Abstract
Heteroresistance may pose a threat to the prognosis of patients following colistin treatment. We investigated colistin heteroresistance in Klebsiella pneumoniae isolates from South Korea. Among 252 K. pneumoniae blood isolates, 231 were susceptible to polymyxins. Heteroresistance to colistin was determined using population analysis profiles, disk diffusion assays, and E-test strip tests for the susceptible isolates. As a result, we identified three colistin-heteroresistant K. pneumoniae isolates belonging to separate clones (ST11, ST461, and ST3217) by multilocus sequence typing analysis. Two colistin-resistant subpopulations were selected from each heteroresistant isolate in either disk diffusion testing or E-testing. Two resistant subpopulations from the same isolate exhibited different amino acid substitutions in the two-component regulatory systems PmrAB and PhoPQ. An in vitro time–kill assay showed that meropenem combined with colistin had a 1× minimum inhibitory concentration bactericidal effect against a multidrug-resistant, colistin-heteroresistant isolate.
Keywords: Klebsiella pneumoniae, colistin, heteroresistance, population analysis profiles, PmrAB, PhoPQ
1. Introduction
Klebsiella pneumoniae is one of the most clinically significant pathogens belonging to the family Enterobacteriaceae. It is an important pathogen in community- and hospital-acquired infections [1]. Carbapenems have been used for infections caused by extended-spectrum β-lactamase (ESBL)-producing K. pneumoniae, and carbapenem-resistant K. pneumoniae first emerged in 1985 [2]. The prevalence of carbapenem-resistant K. pneumoniae has continued to increase globally, and such infections pose a critical threat to human health, with high mortality rates owing to the limited treatment options available [3,4,5]. Colistin and polymyxin B are important therapeutic options for treating infections caused by carbapenem-resistant K. pneumoniae [6,7].
Colistin exerts bactericidal action against Gram-negative pathogens, targeting the lipid A moiety of lipopolysaccharide (LPS) and leading to cell membrane disruption [8]. Unfortunately, colistin resistance has been reported in surveillance studies as well as in clinical case reports [9]. Resistance to colistin generally involves mutations in chromosomal genes. Acquired resistance to polymyxins in strains such as K. pneumoniae and Escherichia coli involves mutations in the two-component regulatory systems PmrAB and PhoPQ or alterations to the negative regulator of PhoPQ, MgrB [10,11].
Antibiotic heteroresistance is a phenomenon where subpopulations of seemingly isogenic bacteria exhibit a range of susceptibilities to a particular antibiotic [12]. It has only been reported in a limited number of studies because it cannot be assessed using ordinary minimum inhibitory concentration (MIC) testing methods. There have been many studies on the heteroresistance to various antibiotics for specific bacterial species, which also encompass colistin heteroresistance in K. pneumoniae isolates [13,14,15,16,17,18]. Although little is known about the clinical significance of antibiotic heteroresistance, instances of treatment failure associated with colistin heteroresistance in K. pneumoniae have been reported [19]. In addition, clinicians may be prompted to investigate the most efficient use of colistin on multidrug-resistant K. pneumoniae, including antibiotic combinations [20].
In this study, we investigated the incidence rate and genomic variation of colistin heteroresistance in K. pneumoniae blood isolates and examined the in vitro efficacy of colistin and meropenem combination treatment against colistin-heteroresistant K. pneumoniae.
2. Materials and Methods
2.1. Bacterial Strains and Antibiotic Susceptibility Testing
In total, 252 nonduplicated K. pneumoniae blood isolates were collected from January to December 2017 from Samsung Medical Center (Seoul, Korea). Species identification was performed using a VITEK-2 system (BioMérieux, Hazelwood, MO, USA).
