Abstract
Although low levels of genetic structure are expected in highly widespread species, geographical and/or ecological factors can limit species distributions and promote population structure and morphological differentiation. In order to determine the effects of geographical isolation on population genetic structure and wing morphology, 281 individuals of the cosmopolitan odonate Pantala flavescens were collected from four continental (Central and South America) and five insular sites (Polynesian islands and the Maldives). COI sequences and eight microsatellite loci were used to characterize genetic diversity and genetic structure between and within locations. Linear and geometric morphometry were used to evaluate differences in the size and shape of wings. Genetic analysis showed a global genetic difference between the continental and insular sites. American locations did not show genetic structure, even in locations separated by a distance of 5000 km. Easter Island showed the lowest values of genetic diversity (mainly mitochondrial diversity) and the highest values of genetic differences compared to other insular and continental sites. Individuals from Easter Island showed smaller forewings, a different abdomen length to thorax length ratio, and a different configuration of anal loop in the hindwings. Thus, the greater isolation, smaller area, and young geological age seem to have determined the genetic and morphological differences in P. flavescens of Easter Island, where selection could promote a loss of migratory behavior and may improve other life history traits, such as reproduction. This work provides new insight into how microevolutionary processes operate in isolated populations of cosmopolitan species.
Subject terms: Ecological genetics, Evolutionary genetics
Introduction
Dispersal is one of the most important biological traits in determining ecological and evolutionary processes in natural populations, affecting local adaptation, speciation, and the evolution of life history traits (Dieckmann et al. 1999). Dispersal is related to the ability to colonize new habitats and enlarge the distributional range of species. It determines the survival of a species in a patchy landscape, providing flexibility as a response to environmental contingencies, and reduces the probability of extinction of the population (Monaghan et al. 2001; Lester et al. 2007; Tesson and Edelaar 2013). Dispersal can also strongly reduce the extent of genetic differences among populations due to the high interchange of individuals, but can also lead to reproductive isolation and evolutionary independence when dispersion is limited by geographical barriers that prevent the gene flow and/or exchange of individuals (Hughes et al. 2009). For example, genetic differences have been observed in migratory species, such as the monarch butterfly Danaus plexippus (Lyons et al. 2012) and the bumblebee Bombus terrestris (Estoup et al. 1996), owing to the great geographical distances between continental and insular populations. In this sense, habitat isolation arises as one of the most important selective forces in shaping the natural dispersal of organisms (Roff 1990; Denno et al. 2001) and consequently the genetic structure of populations.
Oceanic islands are ideal models in which to observe changes in the populations because of their generally small size, distinct boundaries, and simplified biota (Losos and Ricklefs 2009). In addition, the geographic isolation of oceanic islands has led to increased levels of inbreeding and a greater impact of genetic drift (Furlan et al. 2012), but it has also imposed severe restrictions on dispersal, which may result in adaptation toward weaker flying in organisms (Losos and Ricklefs 2009). Changes in body size and loss of dispersal ability are two well-known ecogeographical patterns among island species (Lomolino 2005; Whittaker and Fernandez-Palacios 2007). The development of wingless forms on oceanic islands is a theory that was discussed in early studies and is based on the hypothesis that natural selection reduces wing size because of the high cost associated with wing maintenance (Darwin 1859; Roff 1994). In this sense, it can be expected that geographically isolated populations would show morphological traits associated with a decreasing ability to migrate over long distances.
Pantala flavescens (Fabricius), or the “wandering glider”, is the most widespread odonate, absent only in Antarctica and a part of Europe (Dijkstra and Clausnitzer 2014). It migrates widely, following the temporary abundance of a habitat provided by rains, and it is among the dragonflies most often found on oceanic islands (May 2012). Recently, Christudhas and Mathai (2014), Troast et al. (2016), and Low et al. (2017) suggested high rates of gene flow and panmictic populations of P. flavescens along vast geographic distances. Nevertheless, the presence of individuals with unusual morphological and behavioral characteristics in a remote island of the South Pacific has suggested genetic divergence in this well-known migratory species (Samways and Osborn 1998). Indeed, P. flavescens is the only dragonfly inhabiting Easter Island or Rapa Nui (Dumont and Verschuren 1991; Moore 1992), the most remote inhabited island in the world, located 3510 km from the American continent (Mieth and Bork 2010). Rapa Nui has relatively depauperate freshwater biota, reflecting its young geological age (0.24–0.11 Mya), small size (171 km2), and great geographic isolation (Segers and Dumont 1993). Samways and Osborn (1998) compared the morphological and behavioral traits among continental (South Africa) and insular (Easter Island) populations of P. flavescens and found that insular individuals have mostly reduced, more asymmetrical, and darker wings, and flew lower to the ground than individuals from the continental population. These authors suggested that strong selective pressures toward nonmigratory traits are acting on the insular population due to the extreme isolation from the mainland or other islands.
Based on the above-mentioned premises, the goal of this research was to evaluate whether geographic isolation determines morphological and genetic structure in populations of the cosmopolitan dragonfly P. flavescens. Sequences of the cytochrome oxidase I (COI) gene and microsatellite loci together with morphological analysis results (linear and geometric morphometrics) were obtained and compared among samples of individuals collected from continental (Central and South America) and insular locations (Easter Island, Tonga, and the Maldives).
Materials and methods
Samples and sampling sites
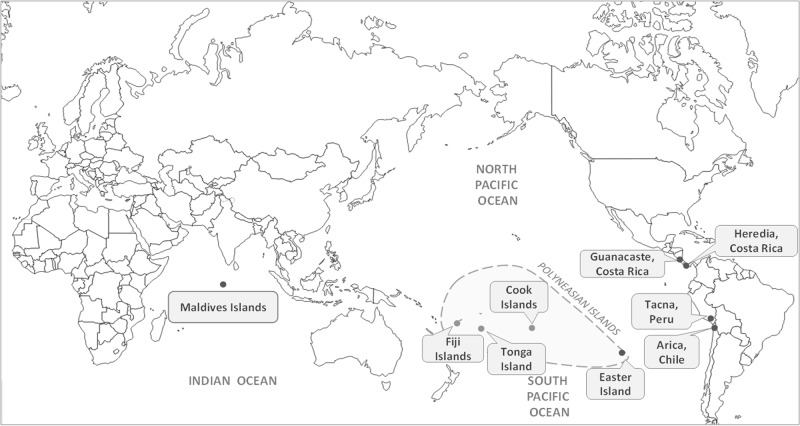
A total of 281 dragonflies were collected for this study (Fig. 1). Samples from Arica (Chile) (n = 34), Tacna, Peru (n = 20), Heredia (n = 29), and Guanacaste (n = 17) in Costa Rica were collected manually using an entomological net during 2015 and 2016; all these samples were used in Alvial et al. (2017). New samples collected from insular sites were obtained in different ways. Samples from Easter Island (n = 82) and Tonga Island (n = 40) were collected in 2017 using an entomological net. Twelve individuals from the Maldives were donated by Charles Anderson, and samples from Cook Islands (n = 29) and Fiji Islands (n = 18) were obtained from the New Zealand Arthropod Collection at the Landcare Research Institute (Auckland, New Zealand).
Fig. 1.
Sampling sites of Pantala flavescens. Modified from www.d-maps.com
While 220 individuals were used in the genetic analysis, the morphological analysis was performed with 236 individuals. This difference was due to the low DNA quality obtained from samples from Cook Islands and Fiji Islands; these individuals were used only in the morphological and morphometric analyses. The sample size used for each analysis is summarized in Table 1.
Table 1.
