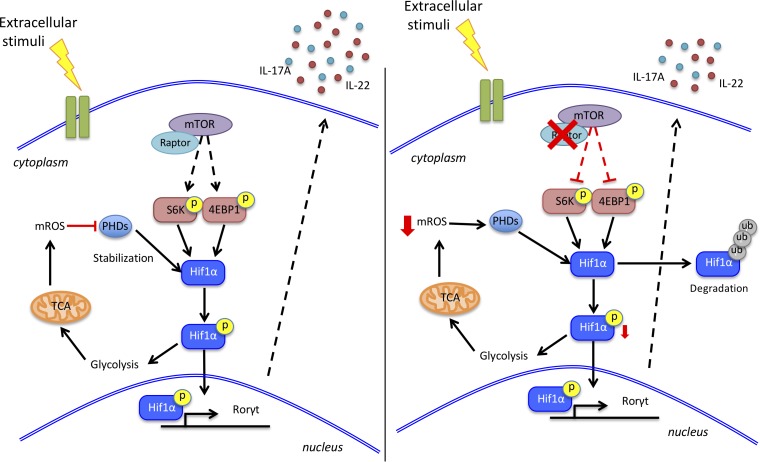
ILC3s are innate immune cells in the intestine that contribute to mucosal barrier integrity and support antimicrobial responses. Di Luccia et al. show that an mTORC1-HIF1α–mediated metabolic program sustains ILC3 numbers and activation in both mouse and human.
Abstract
Group 3 innate lymphoid cells (ILC3s) are the innate counterparts of Th17 that require the transcription factor RORγt for development and contribute to the defense against pathogens through IL-22 and IL-17 secretion. Proliferation and effector functions of Th17 require a specific mTOR-dependent metabolic program that utilizes high-rate glycolysis, while mitochondrial lipid oxidation and production of reactive oxygen species (mROS) support alternative T reg cell differentiation. Whether ILC3s employ a specific metabolic program is not known. Here, we find that ILC3s rely on mTOR complex 1 (mTORC1) for proliferation and production of IL-22 and IL-17A after in vitro activation and Citrobacter rodentium infection. mTORC1 induces activation of HIF1α, which reprograms ILC3 metabolism toward glycolysis and sustained expression of RORγt. However, in contrast to Th17, ILC3 activation requires mROS production; rather than inducing an alternative regulatory fate as it does in CD4 T cells, mROS stabilizes HIF1α and RORγt in ILC3s and thereby promotes their activation. We conclude that ILC3 activation relies on a metabolic program that integrates glycolysis with mROS production.
Graphical Abstract
Introduction
Group 3 innate lymphoid cells (ILC3s) are innate counterparts of T helper 17 cells (Th17s; Eberl et al., 2015; Klose and Artis, 2016; Colonna, 2018). They require retinoic acid receptor–related orphan nuclear receptor γt (RORγt) for development and secrete IL-22 and IL-17. Human ILC3s express the markers NKp44 and CCR6 and preferentially produce IL-22. Several subsets of mouse ILC3s are distinguished based on the expression of cell surface CCR6 and NKp46 as well as the transcription factor T-bet (Diefenbach et al., 2014). CCR6+NKp46−T-bet− ILC3s secrete both IL-22 and IL-17A, whereas CCR6−NKp46−T-bet+ ILC3s and CCR6−NKp46+T-bet+ ILC3s primarily secrete IL-22. ILC3s are most abundant in the gastrointestinal tract, where they contribute to mucosal barrier integrity and resistance to bacterial insults, such as Citrobacter rodentium infection (Satoh-Takayama et al., 2008; Zheng et al., 2008; Sonnenberg et al., 2011; Muñoz et al., 2015; Song et al., 2015; Rankin et al., 2016). Furthermore, ILC3 production of IL-17 supports antifungal and antiviral responses (Gladiator et al., 2013; Hernández et al., 2015). However, inappropriate activation of ILC3s may contribute to inflammatory bowel disease (IBD) and cancer. Many DNA polymorphisms associated with IBD impact genes expressed in ILC3s as well as Th17s (Jostins et al., 2012; Björklund et al., 2016; Koues et al., 2016). Moreover, ILC3s cause gastrointestinal pathology in mouse models of IBD and cancer (Kirchberger et al., 2013; Song et al., 2015; Pearson et al., 2016).
Although ILC3 and Th17 functions largely overlap, the two are differentially regulated, since Th17 cytokine secretion is triggered through the TCR, while ILC3s respond to specific cytokines and other soluble factors in the tissue microenvironment (Bando and Colonna, 2016; Huntington et al., 2016). Metabolism plays a key role in regulation in Th17s, which rely on a marked increase in aerobic glycolysis for energy and biosynthetic precursors (Wang and Green, 2012; Chang et al., 2013; Pearce et al., 2013; Buck et al., 2015). This specific metabolic program is controlled by mechanistic target of rapamycin (mTOR) and its related complex 1 (mTORC1; Pollizzi and Powell, 2014; Jones and Pearce, 2017), which induce expression of the transcription factor hypoxia-inducible factor 1α (HIF1α). HIF1α orchestrates glycolysis (Düvel et al., 2010) and, in addition, directly promotes the transcription of RORγt (Dang et al., 2011; Shi et al., 2011). In contrast to Th17s, T regulatory (T reg) cells exact less energy from glycolysis and rely on an alternative metabolic program that requires mitochondrial oxidation of lipids and pyruvate (Delgoffe et al., 2009, 2011; Michalek et al., 2011; Gerriets et al., 2015; Newton et al., 2016). Mitochondrial metabolism results in the production of mitochondrial ROS (mROS) that attenuate glycolysis by inhibiting pyruvate dehydrogenase kinase, a key checkpoint of glycolysis (Gerriets et al., 2015).
To determine whether the metabolic program regulating ILC3s parallels that of Th17s, we examined the impact of the mTORC1-HIF1α axis on ILC3s at steady state, after in vitro stimulation with cytokines and following in vivo activation by bacterial infection. Using conditional mTORC1 (Rptor) knockout mice and mTORC1-HIF1α pharmacological inhibitors, we demonstrated a major role for the mTORC1-HIF1α axis in sustaining ILC3 numbers, proliferation, and cytokine secretion through induction of both glycolysis and RORγt expression. However, in contrast to Th17s, ILC3 activation also markedly increased mitochondrial respiration and generation of mROS, which consolidated ILC3 glycolysis, proliferation, and effector functions rather than inducing an alternative cell fate. ILC3 reliance on both glycolysis and mROS was conserved in humans. These results reveal an unanticipated metabolic regulation of ILC3s, which integrate glycolysis and mROS production to fulfill the metabolic demands needed for activation.
