Abstract
Tripartite motif containing 11 (TRIM11) plays important roles in the regulation of lung cancer behaviors. However, the mechanisms of action of TRIM11 in tumor angiogenesis remain unclear. In this study, we found that TRIM11 expression is higher in lung adenocarcinoma (ADC) than in normal lung tissues. High TRIM11 expression was found to be associated with advanced progression and a poor prognosis of lung ADCs. Functional assays demonstrated that TRIM11 promoted tumor growth and angiogenesis in vivo and enhanced migration of (and tube formation by) human umbilical vein endothelial cells (HUVECs). Mechanistically, TRIM11 was found to regulate angiogenesis through the signal transducer and activator of transcription 3 (STAT3)/vascular endothelial growth factor A (VEGFA) pathway. Moreover, in clinical samples, VEGFA expression was much higher in cancer tissue samples and positively correlated with TRIM11 expression. TRIM11-overexpressing samples showed higher CD31 staining and microvessel density. Thus, we provide evidence that TRIM11 is a proangiogenic factor in lung ADC and may serve as a therapeutic target for lung ADC treatment.
Keywords: TRIM11, angiogenesis, STAT3/VEGFA, lung adenocarcinoma
Introduction
Lung cancer is one of the most common causes of cancer-related deaths worldwide [1]. Lung adenocarcinoma (ADC) is the major subtype of non-small cell lung cancer, which represents ~85% of all lung cancer cases. Great progress has been made in lung ADC treatment; however, patients with lung ADC still have a high mortality rate [2]. Therefore, improving the understanding of the mechanism of lung ADC progression is urgently needed for better lung ADC treatment.
Tripartite motif containing 11 (TRIM11) is one of the TRIM family proteins, which are characterized by evolutionarily conserved really interesting new gene (RING) finger domain at the N terminus, one or two B-box motifs, and a coiled-coil (RBCC) region [3]. Due to the RING finger, most TRIM family members function as E3 ubiquitin ligases [4]. Moreover, accumulating evidence has demonstrated that TRIM family members play crucial roles in tumor progression [5]. TRIM11 has been reported to be associated with a poor prognosis among patients with hepatocellular carcinoma (HCC); to promote proliferation, invasiveness, and epithelial-mesenchymal transition of HCC cell lines through the PI3K/AKT signaling pathway, and to enhance HCC progression by inhibiting p53 expression both in vitro and in vivo [6-8]. Similarly, TRIM11 serves as an oncogene in breast, prostate, ovarian, and colon cancers [9-12].
Angiogenesis is one of the cancer hallmarks and is widely believed to promote tumorigenesis and tumor progression [13]. Tumor angiogenesis is associated with the interactions among tumor cells and tumor microenvironment and with various related signaling pathways [14]. Growth factors such as vascular endothelial growth factor A (VEGFA) secreted by tumor cells stimulate endothelial cells to form new vasculature and eventually promote angiogenesis [15]. The supporting roles of angiogenesis in tumor progression are 1) effective evacuation of waste and 2) delivery of nutrients to the tumor. High levels of VEGF in tumors and in the blood as well as tumor angiogenesis expressed as mean vascular density (MVD, by CD31 staining) are indicators of poor prognosis for patients with non-small cell lung cancer [16].
In the present study, we explored the mechanisms of action of TRIM11 on lung ADC angiogenesis. TRIM11 expression was found to be higher in lung ADC and was associated with faster progression and a poor prognosis of lung ADCs. TRIM11 promoted tumor growth and angiogenesis in vivo and enhanced migration of (and tube formation by) human umbilical vein endothelial cells (HUVECs) partially through STAT3/VEGFA pathway. Thus, we provide evidence that TRIM11 is a novel oncogene in lung ADC and may serve as a new theranostic biomarker for lung ADC treatment.
Materials and methods
Patients and clinical tissue samples
Formalin-fixed paraffin-embedded cancer tissues and paired normal lung tissues from 46 lung ADC patients were obtained from Shanghai Punan Hospital of Pudong New District (Shanghai, China). Twenty fresh lung ADC tissue samples and paired adjacent normal lung tissue samples were also obtained from Shanghai Punan Hospital of Pudong New District; after surgical resection, these samples were frozen immediately in liquid nitrogen for further analysis. Written informed consent was obtained from all the participating patients. The study protocol was approved by the Ethics Committee of Shanghai Punan Hospital of Pudong New District.