Locations, geographic coordinates, altitude range, date of sampling, and number of individuals/samples collected from nine study sites
| Location | Geographic coordinates | Date of sampling | N | N gen | N lm | N gm |
|---|---|---|---|---|---|---|
| Guanacaste, Costa Rica | 10°50’N, 85°37’W | April–May 2016 | 17 | 17 | 17 | 10 |
| Heredia, Costa Rica | 10°00’N, 84°06’W | April–May 2016 | 29 | 22 | 29 | 10 |
| Arica, Chile | 18°31’S, 70°10’W | January 2015–March 2016 | 34 | 29 | 34 | 28 |
| Tacna, Peru | 18°06’S, 70°20’W | March 2016 | 20 | 19 | 20 | 17 |
| Easter Island, Chile | 27°07’S, 109°22’W | August–September 2015 | 82 | 82 | 47 | 35 |
| Nukualofa, Tonga Island | 21°07’S, 175°13’W | September 2016 | 40 | 40 | 30 | 28 |
| Rarotonga, Cook Islands | 21º13’S, 159º46’W | March 2015 | 29 | – | 29 | |
| Viti Levu, Fiji Islands | 17º49’S, 178º0’E | February 2015 | 18 | – | 18 | |
| South Malé Atoll, Maldives Island | 2º32’N, 72º59’E | November 2015 | 12 | 11 | 12 | 12 |
| Total of samples | 281 | 220 | 236 | 140 | ||
N total number of samples per site, N gen number of samples used in genetic analysis, N lm number of samples used in linear morphology analysis, N gm number of samples used in geometric morphometry analysis
Genetic analysis
As previously mentioned, 220 adult specimens of Pantala flavescens obtained from seven geographic locations across Central and South America, the Maldives, and other South Pacific islands were used for the genetic analysis. Individuals were dissected, and 1 mm3 of thoracic muscle was obtained for DNA extraction using the salt extraction method described by Aljanabi and Martinez (1997). To describe the geographic genetic structure of P. flavescens, a partial region of the COI gene and eight microsatellites described for this species by Cao et al. (2015) were amplified.
COI analysis
A section of the COI gene was amplified using the universal primers N2191 (CCG GTA AAA TTA AAA TAT AAA CTT C) and J1718 (GGA GGA TTT GGA AAT TGA TTA GTT CC), described by Simon et al. (1994). PCR was performed following a modified protocol based on Simon et al. (1994) that uses 2 µL of DNA (50 ng/µL), 2.5 µL of 10 × PCR buffer (Invitrogen), 1.6 µL of MgCl2 (50 mM) (Invitrogen), 2 µL of dNTPs (2.5 mM) (Invitrogen), 14.6 µL of water, 1 µL of forward and reverse primers (50 ng/µL), and 0.3 µL of Taq polymerase (Invitrogen). The PCR procedure started at 95 °C for 2 min, followed by 35 cycles of 95 °C for 30 s, 50 °C for 30 s, and 72 °C for 30 s, and a final step of 72 °C for 5 min. Amplifications were verified on a 1.5% agarose gel, and forward and reverse strands were sequenced using an ABI 3730xl Analyzer by Macrogen Inc. (Korea). Sequences were edited and aligned in PROSEQ 2.91 (Filatov 2002). Molecular diversity indices, such as the number of polymorphic sites (S), haplotype diversity (h), and nucleotide diversity (π), were calculated using DnaSP 4.9 software (Rozas et al. 2003). Genealogical relationships among haplotypes were further assessed using a median-joining network algorithm constructed in PopART v 1.7 (Leigh and Bryant 2015).
To test for phylogeographic structure, the observed genetic differentiation coefficients GST and NST were compared using the software PERMUT v 1.2.1 (Ponds and Petit 1996). This method identifies evidence of a phylogeographic structure when NST is statistically greater than the GST index. Furthermore, the FST index was estimated for pairs of populations using ARLEQUIN v 3.5 (Excoffier and Lischer 2010), and the significance was tested using 1,000 permutations.
Microsatellite analysis
Samples from seven locations were genotyped for eight microsatellite loci described for P. flavescens by Cao et al. (2015) using a modified PCR protocol described by the same authors. The PCR conditions used were as follows: 1.5 µL of DNA (50 ng/µL), 1.5 µL of MgCl2 (50 mM) (Invitrogen), 0.5 µL of 1% BSA, 2 µL of 10 × PCR buffer (Invitrogen), 2 µL of dNTPs (2.5 mM) (Invitrogen), 4.15 µL of water, 0.5 µL of each primer, 0.5 of M13, and 0.15 µL of Taq polymerase (Invitrogen). For the PCR procedure, a touchdown protocol starting at 10 °C above the annealing temperature of each microsatellite was used. PCR products were run in an ABI-PRISM 3010 sequencer (Perkin Elmer) using 500 ROX Size Standard (Applied Biosystems) at the Pontificia Universidad Católica de Chile. The allelic data matrix was obtained using GENEMARKER software (Softgenetics Inc.). To calculate the genotyping error rate, 44 samples (20%) were chosen randomly to be genotyped blindly a second time following the same protocol, and then the genotypes obtained were compared to the original runs, and the number of allelic mismatches was counted according to the method of Morin et al. (2010).
To control the data quality, MICROCHECKER v 2.2 (Van Oosterhout et al. 2004) was used to detect possible genotyping errors and the presence of null alleles in the allelic data. The number of alleles per locus and the expected (HE) and observed (HO) heterozygosity were estimated using GENETIX v 4.05 (Belkhir et al. 2000). This same software was used to test for linkage disequilibrium and for departures from Hardy–Weinberg equilibrium (HWE); 5000 permutations on mono locus genotypes were used to test for statistical significance. Allelic richness (AR) and the inbreeding coefficient (FIS) were estimated with FSTAT v 2.9.3 (Goudet 1995). To ensure that the analysis was performed with a significant number of unrelated individuals, the rxy index (Queller and Goodnight 1989) was estimated with IDENTIX v 1.1 (Belkhir et al. 2001).
Population structure was evaluated using three different methods: (i) computing pairwise FST using GENETIX software (Belkhir et al. 2000), where 1000 permutations were used to test for statistical significance among pairs of sites; (ii) a Bayesian approximation conducted in STRUCTURE v 2.3.4 (Pritchard et al. 2000); and (iii) an iterative reassignment of individuals implemented in FLOCK v 2.0 (Duchesne and Turgeon 2012). The analysis in STRUCTURE was performed using the admixture model and the correlated allele frequencies model. Using the hierarchical method (see Vähä et al. 2007), we ran STRUCTURE software over each cluster predicted in the precedent run until the analysis did not detect evidence of geographical partition in the data. The procedure was run five times for each K estimation (from K = 1 to K = 8), with a burn-in of 200,000 MCMC iterations. The LnP(D) values obtained for each K value were compared using STRUCTURE HARVESTER (Eare and Von Holdt 2012). The graphical displays of the STRUCTURE results were generated using CLUMPAK online software (Kopelman et al. 2015). In contrast, FLOCK assigns genetically similar individuals to K partitions using a positive feedback mechanism where the number of individuals assigned to each of K groups grows with each reallocation. The analysis involved a total of 30 reallocations and 50 runs for K = 2–6, with log-likelihood values ranging from 0 to 0.6.
Morphological analysis
Traditional morphometry
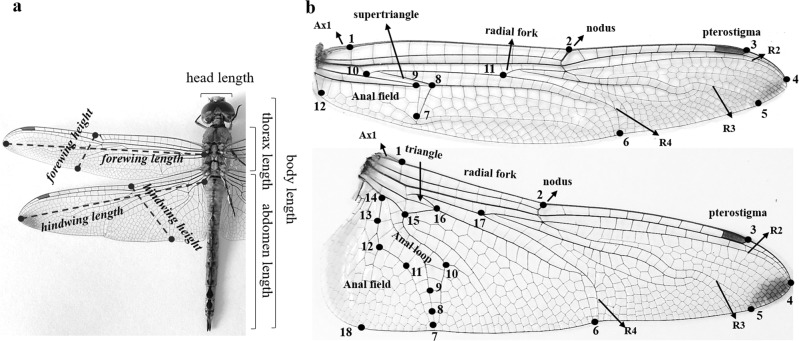
Morphological measures were obtained from 236 individuals (79 females and 157 males) from nine sites (Table 1). All individuals were measured using a digital caliper under a stereoscopic microscope at ×10. Eight morphological measures were obtained, namely, body length (BD, distance between the anterior end of the postclypeus and the tip of the anal appendages), abdomen length (AL, distance between the anterior end of the first abdominal segment and the tip of the anal appendages), thorax length (TL, distance from the anterior margin of the prothorax to the posterior margin of the metathorax), head width (HW), forewing length (FWL, distance from the costal vein base to the end of vein R2), forewing height (FWH, wing width from the nodus to the end of vein R4), hindwing length (HWL, distance from the base of the costal vein to the end of vein R2), and hindwing height (HWH, distance from the first ante nodal cross-vein AX1 to the end of the anal loop) (Fig. 2a).
