Abstract
During periods of reduced O2 supply, the most profound changes in gene expression are mediated by hypoxia-inducible factor (HIF) transcription factors that play a key role in cellular responses to low-O2 tension. Using target-enrichment sequencing, we tested whether variation in 26 genes in the HIF signaling pathway was associated with high altitude and therefore corresponding O2 availability in three duck species that colonized the Andes from ancestral low-altitude habitats in South America. We found strong support for convergent evolution in the case of two of the three duck species with the same genes (EGLN1, EPAS1), and even the same exons (exon 12, EPAS1), exhibiting extreme outliers with a high probability of directional selection in the high-altitude populations. These results mirror patterns of adaptation seen in human populations, which showed mutations in EPAS1, and transcriptional regulation differences in EGLN1, causing changes in downstream target transactivation, associated with a blunted hypoxic response.
Subject terms: Evolutionary genetics, Evolutionary biology
Introduction
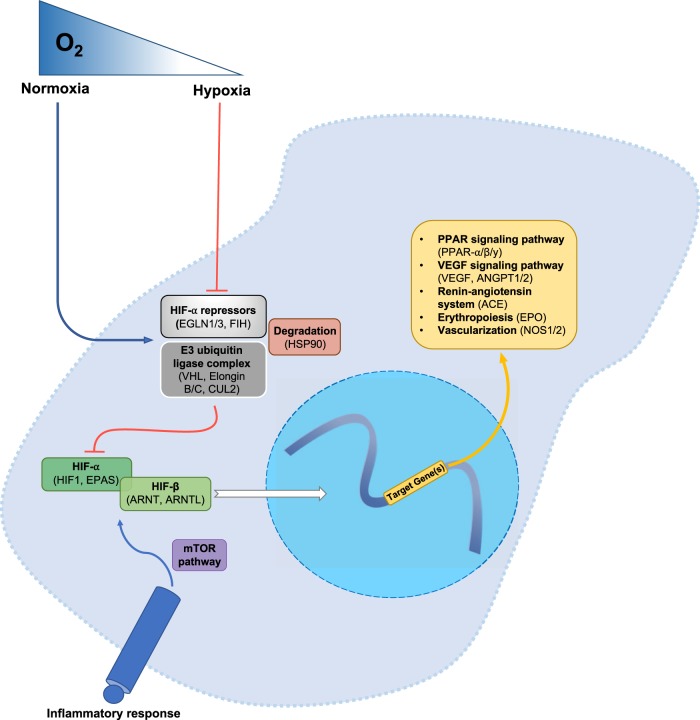
Convergent evolution of adaptive traits in distantly related organisms under the same selective pressures is common, at the genomic, cellular, and phenotypic level (Conte et al. 2012; Gompel and Prud’homme 2009; Losos 2011; Stern and Orgogozo 2009). There are multiple levels, at which the underlying genetic mechanisms for adaptive traits can occur, including different genes in the same pathway, gene, functional region, amino acid, or nucleotide (Storz 2016). Hypoxia (or low O2) is one selective pressure that stimulates a similar physiological response across metazoans allowing organisms to match O2 supply and demand (Semenza 2007b). During reduced O2 supply, changes in gene expression are mediated by a specific transcription factor family that is considered a “master regulator” of O2 homeostasis, known as hypoxia-inducible factors (HIF) (Lisy and Peet 2008; Semenza 2007a; Webb et al. 2009). In mammalian cells, HIF transcription activity is regulated post-translationally through degradation under normoxic conditions. However, under hypoxic conditions, such degradation of the HIFA (HIF1α, HIF2α/EPAS1) subunits does not occur, therefore allowing the HIF heterodimers to enter the nucleus where they recognize hypoxia-response elements (HREs) within the promoters of a large number of genes, effecting changes in transcriptional activity (Fig. 1) (Wenger et al. 2005). Many of these target genes increase O2 transport to hypoxic tissues by promoting red blood cell maturation and angiogenesis/vasomotor control (Haase 2013; Majmundar et al. 2010).
Fig. 1.
A simplified version of the HIF pathway in a cell, with a focus on specific aspects (repressor complexes, HIF transcription factors, downstream target genes) included in the 26 genes assayed (also see Supp. Fig. 1; Kanehisa & Goto, 2000)
Therefore, it is not surprising that in many organisms, HIF transcription factors, and a number of their downstream targets, have been implicated in adaptation to high-altitude environments (Hanaoka et al. 2012; Li et al. 2014; Qiu et al. 2012; Qu et al. 2013; Wang et al. 2014), as well as in other O2 depleted environments (Aggarwal et al. 2010; Ge et al. 2012; Peng et al. 2011; Rytkönen et al. 2007; Terova et al. 2008; Xing et al. 2013). Most examples of convergent evolution in the HIF pathway have been documented in high-altitude human populations (Scheinfeldt and Tishkoff 2010; Simonson et al. 2012). In Andean people, VEGF, NOS2A, and EGLN1 are the strongest-supported candidates (Bigham et al. 2010; Bigham et al. 2009), whereas in Tibetans, EPAS1 and EGLN1 have been identified (Beall et al. 2010; Jeong et al. 2014; Lorenzo et al. 2014; Peng et al. 2011; Simonson et al. 2012; Simonson et al. 2010; Xu et al. 2011; Yi et al. 2010). However, studies focused on Ethiopian highlanders suggest the involvement of a different combination of HIF genes (Alkorta-Aranburu et al. 2012; Huerta-Sánchez et al. 2013; Scheinfeldt et al. 2012).
High-altitude species offer an unparalleled opportunity to understand the molecular and physiological bases of adaptation to hypoxic environments. Therefore, because of the degree to which the HIF pathway is a frequent target of selection for adaptation to hypoxia, we tested whether genetic changes in various parts of the HIF signaling pathway might be statistically associated with high-altitude adaptations in Andean waterfowl species.
Study organisms
Our study includes three species that have low-altitude populations, in addition to high-altitude populations that are continuous year-round residents at > 4000 meters; at this altitude, the O2 partial pressure (pO2) is ~ 60% of that at sea level. These include the yellow-billed pintail (Anas georgica), cinnamon teal (Anas cyanoptera), and the speckled teal (Anas flavirostris flavirostris/Anas f. oxyptera), which were the focus of previous studies examining adaptive changes in hemoglobin (Hb) function (McCracken et al. 2009a; McCracken et al. 2009b; McCracken et al. 2009c; Natarajan et al. 2015). The benefits of using these species is that each of these duck lineages are distantly related by several millions of years of divergence (Johnson and Sorenson 1999) and each species independently colonized the same high-altitude wetlands and puna grasslands of the Andean Plateau (Altiplano), with ancestral low-altitude populations colonizing high-altitude regions and not the reverse (Fjeldså 1985; McCracken et al. 2009a). Although the overall genomic differentiation between populations is still very low for each species (e.g., FST < 0.06), Hb differentiation was markedly higher, with FST ranging from 0.89 to 1.00 for the amino-acid polymorphisms that have been experimentally validated in influencing Hb–O2-binding affinity (Natarajan et al. 2015). This suggests that even though historical gene flow has been relatively high for each of these species, selection on genes related to high-altitude function have led to strong patterns of differentiation, producing unambiguous genomic outliers with measurable functional effects. Generally speaking, this type of pattern occurs through the process of migration-selection balance, when the strength of selection is strong enough to overcome homogenizing forces of gene flow, i.e., if the selection coefficient s is much greater than the migration rate m (Charlesworth et al. 1997; Slatkin 1987; Yeaman and Whitlock 2011). These three Andean duck species are thus ideal for assessing targets of local adaptation in the HIF signaling pathway through outlier analyses, i.e., genomic scans.
Objectives
Here, using target-enrichment sequencing, we test whether variation in 26 genes in the HIF signaling pathway is associated with high altitude in three duck species that colonized the Andes from ancestral low-altitude habitats in South America. Analysis of these three species allowed us to identify whether (a) polymorphism in HIF pathway genes is statistically associated with occupancy of high-altitude hypoxic environments, and (b) if so, whether the same genes functional region, amino acids, or nucleotide substitutions converged in each duck lineage. Here, we focus on the role of exonic variation (i.e., nonsynonymous changes) owing to our ability to better extrapolate its ability to have a direct physiological effect, though we do briefly discuss the potential for intronic variation to also affect a functional change. Ultimately, we found strong support for convergent evolution on the HIF pathway (EPAS1, EGLN1), with the same exonic regions of EPAS1, and intronic regions of EGLN1, exhibiting sharply demarcated outliers with a high probability of directional selection in high-altitude populations of two of the three duck species.