Cell lines and cell culture
Human lung ADC cell lines H1299, A549, H1975, and PC-9 and BEAS-2B bronchial epithelial cells were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All the cells were cultured in the RPMI 1640 medium (Gibco, Grand Island, NY, USA) supplemented with 10% of fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. All the cell lines were maintained in a humidified incubator at 37°C and 5% CO2.
Immunohistochemical (IHC) staining
IHC analysis was performed as described previously [17]. Briefly, tissue sections fixed in formalin and embedded in paraffin were deparaffinized, rehydrated, and then subjected to incubation with citrate buffer (pH 6.0) for antigen retrieval. Then, the tissue slides were incubated with primary antibodies at 4°C overnight, followed by secondary-antibody incubation at room temperature for 60 min. The Dako ChemMateTM Detection Kit (DaKo, Denmark) was next applied to detect the bound primary antibodies. Integrated optical density was determined by means of Image-Pro Plus 6.0 (IPP).
Lentivirus and transduction
A TRIM11-overexpressing lentivirus, TRIM11 knockdown lentivirus (expressing short hairpin RNA; shRNA; target sequence: 5’-CUA UUC AUC UUU CCC GAG A-3’), and the corresponding negative control lentivirus (shNC sequence: 5’-AGC AAT GTC CAC ACT ATA C-3’) were purchased from GenePharma (Shanghai, China). H1299 cells were transduced with the TRIM11 shRNA lentivirus, and A549 cells were transduced with the TRIM11-overexpressing lentivirus using the Lipofectamine 2000 reagent (Invitrogen). The cells were collected 48 h after the transduction.
Real-time PCR
Total-RNA samples were isolated from the tissues or cells using TRIzol. Reverse transcription was performed with the iScriptTM cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA). The primers were as follows: TRIM11: Forward: 5’-GTG CCT ATG GAG CTG AGG AC-3’, Reverse: 5’-CAG GAT CAG CTC AGG GTT G-3’; VEGFA: Forward: 5’-CGC AGC TAC TGC CAT CCA AT-3’, Reverse: 5’-GTG AGG TTT GAT CCG CAT AAT CT-3’. Relative expression levels were determined by the 2-ΔΔCt method. GAPDH expression served as a control.
Western blotting
A western blotting assay was performed as previously described [18]. Cells were lysed with RIPA buffer, and the protein concentrations were determined with the BCA Assay Kit (Beyotime, Haimen, China). Proteins were separated by SDS-PAGE in a 10% gel, followed by transfer to a nitrocellulose membrane (Bio-Rad). After that, the membranes were blocked with 5% milk, and then incubated with the following primary antibodies overnight at 4°C: anti-TRIM11 (Abcam, Cambridge, MA, USA), anti-VEGFA, anti-phospho- (p-)STAT3, anti-STAT3, and anti-β-actin (Cell Signaling Technology, Danvers, MA, USA). β-actin served as a control. Finally, the membranes were incubated with a horseradish peroxidase-conjugated secondary antibody at room temperature for 1 h. The protein bands were detected by means of an ECL Plus Developing System.
Enzyme-linked immunosorbent assay (ELISA)
Human VEGFA ELISA Kit (R&D Systems, Minneapolis, MN, USA) was used to determine the concentrations of VEGFA in the culture supernatant from infected ADC cells.
Transwell migration assays
H1299 and A549 cells transfected with the plasmids were seeded in the upper chamber containing Matrigel in a serum-free medium, while the lower chamber was covered with the complete medium that contained 10% of fetal bovine serum as a chemoattractant. After incubation at 37°C for 12 h, the migratory cells that attached to the lower surface of the membrane were stained with crystal violet. The number of migratory cells was determined under a microscope.
A tube formation assay
HUVECs at 104 cells/well were seeded in a Matrigel-coated 96-well plate and cultured with the serum-free medium for 6 h. Then, the cells were cultured with various conditioned media from H1299 and A549 cells for 10 h. The tube length was calculated from images in the ImageJ software.
