Etzerodt et al. show that the specific targeting of CD163+ macrophages in melanoma drives inflammatory monocyte influx and promotes antitumor immunity, illustrating the importance of selective targeting of tumor-associated myeloid cells for achieving optimal therapeutic responses.
Abstract
Tumor-associated macrophages (TAMs) play critical roles in tumor progression but are also capable of contributing to antitumor immunity. Recent studies have revealed an unprecedented heterogeneity among TAMs in both human cancer and experimental models. Nevertheless, we still understand little about the contribution of different TAM subsets to tumor progression. Here, we demonstrate that CD163-expressing TAMs specifically maintain immune suppression in an experimental model of melanoma that is resistant to anti–PD-1 checkpoint therapy. Specific depletion of the CD163+ macrophages results in a massive infiltration of activated T cells and tumor regression. Importantly, the infiltration of cytotoxic T cells was accompanied by the mobilization of inflammatory monocytes that significantly contributed to tumor regression. Thus, the specific targeting of CD163+ TAMs reeducates the tumor immune microenvironment and promotes both myeloid and T cell–mediated antitumor immunity, illustrating the importance of selective targeting of tumor-associated myeloid cells in a therapeutic context.
Graphical Abstract
Introduction
Tumor-associated macrophages (TAMs) are the most abundant immune cells found in solid tumors, and their important contributions to tumor progression are well documented (Noy and Pollard, 2014). Besides their trophic functions supporting angiogenesis, invasion, and metastasis, TAMs have also been suggested to inhibit T cell proliferation and activation by various mechanisms. This includes release of the immune-suppressive cytokine IL-10 (Jiang et al., 2015) and the local depletion of arginine (Rodriguez et al., 2004) and tryptophan (Munn and Mellor, 2007), on which T cells are highly dependent. The important functions of macrophages in relation to tumor progression has led to a substantial interest in developing new therapeutic strategies for targeting TAMs. Strategies currently in development include blocking of the chemokine CCL2 or its receptor, CCR2, which inhibits TAM recruitment by neutralizing the mobilization of bone marrow–derived monocytes (MNs), and targeting the macrophage growth factor receptor CSF1R (macrophage colony-stimulating factor receptor, c-FMS, and CD115; Yang and Zhang, 2017). While CCL2/CCR2 blockade only targets the recruitment of MN-derived macrophages, CSF1 plays an essential role in the survival and differentiation of tissue-resident macrophages as well as MN-derived macrophages (Wynn et al., 2013). Although clinical data are still limited, reduced numbers of TAMs have been reported after treatment with an mAb against CSF1R, with potentially interesting therapeutic effects in tenosynovial giant cell tumors (Ries et al., 2014; Cassier et al., 2015). However, despite a strong association between TAM accumulation and poor clinical outcome in the majority of clinical studies, in some cases, accumulation of specific TAM subsets can be associated with a good prognosis. One example is the frequency of HLA-DR+ TAMs, which has been associated with beneficial outcomes in several studies (de Vos van Steenwijk et al., 2013; Ino et al., 2013), possibly reflecting the role of macrophages in orchestrating protective immune responses (Mantovani and Allavena, 2015). In fact, recent studies using paired single-cell analysis by mass cytometry and RNA sequencing have revealed an unprecedented level of diversity within the TAM compartment in lung adenocarcinoma and renal cell carcinoma patients (Chevrier et al., 2017; Lavin et al., 2017). In the case of renal cell carcinoma, 17 distinct TAM phenotypes were documented (Chevrier et al., 2017). We still lack fundamental knowledge of the functions of different TAM subsets and their respective contributions to tumor progression. However, it is tempting to speculate that a selective targeting of TAM subsets with protumor functions, while preserving the immune functions of other TAM subsets, could offer significant clinical benefits.
Expression of CD163 by TAMs has been shown to be a particularly strong indicator of poor prognosis in several human cancers (Komohara et al., 2014), including melanoma (Jensen et al., 2009; Bronkhorst et al., 2011; Lee et al., 2018). CD163 is a macrophage- and MN-specific transmembrane protein that functions as a scavenger receptor for haptoglobin–hemoglobin complexes, formed upon intravascular hemolysis (Kristiansen et al., 2001). Expression of CD163 is induced by tumor-promoting cytokines such as IL-6 and IL-10, whereas inflammatory stimuli, including LPS, TNFα, and IFNγ, lead to a rapid down-regulation of expression and removal of membrane-bound CD163 via proteolytic shedding (Etzerodt et al., 2010; Etzerodt and Moestrup, 2013). This, together with the generation of anti-inflammatory heme metabolites from hemoglobin scavenging, has led to the association of CD163+ macrophages with anti-inflammatory functions (Etzerodt and Moestrup, 2013). However, the link between CD163+ TAM accumulation and tumor progression is based exclusively on correlations with clinical progression, and experimental evidence for specific tumor-promoting functions is still lacking.
The recent development of immune checkpoint inhibitors (ICIs), such as anti–PD-1, has had an enormous impact on cancer therapy, particularly in malignant melanoma (Robert et al., 2015; Ugurel et al., 2017). The increased expression of PD-1 ligand (PD-L1) on cancer cells suppresses the activation of PD-1–expressing CD8+ CTLs (Pardoll, 2012). Blocking PD-1/PD-L1 signaling using anti–PD-1 or PD-L1 antibodies thus leads to increased CTL activation, ultimately resulting in unprecedented rates of tumor regression (Tumeh et al., 2014). Unfortunately, only a minority of patients respond to ICI therapy, and the reasons for this are currently an area of intense research. In addition, a major limitation of ICI therapy is the indiscriminate activation of T cells, which can lead to immune-related adverse events that can require cessation of treatment (Larkin et al., 2015; Michot et al., 2016). Thus, new therapeutic strategies to enhance antitumor immunity that can overcome ICI resistance or ameliorate the severe adverse side effects are desperately needed.
Here, we have conducted an in-depth characterization of TAM subsets in a clinically relevant mouse model of melanoma that is resistant to anti–PD-1 therapy. We show that the specific targeting of a minor subset of TAMs expressing CD163 is sufficient to induce tumor regression in this model. Importantly, specific depletion of CD163+ TAMs leads to increased recruitment of effector T cells and CCR2-dependent inflammatory MNs, which both contribute to tumor regression. These studies are the first to demonstrate the selective targeting of CD163+ TAMs and their specific contribution to tumor progression. Furthermore, we show that CD163+ TAMs play a dominant role in suppressing antitumor immunity in anti–PD-1–resistant melanoma.
Results
CD163-expressing macrophages infiltrate spontaneous BrafV600E-driven melanomas
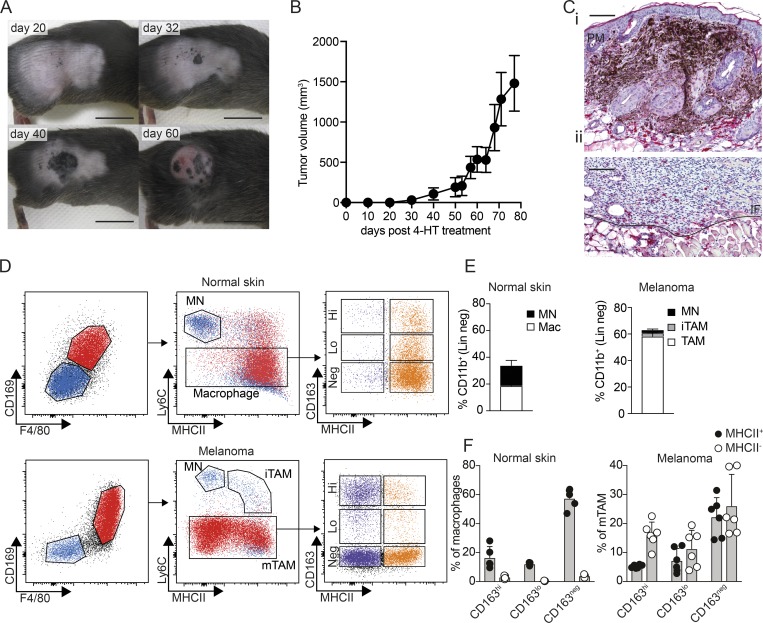
The increased availability of genetically engineered mouse models based on appropriate oncogenic driver mutations has greatly improved the relevance of mouse tumor models to human disease. Activating mutations in BRAF are the most prevalent in human melanoma and are often accompanied by loss of tumor suppressor loci such as PTEN and CDKN2A. The Tyr(CreER, BrafCA, Ptenf/f) mouse model of metastatic melanoma utilizes the melanocyte-restricted tyrosinase promoter to drive expression of a tamoxifen-inducible Cre recombinase, which in turn triggers expression of constitutively active BrafV600E (BrafCA) and deletion of a floxed Pten allele (Ptenf/f; Dankort et al., 2009). In these mice, topical administration of 4-hydroxytamoxifen (4-HT) initially leads to small pigmented lesions within ∼20 d, which progress to amelanotic tumors at around day 40 that subsequently exhibit exponential growth (Fig. 1, A and B). In untreated mice, CD163+ macrophages are evenly dispersed throughout the dermis and adipose tissue (Fig. S1 A). However, after 4-HT treatment of Tyr(CreER, BrafCA, Ptenf/f) mice, we observed accumulation of CD163+ macrophages at the border of pigmented, premelanotic lesions in the dermis (Fig. 1 C, i). When pigmented lesions had transformed into invasive amelanotic tumors, CD163+ macrophages accumulated at the invasive front, whereas only few CD163+ macrophages are present within the tumor (Fig. 1 C, ii). To further examine TAMs in BrafV600E tumors, we performed flow cytometry on single-cell suspensions of both naive skin and tumor tissue. After gating out CD45− cells, lymphocytes and granulocytes (CD45+, CD19−, CD5−, NK1.1−, Siglec F−, Ly6G−, and CD11b+), we found in both normal skin and tumor tissue two major populations of MNs/macrophages based on expression of the mature macrophage markers F4/80 and CD169 (Fig. 1 D). While F4/80 is only expressed by murine macrophages, CD169-expressing TAMs were recently suggested to play an important role in human cancer (Cassetta et al., 2019). The F4/80− CD169− population consisted mainly of Ly6C+ MNs and, in melanomas, also Ly6C+ MHCII+ immature TAMs (iTAMs), as previously described in other models (Movahedi et al., 2010). The larger F4/80+ CD169+ population was negative for Ly6C and showed heterogeneous expression of CD163 and MHCII, suggesting a mature phenotype (Fig. 1 D). Interestingly, mature TAMs (mTAMs) were by far the most abundant subset, making up 60% of the CD11b+ fraction (Fig. 1 E), whereas mature macrophages in the normal skin only accounted for 20% of the CD11b+ fraction. On average, 20% of all tumor-infiltrating leukocytes were mTAMs, which is 100 times more abundant than tumor-infiltrating CD8+ T cells (Fig. S1 B). CD163+ macrophages made up only <25% of all mTAMs, similar to the fraction of CD163+ macrophages in healthy skin. However, whereas the majority of CD163+ and CD163neg macrophages in healthy skin were MHCII+, the majority of mTAMs were MHCII− irrespective of CD163 expression (Fig. 1 F).
Figure 1.
CD163-expressing macrophages infiltrate spontaneous BrafV600E-driven melanomas. (A and B) Development of melanomas in Tyr(CreER, BrafCA, Ptenf/f) mice after application of 4-HT on the right rear flank; scale bars, 1 cm. (C) IHC staining for CD163 (red) on sections from pigmented premelanotic lesions (PM) 32 d after 4-HT (i) and invasive tumors at endpoint (ii). The invasive front (IF) is marked by the solid line. Scale bars, 100 µm. Images are representative of three independent experiments (n = 4). (D) Flow cytometry analysis of macrophages in normal skin or TAMs in late-stage tumors. Myeloid cells, excluding neutrophils, were gated as CD45+, Lin− (CD5, CD19, NK1.1, Siglec F, Ly6G), CD11b+. MNs were gated as F4/80−, CD169−, Ly6C+, MHCII−; iTAMs were F4/80−, CD169−, Ly6C+, MHCII+; and mTAMs were defined as F4/80+, CD169+, Ly6C−, MHCII+/−. The distribution of CD163 and MHCII expression among macrophages in normal skin and mTAMs is shown (see Fig. S1 C for the full gating strategy). (E and F) Relative proportions of MNs, iTAMs, mTAMs, and macrophages among myeloid cells in normal skin or tumor-associated myeloid cells (E) and proportion of CD163hi, CD163lo, and CD163− macrophages and TAMs (F). Data are pooled from three independent experiments (n = 6) and represented as mean ± SEM. Mac, macrophage.
