Healthy tissues can harbor cancer-associated mutations without developing tumors, yet the mechanisms behind this apparent tolerance are unclear. In this work, we demonstrate that the hair follicle skin epithelium uses regeneration as a means of suppressing Ras-driven oncogenic growth.
Abstract
Mutations associated with tumor development in certain tissues can be nontumorigenic in others, yet the mechanisms underlying these different outcomes remains poorly understood. To address this, we targeted an activating Hras mutation to hair follicle stem cells and discovered that Hras mutant cells outcompete wild-type neighbors yet are integrated into clinically normal skin hair follicles. In contrast, targeting the Hras mutation to the upper noncycling region of the skin epithelium leads to benign outgrowths. Follicular Hras mutant cells autonomously and nonautonomously enhance regeneration, which directs mutant cells into continuous tissue cycling to promote integration rather than aberrancy. This follicular tolerance is maintained under additional challenges that promote tumorigenesis in the epidermis, including aging, injury, and a secondary mutation. Thus, the hair follicle possesses a unique, enhanced capacity to integrate and contain Hras mutant cells within both homeostatic and perturbed tissue, demonstrating that in the skin, multiple, distinct mechanisms exist to suppress oncogenic growth.
Introduction
Genetic mutations are a leading cause of cancer. However, recent studies have demonstrated that tissues can harbor cells with known cancer-causing mutations while remaining clinically normal (Ling et al., 2001; Ståhl et al., 2011; Laurie et al., 2012; Martincorena et al., 2015). For instance, although ∼30% of all cancers harbor a Ras mutation (Prior et al., 2012), many of the same Ras mutations have also been identified in noncancerous epithelial tissue (Kumar et al., 1990; Sukumar et al., 1995; Martincorena et al., 2015; Anglesio et al., 2017). One mechanism that the skin uses to deal with mutant cells is elimination, as an activating mutation to β-catenin causes the development of aberrant growths that regress after mutant cells are expelled from the tissue (Brown et al., 2017). It remains unclear, however, whether elimination is generally used to maintain homeostasis in response to any mutation or whether alternative mechanisms exist to suppress tumorigenesis in the skin.
Results and discussion
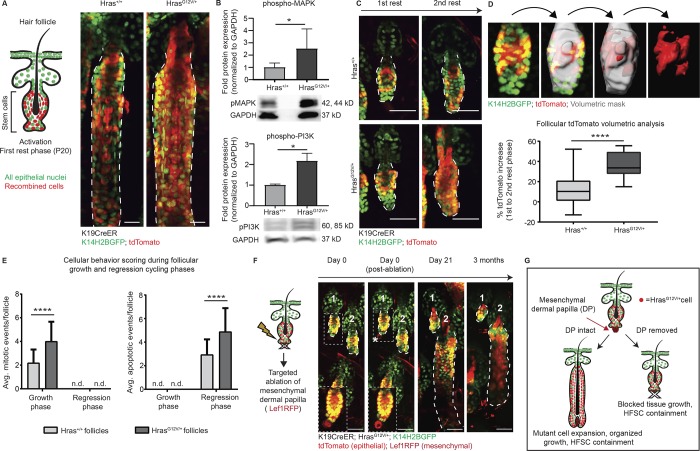
To address this question, we monitored the fate of Hras mutant cells in the skin epithelium through tracking the same cells over time using two-photon live imaging. To activate the mutation, we induced a mutant Hras allele (HrasG12V/+; Chen et al., 2009) specifically in hair follicle stem cells (HFSCs) and their progeny (K19CreER). To visualize all epithelial cells, we labeled their nuclei via a Keratin-14 Histone 2B GFP (K14H2BGFP) mouse line. To identify and track recombined stem cells and their progeny, we also added a cytoplasmic tdTomato Cre-dependent reporter, which approximates the recombination of the mutant cells (Fig. 1 A, left). Revisit imaging revealed that Hras mutant cells persisted in the tissue over time, as indicated by the tdTomato signal (Fig. 1 A, right). Western blot analysis of phospho-MAPK and phosphorylated phosphoinositide 3-kinases (PI3K) confirmed that Ras signaling was elevated in HrasG12V/+ tissue relative to Hras+/+ tissue (Fig. 1 B). These results demonstrate that phenotypically normal tissue can incorporate cells with sustained Ras activation, thus establishing that elimination is not the skin epithelium’s only mechanism for responding to mutant cells.
Figure 1.
Hras mutant cells outcompete wild-type neighbors yet are integrated into normal follicular tissue. All epithelial nuclei are in green (K14H2BGFP), and recombined cells are in red (tdTomato). (A) Left: Cartoon schematic depicting targeted activation to the HFSC compartment using the K19CreER system. Right: Two-photon images of Hras+/+ and HrasG12V/+ follicles during the growth phase. (B) Western blot analysis reveals that Hras signaling is active in the HrasG12V/+ K19CreER skin during the growth phase of follicle cycling as assessed by phospho-MAPK (n = 7 Hras+/+, 8 HrasG12V/+) and phospho-PI3K expression (n = 3 Hras+/+, 4 HrasG12V/+ mice). (C) Two-photon representative images of the same Hras+/+ and HrasG12V/+ follicles from first to second rest phase (∼1 mo) show a significant expansion of tdTomato in the HrasG12V/+ follicles. (D) Top: Imaris tdTomato volumetric analysis schematic showing (from left to right) a two-photon image of the HFSC during rest phase, a volumetric mask of the entire follicle, a volumetric mask of the tdTomato population within the entire follicle, and an isolated volumetric mask of only the tdTomato population. Bottom: The tdTomato expansion in HrasG12V/+ follicles is statistically significantly higher than in Hras+/+ follicles (n = 20 follicles per genotype across four mice per genotype). (E) Analysis of the average number of mitotic (left) and apoptotic (right) events per hair follicle from two-photon images reveals enhanced behaviors in HrasG12V/+ follicles yet proper temporal maintenance of these behaviors within the appropriate cycle phases (mitotic events: n = 64 follicles across 4 Hras+/+ mice, average follicle length 149.6 µm and n = 45 follicles across 5 HrasG12V/+ mice, average follicle length 166.85 µm; apoptotic events: n = 25 follicles across 5 Hras+/+ mice, average follicle length 243.27 µm and n = 40 follicles across 6 HrasG12V/+ mice, average follicle length 251.09 µm). (F) Targeted dermal papilla (DP) ablation during the rest phase locks follicles in permanent quiescence. Follicle 1 is ablated, while follicle 2 is not, and the asterisk denotes the autofluorescent halo produced after ablation (n = 50 follicles across 4 HrasG12V/+ mice, n = 30 follicles across 3 Hras+/+ mice; no ablated follicles ever regrew in either group). Insets are at higher exposure of the red channel to highlight the dermal papilla before and after ablation. (G) Cartoon summary depicting follicular HrasG12V/+ integration and obedience of normal tissue programs. For significance values, *, P < 0.05 and ****, P < 0.0001, as determined by an unpaired, two-tailed t test. Error bars indicate standard deviation and n.d. indicates not detected. All scale bars represent 50 µm.
