Takahashi et al. perform a genome-wide CRISPR screen using the HaloTag-LC3 assay to gain insight into the mechanisms of phagophore closure. They identify a role for VPS37A in coordinating the ESCRT assembly on the phagophore for membrane closure.
Abstract
The process of phagophore closure requires the endosomal sorting complex required for transport III (ESCRT-III) subunit CHMP2A and the AAA ATPase VPS4, but their regulatory mechanisms remain unknown. Here, we establish a FACS-based HaloTag-LC3 autophagosome completion assay to screen a genome-wide CRISPR library and identify the ESCRT-I subunit VPS37A as a critical component for phagophore closure. VPS37A localizes on the phagophore through the N-terminal putative ubiquitin E2 variant domain, which is found to be required for autophagosome completion but dispensable for ESCRT-I complex formation and the degradation of epidermal growth factor receptor in the multivesicular body pathway. Notably, loss of VPS37A abrogates the phagophore recruitment of the ESCRT-I subunit VPS28 and CHMP2A, whereas inhibition of membrane closure by CHMP2A depletion or VPS4 inhibition accumulates VPS37A on the phagophore. These observations suggest that VPS37A coordinates the recruitment of a unique set of ESCRT machinery components for phagophore closure in mammalian cells.
Introduction
Macroautophagy (herein autophagy) is an intracellular degradation process that mediates the clearance of cytoplasmic material, including proteins, organelles, and pathogens (Mizushima et al., 2008). During the catabolic process, membranous structures known as phagophores elongate to enwrap cytoplasmic cargoes within cup-shaped structures that are sealed to form double-membrane autophagosomes and delivered to lysosomes for hydrolytic degradation and nutrient recycling (Lamb et al., 2013). Autophagy is induced in response to a variety of stresses, such as starvation, to maintain cellular and tissue homeostasis (Kroemer et al., 2010). Dysregulation of autophagy has been implicated in various human diseases, including cancer, neurodegeneration, and pathogen infections (Mizushima et al., 2008).
The molecular understanding of autophagy has been built on the genetic studies in budding yeast that have discovered over 30 autophagy-related (ATG) genes (Feng et al., 2014; Ohsumi, 2014). Among the ATGs, 18 have been shown to be essential for autophagosome formation and are categorized into four functional subgroups: the ATG1/ULK complex (ATG1/ULKs, ATG13, ATG17, ATG29, and ATG31) for initiation; the class III phosphoinositide 3-kinase complex (ATG6/BECN1 and ATG14) for nucleation; ATG9 and its recycling system (ATG2, ATG9, and ATG18) for phagophore expansion; and the ATG12 and ATG8/LC3 ubiquitin-like conjugation systems (ATG3, ATG4, ATG5, ATG7, ATG8, ATG10, ATG12, and ATG16) for phagophore elongation and vesicle expansion (Mizushima et al., 2011; Feng et al., 2014). In addition, recent genetic screens in other species, including fission yeast (Sun et al., 2013), worm (Tian et al., 2010), and human (Morita et al., 2018), as well as other nongenetic studies (Mizushima et al., 2011), have resulted in the identification of additional regulators for autophagy. However, the genes required for the process of phagophore closure have remained elusive in these pioneering works, as techniques for distinguishing between unclosed and closed autophagosomal membranes were limited and not adaptable to high-throughput genetic screening. Our recent development of the HaloTag (HT)-LC3 autophagosome completion assay resolves this issue by allowing for the differentiation of phagophores, nascent autophagosomes, and mature autophagic structure in a semiquantitative output (Takahashi et al., 2018).
The endosomal sorting complex required for transport (ESCRT) machinery is an assembly of protein subcomplexes that cooperate together to mediate reverse-topology membrane scission (Schöneberg et al., 2017). The core ESCRT machinery is composed of (1) ESCRT-I and (2) Bro1/ALIX family proteins for the recognition of the sites of action, (3) ESCRT-II for bridging ESCRT-I and ESCRT-III, (4) ESCRT-III for the assembly of filaments to induce membrane remodeling and scission, and (5) the AAA ATPase vacuolar protein sorting 4 (VPS4) complex for the disassembly and recycling of ESCRT-III (McCullough et al., 2013). While the ESCRT membrane scission machinery was originally discovered for its role in ubiquitinated cargo sorting in the multivesicular body (MVB) pathway (Katzmann et al., 2001), recent studies have revealed its involvement in a number of other cellular processes, including virus budding, cytokinesis, microvesicle generation, plasma membrane wound repair, nuclear envelope reformation, nuclear pore complex quality control, and endolysosomal repair (Hurley, 2015; Christ et al., 2017; Skowyra et al., 2018). Most recently, we have used the HT-LC3 assay and demonstrated the requirement of the ESCRT-III component charged MVB protein 2A (CHMP2A) and VPS4 activation in phagophore closure (Takahashi et al., 2018). In this study, we have combined a genome-wide CRISPR library screen with the HT-LC3 assay and identified the ESCRT-I component vacuolar protein sorting-associated protein 37A (VPS37A) as a novel phagophore closure regulator. We show that VPS37A is responsible for the recruitment of the ESCRT machinery for VPS4-mediated membrane scission and closure of the phagophore.
Results
A genome-wide screen using the FACS-based HT-LC3 assay identifies genes involved in phagophore closure
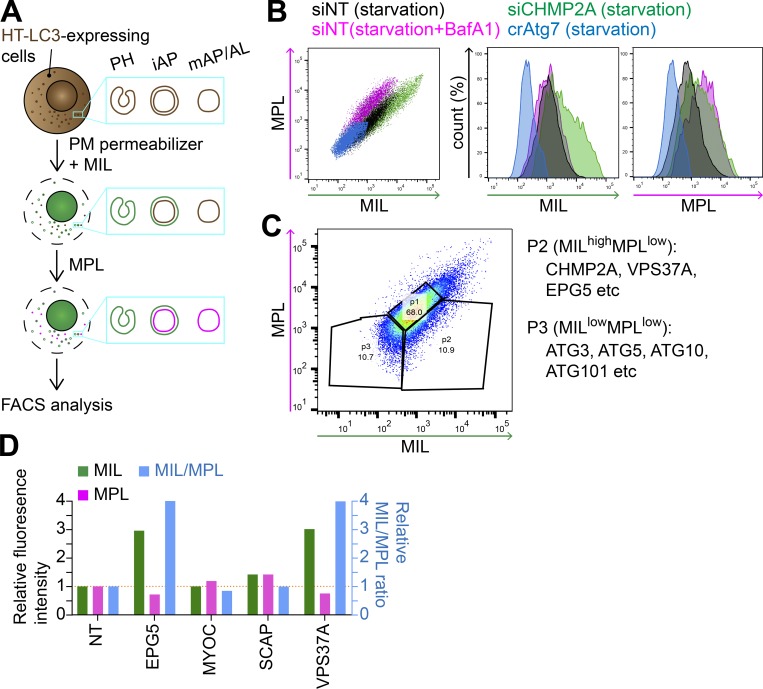
To identify genes involved in phagophore closure, we first adapted the HT-LC3 autophagosome completion assay to a high-throughput workflow using flow cytometry. Similar to the imaging-based assay, HT-LC3–expressing cells were permeabilized using the recombinant cholesterol-dependent pore-forming toxin perfringolysin O (Divakaruni et al., 2013) to release cytosolic HT-LC3-I and then sequentially labeled with a saturating dose of Alexa Fluor 488 (AF488)–conjugated membrane-impermeable ligands (MILs) followed by tetramethylrhodamine (TMR)-conjugated membrane-permeable ligands (MPLs) to detect autophagosomal membrane-unsequestered and sequestered HT-LC3-II, respectively. The resultant cells were then subjected to flow cytometry without fixation (Fig. 1 A). Consistent with our previous report (Takahashi et al., 2018), both MIL and MPL signals were detected during starvation in an Atg7-dependent manner, while impairment of functional autolysosome formation by the lysosomal inhibitor Bafilomycin A1 (BafA1) further accumulated MPL signals (Fig. 1 B and data not shown). Moreover, inhibition of autophagosome completion by ESCRT-III (CHMP2A) depletion substantially increased the MIL intensity while only modestly increasing MPL intensity to reflect an accumulation of cytosol-accessible HT-LC3-II. Collectively, these results validate our high-throughput adaptation of the HT-LC3 assay.
Figure 1.
An HT-LC3 assay–based CRISPR screen identifies VPS37A and EPG5 as potential regulators of autophagosome completion. (A) Schematic strategy of the flow cytometry–based HT-LC3 autophagosome completion assay. HT-LC3–expressing cells are permeabilized and sequentially incubated with AF488-conjugated MILs followed by TMR-conjugated MPLs to label autophagosomal membrane-unsequestered and sequestered HT-LC3-II, respectively. The fluorescence intensities of AF488 and TMR are then analyzed by FACS. iAP, immature autophagosome; mAP/AL, mature autophagosome/autolysosome; PH, phagophore; PM, plasma membrane. (B) HT-LC3–expressing U-2 OS cells transiently transfected with nontargeting siRNA (siNT) or CHMP2A siRNA (siCHMP2A) for 48 h, and HT-LC3–expressing crAtg7 cells were starved in the presence or absence of 100 nM BafA1 for 3 h and subjected to the HT-LC3 assay followed by FACS analysis. (C) The human Brunello lentiviral CRISPR pooled library was used to transduce HT-LC3–expressing U-2 OS cells. 7 d after the transduction, the resultant cells were starved for 3 h and subjected to the HT-LC3 assay followed by FACS analysis. The MILhighMPLlow and MILlowMPLlow populations indicated by the ROIs were sorted and analyzed by next-generation sequencing. Data shown are representative of four independent screens. (D) The selected hits from P2 of the primary screen in C were subjected to secondary screening. Three lentiviruses encoding different gRNAs against a gene were pooled and transduced into the HT-LC3–expressing U-2 OS cell for 2 d. Cells were then selected with 1 µg/ml puromycin for 3 d and then cultured at least another 3 d in complete medium. The resultant stable transfectants were starved in the presence of 100 nM BafA1 for 3 h and subjected to the HT-LC3 assay followed by FACS analysis. The AF488 and TMR intensities relative to those of control crNT cells are shown (n = 1).
We next combined the flow cytometry–based HT-LC3 assay with a genome-wide screen using the human Brunello CRISPR library (Doench et al., 2016). Briefly, HT-LC3–expressing U-2 OS cells were transduced with pooled lentivirus containing 76,441 single guide RNAs (sgRNAs) targeting 19,114 genes. Cells were transduced at low multiplicity of infection, selected by puromycin, and then starved for 3 h before performing the HT-LC3 assay and FACS analysis. Populations were isolated and sorted based on HT-LC3 labeling by MIL and MPL with P1 and P3 (MILlowMPLlow) populations gated as control populations containing autophagy-competent cells or capturing genes required for LC3 conjugation, respectively (Fig. 1 C). The P2 population (MILhighMPLlow) served as our population of interest for the identification of novel candidates that accumulated phagophores due to defects in autophagosome completion. Sorted pools were subjected to genomic DNA extraction followed by next-generation sequencing to determine sgRNA abundance. The amount of each sgRNA in P2 and P3 was normalized to the respective sgRNA values in the unsorted population (first and second screenings) or P1 (third and fourth screenings).