A xenograft tumor model
A total of 106 H1299 or A549 cells were subcutaneously injected into the flanks of 6-week-old female nude mice. After four weeks, all the mice were euthanized to excise the resultant tumors. The tumor volume was calculated via the following formula: ½ × Length × Width2. Tumors were fixed with formalin and embedded in paraffin for IHC staining. All the animal experiments were approved by the Animal Care and Use Committee of Fudan University.
Statistical analysis
All data were expressed as mean ± standard deviation. Statistical analyses were performed in SPSS 13.0 statistical software. Differences between two groups were assessed by unpaired Student’s t test. Kaplan-Meier analyses with the logrank test were conducted to evaluate overall survival. Data with P values < 0.05 were considered significant.
Results
High expression of TRIM11 is associated with faster progression and poor prognosis of lung ADCs
Previously, TRIM11 has been reported to be upregulated at the transcriptional level in lung cancer tissues compared with normal tissues [19]. Similarly, analysis of the data available in Oncomine showed that TRIM11 mRNA levels were significantly higher in human tumor samples as compared with the corresponding normal tissue samples, with the highest levels in lung ADCs (Figure 1A). We then performed IHC analysis to confirm the higher expression of TRIM11 in lung ADCs. As expected, a significantly increased intensity of TRIM11 staining was observed in 46 lung ADC tissues than in paired adjacent normal lung tissues (Figure 1B). The correlation of TRIM11 expression and lung ADC clinical characteristics was studied next. Notably, TRIM11 expression was found to be positively correlated with TNM stages of ADCs (Figure 1C). Using The Cancer Gene Atlas (TCGA) database, we found that lung ADC patients with high tumorous TRIM11 expression show worse overall survival than patients with low TRIM11 expression (Figure 1D). Consistently with these data, among the 46 lung ADCs in this study, high TRIM11 expression negatively correlated with overall survival of the patients (Figure 1E). These results indicated that TRIM11 is associated with faster progression and a poor prognosis of lung ADCs.
Figure 1.
Overexpression of TRIM11 is associated with faster progression and a poor prognosis of lung ADCs. A. Analysis of TRIM11 expression in different types of lung cancer in the Oncomine database. B. Representative IHC staining of TRIM11 in 46 lung ADC samples and paired nontumorous tissue samples. Normalized integrated optical density (IOD) of TRIM11 is shown on the right. C. TRIM11 expression at different clinical stages of lung ADCs. D. Kaplan-Meier analysis of survival of lung ADC patients with distinct expression levels of TRIM11 in the tumor (data from database TCGA). E. Kaplan-Meier analysis of survival of lung ADC patients with distinct expression levels of TRIM11 in 46 tumors. *P < 0.05, **P < 0.01.
TRIM11 promotes tumor growth and angiogenesis in vivo
We then examined the expression of TRIM11 in multiple human lung ADC cell lines. As compared to BEAS-2B bronchial epithelial cells, the mRNA and protein levels of TRIM11 were significantly higher in lung ADC cell lines including H1299, A549, H1975, and PC-9 (Figure 2A, 2B). Next, H1299 cells were infected with the TRIM11 shRNA lentivirus (H1299/shTRIM11) to construct a loss-of-function model, and A549 cells were infected with the TRIM11-overexpressing lentivirus (A549/TRIM11) to construct a gain-of-function cellular model (Figure 2C). These cellular models were then investigated to determine the effect of TRIM11 on tumor growth and angiogenesis in vivo. After continuous monitoring of the nude mice for 4 weeks, subcutaneous tumors formed from H1299/shTRIM11 cells grew much more slowly than did the tumors derived from the corresponding control cells. Conversely, TRIM11 overexpression (A549/TRIM11) resulted in accelerated growth of the xenograft tumors. Furthermore, fewer Ki67+ cells in H1299/shTRIM11-derived tumors, and more Ki67+ cells in A549/TRIM11-derived tumors were noted as compared with their respective controls (Figure 2F), indicating that TRIM11 promoted the proliferation of lung ADC cells. Moreover, TRIM11 downregulation inhibited, whereas TRIM11 upregulation enhanced vascular endothelial cell marker CD31 expression and microvessel density (MVD; Figure 2G). These results indicated that TRIM11 stimulated angiogenesis and promoted tumor growth; these effects may represent a novel mechanism of TRIM11-mediated lung ADC progression.
