Lipolysis and lipophagy are thought to be distinct mechanisms for lipid droplet catabolism. In hepatocytes, Schott et al. report that these pathways may operate synergistically: the lipolysis enzyme ATGL targets large LDs upstream of lipophagy, which is restricted to small LDs.
Abstract
Lipid droplet (LD) catabolism in hepatocytes is mediated by a combination of lipolysis and a selective autophagic mechanism called lipophagy, but the relative contributions of these seemingly distinct pathways remain unclear. We find that inhibition of lipolysis, lipophagy, or both resulted in similar overall LD content but dramatic differences in LD morphology. Inhibition of the lipolysis enzyme adipose triglyceride lipase (ATGL) resulted in large cytoplasmic LDs, whereas lysosomal inhibition caused the accumulation of numerous small LDs within the cytoplasm and degradative acidic vesicles. Combined inhibition of ATGL and LAL resulted in large LDs, suggesting that lipolysis targets these LDs upstream of lipophagy. Consistent with this, ATGL was enriched in larger-sized LDs, whereas lipophagic vesicles were restricted to small LDs as revealed by immunofluorescence, electron microscopy, and Western blot of size-separated LDs. These findings provide new evidence indicating a synergistic relationship whereby lipolysis targets larger-sized LDs to produce both size-reduced and nascently synthesized small LDs that are amenable for lipophagic internalization.
Introduction
Lipid droplets (LDs) are ubiquitous fat-storage organelles that serve as readily accessible reservoirs of high-energy substrates used for β-oxidation within mitochondria. In the parenchymal cells of the liver (hepatocytes), the aberrant accumulation of LDs is the hallmark of steatosis, a key pathological feature of nonalcoholic fatty liver disease, obesity, and metabolic syndrome. This steatosis is viewed as an imbalance between the process of lipid storage and utilization. Thus, an understanding of the cellular machinery required to synthesize and catabolize these organelles is of great interest and an area of intense study. Currently, there are two central processes known to mediate the breakdown of triacylglycerol (TAG) stored within LDs for subsequent oxidation within mitochondria: cytosolic lipolysis and autophagy. In the process of lipolysis, cytosolic lipases including adipose triglyceride lipase (ATGL), hormone-sensitive lipase (HSL), and monoglyceride lipase (MGL) act sequentially to catalyze the liberation of the three fatty acid (FA) moieties comprising the parent TAG molecule (Vaughan et al., 1964; Jenkins et al., 2004; Villena et al., 2004; Zimmermann et al., 2004). The free FAs (FFAs) released by this lipolytic process are presumed to provide substrates for mitochondrial β-oxidation or act as potent signaling molecules for a variety of cellular processes; alternatively, these FAs can be reesterified back into TAG for storage (Kennedy and Lehninger, 1949; Edens et al., 1990; Ong et al., 2011; Khan et al., 2015).
In addition to the actions of the cytoplasmic lipases, it is now established that the catabolic process of autophagy can be used to mobilize LDs during periods of nutrient stress (Singh et al., 2009). Autophagy involves a highly orchestrated network of proteins that act in concert to selectively sequester intracellular contents within double-membrane structures known as autophagosomes. Fusion of autophagosomes with components of the terminal endocytic pathway (e.g., lysosomes) results in the recycling of autophagic cargo into macromolecular components within structures known as autolysosomes. In a highly selective form of LD-targeted autophagy, referred to as “lipophagy,” the specific turnover of LDs occurs through the action of acid lipases deposited into the autolysosome (Kaur and Debnath, 2015). Lipophagy thus represents an alternative to conventional cytosolic lipase-driven LD breakdown (Weidberg et al., 2009; Singh and Cuervo, 2012; Liu and Czaja, 2013; Schulze et al., 2017).
The relative utilization of lipolysis versus lipophagy by hepatocytes and other cells is presently unclear, as manipulation of either of these catabolic processes in mouse models can ultimately result in fatty liver (Singh et al., 2009; Ong et al., 2011). Whether lipolysis and lipophagy occur independently of each other or in tandem is an area of current investigation; indeed, an understanding of the crosstalk occurring between these pathways is only now beginning to emerge.
Evidence suggests that the size of the cargo targeted for degradation may be an important determinant in the capacity of the autophagic machinery to degrade entire organelles; this was recently demonstrated to be the case during a mitochondria-selective form of autophagy known as mitophagy (Gomes et al., 2011). In hepatocytes, LDs have diameters ranging from 60 nm to well over 5 µm in steatotic conditions. We therefore asked whether LD size might play an equally important role in dictating the order and/or prevalence of the catabolic processes used by cells for LD breakdown.
In this study, we find that ATGL, the rate-limiting cytoplasmic lipase, preferentially operates on the largest LDs within the hepatocyte, whereas the lipophagic machinery is restricted in its targeting to only the smallest populations of cytoplasmic LDs (i.e., only those with diameters of <1 µm). We therefore propose that LD size itself represents a fundamental physical parameter dictating the mechanistic processes used for cellular TAG catabolism. Inhibition of neutral lipase activity was shown to result in a significant increase in the average diameter of hepatic LDs. In contrast, the pharmacological or genetic inhibition of lysosomal acid lipase (LAL) led to a two- to threefold accumulation of LDs with diameters averaging <1 µm in size. Importantly, this accumulation of very small LDs by LAL inhibition was completely abrogated by joint inhibition of ATGL, implying the necessity of upstream cytosolic lipase function on large LDs. Differential centrifugation was used to isolate size-based subpopulations of LDs, revealing a preference for ATGL on large LDs and an enrichment of autophagic markers on small LDs. The existence of two independently regulated triglyceride disposal pathways presumably allows the cell to exert tight control over the process of lipid catabolism under conditions of nutritional stress.
Results
Differential LD morphologies by inhibition of lipolysis or lipophagy
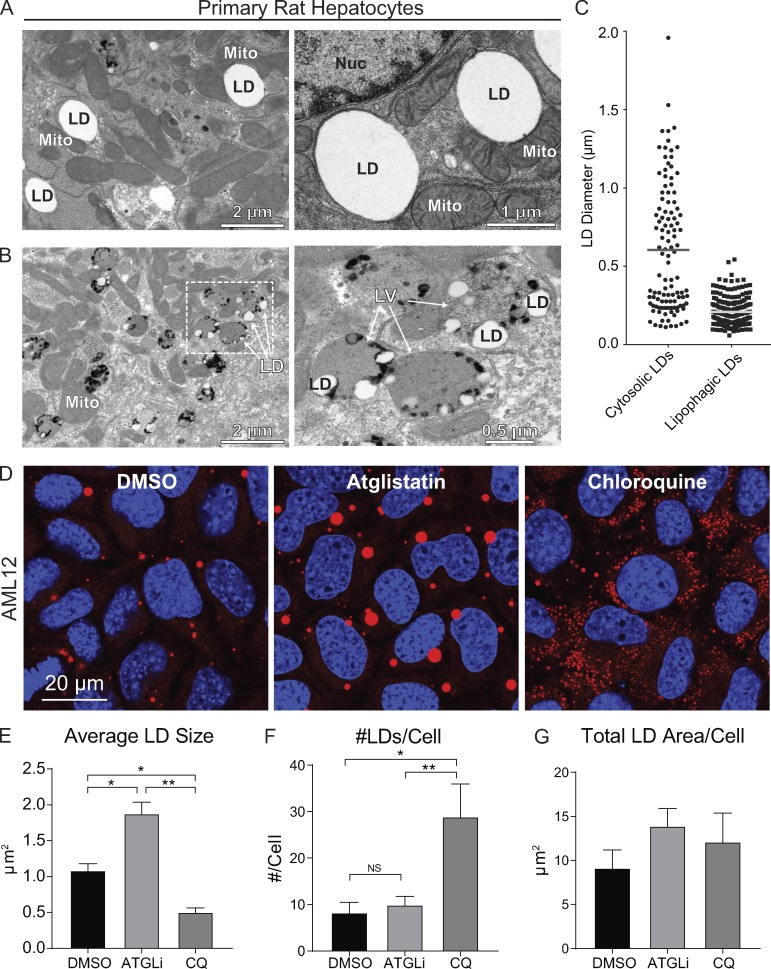
As mentioned above, cargo size is believed to play a role in the selective autophagy of organelles such as mitochondria (Gomes et al., 2011). This premise is consistent with our initial findings from transmission EM of cultured primary rat hepatocytes showing two distinct size-based populations of LDs. As shown in Fig. 1 A, transmission EM performed on freshly isolated primary rat hepatocytes revealed a substantial population of large, cytosolic LDs that interact with adjacent cellular organelles such as mitochondria and ER. In addition to these large LDs, we observed numerous small LDs encased within electron-dense degradative vesicles characteristic of autolysosomes or late endosomes (Fig. 1 B and Fig. S5). Manual quantification of LD sizes from these electron micrographs (n = 4 cells) showed that cytosolic LDs range between 0.1 and 2.0 µm in diameter, with an average diameter of 0.7 µm. In contrast, lipophagic LDs exhibit a more restricted size range between 60 and 500 nm in diameter, with an average diameter of 220 nm (Fig. 1 C). Since the majority of these lipophagic LDs are considerably larger than extracellular lipoproteins that range between 7 and 50 nm in diameter (Rye et al., 1999), we reasoned that the LDs in Fig. 1 B were likely internalized as cytoplasmic LDs. These data indicate that the lipophagy machinery is restricted to degrading small LDs, and suggest that large LDs are perhaps preferentially degraded by lipolysis via ATGL, the rate-limiting enzyme in this process (Smirnova et al., 2006).
Figure 1.
Hepatocytes possess size-based LD subpopulations. (A) Electron micrographs of primary rat hepatocytes showing a subpopulation of large, cytosolic LDs in intimate proximity to ER and mitochondria. (B) These cells also possess a second population of much smaller LDs encased within degradative lipophagic vesicles resembling autolysosomes/late endosomes. Numerous dark granules indicative of lysosomal content are observed in close contact to the LDs. (C) Quantification of LD diameters from EM images shows size-restriction on LDs within lipophagic vesicles (n = 4). (D) To test the hypothesis that large and small LDs are targeted by lipolysis and lipophagy, respectively, confocal micrographs of AML12 hepatocytes stained for LDs with Oil Red O show that a 24-h treatment with ATGLi (20 µM) results in the accumulation of large LDs, whereas the lysosome inhibitor chloroquine (100 µM) leads to the accumulation of numerous small LDs. (E) Quantification of these treated cells reveals a 1.7-fold increase in average LD size by ATGLi and a >50% decrease by chloroquine (CQ) compared with DMSO controls. (F and G) Accordingly, chloroquine-treated cells show a 3.5-fold increase in LD number compared with DMSO or ATGLi treatment (F), while total LD area per cell was not significantly increased after 24 h (G). Asterisks denote statistical significance by one-way ANOVA and Tukey’s post hoc test (*, P < 0.05; **, P < 0.01). Graphs depict mean and SEM from n = 4 experiments.