Characterization of CD163-expressing TAMs in subcutaneous BrafV600E-driven melanomas
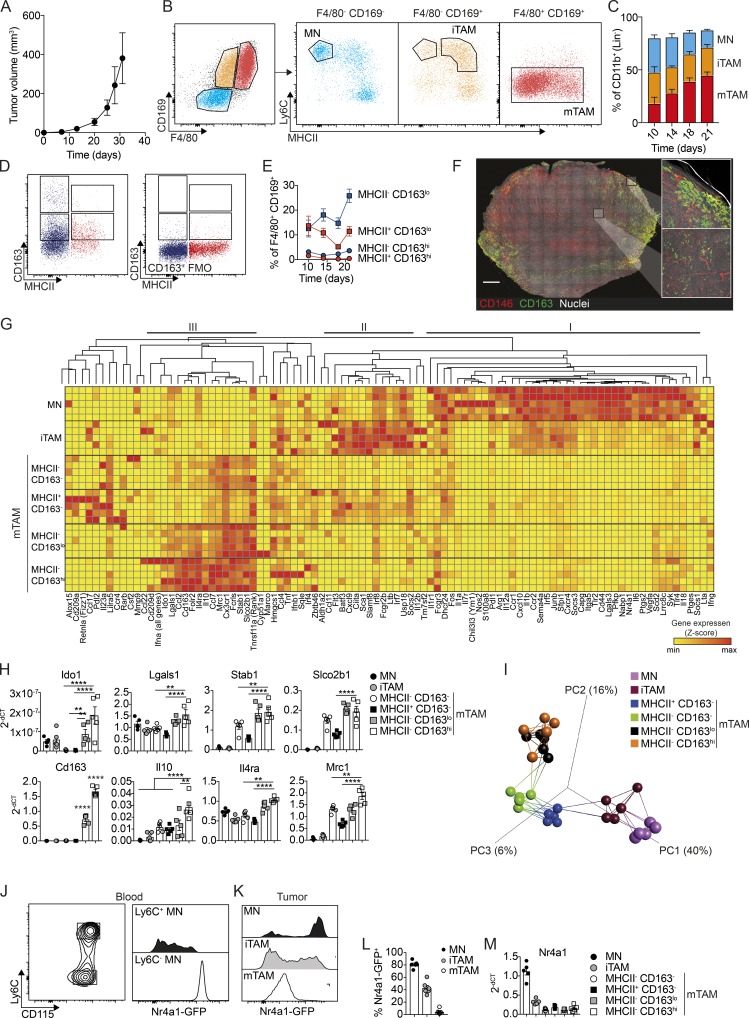
Using cell lines derived from the spontaneous BrafV600E-driven mouse model that mimic the amelanotic stage of tumor growth (Meeth et al., 2016), we sought to determine if implanted subcutaneous tumors with comparable driver mutations would give rise to similar TAM populations found in spontaneous amelanotic tumors. Subcutaneous injection of YUMM1.7 cells, derived from a spontaneous Tyr (CreER, BrafCA, Ptenf/f, and Cdkn2af/f) melanoma, gave rise to tumors with similar growth characteristics as spontaneous tumors (Figs. 1 B and 2 A). Subsequent flow cytometry analysis revealed a similar profile of tumor-associated MN/macrophage populations, based on F4/80 and CD169 expression. However, whereas F4/80− CD169− and F4/80+ CD169+ populations seemed identical in the two models at late stages of tumor growth (Fig. S2 A), there was a pronounced population of F4/80− CD169+ cells in early-stage subcutaneous tumors (Fig. 2 B). This population consisted mainly of the iTAMs, as described in the spontaneous model (Fig. 1 D), in addition to a few Ly6C+ MNs (Fig. 2 B). Accordingly, a higher proportion of MNs (F4/80− CD169−) and iTAMs were present in early tumors, whereas the frequency of mTAMs (F4/80+ CD169+) steadily increased to become the most abundant in late-stage tumors (Figs. 2 C and S2 B). Interestingly, although the frequency of CD163hi and CD163lo mTAMs changed slightly compared with the spontaneous model, the subcutaneous tumors were similarly infiltrated by mostly MHCII-negative TAMs (Fig. 2, D and E; and Fig. S2 C). Immunohistochemistry (IHC) analysis suggested that CD163hi/lo mTAM differed in their spatial distribution; CD163hi cells were mainly localized at the tumor margin, whereas CD163lo cells were located within the tumor stroma (Fig. 2 F). In addition, the proportion of MHCII− CD163lo TAMs increased with tumor progression, whereas the proportion of MHCII− CD163hi cells remained constant (Fig. 2 E).
Figure 2.
Characterization of CD163-expressing TAMs in subcutaneous BrafV600E-driven melanomas. (A) Kinetics of tumor development after s.c. injection of 106 YUMM1.7 cells. (B) Flow cytometry analysis of TAMs in YUMM1.7 tumors. Myeloid cells, excluding neutrophils, were gated as CD45+, Lin− (CD5, CD19, NK1.1, Siglec F, Ly6G), CD11b+. MNs were gated as F4/80−, CD169−, Ly6C+, MHCII−; iTAMs as F4/80−, CD169−, Ly6C+, MHCII+; and mTAMs as F4/80+, CD169+, Ly6C−, MHCII+/−. See Fig. S1 C for the full gating strategy. (C) Proportions of MNs, iTAMs, and mTAMs within the myeloid cell compartment at the indicated time points. (D) The distribution of CD163 and MHCII expression among mTAMs; CD163+ TAM (left) were gated based on the fluorescence minus one (FMO) control for CD163 staining (right). (E) Relative proportion of CD163+ TAMs among mTAMs in YUMM1.7 tumors at the indicated time points; data are represented as mean ± SEM of n = 4. (F) Immunofluorescence staining for CD163 (green) and CD146 (indicating blood vessels, red) and nuclei (Hoechst, white) in a 200-µm-thick vibratome cross section of an entire tumor. Image was acquired as a tile-scan with Z-stacks using 10× objective and subsequent processed in 3D (left). High-resolution images was acquired as single Z-stack and processed in 3D (right). Scale bar, 1,000 µm. (G) High-throughput gene expression analysis of MN, iTAM, and mTAM subsets: (1) MHCII− CD163−, (2) MHCII+ CD163−, (3) MHCII− CD163lo, and (4) MHCII− CD163hi (see Fig. S2 D for full gating strategy). Z-scores were calculated by subtracting the ΔCT(sample) from the ΔCT(mean) across all samples. (H) Relative gene expression (2−ΔCT) of Cd163, Il10, Il4ra, Mrc1, Stab1, Slco2b1, Ido1, and Lgals in MN and TAM subsets. (I) PCA and network analysis of gene expression data highlighting the five nearest neighbors. PCA analysis was performed on normalized data (mean = 0 and variance = 1) generating a correlation based PCA plot. Network analysis connects k nearest neighbors (k = 5) based on similarity calculated by Pearson correlation. (J–L) Flow cytometry analysis of blood MNs (J) and tumor-associated MNs, iTAMs, and mTAMs (K) from Nr4a1GFP mice; histograms show GFP expression in MNs, iTAMs, and mTAMs and the proportion of GFP+ cells (L). (M) Relative Nr4a1 gene expression in MN, iTAM, and mTAM subsets. Data are represented as mean ± SEM of n = 5. All statistically significant differences were calculated using Kruskal–Wallis one-way ANOVA followed by Dunn’s multiple comparisons test; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. All data are representative of a minimum of two independent experiments.
To further characterize the different MN/macrophage populations in this model, we isolated MN, iTAM, and four mTAM populations by flow cytometry and performed high-density quantitative PCR analysis of gene expression using the Fluidigm Biomark system (Fig. 2 G). Gene expression data were analyzed by hierarchical clustering, which revealed a specific cluster of genes up-regulated in CD163-expressing mTAM (cluster III), including Il4ra, Mrc1, Stab1, and Slco2b1 (Fig. 2, G and H). Interestingly, especially in the CD163hi subset, there was also an up-regulation of genes that are known to be associated with negative regulation of CD8+ T cell function and immune suppression, such as Il10, Ido1, and Lgals1 (Fig. 2, G and H). We also observed distinctive clusters of inflammatory genes up-regulated in tumor-associated MNs (cluster I: Cxcl10, Il1b, Irf5, Ccr2, and Il18) and iTAM (cluster II: Cxcl9, Ciita, and Irf7); these genes were down-regulated in mTAMs coincident with the increased expression of genes in cluster III. Many of the genes represented in clusters I and II are IFN responsive and reflect an immune-stimulatory phenotype. We then performed principal component analysis (PCA) and network analysis of gene expression data highlighting the five nearest neighbors to described the relationship between these different populations. PCA was performed on normalized data (mean = 0 and variance = 1) generating a correlation based PCA plot. Network analysis connects k nearest neighbors (k = 5) based on similarity calculated by Pearson correlation (Fig. 2 I). This analysis, in combination with hierarchical clustering (Fig. 2 G), suggested that recruited inflammatory MNs are progressively polarized toward an immune-suppressive mTAM phenotype. We observed high expression of Nr4a1 in tumor-infiltrating MNs (Fig. 2 G). The nuclear receptor Nr4a1 (Nur77) is generally regarded as a marker for so-called patrolling or nonclassical MNs, as high expression is normally only detected on Ly6C− MNs in the circulation (Fig. 2 J). To further investigate Nr4a1 expression in tumor-associated MNs, we analyzed YUMM1.7 tumors from Tg(Nr4a1-GFP) mice; flow cytometry analysis showed comparable levels of GFP expression in Ly6C+ tumor-associated MNs and patrolling (Ly6Clo) MNs in the blood (Fig. 2, J and K), which was progressively reduced in iTAMs and mTAMs, respectively (Fig. 2, L and M), suggesting that tumor-associated MNs may be derived from a population of nonclassical MNs in the circulation, which down-regulate Nr4a1 expression as they differentiate into mTAMs.
Specific depletion of CD163-expressing TAMs promotes tumor regression
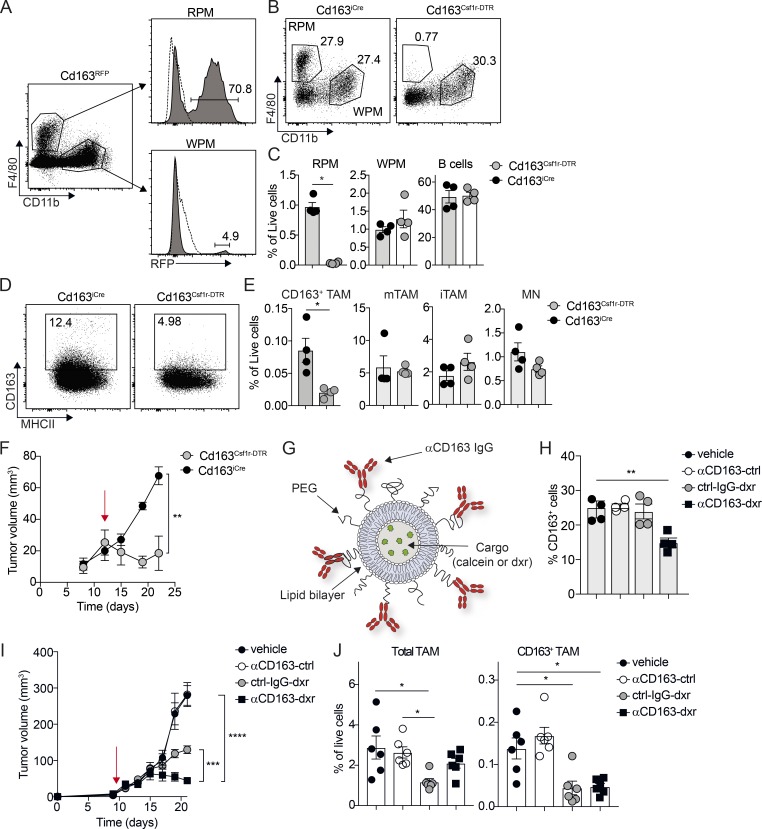
To test the specific contribution of CD163-expressing TAMs to tumor progression, we generated knock-in mice expressing iCre recombinase from the endogenous Cd163-promoter (Cd163-iCre). To validate Cre activity, we crossed Cd163-iCre with Rosa26-LSL-RFP reporter mice (Luche et al., 2007) and analyzed RFP expression in the spleen by flow cytometry (Fig. 3 A and Fig. S3, A–C). As expected, we observed RFP expression mainly in red-pulp macrophages (RPMs), which are known to express high levels of CD163, and to a lower extent in the myeloid compartment of white-pulp macrophages (WPMs; Fig. 3 A). We then crossed these mice to transgenic mice expressing diphtheria toxin receptor (DTR) under control of the Csf1r-promoter, with an upstream lox-stop-lox cassette (Csf1r-LSL-DTR). Thus, Cd163-iCre would trigger DTR expression only in CD163 and Csf1r-expressing myeloid cells and render these cells susceptible for ablation following treatment with diphtheria toxin (DT). Flow cytometry analysis of spleen from Cd163-iCre:Csf1r-LSL-DTR (Cd163Csf1r-DTR) mice 24 h after a single injection of DT (4 ng/kg), showed specific ablation of RPM, without effects on WPM or B cells (Fig. 3, B and C). We next grafted cohorts of Cd163Csf1r-DTR and Csf1r-LSL-DTR mice with YUMM1.7 cells and injected DT into mice with established tumors. 24 h after DT administration, flow cytometry analysis of tumors from Cd163Csf1r-DTR mice showed a specific depletion of >50% of CD163+ TAMs. In contrast, CD163− mTAMs, iTAMs, and MNs were unaffected (Fig. 3, D and E). Next, we attempted to achieve sustained depletion of CD163+ TAMs by repeated DT injections, but despite significantly reduced tumor growth in Cd163Csf1r-DTR mice compared with Csf1r-LSL-DTR mice (Fig. 3 F), this was associated with very severe side effects. In another approach, we developed a method for specific ablation of CD163+ macrophages using antibody-conjugated lipid nanoparticles (LNPs; αCD163-LNP; Etzerodt et al., 2012; Fig. 3 G). These LNPs contain 5% polyethylene glycol (2,000 mw) that minimizes nonspecific phagocytic uptake and increases the specificity of targeting to CD163+ cells (Fig. S3, D and E). To evaluate the targeting of αCD163-LNP to CD163+ TAMs in our melanoma model, calcein-loaded αCD163-LNPs and nontargeted control LNPs were injected i.v. in tumor-bearing mice. Subsequent in vivo fluorescence imaging and flow cytometry analysis showed increased accumulation of targeted LNPs in tumors by specific CD163+ TAM uptake (Fig. S3 F). We next used cytotoxic LNPs to specifically deplete CD163+ TAMs. We loaded LNPs with the DNA damaging agent doxorubicin (dxr) and injected mice i.v. with a single dose of vehicle, empty αCD163-LNPs (αCD163-ctrl), or dxr-loaded targeted and nontargeted LNPs (αCD163-dxr or IgG-dxr, respectively). 24 h later, we analyzed CD163+ RPMs in the spleen by flow cytometry to measure efficacy of CD163+ macrophage depletion and found that a single injection of αCD163-dxr specifically reduced the number of CD163+ RPMs by ∼50%, compared with control (Fig. 3 H). Next, we tested the effects of CD163+ TAM depletion on tumor growth in melanoma-bearing mice. To achieve an efficient and sustained depletion of CD163+ TAMs, mice with palpable tumors were randomized and treated every second day for 2 wk with αCD163-dxr or appropriate controls. Although treatment with nontargeted cytotoxic LNPs (IgG-dxr) was able to slow tumor growth, mice treated with CD163-targeted LNPs (αCD163-dxr) showed almost complete tumor regression after 2 wk (Fig. 3 I). Interestingly, subsequent flow cytometry analysis of tumors showed a reduction of total TAM numbers in mice treated with IgG-dxr (Fig. 3 J), suggesting indiscriminate targeting of TAM subsets. However, CD163-targeted LNPs only depleted the minor fraction of CD163+ TAM, having little impact on total TAM numbers (Fig. 3 J). We also validated the effects of CD163+ TAM depletion in spontaneous BrafV600E-driven melanomas, where αCD163-dxr treatment of tumor-bearing mice resulted in a complete inhibition of tumor growth compared with control-treated mice (Fig. S3 G). It has previously been suggested that CD163 expression can be acquired by tumor cells via cell fusion with CD163+ macrophages (Maniecki et al., 2012). However, we verified that YUMM1.7 cells did not express CD163 after passage in vivo (Fig. S4 A). Furthermore, incubation of YUMM1.7 cells in vitro with αCD163-dxr for 24 h did not affect cell viability (Fig. S4 B), confirming that αCD163-dxr treatment does not target tumor cells.
