Invasive cancer cells degrade and invade into the extracellular matrix by expressing the matrix metalloproteinase MT1-MMP at invadopodia. Miyagawa et al. show that MT1-MMP uses the ER-Golgi SNARE Bet1 to facilitate its own transport to the plasma membrane through their interaction in a cholesterol-rich milieu.
Abstract
Metastasis is a major cause of cancer-related death. Membrane type 1–matrix metalloproteinase (MT1-MMP) is a critical protease for local invasion and metastasis. MT1-MMP is synthesized in the endoplasmic reticulum (ER) and transported in vesicles to invadopodia, specialized subdomains of the plasma membrane, through secretory and endocytic recycling pathways. The molecular mechanism underlying intracellular transport of MT1-MMP has been extensively studied, but is not fully understood. We show that MT1-MMP diverts the SNARE Bet1 from its function in ER-Golgi transport, to promote MT1-MMP trafficking to the cell surface, likely to invadopodia. In invasive cells, Bet1 is localized in MT1-MMP–positive endosomes in addition to the Golgi apparatus, and forms a novel SNARE complex with syntaxin 4 and endosomal SNAREs. MT1-MMP may also use Bet1 for its export from raft-like structures in the ER. Our results suggest the recruitment of Bet1 at an early stage after MT1-MMP expression promotes the exit of MT1-MMP from the ER and its efficient transport to invadopodia.
Introduction
Metastasis, which includes many complex processes such as invasion and dissemination to distant tissues, is a major cause of cancer-related death (Chaffer and Weinberg, 2011). Invasive cancer cells can degrade the ECM and migrate into the surrounding tissues. During these steps, the cells form protrusions of the plasma membrane called invadopodia, which are actin-enriched structures with the ability to degrade the ECM (Linder et al., 2011; Eddy et al., 2017; Paterson and Courtneidge, 2018). Extracellular stimuli such as growth factors and cellular adhesion to the ECM through integrins initiate invadopodia formation by activating several protein and lipid kinases such as Src tyrosine kinase and phosphatidylinositol 3-kinase (Hoshino et al., 2013; Eddy et al., 2017). This activation results in the recruitment of invadopodia-related proteins, such as cortactin (Clark et al., 2007) and neural Wiskott-Aldrich syndrome protein (Yamaguchi et al., 2005), to invadopodia formation sites, leading to actin polymerization and thereby inducing the protrusions of the plasma membrane. Once invadopodia are formed, microtubules extend to and elongate them (Schoumacher et al., 2010) and matrix metalloproteinases (MMPs), including the soluble/secreted MMPs MMP2 and MMP9 (Linder, 2007) and a membrane-bound membrane type 1–MMP (MT1-MMP), a master regulator of invadopodia function (Castro-Castro et al., 2016), are delivered to invadopodia via trafficking vesicles and/or tubulovesicular transport carriers for their maturation (Schoumacher et al., 2010; Jacob et al., 2013; Marchesin et al., 2015).
Intracellular trafficking of MT1-MMP is a complex process. MT1-MMP is synthesized and integrated into the ER membrane as an inactive precursor form (Seiki and Yana, 2003). The ER-integrated MT1-MMP precursor is transported to the Golgi apparatus and then to post-Golgi compartments, where it is cleaved by proprotein convertases such as furin into an active mature form (Yana and Weiss, 2000). After reaching the plasma membrane, MT1-MMP is endocytosed and recycled back to invadopodia of the plasma membrane so that the ECM degradation activity in the invadopodia is optimized. Late endosomes are a major storage compartment for intracellular MT1-MMP (Castro-Castro et al., 2016). A protein complex comprising kinesin-1/2, Arf6, and JIP3/4 delivers MT1-MMP from this storage compartment to invadopodia with tubulovesicular carriers (Marchesin et al., 2015). The transport carriers are tethered to and fused with the plasma membrane at invadopodia in a manner dependent on the tethering complex exocyst and the SNARE VAMP7 (Sakurai-Yageta et al., 2008).
SNAREs are single membrane-spanning or lipid-modified, membrane-anchored proteins that mediate membrane fusion between transport vesicles/carriers and target membranes (Jahn and Scheller, 2006). At least 39 genes encoding SNARE proteins exist in the human genome, and all SNARE proteins have one or two SNARE motifs, which are evolutionarily conserved ∼70–amino acid stretches with α-helical structures. Three or four SNAREs on opposing membranes (one located on transport vesicles and the other two or three on the target membrane) form a four-helical bundle complex through their SNARE motifs to drive membrane fusion. Each SNARE is localized in a unique organelle and forms specific complexes with cognate SNAREs to ensure membrane fusion specificity. VAMP7 was identified as the first SNARE protein that functions in the delivery of MT1-MMP to invadopodia (Steffen et al., 2008). Later, syntaxin 4 (STX4) and SNAP23 were defined as cognate SNAREs for VAMP7 in the plasma membrane at invadopodia (Williams et al., 2014). In addition, another SNARE complex comprised of STX13, SNAP23, and VAMP3 has been proposed to participate in MT1-MMP trafficking to the cell surface and ECM degradation (Kean et al., 2009).
Given the complexity of MT1-MMP trafficking in invasive cancer cells, it is reasonable to assume more SNAREs and their complexes may regulate MT1-MMP trafficking. We therefore screened all SNAREs (32 proteins) expressed in human invasive breast cancer cell line MDA-MB-231 for their possible involvement in MT1-MMP trafficking by means of dominant-negative SNARE mutants. We found that Bet1, a SNARE protein that was previously acknowledged to act in anterograde trafficking from the ER to the Golgi apparatus, is localized in MT1-MMP–positive late endosomes, as well as the Golgi, in invasive cancer cells, and participates in efficient MT1-MMP transport to invadopodia and ECM degradation. Strikingly, MT1-MMP appears to recruit Bet1 to post-Golgi compartments through their interaction in a cholesterol-rich membrane subdomain. Our results reveal a novel mechanism for MT1-MMP delivery to invadopodia: it highjacks the Bet1 function for its own transport.
Results
Bet1 is required for efficient ECM degradation
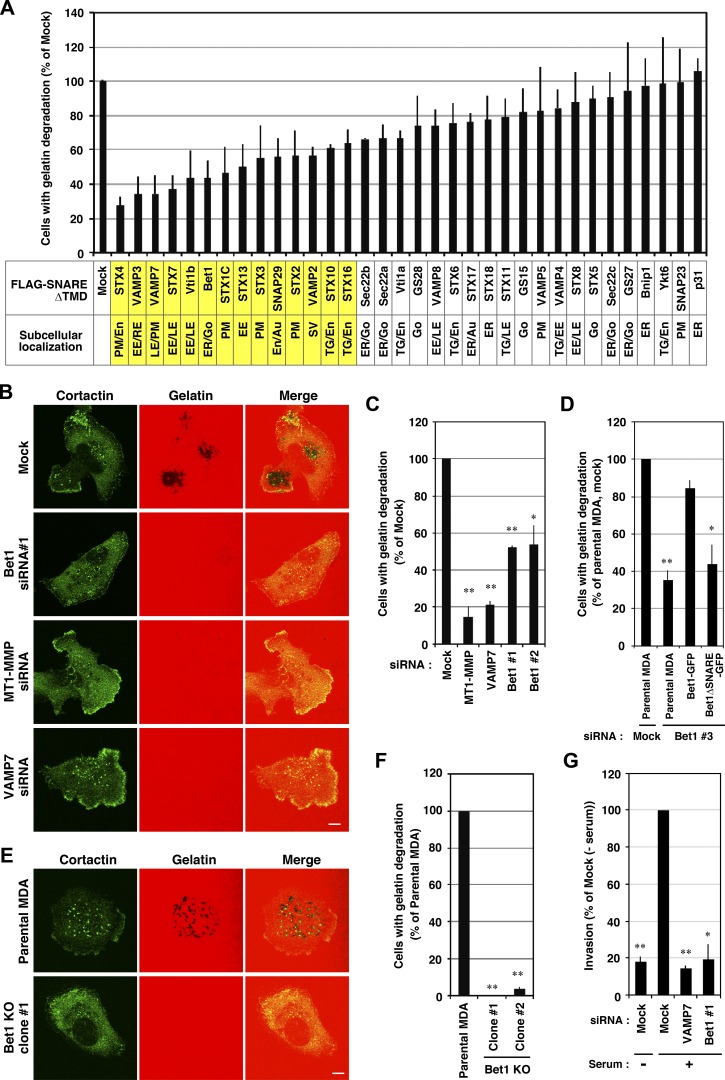
To further identify SNARE proteins involved in ECM degradation, we first surveyed the expression of SNAREs in human invasive breast cancer cell line MDA-MB-231 by RT-PCR. Of the 39 SNARE genes, the expression of 32 was clearly detected, whereas that of the other 7 (STX1B, STX11, STX19, SNAP25, SNAP47, VAMP1, and VAMP2) was not or only very slightly detected (Fig. S1 A). We then examined gelatin degradation by MDA-MB-231 cells, in which putative dominant-negative mutants of SNAREs are transiently overexpressed. Consistent with previous studies (Miyata et al., 2004; Steffen et al., 2008; Kean et al., 2009; Williams et al., 2014), the dominant-negative mutants of STX4, VAMP3, and VAMP7 strongly inhibited gelatin degradation (Fig. 1 A), emphasizing the validity of this screening method, although one of the SNAREs, SNAP23, that has been reported to be involved in MT1-MMP transport (Kean et al., 2009; Williams et al., 2014) was not identified in the screening, probably due to the low level expression of the dominant-negative mutant, suggesting the limitation of our screening.
Figure 1.
Bet1 is required for efficient ECM degradation. (A) Screening of SNARE proteins involved in invadopodium-mediated gelatin degradation by overexpression of dominant-negative SNAREs. MDA-MB-231 cells transiently expressing FLAG-tagged, TMD-deleted SNAREs were cultured on TRITC-gelatin for 7 h, fixed, and stained for FLAG or cortactin, and then the cells exhibiting gelatin degradation were counted under a fluorescence microscope. (B and C) siRNA-mediated knockdown of Bet1 reduces gelatin degradation. MDA-MB-231 cells transfected with the indicated siRNAs were subjected to gelatin degradation assaying as in A. Representative pictures of gelatin degradation (B) and quantitation of the ratio of the cells exhibiting gelatin degradation (C) are shown. (D) WT Bet1, but not its ΔSNARE mutant, rescues the reduced gelatin degradation with Bet1 knockdown. MDA-MB-231 cells stably expressing WT Bet1-GFP or its ΔSNARE mutant were transfected with Bet1 siRNA#3, which targets the 3′-UTR of Bet1 mRNA and depletes endogenous Bet1, but not Bet1-GFP or its ΔSNARE mutant, and then gelatin degradation was examined as in A. (E and F) CRISPR-mediated KO of Bet1 genes abolishes gelatin degradation. MDA-MB-231 cells in which the Bet1 gene was disrupted were subjected to gelatin degradation assaying as in A. Representative pictures of gelatin degradation (E) and quantitation of the ratio of cells exhibiting gelatin degradation (F) in the Bet1-KO cells are shown. (G) Bet1 knockdown impairs invasion through a Matrigel matrix. Invasion activity of Bet1-depleted MDA-MB-231 cells was assessed by Matrigel-coated Transwell assay using 10% FBS as an attractant. Scale bar: 10 μm. *, P < 0.05; **, P < 0.01; vs. mock in C; vs. parental MDA, mock in D; vs. parental MDA in F; vs. mock (+ serum) in G.
