In mitosis, the outer kinetochore is assembled to signal the spindle assembly checkpoint and to attach chromosomes to spindle microtubules. Bonner et al. demonstrate that outer kinetochore assembly requires the enrichment of Aurora B kinase at the inner kinetochore by the conserved central region of INCENP.
Abstract
Outer kinetochore assembly enables chromosome attachment to microtubules and spindle assembly checkpoint (SAC) signaling in mitosis. Aurora B kinase controls kinetochore assembly by phosphorylating the Mis12 complex (Mis12C) subunit Dsn1. Current models propose Dsn1 phosphorylation relieves autoinhibition, allowing Mis12C binding to inner kinetochore component CENP-C. Using Xenopus laevis egg extracts and biochemical reconstitution, we found that autoinhibition of the Mis12C by Dsn1 impedes its phosphorylation by Aurora B. Our data indicate that the INCENP central region increases Dsn1 phosphorylation by enriching Aurora B at inner kinetochores, close to CENP-C. Furthermore, centromere-bound CENP-C does not exchange in mitosis, and CENP-C binding to the Mis12C dramatically increases Dsn1 phosphorylation by Aurora B. We propose that the coincidence of Aurora B and CENP-C at inner kinetochores ensures the fidelity of kinetochore assembly. We also found that the central region is required for the SAC beyond its role in kinetochore assembly, suggesting that kinetochore enrichment of Aurora B promotes the phosphorylation of other kinetochore substrates.
Graphical Abstract
Introduction
Equal segregation of chromosomes depends on the kinetochore, a complex protein structure that connects chromosomes to spindle microtubules (Cheeseman, 2014). By EM, the vertebrate kinetochore appears as discrete “plates” represented by the centromeric chromatin-binding inner kinetochore and the microtubule-binding outer kinetochore (Musacchio and Desai, 2017). The inner kinetochore, composed of the 16-subunit constitutive centromere-associated network (CCAN), is assembled on centromeric nucleosomes and persists throughout the cell cycle. The outer kinetochore contains the 10-subunit Knl1–Mis12–Ndc80 (KMN) network, which links the inner kinetochore to microtubules. In higher eukaryotes, the four-subunit Mis12 complex (Mis12C; Mis12, Dsn1, Nsl1, and Pmf1) serves as a hub for outer kinetochore assembly by interacting with both CENP-C and CENP-T at the inner kinetochore (Gascoigne et al., 2011; Przewloka et al., 2011; Screpanti et al., 2011; Rago et al., 2015; Huis In ’t Veld et al., 2016; Hara et al., 2018) and the Ndc80 complex and Knl1 at the outer kinetochore (Cheerambathur and Desai, 2014). Once the KMN network is assembled, attachments with microtubules are made primarily through Ndc80. If erroneous attachments occur or attachments are lost, the spindle assembly checkpoint (SAC) is activated at the outer kinetochore and checkpoint proteins such as BubR1 and Mad2 are recruited to halt the cell cycle.
Aurora B, a major mitotic kinase, is required for the biorientation of sister chromatids through correction of erroneous kinetochore–microtubule attachments, SAC activation, and, in oocytes, assembly of acentrosomal spindles (Carmena et al., 2012). Aurora B phosphorylates the N-terminal tail of Ndc80 in response to lowered tension across kinetochores to weaken kinetochore–microtubule attachments to promote biorientation. In the SAC, Aurora B is thought to act both upstream and downstream of the checkpoint protein recruitment pathway in human cells and Xenopus laevis egg extracts (Emanuele et al., 2008; Maldonado and Kapoor, 2011; Kim and Yu, 2015). Aurora B is also required for outer kinetochore assembly (Emanuele et al., 2008; Yang et al., 2008; Akiyoshi et al., 2013; Kim and Yu, 2015; Rago et al., 2015). In yeast, chicken, and human cells, phosphorylation of two conserved serines on the Dsn1 subunit of the Mis12C by Aurora B (Ipl1 in yeast) promotes the interaction of Mis12C with inner kinetochore components (Akiyoshi et al., 2013; Kim and Yu, 2015; Rago et al., 2015; Zhou et al., 2017; Hara et al., 2018). These phosphosites lie in an autoinhibitory region of Dsn1 that competes with the binding of inner kinetochore proteins such as CENP-C (Dimitrova et al., 2016; Petrovic et al., 2016). Current models posit that Aurora B phosphorylation regulates Mis12C loading by relieving the autoinhibitory interaction of Dsn1. However, it is unclear how Dsn1 phosphorylation is regulated. In yeast, chicken, and human cells, Mis12C loading and kinetochore assembly can occur through redundant Aurora B–independent mechanisms (Akiyoshi et al., 2013; Kim and Yu, 2015; Rago et al., 2015; Hara et al., 2018), thus making it difficult to study the regulation of Dsn1 phosphorylation in these systems. In contrast, outer kinetochore assembly is completely dependent on Aurora B kinase in Xenopus egg extracts (Emanuele et al., 2008; Haase et al., 2017).
Aurora B is part of the four-subunit chromosomal passenger complex (CPC), along with the proteins INCENP, Survivin, and Borealin (Dasra). INCENP, a large scaffolding protein with highly conserved domains, orchestrates the localization and activity of the CPC throughout mitosis. The C-terminal INbox domain of INCENP binds to and promotes the activation of Aurora B kinase (Bishop and Schumacher, 2002; Sessa et al., 2005; Kelly et al., 2007; Wang et al., 2011). Phosphorylation of the INbox motif by another Aurora B–INCENP complex in trans, regulated through local concentration at centromeres or microtubules, is thought to allow for full kinase activation (Sessa et al., 2005; Kelly et al., 2007; Zaytsev et al., 2016). The CEN domain confers interaction with Survivin and Borealin, which target the CPC to histone modifications present on the chromosome arms and at inner centromeres (Kelly et al., 2010; Wang et al., 2010; Yamagishi et al., 2010). The CEN domain of the INCENP is required for error correction in human cells and Xenopus egg extracts (Haase et al., 2017; Hengeveld et al., 2017), but not for biorientation in budding yeast (Campbell and Desai, 2013).
The central region of INCENP contains two domains (phosphoregulatory domain [PRD] and single α-helical [SAH] domain) that have been implicated in CPC association with microtubules (Vader et al., 2007; Tseng et al., 2010; Wheelock et al., 2017). The INCENP SAH domain forms an α-helix that lacks tertiary structure and directly binds microtubules (Samejima et al., 2015), while phosphorylation of the PRD regulates the microtubule affinity of the SAH domain (Kang et al., 2001; Pereira and Schiebel, 2003; Wheelock et al., 2017). The SAH domain is important for CPC function in eukaryotes (Sandall et al., 2006; Kelly et al., 2007; Vader et al., 2007; Tseng et al., 2010; Samejima et al., 2015; van der Horst et al., 2015; Fink et al., 2017; Wheelock et al., 2017), and its interaction with microtubules has been proposed to underlie its role by targeting Aurora B to microtubules to phosphorylate microtubule-associated proteins. Here, we demonstrate the autoinhibition of the Mis12C impedes Dsn1 phosphorylation and that interplay between CENP-C and Aurora B drives the phosphorylation of Dsn1 and kinetochore assembly. We reveal a microtubule-independent role for the SAH domain and the central region in kinetochore assembly and the SAC through the enrichment of Aurora B at inner kinetochores. Together, our data demonstrate that the INCENP central region ensures the proper assembly of the kinetochore upon entry into mitosis.
Results
Outer kinetochore assembly is entirely dependent on the phosphorylation of Dsn1 by Aurora B kinase in Xenopus egg extracts
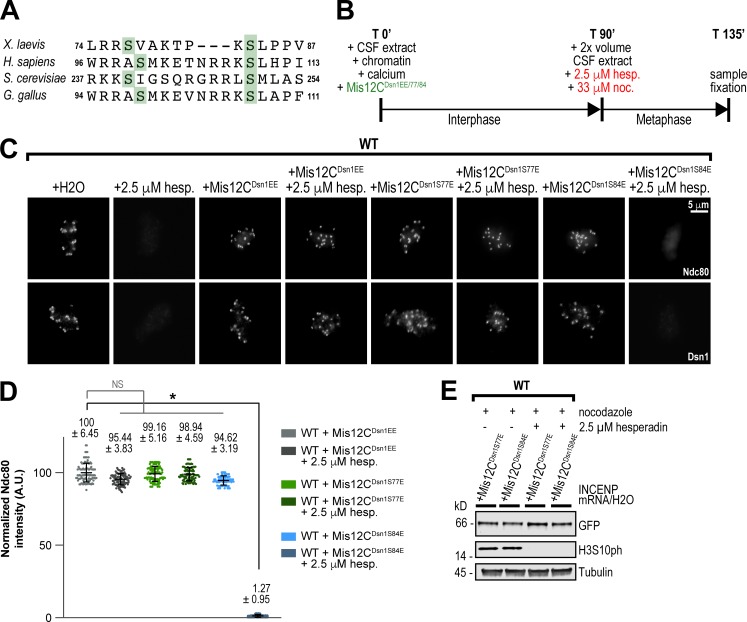
In Xenopus egg extracts, the assembly and maintenance of kinetochores is completely dependent on the mitotic kinase Aurora B, but the underlying phosphosubstrates are unclear (Emanuele et al., 2005, 2008; Haase et al., 2017). In human, chicken, and yeast cells, Aurora B phosphorylation of multiple residues within the Dsn1 subunit of the Mis12C plays a partial role in kinetochore assembly (Fig. 1 A; Akiyoshi et al., 2013; Kim and Yu, 2015; Rago et al., 2015; Zhou et al., 2017; Hara et al., 2018). To investigate the role of Dsn1 phosphorylation in kinetochore assembly in egg extracts, we first mapped Aurora B–dependent phosphorylation sites of the Mis12C immunoprecipitated from egg extracts by mass spectrometry (Fig. S1, A–E). Our analysis revealed numerous phosphorylations within the Dsn1 subunit of the Mis12C, four of which were dependent on Aurora B activity. Two of these Aurora B–dependent phosphorylations, at S77 and S84 of Dsn1, are highly conserved and are important for kinetochore assembly in other species (Fig. 1 A). Indeed, addition of a recombinant phosphomimetic Dsn1 (S77E and S84E) Mis12C (Mis12CDsn1EE) complex to egg extracts treated with the Aurora B inhibitor hesperadin, or immunodepleted of the CPC, restored the assembly of the KMN network of proteins (as detected by KNL1, Dsn1, and Ndc80 staining) at kinetochores to WT levels (Fig. 1, B–E; and Fig. S1, F–J). Addition of S77E phosphomimetic Mis12C, but not S84E, to extracts restored assembly of Dsn1 and Ndc80 at kinetochores (Fig. 1, B–E). Thus, Aurora B–mediated phosphorylation of a single site within the Mis12C controls kinetochore assembly of the KMN network in Xenopus egg extracts, making it an ideal system to study the role of the CPC in outer kinetochore assembly.
Figure 1.
Outer kinetochore assembly is completely dependent on the phosphorylation of Dsn1 by Aurora B in Xenopus egg extracts. (A) Alignment of the Dsn1 autoinhibitory basic region from indicated species showing conserved Aurora B phosphorylation sites. (B) Schematic of kinetochore assembly experiments in Xenopus egg extract. Sperm chromatin was added to WT or ΔCPC CSF extract. To express mutants, recombinant protein or mRNA was added to CSF extract at this point. Here, shown in green, mRNA was added to express the Mis12C or its variants (Mis12CDsn1EE/S77E/S84E), composed of Nsl1, Mis12, Pmf1, and Dsn1-LAP. For all figures, unless recombinant (r) protein is noted in text or figure legend, all mutants were expressed from mRNA. Calcium chloride was added to cycle extract into interphase. After 90 min, 2× volume CSF extract (WT or ΔCPC) was added to cycle the extract into metaphase in addition to drugs shown in red. After a total of 135 min, samples were taken for immunofluorescence and Western blots. (C) Representative immunofluorescence images of replicated chromosomes in WT Xenopus metaphase extracts with the Mis12C expressed from mRNA in extracts and treated with hesperadin in indicated conditions. Chromosomes were stained for Ndc80 and Dsn1. (D) Quantification of fluorescence intensity of Ndc80 on replicated chromosomes in metaphase extracts in indicated conditions shown in C, normalized to WT. n = 96 kinetochores. Error bars represent SD unless otherwise noted, and asterisks indicate a statistically significant difference (*, P < 0.001). A.U., arbitrary units. (E) Western blot for Dsn1-LAP, histone H3 phosphorylation (H3S10ph), and tubulin for samples shown in C and D.
The central region of INCENP promotes outer kinetochore assembly
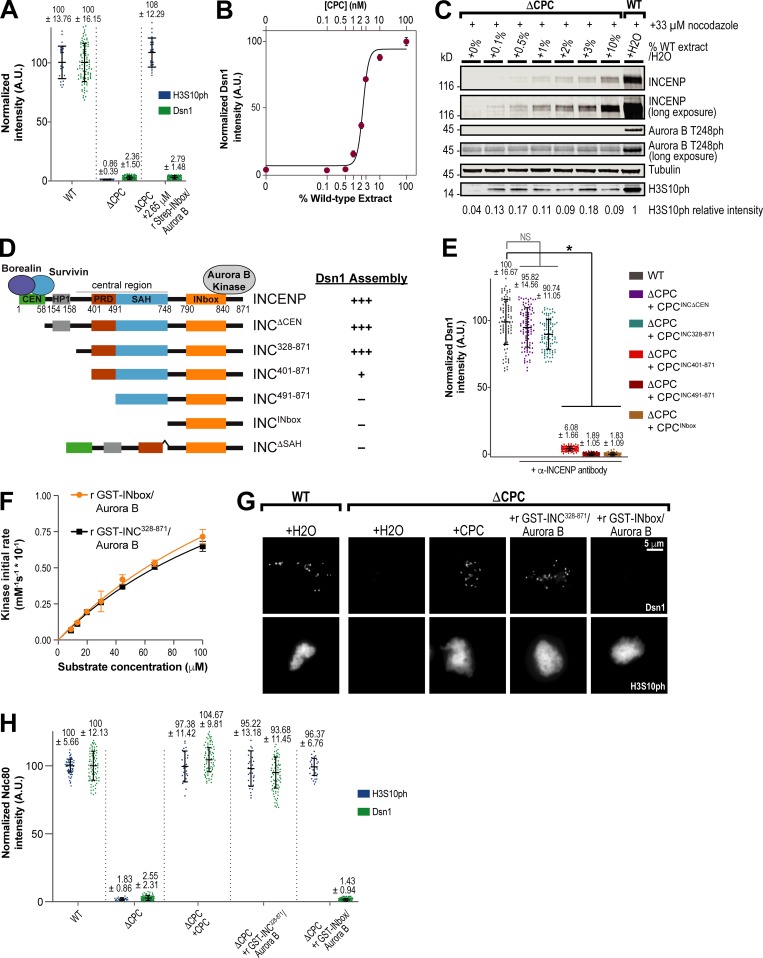
Dsn1 phosphorylation enables Mis12C binding to CCAN proteins such as CENP-C, and current models posit that phosphorylation by Aurora B relieves autoinhibitory contacts between Dsn1 and the CENP-C–binding site on the Mis12C to drive outer kinetochore assembly (Kim and Yu, 2015; Rago et al., 2015; Dimitrova et al., 2016; Petrovic et al., 2016; Hara et al., 2018). A prediction of this model is that active Aurora B kinase should be sufficient to drive kinetochore assembly. To address this, we purified a subcomplex of Aurora B and the INbox domain of INCENP (Aurora B–INbox) and subsequently added high concentrations (∼26-fold higher than endogenous levels) to CPC-depleted extracts. The Aurora B–INbox complex rescued histone H3S10 phosphorylation levels on chromosomes, but not Dsn1 levels (Fig. 2 A and Fig. S2, A–C). Thus, Aurora B kinase activity alone is not sufficient for proper Dsn1 phosphorylation and kinetochore assembly. Indeed, we found that kinetochore assembly of Dsn1 can be achieved even at low concentrations of WT CPC, containing all four subunits (Fig. 2, B and C), suggesting that another part of the CPC is required to promote assembly.
