Abstract
The main objective of this study was to evaluate the potential genetic effects of SEPT14 on male infertility through sequencing the SEPT14 coding region. To address this research gap, 254 men with sperm abnormalities and 116 normozoospermic men were recruited, and the whole-coding regions of SEPT14 were sequenced. Two heterozygous mutations, p.Ala123Thr (3/254 vs. 0/116) and p.Ile333Thr (3/254 vs. 0/116), were identified in these cases. A high percentage of defective sperm heads was found in sperm with mutated SEPT14. Both mutations are highly evolutionarily conserved among vertebrates. The results of a fine morphological and chromatin structural analysis indicated severely malformed sperm heads with abnormal chromatin packaging through transmission electron microscopy and Toluidine blue staining. Compared with controls, high DNA fragmentation was demonstrated in sperm from cases carrying SEPT14 mutations using the comet assay. In addition, these two mutations in SEPT14 affected its polymerization ability in vitro. These data revels that the two SEPT14 missense mutations impaired sperm head morphology and induced DNA damage. Our study suggests that genetic variant of SEPT14 is one of the effects for human sperm formation and male fertility.
Keywords: male infertility, teratozoospermia, septin, SEPT14, DNA damage
1. Introduction
1.1. Male Infertility and Sperm DNA Damage
Approximately 9% of couples are affected by infertility worldwide and male infertility accounts for 30% of couple infertility cases [1,2,3]. And, one of the major causes of male infertility is teratozoospermia, which is frequently accompanied with sperm DNA defects [4]. As well, sperm with high levels of DNA damage or abnormal DNA packaging has negative effects on embryo development and pregnancy outcomes, including pregnancy loss, recurrent spontaneous abortion, and lower live birth rates [5,6,7,8,9,10,11]. According to the various levels of sperm nuclear abnormalities, sperm DNA damage can be categorized as follows: (a) Damage to the actual DNA; (b) nuclear-related gene mutation or protein loss causes decreased DNA compaction; and (c) irregular chromatin structure [12,13,14,15]. To date, only mutations in certain genes have been linked to sperm DNA damage in a clinical aspect (i.e., PROTAMINE, AURKC, SPATA16, PICK1, DPY19L2, and SEPT12) [16,17,18,19].
1.2. Septins
Septins (SEPTs) are highly evolutionarily conserved GTP-binding proteins and are defined as the fourth component of the cytoskeleton [20,21]. All SEPTs polymerize as hetero-oligomeric structures, such as filaments and rings, and are involved in the regulation of cellular processes, including cytokinesis, cell polarity, and membrane dynamics, through cooperation with diverse cytoskeletons [20,21,22,23,24]. The dysfunction of human SEPTs is associated with the pathology of conditions such as cancer, neurological diseases, and male infertility [25,26]. SEPT4 can be a marker used for the diagnosis of human asthenozoospermia [27,28]. Moreover, Sept4-null mice are infertile because of defective morphology and immotile sperm [29,30]. Our previous studies have demonstrated that SEPT12 is a testis-specific expressed gene. In both humans and mice, mutated SEPT12 causes impaired sperm head morphology, bent tail, and DNA damage [18,31,32]. In addition, embryo development in mice fertilized with Sept12-deficient sperm through intracytoplasmic sperm injection ICSI arrests at the early morula stage [33].
1.3. SEPT14
SEPT14 was originally identified as a novel testis-expressed protein [34]. Furthermore, SEPT14 participates in cortical neuronal migration through cooperation with SEPT4 [35]. In addition, two SEPT14 genetic variants have been identified as reduced risk sites of Parkinson disease (PD) through the screening of 720 cases with PD and 740 controls [36]. In male reproduction, the expressional levels of SEPT14 transcripts are required for human spermatogenesis through evaluating the testicular tissues of men with normal and defective, including hypospermatogenesis, maturation arrest, and sertoli cell–only syndrome [37]. Moreover, SEPT14 is mainly localized in the front of the acrosome, neck, and tail [38]. The main objective of this study was to evaluate the potential genetic effects of SEPT14 on male infertility through sequencing the SEPT14 coding region.
2. Materials and Methods
2.1. Participants and Semen Analysis
This study was approved by the Ethics Committee of the Cathay General Hospital (IRB Approval No.: CGH-P102031). The participants were clinically diagnosed by semen analysis and further divided into two groups: controls with normozoospermia (n = 116; semen samples with a sperm concentration of ≥ 15 × 106/mL, a progressive motility of >32%, and a normal morphology of >14%) and patients with abnormal semen parameter (n = 254). All participants signed a written informed consent form. Semen samples were obtained by masturbation after 3–5 days of sexual abstinence. After liquefying the semen at room temperature, routine semen analysis was performed according to the WHO laboratory manual for the examination and processing of human semen, 2010 (World Health Organization, 2010). Sperm morphology examinations were conducted in accordance with the Krueger strict criteria (normal morphology >14%).
2.2. Sperm Genomic DNA Extraction, Polymerase Chain Reaction, and Sequencing
A Wizard Genomic DNA Purification Kit (A1125, Promega, Madison, WI, USA) was used to extract genomic DNA from sperm. Sperm was washed in phosphate-buffered saline (PBS) and centrifuged at 300 g for 5 min twice. The sperm pellet was resuspended and incubated in lysis buffer, to which proteinase K (200 μg/mL) and dithiothreitol (150 mM) were added and left for 2–3 h at 56 °C. Genomic DNA was precipitated with isopropanol and washed in 70% ethanol twice. After centrifugation, DNA pellets were then air-dried and resuspended in nuclease-free ddH2O. The primers used in this study were designed for whole-coding regions and exon/intron boundaries of SEPT14 (GenBank: NM_207366.2) and are listed in Table S1. After polymerase chain reaction (PCR), the nucleotide variants of the SEPT14 coding region were analyzed through Sanger sequencing.