Figure 3.
Specific depletion of CD163-expressing TAMs promotes tumor regression. (A) Flow cytometry analysis of spleen from Cd163iCre/+ × Rosa26lsl-RFP/+ mice (Cd163RFP); RPMs were gated as CD45+, Lin− (CD3, CD19, NK1.1, Ly6G) F4/80+, CD11b− and WPMs as CD45+, Lin− (CD3, CD19, NK1.1, Ly6G) F4/80−, CD11b+. Histograms show relative RFP expression in RPMs and WPMs. (B and C) Flow cytometry analysis of spleen from Cd163iCre/+ × Csf1rlsl-DTR/+ mice (Cd163Csf1r-DTR). RPMs, WPMs, and B cells were analyzed in spleen 24 h after a single injection of 4 ng/kg DT; Cd163iCre/+ littermate control mice injected with DT were used as controls. Representative FACS plots for RPM/WPM analysis (B) and quantification of RPM, WPMs, and B cells in spleen (C) are shown. (D and E) Depletion of CD163+ TAMs in YUMM1.7 melanomas from Cd163Csf1r-DTR mice upon a single injection of 4 ng/kg DT. Representative FACS plots for CD163+ TAMs (D) and quantification of CD163+ TAMs and total TAM, iTAM, and MN in tumors from treated mice (E) are shown. Statistically significant differences were calculated using the Mann–Whitney U test; *, P < 0.05. (F) Tumor growth after sustained depletion of CD163+ TAMs with repeated DT injection; cohorts of Cd163Csf1r-DTR and Cd163iCre/+ mice bearing palpable tumors were treated with 4 ng/kg DT twice a week for 2 wk and tumor volume measured (red arrow indicates initiation of treatment). Statistically significant differences were calculated using a two-way ANOVA followed by Tukey post hoc test; **, P < 0.01. (G) Schematic illustration of αCD163 antibody–conjugated LNPs. PEG, polyethylene glycol. (H) Depletion of CD163-expressing macrophages in spleen after one injection of αCD163-LNP loaded with dxr; mice were injected with dxr-loaded αCD163-LNP (αCD163-dxr), IgG control-LNP (ctrl-IgG-dxr), empty αCD163-LNP (αCD163-ctrl), or PBS alone (vehicle). After 24 h, RPMs were analyzed in spleen by flow cytometry and the proportion of CD163+ RPMs determined. (I) Mice bearing palpable tumors were randomized into groups and treated as in H every second day for 2 wk and tumor volume measured (red arrow indicates initiation of treatment). (J) At endpoint, total TAM and CD163+ TAMs were analyzed by flow cytometry and frequency of live cells calculated. Data in graphs are represented as mean ± SEM of n = 4–6, and results are representative of three independent experiments. Statistically significant differences were calculated using a Kruskal–Wallis one-way ANOVA followed by Dunn’s multiple comparisons test; *, P < 0.05; **, P < 0.01; ***, P < 0.001; **** P < 0.0001.
The profound effects of specific depletion of CD163+ TAMs with targeted cytotoxic LNPs on tumor regression compared with nontargeted LNPs implied that pan-targeting of TAM subsets may in fact abrogate the therapeutic effects conferred by selective targeting of CD163+ TAMs, suggesting that other TAM subsets contribute to tumor regression after depletion CD163+ macrophages.
Targeted depletion of CD163+ TAM reeducates tumor-infiltrating myeloid (TIM) cells
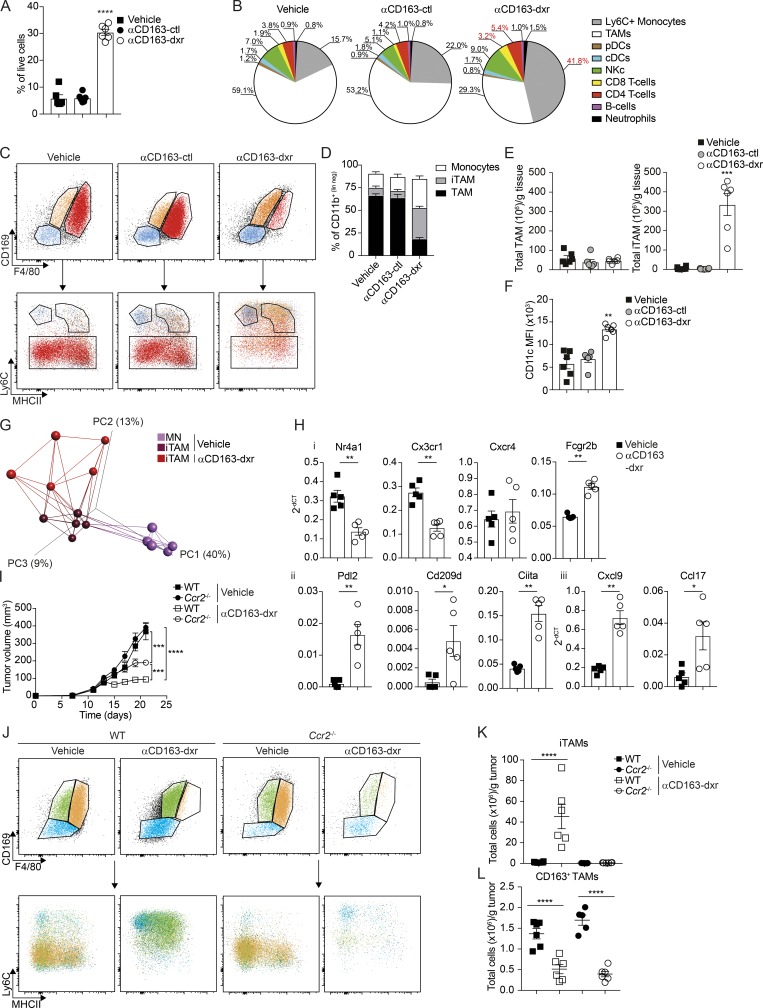
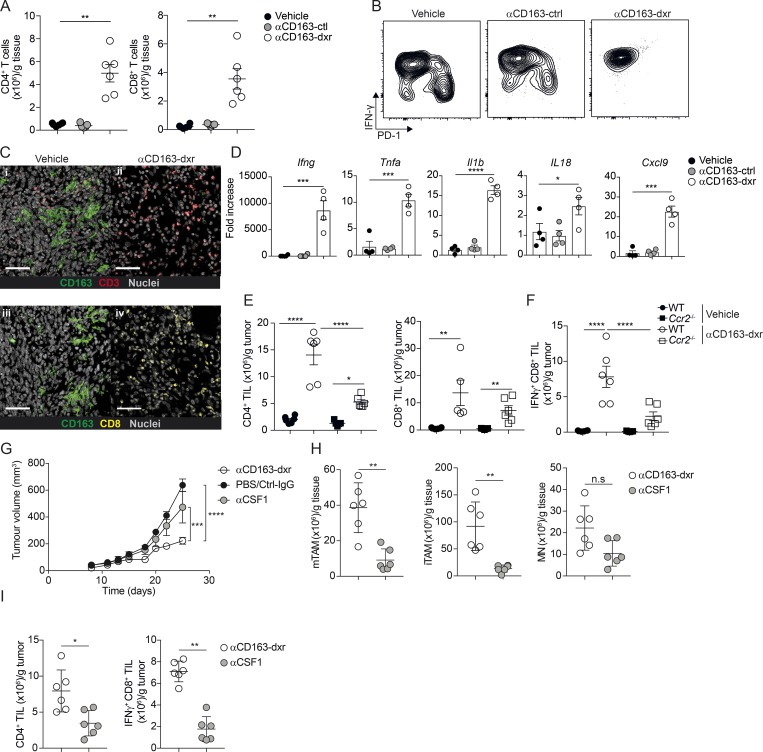
To further analyze the consequence of CD163+ TAM depletion on the tumor immune microenvironment, we performed a high-content immunophenotyping by flow cytometry on tumors after treatment with αCD163-dxr compared with empty CD163-targeted LNP (αCD163-ctrl)– and vehicle-treated mice. The depletion of CD163+ TAMs was associated with a highly significant overall increase in the tumor-infiltrating leukocyte compartment, increasing from 5 to 30% of all cells (Fig. 4 A). Analysis of different immune cell types revealed this was mainly due to increased numbers of tumor-infiltrating lymphocytes (TIL), including both CD4+ and CD8+ T cells, and especially Ly6C+ MNs (Fig. 4 B). In contrast, other immune cell subsets such as B cells, dendritic cells, or natural killer (NK) cells were unaffected (Fig. S4 D).
Figure 4.
Targeted depletion of CD163+ TAM reeducates TIM cells. Mice bearing palpable tumors were randomized into groups and treated as in Fig. 3; tumors were collected at endpoint for analysis by flow cytometry. (A and B) Total leukocyte infiltration (A; CD45+ cells) and high-content unsupervised immunophenotyping analysis after therapeutic depletion of CD163+ TAMs (B; see Fig. S4 C for definitions of populations). cDCs, conventional dendritic cells; pDCs, plasmacytoid dendritic cells. (C and D) Flow cytometry analysis of MNs, iTAMs, and mTAMs after CD163+ TAM depletion (C) and as proportion of tumor-associated CD11b+ cells (D). (E and F) Total number of TAMs (left) and iTAMs (right) per gram of tissue (E) and expression of CD11c, expressed as mean fluorescence intensity (MFI; F). Data are represented as mean ± SEM of n = 6. Statistically significant differences were calculated using a Kruskal–Wallis one-way ANOVA followed by Dunn’s multiple comparisons test; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (G) PCA and network analysis of gene expression in iTAMs from αCD163-dxr treated mice compared with vehicle control. PCA analysis was performed on normalized data (mean = 0 and variance = 1) generating a correlation-based PCA plot. Network analysis connects k nearest neighbors (k = 5) based on similarity calculated by Pearson correlation (see Fig. S4 D for a heatmap of differentially expressed genes). (H) Relative expression of specific genes in iTAMs (2−ΔCT) with and without CD163+ TAM depletion; data are represented as mean ± SEM of n = 5. Statistically significant differences were calculated using a Mann–Whitney U test; *, P < 0.05; **, P < 0.01. (I) Depletion of CD163+ TAM in WT or Ccr2−/− mice grafted with YUMM1.7 tumors; cohorts of mice bearing palpable tumors were treated with either αCD163-dxr or vehicle alone, every second day for 2 wk (n = 6). Statistically significant difference was calculated using two-way ANOVA followed by Tukey post hoc test; ***, P < 0.001; ****, P < 0.0001. (J–L) Flow cytometry analysis of MN, iTAM, and mTAM in tumors from WT and Ccr2−/− mice at endpoint (J); total numbers of iTAM (K) and CD163+ TAM (L) were calculated and expressed per gram of tissue. Data are represented as mean ± SEM of n = 6. Statistically significant differences were calculated using a Kruskal–Wallis one-way ANOVA followed by Dunn’s multiple comparisons test; ****, P < 0.0001. All data are representative of a minimum of two independent experiments.