Figure 2.
The central region of INCENP is required for kinetochore assembly. (A) Quantification of fluorescence intensity of Dsn1 and H3S10ph on replicated chromosomes in WT or CPC-depleted (ΔCPC) metaphase extracts with indicated CPC conditions, normalized to WT. Purified, preactivated, recombinant (r) Strep-INbox–His6-Aurora B was added to ΔCPC extract at 2.65 µM. Samples contained 33 µM nocodazole. n = 96 kinetochores for Dsn1, and n = 50 spindles for H3S10ph. A.U., arbitrary units. (B) Mean integrated fluorescence intensity of Dsn1 was quantified at indicated percentage mixture of CPC-depleted and mock-depleted extracts and normalized to mock-depleted levels (WT; 100% CPC). Estimated CPC concentration is indicated. Solid line represents fit to the Hill equation. n = 50 kinetochores per condition. Error bars represent SEM. (C) Western blot for INCENP, Aurora B phosphorylation (T248ph), and tubulin for samples shown in B. WT extract was mixed with CPC-depleted (ΔCPC) extract at various concentrations to determine how much CPC was necessary to enable outer kinetochore assembly. CPC-depleted extracts with >1% CPC remaining led to detectable kinetochore assembly. (D) Schematic of INCENP constructs used in this study. These constructs were expressed from mRNA in extracts, along with full-length Borealin, Survivin, and Aurora B, to reconstitute ΔCPC extracts, unless otherwise noted. To achieve full activation of Aurora B kinase when expressing mRNAs lacking the CEN domain, either beads bound with anti-INCENP antibodies or substoichiometric amounts of clustering anti-INCENP antibodies were added. Extent of Dsn1 assembly at kinetochores for each condition is indicated. (E) Quantification of fluorescence intensity of Dsn1 on replicated chromosomes in mock-depleted (WT) and ΔCPC metaphase extracts with indicated CPC conditions with anti-INCENP antibody added where specified, normalized to WT. n = 96 kinetochores. Error bars represent SD. *, P < 0.001. (F) Initial rate of chemosensor phosphorylation by indicated preactivated recombinant GST-INCENP–His6-Aurora B complexes as a function of chemosensor concentration. Solid lines represent fit by the Michaelis–Menten equation. Graphs represent data from two independent experiments. Error bars represent SEM. (G) Representative immunofluorescence images of replicated chromosomes in WT and ΔCPC metaphase extracts reconstituted with indicated CPC condition, preactivated recombinant GST-INCENP328–871–His6-Aurora B or preactivated recombinant GST-INbox–His6-Aurora B, and treated with nocodazole. Chromosomes were stained for Dsn1 or histone H3S10ph. (H) Quantification of fluorescence intensity of Dsn1 and H3S10ph on replicated chromosomes in WT or ΔCPC metaphase extracts reconstituted with indicated CPC conditions shown in G, normalized to WT. n = 50 kinetochores per condition. Error bars represent SD.
As described previously, a CPC subcomplex containing INCENP lacking its centromere-targeting CEN domain (residues 58–871) and Aurora B kinase can support outer kinetochore assembly if Aurora B is activated by substoichiometric amounts of clustering anti-INCENP antibodies, mimicking Aurora B clustering at centromeres (Haase et al., 2017; Fig. 2, D and E; and Fig. S2, D and E). To determine the minimal region of INCENP required for kinetochore assembly, further N-terminal INCENP truncations were evaluated for assembly of Dsn1 at kinetochores in the presence of Aurora B and activating antibodies (Fig. 2, D and E; and Fig. S2, D and E). Truncation of the HP1-binding domain, implicated in the localization and modulation of Aurora B activity (Abe et al., 2016; Ruppert et al., 2018), did not perturb Dsn1 levels. However, removal of residues 328–400 from the conserved central region of INCENP (residues 328–747) significantly perturbed kinetochore assembly, and truncation of the CDK-phosphorylated PRD (residues 401–490) completely abrogated assembly. Removal of the SAH domain from an INCENP construct lacking the CEN domain also blocked kinetochore assembly (Fig. S2 F). Thus, our results indicate that the central region of INCENP is important for the Aurora B–dependent assembly of the outer kinetochore.
Although the Aurora B–INbox complex could not rescue kinetochore assembly, it still supported full Aurora B activity toward histone H3 (Fig. 2 A and Fig. S2, A and C). However, the concentration of histone H3 in extracts is up to 340-fold higher than many kinetochore substrates such as Dsn1 (Emanuele et al., 2005; Wühr et al., 2014). Kinetochore assembly may not be rescued by the Aurora B–INbox subcomplex because the INCENP central region is required to enhance the kinetics of substrate phosphorylation, which could affect its ability to phosphorylate lower abundance proteins. We purified recombinant Aurora B–GST-INCENP328–871 complex containing the central region and the INbox domain (Fig. S2 G) and compared its activity to the Aurora B–GST-INbox complex using an established in vitro fluorescence-based assay with an Aurora B consensus site peptide (González-Vera et al., 2009; Zaytsev et al., 2016). Both complexes were highly active and likely fully phosphorylated during purification, displaying no substrate phosphorylation lag time (Fig. S2 H). Furthermore, both subcomplexes displayed nearly identical catalytic efficiencies (kcat/KM [see Material and methods]; Fig. 2 F; kcat/KM for Aurora B–INCENP328–871 = 0.22 µM−1s−1; kcat/KM for Aurora B–INbox = 0.21 µM−1s−1). In CPC-depleted extracts, endogenous concentrations of both complexes rescued H3S10ph levels on chromosomes. Importantly, the recombinant Aurora B/GST-INCENP328–871 complex rescued kinetochore assembly, whereas the Aurora B–GST-INbox complex did not (Fig. 2, G and H). Our results indicate that the requirement for the central region in kinetochore assembly is not due to a general alteration of Aurora B kinase activity but rather that it affects the regulation of Dsn1 phosphorylation.
The role of the SAH domain in kinetochore assembly is independent of its microtubule- and chromatin-binding functions
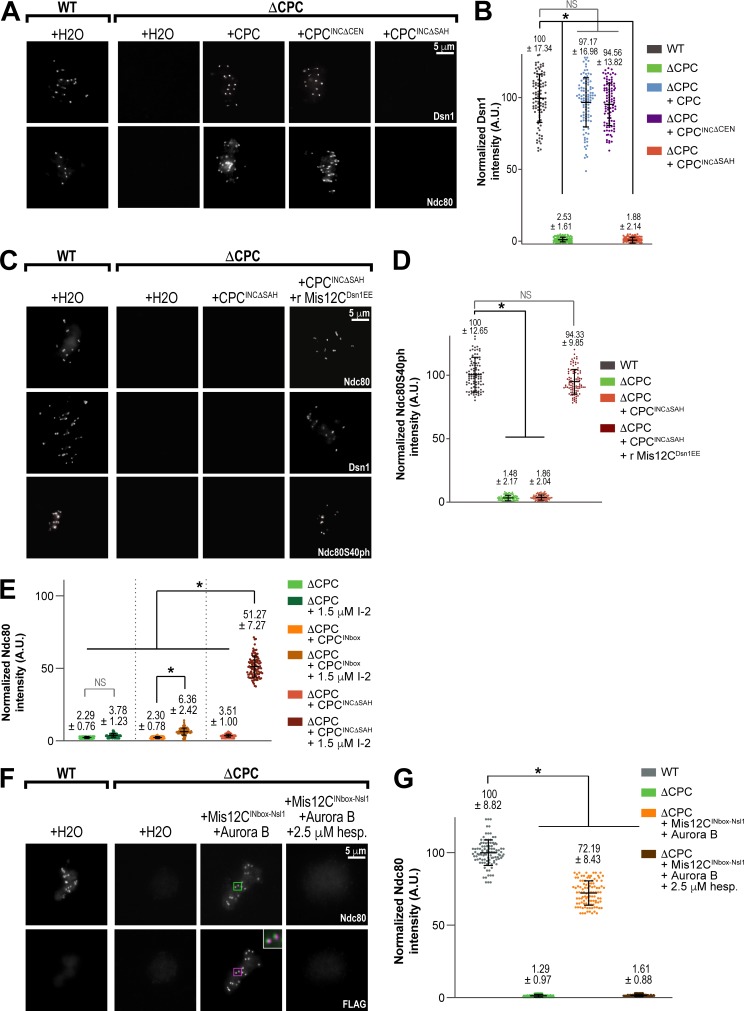
The INCENP central region controls the interaction of the CPC with microtubules in Xenopus egg extracts, yeast, and human cells (Tseng et al., 2010; van der Horst et al., 2015; Fink et al., 2017; Wheelock et al., 2017), and the SAH domain has been shown to directly bind microtubules in vitro (Samejima et al., 2015). We found that deleting the highly conserved SAH domain abrogated Dsn1 and Ndc80 levels at kinetochores (Fig. 3, A and B; and Fig. S3 A). However, replacement of the SAH domain with the microtubule-binding domain of PRC1 (INCΔSAH+PRC1), which substitutes for the spindle assembly function of the SAH domain in egg extracts (Fig. S3 B; Tseng et al., 2010), did not rescue kinetochore assembly (Fig. S3 B), and nocodazole treatment did not affect kinetochore assembly (Fig. S3 B, bottom). Furthermore, phosphomimetic and phospho-null mutations within the conserved CDK sites of the PRD, previously shown to alter CPC affinity for microtubules (Wheelock et al., 2017), did not alter kinetochore assembly (Fig. S3, D and E). Thus, the interaction of the CPC with microtubules is neither necessary nor sufficient for outer kinetochore assembly, suggesting another function of this region. Interestingly, deletion of INCENP residues 134–400, comprising both the HP1-binding site and the 328–400 region important for kinetochore assembly in the absence of the CEN module, did not affect kinetochore assembly (Fig. S3 C). Thus, our results indicate centromere targeting of the CPC can bypass certain central region mutants but that the SAH domain is essential for kinetochore assembly.
Figure 3.
The SAH domain of INCENP is critical for kinetochore assembly. (A) Representative immunofluorescence images of replicated chromosomes in WT and ΔCPC metaphase extracts reconstituted with CPC containing the indicated INCENP construct with anti-INCENP antibody added for the INCΔCEN sample to ensure kinase activation. Samples contained 33 µM nocodazole. Chromosomes were stained for Dsn1 and Ndc80. (B) Quantification of fluorescence intensity of Dsn1 on replicated chromosomes in WT or ΔCPC metaphase extracts reconstituted with indicated CPC conditions shown in A, normalized to WT. n = 96 kinetochores per condition. A.U., arbitrary units. (C) Representative IF images of replicated chromosomes in WT and ΔCPC metaphase extracts reconstituted with CPC containing the indicated INCENP construct and recombinant Mis12CDsn1EE as indicated. Chromosomes were stained for Ndc80, Dsn1, and Ndc80 phosphorylation (Ndc80S40ph). (D) Quantification of fluorescence intensity of Ndc80S40ph on replicated chromosomes in WT or ΔCPC metaphase extracts reconstituted with indicated CPC conditions shown in C, normalized to WT. n = 96 kinetochores per condition. (E) Quantification of fluorescence intensity of Ndc80 in WT and ΔCPC metaphase extracts with indicated CPC conditions and anti-INCENP antibody added to the INbox sample, treated as indicated with I-2, and normalized to WT. n = 96 kinetochores per condition. (F) Representative immunofluorescence images of replicated chromosomes in WT and ΔCPC metaphase extracts with indicated CPC conditions, treated as indicated with hesperadin. Chromosomes were stained for Ndc80 (kinetochore pairs indicated with green boxes) and FLAG (for 3XFLAG-INbox-Nsl1 construct; kinetochore pairs indicated with magenta boxes). The inset white boxes show a merged, 3X zoom image of colocalization of Ndc80 (green) and FLAG (magenta). (G) Quantification of fluorescence intensity of Ndc80 shown in F, normalized to WT. n = 96 kinetochores per condition. Error bars represent SD. *, P < 0.001.
The SAH domain has been reported to interact weakly with chromatin and decrease the dynamics of the CPC at the inner centromere (Wheelock et al., 2017), raising the possibility that it contributes to Aurora B activity at kinetochores by strengthening the CPC interaction with the inner centromere. Although deletion of the SAH domain did not significantly perturb total CPC levels on chromosomes (Fig. S3, F and G) or alter Aurora B activity toward itself or histone H3S10 (Fig. S3, F and H), it is possible that the SAH domain is required for Aurora B activity toward kinetochore substrates, beyond its role in kinetochore assembly. To test this, we measured levels of Aurora B–dependent Ndc80 phosphorylation when stimulated by the depolymerization of kinetochore-attached microtubules, maximal levels of which require the centromeric localization of the CPC in egg extracts (Haase et al., 2017). However, as expected, deletion of the SAH domain abolished Ndc80 phosphorylation due to the lack of kinetochore assembly (Fig. 3, C and D; and Fig. S4, A and B). To circumvent this, we measured the phosphorylation of kinetochore-localized Ndc80 by Aurora B in the presence of recombinant Mis12CDsn1EE, which completely rescued kinetochore assembly of Dsn1 and Ndc80 in INCΔSAH extracts (Fig. 3, C and D; and Fig. S4, A and B). Addition of Mis12CDsn1EE to INCΔSAH extracts containing nocodazole restored Ndc80 phosphorylation levels to WT (Fig. 3, C and D). These results demonstrate that the SAH domain is not required for Ndc80 phosphorylation beyond its role in kinetochore assembly and strongly suggest that it is not required for functional centromere targeting of the CPC. In total, our results demonstrate a specific and essential role for the SAH domain in outer kinetochore assembly through Dsn1 phosphorylation, independent of microtubule and chromatin binding.
Inhibition of PP1 phosphatase partially rescues kinetochore assembly in the absence of the SAH domain
Our results indicate that the SAH domain and the rest of the central region regulate the phosphorylation of Dsn1 to promote kinetochore assembly. The phosphatase PP1 opposes Aurora B kinase in the establishment and maintenance of kinetochores in Xenopus (Fig. S4, C and D; Emanuele et al., 2008). Thus, the SAH domain could function to overcome PP1-mediated dephosphorylation to establish Dsn1 phosphorylation. Treatment of INCΔSAH extracts with the phosphatase inhibitors I-2 or okadaic acid significantly rescued kinetochore assembly of Ndc80, but neither inhibitor could significantly rescue Ndc80 levels in CPC-depleted extracts (Fig. 3 E and Fig. S4, E and F). If the main function of the SAH domain in kinetochore assembly is in regulating PP1 activity, we reasoned that PP1 inhibition should rescue kinetochore assembly as long as Aurora B was active. To test this, we measured Ndc80 assembly at kinetochores in the presence of globally active Aurora B–MBP-INbox subcomplex expressed from mRNAs with and without the PP1 inhibitor I-2. We found that even in the presence of I-2, the Aurora B–MBP-INbox complex could not substantially rescue kinetochore assembly, although weak Ndc80 staining could be observed at kinetochores (∼6% of WT vs. ∼4% for CPC-depleted extracts; Fig. 3 E and Fig. S4, E and F). However, fusion of the INbox directly to the N-terminus of Mis12C subunit Nsl1 (which positions Aurora B close to the Dsn1 N-terminal tail) promoted robust Aurora B–dependent kinetochore assembly, suggesting that high local concentrations of Aurora B around Mis12Cs are required for assembly (Fig. 3, F and G; and Fig. S4 G). Activation of the Aurora B–MBP-INbox complex by beads containing anti-INCENP antibodies, which provide very high local concentrations of Aurora B on the bead surface, did not drive kinetochore assembly in the presence of I-2 (data not shown). Thus, we conclude that the main function of the SAH domain in kinetochore assembly is promoting Aurora B activity toward Dsn1 through local concentration near Mis12Cs.
