Tight junctions (TJs) are a key component of barriers formed by polarized epithelial cells. Otani et al. found that the scaffolding proteins ZO-1/ZO-2 are essential for TJ formation and epithelial polarity. Claudins regulate the TJ strand and electrolyte permeability barrier, while JAM-A regulates membrane apposition and the macromolecule permeability barrier. Claudins and JAM-A act redundantly in regulating epithelial polarity.
Abstract
Tight junctions (TJs) establish the epithelial barrier and are thought to form a membrane fence to regulate epithelial polarity, although the roles of TJs in epithelial polarity remain controversial. Claudins constitute TJ strands in conjunction with the cytoplasmic scaffolds ZO-1 and ZO-2 and play pivotal roles in epithelial barrier formation. However, how claudins and other TJ membrane proteins cooperate to organize TJs remains unclear. Here, we systematically knocked out TJ components by genome editing and show that while ZO-1/ZO-2–deficient cells lacked TJ structures and epithelial barriers, claudin-deficient cells lacked TJ strands and an electrolyte permeability barrier but formed membrane appositions and a macromolecule permeability barrier. Moreover, epithelial polarity was disorganized in ZO-1/ZO-2–deficient cells, but not in claudin-deficient cells. Simultaneous deletion of claudins and a TJ membrane protein JAM-A resulted in a loss of membrane appositions and a macromolecule permeability barrier and in sporadic epithelial polarity defects. These results demonstrate that claudins and JAM-A coordinately regulate TJ formation and epithelial polarity.
Introduction
Epithelia act as barriers to segregate the external environment from the internal body. Epithelial cells are highly polarized, and the asymmetric distribution of their plasma membrane proteins is essential for epithelial transport (Cereijido et al., 1989). Tight junctions (TJs) are epithelial cell junctions that form at the most apical region of intercellular junctions (Farquhar and Palade, 1963). On ultrathin sections, TJs appear as a region where two plasma membranes are closely apposed to one another and contain membrane “kissing points” where adjacent plasma membranes appear to fuse with one another and completely seal the intercellular space (Farquhar and Palade, 1963). TJs are visualized as anastomosing linear fibrils (TJ strands) on freeze-fracture replica EM, and the strands correspond to the membrane kissing points observed on ultrathin sections (Staehelin, 1973).
TJs act as a permeability barrier to restrict free diffusion of solutes through the intercellular space and play central roles in regulating paracellular permeability (“gate” function; Anderson and Van Itallie, 2009; Shen et al., 2011; Zihni et al., 2016). TJs are also thought to act as a membrane fence that prevents intramembrane diffusion of membrane proteins and lipids between the apical and basolateral cell surfaces, thereby maintaining epithelial polarity (“fence” function; De Camilli et al., 1974; Hoi Sang et al., 1979; Dragsten et al., 1981; van Meer and Simons, 1986). In addition, polarity signaling molecules including the Par-3–Par-6–atypical PKC (aPKC) complex localize at TJs (Izumi et al., 1998; Roh et al., 2002). However, recent studies have provided conflicting views on the roles of TJs in epithelial polarity (Umeda et al., 2006; Ikenouchi et al., 2012; Phua et al., 2014), and it remains unclear whether TJs are required for epithelial polarity.
Claudin family genes (27 members in mammals) encode the major integral membrane proteins that constitute TJ strands. These proteins have four transmembrane regions, a short N-terminal cytoplasmic region, and a long C-terminal cytoplasmic region (Furuse et al., 1998a; Günzel and Yu, 2013). When expressed in fibroblasts, claudins can induce cell–cell adhesion activity and reconstitute TJ strand structures (Furuse et al., 1998b). The C-terminal tails of claudins harbor a PDZ-binding motif and interact with the cytoplasmic scaffolding proteins ZO-1, ZO-2, and ZO-3 (Itoh et al., 1999a). ZO-1 and ZO-2 were shown to be essential for TJ strand assembly, because ZO-1 knockout (KO) and ZO-2 depletion by RNAi in the mouse mammary epithelial cell line EpH4 resulted in a loss of TJ strands (Umeda et al., 2006). In addition to claudins, other integral membrane proteins such as tetraspanning membrane protein occludin and immunoglobulin superfamily proteins including junctional adhesion molecules (JAMs) localize to TJs (Furuse et al., 1993; Martìn-Padura et al., 1998). Although it is well established that claudins play pivotal roles in TJ strand formation and regulation of paracellular permeability (Van Itallie and Anderson, 2006; Günzel and Yu, 2013), how claudins and other integral membrane proteins coordinately organize the TJ structure and function remains to be clarified.
Here we tested the roles of TJs in epithelial polarity by systematically knocking out TJ components and provide evidence that TJ is required for epithelial polarity. Furthermore, our results suggest that claudins and JAM-A have overlapping and distinct functions in organizing the TJ structure and function.
Results
ZO-1/ZO-2 regulates TJ assembly
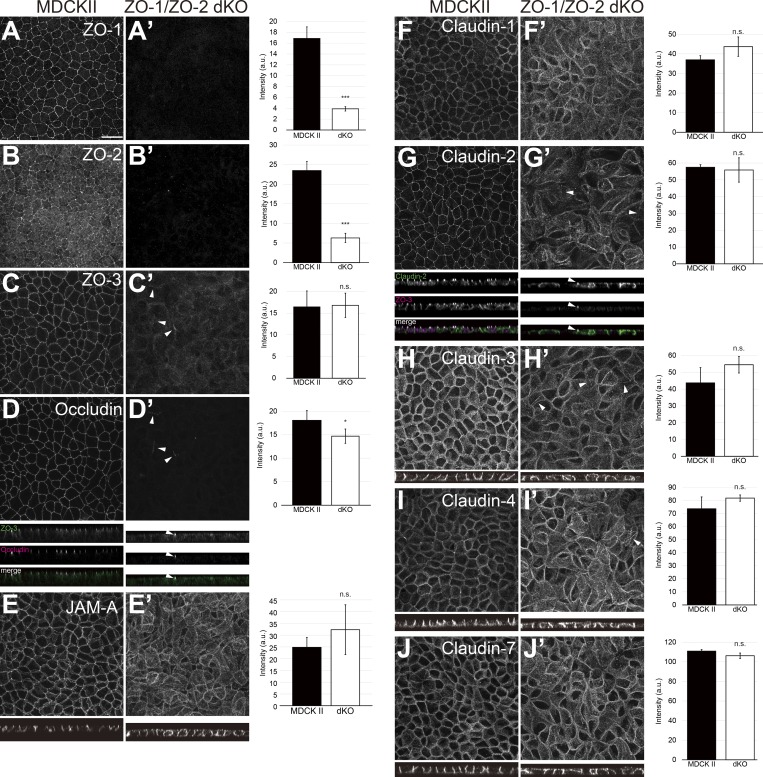
To clarify the roles of TJs in epithelial polarity, we generated ZO-1/ZO-2 double KO (dKO) cells. MDCK II cells derived from canine kidney epithelium were used as a model system, because TJs and epithelial polarity have been extensively investigated in this cell line (Cereijido et al., 1978, 1980). ZO-1/ZO-2 dKO cells were generated by sequential genome editing, and successful KO was confirmed by Sanger sequencing (Fig. S1 A). Three independent clones were isolated, and rescue cell lines expressing ZO-1–GFP were generated for all three cell lines (Fig. S3 H). ZO-1 or ZO-2 single KO cells did not show obvious epithelial barrier or polarity phenotypes except for an increase in the paracellular permeability of 4-kD fluorescent dextran in ZO-1 KO cells (Tokuda et al., 2014; data not shown). Western blotting demonstrated complete loss of ZO-1 and ZO-2, while ZO-3 was slightly reduced (Fig. S1 B). Expression of other TJ proteins was not grossly altered, although the protein level of afadin, an adherens junction (AJ) protein, was significantly reduced for unknown reasons (Fig. S1 B). Immunofluorescence analyses confirmed the loss of ZO-1 and ZO-2 (Fig. 1, A and B). TJ proteins including ZO-3, occludin, JAM-A, and claudins delocalized from the apical junctions in ZO-1/ZO-2 dKO cells, consistent with previous observations (Umeda et al., 2006; Fig. 1, C–J), and JAM-A and claudins diffusely localized along the lateral and apical plasma membrane (Fig. 1, E–J). Occasionally, occludin and claudins, but not JAM-A, were found to localize at apical cell junctions together with ZO-3 in ZO-1/ZO-2 dKO cells (arrowheads in Fig. 1, C′–J′). All phenotypes were rescued by expression of ZO-1–GFP (data not shown). These results demonstrate that ZO-1 and ZO-2 are required for the apical junction localization of TJ proteins.
Figure 1.
ZO-1/ZO-2 are required for apical junction localization of TJ proteins. (A–J) Immunofluorescence analyses of parental MDCK II cells (A–J) and ZO-1/ZO-2 dKO cells (A′–J′). (A and B) ZO-1 (A) and ZO-2 (B) expression was abolished in ZO-1/ZO-2 dKO cells. (C–J) TJ markers including ZO-3 (C), occludin (D), JAM-A (E), claudin-1 (F), claudin-2 (G), claudin-3 (H), claudin-4 (I), and claudin-7 (J) were not concentrated to the apical junctions in ZO-1/ZO-2 dKO cells, and JAM-A (E) and claudins (F–J) were diffusely localized along the lateral and apical plasma membrane. Occasional apical junction accumulation of occludin and claudins, but not JAM-A was observed (arrowheads), colocalizing with ZO-3 (z-sections in D and G). Graphs are quantitation of the fluorescence intensity and represent mean ± SD (n = 2–9). *, P < 0.05; ***, P < 0.0005, compared by t test. Scale bar: 20 µm. n.s., not significant.
Generation of claudin quintuple KO (quinKO) cells
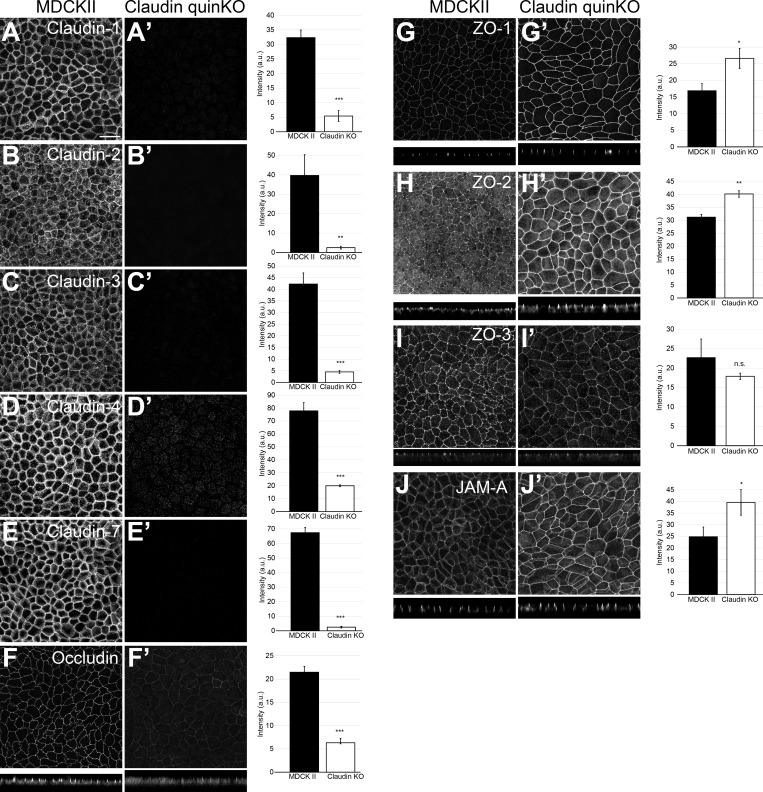
To clarify the roles of claudin-based TJ strands in epithelial polarity, we generated claudin quinKO cells by sequentially knocking out the five major claudins expressed in MDCK II cells (claudin-2, claudin-4, claudin-3, claudin-7, and claudin-1, in that order, based on the RNA sequencing dataset reported by Shukla et al., 2015). Two independent claudin quinKO cell clones were generated, and Sanger sequencing revealed that claudin quinKO cells harbored frameshift mutations for claudin-2, claudin-4, claudin-3, and claudin-7 (Fig. S2 A; Tokuda et al., 2017). Clone 1 contained a frameshift mutation for claudin-1, while clone 2 had a single amino acid insertion in the first transmembrane region of claudin-1 (Fig. S2 A), resulting in intracellular accumulation and degradation of claudin-1 as confirmed by Western blotting (Fig. S2 B) and immunofluorescence (data not shown). Data from clone 1 are shown as representative results throughout the article, although all phenotypes were confirmed in both clones. The Western blotting analyses revealed loss of claudins in claudin quinKO cells, while the expression of JAM-A and tricellulin were slightly increased (Fig. S2 B). The expression of other TJ proteins was not greatly altered (Fig. S2 B). Immunofluorescence analyses confirmed the loss of claudins (Fig. 2, A–E). However, expression of other claudins was not comprehensively examined, and residual expression of other claudins cannot be excluded. The phenotypes of claudin quinKO cells should thus be interpreted as loss of claudin-based TJ strands (see below) and not loss of claudin family genes. ZO-1 and ZO-2, but not ZO-3, were more concentrated at apical cell junctions in claudin quinKO cells (Fig. 2, G–I). Apical cell junction localization of occludin was reduced (Fig. 2 F), while JAM-A accumulation was increased, in claudin quinKO cells (Fig. 2 J).
Figure 2.
Localization of TJ proteins in claudin quinKO cells. (A–J) Immunofluorescence analyses of parental MDCK II cells (A–J) and claudin quinKO cells (A′–J′). (A–E) Claudin-1 (A), claudin-2 (B), claudin-3 (C), claudin-4 (D), and claudin-7 (E) expression was abolished in claudin quinKO cells. (F) Occludin localization to apical junctions was reduced. (G–I) ZO-1 (G) and ZO-2 (H) were more concentrated at apical junctions, while ZO-3 (I) localization was not altered in claudin quinKO cells. (J) JAM-A was more concentrated at apical junctions in claudin quinKO cells. Graphs are quantitation of the fluorescence intensity and represent mean ± SD (n = 3 each). *, P < 0.05; **, P < 0.005; ***, P < 0.0005, compared by t test. Scale bar: 20 µm. n.s., not significant.
Claudin quinKO cells lack TJ strands but retain membrane appositions
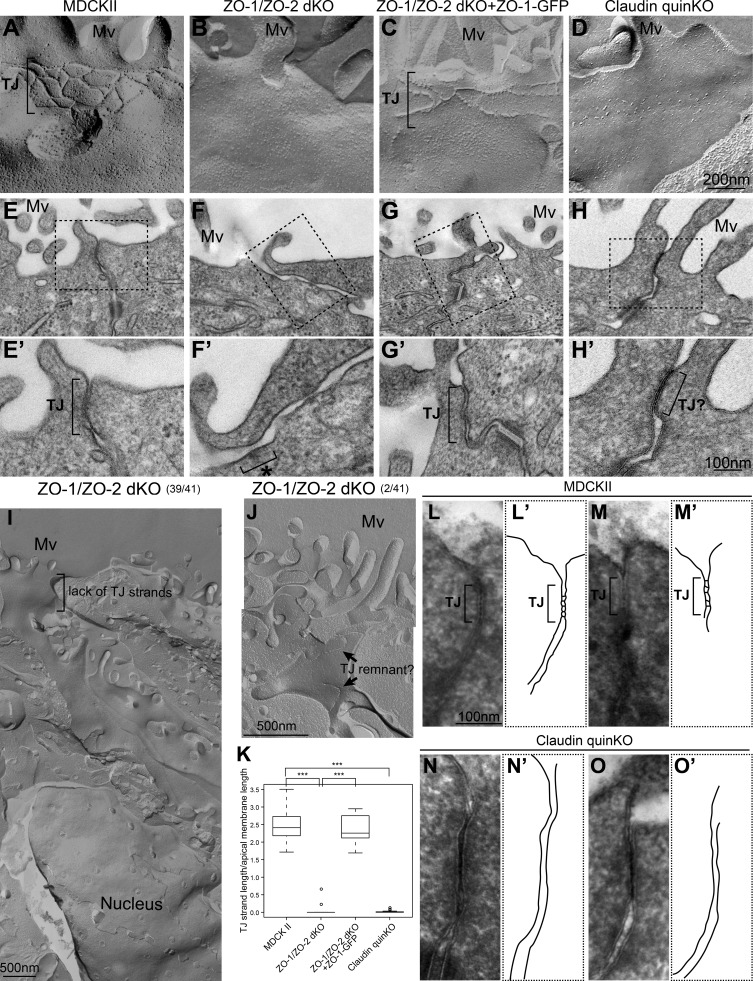
To understand the roles of claudins in TJ assembly, we examined TJ morphology by freeze-fracture replica EM. In parental MDCK II cells, typical TJ strands were observed beneath the apical microvilli (Fig. 3 A). However, ZO-1/ZO-2 dKO cells lacked TJ strands in most cells (39 of 41 junctions observed; Fig. 3, B and I), consistent with previous observations (Umeda et al., 2006). TJ strand-like structures were occasionally seen in ZO-1/ZO-2 dKO cells (2 of 41 junctions observed), but the strands were discontinuous and did not form the zonula occludens (Fig. 3 J). These structures may have corresponded to the occasional apical junction localization of TJ markers observed by immunofluorescence. TJ strand assembly was restored by expression of ZO-1–GFP in ZO-1/ZO-2 dKO cells (Fig. 3 C). Claudin quinKO cells also lacked TJ strands, although accumulation of intramembrane particles was sometimes observed beneath the apical microvilli (Figs. 3 D and S3 A). Quantitative analyses confirmed the loss of TJ strands in ZO-1/ZO-2 dKO cells and claudin quinKO cells (Fig. 3 K). These results suggest that ZO-1, ZO-2, and claudins are required for TJ strand formation.