2.3. Transmission Electron Microscopy
Sperm was prepared for transmission electron microscopy (TEM) (JEM-1400; JEOL, Tokyo, Japan) as previously described [39]. Briefly, sperm was washed in PBS and centrifuged at 300 g for 5 min; this process was repeated twice. The sperm pellet was extremely gently mixed with 0.1% glutaraldehyde at 4 °C overnight. The fixed sperm pellet was rinsed with 0.1 M phosphate buffer (pH 7.2) and then treated with 1% osmium tetroxide at room temperature for 2 h. It was then rinsed again with phosphate buffer and progressively dehydrated through a series of ethanol treatments at increasing concentrations. The Spurr’s resin kit (cat-14300; Electron Microscopy Sciences, PA, USA) was used to embed the sperm pellet at room temperature overnight. Finally, 75-nm thin sections of the embedded samples were cut using an ultramicrotome (EM-UC7, Leica microsystems, Wetzlar, Germany) and were mounted on copper grids. Counter-staining was performed with uranyl acetate and lead citrate. An ultramicrograph was acquired using TEM (JEM-1400; JEOL, Tokyo, Japan) at 100 Kva.
2.4. Toluidine Blue Staining
Toluidine blue (TB) staining is a well-established method for examining chromatin integrity, and in this study, it was performed according to the protocol detailed in relevant studies [40]. TB is a basic metachromatic dye with a high binding ability to DNA phosphate residues. It is incorporated in the incomplete and poor packaging DNA structure of sperm and subsequently produces a blue to deep violet (purple) color. In this study, smears were prepared and fixed in freshly prepared 96% ethanol–acetone (1:1) at 4 °C from 30 min to 12 h before being air-dried. Subsequently, the smears were hydrolyzed in 0.1 N HCl at 4 °C for 5 min and rinsed three times in distilled water for 2 min. Finally, each smear was stained with 0.05% TB (89640, Sigma, St. Louis, MO, USA) in 50% McIlvaine’s citrate phosphate buffer (pH 3.5) for 5 min and then washed with distilled water. TB images were acquired through light microscopy, and for 150 sperm samples, various areas of each slide were examined under oil immersion with ×1000 magnification. Normal sperm with satisfactory chromatin levels was unstained or exhibited pale blue staining, and sperm with abnormal chromatin integrity was stained deep violet. The mean percentage of sperm with both head defects and diminished chromatin integrity in the control group was used as a reference to calculate the fold change in patients with SEPT14 A123T or I333T mutations.
2.5. Comet Assay
Sperm DNA integrity was evaluated using the neutral comet assay [41]. After sperm was washed, the sperm suspension was adjusted to a concentration of 1 × 106/mL in PBS and was mixed with 1% (w/v) low-melting-point agarose (Type VIIA; Sigma-Aldrich, MO, UK) at a ratio of 1:10 (v/v). The mixture was layered onto the surface of two microscope slides to form two microgels, and the slides were transferred to a refrigerator at 4 °C for 10 min. Microgels were submersed in a precooled lysis buffer (2.5 M NaCl, 100 mM EDTA, 10 mM Tris, 1% Triton X-100, and 40 mM dithiothreitol) for 1 h at room temperature and protected from light exposure. Following this initial lysis buffer submersion period, proteinase K (10 μg/mL) was added to the lysis solution, and the microgels underwent additional lysis at 37 °C for 2.5 h. Before electrophoresis, the slides were washed with ddH2O and placed in a horizontal electrophoresis unit, where they were equilibrated for 20 min with TBE buffer. After electrophoresis, the slides were washed twice with ddH2O and stained with SYBR staining solution. After staining, the slides were rinsed with ddH2O and then air-dried. A total of 100 sperm cells per semen sample were randomly selected from the two slides before analysis. Comet contents, including percentage of DNA in the tail (% DNA in the tail) and tail length, were measured using CometScore 2.0. The %DNA in tail measure indicates the proportion of total DNA that has migrated to the tail. Tail length refers to the distance from the fragmented DNA to the head.
2.6. Cloning, Transfection, and Immunofluorescence Staining
Full lengths of human SEPTIN14 were reverse transcription-polymerase chain reaction (RT-PCR)-amplified from a human RNA panel and cloned into pFLAG-CMV2, as described previously [42]. The mutation constructs were prepared using QuikChange Site-directed Mutagenesis Kits (Stratagene, La Jolla, CA, USA). All constructs were confirmed by performing DNA sequencing. After HeLa cells were transfected with plasmids using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), the cells were subjected to immunofluorescence staining. The immunofluorescence staining process is described previously [32]. The cells were exposed to the anti-FLAG antibody (Sigma, F1804, St. Louis, MO, USA) at 4 °C overnight. After washing with PBST, the slides were incubated with secondary antibodies and stained with phalloidin and 4’,6-diamidino-2-phenylindole (DAPI). Subsequently, the slides were mounted on Dako fluorescence mounting medium.
2.7. Statistical Analysis
The quantification results are presented as mean ± standard error of the mean. The Student’s t test (two-tailed) was used to evaluate the significance between the two groups. Differences were considered statistically significant at p < 0.05.