Previous studies have demonstrated that clonal mutations to noncycling epithelial tissues, such as the skin epidermis or the esophageal epithelium, result in robust cellular competition that can ultimately influence the behavior and fate of their wild-type neighbors (Klein et al., 2010; Alcolea and Jones, 2015; Frede et al., 2016; Murai et al., 2018). To better understand how the Hras mutant cells are integrated into the tissue and whether their integration influences the fate of their wild-type neighbors, we performed revisit imaging of follicles over time. Hair follicles go through cyclical and stereotypical phases of rest, growth, and regression throughout the postnatal mouse life. We began tracking the hair follicles during their first rest phase after the initial Cre activation and monitored the starting amount of recombined cells (GFP+/tdTomato+). We imaged the same follicles again 1 mo later, after completing a full hair cycle, and discovered that there was a substantial expansion of tdTomato signal in HrasG12V/+ follicles compared to Hras+/+ follicles (Fig. 1 C). To quantify this, we used image analysis software, IMARIS, to create a 3D volumetric mask of the entire HFSC compartment, isolated the tdTomato population, and then expressed the tdTomato volume as a percentage of total follicle volume (Fig. 1 D, top). By comparing the expansion of tdTomato in HFSCs from first to second rest phase, we observed that there is a statistically significant increase in the number of tdTomato+ cells in HrasG12V/+ follicles (36.06%) compared with Hras+/+ follicles (12.22%) over the same 1-mo period (Fig. 1 D, bottom). These data indicate that the mutant cells are outcompeting wild-type neighbors over time.
Interestingly, despite outcompeting their wild-type neighbors, the HrasG12V/+ cells remained contained within the HFSC compartment, suggesting that the mutant cells are still controlled by normal homeostatic tissue. To test this, we first scored for mitotic events, which occur as follicles are in the growth phase, and apoptotic events, which occur as follicles are in the regression phase. We reasoned that if mutant cells were behaving independently from homeostatic mesenchymal tissue cues, then these cellular behaviors would not occur exclusively during the appropriate phases; rather, we might see apoptosis during growth and mitosis during regression. Interestingly, despite an enhancement of mitotic and apoptotic events in HrasG12V/+ follicles, mitotic events exclusively occurred during the growth phase, and apoptotic events exclusively occurred during the regression phase (Fig. 1 E and Video 1). These data suggest that mutant cells remain responsive to normal tissue cycling cues.
To evaluate whether HrasG12V/+ cells are truly behaving within normal tissue constraints, we sought to halt follicle regeneration. At the bottom of the hair follicle, there is a mesenchymal component, the dermal papilla, which is key for promoting the cycling of the hair follicle epithelium. Previous work demonstrated that targeted deletion of the mesenchymal dermal papilla and its stem cells during the hair follicle resting phase blocks hair cycling (Rompolas et al., 2012). We hypothesized that if HrasG12V/+ cells are truly behaving within normal tissue constraints, then they would not be able to initiate tissue growth without the dermal papilla. In contrast, tissue growth in the absence of the dermal papilla would indicate that HrasG12V/+ cells can override homeostatic tissue programs. We observed that HrasG12V/+ follicles remained locked in rest phase after ablation, identical to what is observed in wild-type follicles (Fig. 1 F). Interestingly, a previous study demonstrated that dermal papilla ablations of mutant β-catenin follicles during telogen does not stop them from growing, indicating that mutant Hras cells are more obedient to homeostatic tissue cues within the hair follicle (Deschene et al., 2014). Altogether, these results indicate that the programs that dictate follicular tissue function are dominant over the influence of constitutive, ligand-independent activation of Hras in this model (Fig. 1 G).
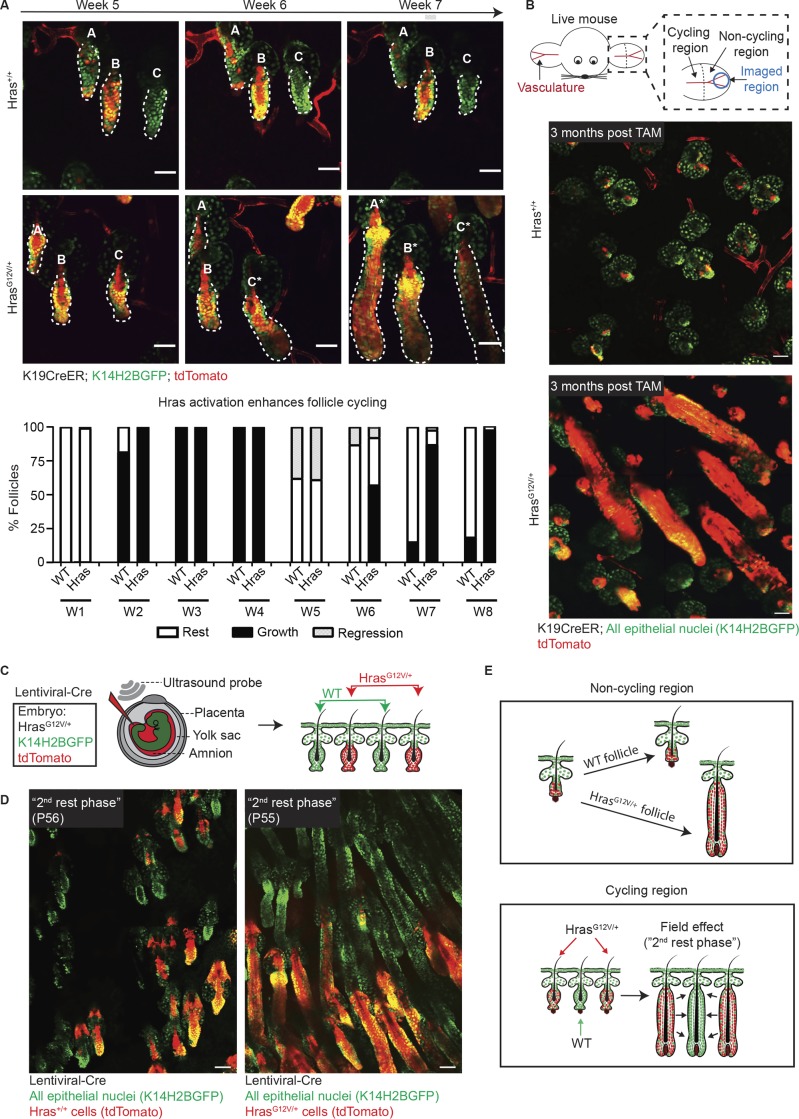
The observation that there were more mitotic and apoptotic cellular events in HrasG12V/+ follicles (Fig. 1 E) caused us to question whether Hras activation has any consequences on the frequency of follicular tissue regeneration. To test this hypothesis, we first tracked follicular cycling in Hras+/+ and HrasG12V/+ mice over an 8-wk period to understand whether Hras activation impacts normal follicular regeneration. Interestingly, as we tracked the same follicles over time via two-photon live imaging, we found that HrasG12V/+ follicles had an abbreviated rest phase as they rapidly regrew after reaching the second rest phase at week 5, which typically lasts for 3–4 wk (Figs. 2 A and S1 A). The increased frequency of follicle cycling also resulted in more rapid hair growth at the macroscopic level (Fig. S1 B). This continuous regeneration implied a precocious mesenchymal activation in the presence of the Hras mutant epithelium, which raised the question as to whether we would observe any phenotype in skin regions where dormancy persists throughout adulthood in the mouse. To accomplish this, we imaged the tip of the ear, which contains follicles that do not normally cycle under wild-type adult conditions, and saw that HrasG12V/+ expression in these follicles promoted them to cycle (Fig. 2 B). Since follicular cycling is dependent on cues from the mesenchyme (Jahoda et al., 1984; Greco et al., 2009; Sennett and Rendl, 2012) and Hras is not sufficient to drive cycling without the mesenchyme, the more frequent and forced tissue cycling indicates a nonautonomous impact of the mutant epithelial cells on the surrounding mesenchyme.