To test this concept, we measured LD morphology in AML12 mouse hepatocytes treated for 24 h with the ATGL inhibitor atglistatin (ATGLi; 20 µM) or the lysosome inhibitor chloroquine (100 µM). We predicted that lipolysis inhibition would result in cells containing large LDs, whereas lysosome inhibition would cause the accumulation of many small LDs trapped within autophagic vesicles. Indeed, as displayed in the confocal micrographs in Fig. 1 D, treatment with these pharmacological inhibitors resulted in dramatic differences in LD morphology as cells treated with the ATGL inhibitor accumulated very large LDs that were nearly twofold larger than DMSO controls, whereas lysosomal perturbation by chloroquine led to the accumulation of numerous small LDs that were roughly half the size of control LDs (Fig. 1 E). In addition, chloroquine treatment caused a nearly fourfold increase in LD number per cell compared with DMSO controls (Fig. 1 F), whereas the total LD area per cell was not significantly increased after 24 h (Fig. 1 G). These observations suggest that ATGL-driven lipolysis preferentially targets larger-sized LDs, whereas small LDs are preferentially catabolized by acidic lipases within lysosomes and/or late endosomes.
Evidence for sequential lipolysis and lipophagy pathways
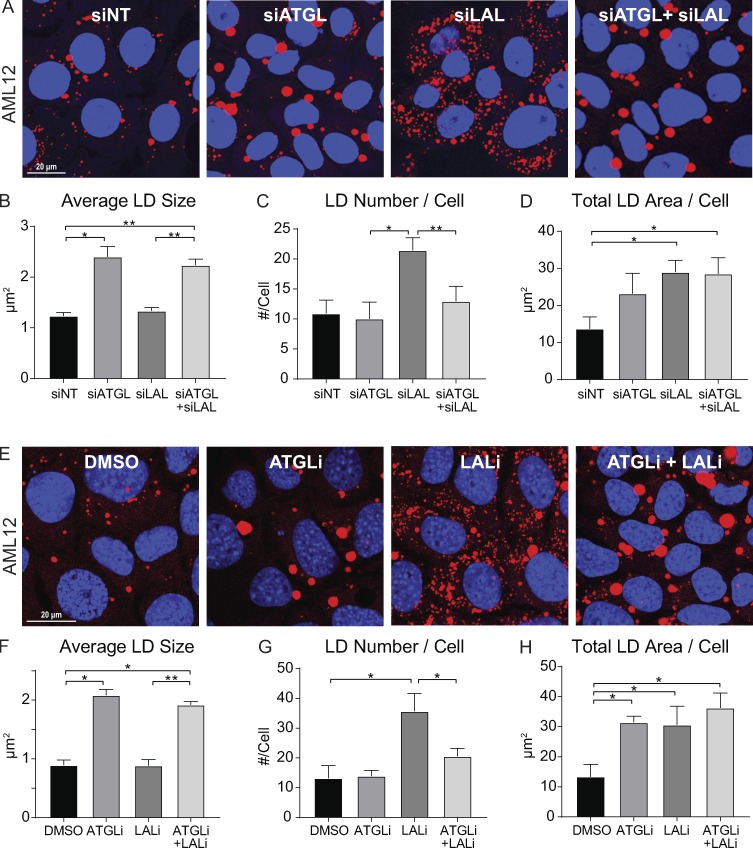
Based on the distinct changes in LD morphology following ATGL versus lysosome inhibition, we tested whether lipolysis and lipophagy operate independently, or if these processes operate in tandem. To test this, we assessed LD morphology in AML12 hepatocytes treated with siRNA against ATGL, LAL, or both. Validation of ATGL and LAL knockdown levels by Western blot and quantitative PCR (qPCR) are shown in Fig. S1 A. As expected, ATGL knockdown resulted in a twofold increase in average LD size over nontargeting siRNA controls (Fig. 2, A and B), and LAL knockdown caused a substantial accumulation of smaller-sized LDs (Fig. 2, A and C), similar to our previous observations using ATGLi and chloroquine in Fig. 1. However, dual knockdown of both ATGL and LAL caused the accumulation of large LDs similar to knockdown of ATGL alone, as LD size was similarly increased about twofold over control siRNA, and the accumulation of small LDs by LAL knockdown was completely reversed (Fig. 2, A–C). Similar increases in total LD area per cell were observed following knockdown of ATGL, LAL, or both (Fig. 2 D), suggesting that these two processes are not independent but operate in tandem.
Figure 2.
Evidence for lipolysis acting upstream of lipophagy on differently sized LDs. (A) Confocal micrographs of AML12 cells stained for LDs with Oil Red O show alterations in LD morphology following 72-h treatment with nontargeting control siRNA (siNT), siATGL, siLAL, or siATGL+siLAL. (B) ATGL knockdown resulted in a twofold increase in average LD size (μm2) and was mimicked by LAL+ATGL double knockdown. (C) Knockdown of LAL caused a greater than twofold increase in the number of LDs/cell, but this increase was blocked by ATGL+LAL knockdown. (D) Quantification of total LD area/cell under each knockdown condition. (E) Similar results were observed in AML12 cells preloaded for 2 h with 150 µM OA and then washed and chased for 48 h in an insulin-free medium containing 2% FBS and treated with DMSO, ATGLi (20 µM), LALi (20 µM), or ATGLi+LALi. (F) ATGLi caused a twofold increase in average LD size which was also observed in the presence of both inhibitors. (G) LALi caused a 2.5-fold increase in the number of LDs per cell, but this was reversed in cells treated with both inhibitors. (H) Quantification of total LD area per cell revealed significant increases by ATGLi, LALi, or both over DMSO control. Asterisks denote statistical significance by one-way ANOVA and Tukey’s post hoc test (*, P < 0.05; **, P < 0.01). Graphs depict mean and SEM. (A–D) n = 3 experiments. (E–H) n = 4 experiments.
As seen in confocal micrographs in Fig. 2 E, similar results were also observed using the ATGL inhibitor ATGLi (20 µM), the LAL inhibitor LAListat (LALi; 50 µM) or both in AML12 cells loaded with oleic acid (OA; 150 µM, 2 h) and chased for 48 h in insulin-free medium with reduced serum (2% FBS) to stimulate LD breakdown. Similar to siRNA knockdown experiments, a twofold increase in LD size was observed in in ATGLi- or ATGLi+LALi-treated cells (Fig. 2 F), and inhibition of LAL caused a more than twofold increase in LD number per cell, which was largely reversed by dual inhibition of ATGL+LAL (Fig. 2 G). These effects were specific for ATGL activity, as inhibition of other cytosolic lipases (HSL and MGL) using CAY10499 increased both large and small LDs and did not reverse the LALi-induced accumulation of small LDs (Fig. S2). However, consistent with siRNA knockdown of ATGL and LAL, the increase in overall LD content was similar with ATGLi, LALi, or both as measured by confocal microscopy (Fig. 2 H) and biochemical TAG analysis (Fig. S1 B). Together, these results suggest that the accumulation of small LDs following lipophagy inhibition is dependent on upstream ATGL-driven lipolysis.
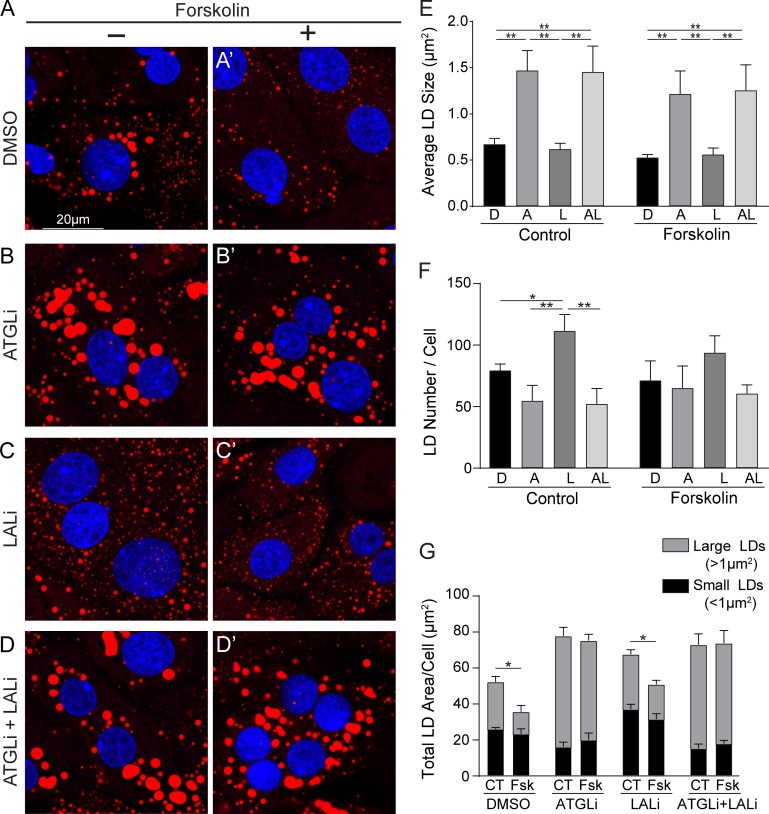
To extend these observations made in the AML12 cells, we also tested the impact of lipolysis and lipophagy inhibition using freshly isolated primary rat hepatocytes treated for 24 h with 20 µM ATGLi, 50 µM LALi, or both. As seen in confocal images and quantification in Fig. 3, the inhibition of ATGL dramatically increased LD size over control hepatocytes (Fig. 3, A, B, and E), whereas LAL inhibition increased the LD number (Fig. 3, C and F). Consistent with the AML12 cells, dual inhibition of both ATGL and LAL resulted in large LDs similar to ATGLi alone (Fig. 3, D and G).
Figure 3.
Primary hepatocytes use sequential lipolysis and lipophagy pathways. (A–D) Confocal micrographs of ORO-stained primary rat hepatocytes showing distinct effects of LD size following 24-h treatment with (A) DMSO, (B) 20 µM ATGLi, (C) 50 µM LALi, or (D) ATGLi+LALi. (A′–D′) These differential effects were also observed in primary hepatocytes stimulated for lipolysis by the cAMP-elevating agent forskolin (10 µM, 24 h). (E) Quantification of LD size revealed a less than twofold increase in LD size following ATGLi or ATGLi+LALi in control and lipolysis-stimulated cells. (F) Accumulation of small LDs was increased 1.4-fold by LALi in unstimulated hepatocytes, but this increase was completely reversed when cotreated with ATGLi+LALi. (G) Quantification of total LD area/cell reveals a significant decrease by forskolin (Fsk) treatment over control (CT) in both DMSO and LALi-treated cells that was almost exclusively due to a reduction in large LDs. ATGLi or ATGLi+LALi completely prevented a reduction in LD area/cell by forskolin. Asterisks denote statistical significance by one-way ANOVA and Tukey’s post hoc test (*, P < 0.05; **, P < 0.01). Graphs depict mean and SEM. n = 3 experiments. A, ATGLi; AL, ATGLi+LALi; D, DMSO; L, LALi.
