Abstract
Glioblastoma (GBM) is a malignant tumor of the central nervous system with high mortality rates. Gene expression profiling may determine the chemosensitivity of GBMs. However, the molecular mechanisms underlying GBM remain to be determined. To screen the novel key genes in its occurrence and development, two glioma databases, GSE122498 and GSE104291, were analyzed in the present study. Bioinformatics analyses were performed using the Database for Annotation, Visualization and Integrated Discovery, the Search Tool for the Retrieval of Interacting Genes, Cytoscape, cBioPortal, and Gene Expression Profiling Interactive Analysis softwares. Patients with recurrent GBM showed worse overall survival rate. Overall, 341 differentially expressed genes (DEGs) were authenticated based on two microarray datasets, which were primarily enriched in ‘cell division’, ‘mitotic nuclear division’, ‘DNA replication’, ‘nucleoplasm’, ‘cytosol, nucleus’, ‘protein binding’, ‘ATP binding’, ‘protein C-terminus binding’, ‘the cell cycle’, ‘DNA replication’, ‘oocyte meiosis’ and ‘valine’. The protein-protein interaction network was composed of 1,799 edges and 237 nodes. Its significant module had 10 hub genes, and CDK1, BUB1B, NDC80, NCAPG, BUB1, CCNB1, TOP2A, DLGAP5, ASPM and MELK were significantly associated with carcinogenesis and the development of GBM. The present study indicated that the DEGs and hub genes, identified based on bioinformatics analyses, had significant diagnostic value for patients with GBM.
Keywords: glioblastoma, bioinformatics analysis, hub genes, microarray datasets, differentially expressed genes
Introduction
Gliomas are one of the common primary neoplasms of the brain, and it is caused by carcinogenesis of the brain and spinal glial cells (1). The annual incidence is 3–8 cases per 100,000. Just as with other types of tumor (such as gastric, breast and colorectal cancer), gliomas are also induced by the interaction between genetic high-risk factors and environmental factors, including air pollution and ionizing radiation (2). Certain known diseases are also genetic susceptibility factors of gliomas, such as neurofibromatosis (type I) and tuberculous sclerotic diseases (3). According to the grading system set by the World Health Organization (WHO) (4), gliomas are classified into four grades: Grade 1, the slightest with the most favorable prognosis; to grade 4, the most severe with the worst prognosis of all grades. The anaplastic gliomas, in terms of traditional cytopathology, are classified as grade 3, and glioblastoma (GBM) is classified as grade 4 (5).
GBM is the most common and lethal malignant primary brain tumor in adults, and is a member of a group of tumors known as gliomas (6). GBM is evolved from astroid neuroglial cell lineage that supports neurocytes, and it accounts for 12–15% of intracranial tumors, and 50–60% of astrocytic tumors (7). The molecular mechanism underlying GBM progression remains unclear, however there is an increasing number of studies suggesting genetic mutations (8–10).
Numerous previous studies have performed bioinformatic analyses to investigate differentially expressed genes (DEGs) in patients with GBM, as well as their roles in different pathways, molecular functions and biological processes (11–13). The overall survival rate is different in patients with GBM to those with different mutation statuses of isocitrate dehydrogenase (IDH), and a previous study has demonstrated that patients with GBM that possess mutated IDH1 have an improved prognosis (14). A total of 23 differently expressed microRNAs (miRNAs) were selected in patients with GMB that possessed mutated and wild IDH1, and these miRNAs were identified as IDH1 mutation miRNAs (11). A molecular marker consisting of 10 miRNAs was identified and validated in the GBM in a previous study; among these, 7 were considered dangerous miRNAs (mir-31, mir-222, mir-148a, mir-221, mir-146b, mir-200b and mir-193a), while the other 3 were considered protective (mir-20a, mir-106a and mir-17-5p) (12). The study screened 3 prognostic genes, including formyl peptide receptor 3, IKBKB interacting protein and S100 calcium binding protein A9, in the mRNA expression profile of a Coarse Grained Parallel Genetic Algorithm, and the biomarker composed of these 3 genes was indicated to serve a prognostic value in patients with GBM with promoter methylation of O(6)-methylguanine DNA methyltransferase (MGMT) (13). A previous study aimed to predict the prognosis of patients with GBM by screening immune-associated molecular markers (15). Arimappamagan et al (16) identified 14 prognostic genes in patients with GBM, and through pathway analysis of the Database for Annotation, Visualization and Integrated Discovery (DAVID), it was revealed that these differential genes were gathering in the inflammatory and immune response pathways (16).
Microarray technology allows simultaneous analysis of changes in the expression of multiple genes to obtain gene sets that could predict GBM (17). DEGs are associated with the grade of tumor and the prognosis of patients with glioma (18). Key molecular markers may serve as independent impact factors (19). Further studies should investigate the underlying mechanisms associated with the abnormally expressed genetic molecular markers. These genetic molecular markers have an impact on the occurrence and malignant progression of GBM, and could serve as therapeutic targets (20). Therefore, the detection and analysis of reliable gene targets of GBM is required (21,22).
The present study aimed to analyze two microarray databases of human gene sets from public datasets, and identify DEGs between patients with GBM and healthy individuals. Subsequently, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology (GO) analyses were performed. In addition, protein-protein interaction (PPI) network analyses, and co-expression network analyses were conducted to help demonstrate molecular targets underlying carcinogenesis of GBM. Overall, 10 hub genes and 341 DEGs were authenticated, which may serve as potential molecular biomarkers for GBM.
Materials and methods
Access to public data
The Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) is an open platform to store genetic data (23). Two expression profiling datasets [GSE122498 (GPL570 platform) (24) and GSE104291 (GPL570 platform) (25)] were obtained from the GEO. The datasets of GSE122498 contained 1 normal sample and 16 GBM samples. Similarly, GSE104291 consisted of 2 normal samples and 24 GBM samples.
DEGs identified by GEO2R
GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) is an interactive online tool to identify DEGs from GEO series (26). GEO2R could be applied to distinguish DEGs between normal brain tissue and GBM tissue samples. According to the method by Benjamini and Hochberg (false discovery rate) (27), the tool could alter the P-values, so as to obtain the adjusted P-values (adj. P), and to maintain a balance between the possibility of false-positives and detection of statistically significant genes. If one probe set does not have the homologous gene, or if one gene has numerous probe sets, the data will be removed. The rule of statistical significance is that adj. P≤0.01 and fold change (FC) ≥2 were considered to indicate a statistically significant result.
Functional annotation for DEGs with the KEGG and GO analysis
DAVID (https://david.ncifcrf.gov/home.jsp; version 6.8), is an online analysis tool suite with the function of Integrated Discovery and Annotation (28). GO is an ontology tool widely used in bioinformatics, which covers three aspects of biology, including ‘biological process (BP)’, ‘cellular component (CC)’ and ‘molecular function (MF)’ (29). KEGG (www.kegg.jp), is one of the most commonly used biological information databases in the world (30). To analyze GO and the biological pathway information of the DEGs, the DAVID online tool was implemented. P<0.05 was considered to indicate a statistically significant result. The UCSC Genome Browser (genome.ucsc.edu) is a graphical viewer for exploring genome annotations and was used to hierarchically cluster key genes.
