Enterobacteriaceae family members such as E. coli exacerbate development of intestinal malignancy. Zhu et al. report that targeting the metabolism of protumoral Enterobacteriaceae by tungstate prevents tumor development in murine models of colitis-associated colorectal cancer.
Abstract
Chronic inflammation and gut microbiota dysbiosis, in particular the bloom of genotoxin-producing E. coli strains, are risk factors for the development of colorectal cancer. Here, we sought to determine whether precision editing of gut microbiota metabolism and composition could decrease the risk for tumor development in mouse models of colitis-associated colorectal cancer (CAC). Expansion of experimentally introduced E. coli strains in the azoxymethane/dextran sulfate sodium colitis model was driven by molybdoenzyme-dependent metabolic pathways. Oral administration of sodium tungstate inhibited E. coli molybdoenzymes and selectively decreased gut colonization with genotoxin-producing E. coli and other Enterobacteriaceae. Restricting the bloom of Enterobacteriaceae decreased intestinal inflammation and reduced the incidence of colonic tumors in two models of CAC, the azoxymethane/dextran sulfate sodium colitis model and azoxymethane-treated, Il10-deficient mice. We conclude that metabolic targeting of protumoral Enterobacteriaceae during chronic inflammation is a suitable strategy to prevent the development of malignancies arising from gut microbiota dysbiosis.
Introduction
Colorectal cancer (CRC) is the third leading cause of cancer-related deaths worldwide (Arnold et al., 2017). Multiple risk factors influence the development of CRC, including age, genetics, lifestyle, diet, and environmental factors (Sears and Garrett, 2014; Brennan and Garrett, 2016). Unlike many other types of cancers that develop in a relatively sterile environment, CRC occurs at the largest interface between host and gut microbial communities and is often associated with an inflammatory response. Chronic inflammatory diseases of the intestinal tract, such as inflammatory bowel disease (IBD), increase the risk for the development of CRC, an association that has been termed colitis-associated CRC (CAC; Grivennikov, 2013). Inflammation enhances the development of CAC by various mechanisms, including the development of a genotoxic environment, as well as expression of proinflammatory cytokines and prostaglandins that promote cellular survival, angiogenesis, and other aspects of the neoplastic process (Brennan and Garrett, 2016; Lasry et al., 2016).
Several studies have reported an association between CRC and changes of the overall community structure of the gut microbiota (Wang et al., 2012a; Wu et al., 2013; Feng et al., 2015). Furthermore, distinct commensal bacteria, such as Fusobacterium spp., can be found in the microenvironment of tumors, and have been shown in experimental models to promote tumor development in genetically predisposed animals (Chen et al., 2012; Kostic et al., 2012; Tjalsma et al., 2012; Sears and Garrett, 2014; Viljoen et al., 2015). These observations have generated interest in investigating how gut microbes are involved in CRC development.
The carcinogenic effects of commensal gut bacteria can involve direct effects of bacteria-derived products. An example of this mechanism involves a small number of bacteria that have been shown to secrete carcinogenic compounds, such as colibactin, a polyketide-derived genotoxin produced by Enterobacteriaceae (Putze et al., 2009; Buc et al., 2013; Prorok-Hamon et al., 2014; Raisch et al., 2014; Wilson et al., 2019). While Enterobacteriaceae are considered to be a part of the “core microbiota” shared by the majority of humans (Mitsuoka and Hayakawa, 1973; Penders et al., 2006; Lozupone et al., 2012), specific members of the Enterobacteriaceae family, in particular Escherichia coli (phylogroups B2 and D), Klebsiella spp., and Proteus mirabilis are frequently detected and overrepresented in the microbiota of CRC patients (Darfeuille-Michaud et al., 1998; Swidsinski et al., 1998; Martin et al., 2004). Exposure of mammalian cells to colibactin induces interstrand DNA cross-linking (Vizcaino and Crawford, 2015; Bossuet-Greif et al., 2018; Wilson et al., 2019) and triggers the ataxia telangiectasia mutated signaling pathway (Nougayrède et al., 2006; Maddocks et al., 2013; Cougnoux et al., 2014). Activation of this damage response results in G2-M cell cycle arrest (Grasso and Frisan, 2015). Expression of colibactin biosynthesis genes is highly induced in biopsies from human CRC patients (Dutilh et al., 2013). Consistent with a role of colibactin in tumor induction, experimental administration of a colibactin-producing E. coli strain favors tumor development in an animal model of inflammation-associated CRC, while E. coli mutants deficient for producing colibactin fail to do so (Arthur et al., 2012; Raisch et al., 2015).
In addition to the release of genotoxin, the gut microbiota can enhance CRC development through its role on tumor-associated inflammation, with cytokines and other inflammatory products magnifying the neoplastic process. Bacteria are known to penetrate tumor tissues and directly enhance inflammation (Dejea et al., 2018). Furthermore, intestinal inflammation promotes dysbiotic changes in the microbiota composition. In particular, Enterobacteriaceae populations expand during bouts of inflammation (Lupp et al., 2007; Garrett et al., 2010). The outgrowth of these pathobionts in turn exacerbates intestinal inflammation, epithelial cell damage, and wound repair responses (Carvalho et al., 2012; Gronbach et al., 2014; Kang and Martin, 2017; Pickard et al., 2017), thus amplifying known risk factors of CAC development.
E. coli and other members of the Enterobacteriaceae family are among the most overrepresented bacteria in IBD and CAC (Swidsinski et al., 1998; Martin et al., 2004; Mukhopadhya et al., 2012). Currently there are few experimental approaches that attempt to restrict colonization with genotoxin-producing Enterobacteriaceae members. Oral antimicrobial therapy with broad-spectrum antibiotics is suitable to temporarily reduce the abundance of certain protumoral bacterial species. However, broad-spectrum antimicrobial therapy also removes beneficial microbes and increases the risk of pathogen expansion (Stecher et al., 2007; Collins et al., 2009; Faber et al., 2016).
To overcome the limitations that broad-spectrum antibiotics have in this setting, we recently developed a strategy that targets the unique metabolic capabilities of Enterobacteriaceae (Winter et al., 2013; Hughes et al., 2017). During episodes of inflammation, Enterobacteriaceae rely on nitrate respiration and formate oxidation, while these metabolic pathways are inactive during homeostatic conditions (Winter et al., 2013; Hughes et al., 2017). E. coli expresses three nitrate reductases and two respiratory formate dehydrogenases to facilitate this disease-associated metabolism (Unden and Dünnwald, 2008). All of these enzymes are metalloenzymes with a molybdenum cofactor (MoCo) in their active site (Enoch and Lester, 1972). In the presence of excess amounts of tungsten ions (W), tungsten is incorporated into the molybdopterin cofactor (Nichols and Rajagopalan, 2005). This tungsten-substituted cofactor is inactive in Enterobacteriaceae and thus MoCo-dependent enzymes are selectively inhibited by tungsten in vitro (Enoch and Lester, 1972; Gates et al., 2003). Administration of tungstate, a water-soluble tungsten salt, ameliorates dysbiosis and intestinal inflammation in acute colitis models (Zhu et al., 2018). Here, we investigated whether administration of tungstate could reduce E. coli–driven tumorigenesis using mouse models of chronic inflammation and CAC.
Results
Tungstate treatment blunts expansion of commensal E. coli in a mouse model of chronic inflammation
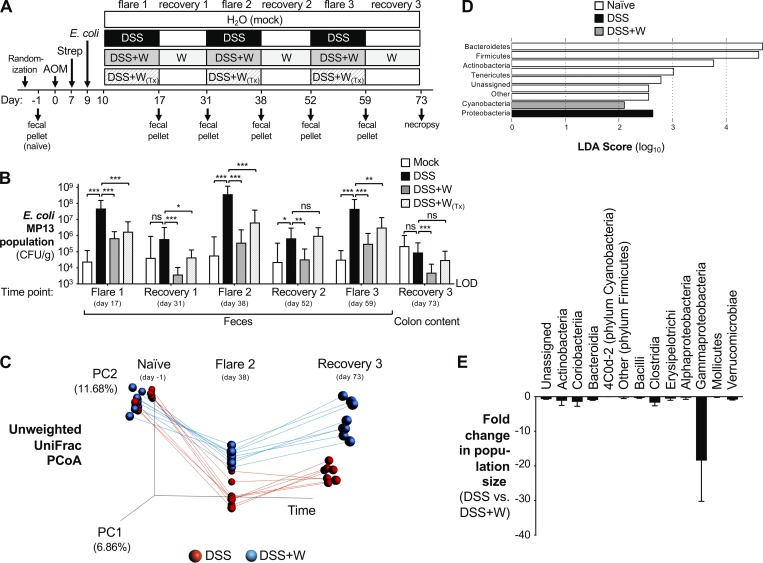
To investigate the role of commensal Enterobacteriaceae in the development of CAC, we experimentally introduced the E. coli strain MP13 into the azoxymethane (AOM)/dextran sulfate sodium (DSS) model of CAC (Fig. 1 A). MP13 is a tetracycline-resistant derivative of the murine commensal E. coli strain MP1 (Lasaro et al., 2014) and was used to facilitate culture-dependent quantification in the intestinal content and the feces. The MP1 genome does not encode the genes required for biosynthesis of colibactin (Lasaro et al., 2014). C57BL/6 mice were treated with a one-time i.p. injection of AOM (20 mg/kg). No Enterobacteriaceae family members were detected in the feces of these animals as determined by growth on MacConkey agar plates. 1 wk later, mice received oral streptomycin treatment to facilitate engraftment of E. coli MP13, and after 48 h, mice were intragastrically inoculated with E. coli. Mice were then treated with 2% DSS in the drinking water for 1 wk or mock treated with water, followed by a recovery phase of 2 wk on regular drinking water. The injury was repeated a total of three times. Fecal samples were obtained after DSS-induced colitis episodes (“flares”) and at the completion of the recovery period. Animals were euthanized after the third recovery phase, and the bacterial load of MP13 in the feces was determined (Fig. 1 B). E. coli MP13 colonized mock-treated mice at a consistently low level throughout the time course. In contrast, induction of gut inflammation by DSS resulted in a pronounced expansion of the E. coli MP13 population, reaching more than three orders of magnitude compared with mock-treated mice during each flare. This population contracted during recovery phases and expanded upon repeated induction of inflammation (Fig. 1 B).
Figure 1.
Impact of tungstate treatment on gut microbial communities during chronic inflammation. Groups of C57BL/6 mice were treated i.p. with 20 mg/kg AOM. After 7 d, animals received streptomycin (Strep; 2 mg/ml in the drinking water) and were intragastrically inoculated with E. coli MP13 2 d later. Mice were treated for 1 wk with 2% DSS (n = 15), 2% DSS supplemented with 0.2% sodium tungstate (W, n = 18), or filtered drinking water (mock, n = 5). Animals were allowed to recover for 2 wk, with the tungsten-treated group receiving 0.2% sodium tungstate throughout the experiment. Injury was repeated two more times. One group of animals received tungstate exclusively during DSS treatment (W(Tx), n = 9). Another group received DSS but was not colonized by MP13 (n = 6). At predetermined time points, feces were collected, and tissues were harvested 73 d after AOM treatment. Two independent experiments were performed and are shown in B; C–E show results from one of these experiments. (A) Schematic representation of the experimental design. (B) Burden of E. coli MP13 in the feces and colon content at the indicated time points determined by plating on selective media. LOD, limit of detection. Bars represent the geometric mean ± 95% confidence interval. P values were calculated by two-way ANOVA and a post hoc Tukey’s multiple comparison test on log-transformed data. (C) Principal coordinate analysis (PCoA) of the gut microbiota composition, as determined by 16S rDNA amplicon sequencing. DSS-treated, red circles; DSS+W–treated, blue circles. Data from each mouse are linked by a colored line. (D) Bacterial phyla that were significantly enriched in indicated groups identified by linear discriminant analysis (LDA) effect size measurements. (E) Comparison of gut microbiota composition at the class level in DSS-treated and DSS+W–treated mice. Bars represent the mean ± 95% confidence interval. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not statistically significant.
