Abstract
Bifidobacteria commonly constitute the most abundant group of microorganisms in the healthy infant gut. Their intestinal establishment is believed to be maternally driven, and their acquisition has even been postulated to occur during pregnancy. In the current study, we evaluated bifidobacterial mother-to infant transmission events in a rat model by means of quantitative PCR (qPCR), as well as by Internally Transcribed Spacer (ITS) bifidobacterial profiling. The occurrence of strains supplied by mothers during pregnancy to their corresponding newborns was observed and identified by analysis immediately following C-section delivery. These findings provide intriguing support for the existence of an unknown route to facilitate bifidobacterial transfer during the very early stages of life.
Keywords: bifidobacteria, infant gut microbiota, microbe-host coevolution, metagenomics
1. Introduction
Bifidobacteria are dominant members of the infant gut microbiota, although their numbers are known to decrease during weaning and adolescence, with a further decline in relative abundance in the elderly population [1,2]. Their role as natural modulators of the host immune system, as well as their impact on the physiology and metabolism of the human large intestine are subject to significant scientific scrutiny [1,3]. Due to these purported health benefits it is no surprise that bifidobacteria are frequently incorporated as functional ingredients in various (functional) foods and pharmaceutical products. Bacterial colonization of the human gut represents a very active field of fundamental and applied research, where bifidobacteria represent a key target group, and where strategies to influence the development, composition, and activities of the infant bifidobacterial members of the gut microbiota by means of nutraceutical products (e.g., probiotics and/or prebiotics) are very topical.
Various publications have described that the first microbial consortia in the gut of newborns may harbor bacteria that are vertically transmitted from the mother [4,5,6,7]. In this context, it has been discovered that amongst these maternally inherited bacteria, bifidobacteria represent an important group of gut commensals [8,9,10]. A recent study focusing on the identification of vertically acquired bifidobacterial strains based on a combination of shotgun metagenomics data, Internally Transcribed Spacer (ITS) bifidobacterial profiling analysis and a cultivation approach, described the finding of several bifidobacterial strains, including members of the Bifidobacterium breve, Bifidobacterium longum subsp. longum and Bifidobacterium bifidum species, which were shared between mother-infant dyads [9]. The identification of vertically transmitted bifidobacterial strains represents a key example of microbe-host co-evolution, and is a phenomenon that was also identified in other primate and non-primate mammals, where bifidobacteria were found to be commonly inherited from mother to her offspring, and where mother’s milk appears to represent an important route to facilitate such transfer events [11]. Furthermore, a recent study revealed that the composition of human milk is able to influence the gut microbiota of newborns, with potential implications for infant development and health [12].
The heretofore accepted dogma that the infant gut is sterile before birth has in recent years been challenged by different studies which suggest the existence of an intra-uterine and placenta bacterial community [13,14,15], though the presence and role of a genuine pre-birth microbiota is still controversial and subject to debate [14,16,17].
In the current study, we investigated vertical transmission events of bifidobacteria employing an in vivo model (Rattus norvegicus), resulting in a different bifidobacterial inheritance outcome when pregnant animals were treated with a single Bifidobacterium strain as compared to a mix of bifidobacterial strains belonging to three different Bifidobacterium species, i.e., B. bifidum, B. breve and B. longum.
2. Results and Discussion
2.1. Evaluation of Vertical Transmission of Bifidobacteria under in Vivo Conditions
As previously shown by metagenomic attempts, identical bifidobacterial strains have been found in the fecal samples of mother-newborn dyads [7,9]. In order to evaluate possible microbial transfer by pregnant rats treated with a bifidobacterial strain mix consisting of B. bifidum PRL2010, B. breve 1895B and B. longum subsp. longum 1886 strains, or rats treated with B. bifidum strain PRL2010 only, animals were divided in two different groups. The first group consisted of three female rats, which were treated with the bifidobacterial mix (Mix Colonization Group—MCG), whereas the second group encompassed nine female rats, to which only PRL2010 was administered (PRL2010 Group—PG). Notably, the bifidobacterial strains used were, previously, isolated from fecal samples from healthy breast-fed infants [11,18,19]. However, even if these bacterial strains were not belonging to the indigenous microbiota of rat, when administered to these animals during pregnancy, they are able to colonize the gut of rat (see below). A similar result has been described previously by Jimenez et al., where a human infant gut commensal, Enterococcus faecium, was able to be vertically transmitted in pregnant mice.