3. Results
3.1. Two Novel Missense Mutations in the SEPT14 Gene
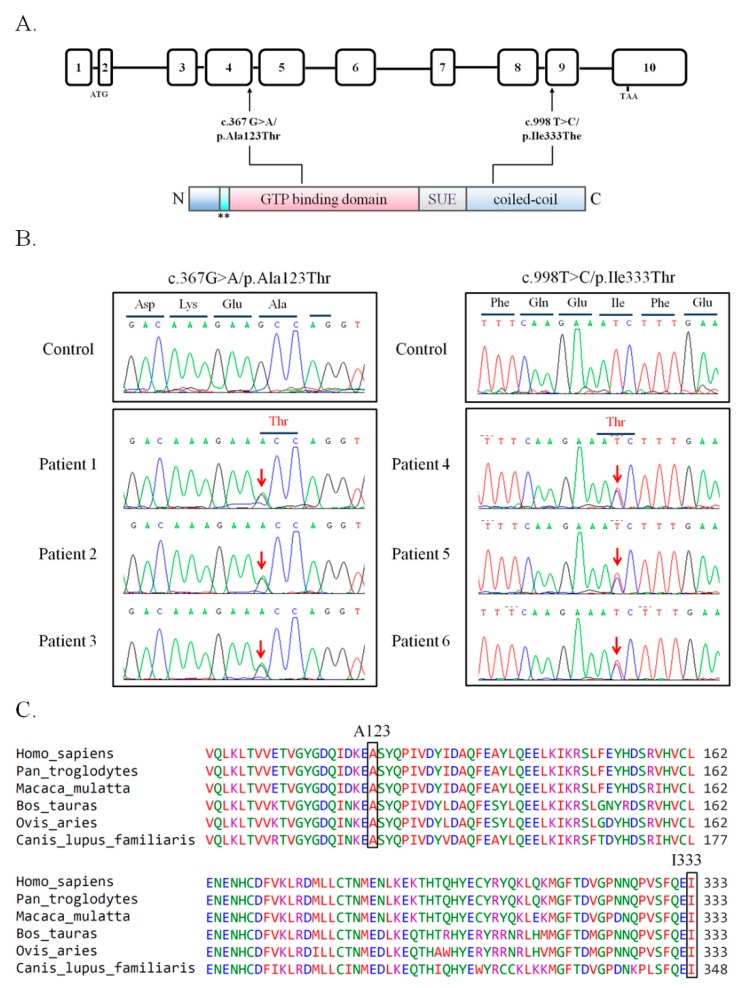
To screen the possible genetic effects of SEPT14 on male fertility, we collected semen samples from 254 male patients with abnormal semen parameter and 116 controls with normozoospermia. We designed the primers (Table S1) targeted to the whole-coding regions of SEPT14 and performed Sanger sequencing to characterize unknown genetic variants of male infertility. Several variants located in exons and exon/intron boundaries were observed (Tables S2 and S3). Notably, two novel missense mutations (c.367G > A, n = 3; c.998T > C, n = 3) were noted in six cases with abnormal semen parameter (Table 1). The c.367G > A mutation was located in exon 4 and caused a substitution of alanine to threonine at position 123 (p.A123T) within the GTP-binding domain of SEPT14 (Figure 1A,B). The other mutation, c.998T > C, was located in exon 9 and induced an amino acid change from isoleucine to threonine at position 333 (p.I333T) within the coiled-coil domain of SEPT14 (Figure 1A,B). Both residues, A123 and I333, are evolutionarily highly conserved among vertebrates (Figure 1C). The clinical data of the six cases are summarized in Table 1. All cases carrying SEPT14 mutations presented with teratozoospermia (91.5% ± 2.88% abnormal sperm), and morphological abnormalities were attributed to head defects (90% ± 4%). Therefore, we speculated that SEPT14 mutations may disturb the morphology of sperm heads and cause male infertility.
Table 1.
Clinical characteristics of infertile patients with c.367G > A/p.Ala123Thr and c.998T > C/p.Ile333Thr SEPTIN14 mutations.
| Nucleotide Sequence/Amino Acid Variation | ||||||
|---|---|---|---|---|---|---|
| c.367G>A/p.Ala123Thr | c.998T>C/p.Ile333Thr | |||||
| Ratio | Patients: 3/254; Controls: 0/116 | Patients: 3/254; Controls: 0/116 | ||||
| Exon | 4 | 9 | ||||
| Case No. | Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 |
| Clinical feature | Oligoteratozoospermia | Teratozoospermia | Teratozoospermia | Teratozoospermia | Teratozoospermia | Teratozoospermia |
| Abnormal morphology (%) (Krueger criteria) | 90 | 88 | 95 | 91 | 90 | 95 |
| Head defect (%) | 91 | 83 | 91 | 88 | 93 | 94 |
| Neck defect (%) | 1 | 9 | 3 | 9 | 4 | 3 |
| Tail defect (%) | 9 | 11 | 10 | 11 | 5 | 18 |
| Immature (%) | 11 | 13 | 1 | 3 | 3 | 5 |
| Sperm concentration (106/ml) (>15 × 106/mL) | 13 | 145 | 70 | 28 | 68 | 20 |
| Progressive motility (%) (>32%) | 49 | 55 | 37 | 48 | 54 | 45 |
| Non-progressive motility (%) | 15 | 11 | 16 | 17 | 14 | 30 |
| Karyotype | 46, XY | 46, XY | 46, XY | 46, XY | 46, XY | 46, XY |
| Age (years) | 31 | 39 | 35 | 32 | 35 | 38 |
| Cigarette smoking, alcohol consumption, genital disease | No | No | No | No | No | No |
Figure 1.
Identification of novel SEPT14 missense mutations associated with male infertility. (A) Schematic of the SEPT14 gene (upper panel) and protein (lower panel). Two missense mutations, A123T and I333T, were observed in patients with teratozoospermia. A123T was located in the GTP-binding domain, and I333T was located in the C-terminal coiled-coil domain. The 10 exons of the human SEPT14 were numbered, and the start and stop codons were annotated. N = N-terminal; ** = phosphoinositide-binding polybasic region; SUE = septin unique element; C = C-terminal. (B) Sanger sequencing chromatograms of SEPT14 mutations. Chromatograms display the sequence corresponding to WT and mutated alleles (upper and lower panels, respectively). These two mutations were noted in three patients and were heterozygous in each patient. Red arrows indicate nucleotide changes. (C) Evolutionary conservation of the p.A123 and p.I333 residues of the SEPT14 protein. Both A123 and I333 residues are highly conserved in various mammalian species. Alignment was performed with Clustal Omega (EMBL-EBI). Key residues, namely A123 and I333, are indicated by black squares. The numbers on the left indicate the amino acid number of the SEPT14 protein sequence.
