Lo and colleagues show that CEP83 is a TTBK2 substrate and further demonstrate that TTBK2-dependent CEP83 phosphorylation is important for the early stages of ciliogenesis, including ciliary vesicle docking and CP110 removal.
Abstract
Primary cilia are microtubule-based organelles that play important roles in development and tissue homeostasis. Tau-tubulin kinase-2 (TTBK2) is genetically linked to spinocerebellar ataxia type 11, and its kinase activity is crucial for ciliogenesis. Although it has been shown that TTBK2 is recruited to the centriole by distal appendage protein CEP164, little is known about TTBK2 substrates associated with its role in ciliogenesis. Here, we perform superresolution microscopy and discover that serum starvation results in TTBK2 redistribution from the periphery toward the root of distal appendages. Our biochemical analyses uncover CEP83 as a bona fide TTBK2 substrate with four phosphorylation sites characterized. We also demonstrate that CEP164-dependent TTBK2 recruitment to distal appendages is required for subsequent CEP83 phosphorylation. Specifically, TTBK2-dependent CEP83 phosphorylation is important for early ciliogenesis steps, including ciliary vesicle docking and CP110 removal. In summary, our results reveal a molecular mechanism of kinase regulation in ciliogenesis and identify CEP83 as a key substrate of TTBK2 during cilia initiation.
Introduction
The primary cilium is a membrane-bound structure with a microtubule-based core called the axoneme and is present in the majority of cells in our body. In many types of cells, the primary cilium serves as a sensory hub for the transduction of extracellular signals to promote intracellular responses, playing important regulatory roles during proliferation and tissue homeostasis (Bisgrove and Yost, 2006; Fliegauf et al., 2007; Gerdes et al., 2009; Singla et al., 2010). Many genetic disorders linked to dysfunctional cilia that are classified as ciliopathies reveal the importance of this structure (Reiter and Leroux, 2017).
The primary cilium originates from the centriole distal end. To faithfully execute cilia formation, the centriole is modified with distal and subdistal appendages that project radially at its distal end. Ciliogenesis is tightly coupled to the cell cycle and occurs when cells are in G0 or early G1 phase. It follows a series of highly ordered steps (Sorokin, 1962; Gilula and Satir, 1972; Carvalho-Santos et al., 2011). In the initiating stage, the distal end of the centriole docks to Golgi-derived membrane vesicles. Images from transmission electron microscopy suggest that this docking process is mediated by a pinwheel-like structure named the centriole distal appendage (DA; Schmidt et al., 2012; Tanos et al., 2013). After the centriole docks to membrane vesicles, a short axoneme bud extends from the centriole distal end that promotes the assembly of the transition zone, which is a specialized structure that forms a gate at the ciliary base. Once the transition zone has been established, a fully mature axoneme is then developed and supported by the intraflagellar transport machinery (Pedersen and Rosenbaum, 2008; Ishikawa and Marshall, 2011). More and more factors involved in ciliogenesis have now been identified, including proteins that are recruited to the cilium base during ciliogenesis, as well as proteins that locate at the centriole distal end (Sánchez and Dynlacht, 2016; Ishikawa and Marshall, 2017).
The importance of DAs in ciliogenesis is well known, since depletion of proteins at DAs affect ciliogenesis (Schmidt et al., 2012; Joo et al., 2013; Tanos et al., 2013; Lu et al., 2015; Kurtulmus et al., 2018). Although it has been proven that DAs mediate membrane docking (Tanos et al., 2013), the molecular regulation remains unclear. Several DA proteins (DAPs) have been identified so far, including CEP164, SCLT1, CCDC41/CEP83, CCDC123/CEP89, FBF1, ANKRD26, tau-tubulin kinase-2 (TTBK2), and LRRC45 (Graser et al., 2007; Schmidt et al., 2012; Joo et al., 2013; Tanos et al., 2013; Ye et al., 2014; Lu et al., 2015; Kurtulmus et al., 2018; Bowler et al., 2019). Their recruitment to the mother centriole is known to be hierarchical. CEP83 is first recruited to centrioles by C2CD3 and is required for the recruitment of SCLT1 and CEP89 (Tanos et al., 2013; Ye et al., 2014). SCLT1 is necessary for the subsequent recruitment of CEP164 and LRRC45. FBF1 is recruited to DAs by LRRC45. Using direct stochastic optical reconstruction microscopy (dSTORM), CEP83, CEP89, SCLT1, and CEP164 have been shown to form the backbone of DAs. CEP83 is located at the root of the pinwheel-like structure, and CEP164 is at the periphery (Yang et al., 2018).
TTBK2 is a serine/threonine protein kinase originally identified as a microtubule-associated protein phosphorylating tau and tubulin (Takahashi et al., 1995; Liachko et al., 2014). Mutations in the TTBK2 gene that cause TTBK2 C-terminal truncations are associated with spinocerebellar ataxia type 11, which is a neurodegenerative disease characterized by progressive ataxia and cerebellum degeneration (Houlden et al., 2007; Edener et al., 2009). In addition, TTBK2 is also known to be a crucial factor in initiating ciliogenesis (Goetz et al., 2012). Previous studies have demonstrated that TTBK2 controls the removal of CP110 from the mother centriole, a process that relies on the kinase activity of TTBK2 (Goetz et al., 2012), relieving its negative effect on ciliogenesis. The recruitment of TTBK2 to mother centrioles is also important for TTBK2’s role in ciliogenesis. Biochemical analyses demonstrate that CEP164 via its N-terminal domain interacts with the C-terminal region of TTBK2 that brings TTBK2 to the DAs and then promotes cilia formation (Čajánek and Nigg, 2014; Oda et al., 2014). Recent studies have shown that TTBK2 phosphorylation of M-phase phosphoprotein 9 (MPP9) at the centriole distal end is important to initiate ciliogenesis (Huang et al., 2018). The presence of MPP9 at the centriole distal end recruits the CP110–CEP97 complex that blocks cilia initiation. When TTBK2 is accumulated at the basal body, the TTBK2-dependent MPP9 phosphorylation promotes MPP9 degradation through the ubiquitin-dependent proteasome pathway. The degradation of MPP9 then facilitates CP110 removal and subsequent cilia formation.
In this study, we provide a new narrative regarding ciliogenesis supported by the interplay between TTBK2 and CEP83 at DAs. We identify CEP83 as a novel substrate of TTBK2 and show that the TTBK2-dependent phosphorylation of CEP83 plays important roles at the initial stages of ciliogenesis, including the removal of CP110 from the centriole distal end and the process of ciliary vesicle docking.
Results
TTBK2 is enriched toward the axoneme upon serum starvation
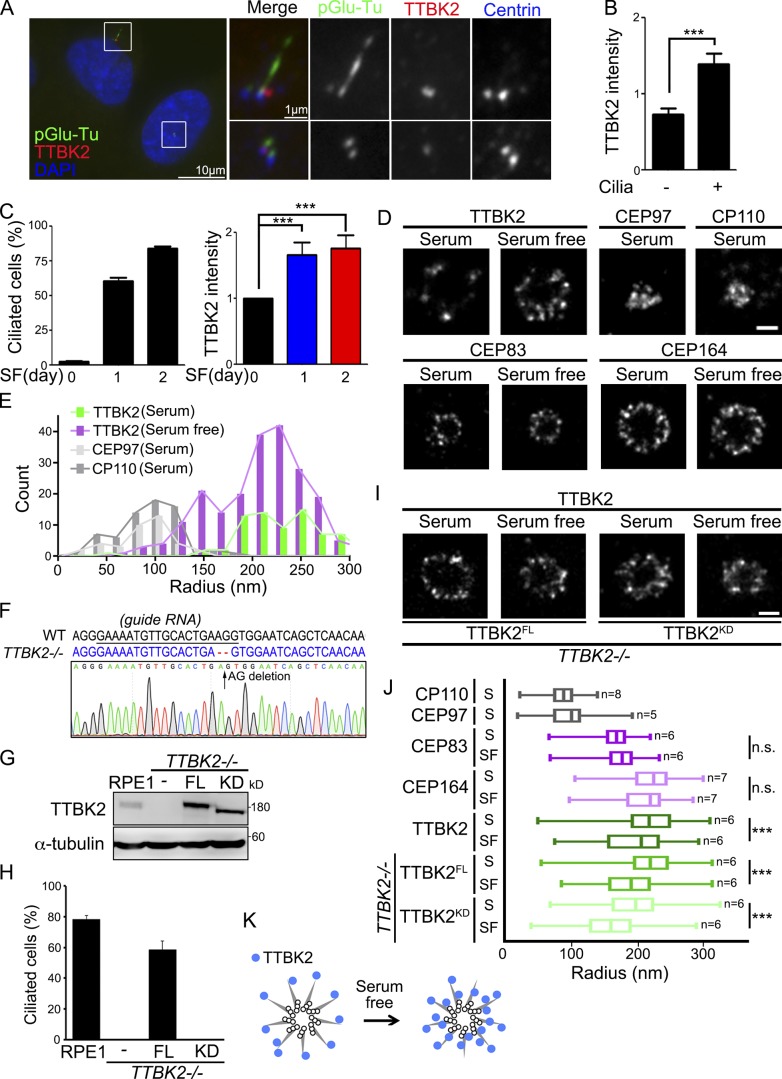
TTBK2 plays a critical role in initiating cilia formation. However, where exactly TTBK2 is localized to regulate ciliary initiation remains largely unclear. By examining endogenous TTBK2 localization at the centrioles in unsynchronized cells, we observed that TTBK2 was more enriched at the centrioles in ciliated cells than in nonciliated cells (Fig. 1, A and B). In addition, TTBK2 was more populated at the centrioles when cells were induced to form cilia by serum starvation (Fig. 1 C). The dSTORM was further used to examine the localization of TTBK2 at the centrioles in detail during ciliogenesis. The axial view of dSTORM images revealed that TTBK2 signal was sporadically scattered at the distal portion of mother centrioles in proliferating cells (Fig. 1 D). In contrast, TTBK2 redistributed and formed a smaller ring around the mother centrioles upon serum starvation (Fig. 1 D; Yang et al., 2018). By comparing with the radial localization of proteins at the centriole distal end, we found that serum starvation shifted the population of TTBK2 signal toward the axoneme closer to CP110 and CEP97 (Fig. 1, D and E). The distribution of CEP83 and CEP164 during ciliogenesis was also analyzed. The dSTORM images showed that the distributions of CEP164 and CEP83 at DAs were not altered upon serum withdrawal, indicating that the structure of DAs was not changed (Fig. 1, D and I). This also suggests that the redistribution of TTBK2 upon serum starvation is not caused by a change in DA structure. TTBK2 knockout cells were then generated to further examine the influence of TTBK2 kinase activity in its redistribution during ciliogenesis (Fig. 1 F). The requirement of TTBK2 kinase activity in cilia formation was confirmed by reexpressing full-length TTBK2 (TTBK2FL) or TTBK2 catalytically inactive mutant (TTBK2KD) in TTBK2 knockout cells (Fig. 1, G and H). The dSTORM images showed that both TTBK2FL and TTBK2KD formed smaller rings around the mother centrioles upon serum starvation, revealing that TTBK2 kinase activity did not affect TTBK2 redistribution at DAs during ciliogenesis (Fig. 1, I and J). The changes of TTBK2 distribution upon serum starvation also suggested that TTBK2 redistribution might be related to its function associated with ciliogenesis (Fig. 1 K).