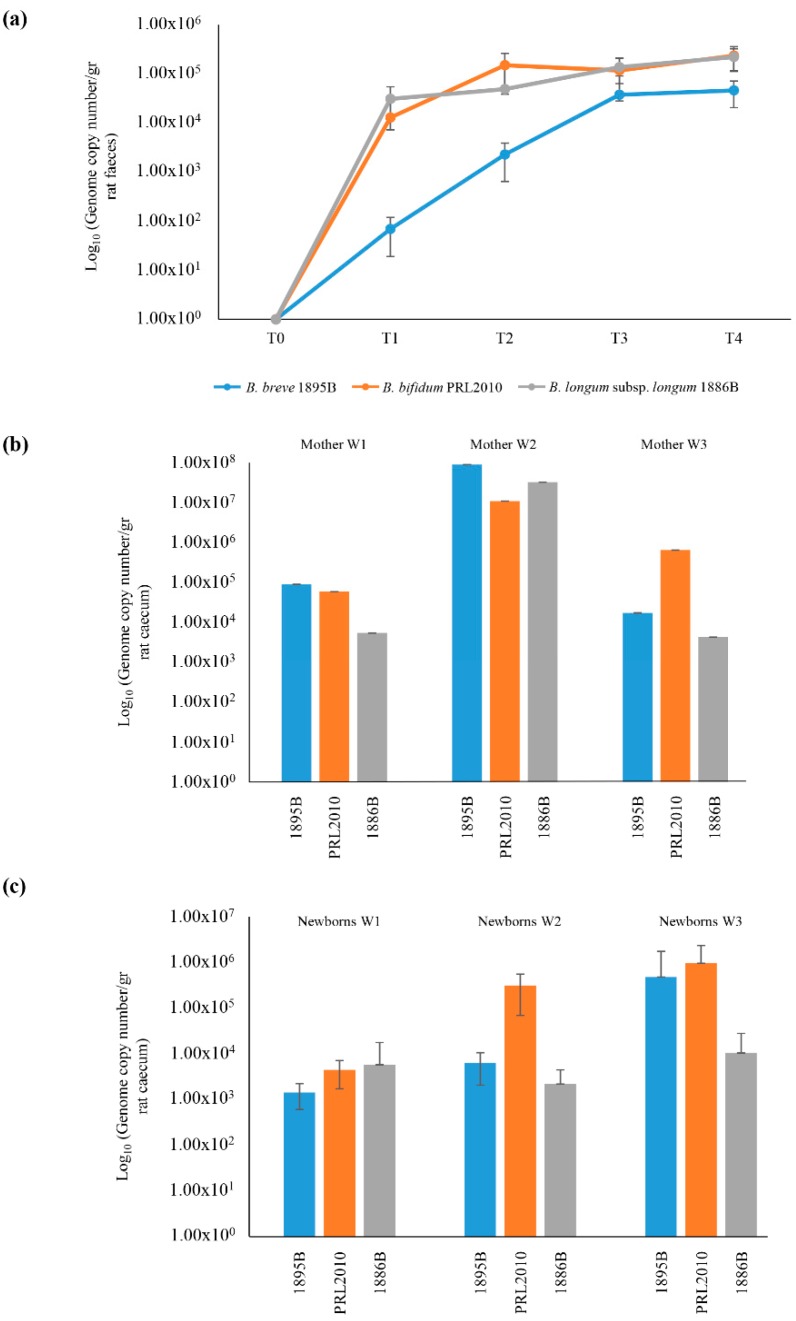
At the start of the experiment, rats were checked for the presence of strains PRL2010, 1895B and/or 1886B in fecal samples by means of PCR using strain-specific primer pairs, revealing that, as expected, these bifidobacterial strains were absent in the animals enrolled in this study and prior to them being fed any of the strains. Dams were administered a daily dose of 109 colony forming units (CFU) of B. bifidum PRL2010 or a mix of B. bifidum PRL2010, B. longum subsp. longum 1886B and B. breve 1895B for 21 days. Presence and clearance of B. bifidum PRL2010, B. longum subsp. longum 1886B and B. breve 1895B were monitored during the gestation period using a qPCR approach based on strain-specific primers (Figure 1). Following Caesarian delivery of the pups, rats were sacrificed and their caecum was removed and assayed for the presence of PRL2010, 1886B and 1895B by means of qPCR. Interestingly, the microbial density estimated by qPCR of B. bifidum PRL2010 in the caecal samples of mothers ranged from 104 to 107 CFU, whereas in caecal samples of pups, the abundance ranged from 103 to 106 CFU (Figure 1). Similarly, the qPCR estimated abundance of B. breve 1895B in the caecum of mothers and of newborns ranged from 104 to 107 and 102 to 106, respectively (Figure 1). The cell load determined by qPCR of B. longum subsp. longum 1886B in the mother’s gut and in pup’s caecum varies from 103 to 107 and 102 to 104, respectively (Figure 1).
Figure 1.
Schematic representation of the vertical transmission of the bifidobacterial mix in rat model treated with B. breve 1895B, B. bifidum PRL2010 and B. longum subsp. longum 1886B strains (Mix Colonization Group—MCG). Panel (a) shows the average of DNA presence of the three strains observed during the bifidobacterial administration. Each point represents the average of the log-population size ± standard deviation for three rats. Panel (b) displays the presence of B. breve 1895B, B. bifidum PRL2010 and B. longum subsp. longum 1886B in the caecum of female rats. Panel (c) exhibits bifidobacterial retrieval from puppies’ caecum. Each pillar represents the average presence for each Bifidobacterium strain ± standard deviation.
In addition, we isolated viable PRL2010/1895B/1897B cells from the caecal samples of the mothers by means of direct cultivation of caecal contents of animals on mMRS followed by the precise strain-assignment of the isolated cells using a PCR approach based on strain-specific primers.
2.2. Maternal Inheritance of B. bifidum PRL2010 Strain
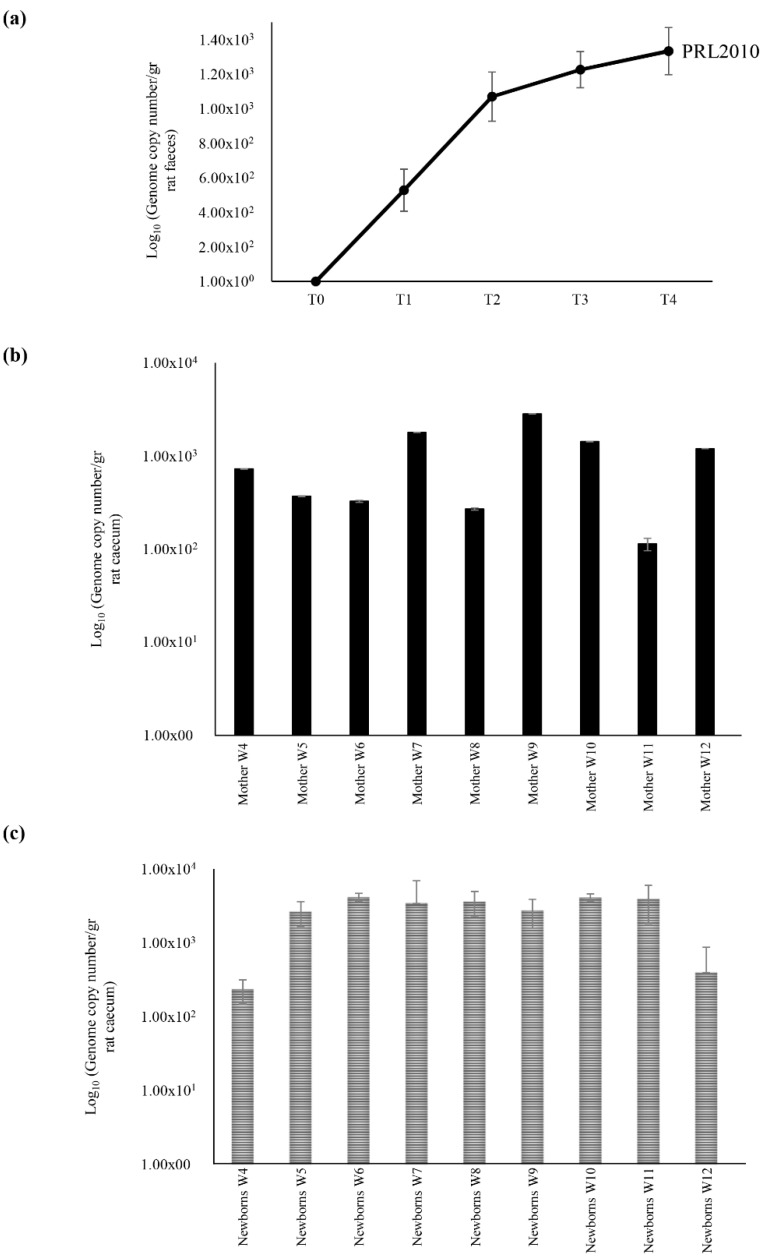
Since B. bifidum strain PRL2010 has been shown to represent a model infant gut commensal [9,18,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34], and as described above for the Mix Colonization Group—MCG, this strain displayed the highest level of vertical transmission from mother to newborns, we decided to further examine the maternal inheritance of strain PRL2010. Bifidobacterial inheritance of nine pregnant rats receiving a daily dose of 109 CFU of B. bifidum PRL2010 for three weeks was monitored by qPCR (Figure 2). These rats delivered their puppies by Caesarian section and PRL2010 levels were evaluated by qPCR using strain-specific primers targeting both the caecal sample of the mother and