Results and discussion
RptorΔRorc mice have reduced ILC3 numbers at steady state
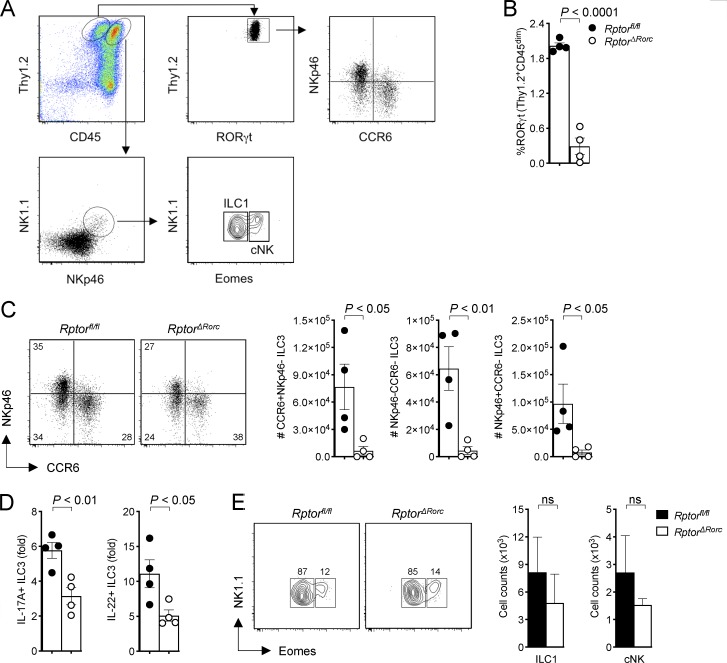
To determine whether ILC3s rely on mTOR signaling for metabolic adaptation to environmental signals, we crossed mice in which the gene for the mTORC1 subunit Raptor is flanked by loxP sites (Rptorfl/fl) with mice expressing a BAC transgene in which the RORγt promoter drives the Cre recombinase (Rorc-Cre). In these conditional knockout mice (called RptorΔRorc mice hereafter), the Raptor gene is deleted in ILC3s as well as in all T cells, because RORγt is transiently expressed during thymic T cell development (Eberl and Littman, 2004). RptorΔRorc mice had significantly fewer small intestinal RORγt+ ILC3s, including the CCR6+NKp46−, NKp46−CCR6−, and NKp46+CCR6− subsets, than did Rptorfl/fl mice (Fig. 1, A–C). Additionally, the residual ILC3s in RptorΔRorc mice produced relatively little IL-17A and IL-22 ex vivo in response to IL-1β plus IL-23 (Fig. 1 D). In contrast to the marked phenotypic changes noted in ILC3s, NK1.1+Eomes− ILC1s and NK1.1+Eomes+ conventional natural killer (cNK) cell numbers were comparable in RptorΔRorc and Rptorfl/fl mice (Fig. 1 E). Thus, mouse intestinal ILC3s require mTORC1 signaling for development and function. This result is further corroborated by the demonstration that deletion of mTORC1 in NKp46+ cells reduces the number of NKp46+ ILC3s, among others (Marçais et al., 2014).
Figure 1.
RptorΔRorc mice have lower numbers of ILC3 at steady state. (A) Gating strategy of ILC3 subsets in RptorΔRorc and Rptorfl/fl small intestine lamina propria. (B) Percentages of small intestine lamina propria ILC3 expressing RORγt in RptorΔRorc and Rptorfl/fl mice (n = 4). (C) Representative FACS plots and numbers of CCR6+NKp46−, CCR6−NKp46− and CCR6−NKp46+ of RORγt+ILC3 in RptorΔRorc and Rptorfl/fl mice. (D) Fold change in cells producing IL-17A and IL-22 within RORγt+ILC3 stimulated with IL-1β + IL-23 ex vivo (n = 4). (E) Representative FACS plots and numbers of ILC1 (NK1.1+Eomes−) and cNK (NK1.1+Eomes+) cells in RptorΔRorc and Rptorfl/fl small intestine lamina propria (n = 4). ns, not significant. Data are pooled from two independent experiments (mean ± SEM; Student's t test; B–E).
Inhibition of mTORC1 signaling impairs the ILC3 response to bacterial infection
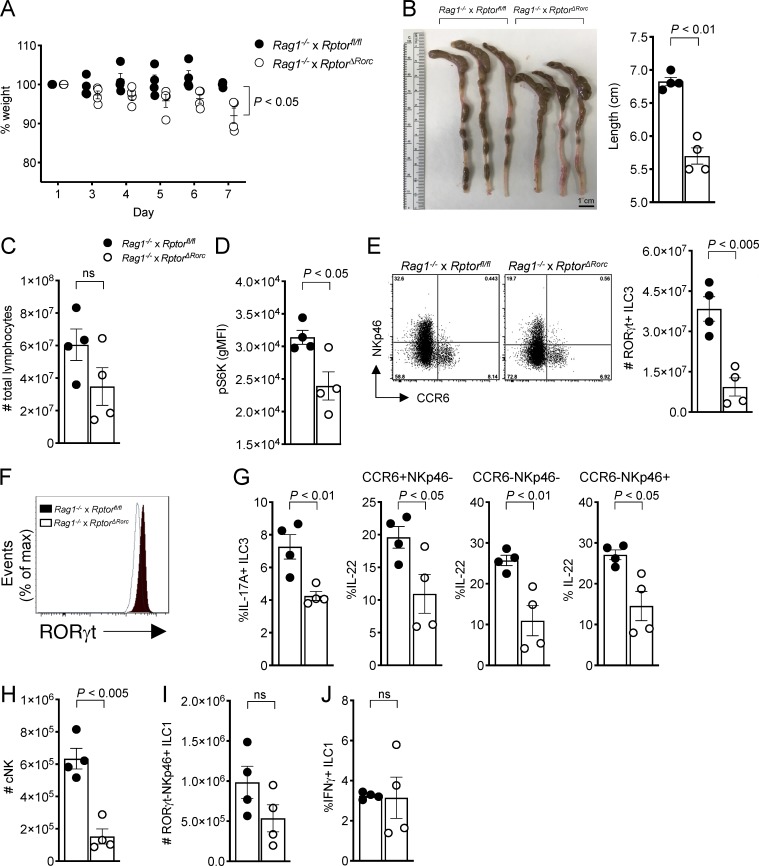
We next sought to determine whether mTORC1 deficiency impacts the ILC3 response to intestinal bacterial infection in vivo. Since RptorΔRorc mice lack the mTORC1 complex in both ILC3s and T cells, we generated Rag1−/− × Rptorfl/fl and Rag1−/− × RptorΔRorc animals, in which ILC3s are the only source of IL-22 and IL-17, and infected mice with C. rodentium by gavage for 7 d. We also treated Rag1−/− mice with rapamycin (Rapa) by i.p. injection to inhibit mTORC1 or with DMSO vehicle, then infected them with C. rodentium (Fig. S1 A). Uninfected Rag1−/− mice treated with Rapa did not show any obvious changes in body weight and colon or cecum length compared with mice injected with DMSO vehicle (Fig. S1 B). Upon C. rodentium infection, Rag1−/− mice lacking mTORC1 in ILC3s or treated with Rapa showed a significant loss of body weight (Figs. 2 A and S1 C), had marked reduction of colon and/or cecum length (Figs. 2 B and S1 D), and had fewer lymphocytes within the lamina propria (Figs. 2 C and S1 E) compared with control mice. Moreover, Rag1−/− × RptorΔRorc significantly impaired phosphorylation of the translational regulator S6 kinase (S6K) in lamina propria ILC3s during the infection (Fig. 2 D). Rag1−/− × RptorΔRorc and Rag1−/− mice treated with Rapa had a twofold reduction of all ILC3s (Figs. 2 E and S1 F); the remaining ILC3s had less intracellular RORγt (Figs. 2 F and S1 G) and failed to produce large amounts of IL-17A and IL-22 during infection (Figs. 2 G and S1 H). Also, we observed a marked decrease in cNK cells from the Rag1−/− × RptorΔRorc infected mice (Fig. 2 H), which could be due to the Rag deficiency that diminished NK cell development and fitness (Karo et al., 2014). In contrast, inhibition of mTORC1 in ILC3s had no detectable impact on the numbers of NKp46+RORγt− ILC1s (Figs. 2 I and S1 I) or their production of IFNγ (Figs. 2 J and S1 J). Thus, our data in Rag1−/− × RptorΔRorc and Rag1−/− mice treated with Rapa strongly supported that mTORC1 is required to sustain ILC3 numbers, expression of RORγt, and production of IL-17A and IL-22 during C. rodentium infection and that mTORC1 acts mainly in a cell-intrinsic fashion.
Figure 2.