Figure 1.
TTBK2 is enriched toward axoneme during ciliogenesis. (A) RPE1 cells were stained with TTBK2 (red), pGlu-Tu (green), and centrin (blue) antibodies. DNA was stained with DAPI (blue). (B) TTBK2 intensity at the centrioles was quantified in ciliated and nonciliated cells. (C) Cells were serum starved (SF) for a different number of days. The percentage of ciliated cells and TTBK2 intensity at the centrioles were quantified. (D) Cells were serum starved for 2 d. The dSTORM images represent the distribution of proteins in radial view at the centrioles in proliferating and serum-starved cells. Scale bar, 200 nm. (E) Histogram analysis of TTBK2, CP110, and CEP97 showing that serum starvation shifted the population of TTBK2 toward the center of centrioles close to CP110 and CEP97. Results are presented for 13, 7, 5, and 8 centrioles for TTBK2 (serum), TTBK2 (serum free), CEP97 (serum), and CP110 (serum), respectively. (F) Sequences analysis of TTBK2 alleles in WT and TTBK2−/− cells. The sequence of WT and edited alleles is in black and blue respectively. The dashed lines represent lesions (AG deletion). (G) FH-TTBK2FL and FH-TTBK2KD were stably expressed in TTBK2−/− cells. Western blot analysis was performed with antibodies against TTBK2 and α-tubulin. (H) Cells were serum starved for 2 d, and the percentage of ciliated cells was quantified. In B, C, and H, >150 cells were analyzed for each independent experiment. Error bars represent mean ± SEM; n = 3. ***, P < 0.001 (Student’s t test). (I) The dSTORM images represent the distribution of TTBK2FL and TTBK2KD in radial view at the centrioles in proliferating cells and serum-starved cells. Scale bar, 200 nm. (J) Quantification of diameters with proteins at the centrioles from D and I. Total cell counts are shown in the figure as n. Error bars represent SD. n.s., not significant. ***, P < 0.001 (Student’s t test). (K) The diagram shows the TTBK2 distribution at centriole DAs during serum starvation.
CEP83 is phosphorylated by TTBK2
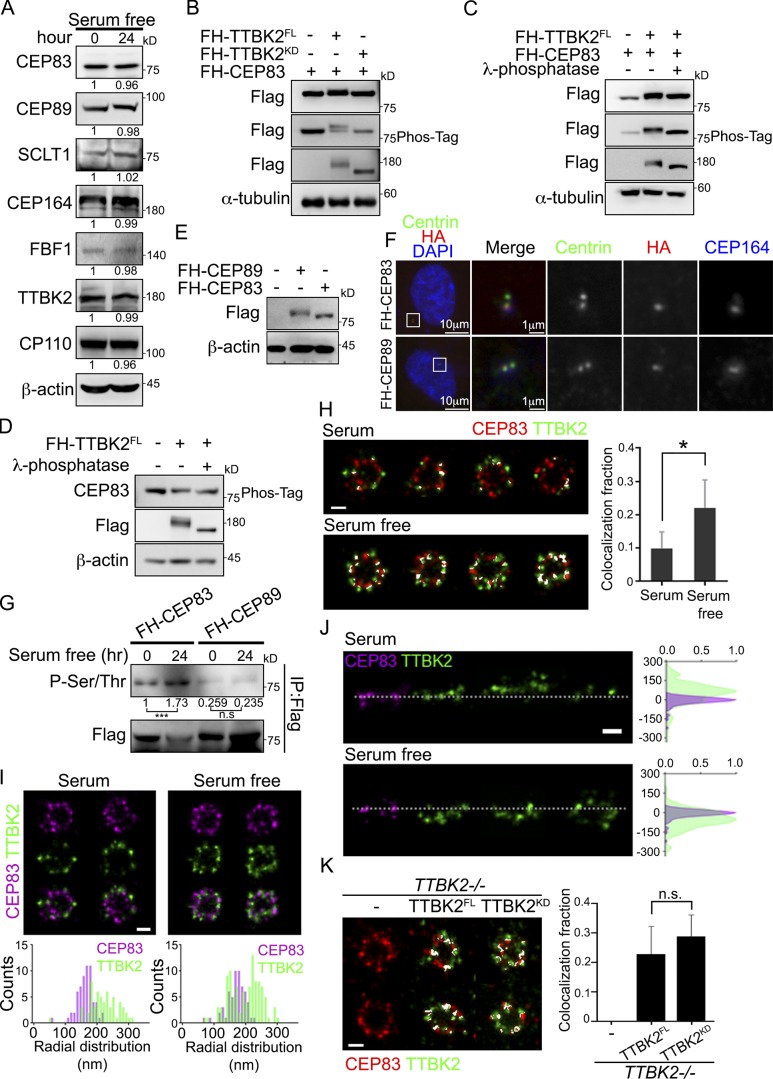
To identify proteins that might respond to TTBK2 redistribution at the onset of ciliogenesis, we first analyzed the expression of proteins at centriole distal end upon serum starvation. Our results indicated that serum starvation did not affect the amount of centriolar proteins (Fig. 2 A). We then checked whether TTBK2 redistribution promoted protein phosphorylation at DAs. A phosphorylation-induced gel mobility shift assay was performed to identify candidate substrates of TTBK2. CEP83 showed an upward gel-mobility shift on both reducing gel and Phos-tag gel when TTBK2FL instead of TTBK2KD was coexpressed (Fig. 2 B). Furthermore, this TTBK2-dependent upward gel-mobility shift of CEP83 was not seen when cell lysate was treated with alkaline phosphatase before gel analysis (Fig. 2, C and D). In addition to CEP83, we also observed upward electrophoretic-mobility shifts of CEP89 and CEP164 (Fig. S1). To investigate whether CEP83 phosphorylation correlated with ciliogenesis, cells were serum deprived to induce cilia formation and CEP83 phosphorylation was examined with an antibody against phosphorylated serine and threonine residues (Fig. 2, E–G). Our results showed that CEP83 phosphorylation dramatically increased during ciliogenesis (Fig. 2 G). The dSTORM images also showed that more colocalization of TTBK2 and CEP83 in serum-starved cells than in proliferating cells (Fig. 2, H and I). In addition, both TTBK2FL and TTBK2KD expressing cells showed similar localization patterns to TTBK2 and CEP83 signals upon serum withdrawal, indicating that the redistribution of TTBK2 to the vicinity of CEP83 did not depend on TTBK2 kinase activity. In brief, our results suggest that CEP83 is a potential substrate of TTBK2 associated with its function in ciliogenesis.
Figure 2.
CEP83 undergoes TTBK2-dependent phosphorylation. (A) Western blot analyses of proteins at the centriole distal end in proliferating and serum-starved cells with the indicated antibodies. Relative amounts of proteins were calculated and normalized to β-actin. (B) 293T cells ectopically expressed FH-TTBK2FL or FH-TTBK2KD together with FH-CEP83. The phosphorylation-induced mobility shift was checked using the reducing and Phos-tag gels followed by Western blot analysis. (C and D) Lysates of 293T cells transfected as indicated were incubated with DMSO (left and middle panel) or alkaline phosphatase (right panel) at 37°C for 1 h. The phosphorylation-induced mobility shift was checked using the reducing and Phos-tag gels followed by Western blot analysis. (E and F) FH-CEP83 or FH-CEP89 is stably expressed in RPE1 cells. Protein expression was confirmed by Western blots (E). Cells were stained with centrin (green), HA (red), and CEP164 (blue) to confirm the localization of CEP83 and CEP89 (F). DNA was stained with DAPI (blue). (G) RPE1 cells stably expressing FH-CEP83 or FH-CEP89 were serum starved for 24 h, and immunoprecipitation (IP) was performed to purify FH-CEP83 or FH-CEP89 using M2 beads. The pulled down complex was immunoblotted with anti-phospho-(Ser/Thr) antibody, and gel bands were quantified. ***, P < 0.001 (Student’s t test). (H) The dSTORM images of TTBK2 and CEP83 in proliferating (serum, n = 6) and serum-starved (serum free, n = 7) cells. Cells were processed for immunofluorescence with anti-TTBK2 and CEP83. The colocalization fraction of TTBK2 and CEP83 is quantified on the right. Error bars represent mean ± SD. *, P < 0.05 (Student’s t test). (I) The radial-view dSTORM images show the distribution of TTBK2 and CEP83 at the centrioles in proliferating and serum-starved cells. (J) The two-color axial-view dSTORM images for TTBK2 and CEP83 illustrate their relative longitudinal positions. (K) Cells were serum starved for 2 d, and the radial-view dSTORM images show the distribution of CEP83 with TTBK2FL or TTBK2KD at centrioles. The results are presented for seven, six, and six centrioles in control, TTBK2FL-, and TTBK2KD-expressing cells, respectively. The colocalization fraction of TTBK2 and CEP83 in each condition is quantified on the right. Error bars represent SD. n.s., not significant. Scale bars, 200 nm (H–K).
TTBK2 phosphorylates CEP83 both in vivo and in vitro
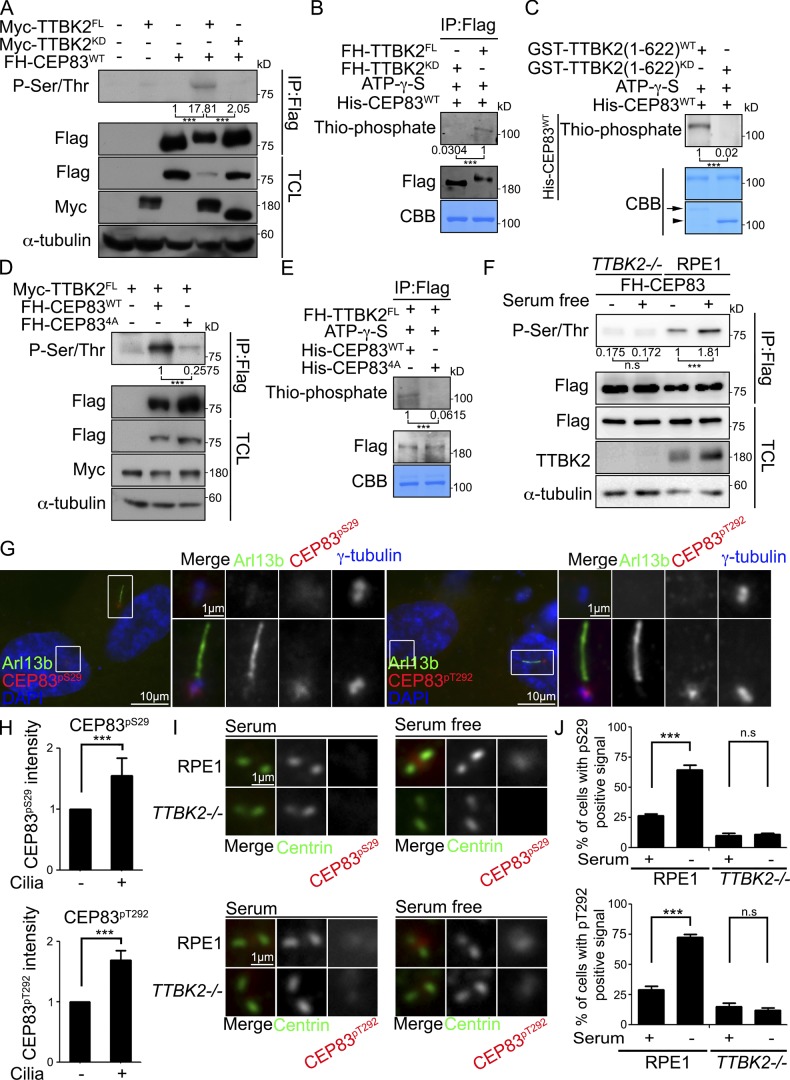
To explore the kinase–substrate relationship between TTBK2 and CEP83, CEP83 was ectopically expressed in 293T cells and immunoprecipitated from cell lysates. According to our results, coexpression of TTBK2, but not its catalytically inactive mutant, led to a significant increase of CEP83 serine and threonine phosphorylation (Fig. 3 A). To examine whether TTBK2 could directly phosphorylate CEP83, we performed in vitro kinase assays using purified TTBK2 either from 293T cells or from bacteria and recombinant CEP83 purified from bacteria as the substrate. TTBK2 was able to phosphorylate CEP83 in the in vitro kinase assay, suggesting that CEP83 is a direct substrate of TTBK2 (Fig. 3, B and C).