Fig. 2.
a Morphological linear measures selected for wing size and corporal size analysis (R2 and R4 = veins); b forewing (above) and hindwing (below) landmarks recorded for each Pantala flavescens. Wing landmarks are: 1, costal vein base (AX1); 2, nodus; 3, end of pterostigma; 4, end of vein R2; 5, end of vein R3; and 6, end of R4. Additional landmarks in the forewing are 7–9, in the supertriangle (t) area; 10, in the arculus; 11, in the radial fork; and 12, at the margin of the anal field. Additional landmarks in the hindwing are 7–15, in the anal loop area; 16, at the extreme of the triangle area, 17, in the radial fork; and 18, at the lower margin of the anal field
These eight morphological measures allowed us to obtain seven ratios, which were included in statistical analysis: abdomen to thorax (AL.TL), thorax to body length (TL.BL), head width to body length (HW.BL), forewing length to body length (FWL.BL), forewing height to body length (FWH.BL), hindwing length to body length (HWL.BL), and hindwing height to body length (HWH.BL). Ratios were used for scaling morphometric variables to remove variation in general body size, as proposed by Daly (1985) and Marinov and Mchugh (2010). To detect possible differences between sexes and among populations based on the seven morphometric ratios, a nonparametric permutation-based multivariate analysis of variance (MANOVA) with 999 permutations was performed. Further, a linear discriminant analysis (LDA) was used to identify the combination of variables that best explained the differences between sexes and among populations. With the most important explanatory variables, LD1 and LD2, a generalized linear model (GLM) with Gaussian link function was used to find differences between localities and sexes. The a posteriori test was conducted using the “lsmeans” package for the significant source of variation in the GLM. All statistical analyses were performed in R v 3.4.3 (R Core Team 2017). The “vegan” library was used for the permutation MANOVA (Oksanen et al. 2018), and the “MASS” library was used to perform the discriminant analysis (Venables and Ripley 2002).
Geometric morphometry
To detect differences in wing shape, a landmark-based geometric morphometric analysis was performed. Morphometric measures were taken from 280 wings corresponding to 140 individuals from seven localities (Table 1). Each wing (left and right wings) was photographed at a fixed position using a digital camera (14-megapixel camera Olympus SP-810UZ). The wings were kept flat by sandwiching between two glass slides while taking the photographs. To capture the shape of the wings, 30 landmarks (12 in the forewing and 18 in the hindwing) were placed on the digital images (Fig. 2b). Landmark selection was performed following Johansson et al. (2009) and Suarez-Tovar and Sarmiento (2016), who found that using vein nodes as landmarks adequately captures wing shape and facilitates comparative analyses. Landmark coordinates were then rotated, scaled, and translated into alignment using a generalized least-squares superimposition method based on the generalized Procrustes analysis (Rohlf and Slice 1990). This procedure removes size differences among individuals and leaves purely allometric shape variation for subsequent analysis. Then, a principal component analysis (PCA) was performed for the relative warp to visualize shape variations; deformation grids were produced by regression of the shape variables on the canonical variables. To assess differences in wing shape configuration, a Procrustes ANOVA (Goodall 1991) was performed with the Procrustes distances among individuals of P. flavescens. This analysis was performed with the “Geomorph” library (Adams and Otarola 2013) implemented in R 3.4.3 software (R Core Team 2017).
Results
Genetics analysis
COI analysis
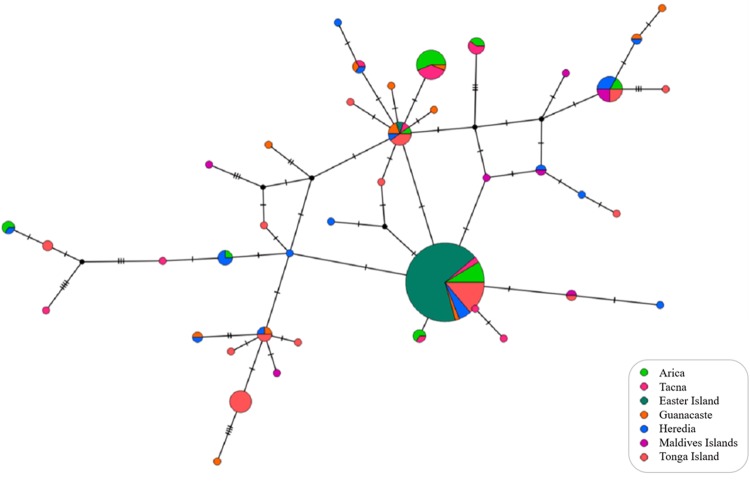
A total of 41 haplotypes were obtained from the 220 individuals of P. flavescens sequenced for a 439-bp segment of the COI gene (GenBank Accession Numbers: KY200583-KY200609, and KY934249-KY934261). No insertions or deletions were detected, indicating that the alignment was straightforward. The number of polymorphic sites ranged from 1 (Easter Island) to 18 (Tacna and Guanacaste). As shown in Table 2, genetic diversity (h) varied from 0.025 (Easter Island) to 0.971 (Guanacaste), and nucleotide diversity (π) varied from 0.0001 (Easter Island) to 0.009 (Guanacaste and Maldives). Two common haplotypes were present in most locations, with the exception of the Maldives. Interestingly, these two haplotypes represent the whole COI diversity in Easter Island. Qualitatively, the haplotype network did not show any apparent geographical structure (Fig. 3).
Table 2.
Summary of genetic diversity indices in the COI gene and eight microsatellite loci for Pantala flavescens
| Location | N COI/microsatellites | nh | np | h (± SD) | π (± SD) | A R | H E | H O |
|---|---|---|---|---|---|---|---|---|
| Maldives | 10/11 | 8 | 5 | 0.933 ± 0.077 | 0.009 ± 0.005 | 2.015 | 0.586 | 0.403 |
| Tonga | 43/46 | 13 | 8 | 0.815 ± 0.045 | 0.006 ± 0.003 | 2.536 | 0.671 | 0.471 |
| Easter Island | 80/82 | 2 | 0 | 0.025 ± 0.024 | 0.0001 ± 0 .0002 | 1.899 | 0.452 | 0.407 |
| Guanacaste | 17/17 | 13 | 6 | 0.971 ± 0.027 | 0.009 ± 0.005 | 2.086 | 0.508 | 0.345 |
| Heredia | 22/21 | 13 | 5 | 0.909 ± 0.045 | 0.007 ± 0.004 | 1.973 | 0.472 | 0.379 |
| Tacna | 19/14 | 10 | 4 | 0.877 ± 0.056 | 0.008 ± 0.005 | 1.953 | 0.475 | 0.346 |
| Arica | 29/29 | 8 | 0 | 0.791 ± 0.051 | 0.006 ± 0.004 | 1.994 | 0.498 | 0.505 |
N Number of sequences, nh number of haplotypes, np private haplotypes, h haplotype diversity, π nucleotide diversity, AR allelic richness, and HO and HEobserved and expected heterozygosities, are shown for each sampling location
Fig. 3.
Median-joining network showing the relationship among COI haplotypes in Pantala flavescens. Circles represent haplotypes, with sizes proportional to their respective frequencies. Colors indicate the population origin of haplotypes. Tick marks represent the deduced number of nucleotide substitutions along each branch
The FST comparisons between pairs of locations are shown in Table 3. In this analysis, most of the largest FST values were observed for paired comparisons between island locations (Maldives, Tonga, and Easter Island), versus locations in the America (Guanacaste, Heredia, Tacna, and Arica), and ranged from 0.10 to 0.52. Most comparisons showed significant differences (P-value < 0.01), except for the comparisons of the Maldives and Tonga with Guanacaste and Heredia. Pairwise comparisons between locations in the Americas presented low FST values (<0.06), showing no significant differences among these locations (P-value > 0.05). On the other hand, pairwise comparisons between island locations (Maldives, Tonga, and Easter Island) showed FST values ranging from 0.15 to 0.72, all showing significant differences (P-value < 0.01). Finally, Easter Island showed the highest and significant FST values (>0.25, P-value < 0.001) for all the sites.
Table 3.