Including the above three, i.e., STX4, VAMP3, and VMAP7, 13 SNAREs whose mutants strongly or moderately inhibited gelatin degradation (highlighted in yellow in Fig. 1 A) were endosomal and plasma membrane SNAREs, consistent with the notion that the endocytic pathway principally regulates the appearance of MT1-MMP at invadopodia (Castro-Castro et al., 2016). The only exception is Bet1 (Fig. 1 A and Fig. S1 B, right, bottom row), an ER-Golgi SNARE (Zhang et al., 1997; Cosson et al., 2005; Inoue et al., 2016). We therefore further explored the involvement of Bet1 in ECM degradation. siRNAs targeting Bet1, MT1-MMP, and VAMP7 effectively knocked down their expression (Fig. S1 C) and significantly inhibited gelatin degradation in MDA-MB-231 cells (Fig. 1, B and C). This phenotype was compensated for by the expression of C-terminally GFP-tagged Bet1 (Bet1-GFP), but not a Bet1-GFP mutant (ΔSNARE) lacking the SNARE domain (Fig. 1 D). The involvement of Bet1 in gelatin degradation was confirmed in Bet1 knockout (KO) cells generated by CRISPR/Cas9-mediated gene editing (Fig. 1, E and F; and Fig. S1 D). In addition, Bet1 depletion impaired invasion activity (Fig. 1 G). These results unequivocally demonstrated that Bet1 is required for ECM degradation.
Bet1 is localized in MT1-MMP–positive endosomes as well as the Golgi apparatus
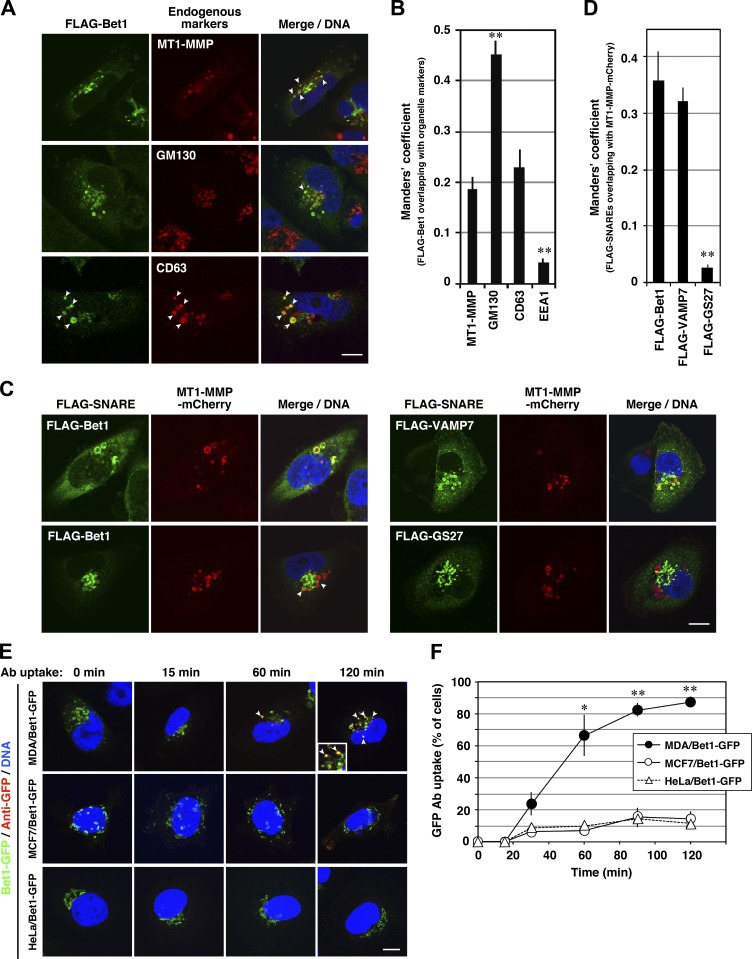
Because an antibody against Bet1 useful for immunofluorescence analysis is not available, we looked at the localization of FLAG-Bet1 in invasive cancer cells. To our surprise, FLAG-Bet1 was found to be not only colocalized with the Golgi marker GM130 but also with MT1-MMP (Fig. 2, A and B), the latter of which has been reported to be principally localized in late endosomes in MDA-MB-231 cells (Steffen et al., 2008; Marchesin et al., 2015). The presence of FLAG-Bet1 in late endosomes was confirmed by the colocalization of FLAG-Bet1 with CD63 (Fig. 2, A and B). Similar colocalization was observed between Tet-inducible N-terminally triple FLAG–tagged Bet1 (3xFLAG-Bet1) expressed at a level comparable to that of endogenous Bet1 and endogenous MT1-MMP (data not shown), and also between FLAG-Bet1 and stably expressed mCherry-tagged MT1-MMP (MT1-MMP-mCherry; Fig. 2, C and D). Consistent with previous studies (Steffen et al., 2008; Marchesin et al., 2015), MT1-MMP-mCherry was found to be localized in late endosomes visualized with FLAG-VAMP7 (Fig. 2, C and D). A similar distribution was observed in other invasive breast cancer cell lines, BT549 and Hs578T (Fig. S2). In contrast to FLAG-Bet1, FLAG-GS27, another ER-Golgi SNARE whose dominant-negative form did not inhibit ECM degradation (Fig. 1 A), did not show any colocalization with MT1-MMP-mCherry (Fig. 2, C and D).
Figure 2.
Bet1 is localized in MT1-MMP–positive endosomes and reaches and is endocytosed from the plasma membrane. (A–D) Bet1 is colocalized with MT1-MMP in endosomal structures. MDA-MB-231 (A) and MDA-MT1-mCh (C) cells transiently expressing FLAG-Bet1 were fixed and then visualized with endogenous MT1-MMP, GM130, or CD63 (A) or MT1-MMP-mCherry (C) by confocal microscopy. In C, FLAG-VAMP7 and FLAG-GS27 were analyzed as FLAG-Bet1. Their colocalization was evaluated with the Manders’ overlap coefficient (B and D). (E) Bet1 reaches the plasma membrane and is endocytosed in MDA-MB-231 cells. MDA-MB-231, MCF7, and HeLa cells stably expressing Bet1-GFP were cultured in the complete medium containing anti-GFP antibodies for the indicated times, fixed, permeabilized, stained with secondary antibodies for anti-GFP, and then visualized by confocal microscopy. (F) Quantitation of the ratio of the cells that take up anti-GFP. Scale bar: 10 μm. *, P < 0.05; **, P < 0.01; vs. MT1-MMP in B; vs. FLAG-Bet1 in D; vs. MCF7/Bet1-GFP and HeLa/Bet1-GFP in F.
To exclude the possibility that the endosomal localization of FLAG-Bet1 is an artifact due to overexpression, we expressed FLAG-Bet1 in noninvasive cells. When expressed in HeLa cells and noninvasive breast cancer cell lines MCF7, SK-BR-3, and T47D, FLAG-Bet1 was found to be well colocalized with GM130 (Fig. S2, B and E), but not markedly with CD63 (Fig. S2, C and F).
Bet1 reaches the plasma membrane and is endocytosed to intracellular compartments
Given that Bet1 is localized in late endosomes, it is tempting to speculate that Bet1 is involved in the trafficking of MT1-MMP from late endosomes to the plasma membrane. If this is the case, Bet1 must reach the plasma membrane in invasive cells. To address this possibility, we adopted a GFP antibody uptake assay. Because Bet1 is a type II membrane protein, the GFP portion of Bet1-GFP is supposed to be exposed to the extracellular space when it arrives at the plasma membrane. Therefore, an antibody against GFP added to the culture medium should be incorporated into cells. When MDA-MB-231 cells stably expressing Bet1-GFP were incubated in GFP antibody–containing medium up to 120 min, the anti-GFP antibody was endocytosed and accumulated in endosomal compartments over time (Fig. 2, E and F). On the other hand, in noninvasive MCF7 and HeLa cells, in which Bet1 was found to be exclusively localized in the Golgi (Fig. S2), uptake and accumulation of an anti-GFP antibody were rarely observed (Fig. 2, E and F). These data suggest that Bet1 can reach the plasma membrane only in invasive cells.
Bet1 is required for MT1-MMP trafficking to the plasma membrane
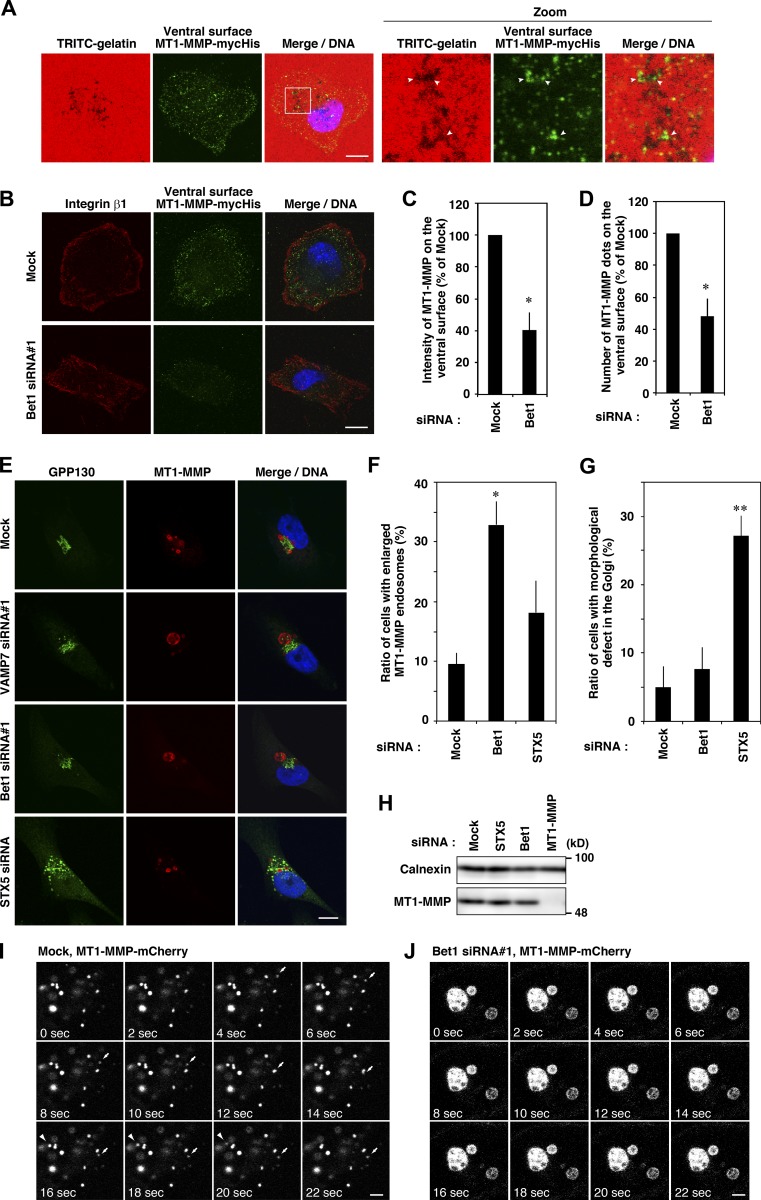
Given the finding that Bet1 is localized in late endosomes and can reach the plasma membrane, and that its depletion inhibits ECM degradation, we next examined whether Bet1 depletion affects the level of MT1-MMP on the ventral cell surface in contact with ECM. To this end, we used cells (MDA-MT1-mycHis) stably expressing C-terminally mycHis-tagged MT1-MMP (MT1-MMP-mycHis) and an antibody that reacts with the extracellular domain of MT1-MMP. In nonpermeabilized cells, mycHis-tagged MT1-MMP gave punctate structures, some of which clearly overlapped with the degraded areas of TRITC-gelatin (Fig. 3 A). Bet1 depletion significantly decreased the MT1-MMP level on the ventral surface marked by integrin β1 (Destaing et al., 2010; Fig. 3, B–D), suggesting that Bet1 is required for efficient delivery of MT1-MMP to the ventral cell surface. In contrast, depletion of Bet1 did not affect the secretion of MMP2 or MMP9 (Fig. S3, A and B), suggesting that Bet1 specifically supports MT1-MMP trafficking, but not that of other ECM-degrading MMPs. In addition, the formation of cortactin-, F-actin–, Tks5-positive structures at the ventral cell surface was not affected by Bet1 depletion (Fig. S3, C and D). Taken together, these results suggest that Bet1 supports the trafficking of MT1-MMP to the cell surface, but not the formation of the actin-based invadopodia-like structure.
