Abstract
The Almeria–Oran Front (AOF) is a recognised hotspot of genetic differentiation in the sea, with genetic discontinuities reported in more than 50 species. The AOF is a barrier to dispersal and an ecological boundary; both can determine the position of these genetic breaks. However, the maintenance of genetic differentiation is likely reinforced by genetic barriers. A general drawback of previous studies is an insufficient density of sampling sites at the transition zone, with a conspicuous lack of samples from the southern coastline. We analysed the fine-scale genetic structure in the mussel Mytilus galloprovincialis using a few ancestry-informative loci previously identified from genome scans. We discovered a 600-km-wide mosaic hybrid zone eastward of the AOF along the Algerian coasts. This mosaic zone provides a new twist to our understanding of the Atlantic–Mediterranean transition because it demonstrates that the two lineages can live in sympatry with ample opportunities to interbreed in a large area, but they hardly do so. This implies that some form of reproductive isolation must exist to maintain the two genetic backgrounds locally cohesive. The mosaic zone ends with an abrupt genetic shift at a barrier to dispersal in the Gulf of Bejaia, Eastern Algeria. Simulations of endogenous or exogenous selection in models that account for the geography and hydrodynamic features of the region support the hypothesis that sister hybrid zones could have been differentially trapped at two alternative barriers to dispersal and/or environmental boundaries, at Almeria in the north and Bejaia in the south. A preponderantly unidirectional north–south gene flow next to the AOF can also maintain a patch of intrinsically maintained genetic background in the south and the mosaic structure, even in the absence of local adaptation. Our results concur with the coupling hypothesis that suggests that natural barriers can explain the position of genetic breaks, while their maintenance depends on genetic barriers.
Subject terms: Speciation, Genetic variation
Introduction
Natural populations are often subdivided into discrete patches within which an apparent panmixia is observed, or where genetic differentiation increases proportionally with geographic distance (Charlesworth et al. 2003). These roughly homogeneous genetic patches are bordered by abrupt genetic discontinuities that are maintained by “barriers to gene flow” (Arnold 2006; Slatkin 1987; Hewitt 1989). Barriers to gene flow allow divergence between populations and possibly the evolution of reproductive isolation and speciation (Barton and Bengtsson 1986; Palumbi 1994; Charlesworth et al. 2003). They also maintain the beta component (i.e. between-populations component) of genetic and species diversity. Barriers to gene flow are therefore crucial in structuring and maintaining biodiversity. The study of their origin, location and the processes involved in their functioning is therefore key in evolutionary biology and ecology.
When genetic discontinuities are spatially concordant among different species, a parsimonious explanation is that the same factors contribute to the barrier across species. For instance, species can share a common history (e.g. shared zones of secondary contact, sometimes called suture zones; Remington 1968; Hewitt 1996; Avise 2000; Hewitt 2000; Swenson and Howard 2005), a comparable mode of dispersal (e.g. planktonic dispersers should be similarly affected by marine connectivity; Pelc et al. 2009) or a similar ecological niche (e.g. adaptation to the same habitat, such as brackish water in the Baltic Sea; Johannesson and Andre 2006). In the marine realm, decades of research in population genetics have allowed researchers to identify and describe hotspots of genetic differentiation (Rocha and Bowen 2008; Hellberg 2009; Riginos and Liggins 2013; see Table 1). Probably because many marine species have large population sizes and high dispersal potential via the larval phase, genetic homogeneity is almost always found within patches delineated by these barriers, and the concentration of genetic breaks at some locations is obvious in the sea (Hellberg 2009; Riginos et al. 2011; Gagnaire et al. 2015). Barriers to gene flow in the marine environment are found at locations of strong environmental gradients or oceanographic currents, which prevent dispersal along shores (Hellberg et al. 2002; Teske et al. 2014). The factors involved in creating such barriers can be physical (oceanic fronts, local and global oceanic currents; Patarnello et al. 2007), ecological (temperature, salinity and tide level; Johannesson et al. 2010; Stanley et al. 2018), historical (secondary contact; Quesada et al. 1995a; Avise 2000) or genetic (reproductive isolation; Bierne et al. 2011). While barriers to gene flow are more likely to result from a combination of factors acting synergistically (Barton and Hewitt 1985; Bierne et al. 2011; Cordero et al. 2014), a single factor is often emphasised to explain the genetic structure in a given case. In addition, the origin (primary divergence vs. secondary contact), localisation (ecotone vs. natural barrier vs. suture zone) and maintenance (dispersal barrier vs. local adaptation vs. reproductive isolation) of genetic differentiation need to be treated as separate questions. For instance, the position of a genetic cline at a barrier to dispersal does not necessarily mean that the genetic differentiation is due to low dispersal at this location; it is equally likely to be the legacy of divergence during past vicariance and/or the additional effect of a genetic barrier trapped by the natural barrier (Barton 1979). Demographic reconstruction can provide insight into the origin of the differentiation that a simple description of the contemporary genetic structure cannot reveal. However, to better understand the maintenance of genetic breaks, genetic analysis of samples far from the transition zone is hardly sufficient, even with high-throughput genomic data (Cruickshank and Hahn 2014; Meirmans 2015; Harrison and Larson 2016). Such genome-wide analyses allow reconstructing the long-term history of populations and identifying highly differentiated loci or genomic islands (Tine et al. 2014; Fraïsse et al. 2016), but tell little about how genetic lineages interbreed at the boundary. A major shortcoming of many genome-wide studies is that they do not undertake detailed sampling at these points of geographic transition, though such sampling is critical for teasing apart competing explanations. Moreover, one also needs to conduct experiments in the laboratory and in the field to obtain direct evidence of the isolation mechanisms at play (Harrison 1993).
Table 1.
Examples of marine hotspots of genetic differentiation
| Barrier to gene flow | Potential processes involved Vicariance (V) Temperature (T) Salinity (S) Hydrography (H) |
References |
|---|---|---|
| Øresund and Danish Belts | S | Johannesson and Andre (2006) |
| Almeria Oran Front | V, H, S, T | Patarnello et al. (2007); Galarza et al. (2009) |
| Balearic Front | H | Galarza et al. (2009) |
| Siculo–Tunisian Strait | V, H, S, T | Bahri Sfar et al. (2000) |
| Cape Cod (Massachusetts) | V, H | Haney et al. (2007) |
| Cape Canaveral (Florida) | V, H, T | Avise (1992); Pelc et al. (2009) |
| Nova Scotia 44–45°N | T | Stanley et al. (2018) |
| Cape Mendocino (California) | C, H | Sivasundar and Palumbi (2010) |
| Point Conception (California) | V, H | Burton (1998); Pelc et al. (2009). |
| Cape Agulhas (South Africa) | H | Teske et al. (2014) |
| 30°S Chilean biogeographic break | H, T | Ewers‐Saucedo et al. (2016) |
The Almeria–Oran Front (AOF) is a well-known marine barrier to gene flow between populations of the Atlantic Ocean and the Mediterranean Sea (Quesada et al. 1995b; Borsa et al. 1997; Borrero-Pérez et al. 2011; Chevolot et al. 2006; Patarnello et al. 2007; Palero et al. 2008; Schunter et al. 2011; Tine et al. 2014). The front is generated by the convergence of Mediterranean and Atlantic waters. Atlantic waters enter the Alboran Sea through the strait of Gibraltar and produce anticyclonic gyres westward of the AOF, the Western and Eastern Alboran Gyres and a north–south unidirectional current at the AOF (see Fig. 1). In addition, part of the flow of the Eastern Alboran Gyre is trapped at the AOF to contribute to the Algerian current that flows eastward of Oran along the Algerian coastline (Viúdez and Tintoré 1995).
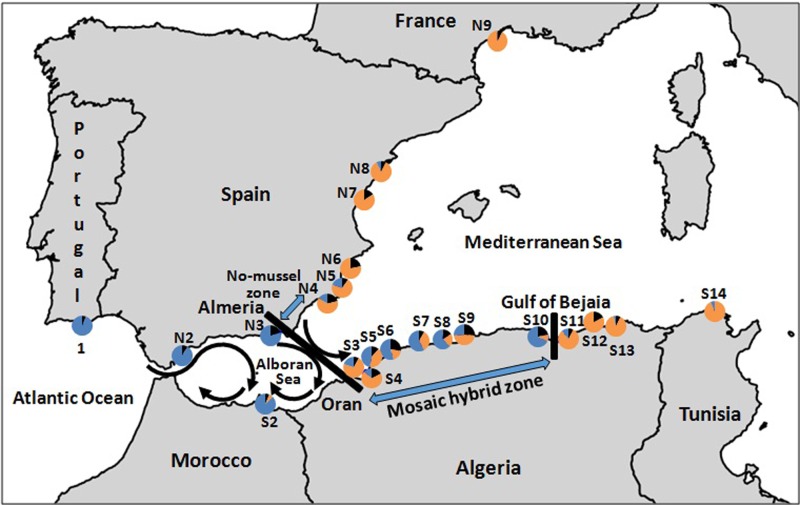
Fig. 1.