The central region of INCENP indirectly promotes Dsn1 phosphorylation by Aurora B
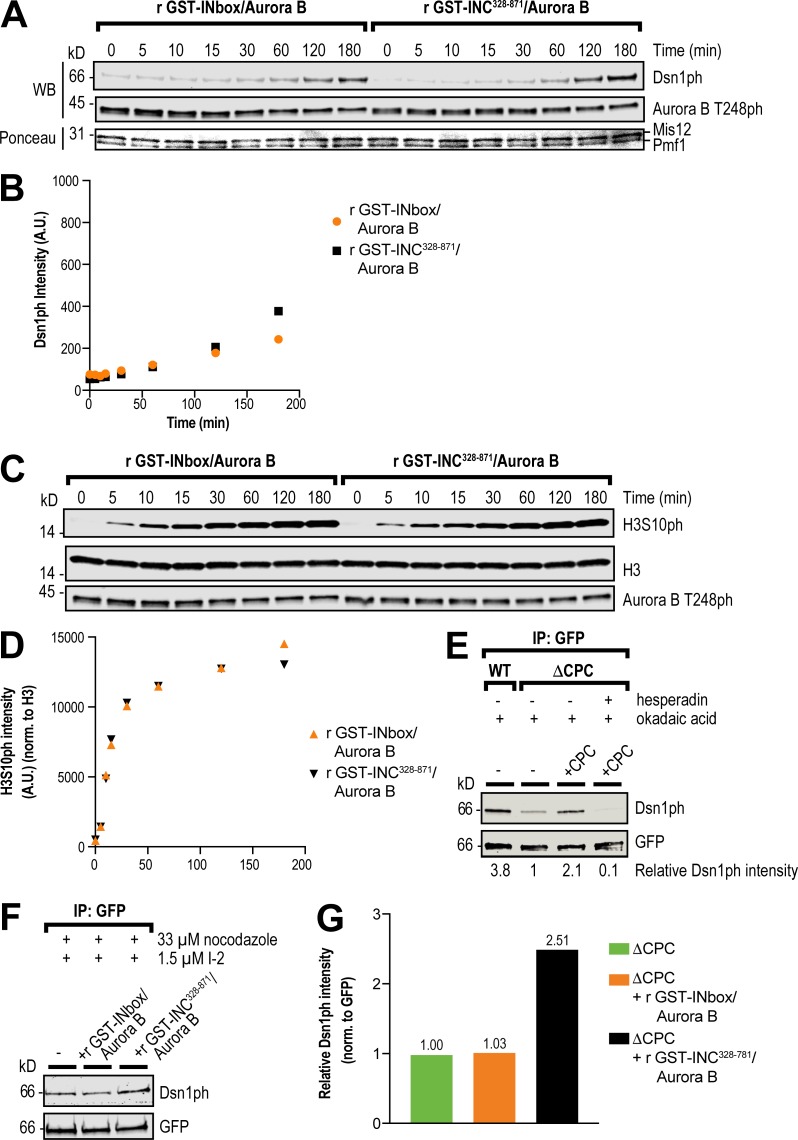
To test whether the central region directly promotes the phosphorylation of the Dsn1 N-terminus by Aurora B, we performed kinase assays in vitro using full-length Mis12C and various Aurora B–INCENP complexes. We generated a phospho-specific antibody against phospho-S77 of Dsn1 (Fig. S5, A and B) and compared the phosphorylation of Dsn1-S77 and of histone H3S10 by the preactivated recombinant Aurora B–GST-INCENP328–871 and Aurora B–GST-INbox complexes. Both complexes exhibited similar phosphorylation kinetics toward Dsn1 (Fig. 4, A and B) and H3S10 (Fig. 4, C and D). Thus, the central region does not directly enhance Aurora B activity or the phosphorylation of Dsn1 in vitro. Because recombinant Aurora B–GST-INCENP328–871 protein could rescue Mis12C assembly in ΔCPC extracts while the Aurora B–GST-INbox complex could not (Fig. 2, G and H), our results indicate that the SAH-domain–dependent promotion of Dsn1 phosphorylation is not direct and requires other factors present in extracts.
Figure 4.
The central region promotes Dsn1 phosphorylation by Aurora B. (A) In vitro kinase assay for Dsn1 phosphorylation. Purified Xenopus recombinant (r) Mis12C was incubated with preactivated recombinant GST-INbox–His6-Aurora B kinase or recombinant GST-INC328-871–His6-Aurora B for the indicated times, and phosphorylation was assessed by Western blot (WB) using a phospho-specific antibody to Ser77 of Dsn1. (B) Quantification of kinase assay for Dsn1 phosphorylation shown in A. A.U., arbitrary units. (C) In vitro kinase assay for histone H3 phosphorylation (H3S10ph). Purified Xenopus recombinant H3–H4 was incubated with preactivated recombinant GST-INbox–His6-Aurora B kinase or recombinant GST-INC328-871–His6-Aurora B for the indicated times, and phosphorylation was assessed by Western blot using a phospho-specific antibody for H3S10ph. (D) Quantification of kinase assay for H3S10 phosphorylation shown in C. (E) Western blot for phosphorylation of Dsn1 in samples of immunoprecipitated (IP) recombinant Mis12Dsn1-LAP complex from WT and ΔCPC metaphase extracts, in indicated CPC conditions, in the presence of nocodazole, okadaic acid, and hesperadin as specified. Normalized phosphorylation levels are indicated below. (F) Western blot for phosphorylation of Dsn1 in samples of immunoprecipitated Mis12Dsn1-LAP complex expressed from mRNAs in ΔCPC metaphase extracts, in indicated CPC conditions. Recombinant GST-INbox–His6-Aurora B or recombinant GST-INC328–871–His6-Aurora B were activated with anti-INCENP beads in the presence of nocodazole and I-2. (G) Quantification of phosphorylation of Dsn1 from samples shown in C, normalized to GFP.
We next sought to measure the phosphorylation level of total Dsn1 in extracts under conditions of kinetochore assembly. However, our antibody was unable to detect endogenous levels of Dsn1-S77ph in whole extracts (data not shown). To circumvent this, we added near-endogenous levels of purified Mis12C, tagged with GFP at the Dsn1 C-terminus, to extracts and immunoprecipitated all of the Mis12C at metaphase after incubation in the presence of replicated chromosomes and different forms of the CPC. Inhibition of PP1 by either okadaic acid or I-2 was required during the incubation and subsequent wash steps to ensure detectable signal (data not shown). We observed robust Dsn1 phosphorylation in extracts containing WT CPC, and Dsn1 phosphorylation was sensitive to both CPC depletion and Aurora B inhibition (Fig. 4 E). However, CPC depletion was not as effective as Aurora B inhibition at preventing Dsn1 phosphorylation, suggesting that very small amounts of residual CPC can support Dsn1 phosphorylation, in line with our observation that low concentrations of the CPC can support kinetochore assembly (Fig. 2, B and C). Nonetheless, the Dsn1 phosphorylation levels observed in CPC-depleted extracts could not support kinetochore assembly (Fig. 2, G and H). Thus, Dsn1 phosphorylation levels as measured by our assay are correlated with kinetochore assembly in egg extracts.
Because the Aurora B–INbox complex did not substantially rescue kinetochore assembly, even when PP1 was inhibited (Fig. S4 F), it should not be able to phosphorylate Dsn1 as well as a complex containing the central region. To test this, we measured Dsn1 phosphorylation in immunoprecipitated samples with recombinant Aurora B–GST-INbox and Aurora B–GST-INCENP328–871 complexes in the presence of PP1 inhibition as above. We saw no detectable increase in Dsn1 phosphorylation upon addition of Aurora B–GST–INbox complex to CPC-depleted extracts, suggesting that Dsn1 phosphorylation is tightly controlled in vivo (Fig. 4, F and G). However, we observed a 2.5-fold increase in Dsn1 phosphorylation in the presence of the Aurora B–GST-INCENP328–871 complex (Fig. 4, F and G). Therefore, our results indicate that the central region promotes Dsn1 phosphorylation indirectly in egg extracts to drive kinetochore assembly.
The spatial control of CPC activity is required for outer kinetochore assembly
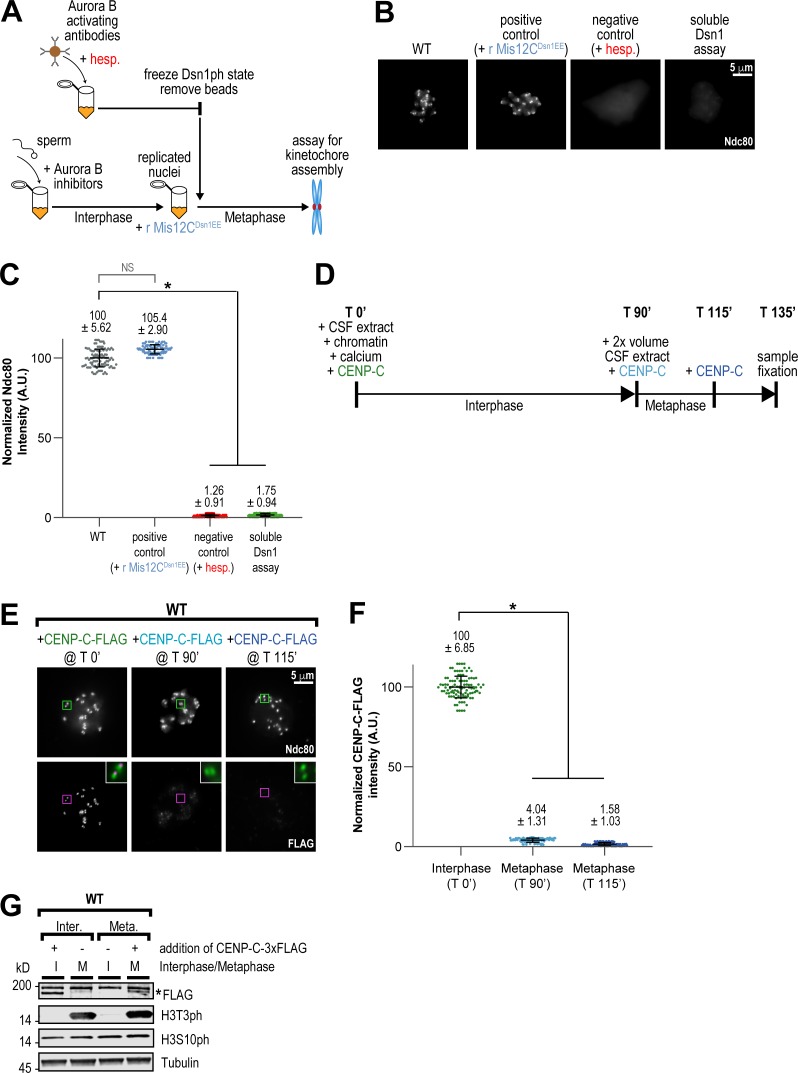
The central region may act through either soluble or kinetochore-bound proteins to promote Dsn1 phosphorylation and kinetochore assembly. To differentiate between these possibilities, we developed an assay to test the spatial requirements of CPC localization and activity to promote assembly (Figs. 5 A and S5 C). First, in the absence of chromosomes, we fully activated Aurora B to phosphorylate substrates by adding in beads coated with anti-INCENP antibodies to cytostatic factor (CSF)–arrested extracts (Kelly et al., 2007; Haase et al., 2017). After removing the beads, we inhibited both Aurora B and PP1 to “freeze” Dsn1 phosphorylation and then added these extracts to interphase extracts containing replicated chromosomes, in which Aurora B was inhibited, to drive them into mitosis. After 45 min, we then assessed kinetochore assembly by immunofluorescence. We found that restriction of Aurora B activity to only soluble CPC molecules before the introduction of chromosomes did not support kinetochore assembly, even in the presence of an intact central region (Fig. 5, B and C). This result suggests that the central region acts through proteins at the kinetochore to promote Dsn1 phosphorylation and Mis12C assembly.
Figure 5.
The CPC promotes kinetochore assembly only at kinetochore-bound CENP-C. (A) Schematic of soluble Dsn1 Assay to test if Mis12C phosphorylated away from chromatin promotes kinetochore assembly (see Fig. S5 C and Materials and methods for more details). In brief, Aurora B was activated by anti-INCENP beads in CSF extract lacking chromosomes and given time to phosphorylate Mis12C. Kinase and phosphatase inhibitors were added to preserve the phosphorylation state of Mis12C, and beads were then removed. This extract, containing soluble phosphorylated Mis12C, was then added to interphase extract containing Aurora B inhibitors and chromatin to drive the reaction into metaphase. Positive and negative control reactions are indicated with blue and red, respectively. (B) Representative immunofluorescence images of replicated chromosomes from the soluble Dsn1 assay outlined in A. Chromosomes were stained for Ndc80. (C) Quantification of fluorescence intensity of Ndc80 for samples in Soluble Dsn1 assay, normalized to WT. n = 96 kinetochores per condition. A.U., arbitrary units. (D) Schematic of kinetochore assembly experiment to test whether CENP-C can be incorporated at kinetochores when added at different points in the cell cycle. CENP-C–3XFLAG from an in vitro translation reaction was added at the beginning of interphase, at the beginning of metaphase (T 90’), and at 115 min. Samples were fixed for immunofluorescence staining at 135 min. (E) Representative IF images of replicated chromosomes in WT extracts with CENP-C–3XFLAG added to extract at indicated time points. Chromosomes were stained for Ndc80 (kinetochore pairs indicated with green boxes) and FLAG (kinetochore pairs indicated with magenta boxes). The inset white boxes show a merged, 3× zoom image of colocalization of Ndc80 (green) and FLAG (magenta) for each time point. (F) Quantification of fluorescence intensity of CENP-C–3XFLAG shown in E, normalized to the T 0’ condition. n = 96 kinetochores per condition. (G) Western blot for FLAG, histone H3 phosphorylation (H3T3ph and H3S10ph), and tubulin for samples shown in E and F. Asterisk denotes CENP-C–3XFLAG band, under the background band. Error bars represent SD. *, P < 0.001.
Restriction of CENP-C dynamics in mitosis provides an additional barrier to kinetochore assembly
The requirement for CPC proximity to kinetochores suggests that productive assembly only occurs at kinetochore-bound receptors (Fig. 5 C). Indeed, in human cells, centromere-bound CENP-C exchanges rapidly in interphase and then ceases to exchange in mitosis (Hemmerich et al., 2008). To test if CENP-C dynamics are similar in egg extracts, we measured the ability of exogenously added FLAG-tagged CENP-C to load onto kinetochores at different points of the cell cycle (Fig. 5 D). We found that while CENP-C-FLAG could load if added to extracts in interphase, CENP-C loading was blocked by early mitosis (Fig. 5, E–G). Thus, our results demonstrate that productive assembly of the Mis12C is restricted to prebound CENP-C molecules in mitosis.