Matrix of pairwise FST values between populations based on COI gene (above the diagonal) and microsatellite loci (below the diagonal)
| Maldives | Tonga | Easter Island | Guanacaste | Heredia | Tacna | Arica | |
|---|---|---|---|---|---|---|---|
| Maldives | 0.158* | 0.721* | 0.046 | 0.024 | 0.104* | 0.116* | |
| Tonga | 0.109* | 0.254* | 0.068 | 0.040 | 0.131* | 0.115* | |
| Easter Island | 0.119* | 0.199* | 0.520* | 0.355* | 0.450* | 0.377* | |
| Guanacaste | 0.344* | 0.248* | 0.410* | 0.036 | 0.053 | 0.052 | |
| Heredia | 0.407* | 0.273* | 0.444* | 0.003 | 0.065 | 0.051 | |
| Tacna | 0.359* | 0.244* | 0.405* | 0.002 | 0.003 | 0.022 | |
| Arica | 0.360* | 0.252* | 0.408* | 0.022 | 0.041 | 0.011 |
Significant values after a Benjamini–Yekutieli correction (Benjamini and Yekutieli 2001) based on the false discovery rate approach (*P-value < 0.01)
The PERMUT results showed a significantly larger NST (0.248) than GST (0.149) values across all populations for the mtDNA sequence, which indicated that the genetic variability of P. flavescens was geographically structured across its global distribution (P-value = 0.014). Nevertheless, the analysis performed for the American sites (NST = 0.038, GST = 0.033, P-value = 0.655) and the island locations (NST = 0.335, GST = 0.296, P-value = 0.289) did not detect any separate phylogeographic structure, showing a phylogeographical signal only at a large geographical scale.
Microsatellite analysis
Summary statistics for the microsatellite loci in each location are shown in Table 2; detailed information is provided in Supplementary Appendix S1. Analysis using MICROCHECKER suggested the presence of null alleles by general excess of homozygotes in three of the eight loci studied: SSR24 (Heredia, Easter Island, and Tonga), SSR2 (Tacna, Guanacaste, and Maldives island), and SSR14 (Arica, Easter Island, and Tonga). The genotyping error rate was low, at < 1% for the 20% of reanalyzed samples. Five out of the 56 comparisons showed evidence of linkage disequilibrium (P-value < 0.01); however, this disequilibrium was not observed in all locations. Departures from HWE were detected in seven of eight microsatellite loci (P-value < 0.01), but the deviations were not associated with any specific locus or location.
A total of 56 alleles were detected across loci, with five alleles at locus SSR10 and nine alleles at locus SSR24. The average AR value was the highest in Tonga (2.536) and the lowest in Easter Island (1.899). The HE and HO per site varied, from 0.452 (Easter Island) to 0.671 (Tonga) and from 0.345 (Guanacaste) to 0.586 (Tonga), respectively (Table 2). The mean rxy values showed statistical evidence (permutation test, P-value > 0.01) of highly related individuals only in Easter Island (see Supplementary Appendix S1). Pairwise FST values based on allelic frequencies ranged from 0.002 to 0.444 (Table 3). Similar to the results obtained with COI, western locations (Maldives, Tonga, and Easter Island), compared to the eastern locations (Guanacaste, Heredia, Tacna, and Arica), showed higher and statistically significant FST values (FST > 0.24, P-value < 0.01). Furthermore, pairwise comparison between locations in America (Guanacaste, Heredia, Tacna, and Arica) showed lower and no significant FST values (FST < 0.04, P-value > 0.05), while paired comparisons between island locations (Maldives, Tonga, and Easter Island) showed high and statistically significant FST values (FST > 0.10, P-value < 0.01).
Initial partitioning of the data led to a clustering of K = 2, which segregated continental from island sites (Fig. 4, Supplementary Appendix S2). The second round was carried out for each of these groups separately. In the case of the continental group, STRUCTURE software produced a clustering of K = 2; however, this clustering did not show any pattern of geographical differentiation. The Island group produced a clustering of K = 2, which clearly separated Easter Island from Maldives–Tonga. In the third round, the Maldives–Tonga cluster led to a subsequent clustering of K = 4 and the Easter Island cluster to a clustering of K = 2; neither analysis evidenced a clear pattern justifying a new level of the hierarchical analysis. Finally, FLOCK software suggested K = 2 as the most likely number of genetic groups (American vs. island locations).
Fig. 4.
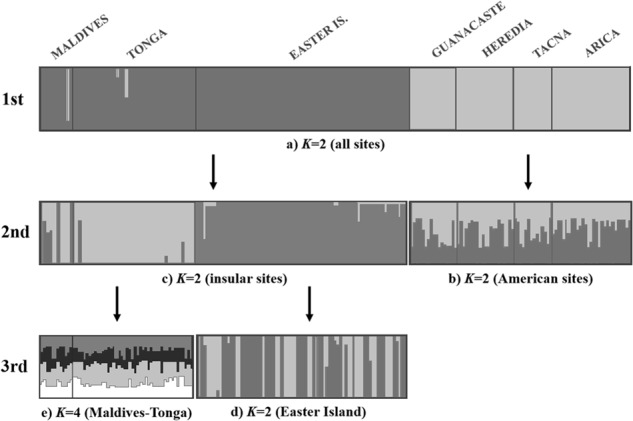
Hierarchical STRUCTURE analysis with three levels of subsets of populations separated and re-analyzed. Vertical lines separate individuals from seven sampling sites (labeled above). Each individual is represented by a thin horizontal line, which is partitioned into K-colored segments representing an individual’s estimated membership fractions in K clusters
Morphological analysis
Linear morphometry
The permutation MANOVA showed a significant difference between sexes and among populations based on the seven morphological ratios (Λ = 0.01042, F = 2.9956, P-value = 0.006). Linear discriminant analysis (LDA) showed that the following ratios explain most differences among populations and between sexes: forewing length:body length ratio (FWL.BL), forewing height:body length ratio (FWH.BL), and abdomen:thorax ratio (AL.TL) (Fig. 5). In this analysis, the linear discriminant 1 (mainly explained by FWL.BL and FWH.BL) segregated all samples from Easter Island from all other sites; while linear discriminant 2 (mainly explained by AL.TL) segregated males and females of each geographic site with an inverse tendency in individuals from Easter Island.
Fig. 5.
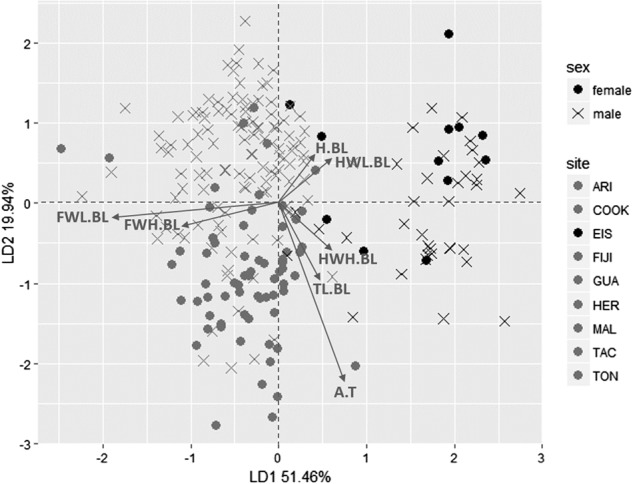
Linear discriminant analysis (LDA) showing a combination of variables that best segregate sex and populations of Pantala flavescens
The GLM showed significant differences in the interaction localities and sex for the aforementioned three ratios (Supplementary Appendix S3). In the case of the abdomen:thorax ratio (AL.TL), the a posteriori test showed that males presented the lowest values of this ratio when compared with females, except for Easter Island, which showed an inverse pattern (Fig. 6a). In other words, the analysis showed that females had a larger abdomen than males, except on Easter Island, where the relationship was inverted. The results showed a similar pattern of change in FWL.BL and FWH.BL, with the lowest values of these ratios in Easter Island compared with all other locations (Fig. 6b, c). Thus, this analysis showed shorter forewings in Easter Island compared with all other sites; the individuals with the longest forewings were observed on Cook Island, the Maldives, and Tonga. Similarly, individuals from Easter Island showed thinner forewings (lower forewing height) than the individuals of other localities; individuals with the thickest forewings were observed in Guanacaste, Heredia, and the Maldives (Fig. 6c).
Fig. 6.