Figure 2.
The Hras mutant epithelium nonautonomously impacts the mesenchyme and follicular regeneration. All epithelial nuclei are in green (K14H2BGFP), and recombined cells are in red (tdTomato). (A) Top: Two-photon revisits reveal that K19CreER;HrasG12V/+ follicles reenter the growth phase more rapidly than K19CreER;Hras+/+ controls. Follicles are labeled A–C for ease of recognition, and cycling follicles are labeled with an asterisk. Bottom: Quantification of follicle staging over an 8-wk (W1–W8) period after tamoxifen induction (n = 1,704 follicles across 4 Hras+/+ mice and n = 1,413 follicles across 3 HrasG12V/+ mice). (B) Top: Cartoon schematic depicting the cycling and noncycling regions of the ear skin and imaged region. Bottom: Two-photon images of the hair follicles in the permanently quiescent region of skin at the tip of the ear in Hras+/+ and HrasG12V/+ mice show that while wild-type follicles do not cycle, HrasG12V/+ follicles do (observed in five K19CreER;HrasG12V/+ mice). TAM, tamoxifen. (C) In utero injection cartoon that depicts targeted lentiviral-Cre delivery to the amniotic sack at embryonic day 9.5. Injection with lentiviral-Cre into the HrasG12V/+;K14H2BGFP;tdTomato system directly tags and activates HrasG12V/+ cells in a mosaic fashion. (D) Two-photon images of Hras+/+ (left) and HrasG12V/+ (right) hair follicles during the “second rest phase” (P55). Mosaic activation of Hras using lentiviral-Cre delivery demonstrates that wild-type follicles (green) within the HrasG12V/+ skin reenter the growth phase precociously along with their Hras mutant (red) neighbors while follicles in the Hras+/+ mouse remain appropriately in rest phase (observed in n = 2 LV-Cre HrasG12V/+ mice compared with n = 3 LV-Cre Hras+/+ mice). (E) Cartoon summary depicting the enhanced regenerative power of HrasG12V/+ follicles in both cycling and noncycling skin. All scale bars represent 50 µm.
Since epithelial Hras activation nonautonomously impacts stromal cells, we next questioned whether these mutant epithelial cells could also promote nonautonomous changes in surrounding wild-type epithelial cells. To accomplish this, we aimed to create a mosaic condition where HrasG12V/+ follicles are interspersed with wild-type ones. Through the previously described CreER approach, tamoxifen injection results in activation throughout almost all follicles, even at lower doses, and does not create a heterogeneous field of individual mutant and wild-type follicles. To achieve this type of mosaicism, we established an approach to activate and directly fluorescently tagged the HrasG12V/+ mutant cells with tdTomato using a low titer of constitutive Cre delivered via in utero lentiviral injection (Figs. 2 C and S2). Through this approach, during a period of time when wild-type follicles are typically in a prolonged rest phase (the second rest phase), we observed that both the HrasG12V/+ follicles (GFP+/tdTomato+) and neighboring wild-type follicles (GFP+/tdTomato−) cycled (Fig. 2 D). The enhanced cycling of wild-type neighboring follicles indicates that Hras activation creates a field effect of continuous regeneration in cycling regions (Fig. 2 E).
Fibroblasts are critical to the orchestration of follicular cycling, which caused us to question whether the altered cycling within the Hras model was accompanied by molecular alterations to the fibroblasts. Importantly, RNA sequencing (RNA-seq) analysis of sorted fibroblasts revealed that despite genetically restricting Hras activation to the epithelial cells, sorted fibroblasts from HrasG12V/+ mice had 105 genes that were significantly differentially expressed compared with Hras+/+ mice. This indicates that there is likely a distinct subpopulation of fibroblasts affected by the nonautonomous impact of the mutant epithelium (Fig. S3, A–D). Notably, Hras itself was strongly up-regulated in the fibroblasts surrounding the HrasG12V/+ epithelium (Fig. S3 D). Furthermore, gene set enrichment analysis revealed a highly significant positive enrichment in the chemokine signaling pathway within fibroblasts sorted from the HrasG12V/+ mice (Fig. S3 E). Altogether, these results suggest that follicular epithelial HrasG12V/+ activation nonautonomously activates the surrounding mesenchyme through alterations to both molecular signatures and cellular behaviors, even in the absence of detectable macroscopic phenotypes.
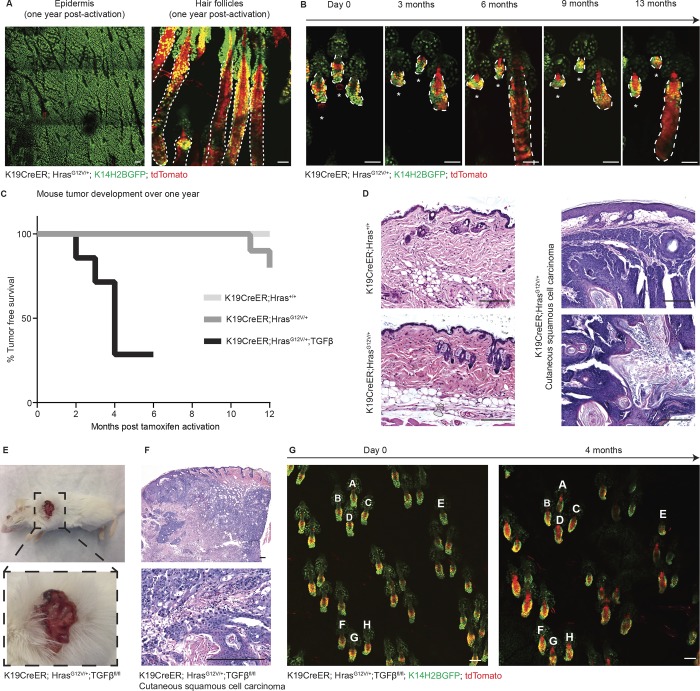
We considered that the observed tolerance to the expanded HrasG12V/+ pool into homeostatic follicular programs might be lost over time. To address this, we tracked mice for 1 yr after activation of HrasG12V/+ and discovered that, surprisingly, the skin remained normal, both at the macroscopic and microscopic levels. HrasG12V/+ cells remained present yet restricted to the HFSC compartment, where they were initially activated, and both the follicles and epidermis retained their normal structures (Fig. 3 A). We next questioned whether the targeted dermal papilla ablations in rest phase follicles described in Fig. 1 E might ultimately lead to aberrant growth given enough time. To investigate this, we tracked these ablated follicles for 1 yr and discovered that they never developed aberrant growth or even cycled despite the persistent presence of Hras mutant cells (Fig. 3 B). Strikingly, the follicles remained normally structured, and the mutant cells remained restricted to the HFSC niche where they were initially activated. Despite their capacity for enhanced proliferation, Hras mutant cells are not sufficient to promote follicular growth without mesenchymal cues, yet they are able to remain latent in the tissue, even 1 yr after activation and ablation.
Figure 3.