Since E. coli and other Enterobacteriaceae rely on MoCo-dependent metabolic pathways, such as nitrate respiration, to bloom during intestinal inflammation (Winter et al., 2013; Hughes et al., 2017), we examined whether these pathways played a role in E. coli population expansion in the AOM/DSS colitis model. We inoculated mice with an equal ratio of the E. coli (Nissle 1917) wild-type strain and an isogenic mutant deficient in MoCo biosynthesis (moaA mutant) as indicators of microbial metabolism (Fig. S1 A), a strategy we had validated in previous reports (Winter et al., 2013; Hughes et al., 2017). In the absence of inflammation, the wild-type strain and the moaA mutant were recovered at comparable levels on day 17 (Fig. S1 B). In contrast, the wild-type strain outcompeted the moaA mutant in DSS-treated mice. Similar results were obtained for a mutant deficient for all three nitrate reductase enzymes (Fig. S1 B). These findings provide support for the idea that MoCo-dependent processes are dispensable under homeostatic conditions, but are critical for E. coli fitness during episodes of gut inflammation.
Enterobacterial molybdoenzymes can be inhibited by soluble tungsten salts (Zhu et al., 2018). Consistent with a role for these metabolic pathways in E. coli expansion, administration of 0.2% sodium tungstate in the drinking water blunted the expansion of E. coli MP13 in all flare and recovery phases throughout the 73-d time course (Fig. 1 B). Moreover, the Nissle 1917 wild-type strain and the moaA mutant were recovered at similar levels on day 17 when mice were concurrently treated with DSS and 0.2% sodium tungstate in the drinking water (Fig. S1 B), suggesting that our experimental administration of tungstate was sufficient to inhibit bacterial molybdoenzymes in the intestinal tract. No overt negative effects of long-term tungstate treatment on the murine host were noted. We had previously shown that mice exposed to 0.2% tungstate in the drinking water do not exhibit liver damage (Zhu et al., 2018). Oral administration of 100 mg sodium tungstate twice daily for the duration of 8 wk as part of a clinical trial did not reveal any overt signs of toxicity in humans (Hanzu et al., 2010).
Tungstate treatment specifically targets Gammaproteobacteria in the dysbiotic gut microbiota
We next sought to explore the impact of long-term oral tungstate treatment on gut microbial communities. We extracted DNA from feces collected before AOM treatment (naive) and at the end of the second flare (day 38) and from colon content at the end of the experiment (day 73), and determined the composition of the bacterial microbiota by 16S rDNA amplicon sequencing (Fig. 1, C–E; and Fig. S1, C–H). Communities in both treatment groups were very similar at the onset of the experiment, as indicated by principal coordinate analysis (Fig. 1 C). Beta diversity analysis revealed that repeated DSS treatment irreversibly shifted the microbiota structure toward a dysbiotic state (Figs. 1 C and S1 C). Population analysis of similarity (ANOSIM) showed significant differences of communities in DSS-treated mice between the naive and in-flare time points (R = 0.89, P = 0.002) and naive and recovery time points (R = 0.90, P = 0.001; Fig. 1 C). This finding was further supported by UniFrac distance analysis (Fig. S1 C). Linear discriminative analysis effect size of communities revealed a marked overrepresentation of Proteobacteria in DSS-treated mice (Fig. 1 D). When mice were treated simultaneously with DSS and tungstate, microbial dysbiosis was significantly reduced compared with DSS-only treatment (DSS+W treatment naive vs. flare, ANOSIM R = 0.69, P = 0.001; Figs. 1 C and S1 C). Similarly, tungstate treatment reduced the severity of dysbiosis during the recovery phase (DSS+W treatment naive vs. recovery; ANOSIM R = 0.53, P = 0.001; Figs. 1 C and S1 C). Oral tungstate administration blunted the expansion of facultative anaerobic Gammaproteobacteria during chronic inflammation and led to a limited compensatory increase in the relative abundance of facultative anaerobic Bacilli (Figs. 1 E and S1, D–H). We had previously shown that oral administration of 0.2% tungstate does not alter the composition of the gut microbiota in the absence of inflammation (Zhu et al., 2018). Collectively, these data demonstrate that the inflammation-associated bloom of Gammaproteobacteria in the AOM/DSS colitis model can be selectively inhibited by oral administration of tungstate.
Oral administration of sodium tungstate treatment reduces intestinal inflammation and tumorigenesis
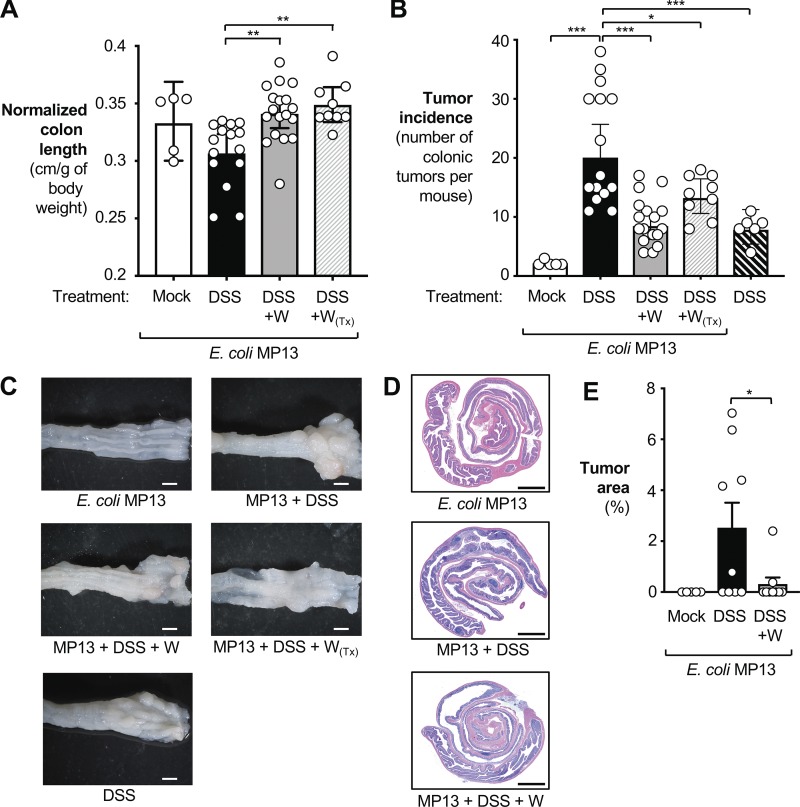
We reasoned that by controlling repeated blooms of Enterobacteriaceae, tungstate treatment could improve chronic intestinal inflammation and thus prevent tumor formation. We therefore analyzed normalized colon length, mRNA levels of a panel of proinflammatory markers in the distal colon, animal body weight, and pathological changes in the colonic tissue (Figs. 2 A and S2, A–D). DSS-treated mice, precolonized with MP13, exhibited shorter normalized colon length and increased mRNA levels of proinflammatory markers in the colon tissue compared with mock-treated, MP13-colonized mice (Figs. 2 A and S2, A–D). Mice that were only treated with AOM and E. coli MP13 exhibited a small number of tumors, while induction of colitis with DSS resulted in significantly increased tumor incidence (Fig. 2, B and C). When we treated mice that were free of any detectable Enterobacteriaceae family members with AOM and DSS alone, the number of tumors was significantly lower compared with E. coli MP13-colonized mice treated with AOM/DSS (Fig. 2, B and C). Since the E. coli MP13 genome does not encode genes required for colibactin biosynthesis, this finding suggests that commensal E. coli strains could aggravate tumorigenesis through colibactin-independent processes.
Figure 2.
Analysis of tumorigenesis in the AOM/DSS colitis model. Tissue was obtained from the animals described in Fig. 1 (73 d after AOM treatment). Animals were initially treated with AOM and colonized with the E. coli MP13 wild-type strain, as indicated. Groups received filtered drinking water (mock; white bars, n = 5), 2% DSS (black bars, n = 15), or 2% DSS + 0.2% sodium tungstate (gray bars, n = 18). Subsets of the animals received tungstate exclusively during DSS-induced flares (W(Tx), n = 9; gray dash pattern) or received DSS but were not colonized by MP13 (black dash pattern, n = 6). Data from two independent experiments are shown. (A) Colon length was normalized to the total body weight of each animal. White dots represent data from individual animals. (B) Tumor incidence in the large intestine. (C) Gross pathology of the distal colon. Scale bars represent 1 mm. (D) Representative images of H&E-stained colonic sections. Scale bars represent 250 µm. (E) Tumor area as determined by a morphometric analysis. White dots represent data from individual animals. Bars represent the geometric mean ± 95% confidence interval. P values were calculated by unpaired, two-tailed Student’s t test on log-transformed data. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Oral administration with sodium tungstate during flares and recovery phases consistently reduced markers of intestinal inflammation in the colon tissue (Figs. 2 A and S2, A–D), although in some cases these trends did not reach statistical significance. Importantly, coadministration of AOM/DSS and tungstate to MP13-colonized mice significantly reduced tumor incidence (Fig. 2, B and C) and tumor area (Fig. 2, D and E).
In the previous set of experiments, tungstate was administered throughout all flare and recovery phases. To better mimic a potential treatment regimen in patients with reoccurring flare-ups, we administered 0.2% sodium tungstate only during the DSS-induced flares (W(Tx); Fig. 1 A). Therapeutic administration of tungstate was effective in blunting expansion of MP13 (Fig. 1 B), in improving colon length shortening (Fig. 2 A), and in reducing the tumor incidence in the large intestine (Fig. 2 B).
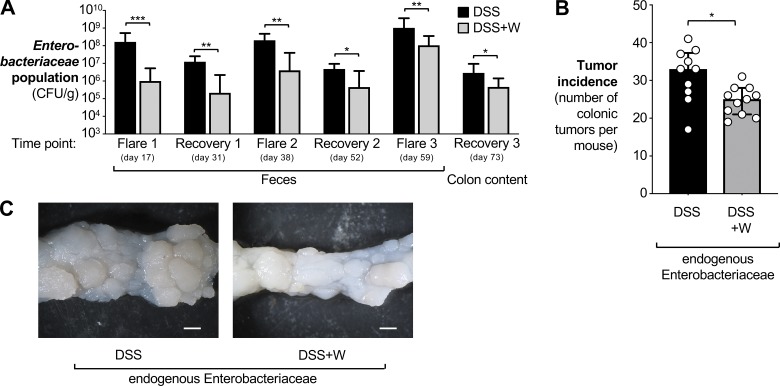
We also considered whether tungstate could prevent tumorigenesis in mice with a native microbiota (Fig. 3 and Fig. S2 E). We thus purchased C57BL/6 mice from a commercial vendor (Charles River Laboratories). These animals naturally harbor Enterobacteriaceae family members (Hughes et al., 2017). Groups of AOM-treated animals received either DSS or DSS + 0.2% sodium tungstate (Fig. 1 A). Colonization by Enterobacteriaceae family members was quantified by plating fecal material and colon content (day 73) on MacConkey agar plates (Fig. 3 A). Akin to our previous findings, tungstate treatment reduced the overall burden of Enterobacteriaceae family members (Fig. 3 A) and tumor incidence in the large intestine (Fig. 3, B and C).
Figure 3.
Contribution of the native gut microbiota to tumorigenesis in the AOM/DSS colitis model. Mice harboring endogenous Enterobacteriaceae were treated with AOM and DSS as described in Fig. 1, except that the antibiotic treatment and colonization with E. coli was omitted. Tissue was obtained 73 d after AOM treatment. Groups received either 2% DSS (n = 10, black bars) or 2% DSS + 0.2% sodium tungstate (n = 11, gray bars). Data from one experiment are shown. (A) The burden of Enterobacteriaceae in the feces and in the colon content at the indicated time points was determined by plating on MacConkey agar plates. Bars represent the geometric mean ± 95% confidence interval. P values were calculated by two-way ANOVA and a post hoc Tukey’s multiple comparison test on log-transformed data. (B) Tumor incidence in the colon. Bars represent the median ± interquartile range. P values were calculated by two-tailed Mann–Whitney U test. (C) Representative images of the distal colon. Scale bars represent 1 mm. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Taken together, these experiments demonstrate that manipulation of the gut microbiota composition by sodium tungstate reduces gut microbiota dysbiosis, dysbiosis-associated inflammation, and tumorigenesis in the AOM/DSS colitis model.