Sampling localities of Mytilus galloprovincialis in the north-eastern Atlantic and the Mediterranean Sea. Proportion of individuals assigned to the Atlantic cluster (in red), the Mediterranean cluster (in green) and unassigned (in black), based on a DAPC analysis with the four loci (COIII, Precol-D, EFbis and EF2). Sample names and their GPS positions are given in supplementary Table S1
Among the examples of genetic breaks at the AOF, one among those described first was for the marine mussel Mytilus galloprovincialis (Quesada et al. 1995a; 1995b, Sanjuan et al. 1994). Since the early reports, the genetic break has been considered a secondary contact between two lineages isolated in Mediterranean and Atlantic glacial refugia (Quesada et al. 1995a), as for other species such as sea bass (Lemaire et al. 2005; Tine et al. 2014; Duranton et al. 2018). The genetic differentiation between the two mussel lineages proved to be highly heterogeneous among loci (Gosset and Bierne 2013; Fraïsse et al. 2016). Heterogeneous differentiation is expected because of a semipermeable genetic barrier maintained by partial reproductive isolation (Barton and Hewitt 1985; Harrison 1993) or because of heterogeneous divergence caused by linked selection (Cruickshank and Hahn 2014; Ravinet et al. 2017), or both (Duranton et al. 2018). To date, only the northern coast near Almeria in Spain has been intensively sampled, and contains an abrupt shift in allele frequency on each side of a 290-km-wide no-mussel zone between Almeria and Alicante (Quesada et al. 1995a). Although some studies have identified weak genetic differentiation between Europe and the North African region east of the AOF (Ouagajjou and Presa 2015; Lourenço et al. 2015), the geographic structure of the transition zone along the southern coast in Algeria was not well characterised. This is an important sampling gap to fill, especially considering that Atlantic waters enter the Mediterranean Sea through the south through the Algerian current described above. The southern side of the transition is indeed a ubiquitous sampling lacuna in the study of the Atlantic–Mediterranean genetic divide.
In the present study, we aimed to advance our understanding of the marine transition zone at the AOF by analysing four ancestry-informative loci (one mitochondrial and three nuclear) at a fine-grained spatial scale. These loci were previously identified to be among the 10 most differentiated loci in two genome scans: one with 388 AFLP loci (Gosset and Bierne 2013) and another with 1269 targeted contigs (~50,000 SNPs, Fraïsse et al. 2016). Our aim was not to infer the history of gene flow between populations, nor the genomic heterogeneity of differentiation, which requires the analysis of a high number of loci and has been treated elsewhere (Fraïsse et al. 2016). Our objective was to investigate the spatial structure of the transition zone and how the two genetic backgrounds meet and interact when in contact, which requires analysing an extensive geographic sample with informative loci (Vines et al. 2016). We accomplished this in three steps. First, as a precaution, we re-analysed the shift in allele frequency in South-Eastern Spain that was reported 20 years ago. We observed the same position of the break in Spain as in earlier studies, confirming the results from Diz and Presa (2008). Second, we sampled a new area along the Algerian coast between Oran and Tunisia, over a distance of 1330 km. We report a 600-km-wide mosaic hybrid zone with populations of mixed ancestry. Third, we explored simple stepping-stone simulation models, which show that the mosaic structure observed can be explained by the local geographic and hydrodynamic characteristics of the coastlines. We used a Y-shaped bi-unidimensional structure of the coastlines to reflect the shape of the coastlines that allows north–south gene exchange west of the AOF, but only along coastlines to the east of the AOF because the distance between Algerian and Spanish coasts is too large for larval dispersal. We also included two alternative barriers to larval dispersal and/or environmental boundaries (one at the AOF and one in the Gulf of Bejaia), and a preponderantly unidirectional north–south gene flow at the AOF.
Materials and methods
Sampling and molecular markers
Mytilus galloprovincialis samples were collected from 13 locations on the southern coast of the Alboran and Mediterranean Seas from Nador (Morocco) to Bizerte (Tunisia), and from 7 locations on the Northern coast from Manilva (Spain) to Peniscola (Table S1, Fig. 1). Mitochondrial DNA sequences proved that all the samples were M. galloprovincialis (see below), as expected in the study area. We also analysed reference samples already reported in previous studies (Gosset and Bierne 2013) in Faro (Portugal) and Sète (France). DNA was extracted from gills using the QIAGEN DNeasy Blood & Tissue Kit, following the manufacturer's instructions. DNA concentration was measured for each sample using a NanoDrop8000 Spectrophotometer (Thermo Scientific) and standardised to a DNA concentration of 100 ng µL–1.
A 350-bp fragment of the F-mtDNA cytochrome oxidase subunit III was PCR amplified and sequenced with primers FOR1 (5′-TATGTACCAGGTCCAAGTCCGTG-3′) and REV1 (5′-TGCTCTTCTTGAATATAA GCGTA-3′) (Zouros et al. 1994). Sequence reactions were precipitated using a standard EDTA/ethanol protocol, suspended in 15 µl Hi-Di formamide and sequenced on an ABI 3130XL automated sequencer.
Based on previous genome scans, three nuclear markers were chosen because of their strong differentiation between the Atlantic and Mediterranean backgrounds of M. galloprovincialis, and ease of analysis. All three produced a length polymorphism of PCR products. EFbis is a widely used marker in mussels and was amplified with the following two primers: EFbis-F 5′-ACAAGATGGACAATACCGAACCACC-3′ and EFbis-R 5′-CTCAAT CATGTTGTCTCCATGCC-3′ (Bierne et al. 2002). EF2 has been described in Gosset and Bierne (2013) and was amplified with the following two primers: EF2-F 5′-GGAAATCCCATGGGTGATTTAGCGG-3′ and EF2-R 5′-GTCAAATAAATACTGAAACACAGTGACTTC-3′. A contig containing the Precollagen-D gene was identified from a genome scan of 1269 contigs as the most differentiated contig between Atlantic and Mediterranean M. galloprovincialis populations (Fraïsse et al. 2016). Using publicly available sequences (Qin et al. 1997), we designed primers to amplify the fifth intron of the gene with the 3′ end of each primer positioned at the exon–intron junction, which prevented amplification of paralogous sequences of the gene family. The primers used to amplify Precol-D locus were Precol-D-F 5′-GACAAGGACCAGCAGGTACCATATT-3′ and Precol-D-R 5′-GAGTGGTCCGGCTGGTCCTAAGAA-3′. Indel polymorphisms in the intron produced nine length alleles in our samples with strong allele frequency differences between Atlantic and Mediterranean samples. Standard PCR protocols were used with annealing temperatures set at 54 °C for EFbis and EF2, and 49 °C for Precol-D. Two microliters of PCR products was mixed with Hi-Di formamide/ROX 500 size standard (12.8 µl formamide and 0.2 µl ROX), and the mixtures were then loaded on an ABI 3130XL capillary automated sequencer. GeneMapper® v4.5 software (Applied Biosystems) was used to read the resulting chromatograms.
Data analysis
COIII sequences were aligned using the ClustalW multiple alignment algorithm in BIOEDIT V7.1.3.0 and revised manually. Aligned sequences were then used to construct a Neigbour-Joining tree with MEGA software v6 (Tamura et al. 2013). Allelic and genotyping frequencies were computed using Genetix software v4.0.5.2 (Belkhir et al. 2002). The number of haplotypes (h), nucleotide diversity (π), average number of nucleotide substitutions per site between populations (Dxy) and net number of nucleotide substitutions per site between populations (Da) were calculated using DnaSP V5.10.1 (Librado and Rozas 2009). Individuals were assigned to the Mediterranean and Atlantic lineages with a discriminant analysis of principal components (DAPC) implemented in the adegenet package in R (Jombart and Ahmed 2011). The individual ancestries were examined by a Bayesian clustering method implemented in STRUCTURE 2.3.4 (Falush et al. 2003). Analysis with STRUCTURE was conducted for various numbers of clusters K under the admixture model, assuming that the allele frequencies are correlated across populations, with a burn-in period of 50,000 steps and a run length of 100,000 iterations. We used NewHybrids software (Anderson and Thompson 2002) to determine the hybrid status of individuals in the hybrid zones. We used a model with six categories of genotypes corresponding to two parental lineages and four categories of hybrids: F1 and F2 hybrids, and the two types of backcrosses. Note that there are many more categories of hybrid genotypes in natural hybrid zones. However, this analysis allows investigating the frequency of early-generation hybrids (expected to be rare in natural bimodal hybrid zones). Individual posterior probabilities of belonging to each category were obtained by running NewHybrids with uniform priors, and a burn-in period of 50,000 steps followed by a run length of 100,000 iterations. Because parental lineages tend to be more introgressed locally within a mosaic hybrid zone than outside of the zone in allopatric populations (Bierne et al. 2003), we did not use allopatric populations as references in the analysis. In this context, including allopatric samples would have inflated the posterior probabilities of a locally introgressed parental genotype to that of backcrosses, which is not desired. To depict allele frequency clines, all four loci were transformed into bi-allelic loci by pooling alleles according to their frequencies in reference samples (sample 1 Faro Portugal for the Atlantic lineage and sample N9 Sète France for the Mediterranean lineage, two localities often studied in previous genetic studies of M. galloprovincialis). Finally, the average departure from Hardy–Weinberg equilibrium was measured by Fis and the average pairwise linkage disequilibrium across all loci, D, was estimated using the variance in the hybrid index as described in Barton and Gale (1993). Departure from Hardy–Weinberg and linkage equilibrium was tested by using permutations with the Genetix software, combining p values using Fisher’s method.
Simulations
We used a model of evolution in a metapopulation of 2 × 60 demes arranged in two parallel linear stepping stones (Bierne et al. 2011; 2013). The purpose of the simulations was to account for the geographic and hydrodynamic characteristics of the region and to compare intrinsic reproductive isolation with local adaptation at two different spatial scales (fine- or coarse-grained environmental heterogeneity). At generation zero, two partially reproductively isolated backgrounds meet and start to exchange genes. The auto-recruitment rate was 1–2m, and migration to adjacent demes was m. A weak barrier to dispersal was set between demes 20 and 21 in the northern chain to simulate the AOF, and another barrier to dispersal in the southern chain between demes 40 and 41 to simulate the Gulf of Bejaia Barrier (GBB). The migration rate at the barriers to dispersal was set to a value mbar, with mbar < m. Bidirectional migration was possible between the northern and southern chains for the first 20 demes (left of the first barrier to dispersal, i.e. in the Alboran Sea) with the same rate of migration, m. For a few more demes (from 1 to 10) bidirectional or unidirectional north–south gene flow was possible between the north and the south, and was then stopped for the remaining demes. The aim of the parameterisation was to account for the fact that the Spanish and Algerian coasts east of the AOF are too distant to be connected directly by a single generation of larval dispersal, which is considered to be ~50 km gen–1 in mussels (McQuaid and Phillips 2000, Gilg and Hilbish 2003). Selection acted at a local adaptation locus (exogenous selection) or against recombinant genotypes (intrinsic selection) at two freely recombining reproductive isolation loci (Bierne et al. 2011). Allelic fitnesses at the exogenous locus were W(C) = 1 and W(c) = 1–t in habitat 1, and W(C) = 1–t and W(c) = 1 in habitat 2. The two-loci fitnesses at the incompatible alleles were W(AB) = W(ab) = 1 and W(Ab) = W(aB) = 1–s. Here we did not consider the interaction between the two types of selection (see Bierne et al. 2011), but considered either of them. A neutral marker was positioned at a recombination map distance of 1 cM to a selected locus.