To further characterize the TIM cell compartment after CD163+ TAM depletion, we performed flow cytometry analysis using our previously established gating strategy (Figs. 2 and 4 C). In αCD163-dxr–treated mice, there was a dramatic increase in recruitment of tumor-associated MNs and iTAMs, collectively making up >300 million cells per gram of tissue (Fig. 4, D and E), and consequently a drastic reduction in the proportion of mTAMs (Fig. 4 D). Interestingly, iTAM that infiltrated tumors after CD163+ TAM depletion showed a significant increase in CD11c expression (Fig. 4 F) and displayed a distinct gene expression profile compared with iTAMs from control tumors (Figs. 4 G and S4 E), including increased expression of Ciita and Cxcl9 (Fig. 4 H). This indicated recruited iTAMs had an immune-stimulatory phenotype typical of activated macrophages or MN–derived dendritic cells. Importantly, iTAMs from αCD163-dxrLNP–treated mice showed a significant decrease in expression of genes normally associated with patrolling or nonclassical MNs, such as Nr4a1 and Cx3cr1 (Fig. 4 H, i), which suggested a reeducation of TIM cells via recruitment of more classical inflammatory MNs.
Mobilization of inflammatory MNs (Ly6Chi, Nr4a1−, and Cx3cr1lo) is highly dependent on expression of the chemokine receptor Ccr2 and severely impaired in Ccr2-deficident mice (Ccr2−/−). In contrast, the distribution of patrolling MNs (Ly6Clo, Nr4a1+, Cx3cr1hi) is only slightly affected in Ccr2−/− mice (Tsou et al., 2007). To assess the contribution of Ccr2-dependent MN recruitment to the accumulation of Ly6C+ MNs and iTAMs after CD163+ TAM depletion, we generated cohorts of WT and Ccr2−/− mice bearing melanomas and treated with αCD163-dxr or vehicle, as described above. Interestingly, tumor progression in vehicle-treated mice was unaffected by Ccr2 deficiency (Fig. 4 I). Furthermore, accumulation of tumor-associated MNs and iTAMs was only marginally reduced and mTAMs were unaffected in Ccr2−/− mice (Fig. 4, J–L). However, tumor regression by αCD163-dxr treatment was significantly abrogated in Ccr2−/− mice (Fig. 4 I), which was accompanied by a complete reversal of iTAM recruitment provoked by CD163+ TAM depletion (Fig. 4, K and L). These data showed that depletion of CD163+ TAMs in melanoma-bearing mice results in Ccr2-dependent recruitment of Ly6C+ MNs and accumulation of immune-stimulatory macrophages that significantly contributes to inhibition of tumor progression.
CD163+ TAM depletion promotes anti–PD-1–resistant CTL responses
Immunophenotyping analysis of melanomas after depletion of CD163+ TAMs showed a significant increase in TILs (Fig. 4 B). To further analyze the effects of CD163+ TAM depletion on TIL recruitment and activation, we performed further flow cytometry analysis. Gating on TILs (CD45+, CD19−, NK1.1−, Siglec F−, Ly6G− and CD11b−, CD3e+, and CD5+), we confirmed a profound increase in both CD4+ and CD8+ TILs in melanoma-bearing mice treated αCD163-dxr compared with vehicle or empty αCD163-LNP (αCD163-ctrl; Fig. 5 A). In control-treated mice, CD8+ TILs displayed a heterogeneous expression of IFNγ and PD-1 (Fig. 5 B); however, in mice treated with αCD163-dxr, the vast majority of CD8+ TILs expressed high levels of IFNγ and no PD-1 (Fig. 5 B). We confirmed increased infiltration of CD8+ TILs in melanomas by confocal microscopy in tumor sections (Fig. 5 C), which correlated with the depletion of CD163+ TAMs (Fig. 5C). In accordance with increased infiltration of activated CTL, the tissue-wide expression of IFNγ (Ifng) was dramatically increased in tumors upon depletion of CD163+ TAMs (Fig. 5 D), and this was accompanied by increased expression of other inflammatory cytokines, including TNFα (Tnfa), IL-1β (Il1b), and IL-18 (Il18; Fig. 5 D), as well as by increased expression of the memory T cell–attracting chemokine Cxcl9 (Cxcl9; Fig. 5 D).
Figure 5.
Depletion of CD163+ TAM promotes CD4 and CD8 T cell recruitment. (A) Total numbers of CD8+ and CD4+ T cells in tumor tissue after therapeutic depletion of CD163+ TAMs, analyzed by flow cytometry. T cells were gated as CD45+, Lin− (CD19, NK1.1, Ly6G, CD11b), CD5+, CD3+ and subsequently identified as either CD4+ or CD8+ and numbers per gram of tissue calculated. See Fig. S1 D for the full gating strategy. (B) Flow cytometry analysis of IFNγ and PD-1 expression in tumor-infiltrating CD8+ T cells. Results are representative of three independent experiments with n = 6 per group. (C) Immunofluorescent staining of tumor tissue for CD163 (green), CD3 (red), and CD8 (yellow) in αCD163-dxr– or vehicle-treated mice. Scale bars, 50 µm. (D) Gene expression analysis in tumor tissue from αCD163-dxr– or vehicle-treated mice. Data are represented as mean ± SEM of n = 5. (E and F) Flow cytometry analysis of tumor-infiltrating T cells in WT and Ccr2−/− mice after therapeutic depletion of CD163+ TAMs; total numbers of (E) CD4+ T cells and CD8+ T cells and (F) IFNγ+ CD8+ T cells were calculated and expressed per gram of tissue. Statistically significant differences were calculated using a Kruskal–Wallis one-way ANOVA followed by Dunn’s multiple comparisons test; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (G) Treatment study comparing efficacy of CD163+ TAM depletion with pan-macrophages depletion using αCSF1 blocking antibody. Mice bearing palpable tumors were randomized into groups and treated with either αCD163-dxr (n = 6) or PBS (n = 4) i.v. every second day for 2 wk or αCSF1 (n = 6) or controls (CtrlIgG, n = 6 or PBS, n = 6) i.p. every 5 d. Statistically significant differences were calculated using a two-way ANOVA followed by Tukey post hoc test; ***, P < 0.001; ****, P < 0.0001. (H and I) At endpoint, the total number of mTAM, iTAM, and MN (H) or CD4+ TILs and IFNγ+ CD8+ TILs (I) was analyzed by flow cytometry and calculated from frequency of live cells. Data are represented as mean ± SEM of n = 6. Statistically significant differences were calculated using a Mann–Whitney U test; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. All data are representative of two independent experiments. n.s., not significant.
The global increase in Cxcl9 expression in tumors upon depletion of CD163+ TAMs correlated with a parallel increase in Cxcl9 expression in freshly recruited iTAMs (Fig. 4 H). Cxcl9 is a potent chemoattractant for memory CD8+ T cells, and induction of Cxcl9 expression in APCs by T cell–derived IFNγ has been shown to be critical for the propagation of CTL responses (Spranger et al., 2017). This led us to investigate if the recruitment of Ccr2-dependent iTAMs, induced by CD163+ TAM depletion (Fig. 4 K), was connected to the observed changes in the TIL compartment. We analyzed TILs in melanomas from WT and Ccr2−/− mice treated with αCD163-dxr or vehicle alone by flow cytometry. CD163+ TAM depletion in both WT and Ccr2−/− mice led to an increase in CD4+ and CD8+ TILs (Fig. 5 E); however, the number of CD4+ TILs, particularly IFNγ-producing CD8+ TILs, was significantly reduced in Ccr2−/− mice compared with WT mice (Fig. 5 F). These data suggested that the Ccr2-dependent recruitment of iTAMs after depletion of CD163+ TAMs contributed to the recruitment and activation of CD4+ and CD8+ TILs, respectively.
Previous studies using pan-macrophage depletion strategies, such as CSF1/CSF1R blockade, have shown only moderate effects on tumor growth in several cancer models, including the spontaneous BrafV600E-driven melanoma model (Perry et al., 2018). Since MN-derived iTAMs are also CSF-1 dependent, we next analyzed the effects of pan-macrophage targeting using an anti-CSF1 blocking antibody in comparison to specific depletion of CD163+ TAMs. As seen previously with IgG-dxr, pan-depletion of TAM subsets using anti-CSF1 resulted in only a slight and less pronounced inhibition of tumor growth compared with specific targeting of CD163+ TAMs (Fig. 5 G). As expected, anti-CSF1 treatment was associated with a strong reduction in all TAM subsets, including MNs (Fig. 5 H). Similarly, to Ccr2−/− mice, the reduction of iTAM after anti-CSF1 treatment was associated with a reduced number of tumor-infiltrating CD4+ and IFNγ-producing CD8+ T cells as compared with specific depletion of CD163+ TAMs (Fig. 5, A and I).
To establish the contributions of CD4+ and CD8+ TIL to tumor regression upon depletion of CD163+ TAM, we administrated CD4 and CD8b-depleting antibodies during treatment with αCD163-dxr. Both CD4+ and CD8+ T cell depletion completely reversed the control of tumor growth by αCD163-dxr treatment in melanoma-bearing mice (Fig. 6, A–C). This clearly demonstrated that tumor regression upon depletion of CD163+ TAM was driven by the recruitment and activation of TILs. Interestingly, depletion of CD4+ TILs alone markedly reduced the number of infiltrating iTAMs and IFNγ-producing CD8+ TILs after depletion of CD163+ TAMs (Fig. 6, D and E). In addition, gene expression analysis showed that depletion of both CD4+ and CD8+ T cells reduced global Ifng expression, whereas global expression of Cxcl9 and Il1b was only reduced in CD4+ T cell–depleted mice (Fig. 6 F). To further understand the relationship between iTAM recruitment and accumulation of CD4+ and CD8+ TILs after depletion of CD163+ TAMs, we monitored the kinetics of their recruitment following CD163+ TAM depletion by flow cytometry (Fig. 6 G). This showed that iTAM accumulation occurs rapidly after the initial depletion of CD163+ TAMs and precedes CD8+ TIL recruitment, whereas the number of CD4+ TILs is steadily increased during the time course of tumor growth. Interestingly, significant iTAM recruitment was not seen until 5 d after CD163+ TAM depletion, which coincided with onset of tumor growth inhibition and CD8+ TIL recruitment.
Figure 6.
CD163+ TAM depletion promotes anti–PD-1–resistant antitumor T cell responses. (A) Tumor-bearing mice were randomized into groups and treated with αCD163-dxr or vehicle every second day in combination with either αCD4 or αCD8-depleting antibodies or isotype control antibody twice a week (n = 5). Statistically significant differences were calculated using a two-way ANOVA followed by Tukey post hoc test; **, P < 0.01. (B–E) At endpoint, the relative numbers of tumor-infiltrating (B) CD4+ T cells, (C) CD8+ T cells, (D) IFNγ+ CD8+ T cells, and (E) iTAMs were determined by flow cytometry. Statistically significant differences were calculated using a Mann–Whitney U test (B and C) or Kruskal–Wallis one-way ANOVA followed by Dunn’s multiple comparisons test (D and E); **, P < 0.01. (F) Gene expression analysis on tumor tissue at endpoint. Data are represented as mean ± SEM of n = 5. Statistically significant differences were calculated using a Kruskal–Wallis one-way ANOVA followed by Dunn’s multiple comparisons test; *, P < 0.05; **, P < 0.01; ***, P < 0.001. (G) Flow cytometric analysis of CD4+, CD8+, and iTAM infiltration in YUMM1.7 tumor 1 d after αCD163-dxr treatment. Mice (n = 5 per time point) received one to five doses of 2 mg/kg dxr (red arrow indicates days of i.v. treatment). Total numbers of cells were calculated from the proportion of live cells and expressed per gram of tissue. (H) Mice inoculated with 106 YUMMER1.7 cells on the right flank were randomized into groups and treated from day 10 with 2 mg/kg dxr or control every second day for 2 wk as previously described for YUMM1.7 tumor bearing mice (Fig. 2 I). (I) Mice with palpable tumors were treated with αCD163-dxr or vehicle in combination with αPD-1 antibody or isotype control, as described above. (J) Tumor-bearing mice were treated with αCD163-dxr or vehicle, and 10 d later, treatment with αPD-1 or isotype control antibody was initiated. Data are represented as mean ± SEM of n = 6. Statistically significant difference was calculated using a two-way ANOVA followed by Tukey post hoc test; **, P < 0.01; ***, P < 0.001. All data are representative of a minimum of two independent experiments. n.s., not significant.