Tungstate reduces colibactin-driven tumorigenesis
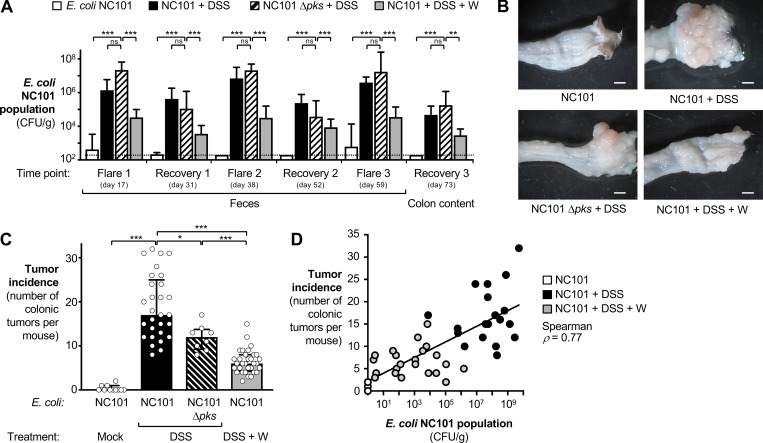
Colibactin-producing E. coli strains potentiate tumorigenesis in mouse models of CAC (Arthur et al., 2012). We investigated whether tungstate treatment would decrease colonization by a murine-adapted, colibactin-producing E. coli (NC101) and thus decrease colibactin-driven CAC. To this end, we colonized groups of AOM-treated C57BL/6 mice with the E. coli NC101 wild-type strain or an isogenic mutant deficient for biosynthesis of colibactin (Δpks mutant), respectively (Fig. 1 A). To assist with quantification in fecal material, both strains carried the low-copy number plasmid pWSK29 encoding a β-lactamase. Animals were treated with DSS or DSS + 0.2% sodium tungstate as described above (Fig. 1 A). The NC101 wild-type strain and the Δpks mutant were recovered from the feces at similar levels throughout the time course, with populations expanding during inflammatory flares and decreasing during recovery (Fig. 4 A). Tumor incidence, determined on day 73, was infrequent in mice that were inoculated with NC101 and received only regular drinking water (Fig. 4, B and C). Repeated induction of colitis (AOM/DSS colitis model) of mice colonized with the NC101 wild-type strain resulted in elevated tumor incidence. In the NC101-colonized as well as the MP1-colonized mice (AOM/DSS colitis model), multifocal neoplastic proliferations in the mucosa, expansive, noninvasive, and nonencapsulated lesions were noted. Neoplastic cells were arranged predominantly in tubules, supported by a moderate amount of lamina propria–like fibrovascular tissue and composed of columnar epithelial cells with well-aggregated pseudostratified appearance. There was moderate cellular anisocytosis and nuclear pleomorphism, with an average of one to two mitotic figures per high-magnification field. Moderate focally extensive inflammation in the stromal tissue, composed of neutrophils and histiocytes, was observed, and most of the epithelium that covered the proliferation was ulcerated with mild neutrophilic infiltration. In some instances, focally extensive areas of enterocyte loss and necrosis, with abundant neutrophilic inflammatory infiltrate and exudation, with debris and regenerative glandular epithelium in adjacent mucosa were noted (Table S1 and Fig. 2 D).
Figure 4.
Effect of oral sodium tungstate administration on genotoxin-producing E. coli populations and tumorigenesis in the AOM/DSS colitis model. Groups of C57BL/6 mice were treated i.p. with AOM. After 7 d, streptomycin-pretreated animals were intragastrically inoculated with the E. coli NC101 wild-type or an isogenic Δpks mutant (n = 8). Groups were then treated for 1 wk with 2% DSS (n = 29), 2% DSS supplemented with 0.2% sodium tungstate (W, n = 32), or filtered drinking water (mock, n = 10) as indicated. Animals were allowed to recover for 2 wk, with the tungsten-treated group receiving 0.2% sodium tungstate throughout the experiment. Injury was repeated two more times. At predetermined time points, feces were collected, and tissues were harvested 73 d after AOM treatment. See Fig. 1 A for a schematic representation. Data from five independent experiments are shown. (A) Burden of NC101 in the feces and the colon content at various time points determined by a culture-dependent method. The dotted line indicates the limit of detection (LOD). Bars represent the geometric mean ± 95% confidence interval. P values were calculated by two-way ANOVA and a post-hoc Tukey’s multiple comparison test on log-transformed data. ns, not significant. (B) Representative images of gross pathology of the distal large intestine. Scale bars represent 1 mm. (C) Tumor incidence in the large intestine. Each dot represents of one animal. Bars indicate the median and the interquartile range. P values were calculated by two-tailed Mann–Whitney U test. (D) Correlation between the NC101 population size (mean of three flares) and the tumor incidence at the end of the experiment. Each dot represents one animal; the color of the dot indicates the treatment. The Spearman’s rank correlation coefficient is indicated, and a linear trendline is shown (P < 0.001). *, P < 0.05; **, P < 0.01, ***, P < 0.001; ns, not statistically significant.
Consistent with a previous report (Arthur et al., 2012), mice colonized with the Δpks mutant exhibited significantly decreased tumor incidence compared with mice colonized with the wild-type strain (P < 0.05; 29% decrease). Tungstate treatment significantly decreased NC101 intestinal burden (Fig. 4 A). Importantly, administration of tungstate significantly lowered tumor incidence in mice colonized with the E. coli NC101 wild-type strain in the AOM/DSS colitis model by ∼64% (P < 0.001; Fig. 4, B and C). NC101 populations in the gut lumen directly correlated with tumor incidence (Fig. 4 D). Notably, the effect size of tungstate treatment was more pronounced (64% reduction) than the effect mediated by genetic ablation of colibactin production (29% reduction), suggesting that tungstate decreases tumor formation by both ameliorating inflammation and reducing the burden of genotoxin-producing bacteria. Tungstate treatment had no discernable effect on tumor size and histological features (Table S1).
Tungstate treatment decreases DNA damage and intestinal inflammation in a model of colibactin-driven CAC
A subset of colibactin-producing E. coli strains, so-called adherent invasive E. coli strains (AIECs), have been isolated from areas of inflamed intestinal tissue in patients with Crohn’s disease (Darfeuille-Michaud et al., 1998). These AIEC strains colonize the mucosa by adhering to the intestinal epithelium (Darfeuille-Michaud et al., 1998), where local release of colibactin can induce DNA damage in host cells (Arthur et al., 2012; Wilson et al., 2019). Furthermore, colibactin production is coregulated with siderophore biosynthesis under iron-limiting conditions, such as intestinal inflammation (Martin et al., 2013; Ganz and Nemeth, 2015; Tronnet et al., 2016).
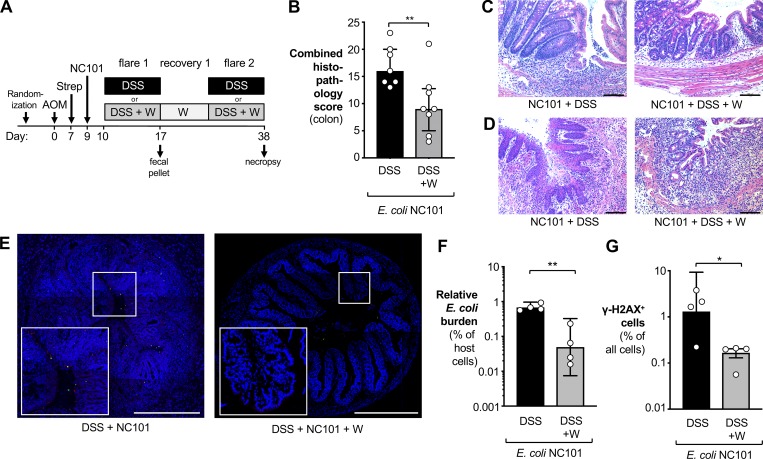
We next investigated whether tungstate treatment would deplete colibactin-producing E. coli strain from the mucosal interface, thus reducing inflammation and DNA damage. Animals were colonized with NC101 and treated with AOM and DSS as described above, and samples were analyzed 38 d after AOM treatment (Fig. 5 A). Mirroring our initial findings with MP13 (Figs. 1 and 2), tungstate treatment of NC101-colonized animals improved intestinal inflammation, as determined by histopathology, mRNA levels of inflammatory marker genes, and colon length (Fig. 5, B–D; and Fig. S3). Tungstate had no discernable effect on animal body weight (Fig. S3 D).
Figure 5.
Analysis of DNA damage and intestinal inflammation. (A) Groups of C57BL/6 mice were treated i.p. with 20 mg/kg AOM. After 7 d, animals received streptomycin (2 mg/ml in the drinking water) and were intragastrically inoculated with the colibactin-producing E. coli NC101 2 d later. Mice were treated for 1 wk with either 2% DSS (black bars) or 2% DSS supplemented with 0.2% sodium tungstate (W; gray bars). Animals were allowed to recover for 2 wk, with the tungsten-treated group receiving 0.2% sodium tungstate throughout the experiment. After 38 d, samples of the colon were obtained. Data from two independent experiments are shown. (B–D) H&E-stained sections were scored by a veterinary pathologist for epithelial damage, polymorphonuclear neutrophil infiltration, submucosal edema, and exudate in the lumen. DSS group: n = 7; DSS+W group: n = 8. (B) Combined histopathology score. Bars indicate the median and the interquartile range. P values were calculated by two-tailed Mann–Whitney U test. (C and D) Representative images of H&E-stained sections of the proximal (C) and distal (D) colon. Scale bars represent 100 µm. (E–G) Transverse sections of colonic tissues were stained for E. coli (red), DNA damage using γ-H2AX foci as marker (green), and cell nuclei (blue). Individual image tiles were assembled to quantify E. coli burden and number of γ-H2AX–positive cells and total mammalian cells. Both groups: n = 4. (E) Representative images. Scale bars represent 500 µm. (F) Burden of E. coli normalized to the number of mammalian cells. (G) Abundance of γ-H2AX–positive cells normalized to the number of all mammalian cells. For F and G, P values were calculated by unpaired, two-tailed Student’s t test on log-transformed data. Bars represent the geometric mean ± 95% confidence interval. *, P < 0.05; **, P < 0.01.
We visualized E. coli via fluorescence in situ hybridization, and colibactin-induced DNA damage was assessed through immunofluorescence staining of phospho-γ-H2AX (Sharma et al., 2012). Transversal sections of colonic tissue were imaged in independent tiles and reassembled as described in Earle et al. (2015) (Fig. 5, E–G). We observed E. coli NC101 bacteria in proximity to the mucosa and notable cellular DNA damage in the gut epithelial tissue in DSS-treated mice. E. coli numbers and DNA damage were reduced in mice receiving both DSS and tungstate (Fig. 5, E–G). Similar observations were made at the 73-d time point (Fig. S4, A–C).
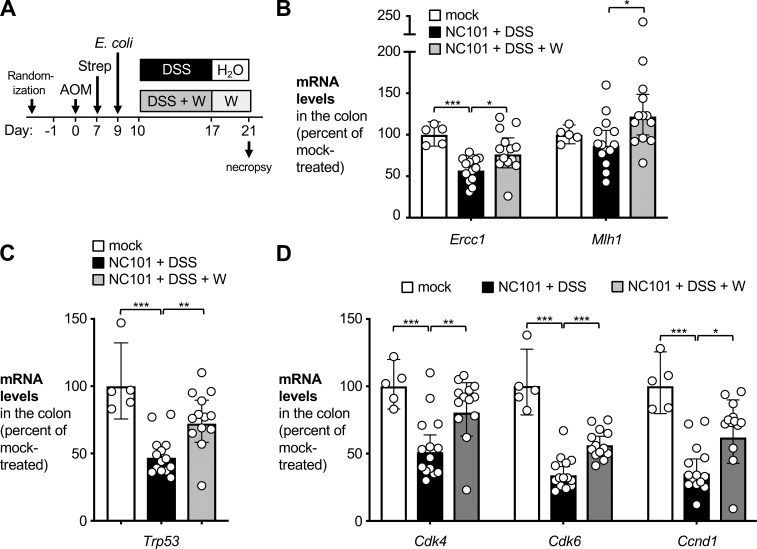
Furthermore, we determined the effect of tungsten on DNA damage response and cell cycle progression ex vivo in intestinal epithelial cells (Fig. 6). Animals were treated as described above (Fig. 6 A), colonocytes were isolated after 21 d, and relative transcription of a panel of genes was determined by quantitative RT-PCR (Fig. 6, B–D). In the AOM/DSS model, transcription of the tumor suppressor p53 (encoded by Trp53) and genes involved in DNA damage response is down-regulated (Zheng et al., 2016; Sharp et al., 2018). Consistent with these previous reports, Trp53, Ercc1, and Mlh1 mRNA levels were decreased in AOM/DSS-treated mice that had been colonized with NC101 animals (Fig. 6, B and C). Importantly, transcription of Ercc1, Mlh1, and Trp53 was rescued in tungsten-treated animals. Similarly, tungstate rescued transcription of genes involved in cell cycle progression, such as Cdk4, Cdk6, and Ccnd1 (Fig. 6 D). Collectively, these data suggest that tungsten impacts tumorigenesis through at least two mechanisms, i.e., by decreasing mucosal inflammation and by reducing DNA damage at early time points.