Figure 3.
TTBK2 phosphorylates CEP83. (A) TTBK2 coexpression promoted CEP83 phosphorylation in 293T cells was detected by immunoprecipitation followed by Western blots with anti-phospho-(Ser/Thr) antibody. (B) FH-TTBK2FL or FH-TTBK2KD was expressed and purified from 293T cells. The TTBK2 in vitro kinase assay was performed with TTBK2 purified from 293T cells and recombinant His-CEP83 purified from bacteria in the presence of ATP-γ-S for 30 min at RT. CEP83 phosphorylation was revealed by Western blot using an antibody against thiophosphate ester. CBB staining indicated the equal amount of His-CEP83WT. (C) Recombinant GST-TTBK2 (1–622)WT, TTBK2(1–622)KD, and His-CEP83WT were expressed and purified from bacteria to perform the TTBK2 in vitro kinase assay. CEP83 phosphorylation was revealed by Western blot using anti-thiophosphate ester antibody. CBB staining indicated the purified GST-TTBK2 (1–622)WT (arrow), GST-TTBK2 (1–622)KD (arrowhead), and His-CEP83WT. (D) FH-CEP83WT or FH-CEP834A was coexpressed with Myc-TTBK2 in 293T cells. FH-CEP83WT or FH-CEP834A phosphorylation was detected by immunoprecipitation (IP) followed by Western blots with anti-phospho-(Ser/Thr) antibody. (E) TTBK2 was purified from 293T cells and incubated with purified CEP83WT or CEP834A from bacteria to perform the TTBK2 in vitro kinase assay. CEP83 phosphorylation was detected by Western blot using an anti-thiophosphate ester antibody. CBB staining indicated the His-tagged CEP83WT and CEP83WT. (F) FH-CEP83 was stably expressed in WT and TTBK2−/− cells. Cells were serum starved for 24 h, and CEP83 phosphorylation was detected by immunoprecipitation followed by Western blots with an anti-phospho-(Ser/Thr) antibody. All of Western blots (A–F) are representative blots from at least three independent experiments. Gel bands were quantified. n.s., not significant. ***, P < 0.001 (Student’s t test). TCL, total cell lysates. (G) RPE1 cells were stained with antibodies against Arl13b, phospho-CEP83Ser29 (CEP83pS29), and phospho-CEP83Thr292 (CEP83pT292). (H) Quantification of phosphorylated CEP83Ser29 and CEP83Thr292 signals at the centrioles in ciliated and nonciliated cells. (I) WT and TTBK2−/− RPE1 cells were serum starved for 2 d and stained with antibodies against centrin, phospho-CEP83Ser29, and phospho-CEP83Thr292. Scale bar, 1 µm. (J) The phosphorylated CEP83Ser29 and CEP83Thr292 signals at the centrioles were quantified. In H and J, 200 cells were analyzed for each independent experiment. Error bars represent mean ± SEM; n = 3. n.s., not significant. ***, P < 0.001 (Student’s t test).
TTBK2 phosphorylates CEP83 at Ser29, Thr292, Thr527, and Ser698
We next attempted to identify the sites on CEP83 that were phosphorylated by TTBK2. CEP83 was ectopically expressed in 293T cells with or without coexpression of TTBK2 followed by CEP83 immunoprecipitation and tandem mass spectrometry (MS/MS) analysis. Our results showed that TTBK2 phosphorylated CEP83 at Ser29, Thr292, Thr527, and Ser698 (Fig. S2, A and B). Notably, all those four peptides were only present in TTBK2 coexpressing samples, suggesting the phosphorylation levels of CEP83 in proliferating cells were low. A phospho-inactive mutant of CEP83 (CEP834A) was then generated to examine the effect of TTBK2 by mutating CEP83 residues Ser29, Thr292, Thr527, and Ser698 to alanine. Coexpression of TTBK2 in 293T cells was unable to promote CEP834A phosphorylation (Fig. 3 D). Finally, an in vitro kinase assay showed that TTBK2 was unable to phosphorylate CEP834A, further supporting the role of TTBK2 in CEP83 phosphorylation (Fig. 3 E). Together, we uncover CEP83 as a TTBK2 substrate with four phosphorylation sites characterized.
TTBK2 mediates serum starvation–induced CEP83 phosphorylation
To examine whether TTBK2 promoted CEP83 phosphorylation during ciliogenesis, CEP83 was stably expressed in WT and TTBK2 knockout cells. CEP83 phosphorylation was then analyzed in proliferating and serum-starved cells. By comparison with WT cells, CEP83 phosphorylation was not induced by serum starvation in TTBK2 knockout cells, indicating that CEP83 phosphorylation was mediated by TTBK2 (Fig. 3 F). The overall changes in endogenous CEP83 phosphorylation were analyzed by 2D gel electrophoresis. In serum-starved cells, CEP83 was resolved by its charge into two main spots that were consistent with the theoretical isoelectric point and molecular weight of CEP83 (Fig. S2 C). However, a right-shifting spot was observed in TTBK2 knockout cells or in cells treated with alkaline phosphatase before gel analysis, suggesting that TTBK2 mediated the serum starvation–induced CEP83 phosphorylation (Fig. S2 C). Two phospho-CEP83 antibodies (anti-phospho-CEP83Ser29 and anti-phospho-CEP83Thr292) were generated to further analyze endogenous CEP83 phosphorylation during ciliogenesis. The specificities of phospho-CEP83 antibodies were confirmed by peptide competition assay in serum-starved RPE1 cells (Fig. S2, D and E). The localization of phosphorylated CEP83Ser29 and CEP83Thr292 signals was also confirmed to colocalize with endogenous CEP83 (Fig. S2 F). By examining CEP83 phosphorylation in unsynchronized cells, we observed that phospho-CEP83 signals were much higher in ciliated cells than in nonciliated cells (Fig. 3, G and H). In addition, CEP83 phosphorylation was increased when cells were serum deprived to promote ciliogenesis, and this was not observed in TTBK2 knockout cells (Fig. 3, I and J). Our results clearly show that CEP83 phosphorylation is induced during ciliogenesis and is mediated by TTBK2.
CEP83 phosphorylation requires the recruitment of TTBK2 to centriole DAs by CEP164
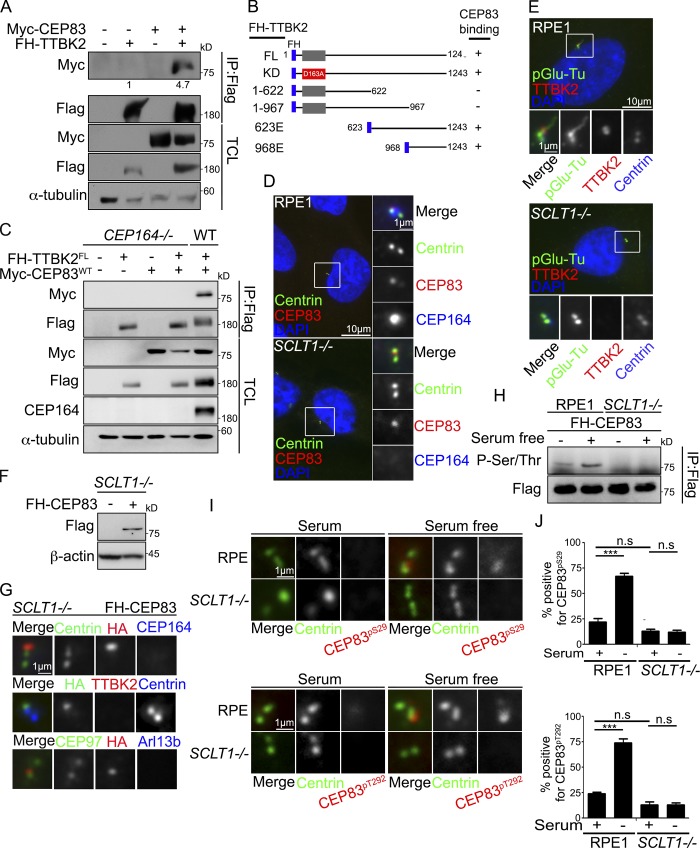
The interaction between TTBK2 and CEP83 was characterized by performing immunoprecipitation followed by Western blot in 293T cells (Fig. 4 A). Their interaction did not require the kinase activity of TTBK2, as expression of TTBK2FL and TTBK2KD showed no difference (Fig. S3 A). TTBK2 deletion mutants were then generated to map its interacting region with CEP83 (Fig. 4 B). The deletion of the TTBK2 C-terminus (TTBK21-967) abrogated its interaction with CEP83, whereas the truncated form of TTBK2 carrying the C-terminal region (TTBK2968E) was able to interact with CEP83, indicating that TTBK2 interacted with CEP83 via its C-terminal region (Figs. 4 B and S3). In addition, CEP834A could be detected in TTBK2 immunoprecipitates, indicating that the TTBK2–CEP83 interaction was independent of CEP83 phosphorylation (Fig. S3 D). Since CEP164 was required for TTBK2 to associate with DAs (Čajánek and Nigg, 2014; Oda et al., 2014), we also tested the involvement of CEP164 in the TTBK2–CEP83 interaction. Using CEP164 knockout 293T cells, we found that TTBK2 did not interact with CEP83 in the absence of CEP164, indicating that the TTBK2–CEP83 interaction required CEP164 (Fig. 4 C).
Figure 4.