Construction of the PPI network and identification of significant modules
Search Tool for the Retrieval of Interacting Genes (STRING; string.embl.de), an online open tool, was applied to construct one the PPI network (31). Cytoscape (version 3.6.1), a free visualization software, was used to present the network (32). A confidence score >0.4 was considered the criterion of judgment. The Molecular Complex Detection (MCODE) (version 1.5.1; a plug-in of Cytoscape) subsequently identified the most important module of the network map (33). The criteria of the MCODE analysis is that the degree of cut-off=2, MCODE scores >5, Max depth=100, node score cut-off=0.2, and k-score=2.
Analysis and identification of hub genes
When the degrees were set (degrees ≥10), the hub genes were excavated. Subsequently, with the KEGG and GO analysis in the DAVID database, functional annotation for the hub genes was performed. One co-expression network of these hub genes and a survival analysis was obtained using cBioPortal (www.cbioportal.org) (34). Furthermore, The Biological Networks Gene Oncology tool (version 3.0.3) was used to analyze and visualize the hub genes’ ‘CC’, ‘BP’ and ‘MF’ (35). The clustering analysis of hub genes was performed using OmicShare (version: 2015–2019; www.omicshare.com/tools/index.php), and the University of California Santa Cruz (UCSC) Xena software (xena.ucsc.edu/welcome-to-ucsc-xena) was used to securely analyze and visualize the hub genes in the scope of public genomic datasets. The expression profiles of these hub genes were analyzed and displayed using the online database Gene Expression Profiling Interactive Analysis (GEPIA; gepia.cancer-pku.cn). The unpaired Student's t-test was used make the comparisons between the normal sample and GBM samples. P<0.05 was used to indicate statistically significant results. The Kaplan-Meier plotter (www.kmplot.com) was used to perform the overall survival rate analysis. The log rank test was used to compare survival curves.
Results
Screening of DEGs in GBM samples
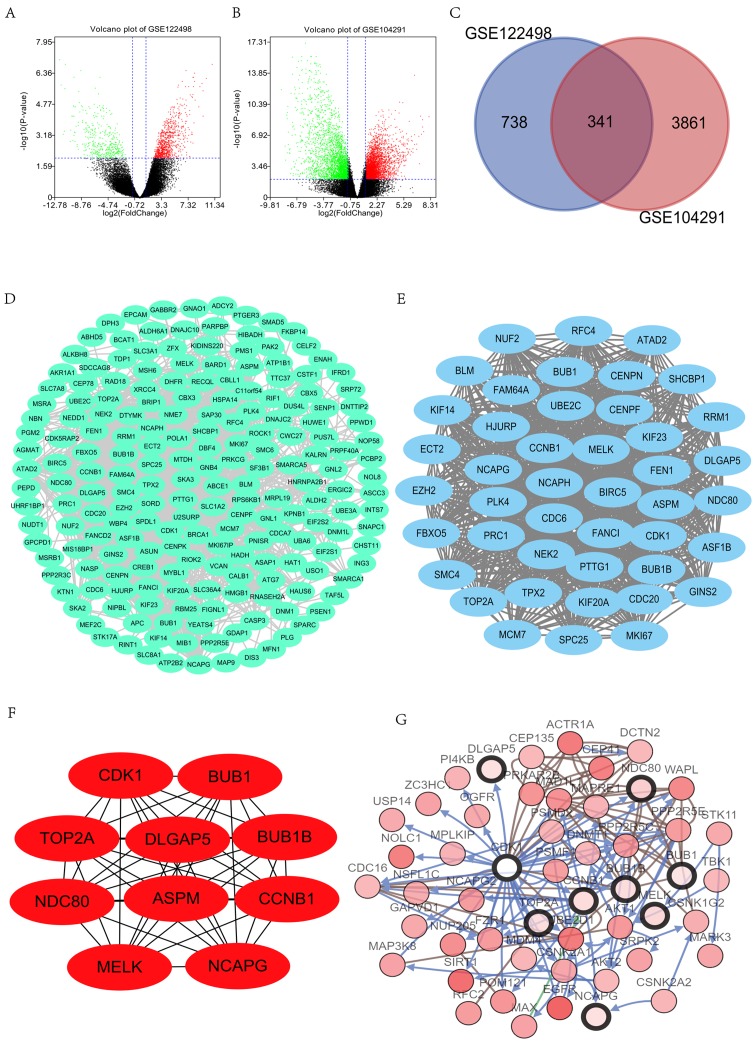
Following the analysis of the datasets (GSE122498 and GSE104291) with GEO2R, the differences between control and GBM tissues were presented in volcano plots (Fig. 1A and B). The analysis of GSE122498 and GSE104291 identified 1,079 and 4,202 DEGs, respectively (Fig. 1C). The Venn diagram revealed that the common part between the 2 datasets included 341 DEGs.
Figure 1.
Identification of DEGs and PPI network construction. (A) Volcano plot presents the difference between non-GBM and GBM tissues after analysis of the datasets GSE122498 with GEO2R. Downregulated genes are presented in green, and upregulated genes are presented in red. (B) Volcano plot presents the difference between non-GBM and GBM tissues after analysis of the datasets GSE104291 with GEO2R. The green presents the down-regulated genes, and the red presents the up-regulated genes. (C) Venn diagram indicated that 341 genes were contained in the GSE122498 and GSE104291 datasets simultaneously. (D) The PPI network of DEGs was constructed using Cytoscape. (E) Most significant module was obtained from PPI network of DEGs using MCODE, including 44 nodes and 884 edges. (F) Network of hub genes. (G) Hub genes and their co-expression genes were analyzed using cBioPortal. Nodes with a bold black outline represent hub genes. Nodes with a thin black outline represent the co-expression genes. *P<0.05. GBM, glioblastoma; DEGs, differentially expressed genes; PPI, protein-protein interaction; GEO, Gene Expression Omnibus; MCODE, Molecular Complex Detection.