To assess the effects of lipolysis and lipophagy inhibition under lipolysis stimulation, primary rat hepatocytes were also treated with forskolin (10 µM, 24 h), a cAMP agonist that activates lipolysis (Schott et al., 2017). As seen in confocal images and corresponding graphs from Fig. 3, A′–D′, inhibition of ATGL, LAL, or both resulted in similar effects on LD morphology between stimulated and unstimulated hepatocytes. When compared with unstimulated cells, forskolin treatment caused a significant decrease in total LD area/cell that was largely due to a reduction in larger-sized LDs (Fig. 3 E, gray bars), whereas no significant decrease in smaller-sized LDs was observed (black bars). Large versus small LDs were defined as greater than or less than 1 µm2, respectively, as this accounted for roughly 50% of the total LD area in each of the two categories in the control (unstimulated) condition. The decrease in LD area per cell by forskolin was completely prevented by ATGL inhibition or by dual inhibition of ATGL+LAL. Taken together, these data suggest that ATGL-driven lipolysis preferentially targets the degradation of large LDs upstream of LAL-driven lipophagy.
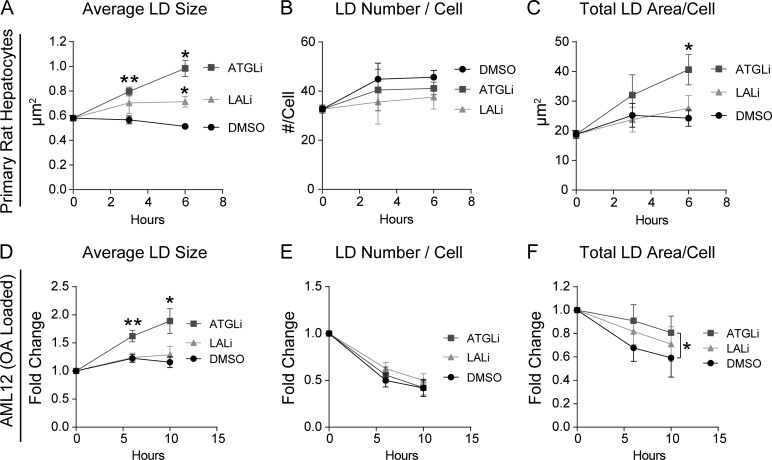
To further define the upstream role of lipolysis relative to lipophagy, we analyzed the kinetics of ATGL versus LAL inhibition in both primary rat hepatocytes and AML12 cells. We predicted that ATGL inhibition would show effects on LD morphology and overall LD content at an earlier time point than LAL inhibition. As seen in Fig. 4 A, ATGL inhibition increased LD size in as little as 3 h in primary rat hepatocytes, while LD number/cell was not affected by ATGL or LAL inhibition during this time (Fig. 4 B). LALi caused an unexpected 20% increase in LD size at t = 6 h (Fig. 4 A), indicating alternative effects of the inhibitor perhaps at acute time points. Nonetheless, LD size was substantially increased 70% by the ATGL inhibitor and was statistically significant within 3 h of treatment. In addition, ATGL inhibition caused a significant increase in total LD area/cell within 6 h over DMSO control (Fig. 4 C), whereas no effect was observed by LAL during this time. As seen in Fig. 4, D–F, similar effects were seen in AML12 cells that were loaded with OA and then washed and chased in OA-free medium. First, ATGL inhibition caused a twofold increase in LD size within 6 h of treatment (Fig. 4 D), while maintaining the total LD area within 10 h (Fig. 4 F). These findings show that ATGL inhibition affects LD size and total area at earlier time points than the LAL inhibitor, which is consistent with the concept that the lipolysis of large LDs precedes the downstream lipophagy of smaller-sized LDs.
Figure 4.
ATGL inhibition alters LDs before LAL inhibition. Time-course experiments were done to test the effects of ATGL-driven lipolysis as an upstream pathway to LAL-driven lipophagy. (A and B) In primary rat hepatocytes, ATGLi caused a significant increase in LD size in as little as 3 h, while no change in LD number per cell was observed. (C) ATGL inhibition increases total LD area in primary rat hepatocytes within 6 h over control hepatocytes (DMSO) or cells treated with LALi (50 µM). (D) In AML12 cells loaded with OA (150 µM, 2 h) and chased in OA-free medium, ATGLi-treated cells showed a significant increase in LD size within 6 h, with no change by LALi over DMSO controls. (E) No change in LD number/cell was observed by 10 h of OA withdrawal. (F) AML12 cells also showed a significant retention in total LD area/cell by ATGLi, but not LALi, within 10 h of OA withdrawal. Asterisks denote statistical significance as determined by Student’s t test (*, P < 0.05; **, P < 0.01). Graphs depict mean ± SEM from n = 4 experiments.
Lipolysis and lipophagy machinery preferentially target size-based LD subpopulations
Based on the findings described above that suggest tandem, size-based lipolysis and lipophagy pathways, we tested if components of these processes might preferentially associate with large and small LDs, respectively, in OA-loaded AML12 cells (150 µM, 16 h). As seen in Fig. 5 A, AML12 hepatocytes display numerous examples of both small and large LDs decorated by endogenous ATGL by immunostaining. However, a subset of small, ATGL-negative LDs was commonly observed, while ATGL fluorescence appeared most prevalent around the largest cellular LDs. Quantification of LD size (Fig. 5 A′) revealed that ATGL-positive LDs were roughly sevenfold larger than ATGL-negative LDs (4.00 versus 0.61 µm2, respectively), suggesting that ATGL preferentially, but not exclusively, localizes to larger-sized LDs. Interestingly, in OA-loaded AML12 cells that were subsequently washed and treated with the lysosome inhibitor chloroquine (50 µM, 24 h), ATGL localization was readily observed on smaller-sized LDs overall (Fig. S3 A, note the numerous small green ATGL puncta that colocalize with Oil Red O–stained LDs) but was nonetheless absent on a subset of small LDs, as indicated by the white arrows in Fig. S3 A. Quantification of LD size revealed that in chloroquine-treated cells, ATGL-negative LDs were 60–70% smaller than ATGL-positive LDs (Fig. S3 B).
Figure 5.
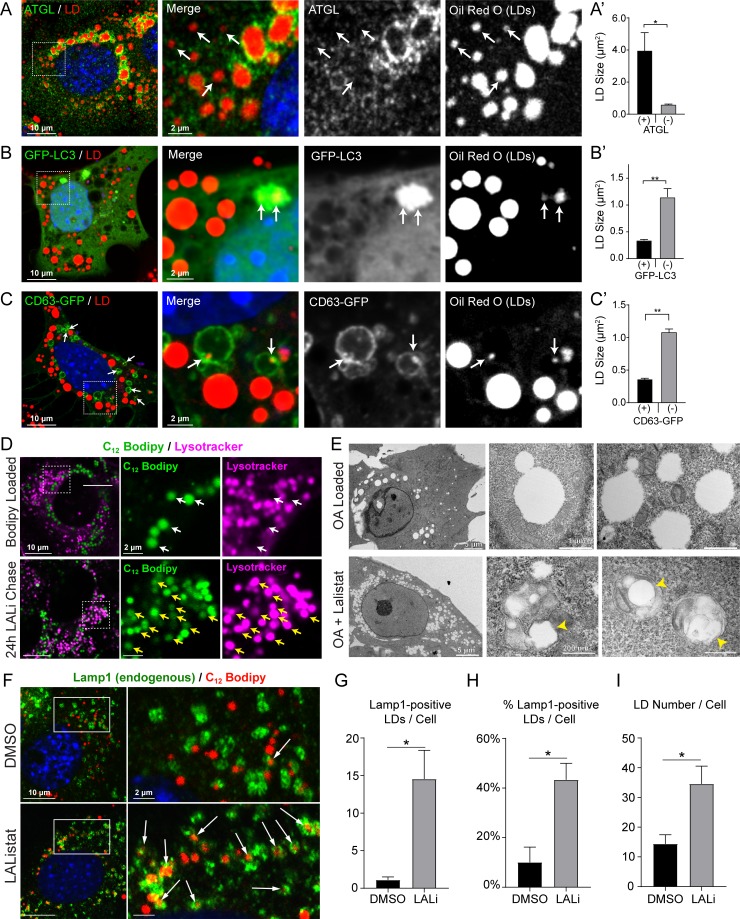
Lipolysis and lipophagy machineries associate with LDs of distinct sizes. (A) Confocal micrograph of OA-loaded AML12 hepatocyte stained with Oil Red O and immunolabeled for ATGL. Inlay images depict endogenous ATGL enriched around larger-sized LDs, whereas some of the smaller LDs lack ATGL staining as indicated by the white arrows. (A′) ATGL-positive LDs are on average sevenfold larger than ATGL-negative LDs. (B) In contrast to ATGL, GFP-LC3 in OA-loaded AML12 cells show preferential association with small LDs. (B′) GFP-LC3–engulfed LDs were 70% smaller than LC3-negative LDs. (C) OA-loaded AML12 cells also revealed numerous examples of small LDs entrapped within CD63-EGFP–positive endo-lysosomes. (C′) Quantification of LD size revealed that CD63-positive LDs were 66% smaller than CD63-negative LDs. (D) Confocal micrograph of AML12 cells pulse labeled with BODIPY C12 show little overlap between LDs and LysoTracker-positive vesicles (white arrows), while LALi treatment (50 µM, 48 h) increased LD accumulation within this compartment as shown by the yellow arrows. (E) Electron micrographs of control OA-loaded AML12 hepatocytes show large cytosolic LDs in contact with mitochondria, whereas treatment with LALi increases the number of small LDs within the cytoplasm and within intracellular vesicles. Yellow arrowheads depict small LDs ∼200 nm in diameter encased within degradative vesicles resembling autolysosomes. (F) Confocal micrograph of LAMP1 immunostained AML12 cells that were loaded for 2 h with red BODIPY C12 (7.5 µM, 2 h) and then washed and chased for 24 h in BODIPY-free medium containing DMSO or 50 µM LALi. White arrows depict LDs that were internalized into LAMP1-positive vesicles (i.e., lysosomes, autolysosomes, and late endosomes). (G and H) Quantification of this internalization revealed a 13-fold increase in the number of LAMP1-positive LDs per cell (G) and a fourfold increase in the percentage of internalized LDs (H). (I) LALi increased the number of BODIPY-labeled LDs per cell more than twofold during this time. Asterisks denote statistical significance as determined by Student’s t test (*, P < 0.05; **, P < 0.01). Graphs depict mean ± SEM. ATGL quantification was done from a total of 120 cells, and LC3/CD63 quantification was from 30 cells each across n = 3 independent experiments. LAMP1 quantification was done from 70–80 cells across n = 3 independent experiments.