Figure 6.
Impact of oral sodium tungstate treatment on cell cycle progression and DNA damage response in the AOM/DSS model. Groups of C57BL/6 mice were treated i.p. with 20 mg/kg AOM. After 7 d, animals received streptomycin (2 mg/ml in the drinking water) and were intragastrically inoculated with the colibactin-producing E. coli NC101 2 d later. Mice were treated for 1 wk with 2% DSS (n = 14), 2% DSS supplemented with 0.2% sodium tungstate (DSS+W, n = 13), or filtered drinking water (mock, n = 5). Animals were allowed to recover for 4 d. Colonocytes were isolated, total RNA was extracted, and relative mRNA levels of marker genes were determined by quantitative RT-PCR. Data from two independent experiments are shown. (A) Schematic representation of the experiment. (B) mRNA levels of Ercc1 and Mlh1, whose gene products are involved in DNA damage response. (C) mRNA levels of Trp53, encoding the tumor suppressor p53. (D) mRNA levels of Cdk4, Cdk6, and Ccnd1, whose products are involved in cell cycle progression. Bars represent the geometric mean ± 95% confidence interval. P values were calculated by unpaired, two-tailed Student’s t test on log-transformed data. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Tungstate treatment reduces polyp formation in an Il10-deficient mouse model, but not in ApcMin+/− mice
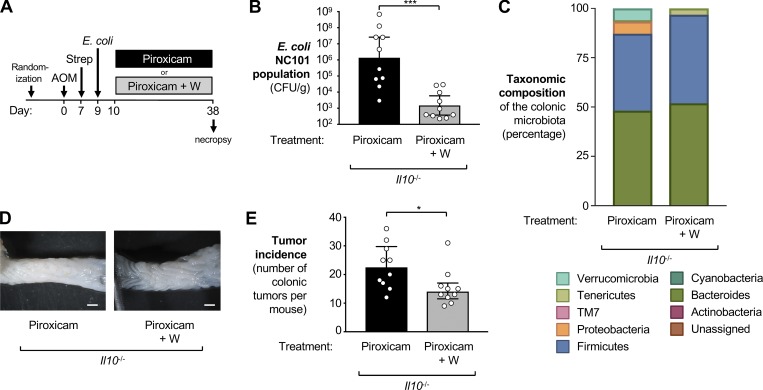
Inflammation in the DSS colitis model is a response to epithelial injury. To exclude the possibility that the observed effects of tungstate on tumorigenesis were unique to DSS-induced colitis, we used a genetic model of gut inflammation. Il10-deficient mice spontaneously develop colitis, a process that can be accelerated by the administration of the nonsteroidal antiinflammatory drug piroxicam (Hale et al., 2005). Groups of Il10-deficient mice were treated i.p. with AOM (20 mg/kg). 1 wk later, streptomycin (2 mg/ml) was administered in the drinking water, followed 2 d later by intragastric inoculation with E. coli NC101. Piroxicam-supplemented diet (100 ppm) was given until the end of the experiment 38 d after the initial AOM injection. One group received normal drinking water while the other group was supplemented with 0.02% sodium tungstate (Fig. 7 A). Tungstate administration significantly decreased the burden of E. coli NC101 in the colon content by three orders of magnitude (Fig. 7 B). The microbiota composition in the two groups (piroxicam vs. piroxicam + tungstate) was significantly different (ANOSIM [unweighted UniFrac]: R = 0.2659, P = 0.003), as determined by 16S rDNA amplicon sequencing (Fig. 7 C). As predicted, tungstate significantly depleted Enterobacteriaceae family members in piroxicam-treated Il10−/− mice. Curiously, we observed a significant decrease in the abundance of members of the phylum Verrucomicrobia (Fig. 7 C). Water-treated mice displayed a large number of colonic polyps (median = 22.5), while tungstate treatment in the drinking water significantly reduced inflammatory markers and polyp formation in this model (median = 14; P < 0.05; Fig. 7, D and E; and Fig. S4 D). This experiment lends further support to the idea that reducing colonization with genotoxin-producing E. coli through oral treatment with tungstate prevents CAC.
Figure 7.
Impact of oral sodium tungstate treatment on E. coli populations and tumorigenesis in AOM-treated Il10-deficient mice. Groups of Il10-deficient mice on the C57BL/6 background were injected i.p. with AOM, treated with streptomycin (Strep), and then colonized with the E. coli NC101 wild-type strain. Inflammation was induced by administration of piroxicam in the rodent diet for 4 wk. One group was treated with 0.02% sodium tungstate (Piroxicam+W, n = 10) in the drinking water while the other received filtered drinking water (Piroxicam, n = 10). Samples were obtained 38 d after AOM injection. Data from two independent experiments are shown. (A) Schematic representation of the experimental design. (B) Burden of E. coli NC101 in the colon content. Bars represent the geometric mean ± 95% confidence interval. P values were calculated by unpaired, two-tailed Student’s t test on log-transformed data. (C) The taxonomic composition of the microbial community in colonic content determined by 16S rDNA amplicon sequencing. (D) Representative images of the colon. Scale bars represent 1 mm. (E) Tumor incidence in the large intestine. Bars indicate the median and the interquartile range. P values were calculated by two-tailed Mann–Whitney U test. *, P < 0.05; ***, P < 0.001.
We also tested the effect of tungstate in ApcMin+/− mice, a model of familial adenomatous polyposis (FAP; Fig. S5). A small subset of gut microbes have been shown to be associated with tumors in this model, such as Fusobacterium nucleatum (Castellarin et al., 2012). In ApcMin+/− mice and in mouse colitis models, colonization with F. nucleatum accelerates tumorigenesis only in mice with preexisting, tumor-promoting mutations (Kostic et al., 2013; Rubinstein et al., 2013). Human FAP patients have predominantly colonic adenomas, while ApcMin+/− mice typically develop numerous polyps in the small intestine and few colonic tumors (Moser et al., 1990; Wu et al., 2009; Kostic et al., 2013; Dolan, 2019). To improve this limitation of the ApcMin+/− mouse model, we modified published protocols (Kostic et al., 2013) and treated ApcMin+/− mice with DSS for 1 wk before colonization with F. nucleatum (Fig. S5 A). One group was treated with 0.02% sodium tungstate in the drinking water throughout the experiment. After 38 d, the number of tumors in the colon was counted. Curiously, we observed ∼23 tumors per animal in the distal portion of the large intestine, yet tungstate treatment had no impact n the number of tumors (Fig. S5, B and C). In a variation of this model, we colonized groups of ApcMin+/− mice with the E. coli strain NC101 to determine whether the presence of this genotoxic E. coli strain could modify tumor development in this model (Fig. S5 D). Tungstate treatment in NC101-colonized ApcMin+/− mice had no effect on the NC101 population size in the colon content (Fig. S5 E) and did not prevent tumor development in the colon (Fig. S5, F and G). Collectively, these findings provide support for our hypothesis that tungstate primarily targets Enterobacteriaceae family members; tumor formation in ApcMin+/− mice is dependent on gut microbiota members such as F. nucleatum and Bacteroides fragilis (Wu et al., 2009; Kostic et al., 2013; Dejea et al., 2018), but not Enterobacteriaceae family members.
Discussion
Bacteria in diverse habitats, including the gut microbiota, express molybdoenzymes (Zhang and Gladyshev, 2010; Zhou et al., 2016). Proteobacteria tend to encode the highest number of molybdoenzymes (Zhang and Gladyshev, 2010). Many enzymes in the enterobacterial electron transport chain, including nitrate, dimethylsulfoxide, and trimethylamine N-oxide reductases, as well as formate dehydrogenases, exhibit a MoCo in their active site. We found that oral tungstate administration during chronic colitis specifically restricts Gammaproteobacteria populations, in particular Enterobacteriaceae family members. This is consistent with the idea that increased availability of respiratory electron acceptors in the gut lumen facilitates a restructuring of the microbial community structure during inflammation (Winter and Bäumler, 2014; Byndloss et al., 2018). In contrast, the effect of tungstate on the abundance of obligate anaerobic gut microbes such as Bacteroidia and Clostridia was very limited in our experimentation.
It is conceivable that tungsten is incorporated into metalloenzymes in obligate anaerobic bacteria; however, either tungsten-inhibited metalloenzymes were dispensable for overall fitness of most obligate anaerobic bacteria or incorporation of tungsten did not affect metalloenzyme activity. Notably, in the Il10/AOM model, tungstate treatment decreased the abundance of members of the phylum Verrucomicrobia. The genome of a member of the phylum Verrucomicrobia was sequenced and encodes at least one putative formate dehydrogenase, a putative molybdenum uptake system (ABC transporter), and genes predicted to be involved in the biosynthesis of a MoCo (Hou et al., 2008). Another sequenced member of the phylum Verrucomicrobia encodes a limited number of genes involved in the biosynthesis of a MoCo (ATCC BAA-835; GenBank accession no. CP001071.1). It is conceivable that erroneous uptake and incorporation of tungsten into a MoCo abolishes formate dehydrogenase activity in Verrucomicrobia members, thus decreasing their abundance in the gut microbiota. However, this does not appear to be a conserved mechanism, as Verrucomicrobia populations were not affected by tungstate in the AOM/DSS model (Fig. 1 E).
Curiously, hyperthermophilic archaea, methanogens, and environmental bacteria express a variety of tungstoenzymes with oxidoreductase activity (Kletzin and Adams, 1996). In these enzymes, tungsten is bound by a molybdopterin cofactor or a molybdopterin derivative (Mukund and Adams, 1991; Schmitz et al., 1992; Johnson et al., 1993). In some instances, tungsten can be functionally replaced by molybdenum. However, hyperthermophilic archaea appear to be strictly dependent on tungsten (Liao, 2013), mirroring the strict dependence of Enterobacteriaceae on molybdenum in their respiratory metalloenzymes.
Microbial dysbiosis frequently occurs as a consequence of mucosal inflammation (Zeng et al., 2017). Conversely, experiments in mouse models suggest that a dysbiotic microbiota can instigate or worsen gut inflammation in some instances. Transfer of a dysbiotic microbiota into genetically susceptible mice is sufficient to trigger colitis (Garrett et al., 2010; Schulfer et al., 2018), and the experimental colonization of mice with human AIEC isolates is sufficient to induce chronic colitis and fibrosis (Carvalho et al., 2009; Small et al., 2013). The molecular mechanisms of how Enterobacteriaceae family members instigate inflammation in the setting of IBD are incompletely understood, but possibly involve a mislocalization of bacteria. AIEC strains frequently form biofilms in close proximity to the epithelium and invade nonphagocytic cells. Ileal epithelial cells in Crohn’s disease patients express higher levels of carcinoembryonic antigen-related cell adhesion molecule 6, which serves as the receptor for type 1 fimbriae expressed by AIEC (Boudeau et al., 2001; Barnich et al., 2007; Barnich and Darfeuille-Michaud, 2010). In our study, we found that tungstate administration not just reduced population levels of the murine AIEC strain NC101 in the gut lumen, but also decreased the number of tissue-associated bacteria and DNA damage in the intestinal epithelium. It should be noted that tungstate treatment did not deplete Gammaproteobacteria entirely from gut microbial communities, but a small population remained throughout the entire experiment. In fact, a complete eradication of Gammaproteobacteria may not be desirable, as these commensals contribute to colonization resistance against enteric pathogens through nutritional competition and release of antibacterial toxins (Kamada et al., 2012; Deriu et al., 2013; Sassone-Corsi et al., 2016).