Figure 3.
Depletion of Bet1 decreases the MT1-MMP level on the ventral cell surface and alters the morphology and dynamics of MT1-MMP–positive endosomes. (A) Some of the MT1-MMP–positive dots on the ventral surface of the cells overlap gelatin degradation areas. MDA-MT1-mycHis cells were seeded onto TRITC-gelatin and then incubated on ice with anti-MT1-MMP antibodies that recognize the extracellular domain before fixation and permeabilization. Then, the cells were fixed, permeabilized, stained with secondary antibodies, and visualized by confocal microscopy. (B) Depletion of Bet1 decreases MT1-MMP dots on the ventral surface. MDA-MT1-mycHis cells were treated with Bet1 siRNAs, and then MT1-MMP–positive dots on the ventral surface of the cells were analyzed as described in A. Antibodies that recognize the extracellular domain of integrin β1 were used to confirm the ventral surface localization of MT1-MMP. (C and D) Determination of the intensity (C) and number (D) of MT1-MMP–positive dots. (E) Bet1 depletion enlarges MT1-MMP–positive endosomes. MDA-MB-231 cells treated with the indicated siRNAs were immunostained for MT1-MMP and GPP130 and then analyzed by confocal microscopy. (F and G) Quantitation of enlarged MT1-MMP–positive endosomes (F) and the dispersed Golgi (G). (H) The protein level of MT1-MMP was not affected by Bet1 knockdown. Total cell lysates were prepared from siRNA-treated MDA-MB-231 cells, and then immunoblotting for MT1-MMP and calnexin (loading control) was performed. (I and J) Enlarged MT1-MMP–positive endosomes were less dynamic. Live cell imaging of MT1-MMP–positive endosomes was performed in mock or Bet1 siRNA-transfected MDA-MT1-mCh cells. Scale bar: 20 μm in A and B; 10 μm in E; 3 μm in I and J. *, P < 0.05; **, P < 0.01; vs. mock in C, D, F, and G.
Bet1 moves together with MT1-MMP, and its depletion causes swelling of endosomes
When Bet1 was depleted, MT1-MMP–positive endosomes became significantly swollen (Fig. 3, E and F). This effect seemed not to be related to Golgi dysfunction because depletion of STX5 caused severer Golgi fragmentation than that caused by Bet1 depletion (Fig. 3, E and G), with a milder effect on MT1-MMP endosome swelling (Fig. 3, E and F). The protein levels of MT1-MMP were comparable in mock, Bet1-, and STX5-depleted cells (Fig. 3 H). Live cell imaging showed that swollen MT1-MMP–positive endosomes in Bet1-depleted cells are less dynamic than smaller endosomes in mock cells (Fig. 3, I and J; Fig. S3 E; and Videos 1 and 2).
Next, we compared the movement of Bet1-GFP with that of MT1-MMP-mCherry in live cells. In live MDA-MB-231 cells stably expressing MT1-MMP-mCherry (MDA-MT1-mCh cells), a part of Bet1-GFP was persistently localized on some MT1-MMP–positive endosomes showing dynamic movement (Fig. S3, F and G; and Video 3). The endosomal Bet1-GFP was associated with degraded gelatin areas, possibly representing ongoing invadopodia maturation (Fig. S3 H and Video 4). Most Bet1-GFP endosomes left the degraded areas within 120 s, while some were associated with them for over 300 s (Fig. S3, H and I; and Video 4). Bet1 endosomes were more stably decorated or associated with cortactin dots (Fig. S3, J and K; and Video 5), which were highly similar to the punctate structures identified in MT1-MMP–positive endosomes in a previous study (Monteiro et al., 2013).
Overexpression of Bet1 increases ECM degradation and the cell surface MT1-MMP level
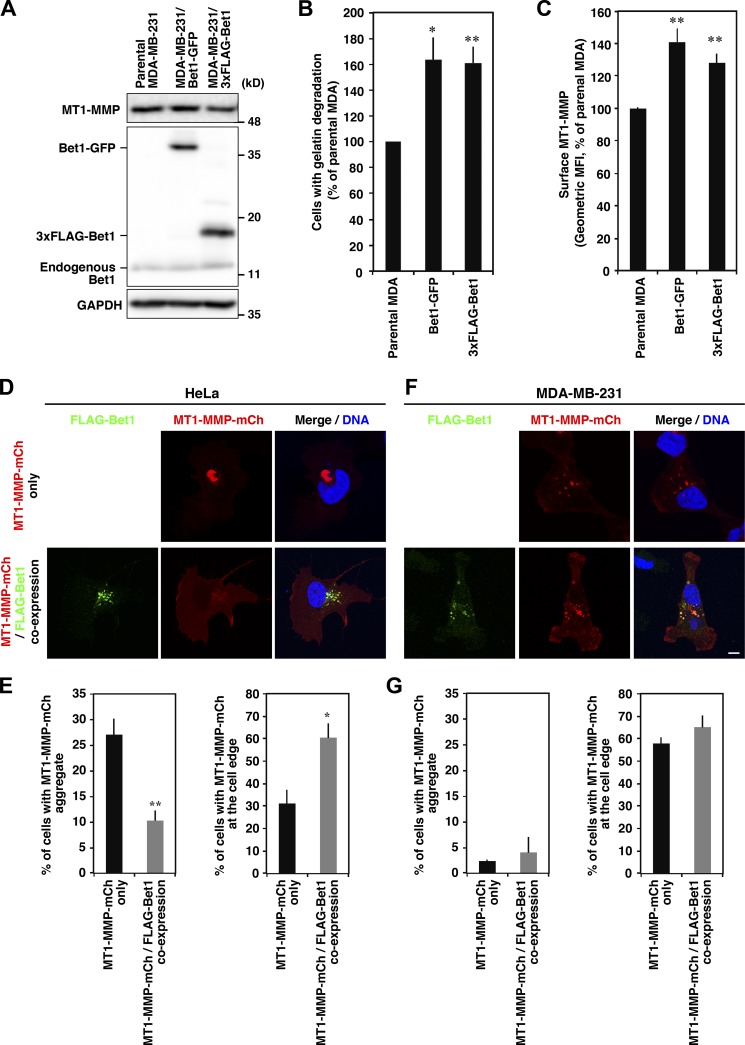
We next examined whether overexpression of Bet1 enhances gelatin degradation. In MDA-MB-231 cells stably expressing Bet1-GFP or 3xFLAG-Bet1, the level of endogenous MT1-MMP was comparable with that in the parental cells (Fig. 4 A). As shown in Fig. 4 B and Fig. S4 A, both cell types showed significantly enhanced gelatin degradation activity compared with the parental cells. In Bet1-GFP and 3xFLAG-Bet1 stable cells, the protein level of MT1-MMP on the ventral cell surface was also elevated, as observed on flow cytometry (Fig. 4 C) and confocal microscopy (Fig. S4, B–D).
Figure 4.
Overexpression of Bet1 elevates gelatin degradation and the protein level of MT1-MMP on the cell surface. (A) Expression levels of MT1-MMP and Bet1 proteins. Endogenous MT1-MMP and endogenous and stably expressed Bet1 proteins in the total cell lysates of parental MDA-MB-231 cells and stable cells expressing Bet1-GFP or 3xFLAG-Bet1 were examined by immunoblotting using specific antibodies. GAPDH was analyzed as a loading control. (B) Quantitation of the ratio of cells exhibiting gelatin degradation. Parental and stable cells were subjected to gelatin degradation assaying as in Fig. 1 A. (C) Flow cytometry of the MT1-MMP level on the cell surface. MT1-MMP on the cell surface was detected with an antibody to the extracellular domain of MT1-MMP without permeabilization and then analyzed by flow cytometry. MFI, mean fluorescence intensity. (D–G) Co-overexpression of Bet1 reduces the aggregation of MT1-MMP and promotes its delivery to the cell edge in HeLa cells. Representative pictures of HeLa (D) and MDA-MB-231 (F) cells transiently transfected with MT1-MMP-mCherry and FLAG-Bet1. Quantitation of the ratio of the cells with MT1-MMP aggregates (left) and at the cell edge (right) of HeLa (E) and MDA-MB-231 (G) cells. Scale bar: 10 μm. *, P < 0.05; **, P < 0.01; vs. parental MDA in B and C; vs. MT1-MMP-mCh only in E.
When MT1-MMP-mCherry was overexpressed in noninvasive HeLa cells, it formed aggregates in the perinuclear region (Fig. 4, D and E). The site of aggregation was likely the ER or an ER subdomain defined by Bap31, but not the Golgi marked by GM130 (data not shown). This aggregation was not observed in invasive MDA-MB-231 cells (Fig. 4, F and G). Notably, coexpression of FLAG-Bet1 in HeLa cells relieved the accumulation of MT1-MMP-mCherry aggregates and increased its level at the cell edge (Fig. 4, D and E). Taken together, these results suggest that Bet1 expression enhances the delivery of MT1-MMP from intracellular compartments to the cell surface, including invadopodia in invasive cancer cells.