Follicular tolerance of Hras is maintained long-term in uninjured tissue. All epithelial nuclei are in green (K14H2BGFP), and recombined cells are in red (tdTomato). (A) Two-photon images of the epidermis (left) and hair follicles (right) of a K19CreER;HrasG12V/+;K14H2BGFP;tdTomato mouse 1 yr after activation reveal that the skin appears grossly normal and the mutant cells (in red) are still present yet contained to the HFSC niche. (B) Two-photon revisit images of the same K19CreER;HrasG12V/+ follicles immediately after dermal papilla ablation (day 0, labeled with an asterisk) and at the indicated time points up to 13 mo. Mutant cells (in red) remain present yet contained to the HFSC niche and the ablated follicles never grow unlike their unablated neighbor. (C) Tumor emergence graph for K19CreER;Hras+/+ (n = 0/9), K19CreER;HrasG12V/+ (n = 2/10, well differentiated cSCCs), and K19CreER;HrasG12V/+;TGFβfl/fl (n = 5/7, poorly differentiated cSCCs) mice monitored over the course of 1 yr after activation. (D) Hematoxylin and eosin images of K19CreER;Hras+/+ (top left) and K19CreER;HrasG12V/+ (bottom left) clinically normal back skin from 1-yr-old mice. On the right are representative images of a tumor developed in a K19CreER;HrasG12V/+ mouse 1 yr after activation. The staining reveals verrucous squamous proliferation with acanthosis, hyperkeratosis and mild atypia consistent with well-differentiated squamous cell carcinoma. (E) Photograph of a cSCC tumor from a K19CreER;HrasG12V/+;TGFβfl/fl mouse 4 mo after tamoxifen activation. (F) Hematoxylin and eosin images revealing islands of atypical keratinocytes within the dermis extending into subcutaneous fat with marked atypia and associated mixed inflammatory infiltrate consistent with moderately differentiated, necrotic invasive squamous cell carcinoma. (G) Two-photon images of the ear skin from the same mouse from E and F at day 0 and 4 mo after tamoxifen activation reveal normal follicular architecture despite the persistent presence of mutant cells (tdTomato+). Follicles are labeled A–H as landmarks. Scale bars in two-photon images represent 50 µm. Scale bars in histology images represent 200 µm.
Although the follicle was normally structured, HrasG12V/+ cells remained active, as demonstrated by follicular hyperthickening during the growth phase at 1 yr, which was continuously corrected as the follicle regressed back into quiescence (Fig. S4, A and B). Persistent follicular tolerance of HrasG12V/+ even 1 yr after activation sharply contrasts the consequence of activating this same mutant allele in the upper noncycling epithelial tissue compartments. Live imaging revisits revealed that similar to in the hair follicle, Hras mutant cells in the epidermis expanded over time and outcompeted their wild-type neighbors (Fig. S5). However, unlike in the hair follicle, we observed that, consistent with previous studies, Ras activation in the epidermis compartment resulted in the development of hyperplasia and ultimately benign skin tumors (i.e., papillomas) within 3–5 mo in 100% of mice (Bailleul et al., 1990; Chen et al., 2009; Lapouge et al., 2011; Ratushny et al., 2012; Beronja et al., 2013; Lowry et al., 2016; Brown et al., 2017; Fig. S4, C and D). In comparison, in mice with follicular activation of Hras, in spite of the persistent presence of active HrasG12V/+ cells, 80% of the mice remained tumor free at 1 yr after activation (Fig. 3, C and D).
These findings demonstrate that the hair follicle has a unique ability to suppress Hras-oncogenic growth when compared with the upper noncycling epithelium; however, we hypothesized that follicular tolerance may be overcome when faced with further insults. To test this, we introduced additional challenges to the HrasG12V/+ model beginning with loss of TGFβ signaling (TGFβfl/fl). When combined with Ras activation, loss of TGFβ signaling has been shown to accelerate tumorigenesis and promote the malignant transformation of skin tumors (Lu et al., 2006; Guasch et al., 2007; Mordasky Markell et al., 2010). Consistent with previous studies, we observed that the combination of both HrasG12V/+ and TGFβfl/fl mutations in the HFSCs ultimately resulted in the development of malignant cutaneous squamous cell carcinoma (cSCC; Fig. 3, C, E, and F). Importantly, while the mutations were activated in hair follicles throughout the entire animal, the incidence of cSCC development was low; mice only developed one tumor each, typically at sites of frequent grooming/scratching, consistent with the findings that wounding promotes Ras-driven tumorigenesis (Bailleul et al., 1990; Cannon et al., 1997; Oki-Idouchi and Lorenzo, 2007; Diez et al., 2009; Ksionda et al., 2013; Berger et al., 2014). Live imaging confirmed that these tumors arose at sites of injury as the epidermis around the tumor was composed of tdTomato+ cells, indicating their exit from their normally restricted follicular niche, behavior that is triggered by epidermal injury (Video 2; Ito et al., 2005; Levy et al., 2005). Meanwhile, the skin away from the tumor appeared clinically normal, demonstrating that a significant level of tolerance is maintained despite additional cell-autonomous cancer-driving lesions (Fig. 3 G). These results demonstrate that while follicles are highly resistant to aberrant growth, a superficial injurious stimulus can promote tumorigenesis.
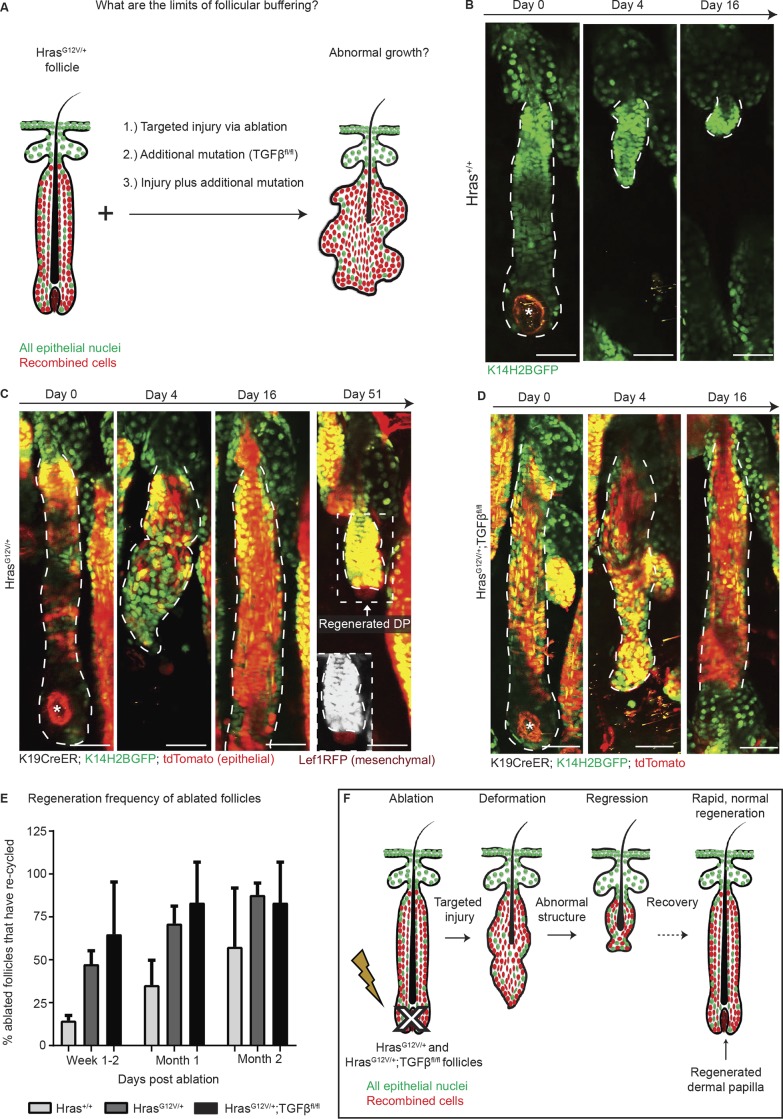
We next questioned whether this breakdown of follicular tolerance observed within our restricted-activation model was due to the injury itself or instead due to the exit of Hras mutant cells from the follicular niche into the epidermis. As previously mentioned, injury of the epidermis has been demonstrated to accelerate Ras-driven tumorigenesis and influence the degree of malignancy of the resultant tumor (Bailleul et al., 1990; Cannon et al., 1997; Oki-Idouchi and Lorenzo, 2007; Diez et al., 2009; Ksionda et al., 2013; Berger et al., 2014). We aimed to determine whether a similar transformation mechanism occurs after follicular injury or if instead the follicular buffering effect of mutant cells extends to recovering from an injurious event. To accomplish this, we performed targeted injury to the growing hair follicle, reasoning that this additional stimulus might prompt HrasG12V/+ cells, which are already proliferating during the growth phase, to proliferate out of homeostatic control (Fig. 4 A). Unlike during the rest phase ablation described in Fig. 1, which specifically targets dermal papilla stem cells, the growth phase ablations