One mechanism of how microbial community members promote tumorigenesis is through the release of genotoxins in the intestinal tract (Arthur et al., 2012). In principle, probiotic bacterial strains could be used to outcompete genotoxic bacteria from the gut ecosystem. A recent report showed that microcins produced by the probiotic E. coli strain Nissle 1917 limit the expansion other Enterobacteriaceae during intestinal inflammation (Sassone-Corsi et al., 2016), and one could speculate that Nissle 1917 could be used to eradicate genotoxin-producing Enterobacteriaceae. However, the Nissle 1917 genome encodes a functional pks synthesis island, and its genotoxicity cannot be disassociated from the probiotic effect (Olier et al., 2012), suggesting that care must be taken when developing bacterial probiotics.
Other strategies have aimed to inhibit proinflammatory and protumoral properties of genotoxic E. coli strains. A recent study reported that inhibition of bacterial quorum sensing in flagellated AIEC strains reduces gut colonization by these strains and improves AIEC-driven intestinal inflammation (Rooks et al., 2017). In addition, small molecule inhibitors of colibactin biosynthesis have been developed (Cougnoux et al., 2016). Administration of these inhibitors significantly reduces colibactin-driven CAC progression in preclinical models (Cougnoux et al., 2016). In our study, we analyzed the effect of tungstate on reducing tumor incidence in a model of CAC in which tumorigenesis was in part driven by colibactin. Interestingly, the effect size of tungstate on tumor numbers was more pronounced than the contribution of colibactin alone (Fig. 3). This outcome is consistent with a scenario in which tungstate decreases tumor formation by acting on chronic inflammation and local colibactin production, both of which are dependent on luminal and mucosa-associated populations of the genotoxin-producing E. coli.
The adenoma-carcinoma sequence in FAP is thought to be an accelerated variant of most sporadic cancers, which are also defective for APC in most instances (Johnson and Fleet, 2013). Enterotoxigenic B. fragilis enhances polyp formation in mouse models of FAP (ApcMin+/− mice; Wu et al., 2009; Chung et al., 2018; Dejea et al., 2018). Furthermore, tumor formation is magnified by F. nucleatum through the recruitment of tumor-infiltrating myeloid cells (Kostic et al., 2013). B. fragilis and F. nucleatum perform only a rudimentary type of anaerobic respiration, i.e., fumarate reduction, and the fumarate reductases expressed by these bacteria are not predicted to require molybdenum or a MoCo (Macy et al., 1978; Kapatral et al., 2002; Nikitina et al., 2015). In line with these considerations, we found that tungstate did not impact polyp formation in ApcMin+/− mice. As such, our strategy of targeting the inflammation-associated bloom of Enterobacteriaceae family members is most applicable to high-risk populations with chronic colitis as one facet of a personalized medicine approach (Sartor and Wu, 2017). In summary, our experiments demonstrate that it is feasible to specifically target the metabolism of malignancy-promoting Enterobacteriaceae in the gut microbiota to reduce the risk of CAC development.
Materials and methods
Bacterial strains
The E. coli strains used in this study are listed in Table S2. NC101 (Patwa et al., 2011) and E. coli MP13 (Lasaro et al., 2014) strains were obtained from Dr. R. Balfour Sartor (University of North Carolina, Chapel Hill, NC) and Dr. Mark Goulian (University of Pennsylvania, Philadelphia, PA), respectively. All strains were routinely propagated in Luria-Bertani (LB) broth or on LB agar plates at 37°C with aeration. When appropriate, the following antibiotics were added: 20 µg/ml tetracycline, 100 µg/ml carbenicillin, and 100 µg/ml kanamycin. To facilitate recovery from fecal material, the NC101 wild-type was transformed with the plasmid pWSK29 (Wang and Kushner, 1991). The resulting strain, NC101(pWSK29), was used for all experiments and is referred to as NC101 throughout the text. Similarly, for the experiment shown in Fig. S1 (A and B), the E. coli Nissle wild-type was carrying pWSK29, while the ΔmoaA mutant (SW930) and the ΔnarZ ΔnarG ΔnapA mutant (SW1029) were marked with pWSK129 (Grozdanov et al., 2004; Spees et al., 2013; Winter et al., 2013; Byndloss et al., 2017; Hughes et al., 2017; Zhu et al., 2018).
Construction of plasmids
Standard molecular cloning techniques (Sambrook et al., 1989) or Gibson Assembly Cloning procedure (New England Biolabs) were used to construct plasmids. The flanking regions of the NC101 pks island were amplified and ligated into pGP706 to give rise to pWZ98. Plasmid inserts were verified by Sanger sequencing. All the plasmids and primers used in this study are listed in Tables S2 and S3.
Construction of mutants by allelic exchange
The pGP706-based suicide plasmid pWZ98 was routinely propagated in DH5α λpir (Pal et al., 2005). pWZ98 was then introduced into the E. coli strain NC101 via bacterial conjugation using S17-1 λpir as the donor (Simon et al., 1983; Winter et al., 2013). LB plates containing appropriate antibiotics were used to select for exconjugants with the suicide plasmid integrated into the chromosome. Sucrose plates (8 g/liter nutrient broth base, 5% sucrose, 15 g/liter agar) were used to select for the second crossover event, which leads to the unmarked deletion of the gene of interest. pWZ98 was used to delete the pks island in E. coli strain NC101, creating WZ140. Deletion of the target gene was confirmed by PCR.
Animal experimentation
All experiments involving mice were approved by the Institutional Animal Care and Use Committee at University of Texas Southwestern Medical Center. Both male and female mice were analyzed, and no significant differences were noted. An approximately equal representation of male and female mice was sought. The number of individual animals per group is noted in each figure. Unless noted otherwise, animals had ad libitum access to autoclaved drinking water and irradiated rodent diet (Teklad global 16% protein 2916; Envigo). Room lighting was on a 12-h light/dark cycle. Animals were housed in individually ventilated cages. All treatments occurred in the designated animal facility. Animals that had to be euthanized for humane reasons before reaching the predetermined time points were excluded from the analysis.
DSS/AOM model and sodium tungstate treatment
C57BL/6J wild-type mice, obtained from the Jackson Laboratory, were bred under specific pathogen–free conditions at University of Texas Southwestern Medical Center. 7–9-wk-old mice were semirandomly assigned into treatment groups before the experiment. AOM (Sigma-Aldrich) was injected i.p. at a dosage of 20 mg/kg. 1 wk after AOM injection, the drinking water was replaced with 2 mg/ml streptomycin solution for 2 d. Mice were then inoculated with 109 CFU of the indicated E. coli strain or remained uninfected. In the competitive colonization experiments, animals were inoculated with an equal mixture of 5 × 108 CFU of wild-type E. coli Nissle 1917 strain and 5 × 108 CFU of indicated mutant. 1 d after inoculation, the drinking water was replaced with filter-sterilized water (mock treatment), a filter-sterilized 2% (wt/vol) DSS solution (relative molecular mass 40,000; Alfa Aesar), or a filter-sterilized solution of 2% (wt/vol) DSS + 0.2% (wt/vol) sodium tungstate solution (Sigma-Aldrich). After 7 d, the drinking solution was replaced with either filter-sterilized water or filter-sterilized 0.2% (wt/vol) sodium tungstate for 2 wk. This treatment was repeated three times in total (see Fig. 1 A). Fecal pellets were collected at the indicated time points. After euthanasia, colon length was measured, and colons were opened longitudinally. Colonic tissue was collected, flash frozen, and stored at −80°C for subsequent mRNA and protein expression analysis. The remaining colonic and cecal tissue was fixed in 40 ml of phosphate-buffered 4% paraformaldehyde (Electron Microscopy Science) at 4°C overnight. Fixed tissue is further washed and stored in 70% ethanol solution. Images of the entire colons were captured using a STEMI 2000-c dissecting microscope (Zeiss) through AxioVision AxioVs40 software (version 4.8.1.0). Fecal material and colonic content were harvested in sterile PBS and the bacterial load of the E. coli strains was enumerated by plating on LB plates supplemented with appropriate antibiotics. In some experiments, fecal samples were not collected to minimize the impact of stress on experimental outcomes. Tissues from all experiments were collected and analyzed for tumor incidence. E. coli NC101 and Nissle 1917 strains were differentially marked with the low-copy-number plasmids pWSK29 or pWSK129 to facilitate bacterial recovery from biological samples (Winter et al., 2013).
AOM/Il10 model
Il10-deficient mice (Kühn et al., 1993), originally obtained from the Jackson Laboratory (B6.129P2-Il10tm1Cgn/J, stock #002251), were bred in the University of Texas Southwestern Medical Center facility. 8–10-wk-old mice were randomly assigned into cages before the experiment. AOM (Sigma-Aldrich) was injected i.p. at a dosage of 20 mg/kg. 1 wk after AOM injection, the drinking water was replaced with 2 mg/ml streptomycin solution for 2 d. Mice were then inoculated with 109 CFU of the indicated E. coli strain. Piroxicam-supplemented diet (100 ppm; Envigo custom diet) was given after inoculation until the end of the experiment (38 d after the initial AOM injection). Sample collection and processing were performed as described above.
APCMin+/− model
7–9-wk-old APCMin+/− mice were semirandomly assigned into treatment groups before the experiment. 1 d after randomization, the drinking water was replaced with a filtered-sterilized 2 mg/ml streptomycin solution for 2 d. Mice were then inoculated with 109 CFU of the E. coli strain NC101. The drinking solution was replaced with either filter-sterilized water (mock-treatment) or filter-sterilized 0.02% (wt/vol) sodium tungstate for 4 wk (see Fig. S5 A). Sample collection and processing were performed as described above.
APCMin+/−/DSS model
7–9-wk-old APCMin+/− mice were semirandomly assigned into treatment groups before the experiment. 1 d after randomization, the drinking water was replaced with a filter-sterilized 2% (wt/vol) DSS solution (relative molecular mass 40,000; Alfa Aesar). 7 d later, the drinking water was replaced with a filtered-sterilized 2 mg/ml streptomycin solution for 2 d. Mice were then inoculated with 109 CFU of the F. nucleatum strain ATCC 23726. The drinking solution was replaced with either filter-sterilized water (mock treatment) or filter-sterilized 0.02% (wt/vol) sodium tungstate for 4 wk (see Fig. S5 D). Sample collection and processing were performed as described above.
Colonocyte isolation
C57BL/6J wild-type mice were injected i.p. with AOM at a dosage of 20 mg/kg. 1 wk after AOM injection, mice were treated with streptomycin and inoculated with 109 CFU of the colibactin-producing E. coli strain NC101. Mice were treated with DSS or DSS + sodium tungstate or left untreated for 7 d and allowed to recover for 4 d. After euthanasia, large intestine was collected, and the colonocytes were harvested using a modified procedure originally described by Roediger and Truelove (1979). Briefly, colon was opened longitudinally and washed extensively to remove luminal content. The colon was then placed in 2 ml of Krebs-Ringer bicarbonate (Ca2+ free) buffer (1.8 g/liter d-glucose, 0.0468 g/liter MgCl2, 0.34 g/liter KCl, 7 g/liter NaCl, 0.1 g/liter Na2HPO4, 0.18 g/liter NaH2PO4, and 1.26 g/liter NaHCO3, pH 7.4) supplemented with 1 mM DTT and 10 mM EDTA (pH 8.0). The tissue was shaken at 220 rpm and 37°C for 40 min. Colonocytes were collected by passing the tissue through a 70-µm cell strainer, and the flow-through cells were centrifuged at 300 g for 5 min. Red blood cells were lysed by incubating cells in ammonium-chloride-potassium lysis buffer (8.02 g/liter NH4Cl, 1 g/liter KHCO3, and 0.037 mg/liter EDTA, pH 7.4) for 1 min. Colonocytes were washed two more times with Ca2+-free Krebs-Ringer bicarbonate buffer before preservation with RNAlater (Qiagen) to stabilize cellular RNA. Total RNA was subsequently extracted using RNeasy Mini Kit (Qiagen), and residual DSS contaminants in RNA samples were removed using Dynabeads mRNA Direct Kit (Life Technologies).