Functional annotation for DEGs with the KEGG and GO analysis
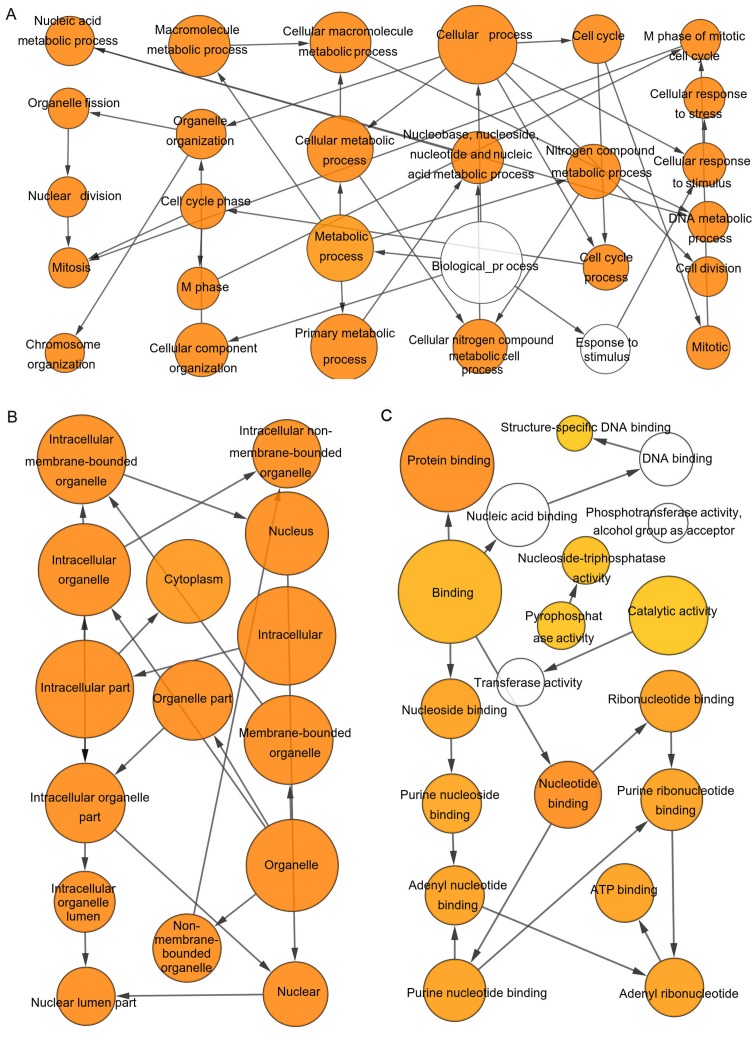
The results of the GO analysis demonstrated that variations in the BP were primarily enriched in ‘cell division’, ‘mitotic nuclear division’ and ‘DNA replication’. Changes in CC were primarily enriched in the nucleoplasm, cytosol and nucleus. The variations in MF were enriched in protein binding, ATP binding and protein C-terminus binding. The KEGG analysis demonstrated that DEGs were prevailingly enriched in the ‘cell cycle’, ‘DNA replication’, ‘oocyte meiosis’, ‘valine, leucine and isoleucine degradation’, ‘fanconi anemia pathway’, ‘GABAergic synapse’, ‘dopaminergic synapse’, and ‘endocrine and other factor-regulated calcium reabsorption’ (Table I).
Table I.
GO and KEGG pathway enrichment analysis of DEGs in glioblastoma samples.
| Term | Description | Count in gene set | P-value |
|---|---|---|---|
| GO:0051301 | Cell division | 30 | 1.16×10−11 |
| GO:0007067 | Mitotic nuclear division | 24 | 1.85×10−10 |
| GO:0006260 | DNA replication | 18 | 3.67×10−9 |
| GO:0005654 | Nucleoplasm | 112 | 2.52×10−18 |
| GO:0005829 | Cytosol | 106 | 1.73×10−10 |
| GO:0005634 | Nucleus | 149 | 2.35×10−10 |
| GO:0005515 | Protein binding | 203 | 1.93×10−8 |
| GO:0005524 | ATP binding | 51 | 7.77×10−6 |
| GO:0008022 | Protein C-terminus binding | 12 | 3.99×10−4 |
| hsa04110 | Cell cycle | 9 | 0.002 |
| hsa03030 | DNA replication | 5 | 0.004 |
| hsa04114 | Oocyte meiosis | 8 | 0.004 |
| hsa00280 | Valine, leucine and isoleucine degradation | 5 | 0.010 |
| hsa03460 | Fanconi anemia pathway | 5 | 0.016 |
| hsa04727 | GABAergic synapse | 6 | 0.019 |
| hsa04728 | Dopaminergic synapse | 7 | 0.030 |
| hsa04961 | Endocrine and other factor-regulated calcium reabsorption | 4 | 0.049 |
GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; DEGs, differentially expressed genes.
Construction of the PPI network and identification of significant module and hub genes
The PPI network was constructed and significant modules were identified, with 1,799 edges and 237 nodes in the PPI network (Fig. 1D), and 884 edges and 44 nodes in the significant module (Fig. 1E). Degrees ≥10 were considered as the criterion of judgment, which was the criterion to determine significance. Overall, 10 genes were identified as hub genes within Cytoscape: CDK1, BUB1B, NDC80, NCAPG, BUB1, CCNB1, TOP2A, DLGAP5, ASPM and MELK (Fig. 1F). Among the hub genes, CDK1 and BUB1B had the highest scores, suggesting that they may play important roles in the occurrence or development of GBM. With DAVID, the KEGG and GO analyses of DEGs involved in hub genes were analyzed. The results revealed that these genes were prevailingly enriched in ‘cell division’, ‘mitotic nuclear division’, ‘cell proliferation’, ‘condensed nuclear chromosome outer kinetochore’, ‘kinetochore’, ‘condensed chromosome kinetochore’, ‘protein kinase activity’, ‘protein serine/threonine kinase activity’ and ‘histone kinase activity’. Analyses of the KEGG pathway indicated that significant genes were primarily enriched in the ‘cell cycle’, ‘progesterone-mediated oocyte maturation’, ‘oocyte meiosis’ and the ‘p53 signaling pathway’ (Table II).
Table II.
GO and KEGG pathway enrichment analysis of DEGs in the most significant module.
| Pathway ID | Pathway description | Count in gene set | P-value |
|---|---|---|---|
| GO:0051301 | Cell division | 6 | 4.50×10−7 |
| GO:0007067 | Mitotic nuclear division | 5 | 5.52×10−6 |
| GO:0008283 | Cell proliferation | 5 | 2.57×10−5 |
| GO:0000942 | Condensed nuclear chromosome outer kinetochore | 3 | 1.30×10−6 |
| GO:0000776 | Kinetochore | 3 | 6.88×10−4 |
| GO:0000777 | Condensed chromosome kinetochore | 3 | 7.94×10−4 |
| GO:0004672 | Protein kinase activity | 4 | 7.28×10−4 |
| GO:0004674 | Protein serine/threonine kinase activity | 4 | 8.33×10−4 |
| GO:0035173 | Histone kinase activity | 2 | 0.002 |
| hsa04110 | Cell cycle | 4 | 5.72×10−6 |
| hsa04914 | Progesterone-mediated oocyte maturation | 3 | 4.70×10−4 |
| hsa04114 | Oocyte meiosis | 3 | 7.66×10−4 |
| hsa04115 | p53 signaling pathway | 2 | 0.029 |
GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; DEGs, differentially expressed genes.