the corresponding pups (Figure 2). The estimated abundance of PRL2010 cells by qPCR ranged from 103 to 104 CFU and from 102 to 104 CFU in mother and newborns samples, respectively (Figure 2). Notably, the PRL2010 colonization was lower in PG mothers respect to MCG mothers (p value < 0.001). Concurrently, the genome copy number of PRL2010 in the newborns from PG group was lower respect those observed in MCG newborns group (p value < 0.001). These findings suggest that microbe-microbe interactions provide an advantage in the vertical transmission efficiency of these species in the infant gut. In addition, such results corroborate previous data regarding the existence of syntrophic interactions between bifidobacteria in the infant gut [9,11,33].
Figure 2.
Schematic representation of vertical inheritance of B. bifidum strain PRL2010, when administered solely in a rat model (PRL2010 Group—PG). Panel (a) shows the population sizes of B. bifidum PRL2010 present in the intestine of female rats. Each point represents the average of the log-population size ± standard deviation for nine rats. Panel (b) displays the B. bifidum PRL2010 retrieval ± standard deviation for each female rat used. Panel (c) exhibits the presence of B. bifidum PRL2010 in the caecum from puppies. Each pillar represents the average colonization ± standard deviation of puppies for each female rat.
However, the bacterial load of PRL2010 in the MCG newborns group resulted lower respect to their mothers. In contrast, the PG newborns group showed a higher abundance of PRL2010 cells when compared to their mothers (Figure S2). Probably, the load of B. bifidum PRL2010 cells in the newborns of PG group is higher respect to their mothers because it has been administered as a mono-strain supplement. Conversely, the simultaneously supplementation of bifidobacterial mix encompassing different strains in MCG group contributed to the decreased level of PRL2010 cells in the newborns.
2.3. Identification of DNA Belonging to PRL2010 in Different Rat Body Sites
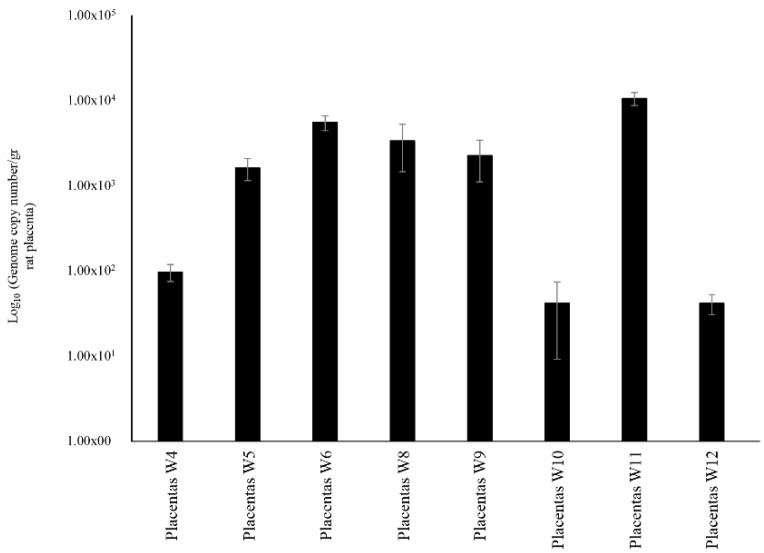
Bifidobacterial DNA occurrence in other body sites of rats such as placenta and blood samples, which was collected from mothers immediately after Caesarian delivery, was investigated by qPCR using PRL2010-specific primers. Remarkably, these experiments revealed the presence of DNA belonging to PRL2010 in the placental tissue but not in blood (Figure 3). Similar results were described previously by Jimenez et al. in a trial examining the vertical transmission of E. faecium HA1 in pregnant mice [35]. Any attempts directed to cultivate PRL2010 cells from placenta samples under mMRS did not result in the isolation of viable cells. These findings suggest that either the rat placenta can only be reached by DNA from lysed PRL2010 cells or that this body compartment only contains dormant PRL2010 cells. These data may also open a novel and intriguing scenario of prenatal colonization of rats by B. bifidum PRL2010.
Figure 3.
Schematic representation of the B. bifidum PRL2010 DNA load observed in placentas’ samples. This graphic displays the level of DNA belonging to B. bifidum PRL2010 present in the placenta of rats. Each pillar represents the average retrieval ± standard deviation of B. bifidum PRL2010 from the placenta of rats.