Kinetochore assembly requires CENP-C–proximal Aurora B kinase
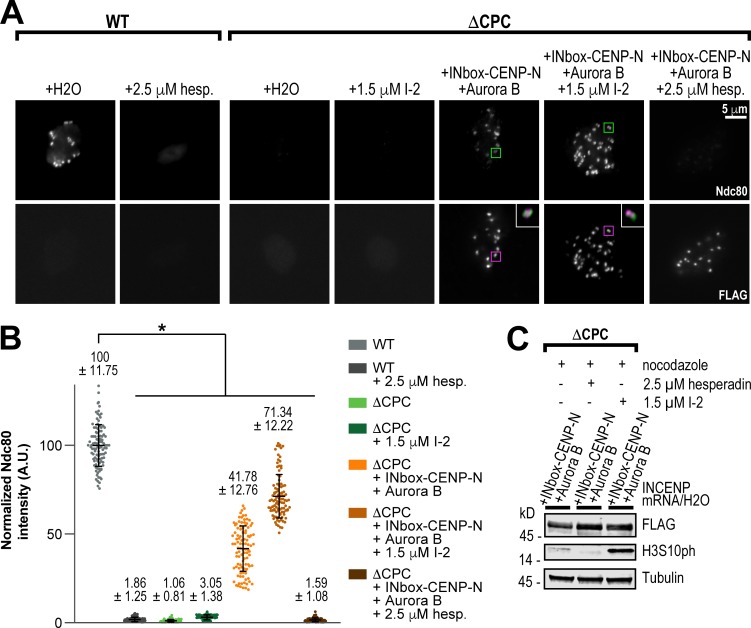
Our results suggest that the central region promotes Dsn1 phosphorylation close to kinetochore-bound CENP-C molecules. Since CPC localized at the inner centromere and on chromosome arms cannot support kinetochore assembly in the absence of the SAH domain (Fig. 3 A and Fig. S3, F and G), we hypothesized that the central region may direct Aurora B activity closer in space to the Mis12C-binding region of CENP-C at kinetochores. In support of this, a study in mammalian cells demonstrated that activated Aurora B first localizes to kinetochores in prophase, shifting to the inner centromere as mitosis progresses (DeLuca et al., 2011), and other works have identified a kinetochore pool of Aurora B as being important for CPC function (Bekier et al., 2015; Krenn and Musacchio, 2015; Hengeveld et al., 2017; Hindriksen et al., 2017). A similar localization pattern of the CPC is observed in budding yeast (Campbell and Desai, 2013), where recent work has implicated the central region of Sli15 (yeast homologue of INCENP) in interactions with the inner kinetochore (Fischböck-Halwachs et al., 2019; García-Rodríguez et al., 2019). To test if kinetochore assembly requires inner kinetochore localization of Aurora B kinase, we fused the INbox domain to the N-terminus of CENP-N (Chittori et al., 2017). Strikingly, we found that inner kinetochore-localized Aurora B supported kinetochore assembly of Ndc80, albeit with reduced Ndc80 levels (∼40% of WT; Fig. 6, A–C). Importantly, this assembly was abrogated by Aurora B inhibition, demonstrating that the fusion itself is not driving assembly. PP1 inhibition enhanced the levels of Ndc80 assembly to nearly the same levels observed in extracts containing the INbox–Mis12C fusion (Figs. 3 G and 6 B). Thus, our results indicate that the central region plays a role in positioning Aurora B to facilitate Dsn1 phosphorylation and outer kinetochore component assembly. Furthermore, that kinetochore assembly was not completely rescued by the INbox–CENP-N fusion suggests that precise positioning of Aurora B kinase to the inner kinetochore is important for outer kinetochore assembly.
Figure 6.
Mis12C assembly requires enrichment of Aurora B at inner kinetochores. (A) Representative immunofluorescence images of replicated chromosomes in WT and ΔCPC metaphase extracts with indicated CPC conditions, treated as indicated with I-2 and hesperadin. Chromosomes were stained for Ndc80 and FLAG (for 3XFLAG-INbox–CENP-N construct). White inset boxes show a merged, 3× zoom image of colocalization of Ndc80 (green) and 3XFLAG-INbox–CENP-N (magenta). (B) Quantification of fluorescence intensity of Ndc80 shown in A, normalized to WT. n = 96 kinetochores per condition. A.U., arbitrary units. (C) Western blot for FLAG, histone H3 phosphorylation (H3S10ph), and tubulin for samples shown in A and B. Error bars represent SD. *, P < 0.001.
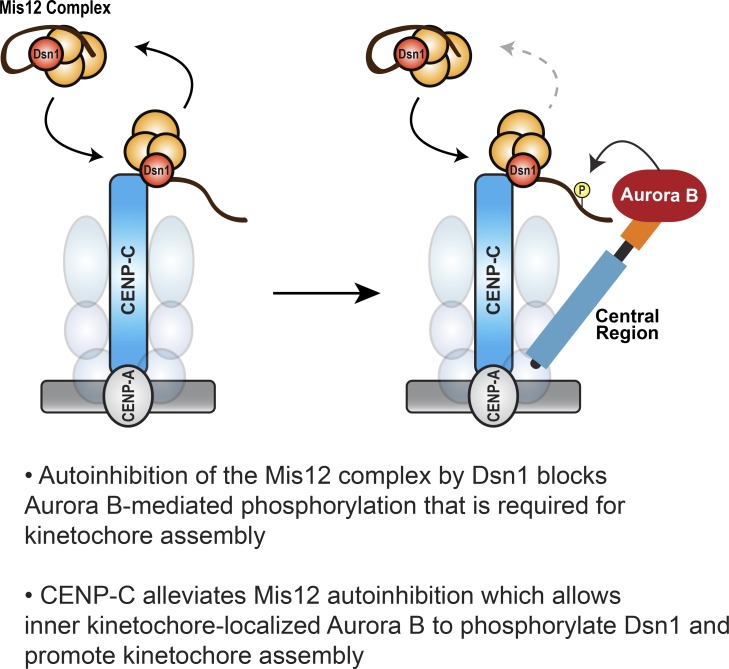
CENP-C relieves autoinhibition of the Mis12C to promote phosphorylation of the Dsn1 N-terminal tail by Aurora B
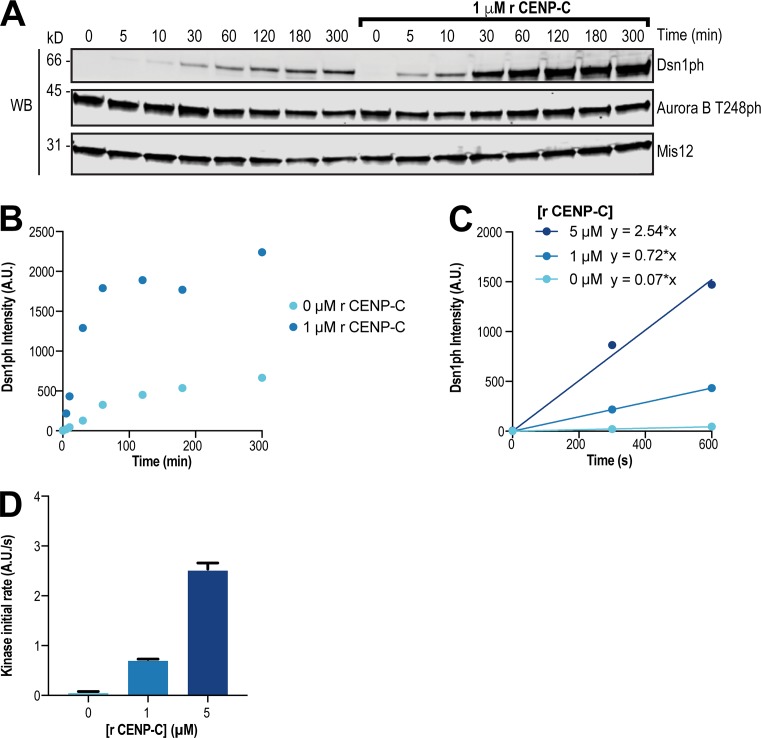
Current models suggest Mis12C autoinhibition is first relieved by phosphorylation of Dsn1 by Aurora B, which then allows CENP-C to bind Dsn1 (Kim and Yu, 2015; Dimitrova et al., 2016; Petrovic et al., 2016). However, our results demonstrate that robust Dsn1 phosphorylation and kinetochore assembly require a high local concentration of Aurora B kinase near CENP-C and suggest that Dsn1 phosphorylation is rate limiting and requires interplay of the CPC with CENP-C. The Dsn1 N-terminal tail shares homology with a portion of the Mis12C-binding domain within the CENP-C N-terminus and competes with CENP-C binding to the Mis12C to inhibit kinetochore assembly (Dimitrova et al., 2016; Petrovic et al., 2016). Since the Aurora B phosphorylation sites are located within the Dsn1 tail (Fig. 1 A), the autoinhibited form of the Mis12C may be resistant to phosphorylation due to reduced substrate access, similar to other autoinhibited proteins (Yu et al., 2010; Saleh et al., 2017). Indeed, in vitro, the kinetics of Dsn1 phosphorylation within autoinhibited Mis12C were slower when compared with the rate of histone H3S10 phosphorylation (Fig. 4, A–D), and robust Dsn1 phosphorylation required long incubations with micromolar concentrations of kinase (Fig. S5 A). Although previous reports indicate that Mis12C autoinhibition by the Dsn1 tail weakens its affinity for CENP-C, it does not completely prevent its binding in vertebrates (Huis In ’t Veld et al., 2016; Petrovic et al., 2016). Therefore, we hypothesized that transient binding events between CENP-C and the Mis12C may expose the S77 phosphorylation site. To test this directly, we measured the activity of the Aurora B–INbox complex toward Dsn1 in the presence and absence of excess amounts of an N-terminal CENP-C fragment (residues 1–55) that competes with the Dsn1 tail for Mis12C binding. We observed a dramatic increase in the kinetics of Dsn1 phosphorylation in the presence of the CENP-C fragment (Fig. 7, A and B), and this response was dose dependent (Fig. 7, C and D). This shows that Mis12C autoinhibition opposes Aurora B phosphorylation of the Dsn1 N-terminus. Thus, our data indicate that Dsn1 phosphorylation is driven by high local concentrations of Aurora B at inner kinetochores and the transient relief of Mis12C autoinhibition by kinetochore-localized CENP-C. We propose a model for kinetochore assembly wherein the coincidence of Aurora B and CENP-C at kinetochores cooperates to promote outer kinetochore assembly.
Figure 7.
Autoinhibition of the Mis12C impedes the phosphorylation of Dsn1 by Aurora B. (A) In vitro kinase assay for Dsn1 phosphorylation. Purified recombinant (r) Mis12C was incubated with preactivated recombinant GST-INbox–His-Aurora B in the presence or absence of recombinant CENP-C2–55–LAP for the indicated times. Western blot shows Dsn1ph, Aurora B T248ph, and Mis12 at indicated time points of assay. (B) Quantification of Dsn1 phosphorylation data from in vitro kinase assay shown in A. A.U., arbitrary units. (C) Quantification of Dsn1 phosphorylation over time used to calculate initial rates. Purified recombinant Mis12C was incubated with preactivated recombinant GST-INbox–His6-Aurora B and indicated concentrations of recombinant CENP-C2–55–LAP. (D) Quantification of initial rates of Dsn1 phosphorylation calculated from data shown in C. Purified recombinant Mis12C was incubated with preactivated recombinant GST-INbox–His6-Aurora B and indicated concentrations of recombinant CENP-C2–55–LAP. Graph represents data from two independent experiments. Error bars represent SEM.
The SAH domain is required for the SAC independently of its roles in kinetochore assembly and microtubule binding
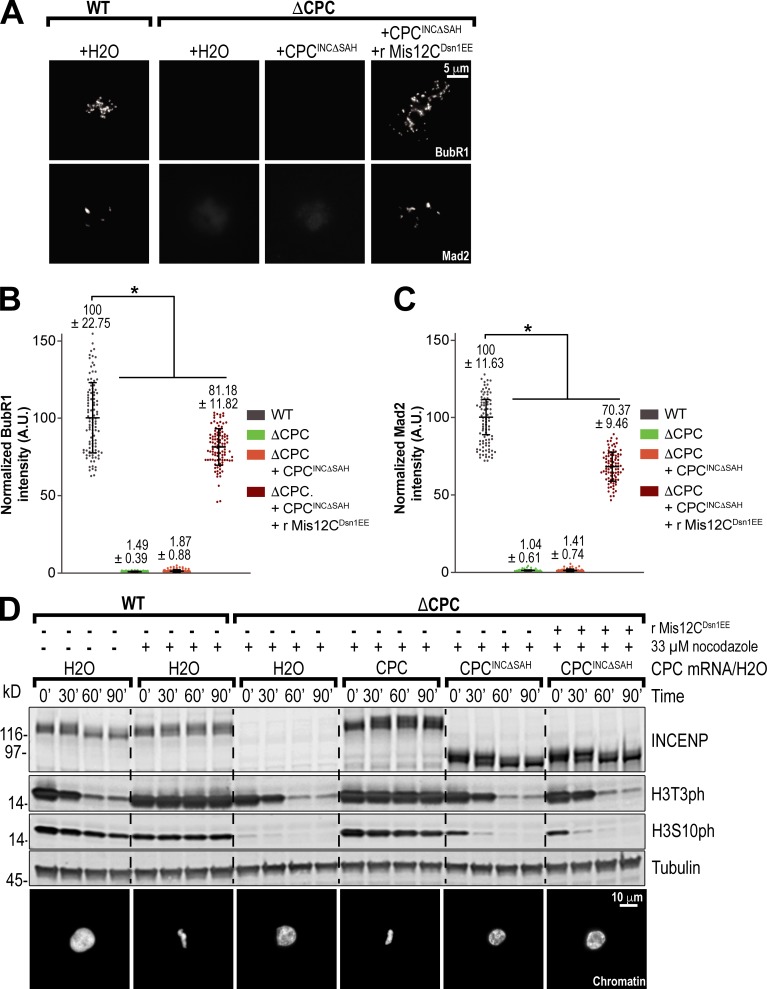
The localization of Aurora B to inner kinetochores may contribute to other CPC-dependent processes. The SAH domain is required for cell cycle arrest in response to nocodazole treatment in Xenopus egg extracts (Wheelock et al., 2017). Indeed, we found that deletion of the SAH domain abrogated recruitment of checkpoint proteins Mad2 and BubR1 to kinetochores in response to nocodazole (Fig. 8, A–C). Significant amounts of Mad2 and BubR1 (70% and 81% of WT, respectively) were recruited to kinetochores upon addition of Mis12CDsn1EE to INCΔSAH extracts (Fig. 8, A–C). Thus, a major role of the SAH domain in the SAC is in mediating the assembly of the outer kinetochore to recruit checkpoint proteins. However, we found that Mis12CDsn1EE could not support a functional SAC arrest in the absence of the SAH or of Aurora B activity, as demonstrated by a lack of condensed nuclei and histone H3T3 and H3S10 phosphorylation, possibly due to incomplete recruitment of SAC proteins (Figs. 8 D and S5 E). Importantly, Mis12CDsn1EE did not perturb the SAC when added to WT extracts (Fig. S5 E). Thus, our results demonstrate that the SAH domain is required for a full SAC response, downstream of its role in kinetochore assembly and independently of its role in microtubule binding, and suggest that positioning Aurora B at inner kinetochores is important for the phosphorylation of other substrates involved in the SAC.
Figure 8.
The SAH domain is required for the SAC independently of its roles in kinetochore assembly and microtubule binding. (A) Representative immunofluorescence images of replicated chromosomes in WT and ΔCPC metaphase extracts with indicated CPC conditions, treated with 33 µM nocodazole and recombinant (r) Mis12CDsn1EE as specified. Chromosomes were stained for BubR1 or Mad2. (B and C) Quantification of fluorescence intensity of BubR1 and Mad2 shown in A normalized to WT. n = 96 kinetochores. A.U., arbitrary units. (D) Western blot for global checkpoint assay for INCENP, histone H3 phosphorylation (H3T3ph and H3S10ph), and tubulin. WT or ΔCPC metaphase extracts with indicated CPC conditions were incubated with sperm nuclei and then challenged with the addition of calcium chloride. Samples were taken at indicated time points after calcium addition, shown with representative images of chromatin morphology at the end of the assay. Error bars represent SD. *, P < 0.001.