Materials and methods
Specimen collection
A total of 60 individuals were used for this study from three different Andean duck species. For the cinnamon teal, individuals from low-altitude populations are the A. c. cyanoptera subspecies (n = 10; elevation range 8–23 m) and from high-altitude are the A. c. orinomus subspecies (n = 10; elevation range 3533–3871 m) (Wilson et al. 2013). For the speckled teal, individuals from low-altitude populations are the A. f. flavirostris subspecies (n = 10; elevation range 77–860 m) and from high-altitude are the A. f. oxyptera subspecies (n = 10; elevation range 3211–4405). For the yellow-billed pintail, individuals from both populations are taxonomically identified as Anas georgica spinicauda. A total of 20 yellow-billed pintails were collected from low- (n = 10; elevation range 292–914 m) and high-altitude (n = 10; elevation range 3332–4070 m). Additional information regarding the samples, including location and dates collected, have been previously described in Graham et al. (2018) and McCracken et al. (2009b).
More detailed information about the phylogeography of these three species, including their population sizes, dispersal behavior across elevational gradients, and gene flow is available in (Wilson et al. 2013), (Graham et al. 2018), and (McCracken et al. 2009a; McCracken et al. 2009c), respectively.
Although these relatively small sample sizes make it impossible to demarcate genomic outliers with low FST such as those experiencing weaker selection, our previous research has shown that these samples can successfully be used to identify extreme outliers with high FST demonstrated to have large phenotypic effects (Natarajan et al. 2015).
HIF pathway gene rationale
HIF pathway genes were chosen based on a combination of (1) being found to be a gene of interest in previous studies of humans using whole-genome scans looking to identify genetic adaptations to high-altitude or hypoxic environments, (2) being a part of the canonical HIF pathway, either through being a known downstream target, or a part of the repression machinery of the pathway, and/or (3) being a part of similar transcription factor families (i.e., bHLH-PAS containing proteins).
A total of 26 genes were able to have probes created for them (Fig. 1, Supp. Fig. 1, Supp. Table 1). These genes represent a wide-breath of coverage across the various parts of the HIF pathway (upstream players, major Transcription Factors/TF, TF repressors, and downstream targets). We were able to capture 100% of the major TFs, and 4/5 of the E3 ubiquitin ligase complex associated with HIF repression; we are unable to capture all known downstream targets of the HIF complex owing to the sheer number of targets it binds to, though we were able to capture all relevant genes in that category (i.e., those which had been major outliers in other high-altitude organisms). Ultimately, we were able to capture a vast majority of the pertinent elements of the HIF pathway incorporating 26 genes, most of which possessed long-coding regions.
Of particular interest were the major HIF pathway TFs (HIF1a, HIF2a/EPAS1); these genes had prior expectations for showing signatures of selection (see ‘Identification of Outlier Loci’ section in the Methods). All HIFα proteins are characterized by the presence of an N-terminal bHLH DNA-binding domain upstream of two PAS domains. In addition, all vertebrate α-subunits include an inhibitory domain called the O2-dependent degradation domain, and an N-terminal transactivation domain (NTAD) (Graham and Presnell 2017). HIF1A and EPAS1 proteins are characterized by the presence of a C-terminal transactivation domain (CTAD) located at the C-terminal end of the protein (Lisy and Peet 2008). These domains are considered critical to the overall function of HIF proteins: the bHLH domain contacts the core nucleotides of HIF-responsive elements (Dinkel et al. 2015), whereas bHLH and PAS domains together mediate both dimerization and sequence specific DNA-binding (Crews and Fan 1999; Ledent and Vervoort 2001). The NTAD is thought to confer target specificity (Hu et al. 2007), whereas the CTAD is required for full HIF activity (Lando et al. 2002) and interactions with co-activators (Carrero et al. 2000; Ema et al. 1999).
DNA extraction and target-enrichment sequencing
Genomic DNA was extracted from muscle tissue using a DNeasy Tissue Kit (Qiagen, Valencia, California, USA) following manufacturer's protocols. In-solution target capture was used to selectively enrich libraries for regions of interest prior to sequencing (Gnirke et al. 2009). All steps of the process were performed by MYcroarray (Ann Arbor, MI), now Arbor Biosciences. A custom MYbaits® biotinylated ssRNA target capture baitset was designed for enriching target sequences. In total, 26 genes associated with the HIF pathway from the duck nuclear genome were selected from the Ensembl A. platyrhynchos v1.0 genome. These 26 sequences were first screened using the web server version of RepeatMasker (http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker) with default settings and selecting A. platyrhynchos as the species, soft-masking all repeats. Next, 120mer probes at 2× tiling density were designed from these soft-masked sequences. All candidate probes were then screened against the duck genome using MYcroarray’s in silico bait analysis software pipeline, in order to filter out any probes that were non-specific in the genome. A final set of 12,062 filtered probes was chosen for 26 genes (Supp. Table 1), which included all probes that (a) did not contain any soft-masked regions and (b) passed MYcroarray’s most liberal filtering threshold. Following hybridization, target regions were purified on magnetic beads followed by post-hybridization amplification to ligate indexing sequences. Sequencing was performed on an Illumina HiSeq platform paired-end (100 bp) with a 250–300 bp insert size.
RAD-seq data
Previously generated RAD-Seq (Restriction Site Associated DNA Sequencing) data were utilized in order to function as a “backdrop” i.e., genomic background, for analyses for the HIF pathway enriched SNP data; the RAD-seq were generated using methods described in (Natarajan et al. 2015) and (Graham et al. 2018). All RAD clusters were then subjected to a BLAST search (database—nr, e value < e−10, annotation cutoff > 50) in Blast2GO (Conesa et al. 2005) using the taxonomy filter for birds (taxa: 8782, Aves) to determine gene identity. Ultimately, SNPs on clusters whose BLAST result was that of a protein-coding gene was then combined with the newly generated target-enrichment data in order to perform selection analyses.
Target enrichment/HIF pathway sequence assembly pipeline
Sequences from the target-enrichment sequencing were received pre-parsed by individual, with adapters trimmed and quality filtered (Q < 30). Additional adapter trimming was performed utilizing fastq-clipper (AGATCGGAAGAGC) and remaining sequences were then filtered by length and quality using fastq-quality filter (reads < 20 bp, and Q < 30) from the FASTX-Toolkit v. 0.0.13 package (Gordon and Hannon 2010). A custom pipeline (scripts located at https://github.com/amgraham07) was created to remove orphan sequences and assemble sequences against the 26 reference genes using BWA v0.7.15 (Li and Durbin 2009). The Samtools package v1.3.1, including BCFtools v.1.3.1 (Li et al. 2009) was then used to create a VCF file, as well as provide assembly statistics (i.e., bp-by-bp coverage). These programs used in the pipeline called SNP variants that were variable against the mallard genome reference, including indels (insertion/deletion); however, the indel information was excluded in the final data set, as the software used does not accommodate indels.
Identification of outlier loci
A combined data set used for detection of outlier loci contained (a) all SNPs associated with the HIF pathway from the target-enrichment sequencing data and (b) SNPs from RAD clusters whose BLAST results hit back to a protein-coding gene (see previous section). The latter provides both a comparison of a priori candidate genes and a genome-wide reference data set for the potential HIF outliers to be analyzed. Ultimately, any outliers described in the results are ones associated with the HIF pathway from target-enrichment sequencing, as the RAD-seq data set only serves as a “backdrop” for analysis.
We tested for signatures of directional selection using three different outlier detection techniques, which minimized the risk of detecting false positives. Outlier loci were considered to be those that were identified by at least two of the three methods as being significant outliers, with the most significant outliers classified as being identified by all three methods.
First, a genomic scan was performed by obtaining pairwise site-by-site FST calculated in Arlequin v. 3.5 (Excoffier and Lischer 2010); these values were then imported into JMP Pro 12 for distribution visualization, as well as for percentile calculations for each species. Candidate loci FST values that exceeded the 99th percentile of the FST values were therefore considered likely targets of selection. Second, MCHEZA, which implements the Dfdist function, was used to demarcate markers putatively under positive directional selection (Antao and Beaumont 2011). MCHEZA analyses were based on the Infinite Alleles Model with 50,000 simulations, a confidence interval of 0.95 and a false-discovery rate of 0.01, using the neutral mean FST and forcing mean FST options. It is important to note here, that MCHEZA was used instead of LOSITAN (Antao et al. 2008) owing to the large size of the data set, and current issues with Java updates (T. Antao, per comm.).