2.4. ITS Bifidobacterial Profiling of the Caecum of Mothers and Newborns
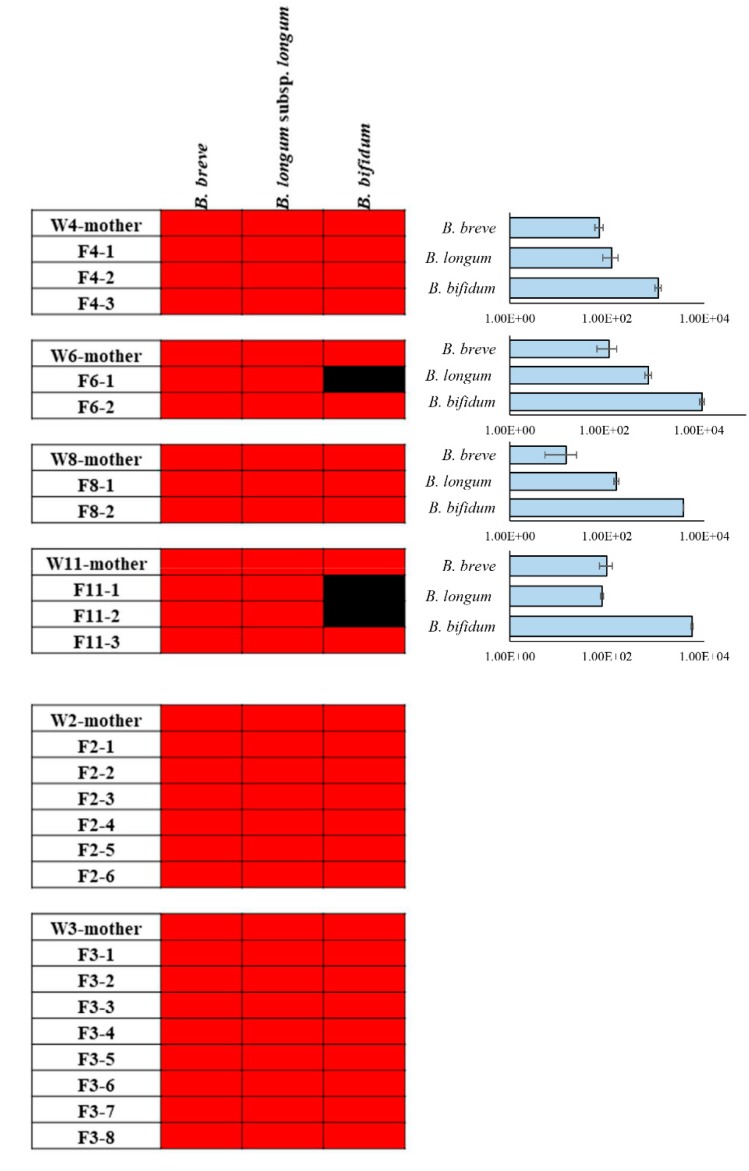
In order to further characterize the bifidobacterial composition of caecal samples from mothers and puppies, ITS bifidobacterial profiling analyses were performed [36] on these 30 samples, producing a total of 212,034 reads with an average of 7068 ± 6457 reads per sample (Table 1). These raw data were processed to identify and classify reads into clusters of identical sequences (OTUs). In detail, we focused our interest on OTUs belonging to the species B. bifidum, B. breve and B. longum subsp. longum. Interestingly, all puppies of the MCG group shared these three OTUs with the corresponding mothers’ samples (Figure 4). Moreover, a comparison between PG mothers with their corresponding puppies showed that in 70% of cases the OTUs belonging to the species B. bifidum, B. breve and B. longum subsp. longum are shared (Figure 4). In order to confirm the presence of B. bifidum, B. breve and B. longum subsp. longum in PG group, we performed a qPCR using species-specific primers using mothers’ caeca (Figure 4). These data revealed that B. breve and B. longum subsp. longum species were present in the gut microbiota of pregnant rats, even if they were not supplemented to the animals (Figure 4).
Table 1.
Filtering table of the analyzed caecal samples.
| Experiment Groups | Samples Name | Input Reads | Final Reads |
|---|---|---|---|
| PG | W4-mother | 3423 | 3126 |
| PG | F4-1 | 2589 | 2386 |
| PG | F4-2 | 8551 | 8012 |
| PG | F4-3 | 4126 | 3776 |
| PG | W6-mother | 2078 | 1970 |
| PG | F6-1 | 2164 | 2036 |
| PG | F6-2 | 5050 | 4852 |
| PG | W8-mother | 1239 | 1201 |
| PG | F8-1 | 6365 | 5797 |
| PG | F8-2 | 4527 | 4050 |
| PG | W11-mother | 1009 | 942 |
| PG | F11-1 | 4106 | 3908 |
| PG | F11-2 | 2133 | 2036 |
| PG | F11-3 | 1692 | 1642 |
| MCG | W2-mother | 22,528 | 22,026 |
| MCG | F2-1 | 12,703 | 11,906 |
| MCG | F2-2 | 32,065 | 29,485 |
| MCG | F2-3 | 11,715 | 10,850 |
| MCG | F2-4 | 17,106 | 15,929 |
| MCG | F2-5 | 13,992 | 12,724 |
| MCG | F2-6 | 10,339 | 9739 |
| MCG | W3-mother | 2686 | 2640 |
| MCG | F3-1 | 4818 | 4620 |
| MCG | F3-2 | 8970 | 8489 |
| MCG | F3-3 | 5346 | 4987 |
| MCG | F3-4 | 5664 | 5373 |
| MCG | F3-5 | 2765 | 2584 |
| MCG | F3-6 | 12,523 | 11,182 |
| MCG | F3-7 | 6129 | 5701 |
| MCG | F3-8 | 8524 | 8065 |
Figure 4.
Heat map of bifidobacterial OTUs belonging to B. bifidum, B. breve and B. longum subsp. longum. Red shading represents presence, and black shading indicates absence. The pillars next to each heat map display the DNA load of these bifidobacterial species in mothers’ caeca of PG group. The x axes represent the genome copy number /gr of cecum samples.