3.2. Severely Malformed Heads and Diminished Chromatin Integrity in Sperm with SEPT14 Mutations
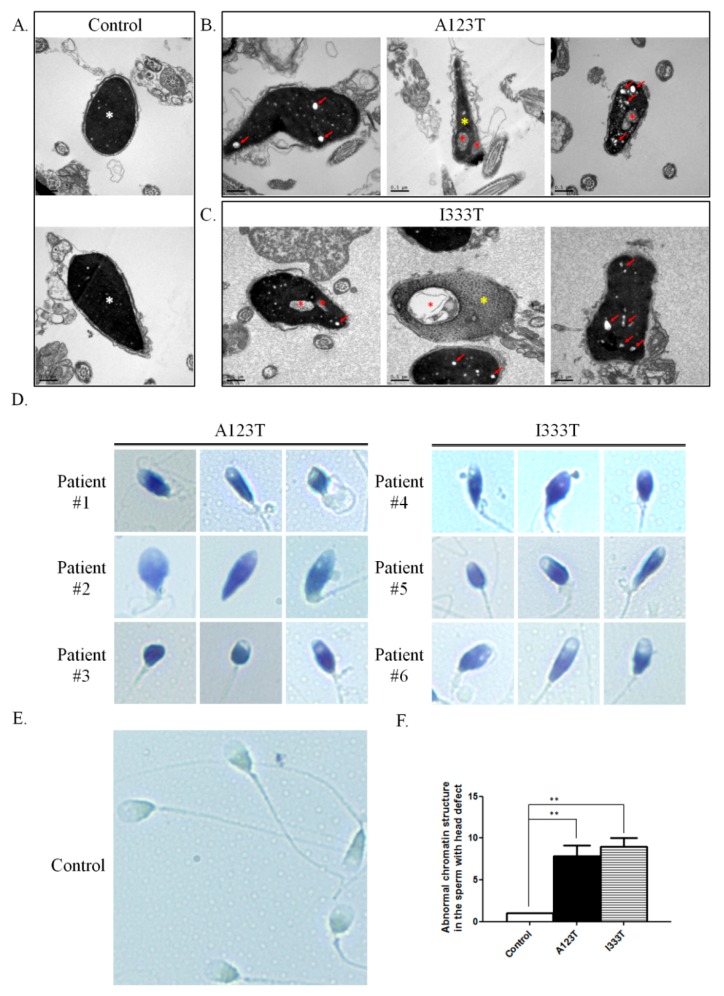
To clarify the detailed morphological effects of SEPT14 mutations on the structure of sperm heads, TEM was used. Compared with controls, sperm from patients carrying A123T or I333T showed prominently malformed heads and contained numerous vacuoles (Figure 2A–C). Furthermore, some sperm samples exhibited abnormal chromatin packaging (Figure 2B, yellow asterisks). Since high chromatin quality is critical for fertilization and achieving successful pregnancy outcomes, we also examined the chromatin integrity of sperm from the patients carrying SEPT14 mutations through TB staining. Based on TB staining, the ratio of the abnormal chromatin structure (deep violet) in sperm with head defects of SEPT14-mutated cases increased significantly compared with controls (light blue) (Figure 2D–F). According to these results, we suggest that SEPT14 mutations impair sperm nuclear morphology and chromatin integrity.
Figure 2.
Sperm from patients with A123T or I333T exhibited severely malformed head shapes and aberrant chromatin integrity. (A–C) Electron microscopic morphology of the sperm heads from controls and cases with SEPT14 mutations. Micrographs of various sections of the sperm heads from controls. A normal sperm head is oval with highly condensed chromatin (white asterisks) (A). Most of the sperm from cases with A123T (B) or I333T (C) exhibited head deformities and numerous nuclear vacuoles (red arrows or asterisks). Moreover, sperm with SEPT14 mutations displayed chromatin packaging failure (yellow asterisk). (D–F) Representative photos of TB staining on sperm from each infertile patient with A123T (D, left panel) or I333T (D, right panel) mutations or controls (E). In sperm from patients with SEPT14 mutations, most of the sperm with head defects was stained in deep violet because of the abnormal chromatin structure (D). In contrast to the abnormal sperm, sperm with satisfactory chromatin structure was unstained or stained pale blue in (E). (F) Quantification of sperm with abnormal chromatin integrity. All quantifications were performed on the control (n = 6), A123T (n = 3), and I333T (n = 3) groups. (** p < 0.01, analyzed using Student’s t test).
3.3. DNA Integrity in Sperm with SEPT14 Mutations
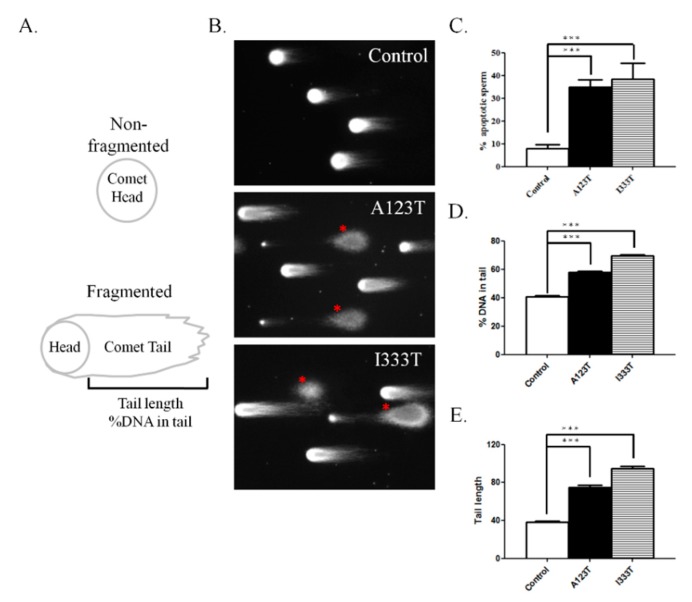
Although a high percentage of sperm with abnormal head morphology and the loss of chromatin packing was found in cases with SEPT14 mutations, the level of DNA damage/fragmentation of sperm remains unknown. To address this concern, the comet assay was used. Two indices of comet assay, namely the percentage of fragmented DNA in the tail and the tail length of comet, were evaluated in the sperm samples through the comet assay (Figure 3A). Figure 3B,C reveal that sperm carrying SEPT14 A123T or I333T is full of DNA fragments, with a high percentage of fragmented DNA in the tail and a longer tail length, compared with controls. Based on these results, we suggest SEPT14 mutations damage DNA integrity in sperm.
Figure 3.
Infertile patients with SEPT14 mutations have significantly higher DNA fragmentation than controls. (A) Illustration of the comet content. DNA fragments migrated out of the heads and resemble a “comet tail” under electrophoresis. The intensity and length of the comet tail reflect the degree of DNA damage. (B) Representative images of DNA damage in sperm obtained from controls and patients with A123T or I333T. (C) Quantification of % apoptotic sperm number. (D and E) Quantification of %DNA in the tail and tail length. Evident DNA damage was observed in sperm from infertile patients with SEPT14 mutations. All quantifications were performed on the control (n = 6), A123T (n = 3), and I333T (n = 3) groups. * Significant difference compared with controls (***, p < 0.001, analyzed using Student’s t test).