In vitro antimicrobial susceptibility testing was performed using the broth microdilution method outlined in the Clinical and Laboratory Standards Institute (CLSI) guidelines [21]. For all isolates, the MICs of four antibiotic agents—imipenem and meropenem (carbapenems) as well as colistin and polymyxin B (polymyxins)—were determined. The MICs of seven other antibiotics (cefotaxime, ceftazidime, cefepime, ciprofloxacin, amikacin, tigecycline, and piperacillin–tazobactam) were also determined for three colistin-heteroresistant isolates and their resistant subpopulations. Most of the antibiotics, except tigecycline, were purchased from Sigma-Aldrich Corp. (St. Louis, MO, USA), and tigecycline (Tygacil® Injection) was provided from Pfizer (Korea). E. coli ATCC 25922 and Pseudomonas aeruginosa ATCC27853 were employed as quality control strains.
2.2. Detection of Colistin-Heteroresistant Isolates
Colistin heteroresistance was detected using a colistin disk diffusion assay (BD BBL™ Sensi-Disc™ antimicrobial susceptibility test disks, colistin 10 μg) or a colistin E-test strip (bioMérieux SA, France) on Mueller–Hinton agar (Difco BBL, USA) plates, where colonies were observed within the clear zone of inhibition. Subpopulations were separated by subculture, and their colistin MIC was assessed by the broth microdilution method and interpreted according to CLSI guidelines [21]. To confirm the presence of colistin heteroresistance, population analysis profiles (PAPs) were obtained. Full 24 h cultures (~108 CFU/mL) were employed. Bacterial cell suspension samples (50 μL) (corresponding to a 0.5 McFarland standard for K. pneumoniae cultures) were plated on Mueller–Hinton agar plates containing 0, 0.5, 1, 2, 4, 6, 8, or 10 mg/L of colistin sulfate (Sigma-Aldrich, St. Louis, MO, USA). After 24 h of incubation at 37 °C, the number of colonies were counted. Colistin heteroresistance was defined as the presence of a colistin-susceptible isolate in which the detectable colistin-resistant subpopulations were able to grow in the presence of ≥10 mg/L of colistin sulfate. The detection limit of colistin-resistant subpopulations was 20 CFU/mL and the lower limit of quantification (LOQ) was 400 CFU/mL (i.e., 2.6 log10 CFU/mL) [16].
2.3. Genotyping and Sequence Analysis of Genes Associated with Colistin Resistance
Multilocus sequence typing (MLST) was performed for three heteroresistant isolates and their resistant subpopulations using a previously described protocol (www.pasteur.fr/recherche/genopole/PF8/mlstKpneumoniae.html) [22]. Genomic DNAs were isolated from overnight cultures in Luria–Bertani agar at 37 °C using the G-spin™ genomic DNA extraction kit for bacteria G-spin™ Genomic DNA Extraction Mini Kit (for Bacteria)G-spin™ Genomic DNA Extraction Mini Kit (for Bacteria) (iNtRON Biotechnology, Korea).
Polymerase chain reaction (PCR) and DNA sequencing were performed to identify nucleotide and resultant amino acid alterations in PhoPQ, PmrAB, and MgrB of parental colistin-heteroresistant isolates and their resistant subpopulations [23]. The presence of the mcr-1 gene was investigated by PCR [24].
2.4. Time–Kill Assays
We examined the time–kill kinetics of colistin and/or meropenem against a colistin-heteroresistant K. pneumoniae blood isolate (S1703-112), which is also multidrug-resistant. Colistin was added to a logarithmic-phase broth culture of approximately 106 CFU/mL to yield concentrations that were 0-, 0.25-, 1-, and 4-fold of the MIC. Samples were collected at 0, 4, 8, 12, 16, 20, and 24 h after adding antibiotics, and a viable cell count was performed by spirally plating the bacterial cell suspension on Mueller–Hinton agar plates after appropriate dilutions. Time–kill curves were constructed by plotting mean colony counts (log10 CFU/mL) versus time. Bactericidal activity was defined as a ≥3 log10 CFU/mL reduction in the total CFU/mL from the original inoculum [25]. Synergy was defined as a ≥2 log10 CFU/mL decrease between the combination and the most efficient agent alone at 24 h [26].