Bet1 forms novel SNARE complexes with endosomal SNAREs in MDA-MB-231 cells
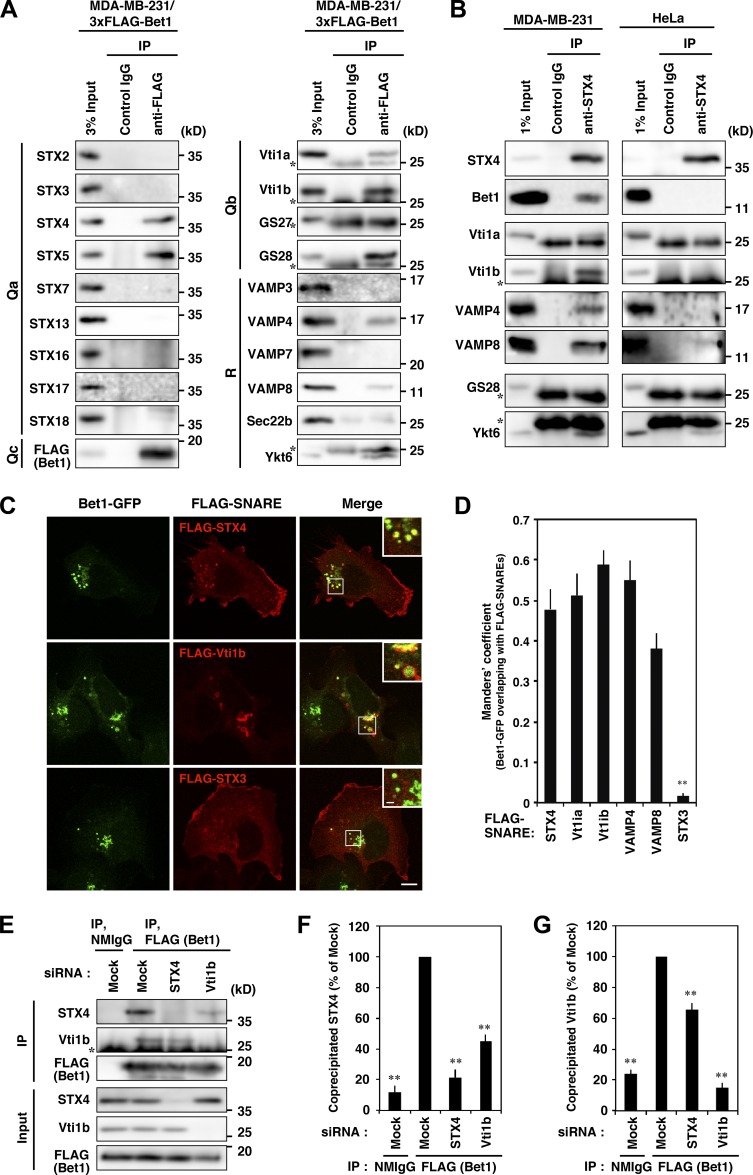
Bet1 has been reported to operate in anterograde transport from the ER to the Golgi as a component of two distinct SNARE complexes, the STX5-GS28-Bet1-Ykt6 and STX5-GS27-Bet1-Sec22b complexes (Xu et al., 2000; Zhang and Hong, 2001). We were interested in whether Bet1 in invasive cancer cells forms different SNARE complexes, in addition to the change in its localization. To examine this, we performed immunoprecipitation using MDA-MB-231 cells stably expressing 3xFLAG-Bet1 and found that it is associated with a number of plasma membrane and endosomal SNAREs (STX4, Vti1a, Vti1b, VAMP4, and VAMP8), as well as conventional Bet1 partners such as STX5, GS28, and Ykt6 (Fig. 5 A). The interaction between endogenous Bet1 and STX4 in MDA-MB-231 cells was verified by immunoprecipitation with an anti-STX4 antibody (Fig. 5 B, left). The immunoprecipitate also contained Vti1a, Vti1b, VAMP4, and VAMP8, as in the case of the immunoprecipitation with 3xFLAG-Bet1. Of note is that no co-precipitation of Bet1 with STX4 was observed in noninvasive HeLa cells (Fig. 5 B, right). These coimmunoprecipitated SNAREs, but not non–co-precipitated SNARE STX3, were also colocalized with Bet1 in endosomal structures (Fig. 5, C and D; and Fig. S4 E).
Figure 5.
Bet1 forms novel SNARE complexes with endosomal SNAREs in MDA-MB-231 cells. (A) 3xFLAG-Bet1 interacts with STX4, Vti1b, and VAMP4 in addition to STX5. MDA-MB-231 cells stably expressing 3xFLAG-Bet1 were lysed, and the lysate was subjected to immunoprecipitation using FLAG-beads, and then the precipitate was subjected to immunoblotting for endogenous SNARE proteins. In the following blots, the brightness and contrast were adjusted so that their input bands became comparable to those of other SNAREs: STX3, STX7, STX16, STX17, STX18, VAMP3, VAMP7, and Vti1b. (B) An endogenous complex comprised of STX4, Bet1, Vti1b, and VAMP4 exists in MDA-MB-231 cells, but not in HeLa cells. Lysates of MDA-MB-231 and HeLa cells were subjected to immunoprecipitation of STX4, and then the co-precipitation of Bet1, Vti1b, and VAMP4 was analyzed. In the following blots, the brightness and contrast were adjusted so that their input bands became comparable to those of other SNAREs: Bet1 and VAMP4 in MDA-MB-231 cells and Bet1, VAMP4, and VAMP8 in HeLa cells. (C and D) Bet1-interacting SNAREs are colocalized with Bet1-GFP in endomembrane compartments. MDA-MB-231 cells stably expressing Bet1-GFP were transfected with FLAG-SNAREs and then subjected to immunofluorescence and confocal microscopy (C). Their colocalization was assessed by using the Manders’ overlap coefficient (D). (E–G) Depletion of STX4 and Vti1b reduces the complex of Bet1 with Vti1b and STX4, respectively. MDA-MB-231 cells stably expressing 3xFLAG-Bet1 were transfected with STX4 and Vti1b siRNAs, and then immunoprecipitation and immunoblotting (E) were performed as in A. Co-precipitated endogenous STX4 (F) and Vti1b (G) were quantified. Scale bar: 10 μm in a regular image; 2 μm in an inset. **, P < 0.01; vs. STX4 in D; vs. FLAG (Bet1), mock in F and G.
To evaluate the specificity of immunoprecipitation, we examined whether the immunoprecipitation efficiency changes upon depletion of each component of the complex. Indeed, the immunoprecipitated amounts of STX4 and Vti1b with 3xFLAG-Bet1 were significantly reduced by the depletion of Vti1b and STX4, respectively (Fig. 5, E–G). These results suggest that Bet1 forms novel endosomal SNARE complexes with STX4 and Vti1b, and perhaps with Vti1a and VAMP4/8 in MDA-MB-231 cells.
MT1-MMP is localized in close proximity to Bet1 in a cholesterol-rich membrane domain
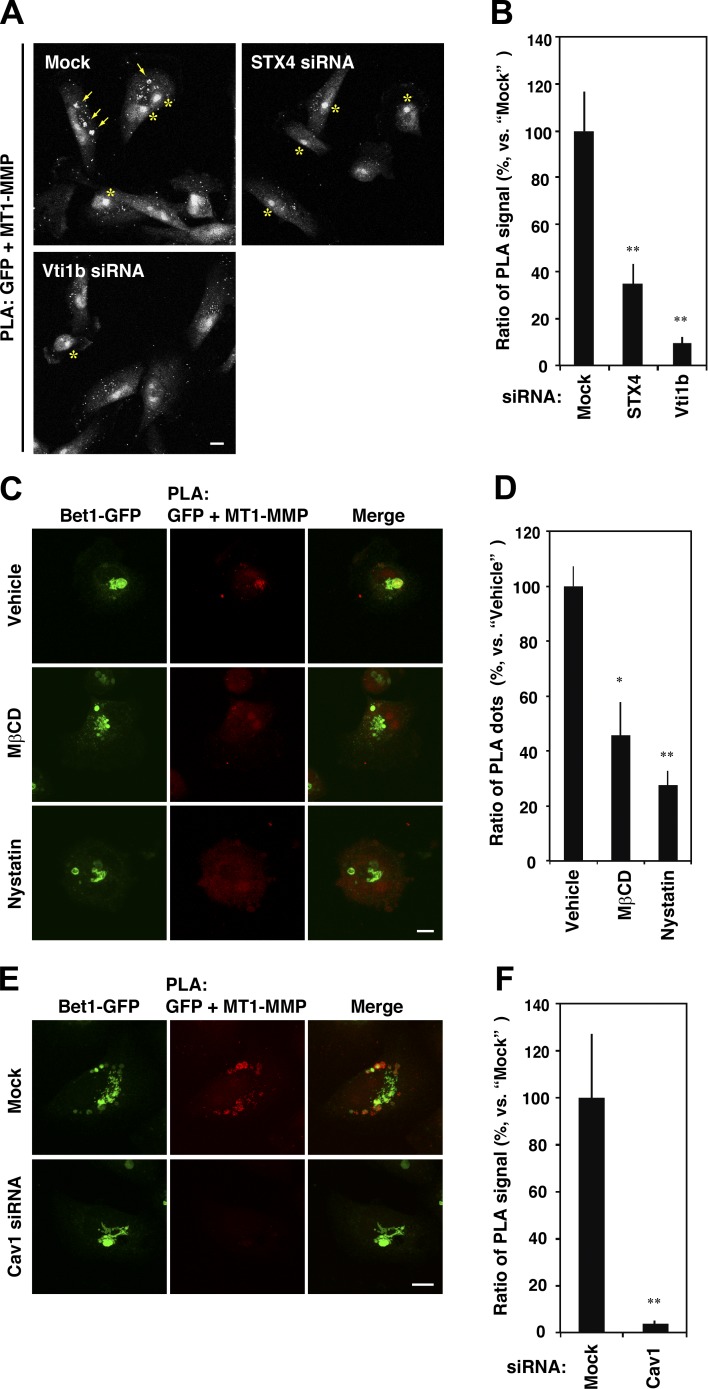
The finding that coexpression of Bet1 with MT1-MMP in noninvasive HeLa cells facilitates the transport of expressed MT1-MMP to the cell surface led us to hypothesize that MT1-MMP itself utilizes and escorts Bet1 for its own transport. Indeed, depletion of MT1-MMP reduced the endosomal localization of Bet1-GFP (Fig. 6, A and B). Moreover, uptake of an anti-GFP antibody by stably Bet1-GFP–expressing cells was markedly inhibited upon MT1-MMP depletion (Fig. 6, C and D). These results suggest that MT1-MMP is a major factor that escorts Bet1 to endocytic compartments and the plasma membrane. A proximity ligation assay (PLA) revealed that MT1-MMP and Bet1-GFP are in close proximity to each other (Fig. 6, E and F). In some cells, the PLA signals strongly accumulated in MT1-MMP–positive endosomes (Fig. 6 G). Bimolecular fluorescence complementation (BiFC) analysis, in which split Venus N- and C-terminal fragments (Vn and Vc) were fused with Bet1 and MT1-MMP, respectively, and expressed in MDA-MB-231 cells, also strongly supports the idea that they interact with each other in the endosomal compartment (Fig. 6 H). Importantly, the PLA signals that were accumulated in endosomes were abrogated on depletion of STX4 or Vti1b (Fig. 7, A and B), suggesting that the assembly of novel endosomal SNAREs is important for the proximity between Bet1-GFP and MT1-MMP.
Figure 6.
