Abstract
Autophagy has an important role in the pathogenesis of plasma cell development and multiple myeloma (MM); however, the prognostic role of autophagy-related genes (ARGs) in MM remains undefined. In the present study, the expression profiles of 234 ARGs were obtained from a Gene Expression Omnibus dataset (accession GSE24080), which contains 559 samples of patients with MM analyzed with 54,675 probes. Univariate Cox regression analysis identified 55 ARGs that were significantly associated with event-free survival of MM. Furthermore, a risk score with 16 survival-associated ARGs was developed using multivariate Cox regression analysis, including ATIC, BNIP3L, CALCOCO2, DNAJB1, DNAJB9, EIF4EBP1, EVA1A, FKBP1B, FOXO1, FOXO3, GABARAP, HIF1A, NCKAP1, PRKAR1A and SUPT20H, was constructed. Using this prognostic signature, patients with MM could be separated into high- and low-risk groups with distinct clinical outcomes. The area under the curve values for the receiver operating characteristic curves were 0.740, 0.741 and 0.712 for 3, 5 and 10 years prognosis predictions, respectively. Notably, the prognostic role of this risk score could be validated with another four independent cohorts (accessions: GSE57317, GSE4581, GSE4452 and GSE4204). In conclusion, ARGs may serve vital roles in the progression of MM, and the ARGs-based prognostic model may provide novel ideas for clinical applications in MM.
Keywords: multiple myeloma, autophagy, autophagy-related genes, survival
Introduction
Autophagy is the process of transporting damaged, denatured or aging proteins, and organelles into lysosomes for digestion and degradation. Under normal physiological conditions, autophagy helps cells to maintain a self-stable state; however, during stress, autophagy prevents accumulation of toxic or carcinogenic damaged proteins and organelles, and inhibits cell carcinogenesis (1–4). Once a tumor is formed, autophagy can be harmful as cells provide additional nutrients and promote tumor growth. Therefore, the role of autophagy in the development of tumors is two-sided (5–9). Since autophagy can regulate cancer formation, proliferation, metastasis and energy metabolism of tumors, antitumor drugs based on regulation of autophagy activity have been used in clinical treatment (10–12). Additionally, inhibition of tumors by improving autophagy activity has become a novel concept for cancer treatment (10–14).
Depending on the way a lysosome accepts the substance to be degraded, autophagy can be divided into macroautophagy, microautophagy and chaperone-mediated autophagy. Macroautophagy, the usual form of autophagy, is the most common type and is characterized by the formation of cup-shaped bilayer membrane structures surrounding the cytoplasmic component, followed by the formation of autophagosomes (15–18). The outer membrane of autophagosomes and the enzymatic fusion form a monolayer membrane structure of autophagosomes, while the inner membrane and contents of autophagosomes are digested (15–18). The aforementioned process is mediated by autophagy-related genes (ARGs). Autophagocytosis is a process in which cells use lysosomes to degrade damaged organelles and macromolecules under regulation of ARGs (19–26). Previous studies have identified 234 ARGs (27). These ARGs have been identified as direct or indirect participants in the process of autophagy; thus, analysis of a list of ARGs can provide a comprehensive overview of the alterations of autophagy in multiple myeloma (MM). Several studies have demonstrated that these ARGs have significant clinical implications for various types of cancer, including glioma, liver cancer and thyroid cancer (23,24,28).
Autophagy has an important role in the pathogenesis of plasma cell development and MM, the incidence rate of which is estimated to be 2–3/100,000, and which mostly affects patients >40 years old (29–31). Generally, autophagy is considered to be involved in pro-survival mechanisms of MM cells and to interact with the ubiquitin-proteasome system to maintain homeostasis of MM cells via degraded and misfolded proteins for energy recovery (32–36). Therefore, inhibiting autophagy may effectively induce MM cell death and can act synergistically using proteasome inhibitors. However, exaggerated activation of autophagy may result in excessive degradation of organelles, which can induce autophagic cell death. Thus, activation of autophagic cell death may represent a promising approach for treatment of MM (32–36). Recent studies have demonstrated that autophagy mediates drug resistance in MM cells and leads to development of clinical complications for MM, while inhibition of autophagy may reverse the response to drugs (37,38). However, the clinical role, particularly the prognostic role of ARGs in MM, has yet to be determined.
In the present study, the expression profiles of ARGs and prognosis data of MM were integrated, and used to develop a risk score to predict the clinical outcome of patients with MM. Previous studies on the role of autophagy in MM tended to focus on a single gene, on the contrary, the present study proposed a novel predictor by integrating several effective indexes, which could provide more effective information concerning autophagy and offer more favorable performance in survival prediction of patients with MM. The present study evaluated the prognostic value of ARGs in MM clinical samples by data mining and bioinformatics analysis of gene expression profiles. Additionally, a risk score was constructed using the prognosis-associated ARGs, which was expected to provide novel ideas for clinical applications in MM.