These results further support the notion of transfer of (bifido)bacterial DNA and/or cells between mother and puppies [11,37] and reinforced the earlier finding that co-existence of multiple bifidobacterial species allows a more efficient transmission of (bifido)bacterial DNA and/or cells from mothers to their newborns [9,11,33].
3. Materials and Methods
3.1. Experiment Design and Bifidobacterial Treatment of Rats
Experiments were performed in accordance with the European Community Council Directive 2010/63/UE and approved by the Italian legislation on animal experimentation (D.L. 04/04/2014, n. 26, authorization no. 370/2018-PR). All efforts were made to reduce sample size and minimize animal suffering. For the purposes of the current study we employed adult (4–6 months) Wistar rats (Rattus norvegicus) which were bred in house under standard conditions [38,39]. The timeline of animal treatment procedures is schematically depicted in supplemental Figure S1. The duration of the animal trial was four weeks in total. Mating was allowed during week 1 and for this purpose, 12 adult female rats (Rattus norvegicus) were coupled with 12 male rats. Pairs were kept in different cages in rooms with controlled temperature (22 ± 2 °C) and humidity (60 ± 10%) and maintained in a 12/12 light/dark cycle (light on from 19: 00 to 7: 00 h), with food and water ad libitum. Following the one week mating period, all female rats were separated and kept in individual cages and were every day orally treated with bifidobacterial strains for three weeks, which represented the gestation period. One group of three female rats (which are here referred to as W1, W2 and W3) was orally inoculated with a mix of three different strains, i.e., B. bifidum PRL2010, B. breve 1895B and B. longum subsp. longum 1886B (Mix Colonization Group–MCG). The second group with the remaining nine female rats (W4, W5, W6, W7, W8, W9, W10, W11 and W12) was orally treated with B. bifidum PRL2010 only (PRL2010 Group—PG). All the strains used in this study were previously isolated from infant stool samples [18,19]. In order to evaluate bifidobacterial transfer, fecal samples were collected at five different time points. The first time point was before the oral administration of bifidobacteria (T0). Then, we collected fecal samples at four time points, i.e., at five, eight, 12 and 17 days (T1, T2, T3 and T4) (Figure S1). On the 19th day of bifidobacterial treatment (i.e., on the day of birth, but prior to labor) female animals were anesthetized with isoflurane (2% in 100% oxygen) and Caesarian delivery was performed under a laminar flow hood and all the surgical instruments used were previously sterilized in order to prevent any microbial contamination. Placentas were harvested and caecum and blood samples were collected from dams and pups.
3.2. Bifidobacterium Strains Growth Conditions
All strains used in this study were cultivated in an anaerobic atmosphere (2.99% H2, 17.01% CO2 and 80% N2) in an anaerobic chamber (Concept 400, Ruskin) on De Man-Rogosa-Sharp (MRS) broth (Scharlau Chemie, Barcelona, Spain) supplemented with 0.05% (w/v) L-cysteine hydrochloride (Sigma-Aldrich, Saint Louis, USA) and incubated at 37 °C. Microbial cultures were harvested by centrifugation (3000 rpm for 8 min), washed and resuspended in 500 µL of 2 % (w/v) sucrose solution. The viable count of each inoculum was determined by retrospective plating on MRS.
3.3. DNA Extraction and qPCR
Bacterial DNA extraction from fecal samples was performed following the manufacturer’s protocol of the QIAamp Fast DNA stool Mini Kit (Qiagen Ltd., Strasse, Germany). Furthermore, bifidobacterial DNA presence was evaluated for the mother’s caecum, placentas, blood samples, and from puppies’ caecal samples. Specifically, the bacterial DNA from puppies and mothers’ caecum and from placentas was extracted following the protocol of Power Viral Environmental RNA/DNA Isolation Kit (Mo Bio, Hilden, Germany), whereas the bacterial DNA from mother’s blood was extracted following the protocol of the DNeasy Blood and Tissue Kit (Qiagen Ltd., Strasse, Germany).
Quantitative PCR (qPCR) was performed as described previously [32]. For B. bifidum PRL2010 were used primers Bbif_0282Fw (5′-GCGAACAATGATGGCACCTA-3′) and Bbif_0282Rv (5′-GTCGAACACCACGACGATGT-3′) [24], for B. breve 1895B were used primers BBR7E_0534_fw (5′-AGCGACGATATGATGCAATG-3′) and BBR7E_0534_rev (5′-CGTGAATACGCTGCACAGTC-3′) and for B. longum subsp. longum 1886B were used primers B1886_0443_fw (5′-AAGCCAAGGACATGTTCGAC-3′) and B1886_0443_rev (5′-TGGTGTATCTGGCGTTCTTG-3′) [19]. For species specific qPCR were used following primer pairs: Bbif1 (5′-CCACATGATCGCATGTGATTG-3′) and Bbif2 (5′-CCGAAGGCTTGCTCCCAAA-3′) for B. bifidum species [29], Bbre1 (5′-CCGGATGCTCCATCACAC-3′) and Bbre2 (5′-ACAAAGTGCCTTGCTCCCT-3′) for B.breve species [40], Blon1 (5′-TTCCAGTTGATCGCATGGTC-3′) and Blon2 (5′-GGGAAGCCGTATCTCTACGA-3′) for B. longum.
3.4. Bifidobacterium Strain Isolation From Mothers’ Caecum
The collected caecum of the mothers was homogenized and serial dilutions (1:10) were performed. All dilutions were cultivated on MRS agar (Scharlau Chemie, Barcelona, Spain) supplemented with 0.05% (w/v) L-cysteine hydrochloride (Sigma-Aldrich) and 50 μg/mL mupirocin (Delchimica, Napoli, Italy). After 48h of incubation in an anaerobic atmosphere (2.99% H2, 17.01% CO2 and 80% N2) in a chamber (Concept 400, Ruskin), morphologically distinct colonies were selected and cultivated in MRS broth (Scharlau Chemie, Barcelona, Spain) supplemented with 0.05% (w/v) L-cysteine hydrochloride (Sigma-Aldrich). Subsequently, after overnight growth, bacterial DNA was extracted as described previously [41]. The identification of specific strains was obtained using a PCR approach based on strain-specific primers (see above).