Consistent with a defect in LC3 conjugation, sgRNAs against genes encoding components of the ULK1 complex (ATG101) and the LC3 ubiquitin-like conjugation systems (ATG3, ATG5, and ATG10; Mizushima et al., 2011) were selectively enriched in the P3 (MILlowMPLlow) population. In contrast, CHMP2A sgRNAs were enriched in the P2 (MILhighMPLlow) population containing candidates with defective phagophore closure, indicating the validity of the screening. Notably, the P2 (MILhighMPLlow) population was also enriched in multiple sgRNAs against genes involved in vesicle trafficking and membrane dynamics (Table S1). Based on the highest probability scores and functional implications in autophagy (Tian et al., 2010; Qiu et al., 2014; Morita et al., 2018; Takahashi et al., 2018; Li et al., 2019), five of these genes (CHMP2A, ectopic P-granules autophagy protein 5 [EPG5], myocilin [MYOC], sterol regulatory element-binding protein cleavage-activating protein [SCAP], and VPS37A) were selected for secondary screening. For the secondary screen, HT-LC3–expressing cells were transduced with three independent lentiviruses encoding distinct sgRNAs against each gene, selected using puromycin, starved in the presence of BafA1, and subjected to the flow cytometry–based HT-LC3 assay. The MIL signal intensity and MIL/MPL ratio were significantly elevated in cells stably expressing sgRNAs against VPS37A (ESCRT-I subunit) and EPG5 compared with the control cells expressing nontargeting (NT) sgRNAs (Fig. 1 D). Despite the detection of CHMP2A sgRNAs among the P2 (MILhighMPLlow) population in the primary screen, CHMP2A CRISPR knockout (crCHMP2A) cells failed to grow and expand, a result that is likely attributed to the roles of CHMP2A in mitosis and cell division (Morita et al., 2010; Olmos et al., 2015).
VPS37A and EPG5 regulate phagophore closure
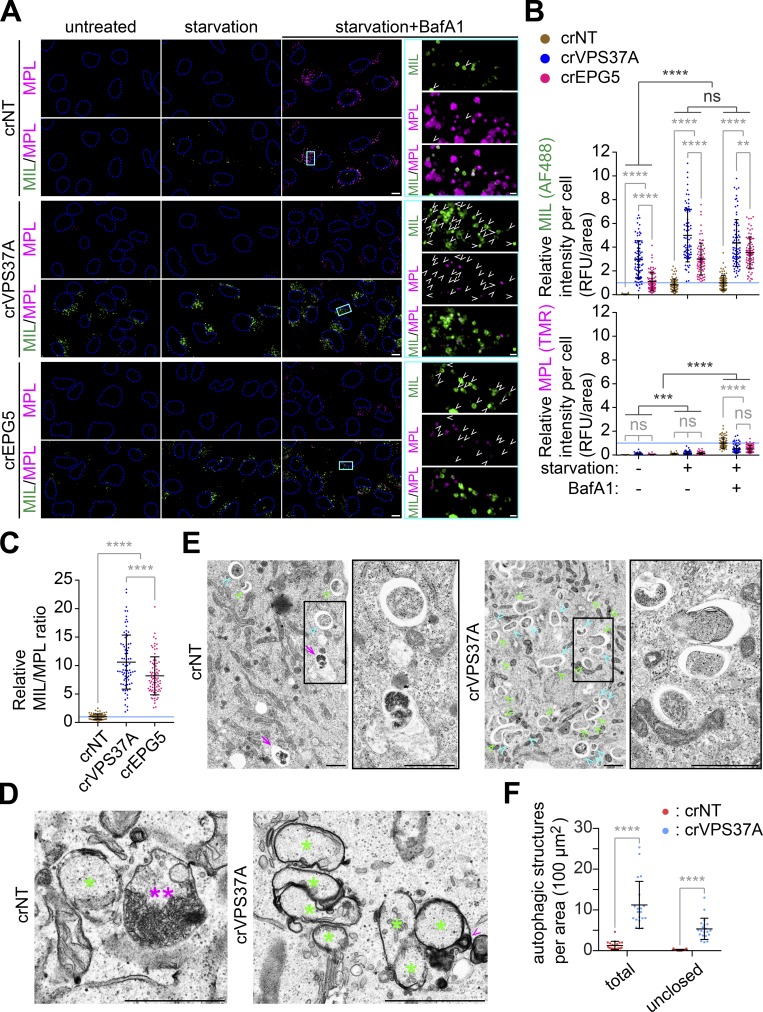
To determine if the MIL signal increases in VPS37A- and EPG5-deficient cells represented phagophore accumulation, HT ligand-labeled VPS37A- and EPG5-deficient cells were analyzed by confocal microscopy. Similar to the FACS analysis, starvation of crNT cells significantly increased both MIL and MPL signals, and the addition of BafA1 further accumulated MPL+ autophagic vacuoles due to defective autolysosome formation (Fig. 2, A and B). Depletion of either VPS37A or EPG5 accumulated MIL+MPL− phagophores, but not MPL+ autophagic vacuoles under the untreated, fed condition. This phenotype was further enhanced by starvation, indicating that VPS37A and EPG5 are dispensable for autophagy induction. Importantly, despite the strong accumulation of MIL signals, loss of VPS37A or EPG5 significantly suppressed the BafA1-induced accumulation of MPL signals to result in an increased MIL-to-MPL ratio, thereby reflecting a defect in the production of closed autophagosomes capable of lysosomal degradation (Fig. 2, A–C). To verify the accumulation of phagophores by VPS37A loss, we next performed electron microscopy. Consistently, an accumulation of cup- or oval-shaped phagophore-like (asterisks in Figs. 2 D and S1), but not autolysosome-like (double asterisks), structures were observed in starved crVPS37A cells. To quantify this observation, we artificially enlarged the intermembrane spaces of phagophores and autophagosomes by preparing samples in the absence of potassium ferrocyanide (Eskelinen, 2008; Takahashi et al., 2018). As expected, in addition to the number of total autophagic structures, the number of clearly unclosed autophagosomal membrane-like structures was significantly increased by VPS37A loss (Fig. 2, E and F). Collectively, these results suggest the importance of VPS37A and EPG5 in autophagosome completion under both basal and induced conditions.
Figure 2.
Loss of VPS37A or EPG5 accumulates unclosed autophagosomal membranes during starvation. (A) HT-LC3–expressing crNT, crVPS37A, and crEPG5 U-2 OS cells were starved in the presence or absence of 100 nM BafA1 for 3 h and subjected to the HT-LC3 assay followed by confocal microscopy. Representative images from three independent experiments are shown. Magnified images in the boxed areas are shown in the right panels. Arrowheads indicate MIL+MPL− phagophores. (B) The cytoplasmic fluorescence intensities of MIL and MPL in each cell in A were quantified and normalized to the respective mean fluorescence intensities of crNT cells starved in the presence of BafA1 (n = 100). RFU, relative fluorescence unit. Statistical significance was determined by two-way ANOVA followed by Tukey’s multiple comparison test. All values are mean ± SD. **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001; ns, not significant. (C) The MIL/MPL ratio for each cell in A is calculated and shown (n = 100). Statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparison test. All values are mean ± SD. ****, P ≤ 0.0001. (D and E) crNT and crVPS37A U-2 OS cells were starved for 3 h and subjected to electron microscopy. The samples were processed in the presence D or absence E of potassium ferrocyanide. In D, magnified images in the boxed areas are shown in the right panels and asterisks, double asterisks, and arrowheads indicate immature autophagic structures including phagophores, an autolysosome-like structure, and endolysosomal contents in the intermembrane space, respectively. In E, green arrowheads, cyan arrowheads, and arrows indicate phagophore-like (clearly opened in 2D micrographs), autophagosome-like, and autolysosome-like structures, respectively. (F) The number of total and unclosed autophagic structures per cytoplasmic area in E was quantified and shown (n = 20). Statistical significance was determined by two-way ANOVA followed by Sidak’s multiple comparison test. All values are mean ± SD. ****, P ≤ 0.0001. The scale bars represent 10 µm in A and 1 µm in the magnified images in A, D, and E.
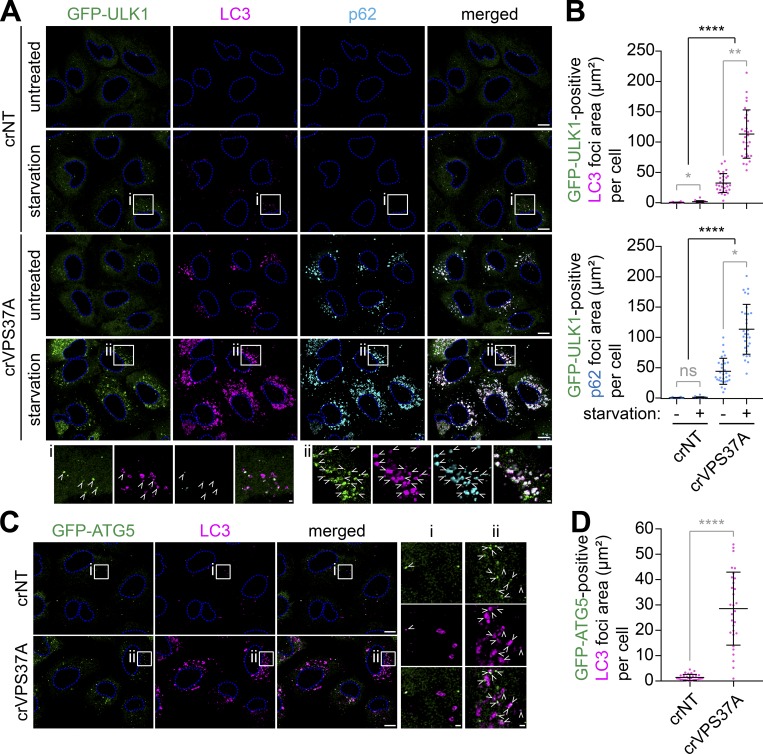
VPS37A deficiency accumulates ULK1, ATG5, and p62 on LC3-positive autophagosomal membranes and impairs autophagic flux
Since our previous study has implicated ESCRT machinery in phagophore closure (Takahashi et al., 2018), we sought to further characterize the role of the ESCRT-I subunit VPS37A in autophagy. We first determined if the phagophore-like structures accumulated in crVPS37A cells are positive for early autophagic makers. Upon the induction of autophagy, the ULK1 complex translocates to ER-associated omegasome precursors to induce phagophore nucleation but dissociates from the membrane upon phagophore closure (Karanasios et al., 2013). Consistently, starvation of crNT cells induced the assembly of omegasome-associated phagophores as detected by GFP-ULK1 cytoplasmic foci partially colocalizing with LC3 (Fig. 3 A, i). In agreement with the HT-LC3 assay results, the number of starvation-induced GFP-ULK1 and LC3 double-positive (ULK1+LC3+) early autophagic structures was dramatically increased by the loss of VPS37A (Fig. 3, A [ii] and B). Moreover, a slight accumulation of ULK1+LC3+ structures was also detected in untreated crVPS37A cells (Fig. 3, A and B) to support the notion that loss of VPS37A impairs phagophore closure under both basal and induced conditions (Fig. 2, A and B). We also observed that the majority of the ULK1+LC3+ structures in crVPS37A cells were positive for p62 (Fig. 3, A and B), indicating that VPS37A is dispensable for the recruitment of autophagic substrates. The accumulation of unsealed LC3-containing autophagosomal membranes by VPS37A depletion was further confirmed using GFP-ATG5 (Fig. 3, C and D), a phagophore marker that is also known to dissociate upon autophagosome completion (Mizushima et al., 2001).
Figure 3.
VPS37A depletion accumulates ULK1, p62, and Atg5 on phagophores. (A) crNT and crVPS37A U-2 OS cells stably expressing GFP-ULK1 were incubated in complete medium or starvation medium for 2 h, stained for LC3 and p62, and analyzed by confocal microscopy. Representative images from three independent experiments are shown. Magnified images in the boxed areas are shown in the bottom panels. Arrowheads indicate GFP-ULK1 and LC3 double-positive immature autophagosomal membranes. (B) The areas of LC3-positive (top) and p62-positive (bottom) autophagic structures that were positive for GFP-ULK1 in each cell in A were quantified and shown (n = 30). Statistical significance was determined by Kruskal–Wallis one-way ANOVA on ranks followed by Dunn’s multiple comparison test. All values are mean ± SD. *, P ≤ 0.05; **, P ≤ 0.01; ****, P ≤ 0.0001; ns, not significant. (C) GFP-ATG5–expressing crNT and crVPS37A U-2 OS cells were starved for 2 h, stained for LC3, and analyzed by confocal microscopy. Representative images from three independent experiments are shown. Magnified images in the boxed areas are shown in the right panels. Arrowheads indicate GFP-ATG5– and LC3 double-positive immature autophagosomal membranes. (D) The areas of GFP-ATG5 and LC3 double-positive autophagic structures in each cell in C are quantified and shown (n = 30). Statistical significance was determined by Student’s t test. All values are mean ± SD. ****, P ≤ 0.0001. The scale bars represent 10 µm in A and C and 1 µm in the magnified images.