Figure 3.
Claudins are required for TJ strand formation but not membrane appositions. (A–D) Freeze-fracture replica EM analyses. (A) TJ strands were observed beneath the apical microvilli in parental MDCK II cells. (B) TJ strands were not found in ZO-1/ZO-2 dKO cells. (C) Expression of ZO-1–GFP restored the TJ strands in ZO-1/ZO-2 dKO cells. (D) Claudin quinKO cells lacked TJ strands, but intramembrane particles were occasionally accumulated beneath the apical microvilli. (E–H) Transmission EM analyses of ultrathin sections. Black squares indicate the regions shown in high-magnification images. (E) TJs with membrane appositions were observed at the most apical cell junctions in MDCK II cells. (F) TJs were absent, and the intercellular space was widened in ZO-1/ZO-2 dKO cells. AJ-like structures associated with actin bundles were observed (asterisk). (G) Expression of ZO-1–GFP restored the formation of TJs in ZO-1/ZO-2 dKO cells. (H) TJ-like structures with membrane appositions were found in claudin quinKO cells. (I) Low-magnification view of a freeze-fracture replica from ZO-1/ZO-2 dKO cells. No TJ strands were found throughout the lateral plasma membrane. (J) An example of fragmented TJ strand-like structures in ZO-1/ZO-2 dKO cells. (K) Quantitation of TJ strand length normalized to the apical surface length of the corresponding fractured region. Graphs represent mean ± SD (n = 15 for MDCK II, n = 24 for ZO-1/ZO-2 dKO, n = 13 for ZO-1/ZO-2 dKO + ZO-1–GFP, n = 22 for claudin quinKO). ***, P < 0.0005, compared by t test. (L–O) TJ membrane kissing points were observed after ferrocyanide-reduced osmium/tannic acid/osmium postfixation. (L and M) TJ kissing points were observed in MDCK cells. (L′ and M′) Tracing of TJs. (N and O) Membranes were closely apposed to one another, but membrane kissing points were not observed in claudin quinKO cells. The most apical cell junctions with membrane appositions were observed. (N′ and O′) Tracing of TJ-like structures. Mv, microvilli. Scale bars: 200 nm (A–D); 100 nm (E–H); 500 nm (I and J); 100 nm (L–O).
Next, we examined the apical cell junction morphology in ultrathin sections by transmission EM. In parental MDCK II cells, close appositions of neighboring plasma membranes, a typical feature of TJs, were observed at the most apical cell junctions (Fig. 3 E). It was difficult to distinguish the TJs and AJs in ultrathin sections of MDCK II cells. In contrast, the intercellular space remained open, and no membrane appositions were observed at the most apical cell junctions in ZO-1/ZO-2 dKO cells, although some AJ-like structures were present (Fig. 3 F). Some ZO-1/ZO-2 dKO cells lacked the AJ-like structures (Fig. S3 B). Expression of ZO-1–GFP restored the TJ structures (Fig. 3 G). Intriguingly, in claudin quinKO cells, TJ-like membrane appositions were observed at the most apical cell junctions, despite the lack of TJ strands (Fig. 3 H). This prompted us to examine the morphology of TJs in more detail by improving the contrast with ferrocyanide-reduced osmium/tannic acid/osmium postfixation. This protocol allowed tracing of the extracellular spaces with an electron-opaque substance and increased the membrane contrast. TJ membrane kissing points were readily observed at the most apical cell junctions in parental MDCK II cells using this method (Fig. 3, L and M). In contrast, although the neighboring plasma membranes at the most apical cell junctions in claudin quinKO cells were closely apposed to one another at ∼6–7-nm distance, membrane kissing points were not observed (Fig. 3, N and O). These results demonstrate that claudins are required for TJ strand formation and suggest that the membrane appositions can form independently of claudin-based TJ strand assembly.
Claudin-based TJ strands are essential for electrolyte, but not macromolecule, permeability barrier formation
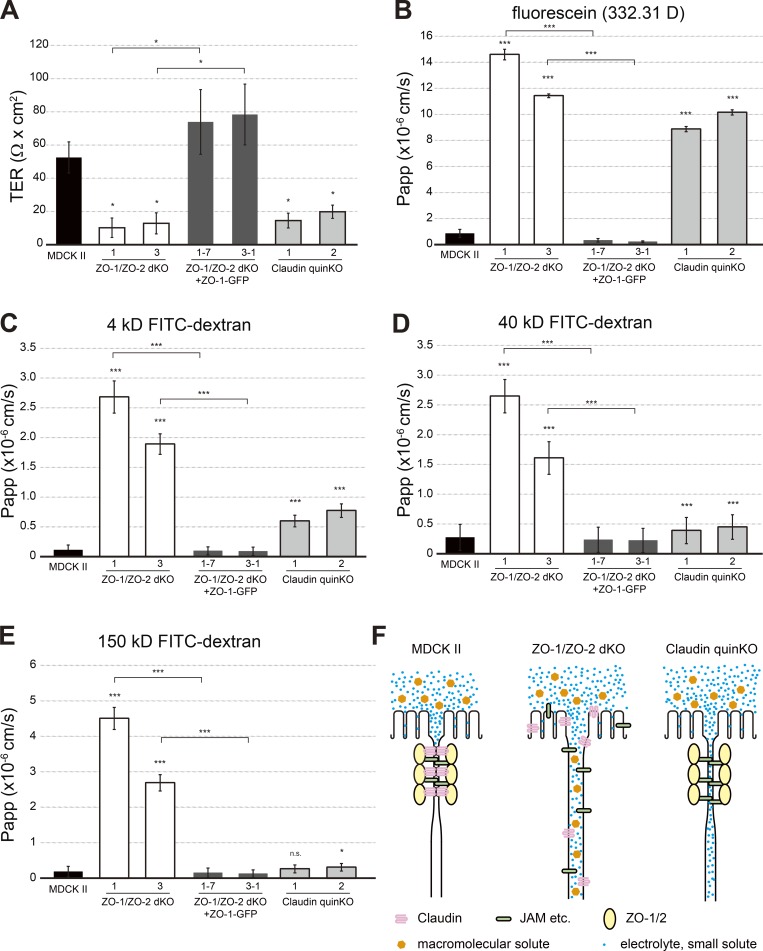
To understand the roles of claudins in epithelial barrier formation, transepithelial electric resistance (TER) was measured. TER reflects the transepithelial movement of ions and reflects the epithelial barrier function against electrolytes. The unit area resistance in parental MDCK II cells was 52.5 ± 9.4 Ω · cm2, while that in ZO-1/ZO-2 dKO cells was decreased to 10.2 ± 5.9 Ω · cm2 in clone 1 and 12.9 ± 6.3 Ω · cm2 in clone 3. Expression of ZO-1–GFP restored the TER value to 73.9 ± 19.5 Ω · cm2 in clone 1/rescue 7 and 78.4 ± 18.3 Ω · cm2 in clone 3/rescue 1. In claudin quinKO cells, the value was decreased to 14.5 ± 4.6 Ω · cm2 in clone 1 and 19.8 ± 4.0 Ω · cm2 in clone 2 (Fig. 4 A). These results suggest that ZO-1, ZO-2, and claudins are essential for establishment of the permeability barrier against electrolytes.
Figure 4.
Size-dependent permeability barrier defects in claudin quinKO cells. (A) TER measurements. Unit area resistance was markedly reduced in ZO-1/ZO-2 dKO and claudin quinKO cells. (B–E) Paracellular flux measurements. (B) Apical-to-basal permeability of fluorescein (332.31 D) was dramatically increased in ZO-1/ZO-2 dKO and claudin quinKO cells. (C) Apical-to-basal permeability of 4-kD FITC-dextran was markedly increased in ZO-1/ZO-2 dKO cells and significantly increased in claudin quinKO cells. (D) Apical-to-basal permeability of 40-kD FITC-dextran was dramatically increased in ZO-1/ZO-2 dKO cells, but only moderately increased in claudin quinKO cells. (E) Apical-to-basal permeability of 150-kD FITC-dextran was dramatically increased in ZO-1/ZO-2 dKO cells, but not in claudin quinKO cells. (F) Model illustrating the barrier defects in ZO-1/ZO-2 dKO cells and claudin quinKO cells. In MDCK II cells, TJs are formed, and paracellular passage of electrolytes and macromolecules is restricted. In ZO-1/ZO-2 dKO cells, the intercellular space is widened, and electrolytes and macromolecules can diffuse along the intercellular space. In claudin quinKO cells, membrane kissing points are lost, but neighboring cell membranes are closely apposed to one another, allowing paracellular diffusion of electrolytes, but not macromolecules. Graphs represent mean ± SD (n = 2 for A; n = 3 for B–E). *, P < 0.05; ***, P < 0.0005, compared by t test. n.s., not significant.
To further characterize the epithelial barrier properties of ZO-1/ZO-2 dKO and claudin quinKO cells, we measured the permeability of fluorescent tracer molecules. In ZO-1/ZO-2 dKO cells, a massive increase in the apical-to-basal flux of fluorescent tracers was observed regardless of the molecular size, indicating disruption of the epithelial barrier function (Fig. 4, B–E). Expression of ZO-1–GFP restored the epithelial barrier function (Fig. 4, B–E). In contrast, although a large increase in permeability of fluorescein (332.31 D) was observed in claudin quinKO cells, the diffusion of larger tracers was progressively restricted in a size-dependent manner (Fig. 4, B–E). As claudin quinKO cells lacked TJ strands, these results indicate that the macromolecule permeability barrier can form in the absence of TJ strands (Fig. 4 F). These results suggest that claudin-based TJ strands are required for the permeability barrier against electrolytes but are not essential for the permeability barrier against macromolecules.
ZO family proteins regulate epithelial polarity independently of TJ strand formation
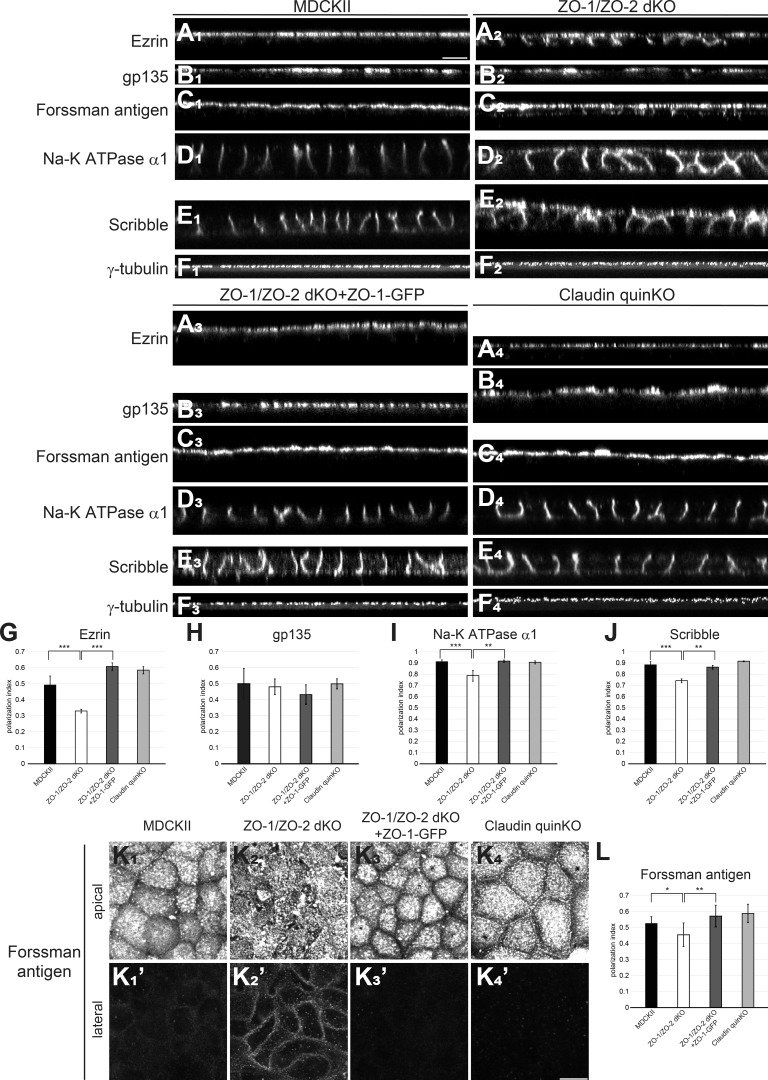
It remains controversial whether TJs are important for epithelial polarity (Umeda et al., 2006; Ikenouchi et al., 2012; Phua et al., 2014). Thus, we examined the epithelial polarity phenotypes of ZO-1/ZO-2 dKO cells and claudin quinKO cells. Parental MDCK II cells were well polarized, with apical marker (ezrin, gp135, and Forssman antigen) localization restricted to the apical membrane (Fig. 5, A1–C1 and K1), and basolateral markers (Na-K ATPase α1 subunit, and Scribble) localized to the basolateral membrane (Fig. 5, D1 and E1), and centrosomes were aligned at the apical cytoplasm (Fig. 5 F1). Intriguingly, epithelial polarity was disorganized in ZO-1/ZO-2 dKO cells. Ezrin and Forssman antigen were localized to both the apical and basolateral membranes (Fig. 5, A2, C2, and K2), while Na-K ATPase α1 subunit and Scribble were detected on the apical membrane in addition to their basolateral localization (Fig. 5, D2 and E2). In contrast, gp135 localization was restricted to the apical plasma membrane (Fig. 5 B2), and centrosomes were aligned at the apical cytoplasm (Fig. 5 F2), suggesting that epithelial polarity is retained to some extent in ZO-1/ZO-2 dKO cells. Expression of ZO-1–GFP rescued the epithelial polarity phenotypes of ZO-1/ZO-2 dKO cells (Fig. 5, A3–F3 and K3). In contrast, no epithelial polarity defects were detected in claudin quinKO cells (Fig. 5, A4–F4), although Forssman antigen was detected along the apical cell junctions, in contrast to MDCK II cells (Fig. 5 K4). The polarity phenotypes were confirmed by quantitative analyses (Fig. 5, G–J and L). These results indicate that ZO-1/ZO-2 regulate epithelial polarity in MDCK II cells.
Figure 5.
ZO-1/ZO-2 regulates epithelial polarity. (A–D) Immunofluorescence analyses of polarity markers. (A1–F1) In MDCK II cells, ezrin (A1), gp135 (B1), and Forssman antigen (C1) were selectively localized to the apical membrane, while Na-K ATPase α1 subunit (D1) and Scribble (E1) were restricted to the basolateral membrane. γ-Tubulin staining (F1) shows that centrosomes are aligned in the apical cytoplasm. (A2–F2) In ZO-1/ZO-2 dKO cells, epithelial polarity was disorganized, and ezrin (A2), Forssman antigen (C2), Na-K ATPase α1 subunit (D2), and Scribble (E2) were detected on both the apical and basolateral membranes, while gp135 (B2) or γ-tubulin (F2) localization was not severely perturbed. (A3–F3) Expression of ZO-1–GFP rescued the epithelial polarity phenotypes of ZO-1/ZO-2 dKO cells. (A4–F4) No epithelial polarity defects were observed in claudin quinKO cells. (G–J) Quantitation of polarization index of ezrin (G), gp135 (H), Na-K ATPase α1 subunit (I), and Scribble (J). Graphs represent mean ± SD (n = 3–9). (K) Forssman antigen localization in apical (K1–K4) and lateral (K′1–K′4) confocal sections. (L) Quantitation of polarization index of Forssman antigen. Graphs represent mean ± SD (n = 6–15). All cells were cultured on Transwell filters for 5–7 d. *, P < 0.05; **, P < 0.005; ***, P < 0.0005, compared by t test. Scale bars: 10 µm.