16S rDNA amplicon sequencing and analysis
Fecal pellets or colonic contents were collected, and DNA extraction was performed using the MoBio PowerFecal kit (MoBio Laboratories), followed by potassium chloride precipitation to remove residual DSS contaminants (Hughes et al., 2017). The V4 region of 16S rDNA of the purified DNA was amplified with the 515f-806R primer pair. The library was purified and quantified with the 2200 TapeStation (Agilent) and sequenced with the Illumina MiSeq platform, generating 250-bp (AOM/DSS model) or 300-bp (Il10/AOM model) paired-end reads. Sequence alignment, operational taxonomic units picking against the Silva database (version 128, released on February 6, 2017; Quast et al., 2013), clustering, phylogenetic and taxonomic profiling, ANOSIM analysis, and the analysis of β diversity (principal component analysis) on the demultiplexed sequences were performed with the Quantitative Insights into Microbial Ecology software package (version 1.91; Caporaso et al., 2010; Vázquez-Baeza et al., 2013). The bacterial 16S rDNA and metagenomics sequencing reads generated and analyzed during the current study are available at the European Bioinformatics Institute repository under accession nos. PRJEB29047 and PRJEB32761.
Immunofluorescence staining and quantification
Paraffin-embedded, transverse sections of colon tissue were hybridized overnight with 2 µM fluorescence in situ hybridization probe specific for Enterobacteriaceae members at 55°C in hybridization buffer (Kempf et al., 2000). The slides were subsequently washed three times with fluorescence in situ hybridization wash buffer and blocked with 4% bovine serum albumin in PBS for 30 min at RT. Blocked slides were stained with anti-phosphorylated γ-H2AX antibody (#9718S; Cell Signaling) and subsequently with Alexa Fluor 488–conjugated anti-rabbit antibody (#4412; Cell Signaling). Tile images were captured using an LSM780 confocal microscope (Zeiss) and stitched using Imaris software 8.2 (Bitplane AG). Quantification of tissue-associated E. coli burden as well DNA damage was performed using the spot and surface function of Imaris software 8.2. In each tissue, 6,688 to 23,809 cells were counted, and only fluorescent objects with a diameter >6 µm (average diameter of mammalian cell nucleus) were counted to eliminate contamination and nonspecific staining.
Quantification of mRNA levels in intestinal tissue
The relative transcription levels of Nos2, Cxcl1, Cxcl2, Il17, Ifng, and Tnfa genes were determined by quantitative RT-PCR as described previously (Winter et al., 2009). Briefly, colonic tissue was homogenized using a Mini beadbeater (Biospec Products). The TRI reagent method was performed to extract RNA from the homogenized tissue (Molecular Research Center). Residual DSS contaminants in RNA samples were removed using Dynabeads mRNA Direct Kit (Life Technologies). TaqMan reverse transcription reagents (Life Technologies) were used to generate cDNA and real-time PCR was performed using SYBR Green (Life Technologies). Data were acquired in a QuantStudio 6 Flex instrument (Life Technologies) and analyzed using the comparative Ct method. The primers of the gene of interest (listed in Table 3; Overbergh et al., 2003; Godinez et al., 2008; Wilson et al., 2008; Wang et al., 2012b) were added at a final concentration of 250 nM. The respective levels of Gapdh mRNA were used for target gene transcription normalization.
Quantification and statistical analysis
Unless noted otherwise, data analysis was performed in GraphPad Prism v7.0c. Values of bacterial population sizes, competitive indices, fold changes in mRNA levels, and normalized colon length were transformed by the natural logarithm before statistical analysis. Time course experiments of E. coli populations were analyzed by two-way ANOVA and a post hoc Tukey’s multiple comparison test. A Student’s t test was used for all other log-transformed data, while tumor incidence was compared using the Mann–Whitney U test, as indicated in the figure legends. A P value <0.05 was considered significant.
Online supplemental material
Fig. S1 describes the impact of tungstate treatment on E. coli fitness and the dynamics of gut microbial communities, and Fig. S2 depicts the analysis of colitis markers in the AOM/DSS colitis model. Fig. S3 analyzes the colonization of NC101 and host inflammatory responses in a murine model. Fig. S4 shows host DNA damage and inflammatory responses in a colibactin-driven tumorigenesis model. Fig. S5 explores the impact of tungstate treatment on ApcMin+/− models of tumorigenesis in the large intestine. Scores of individual samples on the intestinal inflammation and tumor histology are included in Table S1. Strains and plasmids used in this study are listed in Table S2. All primers used in this study are listed in Table S3.
Supplementary Material
Acknowledgments
We thank Dr. R. Balfour Sartor (University of North Carolina, Chapel Hill, NC) and Dr. Mark Goulian (University of Pennsylvania, Philadelphia, PA) for providing E. coli strains.
Work in S.E. Winter’s laboratory was funded by the National Institutes of Health (AI118807 and AI128151), the Welch Foundation (I-1969-20180324), the Burroughs Wellcome Fund (1017880), and an American Cancer Society Research Scholar Grant (RSG-17-048-01-MPC). W. Zhu was supported by a Research Fellows Award from the Crohn’s and Colitis Foundation of America (454921). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the funding agencies. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
S.E. Winter is listed as an inventor on patent application WO2014200929A1, which describes a treatment to prevent the inflammation-associated expansion of Enterobacteriaceae. The other authors declare no additional competing financial interests.
Author contributions: W. Zhu, N. Miyata, M.G. Winter, E.R. Hughes, and L. Spiga performed and analyzed the in vivo experiments and experiments involving quantitative imaging. 16S sequencing data were analyzed by W. Zhu and J. Kim. The histopathology analysis was performed by A. Arenales, M.X. Byndloss, P. Gopal, and R.L. Santos. W. Zhu, E.R. Hughes, L. Sifuentes-Dominguez, and P. Starokadomskyy contributed to assessing DNA damage in gut epithelial cells. W. Zhu, E. Burstein, and S.E. Winter designed the experiments, interpreted the data, and wrote the manuscript with contributions from all authors.
References
- Arnold M., Sierra M.S., Laversanne M., Soerjomataram I., Jemal A., and Bray F.. 2017. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 66:683–691. 10.1136/gutjnl-2015-310912 [DOI] [PubMed] [Google Scholar]
- Arthur J.C., Perez-Chanona E., Mühlbauer M., Tomkovich S., Uronis J.M., Fan T.J., Campbell B.J., Abujamel T., Dogan B., Rogers A.B., et al. . 2012. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 338:120–123. 10.1126/science.1224820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnich N., and Darfeuille-Michaud A.. 2010. Abnormal CEACAM6 expression in Crohn disease patients favors gut colonization and inflammation by adherent-invasive E. coli. Virulence. 1:281–282. 10.4161/viru.1.4.11510 [DOI] [PubMed] [Google Scholar]
- Barnich N., Carvalho F.A., Glasser A.L., Darcha C., Jantscheff P., Allez M., Peeters H., Bommelaer G., Desreumaux P., Colombel J.F., and Darfeuille-Michaud A.. 2007. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J. Clin. Invest. 117:1566–1574. 10.1172/JCI30504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossuet-Greif N., Vignard J., Taieb F., Mirey G., Dubois D., Petit C., Oswald E., and Nougayrède J.P.. 2018. The Colibactin Genotoxin Generates DNA Interstrand Cross-Links in Infected Cells. MBio. 9:e02393-17 10.1128/mBio.02393-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudeau J., Barnich N., and Darfeuille-Michaud A.. 2001. Type 1 pili-mediated adherence of Escherichia coli strain LF82 isolated from Crohn’s disease is involved in bacterial invasion of intestinal epithelial cells. Mol. Microbiol. 39:1272–1284. 10.1111/j.1365-2958.2001.02315.x [DOI] [PubMed] [Google Scholar]
- Brennan C.A., and Garrett W.S.. 2016. Gut Microbiota, Inflammation, and Colorectal Cancer. Annu. Rev. Microbiol. 70:395–411. 10.1146/annurev-micro-102215-095513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buc E., Dubois D., Sauvanet P., Raisch J., Delmas J., Darfeuille-Michaud A., Pezet D., and Bonnet R.. 2013. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS One. 8:e56964 10.1371/journal.pone.0056964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byndloss M.X., Olsan E.E., Rivera-Chávez F., Tiffany C.R., Cevallos S.A., Lokken K.L., Torres T.P., Byndloss A.J., Faber F., Gao Y., et al. . 2017. Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science. 357:570–575. 10.1126/science.aam9949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byndloss M.X., Pernitzsch S.R., and Bäumler A.J.. 2018. Healthy hosts rule within: ecological forces shaping the gut microbiota. Mucosal Immunol. 11:1299–1305. 10.1038/s41385-018-0010-y [DOI] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I., et al. . 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 7:335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho F.A., Barnich N., Sivignon A., Darcha C., Chan C.H., Stanners C.P., and Darfeuille-Michaud A.. 2009. Crohn’s disease adherent-invasive Escherichia coli colonize and induce strong gut inflammation in transgenic mice expressing human CEACAM. J. Exp. Med. 206:2179–2189. 10.1084/jem.20090741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho F.A., Koren O., Goodrich J.K., Johansson M.E., Nalbantoglu I., Aitken J.D., Su Y., Chassaing B., Walters W.A., González A., et al. . 2012. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe. 12:139–152. 10.1016/j.chom.2012.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellarin M., Warren R.L., Freeman J.D., Dreolini L., Krzywinski M., Strauss J., Barnes R., Watson P., Allen-Vercoe E., Moore R.A., and Holt R.A.. 2012. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 22:299–306. 10.1101/gr.126516.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Liu F., Ling Z., Tong X., and Xiang C.. 2012. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One. 7:e39743 10.1371/journal.pone.0039743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung L., Thiele Orberg E., Geis A.L., Chan J.L., Fu K., DeStefano Shields C.E., Dejea C.M., Fathi P., Chen J., Finard B.B., et al. . 2018. Bacteroides fragilis Toxin Coordinates a Pro-carcinogenic Inflammatory Cascade via Targeting of Colonic Epithelial Cells. Cell Host Microbe. 23:203–214.e5. 10.1016/j.chom.2018.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S., Verdu E., Denou E., and Bercik P.. 2009. The role of pathogenic microbes and commensal bacteria in irritable bowel syndrome. Dig. Dis. 27(Suppl 1):85–89. 10.1159/000268126 [DOI] [PubMed] [Google Scholar]
- Cougnoux A., Dalmasso G., Martinez R., Buc E., Delmas J., Gibold L., Sauvanet P., Darcha C., Déchelotte P., Bonnet M., et al. . 2014. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut. 63:1932–1942. 10.1136/gutjnl-2013-305257 [DOI] [PubMed] [Google Scholar]
- Cougnoux A., Delmas J., Gibold L., Faïs T., Romagnoli C., Robin F., Cuevas-Ramos G., Oswald E., Darfeuille-Michaud A., Prati F., et al. . 2016. Small-molecule inhibitors prevent the genotoxic and protumoural effects induced by colibactin-producing bacteria. Gut. 65:278–285. 10.1136/gutjnl-2014-307241 [DOI] [PubMed] [Google Scholar]
- Darfeuille-Michaud A., Neut C., Barnich N., Lederman E., Di Martino P., Desreumaux P., Gambiez L., Joly B., Cortot A., and Colombel J.F.. 1998. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn’s disease. Gastroenterology. 115:1405–1413. 10.1016/S0016-5085(98)70019-8 [DOI] [PubMed] [Google Scholar]