3.5. Bifidobacterial ITS PCR Amplification and Sequencing
Following bacterial DNA extraction from caecal samples of mothers and puppies, partial ITS sequences were amplified using primer pair Probio-bif_Uni/Probio-bif_Rev, which targets the spacer region between the 16S rRNA and 23S rRNA genes within the ribosomal RNA (rRNA) locus [36]. At the same time, we prepared a mock community (Mock Bifidobacterial Community), consisting of a pool of known concentrations of 11 different Bifidobacterium strains prepared by combining equal concentration of bacterial DNA. The DNA from the mix was diluted to produce a final DNA concentration of 2 ng/μL, and 4 μL of these dilutions were used in each PCR reaction using primer pair Probio-bif_Uni/Probio-bif_Rev. Illumina adapter overhang nucleotide sequences were added to the partial ITS amplicons, which were further processed employing the 16S Metagenomic Sequencing Library Preparation Protocol (Part #15044223 Rev. B–Illumina). The library preparation was performed as described above for the 16S rRNA microbial profiling analyses.
Following sequencing, the.fastq files were processed using a custom script based on the QIIME software suite [42]. In order to reconstruct the complete Probio-bif_Uni / Probio-bif_Rev amplicons, the paired-end read pairs were assembled. Quality control retained sequences with a length between 100 and 400 bp and mean sequence quality score of >20, while sequences with homopolymers >7 bp in length and mismatched primers were removed. ITS Operational Taxonomic Units (OTUs) were defined at 100% sequence homology using uclust software [43]. All reads were classified to the lowest possible taxonomic rank using QIIME2 [42,44] and a reference dataset, consisting of an updated version of the bifidobacterial ITS database [36].
3.6. Data Deposition
Raw sequences of the bifidobacterial ITS profiling experiments are accessible through SRA study accession numbers PRJNA556137.
4. Conclusions
This study was aimed to investigate the DNA bifidobacterial transmission from female rats to their corresponding pups through a vertical route. Notably, the efficiency/yield of these DNA transmission events may be influenced by the co-presence of different bifidobacterial strains. The above results confirm previous genus-level overviews [37] and underpin the notion that bifidobacterial transmission is characterized by the development of extensive microbe-microbe co-operation [7,11,33]. In fact, our data suggest that bifidobacterial communities have established co-operative behavior between co-colonizers, acting as evolutionary drivers in the mammalian gut microbiota. This assumption is further reinforced by previous studies that have described the occurrence of cross-feeding between specific bifidobacterial species [7,33]. In addition, our findings have provided preliminary insights about the existence of possible bifidobacterial colonization at the preterm level. Nevertheless, the lack of any data about the viability of PRL2010 cells cannot fully support the existence of fetal colonization of bifidobacteria, and further experiments will be needed in order to investigate this intriguing route of bacterial colonization.
Acknowledgments
Part of this research is conducted using the High Performance Computing (HPC) facility of the University of Parma. We thank GenProbio srl for the financial support of the Laboratory of Probiogenomics.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/2076-2607/7/9/293/s1.
Author Contributions
Conceptualization, F.T., M.V. and D.v.S.; Methodology and Formal Analysis, W.M., S.D., L.M., G.L., R.A., L.C., R.S., C.M., G.A.L.; Writing—Original Draft Preparation, W.M., S.D., F.T.; Writing—Review & Editing, M.V., D.v.S. and A.S.
Funding
This work was funded by the EU Joint Programming Initiative—A Healthy Diet for a Healthy Life (JPI HDHL, http://www.healthydietforhealthylife.eu/) to DvS (in conjunction with Science Foundation Ireland (SFI), Grant number 15/JP-HDHL/3280) and to MV (in conjunction with MIUR, Italy). The study is supported by Fondazione Cariparma, under TeachInParma Project. DvS is a member of The APC Microbiome Institute funded by Science Foundation Ireland (SFI), through the Irish Government’s National Development Plan (Grant number SFI/12/RC/2273-P1 and SFI/12/RC/2273-P2).
Conflicts of Interest
The authors declare that they have no competing interests.
References
- 1.Ventura M., Turroni F., Motherway M.O., MacSharry J., van Sinderen D. Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends Microbiol. 2012;20:467–476. doi: 10.1016/j.tim.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Turroni F., Milani C., Duranti S., Ferrario C., Lugli G.A., Mancabelli L., van Sinderen D., Ventura M. Bifidobacteria and the infant gut: An example of co-evolution and natural selection. Cell. Mol. Life Sci. 2018;75:103–118. doi: 10.1007/s00018-017-2672-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steed H., Macfarlane G.T., Blackett K.L., Macfarlane S., Miller M.H., Bahrami B., Dillon J.F. Bacterial translocation in cirrhosis is not caused by an abnormal small bowel gut microbiota. FEMS Immunol. Med. Microbiol. 2011;63:346–354. doi: 10.1111/j.1574-695X.2011.00857.x. [DOI] [PubMed] [Google Scholar]