CEP83 phosphorylation requires the recruitment of TTBK2 to centrioles by CEP164. (A) FH-TTBK2 and Myc-CEP83 were ectopically expressed in 293T cells. 293T cell lysate was analyzed by immunoprecipitation (IP) followed by Western blots with the indicated antibodies. TCL indicated the total cell lysates. Gel bands were quantified. (B) Schematic diagram shows various TTBK2 mutants tagged with Flag and HA. The ability of each construct to interact with CEP83 is also indicated. (C) FH-TTBK2 and Myc-CEP83 are ectopically expressed in WT and CEP164−/− 293T cells. Lysates were analyzed by immunoprecipitation followed by Western blots with the indicated antibodies. TCL, total cell lysates. (D) WT or SCLT1−/− cells were stained with anti-centrin, CEP83, and CEP164. DNA was stained with DAPI (blue). (E) Cells were serum starved for 2 d and stained with anti-TTBK2 and anti-pGlu-Tu antibodies. DNA was stained with DAPI. (F and G) FH-CEP83 is stably expressed in SCLT1−/− cells. Western blot analysis was performed to confirm CEP83 expression (F). Immunofluorescence was performed with antibodies as indicated to confirm the localization of CEP83 at the centrioles (G). (H) Cells were serum starved for 24 h, and immunoprecipitation was performed to pull down FH-CEP83. Western blot analysis was performed to check for CEP83 serine and threonine phosphorylation. Gel bands were quantified and normalized to the amount of FH-CEP83. (I) WT or SCLT1−/− cells were serum starved for 2 d and stained with antibodies against centrin, phospho-CEP83Ser29 (CEP83pS29), and phospho-CEP83Thr292 (CEP83pT292). Scale bar, 1 µm. (J) The phosphorylated CEP83Ser29 and CEP83Thr292 signals at the centrioles were quantified. More than 200 cells were analyzed for each independent experiment. Error bars represent mean ± SEM; n = 3. n.s., not significant. ***, P < 0.001 (Student’s t test).
To examine whether CEP164 was required for the phosphorylation of CEP83 by TTBK2, we used SCLT1 knockout cells, where CEP83 was present while CEP164 was absent from DAs (Fig. 4, D and E; Tanos et al., 2013; Yang et al., 2018). CEP83 was stably expressed in SCLT1 knockout cells, and CEP83 phosphorylation was analyzed in proliferating and serum-starved cells (Fig. 4, F and G). In SCLT1 knockout cells, CEP83 phosphorylation could not be detected upon serum starvation. This suggested that the recruitment of TTBK2 to DAs by CEP164 was required for TTBK2 to phosphorylate CEP83 (Fig. 4 H). The requirement of CEP164 to promote CEP83 phosphorylation was further confirmed by examining the phosphorylation of endogenous CEP83 using phospho-CEP83 antibodies (Fig. 4, I and J). Our results also suggest that the recruitment of TTBK2 to the centrioles by CEP164 is a prerequisite step for TTBK2 relocation toward the root of centriole DAs to phosphorylate CEP83 during ciliogenesis.
CEP83 inactivation blocks the formation of ciliary vesicles
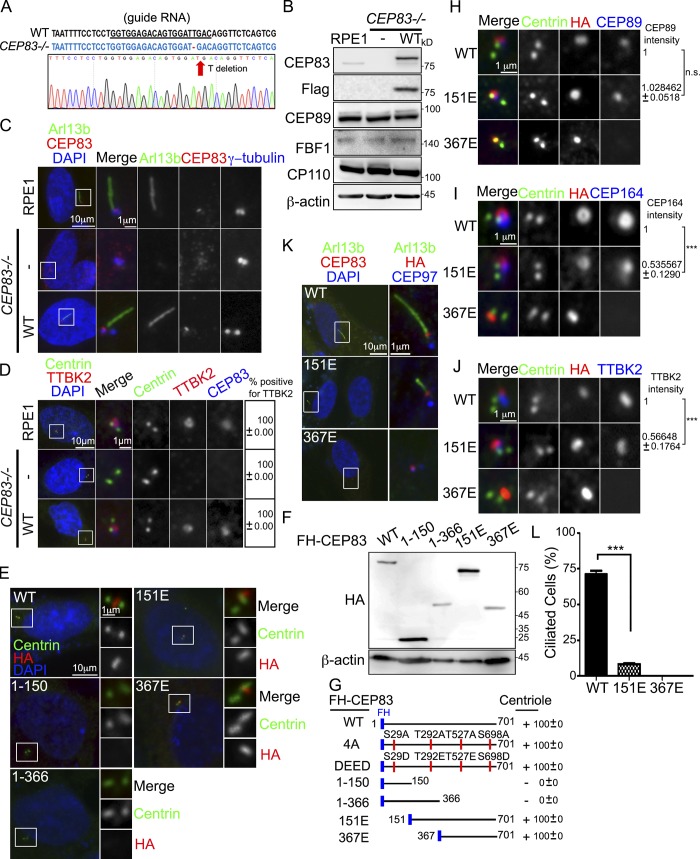
To further study the role of CEP83 in ciliogenesis, we generated CEP83 knockout cells (Figs. 5, A–D). Genotyping showed that a clone with one base pair (T) deletion on both alleles was predicted to produce a truncated products consisting of 27 amino residues only (Fig. 5 A). Western blotting indicated that CEP83 was undetectable without affecting the expression of other centriolar proteins (Fig. 5 B). Since DAs are necessary for ciliogenesis, we confirmed that primary cilia failed to form in CEP83 knockout cells (Figs. 5 C and 6 C). It was known that DA assembly follows a hierarchical relationship (Tanos et al., 2013). Here, we confirmed that loss of CEP83 blocked the recruitment of SCLT1, CEP89, FBF1, and CEP164 to DAs (Fig. S4 A). CEP83 inactivation also blocked the recruitment of important factors for ciliogenesis to the mother centrioles, such as TTBK2 and IFT88 (Figs. 5 D and S4 B; Tanos et al., 2013). Previous reports had shown that DAs were required for docking of membrane vesicles to the mother centrioles. Myosin-Va could be used as a maker to differentiate preciliary vesicles and ciliary vesicles during ciliogenesis (Wu et al., 2018). Here, cells were serum starved for 6 h to induce cilia initiation and a polyglutamylated tubulin (pGlu-Tu) antibody was used to mark the centrioles and ciliary axoneme. Using 3D structured illumination microscopy (SIM), we found that depletion of CEP83 completely blocked the formation of ciliary vesicles, indicating the importance of DAs in the control of ciliary vesicle docking during ciliogenesis (Fig. S4, C and D). Full-length CEP83 was then reintroduced in CEP83 knockout cells, where the formation of primary cilia, DA assembly, as well as TTBK2 and IFT88 localization were all rescued (Fig. 5, C and D; and Fig. S4 B).
Figure 5.
CEP83 inactivation blocks the formation of ciliary vesicles. (A) Sequence analysis of CEP83 alleles in CEP83−/− RPE1 cells. The dashed lines show T deletion. (B) FH-CEP83 was stably expressed in CEP83−/− cells, and Western blot analysis was performed with antibodies as indicated. (C) CEP83 inactivation blocked ciliogenesis. Cells were serum starved for 48 h to induce cilia formation and stained with anti-Arl13b, anti-CEP83, and γ-tubulin antibodies. (D) CEP83 inactivation blocked the recruitment of TTBK2 to the centrioles. The presence of TTBK2 at the centrioles was examined by immunofluorescence using antibodies as indicated. (E and F) CEP83−/− cells stably expressing various FH-CEP83 mutants were analyzed by immunofluorescence and immunoblots using antibodies as indicated. (G) A schematic diagram showing various CEP83 mutants tagged with Flag and HA. The ability of each construct in localizing to centrioles is indicated. (H–J) Immunofluorescence images of CEP83−/− cells stably expressed various FH-CEP83 mutants with indicated antibodies. The intensities of proteins at the centrioles was also quantified. n.s., not significant. (K and L) Cells were serum starved for 2 d to induce cilia formation. The percentage of ciliated cells was quantified. More than 150 cells were analyzed for each independent experiment. Error bars represent mean ± SEM; n = 3. ***, P < 0.001 by Student’s t test (C–E, K, and L). DNA was stained using DAPI (blue).
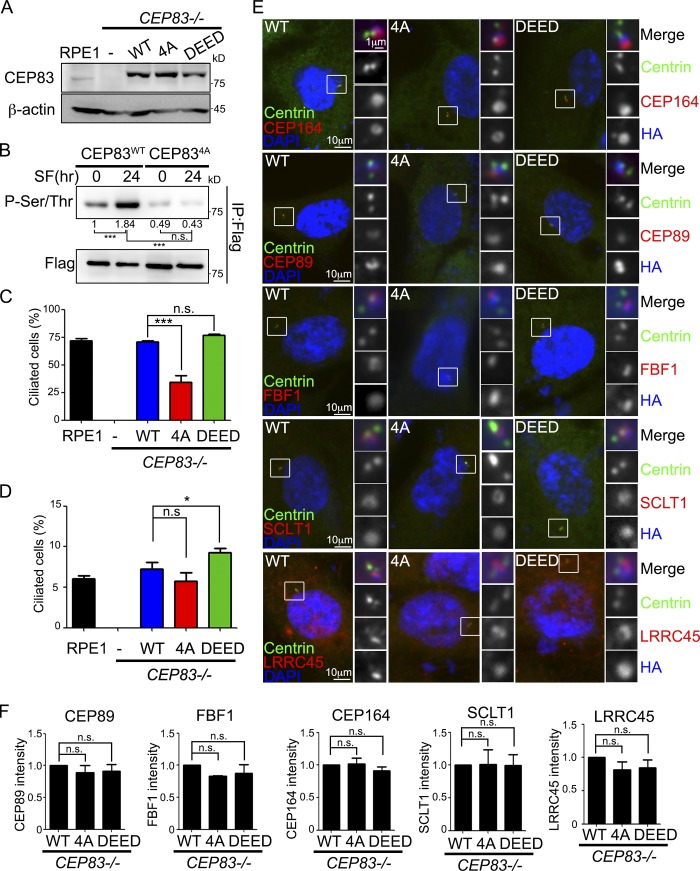
Figure 6.
CEP83 phosphorylation promotes ciliogenesis. (A) CEP83−/− cells were infected with retroviruses carrying various FH-CEP83 mutants. Various FH-CEP83–expressing cell lines were established through clonal propagation from a single cell. Western blots were performed to determine protein expression. (B) CEP83 phosphorylation was detected by immunoprecipitation followed by Western blots with anti-phospho-(Ser/Thr) antibody in proliferating or serum-starved cells. The phosphorylated CEP83 signal was quantified and normalized to total CEP83. (C and D) The percentage of ciliated cells was measured and quantified with an antibody against Arl13b in serum-starved cells (C) or proliferating cells (D). (E) Cells were serum starved for 2 d and analyzed by immunofluorescence with the indicated antibodies. DNA was stained using DAPI (blue). (F) The intensity of CEP89, FBF1, CEP164, SCLT1, and LRRC45 at centrioles was quantified and normalized to the control group (CEP83WT). In C, D, and F, data were collected from three independent experiments (n = 3). More than 150 cells were analyzed for each independent experiment. Error bars represent mean ± SEM; n = 3. n.s., not significant. *, P < 0.05; ***, P < 0.001 by Student’s t test.
Functional domain analysis of CEP83
We cloned and expressed various CEP83 mutants in CEP83 knockout cells in order to map the region responsible for targeting CEP83 to DAs (Figs. 5 F and S4 E). The truncated forms of CEP83 carrying the C-terminal half of CEP83 (CEP83151E and CEP83367E) were able to target to centrioles, whereas other fragments were not, indicating that CEP83 was recruited to centrioles via its C-terminus (Fig. 5, E and G; and Fig. S4 F). Given the role of CEP83 in DA assembly, we also mapped the region of CEP83 responsible for the recruitment of other DAPs. Our results showed that CEP89 was recruited to the centrioles via the middle region of CEP83 (residues 151–366; Fig. 5 H). The recruitment of CEP164 required both the N-terminal and middle regions of CEP83, since CEP164 was not detected at centrioles in CEP83367E-expressing cells and was detected at centrioles in CEP83151E-expressing cells, but at lower levels compared with CEP83WT-expressing cells (Fig. 5 I). The impaired recruitment of CEP164 to centrioles in CEP83151E-expressing cells also reduced the association of TTBK2 with DAs and ciliogenesis (Fig. 5, J–L).