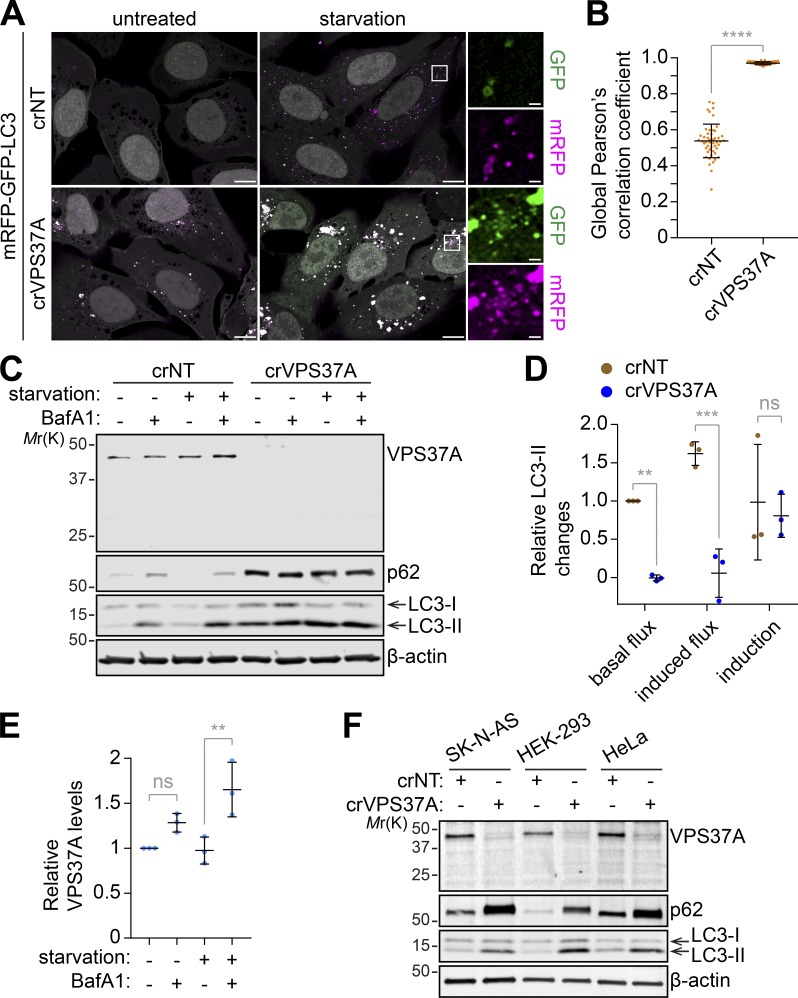
To investigate whether the autophagic defect in crVPS37A results in impaired autophagic degradation, we first monitored autolysosome formation using the tandem fluorescent-tagged LC3 reporter, which contains lysosomal pH-sensitive GFP and pH-resistant mRFP (Kimura et al., 2007). We found that VPS37A loss resulted in the accumulation of mRFP and GFP double-positive LC3 foci, and this was further enhanced by starvation (Fig. 4, A and B), indicating the impairment of functional autolysosome formation, but not autophagy induction. Consistently, LC3-II accumulated in crVPS37A cells under untreated conditions, which was further increased by starvation (Fig. 4, C and D; and Fig. S2). However, in contrast to crNT cells, BafA1 failed to further increase LC3-II or p62 in crVPS37A cells under both untreated (basal) and starvation (induced) conditions, indicating an impairment of autophagic flux. Interestingly, we also observed a significant increase of VPS37A upon starvation in the presence of BafA1 (Figs. 4 E and S2). Importantly, the accumulation of LC3-II and p62 by VPS37A depletion was also observed in several different types of cells including SK-N-AS neuroblastoma, HEK-293 embryonic kidney, and HeLa cervical carcinoma cells (Fig. 4 F). Collectively, these results indicate that VPS37A loss accumulates phagophores leading to impaired autophagic flux.
Figure 4.
VPS37A depletion impairs autophagic flux. (A) crNT and crVPS37A U-2 OS cells were transduced with lentiviruses encoding mRFP-GFP-LC3, incubated in complete medium or starvation medium for 3 h, and subjected to confocal microscopy. Representative images from three independent experiments are shown. Magnified images in the boxed areas are shown in the right panels. The scale bars represent 10 µm in the main images and 1 µm in the magnified images. (B) The acidification of autophagic structures in starved cells in A was assessed by global Pearson’s correlation coefficient (n = 50). Statistical significance was determined by Mann–Whitney nonparametric t test. (C) The indicated U-2 OS cells were incubated in complete medium or starvation medium in the presence or absence of 100 nM BafA1 for 3 h and subjected to IB using the indicated antibodies. Representative blots from three independent experiments are shown. (D) Autophagic flux under untreated (basal flux) and starvation (induced flux) conditions and autophagy induction in C were calculated as described in Materials and methods and normalized to the value of basal flux (n = 3). (E) Fold changes in VPS37A levels relative to untreated control in C are quantified and shown (n = 3). (F) crNT and crVPS37A SK-N-AS, HEK-293, and HeLa cells were subjected to IB using the indicated antibodies. All values are mean ± SD. **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001; ns, not significant.
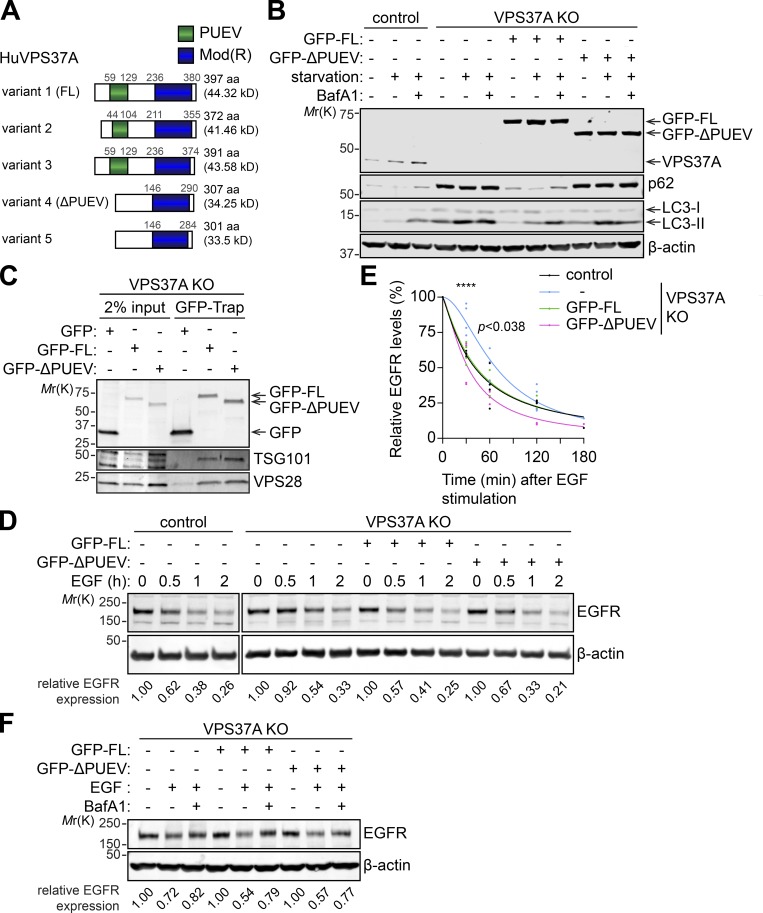
The PUEV domain of VPS37A is required for autophagy but dispensable for EGF receptor (EGFR) degradation
We next examined if the defect in autophagy observed in crVPS37A cells can be rescued by the restoration of VPS37A expression. A National Center for Biotechnology Information database search showed that there are at least five alternative splicing variants of VPS37A expressed in human (Fig. 5 A). While all of the variants contain the C-terminal modifier of rudimentary (Mod(r)) domain, VPS37A variants 1–3 contain an N-terminal putative ubiquitin E2 variant (PUEV) domain that is absent from variants 4 and 5. Based on the predicted molecular weights and using an antibody that detects VPS37A independent of the PUEV domain (Fig. 5, A and B), the PUEV-containing variants appeared to be predominantly expressed in the cell lines we tested (Fig. 4, C and F). Of note, the VPS37A PUEV domain is unique among the four mammalian VPS37 homologues (VPS37A–VPS37D; Stuchell et al., 2004), but its function remains uncharacterized. To determine if the PUEV domain is important for autophagy, we performed rescue experiments using PUEV-containing variant 1 (full length [FL]) and PUEV-lacking variant 4 (ΔPUEV). To avoid sgRNA-mediated silencing of exogenously introduced VPS37A constructs in crVPS37A cells, we generated VPS37A-deficient U-2 OS cells by transient expression of VPS37A sgRNAs in Cas9-expressing U-2 OS cells (indicated as VPS37A knockout [KO] cells). The resultant cells were then transduced with either GFP-tagged FL or ΔPUEV VPS37A variant, and starvation-induced autophagic flux was determined. We observed that the expression of GFP-FL, but not GFP-ΔPUEV, rescued the defect in lysosomal degradation of LC3-II and p62 in VPS37A KO cells (Fig. 5 B), indicating the importance of the PUEV domain in autophagy. Notably, PUEV domain disruption failed to alter the association of VPS37A with other ESCRT-I components, as both GFP-FL and GFP-ΔPUEV coimmunoprecipitated with ESCRT-I subunits TSG101 and VPS28 (Fig. 5 C); this result is consistent with previous data demonstrating that the Mod(r) domain of VPS37A interacts with other ESCRT-I components (Bache et al., 2004). Since VPS37A has been shown to mediate the MVB sorting and lysosomal degradation of EGFR (Bache et al., 2004), we next examined the importance of the PUEV domain in the endocytic degradation of EGFR. As expected, depletion of VPS37A moderately delayed lysosomal degradation of EGFR upon EGF stimulation (Fig. 5, D and E). Interestingly, this phenotype was reversed by the expression of not only GFP-FL but also GFP-ΔPUEV in a manner that was dependent on functional lysosomes (Fig. 5, D–F), suggesting that the Mod(r) domain of VPS37A is sufficient for functionality in the MVB pathway. Collectively, these results indicate that the PUEV domain of VPS37A is required for autophagy but is dispensable for the endocytic degradation of EGFR.
Figure 5.
The PUEV domain of VPS37A is required for autophagy but dispensable for endocytic degradation of EGFR. (A) Schematic domain structures of human VPS37A variants. (B) Cas9-expressing U-2 OS cells (control) were transiently transfected with the plasmid encoding GFP and three different gRNAs against VPS37A for 48 h, followed by flow cytometry sorting for GFP-positive cells. The resultant cells were cultured for 2 wk, and a GFP-negative clone was isolated and expanded to obtain VPS37A-deficient (VPS37A KO) cells. The VPS37A KO cells were then transduced with lentiviruses encoding GFP-VPS37A variant 1 (GFP-FL), variant 4 (GFP-ΔPUEV), or control GFP; starved in the presence or absence of 100 nM BafA1 for 3 h; and subjected to IB using the indicated antibodies. Representative blots from two independent experiments are shown. (C) Cells were starved for 2 h and subjected to GFP-trap followed by IB using the indicated antibodies. Representative blots from three independent experiments are shown. (D) Cells were serum starved for 16 h, coincubated with 100 ng/ml EGF and 10 µg/ml cycloheximide for the indicated durations, and subjected to IB using the indicated antibodies. EGFR levels in each lane were quantified, normalized to β-actin and shown at the bottom. Representative blots from five independent experiments are shown. (E) The EGFR levels relative to respective β-actin in E were quantified and normalized to the value of respective cells at time 0 (n = 5). Nonlinear regression curves were generated and shown. Statistical significance was determined by two-way ANOVA followed by Dunnett’s multiple comparison test. ****, P ≤ 0.0001. (F) Cells were serum starved for 16 h, coincubated with EGF and cycloheximide in the presence or absence of 100 nM BafA1 for 30 min and subjected to IB using the indicated antibodies. EGFR levels in each lane were quantified, normalized to β-actin and shown at the bottom. Representative blots from two independent experiments are shown.