From these findings, it became relevant to test if lipophagic vesicles such as autophagosomes and endo-lysosomes exhibit a preferential association with small versus large LDs. As seen in the confocal micrographs of OA-loaded AML12 cells in Fig. 5 B, small LDs were observed within GFP-LC3 autophagosomes that were 70% smaller than LC3-negative LDs. Similar targeting of small LDs by LC3 was also observed in OA-loaded AML12 cells treated with chloroquine (Fig. S3, B and C). Notably, we found few examples of LDs that were totally engulfed within LC3-positive autophagosomes, which is consistent with previous observations (Ding et al., 2010). However, the internalization of cytosolic LDs was substantially more prevalent within endo-lysosomal vesicles marked by the tetraspanin CD63-EGFP. Numerous examples of small LDs were found within these vesicles in OA-loaded cells both with and without chloroquine treatment. Like LC3, LD size quantification revealed that LDs within CD63 vesicles were ∼70% smaller than LDs that were not associated with these vesicles (Figs. 5 C and S3 C). Taken together, these data suggest that lipolysis and lipophagy machinery exhibit preferential targeting to large and small LDs, respectively.
To further define the terminal degradative compartment of small LDs, pulse-chase lipid analysis was performed using fluorescent BODIPY C12 FA in AML12 cells treated 24 h with DMSO or LALi (50 µM). We predicted that cells treated with lysosome inhibitors might accumulate substantial numbers of small LDs within the acidic vesicular compartments compared with control cells. As seen in Fig. 5 D, white arrows depict little or no colocalization between green fluorescent BODIPY C12 and LysoTracker in OA-loaded AML12 cells (16 h, 150 µM OA + 1 µM BODIPY C12). However, LAL inhibition (50 µM, 24 h) following OA load caused a substantial overlap between BODIPY fluorescence and LysoTracker vesicles, indicating the accumulation of small cytosolic LDs within autolysosomal or late endosomal vesicles. To further define the ultrastructure of these vesicles, transmission electron micrographs were taken of OA-loaded AML12 cells treated with or without LALi (50 µM, 24 h). As seen in Fig. 5 E, OA-loaded control cells contained large, cytosolic LDs in contact with other organelles such as mitochondria, whereas LALi-treated cells accumulated numerous cytoplasmic LDs and a subset of small LDs ∼200 nm in diameter within degradative vesicles resembling autolysosomes (yellow arrows).
To determine the relative proportion of small LDs that are incorporated into lipophagic vesicles, AML12 cells were immunostained for endogenous LAMP1, a lysosomal/late endosomal marker, following pulse-chase analysis using fluorescent BODIPY C12. In these experiments, cells were pulse labeled for 2 h with 150 μM OA + 7.5 μM BODIPY C12 558/568, then chased for 24 h in regular growth medium (10% FBS; ITS) containing DMSO or LALi (50 µM). As seen in Fig. 5 F, the accumulation of small LDs within LAMP1-positive vesicles was greatly increased by LAL inhibition. Quantification revealed a 13-fold increase in the number of LAMP1-positive LDs/cell (70–80 cells per condition, n = 3 experiments; Fig. 5 G). Strikingly, as much as 40% of the LDs in each cell were internalized by LAMP1-positive vesicles following LAL treatment (Fig. 5 H) even as the number of LDs increased 2.4-fold during this time (Fig. 5 I). These data suggest that a substantial number of small LDs are terminally degraded by lipophagy within acidic organelles such as lysosomes, autolysosomes, and late endosomes.
Large and small LDs possess unique protein and lipid signatures
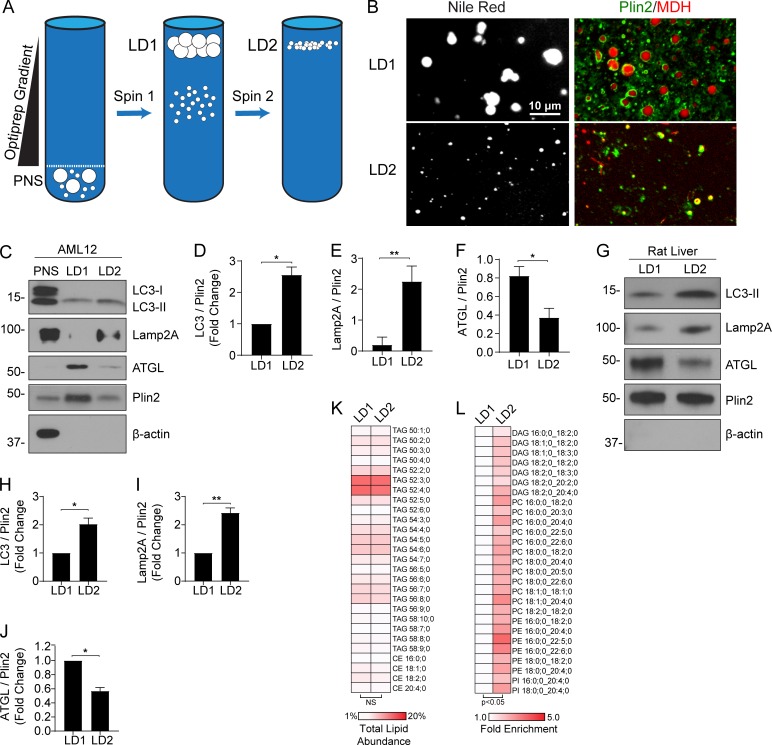
As an extension to the morphological-based studies described above, biochemical criteria were applied to isolated LDs of different sizes. To generate purified LD fractions enriched in small versus large LDs, a two-step differential centrifugation technique was used as depicted in Fig. 6 A (adapted from Brasaemle and Wolins, 2016; Zhang et al., 2016). This method promoted a size separation of large, buoyant LDs in an “LD1” fraction and smaller, less buoyant LDs within an “LD2” fraction that were then assessed by both immunofluorescence and Western blot analyses. As seen in Fig. 6 B, both fractions contained LDs that were positive for Perilipin 2 (PLIN2) immunostaining as well as Nile Red and monodansylpentane (MDH) lipid stains, and the LD1 fraction contained LDs that were substantially larger than that of LD2. Western blot analysis indicated that these LD fractions were not enriched in organelle contaminants such as Golgi, ER, and mitochondria (Fig. S4 A). Most interesting are the findings indicating a marked enrichment of ATGL in the LD1 fraction containing large LDs (more than twofold) while autophagic (LC3, 2.5-fold) and lysosomal markers (LAMP2A, 10-fold) were substantially enriched in the smaller LD2 fraction (Fig. 6, C–F). Confocal microscopy of LD1 and LD2 fractions supported these findings, as ATGL immunofluorescence was abundant on large LDs, whereas LysoTracker staining colocalized with small LDs in the LD2 fraction (Fig. S4 B). These findings are consistent with a size-based preference for components of the lipolysis and lipophagy machineries.
Figure 6.
Biochemical separation of large versus small LDs reveals distinct protein profiles. (A) Cartoon of the two-step centrifugation protocol used to isolate large, buoyant LDs (LD1) and small, less-buoyant LDs (LD2). (B) Confocal images of purified LD1 and LD2 fractions from AML12 cells stained with Nile Red, or stained with MDH and immunolabeled with a PLIN2 antibody. (C) Representative Western blot from oleate-loaded AML12 cells showing post-nuclear supernatant (PNS), LD1, and LD2 fractions. Note the enrichment of autophagic and lysosomal machinery (LC3-II, LAMP2A) in the small LD2 fraction, whereas ATGL is enriched in the large LD1 fraction relative to PLIN2 loading control (n = 3). (D and E) Density quantification revealed the LD2 fraction contained 2.5-fold enrichment of LC3B (D) and a near 10-fold enrichment in LAMP2A (E). (F) Conversely, ATGL was greater than twofold more abundant in the large LD1 fraction. (G–J) Representative Western blot (G) of LD fractions from rat whole liver and quantification show similar enrichment of lipophagic markers LC3B (H) and LAMP2A (I) in the small LD2 fraction, whereas ATGL (J) was enriched in the LD1 fraction. (K and L) Shotgun lipidomics revealed similar levels of TG and CE species in LD1 and LD2 fractions from whole rat liver (n = 4; K), with significant enrichment in several species of DAG, PC, PE, and PI contained in the LD2 fraction (L). Asterisks denote statistical significance by Student’s t test (*, P < 0.05; **, P < 0.01). Graphs depict mean and SEM.
As large and small LDs appear to possess distinct protein signatures (Fig. 6, A–G), we sought to assess whether distinct lipid species were contained within these two subpopulations. Based on the concept that ATGL acts to reduce the size of larger LDs, we predicted that smaller-sized LDs would accumulate remnant lipid species resulting from triglyceride (TAG) lipolysis such as diacylglycerol (DAG) or cholesterol esters (CEs) for which ATGL has lower affinity. Prior to lipidomic analyses, LD1 and LD2 fractions were isolated from whole rat livers (n = 4) using a slightly modified differential centrifugation technique than that of the AML12 cells (described in the Materials and methods section), and confocal microscopy confirmed the size separation of LDs within these fractions (Fig. S4 C). Additional Western blot analysis confirmed the enrichment of LC3B and LAMP2A in the small LD2 fraction, and ATGL enrichment in the large LD1 fraction (Fig. 6, G–J). Surprisingly, lipidomic analysis of large and small LDs revealed near equivalent levels of triglycerides and CEs as a percentage of total lipid abundance (Fig. 6 K and Fig. S4, D and E). Fig. 6 K depicts a heatmap of the 25 most abundant lipid species representing TAGs and CEs that exhibited a similar abundance between the two LD subpopulations. However, as seen in Fig. 6 L, several lipid species were significantly enriched in the LD2 fraction, most notably several DAGs (approximately twofold) and phospholipids phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylinositol (PI; approximately threefold). No lipid classes or individual lipid species were markedly enriched in the LD1 fraction relative to LD2. The enrichment in PC, PE, and PI in LD2 indicates the presence of smaller-sized LDs with higher surface-to-volume ratios, whereas the enrichment in DAG is likely the result of upstream ATGL-driven lipolysis of larger-sized LDs.
Synthesis-dependent and -independent accumulation of small LDs
The observations depicted in Figs. 1, 2, 3, 4, 5, and 6 are consistent with the premise of a tandem stepwise pathway whereby ATGL-driven lipolysis acts on large LDs (>1 µm2) to create a population of small LDs (<1 µm2) that are more amenable for LAL-driven lipophagy. A previous report using cultured adipocytes showed that lipolysis activation forms a nascent population of smaller LDs generated by reesterification of FFAs (Paar et al., 2012). In addition, the contribution of extracellular lipoproteins internalized by clathrin-mediated endocytosis may also provide an alternative source for the generation of small LD accumulation following LAL and lysosome inhibition. To further measure the importance of LD synthesis or exogenous lipids in lipolysis and lipophagy synergy, cells were incubated in the presence of DGAT1 (10 µM PF-06424439) and DGAT2 (10 µM PF-04620110) inhibitors in regular medium and lipoprotein-depleted medium (LDM). Thus, by removing either exogenous lipids or by inhibiting nascent LD synthesis, we tested the relative contributions of these pathways in the accumulation of LDs by lipolysis and lipophagy machinery.