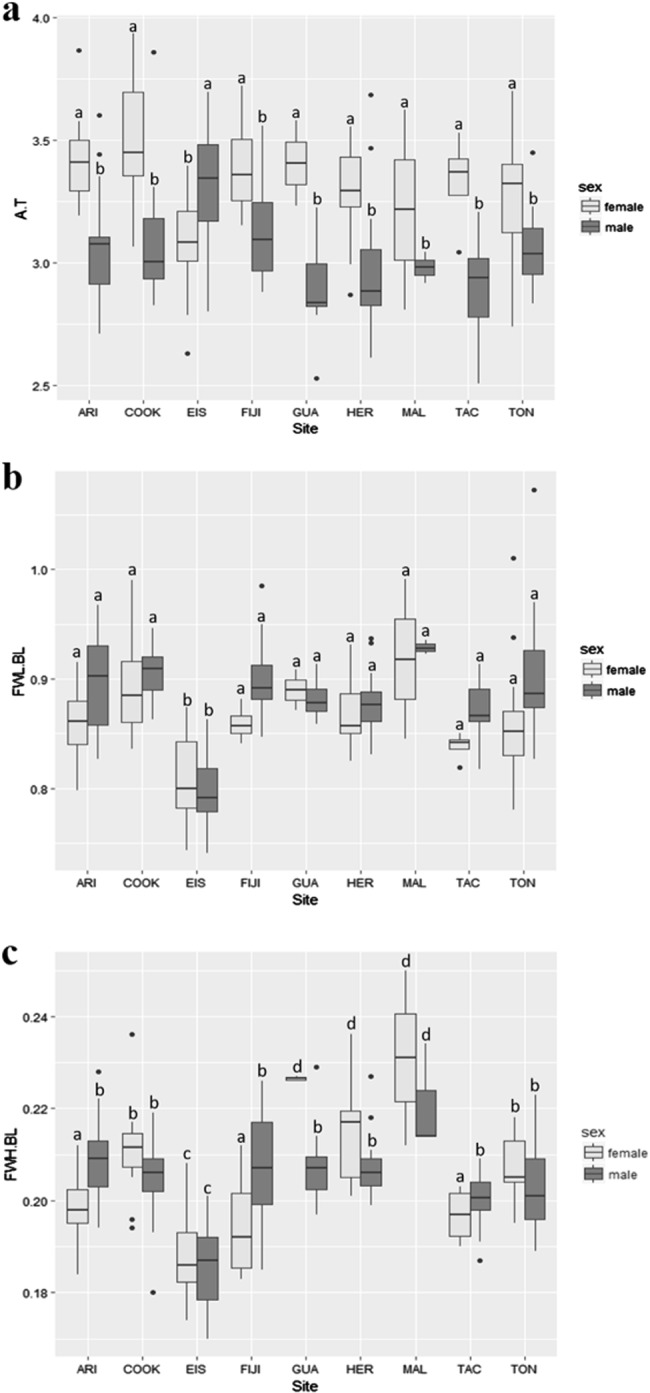
Boxplot of the ratio showing significant statistical differences among locations and sex for Pantala flavescens: a abdomen:thorax ratio, b forewing height:body length ratio, and c forewing height:body length ratio. Different letters in the box represent statistical differences (P-value > 0.05)
Geometric morphometry
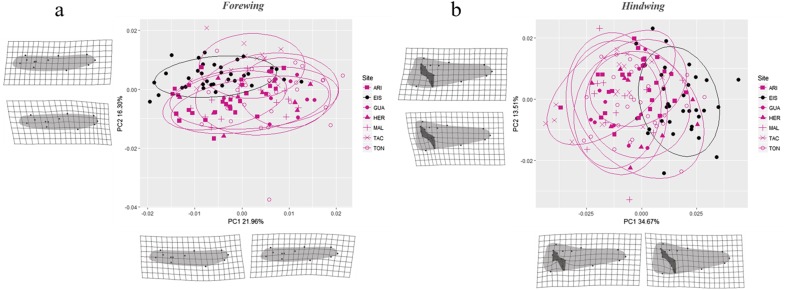
The geometric morphometry analysis revealed significant differences in the configuration of the forewings and the hindwings among individuals of P. flavescens collected in the seven study locations (Procrustes ANOVA, P-value = 0.001). Hindwings explained more of the variation among localities (PC1 = 34.67%, PC2 = 13.51%) than did forewings (PC1 = 21.96%, PC2 = 16.30%) (Fig. 6). Most variation in forewings was explained by landmarks 9 and 10, suggesting that the forewings in the individuals from Easter Island were thinner, curved slightly upward, and had a smaller supertriangle region compared with those of individuals from the other localities (Fig. 7a). In the case of the hindwings, most variation was explained by landmarks 10, 11, and 14, indicating different configurations in the anal loop (Fig. 7b). Individuals from Easter Island showed a more elongated anal loop than individuals from the other locations.
Fig. 7.
PCA plots and deformation grids of the shape variables in the forewings (a) and hindwings (b) of Pantala flavescens
Discussion
Generally, cosmopolitan species have shown great genetic similarity or a lack of genetic structure along their geographical distribution (Freeland et al. 2003); however, the existence of oceanic barriers or terrestrial discontinuities may limit the movement of highly dispersed species, affecting their spatial genetic structure. In the case of P. flavescens, the existence of long continental extensions suggests high gene flow and panmictic populations along vast geographic distances (Christudhas and Mathai 2014; Troast et al. 2016; Alvial et al. 2017; Low et al. 2017). Nevertheless, the absence of continental bridges (e.g., to remote islands) could test these dispersal capabilities and promote a population structure in this cosmopolitan species.
In the present study, based on COI and microsatellite loci variability, the analysis suggested two genetic groups (continental vs. insular), with low connectivity between them. This pattern seems to be of historical origin, given the presence of a phylogeographical signal revealed by the COI haplotypes. The hypothesis of a terrestrial continuum versus an oceanic barrier was evidenced when differentiation indices (FST) were compared among and within both geographic regions. In this way, the highest and most significant values of FST were reported among continental and insular sites. As also evidenced by Alvial et al. (2017), the American sites presented nonsignificant FST values (FST < 0.06; P-value > 0.05), showing that only one cluster composed all these sites in both STRUCTURE and FLOCK software, suggesting high mobility of Pantala flavescens along the American continent. Other studies have shown a null population structure in P. flavescens. For example, four geographical populations of P. flavescens in Peninsular Malaysia did not reveal any population structure (Low et al. 2017), and Troast et al. (2016) described high rates of gene flow among populations of P. flavescens in six distant geographical regions (Guyana, Japan, India, United States, Canada, and Korea).
Conversely, the analysis showed higher and more significant FST values (0.15–0.72; P-value < 0.01) when this index was estimated for between insular populations. Other authors also observed population structure in this dragonfly; Pfeiler and Markow (2017) detected a significant structure when comparing samples from Malaysia and Guyana/Japan. This evidence corroborates the results found in our study, revealing that P. flavescens could have a spatial genetic structure.
Finally, as revealed by the FST index and the cluster analysis, Easter Island showed the highest and most significant differences with all sites. Specifically, the P. flavescens population there showed the lowest haplotypic diversity described for this species (h = 0.025). For example, all other sites analyzed in our study showed h > 0.79, in the Maldives (h = 0.933) and Tonga islands (h = 0.815). Low et al. (2017) described haplotypic diversity ranging from 0.889 to 0.996 in the Malaysia Peninsula, and Pfeiler and Markow (2017) reported a high haplotypic diversity (h = 0.838) when data from Troast et al. (2016) were re-analyzed. Overall, the high differentiation and the lowest values of diversity (haplotype and allelic diversity) reported on this island reflect the effect of the great geographic isolation in this remote South Pacific island.
A similar pattern of a population structure along continental sites versus insular locations was described by Lyons et al. (2012) for the monarch butterfly Danaus plexippus. These authors showed a pattern of decreasing genetic diversity and allelic richness with increasing distance from North America toward Hawaii and New Zealand. Similarly, Estoup et al. (1996) evaluated the differences among continental and insular populations of the bumblebee (Bombus terrestris Linnaeus) and showed genetic homogeneity and large gene flow at the scale of the European continent, but a strong and significant genetic structure in insular Mediterranean populations.
Is there genetic isolation in Easter Island?