spare the dermal papilla stem cells, which at this point in cycling reside outside the follicle. Surprisingly, after broad ablation of the hair follicle bulb, which contains both mesenchymal dermal papilla cells and surrounding epithelial matrix progenitor cells, HrasG12V/+ follicles do not aberrantly grow; rather, they recover and regenerate a properly structured follicle and dermal papilla. Strikingly, this regeneration of the epithelium and mesenchyme occurs even more rapidly than in wild-type follicles subjected to the same injury, despite the fact that mutant cells remain predominant in the resolved follicle (Fig. 4, B and C), further supporting the model of enhanced mesenchymal activity observed in Fig. 2. Surprisingly, targeted follicular injury in double-mutant follicles also resulted in rapid, normal regeneration and no aberrant growth, similar to the HrasG12V/+ single mutants (Fig. 4, D and E). These results demonstrate that the Hras/TGFβ mutant follicles retain the remarkable ability to properly regenerate even in the face of targeted follicular injury, while the same mutations can promote tumorigenesis if subjected to superficial injury (Fig. 4 F).
Figure 4.
Follicles are resistant to oncogenic growth, even in the face of additional tumor promoting insults. All epithelial nuclei are in green (K14H2BGFP), and recombined cells are in red (tdTomato). (A) Cartoon schematic detailing the various approaches for testing the buffering effect. (B–D) Two-photon revisits show that targeted ablation of the bulb region in Hras+/+ (B), HrasG12V/+ (C), and HrasG12V/+;TGFβfl/fl (D) follicles during the growth phase results in follicular deformation and subsequent regeneration. (E) Mutant follicles rapidly regenerate after injury, and within 1 mo after ablation, ∼75% of mutant follicles are cycling again compared with only 35% of wild-type follicles (n = 23 Hras+/+ follicles across two mice, n = 34 HrasG12V/+ across four mice, n = 19 HrasG12V/+;TGFβfl/fl follicles across three mice). All error bars indicate standard deviation. (F) Cartoon summary depicting rapid, normal regeneration of HrasG12V/+ after injury. All scale bars represent 50 µm.
Altogether, our results indicate that the hair follicle has a unique ability to cope with Ras-activated cells. This organ is able to integrate the mutant epithelial cells while remaining clinically normal in spite of the higher regenerative potential of both the epithelium and the mesenchyme. Furthermore, follicular tolerance is maintained even in the face of additional insults that increase the oncogenic potential of the epithelium. In contrast, tolerance breaks down when mutant cells exit the follicular niche, such as after superficial injury. Once out of the follicular niche, Hras mutant cells can no longer be controlled and contained through hair regeneration programs. As we observed, the same mutant cells that are normally channeled into functional growth (enhanced tissue cycling) in the hair follicle cause aberrant growth (skin tumors) in the epidermis. Cellular and structural differences as well as differences regarding the mesenchymal niche are likely at the root of the dramatically different outcomes between the consequences of Hras activation in these two niches. Thus, the hair follicle emerges as a paradigm for principles that could be applied to other organs that have a lesser ability to tolerate mutant cells. Future studies may be used to investigate whether the nonautonomous impact of the mutant cells on the surrounding nonepithelial cellular populations influence these differing outcomes. This will enhance our understanding about whether the manipulation of certain cell types or signaling pathways can enable and/or enhance the ability of other epithelial tissues to also suppress oncogenic growth.
Materials and methods
Mice
K19CreER (Means et al., 2008) or K14CreER mice (Vasioukhin et al., 1999) were bred to HrasG12V/+ mice (Chen et al., 2009) to generate K19CreER;HrasG12V/+ and K14CreER;HrasG12V/+ mice, respectively. To generate K19CreER;HrasG12V/+;TGFβfl/fl mice, K19CreER;HrasG12V/+ mice were crossed with TGFβfl/fl mice, which were obtained from V. Kaartinen (University of Michigan, Ann Arbor, MI; Larsson et al., 2003). K14H2BGFP (Tumbar et al., 2004) transgenic mice were used for visualization of epithelial cells with the two-photon microscope. The reporter line used to visualize Cre recombination, Gt(ROSA)26Sortm14(CAG-tdTomato) (tdTomato reporter line; Madisen et al., 2010), was obtained from The Jackson Laboratory. To label and visualize mesenchymal cells of the skin, including the dermal papillae and fibroblasts, lymphoid enhancer-binding factor 1–RFP mice (Tg(Lef1RFP); Rendl et al., 2005) were used. Mice of either gender from experimental and control groups were randomly selected for live imaging experiments. No blinding was done. All procedures involving animal subjects were performed under the approval of the Institutional Animal Care and Use Committee of the Yale School of Medicine. All mice that developed tumors were euthanized when maximal tumor dimensions of 1 cm3, the size permitted by the committee, were reached.
Tamoxifen induction of mice
K19CreER;HrasG12V/+, K19CreER;HrasG12V/+;TGFβfl/fl, and K14CreER;HrasG12V/+ mice were administered a single dose of 2 mg tamoxifen at postnatal day 20 (P20) for all experiments. All time courses began 3 d after tamoxifen induction (day 0).
Lentiviral production, in vitro infections, and in vivo viral transductions
Lentiviral production, concentration, and in vivo viral transduction were performed as previously described (Endo et al., 2008; Beronja et al., 2010, 2013).
In vivo imaging
Imaging procedures followed those previously described (Rompolas et al., 2012; Deschene et al., 2014; Mesa et al., 2015; Pineda et al., 2015; Brown et al., 2017). A LaVision TriM Scope II (LaVision Biotec) microscope equipped with a Chameleon Vision II (Coherent) two-photon laser (940 nm for live imaging of GFP and 880 nm for whole mounts) and a Chameleon Discovery (Coherent) two-photon laser (1,120 nm for live imaging of tdTomato) was used to acquire z-stacks of ∼90–200 µm (3-µm serial optical sections) using a 20× water-immersion lens (numerical aperture 1.0; Olympus), scanned with a field of view of 0.3–0.5 mm2 at 800 Hz. Mice were imaged at time points after tamoxifen treatment, viral transduction, or laser ablation as indicated. To revisit the same hair follicles over time, organizational clusters of hair follicles and vasculature were used as landmarks to reidentify the region.