Third, a Bayesian approach as implemented in BayeScan v. 2.1 was used to again identify putatively selected loci. BayeScan uses a logistic regression model to separate locus-specific effects of selection from demographic differences (Foll and Gaggiotti 2008). For each SNP, BayeScan estimates the posterior distribution under neutrality α = 0 and separately allowing for selection α ≠ 0 and computes the posterior odds ratio (PO) as a measure of support for the model of local adaptation relative to neutral demography. Foll (2012) proposed a logarithmic scale for the posterior odds defined as: Bayes Factor (BF) 3–10 substantial (log10PO > 0.5–1.0); BF 10–32 strong (log10PO > 1–1.5); BF 32–100 very strong (log10PO > 1.5–2.0); and BF 100–∞ decisive (log10PO > 2.0–∞) evidence for accepting a model. In the genome scans, a threshold for log10PO > 0.5 (substantial), representing a Bayes Factor of 3.0 and posterior probability of 0.76, was used for a marker to be considered under selection. Therefore, loci identified as “substantial” probability of being under selection were classified as being significant outliers, under this approach.
Results
HIF pathway assembly information and population divergence estimates
The total number of bp for the 26 genes in the target-enrichment data set was 839,657 bp per individual, with the goal of ~ 50× coverage per gene in each individual (Supp. Tables 1 and 2). Across all individuals of the three species, the final coverage for each gene was between 74 × (P4HA3) and 584 × (CUL2), with an average across all genes of 414×. This coverage included genes whose total regions covered 81–100% of the reference mallard sequence, with an average of 98.1% ( ± 3.9%) (Supp. Table 2). Within each species, there were a total of 16,339 HIF pathway SNPs for the cinnamon teal, 21,674 SNPs for speckled teal, and 26,484 SNPs for the yellow-billed pintail, which likely reflects apparent differences in the effective population sizes (Ne) of these three species (Graham et al. 2018; McCracken et al. 2009c; Wilson et al. 2013).
The RAD-Seq yielded a total of 18,145 SNPs from cinnamon teal, 47,731 SNPs from speckled teal, and 49,670 SNPs for the yellow-billed pintail (See Natarajan et al. 2015) for other summary statistics concerning the RAD-seq data sets). Among these, 2762 SNPs for cinnamon teal, 6280 SNPs for speckled teal, and 6523 SNPs for yellow-billed pintail mapped to a gene region, representing 13–15% of the total RAD-Seq clusters. Of the subset of those RAD-seq clusters that mapped to a gene, 1441 clusters with an average of 1.9 SNPs per cluster for the cinnamon teal at an average of 121× coverage, 2600 clusters with an average of 2.4 SNPs per cluster for the speckled teal at an average of 85× coverage, and 2619 clusters with an average of 2.5 SNPs per cluster for the yellow-billed pintail at an average of 112× coverage.
Initially, FST was calculated for three separate subsets of the data between each pair of low- and high- altitude populations: (1) target-enrichment/HIF pathway only, (2) RAD-Seq gene-only, and from the (3) combined HIF pathway and RAD-Seq gene-only data sets. All species showed similar average FST values, except for the HIF pathway, which had slightly greater overall divergence than the RAD-Seq data set. These estimates mirrored previous calculations with nuclear loci in these species (Table 1) (McCracken et al. 2009a; McCracken et al. 2009c; Wilson et al. 2013).
Table 1.
Estimates of divergence (FST) for the three species across the different data sets, including (1) the variants associated with the HIF pathway (i.e., 26 genes), (2) variants associated with the RAD-seq data set whose sequence had a significant BLAST hit to any protein-coding gene, and (3) the combined variant data set including both (1 and 2)
| F ST | |||
|---|---|---|---|
| Species | HIF pathway enriched | RAD-seq (genes only) | Combined |
| Cinnamon teal | 0.051 | 0.042 | 0.04 |
| Yellow-billed pintail | 0.045 | 0.013 | 0.02 |
| Speckled teal | 0.124 | 0.058 | 0.06 |
Identifying HIF pathway gene regions with high FST
HIF pathway gene regions with high FST were examined using three different methods (percentile, Dfdist, Bayesian) for cross-comparison among species. These analyses were performed with a combined data set of SNPs from both the HIF pathway enriched sequence data, as well as the RAD-seq gene-only sequence data.
There was general evidence of convergent evolution for the HIF pathway across high-altitude populations of all three duck species (Supp. Table 3–5). For cinnamon teal, the results from the Dfdist analyses showed seven genes with outliers, with EPAS1 (36 SNPs) and NOS1 (10 SNPs) having the most hits, followed by EGLN1 (4 SNPs), CLOCK (2 SNPs), and PPARA1, MTOR, and P4HA3 (one SNP each); however, none were significant in the corresponding Bayesian analysis.
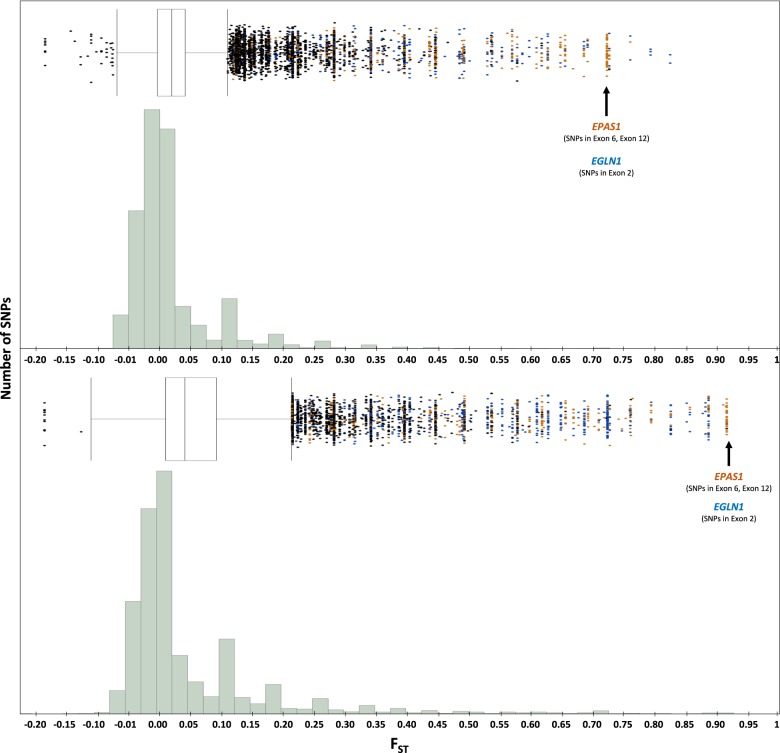
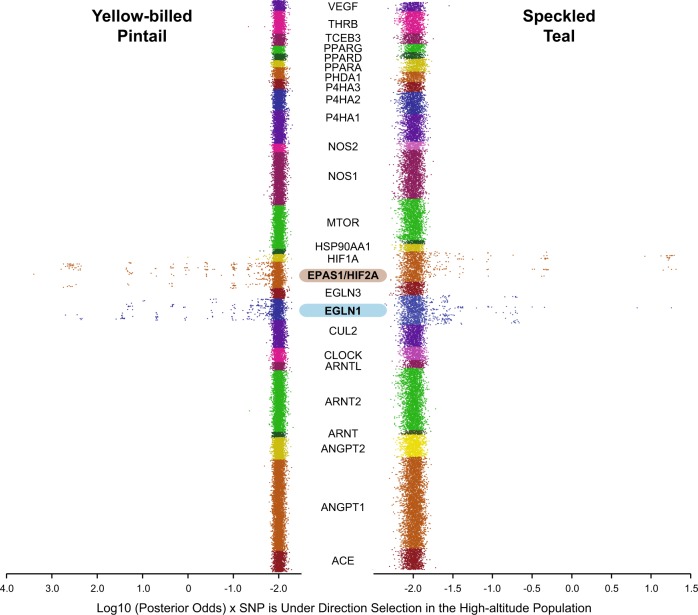
The strongest outliers in both speckled teal and yellow-billed pintail were EGLN1 and EPAS1 and were highly significant in both the Bayesian and Dfdist analyses for both species (Figs 1 and 2, Supp. Table 4, 5). For the HIF pathway SNPs, although most of the SNPs associated with these two genes were located in intronic/noncoding regions, both speckled teal and yellow-billed pintail contained highly significant FST outliers in various exonic regions of EPAS1 and EGLN1—specifically, exon 6 (YBP) and exon 12 (ST, YBP) of EPAS1, and in exon 2 (YBP) of EGLN1 (Figs 3 and 4; Supp. Fig. 2). All exonic SNP variants of particular interest in the low-altitude populations were identical to the presumably ancestral alleles found in the mallard reference, whereas the SNP variants in high-altitude populations were derived. None of the outliers discussed were fixed (i.e., FST = 1.0) between high- and low-altitude populations for any of the three species.