Genetic variation in insular ecosystems depends on three factors: (i) net effects of loss at foundation, (ii) subsequent loss in a finite population, and (iii) gains arising from secondary immigration and new mutations (Frankham 1997). In this way, dispersal frequency is a major determinant of the persistence of the population and the emergence of new variation in the short term (Paulay and Meyer 2002). In the case of low dispersal between populations, genetic drift could be the main force that changes the gene pool of the isolated populations. This seems to be the scenario for Easter Island, where the greatest genetic differences and the lowest genetic diversity were found. It is important to note that mitochondrial diversity was comparatively lower than microsatellite diversity on this island when compared with all other sites. This divergence among mitochondrial and nuclear diversity may be the result of a recent population bottleneck (one that has not yet impacted the genetic diversity of the nuclear genome) and the extreme geographic isolation, which prevents the arrival of new immigrants. In contrast with other Pacific Islands, such as the Galápagos or Hawaiian Islands, which show examples of adaptation and diversification (Roderick and Gillespie 1998; Schmitz et al. 2007), Easter Island is geologically recent (0.24–0.11 Mya), and its freshwater fauna and flora are composed of mainly cosmopolitan species probably transported by humans (Campos and Peña 1973).
However, there are a number of other factors that may impact mitochondrial diversity; for example, the effect of endosymbionts. Wolbachia is known to infect the reproductive tissues of a wide range of arthropods, including odonates (Thipaksorn et al. 2003). This fact has contributed to phylogenetically discordant patterns between mtDNA and nDNA (Hurst and Jiggins 2005; Jiang et al. 2014; Cariou et al. 2017).
Evidence of morphological differences in Easter Island
The morphological analysis clearly suggested differences in forewings and hindwings among individuals from Easter Island and individuals from the other localities analyzed in this study. Thus, P. flavescens collected in Easter Island showed forewings that were smaller, thinner, and slightly curved upward, and hindwings with a different anal loop configuration. This pattern was suggested before by Dumont and Verschuren (1993) and Samways and Osborn (1998). Samways and Osborn (1998) suggested that the great distance from the Easter Island to any mainland, and the infrequent arrival of migrants, promotes a reduction in genetic variability in the population of P. flavescens inhabiting Easter Island.
Other research on flight morphology in dragonflies have suggested that wing size strongly determines flight performance, with narrow-based wings being generally associated with slower flight, broad-based wings benefitting insects that make frequent use of fast-forward motion, and hindwings mainly benefitting insects with the habit of frequent gliding (Hankin 1921; Wootton 1991). Furthermore, dragonflies that migrate on continental scales generally have larger wings and a wider basal portion of the hindwings (Samways and Osborn 1998; Corbet 1999). Effectively, significant differences in the morphological characteristics of the wing have been reported among species that differ in their dispersal behavior (e.g., migrant vs. nonmigrant dragonflies), with broader-based hindwings more typically observed in migrant dragonflies (Johansson et al. 2009; McCauley 2013; Suarez-Tovar and Sarmiento 2016).
During field work, we observed that P. flavescens flies very slowly at low altitude (<2 m) on Easter Island, which contrasts with our observations in Arica and Tacna, where individuals of P. flavescens fly at a great speed at heights of over 20 m. In this sense, the smaller forewings observed in Easter Island could indicate a character of stable differentiation in time, associated mainly with the loss of capability to migrate long distances due to long isolation from a mainland or other islands, corroborating the previous inference made by Dumont and Verschuren (1993) and Samways and Osborn (1998).
However, when Johansson et al. (2009) compared forewing and hindwing shapes between migrating and nonmigrating species of dragonflies, they observed that forewings showed only slight differences in comparison to the differences described in hindwings. That finding suggested that the shape of the hindwings is more important for long-distance flying than the shape of the forewings. Nevertheless, our results did not show significant differences in hindwings. Conversely, we found that forewing size determines the main differences among individuals of P. flavescens. Moreover, we found some broader-based hindwings in the Easter Island individuals, with a more elongated anal loop region with more veins inside. This is contrary to our expectation of narrow hindwings and a more compressed anal loop. Nevertheless, divergence in wing shape could highlight a functional difference that may be influenced by environmental (e.g., habitat, temperature, precipitation, or winds), geographic (e.g., altitude and latitude), or/and internal factors (i.e., genetic diversity, body size, or developmental factors) at intra-species levels (Dellicour et al. 2017). In addition, Outomuro et al. (2012) described that sexual selection could be the main selective agent underlying the greater evolutionary divergence among forewings and hindwings in Calopteryx damselflies. Therefore, we suggest that maneuverability, in response to local factors such as reproductive behavior (mate guarding) and/or climate (strong winds), may influence hindwing shape, while the reduction of the long-distance flight capacity, resulting from geographic isolation in Easter Island, may define smaller-sized forewings.
Another important finding was the difference in abdomen and thorax length between females and males among all locations. Males of all locations showed a longer thorax, with the exception of males of Easter Island, which showed a longer abdomen than females. This also may be a response to an improved maneuverability in mate guarding and/or to a reduced use of flight musculature. In effect, the thorax houses the musculature that powers flight, and relative thorax size can strongly affect flight performance (Schilder and Marden 2004; Marinov and McHugh 2010). Thus, a longer thorax would confer better flight performance in males of all localities except in Easter Island; in addition, the loss of migratory behavior in island populations may facilitate other life history traits, e.g., reproduction, as was reported by Langerotto et al. (2000) and Denno et al. (2001) for insular ecosystems. Langerotto et al. (2000) observed a trade-off between flight capability and reproduction in males of the salt-marsh-inhabiting planthopper Prokelisia dolus, with flightless males mating more successfully and producing more offspring than macropterous males. Similarly, Denno et al. (2001) observed that flightless females of the delphacid planthopper Toya venilia showed higher fecundity, reproduced at an earlier age and produced larger progeny than their flight-capable counterparts. In this way, a larger abdomen in males from Easter Island may facilitate the adoption of the tandem position and increase the success and frequency of copulation. In effect, among dragonflies, larger males have shown a higher mating probability and greater lifetime mating success than smaller males (Tsubaki and Ono 1987; Michiels and Dhondt 1991).
According to the results obtained in this research, habitat isolation would be an important factor leading to genetic and morphological differences in individuals of P. flavescens in Easter Island. As noted by Corbet (1962), in isolated, permanent habitats, dragonflies show specializations that prevent or reduce dispersal; for this reason, islands are extreme examples of situations where dispersal power is reduced by natural selection. Our results suggested that selection and random evolutionary processes, including the founder effect and genetic drift, drive the changes in genetic patterns and wing shape observed in P. flavescens of Easter Island. Overall, we can hypothesize that there is a trade-off between the great capacity for migration and the establishment in an isolated geographical location. In this context, the individuals with lower dispersal capacity could have remained on the island, thus founding an isolated population.
Remote islands are ideal models in which to study microevolutionary processes and provide many opportunities for regional and comparative studies of general ecological and evolutionary biology. Finally, the results exposed in this research provide sufficient reasons for promoting the conservation and preservation of the insular population of P. flavescens. However, further studies are necessary to elucidate the mystery around the arrival of the wandering glider on Easter Island.
Data archiving
Sequences have been archived in Genbank and can be found under the accession numbers KY200583-KY200609 and KY934249-KY934261.
Electronic supplementary material
Acknowledgements
The authors thank doctoral fellowship Nº21130053 (CONICYT), Millennium Nucleus for Ecology and Sustainable Management of Oceanic Islands (ESMOI), Basal Grant PFB 023, ICM P05-002, and CONICYT PIA Apoyo CCTE AFB170008. Thanks to CONICYT–FONDEQUIP EQM150077. The authors thank the staff of the Corporación Nacional Forestal (CONAF) in Easter Island: E Tuki, H Pate, P Hito, G Arévalo, and P Lazo. We thank Ch. Anderson for providing samples from Maldives Islands, D. Ward of Landcare Research, New Zealand for support provided in collections from the South Pacific, P. Duchesne for advising on analysis, N. Rojas-Hernandez for lab assistance, and T. Walter for edition.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
I. E. Alvial, Email: ingrid.alvial@gmail.com
D. Véliz, Phone: +56-229789882, Email: dveliz@uchile.cl
Electronic supplementary material
The online version of this article (10.1038/s41437-018-0165-z) contains supplementary material, which is available to authorized users.