In order to simulate the trapping of a hybrid zone by a barrier to dispersal or an environmental boundary, we need clines to overlap with barriers at their initial position (Goldberg and Lande 2007) or to move towards the barrier (Barton and Turelli 2011). We could have chosen the initial conditions and the parameter values such that the northern cline overlapped with the AOF (between demes 20 and 21) and the southern cline overlapped with the GBB (between demes 40 and 41 of the southern chain). However, we chose to exemplify the arguably more realistic situation of initially moving hybrid zones. Local adaptation clines move to environmental boundaries driven by local selection. However, tension zones maintained by selection against recombinant genotypes stay at their initial contact position in a purely deterministic genetic model with equal fitnesses of parental genotypes and isotropic migration. There are three reasons why tension zones could move. First, they are expected to move down gradients of population density and to be trapped by density troughs (Abbott et al. 2013; Barton 1979; Hewitt 1975). This demographic effect is not accounted for in our purely genetic model, although the tension zone can be trapped by barriers to dispersal, given that it moves towards the barriers for other reasons. Second, tension zones can move because one parental genotype is fitter. These moving tension zones can nonetheless easily be halted by density troughs or barriers to dispersal owing to their bi-stable nature (Barton and Turelli 2011). Random drift is expected to ultimately free such tension zones (Barton 1979, Piálek and Barton 1997), but the trapping can last for a very long time. Finally, tension zones can move haphazardly because of stochastic processes—genetic drift within populations and stochastic dispersal between populations. Here we used these two processes to simulate tension zone movement. We used a slight fitness advantage, a (W(AB) = 1 + a), of the Mediterranean parental genotype starting from a contact within the Mediterranean Sea (at deme 50), or a slight fitness advantage of the Atlantic parental genotype (W(ab) = 1 + a) starting from a contact on the Atlantic side (at deme 10). Alternatively, we introduced random drift by using multinomial sampling of genotypes within each deme at each generation (N = 500 per deme). The latter way of simulating tension zone movement yields various outcomes from the same starting position, because the movement is not directional. We used this alternative only to show that the results are qualitatively similar regardless of the “engine” of the tension zone movement, and we did not intend to complete an extensive analysis of the effect of drift on the outcomes, as this is far from the purpose of the present work.
Results
Analysis of genetic diversity
The analysis of 568 COIII sequences revealed 150 haplotypes and a high nucleotide diversity (π = 0.023). The average and net nucleotide divergence measures between Atlantic and Mediterranean populations were also high (Dxy = 0.029, Da = 0.011). The NJ tree is presented in Supplementary Figure S1. Two clades of haplotypes could be defined according to their phylogenetic relationships and their frequency in Atlantic and Mediterranean samples (Figure S1). The clade nearly fixed in Atlantic samples is known to be due to introgression of alleles from the sister species M. edulis (Quesada et al. 1998). We used phylogenetic relationships to define the Mediterranean and the Atlantic haplogroups. Nine size alleles were detected at the new locus Precol-D and allele frequencies are presented in Table S2. EF2 was found to be bi-allelic, as in Gosset and Bierne (2013). The use of capillary electrophoresis allowed us to distinguish EFbis size alleles with 1-bp differences that were not detected previously with acrylamide gels. We observed 22 alleles, and the correspondence with previous allele names is given in Table S2. Clustering analysis was performed with multi-allelic data. In order to represent allele frequency clines, we transformed multi-allelic loci into bi-allelic loci by pooling alleles according to their frequency in reference samples, sample 1 (Faro, Portugal) for the Atlantic lineage and sample N9 (Sète, France) for the Mediterranean lineage (see Table S2).
Spatial genetic structure
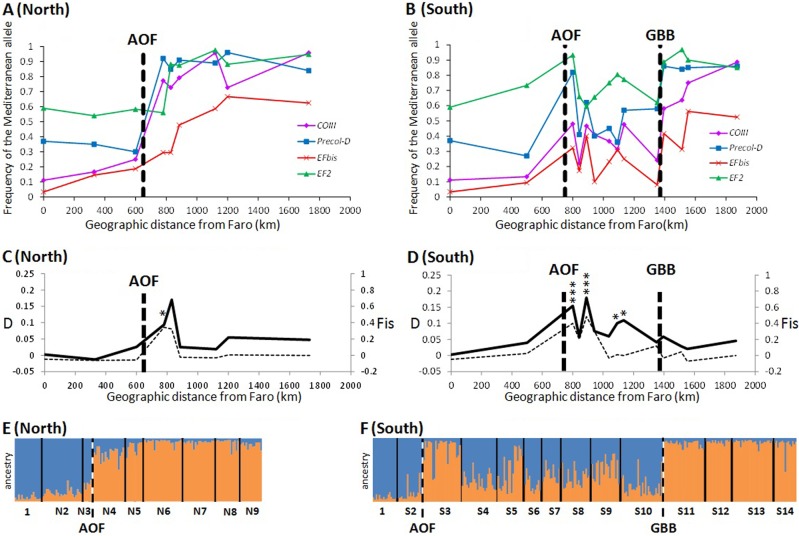
The frequencies of the Mediterranean allele at the mitochondrial COIII locus and the three nuclear markers (Precol-D, EFbis and EF2) in each sample are presented in Fig. 2a, b. Above K=2 genetic clusters, clustering methods did not provide meaningful results(no improvement in the likelihood in the STRUCTURE analysis, low contribution of secondary axes in multivariate analyses and lack of spatial structure of additional clusters). We provide the results for K = 2. As reported first 20 years ago (Quesada et al., 1995b, Sanjuan et al. 1994) and then 10 years later (Diz and Presa 2008), the northern transect along the Spanish coast contained an abrupt change in allele frequencies in the zone of transition between Almeria and Cartagena with all four loci. The three samples collected in Atlantic waters (1-Faro, N2-Manilva and N3-Almeria) have high frequencies of the Atlantic alleles (Fig. 2a), uniformly high Atlantic cluster membership probabilities in the DAPC analysis (Fig. 1, Supplementary File S1) and a high proportion of Atlantic ancestry in the STRUCTURE analysis (Fig. 2e). Conversely, samples from Mediterranean waters (N6-Tabarca, N7-Nules, N8-Peñíscola del Pinatar and N9-Sète) have high frequencies of Mediterranean alleles (Fig. 2b), uniformly high Mediterranean cluster membership probabilities in the DAPC analysis (Fig. 1) and a high proportion of Mediterranean ancestry in the STRUCTURE analysis (Fig. 2f). Two populations (N4-Cartagena and N5-San Pedro) were obtained from the region reported to be devoid of mussels 20 years ago. These samples were predominantly of Mediterranean ancestry, but some individuals were assigned to the Atlantic genetic cluster with high confidence and a few mussels with a genome of mixed ancestry were also present (Figs. 1 and 2e). Departure from Hardy–Weinberg and linkage equilibrium was maximal in these intermediate samples (Fig. 2c), as expected from hybrid zone theory (Barton and Gale 1993), and the departure was significantly different from zero in the sample N4-Cartagena. These samples are crucial for our understanding of the functioning of the AOF barrier to gene flow, as they show that Atlantic mussels are found in observable numbers east of the AOF within the Mediterranean Sea.
Fig. 2.
Mytilus galloprovincialis Mediterranean allele frequencies at the four semi-diagnostic loci analysed (COIII, Precol-D, EFbis and EF2) for a the 9 samples of the northern coastline and b the 14 samples of the southern coastline. AOF: Almeria Oran Front, GBB: Gulf of Bejaïa Barrier. c Average multilocus linkage disequilibrium (D, bold line, left axis) and Hardy–Weinberg disequilibrium (Fis, dashed line, right axis) in samples of the northern coastline and d samples of the southern coastline. Bar plot of the estimated ancestry proportions (Q values) estimated by STRUCTURE with the three nuclear markers (Precol-D, EFbis and EF2) for e samples of the northern coastline and f the southern coastline
The southern transect along the Algerian coast revealed a complex structure on a much wider geographic scale, consisting of a mosaic pattern of alternation between mostly Atlantic, mostly Mediterranean and intermediate samples (Figs. 1 and 2). Individuals assigned to the Atlantic cluster predominated in the sample from S2-Nador (Morocco) situated in the Alboran Sea (Figs. 1 and 2). The Mediterranean cluster predominated in three samples from eastern Algeria (S11-Zaima Mansouriah, S12-Collo and S13-Skikda) and Tunisia (S14-Bizerta) (Figs. 1 and 2). From S3-Oran to S10-Bejaia a mosaic hybrid zone was observed, with a tendency for the Mediterranean ancestry to decrease eastward along the zone. The zone ends with an abrupt genetic shift in the Gulf of Bejaia (Gulf of Bejaia Barrier, GBB), west of which are preponderantly Atlantic samples and east of which is populated by Mediterranean mussels. Within the mosaic zone, samples were found to be a mixture of individuals belonging to both clusters and with mixed ancestry (Figs. 1 and 2f). Again, strong departures from Hardy–Weinberg and linkage equilibrium (Fig. 2d) attest to a strong deficit of hybrids relative to random mating. The departure was strongly significant in S3-Oran and S5-Sidi Lakdher, and significant in S8-Tipaza and S9-Algiers (Fig. 2d). We did not obtain support for the existence of early-generation hybrids in Algerian populations, as none of the 183 individuals sampled in the hybrid zone obtained a cumulative posterior probability >0.8 to be F1, F2 or a backcross in NewHybrids (Supplementary Table S3). This is not surprising as early-generation hybrids must be extremely rare in nature for the genetic differentiation to be so efficiently maintained between two backgrounds. Evidence for hybridisation nonetheless comes from allele frequencies and the comparison of ancestry values that shows that Atlantic mussels from the hybrid zone have a higher fraction of Mediterranean ancestry and Mediterranean mussels a higher fraction of Atlantic ancestry than mussels from peripheral allopatric populations (Fig. 2f). This local introgression of parental backgrounds within the mosaic hybrid zone also explains the lack of power to characterise hybrid genotypes despite the fact that our markers are strongly differentiated between reference samples well outside of the hybrid zone.