Materials and methods
Dataset and data processing
Microarray expression profiles were obtained from the Gene Expression Omnibus (GEO) database (ncbi.nlm.nih.gov/geo/) using the accession number GSE24080, which contained 559 samples of patients with MM (39). GSE24080 belongs to the MAQC-II Project: Multiple myeloma (MM) dataset. All patients had complete data for event-free survival (EFS), which included the recurrence of MM or the onset of certain symptoms associated with MM. Additionally, the platform was [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array (GPL570), which contains 54,675 probes (Affymetrix; Thermo Fisher Scientific, Inc.). To generate the gene expression profile, the expression matrix and microarray platform annotation file were downloaded. When more than one probe detected the same gene expression value, the average value was considered as the gene expression.
For further analysis, a total of 234 human genes and proteins involved in autophagy were acquired from the human autophagy database (HADb; autophagy.lu/). HADb is the first human autophagy-dedicated database, and is a public repository that contains annotation information associated with the up-to-date human genes the have been reported to be involved in autophagy. By October 2018, there were 234 ARGs included in the dataset (27).
Survival analysis and functional characteristics
A total of 234 ARGs were selected and their prognostic values were assessed. ARGs that were significantly associated with the EFS of MM were identified using univariate Cox analysis. Kaplan-Meier plots were used to further analyze the potential of ARGs as prognostic factors in patients. In order to determine the best cut-off value for grouping the patients to observe significant difference in outcome, all gene expression values from the 20 to 80th percentiles were considered. The cut-off with the lowest log-rank P-value was selected to group the patients. Survival analysis was conducted using the ‘survival’ package of R software (version 3.5.1; http://CRAN.R-project.org/package=survival). P<0.05 was considered to indicate a statistically significant difference (40). To reveal molecular functional characteristics, in addition to the autophagy of these prognostic ARGs, functional enrichment analysis was conducted for the Gene Ontology (GO; http://geneontology.org/) and Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.kegg.jp/) databases using the R package ‘clusterProfiler’ (version 3.12.0) (41,42). The protein-protein interaction (PPI) network was generated to display the associations between the prognosis-associated ARGs, by using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (https://string-db.org/; version 10.0).
Risk score construction
To develop a risk score using independent factors with ARGs, least absolute shrinkage and selection operator (LASSO) multivariate Cox regression analysis was performed. Subsequently, for each patient, the risk score was derived by multiplying the expression level of prognosis-associated ARGs and its corresponding coefficient as follows: Risk score=ΣniARGi * βi; in detail, LASSO Cox analysis selected the eligible ARGs for the risk score based on the expression levels of each sample and generated the corresponding coefficients for each of them. Accordingly, a risk score formula consisting of 16 ARGs weighted by the coefficients from LASSO penalized regression was established, where ‘β’ is the coefficient, ‘i’ refers to each ARG and ‘n’ is the number of the prognostic ARGs included in the calculation. The risk score for each patient was calculated, and all patients were divided into high- or low-risk groups based on the median level of the risk score. The performance of the risk score was assessed using the ‘survival receiver-operator characteristic (ROC)’ package for R software (https://CRAN.R-project.org/package=survivalROC; version 1.0.3), which provides an effective approach for evaluating time-dependent ROC using censored data. To quantitatively evaluate the prognostic value, the area under the curve (AUC) of the ROC curves was calculated (24,43–48).
Gene set enrichment analysis
In order to explore the pathways that are affected in the high- or low-risk group, gene set enrichment analysis (GSEA) (http://software.broadinstitute.org/gsea/index.jsp; version 3.0) was performed (49–52). Using GSEA, the present study tested whether the activated/repressed gene signatures were enriched for high-risk vs. low-risk cases. The pre-defined hallmarks were calculated using a normalized enrichment score (NES) and false discovery rate (FDR). Pathways with NES>1 and FDR<0.05 were considered to be significant.