References
- Adams DC, Otarola C. Geomorph: A R package for the collection and analysis of geometric morphometric shape data. Methods Ecol Evol. 2013;4:393–399. doi: 10.1111/2041-210X.12035. [DOI] [Google Scholar]
- Aljanabi S, Martinez I. Universal and rapid salt-extraction of high quality genomic DNA for PCR based techniques. Nucleic Acids Res. 1997;25:4692–4693. doi: 10.1093/nar/25.22.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvial IE, Veliz D, Vargas H, Esquivel C, Vila I. Lack of genetic structure in Pantala flavescens among Central and South American localities (Odonata: Libellulidae) Odonatologica. 2017;46:67–82. [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. GENETIX 4.05, Logiciel sous Windows pour la Genetique des Populations. France: Laboratoire Genome, populations, interactions, CNRS UMR 5000, Université de Montpellier II, Montpellier; 2000. [Google Scholar]
- Belkhir K, Castric V, Bonhomme F. IDENTIX, a software to test for relatedness in a population using permutation methods. Mol Ecol Notes. 2001;2:611–614. doi: 10.1046/j.1471-8286.2002.00273.x. [DOI] [Google Scholar]
- Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29:1165–1188. doi: 10.1214/aos/1013699998. [DOI] [Google Scholar]
- Campos L, Peña E. Los insectos de Isla de Pascua. Rev Chil Entomol. 1973;7:217–229. [Google Scholar]
- Cao L, Fu X, Wu K. Development of 10 microsatellite markers from Pantala flavescens and their applicability in studying genetic diversity. Mol Biol Rep. 2015;42:1275–1279. doi: 10.1007/s11033-015-3868-8. [DOI] [PubMed] [Google Scholar]
- Cariou M, Durst L, Charlat S. The global impact of Wolbachia on mitochondrial diversity and evolution. J Evol Biol. 2017;30:2204–2210. doi: 10.1111/jeb.13186. [DOI] [PubMed] [Google Scholar]
- Christudhas A, Mathai M. Genetic variation of a migratory dragonfly characterized with random DNA marker. J Entomol Zool Stud. 2014;2:182–184. [Google Scholar]
- Corbet P. A biology of dragonflies. London: HF & G Witherby Ltd; 1962. p. 247. [Google Scholar]
- Corbet PS. Dragonflies: behaviour and ecology of Odonata. Colchester: Harley; 1999. [Google Scholar]
- Daly HV. Insect Morphometrics. Annu Rev Entomol. 1985;30:415–438. doi: 10.1146/annurev.en.30.010185.002215. [DOI] [Google Scholar]
- Darwin C. On the origin of species. London: Murray; 1859. [Google Scholar]
- Dellicour S, Gerard M, Prunier J, Dewulf A, Kuhlmann M, Michez D. Distribution and predictors of wing shape and size variability in three sister species of solitary bees. PLoS One. 2017;12:e0173109. doi: 10.1371/journal.pone.0173109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denno R, Hawthorne D, Thorne B, Gratton C. Reduced flight capability in British Virgin island populations of a wing dimorphic insect: the role of habitat isolation, persistence and structure. Ecol Entomol. 2001;26:25–36. doi: 10.1046/j.1365-2311.2001.00293.x. [DOI] [Google Scholar]
- Dieckmann U, O´Hara B, Weisser W. The evolutionary ecology of dispersal. Trends Ecol Evol. 1999;14:88–90. doi: 10.1016/S0169-5347(98)01571-7. [DOI] [Google Scholar]
- Dijkstra K, Clausnitzer V. The Dragonflies and Damselflies of Eastern Africa: handbook for all Odonata from Sudan to Zimbabwe. Stud Afrotropical Zool. 2014;298:1–260. [Google Scholar]
- Duchesne P, Turgeon J. FLOCK attractors provide reliable solutions to the “number of populations” problem. J Hered. 2012;103:734–743. doi: 10.1093/jhered/ess038. [DOI] [PubMed] [Google Scholar]
- Dumont H, Verschuren L. Atypical ecology of Pantala flavescens (Fabr.) on Easter Island (Anisoptera: Libellulidae) Odonalologica. 1993;20:45–51. [Google Scholar]
- Eare D, Von Holdt B. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 2012;4:359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- Estoup A, Solignac M, Cornuet J, Goudet J, Scholls A. Genetic differentiation of continental and island populations of Bombus terrestris (Hymenoptera: Apidae) in Europe. Mol Ecol. 1996;5:19–31. doi: 10.1111/j.1365-294X.1996.tb00288.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HL. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Filatov DA. ProSeq: a software for preparation and evolutionary analysis of DNA sequence data sets. Mol Ecol Notes. 2002;2:621–624. doi: 10.1046/j.1471-8286.2002.00313.x. [DOI] [Google Scholar]
- Frankham R. Do island populations have less genetic variation than mainland populations? Heredity. 1997;78:311–327. doi: 10.1038/hdy.1997.46. [DOI] [PubMed] [Google Scholar]
- Freeland J, May M, Lodge R, Conrad K. Genetic diversity and widespread haplotypes in a migratory dragonfly, the common green darner Anax junius. Ecol Entomol. 2003;28:413–421. doi: 10.1046/j.1365-2311.2003.00521.x. [DOI] [Google Scholar]
- Furlan E, Stoklosa J, Griffiths J, Gust N, Ellis R, Huggins R, Weeks A. Small population size and extremely low levels of genetic diversity in island populations of the platypus, Ornithorhyn chusanatinus. Ecol Evol. 2012;2:844–857. doi: 10.1002/ece3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall CR. Procrustes methods in the statistical analysis of shape. J R Stat Soc Ser A Stat Soc. 1991;53:285–339. [Google Scholar]
- Goudet J. FSTAT (version 1.2) a computer program to calculate F-statistics. J Hered. 1995;86:485–486. doi: 10.1093/oxfordjournals.jhered.a111627. [DOI] [Google Scholar]
- Hankin MA. The soaring flight of dragonflies. Proc Camb Philos Soc. 1921;20:460–465. [Google Scholar]
- Hughes J, Schmidt D, Finn D. Genes in stream: using DNA to understand the movement of freshwater fauna and their riverine habitat. BioScience. 2009;59:573–583. doi: 10.1525/bio.2009.59.7.8. [DOI] [Google Scholar]
- Hurst G, Jiggins F. Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: the effects of inherited symbionts. Proc Biol Sci. 2005;272:1525–1534. doi: 10.1098/rspb.2005.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Zhu J, Chen M, Yang Q, Du X, Chen S, Zhang L, Yu Y. Wolbachia infection status and genetic structure in natural populations of Polytremis nascens (Lepidoptera: Hesperiidae) Infect Genet Evol. 2014;29:202–211. doi: 10.1016/j.meegid.2014.07.026. [DOI] [PubMed] [Google Scholar]
- Johansson F, Soderquist M, Bokma F. Insect wing shape evolution: independent effects of migratory and mate guarding flight on dragonfly wings. Biol J Linn Soc. 2009;97:362–372. doi: 10.1111/j.1095-8312.2009.01211.x. [DOI] [Google Scholar]
- Kopelman N, Mayzel J, Jakobsson M, Rosenberg N, Mayrose I. CLUMPAK: a program or identifying clustering modes and packaging population structure inference across K. Mol Ecol Resour. 2015;15:1179–1191. doi: 10.1111/1755-0998.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langerotto GA, Denno RF, Ott JR. A trade-off between flight capability and reproduction in males of a wing-dimorphic insect. Ecology. 2000;81:865–875. doi: 10.1890/0012-9658(2000)081[0865:ATOBFC]2.0.CO;2. [DOI] [Google Scholar]
- Leigh JW, Bryant D. popart: full‐feature software for haplotype network construction. Methods Ecol Evol. 2015;6:1110–1116. doi: 10.1111/2041-210X.12410. [DOI] [Google Scholar]
- Lester SE, Ruttenberg BI, Gaines SD, Kinlan BP. The relationship between dispersal ability and geographic range size. Ecol Lett. 2007;10:745–758. doi: 10.1111/j.1461-0248.2007.01070.x. [DOI] [PubMed] [Google Scholar]
- Lomolino MV. Body size evolution in insular vertebrates: generality of the island rule. J Biogeogr. 2005;32:1683–1699. doi: 10.1111/j.1365-2699.2005.01314.x. [DOI] [Google Scholar]
- Losos JB, Ricklefs RE. Adaptation and diversification on islands. Nature. 2009;457:830–836. doi: 10.1038/nature07893. [DOI] [PubMed] [Google Scholar]