MT1-MMP recruits Bet1 to endocytic compartments, where they are in proximity to each other. (A) Depletion of MT1-MMP decreases the endosomal localization of Bet1. MDA-MB-231 cells stably expressing Bet1-GFP were transfected with MT1-MMP siRNA, fixed, and immunostained, and then Bet1-GFP and CD63 were visualized by confocal microscopy. (B) Quantitation of colocalization of Bet1-GFP with CD63 by using the Manders’ overlap coefficient. (C) Bet1-GFP reaching the plasma membrane is reduced through MT-MMP depletion. MDA-MB-231 cells stably expressing Bet1-GFP were cultured in the medium containing GFP antibodies for the indicated periods of time, fixed, and stained, and then Bet1-GFP and the endocytosed GFP antibodies were visualized by confocal microscopy. (D) Quantitation of the level of the endocytosed GFP antibodies. (E) MT1-MMP is in proximity to Bet1. PLA was performed with antibodies to MT1-MMP and GFP in parental MDA-MB-231 cells and Bet1-GFP stable cells. (F) Quantitation of the ratio of the number of PLA dots. (G) Representative pictures of the cells with PLA signals on Bet1-GFP–positive endosomes. (H) Interaction between MT1-MMP and Bet1 in endomembrane compartments is detected by BiFC. Bet1 and MT1-MMP fused with the N- and C-terminal fragments of split Venus, Vn-Bet1 and MT1-MMP-Vc, respectively, were transiently transfected to MDA-MB-231 cells, and the cells were spread on a fibronectin-coated coverslip, fixed, and counterstained with antibodies to CD63 or GFP, and the BiFC signals, and the counterstained proteins were visualized by confocal microscopy. Of note is that the BiFC signal was observed only when Vn-Bet1 and MT1-MMP-Vc were coexpressed, i.e., not when each of them was expressed together with a Vc or Vn fragment. Scale bar: 10 μm. **, P < 0.01; vs. mock in B; vs. mock at each time point in D; vs. GFP + MT1-MMP in F.
Figure 7.
MT1-MMP is located in proximity to Bet1 in a raft-like, cholesterol-rich membrane domain. (A) Depletion of STX4 and Vti1b reduces the PLA signals between MT1-MMP and Bet1-GFP. MDA-MB-231 cells stably expressing Bet1-GFP were transfected with STX4 and Vti1b siRNAs, and the PLA was performed as in Fig. 6 E. Arrows indicate accumulated PLA signals in endosomes, whereas asterisks indicate nonspecific staining in nuclei. (B) Quantitation of PLA signals between MT1-MMP and Bet1-GFP as in A. (C) Cholesterol depletion weakens the interplay between MT1-MMP and Bet1. MDA-MB-231 cells stably expressing Bet1-GFP were spread on a fibronectin-coated coverslip for 7 h and then treated with 0.1% DMSO (vehicle), 5 mM methyl-β-cyclodextrin (MβCD) or 50 µg/ml nystatin for 30 min. After that, the cells were fixed and then subjected to PLA for MT1-MMP and Bet1-GFP. (D) Quantitation of PLA signals between MT1-MMP and Bet1-GFP as in C. (E) Depletion of Cav1 abolishes the proximity between MT1-MMP and Bet1-GFP. The experiments were performed as in A except for the use of Cav1 siRNA. (F) Quantitation of PLA signals between MT1-MMP and Bet1-GFP as in E. Scale bar: 10 μm. *, P < 0.05; **, P < 0.01; vs. mock in B and F; vs. vehicle in D.
Because MT1-MMP was not coimmunoprecipitated with Bet1 (data not shown), we assumed that the interaction between Bet1 and MT1-MMP is transient and/or dependent on the milieu surrounding MT1-MMP. As MT1-MMP is localized in lipid rafts (Yamaguchi et al., 2009), we examined whether cholesterol depletion affects the interaction between Bet1-GFP and MT1-MMP. As expected, depletion of cholesterol with methyl-β-cyclodextrin (Ohtani et al., 1989) or nystatin (Rothberg et al., 1992) abrogated the proximity between Bet1-GFP and MT1-MMP (Fig. 7, C and D). Depletion of caveolin-1 (Cav1), a major component of cholesterol rafts, also nearly abolished their proximity (Fig. 7, E and F).
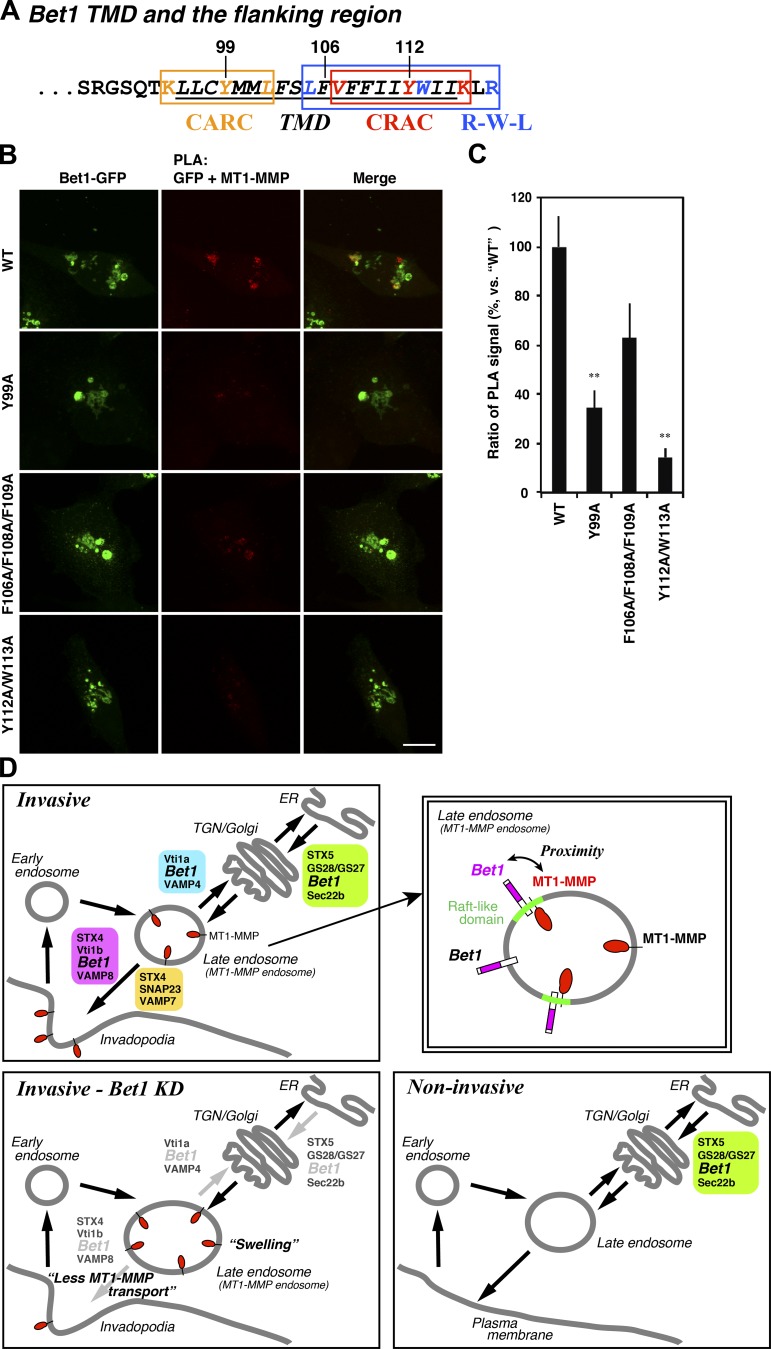
Some membrane proteins that interact with cholesterol in the plasma membrane and endosomal compartments, including G protein–coupled receptors and SNAREs, have cholesterol recognition/interaction motifs called CRAC (cholesterol recognition/interaction amino acid consensus; Fantini and Barrantes, 2013), CARC (inverted CRAC; Fantini and Barrantes, 2013), and R-W-L (a sequence with a combination of basic [R], aromatic [W], and aliphatic [L/V] residues, which is a less stringent CARC motif; Hanson et al., 2008). We found that Bet1 and MT1-MMP have sequences similar to the cholesterol recognition/interaction motifs (Fig. 8 A and Fig. S5 A), and speculated that these motifs might mediate the interaction between Bet1 and MT1-MMP in cholesterol-enriched membrane domains. Indeed, some mutations in these motifs clearly reduced the proximity between Bet1 and MT1-MMP (Fig. 8, B and C; and Fig. S5, B and C). These results suggest that MT1-MMP is in proximity to Bet1 in a cholesterol-rich membrane domain.
Figure 8.
CRAC/CARC/R-W-L motifs in Bet1 are required for the proximity of MT1-MMP and Bet1. (A) Putative CRAC/CARC/R-W-L motifs in Bet1. (B) Bet1 CRAC/CARC/R-W-L motif mutants exhibit reduced PLA signals to MT1-MMP. Bet1 mutants in which consensus residues were replaced with alanine were transfected into MDA-MB-231 cells, which were subjected to PLA reaction with endogenous MT1-MMP as in Fig. 6 E. (C) Quantitation of PLA signals between MT1-MMP and Bet1-GFP as in B. (D) A hypothetical model of the Bet1 function in invasive and noninvasive cancer cells. Scale bar: 10 μm. **, P < 0.01; vs. WT in C.
Discussion
In the present study, we found that in invasive breast cancer cells, Bet1, a SNARE protein known to be localized and to function in the early secretory pathway in noninvasive cells, is localized in MT1-MMP–positive endosomes (Fig. 2, A–D; and Fig. S2, A and D), reaches the plasma membrane (Fig. 2, E and F), and supports the transport of MT1-MMP to plasma membrane–associated invadopodia (Fig. 3, A–D; Fig. 4 C; and Fig. S4, B–D), in addition to its localization and basal function in the ER and Golgi. Moreover, coexpression of Bet1 with MT1-MMP ameliorated the aggregation of expressed MT1-MMP in noninvasive cells and facilitated its transport to the plasma membrane (Fig. 4, D and E). These results suggest that MT1-MMP “hijacks” Bet1 and forces it to assist in the transport of MT1-MMP to invadopodia. Our results also suggest that MT1-MMP itself is a key factor for the change in the Bet1 function. Depletion of MT1-MMP from MDA-MB-231 cells caused redistribution of Bet1 from endosomes to the Golgi apparatus (Fig. 6, A and B) and abrogated the ability of Bet1 to reach the plasma membrane (Fig. 6, C and D). These results and our current working hypothesis are also summarized in Fig. 8 D.
The interaction/proximity between MT1-MMP and Bet1 depends on Bet1-interacting SNAREs, i.e., STX4 and Vti1b (Fig. 7, A and B), a cholesterol-rich milieu, and Cav1 (Fig. 7, C–F). It is of note that STX4 and Vti1b are localized in cholesterol-enriched membranes (Predescu et al., 2005; Reverter et al., 2011) and interact with cholesterol (Hulce et al., 2013; Enrich et al., 2015). Although association of Bet1 with cholesterol has not been demonstrated, it possesses consensus motifs for cholesterol recognition and/or interaction—CRAC, CARC, and R-W-L motifs (Fig. 8 A)—predicting its potential to bind to cholesterol. One of the Bet1-interacting SNAREs, Vti1b, also has the CRAC motif (Enrich et al., 2015). Moreover, MT1-MMP is also localized in cholesterol-enriched lipid rafts (Yamaguchi et al., 2009) and possesses the CRAC motif (Fig. S5 A). As MT1-MMP is a key factor that causes Bet1 recruitment to endosomes, it is tempting to speculate that the expression of MT1-MMP per se creates a cholesterol-rich microdomain by recruiting cholesterol through its CRAC motif. Indeed, some mutations in the cholesterol recognition and/or interaction motifs of Bet1 and MT1-MMP abolished the proximity between Bet1 and MT1-MMP (Fig. 8, B and C; and Fig. S5, B and C).