Laser ablations
Targeted two-photon laser ablations were performed as previously described (Mesa et al., 2015; Pineda et al., 2015; Brown et al., 2017). Briefly, during imaging, a 900-nm laser beam was used to scan the target bulb region and 60–80% laser power was applied for ∼1 s. Ablation parameters were adjusted accordingly depending on the depth of the targeted follicle. For the rest phase ablations, the dermal papilla stem cells were directly and selectively targeted. For the growth phase ablations, the bulb region, consisting of the dermal papilla and surrounding epithelial matrix cells and progenitors, were targeted.
Image analysis
Raw image stacks were imported into ImageJ (National Institutes of Health) or Imaris (BitPlane Scientific Software) for analysis. To quantify follicular bulge width, the average of five measurements randomly taken across the bulge of each follicle was calculated using ImageJ. For tdTomato follicle volumetric analysis, two-photon images were imported into Imaris software. A 3D volumetric mask was created around the entire HFSC compartment by using the draw tool to trace around the follicle through each Z-step. Next, a mask was specifically isolated around the red (tdTomato) channel, and the voxel was adjusted to eliminate noise. The tdTomato mask was then expressed as a percentage of the total follicle volume. For cell behavior analysis, mitotic events and apoptotic events were identified based on cellular morphology from two-photon images of follicles. To perform follicular cycling staging, follicles were analyzed and classified as being in a particular stage of the hair cycle based on stereotypical tissue stage morphology using two-photon images.
Western blot analysis
Western blots were performed on whole skin tissue according to standard protocols using BioRad equipment and reagents. The following rabbit primary antibodies were used at the given concentrations: phospho-p44/42 MAPK (Erk1/2; Thr202/Tyr204; 1:1,000, 9101; Cell Signaling), phospho-PI3K (1:300, 4228; Cell Signaling), and GAPDH (1:1,000, 2118; Cell Signaling). An anti-rabbit IgG HRP (1:1,000, 7074; Cell Signaling) secondary antibody was used.
Hair growth assay
The backs of three K19CreER;Hras+/+ and four K19CreER;HrasG12V/+ mice were shaved 5 wk after tamoxifen activation and monitored for 3 wk to examine the timing of hair regrowth.
Histology
Back skin and skin tumors were 10% formalin fixed, paraffin embedded and used for histological analysis. Paraffin-embedded skin tissues were cut at 5 µm. To display skin morphology, sections were stained with hematoxylin and eosin using standard protocols. Images were taken using an Olympus BX61 microscope equipped with a SPOT flex 15.2 64-Mp shifting pixel camera; 4×, 10×, 20×, and 40× objectives; and SPOT v 5.2 software.
Cell sorting and preparation for RNA-seq
Three K19CreER;Hras+/+;Td tomato+/−/PDGFH2BGFP and five K19CreER;HrasG12V/+;Td tomato+/−;PDGFH2BGFP mice were euthanized for the cell sorting experiment 2 mo after P20 tamoxifen activation. Skin was harvested, digested using both dispase and collagenase, filtered, stained with a CD45–Alexa Fluor 647 antibody (1:200, 103124; BioLegend) and resuspended in 1% FBS in 1× PBS for the sort. Fibroblasts were sorted using a BD Biosciences FACS Aria II based on being CD45–Alexa Fluor 647 negative (immune cells), tdTomato negative (epithelial cells), and GFP positive (fibroblasts) straight into RNA lysis buffer. RNA was prepared from sorted cells and isolated using RNAeasy Micro Kit (74004; Qiagen) according to the manufacturer’s instructions.
RNA-seq quality control
Total RNA quality was determined by estimating the A260/A280 and A260/A230 ratios by nanodrop. RNA integrity was determined by running an Agilent Bioanalyzer gel to measures the ratio of the ribosomal peaks.
RNA-seq library preparation
mRNA was purified from ∼200 ng of total RNA with oligo-dT beads and sheared by incubation at 94°C (Kapa mRNA Hyper Prep). Following first-strand synthesis with random primers, second-strand synthesis was performed with dUTP for generating strand-specific sequencing libraries. The cDNA library was then end repaired and A tailed, adapters were ligated, and second-strand digestion was performed by uracil-DNA-glycosylase. Indexed libraries that met appropriate cutoffs for both were quantified by quantitative RT-PCR using a commercially available kit (KAPA Biosystems) and insert size distributions were determined with the LabChip GX or Agilent Bioanalyzer. Samples with a yield of ≥0.5 ng/µl were used for sequencing.
Flow cell preparation and sequencing
Sample concentrations were normalized to 10 nM and loaded onto an Illumina NovaSeq flow cell at a concentration that yielded 25 million passing filter clusters per sample. Samples were sequenced using 100-bp paired-end sequencing on an Illumina HiSeq NovaSeq according to Illumina protocols. The 8-bp dual index was read during additional sequencing reads that automatically followed the completion of read 1. Data generated during sequencing runs were simultaneously transferred to the Yale Center for Genome Analysis high-performance computing cluster. A positive control (prepared bacteriophage Phi X library) provided by Illumina was spiked into every lane at a concentration of 0.3% to monitor sequencing quality in real time.
RNA-seq data analysis
Samples were analyzed using Partek Flow software. In short, mRNA contaminants were filtered using Bowtie 2, and reads were aligned using STAR. The samples were then normalized to generate counts per million, which were then transformed using DESeq2 (Love et al., 2014) and used for downstream analyses. A heatmap was generated from a filtered list of features with a false discovery rate (FDR)–adjusted P value of <0.1, followed by hierarchical clustering. A volcano plot was created from the unfiltered feature list, with dotted lines denoting an FDR-adjusted P value of 0.1. Fold change of >1.5 or <1.5 is also depicted on the graph. Gene set enrichment analysis was performed on a ranked gene list calculated as the sign (either + or −) of log2(fold change) × −log10(FDR-adjusted P value) and performed using default parameters (Subramanian et al., 2005). This calculated ranked gene list was compared with a list of KEGG pathway gene sets (Kanehisa and Goto, 2000; Kanehisa et al., 2017, 2019) for determination of pathway enrichment, with q value scores of <0.05 deemed as significant.
No statistical methods were used to predetermine sample size. The experiments were not randomized. All investigators were not blinded to allocation during experiments and outcome assessment.
Data availability statement
The authors declare that the main data supporting the findings of this study are available within the paper and its supplemental information.
Online supplemental material
The supplemental figures provide additional data supporting the impact of Hras activation on hair growth (Fig. S1), the viral approach (Fig. S2), the nonautonomous impact of epithelial Hras on the surrounding mesenchyme (Fig. S3), the comparison in phenotypes between Hras activation in the epidermis versus the hair follicle (Fig. S4), and the competition between wild-type and Hras mutant cells in the epidermis (Fig. S5). The supplemental videos include high-resolution z-stacks of mitotic figures in the hair follicle (Video 1) and a tumor at a site of injury (Video 2).
Supplementary Material
Acknowledgments
We thank M. Manning, D. May, and A. Andersen for critical feedback on the manuscript. We thank G. Gu (Vanderbilt University, Nashville, TN) for the K19CreER mice and E. Fuchs (Rockefeller University, New York, NY) for the K14H2BGFP mice. We thank K. Bilguvar, S. Mane, and the Yale Center for Genome Analysis for library preparation and deep sequencing. We thank R. Sundaram for her experimental guidance.