Inhibition of mTORC1 signaling impairs ILC3 response to a bacterial infection. (A) Changes (%) in body weights of C. rodentium–infected Rag1−/− × Rptorfl/fl and Rag1−/− × RptorΔRorc mice (n = 4). (B) Images and lengths of colon and cecum from C. rodentium–infected mice (day 7). (C) Total numbers of intestinal lymphocytes in C. rodentium–infected Rag1−/− × Rptorfl/fl and Rag1−/− × RptorΔRorc mice (day 7, n = 4). (D–F) Expression of pS6K (D), representative FACS plots and numbers of RORγt+ ILC3s (E), and expression of RORγt (F) in C. rodentium–infected Rag1−/− × Rptorfl/fl and Rag1−/− × RptorΔRorc mice (day 7, n = 4). (G) Expression of IL-17A and IL-22 within small intestine ILC3s in C. rodentium–infected mice (day 7, n = 4). (H–J) Numbers of cNK cells (H) and RORγt−NKp46+ ILC1 (I) and percentages of IFNγ+ ILC1 (J) in C. rodentium–infected Rag1−/− × Rptorfl/fl and Rag1−/− × RptorΔRorc mice (day 7, n = 4). ns, not significant. Data are from one experiment representative of two independent experiments (mean ± SEM of n = 4; Student’s t test). gMFI, geometric mean fluorescence intensity.
mTORC1 sustains RORγt expression and cytokine production through HIF1α
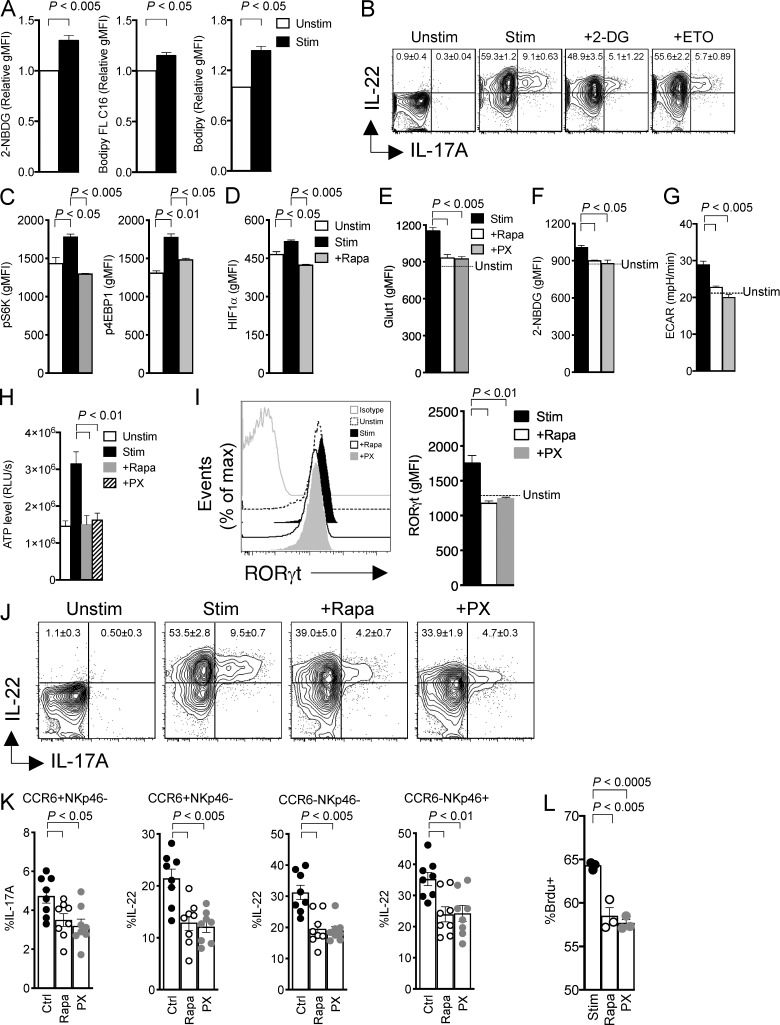
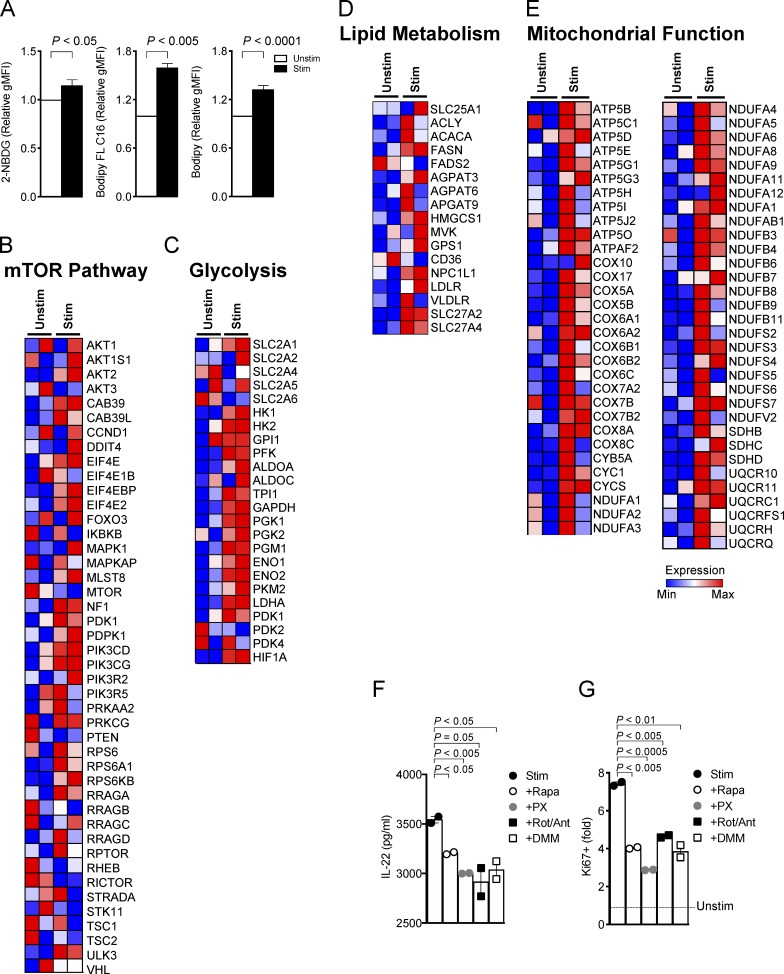
Given that mTORC1 drives anabolic and energetic metabolism, we next asked: what metabolic changes occur during ILC3 activation? We addressed this question in the ILC3 cell line MNK3, which retains phenotypic and functional features characteristic of murine primary ILC3s, including production of IL-17A and IL-22 (Allan et al., 2015). MNK3 cells activated with IL-23 plus IL-1β took up more extracellular glucose than did resting cells, as measured by staining with 2-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-2-deoxyglucose (2-NBDG; Fig. 3 A), which suggests that ILC3 activation requires glucose consumption and glycolysis. Accordingly, restricting glucose by exposing MNK3 cells to 2-deoxy-d-glucose (2-DG) markedly lowered production of IL-17A and IL-22 (Fig. 3 B). MNK3 activation was not entirely associated with glycolysis, as we also detected an increase in fatty acid uptake by tracking the fluorescent long-chain fatty acid analogue boron-dipyrromethene (Bodipy) FL C16 (Fig. 3 A); moreover, inhibiting fatty acid oxidation by treatment with etomoxir (ETO) also curbed production of IL-17A and IL-22 (Fig. 3 B), suggesting that activated MNK3 cells maintain the capacity to use lipids for fuel. Bodipy staining of activated MNK3 cells also revealed an increase in intracellular neutral lipid stores (Fig. 3 A), which may reflect de novo lipogenesis, a metabolic process also reported to distinguish Th17s from T reg cells (Berod et al., 2014).