Human melanomas are characterized by relatively high level of somatic mutation potentially increasing the generation of neoantigens and immunogenicity of the tumor. In keeping with this concept, UVB irradiation of YUMM1.7 cells was used to generate a highly immunogenic cell line called YUMMER1.7 (Wang et al., 2017). In contrast to tumors generated with the parental YUMM1.7 cell line, YUMMER1.7 tumors are characterized by a high T cell infiltration, sensitivity to anti–PD-1 treatment, and frequently spontaneous rejection (Wang et al., 2017). However, despite their high immunogenicity, pan-macrophage depletion using CSF1R blockade showed no effect on growth of YUMMER1.7 tumors (Neubert et al., 2018). Strikingly, specific depletion of CD163+ TAMs resulted in complete rejection of YUMMER1.7 tumors in all treated mice (Fig. 6 H), further highlighting the strong immunosuppressive role of CD163+ TAMs.
Given that YUMM1.7 tumors have previously been reported to be refractory to the effects of PD-1 checkpoint inhibition alone, we sought to investigate if the effects of PD-1 blockade in combination with depletion of CD163+ TAMs. We administered anti–PD-1 antibody to tumor-bearing mice either alone in combination with αCD163-dxr for 10 d (Fig. 6 I) and following αCD163-dxr treatment (Fig. 6 J). As expected, anti–PD-1 treatment alone had no effect on the growth of YUMM1.7 tumors (Fig. 6 J). Furthermore, while αCD163-dxr efficiently controlled tumor growth with or without anti–PD-1 treatment (Fig. 6 I), when αCD163-dxr was substituted for anti–PD-1 alone, tumors quickly relapsed (Fig. 6 J), demonstrating that CD163+ TAM depletion mediates tumor regression independently of PD-1/PD-L1–mediated immune suppression.
To explore the possible clinical significance of CD163+ TAM on responsiveness to anti–PD-1 checkpoint inhibition in human melanoma, we examined the expression of CD163, IL10, LGALS1, and IDO1 in pretreatment tumor biopsy specimens from metastatic melanoma patients after anti–PD-1 therapy (Hugo et al., 2016; Fig. S5). Overall, there was a significant correlation between IL10 and CD163 expression although only IL10 was significantly overexpressed in nonresponding melanoma patients. However, further stratifying the tumors based on confirmed BRAF or NRAS mutations revealed a significant correlation between expression of CD163 and IL10 and response to anti–PD-1 checkpoint therapy.
Discussion
There is now ample experimental and clinical evidence highlighting the important functions of TAMs in progression of cancer. Consequently, the interest in developing novel therapeutic strategies to target TAMs is increasingly high. To date, most strategies have focused on the blockade of CSF-1/CSF1R-signaling, which regulates macrophage differentiation and survival or the CCL2/CCR2 axis, which regulates MN mobilization and recruitment. However, these strategies have shown limited effects in some experimental models and clinical trials (Yang and Zhang, 2017). Recent research has exposed an extensive heterogeneity among TAM subsets in human cancers, which may have important implications for clinical progression (Kiss et al., 2018). Indeed, several clinical studies have shown that certain TAM subsets can be associated with good prognosis in patients (de Vos van Steenwijk et al., 2013; Ino et al., 2013). Hence, more data are needed to further understand the function of specific TAM subsets to aid the future development of more targeted therapies.
Despite their intrinsic immune-stimulatory potential, one of the major tumor-promoting functions of TAMs is thought to be immune suppression. It has been suggested that the tumor microenvironment polarizes macrophages toward an alternative activation state, associated with suppression of TIL function, rather than activation (Ruffell et al., 2014). ICIs, which trigger activation of TILs, have made an unprecedented impact on the treatment of certain cancers, particularly malignant melanoma (Ugurel et al., 2017). However, the majority of patients still do not respond to existing ICI therapies, and the lack of response to ICIs often correlates with low TIL recruitment in primary tumors (Tumeh et al., 2014).
We chose to investigate the role of TAMs in a clinically relevant mouse model of melanoma, which is resistant to the current leading ICI therapy, anti–PD-1. We first characterized the TIM compartment in both autochthonous and subcutaneous melanoma models; TAMs constituted ≤60% of CD11b+ leukocytes in tumors, and on average, 20% of all tumor-infiltrating leukocytes were TAMs, which was 100 times more than the number of CD8+ TILs. Clinical data have strongly linked expression of CD163 by TAMs and poor prognosis in a range of cancers, including melanoma (Jensen et al., 2009; Bronkhorst et al., 2011). However, the functional relevance of these cells in tumor progression is still unclear. CD163+ TAMs represented only a minor fraction of all TAMs in mouse melanomas (<25%). Gene expression analysis of CD163+ TAMs revealed the up-regulation of a cluster of genes associated with inhibition of T cell activation, including Il10, Ido1, and Lgals1. This was in contrast to distinctive clusters of inflammatory genes up-regulated in tumor-infiltrating MNs (such as Cxcl10, Il1b, Irf5, Ccr2, and Il18) and iTAMs (Cxcl9, Ciita, and Irf7); these genes were down-regulated in CD163+ TAMs, coincident with the up-regulation of genes associated with immune suppression. Many of the genes up-regulated in MNs and iTAMs were IFN responsive and reflect immune-stimulatory activity and potential antitumor functions. This suggests recruited MNs are progressively polarized toward an immune-suppressive TAM phenotype.
To assess the specific contribution of CD163+ TAMs to tumor progression, we initially developed a genetic depletion strategy using the Cd163Csf1r-DTR mice, where expression of the simian DTR was restricted to CD163 and Csf1r-expressing macrophages. In these mice, a single injection of DT efficiently depleted CD163+ TAMs. However, YUMM1.7 tumors were rapidly repopulated by MN-derived cells. Although sustained depletion by repeated DT injection showed a convincing effect on tumor growth, mice suffered from severe side effects, including rapid weight loss and hematuria. To overcome the toxicity of sustained DT-mediated CD163+ macrophage depletion, we established a therapeutic approach based on targeted depletion of CD163-expressing TAMs using antibody-conjugated cytotoxic LNPs. Due to the relatively large size of LNPs compared with antibodies, for example, they are believed to preferentially accumulate in tumor tissue via leaky vasculature, while being less able to penetrate deep into healthy tissue (Torchilin, 2011; Ngoune et al., 2016). CD163-targeted LNPs efficiently depleted the minor fraction of CD163+ TAMs, having little impact on total TAM numbers. However, the selective depletion of CD163+ TAMs profoundly reduced tumor growth in both tumor models. Interestingly, we observed that nontargeted cytotoxic LNPs significantly reduced total TAM numbers but were not as effective as CD163-targeted LNPs in reducing tumor growth. This implied that pan-targeting of TAM subsets may in fact abrogate the therapeutic effects conferred by the specific depletion of CD163+ TAMs. This suggests that other TAM subsets could contribute to tumor regression upon CD163+ TAM depletion. To explore this hypothesis, we assessed the impact of CD163+ TAM depletion on the TIM compartment in melanomas. We observed that iTAMs, which infiltrated tumors after CD163+ TAM depletion, were CD11chi and had increased expression of Ciita, Cxcl9, and CD209d, indicating an immune-stimulatory phenotype typical of MN-derived dendritic cells (Menezes et al., 2016). Recruitment of CD11chi iTAM was blocked in Ccr2-deficient mice, which abrogated the reduction in tumor growth induced by CD163+ TAM depletion. Thus, the mobilization of Ccr2-dependent inflammatory MNs significantly contributed to tumor regression. CD163+ TAM depletion also increased the numbers of CD4+ and CD8+ TILs in melanomas, both of which were required for controlling tumor growth. Kinetic analysis showed that the number of iTAMs was rapidly increased after initial CD163+ TAM depletion, followed by a sharp increase in CD8+ TILs that coincided with the onset of tumor regression. In contrast, recruitment of CD4+ TILs only gradually increased over time. Interestingly, both CD4+ and CD8+ TIL recruitment was Ccr2 dependent, indicating a role for the recruitment of inflammatory MNs and likely the accumulation of CD11chi iTAMs. This dependency could be explained by the increased expression of Cxcl9 in iTAMs, a critical chemokine for recruitment of memory T cells, as well as enhanced APC activity due to increased MHCII expression via Ciita (Fig. 7). This would also explain the lack of tumor growth inhibition and T cell infiltration observed upon pan-macrophage depletion by anti-CSF1 blockade.
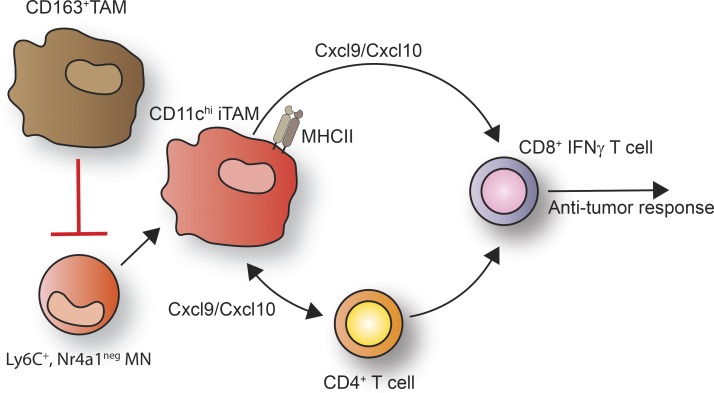
Figure 7.
Schematic representation of the immune-suppressive function of CD163+ TAMs in melanoma. CD163+ TAMs block the recruitment antitumor CD8 T cells by blocking the accumulation of Ly6C+, Nr4a1− MNs and CD11chi iTAMs. Upon CD163+ TAM depletion, Ly6C+, Nr4a1neg MNs are rapidly mobilized, leading to the accumulation of CD11chi iTAMs that in connection with CD4+ T cells drive the recruitment and activation of antitumor CD8+ T cells.
Inhibition of CD4+ TIL accumulation after depletion of CD163+ TAMs markedly reduced the number IFNγ-producing CD8+ TILs, in keeping with the role of CD4+ T cell help in CD8+ TIL activation. However, depletion of CD4+ TILs, but not CD8+ TILs, also significantly reduced the number of infiltrating CD11chi iTAMs, suggesting that CD4+ TILs contribute to inflammatory mobilization, which may in turn promote the recruitment and activation of CD8+ TILs.
In summary, our studies demonstrate a profound immune-suppressive function for CD163+ TAM in melanoma that is independent of the PD-1/PD-L1 axis. Importantly, these findings may be relevant to human melanoma, where we found an increased expression of CD163 and IL10 in pretreatment biopsy specimens from metastatic melanoma patients who were resistant to anti–PD-1 therapy. Furthermore, although we are unable to propose an exact mechanism, our data show that the specific depletion of CD163+ TAMs allows the reprogramming of tumor-infiltrating MNs toward immune-stimulatory functions and tumor regression. These data not only present a new therapeutic strategy based on the specific targeting of TAM subsets but also may explain the lack of efficacy with therapeutic approaches currently in clinical trials, such as CSF1/CSF1R or CCL2/CCR2 blockade, which indiscriminately target all MN-derived macrophages. Thus, the requirement for inflammatory MNs to propagate TIL recruitment and activation may limit the utility of pan-macrophage targeted therapies. Furthermore, targeting TAM subsets could represent a complementary therapeutic approach toward immunologically “cold” tumors that do not respond to current immune-checkpoint therapies.
Materials and methods
Mouse breeding and initiation of mouse melanoma models
Tg (Nr4a1-EGFP; Moran et al., 2011) and Tg(Csf1r-HBEGF; Schreiber et al., 2013) mice were obtained from The Jackson Laboratory. C57BL/6J mice were obtained from Janvier Labs. Cd163-iCre mice were generated from modified embryonic stem cells on a C57BL/6 background. In brief, a FlpO-NeoR cassette encoding IRES-iCre was inserted in the 3′ untranslated region of the CD163 gene using homologous recombination and used to generate chimeric mice that were subsequently crossed to Flp deleter mice to facilitate removal of the NeoR cassette. To induce spontaneous melanoma formation, 5-wk-old mice carrying conditional alleles BRAFCA/+, PTENlox4-5/lox4-5, and Tg(Tyr-CreERT2) (Dankort et al., 2009) were shaved and treated with 1 µl of 7.8 mg/ml 4-HT (Sigma-Aldrich) on the right flank. Subcutaneous tumors were established in 8-wk-old mice by s.c. injection of 106 YUMM1.7 cells (Meeth et al., 2016) or 106 YUMMER1.7 cells (Wang et al., 2017) in 100 µl sterile PBS, pH 7.4, on the right flank. Tumor size was measured using a digital caliper in x, y, and z and volume calculated using the equation for volume of an ellipsoid (volume = 0.5233xyz). For treatment with LNPs, when tumors reached palpable size (∼10 mm3), mice were injected with 100 µl LNPs by retroorbital injection. For CD4+ and CD8+ T cell depletion and anti–PD-1 antibody treatment, mice received 250 µg αCD4 (Clone GK1.5), αCD8b (clone 53–5.8), αPD-1 (clone RMP1-14), or isotype control IgG (IgG1 or IgG2a; BioXCell) by i.p. injection twice a week, with first injection 1 d before treatment with LNPs. For CSF1 blocking antibody treatments, mice received 1 mg anti-Csf1 (clone 5A1) or rat IgG1 isotype control (clone HRPN; both BioXCell) i.p. for the initial treatment followed by 0.5 mg i.p. every 4 d. All mice were housed at the animal facility at Centre d’immunologie Marseille-Luminy with water and food ad libitum and 12-h/12-h night/daylight cycle. All animal experiments were approved and performed in accordance with the limiting principles for using animal in testing (the three Rs: replacement, reduction, and refinement) and approved by the French Ministry of Higher Education and Research.