CEP83 phosphorylation promotes ciliogenesis
To examine the effects of CEP83 phosphorylation in ciliogenesis, we generated cell lines where CEP83 residues Ser29, Thr292, Thr527, and Ser698 were changed to mimic the phosphorylated CEP83 (CEP83DEED). CEP83WT, CEP834A, or CEP83DEED were stably expressed in CEP83 knockout cells and derived using single-cell cloning. Western blot analyses were used to confirm equal expression (Figs. 6 A and S5 A). The phosphorylation levels of CEP83WT and CEP834A were analyzed in proliferating and serum-deprived cells. While serum starvation promoted CEP83WT phosphorylation, the phosphorylation of CEP834A did not change upon serum starvation (Fig. 6 B). We then checked the effects of CEP83 phosphorylation in promoting ciliogenesis. The ciliated frequency of CEP834A-expressing cells was dramatically reduced compared with CEP83WT-expressing, but not CEP83DEED-expressing, cells (Figs. 6 C and S5 C). The ciliated frequency was higher in CEP83DEED-expressing cells than in CEP83WT-expressing cells in proliferating cells, further supporting that CEP83 phosphorylation promoted ciliogenesis (Fig. 6 D).
CEP83 phosphorylation promotes cilia assembly at the initial steps
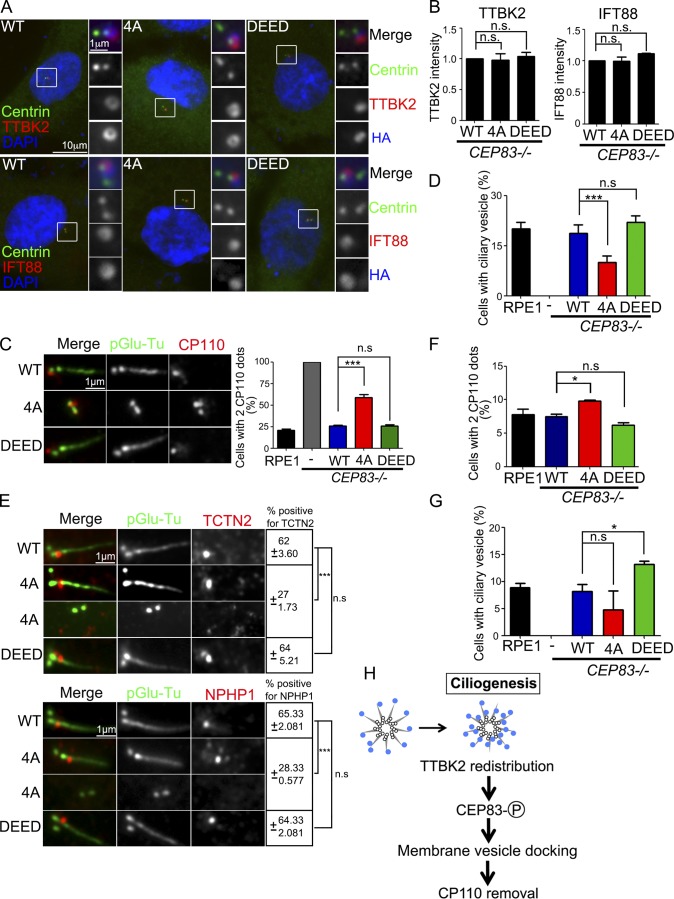
To understand how CEP83 phosphorylation contributed to ciliogenesis, we analyzed the key steps during ciliogenesis. We first asked whether CEP83 phosphorylation affected DA assembly and structure (Schmidt et al., 2012; Tanos et al., 2013; Čajánek and Nigg, 2014; Kurtulmus et al., 2018). Ciliogenesis was induced by serum starvation, and the fluorescent intensities of CEP164, CEP89, FBF1, SCLT1, and LRRC45 showed no difference in CEP83WT-, CEP834A-, or CEP83DEED-expressing cells, indicating that DA assembly and structure were not affected by CEP83 phosphorylation (Fig. 6, E and F). In addition, the recruitment of TTBK2 and IFT88 to the mother centrioles was also not affected by CEP83 phosphorylation (Fig. 7, A and B). Since the removal of CP110 from the mother centriole relied on TTBK2 kinase activity (Goetz et al., 2012), we examined the effect of CEP83 phosphorylation on CP110 removal. Most CEP834A-expressing cells still showed two CP110 foci at the centrioles, indicating that the removal of CP110 was promoted by CEP83 phosphorylation (Fig. 7 C). Since DAs have been shown to mediate docking of membrane vesicles to centrioles, we examined whether CEP83 phosphorylation affected this process. Most of CEP834A-expressing cells contained less ciliary vesicles when compared with the CEP83WT- or CEP83DEED-expressing cells, indicating that CEP83 phosphorylation affected ciliary vesicle docking (Fig. 7 D). The removal of CP110 from the mother centrioles and the docking of membrane vesicles to the mother centrioles promoted transition zone assembly. Our results also showed that CEP83 phosphorylation affected the recruitment of transition zone proteins TCTN2 and NPHP1, thus compromising the establishment of the transition zone (Fig. 7 E). We also performed immunostaining of CP110 and myosin-Va in proliferating cells. CEP83DEED-expressing cells showed more CP110 removed from mother centrioles and contained more ciliary vesicles than CEP83WT-expressing cells, further suggesting a role for CEP83 phosphorylation in promoting centriole to membrane vesicle docking and CP110 removal (Fig. 7, F and G). It is known that docking of membrane vesicles to DAs occurs before CP110 removal during ciliogenesis (Schmidt et al., 2012; Tanos et al., 2013; Lu et al., 2015). Thus, our results suggest a model where TTBK2-dependent CEP83 phosphorylation is important for ciliogenesis by regulating membrane vesicle docking that promotes CP110 removal from the mother centrioles (Fig. 7 H).
Figure 7.
CEP83 phosphorylation promotes cilia formation at the initial stage. (A) Cells were serum starved for 2 d and analyzed by immunofluorescence with the indicated antibodies. DNA was stained using DAPI (blue). (B) The intensity of TTBK2 and CEP83 signals at the centrioles was quantified. (C) Cells were serum starved for 2 d and processed for immunostaining with antibodies against pGlu-Tu and CP110. Scale bar, 1 µm. Graph showed the percentage of cells with two CP110 dots at the centrioles. (D) Cells were serum starved for 6 h. The percentage of cells contained ciliary vesicles was measured with an antibody against myosin-Va. (E) Cells were serum starved for 2 d and processed for immunostaining with antibodies as indicated. Scale bar, 1 µm. The percentage of cells with positive TCTN2 and NPHP1 signals at the centrioles was quantified. (F and G) In proliferating cells, the percentage of cells with two CP110 dots at the centrioles and the percentage of cells containing ciliary vesicles were quantified. (H) The diagram shows that TTBK2 phosphorylates CEP83 in promoting cilia initiation. In B–G, 200 cells were analyzed for each independent experiment. Error bars represent mean ± SEM; n = 3. n.s., not significant; *, P < 0.05; ***, P < 0.001 by Student’s t test.
Discussion
In this study, we observe that TTBK2 is redistributed at DAs from the periphery to the central CEP83-populated area during ciliogenesis. We also uncover the mechanistic basis of TTBK2 function in ciliogenesis, which depends on its kinase activity toward CEP83 upon recruitment by CEP164. Specifically, we find that TTBK2-dependent CEP83 phosphorylation regulates ciliary vesicle docking, CP110 removal, and cilia initiation. The removal of CP110 from the mother centrioles is a critical step for cilia initiation, and CP110 removal occurs after docking of membrane vesicles to the mother centrioles. The docking process is mediated by DAs, and CEP83 is a DAP, suggesting that CEP83 phosphorylation controls the docking of membrane vesicles to mother centrioles, which promotes CP110 removal and cilia initiation (Fig. 7 H).
Docking of membrane vesicles to the mother centrioles is the initial step of ciliogenesis that requires DAs. Although several DAPs have been discovered, CEP83 seems to play a critical role. First, DA assembly is known to be hierarchical (Tanos et al., 2013; Kurtulmus et al., 2018). The presence of CEP83 at centrioles is required for the recruitment of other known DAPs. Superresolution images clearly show that CEP83 locates to the root of centriole DAs (Yang et al., 2018). Results from our work further explain the relationship between CEP83 and other DAPs. Domain mapping experiments indicate that the C-terminal region of CEP83 is required for targeting CEP83 to centrioles and that the following recruitment of other DAPs depends on the N-terminal or middle regions of CEP83 (Fig. 5, E–I). The middle region of CEP83 is sufficient for CEP83 to recruit CEP89 (Fig. 5 H). To efficiently recruit CEP164 to mother centrioles, both the N-terminal and middle regions of CEP83 are required (Fig. 5 I). Our conclusion is also supported by the interaction network of centriole DAPs in a yeast two-hybrid assay (Kurtulmus et al., 2018). Results obtained from the yeast two-hybrid assay also show that CEP83 interacts with the DAP LRRC45 via its N-terminal region (residues 1–160). Since LRRC45 also contributes to ciliogenesis, the dramatic reduction of ciliated frequency in CEP83151E-expressing cells may be due to the failure to recruit LRRC45 to centriole DAs. Results from our mass spectrometry analysis map one TTBK2-dependent CEP83 phosphorylation site at Ser29 (Fig. S2 A). Whether CEP83 phosphorylation at Ser29 affects the CEP83–LRRC45 interaction is not known and can be further explored.
The kinase activity of TTBK2 is necessary for ciliogenesis, thus highlighting the importance of finding TTBK2 substrates associated with this role (Goetz et al., 2012). A recent study identifies MPP9 as a TTBK2 substrate (Huang et al., 2018). At the onset of ciliogenesis, TTBK2-dependent MPP9 phosphorylation promotes MPP9 degradation through the ubiquitin-proteasome system. The degradation of MPP9 then facilitates CP110 removal and subsequent cilia initiation. Although MPP9 is known to be degraded by the ubiquitin-proteasome system pathway, the molecular regulation of MPP9 degradation remains unclear. In our study, we identify CEP83 is a novel TTBK2 substrate for ciliogenesis. By analyzing the protein sequence of CEP83, we find that it contains properties of the intrinsically disordered proteins (Babu, 2016). An intrinsically disordered protein is a protein that lacks a fixed or ordered 3D structure. Therefore, it forms a variety of conformations under different physiological conditions. It is possible that the TTBK2-dependent CEP83 phosphorylation causes a change in the conformation of CEP83. This change could then recruit cilia-specific E3 ligases to the mother centrioles, promoting the degradation of MPP9 and subsequent CP110 removal.
