Microglia are a heterogeneous population whose identity and function are dictated by signals from their microenvironment. Kana et al. show CSF-1 signaling is critical for cerebellar microglial transcriptional identity and homeostasis, and that altering the CSF-1–CSF-1R axis leads to motor and behavioral defects.
Abstract
Microglia, the brain resident macrophages, critically shape forebrain neuronal circuits. However, their precise function in the cerebellum is unknown. Here we show that human and mouse cerebellar microglia express a unique molecular program distinct from forebrain microglia. Cerebellar microglial identity was driven by the CSF-1R ligand CSF-1, independently of the alternate CSF-1R ligand, IL-34. Accordingly, CSF-1 depletion from Nestin+ cells led to severe depletion and transcriptional alterations of cerebellar microglia, while microglia in the forebrain remained intact. Strikingly, CSF-1 deficiency and alteration of cerebellar microglia were associated with reduced Purkinje cells, altered neuronal function, and defects in motor learning and social novelty interactions. These findings reveal a novel CSF-1–CSF-1R signaling-mediated mechanism that contributes to motor function and social behavior.
Introduction
Microglia, the resident macrophages of the central nervous system (CNS), form a continuous cellular network throughout the CNS that controls brain immune and inflammatory responses and contributes to the regulation of neuronal homeostasis (Li and Barres, 2018). Recent studies have revealed that the brain microenvironment is heterogeneous throughout the various brain regions, and that differences in neuronal subtypes, metabolism, and function could influence or be influenced by the microglia that reside in these regions (Grabert et al., 2016; Tay et al., 2017; Ayata et al., 2018), hence the need to study microglia in their regional context.
The cerebellum is an ancient part of the CNS known to control motor coordination and function (Manto et al., 2012), and is increasingly appreciated as an important regulator of higher cognitive functions (Wagner et al., 2017). However, most studies on neuronal–microglial interactions have focused on the forebrain (Parkhurst et al., 2013; Squarzoni et al., 2014; Schafer and Stevens, 2015), and very little is known about the regulation of the microglial network in the cerebellum, prompting us in this study to probe the mechanisms that regulate microglia homeostasis in the cerebellum.
Signaling through the CSF-1 receptor (CSF-1R) is required for microglia homeostasis (Stanley and Chitu, 2014), as suggested by the profound microglia depletion observed in Csf1r-deficient mice (Dai et al., 2002; Ginhoux et al., 2010), but surprisingly, mice that lack the main CSF-1R ligand, CSF-1 (Yoshida et al., 1990; Ginhoux et al., 2010), show only a moderate reduction of microglia (Wegiel et al., 1998). This discrepancy was explained in subsequent reports by the identification of an alternate CSF-1R ligand named IL-34 (Lin et al., 2008), which is produced by CNS populations in the forebrain and shown to control microglial maintenance in the forebrain, but not in the cerebellum (Greter et al., 2012; Stanley and Chitu, 2014). These results prompted us to revisit the distribution of CSF-1R ligands in the cerebellum and their contribution to cerebellar microglial maintenance.
Using a mouse model in which CSF-1 was specifically depleted from Nestin (Nes)+ cells, along with extensive profiling of microglia in different brain regions of the mouse and human brain, we found that the CSF-1R ligand, CSF-1, shaped cerebellar microglia identity and was uniquely required for microglial homeostasis in the cerebellum. Strikingly, disruption of cerebellar microglia in CSF-1–deficient brains was associated with profound alterations of Purkinje cell (PC) morphology and function, and with motor learning and social interaction defects.
Results
IL-34 and CSF-1 have nonoverlapping distribution in the forebrain and cerebellum, where they shape microglia numbers and phenotype
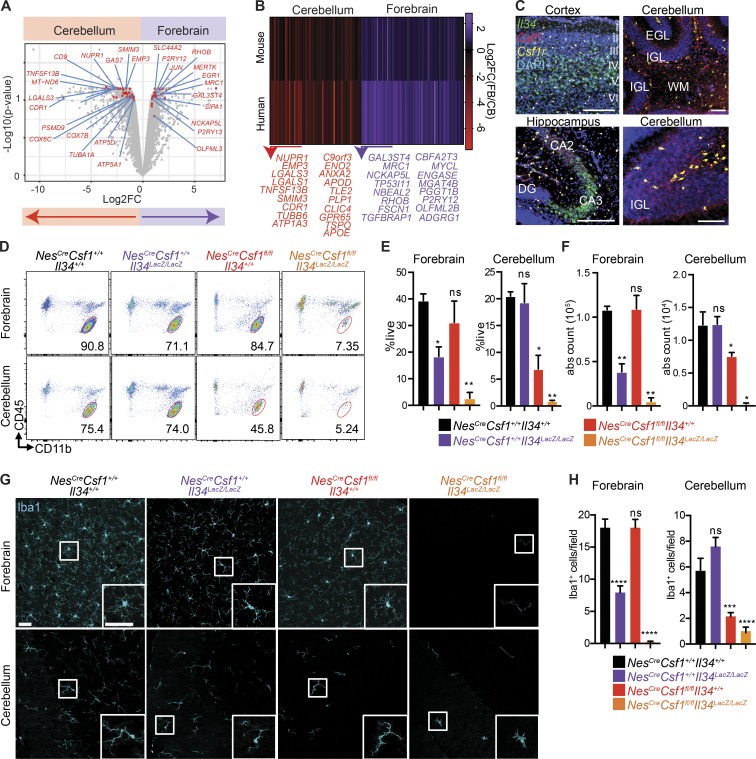
NextGen sequencing of microglia isolated from different human brain regions revealed that human cerebellar microglia expressed a transcriptional program distinct from that of forebrain microglia (Table S1 and Fig. 1, A and B), and were particularly enriched in metabolism and energy genes (COX7B, ATP5A1, and PSMD9), a conserved functional network previously described in mouse cerebellar microglia (Grabert et al., 2016; Ayata et al., 2018). Many orthologous genes more highly expressed in human cerebellar compared with forebrain microglia were also more highly expressed in mouse cerebellar microglia (NUPR1, EMP3, CDR1, LGALS3, and APOE; Fig. 1 B), suggesting that similar tissue factors might imprint the cerebellar microglial program in human and mouse brains.
Figure 1.
Cerebellar microglia depend on CSF-1 growth factor for their maintenance in tissue. (A) Volcano plot showing select DEGs between human cerebellar and forebrain (superior temporal gyrus) microglia. n = 2 matched patient samples/brain region. (B) Heat map and representative genes of adult human and mouse orthologous genes with conserved fold change in cerebellar and forebrain cortical microglia. (C) Representative images from single-molecule RNA in situ hybridization for Il34, Csf1, and Csf1r in WT P8 cerebral cortex, hippocampus, and cerebellum. I–VI, cortical layers; CA, cornu ammonis; DG, dentate gyrus; EGL, external granular layer; IGL, internal granular layer; WM, white matter. Scale bars, 100 µm. (D–F) Representative flow-cytometric plot (D) and quantification of the percent of live (E) and absolute (abs) count (F) of forebrain and cerebellar microglia (defined as doublet−DAPI−CD11b+CD45int). n = 3–7 adult mice/group. Data are pooled from five independent experiments. ns, not significant. (G) Representative confocal images of Iba1 (cyan) immunofluorescence stainings. Images representative of n = 3 adult mice/group. Scale bar, 20 µm. (H) Quantification of Iba1+ microglia/field in cortex and cerebellum. n = 3 adult mice/group, n = 3 or 4 fields/region/mouse. Data are a representative of two independent experiments. DEGs in A defined as Benjamini–Hochberg adjusted P-value < 0.1. Conserved genes in B defined as one to one orthologues expressing >5 TPM in all replicates of at least one brain region in both species, and having an unadjusted P-value < 0.05 and a log2FC > 0.5 in the same direction in both species (log2FC > 0.75 in the same direction for listed genes). Graphs show mean ± SEM. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P < 0.0001 using multiple Student’s t tests (E and F) or ANOVA with Tukey’s post hoc test (H).
Although IL-34 and CSF-1 signal through the same CSF-1R and can both promote macrophage survival, they have distinct distributions in the brain (Stanley and Chitu, 2014). Using RNA in situ hybridization (Fig. 1 C) and cell type–specific translating ribosome affinity purification (TRAP; Fig. S1 A), we found that Il34 expression dominated in forebrain regions, such as the cerebral cortex and the CA3 region of the hippocampus, while Csf1 was most strongly expressed in the cerebellum (Fig. 1 C). The cellular source of Il34 and Csf1 also differed, as glial cells highly expressed Csf1, while Il34 was mainly produced by neurons (Fig. S1 A). Csf1r expression, however, was prominent in microglia throughout the brain (Fig. 1 C). As in mouse brains, human forebrain regions mainly expressed IL34, whereas CSF1 predominated in the cerebellum (Fig. S1 B).
Given the distinct spatial distribution of Csf1 and Il34 in the brain parenchyma, we tested how the specific depletion of each ligand affected microglia in different brain regions. Because Csf1op/op mice, which bear a spontaneous CSF-1 null mutation, have bone and musculoskeletal developmental defects that may indirectly affect brain development (Yoshida et al., 1990), we crossed NesCre/+ mice to Csf1fl/fl mice (NesCreCsf1fl/fl) and significantly reduced Csf1 specifically in the cerebellum, while Il34 expression remained unaffected (Fig. S1 C). In the brain, Nes+ cells mostly consist of CD45− cells, including neuronal and glial progenitor cells, but not microglial cells (Fig. S1 D). Generation of NesCreCsf1fl/fl mice allowed for Csf1 depletion from all cell types deriving from neuroectodermal stem cells, including astrocytes, one of the main sources of Csf1 (Fig. S1 A), as well as oligodendrocytes, and neurons, thus circumventing the development defects observed in Csf1op/op mice.
In contrast to Il34LacZ/LacZ-deficient mice, which have reduced microglia in forebrain regions, including the cerebral cortex and hippocampus (Greter et al., 2012; Wang et al., 2012), NesCreCsf1fl/fl mice had normal forebrain microglia frequencies and numbers, but significantly reduced microglia in the cerebellum (Fig. 1, D–H), thus mirroring the expression pattern of Il34 and Csf1 in their respective brain regions. Importantly, CSF-1 depletion from microglia in Cx3cr1CreCsf1fl/fl mice had no defects in the cerebellar microglial compartment (Fig. S1, E and F), establishing that a microglial CSF-1 source is not required for microglial homeostasis in the cerebellum, and excluding a relevant impact of transient Cx3cr1 expression during neuronal development (Haimon et al., 2018) in Cx3cr1CreCsf1fl/fl mice. We also generated knockout mice that lacked both CSF-1 and IL-34 (NesCreCsf1fl/flIl34Lacz/Lacz) and found almost a complete depletion of microglia within all regions of the brain (Fig. 1, D–H), establishing the critical and nonredundant roles for CSF-1 and IL-34 in microglial homeostasis, as well as the significant contribution of CSF-1 to the maintenance of cerebellar microglia.