Figure 3.
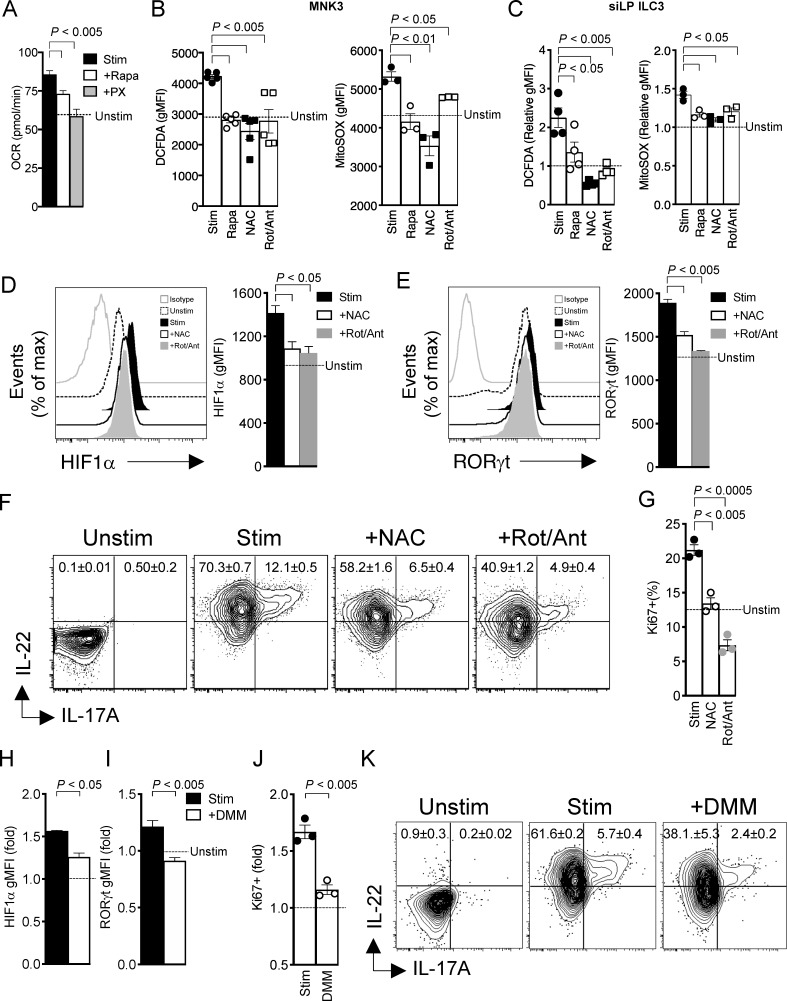
mTORC1 maintains RORγt expression and cytokine production through HIF1α. (A) Uptake of 2-NBDG, Bodipy FL C16, and Bodipy in MNK3 unstimulated (Unstim) or stimulated (Stim) with IL-1β + IL-23 for 16 h, measured by flow cytometry. (B) Intracellular content of IL-17A and IL-22 in MNK3 measured by flow cytometry. Cell were unstimulated or stimulated with IL-1β + IL-23 for 6 h, either without or with 2-DG or ETO. (C) Flow cytometry analysis of phosphorylation of S6K and 4EBP1 in MNK3 unstimulated or stimulated with IL-1β + IL-23 for 16 h without or with Rapa. (D) Expression of HIF1α by flow cytometry analysis in MNK3 treated as in C. (E and F) Expression of Glut1 (E) and uptake of 2-NBDG (F) measured by flow cytometry in MNK3 stimulated with IL-1β + IL-23 for 16 h without or with Rapa or PX. (G and H) Basal ECAR (G) and ATP levels (H) in MNK3. Cells were unstimulated (dotted line or Unstim) or stimulated with IL-1β + IL-23 for 6 h without or with Rapa or PX. RLUs, relative light units. (I) Expression of RORγt in MNK3 stimulated with IL-1β + IL-23 for 6 h in the presence or absence of Rapa or PX, assessed by flow cytometry. A representative histogram and quantification of mean fluorescence intensity (MFI) are shown. (J) Expression of IL-17A and IL-22 in MNK3 treated as in G, measured by flow cytometry. (K) Expression of IL-17A and IL-22 in lamina propria ILC3 stimulated with IL-1β + IL-23 for 3 h without or with Rapa or PX. Ctrl, control. (L) Percentage of BrdU+ MNK3 stimulated for 16 h with or without Rapa and PX, measured by flow cytometry. Data in A–J and L are from one experiment representative of two to three independent experiments, and data in K are pooled from three independent experiments (mean ± SEM of n = 3–8; one-way ANOVA or Student’s t test). gMFI, geometric MFI.
The changes in glucose and fatty acid metabolism noted upon MNK3 cell activation were associated with mTORC1 triggering, as indicated by phosphorylation of two mTORC1 targets, S6K and the eukaryotic translation initiation factor 4E binding protein 1 (4EBP1; Saxton and Sabatini, 2017), which was blocked by Rapa (Fig. 3 C). Since mTORC1 drives glycolytic metabolism by activating transcription of the gene for HIF1α (Düvel et al., 2010), we further investigated the impact of ILC3 activation on HIF1α expression. Stimulation of MNK3 cells with IL-1β plus IL-23 induced HIF1α protein expression, which was abolished by Rapa (Fig. 3 D). Corroborating the role of mTORC1-HIF1α in inducing glycolysis during ILC3 activation, the HIF1α inhibitors PX-478 (PX) and Rapa were equally effective in limiting MNK3 glucose transporter 1 (Glut1) expression, glucose uptake, and the extracellular acidification rate (ECAR), which depends on the generation of lactic acid, the end product of glycolysis (Fig. 3, E–G). Altogether, mTORC1-HIF1α–mediated metabolic changes in activated MNK3 cells resulted in a marked increase in ATP production compared with unstimulated cells, which was blocked by Rapa and PX (Fig. 3 H).
In addition to impacting ILC3 metabolism, mTORC1-HIF1α promoted RORγt expression. Inhibition of mTORC1-HIF1α by Rapa and PX curtailed the expression of RORγt in primary ILC3s (Figs. 3 I and S2). This result was consistent with a direct role of HIF1α in activating the transcription of Rorc previously reported in Th17 cells (Dang et al., 2011; Shi et al., 2011). Ultimately, inhibition of mTORC1-HIF1α by PX and Rapa suppressed IL-17A and IL-22 secretion by MNK3 cells (Fig. 3 J) and all subsets of primary intestinal ILC3s (Fig. 3 K), as well as proliferation of primary ILC3s (Fig. 3 L). We conclude that the mTORC1-HIF1α axis sustains ILC3 activation, cytokine production, and proliferation by promoting both glycolytic metabolism and expression of RORγt.
ILC3 activation relies on mROS
It has been shown that Th17s, while relying on glycolysis, have a low rate of mitochondrial oxidative metabolism, as HIF1α represses mitochondrial function through pyruvate dehydrogenase kinase (Gerriets et al., 2015). Thus, we next tested whether this was also the case for ILC3s. However, stimulation of MNK3 cells augmented the mitochondrial oxygen consumption rate (OCR), which was markedly inhibited by PX and Rapa (Fig. 4 A); this suggests that the mTORC1-HIF1α pathway activates mitochondrial metabolism in MNK3 cells. A consequence of mitochondria activity is the production of mROS through the electron transport chain. Accordingly, increased intracellular content of mROS was evident in activated MNK3 cells, as measured by staining for 2′,7′-dichlorofluorescein diacetate (DCFDA) and MitoSOX; accumulation of intracellular mROS was curbed by Rapa, ROS scavengers (N-acetyl cysteine [NAC] and MitoTEMPO), and the inhibitors of complex I and III, rotenone (Rot) and antimycin A (Ant; Figs. 4 B and S3 A). Similar results were observed in mouse intestinal ILC3s stimulated ex vivo with IL-1β plus IL-23 (Figs. 4 C and S3 A).