The recruitment of TTBK2 to the mother centrioles by CEP164 is important for its role in ciliogenesis (Čajánek and Nigg, 2014; Oda et al., 2014). Our study shows that TTBK2 is redistributed from the periphery toward the root of DAs close to CEP83 during ciliogenesis and how this is important for CEP83 phosphorylation and cilia initiation. Sequence analysis of TTBK2 reveals that TTBK2 is highly enriched in serine residues at its C-terminus, which is also the region that associates with CEP164 (Čajánek and Nigg, 2014). Recent studies have shown that the phosphorylation of serine residues increases the intrinsic propensity of local backbone structure to form the polyproline II helix (He et al., 2016). Thus, it is possible that serum starvation induces a conformational change in TTBK2 by phosphorylating/dephosphorylating the enrichment of serine residues at its C-terminus. The change of TTBK2 local conformation could dissociate TTBK2 from CEP164 toward CEP83. Since ciliogenesis is tightly coupled with cell cycle progression, it is important to know how TTBK2 senses cell cycle signals and then changes its conformation and localization at DAPs to promote ciliogenesis. Thus, identification of upstream regulators that promote TTBK2 phosphorylation or redistribution will be an important issue to understand the cell cycle mechanism that regulates cilia formation.
Ciliopathies are genetic disorders associated with defects in cilia structure or function. In a recent study, mutations of CEP83 were associated with infantile nephronophthisis (NPHP18). All affected individuals showed early onset nephronophthisis (Failler et al., 2014). By examining cells extracted from affected individuals, cells show ciliary defects. Our studies reveal the detailed regulation of CEP83 in controlling DA assembly. We also demonstrate the important roles of CEP83 phosphorylation in controlling cilia initiation. Our results can be applied to study the relationship between CEP83 mutations and NPHP18. Based on our finding regarding the CEP83 phosphorylation by TTBK2, it will be also interesting to explore whether TTBK2 mutations are involved in nephronophthisis-related ciliopathies. In addition, it is known that TTBK2 mutations are genetically linked to spinocerebellar ataxia 11. Mouse studies also indicate that TTBK2 mutations show neuronal tube and sonic hedgehog signaling defects (Goetz et al., 2012). In the future, it will be interesting to examine potential roles of CEP83 in the control of neuronal function and sonic hedgehog signaling.
Materials and methods
Cell culture and reagent
293T, 293FT, and U2OS were cultured in DMEM supplemented with 10% FBS and 1% penicillin–streptomycin. Human telomerase-immortalized retinal pigment epithelial cells (hTERT-RPE1 or RPE1) were cultured in DMEM/F-12 (1:1) supplemented with 10% FBS and 1% penicillin–streptomycin.
Plasmids
Human TTBK2 cDNA was obtained from Open Biosystems, and CEP83 cDNA was directly amplified from an HeLa cDNA library. To generate epitope-tagged TTBK2 and CEP83, fragments were amplified by PCR and cloned into a pcDNA3-FH or pRK5M vector. The pRK5M vector contained sequence for expression of Myc-epitope at the protein C-terminus (Feng et al., 1995). Various CEP83 mutants were cloned to the pRK5M vector and used to analyze the TTBK2–CEP83 interaction (Fig. 4, A and C; and Fig. S3). Various TTBK2 mutants were also cloned to the pRK5M vector and used to analyze the CEP83 interaction (Figs. 3 A and 4 D). The pcDNA3-FH vector was derived from pcDNA3 (Thermo Fisher Scientific) but contained sequences for Flag and HA epitope tag between the HindIII and BamHI cloning sites. Thus, the expressing proteins expressed Flag and HA epitope tag at the N-terminus. The epitope-tagged TTBK2 and CEP83 fragments were also subcloned into a pBabe-puro3 vector (Morgenstern and Land, 1990) so that proteins could be stably expressed in RPE1 cells. For site-directed mutagenesis, we used the QuikChange Lightning Multi Site-Directed Mutagenesis kit (Agilent). CEP83, TTBK2, and their related mutants were also cloned into the pET32a (69015; Novagen) or pGEX-4T-1 (GE Healthcare Life Science) vector for producing recombinant proteins in bacteria.
Primary antibodies
Primary antibodies were obtained from the following sources and used according to the manufacturers’ instructions: mouse anti-Arl13 (immunofluorescence [IF] 1:1,000; ab136648; Abcam), rabbit anti-CP110 (IF 1:1,000; WB 1:1,000; 12780-1-AP; Proteintech), rabbit anti-CEP164 (IF 1:1,000; Western blot [WB] 1:1,000; 45330002; Novus Biologicals), goat anti-CEP164 (IF 1:250; sc-240226; Santa Cruz), mouse anti-HA (IF 1:1,000; WB 1:1,000; 901503; BioLegend), rabbit anti-IFT88 (IF 1:500; 13967-1-AP; Proteintech), mouse anti–pGlu-Tu (IF 1:3,000; 901501; AdipoGen), mouse anti–α-tubulin (WB 1:5,000; T6199; Sigma-Aldrich), mouse anti–γ-tubulin (IF 1:250; sc-51715; Santa Cruz), mouse anti-centrin (IF 1:1,000; 04-1624; Millipore), mouse anti-Flag (WB 1:5,000; F3165; Sigma-Aldrich), rabbit anti-CEP83 (IF 1:200; WB 1:1,000; HPA038161; Atlas Antibodies), rabbit anti-phospho-(Ser/Thr) (WB 1:1,000; 17464; Abcam), rabbit anti-TTBK2 (IF 1:2,000; WB 1:500; HPA018113; Atlas Antibodies), rabbit anti-CEP89 (IF 1:500; WB 1:1,000; ab157609; Abcam), rabbit anti-FBF1 (IF 1:500; WB 1:500; 11531-1-AP; Proteintech), rabbit anti-CEP97 (IF 1:1,000; 22050-1-AP; Proteintech), mouse anti-Myc (WB 1:1,000; 2278; Cell Signaling Technology), mouse anti–β-actin (WB 1:5,000; NB600-501; Novus Biologicals), rabbit anti-thiophosphate ester (WB 1:1,000; ab92570; Abcam), rabbit anti–myosin-Va (IF 1:200; NBP1-92156; Novus Biologicals), rabbit anti-TCTN2 (IF 1:200; 17053-1-AP; Proteintech), goat anti-NPHP1 (IF 1:200; sc-20204; Santa Cruz), rabbit anti-SCLT1 (IF 1:1,000; WB 1:500; 14875-1-AP; Proteintech), rat anti-CEP83 (IF 1: 1,000; Tanos et al., 2013), and rat anti-SCLT1 (IF 1: 1,000; Tanos et al., 2013). Ovalbumin-conjugated SGLTGSQEFQKMLC and CSSSEQNFLINKLH phosphopeptides were used to generate phospho-CEP83Ser29 and phospho-CEP83Thr292 antibodies, respectively in LTK BioLaboratories.
Transient transfection
Transient transfections were performed using T-Pro NTR II transfection reagents (T-Pro Biotechnology). 3 × 106 293T or U2OS cells were plated on a 100-mm plate overnight. Cells were transfected with 10 µg expression constructs according to the manufacturer’s instructions. For transient transfection, cells were harvested 48 h after transfection.
Retrovirus production and infection
5 × 105 293FT cells were plated on a 60-mm dish using T-Pro NTR II transfection reagents with the following plasmids: 1.5 µg V-SVG, 2.5 µg pCMV-gag-pol, and 4 µg of the pBabe-puro3–based constructs. The supernatant containing viral particles was harvested 48 h after transfection. Virus containing media was centrifuged at 1,000 rpm for 5 min and passed through a 0.45-µm filter (Sarstedt). For the infection of RPE1 cells, 5 × 105 RPE1 cells were seeded onto a 60-mm plate the night before infection and incubated with 3 ml viral stock. The medium was changed to fresh culture medium 18–24 h after infection. 2 d after infection, cells were selected and maintained in culture medium containing 2 µg/ml puromycin (Sigma-Aldrich).
Generation of CEP83, TTBK2, and CEP164 knockout RPE1 cells
RNA-guided DNA endonuclease was performed to edit genes through coexpression of the Cas9 protein with gRNAs (http://www.addgene.org/crispr/church/). The targeting sequence for CEP83, TTBK2, and CEP164 was at exon 1 of CEP83 (5′-GTGGAGACAGTGGATTGAC-3′), exon 3 of TTBK2 (5′-GAAAATGTTGCACTGAAGG-3′), and exon 9 of CEP164 (5′-GCTGTTGGCAAAGGGCGACA-3′), respectively. The target sequences were cloned into the gRNA cloning vector (plasmid 41824; Addgene) via the Gibson assembly method (New England Biolabs) as described previously (Mali et al., 2013). Knockout cells were all obtained through clonal propagation from a single cell. For genotyping, the following PCR primers were used: 5′-GTATAATTTGGCTTCCTGAG-3′ and 5′-CCACTTCTAGATTTAAGTAA-3′ for CEP83 alleles, 5′-GATGCTGGCTGTTGACCAAA-3′ and 5′-TATACCCGGCTGACACCAGT-3′ for TTBK2 alleles, and 5′-CTGGGTGATTGATAACCATTGGG-3′ and 5′-CGCAAATGAAGCTCCTGACTCAGT-3′ for CEP164 alleles. PCR products were cloned and sequenced.
Peptide competition assay
The specificity of the two phospho-CEP83 antibodies was validated by peptide competition assay. Phospho-CEP83 antibodies were diluted in 2 ml PBS buffer that contained 3% bovine serum albumin (wt/vol) and preincubated with the phosphorylated or nonphosphorylated peptide (5 µg/ml) overnight at 4°C under gentle rotation before performing immunostaining.
Immunostaining
Cells were grown on 0.1 mg/ml poly-L-lysine–coated coverslips and fixed with methanol at −20°C for 15 min. Cells were then washed three times with PBS and incubated in the blocking buffer that contained 3% bovine serum albumin (wt/vol) and 0.1% Triton X-100 in PBS for 30 min at RT. Primary antibodies were all diluted in blocking buffer and incubated for 2 h at RT. Alexa Fluor 488–, 594–, or 680–conjugated goat secondary antibodies were used at 1:500 dilution (Molecular Probes) and incubated for 1 h at RT. DNA was visualized using DAPI (Thermo Fisher Scientific). Coverslips were mounted on the slides with mounting medium (ProLong Gold Antifade; Thermo Fisher Scientific). Fluorescent images were obtained using an upright microscope (Axio Imager M2 ApoTome2 system; Carl Zeiss) with a Plan-NEOFLUAR 100× (1.3 NA) oil-immersion objective and an Axiocam 702 mono charge-coupled device camera. Images were acquired and processed by ZEN software (Carl Zeiss) or ImageJ software (National Institutes of Health).
Immunoblotting
Cells were washed with ice-cold PBS twice and lysed in ice-cold RIPA lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, and 0.1% SDS) that contained protease inhibitors (Thermo Fisher Scientific) and, where necessary, phosphatase inhibitors (Phospho STOP tablets; Roche). Cell debris was removed by centrifugation at 14,000 g for 15 min at 4°C. Protein concentrations were determined by the Bradford protein assay (Bio-Rad). Equal amounts of proteins were mixed with SDS sample buffer, boiled at 95°C for 5 min, and separated by SDS-PAGE. The resolved proteins were then transferred on polyvinylidene difluoride (PVDF) membranes (0.45 µm Hybond P; GE Healthcare Life Science). Blots were blocked for 1 h at RT with 5% nonfat milk in TBS-T (20 mM Tris, pH 7.6, 137 mM NaCl, and 0.1% Tween-20) and incubated overnight at 4°C with primary antibodies in blocking solution. Membranes were washed three times with TBS-T and incubated with HRP-conjugated anti-mouse or anti-rabbit secondary antibodies for 1 h at RT (Jackson ImmunoResearch). After washing three times with TBS-T, proteins were visualized with ECL Western blotting substrate.