Figure 2.
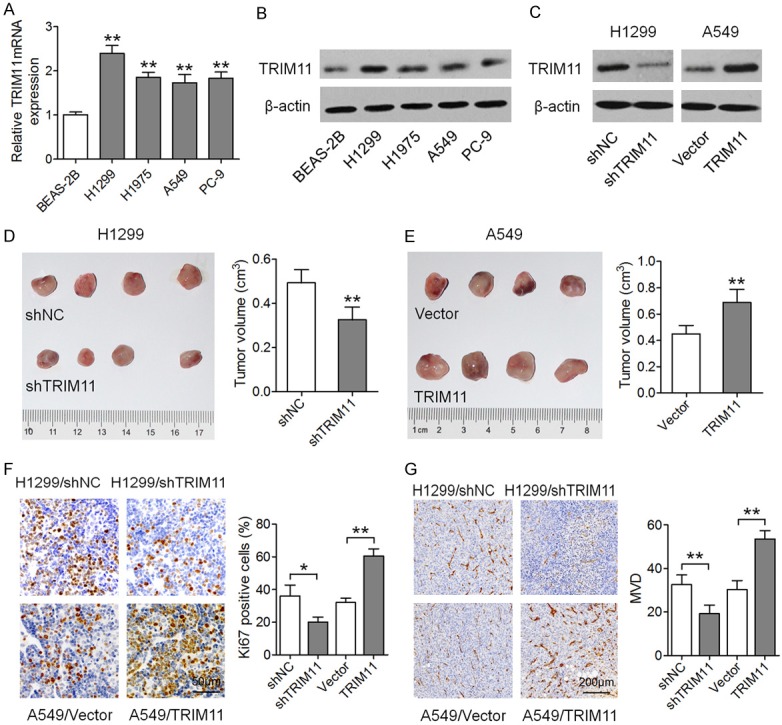
TRIM11 promotes tumor growth and angiogenesis in vivo. (A) Real-time PCR analysis of TRIM11 mRNA levels in the lung ADC cell lines and in bronchial epithelial cell line BEAS-2B. The protein levels of TRIM11 in these cells were determined by western blotting and are depicted in (B). (C) H1299 and A549 cells were infected with the TRIM11 shRNA lentivirus and TRIM11-overexpressing lentivirus, respectively, and western blotting was performed to determine TRIM11 expression. (D) Xenograft tumors derived from H1299/shTRIM11 cells. (E) Xenograft tumors derived from A549/TRIM11 cells. (F) IHC analysis for determining the number of Ki67+ cells in tumor xenografts from the H1299/shTRIM11 and A549/TRIM11 groups. (G) IHC analysis with an anti-CD31 antibody. *P < 0.05, **P < 0.01.
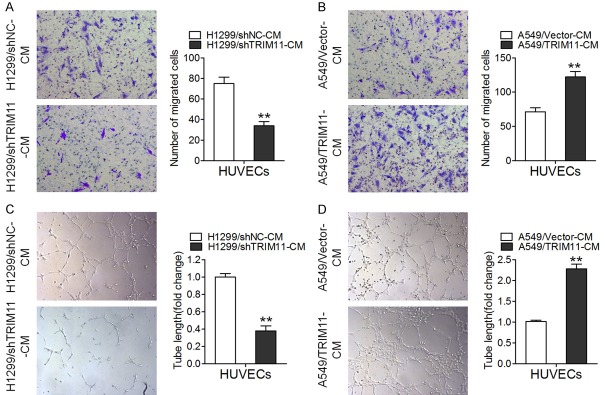
TRIM11 promotes migration of and tube formation by HUVECs
Given that endothelial-cell migration is crucial for angiogenesis [20], we performed Transwell assay to detect the influence of TRIM11 on the migration of HUVECs. As depicted in Figure 3A, the conditioned medium from TRIM11 knockdown H1299 cells (H1299/shTRIM11-CM) obviously inhibited HUVEC migration as compared with the negative control group (H1299/shNC-CM). Not surprisingly, the conditioned medium from TRIM11-overexpressing A549 cells (A549/TRIM11-CM) dramatically enhanced cell migration (Figure 3B). Moreover, tubule formation by HUVECs was inhibited by H1299/shTRIM11-CM as compared with H1299/shNC-CM (Figure 3C). In agreement with these data, A549/TRIM11-CM enhanced tubule formation by HUVECs when compared with A549/Vector-CM (Figure 3D). These results suggested that TRIM11 promoted the migration of (and tube formation by) HUVECs.