Hub genes analysis
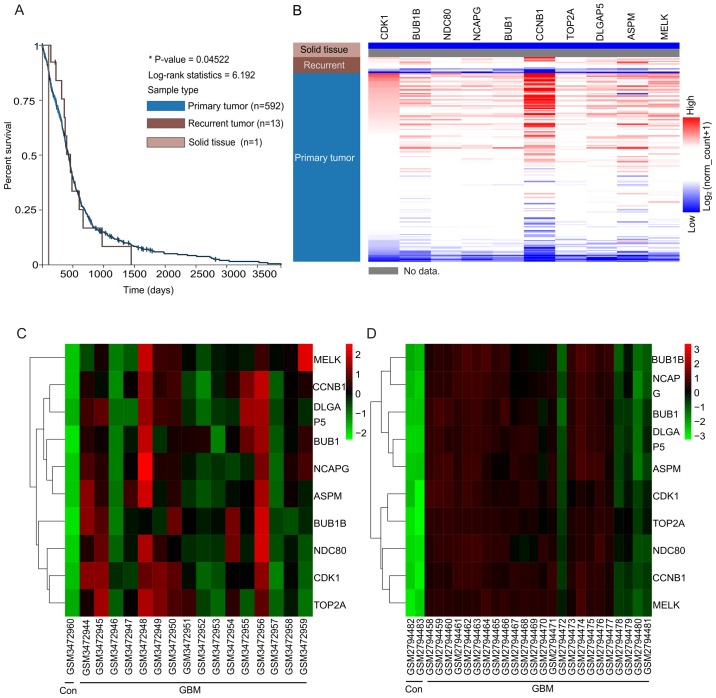
According to the Gene cards, summaries for the function of the 10 hub genes were obtained (Table III). A co-expression network of these significant genes was obtained using cBioPortal (Fig. 1G). The BP, CC and MF analysis for these genes is presented in Fig. 2A-C. Then, a Kaplan-Meier plotter was used to perform the survival analysis. Patients with recurrent GBM demonstrated worse overall survival rate (Fig. 3A). According to the UCSC analysis, hierarchical clustering indicated that these hub genes may differentiate those individuals with GBM from the normal individuals (Fig. 3B). The hub genes were identified between non-GBM samples and GBM samples. It was demonstrated that the expressions of hub genes were upregulated in the GBM (including recurrent and primary GBM), when compared with the solid tissue, non-GBM samples (Fig. 3B). Therefore, the expression patterns of hub genes did not seem to demonstrate any significant difference between primary tumors and recurrent tumors, although relapsed tumors often demonstrated higher aggressiveness compared with primary tumors. The authors suggested that if a differentiation between the recurrent tumor and the primary tumor is required, the initiatory groups should be set as recurrent and primary GBM in the GEO database. Heat maps revealed that the hub genes could differentiate the GBM samples from the non-GBM samples (Fig. 3C and D). Fig. 3C primarily presents the expression levels of hub genes in the GSE122498, but it could also demonstrate that the expression levels of all hub genes in the non-GBM samples were downregulated when compared with the GBM samples. This study may further verify the aforementioned differences in the expression levels of hub genes between non-GBM samples and GBM samples (Fig. 3D).
Table III.
Summaries for the function of 10 hub genes.
| No. | Gene symbol | Full name | Function |
|---|---|---|---|
| 1 | CDK1 | Cyclin dependent kinase 1 | Plays a key role in the control of the eukaryotic cell cycle by modulating the centrosome cycle as well as mitotic onset; promotes G2-M transition, and regulates G1 progress and G1-S transition via association with multiple interphase cyclins. |
| 2 | BUB1B | BUB1 mitotic checkpoint serine/threonine kinase B | Implicated in triggering apoptosis in polyploid cells that exit aberrantly from mitotic arrest. May play a role for tumor suppression. |
| 3 | NDC80 | NDC80, kinetochore complex component | Acts as a component of the essential kinetochore-associated NDC80 complex, which is required for chromosome segregation and spindle checkpoint activity. |
| 4 | NCAPG | Non-SMC condensin I complex subunit G | Regulatory subunit of the condensin complex, a complex required for conversion of interphase chromatin into mitotic-like condense chromo somes. |
| 5 | BUB1 | BUB1 mitotic checkpoint | Serine/threonine-protein kinase that performs 2 crucial functions during mitosis: It is essential for spindle-assembly checkpoint signaling and for correct chromosome alignment. |
| 6 | CCNB1 | Cyclin B1 | Essential for the control of the cell cycle at the G2/M (mitosis) transition. |
| 7 | TOP2A | Topoisomerase | Diseases associated with TOP2A include Female Breast Cancer and |
| (DNA) II alpha | Malignant Peripheral Nerve Sheath Tumor. Among its related pathways are Cell Cycle, Mitotic and SUMOylation. | ||
| 8 | DLGAP5 | DLG associated protein 5 | Potential cell cycle regulator that may play a role in carcinogenesis of cancer cells. Mitotic phosphoprotein regulated by the ubiquitin- proteasome pathway. |
| 9 | ASPM | Abnormal spindle microtubule assembly | Involved in mitotic spindle regulation and coordination of mitotic. processes |
| 10 | MELK | Maternal embryonic leucine zipper kinase | Diseases associated with MELK include Uterine Corpus Endometrial Carcinoma. Among its related pathways are Neuroscience. |
Figure 2.
Enrichment analysis via The Biological Networks Gene Oncology tool. (A) Biological process, (B) cellular components, and (C) molecular function analysis of hub genes was constructed. The color depth of nodes refers to the corrected P-value of ontologies, and the deepest shade represented the most significant enrichment term. The size of nodes refers to the numbers of genes that are involved in the ontologies. P<0.01 was considered to indicate a statistically significant difference.
Figure 3.
University of California Santa Cruz analysis, and heatmaps of the expression levels of hub genes. (A) Overall survival rate analysis of patients with GBM. P<0.05 was considered statistically significant. (B) Hierarchical clustering of hub genes. The samples under the pink bar are normal samples, the samples under the brown bar are recurrent GBM samples and the samples under the blue bar are primary GBM samples. Red, upregulation of genes; blue, downregulation of genes. (C) Hierarchical clustering showed that the hub genes could differentiate the non-GBM samples from the GBM samples in the GSE122498. (D) Hierarchical clustering showed that the hub genes could differentiate the non-GBM samples from the GBM samples in the GSE104291 dataset. Red, upregulation of genes; green, downregulation of genes; GBM, glioblastoma.