Discussion
The coincidence of CENP-C and Aurora B at inner kinetochores drives outer kinetochore assembly
The outer kinetochore is assembled and disassembled every cell cycle, and loss of this regulation can lead to errors in chromosome segregation (Gascoigne and Cheeseman, 2013; Hara and Fukagawa, 2018). Kinetochore assembly must be rapid and occur specifically at the centromere to allow for proper attachment to spindle microtubule and activation of the mitotic checkpoint. Here, we show that multiple factors synergize to ensure the specificity of kinetochore assembly. First, we find that centromere-bound CENP-C molecules become static in mitosis and do not exchange with the soluble pool (Fig. 5, E and F), as observed in human cells (Hemmerich et al., 2008). This shift in CENP-C dynamics restricts productive formation of Mis12C–CENP-C complexes to kinetochore-localized CENP-C. Second, we show that Mis12C autoinhibition serves as a barrier to Aurora B kinase phosphorylation (Fig. 7 A). Our data indicate that substrate access is enhanced through transient interactions of the Mis12C with kinetochore-bound CENP-C (Fig. 7, A–D) and that the Dsn1 phosphorylation rate is increased by high local concentrations of Aurora B kinase proximal to CENP-C (Fig. 6, A and B). Thus, we propose that the coincidence of CENP-C and Aurora B at inner kinetochores is required for proper kinetochore assembly only at centromeres (Fig. 9, A and B). Interestingly, overexpression of CENP-A in cancer cells causes the mislocalization of CENP-A along chromosome arms, which in turn causes ectopic enrichment of CENP-C (Lacoste et al., 2014). However, these sites fail to form outer kinetochores, suggesting that the additional requirement of high local concentrations of Aurora B prevents the formation of functional neocentromeres.
Figure 9.
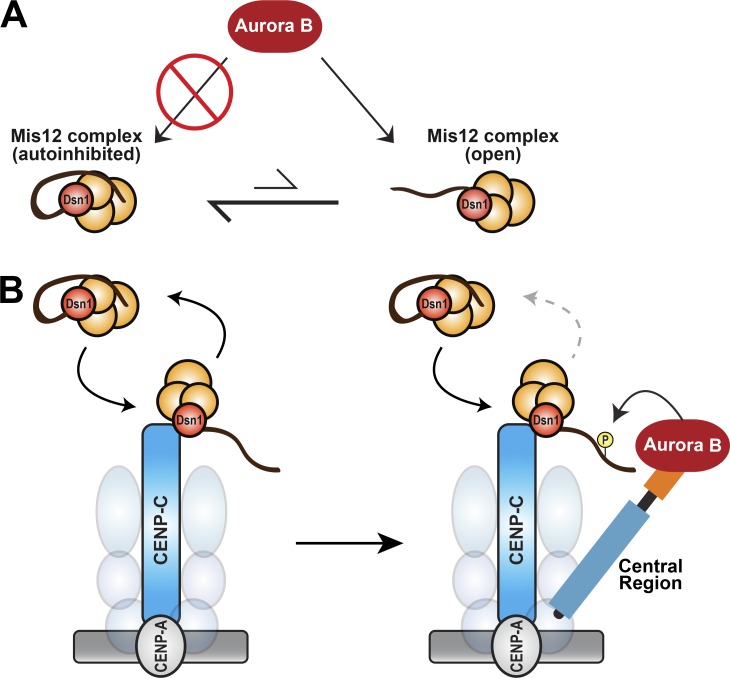
Kinetochore-localized Aurora B is required for kinetochore assembly and function. (A) Mis12C autoinhibition prevents phosphorylation by Aurora B. Away from kinetochores, the autoinhibited form of the Mis12C (orange spheres) predominates and prevents Aurora B phosphorylation of Dsn1 through burial of Dsn1S77. (B) Model for kinetochore assembly through the cooperative actions of CENP-C, the INCENP central region, and Aurora B. At the inner kinetochore, the Mis12C transiently binds to prebound CENP-C (and likely CENP-T) molecules, which relieves Mis12C autoinhibition and exposes the Dsn1 tail. In early mitosis, Aurora B is enriched at inner kinetochores by the central region of INCENP. This high local concentration of Aurora B rapidly phosphorylates the exposed Dsn1 tail to prevent dissociation of the Mis12C. Thus, the coincidence of CENP-C and the CPC at inner kinetochores stabilizes the Mis12C–CENP-C interaction to drive outer kinetochore assembly. Our data indicate that enrichment of Aurora B at the inner kinetochore is also involved in SAC signaling through the phosphorylation substrates other than Dsn1.
Our study identifies the relief of autoinhibition of Mis12C by the Dsn1 tail as the rate-limiting step in phosphorylation by Ser/Thr kinase Aurora B. Prior work has shown that burial of tyrosine kinase substrates in autoinhibited states are barriers to activation and priming phosphorylations at other sites must occur to allow access to the buried substrate (Yu et al., 2010; Saleh et al., 2017). Priming phosphorylation may increase the number of uninhibited Mis12C molecules, since we and others observe other phosphorylated residues (both Aurora B and CDK sites) in the N-terminal tail of Dsn1 (Fig. S1 C; Welburn et al., 2010). However, our data suggest that these phosphorylations are not sufficient to prime phosphorylation of S77 in Dsn1 by Aurora B, since Dsn1 is a poor substrate in vitro and global activation of Aurora B in the absence of PP1 did not support robust kinetochore assembly in extracts (Figs. 3 E and 4 A). Instead, our data indicate that competition from inner kinetochore proteins, such as CENP-C, can alleviate autoinhibition to promote substrate access (Fig. 7). Given the high local concentration of CENP-C at kinetochores, and that the Mis12C-binding domain of human CENP-C binds autoinhibited Mis12C with a Kd of ∼50 nM (Petrovic et al., 2016), we propose that CENP-C can help to locally increase the amount of uninhibited Mis12C for nearby Aurora B to act on.
In other species, CENP-T is thought to form a distinct CDK-dependent kinetochore assembly pathway that works in parallel to the CENP-C assembly pathway to recruit the Ndc80 and Mis12Cs (Akiyoshi et al., 2013; Kim and Yu, 2015; Rago et al., 2015; Hara et al., 2018; Lang et al., 2018). In Xenopus egg extracts, both CENP-T and CENP-C contribute to outer kinetochore assembly, which suggests a similar dichotomy (Krizaic et al., 2015). However, we demonstrate that Dsn1 phosphorylation is necessary and sufficient for kinetochore assembly in egg extracts (Fig. 1, C and D), indicating that the CENP-T pathway is ultimately controlled by Aurora B in egg extracts. In humans, the CENP-T–binding site on the Mis12C partially overlaps with the CENP-C–binding site (Huis In ’t Veld et al., 2016), suggesting that autoinhibition by Dsn1 may also regulate the CENP-T–Mis12C interaction. Indeed, we found that CENP-T only stably interacts with the Mis12C in extracts when Aurora B is activated (Fig. S1 D). Furthermore, we showed that Mis12CDsn1EE, but not Mis12CDsn1WT, can pull down CENP-T from extracts in the presence of the Aurora B inhibitor hesperadin (Fig. S1 E). Interestingly, although Xenopus CENP-T shares high homology with human CENP-T, it lacks the two CDK phosphorylation sites within the Mis12C-binding region required for interaction in humans (Rago et al., 2015; Huis In ’t Veld et al., 2016). This suggests that in Xenopus, the stable interaction of CENP-T with the Mis12C is controlled by Aurora B–mediated phosphorylation of Dsn1. Furthermore, our data suggest that this interaction also requires the central region of INCENP and a high local concentration of Aurora B at inner kinetochores. Together, CENP-C and CENP-T may aid each other by increasing the number of transient interactions of the Mis12C with the inner kinetochore that relieve Mis12C autoinhibition to promote Aurora B–mediated phosphorylation of Dsn1.
The role of the INCENP central region in kinetochore assembly and function
Here, we establish a microtubule-independent role for the INCENP central region in positioning Aurora B kinase at inner kinetochores to promote kinetochore assembly. However, the inner kinetochore receptor for the central region in Xenopus and other vertebrates remains undetermined. Recent work in budding yeast demonstrated that the central region makes multiple contacts with the essential COMA complex, in particular with COMA subunits Ctf19 and Mcm21, which are important for accurate chromosome segregation (Fischböck-Halwachs et al., 2019; García-Rodríguez et al., 2019). Because Dsn1 phosphorylation by Aurora B plays a conserved role in kinetochore assembly and stability in budding yeast (Akiyoshi et al., 2013; Lang et al., 2018), our results suggest that the defects observed in central region mutants in yeast may be caused in part by defects in kinetochore stability. Interestingly, the yeast COMA subunit Ame1 directly binds to the Mis12C and also competes with the Dsn1 N-terminal tail (Dimitrova et al., 2016). That Ipl1 (yeast Aurora B) is recruited to the Ame1-containing COMA complex by the central region aligns with our findings that Dsn1 phosphorylation and kinetochore assembly require precise positioning of the kinase at the inner kinetochore (Figs. 5 C and 6 B).
The less well understood vertebrate CENP–OPQUR complex is related to the COMA complex, and the CENP–O/P subunits share high homology with the COMA subunits Ctf19 and Mcm21 (Hori et al., 2008; Perpelescu and Fukagawa, 2011; McKinley et al., 2015; Pesenti et al., 2018). Recent EM analyses demonstrate a compact assembly of both yeast and human CCAN complexes (Pesenti et al., 2018; Hinshaw and Harrison, 2019). CENP-C directly binds to the CENP–HIKM and CENP–NL complexes, which recruit the CENP–TW and CENP–OPQUR complexes, using a domain that is C-terminal to its Mis12C-binding domain (Hara and Fukagawa, 2018). Thus, the central region could interact with the CENP–OPQUR complex, another CCAN complex, or along the entire CCAN to position Aurora B close to the CENP-C–Mis12C interaction site. Within the central region, we find that the SAH domain is an essential factor in kinetochore assembly in Xenopus egg extracts, and our data suggest that it plays an important role in targeting Aurora B to inner kinetochores (Figs. 3 E and 6 B). Recent work has established that SAH domains can be protein–protein interaction interfaces spanning large proteinaceous assemblies (des Georges et al., 2015; Ulrich et al., 2016), and we propose that the SAH domain may play a similar role for the CPC with the CCAN. Future studies will be required to identify the inner kinetochore-binding sites for the SAH domain and the rest of the central region.
The SAH domain is required for the SAC in egg extracts, and we demonstrate that the SAH domain contributes to the SAC by both assembling the kinetochore and regulating checkpoint protein recruitment (Fig. 8, A–D; and Fig. S5 E; Haase et al., 2017). We propose that other substrates within the KMN network that regulate checkpoint protein recruitment also require a high local concentration of Aurora B at inner kinetochores. Phosphorylation of the MELT repeats within KNL1 by Mps1 kinase is required for SAC activation and triggers BubR1 and Mad2 recruitment to kinetochores (London et al., 2012; Shepperd et al., 2012; Yamagishi et al., 2012; Krenn et al., 2014). Although the mechanism by which Aurora B regulates Mps1 kinetochore localization is not fully understood, the phosphorylation of the Ndc80 N-terminal tail has been proposed to promote Mps1 localization (Hiruma et al., 2015; Ji et al., 2015). However, under checkpoint conditions, we observe that Ndc80 phosphorylation is not affected by the absence of the SAH domain when kinetochore assembly is bypassed (Fig. 3 D). Another possible Aurora B substrate is the N-terminus of KNL1, phosphorylation of which prevents SAC silencing through the recruitment of PP1, which dephosphorylates MELT repeats (Liu et al., 2010; Rosenberg et al., 2011). However, the N-termini of both KNL1 and Ndc80 are both oriented away from the CENP-C–Mis12C interaction (Wan et al., 2009). Since we observe that the Mis12C assembly is sensitive to Aurora B positioning (Fig. 6 B), the candidate substrate may be closer to the inner kinetochore. In support of this, BubR1 has been reported to localize to both the inner and outer kinetochore when the SAC is activated in human cells (Huang and Yen, 2009), suggesting that an unknown Aurora B substrate that is required for BubR1 recruitment resides at or binds to inner kinetochores.
Aurora B kinase is also required for the correction of erroneous kinetochore–microtubule attachments (Krenn and Musacchio, 2015). However, the exact roles of the central region in error correction are not fully understood (Vader et al., 2007; Fink et al., 2017; Fischböck-Halwachs et al., 2019; García-Rodríguez et al., 2019), likely due to the fact that the central region also regulates kinetochore assembly, the SAC, and microtubule assembly (current study; Tseng et al., 2010; Wheelock et al., 2017). Future efforts are required to identify additional kinetochore substrates that require high local concentrations of Aurora B.
Materials and methods
Immunodepletion of Xenopus egg extracts
Xenopus egg extract was prepared as previously described (Kelly et al., 2007). To prevent translation of endogenous RNAs, RNase A (Ambion) was added at a final concentration of 100 ng/ml to extract for 15 min at 12°C. RNase inhibitor (RNasin Plus RNase Inhibitor; Promega) was added at a 1:50 dilution for 5 min at 12°C. Yeast tRNAs (50 µg/ml) were added, and extract was kept on ice. Immunodepletions of the CPC from the extract were performed as previously described (Haase et al., 2017).
Reconstitution of immunodepleted extracts
To express the CPC in immunodepleted extract, we pooled CPC mRNAs (consisting of INCENP, Aurora B, Borealin [Dasra A], and Survivin, unless otherwise indicated) and added them to extracts at the onset of interphase using previously described methods (Haase et al., 2017). Briefly, the CPC members were in vitro transcribed using the mMessage Machine SP6 kit (Thermo Fisher Scientific), pooled together, and then precipitated in the presence of 20 µg glycogen. The capped mRNA pools were then resuspended in RNase-free water so that 1 µl of pooled mRNAs reconstituted 60 µl of CPC-depleted extract.
Kinetochore/spindle assembly in Xenopus egg extracts
To assess kinetochore assembly and checkpoint protein recruitment, sperm chromatin was replicated in extracts, and spindles were assembled according to previously published methods (Haase et al., 2017). Briefly, 1 µl of pooled mRNAs was added to 20 µl of RNase-treated control or immunodepleted extract with sperm nuclei (final concentration of 500/µl) and calcium chloride (0.3 mM). Rhodamine tubulin (Cytoskeleton, Inc.) was added at a 1:200 dilution to observe the progress of spindle assembly. Extract reactions were incubated at 20°C for 80 min to cycle into interphase. To drive the extract into metaphase, 40 µl CSF extract (control or immunodepleted) was added. At the onset of metaphase, nocodazole was added as needed to a final concentration of 33 µM, okadaic acid was added as needed to a final concentration of 8 µM, hesperadin was added as needed to a final concentration of 2 or 2.5 µM, and the phosphatase inhibitor I-2 (NEB) was added to a final concentration of 1.5 µM. In one experiment, to test whether 8 µM okadaic acid or 1.5 µM could successfully oppose inhibition of Aurora B kinase to maintain kinetochores, hesperadin and either okadaic acid or I-2 were added 10 min into metaphase. After 45 min at 20°C, metaphase spindle assembly was assayed by fixing 1-µl samples with Hoechst (10 µg/ml; Hoechst 33258; Invitrogen) and imaging tubulin and DNA. Samples for Western blot and immunofluorescence were taken once metaphase was successfully achieved.