Disruption of CSF-1–CSF-1R signaling predominantly affects cerebellum homeostasis
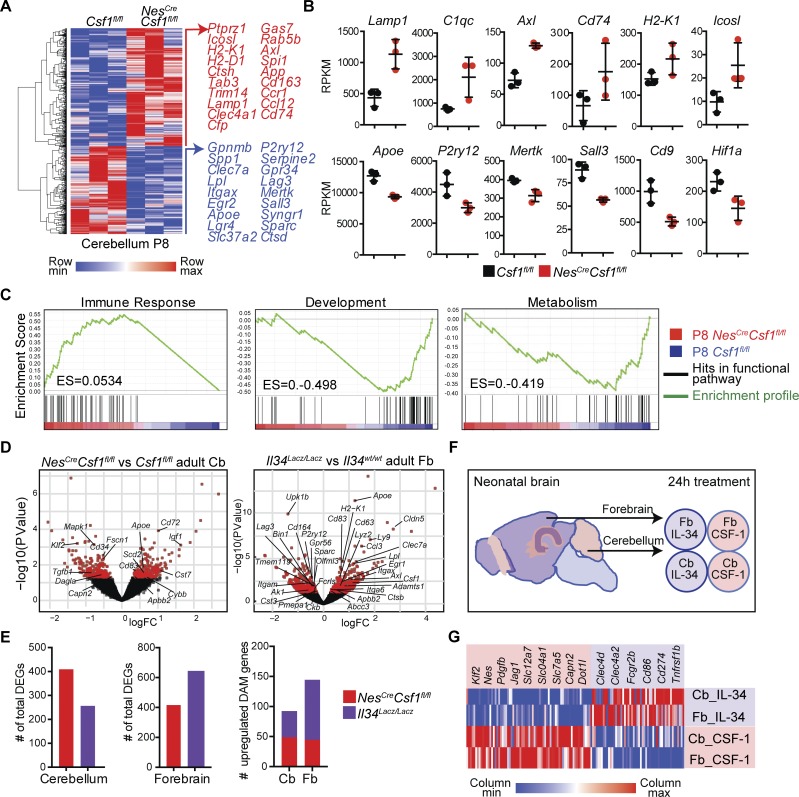
NextGen profiling of the few cerebellar microglia that persisted in NesCreCsf1fl/fl mice at postnatal day 8 (P8), the peak of cerebellar foliation and lengthening of cerebellar folia (Sudarov and Joyner, 2007; Fig. 2 A and Fig. S2, A–C) revealed a significantly altered transcriptional program (Table S2 and Fig. 2, A–C), including down-regulation of homeostatic and sensome genes (Mertk, Gpr34, and P2ry12; Matcovitch-Natan et al., 2016), growth and development genes (Egr2, Spp1, and Cd9), and metabolism genes (Hif1a and Slc37a2; Fig. 2, A and C). Many down-regulated genes (Apoe, Lpl, Spp1, Itgax, Gpnmb, and Clec7a) are also known to be developmentally regulated in microglia (Matcovitch-Natan et al., 2016). There was also a striking up-regulation of immune response genes (Axl, Lamp1, and Clec4a1; Fig. 2, A–C). Down-regulation of growth, development, and metabolism genes (Klf2, Notch1, Tgf1, Dagla, Dotl1, and Capn2) also persisted in adult CSF-1–deficient mice (Fig. 2 D). In contrast, adult forebrain microglia in IL-34–deficient mice acquired a gene signature found in aging and neurodegenerative diseases, also known as a disease-associated microglial (DAM) signature (Keren-Shaul et al., 2017; Fig. 2 D). The DAM signature was more prevalent in adult IL-34–deficient brain compared with adult CSF-1–deficient brain (Fig. 2 E), consistent with prior reports that human Alzheimer’s postmortem brains have reduced IL-34 levels particularly in the forebrain, and not the cerebellum (Walker et al., 2017). Accordingly, we found that ex vivo exposure of neonatal microglia (Fig. 2 F) to CSF-1 or IL-34 drives a common transcriptional program, whether the microglia were isolated from the cerebellum or forebrain (Table S3, Fig. S2 D, and Fig. 2 G), particularly with CSF-1 inducing development and metabolism genes (Klf2, Nes, and Jag1) and IL-34 inducing immune genes (Clec4a2, Cd86, and Tnfrsf1b; Fig. 2 G), indicating that IL-34 and CSF-1 are among the key drivers of regional microglial transcriptional profiles.
Figure 2.
CSF-1 and IL-34 drive distinct microglial programs in the brain tissue. (A) Hierarchical clustering (left) and select representative genes (right) from 860 DEGs between P8 Csf1fl/fl and NesCreCsf1fl/fl cerebellar microglia. (B) Normalized expression (RPKM) of select immune response and homeostatic and metabolism genes in Csf1fl/fl and NesCreCsf1fl/fl cerebellar microglia. n = 3 mice/group. (C) GSEA analysis of P8 NesCreCsf1fl/fl cerebellar microglia showing positive enrichment of genes involved in immune response pathways, and negative enrichment of genes involved in developmental and metabolic pathways. (D) Volcano plots showing select development, metabolism, and DAM genes in adult NesCreCsf1fl/fl cerebellar microglia (left) and select homeostatic and DAM genes in adult Il34Lacz/Lacz forebrain microglia (right). (E) Quantification of total DEGs in adult NesCreCsf1fl/fl and Il34Lacz/Lacz microglia from cerebellum (Cb; left) and forebrain (Fb; middle) and total number of up-regulated intersecting genes from DAM and CSF-1R ligand deficient microglia (right). (F and G) Experimental design (F) used to generate data for heat map (G) of select representative DEGs involved in growth, differentiation, metabolism (red), and immune response (purple) pathways induced in WT neonatal cortical and cerebellar microglia stimulated with either 100 ng/ml IL-34 or 20 ng/ml CSF-1 for 24 h. DEGs defined as read cutoff > 10, P-value < 0.05 (A and E), and LogFC(TPM) > 0.25, Benjamini–Hochberg adjusted P-value < 0.05 (G). Graphs show means ± SEM.
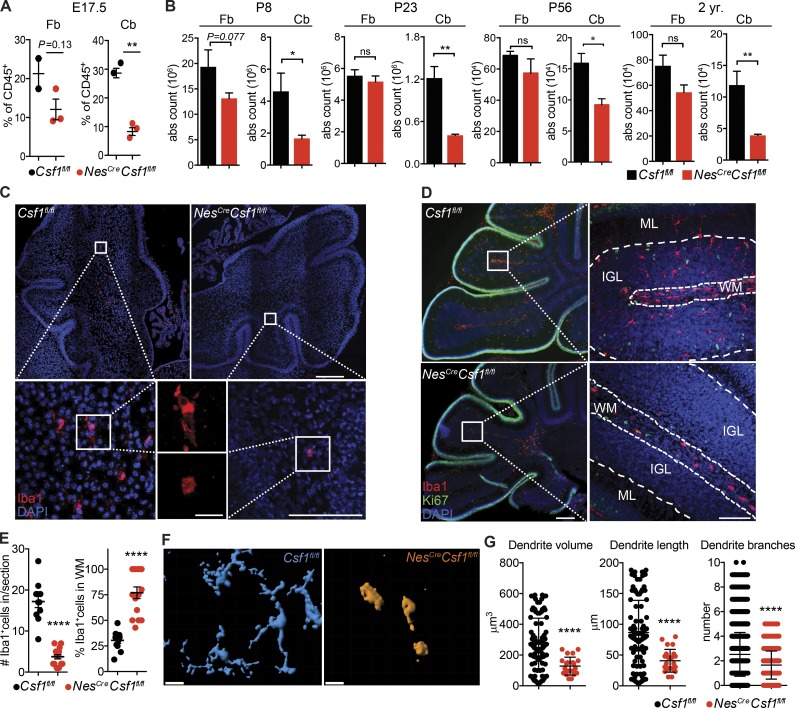
Using congenic parabiotic mice, we confirmed that cerebellar microglia were maintained locally (Fig. S3 A), independent of adult hematopoiesis, as previously described for whole brain microglia (Ajami et al., 2007; Ginhoux et al., 2010). This was confirmed with flow cytometry showing no differences in NesCreCsf1fl/fl cerebellar microglia expression of Tmem119, a microglial marker absent on peripherally derived monocytes and macrophages (Fig. S3 B; Bennett et al., 2016), therefore excluding that peripherally derived monocytes accounted for the remaining microglia in NesCreCsf1fl/fl cerebella. We also confirmed these cells were not replaced by perivascular macrophages, as there were no changes in key genes reported to be specifically up-regulated in these border-associated cells (Mrc1, Cd36, Lyve1, Slc40a1, and Hpgd; Goldmann et al., 2016; Fig. S3 C). Reduction of microglia was already observed at embryonic day 13.5 (E13.5) in NesCreCsf1fl/fl brain rudiments (Hoeffel et al., 2015; Fig. S3, D and E; and Fig. S5), remained low at E17.5 (Fig. S3 F), and was more prominent in the cerebellum than in the forebrain (Fig. 3 A). However, macrophages were not affected in the yolk sac, fetal liver, or fetal limbs of NesCreCsf1fl/fl mice (Fig. S3, G and H). In contrast, Il34LacZ/LacZ mice did not exhibit reduced microglia during embryonic development as previously reported (Greter et al., 2012; Fig. S3 I). Cerebellar microglia remained reduced throughout life in NesCreCsf1fl/fl mice (Fig. 3 B). Interestingly, forebrain microglia in NesCreCsf1fl/fl mice, while reduced in embryos, recovered by P23 (Fig. 3 B), suggesting a developmental switch in growth factor demand and an acquired dependence on the Il34 ligand (Fig. S3, I and J).
Figure 3.
CSF-1 controls microglia morphology, spatial distribution, and development during embryonic and adult life. (A) Flow-cytometric quantification of doublet−DAPI−CD45+Ly6G−CD11bloF4/80hi microglia from E17.5 Csf1fl/fl and NesCreCsf1fl/fl forebrain (Fb) and cerebella (Cb). n = 3–5 mice/group. Data are pooled from two independent experiments. (B) Quantification of doublet−DAPI−CD11b+CD45int microglia from P8, P23, P56, and 2-yr-old Csf1fl/fl and NesCreCsf1fl/fl forebrain and cerebellum. n = 3–5 mice/group. Data are pooled from at least two independent experiments. (C) Representative immunofluorescence stainings of Iba1+ microglia in newborn (P0.5) pups, in control, and NesCreCsf1fl/fl cerebella. Image representative of n ≥ 3 mice/group from two independent litters. Scale bars, 500 μm (upper right), 20 μm (lower middle), and 100 μm (lower right). (D) Representative immunofluorescence stainings showing morphological and spatial distribution of Iba1+ microglia in P8 Csf1fl/fl and NesCreCsf1fl/fl cerebella. Ki67+ cells, proliferating external granule cell layer; ML, molecular layer. Scale bars, 200 µm (left) and 100 µm (right). Image representative of n = 3 mice/group of at least three independent experiments. White segmented lines define borders of layers. (E) Quantification of Iba1+ microglia within P7 Csf1fl/fl and NesCreCsf1fl/fl cerebellar white matter and cortex (left), and percentage of microglia located only in the white matter (right). n = 3 mice/group, four sections/mouse, one representative experiment of two independent experiments. (F and G) Imaris automated 3D reconstruction (F) and process quantification (G) of Iba1+ microglia in P8 Csf1fl/fl (blue) and NesCreCsf1fl/fl (orange) cerebella. Scale bar, 10 µm. n = 3 mice/group, n = 12–34 cells/genotype. Graphs show mean ± SEM. *, P ≤ 0.05; **, P ≤ 0.01; ****, P < 0.0001 using Student’s t test.
Imaging of the cerebellum of NesCreCsf1fl/fl mice at birth (P0.5; Fig. 3 C) and at P6–8 (Fig. 3 D) revealed scarce microglia with a rounded, amoeboid morphology confined to the cerebellar white matter (Fig. 3 E), with significant alterations in microglial process morphology, including volume, length, and number of branches (Fig. 3, F and G). Control microglia, however, were ramified and distributed throughout the white matter, the PC layer, the molecular layer, and the developing granule cell layers (Fig. 3, C–G).