Figure 4.
mTORC1 signaling in ILC3 induces mROS. (A) Basal OCR in MNK3 stimulated (Stim) with IL-1β + IL-23 for 16 h with or without Rapa or PX. (B) DCFDA and MitoSOX staining of MNK3 stimulated with IL-1β + IL-23 for 16 h with or without Rapa, NAC, or Rot/Ant, respectively, assessed by flow cytometry (dotted line indicates unstimulated controls). (C) DCFDA and MitoSOX staining of lamina propria ILC3 stimulated with IL-1β + IL-23 for 3 h in the presence or absence of Rapa, NAC, or Rot/Ant, assessed by flow cytometry (n = 4). Bar graphs show MFI relative to that of naive lamina propria ILC3. (D and E) Expression of HIF1α and RORγt in MNK3 stimulated with IL-1β + IL-23 for 6 h with or without Rapa or PX, assessed by flow cytometry. Representative histograms and quantification of MFI are shown. (F) IL-17A and IL-22 intracellular content of MNK3, unstimulated or stimulated with IL-1β + IL-23 with or without NAC or Rot/Ant, assessed by flow cytometry. (G) Percentage of Ki67+ MNK3 stimulated with IL-1β + IL-23 for 16 h with or without Rapa or PX, assessed by flow cytometry. (H and I) HIF1α and RORγt content of MNK3 stimulated with IL-1β + IL-23 for 6 h with or without DMM, assessed by flow cytometry. Graph bars indicate MFI relative to unstimulated MNK3. (J) Ki67 staining of MNK3 stimulated with IL-1β + IL-23 for 16 h in the presence or absence of DMM, assessed by flow cytometry. Bar graphs indicate fold increase relative to unstimulated MNK3 (dotted line). (K) Expression of IL-17A and IL-22 by MNK3, unstimulated or stimulated with IL-1β + IL-23 for 6 h with or without DMM. Data in A, B, and D–K are from one experiment representative of two or three independent experiments, and data in C are pooled from two independent experiments (mean ± SEM of n = 2–4; one-way ANOVA or Student’s t test).
Since ROS have been shown to stabilize HIF1α protein by preventing its hydroxylation by prolyl hydroxylase domain-containing protein 2 and subsequent proteasomal degradation (Hamanaka and Chandel, 2010), we further tested the impact of mROS on HIF1α activity in ILC3s. We found that both NAC and Rot/Ant treatments curtailed HIF1α expression and also impaired RORγt expression in MNK3 cells (Fig. 4, D and E). Corroborating the concept that mROS are important for ILC3 activation, both NAC and Rot/Ant treatment disrupted IL-17A and IL-22 production (Fig. 4 F) and limited cell division, as assessed by the proliferation marker Ki67 (Fig. 4 G). Similar changes in RORγt, IL-17A, IL-22, and cell division were observed in cells treated with a specific mROS inhibitor, MitoTEMPO (Fig. S3, B–D). We conclude that, upon ILC3 stimulation, mTORC1 signaling activates mitochondrial metabolism and the generation of mROS, which help prolong HIF1α activity and ultimately cytokine secretion and proliferation.
Inhibition of succinate dehydrogenase (SDH) impairs mROS and activation in ILC3
It has been shown that mitochondrial oxidation of succinate by SDH in macrophages leads to reverse electric transport in complex I of the electron transport chain, driving the production of mROS that activate HIF1α (Tannahill et al., 2013; Chouchani et al., 2014; Mills et al., 2016). Therefore, we tested the importance of SDH in ILC3 metabolism and function. Following exposure to the SDH inhibitor dimethyl malonate (DMM), ILC3s had decreased levels of HIF1α and RORγt (Fig. 4, H and I), which consequently reduced ILC3 glucose uptake and lactate production (Fig. S3 E), and failed to proliferate and secrete IL-22 and IL-17A upon stimulation (Fig. 4, J and K). These results suggest that activated ILC3s rely on SDH and reverse electron transport for mROS production and effector functions.
Human ILC3 activation requires mTOR-mediated metabolic reprogramming
We wanted to know whether activated human ILC3s use a metabolic program similar to that observed in mice. We sorted tonsillar CD56+NKp44+CCR6+ ILC3s and stimulated them with a combination of IL-1β and IL-23 ex vivo. We first assessed the metabolic changes of human ILC3s during activation by using 2-NBDG, Bodipy FL C16, and Bodipy staining; we found that activated ILC3s profoundly induced the extracellular glucose uptake, lipid uptake, and cellular lipid content compared with the resting cells (Fig. 5 A). Furthermore, transcriptomes of resting and activated ILC3s were examined for the expression of metabolic genes. ILC3 stimulation significantly increased expression of mTORC1 pathway genes (Fig. 5 B). Moreover, we observed a coordinate increase in expression of genes encoding: (a) key glycolytic mediators, such as Slc2A1 (Glut1), Hk1 and Hk2 (hexokinases), and Hif1a (Fig. 5 C); (b) proteins involved in lipogenesis (e.g., Acaca, Fasn, Hmgcs1, and Gps1) and lipid uptake (e.g., Npc1l1, Ldlr, Slc27a2, and Slc27a4; Fig. 5 D); and (c) proteins involved in mitochondrial respiratory function and ROS generation (Atp5b, Atp5g1, Cox17, Cox5b, Cox8a, Cycs, Sdhb, Sdhc, Sdhd, Ndufa6, and Ndufb8; Fig. 5 E). To directly determine whether mTORC1 impacts human ILC3 effector functions, tonsillar ILC3s were stimulated with IL-23 plus IL-1β in the presence or absence of Rapa, PX, Rot/Ant, or DMM. Inhibition of mTORC1-HIF1α signaling and mitochondrial-derived ROS profoundly inhibited ILC3 production of IL-22 (Fig. 5 F), as well as proliferation (Fig. 5 G), which indicates that cytokine-induced activation of human ILC3s, like mouse ILC3s, relies on metabolic reprogramming.
Figure 5.
Human ILC3 activation requires mTOR-mediated metabolic reprogramming. (A) Uptake of 2-NBDG, Bodipy FL C16, and Bodipy in human ILC3s unstimulated or stimulated with IL-1β + IL-23 for 16 h, measured by flow cytometry. (B–E) Heatmaps of the expression of genes controlling mTOR signaling, glycolysis, lipid metabolism, and mitochondrial function in human tonsillar ILC3, unstimulated (Unstim) or stimulated (Stim) with IL-1β + IL-23 for 16 h. (F) Amount of IL-22 released in culture supernatant by human ILC3, unstimulated or stimulated with IL-1β + IL-23 for 72 h in the presence or absence of Rapa, PX, Rot/Ant, or DMM (n = 2). (G) Ki67 staining of ILC3 stimulated with IL-1β + IL-23 for 72 h as in E, assessed by flow cytometry (mean ± SEM of n = 2; one-way ANOVA or Student’s t test). Graph bars indicate fold increase relative to unstimulated human ILC3s. gMFI, geometric MFI.