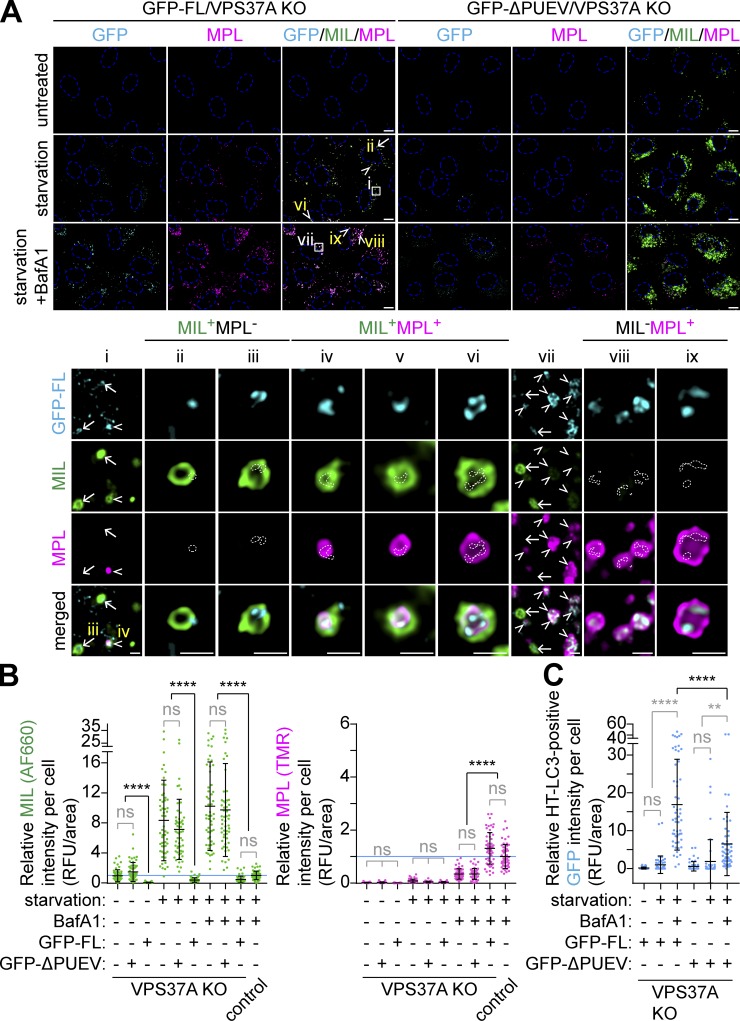
VPS37A translocates to the phagophore through the PUEV domain for the membrane closure
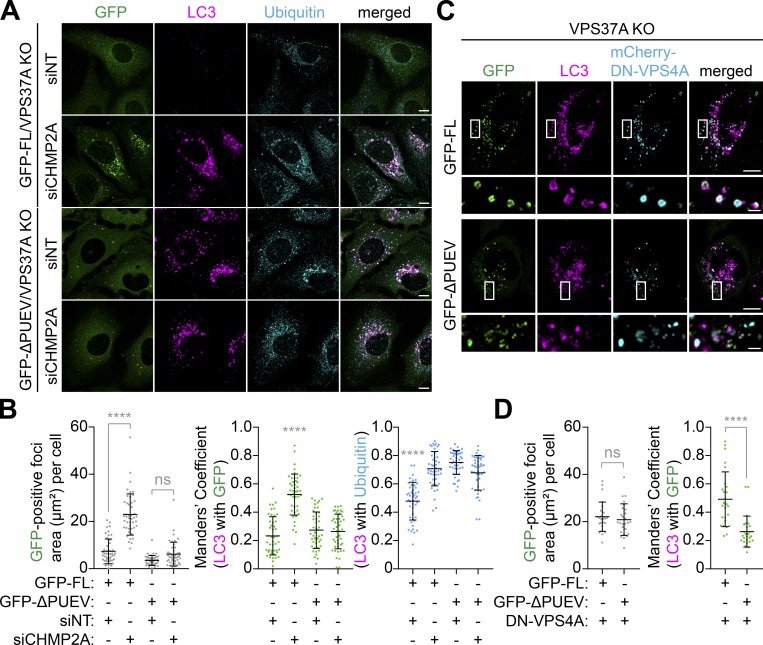
We next examined if the PUEV domain of VPS37A is required for phagophore closure. In agreement with the autophagic flux assay (Fig. 5 B), expression of GFP-FL suppressed the aberrant MIL accumulation in VPS37A-deficient cells under both basal and starvation conditions and increased MPL levels comparable to those in control cells when starved in the presence of BafA1 (Fig. 6, A and B). In stark contrast, GFP-ΔPUEV expression failed to rescue the MIL accumulation and autophagic degradation, indicating that the PUEV domain of VPS37A is important for autophagosome completion and subsequent degradation. Indeed, despite the accumulation of phagophores in GFP-ΔPUEV–expressing VPS37A KO cells (Fig. 6, A and B), GFP-ΔPUEV signals were hardly detected on LC3-positive membranes, and only a minor signal increase was observed in the presence of BafA1 (Fig. 6, A and C). Notably, we detected GFP-FL signals not only on MIL+MPL− phagophores but also in the lumen of MPL+ autophagosomal membranes (Fig. 6 A, i–vi) and found that BafA1 treatment resulted in further accumulation of GFP-FL on MPL+ structures (Fig. 6 A, vii–ix). These results are consistent with our immunoblotting (IB) data that showed an increase of endogenous VPS37A upon lysosomal inhibition (Fig. 4 E) and suggest a possibility that at least a portion of VPS37A located on the phagophore remains on the luminal side of the membrane upon closure and is delivered to lysosomes. We next verified the lysosomal delivery of VPS37A using the mRFP-GFP tandem fluorescence-tagged VPS37A FL and ΔPUEV. Consistent with the observation that GFP-FL is sequestered within autophagosomes upon the membrane closure (Fig. 6 A), we found an accumulation of mRFP+GFP− puncta in starved mRFP-GFP-FL–transduced cells (Fig. S3). In contrast, ΔPUEV foci, which were also induced upon starvation, were positive for both mRFP and GFP. As the starvation-induced ΔPUEV foci appeared to be negative for LC3 (Fig. 7 A), the identity of these structures remains unknown. To further investigate whether the PUEV domain is required for phagophore localization of VPS37A, we stabilized phagophores by depleting the downstream ESCRT-III subunit CHMP2A (Takahashi et al., 2018) and examined the localization of GFP-FL and GFP-ΔPUEV under starvation conditions. As expected, CHMP2A depletion resulted in a strong accumulation of ubiquitin-containing LC3-positive phagophores in both GFP-FL and GFP-ΔPUEV expressing cells (Fig. 7, A and B). Notably, while GFP-FL localized to the accumulated LC3-positive structures in CHMP2A-depleted cells, GFP-ΔPUEV failed to localize on the autophagic structures, even upon CHMP2A depletion. A similar accumulation of GFP-FL on LC3-positive membranes was observed upon the induction of a dominant-negative form of VPS4A (DN-VPS4A, Vps4AE228Q; Fig. 7, C and D). In contrast to CHMP2A depletion, GFP-ΔPUEV formed cytoplasmic foci upon DN-VPS4A expression. However, in stark contrast to GFP-FL, the majority of GFP-ΔPUEV signals were negative for LC3, further supporting that the deletion of the PUEV domain inhibits phagophore localization of VPS37A. Collectively, these results indicate that the PUEV domain is responsible for phagophore localization of VPS37A.
Figure 6.
The PUEV domain of VPS37A is required for autophagosome completion. (A) HT-LC3–expressing U-2 OS cells with the indicated genotypes and transgenes were starved in the presence or absence of 100 nM BafA1 for 3 h and subjected to the HT-LC3 assay using AF660-conjugated MIL- and TMR-conjugated MPL. Representative confocal microscopy images from three independent experiments are shown. Magnified images in the boxed and arrow- or arrowhead-indicated areas are shown in the lower panels. Arrows and arrowheads indicate GFP-VPS37A–positive phagophores (MIL+MPL−) and autophagic vacuoles (MIL+MPL+ or MIL−MPL+), respectively. In the magnified images, dashed-line areas indicate the location of GFP-FL signals. The scale bars represent 10 µm in the main panels and 1 µm in the magnified images. (B and C) The cytoplasmic fluorescence intensities of MIL and MPL (B), and MIL− and/or MPL+ GFP-VPS37A (C) in each cell in A were quantified and normalized to the respective mean fluorescence intensities of control Cas9-expressing U-2 OS cells starved in the presence of BafA1 (n = 55). Statistical significance was determined by Kruskal–Wallis one-way ANOVA on ranks followed by Dunn’s multiple comparison test. All values are mean ± SD. **, P ≤ 0.01; ****, P ≤ 0.0001; ns, not significant.
Figure 7.
The PUEV domain is responsible for phagophore localization of VPS37A. (A) VPS37A KO U-2 OS cells stably expressing GFP-VPS37A FL or ΔPUEV were nucleofected with siNT or siCHMP2A for 48 h, starved for 3 h, stained for LC3 and ubiquitin, and analyzed by confocal microscopy. Representative images from three independent experiments are shown. (B) The areas of GFP-positive foci (left) and colocalization coefficients of LC3 with GFP-VPS37A (middle) and LC3 with ubiquitin (right) per cell in A are quantified and shown (n > 44). Statistical significance was determined by Kruskal–Wallis one-way ANOVA on ranks followed by Dunn’s multiple comparison test. All values are mean ± SD. ****, P ≤ 0.0001; ns, not significant. (C) The indicated U-2 OS cells were transfected with inducible mCherry-VPS4AE228Q (DN-VPS4A) for 16 h, treated with 1 µg/ml doxycycline (Dox) for 6 h, stained for LC3, and analyzed by confocal microscopy. Magnified images in the boxed areas are shown in the bottom panels. (D) The areas of GFP-positive foci (left) and colocalization coefficients of LC3 with GFP-VPS37A (right) per cell in C are quantified and shown (n = 30). Statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparison test. All values are mean ± SD. ****, P ≤ 0.0001. The scale bars represent 10 µm in the main panels and 1 µm in the magnified images in C.
It has recently been reported that the ESCRT machinery can be recruited to damaged lysosomes for the membrane repair that occurs independent of lysophagy induction (Radulovic et al., 2018; Skowyra et al., 2018). To determine if the phagophore localization of VPS37A is distinct from the ESCRT recruitment to damaged membranes, GFP-FL– and HT-LC3–expressing VPS37A KO cells were transfected with the damaged endomembrane marker mCherry-GAL3 (Papadopoulos et al., 2017) and treated with the lysosome-damaging agent l-leucyl-l-leucine O-methyl ester (LLOME) in the presence or absence of the phosphatidylinositol-3-kinase inhibitor wortmannin (WM) followed by labeling with MIL. We observed that nearly all damaged lysosomes labeled with mCherry-GAL3 were enwrapped with MIL+ immature autophagosomal membranes in a WM-sensitive manner (Fig. S4, A and B), indicating the induction of lysophagy. Importantly, LLOME also induced an accumulation of GFP-FL signals on mCherry-GAL3–positive, ruptured lysosome-enwrapped MIL+ membranes, and this was significantly suppressed by WM (Fig. S4, A and B). Moreover, GFP-FL signals were also detected on mCherry-GAL3–negative MIL+ membranes in starved cells (Fig. S4 A). These observations indicate that the phagophore recruitment of VPS37A is, at least in part, dependent on the formation of phosphatidylinositol 3-phosphate or LC3-positive membranes and distinct from the ESCRT recruitment to damaged lysosomal membranes. Interestingly, however, WM treatment did not completely block the translocation of GFP-FL on MIL− damaged lysosomal membranes, supporting the autophagy-independent function of ESCRT-I in lysosome repair (Radulovic et al., 2018).