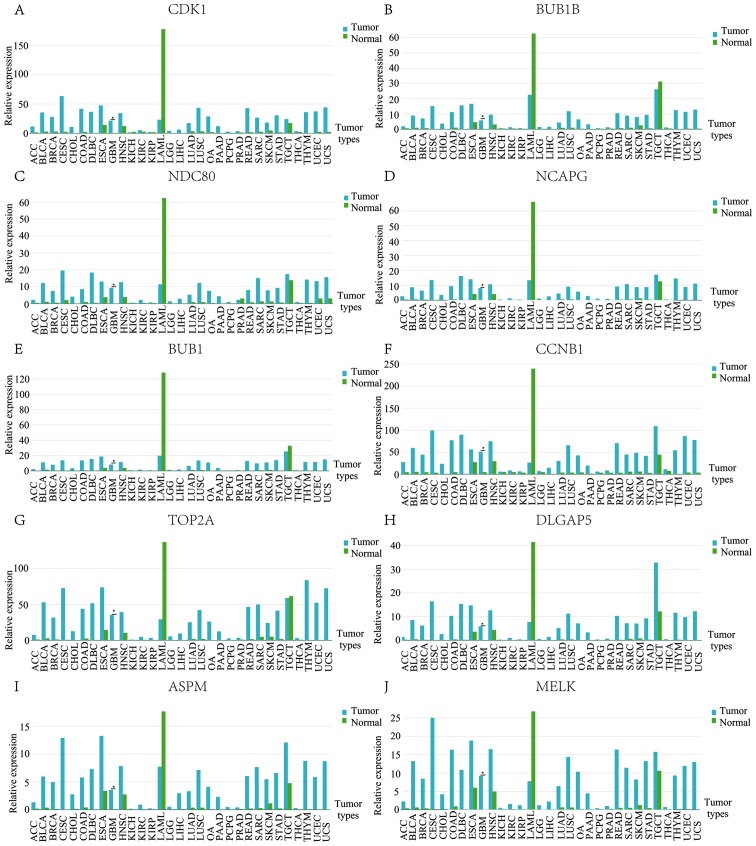
As there were different samples or individuals between the GSE122498 and GSE104291 databases, the names of the different samples are presented at the bottom of Fig. 3C and D. In addition, individual variation exists between the different samples. Therefore, the clustering patterns between datasets GSE122498 and GSE104291 were similar, but not identical. The expression profile of hub genes in human tissue was demonstrated using GEPIA. It was revealed that these genes in GBM were present in higher levels when compared with the matched normal samples (Fig. 4).
Figure 4.
Expression profiles of 10 hub genes in human tissue. (A) CDK1, (B) BUB1B, (C) NDC80, (D) NCAPG, (E) BUB1, (F) CCNB1, (G) TOP2A, (H) DLGAP5, (I) ASPM, and (J) MELK. ACC, Adrenocortical carcinoma; BLCA, Bladder Urothelial Carcinoma; BRCA, Breast invasive carcinoma; CESC, Cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, Cholangio carcinoma; COAD, Colon adenocarcinoma; DLBC, Lymphoid Neoplasm Diffuse Large B-cell Lymphoma; ESCA, Esophageal carcinoma; GBM, Glioblastoma multiforme; HNSC, Head and Neck squamous cell carcinoma; KICH, Kidney Chromophobe; KIRC, Kidney renal clear cell carcinoma; KIRP, Kidney renal papillary cell carcinoma; LAML, Acute Myeloid Leukemia; LGG, Brain Lower Grade Glioma; LIHC, Liver hepatocellular carcinoma; LUAD, Lung adenocarcinoma; LUSC, Lung squamous cell carcinoma; OA, Ovarian serous cystadenocarcinoma; PAAD, Pancreatic adenocarcinoma; PCPG, Pheochromocytoma and Paraganglioma; PRAD, Prostate adenocarcinoma; READ, Rectum adenocarcinoma; SARC, Sarcoma; SKCM, Skin Cutaneous Melanoma; STAD, Stomach adenocarcinoma; TGCT, Testicular Germ Cell Tumors; THCA, Thyroid carcinoma; THYM, Thymoma; UCEC, Uterine Corpus Endometrial Carcinoma; UCS, Uterine Carcinosarcoma.
Discussion
Gliomas are one of the most common primary malignant tumors of the brain, and there are different histological grades and classifications for it (19). According to the WHO, gliomas are classified into four grades: Grade I–IV; and into three pathological types: Astrocytoma, oligodendroglioma and mixed (astrocytoma and oligodendroglioma) gliomas (19). GBM belongs to grade IV glioma with high fatality rate and different severity and histological subtypes (36,37). It is not sensitive to radiotherapy or chemotherapy, and is prone to malignant progression; it lacks clear molecular classification, therapeutic targets and associated targeted drugs (19). The standard treatment for GBM is surgery, followed by radiotherapy, or radiotherapy combined with chemotherapy. If surgery is not practical, radiotherapy or radiotherapy/chemotherapy could be given (38). GBM is capable of extensively invading and infiltrating the normal surrounding brain tissue, making it impossible to completely remove the tumor tissue (5). Following surgery, radiotherapy could kill the remaining tumor cells and prevent recurrence, but it can damage a large number of normal brain cells (38). Even with the best treatment, the recurrence rate of GBM remains high, and was estimated to be 3.20/100,000 worldwide in 2018 (39). Therefore, research into an accurate understanding of the underlying molecular mechanism and reliable therapeutic targets of GBM has generated wide concern.
With the progress of gene-sequencing technology, a large number of DEGs have been identified in a number of other types of tumor (such as gastric, breast and colorectal cancer) (21,22). DEGs may serve a variety of functions in the occurrence and development of diseases, such as transcription, post-transcriptional processing and the regulation of protein expression. The present study aimed to identify the DEGs that play a key role in the occurrence and malignant process of gliomas and that may serve as molecular markers and therapeutic targets for GBM.
CDK1 is a cell cycle regulatory gene (40). According to the results from the present study, the expression levels of CDK1 in GBM tissues were significantly increased compared with normal tissues. The occurrence of tumors is a complex process with multiple damages to normal cell genomes (41). These damages include not only oncogene activation, but also inactivation or deletion of tumor suppressor gene (42). A previous study has revealed that the functional effects of polygene would eventually aggregate into the cell cycle mechanism (43). Among them, the two key checkpoints of the G1/S and G2/M phases of the cell cycle are the primary causes of malignant proliferation (44). However, a compound formed by combining CDK1 with Cyclin B1, the mitotic promoting factor, plays an important role in the G2/M checkpoint of the cell cycle (45). A previous study indicated that the positive degree of CDK1 expression could reflect the malignant degree of tongue squamous cell carcinoma (46). This in turn suggests that CDK1 overexpression may induce genetic mutations and chromosome structural abnormalities, leading to failure of checkpoint regulation of the cell cycle G2/M, which accelerates the progression of the cell cycle and excessive cell proliferation, resulting in tumor development (46). Overexpression of CDK1 was also observed in pancreatic cancer and lung cancer (47,48). Therefore, CDK1 has been indicated to play an important role in tumor occurrence as it may be associated with the occurrence and development of GBM, and the result may provide potential novel insights for further research into the association between GBM and CDK1 expression.