It was reported that ZO-1 KO and ZO-2 depletion by RNAi in mouse mammary epithelial EpH4 cells resulted in loss of TJs, but no epithelial polarity defects (Umeda et al., 2006; Ikenouchi et al., 2012). Therefore, we examined whether the observed epithelial polarity phenotypes in ZO-1/ZO-2 dKO MDCK II cells were caused by cell type differences or incomplete knockdown (KD) of ZO-2 in the previous studies. In parental EpH4 cells, ezrin was restricted to the apical plasma membrane, while Na-K ATPase α1 subunit was restricted to the basolateral membrane (Fig. S4 A). In agreement with the previous studies (Umeda et al., 2006; Ikenouchi et al., 2012), we did not detect epithelial polarity defects in ZO-1 KO/ZO-2 KD EpH4 cells (Fig. S4 A). However, in ZO-1/ZO-2 dKO EpH4 cells, ezrin mislocalized to lateral cell contacts, and Na-K ATPase α1 subunit was also detected on the apical membrane (Fig. S4 A). ZO-2 staining revealed strong reduction of ZO-2 in ZO-1 KO/ZO-2 KD EpH4 cells 2 d after plating (data not shown); however, weak but significant residual expression of ZO-2 was observed in ZO-1 KO/ZO-2 KD EpH4 cells 5 d after plating (Fig. S4 A). In contrast, ZO-2 was lost in ZO-1/ZO-2 dKO EpH4 cells (Fig. S4 A), suggesting that the residual expression of ZO-2 masked the epithelial polarity phenotype in ZO-1 KO/ZO-2 KD EpH4 cells.
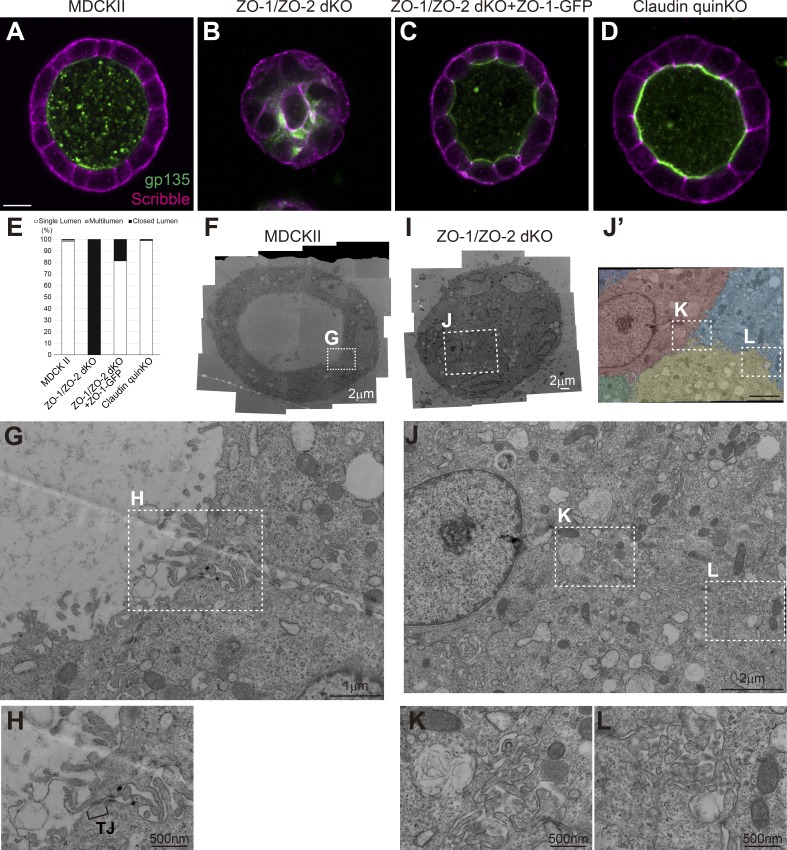
To further investigate the roles of TJs in epithelial polarity, we embedded the cells in collagen gel and examined their ability to form polarized cysts. Parental MDCK II cells formed polarized cysts in collagen gels (Fig. 6, A and E), and TJs were observed at the most apical cell junction (Fig. 6, F–H). In contrast, ZO-1/ZO-2 dKO cells failed to form polarized cysts, and lumen expansion was suppressed (Fig. 6, B and E). The apical marker gp135 and basolateral marker Scribble overlapped with one another, indicating epithelial polarity defects (Fig. 6 B). At the ultrastructural level, microvilli were found along cell–cell contacts, and microlumens that failed to expand were observed between cells (Fig. 6, I–L). Expression of ZO-1–GFP restored the ability to form polarized cysts (Fig. 6, C and E). In contrast to ZO-1/ZO-2 dKO cells, claudin quinKO cells formed polarized cysts (Fig. 6, D and E). Taken together, these results demonstrate that ZO-1 and ZO-2 are required for epithelial polarity. Given that claudins are required for TJ strand formation, these findings suggest that epithelial polarity can be established independently of TJ strand formation.
Figure 6.
ZO-1/ZO-2 regulates polarized cyst formation. (A–D) Localization of apical marker gp135 (green) and basolateral marker Scribble (magenta) in cells embedded in collagen I gels and cultured for 5–7 d. (A) MDCK II cells formed polarized cysts. (B) ZO-1/ZO-2 dKO cells failed to expand their lumens and did not form polarized cysts. Occasional colocalization of gp135 and Scribble was observed. (C) ZO-1/ZO-2 dKO cells were able to form polarized cysts upon expression of ZO-1–GFP. (D) Claudin quinKO cells formed polarized cysts. (E) Quantitation of cyst phenotypes (n = 343 for MDCK II, n = 536 for ZO-1/ZO-2 dKO, n = 311 for ZO-1/ZO-2 dKO + ZO-1–GFP, n = 752 for claudin quinKO). (F–H) Transmission EM observation of cysts formed by MDCK II cells. MDCK II cells formed a polarized epithelium in the collagen gels, and TJs were formed at the most apical region of the intercellular junctions (H). (I–L) Transmission EM observation of ZO-1/ZO-2 dKO cells embedded in collagen I gels. The lumens failed to expand in ZO-1/ZO-2 dKO cells, and microvilli and microlumens were observed between the cells (K and L). The cells were pseudocolored in J′. White squares indicate the regions shown in high-magnification images. Scale bars: 10 µm (A–D); 2 µm (F and I); 1 µm (G and J); 500 nm (H, K, and L).
ZO-1/ZO-2 regulate zonula adherens formation and actomyosin organization
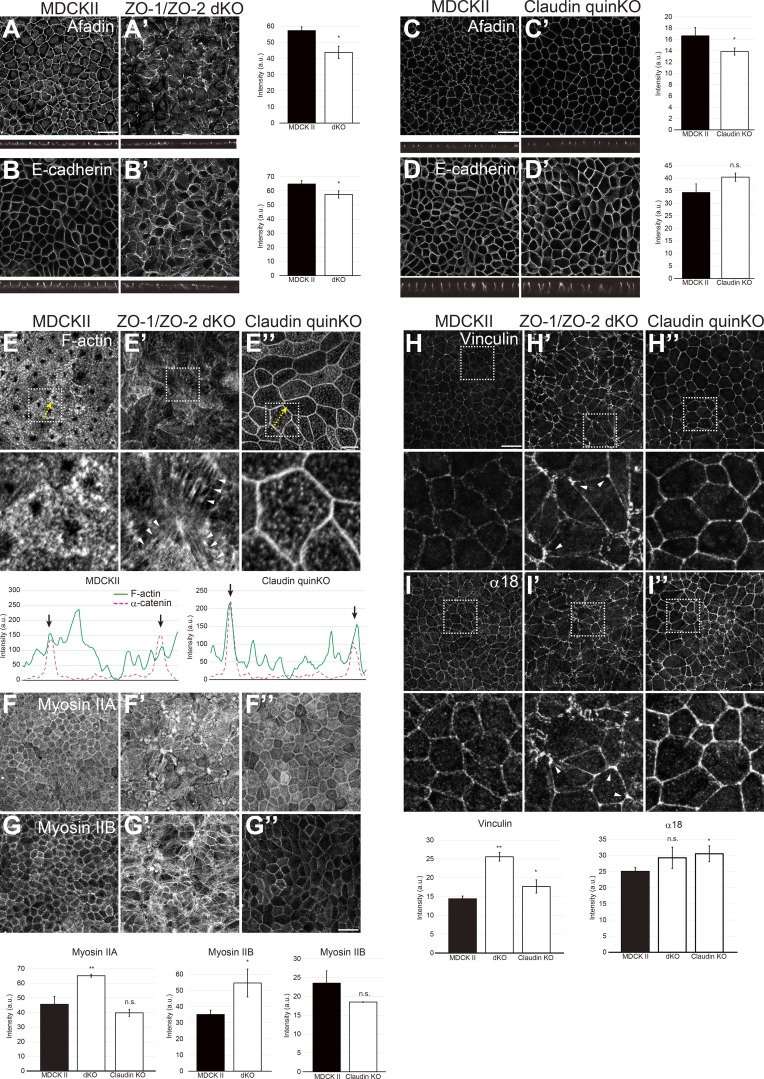
ZO-1 and ZO-2 can localize to AJs in nonepithelial cells (Itoh et al., 1991, 1993, 1999b) and have been implicated in zonula adherens assembly in epithelial cells (Ikenouchi et al., 2007; Yamazaki et al., 2008; Fanning et al., 2012; Itoh et al., 2012; Choi et al., 2016). We examined the localization of AJ markers, including afadin and E-cadherin, in ZO-1/ZO-2 dKO cells and found that although AJ markers were able to concentrate at the apical cell–cell contacts, the localization was discontinuous and fragmented (Fig. 7, A and B), consistent with the transmission EM observations (Figs. 3 F and S3 B). In contrast, in claudin quinKO cells, AJ marker localization was not notably altered (Fig. 7, C and D). These results suggest that ZO-1/ZO-2 are required for AJ assembly.
Figure 7.
ZO-1/ZO-2 is required for zonula adherens formation. (A and B) AJ markers afadin (A) and E-cadherin (B) were apically localized but fragmented in ZO-1/ZO-2 dKO cells. (C and D) Afadin (C) and E-cadherin (D) localizations were not altered, but the junctions appeared to be straighter in claudin quinKO cells. Graphs are quantitation of the fluorescence intensity and represent mean ± SD (n = 3–5). (E–G) F-actin (E), Myosin IIA (F), and IIB (G) organization in ZO-1/ZO-2 dKO cells and claudin quinKO cells. Apical confocal sections are shown in E–G. Actin bundles (E′) and myosin II foci (F′ and G′) were observed in the apical cytoplasm of ZO-1/ZO-2 dKO cells, while F-actin (E″) and Myosin II (F″ and G″) were more enriched at cell junctions in claudin quinKO cells (also see line scan). Line scans represent the fluorescent intensity along the yellow arrows, and black arrows indicate the position of cell junctions. Bar graphs are quantitation of the fluorescence intensity of Myosin II in the apical confocal sections and represent mean ± SD (n = 3–5). (H and I) Tension applied to AJ was monitored by vinculin (H) and α18 (I) localization. Vinculin and α18 localization were increased in ZO-1/ZO-2 dKO and claudin quinKO cells, suggesting that AJs are subjected to increased tension. In ZO-1/ZO-2 dKO cells, vinculin and α18 accumulated at vertices (arrowheads) indicating anisotropy in tension, while vinculin and α18 were increased in an isotropic manner in claudin quinKO cells. Apical confocal sections are shown in H. White squares indicate the regions shown in high-magnification images. Graphs are quantitation of the fluorescence intensity of apical vinculin and total α18 signals and represent mean ± SD (n = 3 each). *, P < 0.05; **, P < 0.005, compared by t test. Scale bar: 20 µm (A–D and F–I); 10 µm (E). n.s., not significant.
ZO-1 and ZO-2 were reported to regulate actomyosin (Otani et al., 2006; Fanning et al., 2012; Tokuda et al., 2014; Choi et al., 2016; Odenwald et al., 2018), and actomyosin plays important roles in AJ organization (Yonemura, 2011). Phalloidin staining revealed that in ZO-1/ZO-2 dKO cells, apical F-actin organization was disorganized, and actin bundles were oriented perpendicular to the cell junctions (Fig. 7 E). In contrast, in claudin quinKO cells, the circumferential actin bundle running parallel to the cell junctions was highly developed (Fig. 7 E″). In ZO-1/ZO-2 dKO cells, Myosin II accumulation was increased and formed foci in the apical cytoplasm localizing along the actin bundles (Fig. 7, F and G), while in claudin quinKO cells, Myosin II was more concentrated at the cell junctions (Fig. 7, F″ and G″).
Contractile actomyosin bundles can apply tension to AJs and induce unfolding of α-catenin that results in recruitment of vinculin (Yonemura et al., 2010). Vinculin localization to AJs was increased in ZO-1/ZO-2 dKO and claudin quinKO cells, suggesting that increased tension is applied to AJs (Fig. 7 H). Staining by α18 monoclonal antibody, which preferentially detects the open conformation of α-catenin (Yonemura et al., 2010), showed similar results (Fig. 7 I). Localization of vinculin and α18 was anisotropic in ZO-1/ZO-2 dKO cells, and increased accumulation at vertices was observed (arrowheads in Fig. 7, H′ and I′), in agreement with previous observations (Choi et al., 2016). In contrast, vinculin and α18 accumulation was isotropic along the cell junctions in claudin quinKO cells (Fig. 7, H″ and I″), resulting in a more linear morphology of the cell junctions (Fig. 10 H).
Figure 10.
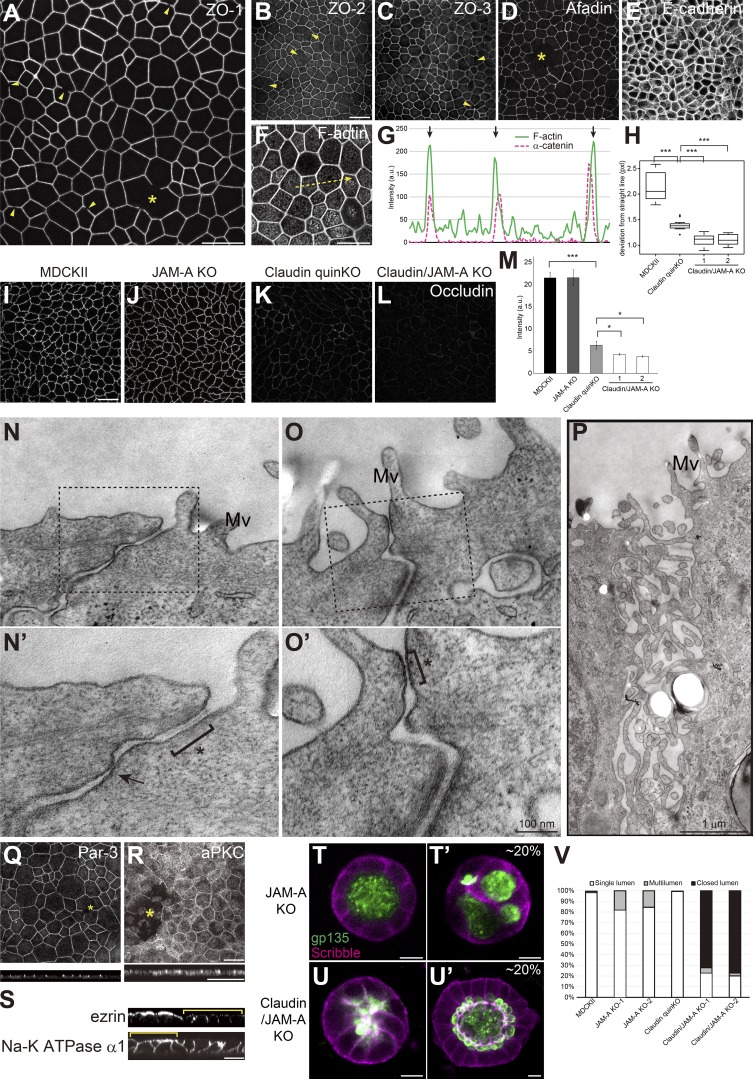
JAM-A regulates membrane appositions and epithelial polarity in claudin quinKO cells. (A–M) Immunofluorescence analyses of cell junction proteins in claudin/JAM-A KO cells. (A) Discontinuity in ZO-1 staining is observed (arrowheads), and large gaps of ZO-1 were occasionally found (asterisk). (B–E) Discontinuity (arrowheads) and gaps (asterisk) were also observed for ZO-2 (B), ZO-3 (C), and Afadin (D) in claudin/JAM-A KO cells. E-cadherin (E) staining was continuous, suggesting that cell–cell contact is maintained. (F and G) F-actin staining shows thickening of circumferential actin bundles. Apical confocal sections are shown. Line scans represent the fluorescent intensity along the yellow arrow in F, and black arrows in G represent the position of cell junctions. (H) Linearity of cell junctions were quantified by measuring the length of deviation from a straight line drawn between the vertices of the corresponding cell junction. Cell junctions of claudin quinKO cells were straighter than MDCK II cells, and extremely straight in claudin/JAM-A KO cells. Graphs represent mean ± SD (n = 15 each). (I–L) Occludin immunostaining was reduced in claudin quinKO cells, and further reduced in claudin/JAM-A KO cells. (M) Quantitation of the fluorescence intensity of occludin. Graphs represent mean ± SD (n = 3 each). Data for MDCK II and claudin quinKO are identical to Fig. 2 F. (N–P) Transmission EM analyses of ultrathin sections of claudin/JAM-A KO cells. Black squares indicate the regions shown in high-magnification images. Intercellular spaces were open in claudin/JAM-A KO cells, and AJ-like structures were found (asterisk). Focal membrane appositions were found in some cases (arrow). Occasionally, apical cell junctions were not formed, and microvilli-like structures were observed along the lateral membranes (P). Mv, microvilli. (Q and R) Large gaps (asterisks) were occasionally observed in Par-3 (Q) and aPKC (R) staining. (S) Sporadic epithelial polarity defects were observed in claudin/JAM-A KO cells (yellow brackets), where ezrin was mislocalized to the lateral cell junctions, and Na-K ATPase α1 subunit was found on both apical and basolateral membranes. (T and U) Localization of apical marker gp135 (green) and basolateral marker Scribble (magenta) in JAM-A KO cells (T) and claudin/JAM-A KO cells (U) embedded in collagen I gels and cultured for 5–7 d. Multilumen phenotypes were observed in ∼20% of the cysts formed by JAM-A KO cells (T′). Majority of the cysts formed by claudin/JAM-A KO cells showed closed lumen phenotypes (U), while ∼20% of them had single lumens, although the lumen morphology was abnormal (U′). (V) Quantitation of cyst phenotypes (n = 343 for MDCK II, n = 398 for JAM-A KO-1, n = 502 for JAM-A KO-2, n = 752 for claudin quinKO, n = 970 for claudin/JAM-A KO-1, n = 1186 for claudin/JAM-A KO-2). Data for MDCK II and claudin quinKO are identical to Fig. 7 E. *, P < 0.05; ***, P < 0.0005. See Figs. 1, 2, 3, 6, 7, and 9 for control images. Scale bars: 20 µm (A–L and Q–S); 100 nm (N and O); 1 µm (P); 10 µm (T and U).