Dephosphorylation assay in cell extract
In some experiments, calf intestinal alkaline phosphatase (CIP; M0290; New England Biolabs) was used to examine protein phosphorylation. After lysing cells by RIPA lysis buffer and determining protein concentrations, 30 µg of cell lysates was suspended in 30 µl CIP buffer and incubated with 30 U CIP at 37°C for 1 h. The control included cell lysate and all reaction components except CIP. Sample buffer was then added to the reaction and boiled at 95°C for 5 min before separating by SDS-PAGE. The Phos-tag gels were made that contained 20 µM Phos-tag (FUJIFILM Wako Chemical Corporation) and 100 µM MnCl2 in the separating gels. Before transferring the gels to PVDF membranes, Phos-tag gels were soaked in the transfer buffer that contains 1 mM EDTA for 20 min with gentle agitation to eliminate the manganese ions in the gel.
SIM and dSTORM imaging and analysis
The 3D-SIM superresolution images were acquired using a ELYRA PS.1 LSM780 system equipped with a Plan Apochromat 63×/NA 1.4 oil-immersion objective (Carl Zeiss). Z stacks with an interval of 110 nm were used to scan whole cells. The raw images were reconstructed using ZEN software under the default parameters.
The dSTORM superresolution images were performed on a modified setup based on a commercial inverted microscope (Eclipse Ti-E; Nikon) with a 100× NA 1.49 oil-immersion objective (Yang et al., 2018). For single-color imaging, a 637-nm laser (OBIS 637 LX 140 mW; Coherent) was used for the excitation of Alexa Flour 647. For dual-color imaging, Alexa Fluor 647 imaging was first performed, and then the second channel imaging of Cy3B was sequentially acquired with a 561-nm light (Jive 561 150 mW; Cobolt). The excitation lights were operated at 3–5 KW/cm2. A weak 405-nm light (100 mW OBIS 405 LX; Coherent) was introduced to convert fluorophores from a dark state to a fluorescence state. The fluorescence emission was spectrally cleaned with bandpass filters (700/75 and 593/40 for Alexa Fluor 647 and Cy3B, respectively; Chroma Technology) before readout by an electron-multiplying charge-coupled device camera (Evolve 512 Delta; Photometrics). Fiducial markers (Tetraspeck, 0.1 µm, T7279; Thermo Fisher Scientific) were used to record the drift during the imaging for the postacquisition linearity correction. Typically, 10,000–20,000 frames were taken at 50 frames per second. The position of each single-molecule event was analyzed using Metamorph Superresolution Module (Molecular Devices), and the final rendered pixel size of dSTORM images was 11.625 nm. For dSTORM, samples was mounted with an imaging chamber (CM-B18-1; Live Cell Instrument) and incubated in the imaging buffer containing 50 mM Tris-HCl and 10 mM NaCl (TN) buffer at pH 8.0 and an oxygen-scavenging system consisting of 100 mM mercaptoethylamine, pH 8.0, 0.5 mg/ml glucose oxidase, 40 µg/ml catalase, and 10% glucose (Sigma-Aldrich). To determine the radial distribution of CEP83/CEP164/TTBK2 in Fig. 2, a center of individual puncta of protein distribution was first fitted, and then their radial positions were described with a distance with respect to the center.
Purification of recombinant proteins
For recombinant protein purification, constructs were transformed into an Escherichia coli Rosetta2 (DE3) strain (Novagen). Cells were cultured in LB media at 37°C until OD600 reached 0.5–0.6. Expression of the recombinant proteins was then induced at 25°C for 16 h by adding IPTG (0.2 mM for GST fusion proteins and 2 mM for His fusion proteins). Purification of the His and GST fusion proteins was performed as described previously (Tsai et al., 2000).
Immunoprecipitation
A 100-mm dish of transfected cells were lysed in a buffer that contained 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Nonidet P-40, and 0.5% sodium deoxycholate together with phosphatase and protease inhibitor. 1 mg of cell lysates was suspended in 1 ml lysis buffer and incubated with 5 µl anti-FLAG M2 beads (Sigma-Aldrich) for 2 h at 4°C under gentle rotation. Beads were then washed three times with the lysis buffer that contained protease and phosphatase inhibitors. The immunocomplex was eluted by SDS sample buffer and separated by gels for Western blot analysis.
TTBK2 in vitro kinase assay
Various FH-TTBK2 proteins were transiently expressed in 293T cells for 48 h and immunoprecipitated by M2 beads. Beads were washed three times with TBS buffer (50 mM Tris-HCl and 150 mM NaCl, pH 7.4) followed by TTBK2 kinase reaction buffer (50 mM Tris, pH 7.4, and 10 mM MgCl2) before performing the TTBK2 kinase assay. The TTBK2 kinase assay was performed by incubating TTBK2 protein on the beads with 5 µg recombinant His-CEP83WT or His-CEP834A for 30 min at RT in 30 µl kinase reaction buffer that contained 100 ng ATP-gamma-S (ab138911; Abcam). After incubation, 1 µl of 50 mM p-nitrobenzyl mesylate (ab138910; Abcam) was added to the kinase reaction for 90 min at RT to stop the kinase reaction (Allen et al., 2007). Proteins were separated by SDS-PAGE and then transferred to the nitrocellulose membranes (GE Healthcare) for Western blot. The immunoreactive bands were detected by Odyssey Image Systems (LI-COR). The gel was also stained with Coomassie brilliant blue (CBB) to ascertain migration of the proteins. For the TTBK2 in vitro kinase assay that used GST-TTBK2 (1–622)WT and GST-TTBK2 (aa 1–622)KD purified from bacteria, 0.5 µg GST-TTBK2 (aa 1–622)WT or GST-TTBK2(aa 1–622)KD and 5 µg CEP83 were added in the 30 µl of kinase reaction buffer that contained 100 ng ATP-gamma-S for 30 min at room temperature.
Quantification of images from immunostaining and immunoblots
For quantification of fluorescent intensity with proteins at the centrioles, all cells were treated the same during the process of immunostaining and image acquisition. ZEN software was used to analyze the image intensity. The same setting was applied to all images. A circle was drawn surrounding the centrioles, and the total pixel value of the marked region was then measured. The signal ratio of the marked region over the proteins at the centrioles was then normalized to the control group. ImageJ software (National Institutes of Health) was used to quantify the immunoblots. A rectangle was drawn surrounding the target band. Results were expressed as density means ± SD by normalizing to the control group. All the quantifications were obtained from at least three independent experiments.
Mass spectrometry
FH-CEP83 was ectopically expressed in 293T cells with or without TTBK2 coexpression. Cells were lysed in RIPA buffer that contained protease and phosphatase inhibitors. CEP83 was affinity purified with M2 magnetic beads. After electrophoresis, CEP83 bands were excised from the gel and digested with trypsin, Lys-C, Arg-C, or Asp-N (Roche). Mass spectrometry was then performed using an LTQ-Orbitrap hybrid tandem mass spectrometer (Thermo Fisher Scientific) in-line with an Agilent 1200 nanoflow HPLC system. File Converter in Xcalibur 2.0.7 (Thermo Fisher Scientific) and in-house programs were used to extract the mass spectrometry and MS/MS signals. The tandem mass spectra collected with liquid chromatography–MS/MS were processed using Sequest/TurboSequest and in house–developed computer programs, which found the best matched peptide sequence in the human protein FASTA database downloaded from the UniProt website. Peptides were verified based on two criteria, namely, TubroSequest XCorr >2.5 and mass error <15 ppm. For searching modified peptides, the saMS approach was used to uncover posttranslational modification–containing peptides (Liu et al., 2015).
2D gel electrophoresis
The protein samples were first concentrated and desalted using methanol-chloroform precipitation. The pellets were dissolved in rehydration buffer that contained 7 M urea, 2 M thiourea, 4% (wt/vol) CHAPS, 40 mM DTT, and 0.5% (vol/vol), pH 4–7, IPG buffer. Samples were then centrifuged at 220,000 g at 22°C for 1 h, and the supernatants were collected for subsequent electrophoreses. Samples were then centrifuged at 220,000 g at 22°C for 1 h, and the supernatants were collected for subsequent electrophoreses. IPG strips for isoelectric focusing were rehydrated for 15 h at 22°C in 450 µl rehydration buffer. Each sample was loaded on strip using the cup-loading method and was focused at 20°C using Ettan IPGphor 3 (GE Healthcare) at 500 V for 3 h, followed by a gradient to 1 kV for 7 h and 8 kV for 3 h, and then maintained at 8 kV for another 12 h. For 2D separation, the strips were incubated in equilibration buffer containing 6 M urea, 30% (vol/vol) glycerol, 2% (wt/vol) SDS, 50 mM Tris-HCl, pH 8.8, and 1% (wt/vol) DTT for 30 min on a rocker at 22°C. The equilibrated strips were sealed onto the top of 1.5-mm-thick 10% discontinuous polyacrylamide gels (16 × 10 cm) with 0.5% (wt/vol) agarose sealing solution. The proteins were separated at 22°C, initially with 80 V for 1.5 h, followed by 160 V until the dye front approached the bottom of the gels.
Statistical data analysis
All data are representing as mean with SEM from at least three independent experiments. Experiment samples and numbers for statistical testing are reported in the corresponding figure legends. Data distribution was assumed to be normal, but this was not formally tested. All P values are from the Student’s t test for two-group comparisons (GraphPad Prism 5; ***, P < 0.001; **, P < 0.01; *, P < 0.05).
Online supplemental material
Fig. S1 shows that TTBK2 coexpression causes phosphorylation-induced mobility shifts of CEP89 and CEP164. Fig. S2 shows that TTBK2 phosphorylates CEP83 at Ser29, Thr292, Thr527, and Ser698 and that the specificities of two phospho-CEP83 antibodies (phospho-CEP83Ser29 and phospho-CEP83Thr292) were confirmed by peptide blocking experiments. Fig. S3 shows that TTBK2 interacts with CEP83 via its C-terminal region. Fig. S4 shows that CEP83 inactivation blocks DA assembly, the recruitment of IFT88 to mother centrioles, and ciliary vesicle docking. Fig. S5 shows that CEP83 phosphorylation promotes cilia formation without affecting DA assembly.
Supplementary Material
Acknowledgments
We thank Dr. Meng-Fu Bryan Tsou (Memorial Sloan-Kettering Cancer Center, New York, NY) for sharing reagents, discussion, and critical comments. We also thank Dr. T.K. Tang (Institute of Biomedical Science, Academia Sinica, Taipei, Taiwan) for helping with the 3D-SIM images and providing critical comments.