Figure 3.
TRIM11 promotes migration of and tube formation by HUVECs. A. The Transwell migration assay was conducted to determine the migratory abilities of HUVECs incubated with the conditioned medium from H1299/shTRIM11 cells. B. The Transwell migration assay of HUVECs incubated with the conditioned medium from A549/TRIM11 cells. C. The capillary tube formation assay of HUVECs incubated with the conditioned medium from H1299/shTRIM11 cells. D. The capillary tube formation assay of HUVECs incubated with the conditioned medium from A549/TRIM11 cells. **P < 0.01.
TRIM11 regulates angiogenesis partially through STAT3/VEGFA pathway
Given that VEGFA is crucial for tumor growth and neovascularization [21,22], we examined the expression levels of VEGFA in H1299/shTRIM11 and A549/TRIM11 cells. Real-time PCR and western blotting analyses revealed that VEGFA mRNA and protein levels were decreased by the TRIM11 knockdown and increased by TRIM11 overexpression (Figure 4A and 4B). Similarly, VEGFA concentrations determined by ELISA in the culture supernatant decreased when TRIM11 was downregulated but increased when TRIM11 was upregulated (Figure 4C). It is worth noting that VEGFA is a target gene of STAT3, which plays a crucial part in tumorigenesis and angiogenesis [23]. We speculated that TRIM11 can upregulate VEGFA through the JAK2/STAT3 pathway, thereby promoting angiogenesis. We found that TRIM11 downregulation reduced p-STAT3 expression, and TRIM11 upregulation increased p-STAT3 expression. In particular, TRIM11 overexpression-mediated VEGFA upregulation was attenuated by STAT3 inhibitor S3I-201 (Figure 4B). In our xenograft tumor model, TRIM11 downregulation decreased VEGFA expression, whereas TRIM11 upregulation increased VEGFA expression (Figure 4D). After that, we tested whether STAT3 activation is required for the effects of TRIM11 on lung ADCs. After inhibition of STAT3 by S3I-201, the conditioned medium from TRIM11-overexpressing A549 cells lost the ability to promote tubule formation by (and migration of) HUVECs (Figure 4E, 4F). These results meant that TRIM11 may regulate lung ADC angiogenesis partially through STAT3/VEGFA pathway.
Figure 4.

TRIM11 regulates angiogenesis partially through STAT3/VEGFA pathway. A. Real-time PCR analysis of VEGFA mRNA levels in H1299/shTRIM11 and A549/TRIM11 cells. B. Western blotting analysis of VEGFA, p-STAT3, and STAT3 in H1299/shTRIM11 cells, A549/TRIM11 cells, and A549/TRIM11 cells treated with the STAT3 inhibitor S3I-201. C. An ELISA was carried out to determine VEGFA concentration in the cell culture supernatant. D. IHC analysis for assessing VEGFA expression in tumor xenografts. E. A capillary tube formation assay of HUVECs incubated with the conditioned medium from A549/TRIM11 cells or from A549/TRIM11 cells treated with S3I-201. F. The Transwell migration assay of HUVECs incubated with the conditioned medium from A549/TRIM11 cells or A549/TRIM11 cells treated with S3I-201. **P < 0.01.
TRIM11 upregulation positively correlates with VEGFA levels in lung ADC samples
Finally, we detected VEGFA mRNA expression levels in 20 fresh lung ADC tissues and in the matched normal tissues. As presented in Figure 5A, VEGFA mRNA levels were much higher in lung ADCs than in normal tissue samples. In addition, there was a positive correlation between TRIM11 and VEGFA expression levels among the collected lung ADC tissue samples (Figure 5B). The association between TRIM11 expression and CD31 staining was evaluated next. Among the 20 collected lung ADC tissues, five samples showed lower TRIM11 expression (hereafter: TRIM11-Low samples) and 11 samples showed higher TRIM11 expression (hereafter: TRIM11-High samples). CD31 expression was much higher in TRIM11-High samples than in TRIM11-Low samples (Figure 5C). Additionally, MVD was dramatically higher in the TRIM11-high samples than in TRIM11-low samples (Figure 5D). These results suggested that TRIM11 upregulation positively correlates with VEGFA overexpression in lung ADC.