- Dejea C.M., Fathi P., Craig J.M., Boleij A., Taddese R., Geis A.L., Wu X., DeStefano Shields C.E., Hechenbleikner E.M., Huso D.L., et al. . 2018. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. 359:592–597. 10.1126/science.aah3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriu E., Liu J.Z., Pezeshki M., Edwards R.A., Ochoa R.J., Contreras H., Libby S.J., Fang F.C., and Raffatellu M.. 2013. Probiotic bacteria reduce salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe. 14:26–37. 10.1016/j.chom.2013.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan S. 2019. Familial Adenomatous Polyposis: Development, Presentation, and Treatment Strategies. Clin. J. Oncol. Nurs. 23:135–138. [DOI] [PubMed] [Google Scholar]
- Dutilh B.E., Backus L., van Hijum S.A., and Tjalsma H.. 2013. Screening metatranscriptomes for toxin genes as functional drivers of human colorectal cancer. Best Pract. Res. Clin. Gastroenterol. 27:85–99. 10.1016/j.bpg.2013.03.008 [DOI] [PubMed] [Google Scholar]
- Earle K.A., Billings G., Sigal M., Lichtman J.S., Hansson G.C., Elias J.E., Amieva M.R., Huang K.C., and Sonnenburg J.L.. 2015. Quantitative Imaging of Gut Microbiota Spatial Organization. Cell Host Microbe. 18:478–488. 10.1016/j.chom.2015.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch H.G., and Lester R.L.. 1972. Effects of molybdate, tungstate, and selenium compounds on formate dehydrogenase and other enzyme systems in Escherichia coli. J. Bacteriol. 110:1032–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber F., Tran L., Byndloss M.X., Lopez C.A., Velazquez E.M., Kerrinnes T., Nuccio S.P., Wangdi T., Fiehn O., Tsolis R.M., and Bäumler A.J.. 2016. Host-mediated sugar oxidation promotes post-antibiotic pathogen expansion. Nature. 534:697–699. 10.1038/nature18597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q., Liang S., Jia H., Stadlmayr A., Tang L., Lan Z., Zhang D., Xia H., Xu X., Jie Z., et al. . 2015. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat. Commun. 6:6528 10.1038/ncomms7528 [DOI] [PubMed] [Google Scholar]
- Ganz T., and Nemeth E.. 2015. Iron homeostasis in host defence and inflammation. Nat. Rev. Immunol. 15:500–510. 10.1038/nri3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett W.S., Gallini C.A., Yatsunenko T., Michaud M., DuBois A., Delaney M.L., Punit S., Karlsson M., Bry L., Glickman J.N., et al. . 2010. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 8:292–300. 10.1016/j.chom.2010.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates A.J., Hughes R.O., Sharp S.R., Millington P.D., Nilavongse A., Cole J.A., Leach E.R., Jepson B., Richardson D.J., and Butler C.S.. 2003. Properties of the periplasmic nitrate reductases from Paracoccus pantotrophus and Escherichia coli after growth in tungsten-supplemented media. FEMS Microbiol. Lett. 220:261–269. 10.1016/S0378-1097(03)00122-8 [DOI] [PubMed] [Google Scholar]
- Godinez I., Haneda T., Raffatellu M., George M.D., Paixão T.A., Rolán H.G., Santos R.L., Dandekar S., Tsolis R.M., and Bäumler A.J.. 2008. T cells help to amplify inflammatory responses induced by Salmonella enterica serotype Typhimurium in the intestinal mucosa. Infect. Immun. 76:2008–2017. 10.1128/IAI.01691-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso F., and Frisan T.. 2015. Bacterial Genotoxins: Merging the DNA Damage Response into Infection Biology. Biomolecules. 5:1762–1782. 10.3390/biom5031762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov S.I. 2013. Inflammation and colorectal cancer: colitis-associated neoplasia. Semin. Immunopathol. 35:229–244. 10.1007/s00281-012-0352-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronbach K., Flade I., Holst O., Lindner B., Ruscheweyh H.J., Wittmann A., Menz S., Schwiertz A., Adam P., Stecher B., et al. . 2014. Endotoxicity of lipopolysaccharide as a determinant of T-cell-mediated colitis induction in mice. Gastroenterology. 146:765–775. 10.1053/j.gastro.2013.11.033 [DOI] [PubMed] [Google Scholar]
- Grozdanov L., Raasch C., Schulze J., Sonnenborn U., Gottschalk G., Hacker J., and Dobrindt U.. 2004. Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J. Bacteriol. 186:5432–5441. 10.1128/JB.186.16.5432-5441.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale L.P., Gottfried M.R., and Swidsinski A.. 2005. Piroxicam treatment of IL-10-deficient mice enhances colonic epithelial apoptosis and mucosal exposure to intestinal bacteria. Inflamm. Bowel Dis. 11:1060–1069. 10.1097/01.MIB.0000187582.90423.bc [DOI] [PubMed] [Google Scholar]
- Hanzu F., Gomis R., Coves M.J., Viaplana J., Palomo M., Andreu A., Szpunar J., and Vidal J.. 2010. Proof-of-concept trial on the efficacy of sodium tungstate in human obesity. Diabetes Obes. Metab. 12:1013–1018. 10.1111/j.1463-1326.2010.01293.x [DOI] [PubMed] [Google Scholar]
- Hou S., Makarova K.S., Saw J.H., Senin P., Ly B.V., Zhou Z., Ren Y., Wang J., Galperin M.Y., Omelchenko M.V., et al. . 2008. Complete genome sequence of the extremely acidophilic methanotroph isolate V4, Methylacidiphilum infernorum, a representative of the bacterial phylum Verrucomicrobia. Biol. Direct. 3:26 10.1186/1745-6150-3-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes E.R., Winter M.G., Duerkop B.A., Spiga L., Furtado de Carvalho T., Zhu W., Gillis C.C., Büttner L., Smoot M.P., Behrendt C.L., et al. . 2017. Microbial Respiration and Formate Oxidation as Metabolic Signatures of Inflammation-Associated Dysbiosis. Cell Host Microbe. 21:208–219. 10.1016/j.chom.2017.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.L., and Fleet J.C.. 2013. Animal models of colorectal cancer. Cancer Metastasis Rev. 32:39–61. 10.1007/s10555-012-9404-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.L., Rajagopalan K.V., Mukund S., and Adams M.W.. 1993. Identification of molybdopterin as the organic component of the tungsten cofactor in four enzymes from hyperthermophilic Archaea. J. Biol. Chem. 268:4848–4852. [PubMed] [Google Scholar]
- Kamada N., Kim Y.G., Sham H.P., Vallance B.A., Puente J.L., Martens E.C., and Núñez G.. 2012. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 336:1325–1329. 10.1126/science.1222195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M., and Martin A.. 2017. Microbiome and colorectal cancer: Unraveling host-microbiota interactions in colitis-associated colorectal cancer development. Semin. Immunol. 32:3–13. 10.1016/j.smim.2017.04.003 [DOI] [PubMed] [Google Scholar]
- Kapatral V., Anderson I., Ivanova N., Reznik G., Los T., Lykidis A., Bhattacharyya A., Bartman A., Gardner W., Grechkin G., et al. . 2002. Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586. J. Bacteriol. 184:2005–2018. 10.1128/JB.184.7.2005-2018.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf V.A., Trebesius K., and Autenrieth I.B.. 2000. Fluorescent In situ hybridization allows rapid identification of microorganisms in blood cultures. J. Clin. Microbiol. 38:830–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kletzin A., and Adams M.W.. 1996. Tungsten in biological systems. FEMS Microbiol. Rev. 18:5–63. 10.1111/j.1574-6976.1996.tb00226.x [DOI] [PubMed] [Google Scholar]
- Kostic A.D., Gevers D., Pedamallu C.S., Michaud M., Duke F., Earl A.M., Ojesina A.I., Jung J., Bass A.J., Tabernero J., et al. . 2012. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 22:292–298. 10.1101/gr.126573.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic A.D., Chun E., Robertson L., Glickman J.N., Gallini C.A., Michaud M., Clancy T.E., Chung D.C., Lochhead P., Hold G.L., et al. . 2013. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 14:207–215. 10.1016/j.chom.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn R., Löhler J., Rennick D., Rajewsky K., and Müller W.. 1993. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 75:263–274. 10.1016/0092-8674(93)80068-P [DOI] [PubMed] [Google Scholar]
- Lasaro M., Liu Z., Bishar R., Kelly K., Chattopadhyay S., Paul S., Sokurenko E., Zhu J., and Goulian M.. 2014. Escherichia coli isolate for studying colonization of the mouse intestine and its application to two-component signaling knockouts. J. Bacteriol. 196:1723–1732. 10.1128/JB.01296-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasry A., Zinger A., and Ben-Neriah Y.. 2016. Inflammatory networks underlying colorectal cancer. Nat. Immunol. 17:230–240. 10.1038/ni.3384 [DOI] [PubMed] [Google Scholar]
- Liao R.Z. 2013. Why is the molybdenum-substituted tungsten-dependent formaldehyde ferredoxin oxidoreductase not active? A quantum chemical study. J. Biol. Inorg. Chem. 18:175–181. 10.1007/s00775-012-0961-5 [DOI] [PubMed] [Google Scholar]
- Lozupone C.A., Stombaugh J.I., Gordon J.I., Jansson J.K., and Knight R.. 2012. Diversity, stability and resilience of the human gut microbiota. Nature. 489:220–230. 10.1038/nature11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupp C., Robertson M.L., Wickham M.E., Sekirov I., Champion O.L., Gaynor E.C., and Finlay B.B.. 2007. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2:119–129. 10.1016/j.chom.2007.06.010 [DOI] [PubMed] [Google Scholar]
- Macy J.M., Ljungdahl L.G., and Gottschalk G.. 1978. Pathway of succinate and propionate formation in Bacteroides fragilis. J. Bacteriol. 134:84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks O.D., Scanlon K.M., and Donnenberg M.S.. 2013. An Escherichia coli effector protein promotes host mutation via depletion of DNA mismatch repair proteins. MBio. 4:e00152–e13. 10.1128/mBio.00152-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin H.M., Campbell B.J., Hart C.A., Mpofu C., Nayar M., Singh R., Englyst H., Williams H.F., and Rhodes J.M.. 2004. Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology. 127:80–93. 10.1053/j.gastro.2004.03.054 [DOI] [PubMed] [Google Scholar]
- Martin P., Marcq I., Magistro G., Penary M., Garcie C., Payros D., Boury M., Olier M., Nougayrède J.P., Audebert M., et al. . 2013. Interplay between siderophores and colibactin genotoxin biosynthetic pathways in Escherichia coli. PLoS Pathog. 9:e1003437 10.1371/journal.ppat.1003437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuoka T., and Hayakawa K.. 1973. [The fecal flora in man. I. Composition of the fecal flora of various age groups]. Zentralbl. Bakteriol. Orig. A. 223:333–342. [PubMed] [Google Scholar]
- Moser A.R., Pitot H.C., and Dove W.F.. 1990. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 247:322–324. 10.1126/science.2296722 [DOI] [PubMed] [Google Scholar]
- Mukhopadhya I., Hansen R., El-Omar E.M., and Hold G.L.. 2012. IBD-what role do Proteobacteria play? Nat. Rev. Gastroenterol. Hepatol. 9:219–230. 10.1038/nrgastro.2012.14 [DOI] [PubMed] [Google Scholar]
- Mukund S., and Adams M.W.. 1991. The novel tungsten-iron-sulfur protein of the hyperthermophilic archaebacterium, Pyrococcus furiosus, is an aldehyde ferredoxin oxidoreductase. Evidence for its participation in a unique glycolytic pathway. J. Biol. Chem. 266:14208–14216. [PubMed] [Google Scholar]
- Nichols J.D., and Rajagopalan K.V.. 2005. In vitro molybdenum ligation to molybdopterin using purified components. J. Biol. Chem. 280:7817–7822. 10.1074/jbc.M413783200 [DOI] [PubMed] [Google Scholar]
- Nikitina A.S., Kharlampieva D.D., Babenko V.V., Shirokov D.A., Vakhitova M.T., Manolov A.I., Shkoporov A.N., Taraskina A.E., Manuvera V.A., Lazarev V.N., and Kostryukova E.S.. 2015. Complete Genome Sequence of an Enterotoxigenic Bacteroides fragilis Clinical Isolate. Genome Announc. 3:e00450-15 10.1128/genomeA.00450-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nougayrède J.P., Homburg S., Taieb F., Boury M., Brzuszkiewicz E., Gottschalk G., Buchrieser C., Hacker J., Dobrindt U., and Oswald E.. 2006. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 313:848–851. 10.1126/science.1127059 [DOI] [PubMed] [Google Scholar]
- Olier M., Marcq I., Salvador-Cartier C., Secher T., Dobrindt U., Boury M., Bacquié V., Pénary M., Gaultier E., Nougayrède J.P., et al. . 2012. Genotoxicity of Escherichia coli Nissle 1917 strain cannot be dissociated from its probiotic activity. Gut Microbes. 3:501–509. 10.4161/gmic.21737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbergh L., Giulietti A., Valckx D., Decallonne R., Bouillon R., and Mathieu C.. 2003. The use of real-time reverse transcriptase PCR for the quantification of cytokine gene expression. J. Biomol. Tech. 14:33–43. [PMC free article] [PubMed] [Google Scholar]