Modelling a Y-shaped bi-unidirectional stepping stone that accounts for the seascape features of the AOF transition zone
Our simple model in a Y-shaped bi-unidimensional stepping stone allowed us to obtain useful information regarding how the principal features of the seascape around the AOF area could produce the mosaic structure observed.
Endogenous selection
It is to be noted that in the tension zone model selection against hybrids maintains the clinal structure, but, in contrast to exogenous selection, the position of the cline is not stabilised by selection. Theoretically, tension zones should be trapped by and should coincide with natural barriers to dispersal, i.e. zones with low population density, such as mountains, rivers or oceanic fronts (Barton 1979). First, we describe the results observed with deterministic simulations at a selected locus in the case of a tension zone moving as a consequence of a fitness advantage of one parental genotype. The objective is not to explore the conditions in which the trapping process occurs, which are well known (Barton and Turelli 2011; Barton 1979), but to illustrate how tension zone trapping can occur in an unusual spatial and connectivity context such as the one in the AOF area. Second, we provide an example of a neutral locus introgressing through the barrier and incorporating genetic drift. Again, we our aim is not to discuss the well-known effects of linkage and random drift (Barton 1986; Polechová and Barton 2011), but to exemplify that the model can retrieve a pattern that resembles the true data.
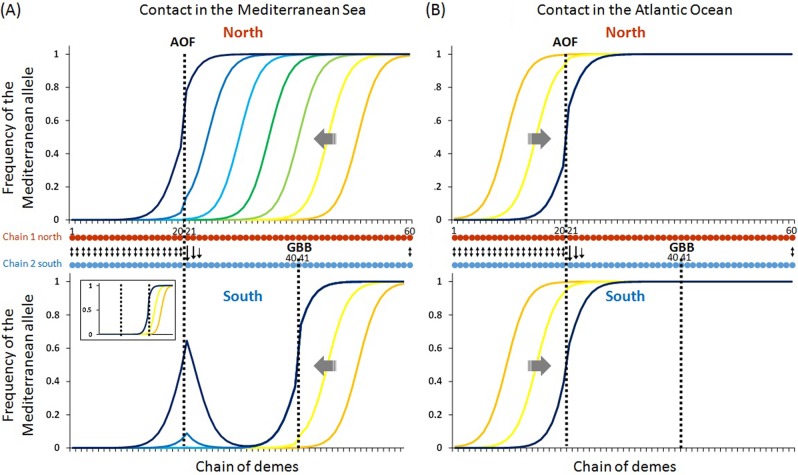
Figure 3 presents the results of the deterministic model: movement of endogenous clines along the northern coast, characterised by a barrier to dispersal between demes 20 and 21 (called AOF in Fig. 3), and the southern coast, characterised by a barrier to dispersal between demes 40 and 41 (called GBB in Fig. 3). We obtained qualitatively similar results when we added a barrier to dispersal between demes 20 and 21 of the southern chain (i.e. Oran), and provide the results with no barrier to exemplify the trapping of the cline even when the barrier is restricted to the northern coast (i.e. Almeria). The most interesting result is a contact within the Mediterranean Sea (between demes 50 and 51) and a westward propagation of the cline owing to a slight fitness advantage of the Mediterranean parental genotype. The clines propagate along the southern and northern coasts, and a barrier to dispersal is first encountered by the southern cline between demes 40 and 41 (GBB), where it is trapped while the northern cline continues to propagate westward (Fig. 3a). The northern cline is subsequently trapped by the AOF barrier between demes 20 and 21 (blue cline). When migration is symmetrical between the north and the south, we end with a pair of shifted clines, one trapped by the AOF in the north and the other trapped by the GBB in the south (see inset in Fig. 3a). In this case the Algerian coast is inhabited by the Atlantic lineage. However, when migration is asymmetrical from the north to the south, the Mediterranean genotype can establish and be maintained in the south, generating a zone of coexistence around the AOF in the south (Fig. 3a bottom panel). This simulation shows that a mosaic hybrid zone can be produced in the south with purely intrinsic reproductive isolation when the major hydrodynamic characteristics of the region are taken into account. We also considered the situation of a contact on the Atlantic side and an eastward propagation of clines owing to a slight fitness advantage of the Atlantic parental genotype. The clines propagate and meet the AOF barrier between demes 20 and 21, where they are trapped (Fig. 3b). The southern cline remains trapped at the AOF, even when the barrier is simulated only in the northern chain (migration between demes 20 and 21 is fixed at mbar in the northern chain, at Almeria, and at m in the southern chain, at Oran). The southern cline does not have the opportunity to meet the barrier to dispersal at the GBB. It is well established that tension zones are expected to be trapped by the first minor barrier encountered (Barton 1979, Barton and Turelli 2011). Despite this prediction, it is usually not well acknowledged that hotspots for hybrid zones (i.e. suture zones) are more likely to be localised at the barrier to dispersal that is closest to an area of frequent contact rather than at the strongest barrier to dispersal across the overall distribution area.
Fig. 3.
a Simulation output obtained with a bi-locus deterministic model with selection against recombinant genotypes (s = 0.1) and a slight advantage of the Mediterranean background (a = 0.01) in a 60-deme Y-shaped stepping-stone model (m = 0.3), including two weak barriers to gene flow (mbar = 0.1) between demes 20 and 21 (AOF) in the northern chain, and between demes 40 and 41 (GBB) in the southern chain of demes, a contact between demes 50 and 51, and unidirectional migration right to the AOF. Above: northern chain, below: southern chain. The inset shows the output when migration is bidirectional between the two chains. Clines are represented for every 500 generations with a rainbow colour code from orange to dark blue. Blue clines superimposed green clines in the southern chain because they remain at the same position (trapping). b Simulation with a contact between demes 10 and 11 and an advantage of the Atlantic background (a = 0.01). Clines are represented for every 500 generations with a rainbow colour code from orange to dark blue. Dark blue clines superimpose light blue and green clines because they remain at the same position (trapping)
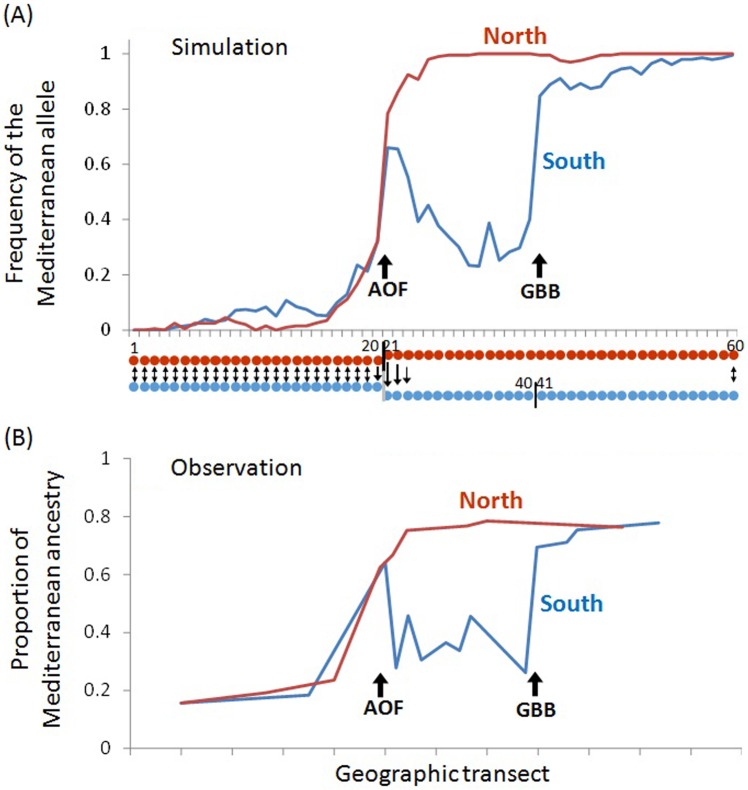
The deterministic model shows that sister tension zones can be differentially trapped at two alternative barriers to larval dispersal, at Almeria in the north and Bejaia in the south, and that a preponderantly asymmetrical north–south migration rate at the AOF can maintain a “pocket” of the Mediterranean background east of Oran in the south. In order to model the true data more closely, we considered a neutral locus linked to a selected locus, which allowed us to account for introgression, and introduced genetic drift. A simulation output is presented in Fig. 4a, with the two coastlines superposed in the same figure and compared to true data in Fig. 4b. In this simulation, the two parental genotypes have the same fitness, which shows that the pocket of Mediterranean background in the south is not due to a fitness advantage but due to the balance between migration from the north and selection against hybrids. A comparison with Fig. 3 shows that introgression of Mediterranean alleles into the Atlantic background and of Atlantic alleles into the Mediterranean background is stronger in the mosaic zone between the two barriers (AOF and GBB) than in peripheral populations. This is expected in mosaic hybrid zones, as already explained for another mussel hybrid zone (Bierne et al. 2003). Small patches of parental populations enclosed within a mosaic hybrid zone and flanked by patches of populations of the alternative genetic background are expected to introgress faster than large and broadly distributed peripheral populations.