Figure 5.
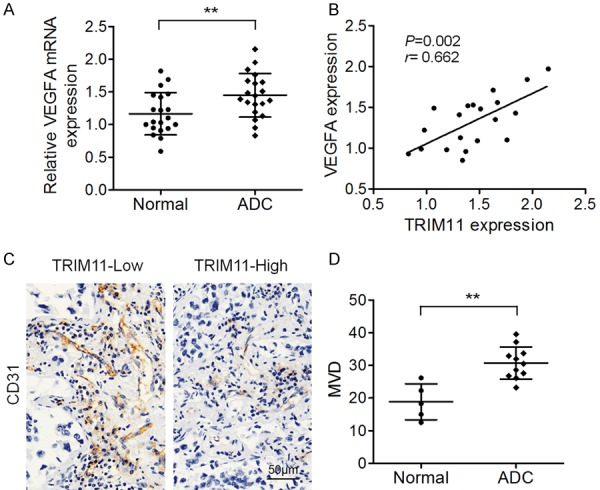
Correlation between TRIM11 and VEGFA expression levels among lung ADC specimens. A. Real-time PCR analysis of VEGFA mRNA levels in 20 lung ADC samples and paired nontumorous tissue samples. B. Pearson’s analysis of correlation between TRIM11 and VEGFA mRNA levels among 20 lung ADCs. C. Representative CD31 staining during IHC analysis of TRIM11-low and TRIM11-high lung ADC tissue samples. D. The average MVD in TRIM11-low and TRIM11-high lung ADC tissue samples. *P < 0.05, **P < 0.01.
Discussion
Lung ADC is still a big threat to public health owing to the high mortality rate [24]. The mechanism that regulates lung ADC progression remains largely unknown. TRIM11 has been reported to be an oncogene in various types of tumors, including lung cancer. Angiogenesis is one of the hallmarks of cancer, and accumulating reports point to the crucial influence of angiogenesis on tumor progression [13,25]. Therefore, we explored the functional mechanisms of action of TRIM11 in tumor angiogenesis. We revealed that upregulated TRIM11 promotes tumor growth and angiogenesis in vivo and enhances migration of and tube formation by HUVECs partially through STAT3/VEGFA pathway. These results provide evidence of a novel mechanism behind TRIM11-mediated lung ADC progression.
TRIM11 is an E3 ubiquitin ligase of the TRIM family of proteins and serves as an oncogene in many types of cancers. TRIM11 mRNA is the direct target of miR-24-3p, and TRIM11 overexpression promotes cell proliferation and colony formation and inhibits apoptosis of colon cancer cell lines. Silencing of TRIM11 inhibits tumor growth in vivo [12]. In HCC clinical tissue samples, higher TRIM11 expression is observed relative to matched nontumorous liver tissues; TRIM11 protein levels are associated with recurrence, postoperative metastasis, and poor overall survival of patients [8]. TRIM11 depletion in ovarian cancer cell lines by small interfering RNA dramatically inhibits cell invasion via matrix metalloproteinase (MMP)-2 and MMP-9 and induces apoptosis via apoptosis-related proteins BCL-2 and BAX [11]. Similarly, TRIM11 turned out to be an oncogene in lung ADC in our present study. TRIM11 expression levels were significantly higher lung ADC patients’ tumor samples. In addition, TRIM11 expression was found to be positively associated with advanced TNM stages and poor prognosis. Functional assays indicated that TRIM11 promotes tumor growth and angiogenesis in vivo and enhances migration of (and tube formation by) HUVECs partially through STAT3/VEGFA pathway.
