Human α/β-tryptase heterotetramer, a previously hidden form of tryptase, explains some of the unusual clinical features of hereditary α-tryptasemia. α/β-Tryptase forms naturally in mast cells and, when secreted, activates clinically relevant proteins, likely impacting a variety of mast cell disorders.
Abstract
Both α-tryptase and β-tryptase are preferentially expressed by human mast cells, but the purpose of α-tryptase is enigmatic, because its tetramers lack protease activity, whereas β-tryptase tetramers are active proteases. The monogenic disorder called hereditary α-tryptasemia, due to increased α-tryptase gene copies and protein expression, presents with clinical features such as vibratory urticaria and dysautonomia. We show that heterotetramers composed of 2α- and 2β-tryptase protomers (α/β-tryptase) form naturally in individuals who express α-tryptase. α/β-Tryptase, but not homotetramer, activates protease-activated receptor-2 (PAR2), which is expressed on cell types such as smooth muscle, neurons, and endothelium. Also, only α/β-tryptase makes mast cells susceptible to vibration-triggered degranulation by cleaving the α subunit of the EGF-like module–containing mucin-like hormone receptor-like 2 (EMR2) mechanosensory receptor. Allosteric effects of α-tryptase protomers on neighboring β-tryptase protomers likely result in the novel substrate repertoire of α/β-tryptase tetramers that in turn cause some of the clinical features of hereditary α-tryptasemia and of other disorders involving mast cells.
Introduction
The tryptase locus on human chromosome 16 includes TPSB2, encoding only β-tryptase, and TPSAB1, encoding either α- or β-tryptase, both homologous to trypsin-like serine proteases and expressed primarily by mast cells (MCs) and, to a lesser extent, basophils. Individuals with only β-tryptase encoded by TPSB2 and TPSAB1, lack α-tryptase. Though a corresponding clinical phenotype for α-tryptase deficiency has yet to emerge (Guida et al., 2000; Soto et al., 2002; Schwartz et al., 2003), prevalence variations among subjects with Asian (10%), European (23%), or African (41%) ancestry suggest a role for natural selection in maintaining α-tryptase (Trivedi et al., 2009). In contrast, no subject lacking a gene encoding an active form of β-tryptase has been reported (Trivedi et al., 2009). Although tryptase has been used clinically as a biomarker for disorders involving MCs such as systemic anaphylaxis and systemic mastocytosis (Schwartz et al., 1987b, 1989, 1995), its biological and pathobiologic functions are not well understood.
Mature tryptase tetramers form in MCs from monomeric protryptase precursors at acidic pH (5.5–6.9) in the presence of heparin (Sakai et al., 1996; Ren et al., 1998) and cathepsins (Le et al., 2011a,b), and then are stored in secretory granules (Schwartz et al., 2003) until released when these cells are activated to degranulate. Heparin stabilizes tryptase tetramers (Schwartz and Bradford, 1986) and orients the protomers in an active conformation (Maun et al., 2018). The active site of each β-tryptase protomer of the planar tetramer faces into a central pore (Pereira et al., 1998; Marquardt et al., 2002), restricting access by all biological inhibitors of serine proteases (Alter et al., 1990). Each β-tryptase protomer allosterically regulates its neighboring protomers, optimizing an active conformation (Maun et al., 2018). In contrast, mature α-tryptase homotetramers exhibit negligible proteolytic activity (Huang et al., 1999; Marquardt et al., 2002), largely explained by a p.G245D substitution (p.G216D in chymotrypsinogen numbering) with the Asp side chain protruding into the substrate binding channel, obstructing potential targets from reaching the catalytic site (Huang et al., 1999; Marquardt et al., 2002; Selwood et al., 2002). So why do most people have a gene for α-tryptase?
Hereditary α-tryptasemia, an autosomal dominant disorder, estimated to affect ∼3% of the total US population, is genetically defined by extra haploid TPSAB1 copies, only when that gene encodes α-tryptase, increasing expression of tryptase mRNA and serum levels of protryptase (Lyons et al., 2014, 2016), while intracellular levels of tryptase in cultured MCs are not affected (Lyons et al., 2016). The elevated serum baseline tryptase level was essentially all protryptase in 18 subjects from 8 different families based on measurements with a mature-specific tryptase immunoassay showing no detectable tryptase (<1 ng/ml; Lyons et al., 2014). The clinical phenotype may include dysautonomia with gastrointestinal dysmotility or postural orthostatic tachycardia, musculoskeletal abnormalities with hyperextensible joints leading to arthritis, chronic pain, and a high prevalence of atopic conditions. Affected patients also complain of symptoms classically associated with MC degranulation, including urticaria, both spontaneous and provoked by physical stimuli such as vibration. How overproduction of α-tryptase might lead to disease pathology, given its apparent lack of enzymatic activity, is unclear.
Murine MC protease (mMCP)–6 and mMPC-7, two homotetrameric tryptases expressed in mice, can form heterotetramers of intermediate binding affinity for heparin (Huang et al., 2000), but human α/β-tryptase heterotetramers have not been reported. We provide evidence that 2α:2β-tryptase heterotetramers form naturally in MCs from individuals with at least one α-tryptase gene, exhibit intermediate physicochemical properties compared with the homotetramers, are increased in individuals with a higher ratio of α-tryptase to β-tryptase–encoding genes, and, most importantly, have unique functional properties that can lead to activation of MCs and other cell types.
Results
α/β-Tryptase heterotetramers form in vitro and in vivo
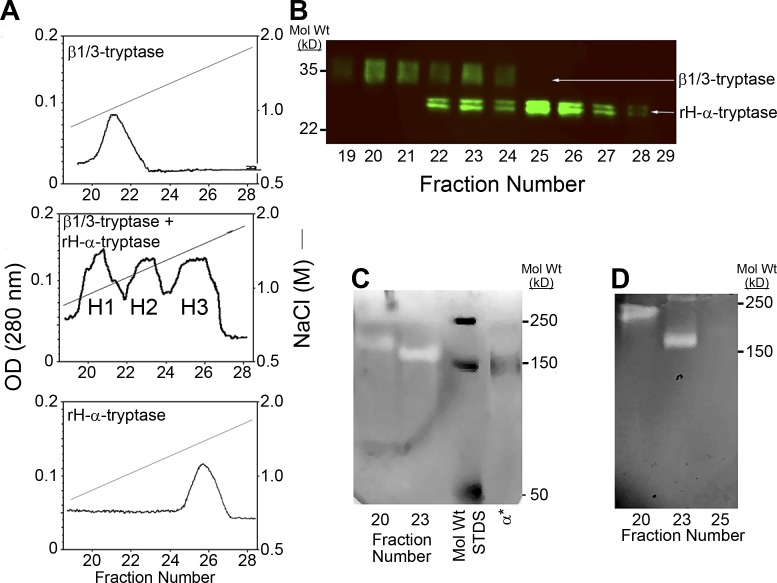
Recombinant human (rH)–α-protryptase (expressed in a baculovirus system; Sakai et al., 1996) and human MC leukemia 1 cell (HMC1)–derived β-protryptases (HMC1 encodes β1 and β3 tryptases, but no α-tryptase; Trivedi et al., 2009) were immunoaffinity purified and processed either together or separately to their mature forms as described (Le et al., 2011b). Activated preparations were then applied to heparin-Sepharose and eluted with a NaCl gradient. When the preparations were processed together three partially overlapping immunoreactive tryptase peaks appeared in the eluate, labeled H1, H2, and H3, eluting at 0.8, 0.9, and 1.1 M NaCl concentrations, respectively (Fig. 1 A). Only H1 and H2 exhibited catalytic activity (not shown). Portions of H1, H2, and H3 were analyzed by Western blotting (Fig. 1 B), which recognizes pro and mature forms of both α and β tryptases. H1 contained primarily HMC1-derived β-tryptase (diffuse tryptase band with an apparent mol wt of ∼37 kD), whereas H3 contained primarily rH-α-tryptase (sharp doublet with an apparent mol wt of ∼28 kD). The marked difference in the electrophoretic mobilities of tryptases in H1 and H3 is largely due to carbohydrate differences, with HMC1 cells producing β-protryptases with large complex N-linked carbohydrate moieties, and insect cells (baculovirus system) producing rH-α-protryptase with small N-linked carbohydrate moieties. In contrast, H2 exhibited the bands observed in both H1 and H3, a mixture of α- and β-tryptases.
Figure 1.
Formation of α/β-tryptase heterotetramers in vitro. (A) Heparin-Sepharose chromatography of homotypic and heterotypic tetramers of natural HMC1-derived β1/3-tryptases and rH-α1-tryptase. Protryptases β1/3 (top), α (bottom), and β1/3 + α (middle) were activated with human CTSB and separated by heparin-Sepharose chromatography with a linear NaCl gradient into three overlapping protein peaks, labeled H1, H2, and H3. Only H1 and H2 were catalytically active. (B) Peak heparin-Sepharose fractions were reduced, denatured, and analyzed by Western blotting with the G3 anti-tryptase mAb. (C) Peak heparin-Sepharose fractions, nonreduced and nondenatured, were analyzed by gelatin zymography along with labeled mol wt standards (STDS) and IRDye 800–labeled α1-tryptase homotetramers (α*). (D) Gelatin zymography after extended electrophoresis to better separate putative α/β-tryptase heterotetramers from β-tryptase homotetramers. Representative of two independent experiments.
To determine whether HII contained α/β-tryptase heterotetramers or a mixture of α-tryptase and β-tryptase homotetramers, H1, H2, and H3 fractions were assessed by gelatin zymography (Fig. 1, C and D). The peak fraction of each H1-3 peak, as well as rH-α-tryptase homotetramers labeled with IRDye 800, were each subjected to gelatin zymography under a condition that permits tryptase tetramers to remain intact (Fig. 1 C). H1 β-tryptase migrated more slowly than H2, each showing a single band of gelatinase activity, whereas α-tryptase showed no gelatinase band (not shown). IRDye 800-α-tryptase exhibited a fluorescent band that migrated faster than tryptase in either H1 or H2 fractions. To better separate such putative heterotetramers, H1 and H2 were subjected to a more prolonged electrophoretic separation (Fig. 1 D), which again showed for each peak fraction a single band of gelatinase activity. These findings indicate that α/β-tryptase heterotetramers can form in vitro when α- and β-protryptases are simultaneously processed together to their mature forms. The single gelatinase band in H2, about equidistant from β-tryptase and fluorescently tagged α-tryptase suggests a single heterotetramer with a 2α:2β composition rather than a mixture of homotetramers or multiple heterotetramers with α:β compositions of 1:3, 2:2, and 3:1.