VPS37A is responsible for ESCRT-I recruitment to the phagophore
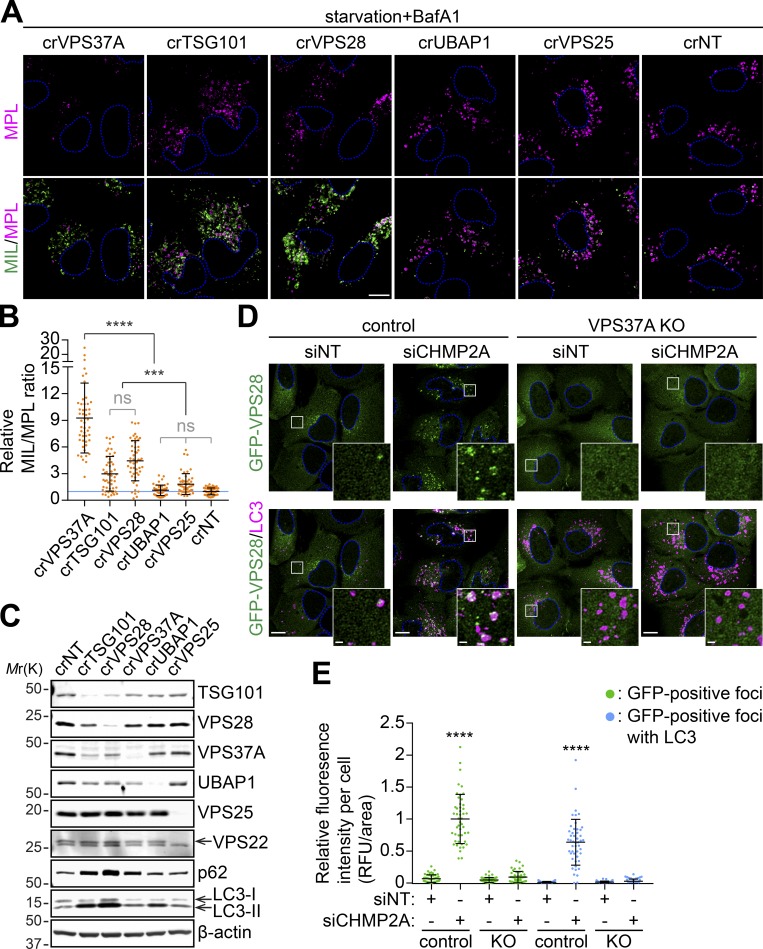
We next determined if VPS37A functions together with other ESCRT-I components and ESCRT-II to regulate phagophore closure. Since several ESCRT components, including TSG101 and VPS28, appeared to be required for cell survival, we performed the experiments after a short-term depletion of the targeted genes. Our data showed that, in addition to crVPS37A, crTSG101 and crVPS28 cells displayed a dramatic accumulation of MIL signals and increased the rates of MIL/MPL upon starvation in the presence of BafA1 (Fig. 8, A and B; and Fig. S5). In contrast, depletion of UBAP1 and VPS25 had minimal effect on the level of MIL and the MIL/MPL ratio. Interestingly, unlike VPS37A depletion, which significantly decreased MPL signals, the levels of MPL in crTSG101 and crVPS28 cells were comparable to that in crNT cells (Fig. S4). Moreover, the levels of LC3-II in crTSG101 and crVPS28 cells under basal, untreated conditions were much higher than that in crVPS37A cells (Fig. 8 C), suggesting the involvement of additional functions of these proteins that may enhance and/or block autophagy. In the IB analysis, we also observed that loss of TSG101 and VPS28, but not VPS37A and UBAP1, strongly decreased the expression of other ESCRT-I components (Fig. 8 C). This observation is in agreement with a previous report (Bache et al., 2004) and indicates that the majority of VPS37A forms a stable complex with TSG101 and VPS28. We next determined if VPS37A is responsible for the phagophore recruitment of the ESCRT-I complex. Similar to VPS37A, GFP-VPS28 signals were occasionally detected on LC3-positive autophagosomal membranes during starvation, and this was significantly increased upon the inhibition of phagophore closure by CHMP2A depletion (Fig. 8, D and E). In contrast, despite accumulation of LC3-positive phagophores that occurred regardless of CHMP2A expression, VPS37A deletion failed to accumulate GFP-VPS28 upon starvation. Collectively, these results indicate that VPS37A is required for ESCRT-I recruitment to the phagophore.
Figure 8.
VPS37A is responsible for ESCRT-I localization to the phagophore. (A–C) Three lentiviruses encoding different gRNAs against a gene were pooled and transduced into the HT-LC3–expressing U-2 OS cell for 2 d. Cells were then selected with 1 µg/ml puromycin for 3 d, cultured another 2 d in complete medium, and subjected to IB using the indicated antibodies (C) or starved in the presence of 100 nM BafA1 for 3 h followed by the HT-LC3 assay (A). In A, representative confocal images from three independent experiments are shown. In B, the MIL/MPL ratio for each cell in A was calculated and shown (n = 50). Statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparison test. All values are mean ± SD. ***, P ≤ 0.001; ****, P ≤ 0.0001; ns, not significant. (D) Control and VPS37A KO U-2 OS cells stably expressing GFP-VPS28 were nucleofected with siNT or siCHMP2A for 48 h, starved for 3 h, stained for LC3, and analyzed by confocal microscopy. Representative images from two independent experiments are shown. Magnified images in the boxed areas are shown as the insets. (E) The fluorescence intensities of total and LC3-associated GFP-VPS28–positive foci per cell in D were quantified and normalized to the total GFP-VPS28 foci value of siCHMP2A-transfected control cells (n = 48). Statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparison test. All values are mean ± SD. ****, P ≤ 0.0001. The scale bars represent 10 µm in the main panels and 1 µm in the insets in D.
VPS37A directs the assembly of CHMP2A for VPS4A recruitment to the phagophore
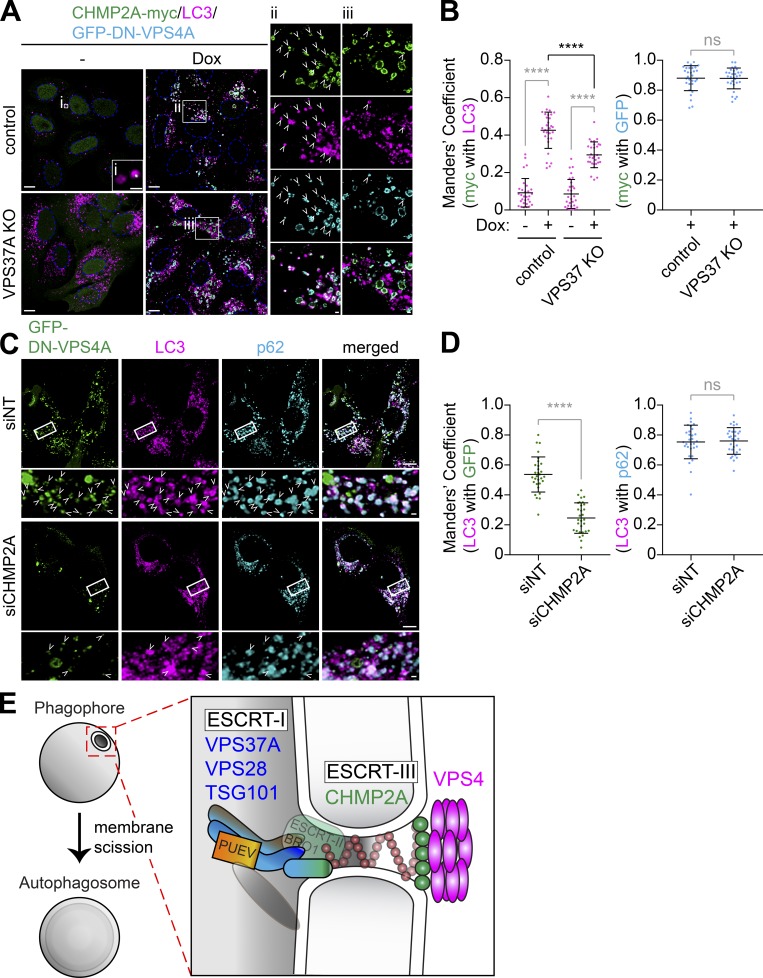
We next examined if VPS37A is required for the recruitment of CHMP2A during autophagosome biogenesis. To minimize the dominant-negative effect of a fluorescence protein–tagged ESCRT-III upon overexpression (Teis et al., 2008), we generated cells that were stably expressing CHMP2A-myc. A small portion of LC3-positive autophagic structures was positive for CHMP2A-myc in starved control cells (Fig. 9 A, i), thereby reflecting its transient recruitment during phagophore closure. In contrast, inhibition of ESCRT disassembly by the induction of DN-VPS4A significantly increased the colocalization of CHMP2A-myc with LC3 (Fig. 9, A [ii] and B), a result consistent with CHMP2A translocation to the phagophore during autophagy (Takahashi et al., 2018). Notably, while VPS37A depletion failed to alter the colocalization of CHMP2A-myc with GFP-DN-VPS4A, the localization of CHMP2A-myc on LC3-positive structures was significantly suppressed in VPS37A-deficient cells (Fig. 9, A [iii] and B). These results indicate that VPS37A plays a critical role in the assembly of CHMP2A on the phagophore. To determine if CHMP2A is required for the phagophore recruitment of VPS4, we depleted cells of CHMP2A and examined the recruitment of GFP-DN-VPS4A to the accumulated LC3-positive phagophores. As expected, p62-containing LC3-positive membranes were accumulated in both siNT and siCHMP2A cells expressing DN-VPS4A due to a defect in AAA ATPase activity and/or ESCRT-III filament assembly. However, our data showed that CHMP2A depletion significantly decreased the recruitment of GFP-DN-VPS4A on LC3-positive membranes (Fig. 9, C and D). Collectively, these data indicate that VPS37A promotes the recruitment of CHMP2A to phagophores for the subsequent CHMP2A-mediated recruitment of VPS4 that is required for phagophore closure.
Figure 9.
VPS37A directs CHMP2A assembly on the phagophore for VPS4 recruitment. (A) Control and VPS37A KO U-2 OS cells stably expressing inducible GFP-DN-VPS4A were cultured in the presence or absence of 1 µg/ml Dox for 4 h, starved in the presence or absence of Dox for 2 h, stained for LC3, and analyzed by confocal microscopy. Magnified images in the boxed areas are shown in the right panels. Arrowheads indicate CHMP2A-myc–positive LC3 foci. (B) Colocalization coefficient of CHMP2A-myc with GFP-DN-VPS4A or LC3 per cell in A were quantified and shown (n = 30). Statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparison test or Mann–Whitney nonparametric t test. All values are mean ± SD. ****, P ≤ 0.0001; ns, not significant. (C) Inducible GFP-DN-VPS4A–expressing U-2 OS cells were nucleofected with the indicated siRNAs (200 pmol) for 26 h, treated with 1 µg/ml Dox for 4 h, starved for 1 h, stained for LC3 and p62, and analyzed by confocal microscopy. Magnified images in the boxed areas are shown in the bottom. (D) Relative fluorescence intensities of GFP-DN-VPS4A and p62 on LC3-positive foci in C were quantified and shown (n = 30). Statistical significance was determined by Mann–Whitney nonparametric t test. All values are mean ± SD. ****, P ≤ 0.0001; ns, not significant. The scale bars represent 10 µm in the main panels and 1 µm in the magnified images. (E) Model for phagophore closure by the ESCRT machinery. ESCRT-I localizes on the phagophore through the VPS37A PUEV domain, which may recognize unidentified proteins on the membrane, to direct the assembly of ESCRT-III for membrane constriction and VPS4 recruitment.