Fig. 2.
RAD-seq cluster distribution against a measurement of population divergence for yellow-billed pintail (top) and speckled teal (bottom): histogram is distribution of all SNP variants (RAD-seq, and HIF pathway genes), with specific genes of interest highlighted: EGLN1 (blue) and EPAS1 (orange), whereas markers with black arrows signify the nonsynonymous/synonymous amino-acid variants in exonic regions of EGLN1 and EPAS1
Fig. 3.
Manhattan scatterplots of each of the 26 HIF pathway gene members listed in alphabetical order, and with their positions in numerical order for both yellow-billed pintail and speckled teal. The two genes with any significant outliers are highlighted (log10PO > 0.5; EGLN1, EPAS1)
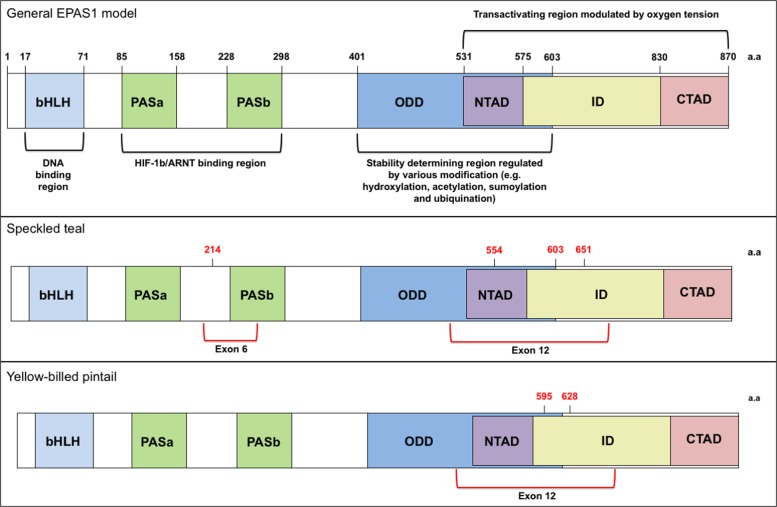
Fig. 4.
General protein model of EPAS1, modified from (Hong et al. 2004), including the canonical domains of the HIF gene family. The variants associated with exonic regions in the gene for both yellow-billed pintail and speckled teal are shown with their associated location/proximity to those domains
Conservation of exons in human and mallard EPAS1 and EGLN1
Further analyses focused on those high FST variants found in exon 6 and 12 in EPAS1 and exon 2 in EGLN1. We compared mallard with human in an explicit test to look for conservation of exon structure. If these exons were not the same, then we could not realistically draw any comparisons from previous literature between the two exons in terms of their potential function. In order to compare the similarity of exon sequence between ducks and humans, the full protein sequences from mallard and human were aligned to each other and exons 6 and 12 were annotated: EPAS1 (human, Q99814; mallard U31HW4), and EGLN1 (human Q9GZT9; mallard, U3J106). Overall, the alignment showed sufficient conservation between sequences, in combination with orthology quality-control statistics for the whole gene from ENSEMBL (GOC—100/100; WGA–91.92/100).
In speckled teal and yellow-billed pintail, four out of the five SNPs resulted in nonsynonymous substitutions in the first and second codon positions (Supp. Fig. 2). Specifically, in EPAS1 there was a Cys → Tyr substitution at amino-acid position 23 in exon 6 and then Arg → His and Tyr → His substitutions at amino-acid positions 32 and 127, respectively, in speckled teal. In yellow-billed pintail, there were two nonsynonymous substitutions at two different positions in exon 12, Pro → Glu at position 71 and Ala → Thr at amino-acid position 104 (Table 2). For EGLN1, the same general approach was taken, by comparing the mallard and human protein sequence, and human and mallard nucleotide sequence for exon 2; however, the observed nucleotide change resulted in a synonymous substitution.
Table 2.
Nonsynonymous changes to exon 12 in EPAS of the speckled teal (ST) and yellow-billed pintail (YBP) between their respective high- and low-altitude populations (also see Fig. 3)
| Species | Low | High | Within exon aa position | Whole protein aa position |
|---|---|---|---|---|
| ST | Cys | Tyr | 26 (of exon 6) | 214 |
| ST | Arg | His | 32 (of exon 12) | 554 |
| ST | Tyr | His | 126 (of exon 12) | 651 |
| YBP | Pro | Glu | 71 (of exon 12) | 595 |
| YBP | Ala | Thr | 104 (of exon 12) | 628 |
Biochemical properties and structural locations of amino-acid replacements in EPAS1
The two nonsynonymous SNPs in EPAS1 exon 12 of yellow-billed pintail resulted in substitutions of amino acids with very different biochemical properties. The Pro → Glu substitution at amino-acid position 71 resulted in a change from a neutral, polar amino acid, to a hydrophobic, nonpolar amino acid in the high-altitude population. The Ala → Thr substitution at amino-acid position 104 resulted in a change from a nonpolar amino acid with a hydrophobic side chain, to a neutral, polar amino acid that has a non-aromatic hydroxyl group attached in the high-altitude population (Supp. Fig. 2).
Of the three nonsynonymous SNPs in EPAS1 exon 12 in speckled teal two resulted in conservative substitutions with similar biochemical properties, Cys → Tyr at position 23 and Arg → His at position 32. Though the Tyr → His at amino-acid position 127, resulted in substitution of a nonpolar aromatic amino acid with a hydrophobic side chain, to a basic, polar amino acid with a positively charged side-chain in the high-altitude population.
Locations of these variants were unable to be placed on existing Protein Data Bank protein models, owing to all available current models containing only the bHLH, and PAS domains, thus lacking any of the ODD/NTAD/CTAD domain regions that are characteristic of the HIF transcription factors (Graham and Presnell 2017). This makes potential functional assessment of the variants in the future problematic.
Discussion
The results of this study show a strong degree of convergent evolution on the HIF pathway in two of the three Andean duck species assayed in this study. Although the cinnamon teal showed evidence of outliers in seven HIF pathway genes, none were significant in the corresponding Bayesian analysis; the evidence for convergence on the HIF pathway as a means of adaptation to high-altitude environments was much stronger in the speckled teal and yellow-billed pintail—specifically, our results suggest that selection acted on the same genes (EPAS1, EGLN1; Figs 2 and 3) and even the same gene regions (exon 12 in EGLN1), though not at the same amino acid positions (Fig. 4; Supp. Fig. 2). It is important to note that all SNP variants for the two species revealed the low-altitude populations being identical to the “ancestral” alleles found in the mallard reference, thus suggesting that such variants are potentially adaptive in the high-altitude populations only.
We focused the results and the discussion more heavily toward those nonsynonymous variants with protein-coding regions because of what prior work has shown regarding those regions (exon 6, and 12 in EPAS1); however, we fully acknowledge the potential for intronic variants (as has been found in human populations) to affect the regulation of gene activity (Hsiao et al. 2016; Park et al. 2018), though it is difficult for us to infer such functions from the sequence data presented in this study. Ultimately, our results also show of convergence with previously studied populations of other organisms living at high-altitudes, specifically humans (which have also shown EPAS1 and EGLN1 as major outliers), extending to the same genes associated with adaptations to low-O2 environments.