Rapid growth of a tumor requires continuous supply of oxygen and nutrients and waste product removal. Angiogenesis is one of the most important phenomena that support tumor growth and is a hallmark of cancers. In a tumor microenvironment, many cytokines have been identified as activators of angiogenesis, e.g., VEGF, basic fibroblast growth factor (bFGF), transforming growth factor (TGF)-α, TGF-β, tumor necrosis factor (TNF)-α, granulocyte colony-stimulating factor, and interleukin 8 [26]. Moreover, VEGF-induced angiogenesis has been reported to promote tumor progression. The monoclonal antibody specific for VEGF suppresses tumor growth and vessel density in nude mice that are injected with human rhabdomyosarcoma cell line, glioblastoma multiforme cell line, or a leiomyosarcoma cell line [27]. In colorectal cancer cell lines, miR-622 overexpression suppresses angiogenesis, whereas CXCR4 overexpression attenuates miR-622-mediated VEGFA inhibition, indicating that miR-622 inhibits colorectal cancer angiogenesis by suppressing the CXCR4-VEGFA axis [28]. Our results revealed that TRIM11 enhances VEGFA expression and promotes HUVEC migration and tube formation partially through STAT3 signing pathway.
It is important to note the limitations of this study. Our data are based on nude mice that were injected with human lung ADC cell lines. The immune system of these mice is ineffective at killing tumor cells, and this situation in turn triggers and accelerates tumor growth. The spontaneously forming lung ADC in mice with the C57BL/6 or BALB/c background and with an intact immune system may be a better way to evaluate the effects of TRIM11 on lung ADC.
In conclusion, our results illustrate higher TRIM11 expression in lung ADC and that TRIM11 promotes lung ADC tumor growth and angiogenesis in vivo. TRIM11 was found to enhance the migration of (and tube formation by) HUVECs partially through STAT3/VEGFA pathway. Therefore, targeting of TRIM11 maybe a novel strategy for lung ADC treatment.
Acknowledgements
This research was supported by the Key Specialized Training Program of Health System in Pudong New District of Shanghai (PWZzk2017-18) and the Development Fund Project of Science and Technology in Pudong New District of Shanghai (PKJ2016-Y39).
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Travis WD. The 2015 WHO classification of lung tumors. Pathologe. 2014;35(Suppl 2):188. doi: 10.1007/s00292-014-1974-3. [DOI] [PubMed] [Google Scholar]
- 3.Ozato K, Shin DM, Chang TH, Morse HC 3rd. TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008;8:849–860. doi: 10.1038/nri2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Versteeg GA, Rajsbaum R, Sanchez-Aparicio MT, Maestre AM, Valdiviezo J, Shi M, Inn KS, Fernandez-Sesma A, Jung J, Garcia-Sastre A. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity. 2013;38:384–398. doi: 10.1016/j.immuni.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer. 2011;11:792–804. doi: 10.1038/nrc3139. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, Rao J, Lou X, Zhai J, Ni Z, Wang X. Upregulated TRIM11 exerts its oncogenic effects in hepatocellular carcinoma through inhibition of P53. Cell Physiol Biochem. 2017;44:255–266. doi: 10.1159/000484678. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Z, Xu C, Zhang X, Huang L, Zheng C, Chen H, Wang Y, Ju H, Yao Q. TRIM11 upregulation contributes to proliferation, invasion, and EMT of hepatocellular carcinoma cells. Oncol Res. 2017;25:691–699. doi: 10.3727/096504016X14774897404770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y, Li L, Qian X, Ge Y, Xu G. High expression of TRIM11 correlates with poor prognosis in patients with hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2017;41:190–196. doi: 10.1016/j.clinre.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 9.Dai X, Geng F, Li M, Liu M. Tripartite motifcontaining 11 regulates the proliferation and apoptosis of breast cancer cells. Oncol Rep. 2019;41:2567–2574. doi: 10.3892/or.2019.7015. [DOI] [PubMed] [Google Scholar]