Fig. 4.
a A simulation output obtained at a neutral locus at a genetic map length of 1 cM to one intrinsic incompatibility after 1000 generations with the Y-shaped bi-unidimensional stepping-stone model, including two weak barriers to gene flow between demes 25 and 26 (AOF) in the northern chain and between demes 40 and 41 (GBB) in the southern chain of demes, and unidirectional migration right to the AOF. b Proportion of Mediterranean ancestry under an admixture model (STRUCTURE Q values). Geographic positions are rescaled so that Almeria superposes to Oran
Exogenous selection
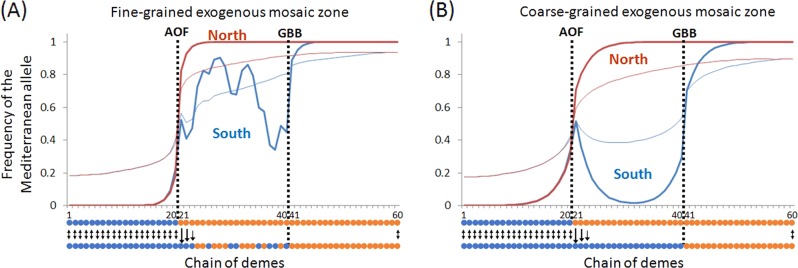
We explored two kinds of seascape: a fine-grained mosaic structure in the hybrid zone, and coarse-grained environmental variation.The first model is a mosaic of habitat 1 (Atlantic-like) that alternates randomly with habitat 2 (Mediterranean-like) in the central area of the southern stepping stone (corresponding to the area between the AOF and the GBB in Algeria). This model corresponds to a fine-grained mosaic of habitats. As expected, notwithstanding that genotypes adapted to habitat 1 and those adapted to habitat 2 flow from peripheral populations, this kind of selection generates a mosaic structure at the selected locus that correlates with habitat variation (Fig. 4a). Despite the strong selection coefficient (s = 0.2) and the recombination rate we used (1 cM) introgression proceeds quickly at the neutral locus in the mosaic zone (Fig. 4a presents the results at generation 1000), and the neutral marker loses the association with both the selected locus and the habitat. We can increase the selection and decrease the recombination to maintain the association for longer periods, but the parameters we used are already strong selection and tight linkage. Our objective here was to briefly verify the well-established result that microgeographic adaptation generates a very weak barrier to neutral gene flow (Flaxman et al. 2012; Thiebert-Plante and Hendry 2010). In the second model we simulated coarse-grained environmental variation (Fig. 3b). As for endogenous selection, we observed two clines at the two environmental boundaries simulated, the AOF in the north and the GBB in the south, when north–south migration was symmetrical. In order to obtain the maintenance of genotypes adapted to habitat 2 (i.e. Mediterranean-like) in the south, asymmetrical north–south migration at the AOF was required. The result obtained is very similar to that obtained with endogenous selection. Introgression proceeds much more slowly than in the fine-grained habitat model, although faster than with endogenous selection (with the same recombination rate of 1 cM). Another difference between the exogenous and endogenous models is that under exogenous selection the habitat 2 genetic background (Mediterranean-like) cannot be maintained without migration from the north, because it is counter-selected in the south. In the endogenous model, however, we can expect a sufficiently big patch of Mediterranean background to persist in the south, at least transiently, as soon as it has been established by an episode of north–south migration.
Discussion
The Almeria–Oran Front (AOF) is arguably one of the most famous hotspots of genetic differentiation in the sea (Patarnello et al. 2007), and Mytilus galloprovincialis one of the iconic species exhibiting a pronounced genetic break at the AOF (Quesada et al. 1995a, b, Sanjuan et al. 1994). We already had indirect evidence that reproductive isolation might have evolved between Mediterranean and Atlantic refugia in order to (i) maintain genetic differentiation upon contact (Quesada et al. 1995a), (ii) maintain a sharp genetic break (Quesada et al. 1995b) and (iii) explain the semi-permeability of the barrier to gene flow (Fraïsse et al. 2016). However, a conclusion could not be reached without more direct evidence that interbreeding opportunities exist at the transition zone, although such opportunities are rare. Perhaps due to this lack of evidence of reproductive isolation, the idea that the Atlantic–Mediterranean divide could simply be explained either by the AOF itself acting as a barrier to larval dispersal or by the ecological differences between Atlantic and Mediterranean waters remains strongly anchored in the marine literature. In addition, given that Atlantic waters enter the Mediterranean Sea through the south by the Algerian current, the description of the southern side of the transition zone was a sampling lacuna that needed to be filled. Sampling that coast revealed far more than one could have expected. The unexplored side of the biogeographic boundary revealed a 600-km-wide mosaic hybrid zone. We argue here that the existence of this large mosaic hybrid zone provides evidence-based support for the existence of partial reproductive isolation, which prevents gene flow when the two lineages live in sympatry/syntopy, with the opportunity to interbreed and exchange genes in the absence of such reproductive isolation. We believe that the existence of partial reproductive isolation between heterogeneously differentiated genetic backgrounds should supplement the disruption of population connectivity and selection against maladaptive migrants to provide a fuller explanation of genetic subdivision in marine (Bierne et al. 2011, Gagnaire et al. 2015, Riginos et al. 2016) as well as terrestrial species (Barton and Hewitt 1985, Hewitt 1989).
The previously reported absence of mussels between Almeria and Alicante (Quesada et al. 1995b) on the Spanish coast suggested that there was no strong opportunity for sympatry and interbreeding between the Atlantic and the Mediterranean linages, in accordance with the theory of AOF as an efficient barrier to dispersal. However, two populations were sampled, south of Alicante in Cartagena and San Pedro (N4 and N5 in Fig. 1), with a low but non-negligible proportion of these individuals assigned to the Atlantic genetic cluster. This is a sufficient proportion of migrant mussels to refute the barrier to dispersal as strong enough to explain the maintenance of the genetic structure. Along with the results from the southern coast presented here, there remains no ambiguity that the Atlantic lineage not only enters the Mediterranean Sea but also co-exists with the Mediterranean lineage, sometimes in balanced proportions, across a widespread area where mussels are numerous. Described as a strong barrier to larval dispersal, the AOF is in fact rather a mix of Atlantic waters and Alboran surface waters, called “Modified Atlantic water” (MAW, Font et al. 1998), in the Eastern Alboran anticyclone and along the Algerian coast, caused by the Algerian current. There are several species without a genetic break at the AOF—e.g. the European flat oyster Ostrea edulis (Launey et al. 2002), the Mediterranean rainbow wrasse Coris julis (Aurelle et al. 2003) and the saddled seabream Oblada melanura (Galarza et al. 2009). These species share similar biological and ecological characteristics with species that do exhibit a break, and this observation was already a strong argument against the hypothesis of a sufficiently reduced dispersal rate at the AOF to explain the genetic differentiation observed in many species.
We now turn to the more complex issue of the nature of the genetic barrier (extrinsic, intrinsic or both). Given the contrasting environments inhabited by the two lineages in the Mediterranean Sea and the Atlantic Ocean, there is little doubt that differential adaptation must exist between the two lineages (including phenotypic plasticity and epigenetic responses). However, the large zone of coexistence in Algeria modifies our view of how the genetic barrier operates. We no longer have to explain adaptation in the two seas, but need to mention that the two lineages are maintained genetically cohesive in sympatry along 600 km of Algerian coasts. As we cannot refute exogenous selection, we argue here that intrinsic pre- or post-zygotic isolation is equally, if not more, likely. We have shown with simple simulations that the mosaic genetic structure observed in Algeria can indeed be obtained with intrinsic reproductive isolation alone (Figs. 3 and 4). It only requires asymmetric north–south dispersal at the AOF, which seems a reasonable assumption. In addition, simulations of exogenous selection to a coarse-grained environment also require the assumption of an asymmetric north–south dispersal at the AOF (Fig. 5b), without which a patch of maladapted Mediterranean mussels cannot be maintained in the south. Adaptation to a patchy fine-grained environment might better explain the mosaic structure, but does not generate a strong barrier to gene flow if neutral markers are not tightly linked to adaptive loci. We probably have not sampled the genome sufficiently densely during our previous genome scans to argue for strong physical linkage between our markers and local adaptation genes (Hoban et al. 2016), and therefore need a process that can generate a barrier to gene flow in large proportions of the genome (Bierne et al. 2011). Selection against migrants that are maladapted to the local environment is often proposed as a process that can generate a genome-wide barrier to gene flow (Marshall et al. 2010; Nosil et al. 2005). However, local selection needs to be extremely strong to generate a sufficiently effective barrier to gene flow (Barton and Bengtsson 1986; Feder and Nosil 2010; Slatkin 1973), thus producing a strong segregation of each ecotype in its own favoured habitat. When lineages coexist in balanced proportions, as observed here in Algeria, some sort of selection against hybrids must exist. It can still be exogenous selection (Kruuk et al. 1999), but hybrids must perform poorly in both habitats. Hybrids are the bridges used by neutral genes to flow across the barrier, and they either need to be produced in low proportion or need to be all unfit for a barrier to gene flow to have broad genomic effects (Barton and Bengtsson 1986). Here, in the Algerian hybrid zone, this is indeed the case, as we observed a strong deficit of intermediate genotypes and strong departures from Hardy–Weinberg and linkage equilibrium among physically unlinked loci (Fig. 2). Finally, the coarse-grained environment we simulated is not very likely. Although Atlantic waters enter the Algerian current, Algerian waters are much more similar to other Mediterranean waters than to Atlantic waters. Fine-grained habitat heterogeneity was also far from evident between our sampling sites in Algeria as we sampled very similar microsites: high-shore rocks were protected from wave action by artificial port breakwaters, with no evident influence of freshwater from nearby estuaries. Altogether, these arguments suggest that intrinsic reproductive isolation, probably pre-zygotic, given the apparent paucity of F1 hybrids, is a parsimonious evidence-based hypothesis that should be considered.