The role of the HIF pathway in adaptations to high-altitude environments
Members of the HIF pathway are consistently among the top genomic outliers in organisms dealing with chronic hypoxic stress, and is especially true for the major transcription factor machinery (EPAS1) and its repression machinery (EGLN1). These genes are frequently targets for selection, because genes that form a hub in a regulatory network between a series of upstream activators and a battery of downstream effector genes are thought to be more likely to become the targets of repeated parallel or convergent evolution (i.e., “hotspot” genes) (Martin and Orgogozo 2013; Rosenblum et al. 2014; Stern and Orgogozo 2009). In addition, such variation within the gene would be more likely to occur in exonic regions/domains that are associated with protein–protein interactions rather than DNA-binding activity (Wagner and Lynch 2008); for example, if they play an essential role in coordinating the expression of target genes in response to multiple input signals in a way that cis-regulatory elements cannot (Wagner and Lynch 2010). Both EPAS1 and EGLN1 fit these criteria because they (1) sit at important junction which ultimately regulates the physiological response through a wide array of downstream targets, and (2) have shown causal variants within protein–protein interaction domains (i.e., ODD/NTAD/CTAD); therefore, these genes are prime candidates to be labeled as “hotspot” genes.
Much of what is known about how the HIF pathway is associated with adaptation to high-altitude environments comes from early genome-wide association studies in humans; these genome-wide scans identified many candidate genes that may contribute to adaptive evolution, including two genes (EPAS1 and EGLN1) that are involved in the HIF pathway, which showed the strongest signals of selective sweeps in Tibetan and Andean humans (Beall et al. 2010; Bigham et al. 2010; Peng et al. 2011; Simonson et al. 2012; Xu et al. 2011), as well as in other high-altitude human populations such as Caucasian/Russians (Pagani et al. 2012), Mongolians (Xing et al. 2013) and others populations in the Himalayas (Arciero et al. 2018; Hackinger et al. 2016). In addition, animals residing in similar high-altitude environments have also had the same HIF pathway members identified as major outliers, including ungulates (Song et al. 2016), birds (Qu et al. 2013), pigs (Ai et al. 2014; Li et al. 2013), and dogs (Gou et al. 2014; Li et al. 2014).
The function of some of these variants has only recently been assessed; these variants are generally associated with the relationship between EPAS1 and EGLN1, so it is important to understand dynamics between the transcription factor (EPAS1) and its repressor (EGLN1). In most organisms, under normoxia, EGLN1 can perform its O2-dependent hydroxylase function on both HIF1A and EPAS1 proteins, which triggers degradation via additional enzymatic complexes. However, under hypoxia, the hydroxylase activity of EGLN1 is suppressed, resulting in the accumulation of HIFAs that can then activate hundreds of downstream target genes, thus inducing numerous physiological responses, including changes in red blood cell (RBC) production (Haase 2013; Hu et al. 2003).
In high-altitude human populations, variants have been shown to alter this relationship. Specifically, EGLN1 has been shown to have both loss-of-function, and gain-of-function mutations, depending on the study population (Song et al. 2014; Xiang et al. 2013). Specifically, these mutations have been shown to affect the ability of EGLN1 to target EPAS1 for further degradation, ultimately activating a broad range of effects orchestrating acclimatization to hypoxia, and potentially leading to a blunted hypoxic response (Lorenzo et al. 2014; Song et al. 2014; Xiang et al. 2013). Unlike EGLN1 variants, many EPAS1 variants are located in the noncoding regions, suggesting that they could affect the regulation of EPAS1 at the transcriptional level (Peng et al. 2017; Peng et al. 2011).
EPAS1 and EGLN1 in high-altitude adaptation in Andean ducks
Functional work on the role of the EPAS1 and EGLN1 variants associated with high-altitude adaptation in humans is the best studied (see previous section); in addition, these genes are highly conserved in nucleotide composition and exon structure across vertebrates, including between ducks and humans (Supp. Fig. 3). Such information is crucial because it allows us to extrapolate from information on well-studied organisms (i.e., humans) to ones that have less information available (i.e., ducks).
Although our results identified no variation in coding regions of EGLN1, the variation found within intronic regions of the high-altitude duck species could still suggest a role for transcriptional regulation of EGLN1 in high-altitude adaptation in the Andean duck species of this study; however, it is hard to hypothesize further about its potential effect in the context of these duck species, beyond speculating. Unlike EGLN1 in our study, we found that EPAS1 contained outliers located within exons: four in speckled teal (one synonymous and three nonsynonymous) and two in yellow-billed pintail (both nonsynonymous; Supp. Table 3, 4). The nonsynonymous variants are located in two specific exons, both of which make-up parts of critical domains of the EPAS1 protein that define HIF function, i.e., the PAS domain (exon 6) and ODD/NTAD domain (exon 12; Fig. 4); therefore, we can hypothesize more thoroughly about possible functional effects on physiological changes that would be associated with adaptation to high-altitude. Specifically, the PAS domains mediate both dimerization and sequence-specific DNA binding (Crews and Fan 1999; Ledent and Vervoort 2001), whereas the ODD is the target of HIF repression machinery during normoxia (i.e., oxygen-dependent degradation), and NTAD is thought to confer target specificity (Hu et al. 2007).
Interestingly, variants in the same exons we identified in EPAS1 (exon 6, exon 12) have also been implicated in being associated with various physiological responses to oxygen stress in humans and other organisms (Buroker et al. 2012; Gale et al. 2008; Newman et al. 2015; Percy et al. 2008a; Percy et al. 2008b; Yi et al. 2010); however, exon structure is not always conserved, owing to exon shuffling and other mechanisms during genome evolution (Keren et al. 2010), so we checked both exons for congruence. Even though humans and birds are separated by hundreds of millions of years of evolution (~ 300 mya), both have retained very similar exon structure; this level of conservation also extends to both exon 6 and exon 12 (e.g., exon 12 humans = exon 12 in ducks; Supp. Fig. 3). Thus, it is appropriate to extrapolate results from what is known about similar variation in the same exons of other organisms, and how it might relate back our outlier variants in the Andean duck species.
Specifically, variation in exon 12 in humans has been implicated with both loss-of-function and gain-of-function mutations associated with RBC production. Such gain-of-function mutations are known to cause erythrocytosis, resulting in an increased number of RBCs through its regulation of erythropoietan, in combination with pulmonary arterial hypertension, in humans (Gale et al. 2008; Percy et al. 2008a; Percy et al. 2008b). Cattle housed at high-altitude also have similar gain-of-function variants in exon 12 associated with pulmonary hypertension (Newman et al. 2015). Although, in both the human and cattle studies, those variants were largely considered deleterious, they demonstrate the ability for genetic variation in exon 12 of EPAS1 to have direct physiological effects similar to those frequently targeted by selection in hypoxic environments. In addition, our results showed that speckled teal had additional outliers in EPAS1 located in exon 6, which has been identified in human high-altitude populations to be responsible for adaptive changes in heart rate and hypertension (Buroker et al. 2012; Yi et al. 2010). Our results mirror variation in EPAS1 in other human populations, in the sense that the gene itself is a target of selection; however, high-altitude adaptation in Tibetan and Andean populations have only shown evidence for variation in non-protein-coding regions of EPAS1, unlike our results that show significant variants in both protein-coding and non-protein-coding regions of EPAS1.
Exactly which mechanistic avenues these Andean duck species are using is unclear, given the current data; yet, it is interesting to note that the speckled teal, like Tibetan humans, are characterized by Hb levels more similar that of their low-altitude counterparts, unlike other some Andean duck populations, like yellow-billed pintail and cinnamon teal (McCracken, unpublished data), who show elevated Hb like Andean people (Beall 2007; Lague et al. 2017). This could suggest a connection between EPAS1, EGLN1, and Hb production, especially based on what we know about Tibetan and Andean populations whose genetic variation in both genes has been implicated in protecting individuals from erythrocytosis/polycythemia, resulting from living at high-altitude (Peng et al. 2017; Peng et al. 2011; Wu and Kayser 2006). With potentially causative exonic variation present in EPAS1 and intronic variation in EGLN1, the relationship between these two genes and potential avenues for high-altitude adaptation is similar to those found in human populations. The ultimate hypothesis from our results is that a combination of function-altering (either gain- or loss-of function) mutations in EPAS1, and transcriptional regulation differences between EGLN1 haplotype variants, may be causing changes in downstream target transactivation, resulting in a blunted hypoxic response. Testing this connection between these variants and such a physiological response would require future functional assessment (next section).
Future functional assessment of EGLN1 and EPAS1 in Andean waterfowl
Despite similar results from other species, it is difficult to say with certainty what effect these variants actually have in the waterfowl species assayed in this study, without functional assessment at the physiological level. The future goal would be to assess these variants in a more causal way than the outlier analyses presented in this study; however, there is some difficulty in performing direct assessment due to the issue of EPAS1 not having a full protein structure available (i.e., lacking the ODD/NTAD/CTAD domains), in addition to the general issues of performing functional genomics in a wild vertebrate species, with no captive caught populations.