Flow cytometry and cell sorting
All tissues were minced and digested in RPMI 1640 medium with 1 mg/ml Collagenase II (Sigma-Aldrich), 50 µg/ml DNaseI (Roche), and 0.1% (wt/vol) BSA for 30 min at 37°C with gentle agitation. Cell suspensions were subsequently passed through a 70-µm cell strainer (BD Biosciences) and collected by centrifugation. For RBC lysis, cell suspensions were incubated with 0.85% NH4Cl for 2 min at room temperature, collected by centrifugation and resuspended in FACS buffer (1×PBS, pH 7.4, 1 mM EDTA, pH 8.0, 3% FCS, and 0.1% NaN3). For flow cytometry and FACS sorting, single-cell suspensions were incubated at 4°C for 10 min with the 2.4G2 antibody followed by the specified antibodies (see Table 1 for details) for 30 min at 4°C. Prior to analysis, cells were incubated with Sytox Blue (Thermo Fisher Scientific) to discriminate dead cells. For IFNγ intracellular staining, surface stained cells were incubated with Live/Dead fixable (Thermo Fisher Scientific) violet for 20 min in PBS to discriminate dead cells and subsequently fixed, permeabilized, and washed with BD Perm/Wash buffer (BD Biosciences) followed by incubation with IFNγ antibodies diluted in Perm/Wash buffer for 30 min at 4°C. Analysis was done on either LSR-2 or Fortessa X-20 flow cytometers equipped with a 350-nm laser (BD Biosciences). Subsequent data analysis was done using FlowJo software V10.4 for Mac (Tree Star). High-content immunophenotyping analysis was performed at Centré d’Immunophénomique (see Fig. S4 C for gating strategies).
Table 1. Antibodies used for flow cytometric analysis.
| Antigen | Clone | Dye | Catalog no. | Supplier |
|---|---|---|---|---|
| CD45.2 | 104 | BUV737 | 564880 | BD Biosciences |
| MHCII | M5/114.15.2 | A700 | 56-5321-82 | eBioscience |
| CD11b | M1/70 | BV395 | 563553 | BD Biosciences |
| F4/80 | BM8 | BV785 | 123141 | BioLegend |
| CD163 | R34 | CF405 | ||
| CD64 | X54-5/7.1 | BV711 | 139311 | BioLegend |
| CD169 | SER-4 | eFluor660 | 50-5755-82 | Invitrogen |
| Ly6C | AL-21 | FITC | 553104 | BD Biosciences |
| TIM4 | 54(RMT4-54) | PerCP-Cy5.5 | 46-5866-82 | eBioscience |
| Siglec F | ES22-10D8 | PE | 130-102-274 | Miltenyi Biotec |
| CD5 | 53-7.3 | BV605 | 563194 | BD Biosciences |
| CD19 | 1D3 | BUV661 | 565076 | BD Biosciences |
| CD11c | HL3 | PE-Cy7 | 558079 | BD Biosciences |
| NK1.1 | PK136 | Allophycocyanin-Cy7 | 108724 | BioLegend |
| Ly6G | 1A8 | Allophycocyanin-Cy7 | 560600 | BD Biosciences |
| CD3 | 17A2 | A700 | 56-0032-82 | eBioscience |
| CD4 | RM4-4 | FITC | 553055 | BD Biosciences |
| CD8 | 53-6.7 | PE-CF594 | 562283 | BD Biosciences |
| PD1 | RMP1-30 | PE-Cy7 | 109110 | BioLegend |
| IFNγ | XMG1.2 | PerCP-Cy5.5 | 45-7311-82 | Invitrogen |
Liposome preparation
Long circulating liposomes encapsulating dxr was prepared and modified for CD163 targeting as previously described (Fritze et al., 2006; Etzerodt et al., 2012). Briefly, liposome formulations were formed using the ethanol-injection method from a mixture of hydrogenated soy phosphatidylcholine, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-(methoxy(polyethylene glycol)-2000), and cholesterol (molar ratio of 55:40:5; Lipoid GmBH and Sigma-Aldrich). Lipids were dissolved in EtOH at 65°C for 15 min followed by hydration (to 10% EtOH) for 1 h at 65°C in aqueous buffer suitable for further downstream applications. Liposomes were sized by extrusion 25 times through a 0.1-µm filter using the Avanti mini-extruder kit (Avanti Polar Lipids) and dialyzed twice against 150 mM NaCl (0.9% NaCl; the second dialysis was overnight at 4°C). Encapsulation of calcein (calLNP) was done by hydrating lipids in a 200 mM calcein (pH 7.4) solution with dialysis repeated five times to remove excess calcein. For remote loading of dxr, lipid was hydrated in 300 mM (NH4)1HPO3. Following extrusion and dialysis, (NH4)1HPO3–containing liposomes were mixed with dxr-HCl for 30 min at 65°C at a dx/lipid ratio of 1:5. Lipid content, drug content, and encapsulation efficiency was subsequently estimated from high-pressure size-exclusion chromatography (UV absorbance 210 nm) using a Dionex Ultimate3000 HPLC system (Thermo Fisher Scientific) equipped with Ascentis C18 column (Sigma-Aldrich). Liposome size was estimated using dynamic light scattering and the DynaPro NanoStar system (Wyatt Technology Europe). Modification of liposomes for CD163 targeting was done as described previously using the postinsertion method of αCD163 antibody, clone 3E10B10 (Torchilin et al., 2001; Etzerodt et al., 2012), or isotype control IgG (BioXCell).
IHC and immunofluorescence
5-mm slices of entire tumors or back skin were fixed in 4% formalin and embedded in agarose for vibratome sectioning, optimal cutting temperature compound for cryostat sectioning, or paraffin for microtome sections. 200-µm vibratome and 10-µm cryostat sections were incubated with anti-CD163-ATTO565 (Etzerodt et al., 2013), anti-CD146-Alexa Fluor 647 (clone ME-9F1; BD Bioscience), CD3e-allophycocyanin (clone 145-2C11; BD Bioscience) and CD8b-FITC (53–5.8; BD Bioscience) together with anti-FITC A488 (A11096; Thermo Fisher Scientific) in 0.1 M Tris, pH 7.2, 1% Triton X-100, and 0.5% BSA for vibratome sections or 1×PBS, 0.1% Triton X-100, and 2% BSA for cryostat sections. Nuclei were visualized with Hoechst 33342 (Sigma-Aldrich). Images were acquired on a Zeiss LSM780 confocal microscope (Carl Zeiss Microscopy) using spectral unmixing with a 20× objective. For IHC, sections were stained with H&E and anti-CD163 and visualized using ImmPRESS-AP anti-rabbit IgG followed by VECTOR Red Alkaline Phosphatase Substrate Kit (Vector Laboratories).
High-throughput gene expression analysis
Total RNA was purified from sorted cell populations using the RNeasy Micro Kit (Qiagen) and concentration determined using the Quant-IT RiboGreen RNA assay kit (Thermo Fisher Scientific). First-strand cDNA synthesis was performed with High Capacity cDNA Reverse transcription Kit (Thermo Fisher Scientific) followed by preamplification of genes of interest using the Fluidigm PreAmp Master Mix (Fluidigm Europe B.V.) using 2.5 ng total RNA and in accordance with the manufacturer’s instructions. Exon-spanning primers to amply genes of interest were designed using Primer-Blast (see Table 2 for details). To increase sensitivity, genes of interest were preamplified by 14 cycles of PCR using pooled assays followed by exonuclease I treatment (New England Biolabs) to remove unincorporated primers. Final preamplified cDNA was diluted 1:5 in Tris-EDTA buffer. High-throughput gene expression analysis was performed using the 96.96 dynamic arrays and Biomark HD system from Fluidigm in accordance with the manufacturer’s instructions and standard settings. Obtained data were analyzed using the Real-Time PCR Analysis Software (Fluidigm), and resulting CT values were normalized to Cph to obtain dCT values. Heatmaps, Z-scores, and hierarchical clustering using the one minus Pearson correlation were generated using Morpheus (https://software.broadinstitute.org/morpheus/). PCA plots were generated using Qlucore Omics Explorer (Qlucore AB).
Table 2. Primer sequences used in high-throughput gene expression analysis.