According to the results of the present study, compared with normal tissues, the expression of BUB1B in GBM tissues increased significantly. Mitosis is the process by which a eukaryotic cell divides into two identical cells, and it plays a crucial role in the evolution and homeostasis of multicellular organisms (49). The mitotic checkpoint is the core regulator during this process, acting as a signal-regulating mechanism that prevents the cell from entering the late stage of mitosis before all chromosomes adhere to the spindle (50). Abnormalities in mitotic checkpoints have been observed in numerous different types of tumor (such as gastric, breast and colorectal cancer), and one of the consequences of abnormalities of mitotic checkpoints is chromosomal instability that make cells more susceptible to malignancy (51). BUB1B is an important constituent protein of the mitotic checkpoint, and is a multidomain protein kinase that responds to centromere tension (52,53). Studies have demonstrated that BUB1B is overexpressed in various different types of tumor, such as renal and breast carcinoma, and its mutation and overexpression are associated with chromosomal instability (54–56). Therefore, further investigation on BUB1B may lead to a greater understanding of its importance in the GBM process, and novel ideas for investigating its molecular mechanisms and establishing more effective treatments.
The NDC80 complex is located on the outer layer of the kinetochore, linking the kinetochore and microtubules (57). It is involved in regulating the normal separation of chromosomes in mitosis, and is also crucial for spindle assembly checkpoints (58). NDC80 is the main component of the NDC80 compound and is highly expressed in actively dividing cells, such as tumor cells. NDC80 plays an important role in normal mitosis, the assembly of kinetochore, the spindle checkpoint, maintenance of chromosomal stability and the occurrence and development of tumors (59,60). According to the results from the present study, the expression levels of NDC80 in GBM tissues were significantly increased compared with normal tissues. Another study indicated that overexpression of NDC80 could result in sustained hyperactivation of mitotic checkpoints and therefore induce tumor formation (61). The expression levels of Mad2 were also significantly increased in mice with high expression of NDC80, and a previous report has demonstrated that overexpression of the Mad2 gene would cause hyperactivation of mitotic checkpoints, resulting in the production of aneuploid chromosomes, which induces tumor formation (62). In summary, the present study revealed that NDC80 is highly expressed in GBM, and high expression levels of NDC80 may play an important role in the occurrence and development of GBM. Studies on the molecular mechanism of NDC80 in the occurrence and development of GBM are useful for investigating the role of NDC80 as a target of intervention for GBM treatment. Future studies could observe tumor changes in a GBM animal model following knockdown of BUB1B (or NDC80). Upregulation of BUB1B expression (or NDC80) could then be observed and variations in the tumor could be recorded. If the knockdown of BUB1B (or NDC80) could decrease the size of GBM and upregulation of BUB1B (or NDC80) could deteriorate the GBM, then it could be suggested that a high expression level of BUB1B (or NDC80) is a risk factor for GBM development. Clinical trials could then be performed in order to verify the curative effect of gene therapy via downregulation of BUB1B (or NDC80) in patients with GBM.
The present study, however, has certain limitations. The screening of 10 key genes is based on bioinformatics analysis, which is an observational study that could only provide clues for further studies on the mechanisms underlying the occurrence and development of GBM. The screened target genes were validated by sequencing databases, such as The Cancer Genome Atlas and GEPIA. In addition, clinical samples need to be collected in order to verify the aforementioned conclusions. However, the present study only provides a potential theory or idea for the mechanism and/or development of a treatment strategy for GBM. Finally, the sample size of this research is small, and it maybe cause the false positive result. Further investigation on the association between mutations in IDHs or the methylation status of the MGMT promoter and the expression levels of hub genes identified in the present study is required.
In conclusion, the present study aimed to identify differentially expressed genes that may be present in the occurrence or development of GBM. Finally, 341 DEGs and 10 hub genes were identified between GBM samples and normal samples, which could be used as diagnostic and therapeutic biomarkers for GBM.
Acknowledgements
The authors would like to acknowledge Dr Ya-lun Dai from the Epidemiology Department of the Beijing Hospital, National Center of Gerontology, for her statistical assistance and suggestions during the submitting process.
Glossary
Abbreviations
- GBM
glioblastoma
- DAVID
Database for Annotation, Visualization and Integrated Discovery
- STRING
Search Tool for the Retrieval of Interacting Genes
- GEPIA
Gene Expression Profiling Interactive Analysis
- DEGs
differentially expressed genes
- WHO
World Health Organization
- IDH
isocitrate dehydrogenase
- MGMT
methylguanine DNA methyltransferase
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- GO
Gene Ontology
- PPI
protein-protein interaction
- GEO
Gene Expression Omnibus
- BP
biological process
- CC
cellular component
- MF
molecular function
- MCODE
Molecular Complex Detection
- UCSC
University of California Santa Cruz
Funding
This study was funded by the Capital Characteristic Clinical Application Research. (grant. no. Z161100000516236).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YFZ and LBM performed the experiment, and were major contributors in writing and submitting the manuscript. XY made substantial contributions to research conception and also drafted the research protocol. ZKH made substantial contributions to analysis and interpretation of data. MJS made substantial contributions to research conception and analysis and interpretation of data. In addition MJS was involved in drafting the manuscript. CHH and DYW were responsible for analyzing the gene expression data and generating the figures. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Laug D, Glasgow SM, Deneen B. A glial blueprint for gliomagenesis. Nat Rev Neurosci. 2018;19:393–403. doi: 10.1038/s41583-018-0014-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amirian ES, Armstrong GN, Zhou R, Lau CC, Claus EB, Barnholtz-Sloan JS, Il'yasova D, Schildkraut J, Ali-Osman F, Sadetzki S, et al. The Glioma international case-control study: A report from the genetic epidemiology of glioma international consortium. Am J Epidemiol. 