It is important to note that, as it relates to functional assessment of variants for either EPAS1 or EGLN1 found in the more prominent human studies, currently none have been directly assessed (i.e., site-directed mutagenesis); instead, they have been indirectly assessed through whole-scale gene knockdowns in mice and tissue culture, and/or measurements of various physiological differences (Peng et al. 2017; Xiang et al. 2013). Even though our extrapolation from previous functional work on the same genes is somewhat circumstantial in respect to the organisms used in this study, we feel that these results from other organisms will potentially help guide future experiments concerning how best to assay the role of the variants in enabling high-altitude living found in this study.
Conclusions
Hypoxia as a stressor in high-altitude environments has been shown to have facilitated a great degree of convergence across many animals, but especially in the case of high-altitude human and Andean duck populations, as demonstrated by the results of this research. Ultimately, we were able to show a potentially high degree of convergence between high-altitude human and duck populations on the HIF pathway itself, but also that convergence included two gene members, EPAS1 and EGLN1. In addition, between the high-altitude populations of speckled teal and yellow-billed pintail, nonsynonymous changes in specific exons (exon 12) in close proximity to protein domains associated with O2-driven protein stability and transactivation suggests evidence of strong molecular convergence. Although the specific molecular mechanisms associated with these variants are currently unknown in this system, previous work from other organisms suggests that these variants are likely resulting in a blunted hypoxic response, potentially through expression of Hb and other downstream targets. Ultimately, we were able to identify a potential molecular mechanism for high-altitude adaptation in Andean duck species, through the HIF pathway; in addition, the results also highlight how frequently natural selection can select for the same genes and pathways in response to a similar selective pressure, resulting in convergent mechanisms of adaptation to similar environments.
Data archiving
Parsed Illumina reads for the RAD-seq data sets are deposited in the NCBI short read archive (SRA PRJEB11624). Raw Illumina reads for the target-enrichment data sets are deposited in the NCBI short reach archive (SRA PRJNA508951). A Dryad digital repository houses additional data files (10.5061/dryad.kd08516). Scripts and other small files associated with analysis are available on GitHub (https://github.com/amgraham07).
Supplementary information
Acknowledgements
We thank the many people and provincial and federal governments in Argentina, Peru, and Bolivia who assisted us with fieldwork for many years. Funding was provided by Alaska EPSCoR (NSF EPS-0092040, EPS-0346770), the National Science Foundation (DEB-0444748 and IOS-0949439), James Kushlan Endowment for Waterbird Biology and Conservation at the University of Miami, and Frank M. Chapman Fund at the American Museum of Natural History. Specimen collections were carried out under University of Alaska-Fairbanks IACUC protocols #02-01 and #05-05. This work was supported in part by the high-performance computing and data storage resources operated by the Research Computing Systems Group at the University of Alaska Fairbanks, Geophysical Institute.
Author contributions
AMG and KGM designed the study; KGM provided funding; AMG performed the research/generated/analyzed the data. AMG and KGM wrote the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41437-018-0173-z) contains supplementary material, which is available to authorized users.
References
- Aggarwal S, Negi S, Jha P, Singh PK, Stobdan T, Pasha MQ, et al. EGLN1 involvement in high-altitude adaptation revealed through genetic analysis of extreme constitution types defined in Ayurveda. PNAS. 2010;107:18961–18966. doi: 10.1073/pnas.1006108107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai H, Yang B, Li J, Xie X, Chen H, Ren J. Population history and genomic signatures for high-altitude adaptation in Tibetan pigs. BMC Genom. 2014;15:834. doi: 10.1186/1471-2164-15-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkorta-Aranburu G, Beall CM, Witonsky DB, Gebremedhin A, Pritchard JK, Di Rienzo A. The genetic architecture of adaptations to high altitude in Ethiopia. PLoS Genet. 2012;8:e1003110. doi: 10.1371/journal.pgen.1003110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antao T, Beaumont MA. Mcheza: a workbench to detect selection using dominant markers. Bioinformatics. 2011;27:1717–1718. doi: 10.1093/bioinformatics/btr253. [DOI] [PubMed] [Google Scholar]
- Antao T, Lopes A, Lopes RJ, Beja-Pereira A, Luikart G. LOSITAN: a workbench to detect molecular adaptation based on a Fst-outlier method. BMC Bioinformatics. 2008;9:323. doi: 10.1186/1471-2105-9-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arciero E, Kraaijenbrink T, Haber M, Mezzavilla M, Ayub Q, Wang W, et al. Demographic history and genetic adaptation in the Himalayan region inferred from genome-wide SNP genotypes of 49 populations. Mol Biol Evol. 2018;35:1916–1933. doi: 10.1093/molbev/msy094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall CM. Two routes to functional adaptation: Tibetan and Andean high-altitude natives. PNAS. 2007;104:8655–8660. doi: 10.1073/pnas.0701985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall CM, Cavalleri GL, Deng L, Elston RC, Gao Y, Knight J, et al. Natural selection on EPAS1 (HIF2α) associated with low hemoglobin concentration in Tibetan highlanders. PNAS. 2010;107:11459–11464. doi: 10.1073/pnas.1002443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigham A, Bauchet M, Pinto D, Mao X, Akey JM, Mei R, et al. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 2010;6:e1001116. doi: 10.1371/journal.pgen.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigham AW, Mao X, Mei R, Brutsaert T, Wilson MJ, Julian CG, et al. Identifying positive selection candidate loci for high-altitude adaptation in Andean populations. Hum Genomics. 2009;4:79. doi: 10.1186/1479-7364-4-2-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buroker NE, Ning XH, Zhou ZN, Li K, Cen WJ, Wu XF, et al. EPAS1 and EGLN1 associations with high altitude sickness in Han and Tibetan Chinese at the Qinghai–Tibetan Plateau. Blood Cells Mol Dis. 2012;49:67–73. doi: 10.1016/j.bcmd.2012.04.004. [DOI] [PubMed] [Google Scholar]
- Carrero P, Okamoto K, Coumailleau P, O’Brien S, Tanaka H, Poellinger L. Redox-regulated recruitment of the transcriptional coactivators CREB-binding protein and SRC-1 to hypoxia-inducible factor 1α. Mol Cell Biol. 2000;20:402–415. doi: 10.1128/mcb.20.1.402-415.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Nordborg M, Charlesworth D. The effects of local selection, balanced polymorphism and background selection on equilibrium patterns of genetic diversity in subdivided populations. Genet Res. 1997;70:155–174. doi: 10.1017/s0016672397002954. [DOI] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Conte G. L., Arnegard M. E., Peichel C. L., Schluter D. The probability of genetic parallelism and convergence in natural populations. Proceedings of the Royal Society B: Biological Sciences. 2012;279(1749):5039–5047. doi: 10.1098/rspb.2012.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews ST, Fan CM. Remembrance of things PAS: regulation of development by bHLH–PAS proteins. Curr Opin Genet Dev. 1999;9:580–587. doi: 10.1016/s0959-437x(99)00003-9. [DOI] [PubMed] [Google Scholar]