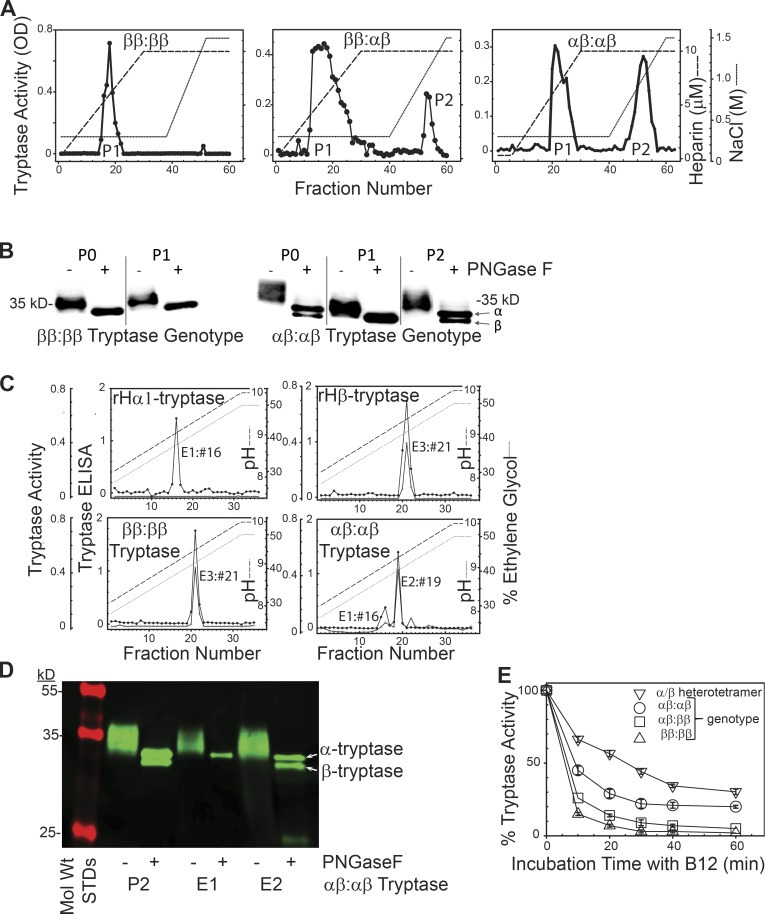
To address whether α/β-tryptase heterotetramers form naturally in MCs, lung tissue samples were tryptase-genotyped (Le et al., 2014; Lyons et al., 2016) and the tryptase formed in vivo and stored in MCs was immunoaffinity purified (P0), and analyzed by phosphocellulose chromatography (Fig. 2 A). The first peak of tryptase activity, P1, was eluted by a heparin glycosaminoglycan gradient, and the second peak, P2, by a NaCl gradient. From a ββ:ββ tryptase-genotyped subject only P1 tryptase activity was detected, whereas those with an αβ:αβ or αβ:ββ tryptase genotype had detectable P2 activity that accounted for more of the total tryptase activity when derived from an αβ:αβ subject than from an αβ:ββ subject.
Figure 2.
Separation and stability of tissue-derived β-tryptase homotetramers and α/β-tryptase heterotetramers. (A) B2-agarose–purified tryptase (P0) from tryptase-genotyped donors and subjected to phosphocellulose chromatography. Representative of tryptase prepared from three individuals with each genotype. (B) Western blotting (G3 anti-tryptase mAb) after SDS-gradient PAGE of P0 and P1 and P2 fractions from A, after being ±N-deglycosylated (PNGase F), reduced (DTT), and heat-denatured (SDS). Representative of experiments from two individuals with each genotype. (C) G5-Sepharose immunoaffinity chromatography of mature rH-α1-tryptase (left upper panel), rH-β2-tryptase (right upper panel), and lung tryptase (ββ:ββ genotype, P0, left lower panel; αβ:αβ genotype, P2, right lower panel). Representative of two individuals with each genotype. (D) SDS-PAGE (10–20%) of P2, E1, and E2 fractions (αβ:αβ genotype) obtained as above from one of the two subjects with an αβ:αβ genotype. (E) Stability of α/β-tryptase heterotetramer (P2, ββ:αααβ genotype), β-tryptase (P0, ββ:ββ genotype), and mixtures of α/β-tryptase and β-tryptase (P0, αβ:ββ or αβ:αβ genotypes) to inhibition by B12 anti-tryptase mAb. Mean ± SD. Data reflect tryptase from three different individuals for each of the three genotypes displayed.
To determine whether P1 contained β-tryptase homotetramers and P2 α/β-tryptase heterotetramers, P0, P1, and P2 fractions were N-deglycosylated, reduced and denatured, separated by SDS-gradient PAGE, and Western blotted. Based on their primary amino acid sequences, mature α-tryptases are slightly larger than β-tryptases. Accordingly, P0 tryptase(s) showed two closely- spaced bands if derived from an αβ:αβ-genotype subject, and a single band from a ββ:ββ-genotype subject (Fig. 2 B). P1 from both the ββ:ββ-genotype and αβ:αβ-genotype specimens showed only the faster-migrating β-tryptase band, while P2 showed both the slower-migrating α-tryptase band and the β-tryptase band, though the band intensity of α-tryptase was greater than the β-tryptase. We therefore considered whether P2 contained a mixture of α/β-tryptase heterotetramers and α-tryptase homotetramers.
To separate α/β-tryptase from α-tryptase tetramers, P2 was subjected to G5 anti-tryptase mAb immunoaffinity chromatography, chosen because G5 binds more slowly to mature α-tryptase than to mature β-tryptase (Fig. S1). Eluting with concurrent pH and ethylene glycol gradients, rH-α1-tryptase homotetramers eluted as a single peak (E1, pH 9.05) and rH-β2-tryptase homotetramers as a single peak (E3, pH 9.35), both detected by a tryptase ELISA, whereas only the β2-tryptase fraction (E3) demonstrated peptidolytic activity (Fig. 2 C). P1 tryptase fractions from lung-derived tryptase from a ββ:ββ-genotyped subject eluted from G5-Sepharose in a single peak at pH 9.35 with both ELISA and peptidolytic activity. However, the P2 tryptase purified from the lungs of αβ:αβ-genotyped subjects eluted in two peaks, one at pH 9.05 detected by ELISA that lacked peptidolytic activity, corresponding to rH-α1-tryptase tetramers (E1), and another at pH 9.23 detected by both ELISA and peptidolytic activity, termed E2. When tryptases from E1 and E2 were Western blotted (Fig. 2 D), E1 migrated with α-tryptase, while E2 displayed both α-tryptase and β-tryptase bands with equal staining intensities. Together, these data support the natural occurrence of catalytically active α/β-tryptase heterotetramers composed of equal amounts of α and β protomers.
An estimate of tryptase activity due to heterotetramers formed in lung MCs was made by calculating the portion of the total tryptase activity present in P2 fractions. As shown in Table 1, this portion increased as the α-tryptase/β-tryptase gene ratio increased from 1:3 to 1:1 in subjects with a normal tryptase genotype. For the two lung specimens found by chance to have a hereditary α-tryptasemia genotype, the heterotetramer portion was higher in the ββ:αααβ than the ββ:ααβ subject, again indicating an α-tryptase gene-dose effect on the heterotetramer portion.
Table 1. Percentages of lung tryptase activity due to α/β-tryptase after purification of tryptase by B2-agarose immunoaffinity chromatography (P0).
| Genotype | Tryptase activity in P2 (%)a | Tryptase activity based on stability to B12 inhibition (%)b | n |
|---|---|---|---|
| αβ:ββ | 15 ± 2 | 15 ± 1 | 3 |
| αβ:αβ | 30 ± 7 | 46 ± 5 | 3 |
| ββ:ααβc | 27 | 27 | 1 |
| ββ:αααβc | 37 | 43 | 1 |
Values are presented as mean ± SD. Data represent two replicate experiments for each of the three different individuals with an αβ:ββ or αβ:αβ tryptase genotype in column 2, triplicate values for each of the three different subjects with an αβ:ββ or αβ:αβ tryptase genotype in column 3, two replicates from the one subject with a ββ:ααβ or a ββ:αααβ tryptase genotype in column 2, and three replicates from the one subject with a ββ:ααβ or a ββ:αααβ tryptase genotype in column 3.
100 × P2/(P1 + P2).
Percentage activity after 20 min compared to β-tryptase (P0, ββ:ββ genotype) and α/β-tryptase (P2, ββ:αααβ genotype).
Hereditary α-tryptasemia genotype.
As homotetramers of α-tryptase are more stable than those of β-tryptase (Marquardt et al., 2002; Selwood et al., 2002), the stabilities of α/β-heterotetramers (P2) and β-tryptase homotetramers (P0, ββ:ββ genotype), formed in vivo, were compared after exposure to the B12 anti-tryptase mAb, which converts heparin-stabilized active tetramers into inactive monomers (Fig. 2 E; Fukuoka and Schwartz, 2006). After a 20 min incubation, the % mean residual activity for β-tryptase was 7 ± 1%, whereas that of α/β-tryptase heterotetramers (P2) was 57 ± 2%, showing that the α/β-tryptase heterotetramers can be distinguished from β-tryptase homotetramers by their higher stability. When P0 tryptase fractions from donors with αβ:αβ, αβ:ββ, ββ:ααβ, or ββ:αααβ tryptase genotypes were compared, stability varied with the α- to β-tryptase gene ratios, allowing an alternative estimate of the portion of tryptase activity accounted for by heterotetramers (Fig. 2 E and Table 1) that corroborates calculations based on P1 and P2 tryptase activities. From here on, α/β-tryptase heterotetramers and β-tryptase homotetramers will be called α/β-tryptase and β-tryptase, respectively.
Because protomers of β-tryptase have allosteric effects on neighboring protomers (Maun et al., 2018), we explored whether differences between α/β-tryptase and β-tryptase would go beyond physicochemical properties to affect substrate repertoire. Accordingly, in the following two sections, we examined whether there were distinct effects of α/β-tryptase versus β-tryptase on two potential targets with clinical implications, EGF-like module-containing mucin-like hormone receptor-like 2 (EMR2), based on its involvement in hereditary vibratory urticaria, and protease-activated receptor-2 (PAR2), based on its stimulatory effects on neuronal, smooth muscle, endothelial, and inflammatory cells.
α/β-Tryptase facilitates vibration-triggered degranulation of human skin MCs by cleaving EMR2α
A severe form of familial vibratory urticaria is caused by a missense mutation (p.C492Y) in adhesion G protein–coupled receptor E2 (ADGRE2). ADGRE2, abundantly expressed in myelocytes, including MCs, encodes EMR2 (CD312), which is autocatalytically cleaved during maturation in the endoplasmic reticulum into two noncovalently associated subunits, dermatan sulfate–binding EMR2α and G protein–coupled receptor EMR2β, which reside on the plasma membrane. EMR2α is thought to inhibit EMR2β unless mechanical force transiently separates the subunits. EMR2-p.C492Y may alter EMR2α:EMR2β binding such that vibration causes EMR2α to be shed from the cell surface, prolonging EMR2β signaling to cause MC degranulation (Boyden et al., 2016).