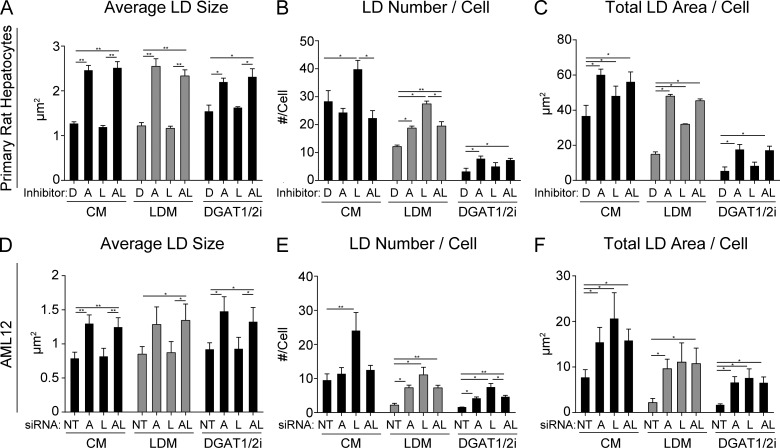
Accordingly, freshly isolated primary hepatocytes were treated for 24 h with DMSO, 20 µM ATGLi, 50 µM LALi, or both (ATGLi+LALi) in control medium (CM), LDM, or CM containing DGAT1/2 inhibitors. First, similar changes in LD size were observed following ATGLi or ATGLi+LALi regardless of exogenous lipids or LD synthesis (Fig. 7 A). In CM, LD number per cell was increased by LALi treatment as expected, and this increase was not observed in the combined presence of ATGLi+LALi (Fig. 7 B), confirming that the increase in LD number caused by lysosome inhibition is first dependent on ATGL-driven lipolysis. In LDM, LALi also caused an increase in LD number per cell, suggesting that exogenous lipids are not required for this increase (Fig. 7 B, gray bars). In the presence of DGAT1/2i, overall LD numbers were reduced with no significant increase by LALi treatment, suggesting that FFA reesterification plays a substantial role in the lipolysis-dependent accumulation of LDs (Fig. 7 B). As seen in Fig. 7 C, similar effects are also observed in total LD area per cell.
Figure 7.
Role of extracellular lipids and triglyceride synthesis in lipolysis and lipophagy. (A–C) Primary rat hepatocytes were treated 24 h with lipase inhibitors (D, DMSO; A, 20 µM ATGLi; L, 50 µM LALi; AL, ATGLi+LALi) in CM (5% FBS), LDM (5% lipoprotein-depleted calf serum), or CM containing DGAT1/2 inhibitors (10 µM each). (A) LD size was increased by ATGLi and ATGLi+LALi regardless of CM, LDM, and DGAT1/2i conditions. (B) LD number per cell was increased by LALi in both CM and LDM in an ATGL-dependent manner, but this was not observed in the presence of DGAT1/2 inhibitors. (C) Total LD area per cell was increased by lipase inhibitors in both CM and LDM, but no increase was observed by LALi in the presence of DGAT1/2i. (D–F) AML12 hepatocytes were treated with 48 h with siRNA as indicated (NT, nontargeting siRNA; A, siATGL; L, siLAL; AL, siATGL+siLAL) and then incubated an additional 18 h in CM, LDM, or DGAT1/2i. Regardless of the presence of lipoproteins or TG synthesis, (D) LD size was increased by siATGL and siATGL+siLAL; (E) LD number increased by LALi; and (F) the total LD area/cell remained elevated by knockdown of ATGL, LAL, or both. Asterisks denote statistical significance by Student’s t test (*, P < 0.05; **, P < 0.01). Graphs depict mean and SEM from n = 3 independent experiments.
The above experiments were also conducted in AML12 cells using siRNA knockdown instead of pharmacological inhibitors. These knockdown variables included nontargeting siRNA, siATGL, siLAL, or siATGL+siLAL. siRNA treatment resulted in a >80% knockdown for both ATGL and LAL as assessed by mRNA and protein levels (Fig. S1 A). Following a 48-h siRNA knockdown, cells were replated for an additional 16 h in CM, LDM, or CM containing DGAT1/2 inhibitors. As shown in Fig. 7 D, LD size was increased in response to ATGL or ATGL+LAL knockdown in all three conditions, similar to primary rat hepatocytes, suggesting that exogenous lipids and FFA reesterification do not impact LD size–based effects of ATGL inhibition. Importantly, the number of LDs per cell was higher in LAL knockdown cells regardless of the culture conditions used (CM, LDM, or DGAT1/2i) as shown in Fig. 7 E, while the overall LD numbers were reduced by roughly 80% when in the absence of exogenous lipids or LD synthesis. Like the primary hepatocytes, exogenous lipids were not required to maintain higher LD numbers or total LD area in LAL-depleted cells (Fig. 7, E and F, gray bars). However, the finding that LD numbers and total LD area were higher in LAL knockdown cells treated with DGAT1/2 inhibitors (Fig. 7, E and F) suggests that lipoproteins and size-reduced cytosolic LDs also contribute to lipid accumulation under these conditions. Taken together, our data suggest that the LD synthesis machinery drives the accumulation of most small LDs that appear following lysosome inhibition, and this requires FAs generated upstream by ATGL-driven lipolysis.
Discussion
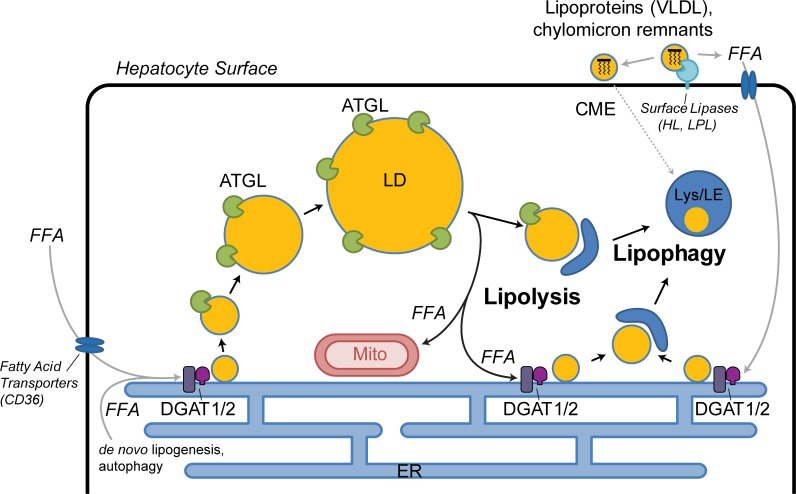
The central findings of this study provide evidence that the hepatocyte is likely to catabolize cytoplasmic LDs using both cytoplasmic lipases and lipophagy. Importantly, this utilization may reflect a stepwise process of small LD production by cytoplasmic lipases such as ATGL followed by autophagic engulfment as depicted in the cartoon working model from Fig. 8. This premise is based on the findings that dual treatment of hepatocytes with inhibitors of ATGL and LAL (Figs. 2, 3, and 7) leads to the accumulation of large LDs, and that lipolysis and lipophagy machinery exhibit size-based targeting of large and small LDs, respectively (Figs. 5 and 6). The LD morphologies resulting from inhibition of lipolysis and lipophagy are consistent with previous studies that have used lipase-defective mouse models that parallel human disease phenotypes. For example, liver-specific ATGL knockout mice exhibit very large hepatocellular LDs by immunohistochemical examination (Ong et al., 2011), and gross macrovesicular steatosis has been reported within periportal zones (Wu et al., 2011). Reciprocally, the accumulation of small LDs following lysosome/LAL perturbation in our cellular models is consistent with microvesicular steatosis in the hepatocytes of patients with LAL deficiencies and other types of lysosomal storage disorders (Bernstein et al., 2013; Reiner et al., 2014). Thus, the combined findings of all these studies support a concept of an epistatic cascade of cytosolic lipase-to-lysosome transition via preferential targeting of size-based LD subpopulations.
Figure 8.
Size-based targeting and LD synthesis dictate lipolysis and lipophagy pathways in hepatocytes. FFAs from extracellular and intracellular sources are packaged into triglycerides and stored within nascent small LDs, some of which may be targeted by lipophagy directly. Maturation into larger-sized LDs will exceed the capacity of the lipophagy machinery and rely on targeting by ATGL-driven lipolysis. Lipolysis produces small LDs both by reducing the size of large LDs and to a greater extent by liberating FFAs, some of which evade mitochondrial β-oxidation and are reesterified at the ER into nascent small LDs by DGATs 1 and 2. Some of the small LDs produced by ATGL-driven lipolysis are subsequently targeted by lipophagic membranes such as autophagosomes and, along with lipoproteins imported via clathrin-mediated endocytosis, are terminally degraded within lysosomes and late endosomes through the action of LAL. CME, clathrin-mediated endocytosis; VLDL, very low density lipoprotein; HL, hepatic lipase; LPL, lipoprotein lipase; Lys/LE, lysosome/late endosome.
Utilization of lipolysis and lipophagy pathways in the hepatocyte
A primary question addressed by this study is defining the source of the many smaller LDs that accumulate in hepatocytes treated with LAL inhibitors (Figs. 1, 2, 3, and 7) and whether they are generated directly from large LDs that are reduced in size, from endocytosed lipids, or from synthesis of nascent LDs. A careful study by others had previously demonstrated that the production of small LDs in adipocytes under lipolytic stimulation can be curtailed by preventing FA esterification and synthesis of nascent LDs (Paar et al., 2012). This suggested that most of the small LDs generated in stimulated cells were not a result of fragmentation or shrinking preexisting LDs but of new synthesis from liberated FAs. While our study also showed that the accumulation of small LDs following LAL inhibition was lipolysis dependent (Figs. 2, 3, and 7), we also tested for alternative lipid trafficking pathways such as lipoprotein endocytosis and FA reesterification.
First, exogenous lipids were removed from the culture medium using lipoprotein-depleted serum to assess the role of endocytosed lipoproteins (Fig. 7). We observe that exogenous lipids contribute to overall LD content but are not required to observe an increase in LD number following LAL inhibition in primary hepatocytes or LAL knockdown in AML12 cells, suggesting that the contribution of endocytosed lipids may be modest. Consistent with this prediction are the many electron micrographs we have viewed, some of which are displayed in Fig. 1 and Fig. S5, that show late endocytic compartments containing large LDs (≥200 nm) that exceed the known diameters of lipoprotein particles ranging between 7 and 50 nm in human blood plasma (Rye et al., 1999). Chylomicron remnants within blood plasma have a somewhat larger diameters (50–150 nm) but are further reduced by hepatic lipase at the hepatocyte surface before endocytic uptake (Cooper, 1997; Crawford and Borensztajn, 1999; Perret et al., 2002). Thus, it is likely the majority of the lipophagic LDs we observe, particularly those ≥200 nm in diameter, are derived from the engulfment of small cytoplasmic LDs.