- Pal D., Venkova-Canova T., Srivastava P., and Chattoraj D.K.. 2005. Multipartite regulation of rctB, the replication initiator gene of Vibrio cholerae chromosome II. J. Bacteriol. 187:7167–7175. 10.1128/JB.187.21.7167-7175.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwa L.G., Fan T.J., Tchaptchet S., Liu Y., Lussier Y.A., Sartor R.B., and Hansen J.J.. 2011. Chronic intestinal inflammation induces stress-response genes in commensal Escherichia coli. Gastroenterology. 141:1842–51.e1: 10. 10.1053/j.gastro.2011.06.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penders J., Thijs C., Vink C., Stelma F.F., Snijders B., Kummeling I., van den Brandt P.A., and Stobberingh E.E.. 2006. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 118:511–521. 10.1542/peds.2005-2824 [DOI] [PubMed] [Google Scholar]
- Pickard J.M., Zeng M.Y., Caruso R., and Núñez G.. 2017. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 279:70–89. 10.1111/imr.12567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prorok-Hamon M., Friswell M.K., Alswied A., Roberts C.L., Song F., Flanagan P.K., Knight P., Codling C., Marchesi J.R., Winstanley C., et al. . 2014. Colonic mucosa-associated diffusely adherent afaC+ Escherichia coli expressing lpfA and pks are increased in inflammatory bowel disease and colon cancer. Gut. 63:761–770. 10.1136/gutjnl-2013-304739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putze J., Hennequin C., Nougayrède J.P., Zhang W., Homburg S., Karch H., Bringer M.A., Fayolle C., Carniel E., Rabsch W., et al. . 2009. Genetic structure and distribution of the colibactin genomic island among members of the family Enterobacteriaceae. Infect. Immun. 77:4696–4703. 10.1128/IAI.00522-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., and Glöckner F.O.. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41(Database issue, D1):D590–D596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisch J., Buc E., Bonnet M., Sauvanet P., Vazeille E., de Vallée A., Déchelotte P., Darcha C., Pezet D., Bonnet R., et al. . 2014. Colon cancer-associated B2 Escherichia coli colonize gut mucosa and promote cell proliferation. World J. Gastroenterol. 20:6560–6572. 10.3748/wjg.v20.i21.6560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisch J., Rolhion N., Dubois A., Darfeuille-Michaud A., and Bringer M.A.. 2015. Intracellular colon cancer-associated Escherichia coli promote protumoral activities of human macrophages by inducing sustained COX-2 expression. Lab. Invest. 95:296–307. 10.1038/labinvest.2014.161 [DOI] [PubMed] [Google Scholar]
- Roediger W.E., and Truelove S.C.. 1979. Method of preparing isolated colonic epithelial cells (colonocytes) for metabolic studies. Gut. 20:484–488. 10.1136/gut.20.6.484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooks M.G., Veiga P., Reeves A.Z., Lavoie S., Yasuda K., Asano Y., Yoshihara K., Michaud M., Wardwell-Scott L., Gallini C.A., et al. . 2017. QseC inhibition as an antivirulence approach for colitis-associated bacteria. Proc. Natl. Acad. Sci. USA. 114:142–147. 10.1073/pnas.1612836114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein M.R., Wang X., Liu W., Hao Y., Cai G., and Han Y.W.. 2013. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 14:195–206. 10.1016/j.chom.2013.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T.. 1989. Molecular Cloning: A Laboratory Manual. Second edition Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 1546 pp. [Google Scholar]
- Sartor R.B., and Wu G.D.. 2017. Roles for Intestinal Bacteria, Viruses, and Fungi in Pathogenesis of Inflammatory Bowel Diseases and Therapeutic Approaches. Gastroenterology. 152:327–339.e4. 10.1053/j.gastro.2016.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassone-Corsi M., Nuccio S.P., Liu H., Hernandez D., Vu C.T., Takahashi A.A., Edwards R.A., and Raffatellu M.. 2016. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature. 540:280–283. 10.1038/nature20557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R.A., Richter M., Linder D., and Thauer R.K.. 1992. A tungsten-containing active formylmethanofuran dehydrogenase in the thermophilic archaeon Methanobacterium wolfei. Eur. J. Biochem. 207:559–565. 10.1111/j.1432-1033.1992.tb17082.x [DOI] [PubMed] [Google Scholar]
- Schulfer A.F., Battaglia T., Alvarez Y., Bijnens L., Ruiz V.E., Ho M., Robinson S., Ward T., Cox L.M., Rogers A.B., et al. . 2018. Intergenerational transfer of antibiotic-perturbed microbiota enhances colitis in susceptible mice. Nat. Microbiol. 3:234–242. 10.1038/s41564-017-0075-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears C.L., and Garrett W.S.. 2014. Microbes, microbiota, and colon cancer. Cell Host Microbe. 15:317–328. 10.1016/j.chom.2014.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Singh K., and Almasan A.. 2012. Histone H2AX phosphorylation: a marker for DNA damage. Methods Mol. Biol. 920:613–626. 10.1007/978-1-61779-998-3_40 [DOI] [PubMed] [Google Scholar]
- Sharp S.P., Malizia R.A., Walrath T., D’Souza S.S., Booth C.J., Kartchner B.J., Lee E.C., Stain S.C., and O’Connor W. Jr. 2018. DNA damage response genes mark the early transition from colitis to neoplasia in colitis-associated colon cancer. Gene. 677:299–307. 10.1016/j.gene.2018.08.016 [DOI] [PubMed] [Google Scholar]
- Simon R., Priefer U., Pühler A., and Engineering V.G.. 1983. A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Nat. Biotechnol. 1:784–791. 10.1038/nbt1183-784 [DOI] [Google Scholar]
- Small C.L., Reid-Yu S.A., McPhee J.B., and Coombes B.K.. 2013. Persistent infection with Crohn’s disease-associated adherent-invasive Escherichia coli leads to chronic inflammation and intestinal fibrosis. Nat. Commun. 4:1957 10.1038/ncomms2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spees A.M., Wangdi T., Lopez C.A., Kingsbury D.D., Xavier M.N., Winter S.E., Tsolis R.M., and Bäumler A.J.. 2013. Streptomycin-induced inflammation enhances Escherichia coli gut colonization through nitrate respiration. MBio. 4:e00430-13 10.1128/mBio.00430-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B., Robbiani R., Walker A.W., Westendorf A.M., Barthel M., Kremer M., Chaffron S., Macpherson A.J., Buer J., Parkhill J., et al. . 2007. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 5:2177–2189. 10.1371/journal.pbio.0050244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidsinski A., Khilkin M., Kerjaschki D., Schreiber S., Ortner M., Weber J., and Lochs H.. 1998. Association between intraepithelial Escherichia coli and colorectal cancer. Gastroenterology. 115:281–286. 10.1016/S0016-5085(98)70194-5 [DOI] [PubMed] [Google Scholar]
- Tjalsma H., Boleij A., Marchesi J.R., and Dutilh B.E.. 2012. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat. Rev. Microbiol. 10:575–582. 10.1038/nrmicro2819 [DOI] [PubMed] [Google Scholar]
- Tronnet S., Garcie C., Rehm N., Dobrindt U., Oswald E., and Martin P.. 2016. Iron Homeostasis Regulates the Genotoxicity of Escherichia coli That Produces Colibactin. Infect. Immun. 84:3358–3368. 10.1128/IAI.00659-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unden G., and Dünnwald P.. 2008. The Aerobic and Anaerobic Respiratory Chain of Escherichia coli and Salmonella enterica: Enzymes and Energetics. Ecosal Plus. 3 10.1128/ecosal.3.2.2 [DOI] [PubMed] [Google Scholar]
- Vázquez-Baeza Y., Pirrung M., Gonzalez A., and Knight R.. 2013. EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience. 2:16 10.1186/2047-217X-2-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viljoen K.S., Dakshinamurthy A., Goldberg P., and Blackburn J.M.. 2015. Quantitative profiling of colorectal cancer-associated bacteria reveals associations between fusobacterium spp., enterotoxigenic Bacteroides fragilis (ETBF) and clinicopathological features of colorectal cancer. PLoS One. 10:e0119462 10.1371/journal.pone.0119462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaino M.I., and Crawford J.M.. 2015. The colibactin warhead crosslinks DNA. Nat. Chem. 7:411–417. 10.1038/nchem.2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R.F., and Kushner S.R.. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 100:195–199. 10.1016/0378-1119(91)90366-J [DOI] [PubMed] [Google Scholar]
- Wang T., Cai G., Qiu Y., Fei N., Zhang M., Pang X., Jia W., Cai S., and Zhao L.. 2012a Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 6:320–329. 10.1038/ismej.2011.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Spandidos A., Wang H., and Seed B.. 2012b PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 40(Database issue, D1):D1144–D1149. 10.1093/nar/gkr1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M.R., Jiang Y., Villalta P.W., Stornetta A., Boudreau P.D., Carrá A., Brennan C.A., Chun E., Ngo L., Samson L.D., et al. . 2019. The human gut bacterial genotoxin colibactin alkylates DNA. Science. 363:eaar7785 10.1126/science.aar7785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R.P., Raffatellu M., Chessa D., Winter S.E., Tükel C., and Bäumler A.J.. 2008. The Vi-capsule prevents Toll-like receptor 4 recognition of Salmonella. Cell. Microbiol. 10:876–890. 10.1111/j.1462-5822.2007.01090.x [DOI] [PubMed] [Google Scholar]
- Winter S.E., and Bäumler A.J.. 2014. Dysbiosis in the inflamed intestine: chance favors the prepared microbe. Gut Microbes. 5:71–73. 10.4161/gmic.27129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter S.E., Thiennimitr P., Nuccio S.P., Haneda T., Winter M.G., Wilson R.P., Russell J.M., Henry T., Tran Q.T., Lawhon S.D., et al. . 2009. Contribution of flagellin pattern recognition to intestinal inflammation during Salmonella enterica serotype typhimurium infection. Infect. Immun. 77:1904–1916. 10.1128/IAI.01341-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter S.E., Winter M.G., Xavier M.N., Thiennimitr P., Poon V., Keestra A.M., Laughlin R.C., Gomez G., Wu J., Lawhon S.D., et al. . 2013. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 339:708–711. 10.1126/science.1232467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N., Yang X., Zhang R., Li J., Xiao X., Hu Y., Chen Y., Yang F., Lu N., Wang Z., et al. . 2013. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb. Ecol. 66:462–470. 10.1007/s00248-013-0245-9 [DOI] [PubMed] [Google Scholar]
- Wu S., Rhee K.J., Albesiano E., Rabizadeh S., Wu X., Yen H.R., Huso D.L., Brancati F.L., Wick E., McAllister F., et al. . 2009. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 15:1016–1022. 10.1038/nm.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng M.Y., Inohara N., and Nuñez G.. 2017. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 10:18–26. 10.1038/mi.2016.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., and Gladyshev V.N.. 2010. General trends in trace element utilization revealed by comparative genomic analyses of Co, Cu, Mo, Ni, and Se. J. Biol. Chem. 285:3393–3405. 10.1074/jbc.M109.071746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Lu Z., Wang R., Chen N., and Zheng P.. 2016. Establishing the colitis-associated cancer progression mouse models. Int. J. Immunopathol. Pharmacol. 29:759–763. 10.1177/0394632016670919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Zhang C., Xiao Q., Zhuang Y., Gu X., Yang F., Xing C., Hu G., and Cao H.. 2016. Effects of Different Levels of Molybdenum on Rumen Microbiota and Trace Elements Changes in Tissues from Goats. Biol. Trace Elem. Res. 174:85–92. 10.1007/s12011-016-0706-3 [DOI] [PubMed] [Google Scholar]
- Zhu W., Winter M.G., Byndloss M.X., Spiga L., Duerkop B.A., Hughes E.R., Büttner L., de Lima Romão E., Behrendt C.L., Lopez C.A., et al. . 2018. Precision editing of the gut microbiota ameliorates colitis. Nature. 553:208–211. 10.1038/nature25172 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.