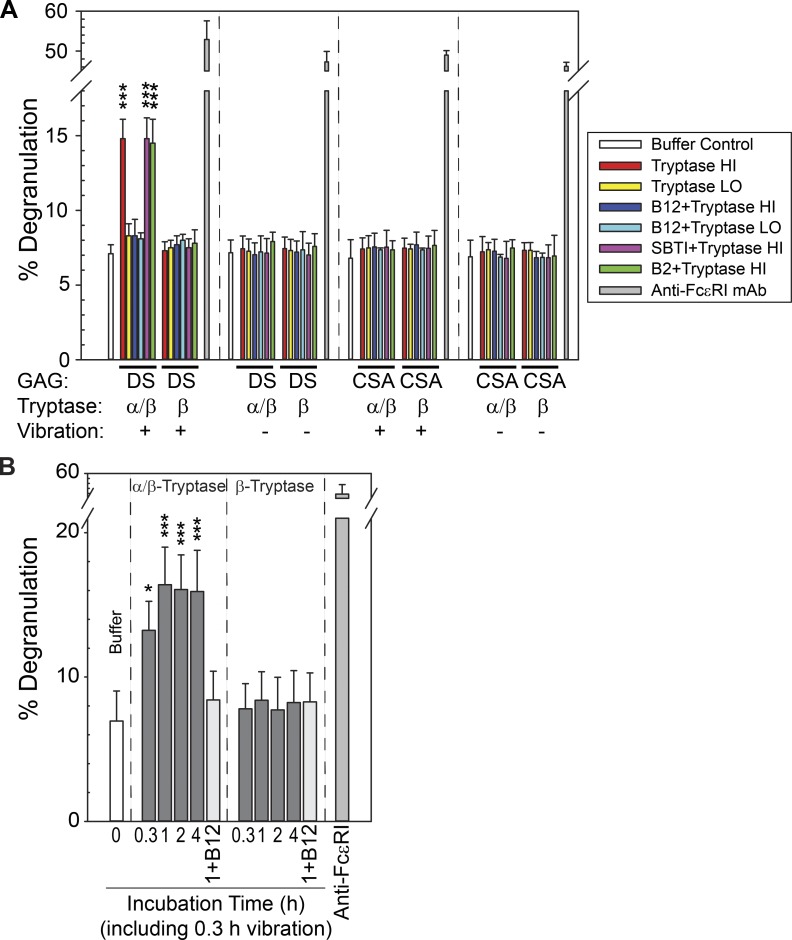
We hypothesized that α/β-tryptase cleaves EMR2α on MCs near the Cys492 site, thereby weakening the association of this resultant EMR2α′ with EMR2β and enabling vibration-triggered degranulation. Accordingly, we incubated skin MCs with heparin-stabilized α/β-tryptase (P2; αβ:αβ genotype) or heparin-stabilized β-tryptase (P0; ββ:ββ genotype) for 1 h, vibrating the mixture for the final 20 min. Vibration-triggered degranulation occurred only after exposure to α/β-tryptase, not to β-tryptase, and only when MCs were vibrated while adherent to dermatan sulfate, not if adherent to chondroitin sulfate A (Fig. 3 A). Degranulation occurred with exposure to α/β-tryptase at 3.5 µg/ml, but not at 0.35 µg/ml. In support of α/β-tryptase being responsible were the findings that vibration-triggered degranulation was inhibited with the B12 anti-tryptase mAb, but not by incubating α/β-tryptase with B2 anti-tryptase or soybean trypsin inhibitor (SBTI; neither of which inhibits tryptase activity). Such activity did not occur in the absence of heparin and was not attenuated by a PAR2 inhibitor (Fig. S2 A). The α/β-tryptase effect was time dependent, as degranulation was less for MCs simultaneously exposed to α/β-tryptase and vibration for 20 min (Fig. 3 B) than if 20-min vibration was initiated after an exposure for ≥40 min with α/β-tryptase.
Figure 3.
α/β-Tryptase but not β-tryptase makes human MCs susceptible to vibration-triggered degranulation. (A) Treatment of skin MCs adherent to dermatan sulfate, but not to chondroitin sulfate A, degranulate when vibrated if pretreated with α/β-tryptase Hi (3.5 µg/ml), but not with α/β-tryptase Lo (0.35 µg/ml) or with β-tryptase at either concentration. Pretreatment of α/β-tryptase Hi with B12 anti-tryptase IgG, but not with SBTI or B2 anti-tryptase IgG, prevents this degranulation. Triplicate data with cells from one subject are shown, representative of two independent experiments. GAG, glycosaminoglycan; DS, dermatan sulfate; CSA, chondroitin sulfate A. (B) Time for incubating α/β-tryptase (3.5 µg/ml) with skin MCs to make them susceptible to vibration-triggered degranulation during the final 20 min of incubation. Triplicate data with cells from one subject are shown, representative of two independent experiments. *, P < 0.05; ***, P < 0.001; ANOVA followed by a Holm–Sidak test compared with the buffer control. Bars show means, and error bars show the SD.
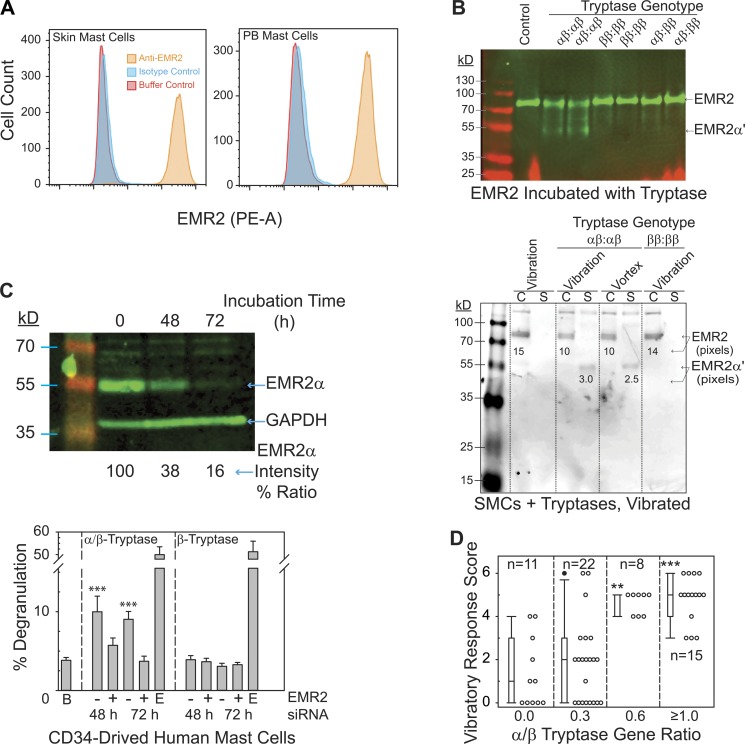
To explore whether α/β-tryptase acted on EMR2, we first confirmed that EMR2 is expressed on skin MCs and on MCs derived in vitro from peripheral blood progenitors (Fig. 4 A). Solubilized EMR2 was purified from HMC1 cells (Fig. S3) and incubated with lung-derived P0 tryptase (n = 2 subjects with each of the tryptase genotypes αβ:αβ, ββ:ββ, or αβ:ββ). Western blotting of untreated EMR2 under nondenaturing/nonreducing conditions with anti-EMR2α mAb revealed one EMR2 band with a mol wt between 70 and 100 kD (Fig. 4 B, upper panel), while a major EMR2α degradation fragment (EMR2α′) with an apparent mol wt of just <55 kD emerged if P0 tryptase preparations from αβ:αβ-genotype donors were used. This fragment was barely visible if P0 tryptase was from αβ:ββ-genotype donors, and was not apparent if tryptase was from ββ:ββ-genotype donors, indicating that EMR2, when in solution, can be selectively targeted by α/β-tryptase.
Figure 4.
α/β-Tryptase targets EMR2α, making MCs susceptible to vibration-triggered degranulation in vitro and urticaria in vivo. (A) EMR2 on skin and CD34-derived MC surfaces by flow cytometry. Histograms representative of two independent experiments. (B) Western blotting of nonreduced/nondenatured, SDS-solubilized EMR2 shows cleavage by P0 α/β-tryptase (αβ:αβ genotype) and negligible cleavage by β-tryptase (αβ:ββ or ββ:ββ genotypes). Purified EMR2 (upper panel, 10–20% acrylamide; Fig. S3) incubated with P0 tryptase (1 µg/ml) from tryptase-genotyped donors or adherent skin MCs (lower panel, 12% acrylamide) incubated with similar tryptase preparations (3.5 µg/ml) for 1 h, and then vibrated or vortexed for 20 min. C, cell extract; S, cell-free medium. Representative of two different subjects with each genotype. (C) EMR2 silencing in CD34+ cell-derived MCs with transfected EMR2 siRNA. These MCs (adherent to dermatan sulfate–coated wells, treated with tryptase × 1 h, and vibrated the last 20 min) were denatured, reduced, and assessed by Western blotting (upper panel, 12% acrylamide; representative of two separate experiments) and for degranulation (lower panel, one experiment performed in triplicate). B, buffer; −, negative-control siRNA-A; +, EMR2 siRNA; E, anti-FcεRI. ANOVA followed by a Holm–Sidak test compared with buffer control. Bars show means, and error bars show the SD. (D) Response scores to a cutaneous vibratory challenge (Fig. S4) were analyzed according to the α/β-tryptase gene ratio by Kruskal–Wallis ANOVA on ranks followed by the Dunn’s method, comparing each α-tryptase gene–containing group to the all β-tryptase gene–containing group. Each symbol represents a different subject tested one time. **, P < 0.01; ***, P < 0.001. Bars show median and 25th/75th percentiles, error bars show the 10th/90th percentiles, and the solid black circle shows the 95th percentile.
To determine whether EMR2 on the MC surface is also a substrate for α/β-tryptase, plate-adherent skin MCs were incubated with P0 tryptase derived from αβ:αβ-genotype or ββ:ββ-genotype donors and either vibrated or vortexed to introduce mechanical stress. Any shed EMR2-derived moieties were separated from cell-associated ones and then analyzed by Western blotting as above (Fig. 4 B, lower panel). A comparable amount of EMR2 (75 kD) was detected in extracts from skin MC pellets that had been incubated either with buffer alone or with P0 tryptase (ββ:ββ-genotype) for which α/β-tryptase was absent. In contrast, incubation of skin MCs with P0 tryptase from αβ:αβ subjects reduced the intensity of the 75-kD band, reflecting less cell-associated EMR2α. Moreover, a new band from the cell supernatant appeared just <55 kD, comparable to the EMR2α′ band seen when EMR2 in solution was incubated with α/β-tryptase. Thus, α/β-tryptase cleaves EMR2α, causing EMR2α′ to be shed with vibration.
MCs derived in vitro from peripheral blood progenitors and attached to dermatan sulfate–coated wells, like primary skin MCs, degranulate when vibrated if attached to dermatan sulfate and preincubated with heparin-stabilized α/β-tryptase, not with heparin-stabilized β-tryptase (Fig. S2 B). Transfecting EMR2 siRNA into these MCs markedly decreased EMR2α protein by 62% after 48 h and 84% after 72 h relative to GAPDH (Fig. 4 C, upper panel). Skin MCs transfected with siRNA or negative-control siRNA-A were then attached to dermatan sulfate–coated wells, incubated with heparin-stabilized α/β-tryptase or β-tryptase for 60 min, and vibrated for the final 20 min (Fig. 4 C, lower panel). Degranulation was apparent only with α/β-tryptase–treated cells that had been exposed to siRNA-A, whereas EMR2 siRNA prevented this response, and none of the transfection conditions affected the ability of MCs to degranulate when exposed to anti-FcεRI IgG.
A common clinical finding of patients with hereditary α-tryptasemia is urticaria, with vibration often being reported as a trigger (Lyons et al., 2014, 2016). To determine whether the α-tryptase gene dosage affects vibration-elicited urticaria, 56 volunteers (35 healthy and 21 with hereditary α-tryptasemia) underwent tryptase genotyping and a cutaneous vibratory challenge as described (Lyons et al., 2014; Boyden et al., 2016). The presence of an α-tryptase gene enhanced the likelihood for vibration-elicited urticaria, and a graded response to increasing α/β tryptase gene ratios was observed for both severity and prevalence, regardless of whether the healthy or the hereditary α-tryptasemia donors were considered together (Fig. 4 D) or separately (Fig. S4). Each subject with a gene ratio of ≥0.6 had a response score ≥3, while the majority of subjects with a gene ratio of 0.0 or 0.3 had response scores ≤2. Each of the subjects who failed to respond to a vibratory cutaneous challenge had no or only one α-tryptase–encoding gene.