CPC titration in Xenopus egg extracts
To determine the levels of CPC required for kinetochore assembly, extracts were immunodepleted of CPC. To these CPC-depleted extracts, mock depleted extract was added in increasing amounts as a volume/volume percentage. CPC-depleted extract was assumed to contain 0% CPC, and mock-depleted extract was assumed to contain 100% CPC. The resulting concentration of the CPC was estimated based on quantitative mass spectrometry measurements (Wühr et al., 2014).
Antibody activation of Aurora B kinase
To activate Aurora B kinase in experiments using N-terminal truncations of INCENP, such as INCENPΔCEN, 1 µl anti-INCENP antibody (15 µg/ml) was added to 40 µl CSF extract, for a final concentration of 0.25 ng/µl. This substoichiometric amount of antibody (relative to CPC concentration in extract) is sufficient to catalyze the activation of Aurora B. After incubation on ice for 20 min, this was added to 20 µl of experimental interphase extract to induce cycling of extract into metaphase.
Cloning of the Mis12C and INbox fusion constructs
We cloned Xenopus Mis12, Nsl1, Pmf1, and Dsn1 into the pCSII vector by Gibson assembly (NEB). Sequences for Mis12, Nsl1, and Pmf1 were obtained from a Xenopus cDNA library (Dharmacon). The sequence for Dsn1 was derived by reverse translating the protein sequence from the Kirschner laboratory mRNA-derived Reference Database (Wühr et al., 2014), and a gene fragment was synthesized (Eurofins Genomics). A localization and affinity purification (LAP) tag (Cheeseman and Desai, 2005) was cloned into the C-terminus of Dsn1. Dsn1 mutations S77E and S84E were generated by Q5 site-directed mutagenesis (NEB) using the primers 5′-ACCCCAAAGGAACTTCCTCCTGTACATCGC-3′ and 5′-TTTGGCCACTTCTCTCCGTAGAGACTTCCTC-3′. For the 3XFLAG-INbox-Nsl1 plasmid, a gene fragment was synthesized (Eurofins Genomics) for a 3XFLAG tag followed by INbox (790–871 of INCENP), and that was cloned into the pCSII vector containing Nsl1 using Gibson Assembly (NEB). Similarly, the 3XFLAG-INbox gene fragment was cloned into a pCSII vector containing CENP-N using Gibson Assembly (NEB).
Expression of xCENP-C–3XFLAG
We cloned a 3XFLAG tag to the C-terminus of Xenopus CENP-C in a pCSII vector (ASP866; a kind gift of A. Straight, Stanford University, Palo Alto, CA; Milks et al., 2009) using Q5 site-directed mutagenesis (NEB). To express xCENP-C–3XFLAG, we used an in vitro transcription and translation (TNT) system (Promega), adding 1 μg plasmid. For the Xenopus extract experiment assessing whether xCENP-C-3XFLAG is incorporated into kinetochores at different cell cycle points, 2 μl of the TNT reaction was added to one reaction at the beginning of interphase, 2 μl of the TNT reaction was added to another reaction upon CSF addition to drive the extract into metaphase, and 2 μl of the TNT reaction was added to another reaction 25 min into metaphase. Samples for Western blot and immunofluorescence were taken once metaphase was successfully achieved (45 min).
Immunoprecipitations
Immunoprecipitations were performed to provide samples for analysis of the phosphorylation sites in Dsn1 and interaction partners. Mis12C mRNAs (1.6 µg/µl complex) were expressed in extracts without chromatin for 40 min at 20°C. To activate Aurora B kinase in the absence of chromatin, anti-INCENP (or an IgG control) antibodies were added to the extracts and incubated for another 80 min at 20°C in the presence of 33 µM nocodazole. Extracts were diluted 1.5 times with lysis buffer (75 mM Hepes, pH 7.7, 1.5 mM EGTA, 1.5 mM MgCl2. 150 mM KCl, 15% glycerol, 0.075% NP-40, 1× Phos-stop [Roche], and 1× Leupeptin/Pepstatin/Chymostatin [Chemicon]). A 25-µl bed volume of twice washed GFP-Agarose beads (Chromotek) was added to each extract (500 μl) condition. Beads and extract were incubated at 4°C with end-over-end rotation. Beads were briefly spun and washed four times with wash buffer (50 mM Hepes, pH 7.7, 1.5 mM EGTA, 1.5 mM MgCl2, 150 mM NaCl, 10% glycerol, 0.05% NP-40, 0.5 mM tris(2-carboxyethyl)phosphine [TCEP], 2× Phos-stop, and 1× LPC [cocktail of protease inhibitors Leupeptin, Pepstatin, and Chymostatin]), changing tubes during washes. Beads were boiled in 2× sample buffer for 5 min at 95°C.
Immunoprecipitations were also performed to provide samples to determine whether the phosphorylation of Dsn1 as measured by our antibody was dependent on the presence of the CPC and Aurora B activity. In control or immunodepleted extract, recombinant Mis12C was added in the presence of 33 µM nocodazole, and 0.3 mM calcium chloride was added to cycle the extract into interphase. After a 75-min incubation at 20°C, CSF extract was added to drive the reaction into metaphase, along with 1 µM okadaic acid. Reactions were incubated for 45 min at 20°C. Input samples were taken for Western blots after the incubation. Reactions were then diluted with lysis buffer (75 mM Hepes, pH 7.7, 1.5 mM MgCl2, 150 mM KCl, 15% glycerol, 0.075% IGEPAL, 1× Phos-Stop, and 1× LPC), and GFP magnetic agarose beads (Chromotek) were added. Beads and extract were incubated at 4°C with end-over-end rotation for 30 min. Beads were captured by magnet and washed four times with wash buffer (50 mM Hepes, pH 7.7, 1.5 mM EGTA, 1.5 mM MgCl2, 150 mM NaCl, 10% glycerol, 0.05% IGEPAL, 0.5 mM TCEP, 1× Phos-Stop, and 1× LPC), changing tubes during washes. Beads were boiled in 2x sample buffer for 5 min at 95°C.
Immunoprecipitations were performed to provide samples to determine whether the phosphorylation of Dsn1 was dependent on the presence of the central region of INCENP. In control or immunodepleted extract, Mis12C mRNAs were expressed according to the same protocol as listed above. After the preexpression period, chromatin was added (final concentration of 7,000/µl) and calcium chloride was added (final concentration of 0.3 mM) to all reactions. GST-INC328–871–His6-Aurora B60–361 or GST-INC788–871/His6-Aurora B60–361 was added to a final concentration of 400 nM in immunodepleted reactions. After all reactions were incubated for 80 min at 20°C, control or immunodepleted CSF extract was added to drive the reactions into metaphase. Nocodazole was added to all reactions (final concentration of 33 µM), the phosphatase inhibitor I-2 was added to all reactions (final concentration of 1.5 µM), and hesperadin was added to one control reaction (final concentration of 2.5 µM). Additionally, anti-INCENP beads were added to the two reactions with recombinant CPC protein to activate Aurora B kinase, and all reactions were incubated for 45 min at 20°C. Western blot samples for inputs were taken. On ice, reactions were then diluted with extraction buffer (10 mM Hepes-KOH, pH 7.7, 750 mM NaCl, 5 mM EDTA, 200 mM sucrose, 0.1 mM TCEP, 0.05% Triton X-100, 0.5 mM spermine, 8 µM okadaic acid, 1× Phos-stop, 2.5 µM hesperadin, and 1× LPC) and incubated for 10 min. The reactions were added to prewashed magnetic agarose GFP-trap beads (Chromotek) and incubated at 4°C with end-over-end rotation for 30 min. Beads were captured by a magnet and washed four times with wash buffer (10 mM Hepes-KOH, pH 7.7, 2 M NaCl, 5 mM EDTA, 0.1 mM TCEP, 0.05% Triton X-100, 1× PhosStop, and 1× LPC), changing tubes during washes. Beads were boiled in 2× sample buffer for 5 min at 95°C.
Soluble Dsn1 assay
To determine whether soluble Mis12C phosphorylated away from chromatin could be incorporated into kinetochores, CSF extract lacking chromatin was incubated with anti-INCENP beads for 60 min at 20°C to fully activate Aurora B kinase for substrate phosphorylation. Then, to stop all phosphatase and kinase activity, okadaic acid was added to a final concentration of 8 µM, the reactions were put on ice, and then hesperadin was added to a final concentration of 2.5 µM. The anti-INCENP beads were removed, and this extract was then added to a separate tube of interphase extract (containing chromatin) in the presence of 2.5 µM hesperadin in order to cycle the reaction into metaphase. Additional okadaic acid was added to maintain a constant concentration of 8 µM. Samples for immunofluorescence were taken at 45 min into metaphase. Western blot samples were taken at four time points during the experiment: at the end of interphase, at the end of the 1-h incubation with anti-INCENP beads, after bead removal, and at 45 min into metaphase. Three control reactions were performed in parallel. One control reaction contained IgG beads to ensure bead manipulation did not affect kinetochore assembly. A positive control reaction contained Mis12CDsn1EE added after bead removal. Finally, a negative control reaction contained hesperadin from the start of the incubation with anti-INCENP beads to prevent any substrate phosphorylation by Aurora B kinase.
Global checkpoint assay
To determine whether extract with various addbacks were competent to hold the SAC, pooled mRNAs were expressed for 30 min at 20°C in control or immunodepleted extract. Sperm nuclei (final concentration of 15,000/µl) and nocodazole (final concentration of 33 µM) were then added and incubated for 45 min at 20°C. Calcium chloride (final concentration of 0.6 mM) was added, and Western blot samples were taken at 0, 30, 60, and 90 min. Chromatin morphology was assessed at 90 min by taking 1-µl samples and staining with Hoechst.
Mass spectrometry
Mass spectrometry was used to determine the phosphorylation sites in Dsn1 and the interacting proteins of the Mis12C. The immunoprecipitated proteins were separated by gel electrophoresis. The members of the Mis12C, as well as other interacting bands, were cut from the gel and in-gel digested with trypsin (Sigma) at 37°C for 16 h, as described previously (Shevchenko et al., 2006). The dried peptides were separated on a 75 µm × 15 cm, 2-µm Acclaim PepMap reverse phase column (Thermo Fisher Scientific) at 300 nl/min using an UltiMate 3000 RSLCnano HPLC (Thermo Fisher Scientific). Peptides were eluted into an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific) using a linear gradient from 96% mobile phase A (0.1% formic acid in water) to 55% mobile phase B (20% water, 80% acetonitrile, and 0.08% formic acid) over 30 min. Parent full-scan mass spectra were collected in the Orbitrap mass analyzer set to acquire data at 120,000 full width half maximum resolution; ions were then isolated in the quadrupole mass filter, fragmented within the higher-energy collisional dissociation cell (higher-energy collisional dissociation normalized energy 32%, stepped ±3%), and the product ions were analyzed in the ion trap. Proteome Discoverer 2.0 (Thermo Fisher Scientific) was used to search the data against Xenopus proteins (Wühr et al., 2014) using SequestHT. The search was limited to tryptic peptides, with maximally two missed cleavages allowed. Cysteine carbamidomethylation was set as a fixed modification, with methionine oxidation and serine/threonine phosphorylation set as variable modifications. The precursor mass tolerance was 10 ppm, and the fragment mass tolerance was 0.6 D. The Percolator node was used to score and rank peptide matches using a 1% false discovery rate. The ptmRS node was used for confidence scoring of phosphorylation sites; all modified spectra were manually validated.
Protein purification
Mis12CDsn1-LAP and Mis12CDsn1EE
The four components of the Xenopus Mis12C, Mis12, Nsl1, Pmf1, and Dsn1–His6–LAP or Dsn1S77E,S84E-His6, were cloned into the protein expression plasmid, pST39, and transformed into BL21(DE3) pLysS Rosetta-2 cells. Cells were induced with 0.3 mM IPTG for 16 h at 18°C. The tagged proteins were purified by a HisTrap column (GE Healthcare), followed by anion exchange chromatography by a MonoQ column and then size exclusion chromatography on a HiLoad 16/600 Superdex 200 column (GE Healthcare). The proteins were concentrated by Amicon Ultra centrifugal filters (Millipore) in 150 mM NaCl, 20 mM Hepes, pH 7.7, 1 mM DTT, and 150 mM sucrose. Glycerol was added to a final concentration of 45%. The proteins were frozen in small aliquots in liquid nitrogen and stored at −80°C. Mis12CDsn1EE was added to the extract at a final concentration of 19 nM.
CENP-C–LAP
His6-WELQ–CENP-C2–55–LAP was cloned into the protein expression plasmid pET28a and transformed into BL21(DE3) cells. Cells were induced with 0.5 mM IPTG for 18 h at 18°C. The tagged protein was purified with Ni-NTA agarose (Qiagen). The His6-tag, which prevents interaction with Mis12C, was cleaved by WELQut protease (Thermo Fisher Scientific). After cleavage, the protein was reapplied to Ni-NTA agarose to remove the His6-tag and protease. The purified protein was concentrated using Amicon Ultra centrifugal filters (Millipore). CENP-C2–55–LAP was frozen in 150 mM NaCl, 20 mM Hepes, pH 7.7, 1 mM DTT, and 150 mM sucrose and stored at −80°C.
Strep-INC788–871–His6-Aurora B
Strep-INC788–871 and His6-Aurora B60–361 were cloned into a modified pCOLADuet-1 dual expression plasmid containing N-terminal Strep II and His6 tags and transformed into BL21(DE3) cells. Cells were induced with 0.5 mM IPTG for 18 h at 18°C. The tagged proteins were purified by Ni-NTA agarose (Qiagen) followed by Strep-Tactin XT resin (iba). Proteins were eluted in 150 mM NaCl, 20 mM Hepes, pH 7.7, and 1 mM TCEP.
GST-INC788–871–His6-Aurora B60–361 and GST-INC328–871–His6-Aurora B60–361
His6-Aurora B60–361 and GST-INC788–871 or GST-INC328–871 were cloned into the protein expression plasmid, pCOLADuet-1, and transformed into Bl21(DE3) cells. Cells were induced with 1 mM IPTG for 22 h at 18°C. The tagged proteins were purified by Glutathione Sepharose 4B resin (GE) followed by Ni-NTA agarose (Qiagen). Proteins were frozen in 300 mM NaCl, 20 mM Hepes, pH 7.7, 1 mM TCEP, 150 mM Imidazole, and 45% glycerol and stored at −80°C.
H3–H4
The protein expression plasmid pET3 containing Xenopus histone H3 or H4 was transformed into BL21(DE3) cells. Cells were induced with 0.2 mM IPTG for 3 h at 37°C. Cells were lysed and centrifuged, and pellets containing inclusion bodies were homogenized with a Dounce homogenizer and then centrifuged. The supernatant was dialyzed against an unfolding buffer and the histones were then purified through cation exchange. After dialysis into 0.1 mM PMSF and 5 mM 2-mercaptoethanol, the sample was lyophilized. Equal amounts of lyophilized H3 and H4 were resuspended, mixed, and dialyzed into a refolding buffer. H3–H4 tetramer was purified by size exclusion chromatography on a Superdex 16/60 column and frozen and stored at −80°C.