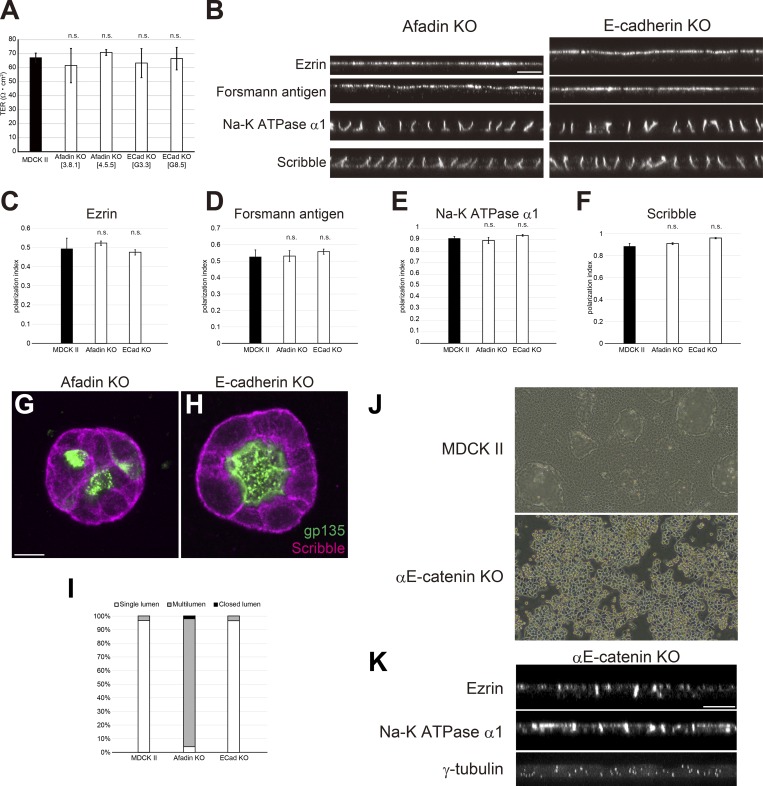
AJ and tension defects cannot fully explain the epithelial polarity defects in ZO-1/ZO-2 dKO cells
As AJs play important roles in epithelial polarity (McNeill et al., 1990; Harris and Peifer, 2004), we considered the possibility that ZO-1 and ZO-2 regulate epithelial polarity through regulation of AJ formation. To test this possibility, we knocked out afadin or E-cadherin (Fig. S4, C and D). Epithelial barrier function was not affected in either afadin KO cells or E-cadherin KO cells (Fig. 8 A). In 2D culture, epithelial polarity was not perturbed in afadin KO cells or E-cadherin KO cells (Fig. 8, B–F). In collagen gels, afadin KO cells exhibited multilumen phenotypes consistent with previous reports (Gao et al., 2017), while E-cadherin KO cells were able to form polarized cysts (Fig. 8, G–I). Although multilumen phenotypes are thought to represent epithelial polarity defects (Monteleon and D’Souza-Schorey, 2012), they were clearly distinct from the lumen expansion defects observed in ZO-1/ZO-2 dKO cells. As afadin or E-cadherin KO may not completely abrogate AJ function, we also generated αE-catenin KO cells (Fig. 8 J). αE-catenin KO cells showed epithelial polarity defects in which ezrin was mislocalized to lateral cell contacts, and Na-K ATPase α1 subunit was also detected on the apical plasma membrane (Fig. 8 K). However, in contrast to ZO-1/ZO-2 dKO cells, centrosome localization was randomized in αE-catenin KO cells (Fig. 8 K), indicating that the polarity defect is more severe. These results suggest that although AJ disorganization may contribute to the epithelial polarity phenotypes in ZO-1/ZO-2 dKO cells, the observed polarity defects in ZO-1/ZO-2 dKO cells and AJ perturbations are distinct from each other.
Figure 8.
The effect of AJs on epithelial polarity is distinct from that of ZO-1/ZO-2. (A) TER measurements showed that the epithelial barrier was not disorganized in afadin KO or E-cadherin KO cells. Graphs represent mean ± SD (n = 3). (B) No epithelial polarity defects were observed in afadin KO and E-cadherin KO cells cultured on Transwell filters. See Fig. 6 A for control images. (C–F) Quantitation of polarization index of ezrin (C), Forssman antigen (D), Na-K ATPase α1 subunit (E), and Scribble (F). Graphs represent mean ± SD (n = 2–9). Data for MDCK II cells are identical to Fig. 6. (G and H) Localization of apical marker gp135 (green) and basolateral marker Scribble (magenta) in afadin KO cells (G) and E-cadherin KO cells (H) embedded in collagen I gels and cultured for 5–7 d. (G) Afadin KO cells showed multilumen phenotypes. (H) E-cadherin KO cells were able to form polarized cysts. See Fig. 7 A for control images. (I) Quantitation of cyst phenotypes (n = 318 for MDCK II, n = 332 for Afadin KO, n = 457 for E-cadherin KO). (J) Phase-contrast images of MDCK II cells and αE-catenin KO cells. Strong cell–cell adhesion is lost in αE-catenin KO cells. (K) Epithelial polarity defects in αE-catenin KO cells. Ezrin and Na-K ATPase α1 subunit were detected on both the apical and basolateral membranes, and centrosome localization was randomized. Scale bars: 10 µm. n.s., not significant.
Myosin II activity was implicated in epithelial polarity (Zihni et al., 2017), and actomyosin was disorganized in ZO-1/ZO-2 dKO cells (Fig. 7, E–G), prompting us to examine the roles of myosin in the epithelial polarity defects of ZO-1/ZO-2 dKO cells. Treatment of ZO-1/ZO-2 dKO cells with myosin II inhibitor blebbistatin did not modify the epithelial polarity phenotypes, although the apical actomyosin bundles characteristic of ZO-1/ZO-2 dKO cells were disrupted (Fig. S4 B). These findings suggest that tension disorganization cannot account for the epithelial polarity phenotypes of ZO-1/ZO-2 dKO cells.
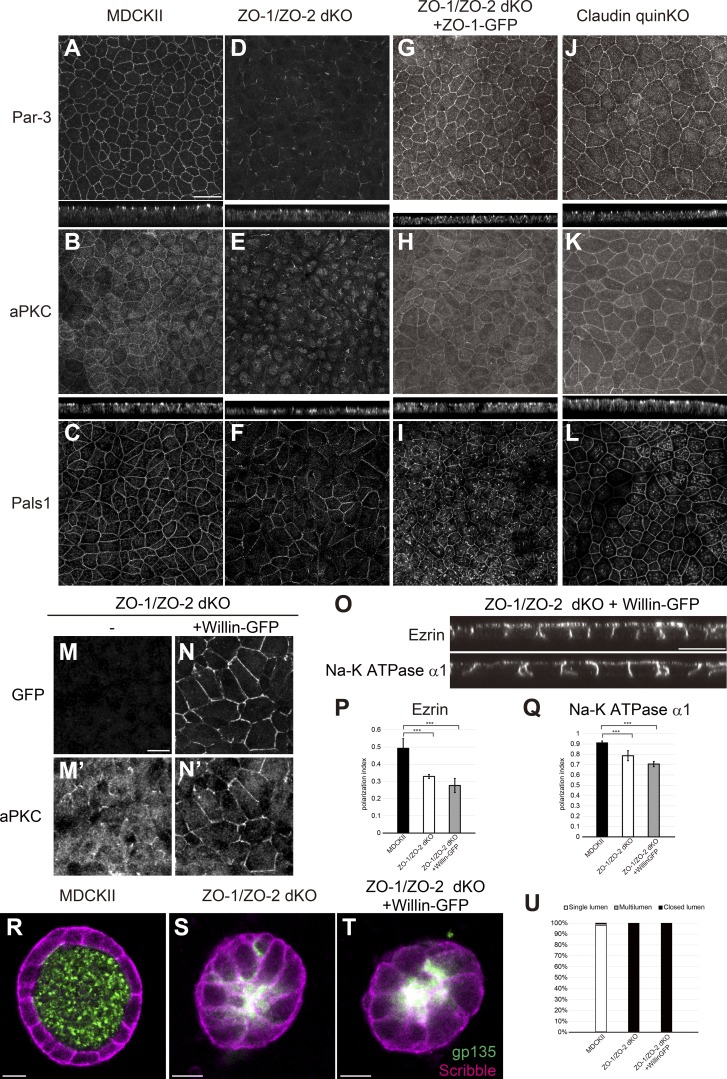
ZO family proteins regulate Par-3–aPKC localization
Polarity signaling molecules including the Par-3–Par-6–aPKC complex localize to TJs (Izumi et al., 1998; Roh et al., 2002). In parental MDCK II cells, Par-3, aPKC, and Pals1 were localized to the apical junctions (Fig. 9, A–C). In contrast, in ZO-1/ZO-2 dKO cells, Par-3 and aPKC localization was fragmented and diffuse, although occasional apical junction localization was observed (Fig. 9, D and E). Pals1 localization was also perturbed, but the defects were less pronounced (Fig. 9 F). Expression of ZO-1–GFP restored the apical junction localization of Par-3, aPKC, and Pals1 (Fig. 9, G–I). The apical junction localization of polarity proteins was not perturbed in claudin quinKO cells (Fig. 9, J–L). These findings suggest that the apical junction localization of polarity signaling molecules requires ZO-1 and ZO-2.
Figure 9.
Polarity signaling molecules are mislocalized in ZO-1/ZO-2 dKO cells. (A–C) Par-3 (A), aPKC (B), and Pals1 (C) were localized to apical junctions in MDCK II cells. (D–F) Par-3 (D) and aPKC (E) were diffuse and fragmented in ZO-1/ZO-2 dKO cells. Pals1 (F) localization was fragmented to a smaller extent in ZO-1/ZO-2 dKO cells. (G–I) Apical junction localization of Par-3 (G), aPKC (H), and Pals1 (I) was restored by expression of ZO-1–GFP in ZO-1/ZO-2 dKO cells. (J–L) Par-3 (J), aPKC (K), and Pals1 (L) were able to localize to apical junctions in claudin quinKO cells. Apical confocal sections are shown for Pals1, and the intracellular signals are nonspecific staining. (M and N) Willin-GFP overexpression promoted apical junction localization of aPKC in ZO-1/ZO-2 dKO cells. (M) aPKC localization was diffuse and fragmented in ZO-1/ZO-2 dKO cells. (N) Willin-GFP overexpression promoted apical junction localization of aPKC. GFP (M and N); aPKC (M′ and N′). (O) Willin-GFP overexpression did not restore the polarized localization of ezrin and Na-K ATPase α1 subunit in ZO-1/ZO-2 dKO cells. (P and Q) Quantitation of polarization index of ezrin (P) and Na-K ATPase α1 subunit (Q). Graphs represent mean ± SD (n = 10 each). ***, P < 0.0005, compared by t test. (R–T) Localization of apical marker gp135 (green) and basolateral marker Scribble (magenta) in MDCK II cells (R), ZO-1/ZO-2 dKO cells (S), and ZO-1/ZO-2 dKO + Willin-GFP cells (T) embedded in collagen I gels and cultured for 5–7 d. Willin-GFP expression did not rescue the lumen phenotype. (U) Quantitation of cyst phenotypes (n = 368 for MDCK II, n = 536 for ZO-1/ZO-2 dKO, n = 1,061 for ZO-1/ZO-2 dKO + Willin-GFP). Data for ZO-1/ZO-2 dKO are identical to Fig. 7 E. Scale bars: 20 µm (A–L and O); 10 µm (M, N, and R–T).
To examine whether mislocalization of Par-3 and aPKC was responsible for the epithelial polarity defects in ZO-1/ZO-2 dKO cells, we overexpressed Willin-GFP in these cells and isolated multiple stable transfectants. Willin protein is recruited to the apical junctional complex by Nectins, a group of cell adhesion molecules localized at AJs, and regulates the apical junction localization of aPKC in parallel to Par-3 (Ishiuchi and Takeichi, 2011, 2012). Overexpression of Willin-GFP in ZO-1/ZO-2 dKO cells promoted the apical junction localization of aPKC (Fig. 9, M and N) but did not rescue the epithelial polarity defects in ZO-1/ZO-2 dKO cells (Fig. 9, O–U). These results suggest that although ZO-1 and ZO-2 regulate the localization of Par-3 and aPKC, the aPKC pathway is not the only mechanism downstream of ZO-1/ZO-2 involved in the regulation of epithelial polarity.
Claudins and JAM-A coordinately regulate structural organization of TJ
The results described so far suggest that although claudins are essential for TJ strands/kissing points and electrolyte permeability barrier formation, other molecules are involved in the formation of the membrane appositions, macromolecule permeability barrier, and epithelial polarity. It is known that in addition to claudins, other integral membrane proteins such as occludin and JAMs localize to TJs (Furuse et al., 1993; Martìn-Padura et al., 1998). In claudin quinKO cells, although occludin localization was reduced (Fig. 2 F), JAM-A was more concentrated to apical cell junctions (Fig. 2 J). As JAM family proteins have been implicated in TJ gate and fence function (Rehder et al., 2006; Laukoetter et al., 2007; Tuncay et al., 2015), we investigated whether JAM-A is involved in the claudin-independent TJ structure and function.
JAM-A KO cells and claudin-2/4/3/7/1/JAM-A sextuple KO (claudin/JAM-A KO) cells were generated (Fig. S5, A–C and I–K). JAM-A KO cells did not show significant change in TJ or AJ marker localization (Fig. S5, D–F). In contrast, in claudin-JAM-A KO cells, discontinuity in ZO-1 staining was observed (Fig. 10 A, arrowheads), and large gaps in ZO-1 were occasionally found (Fig. 10 A, asterisk). Similar results were observed for ZO-2, ZO-3, and Afadin (Fig. 10, B–D). The gaps in ZO-1 were not due to loss of cell–cell contacts, as E-cadherin staining was continuous (Fig. 10 E). The circumferential actin bundle running parallel to the cell junctions was extensively developed (Fig. 10, F and G), resulting in extremely straight cell junctions (Fig. 10 H). Occludin staining was further reduced compared with claudin quinKO cells (Fig. 10, I–M).
Transmission EM revealed widening of the intercellular space at the apical junctions of claudin/JAM-A KO cells (Fig. 10, N and O; and Fig. S3 C). However, focally apposed membranes were observed in some cases (Fig. 10 N′, arrow), suggesting that other adhesion molecules may also contribute to the membrane appositions. AJ-like structures were found in most cases (Fig. 10 N′ and O′, asterisk), although we occasionally observed a complete loss of apical junctional structures where microvilli-like protrusions were observed along the lateral plasma membrane, which may have corresponded to the large gaps in ZO-1 staining (Fig. 10 P). In contrast, no significant change in the TJ structure was observed in JAM-A KO cells (Fig. S3, D–G). These results suggest that JAM-A regulates membrane apposition formation in claudin quinKO cells.