α/β-Tryptase heterotetramers activate PAR2
PAR2 has been considered a potential target for MC tryptase, and PAR2 activation by tryptase in a variety of tissues could offer new insights into the multisystem complaints reported in disorders involving MC activation-degranulation. Human tryptases derived from skin, lung, or HMC1 cells have been reported to activate rH-PAR2 on COS cells (Molino et al., 1997), PAR2 on human endothelial cells or rat ganglion cells (Steinhoff et al., 2000) or on human airway smooth muscle (Berger et al., 2001; Schmidlin et al., 2001). However, heparin-stabilized skin-derived tryptase was unable to activate PAR2 on keratinocytes (Schechter et al., 1998), and heparin-stabilized rH-β1-tryptase was unable to activate PAR2 on fibroblasts (Huang et al., 2001). A potentially unifying study reported that optimal activation of PAR2 by human tryptase requires PAR2 desialation and heparin depletion (Compton et al., 2002a,b), conditions unlikely to predominate in vivo. Importantly, none of these studies considered the human tryptase genotype or presence of α/β-tryptase when using either investigator-prepared or commercial tissue–derived tryptase.
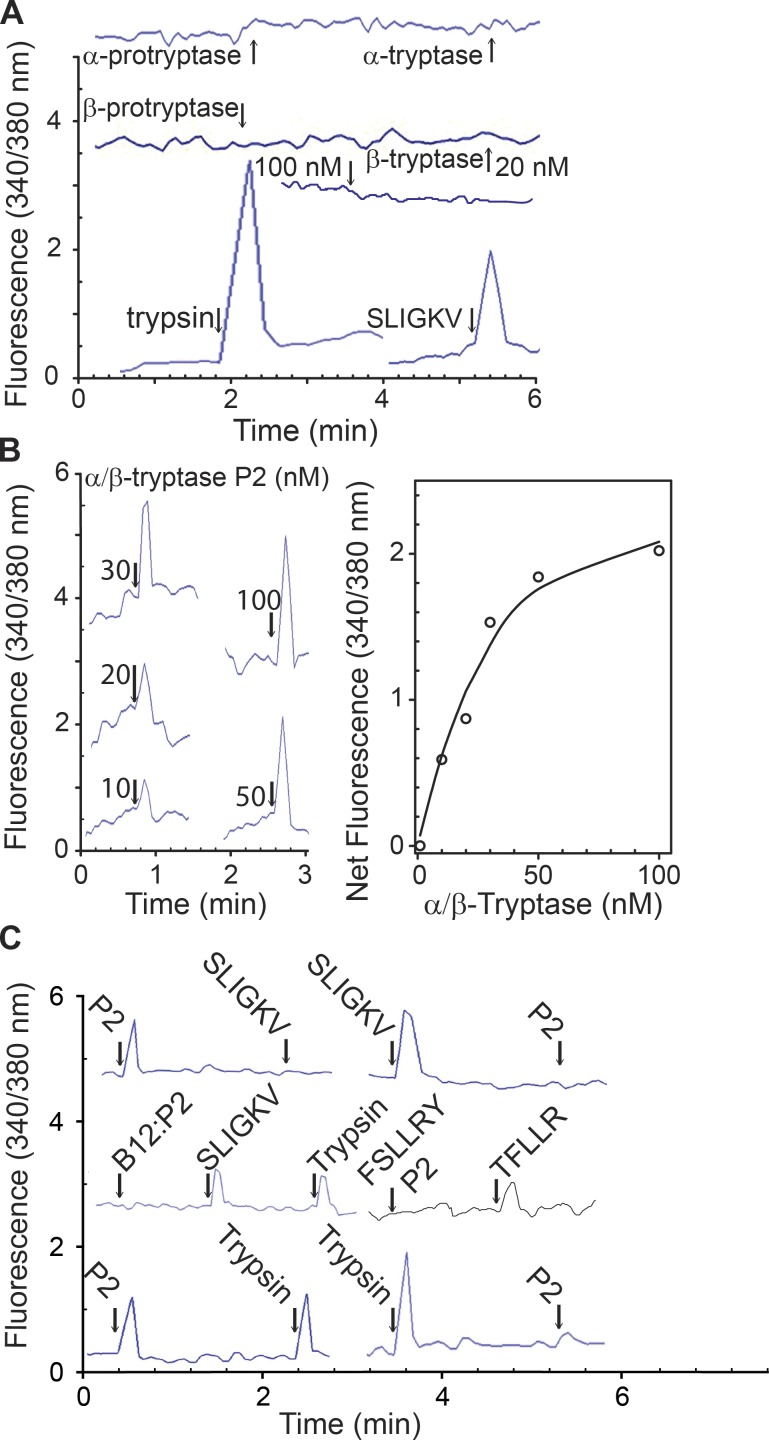
We examined whether activation of naturally expressed PAR2 on Jurkat cells (which express PAR1, PAR2, and PAR3, but not PAR4) might be different for heparin-stabilized α/β-tryptase than β-tryptase. As shown in Fig. 5 A, trypsin (PAR1 and PAR2 agonist) and SLIGKV (PAR2 agonist) each elicited a Ca2+ influx, whereas neither rH-α1-protryptase nor rH-β2-protryptase monomers or their corresponding mature tryptase homotetramers did so. RH-β1 or rH-β3, pro-monomeric or mature tetrameric tryptases, likewise, each failed to alter Ca2+ influx (Fig. S5). In contrast, α/β-tryptase generated a dose-dependent Ca2+ influx with a calculated half-maximal response concentration of 20 nM (Fig. 5 B). SLIGKV completely cross-desensitized the response of Jurkat cells to α/β-tryptase, as did α/β-tryptase to SLIGKV, indicating specificity for PAR2 (Fig. 5 C). Inhibiting α/β-tryptase with the tryptase-specific B12 mAb prevented both α/β-tryptase-elicited Ca2+ influx and cross-desensitization of the SLIGKV response, indicating that PAR2 activation by α/β-tryptase was not due to a contaminant. Pretreating Jurkat cells with FSLLRY (PAR2 antagonist) blocked the response to α/β-tryptase, but not to TFLLR (PAR1 agonist). Moreover, α/β-tryptase partially cross-desensitized the response to trypsin, likely due to trypsin activating PAR1, whereas trypsin completely desensitized the response to α/β-tryptase. Thus, only heparin-stabilized α/β-tryptase activates endogenously expressed and glycosylated PAR2 on Jurkat cells, conditions likely to be present for tryptase and PAR2 in vivo.
Figure 5.
PAR2 on Jurkat cells is activated by α/β-tryptase but not by β-tryptase. (A) Calcium response to rH-α1 protryptase (20 nM) or homotetramer (20 nM protomer), and rH-β2 protryptase (20 nM) or homotetramer (20 or 100 nM protomer), trypsin (10 nM), and SLIGKV (50 nM). Representative of two independent experiments. (B) Dose–response calcium signal by α/β-tryptase. Representative of two independent experiments. (C) Desensitization experiments show that the calcium response to α/β-tryptase (50 nM protomer) is due to PAR2 activation, using trypsin (20 nM) or SLIGKV (50 nM), and is prevented by B12 anti-tryptase (400 nM) or PAR2-specific inhibitor FSLLRY (50 nM), while TFLLR activates PAR1. Each uninterrupted line (A, B [left panel], and C) represents an independent experiment.
Discussion
The current study shows that α/β-tryptase heterotetramers (Fig. 1, C and D; and Fig. 2, C and D) form naturally in MCs in subjects who carry any α-tryptase allele and account for a significant portion of the total tryptase enzymatic activity (Table 1), nearly all of which is stored in MC secretory granules. The apparent α:β protomer equivalency suggests that individual α and β monomers do not combine randomly, but instead form from α and β homodimers, the existence of which was predicted based on the kinetics of monomer-to-tetramer formation (Ren et al., 1998). Further, allosteric effects of α-tryptase protomers on neighboring β-protomers might explain the altered substrate specificity of the heterotetramers, as such interactions have been previously demonstrated between β-tryptase protomers in the homotetramer (Maun et al., 2018). Importantly, the portion of tryptase activity due to α/β-tryptase increases with the α-tryptase gene dosage. Consequently, an α-tryptase gene-dosage effect associated with a particular clinical phenotype, as shown in the current study for a cutaneous response to vibration or as reported for severe Dengue infection (Velasquez et al., 2015) or asthma (Abdelmotelb et al., 2014), might implicate involvement of α/β-tryptase. Also, as α-tryptase gene deficiency varies among individuals with Asian, European, and African backgrounds (Trivedi et al., 2009), affecting α/β-tryptase content, a role for these heterotetramers can now be considered in the context of natural selection.
Although individuals with an αβ:αβ-tryptase genotype are significantly more common than those with hereditary α-tryptasemia, it was the latter disorder that made us consider the possibility of α/β-tryptase heterotetramers. For individuals with hereditary α-tryptasemia, α/β-tryptase likely contributes to some of the associated clinical features. In particular, cleavage of EMR2α by α/β-tryptase likely contributes to the vibratory urticaria responses elicited in nearly all affected patients, perhaps weakening the inhibitory association of this dermatan sulfate–binding subunit with EMR2β, resulting in the EMR2α′ fragment being shed from MCs during vibration, followed by sustained activation of the remaining uninhibited EMR2β G protein–coupled receptor (Fig. 4 B, lower panel). This resembles what happens with the ADGRE2 p.C492Y missense mutation causing familial vibratory urticaria (Boyden et al., 2016), whereby this point mutation rather than EMR2α proteolysis weakens the EMR2α:β association. However, experimental vibration-triggered urticaria occurs in some subjects with a normal tryptase genotype, but the frequency of affected subjects and severity of the reactions rise as the α/β-tryptase gene ratio increases, providing support for the contribution of α/β-tryptase to vibration-triggered urticaria susceptibility in vivo. That some individuals with a ββ:ββ-genotype develop vibration-triggered urticaria indicates that tryptase-independent mechanisms also affect susceptibility to vibration-triggered histamine release. The magnitude of EMR2-dependent MC degranulation could be higher in vivo, where dermatan sulfate in the connective tissue surrounding MCs may engage a higher portion of the EMR2 than in the experimental plate-bound dermatan sulfate.
For α/β-tryptase to make MCs susceptible to vibration-triggered degranulation in vivo, some of this protease must be secreted to have cleaved surface EMR2α before the vibration stimulus. Whether this might happen by low levels of spontaneous secretion via piecemeal or anaphylactic degranulation, by stimulation of activating receptors such as Mas-related G protein–coupled receptor-X2 by endogenous neuropeptides or other cationic proteins, by suboptimal activation of EMR2 or other GPRs, or by secretion in exosomes are possibilities. As cleavage of EMR2α by tryptase is itself irreversible, only replacement of cleaved EMR2 with newly synthesized protein would reverse the tryptase effect, which might be facilitated if extracellular α/β-tryptase activity was inhibited. Some cleavage of EMR2α by α/β-tryptase in vivo must occur well before the vibration-induced trigger, but secretion of additional α/β-tryptase during the reaction may cleave additional EMR2α, further enhancing the response to ongoing vibration. Whether α/β-tryptase also causes EMR2-dependent mechanical activation of MCs in the lungs during coughing or breathing, in the bowel during peristalsis, or in blood vessels during pulsatile blood flow or of EMR2 expressed on myelocytes other than MCs, and how this presents clinically, remains to be determined (I et al., 2017).