- 10.Pan Y, Zhang R, Chen H, Chen W, Wu K, Lv J. Expression of tripartite motif-containing proteactiin 11 (TRIM11) is associated with the progression of human prostate cancer and is downregulated by microRNA-5193. Med Sci Monit. 2019;25:98–106. doi: 10.12659/MSM.911818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y, Sun J, Ma J. Proliferation and invasion of ovarian cancer cells are suppressed by knockdown of TRIM11. Oncol Lett. 2017;14:2125–2130. doi: 10.3892/ol.2017.6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin Y, Zhong J, Li SW, Li JZ, Zhou M, Chen Y, Sang Y, Liu L. TRIM11, a direct target of miR-24-3p, promotes cell proliferation and inhibits apoptosis in colon cancer. Oncotarget. 2016;7:86755–86765. doi: 10.18632/oncotarget.13550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Eilken HM, Adams RH. Dynamics of endothelial cell behavior in sprouting angiogenesis. Curr Opin Cell Biol. 2010;22:617–625. doi: 10.1016/j.ceb.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 15.Folkman J. Role of angiogenesis in tumor growth and metastasis. Semin Oncol. 2002;29:15–18. doi: 10.1053/sonc.2002.37263. [DOI] [PubMed] [Google Scholar]
- 16.Bremnes RM, Camps C, Sirera R. Angiogenesis in non-small cell lung cancer: the prognostic impact of neoangiogenesis and the cytokines VEGF and bFGF in tumours and blood. Lung Cancer. 2006;51:143–158. doi: 10.1016/j.lungcan.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Ramos-Vara JA, Miller MA. Comparison of two polymer-based immunohistochemical detection systems: ENVISION+™ and ImmPRESS™. Journal of Microscopy. 2006;224:135–139. doi: 10.1111/j.1365-2818.2006.01679.x. [DOI] [PubMed] [Google Scholar]
- 18.Ganju RK, Brubaker SA, Meyer J, Dutt P, Yang Y, Qin S, Newman W, Groopman JE. The alpha-chemokine, stromal cell-derived factor-1alpha, binds to the transmembrane G-protein-coupled CXCR-4 receptor and activates multiple signal transduction pathways. J Biol Chem. 1998;273:23169–75. doi: 10.1074/jbc.273.36.23169. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, Shi W, Shi H, Lu S, Wang K, Sun C, He J, Jin W, Lv X, Zou H, Shu Y. TRIM11 overexpression promotes proliferation, migration and invasion of lung cancer cells. J Exp Clin Cancer Res. 2016;35:100. doi: 10.1186/s13046-016-0379-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Lamalice L, Le Boeuf F, Huot J. Endothelial cell migration during angiogenesis. Circ Res. 2007;100:782–794. doi: 10.1161/01.RES.0000259593.07661.1e. [DOI] [PubMed] [Google Scholar]
- 21.Claesson-Welsh L, Welsh M. VEGFA and tumour angiogenesis. J Intern Med. 2013;273:114–127. doi: 10.1111/joim.12019. [DOI] [PubMed] [Google Scholar]
- 22.Dvorak HF. Vascular permeability factor/vascular endothelial growth factor: a critical cytokine in tumor angiogenesis and a potential target for diagnosis and therapy. J. Clin. Oncol. 2002;20:4368–4380. doi: 10.1200/JCO.2002.10.088. [DOI] [PubMed] [Google Scholar]
- 23.Liao XH, Xiang Y, Li H, Zheng L, Xu Y, Xi Yu C, Li JP, Zhang XY, Xing WB, Cao DS, Bao LY, Zhang TC. VEGF-A stimulates STAT3 activity via nitrosylation of myocardin to regulate the expression of vascular smooth muscle cell differentiation markers. Sci Rep. 2017;7:2660. doi: 10.1038/s41598-017-02907-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ Jr, Wu YL, Paz-Ares L. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 25.Bergers G, Benjamin LE. Tumorigenesis and the angiogenic switch. Nat Rev Cancer. 2003;3:401–410. doi: 10.1038/nrc1093. [DOI] [PubMed] [Google Scholar]
- 26.Ribatti D, Crivellato E. Immune cells and angiogenesis. J Cell Mol Med. 2009;13:2822–2833. doi: 10.1111/j.1582-4934.2009.00810.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim KJ, Li B, Winer J, Armanini M, Gillett N, Phillips HS, Ferrara N. Inhibition of vascular endothelial growth factor-induced angiogenesis suppresses tumour growth in vivo. Nature. 1993;362:841–844. doi: 10.1038/362841a0. [DOI] [PubMed] [Google Scholar]
- 28.Fang Y, Sun B, Wang J, Wang Y. miR-622 inhibits angiogenesis by suppressing the CXCR4-VEGFA axis in colorectal cancer. Gene. 2019;699:37–42. doi: 10.1016/j.gene.2019.03.004. [DOI] [PubMed] [Google Scholar]