Fig. 5.
a Simulation output obtained with a deterministic model with exogenous selection in a fine-grained environment between AOF and GBB in the southern coasts (in which habitat type was assigned randomly). Other parameters are the same as in the previous figure. Thick line: selected locus; thin line: neutral locus after 1000 generations (r = 1 cM). b Simulation output obtained with a deterministic model with exogenous selection in a coarse-grained environment. Thick line: selected locus; thin line: neutral locus after 1000 generations (r = 1 cM)
Overall our results provide additional support for the coupling hypothesis, which suggests that genetic breaks are often secondary-contact semipermeable tension zones between heterogeneously differentiated genomes trapped by physical barriers to dispersal or environmental boundaries (Bierne et al. 2011). Physical and environmental factors mostly explain the position of the genetic breaks, but the maintenance of genome-wide genetic differentiation is best explained by reproductive isolation. Our model implies the differential coupling of sister tension zones at two different geographic positions, at the AOF in the north and at the GBB in the south. The abrupt genetic shift at the GBB is a new observation in an understudied area. However, it fits well with the position of a barrier to dispersal identified with oceanographic modelling in three recent analyses (Andrello et al. 2015; Berline et al. 2014; Rossi et al. 2014). It is also of note that these three hydrodynamic studies identified much more efficient barriers to larval dispersal than the AOF within the Mediterranean Sea that are not hotspots of genetic differentiation. Differential coupling of sister tension zones can occur in coastal or river species living in linear landscapes with tree-like reticulation, which are not well described by standard 1D or 2D stepping-stone models (Fourcade et al. 2013). For instance, some hybrid zones secondarily trapped at the entrance of the Baltic Sea are likely to have produced sister hybrid zones that have moved further north along the Norwegian coast to be trapped somewhere in northern Scandinavia (e.g. in Mytilus trossulus, Macoma balthica, Gadus morhua, Platichthys flesus and Gammarus zaddachi; reviewed in Bierne et al. 2011). Recently the Baltic–North Sea contact zone between the strongly divergent mussel species M. trossulus and M. edulis has been investigated in the southern coast area and was identified to be positioned well inside the Baltic Sea in Germany (Stuckas et al. 2017), while it is localised in the Oresund in the north (Väinölä and Hvilsom 1991).
To conclude, the mosaic hybrid zone observed between Oran and Bejaia in M. galloprovincialis mussels contributes to definitively refute the hypothesis that the AOF itself generates a sufficiently strong barrier to dispersal to maintain genetic differentiation in this species. It also opens the debate about the nature of the genetic barrier, which is not necessarily related to differential adaptation to Atlantic and Mediterranean waters but could imply intrinsic pre- or post-zygotic reproductive isolation. The next step will be to conduct laboratory experiments (hybrid crosses) and field studies (settlement and reproduction) to put the alternatives to the test. Our results also call for new genetic studies along Algerian coasts in other marine species. A few samples of sea bass from eastern Algeria proved to be admixed between the Atlantic and Mediterranean lineages (Duranton et al. 2018), although we do not yet know the spatial variation of admixture proportions in this system. Finally, our study reveals how complex the interplay between connectivity (hydrography), environmental variation (seascape) and reproductive isolation (including both extrinsic and intrinsic mechanisms) can be. Without minimising the importance of hydrography and local selection, we believe that the excessive emphasis on oceanographic features for interpreting the genetic structure of marine species sometimes results in an incomplete understanding of the underlying processes explaining its origin and maintenance.
Supplementary information
Acknowledgements
It is a pleasure to thank Marie-Thérèse Augé and Cathy Haag-Liautard for their technical help in the lab, and Marina Albentosa as well as Carlos Saavedra for Spanish samples. We also thank Pierre-Alexandre Gagnaire, Christelle Fraïsse, François Bonhomme, Nathalie Charbonnel, Sophie Arnaud-Haond and Lilia Bahri-Sfar for discussion, as well as Cynthia Riginos, Heiko Stuckas and six anonymous referees for their insightful comments on earlier drafts. Molecular data were produced through the ISEM platform Génomique des Populations Marines (PGPM7) at the Station Marine de Sète (OSU OREME, Observatoire de Recherche Méditerranéen de l’Environnement) and the platform Génomique Environnementale of the LabEx CeMEB (Laboratoire d’Excellence Centre Méditerranéen de l’Environnement et de la Biodiversité). This work was funded by a Languedoc-Roussillon “Chercheur(se)s d’Avenir” grant (Connect7 project). This is article 2018-286 of Institut des Sciences de l’Evolution de Montpellier.
Data archiving
Sequence data have been deposited in GenBank: accession numbers MK259098–MK259664. Genotype data have been deposited in Dryad: 10.5061/dryad.m3t6p5g.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41437-018-0174-y) contains supplementary material, which is available to authorized users.
References
- Abbott R, Albach D, Ansell S, Arntzen JW, Baird SJE, Bierne N, et al. Hybridization and speciation. J Evol Biol. 2013;26:229–246. doi: 10.1111/j.1420-9101.2012.02599.x. [DOI] [PubMed] [Google Scholar]
- Anderson E, Thompson E. A model-based method for identifying species hybrids using multilocus genetic data. Genetics. 2002;160:1217–1229. doi: 10.1093/genetics/160.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrello M, Jacobi MN, Manel S, Thuiller W, Mouillot D. Extending networks of protected areas to optimize connectivity and population growth rate. Ecography. 2015;38:273–282. [Google Scholar]
- Arnold (2006) Evolution through genetic exchange. Oxford University Press, Oxford
- Aurelle D, Guillemaud T, Afonso P, Morato T, Wirtz P, Santos RS, et al. Genetic study of Coris julis (Osteichtyes, Perciformes, Labridae) evolutionary history and dispersal abilities. C R Biol. 2003;326:771–785. doi: 10.1016/j.crvi.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Avise JC. Phylogeography: the history and formation of species. Cambridge: Harvard University Press; 2000. [Google Scholar]
- Avise JC (1992) Molecular population structure and the biogeographic history of a regional fauna: a case history with lessons for conservation biology. Oikos, 62–76
- Bahri-Sfar L, Lemaire C, Hassine OKB, Bonhomme F. Fragmentation of sea bass populations in the western and eastern Mediterranean as revealed by microsatellite polymorphism. Proceedings of the Royal Society of London B: Biological Sciences. 2000;267(1446):929–935. doi: 10.1098/rspb.2000.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton NH. The dynamics of hybrid zone. Heredity. 1979;43:341–359. [Google Scholar]
- Barton NH. The effects of linkage and density-dependant regulation on gene flow. Heredity. 1986;57:415–426. doi: 10.1038/hdy.1986.142. [DOI] [PubMed] [Google Scholar]
- Barton NH, Bengtsson BO. The barrier to genetic exchange between hybridising populations. Heredity. 1986;56:357–376. doi: 10.1038/hdy.1986.135. [DOI] [PubMed] [Google Scholar]
- Barton NH, Gale KS (1993) Genetic analysis of hybrid zones. In: Harrison RG (ed) Hybrid zones and the evolutionary process. Oxford University Press, New York, pp 13–45
- Barton NH, Hewitt GM. Analysis of hybrid zones. Annu Rev Ecol Syst. 1985;16:113–148. [Google Scholar]
- Barton N, Turelli M. Spatial waves of advance with bistable dynamics: cytoplasmic and genetic analogues of Allee effects. Am Nat. 2011;178:E48–E75. doi: 10.1086/661246. [DOI] [PubMed] [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Montpellier, France: Université de Montpellier 2; 2002. [Google Scholar]
- Berline L, Rammou AM, Doglioli A, Molcard A, Petrenko A. A connectivity-based eco-regionalization method of the Mediterranean Sea. PLoS ONE. 2014;9:e111978. doi: 10.1371/journal.pone.0111978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialozyt R, Ziegenhagen B, Petit RJ. Contrasting effects of long distance seed dispersal on genetic diversity during range expansion. J Evol Biol. 2006;19:12–20. doi: 10.1111/j.1420-9101.2005.00995.x. [DOI] [PubMed] [Google Scholar]
- Bierne N, Bonhomme F, Boudry P, Szulkin M, David P. Fitness landscapes support the dominance theory of post-zygotic isolation in the mussels Mytilus edulis and M. galloprovincialis. Proc R Soc B-Biol Sci. 2006;273:1253–1260. doi: 10.1098/rspb.2005.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne N, Borsa P, Daguin C, Jollivet D, Viard F, Bonhomme F, et al. Introgression patterns in the mosaic hybrid zone between Mytilus edulis and M-galloprovincialis. Mol Ecol. 2003;12:447–461. doi: 10.1046/j.1365-294x.2003.01730.x. [DOI] [PubMed] [Google Scholar]