We showed that heparin-stabilized α/β-tryptase activates PAR2 expressed on Jurkat T cells in a dose-dependent manner, while heparin-stabilized β-tryptase lacks this activity, providing a possible explanation for conflicting reports of the effects of tryptases on PAR2 activation. Unlike for EMR2 activation, PAR2 activation would occur immediately upon its cleavage by α/β-tryptase, potentially contributing to clinical phenotypes observed acutely during MC activation. The tryptase concentration (35 µg/ml) used in the current study is likely physiological. Based on MC concentrations in human tissues (Craig et al., 1986; Irani et al., 1990) and the content of tryptase per MC (Schwartz et al., 1987a), mean content of mature tryptase in dermis, lung, and intestinal mucosa, nearly all of which is stored in MCs, ranges from 160 to 270 µg/cm3, suggesting that even with small amounts of degranulation, concentrations of mature tryptase, at least near to where tryptase:heparin complexes are released in vivo, would be within the range observed for activating PAR2 in vitro. Activating PAR2 when MCs degranulate may have many clinical effects. For example, activating PAR2 on vascular endothelium during systemic anaphylaxis might worsen hypotension; on smooth muscle in inflammatory bowel disease (Linden et al., 2001; Lohman et al., 2012) and asthma (Schmidlin et al., 2001; Abdelmotelb et al., 2014) might contract smooth muscle and promote exacerbations; and on epithelial cells in asthma and inflammatory bowel disease and on keratinocytes and sensory nerves in skin might exacerbate inflammation (Frateschi et al., 2011), pruritus (Steinhoff et al., 2003), or hyperalgesia (Vergnolle et al., 2001; Dong and Dong, 2018). Whether activating PAR2 on neurons associated with smooth muscle or blood vessels could promote autonomic dysfunction needs to be considered. Also, activating PAR2 on sensory nerves might lead to neuropeptide secretion, including Substance P and calcitonin gene-related peptide (Steinhoff et al., 2000), which in turn bind Mas-related G protein–coupled receptor-X2 on MCs, stimulating further degranulation (Fujisawa et al., 2014; McNeil et al., 2015), possibly contributing to MC activation syndrome (Sabato et al., 2018).
Based on our study, α/β-tryptase should be considered a mediator that can influence the severity of disorders involving MCs where they are naturally abundant, such as in mucosal tissues, dermis, and perivascular sites, or where they accumulate as part of a disease process, such as in the bronchial smooth muscle of asthmatics (Brightling et al., 2002). Moreover, activation of PAR2 and/or EMR2 by α/β-tryptase may contribute to the pruritus, dysautonomia, and pain symptoms associated with hereditary α-tryptasemia, but α/β-tryptase in patients with a normal tryptase genotype composed of one or two TPSAB1-encoded α-tryptase genes are more common and may affect a wide variety of disorders, including asthma, atopic dermatitis, inflammatory bowel disease, arthritis, and MC activation syndrome. We suspect other differences in substrate specificities between β-tryptase and α/β-tryptase will be found that relate to physiological or pathological situations. Accordingly, a targeted inhibitor of α/β-tryptase may have clinical utility.
Materials and methods
Reagents
Frozen or fresh human adult lung and skin samples, ranging from 15 to 20 g, were purchased through the Collaborative Human Tissue Network; cyanogen bromide–activated Sepharose 4B from GE Healthcare; heparin-Sepharose, Na-benzoyl-dl-arginine 4-nitro-anilide, porcine pancreatic type IX trypsin (13,000–20,000 U/mg), sulphinpyrazone, calcium ionophore (A23187), porcine intestinal mucosa heparin (mean mol wt 16 kD), SBTI, cellulose phosphate (fibrous form), and leupeptin from Sigma-Aldrich; FCS, DMEM, penicillin, streptomycin, and amphotericin from Thermo Fisher Scientific; anti-PAR2 IgG and anti-EMR2 from R&D Systems; and PAR2-activating peptide (SLIGRL-NH2), PAR1-activating peptide (TFLLRN-NH2), PAR2 peptide control LRGILS-NH2, and PAR2 antagonist (FSLLRY-NH2) from Tocris Bioscience. PAR2 agonists or antagonist were diluted in a buffer comprising 20% DMSO and 80% PBS.
Cell cultures
Skin-derived MCs were dispersed from discarded surgical skin obtained from the Collaborative Human Tissue Network and cultured as described (Lotfi-Emran et al., 2018). MCs were dispersed from fresh surgical skin by collagenase and hyaluronidase digestion, partially purified by Percoll density-dependent sedimentation, and cultured in 24-well plates containing X-VIVO 15 serum-free medium containing 100 ng/ml of rH-stem cell factor (rH-SCF). MCs were harvested after ≥6 wk of culture, by which time they were >99% pure and ∼97% viable.
Human peripheral blood buffy coats obtained from the Virginia Blood Bank were used to isolate peripheral blood mononuclear cells, from which CD34+ hematopoietic progenitor cells were purified with the StemCell Technologies EasySep Human CD34 Positive Selection Kit, according to the manufacturer’s directions. In brief, the human buffy coat sample was subjected to isopycnic centrifugation using Ficoll-Paque. The mononuclear cell layer was harvested, washed, and positively selected for CD34+ cells with EasySep CD34+ Positive Selection Kit. These cells were cultured in AIM V serum-free medium with rH-SCF (100 ng/ml), rH-IL-6 (100 ng/ml), and rH-IL-3 (30 ng/ml) for 1 wk, and then cultured in the same medium lacking rH-IL-3. After 6–8 wk, the culture consists of mature human MCs, CD117hi, FcεRI+, and tryptase+ (>95% purity). Such MCs were used after 6 and before 12 wk of culture.
Jurkat T cells were propagated in 75-cm2 T flasks containing RPMI 1640 supplemented with 2 mM l-glutamine, penicillin (100 U/ml), streptomycin (100 µg/ml), 10 mM Hepes, 10% (vol/vol) heat-inactivated FCS, and 1 mM sodium pyruvate. HMC1 cells were maintained in 75-cm2 T flasks containing Iscove’s medium supplemented with 2 mM l-glutamine, penicillin (100 U/ml), streptomycin (100 µg/ml), 10% (vol/vol) heat-inactivated bovine serum, and 0.01% α-thioglycerol.
Tryptase enzymatic activity assays
Tryptase enzymatic activity was monitored in 96-well microtiter plates, using tosyl-Gly-Pro-Lys-p-nitroanilide (TGPK) as the substrate (Le et al., 2011b), except that SBTI (50 µg/ml) was included when proteases other than tryptase were present, such as when using cell extracts. Free p-nitroanilide was measured at a wavelength of 405 nm in a EL312 Kinetic plate reader (Bio-Tek Instruments). One unit of enzyme activity was defined as the amount that cleaves 1 mmol synthetic substrate per min. For inhibition studies, samples were preincubated with various inhibitors at room temperature for 20 min. Protein concentrations were measured with the BCA method using BSA as a standard. G3 mAb, which recognizes both pro- and mature forms of α- and β-tryptases, and mAbs against PAR2 and CD312 (R&D Systems) were used for Western blotting and detected with IRDye-conjugated anti-mouse IgG (Odyssey Infrared Imaging System; LiCor).
Formation of tryptase tetramers from protryptases
rH-protryptases had been cloned as described (Miller et al., 1989, 1990). rH-β1, rH-β2, or rH-β3 or HMC1 β1/3 protryptases (5 µg/ml) were incubated alone or at a 1:1 molar ratio with rH-α1-protryptase and then activated with cathepsin B (CTSB) in the presence of heparin (50 µg/ml) in 50 mM sodium acetate buffer, pH 5.5, containing 150 mM NaCl, 1 mM EDTA, 5 mM l-cysteine, 5% glycerol, and SBTI (50 µg/ml) for 30 min at 37°C (Le et al., 2011b). Activation of β-protryptase, α-protryptases, or mixtures of β- and α- protryptases was measured with TGPK and used in experiments shown in Figs. 1, 5, S1, and S5.
Gel filtration chromatography
Gel filtration studies were performed on a Superose 12 HR 10/30 size-exclusion column (GE Healthcare) using a series Shimadzu High Performance Liquid Chromatography system (PerkinElmer) at a flow rate of 1 ml/min. The column was equilibrated and run in 10 mM 2-mopholinoethanesulfonic acid (MES) buffer, pH 6, containing 1 M NaCl. The column was calibrated with bovine thyroglobulin (669 kD), horse spleen apoferritin (443 kD), sweet potato β-amylase (200 kD), yeast alcohol dehydrogenase (150 kD), BSA (66 kD), and bovine erythrocyte carbonic anhydrase (29 kD).
B2-Sepharose chromatography
Human MC tryptase from human adult lung and skin was immunoaffinity purified as described (Schwartz et al., 1990). Tissue extracts were adjusted to 1 M NaCl by adding a cold 10 mM MES buffer, pH 6.1, containing 4 M NaCl, and then loaded onto a B2 anti-tryptase IgG-Sepharose immunoaffinity column (2.5 × 10 cm). Elution samples containing TGPK-cleaving activity were pooled, concentrated, and dialyzed overnight at 4°C in 10 mM MES, pH 6.1, containing 0.3 M NaCl.
Heparin-agarose chromatography
RH-α1-protryptase and HMC1 β1/β3 protryptases were purified by B2 anti-tryptase mAb-agarose chromatography (Schwartz et al., 1990; Sakai et al., 1996), mixed, and incubated with human CTSB together with heparin (50 µg/ml) for 30 min at 37°C as described (Le et al., 2011b). Reactions were stopped by adding 1 µM E64 to block any remaining CTSB activity, after which heparinase I (2 U/ml) was added and incubated for 30 min at 37°C to remove free heparin. The resulting tryptase complexes were then applied to a 1-ml column of heparin-Sepharose connected to a Shimadzu High Performance Liquid Chromatography system, already equilibrated with 400 mM NaCl and 50 mM sodium acetate, pH 6.0. The column was washed with 10 ml of equilibration buffer; and elution was performed with a 40 ml NaCl linear gradient (0.4–2.0 M) in 50 mM sodium acetate, pH 6.0, at a flow rate of 1 ml/min. To stabilize tryptase tetramers, collected samples were adjusted to 1 M NaCl by the addition of 4 M NaCl. Column fractions were analyzed for tryptase content and activity using immunoblot analysis with the G3 anti-tryptase IgG mAb and gelatin zymography. Measurement of tryptase activity from eluates samples was performed with 0.1 mM TGPK substrate at pH 7.6 in 50 mM Tris-HCl containing 0.15 M NaCl.
Phosphocellulose chromatography
Tryptase samples were dialyzed overnight in 10 mM MES, pH 6.1, containing 0.3 M NaCl and loaded onto a phosphocellulose column (0.5 × 5 cm) equilibrated with 10 mM MES, pH 6.1, containing 0.3 M NaCl. The column was washed with equilibration buffer, followed by a two sequential elution buffers. The first elution was performed with a linear heparin gradient (0–10 µM) in the equilibration buffer containing 0.3 M NaCl, yielding P1. The remaining tryptase was eluted with a linear gradient of 0.3–1.5 M NaCl in equilibration buffer containing 10 µM heparin, yielding P2.
G5 anti-tryptase IgG mAb-Sepharose immunoaffinity chromatography
The G5 IgG anti-tryptase mAb preferentially recognizes mature forms but not proforms of α- and β-tryptase (Schwartz et al., 2003). Fractions from P1 or P2 fractions were separately pooled, concentrated ∼10-fold using Centricon 10 microconcentrators (Amicon), diluted approximately fivefold with 10 mM MES, pH 6.5, to a NaCl concentration of ∼0.4 M, and loaded onto a G5 anti-tryptase IgG mAb-Sepharose column (2.5 × 1 cm) over ∼360 min at 4°C. The column was washed with the same buffer, linear gradients of pH 6–10 and ethylene glycol 0–50% were applied in parallel at 20 ml/h over 6 h at 4°C, and 0.5-ml fractions were collected. Fractions were adjusted to 1 M NaCl, and those with tryptase activity peaks were identified, pooled, concentrated, dialyzed with 10 mM MES, pH 6.5, containing 1 M NaCl and BSA (0.5 mg/ml), and analyzed as described below. rH-β- or rH-α-tryptase homotetramers, used as controls, were subjected to G5-Sepharose immunoaffinity chromatography as above.