| Gene | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) |
|---|---|---|
| Aldh1a2 | TGCAGGCTGGGCTGATAAAA | TGGGCTCGTGTCTTGTGAAA |
| Aox15 | TGCAGCTCTGGCAAGTCA | AACAGCTTGGTCGGTCTTGTA |
| Arg1 | GGATTGGCAAGGTGATGGAA | CGACATCAAAGCTCAGGTGAA |
| Batf3 | CAGAGCCCCAAGGACGATGA | TCAGCTTCGAAATCTCCCTGC |
| Capg | TGAGTTGGGACAGCTTCAACA | TTTCCACCACACCAGGCAAA |
| Ccl11 | CAACAACAGATGCACCCTGAA | CACAGATCTCTTTGCCCAACC |
| Ccl17 | CAGGAAGTTGGTGAGCTGGTA | CTTGCCCTGGACAGTCAGAA |
| Ccl2 | AGCAGCAGGTGTCCCAAA | TTCTTGGGGTCAGCACAGAC |
| Ccl22 | CCTTCTTGCTGTGGCAATTCA | GGCAGCAGATACTGTCTTCCA |
| Ccl4 | CTGTGCTCCAGGGTTCTCA | AGCAAAGACTGCTGGTCTCA |
| Ccl7 | TCTGTGCCTGCTGCTCATA | CATAGCAGCATGTGGATGCA |
| Ccr1 | TCCTCAAAGGCCCAGAAACA | GCTGAGGAACTGGTCAGGAA |
| Ccr2 | TGAGGCTCATCTTTGCCATCA | GGATTCCTGGAAGGTGGTCAA |
| Ccr4 | GACTGTCCTCAGGATCACTTTCA | AGCAGGAGAAGCCAATGAGAA |
| Cd163 | GCCATAACTGCAGGCACAAA | GTTGGTCAGCCTCAGAGACA |
| Cd209a | AAATGGGGAAGAGGCAGCTTC | CAGCCTTCAACTGGGTCAGTTC |
| Cd209d | GAGTGGCACCAGGTATTCTGATA | TTGCGGCTCTGCTTCGTATA |
| Cd44 | TTCCTTCGATGGACCGGTTA | TACTCGCCCTTCTTGCTGTA |
| Chil3l3 | TGGCCCACCAGGAAAGTACA | GACCTCAGTGGTCCTTCATTC |
| Ciita | GCCATCCGGGACCTTAAGAA | ATCTTTGCCAGTGTGGGGAA |
| Ppia | ATGGTCAACCCCACCGTG | TTCTTGCTGTCTTTGGAACTTTGTC |
| Csf2 | AGTCGTCTCTAACGAGTTCTCC | CCGTAGACCCTGCTCGAATA |
| Cx3cr1 | TTCCCATCTGCTCAGGACCTC | AGACCGAACGTGAAGACGAG |
| Cxcl10 | GGGCCATAGGGAAGCTTGAA | GGATTCAGACATCTCTGCTCATCA |
| Cxcl9 | AGCCCCAATTGCAACAAAAC | TCTTCACATTTGCCGAGTCC |
| Cxcr4 | TCAGCCTGGACCGGTACCT | GCAGTTTCCTTGGCCTTTGA |
| Cyp51a1 | TCTTCACCGAGTCCAAGTGC | TGGTGGACTTTTCGCTCCAG |
| Dhcr24 | CGCTCTCGCTCATCTTCGAT | TCCATTCCCGGACCTGTTTC |
| Fcgr2b | GGGAGAAACCCTTCCAGAGG | GGAGGATTGTATGGGCTGCT |
| Fcgr3 | AGAATGCACACTCTGGAAGCC | TCTGCAAAAGCAAACAGCAGC |
| Fcrls | AGTGAGGTGGTAACGCTCAA | CCTCTACGCGTCCTTCACA |
| Flt3 | ATGGAAAACCAGGACGCACT | CACAAGGGTTCCCCCACTTT |
| Folr2 | AGCCTGTCGTACCTCCTTTAC | GCTGTGTTTGGGCACTTGTTA |
| Fos | ATGGGCTCTCCTGTCAACAC | TCTACTTTGCCCCTTCTGCC |
| Hmgcs1 | TCCCCTTTGGCTCTTTCACC | GCCGCCCAATGCAATCATAG |
| Ido1 | ACTTTGTGGACCCAGACACG | GCAGGAGATTCTTTGCCAGC |
| Ifna | TCTGATGCAGCAGGTGGG | AGGGCTCTCCAGACTTCTGCTCTG |
| Ifnb1 | TCCAAGAAAGGACGAACATTCG | TGAGGACATCTCCCACGTCAA |
| Ifng | CCACGGCACAGTCATTGAAA | GCCAGTTCCTCCAGATATCCAA |
| Il10 | GGCGCTGTCATCGATTTCTC | ATGGCCTTGTAGACACCTTGG |
| Il12a | AAACCAGCACATTGAAGACC | GGAAGAAGTCTCTCTAGTAGCC |
| Il12b | GGAAGCACGGCAGCAGAATA | AACTTGAGGGAGAAGTAGGAATGG |
| Il18 | TCTTGGCCCAGGAACAATGG | ACAGTGAAGTCGGCCAAAGT |
| Il1a | AGATGGCCAAAGTTCCTGAC | AGAGATGGTCAATGGCAGAAC |
| Il1b | TGGCAACTGTTCCTGAACTCA | GGGTCCGTCAACTTCAAAGAAC |
| Il1r1 | TCCTGCTCTGGTTTTCTTCC | TCCCAGGGTCTTGGGATAAA |
| Il23a | ACCAGCGGGACATATGAATCT | AGACCTTGGCGGATCCTTTG |
| Il4ra | AACATCTCCAGAGAGGACAACC | CTCAGCCTGGGTTCCTTGTA |
| Il6 | CCAGAAACCGCTATGAAGTTCC | TTGTCACCAGCATCAGTCCC |
| Il7r | GCCAAAAACGAGTCTGAATGTGA | CTGGCTGTGCAGGAAGATCA |
| Irf4 | GACCAGTCACACCCAGAAATCCC | GTTCCTGTCACCTGGCAACC |
| Irf5 | TAGAGGCTACCCAGGAGCAA | GCCCACTCCAGAACACCTTA |
| Irf7 | CTGGAGCCATGGGTATGCA | AAGCACAAGCCGAGACTGCT |
| Irf8 | CCGGCAAGCAGGATTACA | GCTTTGTCTCCCTCTTTAAACTTC |
| Itga5 | TCCAGTGCACCACCATTCAA | TCCTCTCCCTTGGCACTGTA |
| Junb | AGGCAGCTACTTTTCGGGTC | AGGCAGCTACTTTTCGGGTC |
| Lgals1 | GCCAGCAACCTGAATCTCAA | CTTTTCCCAGGTTCAGCACAA |
| Lgals3 | ATCATGGGCACAGTGAAACC | AGTGGAAGGCAACATCATTCC |
| Lilra5 | TGAATCTGGGCCAAGAGACC | ACCAATCCTGGCTGAACACTAA |
| Lrrc8c | TTCTGGGACCACAGATGTTCA | AACTCGGTCACCGGAATCA |
| Lta | GCCTTTCCTGCCTTCGACT | GTCATGTGGAGAACCTGCTGT |
| Ltb | GTTCAACAGCTGCCAAAGGG | CATCCAAGCGCCTATGAGGT |
| Marco | CCAGGACTTTCAGGTGCCAA | TGGCCAGAAGACCCTTTCAT |
| Mrc1 | TCATTGGAAGATCCACTCTGG | CAGCGCTTGTGATCTTCATTATAG |
| Mmp9 | TCCCCAAAGACCTGAAAACC | GGGTGTAACCATAGCGGTAC |
| Nabp1 | ATCGGAGGGCTCATACAGAC | AGTGTCAGGCATCCTTTCCA |
| Nos2 | GCCACCAACAATGGCAACAT | TCGATGCACAACTGGGTGAA |
| Nr4a1 | CAATGCTTCGTGTCAGCACTA | TGTTTGCCAGGCAGATGTAC |
| Pdl1 | CTCGCCTGCAGATAGTTCCC | GTCCAGCTCCCGTTCTACAG |
| Pdl2 | ACCTGCCAGGCTAGAGGTTA | TCCATCCGACTCAGAGGGTC |
| Pfkp | TAAGGAAGCCGTGAAACTCC | CAGGCAGTTTAATGGCAAGAC |
| Ptges | TCTCACTCTCAGTCCCGGTG | CGGGGTTGGCAAAAGCCTTC |
| Ptgs2 | GTTCATCCCTGACCCCCAAG | TTTAAGTCCACTCCATGGCCC |
| Rarb | ACAGATCTCCGCAGCATCA | AGGTGGCATTGATCCAGGAA |
| Retnla | ACTTCTTGCCAATCCAGCTAAC | CAAGCACACCCAGTAGCAGT |
| S100a8 | TTCGTGACAATGCCGTCTGA | TTCTCTCTGAAGGCCTCTGTC |
| Sca1 | TCAGGAGGCAGCAGTTATTGTGGA | TACATTGCAGAGGTCTTCCTGGCA |
| Scd2 | GAGCCTTGTACGGGATCACA | CCAGGGCGCTGATTACATAGTA |
| Sema4a | CTTTGACCCTGTTCACAAGCA | CATGGTGCCGGAATAAAGCA |
| Sfpi1 | CCCACACCGGCCTCAGT | GACAAGGTTTGATAAGGGAAGCA |
| Slamf8 | GGTCAAACCTGGACCCAGAC | GCAGCAGTGAACACTTGAACC |
| Slco2b1 | AGCCTCATGCTACGCCTTTA | ATCTGGGGTCTTTTGGAGTCAA |
| Socs1 | CAACGGAACTGCTTCTTCGC | AGCTCGAAAAGGCAGTCGAA |
| Socs2 | GGTTGCCGGAGGAACAGTC | GAGCCTCTTTTAATTTCTCTTTGGC |
| Socs3 | CCTTCAGCTCCAAAAGCGAG | GCTCTCCTGCAGCTTGCG |
| Sqle | AGTGAACAAACGAGGCGTCC | AGCAAGCTTTTCGGAGCTGA |
| Stab1 | GTACGGTACCACATCTACAACC | TGGTGAGGACACGTCCTTTA |
| Syk | TGGTCACCGGGTGGAATAAT | TTATATTCTGCACAAACTGAGGAC |
| Tlr2 | GGTGCGGACTGTTTCCTTCT | AGATTTGACGCTTTGTCTGAGG |
| Tlr4 | CGCTGCCACCAGTTACAGAT | AGGAACTACCTCTATGCAGGG |
| Tm7sf2 | GTCTTTGCCCTGTGGGCTAT | ACCTCTTAGGTGGACCTGCT |
| Tnf | GGGTGATCGGTCCCCAAA | TGAGGGTCTGGGCCATAGAA |
| Tnrsf11a | GCAGCTCAACAAGGATACGG | GGTGCAGTTGGTCCAAGGTT |
| Usp18 | GACGCAAAGCCTCTGAAAAC | CACATGTCGGAGCTTGCTAA |
| Vegfa | CCAGCACATAGGAGAGATGAG | CTGGCTTTGTTCTGTCTTTCTT |
| Zbtb46 | AAGCGACATACTCTGGTCCAC | GGCTGCAGACATGAACACAC |
Statistical analysis
For treatment studies, statistical analysis was done using two-way ANOVA followed by Tukey post hoc test. For comparison between groups, statistical testing was performed using nonparametric tests such as Mann–Whitney U test or Kruskal–Wallis followed by Dunn’s multiple comparisons test. P values are indicated as *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001. All statistical analysis was performed with GraphPad Prism 7 for Mac.
Online supplemental material
Fig. S1 shows IHC of CD163+ macrophages in naive skin as well as the proportion of leukocytes that are TAMs or CD8+ TILs in spontaneous BrafV600E tumor as well as the gating strategy for TAM and T cells. Fig. S2 shows the flow cytometric analysis of MNs, iTAMs, and TAMs in YUMM1.7 tumors and the sorting strategy used to isolate MNs, iTAMs, and mTAMs for high-throughput gene expression analysis. Fig. S3 shows the gating strategy used for flow cytometric analysis of CD163Cre mice as well as the in vitro and in vivo characterization of αCD163-LNP and the effect of aCD163-dxr on tumor growth in the spontaneous BrafV600E model. Fig. S4 shows the flow cytometric analysis of CD163 expression and toxicity of αCD163-dxr on YUMM1.7 cells as well as the gating strategy used for high-content immunophenotyping, effect of aCD163-dxr on nonmacrophage immune cells, and gene expression analysis of sorted iTAMs after αCD163-dxr treatment. Fig. S5 shows the gene expression analysis of CD163 and IL10 in pretreatment tumor biopsy specimens from metastatic melanoma patients in anti–PD-1 therapy.
Supplementary Material
Acknowledgments
We thank Bernard Malissen (Centre D’immunologie Marseille-Luminy, Marseille, France) for Rosa26-LSL-tdRFP and Ccr2−/− mice and Lionel Chasson (Histology Platform, Centre D'immunologie Marseille-Luminy) for assistance with histology.
These studies were supported by the Novo Nordisk Foundation (grant NNF14OC0008781 to A. Etzerodt), L’Agence Nationale de la Recherche (grants ANR-09-MIEN-029-01 and ANR-10-BLAN-1302-01 to T. Lawrence), and the European Research Council (FP7/2007–2013 grant agreement number 260753 to T. Lawrence) and by institutional funding from the Institut National de la Santé et de la Recherche Médicale, Centre National de la Recherche Scientifique, and Aix-Marseille-Université. Microscopy facilities are supported by L’Agence Nationale de la Recherche (grant ANR-10-INBS-04-01 France Bio Imaging).
S.K. Moestrup and J.H. Graversen are minority shareholders in the small biotech company Affincon Aps, which holds intellectual property rights to CD163 targeting. The remaining authors declare no competing financial interests.
Author contributions: A. Etzerodt and T. Lawrence designed the experiments. A. Etzerodt and K. Tsalkitzi performed the experiments with help from M. Maniecki, W. Damsky, M. Delfini, E. Baudoin, M. Maniecki, J.H. Graversen, N. Auphan-Anezin, and S.K. Moestrup. M. Bosenberg provided Tyr::CreER, BrafCA, Ptenf/f mice and the YUMM1.7 cell line. A. Etzerodt and T. Lawrence wrote the manuscript.