As an extension of these studies, we sought to attenuate synthesis of LDs rather than limiting the external lipid source. This was done using pharmacological inhibitors of DGAT1 and DGAT2 (Fig. 7), blocking the esterification of FAs into triglyceride, a required step in nascent LD synthesis. From these experiments, we found that most of the small LDs that accumulated following LAL inhibition or siRNA knockdown were dependent upon the action of the LD synthesis machinery (Fig. 7, B and E). Thus, it appears that hepatocytes use the lipophagic pathway to not only complete the catabolism of residual LD “remnants” following the action of cytoplasmic lipases such as ATGL, but also to target small newly synthesized LDs resulting from FA reesterification. While the protein blotting of the light and heavy LD fractions are consistent with the premise of ATGL- versus lipophagy-associated subpopulations (Fig. 6), this analysis does not preclude that the LD2 fraction contains a mixture of both nascent and remnant LDs. The extensive lipidomics of these fractions described in Fig. 6 and Fig. S3 does suggest that some conversion of TAG to DAG has taken place preferentially in the heavy LD2 fraction, although this contribution appears minor given the relative homogeneity of lipid classes between the two fractions.
A size-dependent selection of LDs by two central catabolic pathways
An interesting facet of the interplay between cytoplasmic and lysosomal lipases reported here is the concept that LD girth may influence which of these lipases acts first. As discussed above, we predict that the terminal autophagy of LDs is initiated by a size-based mechanism that is dependent on LDs reaching a certain modest diameter, not unlike that observed for mitophagy, as reported elsewhere (Gomes et al., 2011). This prediction is based on the substantial comparative differences observed in LD size phenotypes following the inhibition of either lipolysis or lipophagy. Consistent with this premise, the sizes of LDs were increased following the inhibition of ATGL, suggesting that lipolysis preferentially targets the degradation of large LDs while lipophagic membranes are perhaps unable to enclose such large cargo. Furthermore, primary rat hepatocytes stimulated for lipolysis show a reduction in LD content that appears to be due largely to a reduction in large LDs (Fig. 3 G). Finally, ATGL was observed to be markedly enriched on large LDs both by immunofluorescence microscopy (Fig. 5) and Western blot analysis of size-separated LDs from both AML12 cells and rat livers (Fig. 6). Some ATGL was observed on smaller LDs, however, particularly following lysosomal inhibition (Fig. S3).
The mechanisms that favorably target ATGL to large LDs are yet to be elucidated. It is known that ATGL localizes to LDs through a C-terminal hydrophobic domain (Murugesan et al., 2013) and is thought to be regulated both positively and negatively by phosphorylation at various sites (Mason et al., 2012; Pagnon et al., 2012; Xie et al., 2014). ATGL localization on LDs is also influenced by the ER-Golgi trafficking machinery (Soni et al., 2009), as well as direct binding to the autophagosome marker LC3 (Martinez-Lopez et al., 2016). While it is possible that ATGL phosphorylation or protein interaction dictates its localization to specific LDs, the mechanisms that regulate LD-protein interaction in general are poorly understood. For example, the LD monolayer itself has unique biophysical properties that serve as a gatekeeper for protein interaction. It has been reported that LD monolayers are under high surface tension in comparison to lipid bilayers, and this surface tension restricts many proteins that normally adhere to lipid membranes (Thiam et al., 2013; Prevost et al., 2018). This is likely exacerbated in differently sized LDs with varying degrees of membrane curvature and proteomic contents that could influence the binding of proteins such as ATGL to large versus small LDs. Given the wide heterogeneity of LDs even within individual cells (Hsieh et al., 2012; Zhang et al., 2016; Thiam and Beller, 2017), future studies will need to dissect the contributions of LD diversity, membrane biophysics, and protein–protein interactions in directing ATGL and other proteins to specific LD subpopulations.
The findings that lysosome inhibition increases LD number and incorporation into acidic degradative vesicles (Fig. 5) suggests that, even under basal conditions, the lipophagy pathway is exceptionally active in the catabolism of small LDs (<1 µm2) that may represent remnants of larger-sized LDs as well as a relevant population of newly synthesized LDs. The high basal activity of this process is somewhat surprising and highlights the importance of LD size recognition perhaps over the contributions of nutrient deprivation and other cellular cascades that activate LD–lipophagic interactions. A variety of reports have implicated nutrient deprivation as an activator of this autophagic pathway (reviewed in Schulze et al., 2017) suggesting that lipophagy is a highly regulated process that might work in collaboration with putative sensors of LD size or membrane curvature. Interestingly, LC3 lipidation was reported to occur preferentially on membranes with high curvature due to Atg3 sensing (Nath et al., 2014), raising the possibility that similar curvature-sensing mechanisms could exist for lipophagic targeting of small LDs. It is also likely that the size of degradative vesicles such as lysosomes, autolysosomes, and late endosomes are also limiting to LD internalization and lipid hydrolysis. For example, these vesicles may achieve diameters in excess of 1 µm under certain conditions, while the majority are much smaller in size (Huotari and Helenius, 2011; Xu and Ren, 2015; Su et al., 2016). This range in vesicle size is consistent with the relative size of lipophagic LDs (<1 µm2) that we observe by light and EM (Figs. 1, 5, and S5), and the size-based population of LDs that accumulate following LAL perturbation (Fig. 3 G).
Taken together, these findings support a sophisticated process of hepatocyte LD homeostasis that implicates the action of soluble lipases on large droplets that generate smaller-sized LDs by DGAT1/2 dependent reesterification of FFAs and, to a lesser extent, size-reduction of catabolized LDs. Smaller remnant LDs, generated from the action of ATGL lipolysis and nascent synthesis, attract components of the lipophagic machinery based on a reduced girth (<1 µm2) for degradation within acidic vesicles. Further insights into how the hepatocyte might alter this elaborate process under normal and disease states will prove important.
Lipolysis and lipophagy as tandem pathways for LD breakdown
The stepwise model suggesting size-based LD targeting by lipolysis and lipophagy pathways in the current study is supportive of previous studies showing that ATGL is an important upstream mediator of downstream autophagy. For example, ATGL is well described to regulate SIRT1 to induce autophagy and FA oxidation in hepatocytes via PPARα/PGC-1α signaling (Khan et al., 2015; Sathyanarayan et al., 2017). In agreement with this, we also show that ATGL operates upstream of lipophagy by preferentially targeting large LDs and producing numerous smaller-sized LDs via DGAT1/2 reesterification and, to a lesser extent, reducing the size of existing LDs. Other reports have indicated that autophagy can also act upstream of ATGL/lipolysis by promoting LD synthesis under nutrient-limited conditions (Rambold et al., 2015; Nguyen et al., 2017; Zechner et al., 2017). In this model, macroautophagy facilitates the bulk degradation of cellular organelles, and byproduct membrane lipids are recycled to the ER for LD biogenesis. It is likely that bulk macroautophagy is a separate process from more selective forms of autophagy such as LD-specific lipophagy. Thus, while our data support the hypothesis that ATGL acts upstream of lipophagy, bulk macroautophagy likely plays an important role in LD synthesis further upstream of both lipolysis and lipophagy.
In conclusion, the current study defines lipolysis and lipophagy as tandem pathways in hepatocytes that degrade large and small LDs, respectively. The enrichment of ATGL on larger-sized LDs is consistent with the increase in LD size following ATGL inhibition or siRNA depletion. Conversely, small LDs are preferentially engulfed by lipophagic machinery, and lysosomal inhibition results in a decrease in LD size and the accumulation of numerous small LDs within autophagic and endosomal vesicles. These LD phenotypes correlate with human pathologies of macro- and microvesicular steatosis, which may suggest the need for targeted therapeutic strategies in the treatment of specific fatty liver pathologies.
Materials and methods
Cell culture and reagents
Primary rat hepatocytes were isolated from female Sprague Dawley rats (Envigo) by collagenase perfusion and were cultured in William’s E Medium (5% FBS) as described by previously (Shen et al., 2012). All animals received humane care in accordance with the guidelines established by the American Association for the Accreditation of Laboratory Animal Care, and animal protocols were approved by the Institutional Animal Care and Use Committee at Mayo Clinic. The AML12 cells (CRL-2254; ATCC) are mouse hepatocytes obtained from transgenic MT42 mice (CD1 strain) expressing human transforming growth factor α. AML12 cells were maintained in 1:1 DMEM/F12 medium supplemented with 10% FBS, Pen/Strep, and Insulin-Transferrin-Selenium supplement (41400-05; Gibco; final concentration: 0.01 mg/ml insulin, 0.0055 mg/ml transferrin, and 6.7 ng/ml selenium). The β-actin antibody (A2066), chloroquine diphosphate salt (C6628), DGAT1 inhibitor PF-04620110 (PZ0207), DGAT2 inhibitor PF-06424439 (PZ0233), lipoprotein-depleted serum (S5394), OA (O1008), and Oil Red O (O0625) were from Sigma-Aldrich. The Nile Red (N1142) was from Thermo Fisher Scientific. The MDH was from Abgent (SM1000a). The ATGL antibody (2439) was from Cell Signaling Technology. The PLIN2 antibody (B3121) was from LS Biosciences. The LAMP2A antibody (ab18528) was from Abcam. The LC3 antibody (NB600-1384) was from Novus. The Tsg101 antibody (GTX70255) was from Genetex. The Lamp1 antibody (1D4B) was from the Developmental Studies Hybridoma Bank at the University of Iowa (Iowa City, IA). The ATGL inhibitor ATGLi (#15284) and HSL/MGL inhibitor CAY10499 (10007875) were from Cayman Chemical. The LAL inhibitor LALi (6098) was from Tocris. The C12-BODIPY (FL: D3822; 558/568: D3835) and LysoTracker Deep Red (L12492) were from Thermo Fisher Scientific. The human ATGL-EGFP construct was a kind gift from C. Jackson (Institut Jacques Monod, Paris, France). The EGFP-LC3 was a gift from K. Kirkegaard (Stanford, CA; Addgene plasmid 11546). The CD63-EGFP construct was generated from a human cDNA library (primers: 5′-AAGCTTATGGCGGTGGAAGGAGGA-3′, 5′-GAATTCGGAGACCAGACCCCATCACG-3′), ligated into EGFP N1 vector following restriction endonuclease digestion (5′-HindIII, 3′-EcoRI) and confirmed by sequence analysis. Cells were transfected at 60–80% confluency using Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Fluorescence and live-cell microscopy
Cells were washed in PBS and fixed in 3% formaldehyde as described previously (Cao et al., 1998). To label LDs, fixed samples were washed in 60% isopropanol for 30 s and 60% Oil Red O solution (5 mg/ml in isopropanol) for 1.5 min and then washed in 60% isopropanol for an additional 30 s. Images were acquired using a Zeiss LSM 780 confocal microscope with a 40× oil objective lens (NA = 1.4). Quantification of LD size, number, and total area per cell was done using ImageJ software (Auto Local Threshold tool, Bernsen method).
Triglyceride measurement
Triglycerides were measured from AML12 cells in accordance with a previous protocol (Wang et al., 2017). In brief, lipid was extracted using 1 ml chloroform/methanol (2:1) and vigorous shaking for 1 h at RT. 200 µl double-distilled H2O was added, and samples were vortexed and centrifuged at 3,000 × g for 5 min. The lower lipid phase was collected (∼700 µl), dried overnight, and resuspended in a mixture containing tert-butanol (471712; Sigma-Aldrich), Triton X-114 (X114; Sigma-Aldrich), and methanol (9:4:2). TAG quantification was done in AML12 cells using a colorimetric kit from Pointe Scientific (T7532, T7531-STD) according to manufacturer’s instructions.