N-deglycosylation
Human tryptases were N-deglycosylated with peptide-N-glycosidase F (PNGase F; New England Biolabs) under nondenaturing conditions. PNGase F was incubated with tryptases in 0.01 M Tris-HCl buffer, pH 7.5, at 37°C for 24 h. Deglycosylated tryptase to be subjected to SDS-PAGE under denaturing conditions was first treated with 1% SDS and 80 mM dithiothreitol (DTT) for 5 min in a boiling water bath.
Electrophoresis and zymography
Tryptase tetramers were denatured with 1% SDS and reduced with DTT (10 mM) in a boiling water bath, subjected to SDS-PAGE in a 10–20% polyacrylamide gradient gel or a 12% polyacrylamide gel, and Western blotted with the G3 anti-tryptase IgG, which recognizes both pro- and mature forms of α- and β-tryptases or G5 anti-tryptase IgG, which recognized mature forms of α- and β-tryptases.
Purified human tryptases and their major peak fractions obtained by phosphocellulose or G5-Sepharose chromatography and then N-deglycosylated, were separated by SDS-PAGE using a linear 10–20% acrylamide gradient in a Tris-Tricine buffer system. PageRuler Plus Prestained Protein Ladders (Thermo Fisher Scientific) were used to monitor mol wt. Tryptase bands were analyzed by Western blotting with the G3 anti-tryptase murine IgG mAb. Because α1 (27.701 kD) and α2 (27.743 kD; 99% amino acid sequence identity), are slightly larger than β1 (27.444 kD), β2 (27.446 kD), and β3 (27.581 kD) tryptases (β1, β2, and β3, 98–99% identity), denatured and reduced mature α-tryptases have a slightly slower electrophoretic mobility than β-tryptases. Mature α- and β-tryptase sequences are 92–96% identical.
Tryptase tetramers, stabilized with heparin (50 µg/ml) in 20 mM sodium acetate buffer, pH 6.0, containing 0.15 M NaCl, were concentrated by ultrafiltration to 3.5–5 µg/ml, adjusted to 1% SDS at room temperature, and subjected to gelatin zymography by nondenaturing, nonreducing SDS-PAGE in 8% polyacrylamide gels cast with 0.1% gelatin as described (Le et al., 2011b). SeeBlue Plus2 Pre-Stained Standards (Thermo Fisher Scientific) were used in parallel. After electrophoresis, gels were washed twice with 2.5% Triton X-100 at room temperature to remove SDS; incubated overnight at 37°C in 50 mM Tris-HCl buffer, pH 7.4, containing heparin glycosaminoglycan (50 µg/ml); stained with Coomassie Brilliant Blue G-250 (Thermo Fisher Scientific) for 30 min; washed; and visualized in the Odyssey Image System. As α-tryptase tetramers show negligible proteolytic/gelatinase activity, their migration was assessed after labeling α-protryptase with IRDye 800CW (LiCor), having an N-hydroxysuccinimide reactive group, at a 1:1 molar ratio during an overnight incubation in 100 mM MES buffer, pH 6.0, at 4°C per manufacturer’s instructions.
Inhibition of tryptase by B12 anti-tryptase IgG
Tryptase (5 µg/ml) extracted from human skin MCs from donors having different tryptase genotypes (ββ:ββ, ββ:βα, or βα:βα or with an increased TPSAB1 α-tryptase copy number) and purified by B2-Sepharose immunoaffinity chromatography was incubated with different concentrations of the B12 mAb at 37°C for ≤60 min in 0.1 ml of assay buffer (50 mM Hepes buffer, pH 7.4, containing heparin glycosaminoglycan [50 µg/ml], BSA [0.5 mg/ml], and 0.15 M NaCl). Portions were removed and assessed for enzyme activity with TGPK as substrate.
Cytosolic Ca2+ flux in Jurkat T cells and HMC1 MCs
Jurkat or HMC1 cells were washed with Hepes-buffered HBSS (137 mM NaCl, 4.7 mM KCl, 0.56 mM MgCl2, 2 mM CaCl2, 1.0 mM Na2HPO4, 10 mM Hepes, pH 7.4, 2.0 mM l-glutamine, and 5.5 mM d-glucose) without phenol red, containing 0.1% BSA, resuspended at 5 × 106 cells/ml, and then incubated in the same solution with 4 µM Fura-2/AM and 0.01% Pluronic F-127 for 30 min at room temperature (25°C). Cells then were washed and resuspended in calcium assay buffer (20 mM Hepes, pH 7.4, containing 150 mM NaCl, 3 mM KCl, 1.5 mM CaCl2, 10 mM glucose, and 0.25 mM sulphinpyrazone). Stimulated cell suspensions (0.5 ml) in 1-ml cuvettes were stirred with a magnetic bar and maintained at 25°C. Fluorescence measurements were continuously monitored with a PerkinElmer 650-10S fluorescence spectrometer at excitation wavelengths of 340 and 380 nm and an emission wavelength of 510 nm. The fluorescence ratio of the 510-nm emission responses at the two excitation wavelengths, proportional to the intracellular Ca2+ concentration, was calculated.
PAR2 cross-desensitization studies
PAR2 desensitization analyses were conducted to assess α/β-tryptase specificity for activating PAR2. Various forms of tryptase and known agonists or inhibitors of PAR2 were added serially to Jurkat cells suspended in a 0.5-ml cuvette at 37°C, while the intracellular calcium signal was continuously recorded. After the rise in intracellular calcium triggered by the first PAR2 agonist returned to baseline, desensitizing the response of only this receptor to subsequent triggers, a second test agonist was added, thereby permitting specificity to be assessed.
EMR2 purification
EMR2 was extracted from HMC1 cells by suspending 108 cells in 10 mM triethanolamine HCl buffer, pH 7.8, containing 150 mM NaCl, 5 mM EDTA, 1% NP-40, SBTI (50 µg/ml), and 1× EDTA-free Halt Protease Inhibitor Cocktail (Thermo Fisher Scientific), at 4°C. Solubilized material in the supernatant after centrifugation at 2,000 g for 20 min at 4°C was applied to a MonoQ 5/50 GL anion exchange column (GE Healthcare Life Sciences) equilibrated in 0.05 M Tris-HCl buffer, pH 7.8, containing 0.15 M NaCl, 5 mM CaCl2, and 1 mM MgCl2; washed with this buffer; and then eluted with a 0.12–1.0 M NaCl gradient, followed by step elution with 1.5 M NaCl, each in the equilibration buffer (except for the NaCl concentration and without CaCl2) at a flow rate of 1 ml/min at 4°C, 1 ml/gradient fraction. EMR2-containing fractions identified by Western blotting with anti-EMR2α IgG were combined; dialyzed in the equilibration buffer for dermatan sulfate chromatography; applied to a dermatan sulfate–Sepharose column (2 ml volume) equilibrated in 0.02 M Tris HCl, pH 7.5, containing 0.2 M NaCl, 2.5 mM EDTA, 5 mM CaCl2, and 1 mM MgCl2; washed with this equilibration buffer; and eluted with a 0.2–1-M NaCl linear gradient followed by a 1.5 M NaCl step elution, both in the wash solution without CaCl2. Dermatan sulfate–Sepharose fractions containing the bulk of EMR2 were combined, dialyzed in the equilibration buffer for the subsequent Mono Q chromatography step, and applied to a second Mono Q 5/50 GL column as above, but washed with equilibration buffer containing 0.5 M NaCl before being eluted with a 0.5–1.5 M NaCl gradient in the wash solution without CaCl2. Fractions containing the bulk of EMR2 were combined, concentrated 10–15-fold by ultrafiltration (Amicon Ultra 0.5 ml Centrifugal Filter; 10,000 mol wt cutoff), and subjected to gel filtration chromatography on a Superose 12 HR 10/30 size-exclusion column (GE Healthcare Life Sciences) equilibrated in 0.05 M Tris HCl, pH 7.5, buffer containing 0.15 M NaCl, 2.5 mM CaCl2, 1 mM MgCl2, and 2.5% glycerol. Fractions containing EMR2 by Western blotting were combined, concentrated 10-fold, analyzed by SDS-PAGE, and stained with Coomassie blue. Whether either β-tryptase or α/β-tryptase would cleave this purified, nondenatured, nonreduced EMR2 was further examined in Fig. 4 B (upper panel).
EMR2 silencing and Western blotting
1 d before transfection, CD34-derived MCs (2 × 105 cells/ml) were plated in 1 ml of AIM V serum-free medium containing rH-SCF (100 ng/ml) and rH-IL-6 (100 ng/ml) per well in a 24-well tissue culture plate. Cells were cultured at 37°C in a CO2 incubator for 24 h, after which time the cells were 70–80% confluent, ready to be transfected. EMR2 siRNA (sc-45381) and negative-control siRNA-A (sc-37007) were obtained from Santa Cruz Biotechnology. DNA-siRNA:Lipofectamine 2000 complexes were prepared according to manufacturer’s instructions. siRNA duplexes (8–10 µl containing 80–100 pmol siRNA) were mixed with 100 µl of Opti-MEM I medium without serum. In parallel, Lipofectamine 2000 (10 µl) was mixed with Opti-MEM I medium without serum (100 µl), gently pipetting the solution up and down, and then incubating the mixture for 15–45 min at room temperature. Cells were washed once with 2 ml of Opti-MEM I medium without serum. The wash medium was removed by aspiration, followed immediately by overlaying the transfection solution onto the washed cells, and then incubating these cells for 24, 48, and 72 h at 37°C in a CO2 incubator. Cells were collected, washed with PBS, and used as indicated.
For Western blotting, 50 µl lysis buffer was added to cell pellets, which were then vortexed and centrifuged. Supernatants were collected, and protein concentrations were measured. Supernatants containing 20 µg of protein were mixed with 125 mM Tris-HCl buffer, pH 6.8, containing 4% SDS, such that the final concentration of SDS was 1%. For samples to be reduced and heat-denatured, DTT (80 mM) was added, and samples were heated in a boiling water bath for 5 min. For samples that were not reduced and not heat-denatured, no DTT was added and samples were brought to room temperature before being subjected to SDS-PAGE (12% polyacrylamide). In both cases, PageRuler Plus Prestained Protein Ladder mol wt markers (Thermo Fisher Scientific) were run in parallel to the EMR2 samples. Samples were then transferred by electrophoresis to an Immobilon transfer membrane (Millipore), followed by blocking with Odyssey Blocking Buffer. Membranes were incubated overnight at 4°C with murine IgG mAb against EMR2α (2A1; Thermo Fisher Scientific) and rabbit polyclonal IgG anti-GAPDH (ab9485; Abcam), washed with PBS containing Tween 20, incubated with IRDye 680RD goat anti-mouse IgG (P/N 926-68070; LiCor) together with IRDye 800CW goat anti-rabbit IgG (P/N 926-32211; LiCor), washed, and scanned with an Odyssey CLx Imager.