Claudins and JAM-A coordinately regulate epithelial polarity
To test whether JAM-A is involved in epithelial polarity, we examined the localization of Par-3 and aPKC. Staining of Par-3 and aPKC also revealed large gaps in claudin/JAM-A KO cells, but not in JAM-A KO cells (Fig. 10, Q and R; and Fig. S5 G). Sporadic epithelial polarity defects were observed in claudin/JAM-A KO cells, in which ezrin was mislocalized to the lateral cell membrane and Na-K ATPase α1 subunit was also detected on the apical plasma membrane (Fig. 10 S, brackets). Ezrin and Na-K ATPase α1 subunit localization was not perturbed in JAM-A KO cells (Fig. S5 H). In collagen gels, JAM-A KO cells showed a mild multilumen phenotype, consistent with previous reports (Fig. 10, T and V; Tuncay et al., 2015). Intriguingly, ∼80% of the cysts formed by claudin/JAM-A KO cells showed lumen expansion defects similar to ZO-1/ZO-2 dKO cells (Fig. 10, U and V), while ∼20% of the cysts formed by claudin/JAM-A KO cells formed a single lumen, although the apical surface morphology was disorganized (Fig. 10 U′). These results suggest that claudins and JAM-A coordinately regulate epithelial polarity.
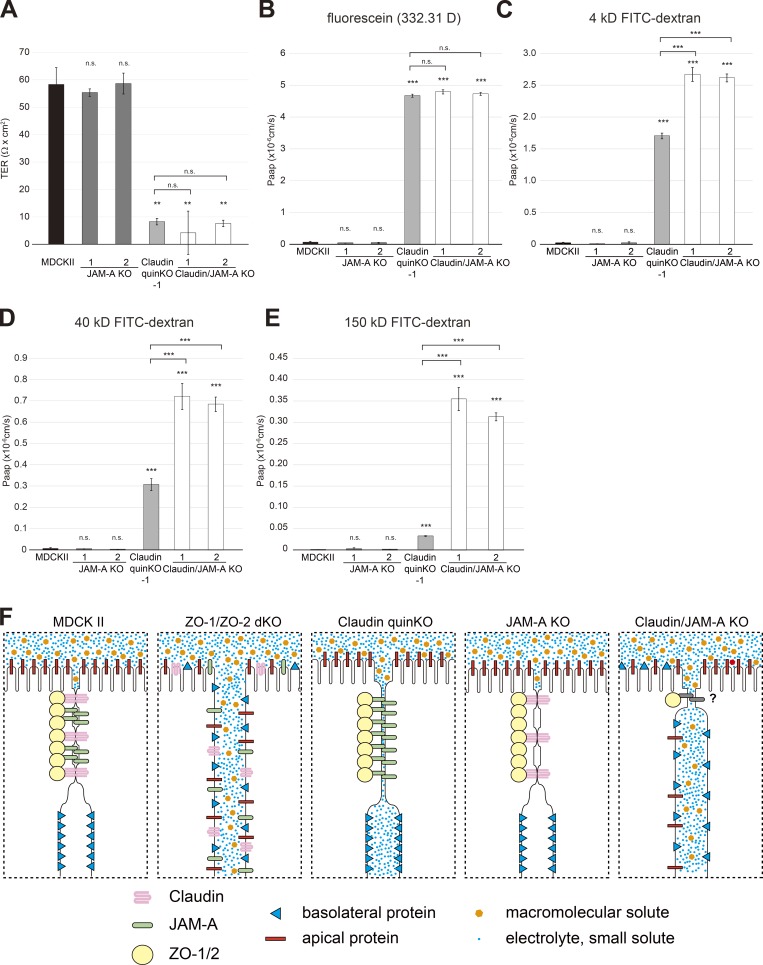
Claudins and JAM-A coordinately regulate the macromolecule permeability barrier formation
Finally, we examined the role of JAM-A in epithelial barrier. JAM-A KO cells did not show any significant epithelial barrier defects (Fig. 11, A–E). In claudin/JAM-A KO cells, the TER values and paracellular permeability of small molecules (FITC) was comparable to that of claudin quinKO cells (Fig. 11, A and B). However, the permeability of molecules larger than 4 kD was significantly increased compared with claudin quinKO cells (Fig. 11, C–E). These results suggest that claudins and JAM-A coordinately regulate the establishment of the macromolecule permeability barrier (Fig. 11 F).
Figure 11.
JAM-A is required for the claudin-independent macromolecule permeability barrier formation. (A) TER measurements. Unit area resistance was markedly reduced in claudin quinKO and claudin/JAM-A KO cells. (B–E) Paracellular flux measurements. (B) Apical-to-basal permeability of fluorescein (332.31 D) was dramatically increased in claudin quinKO and claudin/JAM-A KO cells. (C) Apical-to-basal permeability of 4-kD FITC-dextran was markedly increased in claudin/JAM-A KO cells, and significantly increased in claudin quinKO cells. (D) Apical-to-basal permeability of 40-kD FITC-dextran was dramatically increased in claudin/JAM-A KO cells, but only moderately increased in claudin quinKO cells. (E) Apical-to-basal permeability of 150-kD FITC-dextran was dramatically increased in claudin/JAM-A KO cells, but only modestly in claudin quinKO cells. (F) Summary of the phenotypes of MDCK II, ZO-1/ZO-2 dKO, claudin quinKO, JAM-A KO, and claudin/JAM-A KO cells. In MDCK II cells, claudins and JAM-A are concentrated at the TJs with ZO-1/ZO-2. Membranes closely appose to each other and kissing points are formed, and paracellular diffusion of electrolytes and macromolecules are prohibited. Epithelial polarity is maintained. In ZO-1/ZO-2 dKO cells, claudins and JAM-A are diffusely localized, and membrane appositions and kissing points are lost. Intercellular space is widened, and electrolytes and macromolecules diffuse across the paracellular space. Epithelial polarity is disorganized. In claudin quinKO cells, JAM-A and ZO-1/2 are concentrated at the apical junctions, and membrane appositions are formed despite the lack of kissing points. Although electrolytes can diffuse across the paracellular space, the paracellular diffusion of macromolecules is prohibited. No epithelial polarity defects are observed. In JAM-A KO cells, claudins and ZO-1/2 localize to TJs, and kissing points are formed. Epithelial barrier and polarity are not perturbed. In claudin/JAM-A KO cells, ZO-1 can localize to apical junctions, but discontinuity is observed. Intercellular space is widened although focal membrane appositions are observed in some cases. Electrolytes and macromolecules can diffuse across the paracellular space, and epithelial polarity is disorganized in some regions. Graphs represent mean ± SD (n = 3 each). **, P < 0.005; ***, P < 0.0005, compared with MDCK II cells (or with claudin quinKO cells when notified) by t test. n.s., not significant.
Discussion
TJs play central roles in regulating paracellular permeability (gate function) and are also thought to act as a membrane fence to restrict the intermixing of apical and basolateral membrane lipids and proteins (fence function). However, the roles of TJs in epithelial polarity remain controversial (Umeda et al., 2006; Ikenouchi et al., 2012; Phua et al., 2014), and how claudins and other membrane proteins cooperate to regulate TJ structure and function remains unclear. In the present study, we have demonstrated that ZO-1/ZO-2 are required for epithelial polarity and organize the TJ structure and function by scaffolding the TJ-associated membrane proteins including claudins and JAM-A (Fig. 11 F).
Roles of ZO-1 and ZO-2 in apical junctional complex formation
We have shown that epithelial polarity was disorganized in ZO-1/ZO-2 dKO cells, in contrast to previous KD studies (Umeda et al., 2006; Fanning et al., 2012; Ikenouchi et al., 2012; Odenwald et al., 2018). Moreover, in general, the phenotypes we observed in ZO-1/ZO-2 dKO cells (TJ/AJ structural defects, barrier defects, lumen formation defects, etc.) were more severe compared with the previous KD studies (Van Itallie et al., 2009; Fanning et al., 2012; Choi et al., 2016; Odenwald et al., 2017, 2018). We have shown that in ZO-1 KO/ZO-2 KD EpH4 cells, epithelial polarity was not perturbed, in agreement with previous findings, whereas in ZO-1/ZO-2 dKO EpH4 cells, epithelial polarity was disorganized. Importantly, weak but significant residual ZO-2 expression was found in ZO-1 KO/ZO-2 KD EpH4 cells, while ZO-2 expression was lost in ZO-1/ZO-2 dKO EpH4 cells. Our results are consistent with the idea that even a relatively small amount of residual ZO-1 or ZO-2 can support TJ structure and function to a significant extent, and that it masks some phenotypes. As KD experiments only reduce protein expression and correspond to hypomorphs, it is not surprising that the protein null phenotypes generated by genome editing yield more severe phenotypes.
In ZO-1/ZO-2 dKO cells, the apical junction localization of TJ membrane proteins was disrupted, consistent with the idea that ZO-1 and ZO-2 are essential for TJ assembly. It should be noted that claudins and occludin, but not JAM-A, were occasionally localized to the apical cell junctions together with ZO-3. These results suggest that although ZO-3 is not able to assemble zonula occludens, it can support spontaneous accumulation of claudins and occludin at the apical cell junctions. In contrast to our observations, it has been reported in EpH4 cells that exogenously expressed ZO-3 cannot localize to TJs in ZO-1 KO/ZO-2 KD cells (Umeda et al., 2006). It is possible that ZO-3 or TJ membrane proteins are subjected to differential posttranslational modifications in MDCK II cells and EpH4 cells.
Interestingly, discontinuity in ZO-1 staining was observed in claudin/JAM-A KO cells. These results suggest that while ZO-1 and ZO-2 are essential for TJ assembly, claudins and JAM-A are required for the stable apical junction localization of ZO-1. Although it is unclear how the discontinuity in ZO-1 appears in claudin/JAM-A KO cells, our results suggest that TJ membrane proteins and ZO family proteins may interdependently regulate the formation and/or maintenance of TJs.
In ZO-1/ZO-2 dKO cells, we observed fragmentation of AJs, accompanied with the development of actomyosin bundles in the apical cytoplasm running perpendicular to the cell junctions. In contrast, in claudin quinKO cells or claudin/JAM-A KO cells, the circumferential actin bundles running parallel to the cell junctions were extensively developed, and the cell junctions became linear. It is likely that in ZO-1/ZO-2 dKO cells, the perpendicular orientation of the contractile actomyosin fibers results in an anisotropic increase in tension, an idea supported by the preferential accumulation of tension-sensitive AJ markers at vertices. This likely results in an imbalance in the tension applied to the cell junctions, leading to the fragmentation of AJs. In contrast, in claudin quinKO cells or claudin/JAM-A KO cells, the thickening of the circumferential actomyosin bundles results in an isotropic increase in the tension applied to cell junctions, which probably leads to the more linear morphology of cell junctions. The increase in tension in both ZO-1/ZO-2 dKO cells and claudin quinKO cells is notable, as the latter is accompanied by an increase in junctional ZO-1/ZO-2. Although the molecular mechanisms mediating the actomyosin organization in response to TJ perturbation remain largely unexplored, with the exception of Shroom3 (Choi et al., 2016), it is possible that a mechanism exists to detect dysfunctions of the TJs to optimize the strength of the tension applied to the cell junctions.
Structural organization of TJs
The structural hallmarks of TJs are the close appositions of neighboring plasma membranes, accompanied by membrane kissing points seen on ultrathin sections, and the anastomosing strand structures observed by freeze-fracture replica EM (Farquhar and Palade, 1963; Staehelin, 1973). We showed that claudin-deficient cells lacked TJ strands and membrane kissing points but were able to form membrane appositions. Importantly, further deletion of JAM-A resulted in the widening of intercellular spaces, suggesting that JAM-A plays an important role in membrane apposition formation. These results suggest that TJ strand/kissing point formation and membrane apposition formation can be uncoupled, where claudins play a major role in TJ strand/kissing point formation, while JAM-A can regulate the membrane apposition formation independent of TJ strand assembly. As focal membrane appositions were still observed in claudin/JAM-A KO cells, other adhesion molecules including occludin or JAM-B/C and other related molecules may also contribute to the membrane apposition formation. ZO-1 and ZO-2 were essential for both TJ strand and membrane apposition formation, suggesting that ZO-1/ZO-2 integrate claudins and JAM-A to organize the TJs.
TJ structure and epithelial barrier
Claudin quinKO cells lacked TJ strands, and their permeability barrier against electrolytes and small molecules was disrupted, while diffusion of macromolecules was restricted. These results suggest that claudin-based TJ strands are not absolutely essential for the formation of the macromolecule permeability barrier. Further deletion of JAM-A from claudin quinKO cells disrupted the macromolecule permeability barrier, suggesting that while claudin-based TJ strands form a tight barrier that can restrict the diffusion of electrolytes and small molecules, JAM-A can form a crude barrier independent of TJ strands that restricts diffusion of larger macromolecules. As JAM-A regulates the close appositions of neighboring membranes, it is possible that the membrane appositions can physically preclude the paracellular diffusion of larger molecules in claudin quinKO cells. Occludin and tricellulin have also been implicated in macromolecule permeability barrier formation (Balda et al., 1996; Krug et al., 2009; Al-Sadi et al., 2011; Buschmann et al., 2013), suggesting that multiple TJ proteins coordinately regulate macromolecule permeability barrier formation. Alternatively, JAM-A may regulate macromolecule permeability barrier formation via occludin and tricellulin.
TJs and epithelial polarity
We have shown that ZO-1 and ZO-2 are required for epithelial polarity. Although claudin quinKO cells did not show any epithelial polarity defects, simultaneous deletion of claudins and JAM-A resulted in sporadic epithelial polarity defects, suggesting that claudins and JAM-A coordinately regulate epithelial polarity. These results establish that TJs are required for epithelial polarity.
How do TJs regulate epithelial polarity? We can envisage at least three potential mechanisms. First, ZO-1/ZO-2 may regulate epithelial polarity by establishing a membrane fence at the zonula occludens. This idea is supported by the mislocalization of Forssman antigen, a glycosphingolipid antigen in ZO-1/ZO-2 dKO cells. On the other hand, the localization of some proteins such as gp135 were not affected in ZO-1/ZO-2 dKO cells, which may reflect the differential mobility of membrane proteins on plasma membrane. Interestingly, a recent in vitro study showed that claudin-4 reconstituted in unilamellar vesicles can exclude proteins, but not lipids, from its interface, suggesting that claudins can restrict the intramembrane diffusion of membrane proteins (Belardi et al., 2018). In addition, it has been shown that JAM-A overexpression in fibroblast cells results in an appearance of smooth membrane area on freeze fractures (Itoh et al., 2001), indicating that local accumulation of JAM-A results in exclusion of other membrane particles. When JAM-A is organized in a belt-like fashion in epithelial cells, it may act as a membrane fence by precluding other membrane proteins from diffusing across the zonula occludens. Although the fence function is an attractive possibility, we were unable to directly address the fence function in the present study, because the epithelial barrier was disrupted in ZO-1/ZO-2 dKO cells, and selective labeling of the apical or basolateral plasma membranes with fluorescent lipids was not possible. Thus, whether TJs can act as a membrane fence to restrict intramembrane diffusion of membrane lipids and proteins should be addressed in future studies.
Second, as Par-3 and aPKC localization are disorganized in ZO-1/ZO-2 dKO cells, ZO-1/ZO-2 may regulate epithelial polarity by regulating the spatial organization of the polarity signaling. JAM-A has been reported to bind Par-3 (Ebnet et al., 2001; Itoh et al., 2001), whereas cell–cell junction formation is dependent on the polarity signaling molecules (Suzuki et al., 2001; Hurd et al., 2003; Iden et al., 2012), suggesting that the feedback between TJ/AJ and polarity signaling could be disrupted in ZO-1/ZO-2 dKO cells, leading to a collapse of the epithelial polarity program. Although this is a likely scenario, the failure to rescue the polarity phenotypes by Willin overexpression indicates that this is not the only mechanism downstream of ZO-1/ZO-2 to regulate epithelial polarity. Moreover, the normal localization of gp135 and centrosomes argues that despite the abnormal localization of Par-3 and aPKC, the epithelial polarity is retained to some extent in ZO-1/ZO-2 dKO cells. Third, as ZO-1/ZO-2 regulates the apical junctional complex formation, it is possible that ZO-1/ZO-2 regulates epithelial polarity by regulating the apical junctional complex, which is thought to serve as a polarity landmark (Drubin and Nelson, 1996). For example, the localization of Par-3, which has been shown to act as a receptor for the exocyst complex (Ahmed and Macara, 2017), is perturbed in ZO-1/ZO-2 dKO cells. ZO-1/ZO-2 may regulate polarized transport by establishing the apical cell junctions to form landmarks for polarized delivery of membrane proteins. However, it should be noted that some proteins such as gp135 were localized to the correct plasma membrane domain, indicating that polarized transport is not completely abrogated in ZO-1/ZO-2 dKO cells. In addition, centrosome localization was not affected in ZO-1/ZO-2 dKO cells in contrast to αE-catenin KO cells, suggesting that the epithelial polarity defects of ZO-1/ZO-2 dKO cells are qualitatively distinct from that of a near-complete loss of cell junctions in αE-catenin KO cells.