LD isolation
AML12 cells were grown to 90% confluency in 5 × 15–cm dishes and loaded with OA (150 µM, 16 h). Using a protocol adapted from two previous studies (Brasaemle and Wolins, 2016; Zhang et al., 2016), cells were washed, scraped, and incubated in a hypotonic lysis medium (HLM) before Dounce homogenization. The post-nuclear supernatant was placed at the bottom of a 25% to 0% OptiPrep density gradient (D1556; Sigma-Aldrich) in HLM. Following a 30-min spin at 36,000 × g, the floating fat layer (LD1) was collected and washed for subsequent Western blot analysis. After LD1 was collected, 400 µl of HLM was carefully added to the top of the gradient, and samples were spun for an additional 60 min at 182,000 × g. The LD2 fraction was then collected from the top 200–400 µl. LD2 samples were loaded on SDS-PAGE gels at twice the volume of LD1 samples to account for differences in LD yield, and protein levels were normalized to PLIN2 as a loading control.
To isolate LDs from rat livers, ∼5 g of tissue was surgically extracted from female Sprague Dawley rats (Envigo). Liver tissue was finely minced and washed in ice-cold PBS before incubation in HLM, Dounce homogenization, and layering beneath the OptiPrep gradient as described above. To collect the LD1 fraction from rat whole liver lysates, samples were spun 30 min at 8,000 × g. Following the collection of the floating fat layer (LD1), the samples were spun for 60 min at 182,000 × g to collect the floating LD2 fraction.
siRNA knockdown and qPCR
Cells were transfected with siGENOME pools L/M-048418-01 and M-040220-01 (Dharmacon) targeting murine LIPA and PNPLA2 RNA using the RNAiMAX reagent (Invitrogen) according to manufacturer’s instructions. A nontargeting siRNA duplex from Dharmacon was used in control transfected cells. RNA was isolated from cells using the RNeasy Plus Mini Kit (Qiagen) and was reverse-transcribed with the Super Script III First Strand Kit (Invitrogen) using oligo-(dT) primers. Quantification of gene expression was performed using SYBR green fluorescence on a LightCycler 480 (Roche). Primers for murine LIPA (lysosomal acid lipase; forward: 5′-TGCCCACGGGAACTGTATC-3′ and reverse: 5′-ATCCCCAGCGCATGATTATCT-3′), PNPLA2 (forward: 5′-ATGTTCCCGAGGGAGACCAA-3′ and reverse: 5′-GAGGCTCCGTAGATGTGAGTG-3′), and 18S ribosomal RNA (forward: 5′-CGCTTCCTTACCTGGTTGAT-3′ and reverse: 5′-GAGCGACCAAAGGAACCATA-3′) were synthesized by Integraded DNA Technologies. Expression was calculated using a ΔΔCt method, normalizing to 18S ribosomal RNA levels. Data represent relative expression levels in LIPA and PNPLA2 pooled duplex transfected cells to that of nontargeting siRNA (Dharmacon/GE) control transfected cells.
Lipid extraction for mass spectrometry (MS) lipidomics
MS-based lipid analysis was performed by Lipotype as described previously (Sampaio et al., 2011). Lipids were extracted using a two-step chloroform/methanol procedure (Ejsing et al., 2009). Samples were spiked with internal lipid standard mixture containing cardiolipin 16:1/15:0/15:0/15:0, ceramide 18:1;2/17:0, DAG 17:0/17:0, hexosylceramide 18:1;2/12:0, lyso-phosphatidate 17:0, lyso-PC (LPC) 12:0, lyso-PE (LPE) 17:1, lyso-phosphatidylglycerol 17:1, lyso-PI 17:1, lyso-phosphatidylserine 17:1, phosphatidate 17:0/17:0, PC 17:0/17:0, PE 17:0/17:0, phosphatidylglycerol 17:0/17:0, PI 16:0/16:0, phosphatidylserine 17:0/17:0, CE 20:0, sphingomyelin 18:1;2/12:0;0, and TAG 17:0/17:0/17:0. After extraction, the organic phase was transferred to an infusion plate and dried in a speed vacuum concentrator. First step dry extract was resuspended in 7.5 mM ammonium acetate in chloroform/methanol/propanol (1:2:4, vol:vol:vol) and second step dry extract in 33% ethanol solution of methylamine in chloroform/methanol (0.003:5:1; vol:vol:vol). All liquid handling steps were performed using Hamilton Robotics STARlet robotic platform with the Anti Droplet Control feature for organic solvents pipetting.
MS data acquisition for lipidomics
Samples were analyzed by direct infusion on a QExactive mass spectrometer (Thermo Fisher Scientific) equipped with a TriVersa NanoMate ion source (Advion Biosciences). Samples were analyzed in both positive and negative ion modes with a resolution of Rm/z = 200 = 280,000 for MS and Rm/z = 200 = 17,500 for tandem MS (MSMS) experiments, in a single acquisition. MSMS was triggered by an inclusion list encompassing corresponding MS mass ranges scanned in 1-D increments (Surma et al., 2015). Both MS and MSMS data were combined to monitor CE, DAG, and TAG ions as ammonium adducts; PC and PC O- as acetate adducts; and CL, phosphatidate, PE, PE O-, phosphatidylglycerol, PI, and phosphatidylserine as deprotonated anions. MS only was used to monitor lyso-phosphatidate, LPE, LPE O-, lyso-PI, and lyso-phosphatidylserine as deprotonated anions; and ceramide, hexosylceramide, sphingomyelin, LPC, and LPC O- as acetate adducts.
Lipidomics data analysis and postprocessing
Data were analyzed with in-house developed lipid identification software based on LipidXplorer (Herzog et al., 2011, 2012). Data postprocessing and normalization were performed using an in-house developed data management system. Only lipid identifications with a signal-to-noise ratio greater than five, and a signal intensity fivefold higher than in corresponding blank samples were considered for further data analysis.
Online supplemental material
Fig. S1 shows the validation of ATGL and LAL siRNA knockdown and the accumulation of triglycerides after lipase inhibition. Fig. S2 shows the effect of the HSL/MGL inhibitor CAY10499 on LD size, number, and content in the presence and absence of LALi. Fig. S3 shows the association of lipolysis and lipophagy markers with LDs in AML12 cells treated with the lysosome inhibitor chloroquine. Fig. S4 shows additional biochemical and lipidomic characterization of LD1 and LD2 fractions from AML12 cells and whole rat livers. Fig. S5 shows transmission electron micrographs detailing the ultrastructure of LDs within lipophagic organelles from freshly isolated primary rat hepatocytes.
Supplementary Material
Acknowledgments
We thank the members of the McNiven laboratory, especially Hong Cao, Bridget Mehall, and Katherine Johnson, for helpful discussion and technical assistance.
This work was supported by National Institutes of Health grants R01DK044650 (M.A. McNiven), R01AA020735 (M.A. McNiven and C.A. Casey), T32DK007352 (R.J. Schulze and M.B. Schott), and K99AA026877 (M.B. Schott); the Mayo Clinic Robert and Arlene Kogod Center on Aging (M.B. Schott); the U.S. Department of Veterans Affairs (C.A. Casey); and a Pilot and Feasibility Award to M.B. Schott from the Mayo Clinic Center for Cell Signaling in Gastroenterology (P30DK084567).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare no competing financial interests.
Author contributions: M.B. Schott, S.G. Weller, R.J. Schulze, and M.A. McNiven conceived the study, designed experiments, interpreted data, and wrote the manuscript. M.B. Schott, S.G. Weller, R.J. Schulze, E.W. Krueger, and K. Drizyte-Miller performed and analyzed experiments. C.A. Casey provided vital reagents and critical expertise.