3. Results
Among 252 K. pneumoniae blood isolates, 13 and 12 isolates (5.1% and 4.7%) were resistant to colistin and polymyxin B (MICs, >4 mg/L), respectively. Eight and nine isolates (3.2% and 3.6%) showed intermediate resistance toward colistin and polymyxin B. The others (231 isolates, 91.7%) were susceptible to both colistin and polymyxin B. Only three and one isolates were resistant to meropenem and imipenem, respectively, whereas nine and three isolates exhibited intermediate resistance. The others were susceptible to meropenem and imipenem (240 and 248; 95.2% and 98.4%, respectively).
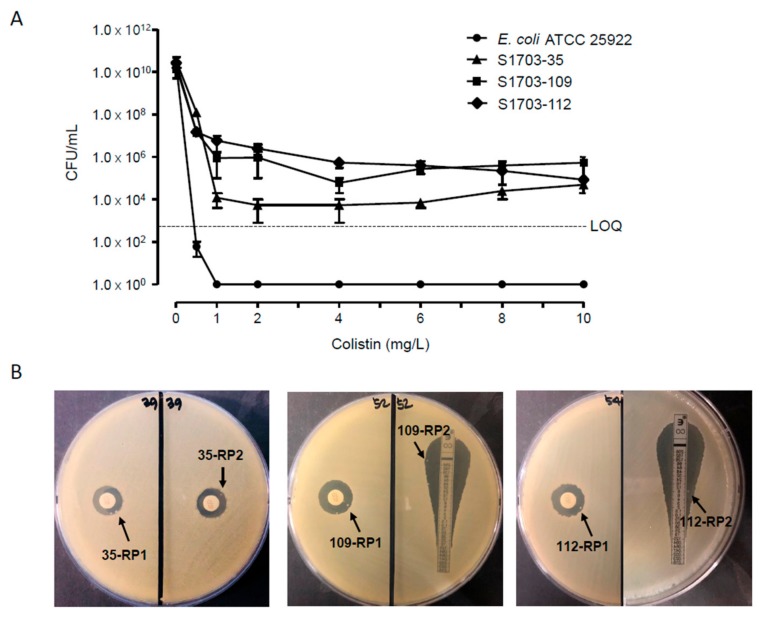
We assayed colistin heteroresistance for 231 susceptible K. pneumoniae isolates. As a result, we identified three isolates (1.3%) being heteroresistant to colistin using a disk diffusion test or E-test. They showed typical bactericidal patterns in population analysis profiling (Figure 1A). For each isolate, we obtained two colonies growing within the zone of inhibition in disk diffusion or E-test (Figure 1B). They were separated by subculture and were named RP1 and RP2 after the isolate number. All the colistin-resistant subpopulations of the three heteroresistant isolates showed colistin MICs of ≥64 μg/mL, whereas the colistin MICs of parental K. pneumoniae isolates were 0.25 or 1 mg/L (Table 1). The colistin MICs of ≥64 μg/mL in the resistant subpopulations persevered after serial subculture in colistin-free media, indicating their stable feature of colistin resistance. The MICs of the other antibiotics tested in this study were not significantly different, except for cefepime and tigecycline in some resistant subpopulations (Table 1). Particularly, the isolate S1703-112 was nonsusceptible to most antibiotics except gentamicin and tigecycline. Thus, we selected this isolate for time–kill assays to investigate the efficacy of a combination of meropenem and colistin. The isolate S1703-112 produced CTX-M-15, an ESBL. According to MLST analysis, the three colistin-heteroresistant K. pneumoniae blood isolates belonged to different clones—ST3217 (S1703-35), ST461 (S1703-109), and ST11 (S1703-112)—which were clones not be strictly associated with colistin resistance. The resistant subpopulations showed the same STs as those of their parental isolates. All isolates were negative for the mcr-1 gene.
Figure 1.
(A) Population analysis profiles of three colistin-heteroresistant Klebsiella pneumoniae blood isolates and Escherichia coli ATCC 25922. LOQ, limit of quantification. The three isolates—S1703-35, S1703-109, and S1703-112—grew in the presence of colistin at concentrations of 4–10 mg/L. (B) The results of disk diffusion test or E-test. Resistant subpopulations (each two in three isolates) analyzed further are indicated; 35-RP, 109-RP, and 112-RP indicate the resistant subpopulations of S1703-35, S1703-109, and S1703-112, respectively. For S1703-35, no resistant colonies were detected in the E-test; thus, we selected resistant colonies in independent disk diffusion tests.