- Dinkel Holger, Van Roey Kim, Michael Sushama, Kumar Manjeet, Uyar Bora, Altenberg Brigitte, Milchevskaya Vladislava, Schneider Melanie, Kühn Helen, Behrendt Annika, Dahl Sophie Luise, Damerell Victoria, Diebel Sandra, Kalman Sara, Klein Steffen, Knudsen Arne C., Mäder Christina, Merrill Sabina, Staudt Angelina, Thiel Vera, Welti Lukas, Davey Norman E., Diella Francesca, Gibson Toby J. ELM 2016—data update and new functionality of the eukaryotic linear motif resource. Nucleic Acids Research. 2015;44(D1):D294–D300. doi: 10.1093/nar/gkv1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ema M, Hirota K, Mimura J, Abe H, Yodoi J, Sogawa K, et al. Molecular mechanisms of transcription activation by HLF and HIF1α in response to hypoxia: their stabilization and redox signal‐induced interaction with CBP/p300. EMBO J. 1999;18:1905–1914. doi: 10.1093/emboj/18.7.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Fjeldså J (1985) Origin, evolution, and status of the avifauna of Andean wetlands. Ornithol Monogr 85–112
- Foll M. BayeScanv2. 1 user manual. Ecology. 2012;20:1450–1462. [Google Scholar]
- Foll M, Gaggiotti O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics. 2008;180:977–993. doi: 10.1534/genetics.108.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale DP, Harten SK, Reid CD, Tuddenham EG, Maxwell PH. Autosomal dominant erythrocytosis and pulmonary arterial hypertension associated with an activating HIF2α mutation. Blood. 2008;112:919–921. doi: 10.1182/blood-2008-04-153718. [DOI] [PubMed] [Google Scholar]
- Ge RL, Simonson TS, Cooksey RC, Tanna U, Qin G, Huff CD, et al. Metabolic insight into mechanisms of high-altitude adaptation in Tibetans. Mol Genet Metab. 2012;106:244–247. doi: 10.1016/j.ymgme.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompel N, Prud’homme B. The causes of repeated genetic evolution. Dev Biol. 2009;332:36–47. doi: 10.1016/j.ydbio.2009.04.040. [DOI] [PubMed] [Google Scholar]
- Gordon A, Hannon G (2010). Fastx-toolkit. Computer program distributed by the author, website http://hannonlabcshledu/fastx_toolkit/index.html
- Gou X, Wang Z, Li N, Qiu F, Xu Z, Yan D, et al. Whole-genome sequencing of six dog breeds from continuous altitudes reveals adaptation to high-altitude hypoxia. Genome Res. 2014;24:1308–1315. doi: 10.1101/gr.171876.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham AM, Lavretsky P, Muñoz-Fuentes V, Green AJ, Wilson RE, McCracken KG. Migration-selection balance drives genetic differentiation in genes associated with high-altitude function in the speckled teal (Anas flavirostris) in the Andes. Genome Biol Evol. 2018;10:14–32. doi: 10.1093/gbe/evx253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham AM, Presnell JS. Hypoxia Inducible Factor (HIF) transcription factor family expansion, diversification, divergence and selection in eukaryotes. PLoS ONE. 2017;12:e0179545. doi: 10.1371/journal.pone.0179545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase VH. Regulation of erythropoiesis by hypoxia-inducible factors. Blood Rev. 2013;27:41–53. doi: 10.1016/j.blre.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackinger S, Kraaijenbrink T, Xue Y, Mezzavilla M, van Driem G, Jobling MA, et al. Wide distribution and altitude correlation of an archaic high-altitude-adaptive EPAS1 haplotype in the Himalayas. Hum Genet. 2016;135:393–402. doi: 10.1007/s00439-016-1641-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka M, Droma Y, Basnyat B, Ito M, Kobayashi N, Katsuyama Y, et al. Genetic variants in EPAS1 contribute to adaptation to high-altitude hypoxia in Sherpas. PLoS ONE. 2012;7:e50566. doi: 10.1371/journal.pone.0050566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SS, Lee H, Kim KW. HIF-1α: a valid therapeutic target for tumor therapy. Cancer Res Treat. 2004;36:343–353. doi: 10.4143/crt.2004.36.6.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao YHE, Bahn JH, Lin X, Chan TM, Wang R, Xiao X. Alternative splicing modulated by genetic variants demonstrates accelerated evolution regulated by highly conserved proteins. Genome Res. 2016;26:440–450. doi: 10.1101/gr.193359.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CJ, Sataur A, Wang L, Chen H, Simon MC. The N-terminal transactivation domain confers target gene specificity of hypoxia-inducible factors HIF-1α and HIF-2α. Mol Biol Cell. 2007;18:4528–4542. doi: 10.1091/mbc.E06-05-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CJ, Wang LY, Chodosh LA, Keith B, Simon MC. Differential roles of hypoxia-inducible factor 1α (HIF-1α) and HIF-2α in hypoxic gene regulation. Mol Cell Biol. 2003;23:9361–9374. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Sánchez E, DeGiorgio M, Pagani L, Tarekegn A, Ekong R, Antao T, et al. Genetic signatures reveal high-altitude adaptation in a set of Ethiopian populations. Mol Biol Evol. 2013;30:1877–1888. doi: 10.1093/molbev/mst089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong C, Alkorta-Aranburu G, Basnyat B, Neupane M, Witonsky DB, Pritchard JK, et al. Admixture facilitates genetic adaptations to high altitude in Tibet. Nat Commun. 2014;5:3281. doi: 10.1038/ncomms4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson Kevin P., Sorenson Michael D. Phylogeny and Biogeography of Dabbling Ducks (Genus: Anas): A Comparison of Molecular and Morphological Evidence. The Auk. 1999;116(3):792–805. [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet. 2010;11:345. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- Lague SL, Chua B, Alza L, Scott GR, Frappell PB, Zhong Y, et al. Divergent respiratory and cardiovascular responses to hypoxia in bar-headed geese and Andean birds. J Exp Biol. 2017;220:4186–4194. doi: 10.1242/jeb.168799. [DOI] [PubMed] [Google Scholar]
- Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Asparagine hydroxylation of the HIF transactivation domain: a hypoxic switch. Science. 2002;295:858–861. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- Ledent V, Vervoort M. The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Res. 2001;11:754–770. doi: 10.1101/gr.177001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Tian S, Jin L, Zhou G, Li Y, Zhang Y, et al. Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat Genet. 2013;45:1431–1438. doi: 10.1038/ng.2811. [DOI] [PubMed] [Google Scholar]
- Li Y, Wu DD, Boyko AR, Wang GD, Wu SF, Irwin DM, et al. Population variation revealed high-altitude adaptation of Tibetan mastiffs. Mol Biol Evol. 2014;31:1200–1205. doi: 10.1093/molbev/msu070. [DOI] [PubMed] [Google Scholar]
- Lisy K, Peet DJ. Turn me on: regulating HIF transcriptional activity. Cell Death Differ. 2008;15:642–649. doi: 10.1038/sj.cdd.4402315. [DOI] [PubMed] [Google Scholar]
- Lorenzo FR, Huff C, Myllymäki M, Olenchock B, Swierczek S, Tashi T, et al. A genetic mechanism for Tibetan high-altitude adaptation. Nat Genet. 2014;46:951–956. doi: 10.1038/ng.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losos JB. Convergence, adaptation, and constraint. Evolution. 2011;65:1827–1840. doi: 10.1111/j.1558-5646.2011.01289.x. [DOI] [PubMed] [Google Scholar]
- Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Orgogozo V. The loci of repeated evolution: a catalog of genetic hotspots of phenotypic variation. Evolution. 2013;67:1235–1250. doi: 10.1111/evo.12081. [DOI] [PubMed] [Google Scholar]
- McCracken KG, Barger CP, Bulgarella M, Johnson KP, Kuhner MK, Moore AV, et al. Signatures of high-altitude adaptation in the major hemoglobin of five species of Andean Dabbling Ducks. Am Nat. 2009;174:631–650. doi: 10.1086/606020. [DOI] [PubMed] [Google Scholar]
- McCracken KG, Barger CP, Bulgarella M, Johnson KP, Sonsthagen SA, Trucco J, et al. Parallel evolution in the major haemoglobin genes of eight species of Andean waterfowl. Mol Ecol. 2009;18:3992–4005. doi: 10.1111/j.1365-294X.2009.04352.x. [DOI] [PubMed] [Google Scholar]
- McCracken KG, Bulgarella M, Johnson KP, Kuhner MK, Trucco J, Valqui TH, et al. Gene flow in the face of countervailing selection: adaptation to high-altitude hypoxia in the beta A hemoglobin subunit of yellow-billed Pintails in the Andes. Mol Biol Evol. 2009;26:815–827. doi: 10.1093/molbev/msp007. [DOI] [PubMed] [Google Scholar]