In vitro kinase assays
Chemosensor assay with CPC subcomplexes
Kinase activity of GST-INC788–871–His6-Aurora B60–361 and GST-INC328–871–His6-Aurora B60–361 was determined by incubating each CPC subcomplex with a generic peptide substrate for Aurora kinases that was conjugated to a Sox fluorescent sensor (ATQ0186; AssayQuant). Phosphorylation was measured via fluorescence at the emission wavelength of 485 nm, generated from an excitation wavelength of 360 nm. Fluorescence was detected by the Synergy Neo2 microplate reader (BioTek), and data were recorded by Gen5 Image Prime v3.04 (BioTek) and analyzed in Prism v7. Kinase assays were performed at 30°C in the kinase assay buffer (54 mM Hepes, pH 7.5, 120 mM NaCl, 10 mM MgCl2, 0.4 mM EGTA, 1 mM ATP, 1.2 mM DTT, and 0.012% Brij-35). A baseline kinase assay was performed in triplicate with 5 nM CPC subcomplex and 24 µM Sox probe substrate. Time points were taken every 45 s for 90 min. Michaelis–Menten kinetic data were generated with 5 nM CPC subcomplex and the following series of Sox probe substrate concentrations in duplicate: 100 µM, 66.67 µM, 44.44 µM, 29.63 µM, 19.75 µM, 13.17 µM, and 8.78 µM. Reaction velocities (relative fluorescence units per second) were converted into initial rates (μM−1s−1) using the slopes of standard curves generated for each substrate concentration. Initial rates (µM−1s−1 × 10−1) were plotted against substrate concentration, and the Michaelis constant (KM) and the maximum initial velocity (Vmax) for each CPC subcomplex was calculated from the fits to the Michaelis–Menten equation. The catalytic rate constant (kcat) was calculated from Vmax.
Kinase assay for Dsn1 phosphorylation with CPC subcomplexes
The kinase assay to assess Dsn1 phosphorylation by GST-INC788–871–His6-Aurora B60–361 or GST-INC328–871–His6-Aurora B60–361 was performed at 25°C with 300 nM Mis12C and 1 nM indicated subcomplex in the kinase assay buffer (20 mM Hepes, pH 7.7, 150 mM NaCl, 10 mM MgCl2, 0.7 mM ATP, 1 mM DTT, and 1× Leupeptin/Pepstatin/Chymostatin; Chemicon). Time points were taken at 0, 5, 10, 15, 30, 60, 120, and 180 min. Phosphorylation of Dsn1 was measured by Western blot with custom phospho-specific antibody to Ser77 of Xenopus Dsn1 (Thermo Fisher Scientific). Antibodies were raised against the peptide CRKSLRR(pS)VAKTPK (New England Peptide), and phospho-specific antibodies were purified from serum by sequential passage over an unmodified and phosphorylated peptide column, followed by elution with 100 mM glycine, pH 2.3.
Kinase assay for Dsn1 phosphorylation with INC788–871–His6–Aurora B and CENP-C
A kinase assay to assess how Dsn1 phosphorylation by the CPC subcomplex is affected by the presence of CENP-C was performed at 25°C with 300 nM Mis12C and 100 nM INC788–871–His6-Aurora B60–361, in the presence or absence of CENP-C2–55–LAP (1 µM or 5 µM), in the kinase assay buffer listed above. Time points were taken at 0, 5, 10, 30, 60, 120, 180, and 300 min. Phosphorylation of Dsn1 was measured by Western blot with the custom phospho-specific antibody to Ser77 of Dsn1. Initial rates for each concentration of CENP-C were calculated using fits to the linear portion of each curve (30 min).
Kinase assay for H3S10 phosphorylation with CPC subcomplexes
A kinase assay to assess the phosphorylation of another Aurora B substrate, histone H3 (Ser10), by each CPC subcomplex was performed at 25°C with 300 nM H3–H4 and 1 nM GST-INC788–871–His6-Aurora B60–361 or GST-INC328–871–His6-Aurora B60–361 in the kinase assay buffer listed above. Time points were taken at 0, 5, 10, 20, 30, 60, and 180 min. Phosphorylation of histone H3S10ph was measured by Western blot with anti-histone H3S10ph antibodies (6G3, 9706; Cell Signaling Technology).
Western blots
Primary antibodies were diluted in Licor blocking solution with a final Tween-20 concentration of 0.1%, except for anti-phospho Aurora and anti-histone H3S10ph, which had no Tween-20. The following antibodies and antibody dilutions were used: anti-INCENP (raised against C-terminal peptide CSNRHHLAVGYGLKY; 5.5 µg/ml), anti-Aurora B (Kelly et al., 2007; 5 µg/ml), anti-Borealin A (Dasra A; Kelly et al., 2007; 5 µg/ml), anti-Survivin (Tseng et al., 2010; 12 µg/ml), anti-phospho Aurora (phospho-Aurora A [Thr288]/Aurora B [Thr 232]/Aurora C [Thr198] 2914, 1:200; Cell Signaling Technology), anti-histone H3 (ab1791, 1:500; Abcam), anti-histone H3S10ph (6G3, 9706, 1:500; Cell Signaling Technology), anti-histone H3T3ph (2162–1, 1:10,000; Epitomics), anti-Mis12 (raised against Mis12199–216 peptide KHKKTKVDGSTLTPICPR; 1:1,000), anti-GFP (11814460001, 1:1,000; Sigma Aldrich), anti-FLAG (F3165, 1:1,000; Sigma Aldrich), anti-α-tubulin (DM1, 1:20,000; Sigma), and custom anti-Dsn1S77ph (1:250). Secondary antibodies from Licor were used (goat anti-rabbit 800 nm and goat anti-mouse 680 nm), as was the Licor imaging system to scan membranes.
Microscopy and fluorescence quantification
To immunostain kinetochores, Xenopus egg extract was fixed for 5 min by ∼20-fold dilution in BRB80, 20% glycerol, 0.5% Triton X-100, and 3.7% formaldehyde at room temperature. Fixed reactions were layered onto a cushion of BRB80 plus 40% glycerol overlaying a poly-L-lysine–coated coverslips (No. 1) placed in a 24-well plate. Nuclei were adhered onto coverslips on plate holders at 4,000 rpm for 15 min at 18°C in a centrifuge (5810R; Eppendorf). Cushions were washed with BRB80, and coverslips were postfixed in ice-cold methanol for 5 min, blocked with Abdil (TBS, 0.1% Tween20 + 2% BSA, and 0.1% sodium azide) overnight at 4°C, and then incubated in primary at room temperature for 1.5 h unless otherwise noted. All washes and antibody dilutions were done with Abdil buffer. Nuclei were stained with Hoechst 33258 before mounting in 80% glycerol plus PBS medium. The following antibodies were used at the indicated dilutions: Dsn1 (Emanuele et al., 2008), 1:500; p-Aurora (2914S; Cell Signaling Technology), 1:500; Ndc80 (McCleland et al., 2003), 1:200; Mad2 (Chen et al., 1996), 1:200; BubR1 (Boyarchuk et al., 2007), 1:100; Borealin (Dasra A), 1:250 (Kelly et al., 2007); CENP-C (Haase et al., 2017), 1:500; Ndc80S44ph (DeLuca et al., 2011), 1:2,000; H3S10ph (9706; Cell Signaling Technology), 1:400; CENP-T (Krizaic et al., 2015), 1:200; Knl1 (Emanuele et al., 2008), 1:200; and anti-FLAG (SLBF6631; Sigma), 1:250.
All immunofluorescence was imaged with a 0.2-µm step size using an Eclipse Ti (Nikon) composed of a Nikon Plan Apo ×100/1.45, oil immersion objective, a Plan Apo 40×/0.95 objective and a Hamamatsu Orca-Flash 4.0 camera. Images were captured and processed using NIS Elements AR 4.20.02 software (Nikon) and analyzed in Fiji ImageJ. The acquired Z-sections of 0.2 µm each were converted to a maximum projection using NIS Elements and Fiji. Kinetochore intensity was measured using Fiji by centering 9 × 9- and 13 × 13-pixel regions over individual kinetochores. Total fluorescence intensity was recorded from each region. To correct for background fluorescence, the difference in intensities between the two regions was determined and then made proportionate to the smaller region. This background value was then subtracted from the smaller region to determine kinetochore intensity with background correction as previously reported (Hoffman et al., 2001). Kinetochore alignment was determined by dividing metaphase spindles longitudinally into three equally sized zones.
Quantification and statistical analysis
All analyses were performed with a minimum number of either 50 spindles or 96 kinetochores for each assay. Sample size was chosen to ensure a high (>90%) theoretical statistical power in order to generate reliable P values. All graphs and statistical analysis were prepared with GraphPad Prism. Fluorescence values from experimental conditions were compared with control conditions using an ordinary one-way ANOVA with Tukey’s multiple comparison tests to determine significance. All graphs show the mean, with error bars representing the SD, unless otherwise indicated.
Online supplemental material
Fig. S1 shows mass spectrometry data for both the phosphorylation sites on Xenopus Dsn1 and the Mis12C interacting proteins. It also shows images and quantification for the rescue of kinetochore assembly by phosphomimetic Mis12CDsn1EE in the presence of hesperadin. Fig. S2 provides additional data showing that kinetochore assembly requires the central region of INCENP. Fig. S3 shows Western blots, images, and quantified data to demonstrate that the SAH domain is required for kinetochore assembly. Fig. S4 includes quantified data to show that phosphomimetic Mis12CDsn1EE rescues kinetochore assembly in the absence of the SAH domain and additional data to show that kinetochore assembly can be partially rescued in the absence of the SAH domain with the addition of phosphatase inhibitor. Fig. S5 shows a Western blot and schematic of an experiment that demonstrated that kinetochore assembly requires localized CPC activity; it also shows that phosphomimetic Mis12CDsn1EE is not sufficient for global SAC in the presence of hesperadin.
Supplementary Material
Acknowledgments
We thank M. Lichten and R. O’Neill for critical reading of the manuscript; members of the Rusan laboratory and the Laboratory of Biochemistry and Molecular Biology for comments and support; and A. Arnaoutov (National Institute of Child Health and Human Development [NICHD], Bethesda, MD), R. Chen (Institute of Biological Chemistry, Academia Sinica, Taipei, Taiwan), M. Dasso (NICHD, Bethesda, MD), J. DeLuca (Colorado State University, Fort Collins, CO), A. Losada (Centro Nacional de Investigaciones Oncológicas, Madrid, Spain), R. Subramanian (Harvard Medical School, Boston, MA), A. Straight (Stanford University, Palo Alto, CA), and P.T. Stukenberg (University of Virginia, Charlottesville, VA) for kindly providing reagents.
This work was supported by the intramural program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health.
The authors declare no competing financial interests.
Author contributions: M.K. Bonner, J. Haase, and A.E. Kelly designed the experiments. M.K. Bonner, J. Haase, H. Halas, J. Swinderman, L.M. Miller Jenkins, and A.E. Kelly performed and analyzed experiments. A.E. Kelly wrote the manuscript with input from all co-authors.
References
- Abe Y., Sako K., Takagaki K., Hirayama Y., Uchida K.S.K., Herman J.A., DeLuca J.G., and Hirota T.. 2016. HP1-Assisted Aurora B Kinase Activity Prevents Chromosome Segregation Errors. Dev. Cell. 36:487–497. 10.1016/j.devcel.2016.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyoshi B., Nelson C.R., and Biggins S.. 2013. The aurora B kinase promotes inner and outer kinetochore interactions in budding yeast. Genetics. 194:785–789. 10.1534/genetics.113.150839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekier M.E., Mazur T., Rashid M.S., and Taylor W.R.. 2015. Borealin dimerization mediates optimal CPC checkpoint function by enhancing localization to centromeres and kinetochores. Nat. Commun. 6:6775 10.1038/ncomms7775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop J.D., and Schumacher J.M.. 2002. Phosphorylation of the carboxyl terminus of inner centromere protein (INCENP) by the Aurora B Kinase stimulates Aurora B kinase activity. J. Biol. Chem. 277:27577–27580. 10.1074/jbc.C200307200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyarchuk Y., Salic A., Dasso M., and Arnaoutov A.. 2007. Bub1 is essential for assembly of the functional inner centromere. J. Cell Biol. 176:919–928. 10.1083/jcb.200609044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell C.S., and Desai A.. 2013. Tension sensing by Aurora B kinase is independent of survivin-based centromere localization. Nature. 497:118–121. 10.1038/nature12057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M., Wheelock M., Funabiki H., and Earnshaw W.C.. 2012. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 13:789–803. 10.1038/nrm3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheerambathur D.K., and Desai A.. 2014. Linked in: formation and regulation of microtubule attachments during chromosome segregation. Curr. Opin. Cell Biol. 26:113–122. 10.1016/j.ceb.2013.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M. 2014. The kinetochore. Cold Spring Harb. Perspect. Biol. 6:a015826 10.1101/cshperspect.a015826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., and Desai A.. 2005. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci. STKE. 2005:pl1 10.1126/stke.2662005pl1 [DOI] [PubMed] [Google Scholar]
- Chen R.H., Waters J.C., Salmon E.D., and Murray A.W.. 1996. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 274:242–246. 10.1126/science.274.5285.242 [DOI] [PubMed] [Google Scholar]
- Chittori S., Hong J., Saunders H., Feng H., Ghirlando R., Kelly A.E., Bai Y., and Subramaniam S.. 2017. Structural mechanisms of centromeric nucleosome recognition by the kinetochore protein CENP-N. Science. 157:2781–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca K.F., Lens S.M.A., and DeLuca J.G.. 2011. Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J. Cell Sci. 124:622–634. 10.1242/jcs.072629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- des Georges A., Dhote V., Kuhn L., Hellen C.U.T., Pestova T.V., Frank J., and Hashem Y.. 2015. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature. 525:491–495. 10.1038/nature14891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova Y.N., Jenni S., Valverde R., Khin Y., and Harrison S.C.. 2016. Structure of the MIND Complex Defines a Regulatory Focus for Yeast Kinetochore Assembly. Cell. 167:1014–1027.e12. 10.1016/j.cell.2016.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele M.J., McCleland M.L., Satinover D.L., and Stukenberg P.T.. 2005. Measuring the stoichiometry and physical interactions between components elucidates the architecture of the vertebrate kinetochore. Mol. Biol. Cell. 16:4882–4892. 10.1091/mbc.e05-03-0239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele M.J., Lan W., Jwa M., Miller S.A., Chan C.S.M., and Stukenberg P.T.. 2008. Aurora B kinase and protein phosphatase 1 have opposing roles in modulating kinetochore assembly. J. Cell Biol. 181:241–254. 10.1083/jcb.200710019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink S., Turnbull K., Desai A., and Campbell C.S.. 2017. An engineered minimal chromosomal passenger complex reveals a role for INCENP/Sli15 spindle association in chromosome biorientation. J. Cell Biol. 216:911–923. 10.1083/jcb.201609123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischböck-Halwachs J., Singh S., Potocnjak M., Hagemann G., Solis-Mezarino V., Woike S., Ghodgaonkar-Steger M., Weissmann F., Gallego L.D., Rojas J., et al. 2019. The COMA complex interacts with Cse4 and positions Sli15/Ipl1 at the budding yeast inner kinetochore. eLife. 8:e42879 10.7554/eLife.42879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Rodríguez L.J., Kasciukovic T., Denninger V., and Tanaka T.U.. 2019. Aurora B-INCENP Localization at Centromeres/Inner Kinetochores Is Required for Chromosome Bi-orientation in Budding Yeast. Curr. Biol. 29:1536–1544.e4. 10.1016/j.cub.2019.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne K.E., and Cheeseman I.M.. 2013. CDK-dependent phosphorylation and nuclear exclusion coordinately control kinetochore assembly state. J. Cell Biol. 201:23–32. 10.1083/jcb.201301006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne K.E., Takeuchi K., Suzuki A., Hori T., Fukagawa T., and Cheeseman I.M.. 2011. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 145:410–422. 10.1016/j.cell.2011.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Vera J.A., Luković E., and Imperiali B.. 2009. A rapid method for generation of selective Sox-based chemosensors of Ser/Thr kinases using combinatorial peptide libraries. Bioorg. Med. Chem. Lett. 19:1258–1260. 10.1016/j.bmcl.2008.12.090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase J., Bonner M.K., Halas H., and Kelly A.E.. 2017. Distinct Roles of the Chromosomal Passenger Complex in the Detection of and Response to Errors in Kinetochore-Microtubule Attachment. Dev. Cell. 42:640–654.e5. 10.1016/j.devcel.2017.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara M., and Fukagawa T.. 2018. Kinetochore assembly and disassembly during mitotic entry and exit. Curr. Opin. Cell Biol. 52:73–81. 10.1016/j.ceb.2018.02.005 [DOI] [PubMed] [Google Scholar]