CSF-1–deficient mice have cerebellar alterations and behavioral defects
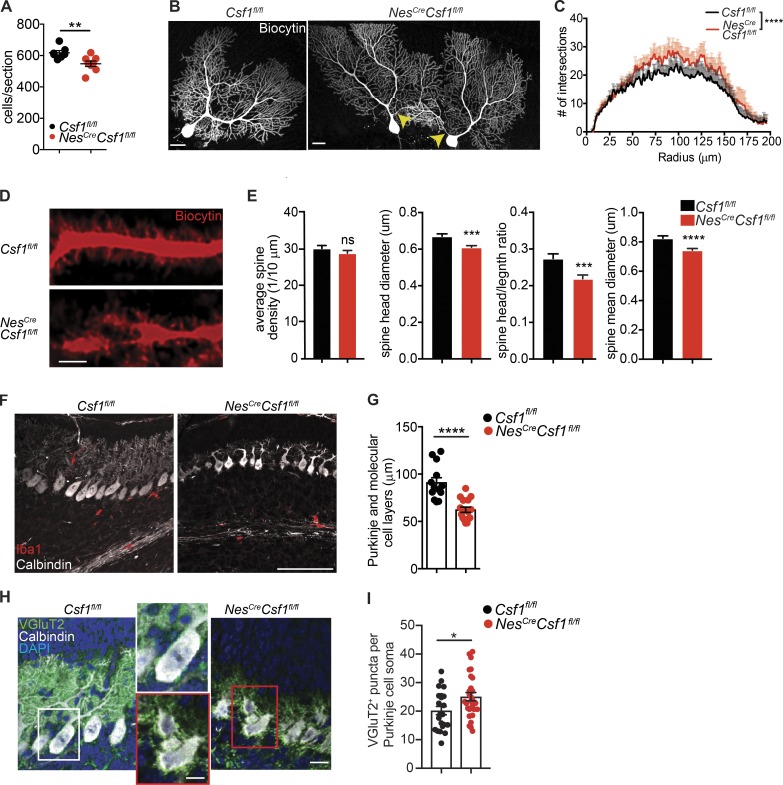
Magnetic resonance imaging (MRI) of NesCreCsf1fl/fl brains revealed increased brain mass, reduced brain size and ventricle volume (Fig. S4, A and B), and increased cerebellar volume (Fig. S4 B), consistent with prior reports in Csf1op/op and Csf1r−/− mice (Nandi et al., 2012). Strikingly, NesCreCsf1fl/fl mice had a significant reduction of PC numbers (Fig. 4 A and Fig. S4 C), which was also confirmed in CSF-1–deficient Csf1op/op mice (Fig. S4 D). In contrast to previous reports (Murase and Hayashi, 2002; Nandi et al., 2012), Csf1r was not expressed by calbindin+ (calb+) PCs (Fig. S4, E and F). We confirmed that Csf1r was mainly expressed in microglial cells using TRAP (Fig. S4 G), as well as with RT-PCR (Fig. S4 H). As in mouse brains, human PCs also lacked CSF1R expression (Fig. S4 I). PCs form a continuous cellular layer within the cerebellar cortex, and their dendritic trees expand into the cerebellar molecular layer. While the main excitatory inputs of the cerebellar cortex originate from climbing and parallel fibers, the only cerebellar output is from PC axons, and their synaptic plasticity is required for successful motor learning (Nguyen-Vu et al., 2013). Imaging of NesCreCsf1fl/fl PCs revealed an increase in aberrant dendrites emerging from PC somas (Fig. 4 B and Fig. S4, J and K), and overall increased branching assessed by dendrite branching complexity quantification. There was no change in dendritic spine density (Fig. 4, D and E), but there was decreased mean spine diameter, spine head diameter, and mean spine head volume-to-length ratio (Fig. 4 E), suggesting a defect in PC neural circuit formation. Climbing fiber elimination is a neurodevelopmental process ensuring that each PC ultimately receives inputs from a single climbing fiber, a process that starts in the first postnatal week (Kano et al., 2018). Developing microglia cluster below the PC layer at P7, and participate in climbing fiber elimination (Nakayama et al., 2018). At P7, NesCreCsf1fl/fl, PC dendrites were shorter, resulting in a thinner molecular cell layer (Fig. 4, F and G), suggesting an altered and delayed PC maturation. PC soma were devoid of microglia interactions (Fig. 3, D and E; and Fig. 4 F) and contained more vesicular glutamate transporter 2 (VGluT2)+ climbing fiber puncta (Fig. 4, H and I), suggesting decreased climbing fiber elimination.
Figure 4.
NesCreCsf1fl/fl mice have morphological PC alterations. (A) Quantification of PC numbers per sagittal section in 6-wk-old Csf1fl/fl and NesCreCsf1fl/fl cerebella, stained with H&E. n = 4 mice/group, one or two sections/mouse, pooled from two independent experiments. (B and C) Representative whole cell imaging (B) and dendrite branching complexity quantification (C) of biocytin-filled and fluorophore-conjugated–streptavidin-stained PC. Arrows point to aberrant dendrites emanating from NesCreCsf1fl/fl PC soma. Scale bars, 20 µm. n = 3 mice/genotype, n = 7 or 8 cells/group, pooled from three independent patch experiments. Only dendrite branching up to 200 µm radii from PC soma center was quantified, as dendrites beyond this distance may not have been reliably filled with biocytin. (D) Representative confocal images of biocytin-labeled PC dendrites and spines from Csf1fl/fl and NesCreCsf1fl/fl cerebella. Scale bars, 2 µm. (E) Imaris quantification of PC dendritic spines in Csf1fl/fl and NesCreCsf1fl/fl cerebella. (D and E) n = 3 mice/genotype, n = 3 neurons/mouse, n = 5 dendrites/neuron, pooled from three independent patch experiments. (F and G) Representative confocal images of calb+ PC and Iba1+ microglia (F) and quantification of PC and molecular cell layer thickness (G) in P7 Csf1fl/fl and NesCreCsf1fl/fl cerebella. n = 3 mice/group, four to six sections/mouse. Scale bar, 100 μm. (H and I) Representative confocal images of VgluT2+ puncta on calb+ PC soma (H) and quantification of VGluT2+ puncta per PC soma (I) in P7 Csf1fl/fl and NesCreCsf1fl/fl cerebella. n = 3 or 4 mice/group, four to six sections/mouse. Scale bars, 10 μm (right panel) and 5 μm (middle panel). Graphs show means ± SEM. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001, using Student’s t test (A, E, G, and I), and two-way ANOVA (C).
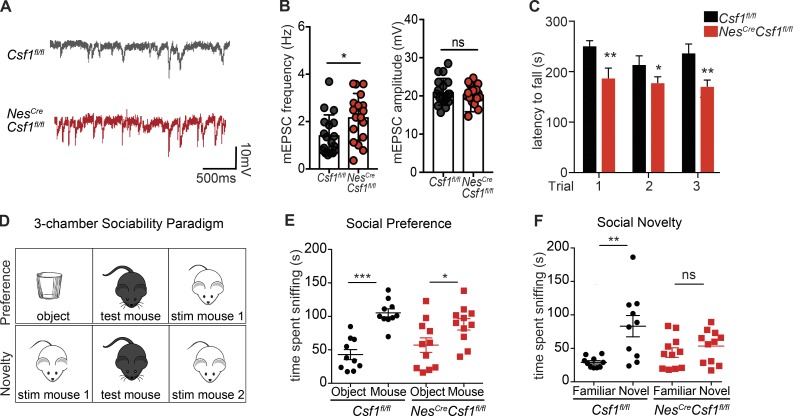
Electrophysiological analysis of PCs in 6-wk-old mice showed increased spontaneous miniature excitatory postsynaptic currents (mEPSCs), with no differences in amplitude (Fig. 5, A and B), consistent with excess dendrite branching observed in the remaining PCs in these mice. Strikingly, NesCreCsf1fl/fl mice showed mild signs of ataxia (Fig. S4 L), and when tested for motor learning capacities during repeated trials on an accelerating rotarod, 5–7-wk-old NesCreCsf1fl/fl mice displayed impairment on the fourth training day (Fig. 5 C), suggesting a motor learning deficit consistent with cerebellar dysfunction. Besides its well-known role in motor coordination and function, the cerebellum is increasingly appreciated as an important regulator of higher cognitive functions (Wagner et al., 2017). We therefore performed two behavioral tests, social preference and social novelty, in NesCreCsf1fl/fl mice using the three-chamber sociability paradigm (Fig. 5 D). Both Csf1fl/fl and NesCreCsf1fl/fl mice showed no defects in the social preference test, both spending significantly more time interacting with the stimulus mouse over the inanimate cup (Fig. 5 E). However, NesCreCsf1fl/fl mice failed to preferentially interact with a novel stimulus mouse (Fig. 5 F), indicating a deficit in social memory, a behavioral defect commonly found in mouse models of neurodevelopmental and neuropsychiatric disorders, such as autism spectrum disorders (ASDs).
Figure 5.
NesCreCsf1fl/fl mice show electrophysiological PC alterations and develop motor and behavioral defects. (A and B) Representative traces (A) and quantification (B) of PC mEPSC frequencies and amplitudes in NesCreCsf1fl/fl PCs compared with controls. n = 3 mice/genotype, n = 6 cells/mouse. Data are a pool of three independent patch experiments. (C) Quantification of the latency for Csf1fl/fl and NesCreCsf1fl/fl mice to fall from accelerating rotating beam. n = 12–15 mice/group, pooled from two independent experiments. (D–F) Schematic (D) and quantification (E and F) of the three-chamber sociability paradigm used to assess defects in social preference (E) and social novelty (F) in Csf1fl/fl and NesCreCsf1fl/fl mice. stim, stimulus. n = 10–12 mice/group, pooled from two independent experiments. Graphs show means ± SEM. ns, not significant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001, using multiple Student’s t tests (A) and one-way ANOVA (C and D).
Discussion
Cerebellar microglia display a unique morphological (Ashwell, 1990; Lawson et al., 1990) and transcriptional (Grabert et al., 2016; Ayata et al., 2018) profile, but little is known about their role in cerebellar development, homeostasis, and function. Here, we explored the role of the macrophage growth factor CSF-1 for microglia development and maintenance in the cerebellum, and the consequences of nervous system CSF-1 depletion for cerebellar development and function. We show that despite both signaling through the same receptor, CSF-1R, the distinct expression patterns of IL-34 and CSF-1 shape microglia numbers and phenotypes in the forebrain and cerebellum. Genetic depletion of Il34 or Csf1 expression resulted in mutually exclusive reductions of postnatal microglia numbers, with IL-34 as the main forebrain cytokine, and CSF-1 as the main cerebellum cytokine. CSF-1–deficient animals, which lack cerebellar microglia, displayed alterations in cerebellar morphology, PC loss with increased branching complexity, and motor learning and sociability defects.
Microglia form a grid-like pattern throughout the brain where each microglia controls its own territory that is not shared with others (Kettenmann et al., 2011), facilitating a fine-tuned microglia–neuron interaction. Strikingly, NesCreCsf1fl/fl microglia were largely confined to central white matter regions of the cerebellum, instead of being evenly distributed throughout the brain, suggesting a migration defect during cerebellar development. The progressive radial distribution of microglia during cerebellar development and growth has been studied in the quail brain (Cuadros et al., 1997), but the mechanism of distribution of microglia throughout the brain during development is largely unknown. Cerebellar astrocytes express Csf1 (Fig. S1 A), a known chemotactic factor for macrophages (Calvo et al., 1998; Pixley and Stanley, 2004). Because cerebellar astrocytes extend their long and ordered cellular processes throughout several cerebellar layers, they might thus serve as guiding structures for developing microglia. Our data, indeed, suggest that microglia may use CSF-1–secreting cells as a scaffold for radial migration into the gray matter, since the absence of neuroectodermal CSF-1 production left remaining microglia stuck in the central white matter of the cerebellum.
Intriguingly, functional pathways induced in postnatal forebrain and cerebellar microglia stimulated with either IL-34 or CSF-1 mirrored the altered biological pathways of microglia from IL-34 or CSF-1–deficient mice, revealing that CSF-1 and IL-34 are among the key drivers of regional microglial transcriptional signatures. Even more interestingly, microglia isolated from mice that underwent maternal immune activation in utero were also shown to up-regulate gene networks involved in embryonic development (Mattei et al., 2017), similar to those described here by CSF-1 treatment, suggesting that increased CSF-1 expressed during maternal immune activation could potentially contribute to this transcriptional phenotype.
Due to their important role in brain development and homeostasis, microglia are involved in virtually all pathological conditions affecting the CNS (Hickman et al., 2018; Li and Barres, 2018). Accordingly, imaging as well as post-mortem studies show that microglia are activated in both multisystem atrophy and bipolar disorder (Watkins et al., 2014; Vieira et al., 2015; Kübler et al., 2019), which affected the microglia donors analyzed here (Fig. 1, A and B). Nevertheless, we found clear, fundamental, and biologically relevant transcriptional differences between human cerebellar and forebrain microglia, regardless of clinical disease. These differences were preserved in mouse forebrain and cerebellar microglia (Fig. 1 C), supporting previous studies reporting regional transcriptional differences in mouse microglia phenotypes (Grabert et al., 2016; Ayata et al., 2018).