- 4.Vaishampayan P.A., Kuehl J.V., Froula J.L., Morgan J.L., Ochman H., Francino M.P. Comparative metagenomics and population dynamics of the gut microbiota in mother and infant. Genome Biol. Evol. 2010;2:53–66. doi: 10.1093/gbe/evp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makino H., Kushiro A., Ishikawa E., Kubota H., Gawad A., Sakai T., Oishi K., Martin R., Ben-Amor K., Knol J., et al. Mother-to-infant transmission of intestinal bifidobacterial strains has an impact on the early development of vaginally delivered infant’s microbiota. PLoS ONE. 2013;8:e78331. doi: 10.1371/journal.pone.0078331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makino H., Kushiro A., Ishikawa E., Muylaert D., Kubota H., Sakai T., Oishi K., Martin R., Ben Amor K., Oozeer R., et al. Transmission of intestinal bifidobacterium longum subsp. Longum strains from mother to infant, determined by multilocus sequencing typing and amplified fragment length polymorphism. Appl. Environ. Microbiol. 2011;77:6788–6793. doi: 10.1128/AEM.05346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milani C., Mancabelli L., Lugli G.A., Duranti S., Turroni F., Ferrario C., Mangifesta M., Viappiani A., Ferretti P., Gorfer V., et al. Exploring vertical transmission of bifidobacteria from mother to child. Appl. Environ. Microbiol. 2015;81:7078–7087. doi: 10.1128/AEM.02037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duranti S., Milani C., Lugli G.A., Turroni F., Mancabelli L., Sanchez B., Ferrario C., Viappiani A., Mangifesta M., Mancino W., et al. Insights from genomes of representatives of the human gut commensal bifidobacterium bifidum. Environ. Microbiol. 2015;17:2515–2531. doi: 10.1111/1462-2920.12743. [DOI] [PubMed] [Google Scholar]
- 9.Duranti S., Lugli G.A., Milani C., James K., Mancabelli L., Turroni F., Alessandri G., Mangifesta M., Mancino W., Ossiprandi M.C., et al. Bifidobacterium bifidum and the infant gut microbiota: An intriguing case of microbe-host co-evolution. Environ. Microbiol. 2019 doi: 10.1111/1462-2920.14705. [DOI] [PubMed] [Google Scholar]
- 10.Milani C., Turroni F., Duranti S., Lugli G.A., Mancabelli L., Ferrario C., van Sinderen D., Ventura M. Genomics of the genus bifidobacterium reveals species-specific adaptation to the glycan-rich gut environment. Appl. Environ. Microbiol. 2016;82:980–991. doi: 10.1128/AEM.03500-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milani C., Mangifesta M., Mancabelli L., Lugli G.A., James K., Duranti S., Turroni F., Ferrario C., Ossiprandi M.C., van Sinderen D., et al. Unveiling bifidobacterial biogeography across the mammalian branch of the tree of life. ISME J. 2017;11:2834–2847. doi: 10.1038/ismej.2017.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moossavi S., Sepehri S., Robertson B., Bode L., Goruk S., Field C.J., Lix L.M., de Souza R.J., Becker A.B., Mandhane P.J., et al. Composition and variation of the human milk microbiota are influenced by maternal and early-life factors. Cell Host Microbe. 2019;25:324–335. doi: 10.1016/j.chom.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 13.Aagaard K., Ma J., Antony K.M., Ganu R., Petrosino J., Versalovic J. The placenta harbors a unique microbiome. Sci. Transl. Med. 2014;6:237ra65. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perez-Munoz M.E., Arrieta M.C., Ramer-Tait A.E., Walter J. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: Implications for research on the pioneer infant microbiome. Microbiome. 2017;5:48. doi: 10.1186/s40168-017-0268-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez J.M., Murphy K., Stanton C., Ross R.P., Kober O.I., Juge N., Avershina E., Rudi K., Narbad A., Jenmalm M.C., et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb. Ecol. Health Dis. 2015;26:26050. doi: 10.3402/mehd.v26.26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jimenez E., Marin M.L., Martin R., Odriozola J.M., Olivares M., Xaus J., Fernandez L., Rodriguez J.M. Is meconium from healthy newborns actually sterile? Res. Microbiol. 2008;159:187–193. doi: 10.1016/j.resmic.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 17.Walker R.W., Clemente J.C., Peter I., Loos R.J.F. The prenatal gut microbiome: Are we colonized with bacteria in utero? Pediatric Obes. 2017;12:3–17. doi: 10.1111/ijpo.12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turroni F., Bottacini F., Foroni E., Mulder I., Kim J.H., Zomer A., Sanchez B., Bidossi A., Ferrarini A., Giubellini V., et al. Genome analysis of bifidobacterium bifidum prl2010 reveals metabolic pathways for host-derived glycan foraging. Proc. Natl. Acad. Sci. USA. 2010;107:19514–19519. doi: 10.1073/pnas.1011100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duranti S., Lugli G.A., Mancabelli L., Armanini F., Turroni F., James K., Ferretti P., Gorfer V., Ferrario C., Milani C., et al. Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome. 2017;5:66. doi: 10.1186/s40168-017-0282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duranti S., Mancabelli L., Mancino W., Anzalone R., Longhi G., Statello R., Carnevali L., Sgoifo A., Bernasconi S., Turroni F., et al. Exploring the effects of colostrononi on the mammalian gut microbiota composition. PLoS ONE. 2019;14:e0217609. doi: 10.1371/journal.pone.0217609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duranti S., Gaiani F., Mancabelli L., Milani C., Grandi A., Bolchi A., Santoni A., Lugli G.A., Ferrario C., Mangifesta M., et al. Elucidating the gut microbiome of ulcerative colitis: Bifidobacteria as novel microbial biomarkers. FEMS Microbiol. Ecol. 2016;92:fiw191. doi: 10.1093/femsec/fiw191. [DOI] [PubMed] [Google Scholar]
- 22.Egan M., Motherway M.O., Kilcoyne M., Kane M., Joshi L., Ventura M., van Sinderen D. Cross-feeding by bifidobacterium breve ucc2003 during co-cultivation with bifidobacterium bifidum prl2010 in a mucin-based medium. BMC Microbiol. 2014;14:282. doi: 10.1186/s12866-014-0282-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferrario C., Duranti S., Milani C., Mancabelli L., Lugli G.A., Turroni F., Mangifesta M., Viappiani A., Ossiprandi M.C., van Sinderen D., et al. Exploring amino acid auxotrophy in bifidobacterium bifidum prl2010. Front. Microbiol. 2015;6:1331. doi: 10.3389/fmicb.2015.01331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turroni F., Serafini F., Foroni E., Duranti S., O’Connell Motherway M., Taverniti V., Mangifesta M., Milani C., Viappiani A., Roversi T., et al. Role of sortase-dependent pili of bifidobacterium bifidum prl2010 in modulating bacterium-host interactions. Proc. Natl. Acad. Sci. USA. 2013;110:11151–11156. doi: 10.1073/pnas.1303897110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Serafini F., Strati F., Ruas-Madiedo P., Turroni F., Foroni E., Duranti S., Milano F., Perotti A., Viappiani A., Guglielmetti S., et al. Evaluation of adhesion properties and antibacterial activities of the infant gut commensal bifidobacterium bifidum prl2010. Anaerobe. 2013;21:9–17. doi: 10.1016/j.anaerobe.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Serafini F., Turroni F., Ruas-Madiedo P., Lugli G.A., Milani C., Duranti S., Zamboni N., Bottacini F., van Sinderen D., Margolles A., et al. Kefir fermented milk and kefiran promote growth of bifidobacterium bifidum prl2010 and modulate its gene expression. Int. J. Food Microbiol. 2014;178:50–59. doi: 10.1016/j.ijfoodmicro.2014.02.024. [DOI] [PubMed] [Google Scholar]