- Bierne N, David P, Boudry P, Bonhomme F. Assortative fertilization and selection at larval stage in the mussels Mytilus edulis and M-galloprovincialis. Evolution. 2002;56:292–298. doi: 10.1111/j.0014-3820.2002.tb01339.x. [DOI] [PubMed] [Google Scholar]
- Bierne N, Gagnaire PA, David P. The geography of introgression in a patchy environment and the thorn in the side of ecological speciation. Curr Zool. 2013;59:72–86. [Google Scholar]
- Bierne N, Welch J, Loire E, Bonhomme F, David P. The coupling hypothesis: why genome scans may fail to map local adaptation genes. Mol Ecol. 2011;20:2044–2072. doi: 10.1111/j.1365-294X.2011.05080.x. [DOI] [PubMed] [Google Scholar]
- Borrero-Pérez GH, González-Wangüemert M, Marcos C, Pérez-Ruzafa A. Phylogeography of the Atlanto-Mediterranean sea cucumber Holothuria (Holothuria) mammata: the combined effects of historical processes and current oceanographical pattern. Mol Ecol. 2011;20:1964–1975. doi: 10.1111/j.1365-294X.2011.05068.x. [DOI] [PubMed] [Google Scholar]
- Borsa P, Naciri M, Bahri L, Chikhi L, Garcia de Leon FJ, Kotoulas G, et al. Zoogéographie infra-spécifique de la mer Méditerranée. Vie Milieu. 1997;47:295–305. [Google Scholar]
- Burton RS. Intraspecific phylogeography across the Point Conception biogeographic boundary. Evolution. 1998;52(3):734–745. doi: 10.1111/j.1558-5646.1998.tb03698.x. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D, Barton NH. The effects of genetic and geographic structure on neutral variation. Annu Rev Ecol Syst. 2003;34:99–125. [Google Scholar]
- Chevolot M, Hoarau G, Rijnsdorp AD, Stam WT, Olsen JL. Phylogeography and population structure of thornback rays (Raja clavata L., Rajidae) Mol Ecol. 2006;15:3693–3705. doi: 10.1111/j.1365-294X.2006.03043.x. [DOI] [PubMed] [Google Scholar]
- Cordero D, Peña JB, Saavedra C. Phylogeographic analysis of introns and mitochondrial DNA in the clam Ruditapes decussatus uncovers the effects of Pleistocene glaciations and endogenous barriers to gene flow. Mol Phylogenet Evol. 2014;71:274–287. doi: 10.1016/j.ympev.2013.11.003. [DOI] [PubMed] [Google Scholar]
- Cruickshank TE, Hahn MW. Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Mol Ecol. 2014;23:3133–3157. doi: 10.1111/mec.12796. [DOI] [PubMed] [Google Scholar]
- Diz AP, Presa P. Regional patterns of microsatellite variation in Mytilus galloprovincialis from the Iberian Peninsula. Mar Biol. 2008;154:277–286. [Google Scholar]
- Duranton M, Allal F, Fraïsse C, Bierne N, Bonhomme F, Gagnaire PA. The origin and remolding of genomic islands of differentiation in the European sea bass. Nature communications. 2018;9(1):2518. doi: 10.1038/s41467-018-04963-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewers-Saucedo C, Pringle JM, Sep£lveda HH, Byers JE, Navarrete SA, Wares JP. The oceanic concordance of phylogeography and biogeography: a case study in N otochthamalus. Ecology and evolution. 2016;6(13):4403–4420. doi: 10.1002/ece3.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder JL, Nosil P. The efficacy of divergence hitchhiking in generating genomic islands during ecological speciation. Evolution. 2010;64:1729–1747. doi: 10.1111/j.1558-5646.2010.00943.x. [DOI] [PubMed] [Google Scholar]
- Flaxman SM, Feder JL, Nosil P. Spatially explicit models of divergence and genome hitchhiking. J Evol Biol. 2012;25:2633–2650. doi: 10.1111/jeb.12013. [DOI] [PubMed] [Google Scholar]
- Fourcade Y, Chaput‐Bardy A, Secondi J, Fleurant C, Lemaire C. Is local selection so widespread in river organisms? Fractal geometry of river networks leads to high bias in outlier detection. Mol Ecol. 2013;22:2065–2073. doi: 10.1111/mec.12158. [DOI] [PubMed] [Google Scholar]
- Font J, Millot C, Salas J, Julià A, Chic O. The drift of Modified Atlantic Water from the Alboran Sea to the eastern Mediterranean. Sci Mar. 1998;62:211–216. [Google Scholar]
- Fraïsse C, Belkhir K, Welch JJ, Bierne N. Local interspecies introgression is the main cause of extreme levels of intraspecific differentiation in mussels. Mol Ecol. 2016;25:269–286. doi: 10.1111/mec.13299. [DOI] [PubMed] [Google Scholar]
- Gagnaire PA, Broquet T, Aurelle D, Viard F, Souissi A, Bonhomme F, et al. Using neutral, selected and hitchhiker loci to assess connectivity of marine populations in the genomic era. Evol Appl. 2015;8:769–786. doi: 10.1111/eva.12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galarza JA, Carreras-Carbonell J, Macpherson E, Pascual M, Roques S, Turner GF, et al. The influence of oceanographic fronts and early-life-history traits on connectivity among littoral fish species. Proc Natl Acad Sci. 2009;106:1473–1478. doi: 10.1073/pnas.0806804106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilg MR, Hilbish TJ. The geography of marine larval dispersal: coupling genetics with fine‐scale physical oceanography. Ecology. 2003;84:2989–2998. [Google Scholar]
- Goldberg EE, Lande R. Species’ borders and dispersal barriers. Am Nat. 2007;170:297–304. doi: 10.1086/518946. [DOI] [PubMed] [Google Scholar]
- Gosset C, Bierne N. Differential introgression from a sister species explains high Fst outlier loci within a mussel species. J Evol Biol. 2013;26:14–26. doi: 10.1111/jeb.12046. [DOI] [PubMed] [Google Scholar]
- Haney RA, Silliman BR, Fry AJ, Layman CA, Rand DM. The Pleistocene history of the sheepshead minnow (Cyprinodon variegatus): non-equilibrium evolutionary dynamics within a diversifying species complex. Molecular phylogenetics and evolution. 2007;43(3):743–754. doi: 10.1016/j.ympev.2006.08.022. [DOI] [PubMed] [Google Scholar]
- Harrison RG. Hybrid zones and the evolutionary process. Oxford: Oxford University Press; 1993. [Google Scholar]
- Harrison RG, Larson EL. Heterogeneous genome divergence, differential introgression, and the origin and structure of hybrid zones. Mol Ecol. 2016;25:2454–2466. doi: 10.1111/mec.13582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellberg ME. Gene flow and isolation among populations of marine animals. Annu Rev Ecol, Evol, Syst. 2009;40:291–310. [Google Scholar]
- Hellberg ME, Burton RS, Neigel JE, Palumbi SR. Genetic assesment of connectivity among marine populations. Bull Mar Sci. 2002;70:273–290. [Google Scholar]
- Hewitt GM. A sex-chromosome hybrid zone in the grasshopper Podisma pedestris (Orthoptera: Acrididae) Heredity. 1975;35:375–387. doi: 10.1038/hdy.1975.108. [DOI] [PubMed] [Google Scholar]
- Hewitt, G. M. (1989). The subdivision of species by hybrid zones. Speciation and its Consequences, 85-110
- Hewitt GM. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc. 1996;58:247–276. [Google Scholar]
- Hewitt GM. The genetic legacy of the quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- Hoban S, Kelley JL, Lotterhos KE, Antolin MF, Bradburd G, Lowry DB, Poss ML, Reed LK, et al. Finding the Genomic Basis of Local Adaptation: Pitfalls, Practical Solutions, and Future Directions. The American Naturalist. 2016;188:379–397. doi: 10.1086/688018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim KM, Nichols RA, Hewitt GM. Spatial patterns of genetic variation generated by different forms of dispersal during range expansion. Heredity. 1996;77:282–291. [Google Scholar]
- Johannesson K, Andre C. Life on the margin: genetic isolation and diversity loss in a peripheral marine ecosystem, the Baltic Sea. Mol Ecol. 2006;15:2013–2029. doi: 10.1111/j.1365-294X.2006.02919.x. [DOI] [PubMed] [Google Scholar]
- Johannesson K, Panova M, Kemppainen P, Andre C, Rolan-Alvarez E, Butlin RK. Repeated evolution of reproductive isolation in a marine snail: unveiling mechanisms of speciation. Philos Trans R Soc Lond B Biol Sci. 2010;365:1735–1747. doi: 10.1098/rstb.2009.0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T, Ahmed I. adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics. 2011;27:3070–3071. doi: 10.1093/bioinformatics/btr521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruuk LEB, Baird SJE, Gale KS, Barton NH. A comparison of multilocus clines maintained by environmental adaptation or by selection against hybrids. Genetics. 1999;153(4):1959–1971. doi: 10.1093/genetics/153.4.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Launey S, Ledu C, Boudry P, Bonhomme F, Naciri-Graven Y. Geographic structure in the European flat oyster, Ostrea edulis L, as revealed by microsatellite polymorphism. J Hered. 2002;93:331–351. doi: 10.1093/jhered/93.5.331. [DOI] [PubMed] [Google Scholar]
- Le Corre V, Machon N, Petit RJ, Kremer A. Colonization with long-distance seed dispersal and genetic structure of maternally inherited genes in forest trees: a simulation study. Genet Res. 1997;69:117–125. [Google Scholar]