Table 1.
Antibiotic susceptibility against three colistin-heteroresistant K. pneumoniae isolates and their resistant populations.
| Antibiotics | MIC (mg/L) a, b | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| S1703-35 | S1703-109 | S1703-112 | |||||||
| P | RP1 | RP2 | P | RP1 | RP2 | P | RP1 | RP2 | |
| Colistin | 1 (S) | 128 (R) | 64 (R) | 0.25 (S) | 64 (R) | 64 (R) | 1 (S) | 256 (R) | 128 (R) |
| Polymyxin B | 1 (S) | 64 (R) | 32 (R) | 0.25 (S) | 64 (R) | 32 (R) | 1 (S) | 64 (R) | 64 (R) |
| Meropenem | 0.06 (S) | 0.125 (S) | 0.125 (S) | 0.06 (S) | 0.06 (S) | 0.06 (S) | 4 (R) | 4 (R) | 2 (I) |
| Imipenem | 1 (S) | 1 (S) | 0.5 (S) | 0.25 (S) | 0.25 (S) | 0.25 (S) | 2 (I) | 1 (S) | 1 (S) |
| Cefotaxime | 0.125 (S) | 0.25 (S) | 0.25 (S) | 0.25 (S) | 0.25 (S) | 0.125 (S) | >128 (R) | >128 (R) | >128 (R) |
| Ceftazidime | 0.5 (S) | 1 (S) | 1 (S) | 1 (S) | 1 (S) | 1 (S) | >64 (R) | >64 (R) | >64 (R) |
| Cefepime | 0.25 (S) | 1 (S) | 1 (S) | 0.125 (S) | 1 (S) | 1 (S) | >64 (R) | >64 (R) | >64 (R) |
| Amikacin | 4 (S) | 4 (S) | 4 (S) | 2 (S) | 2 (S) | 2 (S) | 32 (I) | 32 (I) | 32 (I) |
| Gentamicin | 1 (S) | 1 (S) | 1 (S) | 0.5 (S) | 0.5 (S) | 0.5 (S) | 2 (S) | 2 (S) | 1 (S) |
| Ciprofloxacin | 0.25 (S) | 0.25 (S) | 0.25 (S) | 0.06 (S) | 0.06 (S) | 0.06 (S) | >64 (R) | >64 (R) | >64 (R) |
| Aztreonam | 0.125 (S) | 0.125 (S) | 0.125 (S) | 0.125 (S) | 0.125 (S) | 0.125 (S) | >64 (R) | >64 (R) | >64 (R) |
| Tigecycline | 2 (S) | 1 (S) | 1 (S) | 2 (S) | 0.5 (S) | 0.5 (S) | 1 (S) | 1 (S) | 1 (S) |
| Piperacillin–tazobactam | 16/4 (S) | 8/4 (S) | 8/4 (S) | 8/4 (S) | 8/4 (S) | 8/4 (S) | >256/4 (R) | >256/4 (R) | >256/4 (R) |
a MIC, minimal inhibitory concentration; P, parental; RP, resistant population; S, susceptible; I, intermediate; R, resistant. b Data are underlined when the MIC increased more than 2-fold in the RP compared with the parental isolate (P).
We investigated the amino acid alterations of the two-component regulatory systems PmrAB and PhoPQ, which are known to be associated with colistin resistance in K. pneumoniae (Table 2). We identified amino acid variations in 18 sites, where 11 were likely not associated with colistin resistance because the amino acids in the resistant subpopulations could be found in other parental isolates. As a result, it was assumed that seven amino acid substitutions may be associated with colistin resistance in resistant subpopulations: two in PmrA, one in PmrB, two in PhoP, and two in PhoQ. Of note, two resistant subpopulations from the same parental isolate did not show amino acid variations in PmrAB and PhoPQ. Two variations in PhoP (Arg198His and Lys199Asn) and one in PhoQ (Leu414Agr) were identified in S1703-35-RP1 but not in S1703-35-RP2. Further, Asp152Asn in PhoQ was identified only in S1703-35-RP2. For resistant subpopulations of S1703-109, Ile178Phe in PmrA and Asp150Asn in PmrB were found in different resistant subpopulations. In addition, Leu414Agr in PhoQ was identified in S1703-112-RP2 but not in S1703-112-RP1. No changes were found in MgrB.