- 27.Turroni F., Taverniti V., Ruas-Madiedo P., Duranti S., Guglielmetti S., Lugli G.A., Gioiosa L., Palanza P., Margolles A., van Sinderen D., et al. Bifidobacterium bifidum prl2010 modulates the host innate immune response. Appl. Environ. Microbiol. 2014;80:730–740. doi: 10.1128/AEM.03313-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turroni F., Foroni E., Montanini B., Viappiani A., Strati F., Duranti S., Ferrarini A., Delledonne M., van Sinderen D., Ventura M. Global genome transcription profiling of bifidobacterium bifidum prl2010 under in vitro conditions and identification of reference genes for quantitative real-time pcr. Appl. Environ. Microbiol. 2011;77:8578–8587. doi: 10.1128/AEM.06352-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turroni F., Strati F., Foroni E., Serafini F., Duranti S., van Sinderen D., Ventura M. Analysis of predicted carbohydrate transport systems encoded by bifidobacterium bifidum prl2010. Appl. Environ. Microbiol. 2012;78:5002–5012. doi: 10.1128/AEM.00629-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turroni F., Duranti S., Bottacini F., Guglielmetti S., Van Sinderen D., Ventura M. Bifidobacterium bifidum as an example of a specialized human gut commensal. Front. Microbiol. 2014;5:437. doi: 10.3389/fmicb.2014.00437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turroni F., Serafini F., Mangifesta M., Arioli S., Mora D., van Sinderen D., Ventura M. Expression of sortase-dependent pili of bifidobacterium bifidum prl2010 in response to environmental gut conditions. FEMS Microbiol. Lett. 2014;357:23–33. doi: 10.1111/1574-6968.12509. [DOI] [PubMed] [Google Scholar]
- 32.Turroni F., Ozcan E., Milani C., Mancabelli L., Viappiani A., van Sinderen D., Sela D.A., Ventura M. Glycan cross-feeding activities between bifidobacteria under in vitro conditions. Front. Microbiol. 2015;6:1030. doi: 10.3389/fmicb.2015.01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turroni F., Milani C., Duranti S., Mancabelli L., Mangifesta M., Viappiani A., Lugli G.A., Ferrario C., Gioiosa L., Ferrarini A., et al. Deciphering bifidobacterial-mediated metabolic interactions and their impact on gut microbiota by a multi-omics approach. ISME J. 2016;10:1656–1668. doi: 10.1038/ismej.2015.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turroni F., Milani C., van Sinderen D., Ventura M. Genetic strategies for mucin metabolism in bifidobacterium bifidum prl2010: An example of possible human-microbe co-evolution. Gut Microbes. 2011;2:183–189. doi: 10.4161/gmic.2.3.16105. [DOI] [PubMed] [Google Scholar]
- 35.Jimenez E., Fernandez L., Marin M.L., Martin R., Odriozola J.M., Nueno-Palop C., Narbad A., Olivares M., Xaus J., Rodriguez J.M. Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Curr. Microbiol. 2005;51:270–274. doi: 10.1007/s00284-005-0020-3. [DOI] [PubMed] [Google Scholar]
- 36.Milani C., Lugli G.A., Turroni F., Mancabelli L., Duranti S., Viappiani A., Mangifesta M., Segata N., van Sinderen D., Ventura M. Evaluation of bifidobacterial community composition in the human gut by means of a targeted amplicon sequencing (its) protocol. FEMS Ecol. 2014;90:493–503. doi: 10.1111/1574-6941.12410. [DOI] [PubMed] [Google Scholar]
- 37.Ley R.E., Hamady M., Lozupone C., Turnbaugh P.J., Ramey R.R., Bircher J.S., Schlegel M.L., Tucker T.A., Schrenzel M.D., Knight R., et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolfensohn S., Lloyd M. Handbook of Laboratory Animal Management and Welfare. 3rd ed. Volume 1. Oxford University Press; London, UK: 2003. p. 416. [Google Scholar]
- 39.Carnevali L., Montano N., Statello R., Coude G., Vacondio F., Rivara S., Ferrari P.F., Sgoifo A. Social stress contagion in rats: Behavioural, autonomic and neuroendocrine correlates. Psychoneuroendocrinology. 2017;82:155–163. doi: 10.1016/j.psyneuen.2017.05.017. [DOI] [PubMed] [Google Scholar]
- 40.Alessandri G., Milani C., Duranti S., Mancabelli L., Ranjanoro T., Modica S., Carnevali L., Statello R., Bottacini F., Turroni F., et al. Ability of bifidobacteria to metabolize chitin-glucan and its impact on the gut microbiota. Sci. Rep. 2019;9:5755. doi: 10.1038/s41598-019-42257-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ventura M., Elli M., Reniero R., Zink R. Molecular microbial analysis of bifidobacterium isolates from different environments by the species-specific amplified ribosomal DNA restriction analysis (ardra) FEMS Microbiol. Ecol. 2001;36:113–121. doi: 10.1111/j.1574-6941.2001.tb00831.x. [DOI] [PubMed] [Google Scholar]
- 42.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pena A.G., Goodrich J.K., Gordon J.I., et al. Qiime allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edgar R.C. Search and clustering orders of magnitude faster than blast. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 44.Bokulich N.A., Kaehler B.D., Rideout J.R., Dillon M., Bolyen E., Knight R., Huttley G.A., Gregory Caporaso J. Optimizing taxonomic classification of marker-gene amplicon sequences with qiime 2’s q2-feature-classifier plugin. Microbiome. 2018;6:90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.