We also show that CSF-1–deficient animals, which lack cerebellar microglia, display alterations in cerebellar morphology, PC number and function, and motor learning and sociability defects. Previous studies have shown that microglia serve a key role in proper cerebellar climbing fiber elimination, both shifting the excitatory drive and decreasing the inhibitory drive onto PCs, starting early in cerebellar development and lasting into adulthood (Nakayama et al., 2018). Indeed, NesCreCsf1fl/fl cerebella have increased VGluT2 excitatory inputs onto developing (P7) PC soma compared with controls, suggesting a defect in early-stage climbing fiber elimination (Ichikawa et al., 2016). Moreover, reduced pruning due to microglia depletion in CSF-1–deficient cerebella could be a potential mechanism by which adult NesCreCsf1fl/fl PC branching is increased, and thereby also contributing to the increased firing potential (mEPSC) seen in their PCs. Not only can increased excitatory drive result in neuronal, and particularly PC death (Johnston, 2005; Slemmer et al., 2005), but altered climbing fiber elimination can also lead to cerebellar motor dysfunction and ataxia (Johnson et al., 2007). Lastly, increasing evidence suggests that cerebellar dysfunction may also contribute to a subset of ASD (Fatemi et al., 2012), a heterogeneous group of disorders, which presents a substantial challenge to diagnosis and treatment. Accordingly, postmortem brain studies of patients with ASD have consistently reported reduced PC counts in the cerebellar cortex (Bauman and Kemper, 2005; Fatemi et al., 2012), and cerebellar injury is linked to increased incidence of ASD (Limperopoulos et al., 2007). Moreover, studies in mouse models have shown that genetic disruption of cerebellar PC function can lead to similar social interaction defects as shown in CSF-1–deficient cerebella (Tsai et al., 2012).
Previous reports suggested that PCs express Csf1r (Murase and Hayashi, 2002; Nandi et al., 2012). However, we found no Csf1r or CSF1R expression in mouse and human PCs, respectively, using multiple techniques and different ages, providing evidence for microglial deregulation as the single driver for the neuronal and behavioral perturbations observed in our CSF-1 deficiency model. In addition, Iba1CreCsf1rfl/fl mice showed similar PC perturbations (Nakayama et al., 2018), again supporting a microglial role for PC development and regulation. Further studies must be performed to exclude a transient but relevant PC Csf1r expression during embryonic or early postnatal development.
The distinct distribution of IL-34 and CSF-1, and the distinct regional microglial transcriptional signatures described here, are conserved in both the mouse and human brain, highlighting the importance of this discrete expression and signaling for microglial biology. Altogether these results reveal the CSF-1–CSF-1R axis as a critical regulator of cerebellar development and integrity and identify the CSF-1–CSF1-R axis as a potential therapeutic target to help restore cerebellar microglial and PC function, and to improve developmental motor and sociability disorders.
Materials and methods
Animals
NesCre, Cx3cr1Cre, C57BL/6 CD45.2 and CD45.1, ubiquitin-GFP, and R26-stop-EYFP were purchased from the Jackson Laboratory. Csf1fl/fl mice were provided by Sherry Abboud Werner (University of Texas Health Science Center, San Antonio, TX; Harris et al., 2012). Il34Lacz/Lacz mice were generated by the European Conditional Mouse Mutagenesis Program (International Knockout Mouse Consortium project ID: 33127; provided by Marco Colonna, Washington University School of Medicine, St. Louis, MO; Wang et al., 2012). NesCreCsf1fl/fl and NesCreCsf1fl/flIl34LacZ/LacZ mice were generated in house by crossing NesCre, Csf1fl/fl, and Il34Lacz/Lacz mouse lines. Csf1op/op were originally obtained from the Jackson Laboratory on an outbred C57/BL/J C3Heb/FeJ-a/a CD1 background and backcrossed for ≥10 generations onto the FVB/NJ background. Tg(Aldh1l1-eGFPL10a)JD130, Tg(Pcp2-eGFPL10a)DR168, Tg(NeuroD1-eGFPL10a)JP241, Tg(Drd1-eGFPL10a)CP73, and Tg(Drd2-eGFPL10a)CP101 mice were used for astrocyte-, PC-, granule cell–, D1 neuron–, and D2 neuron–TRAP, respectively (Doyle et al., 2008; Heiman et al., 2008). Eef1a1LSL.eGFPL10a/+ mice, which carry a loxP-flanked STOP cassette (LSL) upstream of the eGFP-L10a gene under the control of eukaryotic translation elongation factor 1 α 1, Eef1a1 (provided by Ana Domingos, University of Oxford, Oxford, UK; Stanley et al., 2013), were crossed with Cx3cr1CreErt2/+(Litt) (provided by Dan Littman, New York University School of Medicine, New York, NY; Parkhurst et al., 2013) to generate a microglia-specific TRAP mouse. Microglia-specific mRNA enrichment was validated (Ayata et al., 2018). To activate tamoxifen-inducible Cre (CreErt2 or Cre/Esr1*), mice were gavaged at 4–6 wk of age with five doses of 100 mg/kg of tamoxifen (T5648; Sigma-Aldrich) in corn oil (C8267; Sigma-Aldrich) with a separation of ≥48 h between doses. For timed embryo studies, male and female mice were bred overnight and vaginal plugs looked for the following morning, with this stage considered E0. Mice were housed at two to five animals per cage with a 12-h light/dark cycle (lights on from 0700 to 1900 h) at constant temperature (23°C) with ad libitum access to food and water. All animal protocols were approved by the Institutional Animal Care and Use Committee at the Icahn School of Medicine at Mount Sinai and were performed in accordance with National Institutes of Health guidelines.
RNAscope in situ hybridization
Expression and localization of Csf1r, Csf1, and Il34 was assessed using RNAscope in situ hybridization technology (ACD) following the manufacturer’s instructions. The multiplexed assay was performed on fixed frozen brain, cut in the sagittal plane at 14-μm thickness. Proprietary probes to Csf1r, Il34, and Csf1 were hybridized to their respective target RNA for 2 h at 40°C and their signal amplified for 15–30 min at 40°C. The last amplification step contained the fluorophores used for visualization, Alexa Fluor 488 for Il34, Atto550 for Csf1, and Atto647 for Csf1r. Slides were then counter-stained with DAPI for 2 min, mounted with ProLong Gold (Invitrogen), and cover-slipped. Hybridized brain slices were visualized using confocal microscopy (Zeiss) and NanoZoomer whole-slide imaging software (Hamamatsu).
Single-molecule RNA in situ hybridization with immunofluorescence
6-wk-old WT mice (n = 5) were anesthetized with ketamine (120 mg/kg) and xylazine (24 mg/kg) and perfused transcardially with 10 ml PBS followed by 40 ml 10% formalin in PBS. The brains were then removed and fixed in 10% neutral-buffered formalin for another 48 h. Fixed brains were embedded in paraffin, sectioned at 5 µm, mounted on Superfrost Plus slides (Thermo Fisher Scientific), baked in a dry oven for 1 h at 60°C, and stored at room temperature until further processing. In situ hybridization was performed using RNAScope custom-designed probes for Csf1r in combination with the RNAScope 2.0 Red kit following the manufacturer’s recommendation (Advanced Cell Diagnostics). After completing in situ hybridization before colorimetric reaction, sections were rinsed with ddH2O, 0.1× PBS, 0.25× PBS, 0.5× PBS, and 1× PBS, blocked in 2% normal goat serum in PBS for 1 h at room temperature, and incubated with calbindin D-28 antibody (dilution 1:5,000; Swant) in 2% normal goat serum in PBS overnight at 4°C. Calbindin signal was amplified with anti-rabbit horseradish peroxidase–conjugated secondary antibody and Alexa Fluor 488 tyramide from the Tyramide Signal Amplification Kit (Life Technologies) following the manufacturer’s instructions, and nuclei were stained with DAPI (0.2 mg/ml). Sections were then rinsed with 1× PBS, 0.5× PBS, 0.25× PBS, 0.1× PBS, and ddH2O; subjected to colorimetric reaction; dried 15 min at 60°C; mounted using EcoMount (EM897L; Biocare Medical); and dried overnight. Single-plane tile scans were taken under a LSM780 Confocal Microscope (Zeiss) with 10× objective for quantification and representation. Csf1r+ calb+ PCs were manually counted using Zen 2011 software and represented as dot plots using GraphPad Prism v5.01.
Immunofluorescence staining
Mice were transcardially perfused with cold 1× PBS and their brains dissected and put into 4% paraformaldehyde overnight. Following fixation, brains were placed into 30% sucrose for two nights before embedding into optimal cutting temperature compound. 20-μm sections were cut from each brain using a cryostat. Sections were then thawed at room temperature, washed in 1× PBS, and incubated with blocking buffer (1× PBS, 0.5% Triton X-100, 2% BSA, and 5% goat serum) for 1 h at 4°C. The following primary antibodies were used overnight at 4°C: Iba1 (1:500; WAKO), Ki-67 (1:100; eBioscience), CD68 (1:250; Serotec), NeuN (1:100; Abcam), calbindin (1:500; CB-955; Abcam), and VGluT2 (1:1,000; AB2251-I; EMD Millipore). Secondary antibodies (all from eBioscience) conjugated to their respective fluorophores were incubated at 1:200 for 2 h at room temperature. Slides were then counterstained with DAPI and mounted with Prolong Gold (Invitrogen) media. Images were acquired using wide-field and confocal microscopy (Zeiss), and analyzed using Fiji (ImageJ) and Imaris (Bitplane). Manual quantifications were performed by an observer blinded to genotypes.
Cell suspension preparation and microglia isolation
Perinatal and adult brains, yolk sac, embryonic limbs, and fetal livers were dissected from mice at specified ages, as previously described (Greter et al., 2012; Lavin et al., 2014; Hoeffel et al., 2015). Brains of P21 and older were perfused with ice-cold PBS before tissue isolation, apart from brains used for microglia sorting and subsequent ultra-low input (ULI) RNA sequencing (RNA-seq), according to the Immgen standardized protocol listed on immgen.org. For the perinatal brain, yolk sac, and limbs, the tissue was enzymatically digested with 0.4 mg/μl collagenase IV (C5138; Sigma-Aldrich) for 10–20 min at 37°C, followed by mechanical trituration with an 18-G blunt-tipped syringe and filtration through a 70-μm filter. Adult brain was digested with 0.4 mg/μl collagenase IV for 30 min. Fetal liver tissue was only mechanically triturated to obtain a homogeneous cell suspension. Cell pellets from all adult brain were further processed by centrifugation in 5 ml of 40% Percoll (Sigma-Aldrich), at 2,300 rpm for 25 min with no brake. Myelin was aspirated, and the cell pellet was washed twice with FACS buffer (PBS without Ca2+ and Mg2+ supplemented with 2% heat-inactivated FBS and 5 mM EDTA) before staining. For parabionts, brain regions specified were dissected, minced, and digested in RPMI supplemented with 10% FCS, 0.2 mg/ml collagenase IV, and 0.06 mg/ml DNaseI, for 1 h at 37°C. Cells were pelleted at 1,350 rpm for 5 min, and resuspended with 7 ml of 30% Percoll. The cell suspension was spun at 3,000 rpm for 10 min with no brake and washed twice with FACS buffer before staining.
Flow cytometry and microglia sorting
Following single-cell suspension preparation, cells were pelleted and subsequently stained with fluorophore-conjugated antibodies against CD11b (clone M1/70), CD45 (clone 30-F11), CD45.1 (clone A20), CD45.2 (clone 104), CX3CR1 (clone SA011F11; BioLegend), F4/80 (clone BM8; BioLegend), Ly6C (clone HK1.4), Ly6G (clone 1A8; BioLegend), Gr-1 (clone RB6-8C5), CD115 (clone AFS98), MHC II (clone M5/114.15.2), CD11c (clone N418), CD64 (clone X54-5/7.1; BioLegend), CD86 (clone GL1; BioLegend), Tmem119 (clone 106–6; Abcam), and either DAPI or propium iodide viability dyes (all from eBioscience if not indicated otherwise). Flow cytometry was performed using a Fortessa analyzer (BD Biosciences), and FACS was performed using a FACS Aria II (BD Biosciences) or LSRII (BD Biosciences). The gating strategies used for flow cytometry analysis of embryonic tissues are adapted from Hoeffel et al. (2015) and shown in Fig. S5. Resident microglia were sorted as doublet−DAPI−CD11b+CD45int (Fig. S2 B). Flow cytometry data analysis was performed using FlowJo (TreeStar) software. For ULI RNA-seq, microglia were double-sorted to reach a purity of >98%, and 1,000 cells were sequenced.