This work was supported by the Ministry of Science and Technology, Taiwan (MOST 105-2628-B-010-004-MY3, MOST 107-2313-B-010-001, MOST 108-2628-B-010-007, MOST 107-2633-B-009-003, and Shackleton Program Grant), the Yen Tjing Ling Medical Foundation (CI-107-17 and CI-108-12), and the Higher Education Sprout Project by the Ministry of Education, Taiwan (107AC-D920). For DNA sequencing, we acknowledge the Clinical and Industrial Genomic Application Development Service of the National Core Facility for Biopharmaceuticals, Taiwan (MOST 107-2319-B-010-002).
The authors declare no competing financial interests.
Author contributions: C.-H. Lo and I.-H. Lin performed most of experiments and contributed to study design and data analysis. T.T. Yang performed the dSTORM imaging and analyzed the data. C.P. Chou quantified the fluorescent intensity. B.E. Tanos generated the CEP83 knockout cells. C.-W. Chang generated the TTBK2 knockout cells. Y.-C. Huang performed the mass spectrometry assay and 2D gel electrophoresis. B.E. Tanos, J.-C. Liao, Y.-G. Tsay, and W.-J. Wang discussed the paper. C.H. Lo wrote the initial draft of manuscript. W.-J. Wang designed the study, interpreted data, and wrote final manuscript with discussion from all authors. All authors helped with editing.
References
- Allen J.J., Li M., Brinkworth C.S., Paulson J.L., Wang D., Hübner A., Chou W.H., Davis R.J., Burlingame A.L., Messing R.O., et al. . 2007. A semisynthetic epitope for kinase substrates. Nat. Methods. 4:511–516. 10.1038/nmeth1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu M.M. 2016. The contribution of intrinsically disordered regions to protein function, cellular complexity, and human disease. Biochem. Soc. Trans. 44:1185–1200. 10.1042/BST20160172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove B.W., and Yost H.J.. 2006. The roles of cilia in developmental disorders and disease. Development. 133:4131–4143. 10.1242/dev.02595 [DOI] [PubMed] [Google Scholar]
- Bowler M., Kong D., Sun S., Nanjundappa R., Evans L., Farmer V., Holland A., Mahjoub M.R., Sui H., and Loncarek J.. 2019. High-resolution characterization of centriole distal appendage morphology and dynamics by correlative STORM and electron microscopy. Nat. Commun. 10:993 10.1038/s41467-018-08216-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čajánek L., and Nigg E.A.. 2014. Cep164 triggers ciliogenesis by recruiting Tau tubulin kinase 2 to the mother centriole. Proc. Natl. Acad. Sci. USA. 111:E2841–E2850. 10.1073/pnas.1401777111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-Santos Z., Azimzadeh J., Pereira-Leal J.B., and Bettencourt-Dias M.. 2011. Evolution: Tracing the origins of centrioles, cilia, and flagella. J. Cell Biol. 194:165–175. 10.1083/jcb.201011152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edener U., Kurth I., Meiner A., Hoffmann F., Hübner C.A., Bernard V., Gillessen-Kaesbach G., and Zühlke C.. 2009. Missense exchanges in the TTBK2 gene mutated in SCA11. J. Neurol. 256:1856–1859. 10.1007/s00415-009-5209-0 [DOI] [PubMed] [Google Scholar]
- Failler M., Gee H.Y., Krug P., Joo K., Halbritter J., Belkacem L., Filhol E., Porath J.D., Braun D.A., Schueler M., et al. . 2014. Mutations of CEP83 cause infantile nephronophthisis and intellectual disability. Am. J. Hum. Genet. 94:905–914. 10.1016/j.ajhg.2014.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X.H., Filvaroff E.H., and Derynck R.. 1995. Transforming growth factor-beta (TGF-beta)-induced down-regulation of cyclin A expression requires a functional TGF-beta receptor complex. Characterization of chimeric and truncated type I and type II receptors. J. Biol. Chem. 270:24237–24245. 10.1074/jbc.270.41.24237 [DOI] [PubMed] [Google Scholar]
- Fliegauf M., Benzing T., and Omran H.. 2007. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 8:880–893. 10.1038/nrm2278 [DOI] [PubMed] [Google Scholar]
- Gerdes J.M., Davis E.E., and Katsanis N.. 2009. The vertebrate primary cilium in development, homeostasis, and disease. Cell. 137:32–45. 10.1016/j.cell.2009.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilula N.B., and Satir P.. 1972. The ciliary necklace. A ciliary membrane specialization. J. Cell Biol. 53:494–509. 10.1083/jcb.53.2.494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz S.C., Liem K.F. Jr., and Anderson K.V.. 2012. The spinocerebellar ataxia-associated gene Tau tubulin kinase 2 controls the initiation of ciliogenesis. Cell. 151:847–858. 10.1016/j.cell.2012.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S., Stierhof Y.D., Lavoie S.B., Gassner O.S., Lamla S., Le Clech M., and Nigg E.A.. 2007. Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 179:321–330. 10.1083/jcb.200707181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He E., Yan G., Zhang J., Wang J., and Li W.. 2016. Effects of phosphorylation on the intrinsic propensity of backbone conformations of serine/threonine. J. Biol. Phys. 42:247–258. 10.1007/s10867-015-9405-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlden H., Johnson J., Gardner-Thorpe C., Lashley T., Hernandez D., Worth P., Singleton A.B., Hilton D.A., Holton J., Revesz T., et al. . 2007. Mutations in TTBK2, encoding a kinase implicated in tau phosphorylation, segregate with spinocerebellar ataxia type 11. Nat. Genet. 39:1434–1436. 10.1038/ng.2007.43 [DOI] [PubMed] [Google Scholar]
- Huang N., Zhang D., Li F., Chai P., Wang S., Teng J., and Chen J.. 2018. M-Phase Phosphoprotein 9 regulates ciliogenesis by modulating CP110-CEP97 complex localization at the mother centriole. Nat. Commun. 9:4511 10.1038/s41467-018-06990-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., and Marshall W.F.. 2011. Ciliogenesis: building the cell’s antenna. Nat. Rev. Mol. Cell Biol. 12:222–234. 10.1038/nrm3085 [DOI] [PubMed] [Google Scholar]
- Ishikawa H., and Marshall W.F.. 2017. Intraflagellar Transport and Ciliary Dynamics. Cold Spring Harb. Perspect. Biol. 9: a021998 10.1101/cshperspect.a021998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo K., Kim C.G., Lee M.S., Moon H.Y., Lee S.H., Kim M.J., Kweon H.S., Park W.Y., Kim C.H., Gleeson J.G., and Kim J.. 2013. CCDC41 is required for ciliary vesicle docking to the mother centriole. Proc. Natl. Acad. Sci. USA. 110:5987–5992. 10.1073/pnas.1220927110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtulmus B., Yuan C., Schuy J., Neuner A., Hata S., Kalamakis G., Martin-Villalba A., and Pereira G.. 2018. LRRC45 contributes to early steps of axoneme extension. J. Cell Sci. 131:jcs223594 10.1242/jcs.223594 [DOI] [PubMed] [Google Scholar]
- Liachko N.F., McMillan P.J., Strovas T.J., Loomis E., Greenup L., Murrell J.R., Ghetti B., Raskind M.A., Montine T.J., Bird T.D., et al. . 2014. The tau tubulin kinases TTBK1/2 promote accumulation of pathological TDP-43. PLoS Genet. 10:e1004803 10.1371/journal.pgen.1004803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N.Y., Lee H.H., Chang Z.F., and Tsay Y.G.. 2015. Examination of segmental average mass spectra from liquid chromatography-tandem mass spectrometric (LC-MS/MS) data enables screening of multiple types of protein modifications. Anal. Chim. Acta. 892:115–122. 10.1016/j.aca.2015.07.032 [DOI] [PubMed] [Google Scholar]
- Lu Q., Insinna C., Ott C., Stauffer J., Pintado P.A., Rahajeng J., Baxa U., Walia V., Cuenca A., Hwang Y.S., et al. . 2015. Early steps in primary cilium assembly require EHD1/EHD3-dependent ciliary vesicle formation. Nat. Cell Biol. 17:228–240. 10.1038/ncb3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., and Church G.M.. 2013. RNA-guided human genome engineering via Cas9. Science. 339:823–826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern J.P., and Land H.. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587–3596. 10.1093/nar/18.12.3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T., Chiba S., Nagai T., and Mizuno K.. 2014. Binding to Cep164, but not EB1, is essential for centriolar localization of TTBK2 and its function in ciliogenesis. Genes Cells. 19:927–940. 10.1111/gtc.12191 [DOI] [PubMed] [Google Scholar]
- Pedersen L.B., and Rosenbaum J.L.. 2008. Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Curr. Top. Dev. Biol. 85:23–61. 10.1016/S0070-2153(08)00802-8 [DOI] [PubMed] [Google Scholar]
- Reiter J.F., and Leroux M.R.. 2017. Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol. 18:533–547. 10.1038/nrm.2017.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez I., and Dynlacht B.D.. 2016. Cilium assembly and disassembly. Nat. Cell Biol. 18:711–717. 10.1038/ncb3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K.N., Kuhns S., Neuner A., Hub B., Zentgraf H., and Pereira G.. 2012. Cep164 mediates vesicular docking to the mother centriole during early steps of ciliogenesis. J. Cell Biol. 199:1083–1101. 10.1083/jcb.201202126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla V., Romaguera-Ros M., Garcia-Verdugo J.M., and Reiter J.F.. 2010. Ofd1, a human disease gene, regulates the length and distal structure of centrioles. Dev. Cell. 18:410–424. 10.1016/j.devcel.2009.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin S. 1962. Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J. Cell Biol. 15:363–377. 10.1083/jcb.15.2.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M., Tomizawa K., Sato K., Ohtake A., and Omori A.. 1995. A novel tau-tubulin kinase from bovine brain. FEBS Lett. 372:59–64. 10.1016/0014-5793(95)00955-9 [DOI] [PubMed] [Google Scholar]
- Tanos B.E., Yang H.J., Soni R., Wang W.J., Macaluso F.P., Asara J.M., and Tsou M.F.. 2013. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev. 27:163–168. 10.1101/gad.207043.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y.T., Su Y.H., Fang S.S., Huang T.N., Qiu Y., Jou Y.S., Shih H.M., Kung H.J., and Chen R.H.. 2000. Etk, a Btk family tyrosine kinase, mediates cellular transformation by linking Src to STAT3 activation. Mol. Cell. Biol. 20:2043–2054. 10.1128/MCB.20.6.2043-2054.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.T., Chen H.Y., and Tang T.K.. 2018. Myosin-Va is required for preciliary vesicle transportation to the mother centriole during ciliogenesis. Nat. Cell Biol. 20:175–185. 10.1038/s41556-017-0018-7 [DOI] [PubMed] [Google Scholar]
- Yang T.T., Chong W.M., Wang W.J., Mazo G., Tanos B., Chen Z., Tran T.M.N., Chen Y.D., Weng R.R., Huang C.E., et al. . 2018. Super-resolution architecture of mammalian centriole distal appendages reveals distinct blade and matrix functional components. Nat. Commun. 9:2023 10.1038/s41467-018-04469-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., Zeng H., Ning G., Reiter J.F., and Liu A.. 2014. C2cd3 is critical for centriolar distal appendage assembly and ciliary vesicle docking in mammals. Proc. Natl. Acad. Sci. USA. 111:2164–2169. 10.1073/pnas.1318737111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.