References
- Bronkhorst I.H.G., Ly L.V., Jordanova E.S., Vrolijk J., Versluis M., Luyten G.P.M., and Jager M.J.. 2011. Detection of M2-macrophages in uveal melanoma and relation with survival. Invest. Ophthalmol. Vis. Sci. 52:643–650. 10.1167/iovs.10-5979 [DOI] [PubMed] [Google Scholar]
- Cassetta L., Fragkogianni S., Sims A.H., Swierczak A., Forrester L.M., Zhang H., Soong D.Y.H., Cotechini T., Anur P., Lin E.Y., et al. 2019. Human Tumor-Associated Macrophage and Monocyte Transcriptional Landscapes Reveal Cancer-Specific Reprogramming, Biomarkers, and Therapeutic Targets. Cancer Cell. 35:588–602.e10. 10.1016/j.ccell.2019.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassier P.A., Italiano A., Gomez-Roca C.A., Le Tourneau C., Toulmonde M., Cannarile M.A., Ries C., Brillouet A., Müller C., Jegg A.-M., et al. 2015. CSF1R inhibition with emactuzumab in locally advanced diffuse-type tenosynovial giant cell tumours of the soft tissue: a dose-escalation and dose-expansion phase 1 study. Lancet Oncol. 16:949–956. 10.1016/S1470-2045(15)00132-1 [DOI] [PubMed] [Google Scholar]
- Chevrier S., Levine J.H., Zanotelli V.R.T., Silina K., Schulz D., Bacac M., Ries C.H., Ailles L., Jewett M.A.S., Moch H., et al. 2017. An Immune Atlas of Clear Cell Renal Cell Carcinoma. Cell. 169:736–749.e18. 10.1016/j.cell.2017.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dankort D., Curley D.P., Cartlidge R.A., Nelson B., Karnezis A.N., Damsky W.E. Jr., You M.J., DePinho R.A., McMahon M., and Bosenberg M.. 2009. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat. Genet. 41:544–552. 10.1038/ng.356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos van Steenwijk P.J., Ramwadhdoebe T.H., Goedemans R., Doorduijn E.M., van Ham J.J., Gorter A., van Hall T., Kuijjer M.L., van Poelgeest M.I.E., van der Burg S.H., and Jordanova E.S.. 2013. Tumor-infiltrating CD14-positive myeloid cells and CD8-positive T-cells prolong survival in patients with cervical carcinoma. Int. J. Cancer. 133:2884–2894. 10.1002/ijc.28309 [DOI] [PubMed] [Google Scholar]
- Etzerodt A., and Moestrup S.K.. 2013. CD163 and inflammation: biological, diagnostic, and therapeutic aspects. Antioxid. Redox Signal. 18:2352–2363. 10.1089/ars.2012.4834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzerodt A., Maniecki M.B., Møller K., Møller H.J., and Moestrup S.K.. 2010. Tumor necrosis factor α-converting enzyme (TACE/ADAM17) mediates ectodomain shedding of the scavenger receptor CD163. J. Leukoc. Biol. 88:1201–1205. 10.1189/jlb.0410235 [DOI] [PubMed] [Google Scholar]
- Etzerodt A., Maniecki M.B., Graversen J.H., Møller H.J., Torchilin V.P., and Moestrup S.K.. 2012. Efficient intracellular drug-targeting of macrophages using stealth liposomes directed to the hemoglobin scavenger receptor CD163. J. Control. Release. 160:72–80. 10.1016/j.jconrel.2012.01.034 [DOI] [PubMed] [Google Scholar]
- Etzerodt A., Kjolby M., Nielsen M.J., Maniecki M., Svendsen P., and Moestrup S.K.. 2013. Plasma clearance of hemoglobin and haptoglobin in mice and effect of CD163 gene targeting disruption. Antioxid. Redox Signal. 18:2254–2263. 10.1089/ars.2012.4605 [DOI] [PubMed] [Google Scholar]
- Fritze A., Hens F., Kimpfler A., Schubert R., and Peschka-Süss R.. 2006. Remote loading of doxorubicin into liposomes driven by a transmembrane phosphate gradient. Biochim. Biophys. Acta. 1758:1633–1640. 10.1016/j.bbamem.2006.05.028 [DOI] [PubMed] [Google Scholar]
- Hugo W., Zaretsky J.M., Sun L., Song C., Moreno B.H., Hu-Lieskovan S., Berent-Maoz B., Pang J., Chmielowski B., Cherry G., et al. 2016. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell. 165:35–44. 10.1016/j.cell.2016.02.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ino Y., Yamazaki-Itoh R., Shimada K., Iwasaki M., Kosuge T., Kanai Y., and Hiraoka N.. 2013. Immune cell infiltration as an indicator of the immune microenvironment of pancreatic cancer. Br. J. Cancer. 108:914–923. 10.1038/bjc.2013.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen T.O., Schmidt H., Møller H.J., Høyer M., Maniecki M.B., Sjoegren P., Christensen I.J., and Steiniche T.. 2009. Macrophage markers in serum and tumor have prognostic impact in American Joint Committee on Cancer stage I/II melanoma. J. Clin. Oncol. 27:3330–3337. 10.1200/JCO.2008.19.9919 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Li Y., and Zhu B.. 2015. T-cell exhaustion in the tumor microenvironment. Cell Death Dis. 6:e1792–e1792. 10.1038/cddis.2015.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss M., Van Gassen S., Movahedi K., Saeys Y., and Laoui D.. 2018. Myeloid cell heterogeneity in cancer: not a single cell alike. Cell. Immunol. 330:188–201. 10.1016/j.cellimm.2018.02.008 [DOI] [PubMed] [Google Scholar]
- Komohara Y., Jinushi M., and Takeya M.. 2014. Clinical significance of macrophage heterogeneity in human malignant tumors. Cancer Sci. 105:1–8. 10.1111/cas.12314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansen M., Graversen J.H., Jacobsen C., Sonne O., Hoffman H.J., Law S.K., and Moestrup S.K.. 2001. Identification of the haemoglobin scavenger receptor. Nature. 409:198–201. 10.1038/35051594 [DOI] [PubMed] [Google Scholar]
- Larkin J., Chiarion-Sileni V., Gonzalez R., Grob J.J., Cowey C.L., Lao C.D., Schadendorf D., Dummer R., Smylie M., Rutkowski P., et al. 2015. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N. Engl. J. Med. 373:23–34. 10.1056/NEJMoa1504030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin Y., Kobayashi S., Leader A., Amir E.D., Elefant N., Bigenwald C., Remark R., Sweeney R., Becker C.D., Levine J.H., et al. 2017. Innate Immune Landscape in Early Lung Adenocarcinoma by Paired Single-Cell Analyses. Cell. 169:750–765.e17. 10.1016/j.cell.2017.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.J., Lee M.H., Kim H.T., Won C.H., Lee M.W., Choi J.H., and Chang S.E.. 2018. Prognostic significance of CD163 expression and its correlation with cyclooxygenase-2 and vascular endothelial growth factor expression in cutaneous melanoma. Melanoma Res.:1 10.1097/CMR.0000000000000549 [DOI] [PubMed] [Google Scholar]
- Luche H., Weber O., Nageswara Rao T., Blum C., and Fehling H.J.. 2007. Faithful activation of an extra-bright red fluorescent protein in “knock-in” Cre-reporter mice ideally suited for lineage tracing studies. Eur. J. Immunol. 37:43–53. 10.1002/eji.200636745 [DOI] [PubMed] [Google Scholar]
- Maniecki M.B., Etzerodt A., Ulhøi B.P., Steiniche T., Borre M., Dyrskjøt L., Orntoft T.F., Moestrup S.K., and Møller H.J.. 2012. Tumor-promoting macrophages induce the expression of the macrophage-specific receptor CD163 in malignant cells. Int. J. Cancer. 131:2320–2331. 10.1002/ijc.27506 [DOI] [PubMed] [Google Scholar]
- Mantovani A., and Allavena P.. 2015. The interaction of anticancer therapies with tumor-associated macrophages. J. Exp. Med. 212:435–445. 10.1084/jem.20150295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeth K., Wang J.X., Micevic G., Damsky W., and Bosenberg M.W.. 2016. The YUMM lines: a series of congenic mouse melanoma cell lines with defined genetic alterations. Pigment Cell Melanoma Res. 29:590–597. 10.1111/pcmr.12498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes S., Melandri D., Anselmi G., Perchet T., Loschko J., Dubrot J., Patel R., Gautier E.L., Hugues S., Longhi M.P., et al. 2016. The Heterogeneity of Ly6Chi Monocytes Controls Their Differentiation into iNOS+ Macrophages or Monocyte-Derived Dendritic Cells. Immunity. 45:1205–1218. 10.1016/j.immuni.2016.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michot J.M., Bigenwald C., Champiat S., Collins M., Carbonnel F., Postel-Vinay S., Berdelou A., Varga A., Bahleda R., Hollebecque A., et al. 2016. Immune-related adverse events with immune checkpoint blockade: a comprehensive review. Eur. J. Cancer. 54:139–148. 10.1016/j.ejca.2015.11.016 [DOI] [PubMed] [Google Scholar]
- Moran A.E., Holzapfel K.L., Xing Y., Cunningham N.R., Maltzman J.S., Punt J., and Hogquist K.A.. 2011. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J. Exp. Med. 208:1279–1289. 10.1084/jem.20110308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movahedi K., Laoui D., Gysemans C., Baeten M., Stangé G., Van den Bossche J., Mack M., Pipeleers D., In’t Veld P., De Baetselier P., and Van Ginderachter J.A.. 2010. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer Res. 70:5728–5739. 10.1158/0008-5472.CAN-09-4672 [DOI] [PubMed] [Google Scholar]
- Munn D.H., and Mellor A.L.. 2007. Indoleamine 2,3-dioxygenase and tumor-induced tolerance. J. Clin. Invest. 117:1147–1154. 10.1172/JCI31178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubert N.J., Schmittnaegel M., Bordry N., Nassiri S., Wald N., Martignier C., Tillé L., Homicsko K., Damsky W., Maby-El Hajjami H., et al. 2018. T cell-induced CSF1 promotes melanoma resistance to PD1 blockade. Sci. Transl. Med. 10:eaan3311 10.1126/scitranslmed.aan3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngoune R., Peters A., von Elverfeldt D., Winkler K., and Pütz G.. 2016. Accumulating nanoparticles by EPR: A route of no return. J. Control. Release. 238:58–70. 10.1016/j.jconrel.2016.07.028 [DOI] [PubMed] [Google Scholar]
- Noy R., and Pollard J.W.. 2014. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 41:49–61. 10.1016/j.immuni.2014.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardoll D.M. 2012. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer. 12:252–264. 10.1038/nrc3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry C.J., Muñoz-Rojas A.R., Meeth K.M., Kellman L.N., Amezquita R.A., Thakral D., Du V.Y., Wang J.X., Damsky W., Kuhlmann A.L., et al. 2018. Myeloid-targeted immunotherapies act in synergy to induce inflammation and antitumor immunity. J. Exp. Med. 215:877–893. 10.1084/jem.20171435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries C.H., Cannarile M.A., Hoves S., Benz J., Wartha K., Runza V., Rey-Giraud F., Pradel L.P., Feuerhake F., Klaman I., et al. 2014. Targeting tumor-associated macrophages with anti-CSF-1R antibody reveals a strategy for cancer therapy. Cancer Cell. 25:846–859. 10.1016/j.ccr.2014.05.016 [DOI] [PubMed] [Google Scholar]
- Robert C., Schachter J., Long G.V., Arance A., Grob J.J., Mortier L., Daud A., Carlino M.S., McNeil C., Lotem M., et al. KEYNOTE-006 investigators . 2015. Pembrolizumab versus Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 372:2521–2532. 10.1056/NEJMoa1503093 [DOI] [PubMed] [Google Scholar]
- Rodriguez P.C., Quiceno D.G., Zabaleta J., Ortiz B., Zea A.H., Piazuelo M.B., Delgado A., Correa P., Brayer J., Sotomayor E.M., et al. 2004. Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res. 64:5839–5849. 10.1158/0008-5472.CAN-04-0465 [DOI] [PubMed] [Google Scholar]
- Ruffell B., Chang-Strachan D., Chan V., Rosenbusch A., Ho C.M.T., Pryer N., Daniel D., Hwang E.S., Rugo H.S., and Coussens L.M.. 2014. Macrophage IL-10 blocks CD8+ T cell-dependent responses to chemotherapy by suppressing IL-12 expression in intratumoral dendritic cells. Cancer Cell. 26:623–637. 10.1016/j.ccell.2014.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber H.A., Loschko J., Karssemeijer R.A., Escolano A., Meredith M.M., Mucida D., Guermonprez P., and Nussenzweig M.C.. 2013. Intestinal monocytes and macrophages are required for T cell polarization in response to Citrobacter rodentium. J. Exp. Med. 210:2025–2039. 10.1084/jem.20130903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spranger S., Dai D., Horton B., and Gajewski T.F.. 2017. Tumor-Residing Batf3 Dendritic Cells Are Required for Effector T Cell Trafficking and Adoptive T Cell Therapy. Cancer Cell. 31:711–723.e4. 10.1016/j.ccell.2017.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchilin V. 2011. Tumor delivery of macromolecular drugs based on the EPR effect. Adv. Drug Deliv. Rev. 63:131–135. 10.1016/j.addr.2010.03.011 [DOI] [PubMed] [Google Scholar]
- Torchilin V.P., Levchenko T.S., Lukyanov A.N., Khaw B.A., Klibanov A.L., Rammohan R., Samokhin G.P., and Whiteman K.R.. 2001. p-Nitrophenylcarbonyl-PEG-PE-liposomes: fast and simple attachment of specific ligands, including monoclonal antibodies, to distal ends of PEG chains via p-nitrophenylcarbonyl groups. Biochim. Biophys. Acta. 1511:397–411. 10.1016/S0005-2728(01)00165-7 [DOI] [PubMed] [Google Scholar]
- Tsou C.-L., Peters W., Si Y., Slaymaker S., Aslanian A.M., Weisberg S.P., Mack M., and Charo I.F.. 2007. Critical roles for CCR2 and MCP-3 in monocyte mobilization from bone marrow and recruitment to inflammatory sites. J. Clin. Invest. 117:902–909. 10.1172/JCI29919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumeh P.C., Harview C.L., Yearley J.H., Shintaku I.P., Taylor E.J.M., Robert L., Chmielowski B., Spasic M., Henry G., Ciobanu V., et al. 2014. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 515:568–571. 10.1038/nature13954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugurel S., Röhmel J., Ascierto P.A., Flaherty K.T., Grob J.J., Hauschild A., Larkin J., Long G.V., Lorigan P., McArthur G.A., et al. 2017. Survival of patients with advanced metastatic melanoma: the impact of novel therapies-update 2017. Eur. J. Cancer. 83:247–257. 10.1016/j.ejca.2017.06.028 [DOI] [PubMed] [Google Scholar]
- Wang J., Perry C.J., Meeth K., Thakral D., Damsky W., Micevic G., Kaech S., Blenman K., and Bosenberg M.. 2017. UV-induced somatic mutations elicit a functional T cell response in the YUMMER1.7 mouse melanoma model. Pigment Cell Melanoma Res. 30:428–435. 10.1111/pcmr.12591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn T.A., Chawla A., and Pollard J.W.. 2013. Macrophage biology in development, homeostasis and disease. Nature. 496:445–455. 10.1038/nature12034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., and Zhang Y.. 2017. Tumor-associated macrophages, potential targets for cancer treatment. Biomark. Res. 5:25 10.1186/s40364-017-0106-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.