Parabiosis
Parabiotic mice were generated by surgically linking age- and size-matched congenic CD45.2 (C57BL/6) and CD45.1 (C57BL/6) mice as previously described (Ginhoux et al., 2009).
Primary neonatal microglia culture
Preparation of primary neonatal microglia culture was adapted from (Bronstein et al., 2013). The cerebellum and cortex from 10–20 newborn pups (P2–3) were dissected and mechanically triturated with a P1,000 (200–1,000 μl) pipette in ice-cold 1× HBSS. Following incubation for 15 min at 37°C in 0.25% trypsin/EDTA (Thermo Fisher Scientific), cells were spun down and washed twice in 1× HBSS. Cells were then resuspended in complete medium (DMEM, 10% FBS, 100 U/ml penicillin, and 100 mg/ml streptomycin) and split evenly into three poly-D-lysine (Sigma-Aldrich)–coated T-75 flasks. Half of the medium was changed 3 d later, and subsequently every other day. Between days 10–14 after plating into flasks, the media were removed from each flask, and the cells were dissociated by incubating in Accutase (Thermo Fisher Scientific) for 10 min at 37°C. Cells were then pelleted, washed, and stained on ice for 30 min before FACS. Sorted DAPI−CD11b+CD45int microglia from the mixed culture were seeded at density of 2 × 105 cells onto poly-D-lysine–coated 12-well plates in complete medium containing either 100 ng/ml IL-34 or 20 ng/ml CSF-1 for 24 h.
Human samples
Brain tissue was provided by the Netherlands Brain Bank (NBB). Informed consent for research purposes was obtained for brain autopsy and the usage of tissue and clinical information. The procedures of the NBB are in accordance with all national laws and regulations and respect human rights (the right to life, liberty, and security of person). The NBB adheres to the standards for quality, safety, and ethics for obtaining and handling of human tissue, as described in BrainNet Europe’s Code of Conduct for brain banking. The NBB’s procedures have been approved by the ethics committee of the VU University Medical Center (Amsterdam, Netherlands), where all autopsies take place. For this study, brain tissues were collected from two female donors with bipolar disorder (aged 45 yr) and multisystem atrophy (aged 60 yr) and processed according to standardized protocols for next-generation sequencing of primary microglia.
Next-generation sequencing
Human microglia
Primary microglia obtained from the NBB were isolated as described before (Melief et al., 2016) with minor modifications. In short, 2–10 g of superior temporal gyrus and cerebellum was collected from two female donors within 8 h after death and stored in Hibernate medium on ice, and the isolation procedure was started within 2 to 24 h after autopsy. A single-cell suspension was generated by dissociating the tissue mechanically through a metal sieve, followed by an enzymatic digestion using collagenase Type I (3,700 U/ml; Worthington) and DNaseI (200 µg/ml; Roche). Microglia were further purified using a Percoll gradient and positive selection using CD11b-conjugated magnetic microbeads (Miltenyi Biotec), which resulted in a 99% pure microglia population as analyzed by flow cytometry. Microglia were lysed in RLT buffer (Qiagen), and RNA was extracted according to the protocol provided by the RNeasy Mini Kit (Qiagen). cDNA synthesis and library preparation were performed using the SMART-Seq v4 Ultra Low Input RNA Kit and Low Input Library Prep Kit v2 (Clontech), respectively. Sequencing was performed using the Illumina NextSeq-500 system. Transcript abundances were quantified with the Ensembl GRCh38 cDNA reference using Kallisto version 0.43.0. Transcript abundances were summarized to gene level using tximport. The expression matrix was filtered only for transcripts with >5 reads per kilobase of transcript per million mapped reads (RPKM) in both replicates of at least one brain region, leaving 9,927 genes. Differential expression statistics between different brain regions were generated using DEseq2. Genes with a Benjamini–Hochberg adjusted P-value <0.1 were called differentially expressed. Gene ontology (GO) term enrichment was performed using the topGO package in R using genes up-regulated in either brain region as input.
Primary adult mouse microglia
103 forebrain and cerebellar microglia (DAPI−CD11b+CD45int) were FACS-sorted (FACS Aria II; BD) according to the Immgen standardized protocol for ULI RNA-seq (Tan et al., 2016). RNA extraction, library preparation, and sequencing were performed at the Broad Technology Labs Boston using the NextSeq-500 system (Illumina). Differentially expressed genes for ULI RNA-seq were determined as having ≥10 reads and a P-value <0.05. Differential expression statistics were calculated with the edgeR package in R. Comparison of CSF-1–deficient microglia RNA-seq data to published data of sorted myeloid populations from adult mice (40) were downloaded from the National Center for Biotechnology Information (Gene Expression Omnibus [GEO] accession no. GSE63340). The dataset was renormalized with the dataset from the present study using the TMM normalization as implemented in the calcNormFactors() function in the edgeR package. Datasets were hierarchically clustered using Spearman correlation distance of the top 1,000 differentially expressed genes (DEGs) among sample types as determined using the ebayes() function in the Limma package. For GO term enrichment analysis, DEGs were input to the Panther Classification System using the statistical enrichment test on the “GO biological process complete” annotation dataset. Gene lists for each GO term were generated in R using the intersection of the DEGs and Bioconductor’s Genome wide annotation for Mouse (org.Mm.egGO2ALLEGS). Panther Classification System and Bioconductor’s Genome wide annotation for Mouse are both sourced by the Gene Ontology database. Gene set enrichment analysis plots were generated using GSEA software (Subramanian et al., 2005) using our DEG values for P8 Csf1fl/fl and NesCreCsf1fl/fl cerebellar microglia and pathway gene set lists curated from our GO term analysis, with 1,000 gene permutations. To compare human and mouse microglia, DEseq2 differential expression tables for human and mouse brain region comparisons were filtered for one-to-one orthologues (determined via Ensembl) that were expressed >5 RPKM in all replicates of at least one brain region in both species, leaving 7,071 genes. To determine conserved brain region expression differences, genes that had an adjusted P-value < 0.1 and a log2FC > 0.5 in the same direction in both species were selected.
Cultured neonatal mouse microglia
Following 24-h incubation with either IL-34 or CSF-1, the supernatant was removed, and the attached microglia were harvested directly in Qiazol (Qiagen). Total RNA was extracted using the miRNeasy Kit (Qiagen) following the manufacturer’s protocol. cDNA synthesis and library preparation were performed using the SMART-Seq v4 Ultra Low Input RNA Kit and Low Input Library Prep Kit v2 (Clontech), respectively. Sequencing was performed using the Illumina NextSeq-500 system. Transcript abundances were quantified with the Ensembl GRCm38 Mouse cDNA and non-coding RNA reference using Kallisto version 0.43.0. Transcript abundances were summarized to gene level using tximport. The expression matrix was filtered only for transcripts with >2 transcripts per kilobase million (TPM) in all replicates of at least one condition, leaving 10,751 genes. Differentially expressed genes for neonatal microglial cultures were determined using a one-way ANOVA and a Benjamini–Hochberg adjusted P-value threshold < 0.01. Differential expression statistics between specific conditions were calculated using the Limma package in R (Ritchie et al., 2015). Hierarchical clustering of genes was done using Pearson correlation distance and average linkage. Principal component analysis for the neonatal cultures was done using Log2(TPM + 1) values for genes determined to be differentially expressed by one-way ANOVA, 675 genes in total. Genes specifically up-regulated for either cytokine were identified by selecting those that had at least a log2FC > 0.25 and an adjusted P-value < 0.05 in one condition compared with all of the other three.
TRAP
6–8-wk-old transgenic Tg(Aldh1l1-eGFPL10a)JD130, Tg(Pcp2-eGFPL10a)DR168, Tg(NeuroD1-eGFPL10a)JP241, Tg(Drd1-eGFPL10a)CP73, Tg(Drd2-eGFPL10a)CP101, and Cx3cr1CreErt2/+(Litt); Eef1a1LSL.eGFPL10a/+ mice (n = 2–6 per genotype) were euthanized with CO2, and brain regions of interest were dissected. Ribosome-associated mRNA from neurons, microglia, or astrocytes was isolated from each region as previously described (Heiman et al., 2008, 2014) where each sample corresponds to a single mouse. RNA clean-up from TRAP samples and 5% of the unbound fractions from TRAP samples was performed using the RNeasy Mini Kit (Qiagen) following the manufacturer’s instructions. RNA integrity was assayed using an RNA Pico chip on a Bioanalyzer 2100 (Agilent), and only samples with RNA integrity number > 9 were considered for subsequent analysis. Double-stranded cDNA was generated from 1–5 ng of RNA using Nugen Ovation V2 kit (NuGEN) following the manufacturer’s instructions. Fragments of 200 bp were obtained by sonicating 500 ng of cDNA per sample using the Covaris-S2 system (duty cycle: 10%, intensity: 5.0, bursts per second: 200, duration: 120 s, mode: frequency sweeping, power: 23 W, temperature: 5.5–6°C; Covaris). Subsequently, these fragments were used to produce libraries for sequencing by the TruSeq DNA Sample kit (Ilumina) following the manufacturer’s instructions. The quality of the libraries was assessed by 2200 TapeStation (Agilent). Multiplexed libraries were directly loaded on NextSeq 500 (Ilumina) with High Output single read sequencing for 75 cycles. Raw sequencing data were processed by using Illumina bcl2fastq2 Conversion Software v2.17. Raw sequencing reads were mapped to the mouse genome (mm9) using the TopHat2 package (v2.1.0; Kim and Salzberg, 2011). Reads were counted using HTSeq-count (v0.6.0; Anders et al., 2015) against the Ensembl v67 annotation. The read alignment, read counting, and quality assessment using metrics such as total mapping rate and mitochondrial and ribosomal mapping rates were done in parallel using an in-house workflow pipeline called SPEctRA (Purushothaman and Shen, 2016). Dow plots representing RPKM of indicated genes were made on GraphPad Prism v5.01.
IL-34 ELISA
Brains were isolated, and indicated regions were dissected, flash-frozen in liquid nitrogen, and stored at −80°C until further use. Proteins were extracted by homogenizing the tissue in ice-cold radioimmunoprecipitation assay buffer (Sigma-Aldrich), and lysates were centrifuged at 16,000 ×g at 4°C. Supernatants containing 70 µg of total protein were used to determine IL-34 concentration using an IL-34 ELISA kit (BioLegend).
Quantitative RT-PCR (qRT-PCR)
RNA was extracted from FACS-isolated GFP+ cells from P6 Atoh1-GFP mice (Chen et al., 2002), FACS-isolated CFP+ cells from P6 Nes-CFP mice (Mignone et al., 2004), FACS-isolated TdTomato+ cells from P6 Pcp2Cre/+;R26LSL-TdTomato/+ mice (Zhang et al., 2004; Madisen et al., 2010), and 6-wk-old Csf1fl/fl and NesCreCsf1fl/fl mice using a miRNeasy Micro Kit (Qiagen), according to the manufacturer’s protocol. cDNA was prepared using the iScript cDNA synthesis kit (Bio-Rad). qRT-PCR was performed using PowerUp Sybr Green Master Mix (Applied Biosystems). Fold-changes in expression were calculated using the ΔΔCt method. The Gapdh gene was used to normalize the results. The following primer pairs were used: Csf1: F, 5′-CATCCAGGCAGAGACTGACA-3′ and R, 5′-CTTCGTGATCCTCCTTCCAG-3′ (Harris et al., 2012); Csf1r: F, 5′-TGTCATCGAGCCTAGTGGC-3′ and R, 5′-CGGGAGATTCAGGGTCCAAG-3′; Il34: F, 5′-CTTTGGGAAACGAGAATTTGGAGA-3′ and R, 5′-GCAATCCTGTAGTTGATGGGGAAG-3′; and Gapdh: F, 5′-CCAAGGTGTCCGTCGTGGATCT-3′ and R, 5′-GTTGAAGTCGCAGGAGACAACC-3′.