Table 2.
Amino acid substitutions in PmrA, PmrB, PhoP, and PhoQ in three colistin-heteroresistant K. pneumoniae isolates and their resistant subpopulations.
| Isolatea | Amino Acid Substitutions in: | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PmrA | PmrB | PhoP | PhoQ | ||||||||||||||||
| 178 | 203 | 43 | 150 | 163 | 185 | 186 | 198 | 199 | 216 | 152 | 154 | 359 | 414 | 421 | 423 | 429 | 430 | ||
| S1703-35 | P | Iso | Arg | Glu | Asp | Arg | Arg | Lys | Arg | Lys | Gln | Asp | Lys | Arg | Leu | Asp | Ala | Val | Phe |
| RP1 | Lys | Asn | Leu | Glu | His | Asn | Lys | Arg | Pro | Ala | Val | ||||||||
| RP2 | Lys | Asn | Glu | Asn | Lys | Pro | Ala | Val | |||||||||||
| S1703-109 | P | Ile | Gly | Arg | Asp | Cys | Thr | Gly | Gly | Cys | Gly | Asp | Ser | Lys | Leu | Gly | Pro | Ala | Val |
| RP1 | Phe | Glu | |||||||||||||||||
| RP2 | Glu | Asn | |||||||||||||||||
| S1703-112 | P | Ile | Arg | Pro | Leu | Glu | Arg | Lys | Arg | Asp | Gln | Lys | Leu | Gly | Pro | Ala | Val | ||
| RP1 | Lys | Arg | Arg | Gln | Lys | Arg | |||||||||||||
| RP2 | Lys | Arg | Arg | Gln | Lys | Arg | Arg | ||||||||||||
a P, parental; RP, resistant population. b Amino acid alterations that are supposed to be associated with colistin resistance are indicated as white letters with a grey background.
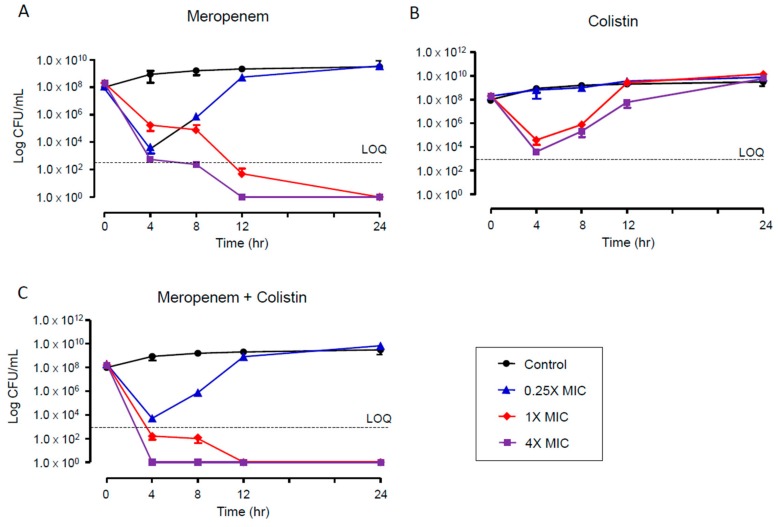
The time–kill assays were performed for the multidrug-resistant and colistin-heteroresistant K. pneumoniae isolate S1703-112. While 4- and 1-fold MICs of meropenem showed complete killing efficacy after 12 and 24 h, respectively (Figure 2A), colistin did not eradicate the colistin-heteroresistant isolate even at 4× MIC (Figure 2B). Although the combination of 0.25× MICs of meropenem and colistin did not kill the heteroresistant isolate, the combination of 1× and 4× MICs demonstrated a rapid killing effect compared with a single regimen of meropenem (Figure 2C).
Figure 2.