Discussion
Although phagophore closure is frequently described as a membrane fusion event, the process bears a similar topology to ESCRT-mediated membrane scission that is proposed to generate two separate lipid bilayers (the outer and inner autophagosomal membranes; Rusten and Stenmark, 2009; Hurley, 2015; Knorr et al., 2015). Indeed, by developing the HT-LC3 autophagosome completion assay, we have recently demonstrated the involvement of the ESCRT-III component CHMP2A and the AAA ATPase VPS4 in phagophore closure (Takahashi et al., 2018). In this study, we use the HT-LC3 assay to perform a genome-wide CRISPR screen and identify the ESCRT-I subunit VPS37A as a critical component for the phagophore localization of ESCRTs to drive membrane closure. Interestingly, our data show that in addition to VPS37A, the core ESCRT-I components TSG101 and VPS28, but not the endosome-specific ESCRT-I component UBAP1, are required for autophagosome completion. Moreover, using an autophagy-defective, yet endocytic receptor sorting-competent VPS37A variant, we also show that VPS37A promotes phagophore closure independent of its function in the endocytic receptor sorting pathway, thereby excluding an enhanced induction of autophagy or accumulation of autophagic structures due to impaired MVB formation (Rusten and Stenmark, 2009; Djeddi et al., 2012). These results build on our previous work (Takahashi et al., 2018) to experimentally demonstrate the significance of ESCRT-mediated membrane scission in phagophore closure (Fig. 9 E).
The FL variant of VPS37A is composed of the N-terminal PUEV domain and the C-terminal Mod(r) domain. Our data show that disruption of the PUEV domain inhibits the phagophore localization of VPS37A upon starvation and impairs autophagosome completion. Interestingly, the ubiquitin E2 variant domain of TSG101 has been shown to interact with ubiquitin as well as the PT/SAP motif (Pornillos et al., 2002). As the phagophores accumulated in VPS37A- or CHMP2A-deficient cells are found to be positive for ubiquitin, the recognition of ubiquitinated proteins on the phagophore through the PUEV domain may be responsible for the recruitment of VPS37A. However, the residues responsible for the interaction with ubiquitin and the PT/SAP motif in the TSG101 ubiquitin E2 variant domain are not conserved in the VPS37A PUEV domain (Stuchell et al., 2004), and unpublished data has been documented that VPS37A fails to interact with ubiquitin (Bache et al., 2004). Since CHMP2A depletion accumulates the PUEV-containing, FL variant of VPS37A, but not the ΔPUEV variant on phagophores, future analyses of the PUEV-binding proteins in CHMP2A-depleted cells may provide molecular insight into the ordered recruitment of ESCRT subcomplexes during autophagy.
In contrast to its role in phagophore closure, we find that the PUEV domain of VPS37A is dispensable for its function in EGFR sorting in the MVB pathway. This observation is in agreement with a previous study demonstrating that while individually dispensable, multiple ubiquitin-binding domains within the ESCRT-I/II components cooperate in the recognition and sorting of ubiquitinated cargos in the MVB pathway (Shields et al., 2009). As the disruption of the PUEV domain does not interfere with the interaction of VPS37A with TSG101 and VPS28, the ability of VPS37A to form a complex with other ESCRT-I components through the Mod(r) domain (Stuchell et al., 2004) may be sufficient to maintain cargo sorting in the MVB pathway. Along this line, despite the absence of the PUEV domain, yeast and worm VPS37 proteins are found to be indispensable for functional MVB formation (Katzmann et al., 2001; Djeddi et al., 2012).
The observation that CHMP2A depletion accumulates VPS28 on phagophores in control, but not VPS37A KO, cells suggests the role of VPS37A in directing the ESCRT machinery to the membrane closure site. Consistently, VPS37A loss significantly suppresses the phagophore accumulation of CHMP2A induced by DN-VPS4A expression. However, unlike VPS28, whose localization to the phagophore appears to be dependent on VPS37A, a significant portion of CHMP2A remains to localize on LC3-positive membranes in the absence of VPS37A. A possible explanation for this is due to the nonproductive, abnormal fusion of a phagophore with endosomes/lysosomes (Takahashi et al., 2018) that may deliver endosomal DN-VPS4–trapped CHMP2A to autophagosomal membranes. Indeed, we occasionally observe autophagic structures in which endolysosomal contents are detected in the intermembrane space in VPS37A KO cells. Alternatively, it would be possible that an unidentified mechanism can partially bypass the requirement of ESCRT-I for CHMP2A recruitment. The ESCRT-binding protein ALIX is such a candidate, as it functions in parallel with ESCRT-I/II to recruit ESCRT-III during cytokinetic abscission (Christ et al., 2016). During the revision of this manuscript, ESCRT-dependent autophagosome closure that is mediated through the Rab5-dependent interaction between the ESCRT-III component Snf7 and Atg17 has been demonstrated in yeast (Zhou et al., 2019). Since Atg17 and its mammalian functional counterpart FIP200 share several features (Hara et al., 2008), it would also be of future interest to determine if FIP200 can interact with CHMP4 and recruit the ESCRT-III complex for phagophore closure.
Our screening also identified EPG5 sgRNAs in the MILhighMPLlow population. Interestingly, EPG5 was originally discovered by a worm genetic screen for mutants with defective autophagic degradation and suggested to be required for autolysosome formation (Tian et al., 2010). The observation that MIL, but not MPL, signals are accumulated in crEPG5 cells suggests that EPG5 may have additional roles in autophagosome completion (e.g., ESCRT recruitment or LC3-II delipidation upon autophagosome maturation) besides its role as a Rab7 effector (Wang et al., 2016).
Materials and methods
Reagents
The following antibodies were used for immunofluorescence (IF) and IB: mouse antibodies against β-actin (IB, A5441, 1:10,000; Sigma-Aldrich), TSG101 (IB, ab83, 1:1,000; Abcam), VPS28 (IB, sc-166537, 1:100; Santa Cruz Biotechnology), VPS37A (IB, sc-376978, 1:100; Santa Cruz Biotechnology); rabbit antibodies against EGFR (IB, 4267, 1:1,000; Cell Signaling), GFP (IB, 2956, 1:1,000; Cell Signaling), MAP1LC3B (IB, NB100-2220, 1:3,000; Novus; IF, 3868, 1:200; Cell Signaling), UBAP1 (IB, 12385–1-AP, 1:1,000; Proteintech), VPS22 (IB, NBP2-15125, 1:1,000; Novus), VPS25 (IB, 15669–1-AP, 1:1,000; Proteintech), and guinea pig antibody against p62 (03-GP62-C, 1:4,000 for IB, 1:400 for IF; American Research Products). ON-TARGETplus SMART Pool nontargeting (D-001810-10) and CHMP2A (L-020247-01) siRNAs were purchased from GE Healthcare. The following plasmids were obtained from Hartmut Land (University of Rochester Medical Center, Rochester, NY), Jay Morgenstern (Imperial Cancer Research Fund, London, UK), Bob Weinberg (Whitehead Institute for Biomedical Research, Cambridge, MA), Feng Zhang (Massachusetts Institute of Technology, Cambridge, MA), Brett Stringer (QIMR Berghofer Medical Research Institute, Brisbane, Australia), Christopher Vakoc (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY), Noboru Mizushima (The University of Tokyo, Tokyo, Japan), Wesley Sundquist (University of Utah School of Medicine, Salt Lake City, UT), Konrad Buessow (Max Planck Institute for Molecular Genetics, Berlin, Germany), and Hemmo Meyer (University of Duisburg-Essen, Essen, Germany) through Addgene: pBabe-Hygro (#1765; Morgenstern and Land, 1990), lentiCas9-Blast (#52962; Sanjana et al., 2014), lentiCRISPRv2 (puro; #98290; Brett Stringer Lab), LRG (Lenti_sgRNA_EFS_GFP, #65656; Shi et al., 2015), pMXs-IP-EGFP-mATG5 (#38196; Hara et al., 2008), pMXs-IP-EGFP-ULK1 (#38193; Hara et al., 2008), pMXs-IP-EGFP-LC3 (#38195; Hara et al., 2008), pEGFP-VPS4-E228Q (#80351; Votteler et al., 2016), pQTEV-VPS28 (#34803; Büssow et al., 2005), and pmCherry-GAL3 (#85662; Papadopoulos et al., 2017). To generate pCDH1-CMV-MCS-SV40-Hygro, the puromycin resistance gene (BamHI–SalI [blunted] in pCDH1-CMV-MCS1-EF1-Puro; #CD510A-1; System Biosciences) was replaced to the hygromycin resistance gene (BanHI–ClaI [blunted]) from pBabe-Hygro. pCDH1-CMV-HT-LC3-SV40-Hygro was generated by subcloning the PCR-amplified (primer set: 5′-GAAGATTCTAGAACCATGGCAGAAATCGG-3′; 5′-TATAGGATCCCTACACTGACAATTTCATCCCG-3′) HT-LC3 cassette in to the XbaI–BamHI site of pCDH1-CMV-MCS-SV40-Hygro. For VPS37A expression constructs, total RNA was extracted from HEK293T cells using Qiagen RNeasy mini kit (catalog number 74104). The first-strand cDNA was synthesized via GoScript ReverseTranscription System (catalog number A500; Promega) according to the manufacturer’s instruction. The VPS37A variant 1 (FL) cDNA was then amplified by PCR using the primers 5′-TCTCGAGCTCAAAGCTGGCTTTTTCCCCTGAC-3′ and 5′-TATAGGATCCCTATAGTGGAGCATGAAATTG-3′ and subcloned into the SacI–BamHI site of pEGFP-C1 (6082–1; Clontech). The VPS37A variant 4 (ΔPUEV) cDNA was amplified by PCR using the primers 5′-TCTCGAGCTCAAGATAAACAAGGAGTGTATG-3′ and 5′-TATAGGATCCCTATAGTGGAGCATGAAATTG-3′, and pEGFP-FL as a template. Finally, the GFP-FL and GFP-ΔPUEV cassettes were amplified by PCR using the primers 5′-GATCGCACCGGTCGCCACCTCTAGAATGGTGAGCAAGGGCGAG-3′ and 5′-TATAGGATCCCTATAGTGGAGCATGAAATTG3′, and subcloned into the XbaI–BamHI site of pCDH1-CMV-MCS1-EF1-Puro to generate pCDH1-CMV-GFP-FL(VPS37A variant 1)-EF1-Puro and pCDH1-CMV-GFP-ΔPUEV(VPS37A variant 4)-EF1-Puro, respectively. To make an inducible dominant-negative VPS4 (VPS4AE228Q) expression plasmid, pCDH1-TetOn-MCS-EF1-puro was generated by subcloning the tetracycline operator and reverse tetracycline-controlled transactivator into the ClaI–XbaI and SalI sites, respectively, of pCDH1-CMV-MCS-EF1-Puro. The EGFP cassette in the AgeI–XhoI site of pEGFP-VPS4-E228Q was replaced with the mCherry cassette and the resultant mCherry-DN-VPS4 cassette was subcloned into the XbaI–NotI site of pCDH1-TetOn-MCS-EF1-puro to generate pCDH1-TetOn-DN-VPS4-EF1-puro. pCDH1-CMV-CHMP2A-myc-SV40-hygro was generated using Gibson Assembly. The linker sequence between CHMP2A and myc are N′-(CHMP2A)-GGGGS-(myc)-C′. To generate a lentiviral vector encoding human VPS28, the VPS37A cDNA at the XhoI–NotI site in pCDH1-CMV-GFP-FL was replaced to the human VPS28 cDNA amplified from the plasmid pQTEV-VPS28 using the primer 5′-TATACTCGAGGATTTCATGGGATCCCAGCC-3′ and 5′-TCGAGCGGCCGCTCAGGCATG-3′. For the tandem fluorescent-tagged VPS37A (RFP-GFP-VPS37A) expression constructs, the EGFP-LC3 cassette in tfLC3 (pCDH-RFP-EGFP-LC3; Tang et al., 2017) was replaced to GFP-FL or GFP-ΔPUEV at the Age1–Bamh1 site. All other reagents were obtained from the following sources: autoMACS Rinsing Solution (130–091-222), Miltenyi Biotec; BafA1 (B-1080), LC Laboratories; BSA (126575), EMD Millipore; MIL (AF488-conjugated, G1001; AF660-conjugated, G8471), Promega; MPL (TMR-conjugated, G8251), Promega; normal goat serum (G9023), Sigma-Aldrich; Nucleofector Kit V (VCA-1003), Lonza; PFA (15710), Electron Microscopy Sciences; recombinant human EGF (PHG0311), Invitrogen; and XF Plasma Membrane Permeabilizer/recombinant perfringolysin O mutant (102504–100), Seahorse Bioscience.