This work is supported by The New York Stem Cell Foundation, the Edward Mallinckrodt Jr. Foundation, the Glenn Foundation for Medical Research, the Howard Hughes Medical Institute Scholar award, and the National Institute of Arthritis and Musculoskeletal and Skin Disease, National Institutes of Health (grants 2R01AR063663-06A1, 1R01AR072668-01, and 5R01AR067755-02). C.M. Pineda was supported by the National Cancer Institute, National Institutes of Health (award number F31CA206419). N.R. Fons was supported by the Yale Cancer Biology Training Program via the Yale Cancer Center and Yale School of Medicine. T. Xin was supported by the James Hudson Brown-Alexander Brown Coxe postdoctoral fellowship. K. Tai was supported by the Yale College First-Year Summer Research Fellowship in the Sciences. E. Marsh was supported in part by the National Institutes of Health (grant T32 GM007499). D.X. Nguyen was funded by the National Institutes of Health (grant R01CA191489) and has also received research funding from AstraZeneca and Leidos Biomedical Research. These funders had no role in this manuscript.
The authors declare no competing financial interests.
Author contributions: C.M. Pineda and V. Greco designed experiments and analyzed data. C.M. Pineda performed two-photon imaging, manipulation and analysis, laser ablations, protein preparation, western blot analysis, mouse genetics, and viral construct generation and injections. D.G. Gonzalez assisted with mouse work and genetics, drug administration, imaging, and Imaris analysis. C. Matte-Martone assisted with drug administration, mouse work, viral injections, and RNA-seq preparatory work and analysis. J. Boucher assisted with mouse genetics and tissue processing. E. Lathrop assisted with mouse genetics. S. Gallini assisted with molecular analysis. N.R. Fons assisted with RNA-seq analysis. T. Xin assisted with molecular analysis and mouse work. K. Tai assisted with molecular analysis. E. Marsh assisted with RNA-seq analyses. D.X. Nguyen assisted with RNA-seq analysis. K.C. Suozzi assisted with clinical diagnosis of histological images. S. Beronja provided viral reagents (Beronja et al., 2010), protocols, and training as well as extended experimental discussions throughout this project development.
References
- Alcolea M.P., and Jones P.H.. 2015. Cell competition: winning out by losing notch. Cell Cycle. 14:9–17. 10.4161/15384101.2014.988027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anglesio M.S., Papadopoulos N., Ayhan A., Nazeran T.M., Noë M., Horlings H.M., Lum A., Jones S., Senz J., Seckin T., et al. . 2017. Cancer-associated mutations in endometriosis without cancer. N. Engl. J. Med. 376:1835–1848. 10.1056/NEJMoa1614814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailleul B., Surani M.A., White S., Barton S.C., Brown K., Blessing M., Jorcano J., and Balmain A.. 1990. Skin hyperkeratosis and papilloma formation in transgenic mice expressing a ras oncogene from a suprabasal keratin promoter. Cell. 62:697–708. 10.1016/0092-8674(90)90115-U [DOI] [PubMed] [Google Scholar]
- Berger F., Geddert H., Faller G., Werner M., and Dimmler A.. 2014. Pattern of TGFbeta receptor 1 expression differs between kras-mutated keratoacanthomas and squamous cell carcinomas of the skin. Pathol. Res. Pract. 210:596–602. 10.1016/j.prp.2014.05.006 [DOI] [PubMed] [Google Scholar]
- Beronja S., Livshits G., Williams S., and Fuchs E.. 2010. Rapid functional dissection of genetic networks via tissue-specific transduction and RNAi in mouse embryos. Nat. Med. 16:821–827. 10.1038/nm.2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beronja S., Janki P., Heller E., Lien W.H., Keyes B.E., Oshimori N., and Fuchs E.. 2013. RNAi screens in mice identify physiological regulators of oncogenic growth. Nature. 501:185–190. 10.1038/nature12464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S., Pineda C.M., Xin T., Boucher J., Suozzi K.C., Park S., Matte-Martone C., Gonzalez D.G., Rytlewski J., Beronja S., and Greco V.. 2017. Correction of aberrant growth preserves tissue homeostasis. Nature. 548:334–337. 10.1038/nature23304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon R.E., Spalding J.W., Trempus C.S., Szczesniak C.J., Virgil K.M., Humble M.C., and Tennant R.W.. 1997. Kinetics of wound-induced v-Ha-ras transgene expression and papilloma development in transgenic Tg.AC mice. Mol. Carcinog. 20:108–114. [DOI] [PubMed] [Google Scholar]
- Chen X., Mitsutake N., LaPerle K., Akeno N., Zanzonico P., Longo V.A., Mitsutake S., Kimura E.T., Geiger H., Santos E., et al. . 2009. Endogenous expression of Hras(G12V) induces developmental defects and neoplasms with copy number imbalances of the oncogene. Proc. Natl. Acad. Sci. USA. 106:7979–7984. 10.1073/pnas.0900343106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschene E.R., Myung P., Rompolas P., Zito G., Sun T.Y., Taketo M.M., Saotome I., and Greco V.. 2014. β-Catenin activation regulates tissue growth non-cell autonomously in the hair stem cell niche. Science. 343:1353–1356. 10.1126/science.1248373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez F.R., Garrido A.A., Sharma A., Luke C.T., Stone J.C., Dower N.A., Cline J.M., and Lorenzo P.S.. 2009. RasGRP1 transgenic mice develop cutaneous squamous cell carcinomas in response to skin wounding: potential role of granulocyte colony-stimulating factor. Am. J. Pathol. 175:392–399. 10.2353/ajpath.2009.090036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M., Zoltick P.W., Peranteau W.H., Radu A., Muvarak N., Ito M., Yang Z., Cotsarelis G., and Flake A.W.. 2008. Efficient in vivo targeting of epidermal stem cells by early gestational intraamniotic injection of lentiviral vector driven by the keratin 5 promoter. Mol. Ther. 16:131–137. 10.1038/sj.mt.6300332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frede J., Greulich P., Nagy T., Simons B.D., and Jones P.H.. 2016. A single dividing cell population with imbalanced fate drives oesophageal tumour growth. Nat. Cell Biol. 18:967–978. 10.1038/ncb3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco V., Chen T., Rendl M., Schober M., Pasolli H.A., Stokes N., Dela Cruz-Racelis J., and Fuchs E.. 2009. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell. 4:155–169. 10.1016/j.stem.2008.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guasch G., Schober M., Pasolli H.A., Conn E.B., Polak L., and Fuchs E.. 2007. Loss of TGFbeta signaling destabilizes homeostasis and promotes squamous cell carcinomas in stratified epithelia. Cancer Cell. 12:313–327. 10.1016/j.ccr.2007.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M., Liu Y., Yang Z., Nguyen J., Liang F., Morris R.J., and Cotsarelis G.. 2005. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat. Med. 11:1351–1354. 10.1038/nm1328 [DOI] [PubMed] [Google Scholar]
- Jahoda C.A.B., Horne K.A., and Oliver R.F.. 1984. Induction of hair growth by implantation of cultured dermal papilla cells. Nature. 311:560–562. 10.1038/311560a0 [DOI] [PubMed] [Google Scholar]