MC vibratory challenge in vitro
Wells in 96-well flat-bottom plates were coated with either dermatan sulfate, chondroitin sulfate, or poly-l-lysine (100 µg/ml) in PBS overnight at 4°C, and then washed three times with PBS alone. MCs (25,000 cells per 100-µl well) in 10 mM Hepes buffer, pH 7.4, containing 1.8 mM CaCl2, 1 mM MgCl2, and 0.04% BSA, were then added to each experimental well, centrifuged for 5 min at 450 g at room temperature, washed three times with this Hepes buffer, and suspended in this Hepes buffer, also containing heparin glycosaminoglycan (10 µg/ml). Various forms of tryptase (10 µl of either 0.3 or 3 µg/ml of 0.02 M Mes, pH 6.0, containing 1 M NaCl and BSA [250 µg/ml]) or control solutions (10 µl) are added to the cells for the time periods specified at 37°C. Attached cells are vibrated (750 rpm; Stat Fax 2200; Awareness Technology) or vortexed (Level 4–5, Vortex-Genie 2 Shaker; Scientific Industries) or incubated without agitation for 30 min at 37°C, and then centrifuged at 450 g at 4°C for 5 min to separate released from retained substances found in the respective supernatant and pellet fractions. Cells in the pellet are resuspended in Tyrode’s buffer containing 5 mM EDTA, SBTI (50 µg/ml), 1× EDTA-free Halt Protease Inhibitor Cocktail (Thermo Fisher Scientific), and 1% Triton X-100, and lysed by sonication (four 15-s pulses at room temperature; Solid State Ultrasonic FS-9 bath; Fisher Scientific), after which β-hexosaminidase levels are measured (Schwartz et al., 1981) and % degranulation calculated as 100 × [supernatant/(supernatant + pellet)].
Vibratory cutaneous challenge
Each subject underwent a vibratory cutaneous challenge and was tryptase genotyped after providing written informed consent on National Institutes of Health–approved research protocols (NCT01164241 and/or NCT00852943; Lyons et al., 2014, 2016), and received one point each for erythema, induration, pruritus/tingling/pain, or warmth and two additional points for either expansion beyond the margins of the vortex and/or systemic symptoms, for a maximal possible response score of six.
Statistical analysis
Data were analyzed using SigmaStat (Systat Software). For normally distributed data, a one-way ANOVA compared with the control was used with a Holm–Sidak post hoc analysis to isolate the groups that were statistically different from the control group. For data not normally distributed, a nonparametric Kruskal–Wallis ANOVA compared with control group was used with a post hoc Dunn’s method to isolate the groups that were statistically different from the control group. Significance levels were as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001. Data in each plot is displayed as mean ± SD if normally distributed or as a box and whisker plot, with median, 25th/75th percentiles, 10th/90th percentiles, and outliers for data not normally distributed.
Online supplemental material
Fig. S1 shows that binding of the G5 mAb to mature rH-β2-tryptase occurs faster than to mature rH-α1-tryptase, which led us to successfully separate α-tryptase tetramers from α/β-tryptase heterotetramers in Fig. 2 C by G5-Sepharose immunoaffinity chromatography, whereby α-tryptase eluted earlier than α/β-tryptase. Fig. S2 shows that heparin is required for α/β-tryptase to cause MCs to become susceptible to vibration-triggered degranulation, that β-tryptase lacks this ability regardless of the presence of absence of heparin, and that the time course for α/β-tryptase to make human MCs derived in vitro from peripheral blood CD34 progenitors is comparable to the time course shown in Fig. 3 B for skin MCs. Fig. S3 shows the purification of EMR2 from HMC1 cells (an MC leukemia cell line), which was then shown in Fig. 4 B, upper panel, to be cleaved by α/β-tryptase but not β-tryptase, producing an EMR2α fragment migrating at a slightly faster electrophoretic mobility than the 55-kD mol wt standard. Fig. S4 examines the data shown in Fig. 4 D based on the α/β tryptase genotype rather than the α/β tryptase gene ratio, showing that having two or more α-tryptase genes, regardless whether the subject has a normal genotype with two α-tryptase genes (one on each parental chromosome 16) or a genotype with extra copies of the α-tryptase gene, would have a higher cutaneous response to vibration than those with either one or zero α-tryptase genes. Fig. S5 shows that neither β1 nor β3 protryptase nor mature tryptase activates PAR2 on Jurkat cells, analogous to what was shown in Fig. 5 A for rH-β2 protryptase and mature tryptase.
Supplementary Material
Acknowledgments
Q.T. Le and L.B. Schwartz thank B. Ward and M. Hicks (Virginia Commonwealth University, Richmond, VA) for sharing MCs derived from peripheral blood progenitors and V. Harlow for performing tryptase ELISAs. J.J. Lyons and J.D. Milner thank J. Chovanec, S. Blackstone, T. DiMaggio, N. Jones, and C. Nelson for assistance with tryptase genotyping and vibration urticarial challenges.
This work was supported by the Charles Thomas Research Fund of the Medical College of Virginia Foundation and the National Institutes of Health, grants U18AI77435 and R0127517 (L.B. Schwartz); and by the Division of Intramural Research of the National Institute of Allergy and Infectious Diseases, National Institutes of Health. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
L.B. Schwartz is a paid consultant for Genentech, Inc., collects licensing fees for the B12 mAb paid to Virginia Commonwealth University by Genentech, Inc., and collects royalties for the clinical tryptase immunoassay paid to Virginia Commonwealth University by Thermo Fisher Scientific Inc.; Q.T. Le collects licensing fees for the B12 mAb paid to Virginia Commonwealth University by Genentech; and R.A. Lazarus is an employee of Genentech Inc. All other authors declare no competing financial interests.
Author contributions: L.B. Schwartz contributed to experimental design and data analysis; Q.T. Le contributed to designing, performing, and analyzing experiments; R.A. Lazarus contributed to data analysis; J.J. Lyons contributed to designing, performing, and analyzing experiments; J.D. Milner contributed to designing and analyzing experiments; A.N. Naranjo and A. Olivera contributed to designing experiments; and D.D. Metcalfe contributed to designing experiments. All coauthors contributed to writing the manuscript.
References
- Abdelmotelb A.M., Rose-Zerilli M.J., Barton S.J., Holgate S.T., Walls A.F., and Holloway J.W.. 2014. Alpha-tryptase gene variation is associated with levels of circulating IgE and lung function in asthma. Clin. Exp. Allergy. 44:822–830. 10.1111/cea.12259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter S.C., Kramps J.A., Janoff A., and Schwartz L.B.. 1990. Interactions of human mast cell tryptase with biological protease inhibitors. Arch. Biochem. Biophys. 276:26–31. 10.1016/0003-9861(90)90005-J [DOI] [PubMed] [Google Scholar]
- Berger P., Perng D.W., Thabrew H., Compton S.J., Cairns J.A., McEuen A.R., Marthan R., Tunon De Lara J.M., and Walls A.F.. 2001. Tryptase and agonists of PAR-2 induce the proliferation of human airway smooth muscle cells. J. Appl. Physiol. 91:1372–1379. 10.1152/jappl.2001.91.3.1372 [DOI] [PubMed] [Google Scholar]
- Boyden S.E., Desai A., Cruse G., Young M.L., Bolan H.C., Scott L.M., Eisch A.R., Long R.D., Lee C.C., Satorius C.L., et al. . 2016. Vibratory Urticaria Associated with a Missense Variant in ADGRE2. N. Engl. J. Med. 374:656–663. 10.1056/NEJMoa1500611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brightling C.E., Bradding P., Symon F.A., Holgate S.T., Wardlaw A.J., and Pavord I.D.. 2002. Mast-cell infiltration of airway smooth muscle in asthma. N. Engl. J. Med. 346:1699–1705. 10.1056/NEJMoa012705 [DOI] [PubMed] [Google Scholar]
- Compton S.J., McGuire J.J., Saifeddine M., and Hollenberg M.D.. 2002a Restricted ability of human mast cell tryptase to activate proteinase-activated receptor-2 in rat aorta. Can. J. Physiol. Pharmacol. 80:987–992. 10.1139/y02-125 [DOI] [PubMed] [Google Scholar]
- Compton S.J., Sandhu S., Wijesuriya S.J., and Hollenberg M.D.. 2002b Glycosylation of human proteinase-activated receptor-2 (hPAR2): role in cell surface expression and signalling. Biochem. J. 368:495–505. 10.1042/bj20020706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig S.S., DeBlois G., and Schwartz L.B.. 1986. Mast cells in human keloid, small intestine, and lung by an immunoperoxidase technique using a murine monoclonal antibody against tryptase. Am. J. Pathol. 124:427–435. [PMC free article] [PubMed] [Google Scholar]
- Dong X., and Dong X.. 2018. Peripheral and Central Mechanisms of Itch. Neuron. 98:482–494. 10.1016/j.neuron.2018.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frateschi S., Camerer E., Crisante G., Rieser S., Membrez M., Charles R.P., Beermann F., Stehle J.C., Breiden B., Sandhoff K., et al. . 2011. PAR2 absence completely rescues inflammation and ichthyosis caused by altered CAP1/Prss8 expression in mouse skin. Nat. Commun. 2:161 10.1038/ncomms1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa D., Kashiwakura J., Kita H., Kikukawa Y., Fujitani Y., Sasaki-Sakamoto T., Kuroda K., Nunomura S., Hayama K., Terui T., et al. . 2014. Expression of Mas-related gene X2 on mast cells is upregulated in the skin of patients with severe chronic urticaria. J. Allergy Clin. Immunol. 134:622–633.e9. 10.1016/j.jaci.2014.05.004 [DOI] [PubMed] [Google Scholar]
- Fukuoka Y., and Schwartz L.B.. 2006. The B12 anti-tryptase monoclonal antibody disrupts the tetrameric structure of heparin-stabilized beta-tryptase to form monomers that are inactive at neutral pH and active at acidic pH. J. Immunol. 176:3165–3172. 10.4049/jimmunol.176.5.3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guida M., Riedy M., Lee D., and Hall J.. 2000. Characterization of two highly polymorphic human tryptase loci and comparison with a newly discovered monkey tryptase ortholog. Pharmacogenetics. 10:389–396. 10.1097/00008571-200007000-00002 [DOI] [PubMed] [Google Scholar]
- Huang C., Li L., Krilis S.A., Chanasyk K., Tang Y., Li Z., Hunt J.E., and Stevens R.L.. 1999. Human tryptases alpha and beta/II are functionally distinct due, in part, to a single amino acid difference in one of the surface loops that forms the substrate-binding cleft. J. Biol. Chem. 274:19670–19676. 10.1074/jbc.274.28.19670 [DOI] [PubMed] [Google Scholar]
- Huang C., Morales G., Vagi A., Chanasyk K., Ferrazzi M., Burklow C., Qiu W.T., Feyfant E., Sali A., and Stevens R.L.. 2000. Formation of enzymatically active, homotypic, and heterotypic tetramers of mouse mast cell tryptases. Dependence on a conserved Trp-rich domain on the surface. J. Biol. Chem. 275:351–358. 10.1074/jbc.275.1.351 [DOI] [PubMed] [Google Scholar]