3.4. SEPT14 Mutations Damage the Forming Ability of the Filamentous Structure
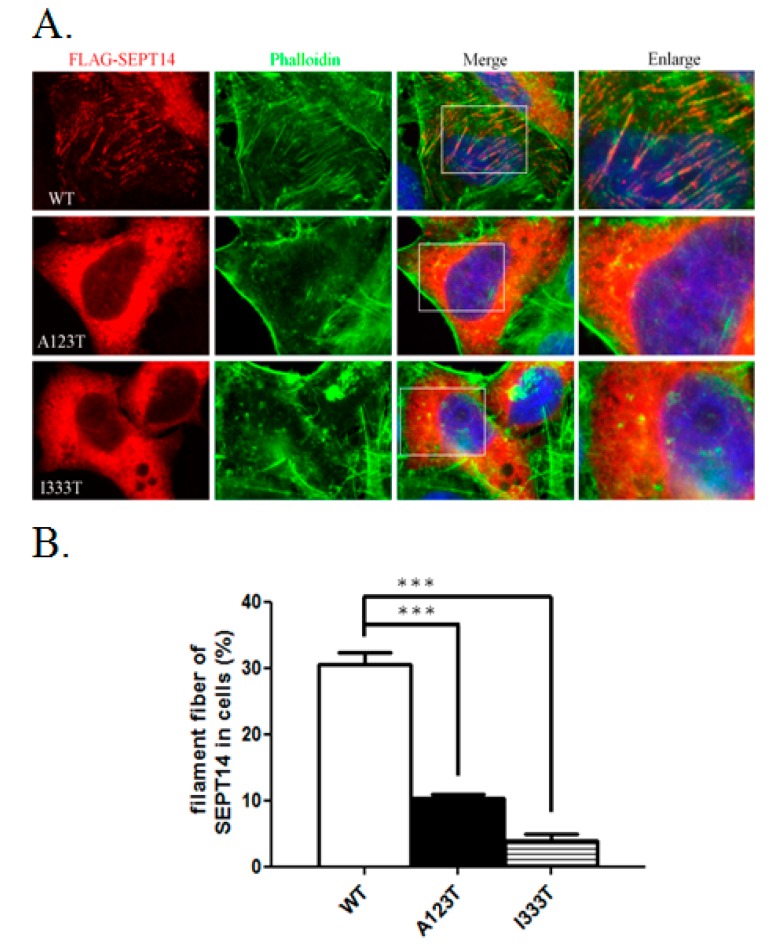
To determine whether SEPT14 mutations affect its cellular function e.g., filamentous formation, wild-type (WT) and mutant SEPT14 constructs were transfected into the cells, followed by immunofluorescence staining (Figure 4). Figure 4A shows that FLAG-SEPT14 (WT) forms the filamentous structure (red) and is colocalized with F-actin (green) in the cell, as revealed by staining with phalloidin. Furthermore, compared with FLAG-SEPT14 (WT), only few of the filament structures observed in the cell were transfected with the mutated SEPT14 (FLAG-SEPT14A123T and FLAG-SEPT14I333T) constructs. Through quantitative estimation from three independent assays, Figure 4B reveals that the percentage of formed filaments of the mutated SEPT14 constructs decreased significantly compared with FLAG-SEPT14 (WT). This result demonstrates that mutated SEPT14 damages the forming ability of the filamentous structure.
Figure 4.
Filament forming ability of WT-SEPT14 and mutant SEPT14. (A) Immunofluorescence staining of the cells transfected with plasmids encoding WT FLAG-SEPT14 (WT panel), FLAG-SEPT14A123T (A123T panel), and FLAG-SEPT14I333T (I333T panel). From left to right: Staining with anti-FLAG antibody (SEPT14), staining with phalloidin (phalloidin), merging with the figures of staining with anti-FLAG antibody, phalloidin, and DAPI (merge), and enlarged figure (enlarge). (B) The quantification bar chart was based on the observation of more than 100 cells for each experiment in Figure 4A. Filament fibers indicate the percentages of cells with SEPT14 filaments. (p < 0.01, Student’s t test comparing filament fibers to others). Data are represented as means ± standard error of three tests.
4. Discussion
SEPT14 is the last identified gene in the SEPTIN family and is abundantly expressed in testes and neurons. In this study, we characterized the genetic effects of SEPT14 on cases with abnormal sperm parameters by sequencing the whole-coding regions of SEPT14. We identified two mutations, A123T and I333T, in teratozoospermia cases. Moreover, sperm from cases carrying SEPT14 mutations not only displayed severe structural defects of the sperm head but also presented with high levels of DNA damage. In addition, these mutations affected the polymerization ability of SEPT14 in vitro. This is the first study to reveal the genetic alternations and spermatogenetic functions of SEPT14 in maintaining human sperm head morphology and nuclear DNA integration.
4.1. SEPTs and Sperm DNA Damage
We previously discovered two heterozygous missense mutations of SEPT12, in which threonine was replaced with methionine at amino acid 89 (T89M; 1/160) and aspartate was replaced with asparagine at amino acid 197 (D197N; 1/160), from screening 160 male patients with infertility [31]. Both mutations were located within the GTPase domain, a critical domain for binding GTP for SEPT polymerization. However, sperm from patients with SEPT12D197N presented a defective annulus between the midpiece and the principal piece of the sperm tail, with a bent tail. Moreover, we discovered a SEPT12 variant (c.474 G > A; 15/160), which affects RNA splicing and creates a truncated C-terminal of SEPT12. It inhibits SEPT12 polymerization considerably. Approximately 88%–99% of sperm from infertile men carrying the c.474 G > A SNP display severe sperm head defects, with DNA damage and nuclear defects [18]. Due to the dissimilar phenotypes between sperm with mutations and c.474 G > A variants, we speculate that SEPT12 has multiple functions during sperm head and sperm tail annulus formation. In this study, two missense mutations (A123T, 3/254; I333T, 3/254) of SEPT14 were observed in treatozoospermia cases with severe sperm head defects (Table 1 and Figure 1). Sperm from these patients exhibited a disrupted ultrastructure of sperm heads and DNA damage (Figure 2 and Figure 3). Moreover, the mutations of SEPT14 decreased its polymerization ability (Figure 4). The present study supports that mutated SEPT14 affects sperm head formation and causes DNA damage through the loss of polymerization ability.