- Hara M., Ariyoshi M., Okumura E.-I., Hori T., and Fukagawa T.. 2018. Multiple phosphorylations control recruitment of the KMN network onto kinetochores. Nat. Cell Biol. 20:1378–1388. 10.1038/s41556-018-0230-0 [DOI] [PubMed] [Google Scholar]
- Hemmerich P., Weidtkamp-Peters S., Hoischen C., Schmiedeberg L., Erliandri I., and Diekmann S.. 2008. Dynamics of inner kinetochore assembly and maintenance in living cells. J. Cell Biol. 180:1101–1114. 10.1083/jcb.200710052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengeveld R.C.C., Vromans M.J.M., Vleugel M., Hadders M.A., and Lens S.M.A.. 2017. Inner centromere localization of the CPC maintains centromere cohesion and allows mitotic checkpoint silencing. Nat. Commun. 8:15542 10.1038/ncomms15542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindriksen S., Lens S.M.A., and Hadders M.A.. 2017. The Ins and Outs of Aurora B Inner Centromere Localization. Front. Cell Dev. Biol. 5:112 10.3389/fcell.2017.00112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw S.M., and Harrison S.C.. 2019. The structure of the Ctf19c/CCAN from budding yeast. eLife. 8:e44239 10.7554/eLife.44239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiruma Y., Sacristan C., Pachis S.T., Adamopoulos A., Kuijt T., Ubbink M., von Castelmur E., Perrakis A., and Kops G.J.P.L.. 2015. Competition between MPS1 and microtubules at kinetochores regulates spindle checkpoint signaling. Science. 348:1264–1267. 10.1126/science.aaa4055 [DOI] [PubMed] [Google Scholar]
- Hoffman D.B., Pearson C.G., Yen T.J., Howell B.J., and Salmon E.D.. 2001. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell. 12:1995–2009. 10.1091/mbc.12.7.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., Okada M., Maenaka K., and Fukagawa T.. 2008. CENP-O class proteins form a stable complex and are required for proper kinetochore function. Mol. Biol. Cell. 19:843–854. 10.1091/mbc.e07-06-0556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., and Yen T.J.. 2009. BubR1 is an effector of multiple mitotic kinases that specifies kinetochore: microtubule attachments and checkpoint. Cell Cycle. 8:1164–1167. 10.4161/cc.8.8.8151 [DOI] [PubMed] [Google Scholar]
- Huis In ’t Veld P.J., Jeganathan S., Petrovic A., Singh P., John J., Krenn V., Weissmann F., Bange T., and Musacchio A.. 2016. Molecular basis of outer kinetochore assembly on CENP-T. eLife. 5:e21007 10.7554/eLife.21007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z., Gao H., and Yu H.. 2015. Kinetochore attachment sensed by competitive Mps1 and microtubule binding to Ndc80C. Science. 348:1260–1264. 10.1126/science.aaa4029 [DOI] [PubMed] [Google Scholar]
- Kang J., Cheeseman I.M., Kallstrom G., Velmurugan S., Barnes G., and Chan C.S.M.. 2001. Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)-related protein Sli15 during chromosome segregation. J. Cell Biol. 155:763–774. 10.1083/jcb.200105029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A.E., Sampath S.C., Maniar T.A., Woo E.M., Chait B.T., and Funabiki H.. 2007. Chromosomal enrichment and activation of the aurora B pathway are coupled to spatially regulate spindle assembly. Dev. Cell. 12:31–43. 10.1016/j.devcel.2006.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A.E., Ghenoiu C., Xue J.Z., Zierhut C., Kimura H., and Funabiki H.. 2010. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science. 330:235–239. 10.1126/science.1189505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., and Yu H.. 2015. Multiple assembly mechanisms anchor the KMN spindle checkpoint platform at human mitotic kinetochores. J. Cell Biol. 208:181–196. 10.1083/jcb.201407074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krenn V., and Musacchio A.. 2015. The Aurora B Kinase in Chromosome Bi-Orientation and Spindle Checkpoint Signaling. Front. Oncol. 5:225 10.3389/fonc.2015.00225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krenn V., Overlack K., Primorac I., van Gerwen S., and Musacchio A.. 2014. KI motifs of human Knl1 enhance assembly of comprehensive spindle checkpoint complexes around MELT repeats. Curr. Biol. 24:29–39. 10.1016/j.cub.2013.11.046 [DOI] [PubMed] [Google Scholar]
- Krizaic I., Williams S.J., Sánchez P., Rodríguez-Corsino M., Stukenberg T., and Losada A.. 2015. The distinct functions of CENP-C and CENP-T/W in centromere propagation and function in Xenopus egg extracts. Nucleus. 6:133–143. 10.1080/19491034.2014.1003509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacoste N., Woolfe A., Tachiwana H., Garea A.V., Barth T., Cantaloube S., Kurumizaka H., Imhof A., and Almouzni G.. 2014. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol. Cell. 53:631–644. 10.1016/j.molcel.2014.01.018 [DOI] [PubMed] [Google Scholar]
- Lang J., Barber A., and Biggins S.. 2018. An assay for de novo kinetochore assembly reveals a key role for the CENP-T pathway in budding yeast. eLife. 7:e37819 10.7554/eLife.37819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Vleugel M., Backer C.B., Hori T., Fukagawa T., Cheeseman I.M., and Lampson M.A.. 2010. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J. Cell Biol. 188:809–820. 10.1083/jcb.201001006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- London N., Ceto S., Ranish J.A., and Biggins S.. 2012. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr. Biol. 22:900–906. 10.1016/j.cub.2012.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado M., and Kapoor T.M.. 2011. Constitutive Mad1 targeting to kinetochores uncouples checkpoint signalling from chromosome biorientation. Nat. Cell Biol. 13:475–482. 10.1038/ncb2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleland M.L., Gardner R.D., Kallio M.J., Daum J.R., Gorbsky G.J., Burke D.J., and Stukenberg P.T.. 2003. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 17:101–114. 10.1101/gad.1040903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley K.L., Sekulic N., Guo L.Y., Tsinman T., Black B.E., and Cheeseman I.M.. 2015. The CENP-L-N Complex Forms a Critical Node in an Integrated Meshwork of Interactions at the Centromere-Kinetochore Interface. Mol. Cell. 60:886–898. 10.1016/j.molcel.2015.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milks K.J., Moree B., and Straight A.F.. 2009. Dissection of CENP-C-directed centromere and kinetochore assembly. Mol. Biol. Cell. 20:4246–4255. 10.1091/mbc.e09-05-0378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A., and Desai A.. 2017. A Molecular View of Kinetochore Assembly and Function. Biology (Basel). 6:5 10.3390/biology6010005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., and Schiebel E.. 2003. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science. 302:2120–2124. 10.1126/science.1091936 [DOI] [PubMed] [Google Scholar]
- Perpelescu M., and Fukagawa T.. 2011. The ABCs of CENPs. Chromosoma. 120:425–446. 10.1007/s00412-011-0330-0 [DOI] [PubMed] [Google Scholar]
- Pesenti M.E., Prumbaum D., Auckland P., Smith C.M., Faesen A.C., Petrovic A., Erent M., Maffini S., Pentakota S., Weir J.R., et al. 2018. Reconstitution of a 26-Subunit Human Kinetochore Reveals Cooperative Microtubule Binding by CENP-OPQUR and NDC80. Mol. Cell. 71:923–939.e10. 10.1016/j.molcel.2018.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic A., Keller J., Liu Y., Overlack K., John J., Dimitrova Y.N., Jenni S., van Gerwen S., Stege P., Wohlgemuth S., et al. 2016. Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores. Cell. 167:1028–1040.e15. 10.1016/j.cell.2016.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przewloka M.R., Venkei Z., Bolanos-Garcia V.M., Debski J., Dadlez M., and Glover D.M.. 2011. CENP-C is a structural platform for kinetochore assembly. Curr. Biol. 21:399–405. 10.1016/j.cub.2011.02.005 [DOI] [PubMed] [Google Scholar]
- Rago F., Gascoigne K.E., and Cheeseman I.M.. 2015. Distinct organization and regulation of the outer kinetochore KMN network downstream of CENP-C and CENP-T. Curr. Biol. 25:671–677. 10.1016/j.cub.2015.01.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg J.S., Cross F.R., and Funabiki H.. 2011. KNL1/Spc105 recruits PP1 to silence the spindle assembly checkpoint. Curr. Biol. 21:942–947. 10.1016/j.cub.2011.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppert J.G., Samejima K., Platani M., Molina O., Kimura H., Jeyaprakash A.A., Ohta S., and Earnshaw W.C.. 2018. HP1α targets the chromosomal passenger complex for activation at heterochromatin before mitotic entry. EMBO J. 37:e97677 10.15252/embj.201797677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh T., Rossi P., and Kalodimos C.G.. 2017. Atomic view of the energy landscape in the allosteric regulation of Abl kinase. Nat. Struct. Mol. Biol. 24:893–901. 10.1038/nsmb.3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samejima K., Platani M., Wolny M., Ogawa H., Vargiu G., Knight P.J., Peckham M., and Earnshaw W.C.. 2015. The INCENP Coil is a Single Alpha Helical (SAH) Domain that Binds Directly to Microtubules and is Important for CPC Localization and Function in Mitosis. J. Biol. Chem. 290:21460–21472. 10.1074/jbc.M115.645317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandall S., Severin F., McLeod I.X., Yates J.R. III, Oegema K., Hyman A., and Desai A.. 2006. A Bir1-Sli15 complex connects centromeres to microtubules and is required to sense kinetochore tension. Cell. 127:1179–1191. 10.1016/j.cell.2006.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screpanti E., De Antoni A., Alushin G.M., Petrovic A., Melis T., Nogales E., and Musacchio A.. 2011. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr. Biol. 21:391–398. 10.1016/j.cub.2010.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa F., Mapelli M., Ciferri C., Tarricone C., Areces L.B., Schneider T.R., Stukenberg P.T., and Musacchio A.. 2005. Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol. Cell. 18:379–391. 10.1016/j.molcel.2005.03.031 [DOI] [PubMed] [Google Scholar]
- Shepperd L.A., Meadows J.C., Sochaj A.M., Lancaster T.C., Zou J., Buttrick G.J., Rappsilber J., Hardwick K.G., and Millar J.B.A.. 2012. Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr. Biol. 22:891–899. 10.1016/j.cub.2012.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Tomas H., Havlis J., Olsen J.V., and Mann M.. 2006. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1:2856–2860. 10.1038/nprot.2006.468 [DOI] [PubMed] [Google Scholar]
- Tseng B.S., Tan L., Kapoor T.M., and Funabiki H.. 2010. Dual detection of chromosomes and microtubules by the chromosomal passenger complex drives spindle assembly. Dev. Cell. 18:903–912. 10.1016/j.devcel.2010.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich A.K.C., Seeger M., Schütze T., Bartlick N., and Wahl M.C.. 2016. Scaffolding in the Spliceosome via Single α Helices. Structure. 24:1972–1983. 10.1016/j.str.2016.09.007 [DOI] [PubMed] [Google Scholar]
- Vader G., Cruijsen C.W.A., van Harn T., Vromans M.J.M., Medema R.H., and Lens S.M.A.. 2007. The chromosomal passenger complex controls spindle checkpoint function independent from its role in correcting microtubule kinetochore interactions. Mol. Biol. Cell. 18:4553–4564. 10.1091/mbc.e07-04-0328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst A., Vromans M.J.M., Bouwman K., van der Waal M.S., Hadders M.A., and Lens S.M.A.. 2015. Inter-domain Cooperation in INCENP Promotes Aurora B Relocation from Centromeres to Microtubules. Cell Reports. 12:380–387. 10.1016/j.celrep.2015.06.038 [DOI] [PubMed] [Google Scholar]
- Wan X., O’Quinn R.P., Pierce H.L., Joglekar A.P., Gall W.E., DeLuca J.G., Carroll C.W., Liu S.-T., Yen T.J., McEwen B.F., et al. 2009. Protein architecture of the human kinetochore microtubule attachment site. Cell. 137:672–684. 10.1016/j.cell.2009.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E., Ballister E.R., and Lampson M.A.. 2011. Aurora B dynamics at centromeres create a diffusion-based phosphorylation gradient. J. Cell Biol. 194:539–549. 10.1083/jcb.201103044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Dai J., Daum J.R., Niedzialkowska E., Banerjee B., Stukenberg P.T., Gorbsky G.J., and Higgins J.M.G.. 2010. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science. 330:231–235. 10.1126/science.1189435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn J.P.I., Vleugel M., Liu D., Yates J.R. III, Lampson M.A., Fukagawa T., and Cheeseman I.M.. 2010. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell. 38:383–392. 10.1016/j.molcel.2010.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheelock M.S., Wynne D.J., Tseng B.S., and Funabiki H.. 2017. Dual recognition of chromatin and microtubules by INCENP is important for mitotic progression. J. Cell Biol. 216:925–941. 10.1083/jcb.201609061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wühr M., Freeman R.M. Jr., Presler M., Horb M.E., Peshkin L., Gygi S., and Kirschner M.W.. 2014. Deep proteomics of the Xenopus laevis egg using an mRNA-derived reference database. Curr. Biol. 24:1467–1475. 10.1016/j.cub.2014.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y., Honda T., Tanno Y., and Watanabe Y.. 2010. Two histone marks establish the inner centromere and chromosome bi-orientation. Science. 330:239–243. 10.1126/science.1194498 [DOI] [PubMed] [Google Scholar]
- Yamagishi Y., Yang C.-H., Tanno Y., and Watanabe Y.. 2012. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat. Cell Biol. 14:746–752. 10.1038/ncb2515 [DOI] [PubMed] [Google Scholar]
- Yang Y., Wu F., Ward T., Yan F., Wu Q., Wang Z., McGlothen T., Peng W., You T., Sun M., et al. 2008. Phosphorylation of HsMis13 by Aurora B kinase is essential for assembly of functional kinetochore. J. Biol. Chem. 283:26726–26736. 10.1074/jbc.M804207200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Martins I.R.S., Li P., Amarasinghe G.K., Umetani J., Fernandez-Zapico M.E., Billadeau D.D., Machius M., Tomchick D.R., and Rosen M.K.. 2010. Structural and energetic mechanisms of cooperative autoinhibition and activation of Vav1. Cell. 140:246–256. 10.1016/j.cell.2009.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaytsev A.V., Segura-Peña D., Godzi M., Calderon A., Ballister E.R., Stamatov R., Mayo A.M., Peterson L., Black B.E., Ataullakhanov F.I., et al. 2016. Bistability of a coupled Aurora B kinase-phosphatase system in cell division. eLife. 5:e10644 10.7554/eLife.10644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Zheng F., Wang C., Wu M., Zhang X., Wang Q., Yao X., Fu C., Zhang X., and Zang J.. 2017. Phosphorylation of CENP-C by Aurora B facilitates kinetochore attachment error correction in mitosis. Proc. Natl. Acad. Sci. USA. 114:E10667–E10676. 10.1073/pnas.1710506114 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.