Cell culture and autophagy induction
HeLa (CCL-2), 293 (CRL-1573), 293T/17 (CRL-11268), and SK-N-AS (CRL-2137) cells were maintained in DMEM supplemented with 10% FBS. U-2 OS cells were maintained in McCoy’s 5A medium, supplemented with 10% FBS and antibiotic-antimycotic (30-004-CI; Corning). All cells were obtained from American Type Culture Collection. To induce autophagy, cells were rinsed three times with Dulbecco’s PBS and incubated with amino acid–free DMEM (048–33575; Wako).
Transfection and viral transduction
To produce recombinant lentiviruses, each lentiviral vector was cotransfected with the Invitrogen ViraPower lentiviral packaging plasmids (pLP1, pLP2, and pLP/VSVG) into 293T/F17 cells using Lipofectamine 3000 (Invitrogen). Viral supernatants were collected 24 and 48 h after transfection followed by filtration (0.45-µm pore). Recombinant retroviruses were generated using the Phoenix Amphotropic system (Nolan Lab, Stanford University, Stanford, CA).
Library amplification and screening
The human Brunello CRISPR library (one-vector system) was amplified in ElecroMAX Stbl4 competent cells (Invitrogen) as described previously (Doench et al., 2016) and sequenced using the Illumina HiSeq 2500 system to ensure sufficient coverage. For screening, 108 U-2 OS cells stably expressing HT-LC3 were transduced with recombinant lentiviruses at the amount to reach up to 40% infection efficiency for 48 h followed by selection with 2 µg/ml puromycin for 72 h. 7 d after the transduction, cells were subjected to the HT-LC3 assay and sorted using the BD FACSAria SORP cell sorter. Approximately 107 cells in P2 or P3, and 3 × 107 cells in P1 or unsorted population were subjected to genomic DNA extraction using the Blood and Cell Culture Mini kit (13323; Qiagen). Genomically integrated sgRNA sequences were amplified by PCR using p5 (5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTTTGTGGAAAGGACGAAACACCG) and barcoded p7 (5′-CAAGCAGAAGACGGCATACGAGATNNNNNNNNGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTCCAATTCCCACTCCTTTCAAGACCT-3′) primers and Herculase II Fusion DNA Polymerase (600677; Agilent Technologies) via the method described by Doench et al. (2016) to generate the Illumina sequencing libraries. The resulting sequencing libraries were purified by gel extraction using QIAquick Gel Extraction Kit (28704; Qiagen) followed by quality analysis via Agilent 2100 Bioanalyzer instrument. The Illumina sequencing libraries were pooled together with other samples that are well diverse in nucleotide composition to avoid the monotemplating during Illumina sequencing, and the pooled libraries were sequenced by a Hiseq 2500 (Illumina) instrument by a single-end 50 cycle (1 × 50) on a Rapid flow cell.
Next-generation sequencing data analysis
Raw sequencing reads were subjected to count the spacer distribution by using the count_spacers.py, a python script, as distributed and described by Joung et al. (2017). Raw sequencing reads were processed based on the protocol described previously (Joung et al., 2017) followed by redundant siRNA activity analysis (König et al., 2007). sgRNA enrichments in the MILhigh population (P2) was assessed by comparing the log2 fold ratio of the abundance of each sgRNA in the P2 population versus the unsorted population (first and second screenings) or versus the P1 population with moderate MIL and MPL signals (third and fourth screenings). Top hits (P < 0.05) that overlapped with at least two biological replicates whose function may be involved in membrane trafficking were chosen for further validation.
CRISPR/Cas9-mediated genome editing
Each sgRNA listed in Table S2 was subcloned into the BsmB1 site of lentiCRISPRv2 or LRG. For the secondary screening, three independent recombinant lentiviruses encoding distinct sgRNAs against each gene were generated using the lentiCRISPRv2 system (Brett Stringer Lab), transduced into HT-LC3-expressing U-2 OS cells for 48 h, and selected with 2 µg/ml puromycin for 3 d. The cells were cultured at least another 3 d in complete medium before use for the experiments. To generate VPS37A KO U-2 OS cells, recombinant lentiviruses encoding Cas9 were generated using the lentiCas9-Blast system (Sanjana et al., 2014) and transduced into U-2 OS cells. The cells were selected by 10 µg/ml blasticidin for 10 d and then transiently transfected with a mixture of LRG plasmids encoding three distinct VPS37A sgRNAs for 48 h followed by flow cytometry sorting for GFP-positive cells. 2 wk after culture in complete medium, a GFP-negative single clone was picked and expanded to obtain VPS37A KO cells.
HT-LC3 autophagosome completion assay
For FACS analysis, HT-LC3–expressing cells were trypsinized, rinsed with PBS, and resuspended in 1× MAS buffer (220 mM mannitol, 70 mM sucrose, 10 mM KH2PO4, 5 mM MgCl2, 2 mM Hepes, and 1 mM EGTA) containing 4 nM XF-PMP and 1 µM MIL-AF488 at 37°C for 15 min. Cells were then rinsed with magnetic-activated cell sorting separation buffer (MACSSB; autoMACS Rinsing Solution containing 0.5% BSA and 2 mM EDTA), resuspended in MACSSB containing 1 µM MPL-TMR, incubated at RT for 5 min, rinsed twice with MACSSB, resuspended in MACSSB, and subjected to flow cytometry. For confocal microscopy, cells were incubated in 1× MAS buffer containing 3 nM XF-PMP and 1 µM MIL-AF488 or 3.5 µM MIL-AF660 at 37°C for 15 min, fixed in 4% PFA for 5 min, washed three times in PBS, and incubated with 5 µM TMR for 30 min. After washing three times in PBS, cells were analyzed by confocal microscopy.
Flow cytometry, confocal microscopy, and electron microscopy
For flow cytometry, cells were resuspended in MACSSB and analyzed on a BD LDRFortessa cell analyzer. Data were analyzed using FlowJo software. For confocal microscopy, cells were grown on Lab-TekII Chambered Coverglass, Chamber Slide (154941; Nunc). For IF, cells were permeabilized and fixed in methanol at −20°C for 10 min, blocked in 10% normal goat serum for 1 h, and incubated with the primary antibodies overnight at 4°C followed by the secondary antibodies for 1 h at RT. Fluorescent images were obtained at RT using a Leica AOBS SP8 laser-scanning confocal microscope (63× water [1.4 numerical aperture] or oil-immersion [1.2 numerical aperture] lens) with the highly sensitive HyD detectors and the Leica image acquisition software LAX, deconvolved using Huygens deconvolution software (Scientific Volume Imaging), and analyzed using Imaris software (Bitplane) and Volocity software (PerkinElmer) without gamma adjustment. PBS was used as imaging medium. For electron microscopy, cells were grown on Thermanox plastic coverslips (174950; Thermo Scientific) overnight, incubated in starvation medium (048-33575; Wako) for 3 h, fixed in 2% PFA/2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.3, for 1.5 h at RT followed by postfixation buffer (1% osmium tetroxide/1.5% potassium ferrocyanide/0.1 M sodium cacodylate, pH 7.3) for 1 h or overnight, dehydrated in a graded series of ethanol, embedded in EMbed 812 resin (14120; Electron Microscopy Sciences), sectioned at a thickness of 70 nm, mounted on mesh copper grids, stained with aqueous uranyl acetate and lead citrate, and analyzed using a JEOL JEM 1400 transmission electron microscope.
IB and coimmunoprecipitation
Total cell lysates were prepared in radio-immunoprecipitation assay buffer (150 mM NaCl, 10 mM Tris-HCl, pH 7.4, 0.1% SDS, 1% Triton X-100, 1% deoxycholate, and 5 mM EDTA, pH 8.0) containing protease inhibitors and subjected to SDS-PAGE followed by IB with the indicated antibodies. The signal intensities were quantified using Image Studio version 5 software (LI-COR Biotechnology; Serfass et al., 2017). To measure changes in basal and induced autophagic flux and autophagy induction, LC3-II levels in each lane were normalized to the respective value of β-actin and normalized LC3-II values were used for calculations as described previously (Tooze et al., 2015). The formulas to calculate each event were as follows (CM: complete medium [DMEM supplemented with 10% FBS]; SM: starvation medium): basal flux = (CM with BafA1) − CM; induced flux = (SM with BafA1) − SM; induction = (SM with BafA1) − (CM with BafA1). For coimmunoprecipitation, total cell lysates were prepared in 0.5% NP-40 lysis buffer (10 mM Tris/Cl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, and 0.5% NP-40) containing protease inhibitors and subjected to immunoprecipitation using GFP-Trap beads (gtma; Chromotek). The resulting immunocomplexes were washed three times with wash buffer (10 mM Tris/Cl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, and 0.05% NP-40) and subjected to IB.
Endocytic EGFR degradation assay
Cells at 75% confluency were serum starved for 16 h, incubated in DMEM containing 10 µg/ml cycloheximide and 100 ng/ml EGF at 37°C for the indicated periods of time, and subjected to IB.
Statistical analyses
Statistical significance was determined using Graph Pad Prism 7.0. Threshold for statistical significance for each test was set at 95% confidence (P < 0.05).
Online supplemental material
Fig. S1 shows lower magnification images for Fig. 2 D. Fig. S2 shows additional immunoblots used for quantification in Fig. 4, D and E. Fig. S3 shows the lysosomal degradation of GFP-VPS37A FL during autophagy. Fig. S4 shows the immature autophagosomal membrane translocation of GFP-VPS37A FL upon lysosomal damage. Fig. S5 shows quantification results for Fig. 8 A. Table S1 shows the primary screen data. Table S2 shows sgRNA sequences used for the experiments.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants GM127954 and CA222349, the Lois High Berstler Research Endowment Fund, and the Four Diamonds. Confocal images were generated using the Leica SP8 microscope (National Institutes of Health Shared Instrumentation grant S10OD010756-01A1) located in the Penn State College of Medicine Microscopy Imaging Core Facility.
The authors declare no competing financial interests.
Author contributions: Y. Takahashi and X. Liang performed the experiments and analyzed the data; T. Hattori, Z. Tang, H. He, M.M. Young, X. Liu, Y. Imamura-Kawasawa, N.J. Buchkovich, H. Chen, and T. Abraham assisted in preparing reagents, collecting and analyzing the data, and preparing the manuscript; and Y. Takahashi, X. Liang, and H.-G. Wang designed the experiments, discussed the results, and wrote the manuscript.