4.2. Sperm DNA Damage and ART
Sperm DNA damage may be attributable to factors such as cigarette smoking, chemotherapy or radiation therapy, environmental toxins, genital tract inflammation, reactive oxygen species, testicular hyperthermia, hormonal factors, and gene mutations [12,43,44]. Many studies indicated high level of sperm DNA damage is related to a high miscarriage rate, decreased pregnancy rate [45,46,47,48,49,50,51]. In our previous study, sperm with the defective Sept12 allele exhibited abnormal sperm heads and increased DNA damage [32,33]. Mouse embryo development through injected oocyte with Sept12-deficient sperm arrests at the early morula stage [33]. In the present study, we also demonstrated that sperm with mutated SEPT14 exhibited abnormal sperm heads and increased DNA damage. We suggest that mutated SEPT14 is one of the causes of sperm DNA damage in infertile men and may be an indicator for the successful rates of assisted reproductive technology (ART).
5. Conclusions
This study is the first to link the genetic changes of SEPT14 to sperm DNA damage and male infertility.
Acknowledgments
This research was funded by grants from the Ministry of Science and Technology of the Republic of China (MOST 106-2320-B-030-003-MY3; MOST 105-2314-B-030-001). We extend our gratitude to Yen-Sheng Wu for his technical support at the Electron Microscope Laboratory of Tzong Jwo Jang, College of Medicine, Fu Jen Catholic University, Taiwan.
Supplementary Materials
The following are available online at https://www.mdpi.com/2077-0383/8/9/1297/s1, Table S1: Sequences of primers used in this study for SEPTIN14 novel variants, Table S2: Variants identified in infertile men in the coding region of SEPTIN14, Table S3: Variants identified in infertile men in the noncoding regions of the SEPTIN14 gene.
Author Contributions
Y.-Y.W. and Y.-H.L. designed the study and proposed the hypothesis. Y.-Y.W., T.-H.L., and M.-F.C. performed the experiments and analyzed the data. Y.-Y.W., H.-L.L., P.-L.K., Y.-H.L. drafted the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Boivin J., Bunting L., Collins J.A., Nygren K.G. International estimates of infertility prevalence and treatment-seeking: Potential need and demand for infertility medical care. Hum. Reprod. 2007;22:1506–1512. doi: 10.1093/humrep/dem046. [DOI] [PubMed] [Google Scholar]
- 2.Vayena E.R.P., Griffin P.D. Medical, Ethical & Social Aspects of Assisted Reproduction Current Practices & Controversies in Assisted Reproduction: Report of a WHO Meeting. WHO; Geneva, Switzerland: 2001. [Google Scholar]
- 3.Brugh V.M., 3rd, Lipshultz L.I. Male factor infertility: Evaluation and management. Med. Clin. N. Am. 2004;88:367–385. doi: 10.1016/S0025-7125(03)00150-0. [DOI] [PubMed] [Google Scholar]
- 4.Brahem S., Mehdi M., Elghezal H., Saad A. Detection of DNA fragmentation and meiotic segregation in human with isolated teratozoospermia. J. Assist. Reprod. Genet. 2011;28:41–48. doi: 10.1007/s10815-010-9482-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palermo G.D., Neri Q.V., Cozzubbo T., Rosenwaks Z. Perspectives on the assessment of human sperm chromatin integrity. Fertil. Steril. 2014;102:1508–1517. doi: 10.1016/j.fertnstert.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Zhao J., Zhang Q., Wang Y., Li Y. Whether sperm deoxyribonucleic acid fragmentation has an effect on pregnancy and miscarriage after in vitro fertilization/intracytoplasmic sperm injection: A systematic review and meta-analysis. Fertil. Steril. 2014;102:998–1005 e8. doi: 10.1016/j.fertnstert.2014.06.033. [DOI] [PubMed] [Google Scholar]
- 7.Osman A., Alsomait H., Seshadri S., El-Toukhy T., Khalaf Y. The effect of sperm DNA fragmentation on live birth rate after IVF or ICSI: A systematic review and meta-analysis. Reprod. Biomed. Online. 2015;30:120–127. doi: 10.1016/j.rbmo.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 8.Wdowiak A., Bakalczuk S., Bakalczuk G. The effect of sperm DNA fragmentation on the dynamics of the embryonic development in intracytoplasmatic sperm injection. Reprod. Biol. 2015;15:94–100. doi: 10.1016/j.repbio.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Agarwal A., Cho C.L., Esteves S.C. Should we evaluate and treat sperm DNA fragmentation? Curr. Opin. Obstet. Gynecol. 2016;28:164–171. doi: 10.1097/GCO.0000000000000271. [DOI] [PubMed] [Google Scholar]
- 10.Zini A., Boman J.M., Belzile E., Ciampi A. Sperm DNA damage is associated with an increased risk of pregnancy loss after IVF and ICSI: Systematic review and meta-analysis. Hum. Reprod. 2008;23:2663–2668. doi: 10.1093/humrep/den321. [DOI] [PubMed] [Google Scholar]
- 11.Evenson D., Wixon R. Meta-analysis of sperm DNA fragmentation using the sperm chromatin structure assay. Reprod. Biomed. Online. 2006;12:466–472. doi: 10.1016/S1472-6483(10)62000-7. [DOI] [PubMed] [Google Scholar]
- 12.Schulte R.T., Ohl D.A., Sigman M., Smith G.D. Sperm DNA damage in male infertility: Etiologies, assays, and outcomes. J. Assist. Reprod. Genet. 2010;27:3–12. doi: 10.1007/s10815-009-9359-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evenson D.P., Larson K.L., Jost L.K. Sperm chromatin structure assay: Its clinical use for detecting sperm DNA fragmentation in male infertility and comparisons with other techniques. J. Androl. 2002;23:25–43. doi: 10.1002/j.1939-4640.2002.tb02599.x. [DOI] [PubMed] [Google Scholar]