Time–kill curves for meropenem (A), colistin (B), and combination of meropenem and colistin (C) against a colistin-heteroresistant K. pneumoniae isolate (S1703-112) that is multidrug-resistant.
4. Discussion
Heteroresistance has been recognized in both Gram-positive and -negative bacteria and is a phenomenon in which a subpopulation of seemingly isogenic bacteria exhibits a range of susceptibilities to a particular antibiotic [16]. Heteroresistance may have an effect on the outcome of clinical infection, particularly because of limitations in detection by routine microbiological susceptibility testing [12]. This study showed that heteroresistance among apparently susceptible isolates forms a reservoir for the emergence of colistin resistance during treatment.
In this study, only a few K. pneumoniae isolates were heteroresistant to colistin. They were clonally unrelated to each other. The rate of colistin heteroresistance found here was lower than that in a previous study [13], in which it was reported that 12 among the 16 colistin-susceptible, carbapenemase-producing K. pneumoniae isolates from Greece were heteroresistant to colistin. The rates of colistin heteroresistance vary according to locality, isolation source, treatment of colistin, and so forth. In addition, undetected colistin heteroresistance has been reported in K. pneumoniae [19,27], suggesting the possibility that the rate of colistin heteroresistance may be higher than that identified in this study.
We identified amino acid alterations that are supposed to be associated with colistin resistance in resistant subpopulations, but it was not known if the genetic changes were induced by colistin treatment. The amino acid alterations have not been previously reported, and it is not known if the changes affect the function of PmrAB or PhoPQ. Of note, two resistant subpopulations from the same isolate showed different amino acid substitutions in the two-component regulatory systems PmrAB and PhoPQ. To our knowledge, variations between colistin-resistant subpopulations have not been reported thus far. However, diverse genetic variations between colistin-resistant K. pneumoniae mutants derived from the same parental strain after treatment have been reported [23]. Our results may indicate that diverse subpopulations with resistance to colistin coexist in the heteroresistant or susceptible isolates, which may develop into resistant strains with diverse mutations associated with colistin resistance.
The combination of meropenem and colistin has been suggested to treat multidrug-resistant K. pneumoniae infections [28]. The results of our time–kill assays showed that monotherapy with colistin may be problematic for the treatment of infections caused by colistin-heteroresistant K. pneumoniae. Although meropenem alone was effective at killing the heteroresistant isolate, the combination of meropenem and colistin allowed rapid eradication at 1× MICs. Meropenem combined with colistin at the appropriate dosage intervals might be a therapeutic option for infections caused by colistin-heteroresistant K. pneumoniae. However, the effectiveness of the combination should be investigated for carbapenemase-producing K. pneumoniae isolates.
5. Conclusions
We identified three colistin-heteroresistant K. pneumoniae isolates. The resistant populations of the same isolate showed different amino acid alterations in PmrAB and PhoPQ. Meropenem combined with colistin would be a suitable therapeutic option for infections caused by multidrug-resistant, colistin-heteroresistant K. pneumoniae isolates.
Acknowledgments
The Klebsiella pneumoniae isolates used in this study were obtained from the Asian Bacterial Bank (ABB) of the Asia Pacific Foundation for Infectious Diseases (APFID) (Seoul, South Korea).
Author Contributions
Conceptualization, H.S.C., S.Y.K., Y.M.W. and K.S.K.; methodology, S.Y.K.; software, S.Y.K. and K.S.K.; validation, Y.M.W. and K.R.P.; formal analysis, H.S.C. and S.Y.K.; investigation, H.S.C. and S.Y.K.; resources, Y.M.W and K.R.P.; data curation, K.S.K.; writing—original draft preparation, H.S.C. and S.Y.K.; writing—review and editing, Y.M.W., K.R.P. and K.S.K.; visualization, K.S.K.; supervision, K.R.P. and K.S.K.; project administration, K.S.K.; funding acquisition, Y.M.W.