- Kanehisa M., and Goto S.. 2000. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Furumichi M., Tanabe M., Sato Y., and Morishima K.. 2017. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45(D1):D353–D361. 10.1093/nar/gkw1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Sato Y., Furumichi M., Morishima K., and Tanabe M.. 2019. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 47(D1):D590–D595. 10.1093/nar/gky962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A.M., Brash D.E., Jones P.H., and Simons B.D.. 2010. Stochastic fate of p53-mutant epidermal progenitor cells is tilted toward proliferation by UV B during preneoplasia. Proc. Natl. Acad. Sci. USA. 107:270–275. 10.1073/pnas.0909738107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksionda O., Limnander A., and Roose J.P.. 2013. RasGRP Ras guanine nucleotide exchange factors in cancer. Front. Biol. (Beijing). 8:508–532. 10.1007/s11515-013-1276-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Sukumar S., and Barbacid M.. 1990. Activation of ras oncogenes preceding the onset of neoplasia. Science. 248:1101–1104. 10.1126/science.2188364 [DOI] [PubMed] [Google Scholar]
- Lapouge G., Youssef K.K., Vokaer B., Achouri Y., Michaux C., Sotiropoulou P.A., and Blanpain C.. 2011. Identifying the cellular origin of squamous skin tumors. Proc. Natl. Acad. Sci. USA. 108:7431–7436. 10.1073/pnas.1012720108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson J., Blank U., Helgadottir H., Björnsson J.M., Ehinger M., Goumans M.J., Fan X., Levéen P., and Karlsson S.. 2003. TGF-beta signaling-deficient hematopoietic stem cells have normal self-renewal and regenerative ability in vivo despite increased proliferative capacity in vitro. Blood. 102:3129–3135. 10.1182/blood-2003-04-1300 [DOI] [PubMed] [Google Scholar]
- Laurie C.C., Laurie C.A., Rice K., Doheny K.F., Zelnick L.R., McHugh C.P., Ling H., Hetrick K.N., Pugh E.W., Amos C., et al. . 2012. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat. Genet. 44:642–650. 10.1038/ng.2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy V., Lindon C., Harfe B.D., and Morgan B.A.. 2005. Distinct stem cell populations regenerate the follicle and interfollicular epidermis. Dev. Cell. 9:855–861. 10.1016/j.devcel.2005.11.003 [DOI] [PubMed] [Google Scholar]
- Ling G., Persson A., Berne B., Uhlén M., Lundeberg J., and Ponten F.. 2001. Persistent p53 mutations in single cells from normal human skin. Am. J. Pathol. 159:1247–1253. 10.1016/S0002-9440(10)62511-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry W.E., Flores A., and White A.C.. 2016. Exploiting mouse models to study ras-induced cutaneous squamous cell carcinoma. J. Invest. Dermatol. 136:1543–1548. 10.1016/j.jid.2016.03.017 [DOI] [PubMed] [Google Scholar]
- Lu S.L., Herrington H., Reh D., Weber S., Bornstein S., Wang D., Li A.G., Tang C.F., Siddiqui Y., Nord J., et al. . 2006. Loss of transforming growth factor-β type II receptor promotes metastatic head-and-neck squamous cell carcinoma. Genes Dev. 20:1331–1342. 10.1101/gad.1413306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R., et al. . 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13:133–140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I., Roshan A., Gerstung M., Ellis P., Van Loo P., McLaren S., Wedge D.C., Fullam A., Alexandrov L.B., Tubio J.M., et al. . 2015. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 348:880–886. 10.1126/science.aaa6806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Means A.L., Xu Y., Zhao A., Ray K.C., and Gu G.. 2008. A CK19(CreERT) knockin mouse line allows for conditional DNA recombination in epithelial cells in multiple endodermal organs. Genesis. 46:318–323. 10.1002/dvg.20397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesa K.R., Rompolas P., Zito G., Myung P., Sun T.Y., Brown S., Gonzalez D.G., Blagoev K.B., Haberman A.M., and Greco V.. 2015. Niche-induced cell death and epithelial phagocytosis regulate hair follicle stem cell pool. Nature. 522:94–97. 10.1038/nature14306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordasky Markell L., Pérez-Lorenzo R., Masiuk K.E., Kennett M.J., and Glick A.B.. 2010. Use of a TGFbeta type I receptor inhibitor in mouse skin carcinogenesis reveals a dual role for TGFbeta signaling in tumor promotion and progression. Carcinogenesis. 31:2127–2135. 10.1093/carcin/bgq191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai K., Skrupskelyte G., Piedrafita G., Hall M., Kostiou V., Ong S.H., Nagy T., Cagan A., Goulding D., Klein A.M., et al. . 2018. Epidermal tissue adapts to restrain progenitors carrying clonal p53 mutations. Cell Stem Cell. 23:687–699.e8. 10.1016/j.stem.2018.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki-Idouchi C.E., and Lorenzo P.S.. 2007. Transgenic overexpression of RasGRP1 in mouse epidermis results in spontaneous tumors of the skin. Cancer Res. 67:276–280. 10.1158/0008-5472.CAN-06-3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda C.M., Park S., Mesa K.R., Wolfel M., Gonzalez D.G., Haberman A.M., Rompolas P., and Greco V.. 2015. Intravital imaging of hair follicle regeneration in the mouse. Nat. Protoc. 10:1116–1130. 10.1038/nprot.2015.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior I.A., Lewis P.D., and Mattos C.. 2012. A comprehensive survey of Ras mutations in cancer. Cancer Res. 72:2457–2467. 10.1158/0008-5472.CAN-11-2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratushny V., Gober M.D., Hick R., Ridky T.W., and Seykora J.T.. 2012. From keratinocyte to cancer: the pathogenesis and modeling of cutaneous squamous cell carcinoma. J. Clin. Invest. 122:464–472. 10.1172/JCI57415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendl M., Lewis L., and Fuchs E.. 2005. Molecular dissection of mesenchymal-epithelial interactions in the hair follicle. PLoS Biol. 3:e331 10.1371/journal.pbio.0030331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P., Deschene E.R., Zito G., Gonzalez D.G., Saotome I., Haberman A.M., and Greco V.. 2012. Live imaging of stem cell and progeny behaviour in physiological hair-follicle regeneration. Nature. 487:496–499. 10.1038/nature11218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sennett R., and Rendl M.. 2012. Mesenchymal-epithelial interactions during hair follicle morphogenesis and cycling. Semin. Cell Dev. Biol. 23:917–927. 10.1016/j.semcdb.2012.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ståhl P.L., Stranneheim H., Asplund A., Berglund L., Pontén F., and Lundeberg J.. 2011. Sun-induced nonsynonymous p53 mutations are extensively accumulated and tolerated in normal appearing human skin. J. Invest. Dermatol. 131:504–508. 10.1038/jid.2010.302 [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumar S., McKenzie K., and Chen Y.. 1995. Animal models for breast cancer. Mutat. Res. 333:37–44. 10.1016/0027-5107(95)00129-8 [DOI] [PubMed] [Google Scholar]
- Tumbar T., Guasch G., Greco V., Blanpain C., Lowry W.E., Rendl M., and Fuchs E.. 2004. Defining the epithelial stem cell niche in skin. Science. 303:359–363. 10.1126/science.1092436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasioukhin V., Degenstein L., Wise B., and Fuchs E.. 1999. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc. Natl. Acad. Sci. USA. 96:8551–8556. 10.1073/pnas.96.15.8551 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the main data supporting the findings of this study are available within the paper and its supplemental information.