Human PC and brain tissue RNA-seq data
Single-cell RNA-seq data from human PCs were extracted from the Encyclopedia of DNA Elements (ENCODE) database. 6-yr-old patient ENCODE listing was ENCSR900JSG, GEO:GSE78474. 20-yr-old patient ENCODE listing was ENCSR8882YA, GEO:GSE78473. IL34 and CSF1 RNA-seq data from human forebrain and cerebellar tissue were extracted from the Human Protein Atlas database (Uhlen et al., 2017) using the Genome Tissue Expression RNA-seq dataset.
Patch-clamp recording
Animals were decapitated under isofluorane anesthesia. Brains were quickly removed and transferred into ice-cold (0–4°C) artificial cerebrospinal fluid (ACSF) of the following composition (in mM): 210.3 sucrose, 11 glucose, 2.5 KCl, 1 NaH2PO4, 26.2 NaHCO3, 0.5 CaCl2, and 4 MgCl2. Acute sagittal slices (350 µm) were prepared from the cerebellum of 4-wk-old mutant and control littermates. Slices were allowed to recover for 40 min at room temperature in the same solution, but with reduced sucrose (105.2 mM) and addition of NaCl (109.5 mM). Following recovery, slices were maintained at room temperature in standard ACSF composed of the following (in mM): 119 NaCl, 2.5 KCl, 1 NaH2PO4, 26.2 NaHCO3, 11 glucose, 2 CaCl2, and 2 MgCl2. Whole-cell patch-clamp recordings (Edwards et al., 1989; Tsai et al., 2012) were taken using borosilicate glass electrodes (3–5 MΩ), from the soma of visually identified PCs located at the ganglionic layer of the cerebellum, which are clearly visible even under 10× objective. Whole-cell voltage-clamp recordings were obtained with the internal solution containing (in mM): 120 Cs-methanesulfonate, 10 Hepes, 0.5 EGTA, 8 NaCl, 4 Mg-ATP, 1 QX-314, 10 Na-phosphocreatine, and 0.4 Na-GTP. mEPSCs were passively recorded in the presence of tetrodotoxin (1 µM; Abcam) in the standard ACSF (as above). mEPSCs were recorded at −60 mV. Data were low-pass filtered at 10 kHz and acquired at 10 kHz using Multiclamp 700B (Axon Instruments) and pClamp 10 (Molecular Devices). PCs for recording were selected at random. Series and membrane resistance was continuously monitored, and recordings were discarded when these measurements changed by >20%. Recordings in which series resistance exceeded 25 MΩ were rejected. Detection and analysis of EPSCs were performed using MiniAnalysis (Synaptosoft). Series resistance was monitored and canceled using a bridge circuit, and pipette capacitance was compensated. Voltage signals were low-pass filtered at 10 kHz.
VGluT2 and PC quantification and structural analysis
PC quantification
Male P7, P21, and 6-wk-old Csf1fl/fl and NesCreCsf1fl/fl littermates and 2-wk-old FVB WT and Csf1op/op mice on FVB background were perfused transcardially with 4% paraformaldehyde, pH 7.4, and brains were incubated overnight in the same. For P7 and P21 Csf1fl/fl and NesCreCsf1fl/fl mice, brains were cut in half sagittally, sectioned at 15 µm, and stained with anti-calbindin, anti-VGluT2 antibody, and DAPI. PCs were counted in the manually determined PC and molecular cell layers using Imaris. VGluT2 puncta on manually outlined PC soma were automatically determined using Imaris Spot Function. For 6-wk-old Csf1fl/fl and NesCreCsf1fl/fl mice, brains were cut in half sagittally and sectioned at 5 µm. Slides were stained with H&E and scanned for digital imaging (Pannoramic 250 Flash III whole-slide scanner, 3DHISTECH). All PCs of all whole cerebellar sections were manually counted by an observer blinded to genotypes. For WT and Csf1op/op mice, 5-µm-thick paraffin-embedded brain sections were stained with H&E and pictures obtained by a Leica microscope. PCs were quantitated from H&E-stained sections from cerebellar lobes IX–X by manual counting.
Intracellular labeling of PCs
Randomly selected PCs that were being passively recorded were filled with 0.4% biocytin (Tocris) using patch-clamp recording pipettes once the recordings were completed (Kang, 2014). Neurons in deeper portions of the PC layer were targeted and filled for 15 min, and then the pipette was slowly withdrawn so that the cell membrane could reseal. Slices (350 mm thick) were then fixed in 4% paraformaldehyde in 0.1 M phosphate buffer for 24 h, rinsed thoroughly in PBS and incubated for 90 min in a PBS solution containing 0.5% Triton X-100, 10% normal goat serum, and streptavidin Alexa Fluor 488 conjugate (1:500; Life Technologies). Slices were then rinsed in PBS, mounted on Superfrost Plus slides (VWR International), air-dried, and coverslipped in Vectashield mounting media (Vector Labs) for the subsequent imaging.
Dendritic spine imaging and characterization
Images were acquired using an Upright LSM 780 Confocal microscope (Carl Zeiss). A nonbiased approach was taken to characterize dendritic spines on secondary dendrites within 200 μm of the soma. To assure sufficient sampling, five dendritic spine segments were taken from each neuron, and three neurons were assessed per animal (Dumitriu et al., 2011). Dendritic segments were imaged using a 100× lens (numerical aperture 1.4; Carl Zeiss) and a zoom of 4.0. Images were taken with a resolution of 300 × 100 pixels, dwell time was 1.58 µm/s, and the line average was set to 8. Pixel size was 0.03 μm in the x-y plane and 0.05 μm in the z plane. To assure consistency of imaging across different confocal sessions and at different depths of the tissue, power was consistently set to 3.0%, and gain was adjusted within the range of 600–800 U to achieve consistent light intensity values within the same set of neurons. Images were de-convolved using a three-dimensional (3D) resolution enhancement with AutoDeblur software (Media Cybernetics; Rodriguez et al., 2006, 2008; Christoffel et al., 2011) and then run through the spot detector in Imaris (Bitplane). PC dendritic spine reconstruction and analysis were performed using Imaris FilamentTracer. Start and end points of dendrites to be segmented were manually selected and traced using the AutoPath method, and automatic thresholds used to determine dendrite diameter and surface rendering. Spines were considered to have a minimum seed point diameter of 0.2 μm and a maximum spine length of 2.0 μm (including branched spines). The seed point threshold was manually decreased to detect the maximum seed points for each dendrite. Automatic thresholds were used for spine diameter rendering. The average values for dendritic spine length, density, diameter, and volume measurements were calculated by the Imaris software and exported from the Statistics tab. All analyses were performed blinded to the group conditions until the analyses were completed.
Whole-cell imaging and dendrite complexity quantification
Images were acquired using an Upright LSM 780 Confocal microscope (Carl Zeiss) using a 63× lens (numerical aperture 1.4; Carl Zeiss) and a zoom of 0.6. Images were taken with a resolution of 1,024 × 1,024 pixels, dwell time was 1.58 µm/s, and the line average was set to 4. Pixel size was 0.22 µm in the x-y plane and 1.00 μm in the z plane. Gain was set to 600–800 U. Dendrite complexity quantification was performed using the Sholl analysis plugin in Fiji (ImageJ).
Behavioral tests
Ataxia assessment
Cerebellar ataxia in Csf1fl/fl and NesCreCsf1fl/fl mice was assessed using a composite score of hind limb clasping, ledge test, gait, and kyphosis, as previously described (Ginhoux et al., 2009).
Rotarod
Motor effects in Il34LacZ/LacZ and NesCreCsf1fl/fl animals, and their respective littermate controls, were tested using the accelerating rotarod paradigm (Brooks and Dunnett, 2009), with additional video recording. Mice were placed on a motorized beam that was accelerated from 4–40 rpm. The latency of the mice to fall was recorded. If an animal clung to the rod and completed a full passive rotation, the timer was stopped for the animal and the time was noted. Prior to the final recorded rotarod session on day 4, with three trials at 5-min intervals each, animals were habituated to the paradigm during three training days with three trials of 5 min each. Differences between groups were statistically evaluated by Student’s t tests.
Sociability assessments
Sociability deficits were assessed in 6–8-wk-old male NesCreCsf1fl/fl mice and their littermate controls. The premise for these behavioral tests lies in the preference of mice spending time around (1) other mice over inanimate objects and (2) novel over familiar subjects (File and Hyde, 1978; Ellegood and Crawley, 2015). To begin, test mice were habituated to an empty three-chambered Plexiglass box for 10 min. Mice were then restricted to the middle chamber by dividers, while a novel inanimate object and a novel stimulus mouse were placed under wire cups in either of the two flanking chambers (social preference). The dividers were then removed and the test mouse was allowed to freely explore the entire box for 10 min. Following this, test mice were again restricted to the middle chamber, while the inanimate object was replaced with a second novel stimulus mouse (social novelty). Test mice were again released from the middle chamber and allowed to explore for 10 min the chambers with the familiar and new stimulus mice. The behavior sessions were recorded using EthoVision (Noldus), and the time each test mouse spent near and/or sniffing the object or stimulus mice was manually recorded. Stimulus mice were age- and sex-matched for these assessments.
MRI
All imaging was performed at the Translational and Molecular Imaging Institute at Mount Sinai Hospital on a Bruker Biospec 70/30 7Tesla scanner with a B-GA12S gradient insert (gradient strength 440 mT/m and slew rate 3,444 T/m/s). A Bruker four-channel mice brain phased array was used for all data acquisition in conjunction with a Bruker volume transmit 86-mm coil. All mice were imaged under isoflurane anesthesia (3% induction and 1.5% maintenance). All mice were imaged on a heated bed, and respiration was monitored continuously. The following protocols were used. After a three-plane localizer, a diffusion tensor imaging protocol was acquired with a Pulsed Gradient Spin Echo echo-planar imaging sequence with the following parameters: repetition time = 5,500 ms, echo time = 22.672 ms, four segments, 30 gradient directions with b-value = 1,000 s/mm2 and 5 B0s, field-of-view = 25 mm, matrix = 128 × 128, slice thickness = 0.5 mm, skip = 0, three averages, and total acquisition time = 38 min. A T2 anatomical scan was obtained with a 3D Rapid Acquisition with Relaxation Enhancement sequence with a Rapid Acquisition with Relaxation Enhancement factor of 8, repetition time = 2,500 ms, effective pulse duration = 62.5 ms, field-of-view = 25.6 mm × 25.6 mm, slice thickness = 0.4 mm, skip = 0, 18 slices, matrix size 256 × 256, 30 averages, and total acquisition time = 31 min. Scans were transferred to offline workstation for processing. In-house software TMII-ROI developed under MATLAB (R2015b; 2000; MathWorks) was used to analyze volume.
Statistical analysis
For all graphical analyses, mean values, SEM values, Student’s t tests (two-tailed, unpaired), and one-way ANOVAs were calculated using Prism 7 (GraphPad).
Data and material availability
All data are available in the main text or the supplementary materials. RNA-seq and ULI RNA-seq data are deposited in GEO under accession nos. GSE133357 (human microglia), GSE133360 (neonatal stimulation microglia), and GSE133362.
Online supplemental material
Fig. S1 shows distinct expression patterns of CSF-1 and IL-34 mRNA in mice and humans. Fig. S2 shows NesCreCsf1fl/fl microglia profile clusters with published microglia profile and distinct clustering of CSF-1– or IL-34–stimulated forebrain and cerebellar neonatal microglia. Fig. S3 shows characterization of cerebellar microglia and embryonic macrophages in control and CSF-1–deficient mice. Fig. S4 shows morphological and behavioral alterations in CSF-1–deficient mice. Fig. S5 shows gating strategies for flow cytometry analysis of embryonic tissues in CSF-1–deficient mice. Table S1 shows GO term pathways for RNA-seq expression data of purified human cerebellar and forebrain microglia. Table S2 shows GO term pathways for RNA-seq expression data of sorted P8 and adult Csf1fl/fl and NesCreCsf1fl/fl, and adult Il34wt/wt and Il34LacZ/LacZ cerebellar and forebrain microglia. Table S3 shows GO term pathways for RNA-seq expression data of cultured and sorted primary neonatal cerebellar and forebrain microglia stimulated with CSF-1 or IL-34.