Funding
This research was funded by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Science and ICT (grant no. 2018R1D1A1B07049433).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Paczosa M., Mecsas J. Klebsiella pneumoniae: Going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 2016;80:629–661. doi: 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knothe H., Antal M., Krcméry V. Imipenem and ceftazidime resistance in Pseudomonas aeruginosa and Klebsiella pneumoniae. J. Antimicrob. Chemother. 1987;19:136–138. doi: 10.1093/jac/19.1.136. [DOI] [PubMed] [Google Scholar]
- 3.Van Duin D., Kaye K.S., Neuner E.A., Bonomo R.A. Carbapenem-resistant Enterobacteriaceae: A review of treatment and outcomes. Diagn. Microbiol. Infect. Dis. 2013;75:115–120. doi: 10.1016/j.diagmicrobio.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geraci D.M., Bonura C., Giuffrè M., Saporito L., Graziano G., Aleo A., Fasciana T., Di Bernardo F., Stampone T., Palma D.M., et al. Is the monoclonal spread of the ST258, KPC-3-producing clone being replaced in southern Italy by the dissemination of multiple clones of carbapenem-nonsusceptible, KPC-3-producing Klebsiella pneumoniae? Clin. Microbiol. Infect. 2015;21:e15–e17. doi: 10.1016/j.cmi.2014.08.022. [DOI] [PubMed] [Google Scholar]
- 5.Bonura C., Giuffrè M., Aleo A., Fasciana T., Di Bernardo F., Stampone T., Giammanco A., MDR-GN Working Group. Palma D.M., Mammina C. An update of the evolving epidemic of blaKPC carrying Klebsiella pneumoniae in Sicily, Italy, 2014: Emergence of multiple non-ST258 clones. PLoS ONE. 2015;10:e0132936. doi: 10.1371/journal.pone.0132936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Potter R.F., D’Souza A.W., Dantas G.D. The rapid spread of carbapenem-resistant Enterobacteriaceae. Drug Resist. Updat. 2016;29:30–46. doi: 10.1016/j.drup.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mammina C., Bonura C., Di Bernardo F., Aleo A., Fasciana T., Sodano C., Saporito M.A., Verde M.S., Tetamo R., Palma D.M. Ongoing spread of colistin-resistant Klebsiella pneumoniae in different wards of an acute general hospital, Italy, June to December 2011. Eurosurveillance. 2012;17:20248. [PubMed] [Google Scholar]
- 8.Poirel L., Jayol A., Nordmann P. Polymyxins: Antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 2017;30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ah Y.M., Kim A.J., Lee J.Y. Colistin resistance in Klebsiella pneumoniae. Int. J. Antimicrob. Agents. 2014;44:8–15. doi: 10.1016/j.ijantimicag.2014.02.016. [DOI] [PubMed] [Google Scholar]
- 10.Poirel L., Jayol A., Bontron S., Villegas M.V., Ozdamar M., Tükoglu S., Nordmann P. The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J. Antimicrob. Chemother. 2015;70:75–80. doi: 10.1093/jac/dku323. [DOI] [PubMed] [Google Scholar]
- 11.Olaitan A.O., Morand S., Rolain J.M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014;5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El-Halfawy O.M., Valvano M.A. Antimicrobial heteroresistance: An emerging field in need of clarity. Clin. Microbiol. Rev. 2015;28:191–207. doi: 10.1128/CMR.00058-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meletis G., Tzampaz E., Sianou E., Tzavaras I., Sofianou D. Colistin heteroresistance in carbapenemase-producing Klebsiella pneumoniae. J. Antimicrob. Chemother. 2011;66:946–947. doi: 10.1093/jac/dkr007. [DOI] [PubMed] [Google Scholar]
- 14.Jayol A., Nordmann P., Brink A., Poirel L. Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrob. Agents Chemother. 2015;59:2780–2784. doi: 10.1128/AAC.05055-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silva A., Sousa A.M., Alves D., Lourenco A., Pereira M.O. Heteroresistance to colistin in Klebsiella pneumoniae is triggered by small colony variants sub-populations within biofilms. Pathog. Dis. 2016;74:ftw036. doi: 10.1093/femspd/ftw036. [DOI] [PubMed] [Google Scholar]