- Huang C., De Sanctis G.T., O’Brien P.J., Mizgerd J.P., Friend D.S., Drazen J.M., Brass L.F., and Stevens R.L.. 2001. Evaluation of the substrate specificity of human mast cell tryptase beta I and demonstration of its importance in bacterial infections of the lung. J. Biol. Chem. 276:26276–26284. 10.1074/jbc.M102356200 [DOI] [PubMed] [Google Scholar]
- I K.Y., Huang Y.S., Hu C.H., Tseng W.Y., Cheng C.H., Stacey M., Gordon S., Chang G.W., and Lin H.H.. 2017. Activation of Adhesion GPCR EMR2/ADGRE2 Induces Macrophage Differentiation and Inflammatory Responses via Gα16/Akt/MAPK/NF-κB Signaling Pathways. Front. Immunol. 8:373 10.3389/fimmu.2017.00373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irani A.A., Garriga M.M., Metcalfe D.D., and Schwartz L.B.. 1990. Mast cells in cutaneous mastocytosis: accumulation of the MCTC type. Clin. Exp. Allergy. 20:53–58. 10.1111/j.1365-2222.1990.tb02775.x [DOI] [PubMed] [Google Scholar]
- Le Q.T., Gomez G., Zhao W., Hu J., Xia H.Z., Fukuoka Y., Katunuma N., and Schwartz L.B.. 2011a Processing of human protryptase in mast cells involves cathepsins L, B, and C. J. Immunol. 187:1912–1918. 10.4049/jimmunol.1001806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Q.T., Min H.K., Xia H.Z., Fukuoka Y., Katunuma N., and Schwartz L.B.. 2011b Promiscuous processing of human alphabeta-protryptases by cathepsins L, B, and C. J. Immunol. 186:7136–7143. 10.4049/jimmunol.1001804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Q.T., Lotfi-Emran S., Min H.K., and Schwartz L.B.. 2014. A simple, sensitive and safe method to determine the human α/β-tryptase genotype. PLoS One. 9:e114944 10.1371/journal.pone.0114944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden D.R., Manning B.P., Bunnett N.W., and Mawe G.M.. 2001. Agonists of proteinase-activated receptor 2 excite guinea pig ileal myenteric neurons. Eur. J. Pharmacol. 431:311–314. 10.1016/S0014-2999(01)01447-9 [DOI] [PubMed] [Google Scholar]
- Lohman R.J., Cotterell A.J., Suen J., Liu L., Do A.T., Vesey D.A., and Fairlie D.P.. 2012. Antagonism of protease-activated receptor 2 protects against experimental colitis. J. Pharmacol. Exp. Ther. 340:256–265. 10.1124/jpet.111.187062 [DOI] [PubMed] [Google Scholar]
- Lotfi-Emran S., Ward B.R., Le Q.T., Pozez A.L., Manjili M.H., Woodfolk J.A., and Schwartz L.B.. 2018. Human mast cells present antigen to autologous CD4+ T cells. J. Allergy Clin. Immunol. 141:311–321.e10. 10.1016/j.jaci.2017.02.048 [DOI] [PubMed] [Google Scholar]
- Lyons J.J., Sun G., Stone K.D., Nelson C., Wisch L., O’Brien M., Jones N., Lindsley A., Komarow H.D., Bai Y., et al. . 2014. Mendelian inheritance of elevated serum tryptase associated with atopy and connective tissue abnormalities. J. Allergy Clin. Immunol. 133:1471–1474. 10.1016/j.jaci.2013.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons J.J., Yu X., Hughes J.D., Le Q.T., Jamil A., Bai Y., Ho N., Zhao M., Liu Y., O’Connell M.P., et al. . 2016. Elevated basal serum tryptase identifies a multisystem disorder associated with increased TPSAB1 copy number. Nat. Genet. 48:1564–1569. 10.1038/ng.3696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt U., Zettl F., Huber R., Bode W., and Sommerhoff C.. 2002. The crystal structure of human alpha1-tryptase reveals a blocked substrate-binding region. J. Mol. Biol. 321:491–502. 10.1016/S0022-2836(02)00625-3 [DOI] [PubMed] [Google Scholar]
- Maun H.R., Liu P.S., Franke Y., Eigenbrot C., Forrest W.F., Schwartz L.B., and Lazarus R.A.. 2018. Dual functionality of β-tryptase protomers as both proteases and cofactors in the active tetramer. J. Biol. Chem. 293:9614–9628. 10.1074/jbc.M117.812016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil B.D., Pundir P., Meeker S., Han L., Undem B.J., Kulka M., and Dong X.. 2015. Identification of a mast-cell-specific receptor crucial for pseudo-allergic drug reactions. Nature. 519:237–241. 10.1038/nature14022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.S., Westin E.H., and Schwartz L.B.. 1989. Cloning and characterization of complementary DNA for human tryptase. J. Clin. Invest. 84:1188–1195. 10.1172/JCI114284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.S., Moxley G., and Schwartz L.B.. 1990. Cloning and characterization of a second complementary DNA for human tryptase. J. Clin. Invest. 86:864–870. 10.1172/JCI114786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molino M., Barnathan E.S., Numerof R., Clark J., Dreyer M., Cumashi A., Hoxie J.A., Schechter N., Woolkalis M., and Brass L.F.. 1997. Interactions of mast cell tryptase with thrombin receptors and PAR-2. J. Biol. Chem. 272:4043–4049. 10.1074/jbc.272.7.4043 [DOI] [PubMed] [Google Scholar]
- Pereira P.J., Bergner A., Macedo-Ribeiro S., Huber R., Matschiner G., Fritz H., Sommerhoff C.P., and Bode W.. 1998. Human β-tryptase is a ring-like tetramer with active sites facing a central pore. Nature. 392:306–311. 10.1038/32703 [DOI] [PubMed] [Google Scholar]
- Ren S., Sakai K., and Schwartz L.B.. 1998. Regulation of human mast cell beta-tryptase: conversion of inactive monomer to active tetramer at acid pH. J. Immunol. 160:4561–4569. [PubMed] [Google Scholar]
- Sabato V., Chovanec J., Faber M., Milner J.D., Ebo D., and Lyons J.J.. 2018. First Identification of an Inherited TPSAB1 Quintuplication in a Patient with Clonal Mast Cell Disease. J. Clin. Immunol. 38:457–459. 10.1007/s10875-018-0506-y [DOI] [PubMed] [Google Scholar]
- Sakai K., Ren S., and Schwartz L.B.. 1996. A novel heparin-dependent processing pathway for human tryptase. Autocatalysis followed by activation with dipeptidyl peptidase I. J. Clin. Invest. 97:988–995. 10.1172/JCI118523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter N.M., Brass L.F., Lavker R.M., and Jensen P.J.. 1998. Reaction of mast cell proteases tryptase and chymase with protease activated receptors (PARs) on keratinocytes and fibroblasts. J. Cell. Physiol. 176:365–373. [DOI] [PubMed] [Google Scholar]
- Schmidlin F., Amadesi S., Vidil R., Trevisani M., Martinet N., Caughey G., Tognetto M., Cavallesco G., Mapp C., Geppetti P., and Bunnett N.W.. 2001. Expression and function of proteinase-activated receptor 2 in human bronchial smooth muscle. Am. J. Respir. Crit. Care Med. 164:1276–1281. 10.1164/ajrccm.164.7.2101157 [DOI] [PubMed] [Google Scholar]
- Schwartz L.B., and Bradford T.R.. 1986. Regulation of tryptase from human lung mast cells by heparin. Stabilization of the active tetramer. J. Biol. Chem. 261:7372–7379. [PubMed] [Google Scholar]
- Schwartz L.B., Lewis R.A., Seldin D., and Austen K.F.. 1981. Acid hydrolases and tryptase from secretory granules of dispersed human lung mast cells. J. Immunol. 126:1290–1294. [PubMed] [Google Scholar]
- Schwartz L.B., Irani A.M., Roller K., Castells M.C., and Schechter N.M.. 1987a Quantitation of histamine, tryptase, and chymase in dispersed human T and TC mast cells. J. Immunol. 138:2611–2615. [PubMed] [Google Scholar]
- Schwartz L.B., Metcalfe D.D., Miller J.S., Earl H., and Sullivan T.. 1987b Tryptase levels as an indicator of mast-cell activation in systemic anaphylaxis and mastocytosis. N. Engl. J. Med. 316:1622–1626. 10.1056/NEJM198706253162603 [DOI] [PubMed] [Google Scholar]
- Schwartz L.B., Yunginger J.W., Miller J., Bokhari R., and Dull D.. 1989. Time course of appearance and disappearance of human mast cell tryptase in the circulation after anaphylaxis. J. Clin. Invest. 83:1551–1555. 10.1172/JCI114051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz L.B., Bradford T.R., Lee D.C., and Chlebowski J.F.. 1990. Immunologic and physicochemical evidence for conformational changes occurring on conversion of human mast cell tryptase from active tetramer to inactive monomer. Production of monoclonal antibodies recognizing active tryptase. J. Immunol. 144:2304–2311. [PubMed] [Google Scholar]
- Schwartz L.B., Sakai K., Bradford T.R., Ren S., Zweiman B., Worobec A.S., and Metcalfe D.D.. 1995. The α form of human tryptase is the predominant type present in blood at baseline in normal subjects and is elevated in those with systemic mastocytosis. J. Clin. Invest. 96:2702–2710. 10.1172/JCI118337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz L.B., Min H.K., Ren S., Xia H.Z., Hu J., Zhao W., Moxley G., and Fukuoka Y.. 2003. Tryptase precursors are preferentially and spontaneously released, whereas mature tryptase is retained by HMC-1 cells, Mono-Mac-6 cells, and human skin-derived mast cells. J. Immunol. 170:5667–5673. 10.4049/jimmunol.170.11.5667 [DOI] [PubMed] [Google Scholar]
- Selwood T., Wang Z.M., McCaslin D.R., and Schechter N.M.. 2002. Diverse stability and catalytic properties of human tryptase alpha and beta isoforms are mediated by residue differences at the S1 pocket. Biochemistry. 41:3329–3340. 10.1021/bi015662v [DOI] [PubMed] [Google Scholar]
- Soto D., Malmsten C., Blount J.L., Muilenburg D.J., and Caughey G.H.. 2002. Genetic deficiency of human mast cell α-tryptase. Clin. Exp. Allergy. 32:1000–1006. 10.1046/j.1365-2222.2002.01416.x [DOI] [PubMed] [Google Scholar]
- Steinhoff M., Vergnolle N., Young S.H., Tognetto M., Amadesi S., Ennes H.S., Trevisani M., Hollenberg M.D., Wallace J.L., Caughey G.H., et al. . 2000. Agonists of proteinase-activated receptor 2 induce inflammation by a neurogenic mechanism. Nat. Med. 6:151–158. 10.1038/72247 [DOI] [PubMed] [Google Scholar]
- Steinhoff M., Neisius U., Ikoma A., Fartasch M., Heyer G., Skov P.S., Luger T.A., and Schmelz M.. 2003. Proteinase-activated receptor-2 mediates itch: a novel pathway for pruritus in human skin. J. Neurosci. 23:6176–6180. 10.1523/JNEUROSCI.23-15-06176.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi N.N., Tamraz B., Chu C., Kwok P.Y., and Caughey G.H.. 2009. Human subjects are protected from mast cell tryptase deficiency despite frequent inheritance of loss-of-function mutations. J. Allergy Clin. Immunol. 124:1099–105.e1: 4. 10.1016/j.jaci.2009.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez C.V., Roman A.D., Lan N.T., Huy N.T., Mercado E.S., Espino F.E., Perez M.L., Huong V.T., Thuy T.T., Tham V.D., et al. . 2015. Alpha tryptase allele of Tryptase 1 (TPSAB1) gene associated with Dengue Hemorrhagic Fever (DHF) and Dengue Shock Syndrome (DSS) in Vietnam and Philippines. Hum. Immunol. 76:318–323. 10.1016/j.humimm.2015.03.009 [DOI] [PubMed] [Google Scholar]
- Vergnolle N., Bunnett N.W., Sharkey K.A., Brussee V., Compton S.J., Grady E.F., Cirino G., Gerard N., Basbaum A.I., Andrade-Gordon P., et al. . 2001. Proteinase-activated receptor-2 and hyperalgesia: A novel pain pathway. Nat. Med. 7:821–826. 10.1038/89945 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.