Supplementary Material
Acknowledgments
We thank the members of the M. Merad and B.D. Brown laboratory and Soyon Hong for extensive discussions and critiques of the manuscript, the Immgen Consortium for sequencing, the ENCODE Consortium and the ENCODE production laboratories for generating the particular datasets, Patrick Hof for advice on PC quantification, Sayan Nandi for help with PC quantification, and the Mount Sinai Flow Cytometry Core and Translational Imaging Core.
This work was supported by National Institutes of Health grants R01CA154947, R01MH104559, and U19AI128949 (M. Merad), an Advanced Postdoc Mobility Fellowship of the Swiss National Foundation (V. Kana), a postdoctoral fellowship from the Human Frontier Science Program Organization (LT000110/2015-L/1; M. Casanova-Acebes), a National Institutes of Health Director New Innovator Award (DP2 MH100012-01; A. Schaefer), National Institutes of Health grant 1RF1 AG054011-01 (A. Schaefer), a Brain and Behavior Research Foundation NARSAD Young Investigator Award (25065; P. Ayata), National Institutes of Health grant T32 AG049688 (A. Baccarini), National Institute of Mental Health grant F30MH111143 (E. Nabel), National Institutes of Health grant R21MH106919 (H. Morishita), National Eye Institute grants R01EY024918, R01EY 026053, and R21EY026702 (H. Morishita), National Institute of Neurological Disorders and Stroke grant R21NS105119 (H. Morishita), the Naito Foundation (K. Yamamuro), the Uehara Memorial Foundation (K. Yamamuro), the Seaver Foundation (E. Nabel), an EMBO Young Investigator Programme award (F. Ginhoux), National Research Foundation Senior Investigatorship grant NFR2016NRF-NRFI001-02 (F. Ginhoux), the Singapore Immunology Network Core Funding (F. Ginhoux and P. See), and National Institutes of Health grant R01 NS091519 (E.R. Stanley).
The authors declare no competing financial interests.
Author contributions: V. Kana, F.A. Desland, M. Casanova-Acebes, P. Ayata, A. Baccarini, E. Nabel, K. Yamamuro, M. Sneeboer, I.-L. Tan, M.E. Flanigan, H. Le Bourhis, E.S. Sweet, N. Tung, A. Wroblewska, Y. Lavin, P. See, and A. Baccarini performed experiments. V. Kana, F.A. Desland, P. Ayata, A. Badimon, E. Nabel, K. Yamamuro, I.-L. Tan, S.A. Rose, C. Chang, A. Leader, H. Le Bourhis, and P. See analyzed data. V. Kana, F.A. Desland, P. Ayata, A. Baccarini, E. Nabel, K. Yamamuro, I.-L. Tan, P. See, F. Ginhoux, V. Chitu, H. Morishita, A. Schaefer, and M. Merad designed experiments. V. Kana, F.A. Desland, M. Casanova-Acebes, P. Ayata, A. Baccarini, E. Nabel, K. Yamamuro, P. See, F. Ginhoux, V. Chitu, E.R. Stanley, S.J. Russo, Z. Yue, B.D. Brown, A.L. Joyner, L.D. De Witte, H. Morishita, A. Schaefer, and M. Merad provided intellectual contributions to the project. V. Kana, F.A. Desland, and M. Merad wrote the paper.
References
- Ajami B., Bennett J.L., Krieger C., Tetzlaff W., and Rossi F.M.. 2007. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat. Neurosci. 10:1538–1543. 10.1038/nn2014 [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., and Huber W.. 2015. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 31:166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwell K. 1990. Microglia and cell death in the developing mouse cerebellum. Brain Res. Dev. Brain Res. 55:219–230. 10.1016/0165-3806(90)90203-B [DOI] [PubMed] [Google Scholar]
- Ayata P., Badimon A., Strasburger H.J., Duff M.K., Montgomery S.E., Loh Y.E., Ebert A., Pimenova A.A., Ramirez B.R., Chan A.T., et al. 2018. Epigenetic regulation of brain region-specific microglia clearance activity. Nat. Neurosci. 21:1049–1060. 10.1038/s41593-018-0192-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman M.L., and Kemper T.L.. 2005. Neuroanatomic observations of the brain in autism: a review and future directions. Int. J. Dev. Neurosci. 23:183–187. 10.1016/j.ijdevneu.2004.09.006 [DOI] [PubMed] [Google Scholar]
- Bennett M.L., Bennett F.C., Liddelow S.A., Ajami B., Zamanian J.L., Fernhoff N.B., Mulinyawe S.B., Bohlen C.J., Adil A., Tucker A., et al. 2016. New tools for studying microglia in the mouse and human CNS. Proc. Natl. Acad. Sci. USA. 113:E1738–E1746. 10.1073/pnas.1525528113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronstein R., Torres L., Nissen J.C., and Tsirka S.E.. 2013. Culturing microglia from the neonatal and adult central nervous system. J. Vis. Exp. 78:50647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks S.P., and Dunnett S.B.. 2009. Tests to assess motor phenotype in mice: a user’s guide. Nat. Rev. Neurosci. 10:519–529. 10.1038/nrn2652 [DOI] [PubMed] [Google Scholar]
- Calvo C.F., Dobbertin A., Gelman M., Glowinski J., and Mallat M.. 1998. Identification of CSF-1 as a brain macrophage migratory activity produced by astrocytes. Glia. 24:180–186. [DOI] [PubMed] [Google Scholar]
- Chen P., Johnson J.E., Zoghbi H.Y., and Segil N.. 2002. The role of Math1 in inner ear development: Uncoupling the establishment of the sensory primordium from hair cell fate determination. Development. 129:2495–2505. [DOI] [PubMed] [Google Scholar]
- Christoffel D.J., Golden S.A., Dumitriu D., Robison A.J., Janssen W.G., Ahn H.F., Krishnan V., Reyes C.M., Han M.H., Ables J.L., et al. 2011. IκB kinase regulates social defeat stress-induced synaptic and behavioral plasticity. J. Neurosci. 31:314–321. 10.1523/JNEUROSCI.4763-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadros M.A., Rodríguez-Ruiz J., Calvente R., Almendros A., Marín-Teva J.L., and Navascués J.. 1997. Microglia development in the quail cerebellum. J. Comp. Neurol. 389:390–401. [DOI] [PubMed] [Google Scholar]
- Dai X.M., Ryan G.R., Hapel A.J., Dominguez M.G., Russell R.G., Kapp S., Sylvestre V., and Stanley E.R.. 2002. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood. 99:111–120. 10.1182/blood.V99.1.111 [DOI] [PubMed] [Google Scholar]
- Doyle J.P., Dougherty J.D., Heiman M., Schmidt E.F., Stevens T.R., Ma G., Bupp S., Shrestha P., Shah R.D., Doughty M.L., et al. 2008. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 135:749–762. 10.1016/j.cell.2008.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitriu D., Rodriguez A., and Morrison J.H.. 2011. High-throughput, detailed, cell-specific neuroanatomy of dendritic spines using microinjection and confocal microscopy. Nat. Protoc. 6:1391–1411. 10.1038/nprot.2011.389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards F.A., Konnerth A., Sakmann B., and Takahashi T.. 1989. A thin slice preparation for patch clamp recordings from neurones of the mammalian central nervous system. Pflugers Arch. 414:600–612. 10.1007/BF00580998 [DOI] [PubMed] [Google Scholar]
- Ellegood J., and Crawley J.N.. 2015. Behavioral and Neuroanatomical Phenotypes in Mouse Models of Autism. Neurotherapeutics. 12:521–533. 10.1007/s13311-015-0360-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi S.H., Aldinger K.A., Ashwood P., Bauman M.L., Blaha C.D., Blatt G.J., Chauhan A., Chauhan V., Dager S.R., Dickson P.E., et al. 2012. Consensus paper: pathological role of the cerebellum in autism. Cerebellum. 11:777–807. 10.1007/s12311-012-0355-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- File S.E., and Hyde J.R.. 1978. Can social interaction be used to measure anxiety? Br. J. Pharmacol. 62:19–24. 10.1111/j.1476-5381.1978.tb07001.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F., Liu K., Helft J., Bogunovic M., Greter M., Hashimoto D., Price J., Yin N., Bromberg J., Lira S.A., et al. 2009. The origin and development of nonlymphoid tissue CD103+ DCs. J. Exp. Med. 206:3115–3130. 10.1084/jem.20091756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F., Greter M., Leboeuf M., Nandi S., See P., Gokhan S., Mehler M.F., Conway S.J., Ng L.G., Stanley E.R., et al. 2010. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 330:841–845. 10.1126/science.1194637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann T., Wieghofer P., Jordão M.J., Prutek F., Hagemeyer N., Frenzel K., Amann L., Staszewski O., Kierdorf K., Krueger M., et al. 2016. Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat. Immunol. 17:797–805. 10.1038/ni.3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabert K., Michoel T., Karavolos M.H., Clohisey S., Baillie J.K., Stevens M.P., Freeman T.C., Summers K.M., and McColl B.W.. 2016. Microglial brain region-dependent diversity and selective regional sensitivities to aging. Nat. Neurosci. 19:504–516. 10.1038/nn.4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greter M., Lelios I., Pelczar P., Hoeffel G., Price J., Leboeuf M., Kündig T.M., Frei K., Ginhoux F., Merad M., and Becher B.. 2012. Stroma-derived interleukin-34 controls the development and maintenance of langerhans cells and the maintenance of microglia. Immunity. 37:1050–1060. 10.1016/j.immuni.2012.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimon Z., Volaski A., Orthgiess J., Boura-Halfon S., Varol D., Shemer A., Yona S., Zuckerman B., David E., Chappell-Maor L., et al. 2018. Re-evaluating microglia expression profiles using RiboTag and cell isolation strategies. Nat. Immunol. 19:636–644. 10.1038/s41590-018-0110-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris S.E., MacDougall M., Horn D., Woodruff K., Zimmer S.N., Rebel V.I., Fajardo R., Feng J.Q., Gluhak-Heinrich J., Harris M.A., and Abboud Werner S.. 2012. Meox2Cre-mediated disruption of CSF-1 leads to osteopetrosis and osteocyte defects. Bone. 50:42–53. 10.1016/j.bone.2011.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M., Schaefer A., Gong S., Peterson J.D., Day M., Ramsey K.E., Suárez-Fariñas M., Schwarz C., Stephan D.A., Surmeier D.J., et al. 2008. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 135:738–748. 10.1016/j.cell.2008.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M., Kulicke R., Fenster R.J., Greengard P., and Heintz N.. 2014. Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP). Nat. Protoc. 9:1282–1291. 10.1038/nprot.2014.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman S., Izzy S., Sen P., Morsett L., and El Khoury J.. 2018. Microglia in neurodegeneration. Nat. Neurosci. 21:1359–1369. 10.1038/s41593-018-0242-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffel G., Chen J., Lavin Y., Low D., Almeida F.F., See P., Beaudin A.E., Lum J., Low I., Forsberg E.C., et al. 2015. C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity. 42:665–678. 10.1016/j.immuni.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa R., Hashimoto K., Miyazaki T., Uchigashima M., Yamasaki M., Aiba A., Kano M., and Watanabe M.. 2016. Territories of heterologous inputs onto Purkinje cell dendrites are segregated by mGluR1-dependent parallel fiber synapse elimination. Proc. Natl. Acad. Sci. USA. 113:2282–2287. 10.1073/pnas.1511513113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E.M., Craig E.T., and Yeh H.H.. 2007. TrkB is necessary for pruning at the climbing fibre-Purkinje cell synapse in the developing murine cerebellum. J. Physiol. 582:629–646. 10.1113/jphysiol.2007.133561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M.V. 2005. Excitotoxicity in perinatal brain injury. Brain Pathol. 15:234–240. 10.1111/j.1750-3639.2005.tb00526.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J. 2014. Biocytin staining of glia and neurons in brain slices. Cold Spring Harb. Protoc. 2014:948–950. 10.1101/pdb.prot083527 [DOI] [PubMed] [Google Scholar]