Another important remaining question is how ZO-1 and ZO-2 become localized to the most apical region of intercellular junctions to induce TJ formation. As TJs are located at the border of the apical and basolateral membranes, it is reasonable to assume that the polarity signaling complexes play important roles in TJ positioning. Further genetic dissection and reconstitution studies should reveal the molecular mechanisms that control junctional complex assembly and epithelial polarity in the future.
Materials and methods
Cell culture
MDCK II cells derived from canine kidney were provided by Masayuki Murata (University of Tokyo, Tokyo, Japan) and cultured in DMEM (low glucose; #05919; Nissui) supplemented with 10% FCS (Furuse et al., 2001). EpH4 cells derived from mouse mammary gland (Reichmann et al., 1992) were provided by Ernst Reichmann (University of Zurich, Zurich, Switzerland) and cultured in DMEM (low glucose) supplemented with 10% FCS. Claudin-2/4 dKO MDCK II cells (Tokuda et al., 2017), ZO-1 KO/ZO-2 KD EpH4 cells (Umeda et al., 2006), and ZO-1/ZO-2 dKO EpH4 cells (Nishimura et al., 2016; provided by Junichi Ikenouchi, Kyushu University, Fukuoka, Japan) were described previously. All cells were cultured at 37°C under 5% CO2.
For immunofluorescence or physiological analyses, cells were cultured on Transwell polycarbonate filters (0.4-µm pore size; #3413; Corning), with 1 × 105 cells seeded per 6.5-mm-diameter filter. For 3D culture, MDCK II cells were embedded in bovine dermis atelocollagen (IPC-50; Koken). Briefly, bovine atelocollagen and cell suspension prepared in ice-cold DMEM supplemented with 10% FCS and 20 mM Hepes (pH 7.4) were mixed at a 4:6 ratio on ice, and 150 µl of the solution was plated on 12-mm-diameter Transwell-Clear polyester filters (0.4-µm pore size; #3460; Corning), with 6 × 104 cells seeded per filter. The gel was allowed to form at 37°C for 10 min, and DMEM supplemented with 10% FCS was added to the upper and lower chambers. Myosin II inhibitor (-)-blebbistatin (#021-17041; Wako) was used at 50 µM for 2 h (Straight et al., 2003). Phase-contrast images were acquired by AdvanCam-HD1080P camera (Advan Vision) mounted on an inverted microscope CKX53 (Olympus Lifescience).
Genome editing and transfection
ZO-1/ZO-2 dKO cells were generated by sequential genome editing using transcription activator-like effector nuclease (TALEN). The ZO-1 and ZO-2 TALENs were described previously (Tokuda et al., 2014). Initially, ZO-2 KO cells were established by transfecting ZO-2 TALEN vectors into parental MDCK II cells and cloning by limited dilution. ZO-1 TALEN vectors were further transfected into parental MDCK II or ZO-2 KO cells and cloned by limited dilution to establish ZO-1 KO cells and ZO-1/ZO-2 dKO cells, respectively.
Claudin KO cells were generated by sequential genome editing using TALENs. Claudin-2/4 dKO cells (Tokuda et al., 2017) and claudin-3/7/1 TALENs were described previously (Tokuda and Furuse, 2015). Initially, claudin-3 TALEN vectors were transfected into claudin-2/4 dKO cells, and claudin-2/4/3 triple KO cells were cloned by limited dilution. Claudin-7 TALEN vectors were further transfected into claudin-2/4/3 triple KO cells, and claudin-2/4/3/7 quadruple KO cells were cloned by limited dilution. Finally, claudin-1 TALEN vectors were transfected into a claudin-2/4/3/7 quadruple KO cell clone, and claudin-2/4/3/7/1 quinKO cells were cloned by limited dilution.
E-cadherin KO cells, afadin KO cells, αE-catenin KO cells, JAM-A KO cells, and claudin/JAM-A KO cells were generated by electroporation of parental MDCK II cells (or claudin quinKO cells for generation of claudin/JAM-A KO cells) with Cas9–gRNA RNP complexes using a CUY21 Pro-Vitro electroporator (Nepagene). CRISPR RNA and trans-activating CRISPR RNA were synthesized by IDT and annealed with one another, and the gRNA duplex was further incubated with Cas9 protein (IDT) at RT for 10 min to form the Cas9–gRNA RNP complex. Cas9 and gRNA duplex were mixed at a 1:1.2 molar ratio, and 100 pmol Cas9 and 120 pmol gRNA duplex were introduced to 1 × 105 to 1 × 106 cells. Electroporation was performed with the following conditions: prepulse, 150 V for 10 ms; postpulses, 10 pulses of 20 V for 50 ms at 50-ms intervals. The target sequences were as follows (PAM sequences are underlined): afadin, 5′-GGATGATAGGCCTTTCCAAGGG-3′; E-cadherin, 5′-GATGACACCCGATTCAAAGTGGG-3′; αE-catenin, 5′-GTTTCTCAAGGAAGAGCTTGTGG-3′; and JAM-A, 5′-CCTATGAGGACCGAGTTACCTTC-3′.
Screening of KO cells was performed by immunofluorescence in glass-bottom 96-well plates (#4580; Corning). αE-catenin KO cells were isolated on the basis of loss of cell–cell adhesion. Loss of protein expression was confirmed by Western blotting. The genomic regions of the targeted genes were amplified by genomic PCR and subcloned into pTAC-1 vector (BioDynamics Laboratory) by TA cloning or digested with EcoRI/SalI or EcoRI/XhoI and ligated into pBS-SK(-) vector digested with EcoRI/SalI. Successful KO was confirmed by Sanger sequencing with a BigDye Terminator v3.1 Sequencing Kit (Thermo Fisher Scientific) and analysis with an Applied Biosystems 3130xl DNA analyzer (Thermo Fisher Scientific). The following primers were used for genomic PCR (restriction enzyme sites are underlined): ZO-1 forward, 5′-AAGGAAGTTCCTGCGTGTAGTTTC-3′, and reverse, 5′-CAGGAATAAAAAGAAAAAGCTAACC-3′; ZO-2 forward, 5′-TTTTGTAATGATTGTTTTGGAGTGG-3′, and reverse, 5′-AACTCAAGGACAGGGGATGCCTGGG-3′; claudin-1 forward, 5′-TTTTCTCGAGCCTGATCCTTCCCAGGGGT-3′, and reverse, 5′-TTTTTGAATTCACCTTGCACTGAATCTGCCC-3′; claudin-3 forward, 5′-AAGCACAGGCAGGTGCAGGCGCTGC-3′, and reverse, 5′-AGCCCGAAGGCGGCCAGCAGGATGG-3′; claudin-7 forward, 5′-GGGGTCGACCCGGCCTTCGCGGATCGCTCTTTGG-3′, and reverse, 5′-CCCGAATTCTCGTACATTTTGCAGCTCATCATGC-3′; afadin forward, 5′-CCCGTCGACCACATGCGAAGTGCACAGAT-3′, and reverse, 5′-AAAAAGAATTCGGTCCAAACAACCAGAAAGG-3′; E-cadherin forward, 5′-CCCGTCGACGGCAGGTTTCTGTTTGCACT-3′, and reverse, 5′-TTTTTGAATTCGGTATTGGCCATGTCTGGAG-3′; and JAM-A forward, 5′-CCCGTCGACCAGCTCATCCAGCTCATCCA-3′, and reverse, 5′-TTTTTGAATTCTTGACGGTGACCTCCCCATA-3′.
pCANw-ZO-1–GFP was generated by subcloning mouse ZO-1 into pCANw-Sal-EGFP (Ichii and Takeichi, 2007). pCANw-Willin-GFP was kindly provided by Masatoshi Takeichi (RIKEN Center for Biosystems Dynamics Research, Kobe, Japan; Ishiuchi and Takeichi, 2011). Transfection of plasmid vectors was performed with Lipofectamine LTX (#15338-100; Thermo Fisher Scientific), according to the manufacturer’s instructions.
Antibodies
The following antibodies were used in this study: mouse monoclonal anti–ZO-1 (clone T-8754; Itoh et al., 1991); rabbit polyclonal anti–ZO-1 (#61-7300; Thermo Fisher Scientific; for Western blotting); rabbit polyclonal anti–ZO-2 (#38-9100; Thermo Fisher Scientific); rabbit polyclonal anti–ZO-3 (#36-4100; Thermo Fisher Scientific); rabbit polyclonal anti-tricellulin/MARVELD2 (#48-8400; Thermo Fisher Scientific); rabbit polyclonal anti-l/s-afadin (#A0224; Sigma-Aldrich); rabbit polyclonal anti-myosin IIA (#M8064; Sigma-Aldrich); rabbit polyclonal anti-myosin heavy chain IIB (#Poly19099; BioLegend); rabbit polyclonal anti–claudin-1 (#51-9000; Thermo Fisher Scientific); mouse monoclonal anti–claudin-2 (clone 12H12; #32-5600; Thermo Fisher Scientific); rabbit polyclonal anti–claudin-3 (#34-1700; Thermo Fisher Scientific); mouse monoclonal anti–claudin-4 (clone 3E2C1; #32-9400; Thermo Fisher Scientific); rabbit polyclonal anti–claudin-7 (#34-9100; Thermo Fisher Scientific); rat monoclonal anti-occludin (clone MOc37; Saitou et al., 1997); rabbit polyclonal anti-occludin (Saitou et al., 1997; for Western blotting); rabbit polyclonal anti-canine JAM-A (Rehder et al., 2006); rat monoclonal anti–E-cadherin (clone ECCD-2; #M108; Takara; Shirayoshi et al., 1986); mouse monoclonal anti–E-cadherin (clone rr1; Developmental Studies Hybridoma Bank; for Western blotting; Gumbiner and Simons, 1986); rabbit polyclonal anti-α-catenin (#C2081; Sigma-Aldrich); rat monoclonal anti-α-catenin (clone α18; provided by Akira Nagafuchi, Nara Medical University, Nara, Japan; Nagafuchi and Tsukita, 1994); mouse monoclonal anti–β-catenin (clone 14; #610153; BD Biosciences); mouse anti-vinculin (clone VIN-11-5; #V4505; Sigma-Aldrich); rabbit polyclonal anti-Scribble (H-300; #sc-28737; Santa Cruz Biotechnology); mouse anti-sodium potassium ATPase α 1 (clone 464.6/6H; Novus Biologicals; Pietrini et al., 1992); mouse monoclonal anti–γ-tubulin (clone GTU88; #T5326; Sigma-Aldrich); mouse monoclonal anti–α-tubulin (clone DM1A; #05-829; Merck; Blose et al., 1984); mouse monoclonal anti–β-actin (clone AC-15; #A1978; Sigma-Aldrich; Gimona et al., 1994); rat monoclonal anti-ezrin (clone M11; provided by Shigenobu Yonemura, Tokushima University, Tokushima, Japan; Takeuchi et al., 1994); rat monoclonal anti-Forssman antigen (clone 12B12; provided by Junichi Ikenouchi; Zinkl et al., 1996); mouse monoclonal anti-gp135 (clone 3F2/D8; Developmental Studies Hybridoma Bank; Ojakian and Schwimmer, 1988); rabbit anti–Par-3 (#07-330; Merck); rabbit anti-aPKC (#sc-216; Santa Cruz Biotechnology); rabbit anti Pals1 (#07-708; Merck); and mouse monoclonal anti-GFP (clones 7.1 and 13.1; Roche Applied Science).
The following secondary antibodies and detection reagents were used: donkey anti-mouse IgG Alexa Fluor 488–conjugated (#A21202; Molecular Probes); donkey anti-rat IgG Alexa Fluor 488–conjugated (#A21208; Molecular Probes); donkey anti-rabbit IgG Alexa Fluor 488–conjugated (#A21206; Molecular Probes); donkey anti-mouse IgG Cy3-conjugated (#715-165-151; Jackson ImmunoResearch Laboratories); donkey anti-rat IgG Cy3-conjugated (#712-165-153; Jackson ImmunoResearch Laboratories); donkey anti-rabbit IgG Cy3-conjugated (#711-165-152; Jackson ImmunoResearch Laboratories); donkey anti-mouse IgG Cy5-conjugated (#715-175-151; Jackson ImmunoResearch Laboratories); donkey anti-rat IgG Cy5-conjugated (#712-175-153; Jackson ImmunoResearch Laboratories); donkey anti-rabbit IgG Cy5-conjugated (#711-175-152; Jackson ImmunoResearch Laboratories); Alexa Fluor 488–conjugated Phalloidin (#A12379; Molecular Probes); sheep anti-mouse IgG HRP-conjugated whole antibody (#NA931V; GE Healthcare); and donkey anti-rabbit HRP-conjugated F(a′b′)2 fragment (#NA9340V; GE Healthcare).
Western blotting
Cells were rinsed with PBS, lysed in Laemmli sample buffer supplemented with 100 mM DTT, and incubated at 95°C for 5–10 min. The obtained samples were separated by SDS-PAGE using standard methods and 6%, 8%, 10%, or 12% polyacrylamide gels. The separated proteins were transferred to 0.45-µm-pore Protran nitrocellulose transfer membranes (#10-401-196; Whatman) and blocked with 5% skim milk in TBS. The primary antibodies were diluted in the blocking buffer or Can Get Signal (#NKB-101; Toyobo Life Science) and incubated with the membranes for 1–2 h at RT. After three washes with 0.1% Tween-20 in TBS for 10 min at RT with agitation, signals were detected by chemiluminescence using an ECL Prime Kit (#RPN2232; GE Healthcare), and images were captured using an LAS3000 mini (Fujifilm). Image processing (brightness and contrast adjustments) was performed using Fiji/ImageJ 1.52f software (National Institutes of Health).
Immunohistochemistry
MDCK II cells cultured on Transwell filters for 5–7 d were fixed with 1% PFA in PBS for 5 min at RT (for ZO-1, myosin IIA, and α18), 4% PFA in PBS for 15 min at RT (for JAM-A, ezrin, vinculin, and phalloidin), 10% TCA for 15 min at 4°C (for ZO-2), or 100% methanol for 15 min at −20°C (for ZO-3, claudins, occludin, afadin, E-cadherin, Myosin IIB, Scribble, Na-K ATPase α1, aPKC, Par-3, Pals1, and GFP). The filters were rinsed with PBS, and the cells were permeabilized with 0.1% Triton X-100 in PBS for 15 min at RT. The filters were excised with scalpels and blocked by incubation with 10% FCS in PBS. The primary antibodies were diluted in the blocking solution and incubated with the filters for 1–2 h at RT. After three washes with 0.1% Triton X-100 in PBS, the filters were incubated with the secondary antibodies for 30–60 min at RT. After three washes with 0.1% Triton X-100 in PBS, the samples were mounted in FluoroSave Reagent (#345789; Calbiochem).
For staining with the anti-Forssman antigen antibody, MDCK II cells cultured on Transwell filters for 5–7 d were fixed with 1% PFA in PBS for 10 min at RT. The filters were rinsed with PBS, and the cells were permeabilized with 0.1% saponin in PBS for 10 min at 4°C. The filters were excised with scalpels and blocked by incubation with 10% FCS in PBS at 4°C. The primary antibodies were diluted in the blocking solution and incubated with the filters for 1–2 h at 4°C. After three washes with PBS at 4°C, the filters were postfixed with 2% PFA in PBS for 10 min at 4°C and rinsed three times with PBS at 4°C. The filters were incubated in the secondary antibodies for 30–60 min at RT. After three washes with PBS, the samples were mounted in FluoroSave reagent.
For staining of cells embedded in collagen gel, MDCK II cells cultured in collagen gel for 5–7 d were rinsed twice with PBS and incubated with collagenase type VII (100 U/ml; #C0773; Sigma-Aldrich) for 15 min at RT. After two rinses with PBS, the cells were fixed with 2% PFA in PBS for 30 min at RT. After another two rinses with PBS, the cells were permeabilized with 0.5% Triton X-100 in PBS for 30 min at RT, rinsed twice with PBS, and blocked with 10% FCS in PBS for 1 h at RT. The primary antibodies were diluted in the blocking solution and incubated with the gels overnight at 4°C with gentle rocking. The gels were rinsed with PBS five times and incubated with the secondary antibodies overnight at 4°C with gentle rocking. After five washes with PBS, the samples were mounted on glass-base dishes (#3970-035; Iwaki) in Vectashield antifade mounting medium (#H-1000; Vector Laboratories).