- 16.Halaby T., Kucukkose E., Janssen A.B., Rogers M.R., Doorduijn D.J., van der Zanden A.G., Al Naiemi N., Vandenbroucke-Grauls C.M., van Schaik W. Genomic characterization of colistin heteroresistance in Klebsiella pneumoniae during a nosocomial outbreak. Antimicrob. Agents Chemother. 2016;60:6837–6843. doi: 10.1128/AAC.01344-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bardet L., Baron S., Leangapichart T., Okdah L., Diene S.M., Rolain J.M. Deciphering heteroresistance to colistin in a Klebsiella pneumoniae isolate from Marseille, France. Antimicrob. Agents Chemother. 2017;61:e00356-17. doi: 10.1128/AAC.00356-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barragán-Prada H., Ruiz-Hueso P., Tedim A.P., González-Candelas F., Galán J.C., Cantón R., Morosini M.I. Emergence and dissemination of colistin-resistnat Klebsiella pneumoniae isolates expressing OXA-48 plus CTX-M-15 in patients not previously treated with colistin in a Spanish university hospital. Diag. Microbiol. Infect. Dis. 2019;93:147–153. doi: 10.1016/j.diagmicrobio.2018.08.014. [DOI] [PubMed] [Google Scholar]
- 19.Band V.I., Satola S.W., Burd E.M., Farley M.M., Jacob J.T., Weiss D.S. Carbapenem-resistant Klebsiella pneumoniae exhibiting clinically undetected colistin heteroresistance leads to treatment failure in a murine model of infection. mBio. 2018;9:e02448-17. doi: 10.1128/mBio.02448-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lenhard J.R., Nation R.L., Tsuji B.T. Synergistic combinations of polymyxins. Int. J. Antimicrob. Agents. 2016;48:607–613. doi: 10.1016/j.ijantimicag.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Susceptibility Testing. CLSI; Wayne, PA, USA: 2018. Twenty-seventh Informational Supplement M100-S28. [Google Scholar]
- 22.Diancourt L., Passet V., Verhoef J., Grimont P.A., Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 2005;43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim S.Y., Choi H.J., Ko K.S. Differential expression of two-component systems, pmrAB and phoPQ, with different growth phases of Klebsiella pneumoniae in the presence or absence of colistin. Curr. Microbiol. 2014;69:37–41. doi: 10.1007/s00284-014-0549-0. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y.Y., Wang Y., Walsh T.R., Yi L.X., Zhang R., Spencer J., Doi Y., Tian G., Dong B., Huang X., et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 25.Pournaras S., Vrioni G., Neou E., Dendrinos J., Dimitroulia E., Poulou A., Tsakris A. Activity of tigecycline alone and in combination with colistin and meropenem against Klebsiella pneumoniae carbapenemase (KPC)-producing Enterobacteriaceae strains by time-kill assay. Int. J. Antimicrob. Agents. 2011;37:244–247. doi: 10.1016/j.ijantimicag.2010.10.031. [DOI] [PubMed] [Google Scholar]
- 26.Vidaillac C., Benichou L., Duval R.E. In vitro synergy of colistin combinations against colistin-resistant Acinetobacter baumannii, Pseudomonas aeruginosa, and Klebsiella pneumoniae isolates. Antimicrob. Agents Chemother. 2012;56:4856–4861. doi: 10.1128/AAC.05996-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wozniak J.E., Band V.I., Conley A.B., Rishishwar L., Burd E.M., Satola S.W., Hardy D.J., Tsay R., Farley M.M., Jacob J.T., et al. A nationwide screen of carbapenem-resistant Klebsiella pneumoniae reveals an isolate with enhanced virulence and clinically undetected colistin heteroresistance. Antimicrob. Agents Chemother. 2019;63:e00107–e00119. doi: 10.1128/AAC.00107-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crémieux A.C., Dinh A., Nordmann P., Mouton W., Tattevin P., Ghout I., Jayol A., Aimer O., Gatin L., Verdier M.C., et al. Efficacy of colistin alone and in various combinations for the treatment of experimental osteomyelitis due to carbapenemase-producing Klebsiella pneumoniae. J. Antimicrob. Chemother. 2019;74:2666–2675. doi: 10.1093/jac/dkz257. [DOI] [PubMed] [Google Scholar]