References
- Bernstein D.L., Hülkova H., Bialer M.G., and Desnick R.J.. 2013. Cholesteryl ester storage disease: review of the findings in 135 reported patients with an underdiagnosed disease. J. Hepatol. 58:1230–1243. 10.1016/j.jhep.2013.02.014 [DOI] [PubMed] [Google Scholar]
- Brasaemle D.L., and Wolins N.E.. 2016. Isolation of Lipid Droplets from Cells by Density Gradient Centrifugation. Curr. Protoc. Cell Biol. 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Garcia F., and McNiven M.A.. 1998. Differential distribution of dynamin isoforms in mammalian cells. Mol. Biol. Cell. 9:2595–2609. 10.1091/mbc.9.9.2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A.D. 1997. Hepatic uptake of chylomicron remnants. J. Lipid Res. 38:2173–2192. [PubMed] [Google Scholar]
- Crawford S.E., and Borensztajn J.. 1999. Plasma clearance and liver uptake of chylomicron remnants generated by hepatic lipase lipolysis: evidence for a lactoferrin-sensitive and apolipoprotein E-independent pathway. J. Lipid Res. 40:797–805. [PubMed] [Google Scholar]
- Ding W.X., Li M., Chen X., Ni H.M., Lin C.W., Gao W., Lu B., Stolz D.B., Clemens D.L., and Yin X.M.. 2010. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology. 139:1740–1752. 10.1053/j.gastro.2010.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edens N.K., Leibel R.L., and Hirsch J.. 1990. Mechanism of free fatty acid re-esterification in human adipocytes in vitro. J. Lipid Res. 31:1423–1431. [PubMed] [Google Scholar]
- Ejsing C.S., Sampaio J.L., Surendranath V., Duchoslav E., Ekroos K., Klemm R.W., Simons K., and Shevchenko A.. 2009. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. USA. 106:2136–2141. 10.1073/pnas.0811700106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes L.C., Di Benedetto G., and Scorrano L.. 2011. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 13:589–598. 10.1038/ncb2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog R., Schwudke D., Schuhmann K., Sampaio J.L., Bornstein S.R., Schroeder M., and Shevchenko A.. 2011. A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol. 12:R8 10.1186/gb-2011-12-1-r8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog R., Schuhmann K., Schwudke D., Sampaio J.L., Bornstein S.R., Schroeder M., and Shevchenko A.. 2012. LipidXplorer: a software for consensual cross-platform lipidomics. PLoS One. 7:e29851 10.1371/journal.pone.0029851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh K., Lee Y.K., Londos C., Raaka B.M., Dalen K.T., and Kimmel A.R.. 2012. Perilipin family members preferentially sequester to either triacylglycerol-specific or cholesteryl-ester-specific intracellular lipid storage droplets. J. Cell Sci. 125:4067–4076. 10.1242/jcs.104943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huotari J., and Helenius A.. 2011. Endosome maturation. EMBO J. 30:3481–3500. 10.1038/emboj.2011.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins C.M., Mancuso D.J., Yan W., Sims H.F., Gibson B., and Gross R.W.. 2004. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J. Biol. Chem. 279:48968–48975. 10.1074/jbc.M407841200 [DOI] [PubMed] [Google Scholar]
- Kaur J., and Debnath J.. 2015. Autophagy at the crossroads of catabolism and anabolism. Nat. Rev. Mol. Cell Biol. 16:461–472. 10.1038/nrm4024 [DOI] [PubMed] [Google Scholar]
- Kennedy E.P., and Lehninger A.L.. 1949. Oxidation of fatty acids and tricarboxylic acid cycle intermediates by isolated rat liver mitochondria. J. Biol. Chem. 179:957–972. [PubMed] [Google Scholar]
- Khan S.A., Sathyanarayan A., Mashek M.T., Ong K.T., Wollaston-Hayden E.E., and Mashek D.G.. 2015. ATGL-catalyzed lipolysis regulates SIRT1 to control PGC-1α/PPAR-α signaling. Diabetes. 64:418–426. 10.2337/db14-0325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., and Czaja M.J.. 2013. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ. 20:3–11. 10.1038/cdd.2012.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Lopez N., Garcia-Macia M., Sahu S., Athonvarangkul D., Liebling E., Merlo P., Cecconi F., Schwartz G.J., and Singh R.. 2016. Autophagy in the CNS and Periphery Coordinate Lipophagy and Lipolysis in the Brown Adipose Tissue and Liver. Cell Metab. 23:113–127. 10.1016/j.cmet.2015.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason R.R., Meex R.C., Lee-Young R., Canny B.J., and Watt M.J.. 2012. Phosphorylation of adipose triglyceride lipase Ser(404) is not related to 5′-AMPK activation during moderate-intensity exercise in humans. Am. J. Physiol. Endocrinol. Metab. 303:E534–E541. 10.1152/ajpendo.00082.2012 [DOI] [PubMed] [Google Scholar]
- Murugesan S., Goldberg E.B., Dou E., and Brown W.J.. 2013. Identification of diverse lipid droplet targeting motifs in the PNPLA family of triglyceride lipases. PLoS One. 8:e64950 10.1371/journal.pone.0064950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath S., Dancourt J., Shteyn V., Puente G., Fong W.M., Nag S., Bewersdorf J., Yamamoto A., Antonny B., and Melia T.J.. 2014. Lipidation of the LC3/GABARAP family of autophagy proteins relies on a membrane-curvature-sensing domain in Atg3. Nat. Cell Biol. 16:415–424. 10.1038/ncb2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.B., Louie S.M., Daniele J.R., Tran Q., Dillin A., Zoncu R., Nomura D.K., and Olzmann J.A.. 2017. DGAT1-Dependent Lipid Droplet Biogenesis Protects Mitochondrial Function during Starvation-Induced Autophagy. Dev. Cell. 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong K.T., Mashek M.T., Bu S.Y., Greenberg A.S., and Mashek D.G.. 2011. Adipose triglyceride lipase is a major hepatic lipase that regulates triacylglycerol turnover and fatty acid signaling and partitioning. Hepatology. 53:116–126. 10.1002/hep.24006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paar M., Jüngst C., Steiner N.A., Magnes C., Sinner F., Kolb D., Lass A., Zimmermann R., Zumbusch A., Kohlwein S.D., and Wolinski H.. 2012. Remodeling of lipid droplets during lipolysis and growth in adipocytes. J. Biol. Chem. 287:11164–11173. 10.1074/jbc.M111.316794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnon J., Matzaris M., Stark R., Meex R.C., Macaulay S.L., Brown W., O’Brien P.E., Tiganis T., and Watt M.J.. 2012. Identification and functional characterization of protein kinase A phosphorylation sites in the major lipolytic protein, adipose triglyceride lipase. Endocrinology. 153:4278–4289. 10.1210/en.2012-1127 [DOI] [PubMed] [Google Scholar]
- Perret B., Mabile L., Martinez L., Tercé F., Barbaras R., and Collet X.. 2002. Hepatic lipase: structure/function relationship, synthesis, and regulation. J. Lipid Res. 43:1163–1169. [PubMed] [Google Scholar]
- Prevost C., Sharp M.E., Kory N., Lin Q., Voth G.A., Farese R.V. Jr., and Walther T.C.. 2018. Mechanism and Determinants of Amphipathic Helix-Containing Protein Targeting to Lipid Droplets. Dev Cell. 44:73–86.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold A.S., Cohen S., and Lippincott-Schwartz J.. 2015. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev. Cell. 32:678–692. 10.1016/j.devcel.2015.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner Ž., Guardamagna O., Nair D., Soran H., Hovingh K., Bertolini S., Jones S., Ćorić M., Calandra S., Hamilton J., et al. 2014. Lysosomal acid lipase deficiency--an under-recognized cause of dyslipidaemia and liver dysfunction. Atherosclerosis. 235:21–30. 10.1016/j.atherosclerosis.2014.04.003 [DOI] [PubMed] [Google Scholar]
- Rye K.A., Clay M.A., and Barter P.J.. 1999. Overview of plasma lipid transport. In Plasma Lipids and their Role in Disease. Barter P.J. and Rye K.A., editors. Harwood Academic Publishers, Sydney: 1–16. 10.4324/9780203304815_chapter_1 [DOI] [Google Scholar]
- Sampaio J.L., Gerl M.J., Klose C., Ejsing C.S., Beug H., Simons K., and Shevchenko A.. 2011. Membrane lipidome of an epithelial cell line. Proc. Natl. Acad. Sci. USA. 108:1903–1907. 10.1073/pnas.1019267108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyanarayan A., Mashek M.T., and Mashek D.G.. 2017. ATGL Promotes Autophagy/Lipophagy via SIRT1 to Control Hepatic Lipid Droplet Catabolism. Cell Reports. 19:1–9. 10.1016/j.celrep.2017.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott M.B., Rasineni K., Weller S.G., Schulze R.J., Sletten A.C., Casey C.A., and McNiven M.A.. 2017. β-Adrenergic induction of lipolysis in hepatocytes is inhibited by ethanol exposure. J. Biol. Chem. 292:11815–11828. 10.1074/jbc.M117.777748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze R.J., Drižytė K., Casey C.A., and McNiven M.A.. 2017. Hepatic Lipophagy: New Insights into Autophagic Catabolism of Lipid Droplets in the Liver. Hepatol Commun. 1:359–369. 10.1002/hep4.1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L., Hillebrand A., Wang D.Q., and Liu M.. 2012. Isolation and primary culture of rat hepatic cells. J. Vis. Exp. (64):3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., and Cuervo A.M.. 2012. Lipophagy: connecting autophagy and lipid metabolism. Int. J. Cell Biol. 2012:282041 10.1155/2012/282041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A.M., and Czaja M.J.. 2009. Autophagy regulates lipid metabolism. Nature. 458:1131–1135. 10.1038/nature07976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E., Goldberg E.B., Makarova K.S., Lin L., Brown W.J., and Jackson C.L.. 2006. ATGL has a key role in lipid droplet/adiposome degradation in mammalian cells. EMBO Rep. 7:106–113. 10.1038/sj.embor.7400559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni K.G., Mardones G.A., Sougrat R., Smirnova E., Jackson C.L., and Bonifacino J.S.. 2009. Coatomer-dependent protein delivery to lipid droplets. J. Cell Sci. 122:1834–1841. 10.1242/jcs.045849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Q.P., Du W., Ji Q., Xue B., Jiang D., Zhu Y., Ren H., Zhang C., Lou J., Yu L., and Sun Y.. 2016. Vesicle Size Regulates Nanotube Formation in the Cell. Sci. Rep. 6:24002 10.1038/srep24002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surma M.A., Herzog R., Vasilj A., Klose C., Christinat N., Morin-Rivron D., Simons K., Masoodi M., and Sampaio J.L.. 2015. An automated shotgun lipidomics platform for high throughput, comprehensive, and quantitative analysis of blood plasma intact lipids. Eur. J. Lipid Sci. Technol. 117:1540–1549. 10.1002/ejlt.201500145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiam A.R., and Beller M.. 2017. The why, when and how of lipid droplet diversity. J. Cell Sci. 130:315–324. 10.1242/jcs.192021 [DOI] [PubMed] [Google Scholar]
- Thiam A.R., Farese R.V. Jr., and Walther T.C.. 2013. The biophysics and cell biology of lipid droplets. Nat. Rev. Mol. Cell Biol. 14:775–786. 10.1038/nrm3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan M., Berger J.E., and Steinberg D.. 1964. Hormone-Sensitive Lipase and Monoglyceride Lipase Activities in Adipose Tissue. J. Biol. Chem. 239:401–409. [PubMed] [Google Scholar]
- Villena J.A., Roy S., Sarkadi-Nagy E., Kim K.H., and Sul H.S.. 2004. Desnutrin, an adipocyte gene encoding a novel patatin domain-containing protein, is induced by fasting and glucocorticoids: ectopic expression of desnutrin increases triglyceride hydrolysis. J. Biol. Chem. 279:47066–47075. 10.1074/jbc.M403855200 [DOI] [PubMed] [Google Scholar]
- Wang L., Zhou J., Yan S., Lei G., Lee C.H., and Yin X.M.. 2017. Ethanol-triggered Lipophagy Requires SQSTM1 in AML12 Hepatic Cells. Sci. Rep. 7:12307 10.1038/s41598-017-12485-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H., Shvets E., and Elazar Z.. 2009. Lipophagy: selective catabolism designed for lipids. Dev. Cell. 16:628–630. 10.1016/j.devcel.2009.05.001 [DOI] [PubMed] [Google Scholar]
- Wu J.W., Wang S.P., Alvarez F., Casavant S., Gauthier N., Abed L., Soni K.G., Yang G., and Mitchell G.A.. 2011. Deficiency of liver adipose triglyceride lipase in mice causes progressive hepatic steatosis. Hepatology. 54:122–132. 10.1002/hep.24338 [DOI] [PubMed] [Google Scholar]
- Xie X., Langlais P., Zhang X., Heckmann B.L., Saarinen A.M., Mandarino L.J., and Liu J.. 2014. Identification of a novel phosphorylation site in adipose triglyceride lipase as a regulator of lipid droplet localization. Am. J. Physiol. Endocrinol. Metab. 306:E1449–E1459. 10.1152/ajpendo.00663.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., and Ren D.. 2015. Lysosomal physiology. Annu. Rev. Physiol. 77:57–80. 10.1146/annurev-physiol-021014-071649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechner R., Madeo F., and Kratky D.. 2017. Cytosolic lipolysis and lipophagy: two sides of the same coin. Nat. Rev. Mol. Cell Biol. 18:671–684. 10.1038/nrm.2017.76 [DOI] [PubMed] [Google Scholar]
- Zhang S., Wang Y., Cui L., Deng Y., Xu S., Yu J., Cichello S., Serrero G., Ying Y., and Liu P.. 2016. Morphologically and Functionally Distinct Lipid Droplet Subpopulations. Sci. Rep. 6:29539 10.1038/srep29539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann R., Strauss J.G., Haemmerle G., Schoiswohl G., Birner-Gruenberger R., Riederer M., Lass A., Neuberger G., Eisenhaber F., Hermetter A., and Zechner R.. 2004. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 306:1383–1386. 10.1126/science.1100747 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.