- 14.Evenson D.P., Darzynkiewicz Z., Melamed M.R. Relation of mammalian sperm chromatin heterogeneity to fertility. Science. 1980;210:1131–1133. doi: 10.1126/science.7444440. [DOI] [PubMed] [Google Scholar]
- 15.Evenson D.P., Jost L.K., Baer R.K., Turner T.W., Schrader S.M. Individuality of DNA denaturation patterns in human sperm as measured by the sperm chromatin structure assay. Reprod. Toxicol. 1991;5:115–125. doi: 10.1016/0890-6238(91)90039-I. [DOI] [PubMed] [Google Scholar]
- 16.De Braekeleer M., Nguyen M.H., Morel F., Perrin A. Genetic aspects of monomorphic teratozoospermia: A review. J. Assist. Reprod. Genet. 2015;32:615–623. doi: 10.1007/s10815-015-0433-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coutton C., Escoffier J., Martinez G., Arnoult C., Ray P.F. Teratozoospermia: Spotlight on the main genetic actors in the human. Hum. Reprod. Update. 2015;21:455–485. doi: 10.1093/humupd/dmv020. [DOI] [PubMed] [Google Scholar]
- 18.Lin Y.H., Wang Y.Y., Chen H.I., Kuo Y.C., Chiou Y.W., Lin H.H., Wu C.M., Hsu C.C., Chiang H.S., Kuo P.L. SEPTIN12 Genetic Variants Confer Susceptibility to Teratozoospermia. PLoS ONE. 2012;7:e34011. doi: 10.1371/journal.pone.0034011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ravel C., Chantot-Bastaraud S., El Houate B., Berthaut I., Verstraete L., De Larouziere V., Lourenco D., Dumaine A., Antoine J.M., Mandelbaum J., et al. Mutations in the protamine 1 gene associated with male infertility. Mol. Hum. Reprod. 2007;13:461–464. doi: 10.1093/molehr/gam031. [DOI] [PubMed] [Google Scholar]
- 20.Mostowy S., Cossart P. Septins: The fourth component of the cytoskeleton. Nat. Rev. Mol. Cell Biol. 2012;13:183–194. doi: 10.1038/nrm3284. [DOI] [PubMed] [Google Scholar]
- 21.Kinoshita M., Noda M. Roles of septins in the mammalian cytokinesis machinery. Cell Struct. Funct. 2001;26:667–670. doi: 10.1247/csf.26.667. [DOI] [PubMed] [Google Scholar]
- 22.Kinoshita M., Field C.M., Coughlin M.L., Straight A.F., Mitchison T.J. Self- and actin-templated assembly of Mammalian septins. Dev. Cell. 2002;3:791–802. doi: 10.1016/S1534-5807(02)00366-0. [DOI] [PubMed] [Google Scholar]
- 23.Bai X., Bowen J.R., Knox T.K., Zhou K., Pendziwiat M., Kuhlenbaumer G., Sindelar C.V., Spiliotis E.T. Novel septin 9 repeat motifs altered in neuralgic amyotrophy bind and bundle microtubules. J. Cell Biol. 2013;203:895–905. doi: 10.1083/jcb.201308068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith C., Dolat L., Angelis D., Forgacs E., Spiliotis E.T., Galkin V.E. Septin 9 Exhibits Polymorphic Binding to F-Actin and Inhibits Myosin and Cofilin Activity. J. Mol. Biol. 2015;427:3273–3284. doi: 10.1016/j.jmb.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall P.A., Jung K., Hillan K.J., Russell S.E. Expression profiling the human septin gene family. J. Pathol. 2005;206:269–278. doi: 10.1002/path.1789. [DOI] [PubMed] [Google Scholar]
- 26.Peterson E.A., Petty E.M. Conquering the complex world of human septins: Implications for health and disease. Clin. Genet. 2010;77:511–524. doi: 10.1111/j.1399-0004.2010.01392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sugino Y., Ichioka K., Soda T., Ihara M., Kinoshita M., Ogawa O., Nishiyama H. Septins as diagnostic markers for a subset of human asthenozoospermia. J. Urol. 2008;180:2706–2709. doi: 10.1016/j.juro.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 28.Lhuillier P., Rode B., Escalier D., Lores P., Dirami T., Bienvenu T., Gacon G., Dulioust E., Toure A. Absence of annulus in human asthenozoospermia: Case report. Hum. Reprod. 2009;24:1296–1303. doi: 10.1093/humrep/dep020. [DOI] [PubMed] [Google Scholar]
- 29.Ihara M., Kinoshita A., Yamada S., Tanaka H., Tanigaki A., Kitano A., Goto M., Okubo K., Nishiyama H., Ogawa O., et al. Cortical organization by the septin cytoskeleton is essential for structural and mechanical integrity of mammalian spermatozoa. Dev. Cell. 2005;8:343–352. doi: 10.1016/j.devcel.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 30.Kissel H., Georgescu M.M., Larisch S., Manova K., Hunnicutt G.R., Steller H. The Sept4 septin locus is required for sperm terminal differentiation in mice. Dev. Cell. 2005;8:353–364. doi: 10.1016/j.devcel.2005.01.021. [DOI] [PubMed] [Google Scholar]
- 31.Kuo Y.C., Lin Y.H., Chen H.I., Wang Y.Y., Chiou Y.W., Lin H.H., Pan H.A., Wu C.M., Su S.M., Hsu C.C., et al. SEPT12 mutations cause male infertility with defective sperm annulus. Hum. Mutat. 2012;33:710–719. doi: 10.1002/humu.22028. [DOI] [PubMed] [Google Scholar]
- 32.Lin Y.H., Lin Y.M., Wang Y.Y., Yu I.S., Lin Y.W., Wang Y.H., Wu C.M., Pan H.A., Chao S.C., Yen P.H., et al. The expression level of septin12 is critical for spermiogenesis. Am. J. Pathol. 2009;174:1857–1868. doi: 10.2353/ajpath.2009.080955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin Y.H., Chou C.K., Hung Y.C., Yu I.S., Pan H.A., Lin S.W., Kuo P.L. SEPT12 deficiency causes sperm nucleus damage and developmental arrest of preimplantation embryos. Fertil. Steril. 2011;95:363–365. doi: 10.1016/j.fertnstert.2010.07.1064. [DOI] [PubMed] [Google Scholar]