Confocal microscopy
Confocal microscopy was performed using a TCS-SPE laser scanning confocal microscope mounted on a DMI 4000 B inverted microscope using HCX PL Fluotar 40×/NA 0.75, HCX PL APO 63×/NA 1.40, and HCX FL APO 100×/NA 1.40 objectives with diode lasers (488/532/635 nm; all from Leica Microsystems). Image acquisition was performed with LAS AF software (Leica Microsystems). Image processing (z-stacking, orthogonal views, brightness and contrast adjustments, merge channels, and median filters) was performed using Fiji/ImageJ 1.52f software.
Fluorescent intensity was quantified using Fiji/ImageJ 1.52f software. In brief, stacked images were generated for the whole cells or for the apical regions, and after background subtraction, mean intensity was measured. The polarization index was quantified by using the z-section images using Fiji/ImageJ 1.52f software. In brief, after background subtraction, a segmented line (5 pixels wide) was overlaid on the apical plasma membrane, and the total intensity of apical plasma membrane was measured. Subsequently, the total intensity of the image was measured. Basolateral intensity was determined by subtracting the apical intensity from total intensity. The polarization index was determined by calculating the ratio of signals located at the correct plasma membrane domains. Junction linearity was quantified by measuring the length of deviation from a straight line drawn between the vertices of the corresponding cell junction. Graphs were generated using Excel (Microsoft) or R (R Foundation). Statistical tests were performed using Excel or R.
EM
MDCK II cells were cultured on Transwell filters for 5–7 d and rinsed once with PBS. Fixative (2% glutaraldehyde and 2% PFA in 0.1 M cacodylate buffer, pH 7.4) was gently added to the upper and lower chambers. After fixation at RT for 30–60 min, the samples were transferred to 4°C. The filters were excised with scalpels, washed three times with 0.1 M cacodylate buffer (pH 7.4), and postfixed with 1% OsO4 in 0.1 M cacodylate buffer (pH 7.4) for 30 min on ice. After three 10-min washes with water, the filters were stained en bloc with 0.5% uranyl acetate for 30 min at RT. After three washes with water, the filters were dehydrated in a graded ethanol series (65%, 75%, and 85%) for 10 min at each concentration, further dehydrated in 95% and 99.5% ethanol for 15 min at each concentration, and transferred to 100% ethanol for two 15-min incubations. The filters were incubated in propylene oxide for 1 min, transferred to a 1:1 mixture of propylene oxide/Quetol 812 resin (Nisshin EM), and incubated overnight. After three transfers of the filters through Quetol 812, the resin was polymerized at 60°C for >48 h.
For analysis of TJ membrane kissing points, MDCK II cells were cultured on Transwell filters for 5–7 d. The same volume of fixative (2% glutaraldehyde and 4% PFA in 0.1 M cacodylate buffer, pH 7.4, supplemented with 20 mM CaCl2) was directly added to the medium in the upper and lower chambers, and the medium/fixative mixture was immediately removed and replaced with fresh fixative. The fixative was further replaced with fresh fixative again to ensure complete replacement of the medium with the fixative. Fixation was performed for 1 h at RT. After four 3-min washes with 0.1 M cacodylate buffer (pH 7.4) supplemented with 20 mM CaCl2, the filters were excised with scalpels and postfixed with 1% OsO4 and 1.5% KFeCN in 0.1 M cacodylate buffer (pH 7.4) for 45 min on ice. After three 3-min washes with water, the filters were incubated with 1% tannic acid for 30 min at RT. After four 3-min washes with water, the filters were further fixed with 2% OsO4 for 30 min at RT. After four 3-min washes with water, the filters were stained en bloc with 2% uranyl acetate for 20 min at 37°C, washed twice with water, and stained en bloc with Walton’s lead aspartate for 30 min at 60°C (Walton, 1979). The filters were washed twice with water, dehydrated on ice in a graded ethanol series (30%, 50%, 60%, 70%, 80%, 90%, and 95%) for 5 min at each concentration, and transferred to 100% ethanol for three 10-min incubations. The filters were incubated in propylene oxide for 1 min, transferred to a 1:1 mixture of propylene oxide/Quetol 812 resin, and incubated for 45 min. The filters were further incubated twice in 100% Quetol 812 for 45 min, and the resin was polymerized at 60°C for >48 h.
Semithin sections (0.5 µm) were cut and stained with toluidine blue to examine the sample preparations. Ultrathin sections (50–80 nm) were cut and mounted on 200-mesh formvar-coated copper grids. The sections were stained with 0.5% aqueous uranyl acetate in the dark for 3 min at RT. After washing with water, the sections were further stained with Sato’s lead solution (Sato, 1968) for 3 min at RT, washed with water, and allowed to dry.
For freeze-fracture replica EM, MDCK II cells cultured on Transwell filters for 5–7 d were rinsed once with 0.1 M phosphate buffer (pH 7.4) and fixed with 2% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) at 4°C overnight. After three 10-min washes with 0.1 M phosphate buffer (pH 7.4), the samples were cryoprotected with 30% glycerol in 0.1 M phosphate buffer (pH 7.4) for 30 min at RT. After excision of the filters with scalpels, the cells were scraped from the filters with scrapers and mounted on gold stubs. After removing the excess buffer, the samples were snap-frozen in liquid N2. The frozen samples were transferred to a freeze-fracture machine (BAF-060; Bal-Tec) and subjected to fracturing at −110°C. Immediately after fracturing, the samples were coated with a thin layer (∼2 nm) of platinum at a 45° angle and a thin layer (∼20 nm) of carbon at a 90° angle. After retrieval from the machine, the samples were coated with collodion and cleaned with domestic bleach. After three 10-min rinses with water, replicas were collected on 200-mesh formvar-coated copper grids.
Samples were observed with a JEM1011 or JEM1010 transmission EM (JEOL) at 100-kV accelerating voltage. Images were captured with a MegaViewG2 or Veleta CCD camera using iTEM software (all from Olympus Soft Imaging Solutions). Cell–cell junctions at the most apical cell contacts were observed.
Image processing (brightness and contrast adjustments) was performed using Fiji/ImageJ 1.52f software, and a pseudocolor image was generated using Illustrator CC2018 (Adobe). TJ strand length was quantified by overlaying segmented lines on the TJ strands and measuring their length using Fiji/ImageJ 1.52f software. When more than three intramembrane particles were aligned at the apical region, they were classified as TJ strands. The strand length was normalized against the apical surface length of the corresponding fractured region. The minimum distance between two plasma membranes was measured using Fiji/ImageJ 1.52f software. Graphs were generated using Excel or R. Statistical tests were performed by using Excel or R.
TER measurement
MDCK II cells were cultured on Transwell polycarbonate filters (0.4-µm pore size; #3413; Corning) for 5–7 d. After cells were equilibrated to RT, the electric resistance between the apical and basolateral chambers was measured using a Millicell ERS-2 electrical resistance system (Merck Millipore). Blank measurements were performed on Transwell filters without cells. After subtraction of the mean blank values, the electric resistance was multiplied by the growth area of the Transwell filter to yield the unit area resistance (Ω · cm2). Graphs were generated using Excel. Statistical tests were performed using Excel.
Paracellular flux measurement
MDCK II cells were cultured on Transwell polycarbonate filters (0.4-µm pore size, 6.5-mm diameter; #3413; Corning) for 5–7 d. The medium was then changed to phenol red–free DMEM (low glucose; #08490-05; Nacalai Tesque) supplemented with 10% FCS. The following fluorescent tracers were added to the apical chamber: 200 µM fluorescein (#16106-82; Nacalai Tesque); 200 µM 4-kD FITC-dextran (#FD4; Sigma-Aldrich); 50 µM 40-kD FITC-dextran (#FD40; Sigma-Aldrich); and 50 µM 150-kD FITC-dextran (#FD150S; Sigma-Aldrich). The volumes of medium in the apical (donor) and basal (acceptor) chambers were 100 and 600 µl, respectively. The cells were incubated for 1 h at 37°C under 5% CO2, and the basal chamber medium was recovered and protected from light. The fluorescence intensity in the recovered medium was measured using a Typhoon 9400 scanner (GE Healthcare) or SpectraMax i3 microplate reader (Molecular Devices). Standard curves were obtained by measuring the fluorescence intensities of serial dilution series of the fluorescent tracers, and blank measurements were performed by measuring the fluorescence intensity of wells without fluorescent tracers. After subtraction of the mean blank values, the apparent permeability (Papp) was determined by the following formula: Papp (cm × s) = {(dQ/dt) × Vacc}/(A × C), where dQ is the amount of tracer transported to the acceptor chamber during time interval dt, Vacc is the acceptor volume, A is the area, and C is the initial tracer concentration. Graphs were generated using Excel. Statistical tests were performed using Excel.
Online supplemental material
Fig. S1 shows the genomic sequence and Western blotting of ZO-1/ZO-2 dKO cells. Fig. S2 shows the genomic sequence and Western blotting of claudin quinKO cells. Fig. S3 shows additional images and quantitation of EM images and Western blotting of ZO-1–GFP rescue cells. Fig. S4 shows the data of EpH4 cells, the lack of effects of myosin II inhibition on epithelial polarity phenotypes, and the genomic sequence of afadin KO and E-cadherin KO cells. Fig. S5 shows the genomic sequence and characterization of JAM-A KO and claudin/JAM-A KO cells.
Supplementary Material
Acknowledgments
We thank the Developmental Studies Hybridoma Bank, Masayuki Murata, Ernst Reichmann, Shigenobu Yonemura, Akira Nagafuchi, Junichi Ikenouchi, and Masatoshi Takeichi for kindly providing reagents; the EM facility in the National Institute for Physiological Sciences, Osamu Nagata, and Sei Saitoh for support in EM; Mika Watanabe and Yuichiro Kano for technical assistance; Motohiro Nishida for allowing us to use the microplate reader; Junichi Ikenouchi and Toyoshi Fujimoto for advice on lipid polarity; and Akira Nagafuchi, Shigenobu Yonemura, and all members of Furuse laboratory for discussions and comments. We also thank Alison Sherwin from Edanz Group for editing a draft of this manuscript.
This work was supported by a Japan Society for the Promotion of Science Grant-in-Aid for Challenging Exploratory Research (16K15226, M. Furuse), Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research (B) (26291043 and 18H02440, M. Furuse), a Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research (C) (18K06234, T. Otani), a Japan Society for the Promotion of Science Grant-in-Aid for Young Scientists (B) (16K18544, T. Otani), a Ministry of Education, Culture, Sports, Science and Technology/Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research on Innovative Areas (17H05627, T. Otani), the National Institute of Natural Science Program for Cross-Disciplinary Study (T. Otani), the Inamori Foundation (T. Otani), and the Takeda Science Foundation (M. Furuse and T. Otani).
The authors declare no competing financial interests.
Author contributions: T. Otani and M. Furuse designed the study. S. Tokuda, K. Ebnet, and M. Furuse contributed key materials. T. Otani performed and analyzed most of the experiments. T.P. Nguyen performed the immunofluorescence analyses of JAM-A KO and claudin/JAM-A KO cells. T. Sugawara and M. Furuse performed and analyzed experiments. K. Furuse performed the freeze-fracture replica EM analysis. K. Sugihara and T. Miura performed junction linearity quantitation. T. Otani and M. Furuse wrote the manuscript. All authors read and commented on the manuscript.
References
- Ahmed S.M., and Macara I.G.. 2017. The Par3 polarity protein is an exocyst receptor essential for mammary cell survival. Nat. Commun. 8:14867 10.1038/ncomms14867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sadi R., Khatib K., Guo S., Ye D., Youssef M., and Ma T.. 2011. Occludin regulates macromolecule flux across the intestinal epithelial tight junction barrier. Am. J. Physiol. Gastrointest. Liver Physiol. 300:G1054–G1064. 10.1152/ajpgi.00055.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J.M., and Van Itallie C.M.. 2009. Physiology and function of the tight junction. Cold Spring Harb. Perspect. Biol. 1:a002584 10.1101/cshperspect.a002584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balda M.S., Whitney J.A., Flores C., González S., Cereijido M., and Matter K.. 1996. Functional dissociation of paracellular permeability and transepithelial electrical resistance and disruption of the apical-basolateral intramembrane diffusion barrier by expression of a mutant tight junction membrane protein. J. Cell Biol. 134:1031–1049. 10.1083/jcb.134.4.1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belardi B., Son S., Vahey M.D., Wang J., Hou J., and Fletcher D.A.. 2018. Claudin-4 reconstituted in unilamellar vesicles is sufficient to form tight interfaces that partition membrane proteins. J. Cell Sci. 132:jcs221556 10.1242/jcs.221556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blose S.H., Meltzer D.I., and Feramisco J.R.. 1984. 10-nm filaments are induced to collapse in living cells microinjected with monoclonal and polyclonal antibodies against tubulin. J. Cell Biol. 98:847–858. 10.1083/jcb.98.3.847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschmann M.M., Shen L., Rajapakse H., Raleigh D.R., Wang Y., Wang Y., Lingaraju A., Zha J., Abbott E., McAuley E.M., et al. 2013. Occludin OCEL-domain interactions are required for maintenance and regulation of the tight junction barrier to macromolecular flux. Mol. Biol. Cell. 24:3056–3068. 10.1091/mbc.e12-09-0688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereijido M., Robbins E.S., Dolan W.J., Rotunno C.A., and Sabatini D.D.. 1978. Polarized monolayers formed by epithelial cells on a permeable and translucent support. J. Cell Biol. 77:853–880. 10.1083/jcb.77.3.853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereijido M., Ehrenfeld J., Meza I., and Martínez-Palomo A.. 1980. Structural and functional membrane polarity in cultured monolayers of MDCK cells. J. Membr. Biol. 52:147–159. 10.1007/BF01869120 [DOI] [PubMed] [Google Scholar]
- Cereijido M., Ponce A., and Gonzalez-Mariscal L.. 1989. Tight junctions and apical/basolateral polarity. J. Membr. Biol. 110:1–9. 10.1007/BF01870987 [DOI] [Google Scholar]
- Choi W., Acharya B.R., Peyret G., Fardin M.A., Mège R.M., Ladoux B., Yap A.S., Fanning A.S., and Peifer M.. 2016. Remodeling the zonula adherens in response to tension and the role of afadin in this response. J. Cell Biol. 213:243–260. 10.1083/jcb.201506115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Camilli P., Peluchetti D., and Meldolesi J.. 1974. Structural difference between luminal and lateral plasmalemma in pancreatic acinar cells. Nature. 248:245–247. 10.1038/248245b0 [DOI] [PubMed] [Google Scholar]
- Dragsten P.R., Blumenthal R., and Handler J.S.. 1981. Membrane asymmetry in epithelia: is the tight junction a barrier to diffusion in the plasma membrane? Nature. 294:718–722. 10.1038/294718a0 [DOI] [PubMed] [Google Scholar]
- Drubin D.G., and Nelson W.J.. 1996. Origins of cell polarity. Cell. 84:335–344. 10.1016/S0092-8674(00)81278-7 [DOI] [PubMed] [Google Scholar]
- Ebnet K., Suzuki A., Horikoshi Y., Hirose T., Meyer Zu Brickwedde M.K., Ohno S., and Vestweber D.. 2001. The cell polarity protein ASIP/PAR-3 directly associates with junctional adhesion molecule (JAM). EMBO J. 20:3738–3748. 10.1093/emboj/20.14.3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning A.S., Van Itallie C.M., and Anderson J.M.. 2012. Zonula occludens-1 and -2 regulate apical cell structure and the zonula adherens cytoskeleton in polarized epithelia. Mol. Biol. Cell. 23:577–590. 10.1091/mbc.e11-09-0791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar M.G., and Palade G.E.. 1963. Junctional complexes in various epithelia. J. Cell Biol. 17:375–412. 10.1083/jcb.17.2.375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M., Hirase T., Itoh M., Nagafuchi A., Yonemura S., Tsukita S., and Tsukita S.. 1993. Occludin: a novel integral membrane protein localizing at tight junctions. J. Cell Biol. 123:1777–1788. 10.1083/jcb.123.6.1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M., Fujita K., Hiiragi T., Fujimoto K., and Tsukita S.. 1998a Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J. Cell Biol. 141:1539–1550. 10.1083/jcb.141.7.1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M., Sasaki H., Fujimoto K., and Tsukita S.. 1998b A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J. Cell Biol. 143:391–401. 10.1083/jcb.143.2.391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M., Furuse K., Sasaki H., and Tsukita S.. 2001. Conversion of zonulae occludentes from tight to leaky strand type by introducing claudin-2 into Madin-Darby canine kidney I cells. J. Cell Biol. 153:263–272. 10.1083/jcb.153.2.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Yang Z., Hiremath C., Zimmerman S.E., Long B., Brakeman P.R., Mostov K.E., Bryant D.M., Luby-Phelps K., and Marciano D.K.. 2017. Afadin orients cell division to position the tubule lumen in developing renal tubules. Development. 144:3511–3520. 10.1242/dev.148908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimona M., Vandekerckhove J., Goethals M., Herzog M., Lando Z., and Small J.V.. 1994. Beta-actin specific monoclonal antibody. Cell Motil. Cytoskeleton. 27:108–116. 10.1002/cm.970270203 [DOI] [PubMed] [Google Scholar]