- Natarajan C, Projecto-Garcia J, Moriyama H, Weber RE, Muñoz-Fuentes V, Green AJ, et al. Convergent evolution of hemoglobin function in high-altitude Andean waterfowl involves limited parallelism at the molecular sequence level. PLoS Genet. 2015;11:e1005681. doi: 10.1371/journal.pgen.1005681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JH, Holt TN, Cogan JD, Womack B, Phillips JA, III, Li C, et al. Increased prevalence of EPAS1 variant in cattle with high-altitude pulmonary hypertension. Nat Commun. 2015;6::6863. doi: 10.1038/ncomms7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani L, Ayub Q, MacArthur DG, Xue Y, Baillie JK, Chen Y, et al. High altitude adaptation in Daghestani populations from the Caucasus. Hum Genet. 2012;131:423–433. doi: 10.1007/s00439-011-1084-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Pan Z, Zhang Z, Lin L, Xing Y. The expanding landscape of alternative splicing variation in human populations. Am J Hum Genet. 2018;102:11–26. doi: 10.1016/j.ajhg.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Cui C, He Y, Zhang H, Yang D, Zhang Q, et al. Down-regulation of EPAS1 transcription and genetic adaptation of Tibetans to high-altitude hypoxia. Mol Biol Evol. 2017;34:818–830. doi: 10.1093/molbev/msw280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Yang Z, Zhang H, Cui C, Qi X, Luo X, et al. Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Mol Biol Evol. 2011;28:1075–1081. doi: 10.1093/molbev/msq290. [DOI] [PubMed] [Google Scholar]
- Percy MJ, Beer PA, Campbell G, Dekker AW, Green AR, Oscier D, et al. Novel exon 12 mutations in the HIF2A gene associated with erythrocytosis. Blood. 2008;111:5400–5402. doi: 10.1182/blood-2008-02-137703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percy MJ, Furlow PW, Lucas GS, Li X, Lappin TR, McMullin MF, et al. A gain-of-function mutation in the HIF2A gene in familial erythrocytosis. N Engl J Med. 2008;358:162–168. doi: 10.1056/NEJMoa073123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Q, Zhang G, Ma T, Qian W, Wang J, Ye Z, et al. The yak genome and adaptation to life at high altitude. Nat Genet. 2012;44:946–949. doi: 10.1038/ng.2343. [DOI] [PubMed] [Google Scholar]
- Qu Y, Zhao H, Han N, Zhou G, Song G, Gao B, et al. Ground tit genome reveals avian adaptation to living at high altitudes in the Tibetan plateau. Nat Commun. 2013;4:2071. doi: 10.1038/ncomms3071. [DOI] [PubMed] [Google Scholar]
- Rosenblum EB, Parent CE, Brandt EE. The molecular basis of phenotypic convergence. Annu Rev Ecol Evol Syst. 2014;45:203–226. [Google Scholar]
- Rytkönen KT, Vuori KA, Primmer CR, Nikinmaa M. Comparison of hypoxia-inducible factor-1 alpha in hypoxia-sensitive and hypoxia-tolerant fish species. Comp Biochem Physiol Part D: Genom Proteom. 2007;2:177–186. doi: 10.1016/j.cbd.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Scheinfeldt LB, Soi S, Thompson S, Ranciaro A, Woldemeskel D, Beggs W, et al. Genetic adaptation to high altitude in the Ethiopian highlands. Genome Biol. 2012;13:R1. doi: 10.1186/gb-2012-13-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinfeldt LB, Tishkoff SA. Living the high life: high-altitude adaptation. Genome Biol. 2010;11:133. doi: 10.1186/gb-2010-11-9-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. Hypoxia-inducible factor 1 (HIF-1) pathway. Sci STKE. 2007;2007:cm8. doi: 10.1126/stke.4072007cm8. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Life with oxygen. Science. 2007;318:62–64. doi: 10.1126/science.1147949. [DOI] [PubMed] [Google Scholar]
- Simonson TS, McClain DA, Jorde LB, Prchal JT. Genetic determinants of Tibetan high-altitude adaptation. Hum Genet. 2012;131:527–533. doi: 10.1007/s00439-011-1109-3. [DOI] [PubMed] [Google Scholar]
- Simonson TS, Yang Y, Huff CD, Yun H, Qin G, Witherspoon DJ, et al. Genetic evidence for high-altitude adaptation in Tibet. Science. 2010;329:72–75. doi: 10.1126/science.1189406. [DOI] [PubMed] [Google Scholar]
- Slatkin M. The average number of sites separating DNA sequences drawn from a subdivided population. Theor Popul Biol. 1987;32:42–49. doi: 10.1016/0040-5809(87)90038-4. [DOI] [PubMed] [Google Scholar]
- Song D, Li Ls, Arsenault PR, Tan Q, Bigham AW, Heaton-Johnson KJ, et al. Defective Tibetan PHD2 binding to p23 links high altitude adaption to altered oxygen sensing. J Biol Chem. 2014;289:14656–14665. doi: 10.1074/jbc.M113.541227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Yao N, Yang M, Liu X, Dong K, Zhao Q, et al. Exome sequencing reveals genetic differentiation due to high-altitude adaptation in the Tibetan cashmere goat (Capra hircus) BMC Genomics. 2016;17:122. doi: 10.1186/s12864-016-2449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V. Is genetic evolution predictable? Science. 2009;323:746–751. doi: 10.1126/science.1158997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF. Causes of molecular convergence and parallelism in protein evolution. Nat Rev Genet. 2016;17:239–250. doi: 10.1038/nrg.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terova G, Rimoldi S, Corà S, Bernardini G, Gornati R, Saroglia M. Acute and chronic hypoxia affects HIF-1α mRNA levels in sea bass (Dicentrarchus labrax) Aquaculture. 2008;279:150–159. [Google Scholar]
- Wagner GP, Lynch VJ. The gene regulatory logic of transcription factor evolution. Trends Ecol Evol. 2008;23:377–385. doi: 10.1016/j.tree.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Wagner GP, Lynch VJ. Evolutionary novelties. Curr Biol. 2010;20:R48–R52. doi: 10.1016/j.cub.2009.11.010. [DOI] [PubMed] [Google Scholar]
- Wang GD, Fan RX, Zhai W, Liu F, Wang L, Zhong L, et al. Genetic convergence in the adaptation of dogs and humans to the high-altitude environment of the Tibetan Plateau. Genome Biol Evol. 2014;6:2122–2128. doi: 10.1093/gbe/evu162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb JD, Coleman ML, Pugh CW. Hypoxia, hypoxia-inducible factors (HIF), HIF hydroxylases and oxygen sensing. Cell Mol Life Sci. 2009;66:3539–3554. doi: 10.1007/s00018-009-0147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger RH, Stiehl DP, Camenisch G. Integration of oxygen signaling at the consensus HRE. Sci STKE. 2005;2005:re12. doi: 10.1126/stke.3062005re12. [DOI] [PubMed] [Google Scholar]
- Wilson RE, Peters JL, McCracken KG. Genetic and phenotypic divergence between low- and high- altitude populations of two recently diverged cinnamon teal subspecies. Evolution. 2013;67:170–184. doi: 10.1111/j.1558-5646.2012.01740.x. [DOI] [PubMed] [Google Scholar]
- Wu T, Kayser B. High altitude adaptation in Tibetans. High Alt Med Biol. 2006;7:193–208. doi: 10.1089/ham.2006.7.193. [DOI] [PubMed] [Google Scholar]
- Xiang K, Peng Y, Yang Z, Zhang X, Cui C, Zhang H, et al. Identification of a Tibetan-specific mutation in the hypoxic gene EGLN1 and its contribution to high-altitude adaptation. Mol Biol Evol. 2013;30:1889–1898. doi: 10.1093/molbev/mst090. [DOI] [PubMed] [Google Scholar]
- Xing J, Wuren T, Simonson TS, Watkins WS, Witherspoon DJ, Wu W, et al. Genomic analysis of natural selection and phenotypic variation in high-altitude Mongolians. PLoS Genet. 2013;9:e1003634. doi: 10.1371/journal.pgen.1003634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Li S, Yang Y, Tan J, Lou H, Jin W, et al. A genome-wide search for signals of high-altitude adaptation in Tibetans. Mol Biol Evol. 2011;28:1003–1011. doi: 10.1093/molbev/msq277. [DOI] [PubMed] [Google Scholar]
- Yeaman S, Whitlock MC. The genetic architecture of adaptation under migration–selection balance. Evolution. 2011;65:1897–1911. doi: 10.1111/j.1558-5646.2011.01269.x. [DOI] [PubMed] [Google Scholar]
- Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZXP, Pool JE, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Parsed Illumina reads for the RAD-seq data sets are deposited in the NCBI short read archive (SRA PRJEB11624). Raw Illumina reads for the target-enrichment data sets are deposited in the NCBI short reach archive (SRA PRJNA508951). A Dryad digital repository houses additional data files (10.5061/dryad.kd08516). Scripts and other small files associated with analysis are available on GitHub (https://github.com/amgraham07).