SNAREs can be classified into v (vesicle)- and t (target)-SNAREs on the basis of their relative positions in opposing membranes, and Qa,b,c- and R-SNAREs (Q: Gln and R: Arg) on the basis of amino acids located in the zero layer of the SNARE complex (Jahn and Scheller, 2006). There is a rough correlation between these two classifications: v-SNAREs, which provide one helix from vesicles, and t-SNAREs, which supply three helices from target membranes, correspond to R- and Q-SNAREs, respectively. Although Bet1 is a Qc-SNARE (Hong, 2005), our results favor the idea that Bet1 may function as a v-SNARE on tubulovesicular carriers. This is not surprising because the liposome fusion assay demonstrated that yeast Bet1 appears to function as a v-SNARE (Parlati et al., 2000), that is, a SNARE on the membrane that provides one helix for membrane fusion.
Although the presence of Bet1 on endosomes has so far not been reported in mammalian cells, a genome-wide interaction study showed that CG14084, the Drosophila melanogaster orthologue of mammalian Bet1, interacts with Drosophila endosomal and plasma membrane SNAREs such as VAMP7, STX1A, and STX4, in addition to ER-Golgi SNAREs (Guruharsha et al., 2011). In addition, GS15, also known as Bet1L, which is an isoform of Bet1, was originally identified as a Golgi-associated SNARE (Xu et al., 2002), but later was shown to act in endosomes as well as the Golgi (Tai et al., 2004). These results suggest that Bet1 potentially functions in endosomes.
Our results revealed that invasive cancer cells exploit Bet1 through routes different from the original ones. This strategy of invasive cancer cells may not be limited to Bet1. Recently, Rab2A, a key small G protein for ER-Golgi transport (Tisdale et al., 1992), was found to facilitate the trafficking of MT1-MMP from late endosomes to the plasma membrane through interaction with VPS39, a critical subunit of the homotypic fusion and vacuole protein sorting complex, in invasive breast cancer cells (Kajiho et al., 2016). As in the case of Bet1, Drosophila Rab2A also functions in the endosome–lysosome pathway (Gillingham et al., 2014; Lőrincz et al., 2017). Therefore, Bet1 and Rab2 have the potential to function in the endocytic pathway as well as the early secretory pathway. Invasive cancer cells exploit this potential for the production of the ECM degradation machinery.
In conclusion, our findings suggest that MT1-MMP changes the Bet1 function for its efficient transport to invadopodia. Bet1 is critical for the formation of functional invadopodia that degrade ECM, and therefore could be a novel target for diagnosis, treatment, and prognosis prediction of the disease.
Materials and methods
Cell culture and stable-cell isolation
Human breast cancer cell lines, i.e., MDA-MB-231, BT549, and Hs578T, were obtained from the American Type Culture Collection and cultured as described previously (Yamaguchi et al., 2009). Other cells were maintained as described previously (Inoue et al., 2015). Stable cells were isolated using plasmids and retrovirus vectors containing a tandem sequence of the internal ribosomal entry site (IRES), and selection markers such as pCI-IRES-bsr, pCX4-bsr, and pMXs-IP (Akagi et al., 2000; Yonekawa et al., 2011). Blasticidin S (Kaken Pharmaceutical Co. Ltd.) and puromycin (Wako Pure Chemicals) were added to the culture medium at 10 μg/ml and 1 μg/ml, respectively. Bet1 gene–disrupted MDA-MB-231 cells were generated by the CRISPR/Cas9-based method using pSpCas9(BB)-2A-GFP (Addgene). The gRNAs for Bet1 were as follows: gRNA1 for clone 1, 5′-GAGTACCTCCTGGCAACTAT-3′, and gRNA2 for clone 2, 5′-AACTATGGCTATGCTAATAG-3′.
DNA constructs, siRNAs, and transfection
Total RNA purified from subconfluent MDA-MB-231 cells was subjected to reverse-transcription reaction to synthesize complementary DNA, and the ORFs encoding SNAREs were then PCR-amplified using specific primers. The amplified DNAs were cloned into pFLAG-CMV-6 (Sigma-Aldrich), pEGFP (Clontech), and the 3xFLAG vector pcDNA4/TO-3xFLAG (donated by Dr. Craig R. Roy, Yale University, New Haven, CT; Ingmundson et al., 2007). The dominant-negative mutants of transmembrane domain (TMD)–containing SNAREs were constructed by deleting the TMD by PCR-based methods. The mutants of TMD-less SNAREs (STX11, SNAP23, SNAP29, and Ykt6) were generated according to previous studies (Scott et al., 2003; Fukasawa et al., 2004; Offenhäuser et al., 2011). For BiFC, Vn (aa 1–172) and Vc (aa 154–236) fragments of Venus were fused with the N-terminus of Bet1 and the C-terminus of MT1-MMP, respectively, in the pCI vector (Promega). Site-directed mutagenesis of Bet1 and MT1-MMP was performed by the QuikChange mutagenesis method (Agilent Technologies). The target sequences of the siRNAs used in this study were as follows: Bet1 siRNA#1, 5′-AAGCAAAGTAACTGCTATAAA-3′; Bet1 siRNA#2, 5′-AAGTTAAAACCCAGAATAAAT-3′; Bet1 siRNA#3, 5′-AACATTCCTAGTGTTCAAATA-3′; STX4 siRNA, 5′-GACAATTCGGCAGACTATTGT-3′; Vti1b siRNA, 5′-AAGAAAAGAAGAAATTGATCA-3′; VAMP7 siRNA, 5′-ACGTACTCACATGGCAATTAT-3′; and Cav1 siRNA, 5′-AACAATTTATGAATTGAATTA-3′. MT1-MMP and STX5 siRNAs were described in previous papers (Steffen et al., 2008; Miyazaki et al., 2012). Plasmids and siRNAs were transfected into cells using Lipofectamine 2000 and RNAiMAX (Thermo Fisher Scientific), respectively, according to the manufacturer’s instructions.
Immunofluorescence and live cell imaging
Immunostaining and confocal microscopy were performed as described previously (Inoue et al., 2008, 2015). Briefly, cells were cultured on gelatin- or fibronectin-coated glass coverslips, fixed with 4% paraformaldehyde in PBS, permeabilized with 0.1% Triton X-100 in PBS, and then stained with the antibodies and fluorescent dyes indicated in each figure. To detect cell surface MT1-MMP, cells were incubated with antibodies to the extracellular domain of MT1-MMP (clone 128527; R&D Systems) in the complete medium for 30 min on ice before fixation and permeabilization. A FluoView 1000 laser-scanning confocal microscope equipped with a 60×, 1.35-numerical aperture objective lens (Olympus) was used to capture images. For live cell imaging, cells were seeded on to gelatin- or fibronectin-coated glass-bottom dishes (AGC Techno Glass), and images were captured at 1-s intervals for 5 min. Colocalization of two channels was quantified with JACoP plugin in ImageJ software (Bolte and Cordelières, 2006).
Gelatin degradation assay
Coverslips coated with TRITC-labeled gelatin were prepared as described previously (Bowden et al., 2001). Cells were seeded onto the coverslips and cultured for 7 h to allow them to degrade fluorescent gelatin. After cells had been fixed and counter-stained with the indicated antibodies, e.g., anti-cortactin or FLAG, the numbers of total cells and cells over the areas of the degraded gelatin were determined under a fluorescence microscope. More than 100 cells were counted in each experiment, and then the ratios of the cells to the degraded gelatin areas were calculated. Alternatively, the degraded areas and whole-cell areas were measured with ImageJ software, and the ratios of the degraded areas were calculated. For each experiment, the results are presented as relative values to the control cells.
Antibody endocytosis assay and FACS
Cells stably expressing Bet1-GFP were spread on fibronectin-coated coverslips and incubated in the complete medium containing antibodies to GFP (clone mFx73; Wako Pure Chemicals) on ice for 30 min, and then heated up to 37°C and cultured for the indicated periods of time. After extensive washing, the cells were fixed, stained with Alexa Fluor 594–labeled anti-mouse IgG and Hoechst 33342, and then analyzed by confocal microscopy as described above. For FACS to detect MT1-MMP on the cell surface, cells were harvested with PBS containing 5 mM EDTA, incubated sequentially with antibodies to the extracellular domain of MT1-MMP and fluorescence-labeled secondary antibodies in PBS containing 0.5% BSA and 5 mM EDTA on ice without fixation or permeabilization, and then analyzed with a flow cytometer (FACSVerse; BD Biosciences).
Immunoprecipitation
Immunoprecipitation was performed as described previously (Inoue et al., 2015). Briefly, subconfluent cells were lysed in lysis buffer comprising 1% Triton X-100; 25 mM Hepes-KOH, pH 7.2; 150 mM KCl; 2 mM EDTA; 1 mM DTT; and protease inhibitors, and then the lysates were reacted with the indicated antibodies or control IgG from nonimmunized animals and Protein G beads (Jackson ImmunoResearch). After centrifugation and extensive washing with the lysis buffer, the precipitated materials were analyzed by immunoblotting using the indicated antibodies.
PLA
The PLA was performed using a Duolink PLA kit (Sigma-Aldrich) according to the manufacturer's instructions and as previously described (Arasaki et al., 2015) except for the use of anti–MT1-MMP (R&D Systems) and anti-GFP (Thermo Fisher Scientific), or anti-mCherry (GeneTex) and anti-FLAG M2 (Sigma-Aldrich). Cholesterol depletion was performed as described previously (Ohtani et al., 1989; Rothberg et al., 1992). Briefly, cells were spread on fibronectin-coated coverslips and left for 7 h, and then treated with 5 μM methyl-β-cyclodextrin (Wako Pure Chemicals) or 50 μg/ml nystatin (Sigma-Aldrich) for 30 min at 37°C. After that, the cells were fixed and subjected to PLA reactions. PLA signals were imaged by confocal microscopy and quantified by using ImageJ software.
Gelatin zymography
Cells were seeded at 0.7 × 105 cells/well in a 12-well plate. After 24 h, siRNAs were transfected as described above, and then the cells were cultured for 24 h. The transfected cells were then split at 1:2 and plated on gelatin-coated 12-well plates with 0.5 ml of serum-free Opti-MEM (Thermo Fisher Scientific) and cultured for 48 h. The conditioned media were collected and mixed with an equal volume of nonreducing sample buffer (250 mM Tris-HCl, pH 6.8; 25% glycerol; 10% SDS; and 0.01% bromophenol blue), and then proteins secreted into the media were separated by 1 mg/ml gelatin-containing SDS-PAGE. The gel was incubated in renaturation buffer (2.5% Triton X-100) for 2 h at 37°C and in substrate buffer (50 mM Tris-HCl, pH 7.4, and 5 mM CaCl2) for 24 h at 37°C and stained with Coomassie Brilliant Blue. Unstained bands, where MMP2 and MMP9 degraded gelatin in the gel, were quantified using ImageJ software.
Transwell invasion assay
Transwell invasion assays were performed as described previously (Shaw, 2005). Briefly, cells were seeded into Transwell chambers (8-μm pores; Corning) coated with Matrigel (BD) with serum-free medium and then cultured for 24 h with culture medium containing 10% FBS as a chemoattractant in the bottom chambers to allow the cells to invade into the Matrigel layer and reach to the lower side of the Transwell porous membrane. The invaded cells were stained with crystal violet and counted by microscopy.