- 34.Peterson E.A., Kalikin L.M., Steels J.D., Estey M.P., Trimble W.S., Petty E.M. Characterization of a SEPT9 interacting protein, SEPT14, a novel testis-specific septin. Mamm. Genome. 2007;18:796–807. doi: 10.1007/s00335-007-9065-x. [DOI] [PubMed] [Google Scholar]
- 35.Shinoda T., Ito H., Sudo K., Iwamoto I., Morishita R., Nagata K. Septin 14 is involved in cortical neuronal migration via interaction with Septin 4. Mol. Biol. Cell. 2010;21:1324–1334. doi: 10.1091/mbc.e09-10-0869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rozenkrantz L., Gan-Or Z., Gana-Weisz M., Mirelman A., Giladi N., Bar-Shira A., Orr-Urtreger A. SEPT14 Is Associated with a Reduced Risk for Parkinson’s Disease and Expressed in Human Brain. J. Mol. Neurosci. MN. 2016;59:343–350. doi: 10.1007/s12031-016-0738-3. [DOI] [PubMed] [Google Scholar]
- 37.Shafipour M., Sabbaghian M., Shahhoseini M., Sadighi Gilani M.A. Comparative expression analysis of Septin 14 in testes of infertile men with normal spermatogenesis and spermatogenic failure. Iran. J. Reprod. Med. 2014;12:205–208. [PMC free article] [PubMed] [Google Scholar]
- 38.Vahabi Barzi N., Hosseinifar H., Nickhah Kalashami Z., Sadighi Gilani M.A., Sabbaghian M. Localization of septin 14 protein in sperm. Iran. J. Reprod. Med. 2015;13:70. [Google Scholar]
- 39.Wang Y.Y., Lin Y.H., Wu Y.N., Chen Y.L., Lin Y.C., Cheng C.Y., Chiang H.S. Loss of SLC9A3 decreases CFTR protein and causes obstructed azoospermia in mice. PLoS Genet. 2017;13:e1006715. doi: 10.1371/journal.pgen.1006715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Erenpreiss J., Bars J., Lipatnikova V., Erenpreisa J., Zalkalns J. Comparative study of cytochemical tests for sperm chromatin integrity. J. Androl. 2001;22:45–53. [PubMed] [Google Scholar]
- 41.Haines G.A., Hendry J.H., Daniel C.P., Morris I.D. Germ cell and dose-dependent DNA damage measured by the comet assay in murine spermatozoaa after testicular X-irradiation. Biol. Reprod. 2002;67:854–861. doi: 10.1095/biolreprod.102.004382. [DOI] [PubMed] [Google Scholar]
- 42.Huang C.Y., Wang Y.Y., Chen Y.L., Chen M.F., Chiang H.S., Kuo P.L., Lin Y.H. CDC42 Negatively Regulates Testis-Specific SEPT12 Polymerization. Int. J. Mol. Sci. 2018;19:2627. doi: 10.3390/ijms19092627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakkas D., Alvarez J.G. Sperm DNA fragmentation: Mechanisms of origin, impact on reproductive outcome, and analysis. Fertil. Steril. 2010;93:1027–1036. doi: 10.1016/j.fertnstert.2009.10.046. [DOI] [PubMed] [Google Scholar]
- 44.Zini A., Libman J. Sperm DNA damage: Clinical significance in the era of assisted reproduction. CMAJ Can. Med. Assoc. J. 2006;175:495–500. doi: 10.1503/cmaj.060218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Avendano C., Franchi A., Duran H., Oehninger S. DNA fragmentation of normal spermatozoa negatively impacts embryo quality and intracytoplasmic sperm injection outcome. Fertil. Steril. 2010;94:549–557. doi: 10.1016/j.fertnstert.2009.02.050. [DOI] [PubMed] [Google Scholar]
- 46.Larson-Cook K.L., Brannian J.D., Hansen K.A., Kasperson K.M., Aamold E.T., Evenson D.P. Relationship between the outcomes of assisted reproductive techniques and sperm DNA fragmentation as measured by the sperm chromatin structure assay. Fertil. Steril. 2003;80:895–902. doi: 10.1016/S0015-0282(03)01116-6. [DOI] [PubMed] [Google Scholar]
- 47.Lopes S., Sun J.G., Jurisicova A., Meriano J., Casper R.F. Sperm deoxyribonucleic acid fragmentation is increased in poor-quality semen samples and correlates with failed fertilization in intracytoplasmic sperm injection. Fertil. Steril. 1998;69:528–532. doi: 10.1016/S0015-0282(97)00536-0. [DOI] [PubMed] [Google Scholar]
- 48.Virro M.R., Larson-Cook K.L., Evenson D.P. Sperm chromatin structure assay (SCSA) parameters are related to fertilization, blastocyst development, and ongoing pregnancy in in vitro fertilization and intracytoplasmic sperm injection cycles. Fertil. Steril. 2004;81:1289–1295. doi: 10.1016/j.fertnstert.2003.09.063. [DOI] [PubMed] [Google Scholar]
- 49.Hamidi J., Frainais C., Amar E., Bailly E., Clement P., Menezo Y. A double-blinded comparison of in situ TUNEL and aniline blue versus flow cytometry acridine orange for the determination of sperm DNA fragmentation and nucleus decondensation state index. Zygote. 2015;23:556–562. doi: 10.1017/S0967199414000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bungum M., Humaidan P., Axmon A., Spano M., Bungum L., Erenpreiss J., Giwercman A. Sperm DNA integrity assessment in prediction of assisted reproduction technology outcome. Hum. Reprod. 2007;22:174–179. doi: 10.1093/humrep/del326. [DOI] [PubMed] [Google Scholar]
- 51.Borini A., Tarozzi N., Bizzaro D., Bonu M.A., Fava L., Flamigni C., Coticchio G. Sperm DNA fragmentation: Paternal effect on early post-implantation embryo development in ART. Hum. Reprod. 2006;21:2876–2881. doi: 10.1093/humrep/del251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.