Concluding remarks
Previous analyses of the metabolic programs employed by Th17 and T reg cells have shown a clear dichotomy. Th17s rely on the mTORC1-HIF1α pathway, which promotes glycolysis and directly induces RORγt expression (Delgoffe et al., 2009, 2011; Dang et al., 2011; Michalek et al., 2011; Shi et al., 2011; Kurebayashi et al., 2012; Gerriets et al., 2015). T reg cells rely on lipid oxidation and generation of mROS in mitochondria (Michalek et al., 2011; Gerriets et al., 2015). Our study demonstrates that although ILC3s are the innate counterparts of Th17s, they have adopted quite distinct metabolic strategies to cope with environmental cues. During ILC3 activation, mTORC1-HIF1α signaling reprograms the metabolism toward glycolysis and augments RORγt expression, as observed in Th17s. Of note, it has been shown that lower oxygen conditions can elevate the activity of the mTORC1-HIF1α axis to promote Th17 differentiation (Ikejiri et al., 2012); thus, a hypoxic environment might also promote ILC3 development. Furthermore, de novo fatty acid synthesis is evident in activated ILC3s; this is a metabolic feature that distinguishes Th17 cells from T reg cells (Berod et al., 2014). However, the metabolic adaptation of ILC3s diverges from that of Th17s in their use of mitochondrial metabolism and the impact of mROS. Th17s maintain low levels of mitochondrial metabolism and generate very little mROS, which can deviate CD4 T cell differentiation toward T reg cells by inhibiting pyruvate dehydrogenase kinase, a key regulator of glycolysis (Gerriets et al., 2015). In contrast, ILC3s produce mROS, which stabilizes HIF1α activity, enhances RORγt expression, and ultimately promotes effector functions.
ILC3s’ reliance on mROS is reminiscent of the metabolic changes observed in LPS-activated macrophages (Mills et al., 2016, 2017). In these cells, glycolysis is associated with a break in the Krebs cycle and activation of SDH; this results in the generation of mROS, which activates HIF1α and glycolysis. Further supporting the parallel between ILC3s and LPS-activated macrophages, we found that SDH inhibition by DMM dampened ILC3 activation. The importance of mROS in ILC3 activation is also consistent with previous studies showing that mROS signaling is needed for T cell activation, at least during antigen-specific expansion (Sena et al., 2013). Moreover, the mTORC1-HIF1α pathway has been shown to promote mitochondrial metabolism in various contexts (Semenza, 2011; Lamming and Sabatini, 2013; Morita et al., 2013). Why mROS enhance ILC3 functions but deviate CD4 T cell differentiation away from Th17s and toward T reg cells remains unclear. It is noteworthy that ILCs include effector subsets corresponding to TH1, TH2, and Th17 cells but lack a regulatory subset expressing FoxP3. Thus, we envision that while ILC3s use and integrate both glycolytic and mROS signals to boost effector functions, CD4 T cells have adopted glycolysis and mROS as separate pathways to control the balance between Th17 and T reg cells. Future studies will examine the mROS downstream targets that are differentially expressed and/or regulated in ILCs and CD4 T cells.
Materials and methods
Mice
B6.129S7-Rag1tm1Mom/J (Rag1−/−; #002216) and B6.Cg-Rptortm1.1Dmsa/J (Rptorfl/fl; #013188) mice were purchased from the Jackson Laboratory. Rorc-Cre mice have been described (Eberl and Littman, 2004). All mice were bred and maintained in specific pathogen–free facilities under protocols approved by the institutional animal care at Washington University School of Medicine and were used at 8–10 wk of age.
C. rodentium infection
8–10-wk-old age- and gender-matched mice were orally administered 2 × 109 C. rodentium, strain DBS100 (ATCC), as previously described (Cella et al., 2009). Body weight of mice was measured until day 7. For Rapa injection, mice were injected i.p. with 200 µl of Rapa (5 mg/kg body weight) or DMSO vehicle before infection.
Tissue dissociation
Cells were isolated from the small intestine lamina propria by treating tissue with EDTA (Corning), digesting with Collagenase IV (Sigma-Aldrich), and then subjecting digests to density gradient centrifugation.
Cell culture and activation
MNK3 cells or single-cell preparations of small intestine lamina propria were cultured in complete RPMI (Corning) with or without a combination of IL-1β (10 ng/ml) and IL-23 (10 ng/ml) in the presence or absence of 20 nM Rapa (Millipore), 20 µM PX (Selleckchem), 0.1 µM Rot (Sigma-Aldrich) + 1 µM Ant (Sigma-Aldrich), 5 mM NAC (Sigma-Aldrich), 50 µM MitoTEMPO (Sigma-Aldrich), 20 mM DMM (Sigma-Aldrich), 1 mM 2-DG (Sigma-Aldrich), and 200 µM ETO (Cayman). Brefeldin A (BD GolgiPlug) was added to cultures 3 h before analysis of intracellular cytokines.
Flow cytometry
Rat anti-human/mouse RORγt (AFKJS-9), rat anti-mouse Eomes (Dan11mag), rat anti-mouse/rat Ki67 (SolA15), anti-mouse NK1.1 (PK136), anti-mouse CD3 (145-2C11), anti-mouse CD5 (53–7.3), rat anti-mouse CD19 (1D3), and rat anti-mouse IL-22 (1H8PWSR) were purchased from eBioscience. Rat anti-mouse IL-17A (TC11-18H10), rat anti-mouse Thy1.2 (30-H12), rat anti-mouse CD196 (CCR6; 140706), and BrdU were purchased from BD Biosciences. Rat anti-mouse CD45 (30-F11) was purchased from BioLegend. Anti-phospho-p70 S6K (pS6K) and anti-phospho-4EBP1 (p4EBP1) were purchased from Cell Signaling. Anti-human/mouse HIF1α (241812) antibody was purchased from R&D Systems. Anti-human/mouse Glut1 antibody was purchased from Metafora. Dead cells were excluded using a Live/Dead Fixable Cell Stain Kit (Thermo Fisher Scientific). Intracellular staining was either the BD Biosciences Fixation/Permeabilization Solution Kit or eBioscience Transcription Factor staining kit. For uptake of 2-NBDG (10 µg/ml; Invitrogen), Bodipy (500 µg/ml; Invitrogen), and Bodipy FL C16 (1 µM; Invitrogen), was performed by incubating cells in RPMI medium for 30–60 min at 37°C and measured by flow cytometry. Detection of ROS using DCFDA (20 µM; Abcam) was performed by incubating cells in RPMI medium 1 h at 37°C or using MitoSOX (5 µM; Invitrogen) by incubating cells in HBSS for 10 min at 37°C and measuring by flow cytometry. Data were acquired on a FACSCanto II flow cytometer (BD Biosciences) and analyzed with FlowJo v.9.5.2 (TreeStar).
Metabolism assays
For real-time analysis of ECARs and OCRs, MNK3 cells (1.5 × 105 cells/well) were analyzed using an XF-96 Extracellular Flux Analyzer (Seahorse Bioscience) as described in detail previously (Huang et al., 2014). ATP assays were measured with an ATP Determination Kit according to the manufacturer’s instructions (Invitrogen).
Cell fractionation and immunoblotting
Cells were lysed in buffer containing 1% Triton X-100 and 0.1% SDS with protease and phosphatase inhibitors. Anti-HIF1α (D1S7W; Cell Signaling), anti-RORγt (Biorbyt), and anti–β-actin (Cell Signaling) were used and detected with peroxidase-linked secondary antibody followed by ECL Western Blotting Detection Reagents (Thermo Fisher Scientific).
Human samples
Cells were sorted from two to three pooled donors and then stimulated with a combination of human IL-1β (10 ng/ml), IL-23 (10 ng/ml), and IL-15 (25 ng/ml) at 37°C for 16 h or as indicated. For microarray analysis, RNA was isolated (RNeasy Plus Micro Kit; Qiagen), amplified, and hybridized to the Affymetrix Human Gene (v.1.0) ST arrays. Data were analyzed with GenePattern software (Broad Institute) and Ingenuity Pathway Analysis (Qiagen) as previously described (Robinette et al., 2015). Secretion of IL-22 by human ILC3 (105) was collected and measured with a human IL-22 Quantikine ELISA kit according to the manufacturer’s instructions (R&D Systems). All human studies were conducted under the approval of Institutional Review Boards of Washington University (IRB#03-1214). Microarray data are deposited in the Gene Expression Omnibus database under accession no. GSE132843.