- Gumbiner B., and Simons K.. 1986. A functional assay for proteins involved in establishing an epithelial occluding barrier: identification of a uvomorulin-like polypeptide. J. Cell Biol. 102:457–468. 10.1083/jcb.102.2.457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günzel D., and Yu A.S.L.. 2013. Claudins and the modulation of tight junction permeability. Physiol. Rev. 93:525–569. 10.1152/physrev.00019.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T.J.C., and Peifer M.. 2004. Adherens junction-dependent and -independent steps in the establishment of epithelial cell polarity in Drosophila. J. Cell Biol. 167:135–147. 10.1083/jcb.200406024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoi Sang U., Saier M.H. Jr., and Ellisman M.H.. 1979. Tight junction formation is closely linked to the polar redistribution of intramembranous particles in aggregating MDCK epithelia. Exp. Cell Res. 122:384–391. 10.1016/0014-4827(79)90315-X [DOI] [PubMed] [Google Scholar]
- Hurd T.W., Gao L., Roh M.H., Macara I.G., and Margolis B.. 2003. Direct interaction of two polarity complexes implicated in epithelial tight junction assembly. Nat. Cell Biol. 5:137–142. 10.1038/ncb923 [DOI] [PubMed] [Google Scholar]
- Ichii T., and Takeichi M.. 2007. p120-catenin regulates microtubule dynamics and cell migration in a cadherin-independent manner. Genes Cells. 12:827–839. 10.1111/j.1365-2443.2007.01095.x [DOI] [PubMed] [Google Scholar]
- Iden S., Misselwitz S., Peddibhotla S.S.D., Tuncay H., Rehder D., Gerke V., Robenek H., Suzuki A., and Ebnet K.. 2012. aPKC phosphorylates JAM-A at Ser285 to promote cell contact maturation and tight junction formation. J. Cell Biol. 196:623–639. 10.1083/jcb.201104143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenouchi J., Umeda K., Tsukita S., Furuse M., and Tsukita S.. 2007. Requirement of ZO-1 for the formation of belt-like adherens junctions during epithelial cell polarization. J. Cell Biol. 176:779–786. 10.1083/jcb.200612080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenouchi J., Suzuki M., Umeda K., Ikeda K., Taguchi R., Kobayashi T., Sato S.B., Kobayashi T., Stolz D.B., and Umeda M.. 2012. Lipid polarity is maintained in absence of tight junctions. J. Biol. Chem. 287:9525–9533. 10.1074/jbc.M111.327064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiuchi T., and Takeichi M.. 2011. Willin and Par3 cooperatively regulate epithelial apical constriction through aPKC-mediated ROCK phosphorylation. Nat. Cell Biol. 13:860–866. 10.1038/ncb2274 [DOI] [PubMed] [Google Scholar]
- Ishiuchi T., and Takeichi M.. 2012. Nectins localize Willin to cell-cell junctions. Genes Cells. 17:387–397. 10.1111/j.1365-2443.2012.01593.x [DOI] [PubMed] [Google Scholar]
- Itoh M., Yonemura S., Nagafuchi A., Tsukita S., and Tsukita S.. 1991. A 220-kD undercoat-constitutive protein: its specific localization at cadherin-based cell-cell adhesion sites. J. Cell Biol. 115:1449–1462. 10.1083/jcb.115.5.1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M., Nagafuchi A., Yonemura S., Kitani-Yasuda T., Tsukita S., and Tsukita S.. 1993. The 220-kD protein colocalizing with cadherins in non-epithelial cells is identical to ZO-1, a tight junction-associated protein in epithelial cells: cDNA cloning and immunoelectron microscopy. J. Cell Biol. 121:491–502. 10.1083/jcb.121.3.491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M., Furuse M., Morita K., Kubota K., Saitou M., and Tsukita S.. 1999a Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J. Cell Biol. 147:1351–1363. 10.1083/jcb.147.6.1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M., Morita K., and Tsukita S.. 1999b Characterization of ZO-2 as a MAGUK family member associated with tight as well as adherens junctions with a binding affinity to occludin and α catenin. J. Biol. Chem. 274:5981–5986. 10.1074/jbc.274.9.5981 [DOI] [PubMed] [Google Scholar]
- Itoh M., Sasaki H., Furuse M., Ozaki H., Kita T., and Tsukita S.. 2001. Junctional adhesion molecule (JAM) binds to PAR-3: a possible mechanism for the recruitment of PAR-3 to tight junctions. J. Cell Biol. 154:491–497. 10.1083/jcb.200103047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M., Tsukita S., Yamazaki Y., and Sugimoto H.. 2012. Rho GTP exchange factor ARHGEF11 regulates the integrity of epithelial junctions by connecting ZO-1 and RhoA-myosin II signaling. Proc. Natl. Acad. Sci. USA. 109:9905–9910. 10.1073/pnas.1115063109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi Y., Hirose T., Tamai Y., Hirai S., Nagashima Y., Fujimoto T., Tabuse Y., Kemphues K.J., and Ohno S.. 1998. An atypical PKC directly associates and colocalizes at the epithelial tight junction with ASIP, a mammalian homologue of Caenorhabditis elegans polarity protein PAR-3. J. Cell Biol. 143:95–106. 10.1083/jcb.143.1.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug S.M., Amasheh S., Richter J.F., Milatz S., Günzel D., Westphal J.K., Huber O., Schulzke J.D., and Fromm M.. 2009. Tricellulin forms a barrier to macromolecules in tricellular tight junctions without affecting ion permeability. Mol. Biol. Cell. 20:3713–3724. 10.1091/mbc.e09-01-0080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laukoetter M.G., Nava P., Lee W.Y., Severson E.A., Capaldo C.T., Babbin B.A., Williams I.R., Koval M., Peatman E., Campbell J.A., et al. 2007. JAM-A regulates permeability and inflammation in the intestine in vivo. J. Exp. Med. 204:3067–3076. 10.1084/jem.20071416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martìn-Padura I., Lostaglio S., Schneemann M., Williams L., Romano M., Fruscella P., Panzeri C., Stoppacciaro A., Ruco L., Villa A., et al. 1998. Junctional adhesion molecule, a novel member of the immunoglobulin superfamily that distributes at intercellular junctions and modulates monocyte transmigration. J. Cell Biol. 142:117–127. 10.1083/jcb.142.1.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill H., Ozawa M., Kemler R., and Nelson W.J.. 1990. Novel function of the cell adhesion molecule uvomorulin as an inducer of cell surface polarity. Cell. 62:309–316. 10.1016/0092-8674(90)90368-O [DOI] [PubMed] [Google Scholar]
- Monteleon C.L., and D’Souza-Schorey C.. 2012. Modeling disease using three dimensional cell culture: multi-lumen and inverted cyst phenotypes. Front. Biosci. (Elite Ed.). 4:2764–2771. [DOI] [PubMed] [Google Scholar]
- Nagafuchi A., and Tsukita S.. 1994. The Loss of the Expression of α Catenin, the 102 kD Cadherin Associated Protein, in Central Nervous Tissues during Development. Dev. Growth Differ. 36:59–71. 10.1111/j.1440-169X.1994.00059.x [DOI] [PubMed] [Google Scholar]
- Nishimura T., Ito S., Saito H., Hiver S., Shigetomi K., Ikenouchi J., and Takeichi M.. 2016. DAAM1 stabilizes epithelial junctions by restraining WAVE complex-dependent lateral membrane motility. J. Cell Biol. 215:559–573. 10.1083/jcb.201603107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odenwald M.A., Choi W., Buckley A., Shashikanth N., Joseph N.E., Wang Y., Warren M.H., Buschmann M.M., Pavlyuk R., Hildebrand J., et al. 2017. ZO-1 interactions with F-actin and occludin direct epithelial polarization and single lumen specification in 3D culture. J. Cell Sci. 130:243–259. 10.1242/jcs.188185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odenwald M.A., Choi W., Kuo W.T., Singh G., Sailer A., Wang Y., Shen L., Fanning A.S., and Turner J.R.. 2018. The scaffolding protein ZO-1 coordinates actomyosin and epithelial apical specializations in vitro and in vivo. J. Biol. Chem. 293:17317–17335. 10.1074/jbc.RA118.003908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojakian G.K., and Schwimmer R.. 1988. The polarized distribution of an apical cell surface glycoprotein is maintained by interactions with the cytoskeleton of Madin-Darby canine kidney cells. J. Cell Biol. 107:2377–2387. 10.1083/jcb.107.6.2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani T., Ichii T., Aono S., and Takeichi M.. 2006. Cdc42 GEF Tuba regulates the junctional configuration of simple epithelial cells. J. Cell Biol. 175:135–146. 10.1083/jcb.200605012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phua D.C.Y., Xu J., Ali S.M., Boey A., Gounko N.V., and Hunziker W.. 2014. ZO-1 and ZO-2 are required for extra-embryonic endoderm integrity, primitive ectoderm survival and normal cavitation in embryoid bodies derived from mouse embryonic stem cells. PLoS One. 10.1371/journal.pone.0099532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrini G., Matteoli M., Banker G., and Caplan M.J.. 1992. Isoforms of the Na,K-ATPase are present in both axons and dendrites of hippocampal neurons in culture. Proc. Natl. Acad. Sci. USA. 89:8414–8418. 10.1073/pnas.89.18.8414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehder D., Iden S., Nasdala I., Wegener J., Brickwedde M.K.M.Z., Vestweber D., and Ebnet K.. 2006. Junctional adhesion molecule-a participates in the formation of apico-basal polarity through different domains. Exp. Cell Res. 312:3389–3403. 10.1016/j.yexcr.2006.07.004 [DOI] [PubMed] [Google Scholar]
- Reichmann E., Schwarz H., Deiner E.M., Leitner I., Eilers M., Berger J., Busslinger M., and Beug H.. 1992. Activation of an inducible c-FosER fusion protein causes loss of epithelial polarity and triggers epithelial-fibroblastoid cell conversion. Cell. 71:1103–1116. 10.1016/S0092-8674(05)80060-1 [DOI] [PubMed] [Google Scholar]
- Roh M.H., Makarova O., Liu C.J., Shin K., Lee S., Laurinec S., Goyal M., Wiggins R., and Margolis B.. 2002. The Maguk protein, Pals1, functions as an adapter, linking mammalian homologues of Crumbs and Discs Lost. J. Cell Biol. 157:161–172. 10.1083/jcb.200109010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M., Ando-Akatsuka Y., Itoh M., Furuse M., Inazawa J., Fujimoto K., and Tsukita S.. 1997. Mammalian occludin in epithelial cells: its expression and subcellular distribution. Eur. J. Cell Biol. 73:222–231. [PubMed] [Google Scholar]
- Sato T. 1968. A modified method for lead staining of thin sections. J. Electron Microsc. (Tokyo). 17:158–159. [PubMed] [Google Scholar]
- Shen L., Weber C.R., Raleigh D.R., Yu D., and Turner J.R.. 2011. Tight junction pore and leak pathways: a dynamic duo. Annu. Rev. Physiol. 73:283–309. 10.1146/annurev-physiol-012110-142150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayoshi Y., Nose A., Iwasaki K., and Takeichi M.. 1986. N-linked oligosaccharides are not involved in the function of a cell-cell binding glycoprotein E-cadherin. Cell Struct. Funct. 11:245–252. 10.1247/csf.11.245 [DOI] [PubMed] [Google Scholar]
- Shukla P., Vogl C., Wallner B., Rigler D., Müller M., and Macho-Maschler S.. 2015. High-throughput mRNA and miRNA profiling of epithelial-mesenchymal transition in MDCK cells. BMC Genomics. 16:944 10.1186/s12864-015-2036-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehelin L.A. 1973. Further observations on the fine structure of freeze-cleaved tight junctions. J. Cell Sci. 13:763–786. [DOI] [PubMed] [Google Scholar]
- Straight A.F., Cheung A., Limouze J., Chen I., Westwood N.J., Sellers J.R., and Mitchison T.J.. 2003. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 299:1743–1747. 10.1126/science.1081412 [DOI] [PubMed] [Google Scholar]
- Suzuki A., Yamanaka T., Hirose T., Manabe N., Mizuno K., Shimizu M., Akimoto K., Izumi Y., Ohnishi T., and Ohno S.. 2001. Atypical protein kinase C is involved in the evolutionarily conserved par protein complex and plays a critical role in establishing epithelia-specific junctional structures. J. Cell Biol. 152:1183–1196. 10.1083/jcb.152.6.1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K., Sato N., Kasahara H., Funayama N., Nagafuchi A., Yonemura S., Tsukita S., and Tsukita S.. 1994. Perturbation of cell adhesion and microvilli formation by antisense oligonucleotides to ERM family members. J. Cell Biol. 125:1371–1384. 10.1083/jcb.125.6.1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuda S., and Furuse M.. 2015. Claudin-2 knockout by TALEN-mediated gene targeting in MDCK cells: claudin-2 independently determines the leaky property of tight junctions in MDCK cells. PLoS One. 10:e0119869 10.1371/journal.pone.0119869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuda S., Higashi T., and Furuse M.. 2014. ZO-1 knockout by TALEN-mediated gene targeting in MDCK cells: involvement of ZO-1 in the regulation of cytoskeleton and cell shape. PLoS One. 9:e104994 10.1371/journal.pone.0104994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuda S., Hirai T., and Furuse M.. 2017. Claudin-4 knockout by TALEN-mediated gene targeting in MDCK cells: Claudin-4 is dispensable for the permeability properties of tight junctions in wild-type MDCK cells. PLoS One. 12:e0182521 10.1371/journal.pone.0182521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuncay H., Brinkmann B.F., Steinbacher T., Schürmann A., Gerke V., Iden S., and Ebnet K.. 2015. JAM-A regulates cortical dynein localization through Cdc42 to control planar spindle orientation during mitosis. Nat. Commun. 6:8128 10.1038/ncomms9128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda K., Ikenouchi J., Katahira-Tayama S., Furuse K., Sasaki H., Nakayama M., Matsui T., Tsukita S., Furuse M., and Tsukita S.. 2006. ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell. 126:741–754. 10.1016/j.cell.2006.06.043 [DOI] [PubMed] [Google Scholar]
- Van Itallie C.M., and Anderson J.M.. 2006. Claudins and epithelial paracellular transport. Annu. Rev. Physiol. 68:403–429. 10.1146/annurev.physiol.68.040104.131404 [DOI] [PubMed] [Google Scholar]
- Van Itallie C.M., Fanning A.S., Bridges A., and Anderson J.M.. 2009. ZO-1 stabilizes the tight junction solute barrier through coupling to the perijunctional cytoskeleton. Mol. Biol. Cell. 20:3930–3940. 10.1091/mbc.e09-04-0320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer G., and Simons K.. 1986. The function of tight junctions in maintaining differences in lipid composition between the apical and the basolateral cell surface domains of MDCK cells. EMBO J. 5:1455–1464. 10.1002/j.1460-2075.1986.tb04382.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton J. 1979. Lead aspartate, an en bloc contrast stain particularly useful for ultrastructural enzymology. J. Histochem. Cytochem. 27:1337–1342. 10.1177/27.10.512319 [DOI] [PubMed] [Google Scholar]
- Yamazaki Y., Umeda K., Wada M., Nada S., Okada M., Tsukita S., and Tsukita S.. 2008. ZO-1- and ZO-2-dependent integration of myosin-2 to epithelial zonula adherens. Mol. Biol. Cell. 19:3801–3811. 10.1091/mbc.e08-04-0352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S. 2011. Cadherin-actin interactions at adherens junctions. Curr. Opin. Cell Biol. 23:515–522. 10.1016/j.ceb.2011.07.001 [DOI] [PubMed] [Google Scholar]
- Yonemura S., Wada Y., Watanabe T., Nagafuchi A., and Shibata M.. 2010. α-Catenin as a tension transducer that induces adherens junction development. Nat. Cell Biol. 12:533–542. 10.1038/ncb2055 [DOI] [PubMed] [Google Scholar]
- Zihni C., Mills C., Matter K., and Balda M.S.. 2016. Tight junctions: from simple barriers to multifunctional molecular gates. Nat. Rev. Mol. Cell Biol. 17:564–580. 10.1038/nrm.2016.80 [DOI] [PubMed] [Google Scholar]
- Zihni C., Vlassaks E., Terry S., Carlton J., Leung T.K.C., Olson M., Pichaud F., Balda M.S., and Matter K.. 2017. An apical MRCK-driven morphogenetic pathway controls epithelial polarity. Nat. Cell Biol. 19:1049–1060. 10.1038/ncb3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkl G.M., Zuk A., van der Bijl P., van Meer G., and Matlin K.S.. 1996. An antiglycolipid antibody inhibits Madin-Darby canine kidney cell adhesion to laminin and interferes with basolateral polarization and tight junction formation. J. Cell Biol. 133:695–708. 10.1083/jcb.133.3.695 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.