- Lemaire C, Versini JJ, Bonhomme F. Maintenance of genetic differentiation across a transition zone in the sea: discordance between nuclear and cytoplasmic markers. J Evol Biol. 2005;18:70–80. doi: 10.1111/j.1420-9101.2004.00828.x. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSPv5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Lourenço CR, Nicastro KR, Serrão EA, Castilho R, Zardi GI. Behind the mask: cryptic genetic diversity of Mytilus galloprovincialis along southern European and northern African shores. J Mollusca Stud. 2015;81:380–387. [Google Scholar]
- M’Gonigle LK, FitzJohn RG. Assortative mating and spatial structure in hybrid zones. Evolution. 2010;64:444–455. doi: 10.1111/j.1558-5646.2009.00838.x. [DOI] [PubMed] [Google Scholar]
- Manel S, Holderegger R. Ten years of landscape genetics. Trends Ecol Evol. 2013;28:614–621. doi: 10.1016/j.tree.2013.05.012. [DOI] [PubMed] [Google Scholar]
- Marshall DJ, Monro K, Bode M, Keough MJ, Swearer S. Phenotype–environment mismatches reduce connectivity in the sea. Ecol Lett. 2010;13:128–140. doi: 10.1111/j.1461-0248.2009.01408.x. [DOI] [PubMed] [Google Scholar]
- McQuaid CD, Phillips TE. Limited wind-driven dispersal of intertidal mussel larvae: in situ evidence from the plankton and the spread of the invasive species Mytilus galloprovincialis in South Africa. Mar Ecol Prog Ser. 2000;201:211–220. [Google Scholar]
- Meirmans PG. Seven common mistakes in population genetics and how to avoid them. Mol Ecol. 2015;24:3223–3231. doi: 10.1111/mec.13243. [DOI] [PubMed] [Google Scholar]
- Nichols RA, Hewitt GM. The genetic consequences of long distance dispersal during colonization. Heredity. 1994;72:312–317. [Google Scholar]
- Nosil P, Vines TH, Funk DJ. Reproductive isolation caused by natural selection against immigrants from divergent habitats. Evolution. 2005;59:705–719. [PubMed] [Google Scholar]
- Ouagajjou Y, Presa P. The connectivity of Mytilus galloprovincialis in northern Morocco: a gene flow crossroads between continents. Estuar, Coast Shelf Sci. 2015;152:1–10. [Google Scholar]
- Palero F, Abelló P, Macpherson E, Gristina M, Pascual M. Phylogeography of the European spiny lobster (Palinurus elephas): influence of current oceanographical features and historical processes. Mol Phylogenet Evol. 2008;48:708–717. doi: 10.1016/j.ympev.2008.04.022. [DOI] [PubMed] [Google Scholar]
- Palumbi SR. Genetic divergence, reproductive isolation, and marine speciation. Annu Rev Ecol Syst. 1994;25:547–572. [Google Scholar]
- Patarnello T, Volckaert FA, Castilho R. Pillars of Hercules: is the Atlantic-Mediterranean transition a phylogeographical break? Mol Ecol. 2007;16:4426–4444. doi: 10.1111/j.1365-294X.2007.03477.x. [DOI] [PubMed] [Google Scholar]
- Pelc RA, Warner RR, Gaines SD. Geographical patterns of genetic structure in marine species with contrasting life histories. J Biogeogr. 2009;36:1881–1890. [Google Scholar]
- Piálek J, Barton NH. The spread of an advantageous allele across a barrier: the effects of random drift and selection against heterozygotes. Genetics. 1997;145:493–504. doi: 10.1093/genetics/145.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polechová J, Barton N. Genetic drift widens the expected cline but narrows the expected cline width. Genetics. 2011;189:227–235. doi: 10.1534/genetics.111.129817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin XX, Coyne KJ, Waite JH. Tough tendons mussel byssus has collagen with silk-like domains. J Biol Chem. 1997;51:32623–32627. doi: 10.1074/jbc.272.51.32623. [DOI] [PubMed] [Google Scholar]
- Quesada H, Beynon CM, Skibinski DOF. A mitochondrial DNA discontinuity in the mussel Mytilus galloprovincialis: pleistocene vicariance biogeography and secondary intergradation. Mol Biol Evol. 1995;12:521–524. doi: 10.1093/oxfordjournals.molbev.a040227. [DOI] [PubMed] [Google Scholar]
- Quesada H, Warren M, Skibinski DOF. Nonneutral evolution and differential mutation rate of gender-associated mitochondrial DNA lineages in the marine mussel Mytilus. Genetics. 1998;149:1511–1526. doi: 10.1093/genetics/149.3.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada H, Zapata C, Alvarez G. A multilocus allozyme discontinuity in the mussel Mytilus galloprovincialis: the interaction of ecological and life-history factors. Mar Ecol Prog Ser. 1995;116:99–115. [Google Scholar]
- Rand DM, Harrison RG. Ecological genetics of a mosaic hybrid zone: mitochondrial, nuclear, and reproductive differentiation of crickets by soil type. Evolution. 1989;43:432–449. doi: 10.1111/j.1558-5646.1989.tb04238.x. [DOI] [PubMed] [Google Scholar]
- Ravinet M, Faria R, Butlin RK, Galindo J, Bierne N, Rafajlović M, et al. Interpreting the genomic landscape of speciation: a road map for finding barriers to gene flow. J Evol Biol. 2017;30:1450–1477. doi: 10.1111/jeb.13047. [DOI] [PubMed] [Google Scholar]
- Remington Charles L. Evolutionary Biology. Boston, MA: Springer US; 1968. Suture-Zones of Hybrid Interaction Between Recently Joined Biotas; pp. 321–428. [Google Scholar]
- Riginos C, Douglas KE, Jin Y, Shanahan DF, Treml EA. Effects of geography and life history traits on genetic differentiation in benthic marine fishes. Ecography. 2011;34:566–575. [Google Scholar]
- Riginos C, Liggins L. Seascape genetics: populations, individuals, and genes marooned and adrift. Geogr Compass. 2013;7:197–216. [Google Scholar]
- Riginos C, Crandall ED, Liggins L, Bongaerts P, Treml EA. Navigating the currents of seascape genomics: how spatial analyses can augment population genomic studies. Current zoology. 2016;62(6):581–601. doi: 10.1093/cz/zow067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha L, Bowen B. Speciation in coral‐reef fishes. J Fish Biol. 2008;72:1101–1121. [Google Scholar]
- Rossi V, Ser‐Giacomi E, López C, Hernández‐García E. Hydrodynamic provinces and oceanic connectivity from a transport network help designing marine reserves. Geophys Res Lett. 2014;41:2883–2891. [Google Scholar]
- Sanjuan A, Zapata C, Alvarez G. Mytilus galloprovincialis and M. edulis on the coasts of the Iberian Peninsula. Mar Ecol Prog. 1994;113:131–46. [Google Scholar]
- Schunter C, Carreras-Carbonell J, Macpherson E, Tintoré J, Vidal‐Vijande E, et al. Matching genetics with oceanography: directional gene flow in a Mediterranean fish species. Mol Ecol. 2011;20:5167–5181. doi: 10.1111/j.1365-294X.2011.05355.x. [DOI] [PubMed] [Google Scholar]
- Sivasundar A, Palumbi SR. Life history, ecology and the biogeography of strong genetic breaks among 15 species of Pacific rockfish, Sebastes. Marine biology. 2010;157(7):1433–1452. [Google Scholar]
- Stanley RR, DiBacco C, Lowen B, Beiko RG, Jeffery NW, et al. A climate-associated multispecies cryptic cline in the northwest Atlantic. Sci Adv. 2018;4:eaaq0929. doi: 10.1126/sciadv.aaq0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin Gene flow and the geographic structure of natural populations. Science. 1987;236:787–792. doi: 10.1126/science.3576198. [DOI] [PubMed] [Google Scholar]
- Slatkin M. Gene flow and selection in a cline. Genetics. 1973;75:733–756. doi: 10.1093/genetics/75.4.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuckas H, Knöbel L, Schade H, Breusing C, Hinrichsen HH, Bartel M, et al. Combining hydrodynamic modelling with genetics: can passive larval drift shape the genetic structure of Baltic Mytilus populations? Mol Ecol. 2017;26:2765–2782. doi: 10.1111/mec.14075. [DOI] [PubMed] [Google Scholar]
- Swenson NG, Howard DJ. Clustering of contact zones, hybrid zones, and phylogeographic breaks in North America. Am Nat. 2005;166:581–591. doi: 10.1086/491688. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teske PR, Papadopoulos I, Barker NP, McQuaid CD, Beheregaray LB. Mitonuclear discordance in genetic structure across the Atlantic/Indian Ocean biogeographical transition zone. J Biogeogr. 2014;41:293–401. [Google Scholar]
- Thibert-Plante X, Hendry A. When can ecological speciation be detected with neutral loci? Mol Ecol. 2010;19:2301–2314. doi: 10.1111/j.1365-294X.2010.04641.x. [DOI] [PubMed] [Google Scholar]
- Tine M, Kuhl H, Gagnaire P-A, Louro B, Desmarais E, Martins RST et al. (2014) The European seabass genome and its variation provide insights into adaptation to euryhalinity and speciation. Nat Commun 5:5770 [DOI] [PMC free article] [PubMed]
- Väinölä R, Hvilsom MM. Genetic divergence and a hybrid zone between Baltic and North Sea Mytilus populations (Mytilidae: Mollusca) Biol J Linn Soc. 1991;43:127–148. [Google Scholar]
- Vines TH, Dalziel AC, Albert AY, Veen T, Schulte PM, Schluter D. Cline coupling and uncoupling in a stickleback hybrid zone. Evolution. 2016;70:1023–1038. doi: 10.1111/evo.12917. [DOI] [PubMed] [Google Scholar]
- Viúdez Aacute, Tintoré J. Time and space variability in the eastern Alboran Sea from March to May 1990. J Geophys Res: Oceans. 1995;100:8571–8586. [Google Scholar]
- Zouros E, Oberhauser Ball A, Saavedra C, Freeman KR. A unusual type of mtDNA inheritance in the blue mussel Mytilus. Proc Natl Acad Sci USA. 1994;91:7463–7467. doi: 10.1073/pnas.91.16.7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data have been deposited in GenBank: accession numbers MK259098–MK259664. Genotype data have been deposited in Dryad: 10.5061/dryad.m3t6p5g.