2016;183:85–91. doi: 10.1093/aje/kwv235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benenemissi IH, Sifi K, Sahli LK, Semmam O, Abadi N, Satta D. Angiotensin-converting enzyme insertion/deletion gene polymorphisms and the risk of glioma in an Algerian population. Pan Afr Med J. 2019;32:197. doi: 10.11604/pamj.2019.32.197.15129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hempel JM, Schittenhelm J, Bisdas S, Brendle C, Bender B, Bier G, Skardelly M, Tabatabai G, Castaneda Vega S, Ernemann U, Klose U. In vivo assessment of tumor heterogeneity in WHO 2016 glioma grades using diffusion kurtosis imaging: Diagnostic performance and improvement of feasibility in routine clinical practice. J Neuroradiol. 2018;45:32–40. doi: 10.1016/j.neurad.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research Networ. Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA, Rheinbay E, Miller CR, Vitucci M, et al. Comprehensive, integrative genomic analysis of diffuse Lower-Grade gliomas. N Engl J Med. 2015;372:2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmid RS, Simon JM, Vitucci M, McNeill RS, Bash RE, Werneke AM, Huey L, White KK, Ewend MG, Wu J, Miller CR. Core pathway mutations induce de-differentiation of murine astrocytes into glioblastoma stem cells that are sensitive to radiation but resistant to temozolomide. Neuro Oncol. 2016;18:962–973. doi: 10.1093/neuonc/nov321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischer U, Struss AK, Hemmer D, Michel A, Henn W, Steudel WI, Meese E. PHF3 expression is frequently reduced in glioma. Cytogenet Cell Genet. 2001;94:131–136. doi: 10.1159/000048804. [DOI] [PubMed] [Google Scholar]
- 8.Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tönjes M, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 10.Ceccon G, Werner JM, Dunkl V, Tscherpel C, Stoffels G, Brunn A, Deckert M, Fink GR, Galldiks N. Dabrafenib treatment in a patient with an Epithelioid Glioblastoma and BRAF V600E Mutation. Int J Mol Sci. 2018;19:E1090. doi: 10.3390/ijms19041090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z, Bao Z, Yan W, You G, Wang Y, Li X, Zhang W. Isocitrate dehydrogenase 1 (IDH1) mutation-specific microRNA signature predicts favorable prognosis in glioblastoma patients with IDH1 wild type. J Exp Clin Cancer Res. 2013;32:59. doi: 10.1186/1756-9966-32-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srinivasan S, Patric IR, Somasundaram K. A ten-microRNA expression signature predicts survival in glioblastoma. PLoS One. 2011;6:e17438. doi: 10.1371/journal.pone.0017438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W, Zhang L, Wang Z, Yang F, Wang H, Liang T, Wu F, Lan Q, Wang J, Zhao J. A three-gene signature for prognosis in patients with MGMT promoter-methylated glioblastoma. Oncotarget. 2016;7:69991–69999. doi: 10.18632/oncotarget.11726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karsy M, Guan J, Cohen AL, Jensen RL, Colman H. New molecular considerations for glioma: IDH, ATRX, BRAF, TERT, H3 K27M. Curr Neurol Neurosci Rep. 2017;17:19. doi: 10.1007/s11910-017-0722-5. [DOI] [PubMed] [Google Scholar]
- 15.Solanki C, Sadana D, Arimappamagan A, Rao KV, Rajeswaran J, Subbakrishna DK, Santosh V, Pandey P. Impairments in quality of life and cognitive functions in long-term survivors of glioblastoma. J Neurosci Rural Pract. 2017;8:228–235. doi: 10.4103/0976-3147.203829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arimappamagan A, Somasundaram K, Thennarasu K, Peddagangannagari S, Srinivasan H, Shailaja BC, Samuel C, Patric IR, Shukla S, Thota B, et al. A fourteen gene GBM prognostic signature identifies association of immune response pathway and mesenchymal subtype with high risk group. PLoS One. 2013;8:e62042. doi: 10.1371/journal.pone.0062042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vastrad B, Vastrad C, Godavarthi A, Chandrashekar R. Molecular mechanisms underlying gliomas and glioblastoma pathogenesis revealed by bioinformatics analysis of microarray data. Med Oncol. 2017;34:182. doi: 10.1007/s12032-017-1043-x. [DOI] [PubMed] [Google Scholar]
- 18.Zhong S, Wu B, Dong X, Han Y, Jiang S, Zhang Y, Bai Y, Luo SX, Chen Y, Zhang H, Zhao G. Identification of driver genes and key pathways of glioblastoma shows JNJ-7706621 as a novel antiglioblastoma drug. World Neurosurg. 2018;109:e329–e342. doi: 10.1016/j.wneu.2017.09.176. [DOI] [PubMed] [Google Scholar]
- 19.Aldape K, Zadeh G, Mansouri S, Reifenberger G, von Deimling A. Glioblastoma: Pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129:829–848. doi: 10.1007/s00401-015-1432-1. [DOI] [PubMed] [Google Scholar]
- 20.Poon CC, Sarkar S, Yong VW, Kelly JJP. Glioblastoma-associated microglia and macrophages: Targets for therapies to improve prognosis. Brain. 2017;140:1548–1560. doi: 10.1093/brain/aww355. [DOI] [PubMed] [Google Scholar]
- 21.Kim SM, Kwon IJ, Myoung H, Lee JH, Lee SK. Identification of human papillomavirus (HPV) subtype in oral cancer patients through microarray technology. Eur Arch Otorhinolaryngol. 2018;275:535–543. doi: 10.1007/s00405-017-4828-9. [DOI] [PubMed] [Google Scholar]
- 22.Cheng Y, Ping J, Chen J. Identification of potential gene network associated with HCV-Related hepatocellular carcinoma using microarray analysis. Pathol Oncol Res. 2018;24:507–514. doi: 10.1007/s12253-017-0273-8. [DOI] [PubMed] [Google Scholar]
- 23.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hilf N, Kuttruff-Coqui S, Frenzel K, Bukur V, Stevanovic S, Gouttefangeas C, Platten M, Tabatabai G, Dutoit V, van der Burg SH, et al. Actively personalized vaccination trial for newly diagnosed glioblastoma. Nature. 2019;565:240–245. doi: 10.1038/s41586-018-0810-y. [DOI] [PubMed] [Google Scholar]
- 25.Bady P, Diserens AC, Castella V, Kalt S, Heinimann K, Hamou MF, Delorenzi M, Hegi ME. DNA fingerprinting of glioma cell lines and considerations on similarity measurements. Neuro Oncol. 2012;14:701–711. doi: 10.1093/neuonc/nos072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: Archive for functional genomics data sets-update. Nucleic Acids Res 41 (Database Issue) 2013:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solari A, Goeman JJ. Minimally adaptive BH: A tiny but uniform improvement of the procedure of Benjamini and Hochberg. Biom J. 2017;59:776–780. doi: 10.1002/bimj.201500253. [DOI] [PubMed] [Google Scholar]
- 28.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID gene functional classification tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa M. The KEGG database. Novartis Found Symp. 2002;247:91–103. doi: 10.1002/0470857897.ch8. 119-128, 244–252. [DOI] [PubMed] [Google Scholar]
- 31.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maere S, Heymans K, Kuiper M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 36.Bian EB, Li J, Xie YS, Zong G, Li J, Zhao B. LncRNAs: New players in gliomas, with special emphasis on the interaction of lncRNAs With EZH2. J Cell Physiol. 2015;230:496–503. doi: 10.1002/jcp.24549. [DOI] [PubMed] [Google Scholar]