References
- Bache K.G., Slagsvold T., Cabezas A., Rosendal K.R., Raiborg C., and Stenmark H.. 2004. The growth-regulatory protein HCRP1/hVps37A is a subunit of mammalian ESCRT-I and mediates receptor down-regulation. Mol. Biol. Cell. 15:4337–4346. 10.1091/mbc.e04-03-0250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büssow K., Scheich C., Sievert V., Harttig U., Schultz J., Simon B., Bork P., Lehrach H., and Heinemann U.. 2005. Structural genomics of human proteins—target selection and generation of a public catalogue of expression clones. Microb. Cell Fact. 4:21 10.1186/1475-2859-4-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ L., Wenzel E.M., Liestøl K., Raiborg C., Campsteijn C., and Stenmark H.. 2016. ALIX and ESCRT-I/II function as parallel ESCRT-III recruiters in cytokinetic abscission. J. Cell Biol. 212:499–513. 10.1083/jcb.201507009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ L., Raiborg C., Wenzel E.M., Campsteijn C., and Stenmark H.. 2017. Cellular Functions and Molecular Mechanisms of the ESCRT Membrane-Scission Machinery. Trends Biochem. Sci. 42:42–56. 10.1016/j.tibs.2016.08.016 [DOI] [PubMed] [Google Scholar]
- Divakaruni A.S., Wiley S.E., Rogers G.W., Andreyev A.Y., Petrosyan S., Loviscach M., Wall E.A., Yadava N., Heuck A.P., Ferrick D.A., et al. . 2013. Thiazolidinediones are acute, specific inhibitors of the mitochondrial pyruvate carrier. Proc. Natl. Acad. Sci. USA. 110:5422–5427. 10.1073/pnas.1303360110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djeddi A., Michelet X., Culetto E., Alberti A., Barois N., and Legouis R.. 2012. Induction of autophagy in ESCRT mutants is an adaptive response for cell survival in C. elegans. J. Cell Sci. 125:685–694. 10.1242/jcs.091702 [DOI] [PubMed] [Google Scholar]
- Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R., et al. . 2016. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34:184–191. 10.1038/nbt.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskelinen E.L. 2008. Fine structure of the autophagosome. Methods Mol. Biol. 445:11–28. 10.1007/978-1-59745-157-4_2 [DOI] [PubMed] [Google Scholar]
- Feng Y., He D., Yao Z., and Klionsky D.J.. 2014. The machinery of macroautophagy. Cell Res. 24:24–41. 10.1038/cr.2013.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J.L., and Mizushima N.. 2008. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181:497–510. 10.1083/jcb.200712064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J.H. 2015. ESCRTs are everywhere. EMBO J. 34:2398–2407. 10.15252/embj.201592484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J., Konermann S., Gootenberg J.S., Abudayyeh O.O., Platt R.J., Brigham M.D., Sanjana N.E., and Zhang F.. 2017. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat. Protoc. 12:828–863. 10.1038/nprot.2017.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanasios E., Stapleton E., Manifava M., Kaizuka T., Mizushima N., Walker S.A., and Ktistakis N.T.. 2013. Dynamic association of the ULK1 complex with omegasomes during autophagy induction. J. Cell Sci. 126:5224–5238. 10.1242/jcs.132415 [DOI] [PubMed] [Google Scholar]
- Katzmann D.J., Babst M., and Emr S.D.. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 106:145–155. 10.1016/S0092-8674(01)00434-2 [DOI] [PubMed] [Google Scholar]
- Kimura S., Noda T., and Yoshimori T.. 2007. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 3:452–460. 10.4161/auto.4451 [DOI] [PubMed] [Google Scholar]
- Knorr R.L., Lipowsky R., and Dimova R.. 2015. Autophagosome closure requires membrane scission. Autophagy. 11:2134–2137. 10.1080/15548627.2015.1091552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König R., Chiang C.Y., Tu B.P., Yan S.F., DeJesus P.D., Romero A., Bergauer T., Orth A., Krueger U., Zhou Y., and Chanda S.K.. 2007. A probability-based approach for the analysis of large-scale RNAi screens. Nat. Methods. 4:847–849. 10.1038/nmeth1089 [DOI] [PubMed] [Google Scholar]
- Kroemer G., Mariño G., and Levine B.. 2010. Autophagy and the integrated stress response. Mol. Cell. 40:280–293. 10.1016/j.molcel.2010.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C.A., Yoshimori T., and Tooze S.A.. 2013. The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 14:759–774. 10.1038/nrm3696 [DOI] [PubMed] [Google Scholar]
- Li D., Chen A., Lan T., Zou Y., Zhao L., Yang P., Qu H., Wei L., Varghese Z., Moorhead J.F., et al. . 2019. SCAP knockdown in vascular smooth muscle cells alleviates atherosclerosis plaque formation via up-regulating autophagy in ApoE-/- mice. FASEB J. 33:3437–3450. 10.1096/fj.201800975RRR [DOI] [PubMed] [Google Scholar]
- McCullough J., Colf L.A., and Sundquist W.I.. 2013. Membrane fission reactions of the mammalian ESCRT pathway. Annu. Rev. Biochem. 82:663–692. 10.1146/annurev-biochem-072909-101058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., and Yoshimori T.. 2001. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152:657–668. 10.1083/jcb.152.4.657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A.M., and Klionsky D.J.. 2008. Autophagy fights disease through cellular self-digestion. Nature. 451:1069–1075. 10.1038/nature06639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T., and Ohsumi Y.. 2011. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27:107–132. 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- Morgenstern J.P., and Land H.. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587–3596. 10.1093/nar/18.12.3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E., Colf L.A., Karren M.A., Sandrin V., Rodesch C.K., and Sundquist W.I.. 2010. Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc. Natl. Acad. Sci. USA. 107:12889–12894. 10.1073/pnas.1005938107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K., Hama Y., Izume T., Tamura N., Ueno T., Yamashita Y., Sakamaki Y., Mimura K., Morishita H., Shihoya W., et al. . 2018. Genome-wide CRISPR screen identifies TMEM41B as a gene required for autophagosome formation. J. Cell Biol. 217:3817–3828. 10.1083/jcb.201804132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y. 2014. Historical landmarks of autophagy research. Cell Res. 24:9–23. 10.1038/cr.2013.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos Y., Hodgson L., Mantell J., Verkade P., and Carlton J.G.. 2015. ESCRT-III controls nuclear envelope reformation. Nature. 522:236–239. 10.1038/nature14503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos C., Kirchner P., Bug M., Grum D., Koerver L., Schulze N., Poehler R., Dressler A., Fengler S., Arhzaouy K., et al. . 2017. VCP/p97 cooperates with YOD1, UBXD1 and PLAA to drive clearance of ruptured lysosomes by autophagy. EMBO J. 36:135–150. 10.15252/embj.201695148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornillos O., Alam S.L., Rich R.L., Myszka D.G., Davis D.R., and Sundquist W.I.. 2002. Structure and functional interactions of the Tsg101 UEV domain. EMBO J. 21:2397–2406. 10.1093/emboj/21.10.2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Shen X., Shyam R., Yue B.Y., and Ying H.. 2014. Cellular processing of myocilin. PLoS One. 9:e92845 10.1371/journal.pone.0092845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radulovic M., Schink K.O., Wenzel E.M., Nähse V., Bongiovanni A., Lafont F., and Stenmark H.. 2018. ESCRT-mediated lysosome repair precedes lysophagy and promotes cell survival. EMBO J. 37:e99753 10.15252/embj.201899753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusten T.E., and Stenmark H.. 2009. How do ESCRT proteins control autophagy? J. Cell Sci. 122:2179–2183. 10.1242/jcs.050021 [DOI] [PubMed] [Google Scholar]
- Sanjana N.E., Shalem O., and Zhang F.. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 11:783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöneberg J., Lee I.H., Iwasa J.H., and Hurley J.H.. 2017. Reverse-topology membrane scission by the ESCRT proteins. Nat. Rev. Mol. Cell Biol. 18:5–17. 10.1038/nrm.2016.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serfass J.M., Takahashi Y., Zhou Z., Kawasawa Y.I., Liu Y., Tsotakos N., Young M.M., Tang Z., Yang L., Atkinson J.M., et al. . 2017. Endophilin B2 facilitates endosome maturation in response to growth factor stimulation, autophagy induction, and influenza A virus infection. J. Biol. Chem. 292:10097–10111. 10.1074/jbc.M117.792747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Wang E., Milazzo J.P., Wang Z., Kinney J.B., and Vakoc C.R.. 2015. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat. Biotechnol. 33:661–667. 10.1038/nbt.3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields S.B., Oestreich A.J., Winistorfer S., Nguyen D., Payne J.A., Katzmann D.J., and Piper R.. 2009. ESCRT ubiquitin-binding domains function cooperatively during MVB cargo sorting. J. Cell Biol. 185:213–224. 10.1083/jcb.200811130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra M.L., Schlesinger P.H., Naismith T.V., and Hanson P.I.. 2018. Triggered recruitment of ESCRT machinery promotes endolysosomal repair. Science. 360:eaar5078 10.1126/science.aar5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuchell M.D., Garrus J.E., Müller B., Stray K.M., Ghaffarian S., McKinnon R., Kräusslich H.G., Morham S.G., and Sundquist W.I.. 2004. The human endosomal sorting complex required for transport (ESCRT-I) and its role in HIV-1 budding. J. Biol. Chem. 279:36059–36071. 10.1074/jbc.M405226200 [DOI] [PubMed] [Google Scholar]
- Sun L.L., Li M., Suo F., Liu X.M., Shen E.Z., Yang B., Dong M.Q., He W.Z., and Du L.L.. 2013. Global analysis of fission yeast mating genes reveals new autophagy factors. PLoS Genet. 9:e1003715 10.1371/journal.pgen.1003715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., He H., Tang Z., Hattori T., Liu Y., Young M.M., Serfass J.M., Chen L., Gebru M., Chen C., et al. . 2018. An autophagy assay reveals the ESCRT-III component CHMP2A as a regulator of phagophore closure. Nat. Commun. 9:2855 10.1038/s41467-018-05254-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Takahashi Y., Chen C., Liu Y., He H., Tsotakos N., Serfass J.M., Gebru M.T., Chen H., Young M.M., and Wang H.G.. 2017. Atg2A/B deficiency switches cytoprotective autophagy to non-canonical caspase-8 activation and apoptosis. Cell Death Differ. 24:2127–2138. 10.1038/cdd.2017.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teis D., Saksena S., and Emr S.D.. 2008. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev. Cell. 15:578–589. 10.1016/j.devcel.2008.08.013 [DOI] [PubMed] [Google Scholar]
- Tian Y., Li Z., Hu W., Ren H., Tian E., Zhao Y., Lu Q., Huang X., Yang P., Li X., et al. . 2010. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell. 141:1042–1055. 10.1016/j.cell.2010.04.034 [DOI] [PubMed] [Google Scholar]
- Tooze S.A., Dooley H.C., Jefferies H.B., Joachim J., Judith D., Lamb C.A., Razi M., and Wirth M.. 2015. Assessing mammalian autophagy. Methods Mol. Biol. 1270:155–165. 10.1007/978-1-4939-2309-0_12 [DOI] [PubMed] [Google Scholar]
- Votteler J., Ogohara C., Yi S., Hsia Y., Nattermann U., Belnap D.M., King N.P., and Sundquist W.I.. 2016. Designed proteins induce the formation of nanocage-containing extracellular vesicles. Nature. 540:292–295. 10.1038/nature20607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Miao G., Xue X., Guo X., Yuan C., Wang Z., Zhang G., Chen Y., Feng D., Hu J., and Zhang H.. 2016. The Vici Syndrome Protein EPG5 Is a Rab7 Effector that Determines the Fusion Specificity of Autophagosomes with Late Endosomes/Lysosomes. Mol. Cell. 63:781–795. 10.1016/j.molcel.2016.08.021 [DOI] [PubMed] [Google Scholar]
- Zhou F., Wu Z., Zhao M., Murtazina R., Cai J., Zhang A., Li R., Sun D., Li W., Zhao L., et al. . 2019. Rab5-dependent autophagosome closure by ESCRT. J. Cell Biol. 218:1908–1927. 10.1083/jcb.201811173 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.