Online supplemental material
Fig. S1 shows that Rapa treatment impairs ILC3s’ response to a bacterial infection. Fig. S2 shows the expression of RORγt and HIF1α in MNK3s by immunoblotting. Fig. S3 indicates that mROS supports ILC3 function.
Supplementary Material
Acknowledgments
The authors thank Dr. Jennifer Bando for helpful discussion, Lydia Raines for assistance with immunoblotting experiments, the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for providing help with genomic analysis, and Dr. Babak Razani (Cardiovascular Division, Washington University School of Medicine, St. Louis, MO) for Rptorfl/fl mice.
This work was supported by the Cleveland Digestive Diseases Research Core Center Pilot Grant and Case Comprehensive Cancer Center, Case Western Reserve University American Cancer Society Pilot Grant to S.C.-C. Huang (1P30DK097948 and IRG-91-022-19) and the National Institutes of Health and Crohn’s and Colitis Foundation of America to M. Colonna.
M. Colonna receives research financial support from Pfizer. The other authors declare no competing financial interests.
Author contributions: S.C.-C. Huang and M. Colonna conceived the study and wrote the manuscript. S.C.-C. Huang, B. Di Luccia, S. Gilfillan, and M. Cella performed experiments and analyzed data.
References
- Allan D.S.J., Kirkham C.L., Aguilar O.A., Qu L.C., Chen P., Fine J.H., Serra P., Awong G., Gommerman J.L., Zúñiga-Pflücker J.C., and Carlyle J.R.. 2015. An in vitro model of innate lymphoid cell function and differentiation. Mucosal Immunol. 8:340–351. 10.1038/mi.2014.71 [DOI] [PubMed] [Google Scholar]
- Bando J.K., and Colonna M.. 2016. Innate lymphoid cell function in the context of adaptive immunity. Nat. Immunol. 17:783–789. 10.1038/ni.3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berod L., Friedrich C., Nandan A., Freitag J., Hagemann S., Harmrolfs K., Sandouk A., Hesse C., Castro C.N., Bähre H., et al. . 2014. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat. Med. 20:1327–1333. 10.1038/nm.3704 [DOI] [PubMed] [Google Scholar]
- Björklund Å.K., Forkel M., Picelli S., Konya V., Theorell J., Friberg D., Sandberg R., and Mjösberg J.. 2016. The heterogeneity of human CD127(+) innate lymphoid cells revealed by single-cell RNA sequencing. Nat. Immunol. 17:451–460. 10.1038/ni.3368 [DOI] [PubMed] [Google Scholar]
- Buck M.D., O’Sullivan D., and Pearce E.L.. 2015. T cell metabolism drives immunity. J. Exp. Med. 212:1345–1360. 10.1084/jem.20151159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella M., Fuchs A., Vermi W., Facchetti F., Otero K., Lennerz J.K.M., Doherty J.M., Mills J.C., and Colonna M.. 2009. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 457:722–725. 10.1038/nature07537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-H., Curtis J.D., Maggi L.B. Jr., Faubert B., Villarino A.V., O’Sullivan D., Huang S.C.-C., van der Windt G.J.W., Blagih J., Qiu J., et al. . 2013. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 153:1239–1251. 10.1016/j.cell.2013.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouchani E.T., Pell V.R., Gaude E., Aksentijević D., Sundier S.Y., Robb E.L., Logan A., Nadtochiy S.M., Ord E.N.J., Smith A.C., et al. . 2014. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature. 515:431–435. 10.1038/nature13909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna M. 2018. Innate Lymphoid Cells: Diversity, Plasticity, and Unique Functions in Immunity. Immunity. 48:1104–1117. 10.1016/j.immuni.2018.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang E.V., Barbi J., Yang H.-Y., Jinasena D., Yu H., Zheng Y., Bordman Z., Fu J., Kim Y., Yen H.-R., et al. . 2011. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 146:772–784. 10.1016/j.cell.2011.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgoffe G.M., Kole T.P., Zheng Y., Zarek P.E., Matthews K.L., Xiao B., Worley P.F., Kozma S.C., and Powell J.D.. 2009. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 30:832–844. 10.1016/j.immuni.2009.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgoffe G.M., Pollizzi K.N., Waickman A.T., Heikamp E., Meyers D.J., Horton M.R., Xiao B., Worley P.F., and Powell J.D.. 2011. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat. Immunol. 12:295–303. 10.1038/ni.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diefenbach A., Colonna M., and Koyasu S.. 2014. Development, differentiation, and diversity of innate lymphoid cells. Immunity. 41:354–365. 10.1016/j.immuni.2014.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Düvel K., Yecies J.L., Menon S., Raman P., Lipovsky A.I., Souza A.L., Triantafellow E., Ma Q., Gorski R., Cleaver S., et al. . 2010. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell. 39:171–183. 10.1016/j.molcel.2010.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl G., and Littman D.R.. 2004. Thymic origin of intestinal alphabeta T cells revealed by fate mapping of RORgammat+ cells. Science. 305:248–251. 10.1126/science.1096472 [DOI] [PubMed] [Google Scholar]
- Eberl G., Colonna M., Di Santo J.P., and McKenzie A.N.J.. 2015. Innate lymphoid cells. Innate lymphoid cells: a new paradigm in immunology. Science. 348:aaa6566 10.1126/science.aaa6566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerriets V.A., Kishton R.J., Nichols A.G., Macintyre A.N., Inoue M., Ilkayeva O., Winter P.S., Liu X., Priyadharshini B., Slawinska M.E., et al. . 2015. Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J. Clin. Invest. 125:194–207. 10.1172/JCI76012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladiator A., Wangler N., Trautwein-Weidner K., and LeibundGut-Landmann S.. 2013. Cutting edge: IL-17-secreting innate lymphoid cells are essential for host defense against fungal infection. J. Immunol. 190:521–525. 10.4049/jimmunol.1202924 [DOI] [PubMed] [Google Scholar]
- Hamanaka R.B., and Chandel N.S.. 2010. Mitochondrial reactive oxygen species regulate cellular signaling and dictate biological outcomes. Trends Biochem. Sci. 35:505–513. 10.1016/j.tibs.2010.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández P.P., Mahlakoiv T., Yang I., Schwierzeck V., Nguyen N., Guendel F., Gronke K., Ryffel B., Hoelscher C., Dumoutier L., et al. . 2015. Interferon-λ and interleukin 22 act synergistically for the induction of interferon-stimulated genes and control of rotavirus infection. Nat. Immunol. 16:698–707. 10.1038/ni.3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.C.-C., Everts B., Ivanova Y., O’Sullivan D., Nascimento M., Smith A.M., Beatty W., Love-Gregory L., Lam W.Y., O’Neill C.M., et al. . 2014. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat. Immunol. 15:846–855. 10.1038/ni.2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntington N.D., Carpentier S., Vivier E., and Belz G.T.. 2016. Innate lymphoid cells: parallel checkpoints and coordinate interactions with T cells. Curr. Opin. Immunol. 38:86–93. 10.1016/j.coi.2015.11.008 [DOI] [PubMed] [Google Scholar]
- Ikejiri A., Nagai S., Goda N., Kurebayashi Y., Osada-Oka M., Takubo K., Suda T., and Koyasu S.. 2012. Dynamic regulation of Th17 differentiation by oxygen concentrations. Int. Immunol. 24:137–146. 10.1093/intimm/dxr111 [DOI] [PubMed] [Google Scholar]