- Low V, Rashid Y, Yusoff A, Siow W, Prakash B, Tan T, Noorhidayah M, Chen C, Azirun M. Pleistocene demographic expansion and high gene flow in the Globe Skimmer dragonfly Pantala flavescens Fabricius (Odonata: Libellulidae) in Peninsular Malaysia. Zool Anz. 2017;266:23–27. doi: 10.1016/j.jcz.2016.10.002. [DOI] [Google Scholar]
- Lyons J, Pierce A, Barribeau S, Sternberg E, Mongue A, Roode J. Lack of genetic differentiation between monarch butterflies with divergent migration destinations. Mol Ecol. 2012;21:3433–3444. doi: 10.1111/j.1365-294X.2012.05613.x. [DOI] [PubMed] [Google Scholar]
- Marinov M, McHugh P. Comparative study of the Chatham Islands Odonata: Morphological variability, behaviour and demography of the endemic Xanthocnemis tuanuii Rowe, 1987. Int Dragonfly Fund-Report. 2010;30:1–44. [Google Scholar]
- May M. A critical overview of progress in studies of migration of dragonflies (Odonata: Anisoptera), with emphasis on North America. J Insect Conserv. 2012;17:1–15. doi: 10.1007/s10841-012-9540-x. [DOI] [Google Scholar]
- McCauley SJ. Relationship between morphology, dispersal and habitat distribution in three species of Libellula (Odonata: Anisoptera) Aquat Insects. 2013;34:195–201. doi: 10.1080/01650424.2013.800557. [DOI] [Google Scholar]
- Michiels N, Dhondt A. Sources of variation in male mating success and female oviposition rate in a nonterritorial dragonfly. Behav Ecol Sociobiol. 1991;29:17–25. doi: 10.1007/BF00164290. [DOI] [Google Scholar]
- Mieth A, Bork H. Humans, climate or introduced rats-which is to blame for the woodland destruction on prehistoric Rapa Nui (Easter Island) J Archaeol Sci. 2010;37:417–426. doi: 10.1016/j.jas.2009.10.006. [DOI] [Google Scholar]
- Monaghan T, Spaak P, Robinson C, Ward J. Genetic differentiation of Baetis alpines Pichet (Ephemeroptera: Baetidae) in fragmented alpine streams. Heredity. 2001;86:395–403. doi: 10.1046/j.1365-2540.2001.00843.x. [DOI] [PubMed] [Google Scholar]
- Moore NW. Behaviour of imaginal Pantala flavescens (Fabr.) on Easter Island (Anisoptera: Libellulidae) Odonatologica. 1992;22:71–76. [Google Scholar]
- Morin P, Martien K, Archer F, Cipriano F, Steel D, Jackson J, Taylor B. Applied conservation genetics and the need for quality control and reporting of genetic data used in fisheries and wildlife management. J Hered. 2010;101:1–10. doi: 10.1093/jhered/esp107. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Blanchet F, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin P, O'Hara R, Simpson G, Solymos P, Stevens M, Szoecs E, Wagner H (2018) Vegan: Community Ecology Package. https://CRAN.R-project.org/package=vegan. Accessed September 4, 2018
- Outomuro D, Bokma F, Johansson F. Hind wing shape evolves faster than front wing shape in Calopteryx damselflies. Evol Biol. 2012;39:116–125. doi: 10.1007/s11692-011-9145-4. [DOI] [Google Scholar]
- Paulay G, Meyer C. Diversification in the Tropical pacific: comparisons between marine and terrestrial systems and the importance of founder speciation. Integr Comp Biol. 2002;42:922–934. doi: 10.1093/icb/42.5.922. [DOI] [PubMed] [Google Scholar]
- Pfeiler E, Markow TA. Population connectivity and genetic diversity in long-distance migrating insects: divergent patterns in representative butterflies and dragonflies. Biol J Linnean Soc. 2017;122:479–486. doi: 10.1093/biolinnean/blx074. [DOI] [Google Scholar]
- Ponds O, Petit RJ. Measuring and testing genetic differentiation with ordered v/s unordered alleles. Genetics. 1996;144:1237–1245. doi: 10.1093/genetics/144.3.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;115:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queller DC, Goodnight KF. Estimating relatedness using genetic markers. Evolution. 1989;43:258–275. doi: 10.1111/j.1558-5646.1989.tb04226.x. [DOI] [PubMed] [Google Scholar]
- R Core Team (2017) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. Accessed September 4, 2018
- Roderick R, Gillespie G. Speciation and phylogeography of Hawaiian terrestrial arthropods. Mol Ecol. 1998;7:519–531. doi: 10.1046/j.1365-294x.1998.00309.x. [DOI] [PubMed] [Google Scholar]
- Roff DA. The evolution of flightlessness: Is history important? Evol Ecol. 1994;8:639–657. doi: 10.1007/BF01237847. [DOI] [Google Scholar]
- Roff DA. The evolution of flightlessness in insects. Ecol Monogr. 1990;60:389–421. doi: 10.2307/1943013. [DOI] [Google Scholar]
- Rohlf FJ, Slice DE. Extensions of the Procrustes method for the optimal superimposition of landmarks. Syst Zool. 1990;39:40–59. doi: 10.2307/2992207. [DOI] [Google Scholar]
- Rozas J, Sanchez-Del Barrio JC, Messeguer X, Rozas R. DnaSP DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Samways M, Osborn R. Divergence in a transoceanic circumtropical dragonfly on a remote island. J Biogeogr. 1998;25:935–946. doi: 10.1046/j.1365-2699.1998.00245.x. [DOI] [Google Scholar]
- Schilder RJ, Marden JH. A hierarchical analysis of the scaling force and power production by dragonfly motors. J Exp Biol. 2004;207:767–776. doi: 10.1242/jeb.00817. [DOI] [PubMed] [Google Scholar]
- Schmitz P, Cibois A, Landry B. Molecular phylogeny and dating of an insular endemic moth radiation inferred from mitochondrial and nuclear genes: the genus Galagete (Lepidoptera: Austostichidae) of the Galápagos Islands. Mol Phylogenetics Evol. 2007;45:180–192. doi: 10.1016/j.ympev.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Segers H, Dumont H. Zoogeography of Pacific Ocean islands: a comparison of the rotifer faunas of Easter Island and the Galapagos archipelago. Hydrobiologia. 1993;255:475–480. doi: 10.1007/BF00025876. [DOI] [Google Scholar]
- Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am. 1994;87:651–701. doi: 10.1093/aesa/87.6.651. [DOI] [Google Scholar]
- Suarez-Tovar CM, Sarmiento CE. Beyond the wing planform: morphological differentiation between migratory and nonmigratory dragonfly species. J Evol Biol. 2016;29:690–703. doi: 10.1111/jeb.12830. [DOI] [PubMed] [Google Scholar]
- Tesson S, Edelaar P. Dispersal in a changing world: opportunities, insights and challenges. Mov Ecol. 2013;14:1–10. doi: 10.1186/2051-3933-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thipaksorn A, Jamnongluk W, Kittayapong P. Molecular evidence of Wolbachia infection in natural populations of tropical odonates. Curr Microbiol. 2003;47:314–318. doi: 10.1007/s00284-002-4010-4. [DOI] [PubMed] [Google Scholar]
- Troast D, Suhling F, Jinguji H, Sahlen G, Ware J. A global population genetic study of Pantala flavescens. PLoS One. 2016;11:e0148949. doi: 10.1371/journal.pone.0148949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubaki Y, Ono T. Effects of age and body size on the male territorial system of the dragonfly, Nannophya pygmaea rambur (Odonata: Libellulidae) Anim Behav. 1987;35:518–525. doi: 10.1016/S0003-3472(87)80276-2. [DOI] [Google Scholar]
- Vähä J, Erkinaro J, Niemelä E, Primmer C. Life-history and habitat features influence the within-river genetic structure of Atlantic salmon. Mol Ecol. 2007;16:2638–2654. doi: 10.1111/j.1365-294X.2007.03329.x. [DOI] [PubMed] [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DP, Shipley P. Microchecker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes. 2004;4:535–538. doi: 10.1111/j.1471-8286.2004.00684.x. [DOI] [Google Scholar]
- Venables W. N., Ripley B. D. Modern Applied Statistics with S. New York, NY: Springer New York; 2002. [Google Scholar]
- Whittaker R, Fernandez-Palacios J (2007) Island biogeography: ecology, evolution and conservation. Second Edition. Oxford University Press, UK
- Wootton RJ. The functional morphology of the wings of Odonata. Adv Odonatol. 1991;5:153–169. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences have been archived in Genbank and can be found under the accession numbers KY200583-KY200609 and KY934249-KY934261.