Antibodies and other reagents
The commercial mouse monoclonal antibodies used here were anti-FLAG M2 (Sigma-Aldrich), anti–MT1-MMP (R&D Systems), anti-cortactin (clone 4F11; EMD Millipore), anti-Bet1 (clone 17; Santa Cruz Biotechnology), anti-calnexin (clone 37; BD), anti-GM130 (clone 35; BD), anti-EEA1 (clone 14; BD), anti-LAMP1 (clone H4A3; Developmental Studies Hybridoma Bank), anti-α-tubulin (clone B-5-1-2; Sigma-Aldrich), and anti-integrin β1 (clone 12G10; Santa Cruz Biotechnology). The commercial rabbit polyclonal antibodies used were anti-FLAG (Sigma-Aldrich), anti–MT1-MMP (EMD Millipore), anti-STX16 (Synaptic Systems), anti-GFP (Thermo Fisher Scientific), and anti-GPP130 (Covance). The rabbit polyclonal antibodies to STX4, STX5, STX7, STX17, and STX18 were as described previously (Hatsuzawa et al., 2000; Mizoguchi et al., 2000; Arasaki et al., 2015; Inoue et al., 2015). Anti-STX2, -STX3, -VAMP7, and -Vti1b polyclonal antibodies were raised by immunizing rabbits with the purified recombinant His-tagged cytoplasmic domains of human STX2 (aa 1–264), STX3 (aa 1–263), VAMP7 (aa 1–188), and Vti1b (aa 1–209), respectively, and then the antisera were affinity-purified. Secondary antibodies labeled with HRP and fluorochrome were purchased from BioRad Laboratories and Life Technologies, respectively.
Statistical analysis
The results of at least three experiments were expressed as means ± SEM. Statistical analysis was performed using a two-tailed, unpaired Student's t test, with P < 0.05 considered to be statistically significant.
Online supplemental material
Fig. S1 shows the expression of 39 SNAREs in MDA-MB-231 cells; dominant-negative effects of STX7, Vti1b, and Bet1 ΔTMD mutants on gelatin degradation; knockdown levels of Bet1, MT1-MMP, and VAMP7 proteins; and Bet1 protein levels in Bet1 KO cell clones. Fig. S2 shows the localization analysis of Bet1 in invasive and noninvasive breast cancer cell lines. Fig. S3 shows gelatin zymography by secreted MMP2 and MMP9 from Bet1-depleted MDA-MB-231 cells; Tks5-, cortactin-, and F-actin–positive invadopodial structure in Bet1-depleted cells; and live cell imaging of Bet1, MT1-MMP, cortactin, and degraded gelatin spots. Fig. S4 shows gelatin degradation and ventral surface MT1-MMP in Bet1-overexpressing MDA-MB-231 cells, and colocalization of Bet1 with Vti1b, VAMP4, and VAMP8. Fig. S5 shows a putative CRAC sequence in the TMD of MT1-MMP, and PLA between Bet1 and MT1-MMP CRAC mutants. Videos 1 and 2 show dynamics of MT1-MMP-mCherry in mock and Bet1-depleted MDA-MB-231 cells, respectively. Video 3 shows dynamics of colocalization between Bet1-GFP and MT1-MMP-mCherry. Video 4 shows dynamic of Bet1-GFP vesicles associated with degraded gelatin spots. Video 5 shows dynamics of associations between mCherry-Bet1 and cortactin-GFP.
Supplementary Material
Acknowledgments
We thank Asami Tsuchida, Ayano Nakada, Haruka Kusukami, Risae Gokita, Yuka Katano, Sanae Kikuchi, Marina Sakaniwa, Kimiko Minegishi, Akira Hamauji, Takuma Yamaguchi, Shiori Yoshida, and Sachiho Kageyama for experimental support.
This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan to H. Inoue (Project Numbers 23570174, 26440064, and 18K06138), K. Arasaki (26713016), Y. Wakana (15K18507), and M. Tagaya. (25291029).
The authors declare no competing financial interests.
Author contributions: T. Miyagawa, K. Hasegawa, Y. Aoki, and T. Watanabe performed the immunofluorescence and gelatin degradation experiments. T. Watanabe cloned and constructed most of the SNARE expression vectors. T. Miyagawa, Y. Aoki, and T. Watanabe performed the screening of SNAREs that are involved in MT1-MMP trafficking. K. Hasegawa conducted the immunoprecipitation experiments. Y. Aoki and Y. Otagiri performed the experiments of live cell imaging. Y. Otagiri conducted the BiFC experiments and constructed CRAC mutants. K. Hasegawa and H. Inoue performed the PLA experiments. K. Arasaki, Y. Wakana, and M. Tagaya provided critical feedback for experiments. H. Yamaguchi provided materials, protocols, and critical feedback for experiments. K. Asano and M. Tanaka assisted with flow cytometry. M. Tagaya and H. Inoue wrote the manuscript. H. Inoue performed most of the experiments and conceived and supervised the project.
References
- Akagi T., Shishido T., Murata K., and Hanafusa H.. 2000. v-Crk activates the phosphoinositide 3-kinase/AKT pathway in transformation. Proc. Natl. Acad. Sci. USA. 97:7290–7295. 10.1073/pnas.140210297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arasaki K., Shimizu H., Mogari H., Nishida N., Hirota N., Furuno A., Kudo Y., Baba M., Baba N., Cheng J., et al. 2015. A role for the ancient SNARE syntaxin 17 in regulating mitochondrial division. Dev. Cell. 32:304–317. 10.1016/j.devcel.2014.12.011 [DOI] [PubMed] [Google Scholar]
- Bolte S., and Cordelières F.P.. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224:213–232. 10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
- Bowden E.T., Coopman P.J., and Mueller S.C.. 2001. Invadopodia: Unique methods for measurement of extracellular matrix degradation in vitro. Methods Cell Biol. 63:613–627. 10.1016/S0091-679X(01)63033-4 [DOI] [PubMed] [Google Scholar]
- Castro-Castro A., Marchesin V., Monteiro P., Lodillinsky C., Rossé C., and Chavrier P.. 2016. Cellular and molecular mechanisms of MT1-MMP-dependent cancer cell invasion. Annu. Rev. Cell Dev. Biol. 32:555–576. 10.1146/annurev-cellbio-111315-125227 [DOI] [PubMed] [Google Scholar]
- Chaffer C.L., and Weinberg R.A.. 2011. A perspective on cancer cell metastasis. Science. 331:1559–1564. 10.1126/science.1203543 [DOI] [PubMed] [Google Scholar]
- Clark E.S., Whigham A.S., Yarbrough W.G., and Weaver A.M.. 2007. Cortactin is an essential regulator of matrix metalloproteinase secretion and extracellular matrix degradation in invadopodia. Cancer Res. 67:4227–4235. 10.1158/0008-5472.CAN-06-3928 [DOI] [PubMed] [Google Scholar]
- Cosson P., Ravazzola M., Varlamov O., Söllner T.H., Di Liberto M., Volchuk A., Rothman J.E., and Orci L.. 2005. Dynamic transport of SNARE proteins in the Golgi apparatus. Proc. Natl. Acad. Sci. USA. 102:14647–14652. 10.1073/pnas.0507394102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destaing O., Planus E., Bouvard D., Oddou C., Badowski C., Bossy V., Raducanu A., Fourcade B., Albiges-Rizo C., and Block M.R.. 2010. β1A integrin is a master regulator of invadosome organization and function. Mol. Biol. Cell. 21:4108–4119. 10.1091/mbc.e10-07-0580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy R.J., Weidmann M.D., Sharma V.P., and Condeelis J.S.. 2017. Tumor cell invadopodia: Invasive protrusions that orchestrate metastasis. Trends Cell Biol. 27:595–607. 10.1016/j.tcb.2017.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enrich C., Rentero C., Hierro A., and Grewal T.. 2015. Role of cholesterol in SNARE-mediated trafficking on intracellular membranes. J. Cell Sci. 128:1071–1081. 10.1242/jcs.164459 [DOI] [PubMed] [Google Scholar]
- Fantini J., and Barrantes F.J.. 2013. How cholesterol interacts with membrane proteins: an exploration of cholesterol-binding sites including CRAC, CARC, and tilted domains. Front. Physiol. 4:31 10.3389/fphys.2013.00031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa M., Varlamov O., Eng W.S., Söllner T.H., and Rothman J.E.. 2004. Localization and activity of the SNARE Ykt6 determined by its regulatory domain and palmitoylation. Proc. Natl. Acad. Sci. USA. 101:4815–4820. 10.1073/pnas.0401183101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham A.K., Sinka R., Torres I.L., Lilley K.S., and Munro S.. 2014. Toward a comprehensive map of the effectors of rab GTPases. Dev. Cell. 31:358–373. 10.1016/j.devcel.2014.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruharsha K.G., Rual J.F., Zhai B., Mintseris J., Vaidya P., Vaidya N., Beekman C., Wong C., Rhee D.Y., Cenaj O., et al. 2011. A protein complex network of Drosophila melanogaster. Cell. 147:690–703. 10.1016/j.cell.2011.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson M.A., Cherezov V., Griffith M.T., Roth C.B., Jaakola V.P., Chien E.Y., Velasquez J., Kuhn P., and Stevens R.C.. 2008. A specific cholesterol binding site is established by the 2.8 Å structure of the human β2-adrenergic receptor. Structure. 16:897–905. 10.1016/j.str.2008.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsuzawa K., Hirose H., Tani K., Yamamoto A., Scheller R.H., and Tagaya M.. 2000. Syntaxin 18, a SNAP receptor that functions in the endoplasmic reticulum, intermediate compartment, and cis-Golgi vesicle trafficking. J. Biol. Chem. 275:13713–13720. 10.1074/jbc.275.18.13713 [DOI] [PubMed] [Google Scholar]
- Hong W. 2005. SNAREs and traffic. Biochim. Biophys. Acta. 1744:493–517. 10.1016/j.bbamcr.2005.03.014 [DOI] [PubMed] [Google Scholar]
- Hoshino D., Branch K.M., and Weaver A.M.. 2013. Signaling inputs to invadopodia and podosomes. J. Cell Sci. 126:2979–2989. 10.1242/jcs.079475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulce J.J., Cognetta A.B., Niphakis M.J., Tully S.E., and Cravatt B.F.. 2013. Proteome-wide mapping of cholesterol-interacting proteins in mammalian cells. Nat. Methods. 10:259–264. 10.1038/nmeth.2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingmundson A., Delprato A., Lambright D.G., and Roy C.R.. 2007. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature. 450:365–369. 10.1038/nature06336 [DOI] [PubMed] [Google Scholar]
- Inoue H., Ha V.L., Prekeris R., and Randazzo P.A.. 2008. Arf GTPase-activating protein ASAP1 interacts with Rab11 effector FIP3 and regulates pericentrosomal localization of transferrin receptor-positive recycling endosome. Mol. Biol. Cell. 19:4224–4237. 10.1091/mbc.e08-03-0290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H., Matsuzaki Y., Tanaka A., Hosoi K., Ichimura K., Arasaki K., Wakana Y., Asano K., Tanaka M., Okuzaki D., et al. 2015. γ-SNAP stimulates disassembly of endosomal SNARE complexes and regulates endocytic trafficking pathways. J. Cell Sci. 128:2781–2794. 10.1242/jcs.158634 [DOI] [PubMed] [Google Scholar]