- Jones R.G., and Pearce E.J.. 2017. MenTORing Immunity: mTOR Signaling in the Development and Function of Tissue-Resident Immune Cells. Immunity. 46:730–742. 10.1016/j.immuni.2017.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L., Ripke S., Weersma R.K., Duerr R.H., McGovern D.P., Hui K.Y., Lee J.C., Schumm L.P., Sharma Y., Anderson C.A., et al. International IBD Genetics Consortium (IIBDGC) . 2012. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 491:119–124. 10.1038/nature11582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karo J.M., Schatz D.G., and Sun J.C.. 2014. The RAG recombinase dictates functional heterogeneity and cellular fitness in natural killer cells. Cell. 159:94–107. 10.1016/j.cell.2014.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchberger S., Royston D.J., Boulard O., Thornton E., Franchini F., Szabady R.L., Harrison O., and Powrie F.. 2013. Innate lymphoid cells sustain colon cancer through production of interleukin-22 in a mouse model. J. Exp. Med. 210:917–931. 10.1084/jem.20122308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose C.S.N., and Artis D.. 2016. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat. Immunol. 17:765–774. 10.1038/ni.3489 [DOI] [PubMed] [Google Scholar]
- Koues O.I., Collins P.L., Cella M., Robinette M.L., Porter S.I., Pyfrom S.C., Payton J.E., Colonna M., and Oltz E.M.. 2016. Distinct Gene Regulatory Pathways for Human Innate versus Adaptive Lymphoid Cells. Cell. 165:1134–1146. 10.1016/j.cell.2016.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurebayashi Y., Nagai S., Ikejiri A., Ohtani M., Ichiyama K., Baba Y., Yamada T., Egami S., Hoshii T., Hirao A., et al. . 2012. PI3K-Akt-mTORC1-S6K1/2 axis controls Th17 differentiation by regulating Gfi1 expression and nuclear translocation of RORγ. Cell Reports. 1:360–373. 10.1016/j.celrep.2012.02.007 [DOI] [PubMed] [Google Scholar]
- Lamming D.W., and Sabatini D.M.. 2013. A Central role for mTOR in lipid homeostasis. Cell Metab. 18:465–469. 10.1016/j.cmet.2013.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais A., Cherfils-Vicini J., Viant C., Degouve S., Viel S., Fenis A., Rabilloud J., Mayol K., Tavares A., Bienvenu J., et al. . 2014. The metabolic checkpoint kinase mTOR is essential for IL-15 signaling during the development and activation of NK cells. Nat. Immunol. 15:749–757. 10.1038/ni.2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalek R.D., Gerriets V.A., Jacobs S.R., Macintyre A.N., MacIver N.J., Mason E.F., Sullivan S.A., Nichols A.G., and Rathmell J.C.. 2011. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J. Immunol. 186:3299–3303. 10.4049/jimmunol.1003613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills E.L., Kelly B., Logan A., Costa A.S.H., Varma M., Bryant C.E., Tourlomousis P., Däbritz J.H.M., Gottlieb E., Latorre I., et al. . 2016. Succinate Dehydrogenase Supports Metabolic Repurposing of Mitochondria to Drive Inflammatory Macrophages. Cell. 167:457–470.e13. 10.1016/j.cell.2016.08.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills E.L., Kelly B., and O’Neill L.A.J.. 2017. Mitochondria are the powerhouses of immunity. Nat. Immunol. 18:488–498. 10.1038/ni.3704 [DOI] [PubMed] [Google Scholar]
- Morita M., Gravel S.-P., Chénard V., Sikström K., Zheng L., Alain T., Gandin V., Avizonis D., Arguello M., Zakaria C., et al. . 2013. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 18:698–711. 10.1016/j.cmet.2013.10.001 [DOI] [PubMed] [Google Scholar]
- Muñoz M., Eidenschenk C., Ota N., Wong K., Lohmann U., Kühl A.A., Wang X., Manzanillo P., Li Y., Rutz S., et al. . 2015. Interleukin-22 induces interleukin-18 expression from epithelial cells during intestinal infection. Immunity. 42:321–331. 10.1016/j.immuni.2015.01.011 [DOI] [PubMed] [Google Scholar]
- Newton R., Priyadharshini B., and Turka L.A.. 2016. Immunometabolism of regulatory T cells. Nat. Immunol. 17:618–625. 10.1038/ni.3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce E.L., Poffenberger M.C., Chang C.-H., and Jones R.G.. 2013. Fueling immunity: insights into metabolism and lymphocyte function. Science. 342:1242454 10.1126/science.1242454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C., Thornton E.E., McKenzie B., Schaupp A.-L., Huskens N., Griseri T., West N., Tung S., Seddon B.P., Uhlig H.H., and Powrie F.. 2016. ILC3 GM-CSF production and mobilisation orchestrate acute intestinal inflammation. eLife. 5:e10066 10.7554/eLife.10066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollizzi K.N., and Powell J.D.. 2014. Integrating canonical and metabolic signalling programmes in the regulation of T cell responses. Nat. Rev. Immunol. 14:435–446. 10.1038/nri3701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin L.C., Girard-Madoux M.J.H., Seillet C., Mielke L.A., Kerdiles Y., Fenis A., Wieduwild E., Putoczki T., Mondot S., Lantz O., et al. . 2016. Complementarity and redundancy of IL-22-producing innate lymphoid cells. Nat. Immunol. 17:179–186. 10.1038/ni.3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinette M.L., Fuchs A., Cortez V.S., Lee J.S., Wang Y., Durum S.K., Gilfillan S., and Colonna M.. Immunological Genome Consortium . 2015. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat. Immunol. 16:306–317. 10.1038/ni.3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh-Takayama N., Vosshenrich C.A.J., Lesjean-Pottier S., Sawa S., Lochner M., Rattis F., Mention J.-J., Thiam K., Cerf-Bensussan N., Mandelboim O., et al. . 2008. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 29:958–970. 10.1016/j.immuni.2008.11.001 [DOI] [PubMed] [Google Scholar]
- Saxton R.A., and Sabatini D.M.. 2017. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 168:960–976. 10.1016/j.cell.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza G.L. 2011. Hypoxia-inducible factor 1: regulator of mitochondrial metabolism and mediator of ischemic preconditioning. Biochim. Biophys. Acta. 1813:1263–1268. 10.1016/j.bbamcr.2010.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena L.A., Li S., Jairaman A., Prakriya M., Ezponda T., Hildeman D.A., Wang C.-R., Schumacker P.T., Licht J.D., Perlman H., et al. . 2013. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity. 38:225–236. 10.1016/j.immuni.2012.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L.Z., Wang R., Huang G., Vogel P., Neale G., Green D.R., and Chi H.. 2011. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J. Exp. Med. 208:1367–1376. 10.1084/jem.20110278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C., Lee J.S., Gilfillan S., Robinette M.L., Newberry R.D., Stappenbeck T.S., Mack M., Cella M., and Colonna M.. 2015. Unique and redundant functions of NKp46+ ILC3s in models of intestinal inflammation. J. Exp. Med. 212:1869–1882. 10.1084/jem.20151403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg G.F., Monticelli L.A., Elloso M.M., Fouser L.A., and Artis D.. 2011. CD4(+) lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity. 34:122–134. 10.1016/j.immuni.2010.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannahill G.M., Curtis A.M., Adamik J., Palsson-McDermott E.M., McGettrick A.F., Goel G., Frezza C., Bernard N.J., Kelly B., Foley N.H., et al. . 2013. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 496:238–242. 10.1038/nature11986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., and Green D.R.. 2012. Metabolic checkpoints in activated T cells. Nat. Immunol. 13:907–915. 10.1038/ni.2386 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Valdez P.A., Danilenko D.M., Hu Y., Sa S.M., Gong Q., Abbas A.R., Modrusan Z., Ghilardi N., de Sauvage F.J., and Ouyang W.. 2008. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 14:282–289. 10.1038/nm1720 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.