- 37.Ostrom QT, Bauchet L, Davis FG, Deltour I, Fisher JL, Langer CE, Pekmezci M, Schwartzbaum JA, Turner MC, Walsh KM, et al. The epidemiology of glioma in adults: A ‘state of the science’ review. Neuro Oncol. 2014;16:896–913. doi: 10.1093/neuonc/nou087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perry JR, Laperriere N, O'Callaghan CJ, Brandes AA, Menten J, Phillips C, Fay M, Nishikawa R, Cairncross JG, Roa W, et al. Short-course radiation plus Temozolomide in elderly patients with Glioblastoma. N Engl J Med. 2017;376:1027–1037. doi: 10.1056/NEJMoa1611977. [DOI] [PubMed] [Google Scholar]
- 39.Peñaranda Fajardo NM, Meijer C, Kruyt FA. The endoplasmic reticulum stress/unfolded protein response in gliomagenesis, tumor progression and as a therapeutic target in glioblastoma. Biochem Pharmacol. 2016;118:1–8. doi: 10.1016/j.bcp.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 40.Sacristan MP, Ovejero S, Bueno A. Human Cdc14A becomes a cell cycle gene in controlling Cdk1 activity at the G2/M transition. Cell Cycle. 2011;10:387–391. doi: 10.4161/cc.10.3.14643. [DOI] [PubMed] [Google Scholar]
- 41.Glousker G, Touzot F, Revy P, Tzfati Y, Savage SA. Unraveling the pathogenesis of Hoyeraal-Hreidarsson syndrome, a complex telomere biology disorder. Br J Haematol. 2015;170:457–471. doi: 10.1111/bjh.13442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rooj AK, Mineo M, Godlewski J. MicroRNA and extracellular vesicles in glioblastoma: Small but powerful. Brain Tumor Pathol. 2016;33:77–88. doi: 10.1007/s10014-016-0259-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liau BB, Sievers C, Donohue LK, Gillespie SM, Flavahan WA, Miller TE, Venteicher AS, Hebert CH, Carey CD, Rodig SJ, et al. Adaptive chromatin remodeling drives glioblastoma stem cell plasticity and drug tolerance. Cell Stem Cell. 2017;20:233–246.e7. doi: 10.1016/j.stem.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamashita T, Nishimura K, Saiki R, Okudaira H, Tome M, Higashi K, Nakamura M, Terui Y, Fujiwara K, Kashiwagi K, Igarashi K. Role of polyamines at the G1/S boundary and G2/M phase of the cell cycle. Int J Biochem Cell Biol. 2013;45:1042–1050. doi: 10.1016/j.biocel.2013.02.021. [DOI] [PubMed] [Google Scholar]
- 45.Castedo M, Perfettini JL, Roumier T, Yakushijin K, Horne D, Medema R, Kroemer G. The cell cycle checkpoint kinase Chk2 is a negative regulator of mitotic catastrophe. Oncogene. 2004;23:4353–4361. doi: 10.1038/sj.onc.1207573. [DOI] [PubMed] [Google Scholar]
- 46.Wada S, Yue L, Furuta I. Prognostic significance of p34cdc2 expression in tongue squamous cell carcinoma. Oral Oncol. 2004;40:164–169. doi: 10.1016/S1368-8375(03)00146-5. [DOI] [PubMed] [Google Scholar]
- 47.Zhang R, Loganathan S, Humphreys I, Srivastava SK. Benzyl isothiocyanate-induced DNA damage causes G2/M cell cycle arrest and apoptosis in human pancreatic cancer cells. J Nutr. 2006;136:2728–2734. doi: 10.1093/jn/136.11.2728. [DOI] [PubMed] [Google Scholar]
- 48.Soria JC, Jang SJ, Khuri FR, Hassan K, Liu D, Hong WK, Mao L. Overexpression of cyclin B1 in early-stage non-small cell lung cancer and its clinical implication. Cancer Res. 2000;60:4000–4004. [PubMed] [Google Scholar]
- 49.Girke P, Seufert W. Compositional reorganization of the nucleolus in budding yeast mitosis. Mol Biol Cell. 2019;30:591–606. doi: 10.1091/mbc.E18-08-0524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guerrero PA, Tchaicha JH, Chen Z, Morales JE, McCarty N, Wang Q, Sulman EP, Fuller G, Lang FF, Rao G, McCarty JH. Glioblastoma stem cells exploit the αvβ8 integrin-TGFβ1 signaling axis to drive tumor initiation and progression. Oncogene. 2017;36:6568–6580. doi: 10.1038/onc.2017.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shing JC, Choi JW, Chapman R, Schroeder MA, Sarkaria JN, Fauq A, Bram RJ. A novel synthetic 1,3-phenyl bis-thiourea compound targets microtubule polymerization to cause cancer cell death. Cancer Biol Ther. 2014;15:895–905. doi: 10.4161/cbt.28881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagarajan P, Curry JL, Ning J, Piao J, Torres-Cabala CA, Aung PP, Ivan D, Ross MI, Levenback CF, Frumovitz M, et al. Tumor thickness and mitotic rate robustly predict melanoma-specific survival in patients with primary vulvar melanoma: A retrospective review of 100 cases. Clin Cancer Res. 2017;23:2093–2104. doi: 10.1158/1078-0432.CCR-16-2126. [DOI] [PubMed] [Google Scholar]
- 53.Jun DY, Lee JY, Park HS, Lee YH, Kim YH. Tumor suppressor protein p53-mediated repression of human mitotic centromere-associated kinesin gene expression is exerted via down-regulation of Sp1 level. PLoS One. 2017;12:e0189698. doi: 10.1371/journal.pone.0189698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pinto M, Vieira J, Ribeiro FR, Soares MJ, Henrique R, Oliveira J, Jerónimo C, Teixeira MR. Overexpression of the mitotic checkpoint genes BUB1 and BUBR1 is associated with genomic complexity in clear cell kidney carcinomas. Cell Oncol. 2008;30:389–395. doi: 10.3233/CLO-2008-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Myrie KA, Percy MJ, Azim JN, Neeley CK, Petty EM. Mutation and expression analysis of human BUB1 and BUB1B in aneuploid breast cancer cell lines. Cancer Lett. 2000;152:193–199. doi: 10.1016/S0304-3835(00)00340-2. [DOI] [PubMed] [Google Scholar]
- 56.Scintu M, Vitale R, Prencipe M, Gallo AP, Bonghi L, Valori VM, Maiello E, Rinaldi M, Signori E, Rabitti C, et al. Genomic instability and increased expression of BUB1B and MAD2L1 genes in ductal breast carcinoma. Cancer Lett. 2007;254:298–307. doi: 10.1016/j.canlet.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 57.Herman JA, Toledo CM, Olson JM, DeLuca JG, Paddison PJ. Molecular pathways: Regulation and targeting of kinetochore-microtubule attachment in cancer. Clin Cancer Res. 2015;21:233–239. doi: 10.1158/1078-0432.CCR-13-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DeLuca JG, Dong Y, Hergert P, Strauss J, Hickey JM, Salmon ED, McEwen BF. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol Biol Cell. 2005;16:519–531. doi: 10.1091/mbc.e04-09-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Iemura K, Tanaka K. Chromokinesin Kid and kinetochore kinesin CENP-E differentially support chromosome congression without end-on attachment to microtubules. Nat Commun. 2015;6:6447. doi: 10.1038/ncomms7447. [DOI] [PubMed] [Google Scholar]
- 60.Shrestha RL, Draviam VM. Lateral to end-on conversion of chromosome-microtubule attachment requires kinesins CENP-E and MCAK. Curr Biol. 2013;23:1514–1526. doi: 10.1016/j.cub.2013.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Diaz-Rodriguez E, Sotillo R, Schvartzman JM, Benezra R. Hec1 overexpression hyperactivates the mitotic checkpoint and induces tumor formation in vivo. Proc Natl Acad Sci USA. 2008;105:16719–16724. doi: 10.1073/pnas.0803504105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sotillo R, Hernando E, Diaz-Rodriguez E, Teruya-Feldstein J, Cordón-Cardo C, Lowe SW, Benezra R. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.