- Inoue H., Tani K., and Tagaya M.. 2016. SNARE-associated proteins and receptor trafficking. Receptors Clin. Investig. 3:e1377 10.14800/rci.1377 [DOI] [Google Scholar]
- Jacob A., Jing J., Lee J., Schedin P., Gilbert S.M., Peden A.A., Junutula J.R., and Prekeris R.. 2013. Rab40b regulates trafficking of MMP2 and MMP9 during invadopodia formation and invasion of breast cancer cells. J. Cell Sci. 126:4647–4658. 10.1242/jcs.126573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R., and Scheller R.H.. 2006. SNAREs—Engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7:631–643. 10.1038/nrm2002 [DOI] [PubMed] [Google Scholar]
- Kajiho H., Kajiho Y., Frittoli E., Confalonieri S., Bertalot G., Viale G., Di Fiore P.P., Oldani A., Garre M., Beznoussenko G.V., et al. 2016. RAB2A controls MT1-MMP endocytic and E-cadherin polarized Golgi trafficking to promote invasive breast cancer programs. EMBO Rep. 17:1061–1080. 10.15252/embr.201642032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kean M.J., Williams K.C., Skalski M., Myers D., Burtnik A., Foster D., and Coppolino M.G.. 2009. VAMP3, syntaxin-13 and SNAP23 are involved in secretion of matrix metalloproteinases, degradation of the extracellular matrix and cell invasion. J. Cell Sci. 122:4089–4098. 10.1242/jcs.052761 [DOI] [PubMed] [Google Scholar]
- Linder S. 2007. The matrix corroded: Podosomes and invadopodia in extracellular matrix degradation. Trends Cell Biol. 17:107–117. 10.1016/j.tcb.2007.01.002 [DOI] [PubMed] [Google Scholar]
- Linder S., Wiesner C., and Himmel M.. 2011. Degrading devices: Invadosomes in proteolytic cell invasion. Annu. Rev. Cell Dev. Biol. 27:185–211. 10.1146/annurev-cellbio-092910-154216 [DOI] [PubMed] [Google Scholar]
- Lőrincz P., Tóth S., Benkő P., Lakatos Z., Boda A., Glatz G., Zobel M., Bisi S., Hegedűs K., Takáts S., et al. 2017. Rab2 promotes autophagic and endocytic lysosomal degradation. J. Cell Biol. 216:1937–1947. 10.1083/jcb.201611027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesin V., Castro-Castro A., Lodillinsky C., Castagnino A., Cyrta J., Bonsang-Kitzis H., Fuhrmann L., Irondelle M., Infante E., Montagnac G., et al. 2015. ARF6-JIP3/4 regulate endosomal tubules for MT1-MMP exocytosis in cancer invasion. J. Cell Biol. 211:339–358. 10.1083/jcb.201506002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T., Ohnishi H., Suzuki J., Yoshikumi Y., Ohno H., Mashima H., Yasuda H., Ishijima T., Osawa H., Satoh K., et al. 2004. Involvement of syntaxin 4 in the transport of membrane-type 1 matrix metalloproteinase to the plasma membrane in human gastric epithelial cells. Biochem. Biophys. Res. Commun. 323:118–124. 10.1016/j.bbrc.2004.08.064 [DOI] [PubMed] [Google Scholar]
- Miyazaki K., Wakana Y., Noda C., Arasaki K., Furuno A., and Tagaya M.. 2012. Contribution of the long form of syntaxin 5 to the organization of the endoplasmic reticulum. J. Cell Sci. 125:5658–5666. 10.1242/jcs.105304 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T., Nakajima K., Hatsuzawa K., Nagahama M., Hauri H.P., Tagaya M., and Tani K.. 2000. Determination of functional regions of p125, a novel mammalian Sec23p-interacting protein. Biochem. Biophys. Res. Commun. 279:144–149. 10.1006/bbrc.2000.3846 [DOI] [PubMed] [Google Scholar]
- Monteiro P., Rossé C., Castro-Castro A., Irondelle M., Lagoutte E., Paul-Gilloteaux P., Desnos C., Formstecher E., Darchen F., Perrais D., et al. 2013. Endosomal WASH and exocyst complexes control exocytosis of MT1-MMP at invadopodia. J. Cell Biol. 203:1063–1079. 10.1083/jcb.201306162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offenhäuser C., Lei N., Roy S., Collins B.M., Stow J.L., and Murray R.Z.. 2011. Syntaxin 11 binds Vti1b and regulates late endosome to lysosome fusion in macrophages. Traffic. 12:762–773. 10.1111/j.1600-0854.2011.01189.x [DOI] [PubMed] [Google Scholar]
- Ohtani Y., Irie T., Uekama K., Fukunaga K., and Pitha J.. 1989. Differential effects of α-, β- and γ-cyclodextrins on human erythrocytes. Eur. J. Biochem. 186:17–22. 10.1111/j.1432-1033.1989.tb15171.x [DOI] [PubMed] [Google Scholar]
- Parlati F., McNew J.A., Fukuda R., Miller R., Söllner T.H., and Rothman J.E.. 2000. Topological restriction of SNARE-dependent membrane fusion. Nature. 407:194–198. 10.1038/35025076 [DOI] [PubMed] [Google Scholar]
- Paterson E.K., and Courtneidge S.A.. 2018. Invadosomes are coming: New insights into function and disease relevance. FEBS J. 285:8–27. 10.1111/febs.14123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Predescu S.A., Predescu D.N., Shimizu K., Klein I.K., and Malik A.B.. 2005. Cholesterol-dependent syntaxin-4 and SNAP-23 clustering regulates caveolar fusion with the endothelial plasma membrane. J. Biol. Chem. 280:37130–37138. 10.1074/jbc.M505659200 [DOI] [PubMed] [Google Scholar]
- Reverter M., Rentero C., de Muga S.V., Alvarez-Guaita A., Mulay V., Cairns R., Wood P., Monastyrskaya K., Pol A., Tebar F., et al. 2011. Cholesterol transport from late endosomes to the Golgi regulates t-SNARE trafficking, assembly, and function. Mol. Biol. Cell. 22:4108–4123. 10.1091/mbc.e11-04-0332r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothberg K.G., Heuser J.E., Donzell W.C., Ying Y.S., Glenney J.R., and Anderson R.G.. 1992. Caveolin, a protein component of caveolae membrane coats. Cell. 68:673–682. 10.1016/0092-8674(92)90143-Z [DOI] [PubMed] [Google Scholar]
- Sakurai-Yageta M., Recchi C., Le Dez G., Sibarita J.B., Daviet L., Camonis J., D’Souza-Schorey C., and Chavrier P.. 2008. The interaction of IQGAP1 with the exocyst complex is required for tumor cell invasion downstream of Cdc42 and RhoA. J. Cell Biol. 181:985–998. 10.1083/jcb.200709076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoumacher M., Goldman R.D., Louvard D., and Vignjevic D.M.. 2010. Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J. Cell Biol. 189:541–556. 10.1083/jcb.200909113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott C.C., Furuya W., Trimble W.S., and Grinstein S.. 2003. Activation of store-operated calcium channels: Assessment of the role of snare-mediated vesicular transport. J. Biol. Chem. 278:30534–30539. 10.1074/jbc.M304718200 [DOI] [PubMed] [Google Scholar]
- Seiki M., and Yana I.. 2003. Roles of pericellular proteolysis by membrane type-1 matrix metalloproteinase in cancer invasion and angiogenesis. Cancer Sci. 94:569–574. 10.1111/j.1349-7006.2003.tb01484.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw L.M. 2005. Tumor cell invasion assays. Methods Mol. Biol. 294:97–105. 10.1385/1-59259-860-9:097 [DOI] [PubMed] [Google Scholar]
- Steffen A., Le Dez G., Poincloux R., Recchi C., Nassoy P., Rottner K., Galli T., and Chavrier P.. 2008. MT1-MMP-dependent invasion is regulated by TI-VAMP/VAMP7. Curr. Biol. 18:926–931. 10.1016/j.cub.2008.05.044 [DOI] [PubMed] [Google Scholar]
- Tai G., Lu L., Wang T.L., Tang B.L., Goud B., Johannes L., and Hong W.. 2004. Participation of the syntaxin 5/Ykt6/GS28/GS15 SNARE complex in transport from the early/recycling endosome to the trans-Golgi network. Mol. Biol. Cell. 15:4011–4022. 10.1091/mbc.e03-12-0876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisdale E.J., Bourne J.R., Khosravi-Far R., Der C.J., and Balch W.E.. 1992. GTP-binding mutants of rab1 and rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the Golgi complex. J. Cell Biol. 119:749–761. 10.1083/jcb.119.4.749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K.C., McNeilly R.E., and Coppolino M.G.. 2014. SNAP23, Syntaxin4, and vesicle-associated membrane protein 7 (VAMP7) mediate trafficking of membrane type 1-matrix metalloproteinase (MT1-MMP) during invadopodium formation and tumor cell invasion. Mol. Biol. Cell. 25:2061–2070. 10.1091/mbc.e13-10-0582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Joglekar A.P., Williams A.L., and Hay J.C.. 2000. Subunit structure of a mammalian ER/Golgi SNARE complex. J. Biol. Chem. 275:39631–39639. 10.1074/jbc.M007684200 [DOI] [PubMed] [Google Scholar]
- Xu Y., Martin S., James D.E., and Hong W.. 2002. GS15 forms a SNARE complex with syntaxin 5, GS28, and Ykt6 and is implicated in traffic in the early cisternae of the Golgi apparatus. Mol. Biol. Cell. 13:3493–3507. 10.1091/mbc.e02-01-0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H., Lorenz M., Kempiak S., Sarmiento C., Coniglio S., Symons M., Segall J., Eddy R., Miki H., Takenawa T., and Condeelis J.. 2005. Molecular mechanisms of invadopodium formation: the role of the N-WASP-Arp2/3 complex pathway and cofilin. J. Cell Biol. 168:441–452. 10.1083/jcb.200407076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H., Takeo Y., Yoshida S., Kouchi Z., Nakamura Y., and Fukami K.. 2009. Lipid rafts and caveolin-1 are required for invadopodia formation and extracellular matrix degradation by human breast cancer cells. Cancer Res. 69:8594–8602. 10.1158/0008-5472.CAN-09-2305 [DOI] [PubMed] [Google Scholar]
- Yana I., and Weiss S.J.. 2000. Regulation of membrane type-1 matrix metalloproteinase activation by proprotein convertases. Mol. Biol. Cell. 11:2387–2401. 10.1091/mbc.11.7.2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekawa S., Furuno A., Baba T., Fujiki Y., Ogasawara Y., Yamamoto A., Tagaya M., and Tani K.. 2011. Sec16B is involved in the endoplasmic reticulum export of the peroxisomal membrane biogenesis factor peroxin 16 (Pex16) in mammalian cells. Proc. Natl. Acad. Sci. USA. 108:12746–12751. 10.1073/pnas.1103283108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., and Hong W.. 2001. Ykt6 forms a SNARE complex with syntaxin 5, GS28, and Bet1 and participates in a late stage in endoplasmic reticulum-Golgi transport. J. Biol. Chem. 276:27480–27487. 10.1074/jbc.M102786200 [DOI] [PubMed] [Google Scholar]
- Zhang T., Wong S.H., Tang B.L., Xu Y., Peter F., Subramaniam V.N., and Hong W.. 1997. The mammalian protein (rbet1) homologous to yeast Bet1p is primarily associated with the pre-Golgi intermediate compartment and is involved in vesicular transport from the endoplasmic reticulum to the Golgi apparatus. J. Cell Biol. 139:1157–1168. 10.1083/jcb.139.5.1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.