- Kano M., Watanabe T., Uesaka N., and Watanabe M.. 2018. Multiple Phases of Climbing Fiber Synapse Elimination in the Developing Cerebellum. Cerebellum. 17:722–734. 10.1007/s12311-018-0964-z [DOI] [PubMed] [Google Scholar]
- Keren-Shaul H., Spinrad A., Weiner A., Matcovitch-Natan O., Dvir-Szternfeld R., Ulland T.K., David E., Baruch K., Lara-Astaiso D., Toth B., et al. 2017. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell. 169:1276–1290.e17. 10.1016/j.cell.2017.05.018 [DOI] [PubMed] [Google Scholar]
- Kettenmann H., Hanisch U.K., Noda M., and Verkhratsky A.. 2011. Physiology of microglia. Physiol. Rev. 91:461–553. 10.1152/physrev.00011.2010 [DOI] [PubMed] [Google Scholar]
- Kim D., and Salzberg S.L.. 2011. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol. 12:R72 10.1186/gb-2011-12-8-r72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kübler D., Wächter T., Cabanel N., Su Z., Turkheimer F.E., Dodel R., Brooks D.J., Oertel W.H., and Gerhard A.. 2019. Widespread microglial activation in multiple system atrophy. Mov. Disord. 34:564–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin Y., Winter D., Blecher-Gonen R., David E., Keren-Shaul H., Merad M., Jung S., and Amit I.. 2014. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 159:1312–1326. 10.1016/j.cell.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson L.J., Perry V.H., Dri P., and Gordon S.. 1990. Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience. 39:151–170. 10.1016/0306-4522(90)90229-W [DOI] [PubMed] [Google Scholar]
- Li Q., and Barres B.A.. 2018. Microglia and macrophages in brain homeostasis and disease. Nat. Rev. Immunol. 18:225–242. 10.1038/nri.2017.125 [DOI] [PubMed] [Google Scholar]
- Limperopoulos C., Bassan H., Gauvreau K., Robertson R.L. Jr., Sullivan N.R., Benson C.B., Avery L., Stewart J., Soul J.S., Ringer S.A., et al. 2007. Does cerebellar injury in premature infants contribute to the high prevalence of long-term cognitive, learning, and behavioral disability in survivors? Pediatrics. 120:584–593. 10.1542/peds.2007-1041 [DOI] [PubMed] [Google Scholar]
- Lin H., Lee E., Hestir K., Leo C., Huang M., Bosch E., Halenbeck R., Wu G., Zhou A., Behrens D., et al. 2008. Discovery of a cytokine and its receptor by functional screening of the extracellular proteome. Science. 320:807–811. 10.1126/science.1154370 [DOI] [PubMed] [Google Scholar]
- Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R., et al. 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13:133–140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manto M., Bower J.M., Conforto A.B., Delgado-García J.M., da Guarda S.N., Gerwig M., Habas C., Hagura N., Ivry R.B., Mariën P., et al. 2012. Consensus paper: roles of the cerebellum in motor control--the diversity of ideas on cerebellar involvement in movement. Cerebellum. 11:457–487. 10.1007/s12311-011-0331-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matcovitch-Natan O., Winter D.R., Giladi A., Vargas Aguilar S., Spinrad A., Sarrazin S., Ben-Yehuda H., David E., Zelada González F., Perrin P., et al. 2016. Microglia development follows a stepwise program to regulate brain homeostasis. Science. 353:aad8670 10.1126/science.aad8670 [DOI] [PubMed] [Google Scholar]
- Mattei D., Ivanov A., Ferrai C., Jordan P., Guneykaya D., Buonfiglioli A., Schaafsma W., Przanowski P., Deuther-Conrad W., Brust P., et al. 2017. Maternal immune activation results in complex microglial transcriptome signature in the adult offspring that is reversed by minocycline treatment. Transl. Psychiatry. 7:e1120 10.1038/tp.2017.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melief J., Sneeboer M.A., Litjens M., Ormel P.R., Palmen S.J., Huitinga I., Kahn R.S., Hol E.M., and de Witte L.D.. 2016. Characterizing primary human microglia: A comparative study with myeloid subsets and culture models. Glia. 64:1857–1868. 10.1002/glia.23023 [DOI] [PubMed] [Google Scholar]
- Mignone J.L., Kukekov V., Chiang A.S., Steindler D., and Enikolopov G.. 2004. Neural stem and progenitor cells in nestin-GFP transgenic mice. J. Comp. Neurol. 469:311–324. 10.1002/cne.10964 [DOI] [PubMed] [Google Scholar]
- Murase S., and Hayashi Y.. 2002. Neuronal expression of macrophage colony stimulating factor in Purkinje cells and olfactory mitral cells of wild-type and cerebellar-mutant mice. Histochem. J. 34:85–95. 10.1023/A:1021308328278 [DOI] [PubMed] [Google Scholar]
- Nakayama H., Abe M., Morimoto C., Iida T., Okabe S., Sakimura K., and Hashimoto K.. 2018. Microglia permit climbing fiber elimination by promoting GABAergic inhibition in the developing cerebellum. Nat. Commun. 9:2830 10.1038/s41467-018-05100-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi S., Gokhan S., Dai X.M., Wei S., Enikolopov G., Lin H., Mehler M.F., and Stanley E.R.. 2012. The CSF-1 receptor ligands IL-34 and CSF-1 exhibit distinct developmental brain expression patterns and regulate neural progenitor cell maintenance and maturation. Dev. Biol. 367:100–113. 10.1016/j.ydbio.2012.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Vu T.D., Kimpo R.R., Rinaldi J.M., Kohli A., Zeng H., Deisseroth K., and Raymond J.L.. 2013. Cerebellar Purkinje cell activity drives motor learning. Nat. Neurosci. 16:1734–1736. 10.1038/nn.3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhurst C.N., Yang G., Ninan I., Savas J.N., Yates J.R. III, Lafaille J.J., Hempstead B.L., Littman D.R., and Gan W.B.. 2013. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell. 155:1596–1609. 10.1016/j.cell.2013.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pixley F.J., and Stanley E.R.. 2004. CSF-1 regulation of the wandering macrophage: complexity in action. Trends Cell Biol. 14:628–638. 10.1016/j.tcb.2004.09.016 [DOI] [PubMed] [Google Scholar]
- Purushothaman I., and Shen L. SPEctRA: First stable release. Zenodo; 2016. [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., and Smyth G.K.. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A., Ehlenberger D.B., Hof P.R., and Wearne S.L.. 2006. Rayburst sampling, an algorithm for automated three-dimensional shape analysis from laser scanning microscopy images. Nat. Protoc. 1:2152–2161. 10.1038/nprot.2006.313 [DOI] [PubMed] [Google Scholar]
- Rodriguez A., Ehlenberger D.B., Dickstein D.L., Hof P.R., and Wearne S.L.. 2008. Automated three-dimensional detection and shape classification of dendritic spines from fluorescence microscopy images. PLoS One. 3:e1997 10.1371/journal.pone.0001997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer D.P., and Stevens B.. 2015. Microglia Function in Central Nervous System Development and Plasticity. Cold Spring Harb. Perspect. Biol. 7:a020545 10.1101/cshperspect.a020545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slemmer J.E., De Zeeuw C.I., and Weber J.T.. 2005. Don’t get too excited: mechanisms of glutamate-mediated Purkinje cell death. Prog. Brain Res. 148:367–390. 10.1016/S0079-6123(04)48029-7 [DOI] [PubMed] [Google Scholar]
- Squarzoni P., Oller G., Hoeffel G., Pont-Lezica L., Rostaing P., Low D., Bessis A., Ginhoux F., and Garel S.. 2014. Microglia modulate wiring of the embryonic forebrain. Cell Reports. 8:1271–1279. 10.1016/j.celrep.2014.07.042 [DOI] [PubMed] [Google Scholar]
- Stanley E.R., and Chitu V.. 2014. CSF-1 receptor signaling in myeloid cells. Cold Spring Harb. Perspect. Biol. 6:a021857 10.1101/cshperspect.a021857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley S., Domingos A.I., Kelly L., Garfield A., Damanpour S., Heisler L., and Friedman J.. 2013. Profiling of Glucose-Sensing Neurons Reveals that GHRH Neurons Are Activated by Hypoglycemia. Cell Metab. 18:596–607. 10.1016/j.cmet.2013.09.002 [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarov A., and Joyner A.L.. 2007. Cerebellum morphogenesis: the foliation pattern is orchestrated by multi-cellular anchoring centers. Neural Dev. 2:26 10.1186/1749-8104-2-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan T.G., Sefik E., Geva-Zatorsky N., Kua L., Naskar D., Teng F., Pasman L., Ortiz-Lopez A., Jupp R., Wu H.J., et al. 2016. Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc. Natl. Acad. Sci. USA. 113:E8141–E8150. 10.1073/pnas.1617460113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay T.L., Mai D., Dautzenberg J., Fernández-Klett F., Lin G., Sagar M., Datta M., Drougard A., Stempfl T., Ardura-Fabregat A., et al. 2017. A new fate mapping system reveals context-dependent random or clonal expansion of microglia. Nat. Neurosci. 20:793–803. 10.1038/nn.4547 [DOI] [PubMed] [Google Scholar]
- Tsai P.T., Hull C., Chu Y., Greene-Colozzi E., Sadowski A.R., Leech J.M., Steinberg J., Crawley J.N., Regehr W.G., and Sahin M.. 2012. Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature. 488:647–651. 10.1038/nature11310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M., Zhang C., Lee S., Sjostedt E., Fagerberg L., Bidkhori G., Benfeitas R., Arif M., Liu Z., Edfors F., et al. 2017. A pathology atlas of the human cancer transcriptome. Science. 357:eaan2507 10.1126/science.aan2507 [DOI] [PubMed] [Google Scholar]
- Vieira B.D., Radford R.A., Chung R.S., Guillemin G.J., and Pountney D.L.. 2015. Neuroinflammation in Multiple System Atrophy: Response to and Cause of α-Synuclein Aggregation. Front. Cell. Neurosci. 9:437 10.3389/fncel.2015.00437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M.J., Kim T.H., Savall J., Schnitzer M.J., and Luo L.. 2017. Cerebellar granule cells encode the expectation of reward. Nature. 544:96–100. 10.1038/nature21726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker D.G., Tang T.M., and Lue L.F.. 2017. Studies on Colony Stimulating Factor Receptor-1 and Ligands Colony Stimulating Factor-1 and Interleukin-34 in Alzheimer’s Disease Brains and Human Microglia. Front. Aging Neurosci. 9:244 10.3389/fnagi.2017.00244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Szretter K.J., Vermi W., Gilfillan S., Rossini C., Cella M., Barrow A.D., Diamond M.S., and Colonna M.. 2012. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat. Immunol. 13:753–760. 10.1038/ni.2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins C.C., Sawa A., and Pomper M.G.. 2014. Glia and immune cell signaling in bipolar disorder: insights from neuropharmacology and molecular imaging to clinical application. Transl. Psychiatry. 4:e350 10.1038/tp.2013.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegiel J., Wiśniewski H.M., Dziewiatkowski J., Tarnawski M., Kozielski R., Trenkner E., and Wiktor-Jedrzejczak W.. 1998. Reduced number and altered morphology of microglial cells in colony stimulating factor-1-deficient osteopetrotic op/op mice. Brain Res. 804:135–139. 10.1016/S0006-8993(98)00618-0 [DOI] [PubMed] [Google Scholar]
- Yoshida H., Hayashi S., Kunisada T., Ogawa M., Nishikawa S., Okamura H., Sudo T., Shultz L.D., and Nishikawa S.. 1990. The murine mutation osteopetrosis is in the coding region of the macrophage colony stimulating factor gene. Nature. 345:442–444. 10.1038/345442a0 [DOI] [PubMed] [Google Scholar]
- Zhang X.M., Ng A.H., Tanner J.A., Wu W.T., Copeland N.G., Jenkins N.A., and Huang J.D.. 2004. Highly restricted expression of Cre recombinase in cerebellar Purkinje cells. Genesis. 40:45–51. 10.1002/gene.20062 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.