Validation of the autophagy-associated risk score in other independent cohorts
MM-related microarray and RNA-sequencing datasets were screened in the GEO (https://www.ncbi.nlm.nih.gov/geo/), ArrayExpress (https://www.ebi.ac.uk/arrayexpress/) and SRA databases (https://www.ncbi.nlm.nih.gov/sra), and the search strategy was as follows: ‘Myeloma AND Homo sapiens’. Subsequently, prognosis-associated datasets were selected for further analysis. Four independent cohorts met the inclusion criteria, including GSE57317, GSE4581, GSE4452 and GSE4204. The expression data of the aforementioned 16 genes [5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase (ATIC), BCL2 interacting protein 3 like (BNIP3L), calcium binding and coiled-coil domain 2 (CALCOCO2), DnaJ heat shock protein family (Hsp 40) member B1 (DNAJB1), DnaJ heat shock protein family (Hsp 40) member B9 (DNAJB9), eukaryotic translation initiation factor 4E binding protein 1 (EIF4EBP1), eva-1 homolog A (EVA1A), FKBP prolyl isomerase 1B (FKBP1B), forkhead box O1 (FOXO1), forkhead box O3 (FOXO3), GABA type A receptor-associated protein (GABARAP), hypoxia-inducible factor 1 subunit α (HIF1A), NCK associated protein 1 (NCKAP1), protein kinase cAMP-dependent type I regulatory subunit α (PRKAR1A), SPT20 homolog SAGA complex component (SUPT20H) and transmembrane 9 superfamily member 1 (TM9SF1)] were extracted from these selected datasets. The risk score was calculated and one-way Cox regression analysis was performed using SPSS version 19.0 (IBM Corp.) and individual hazard ratios (HRs) were calculated. The HR and corresponding 95% confidence interval (CI) estimates were calculated and pooled to determine the association of risk score with clinical outcome. The random-effects model was conducted. Finally, a meta-analysis was carried out to impute the summarized HR combining all five datasets.
Results
Prognostic autophagy-specific gene screening
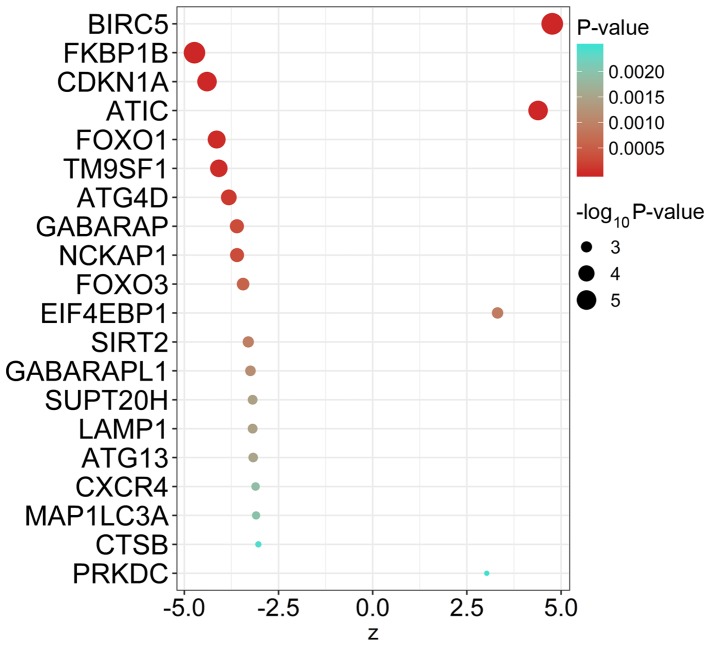
Univariate Cox regression analysis using autophagy-specific gene expression values for MM samples identified 55 ARGs that were significantly associated with EFS of MM (Table I). The top 20 ARGs with significant association with survival are displayed in Fig. 1.
Table I.
Prognosis-associated autophagy-related genes in multiple myeloma.
| Gene | Hazard ratio | Z-score | P-value |
|---|---|---|---|
| BIRC5 | 1.370446 | 4.764858 | 1.89×10−6 |
| FKBP1B | 0.804927 | −4.731650 | 2.23×10−6 |
| CDKN1A | 0.693169 | −4.397310 | 1.10×10−5 |
| ATIC | 1.828409 | 4.392418 | 1.12×10−5 |
| FOXO1 | 0.685527 | −4.142160 | 3.44×10−5 |
| TM9SF1 | 0.607627 | −4.082530 | 4.45×10−5 |
| ATG4D | 0.651409 | −3.819260 | 1.34×10−4 |
| GABARAP | 0.572865 | −3.603840 | 3.14×10−4 |
| NCKAP1 | 0.813461 | −3.598490 | 3.20×10−4 |
| FOXO3 | 0.678902 | −3.439260 | 5.83×10−4 |
| X10IF4X10BP1 | 1.245795 | 3.315612 | 9.14×10−4 |
| SIRT2 | 0.754843 | −3.30136 | 9.62×10−4 |
| GABARAPL1 | 0.760565 | −3.245810 | 1.17×10−3 |
| SUPT20H | 0.645072 | −3.185440 | 1.45×10−3 |
| LAMP1 | 0.756851 | −3.185010 | 1.45×10−3 |
| ATG13 | 0.677256 | −3.172030 | 1.51×10−3 |
| CXCR4 | 0.840891 | −3.106610 | 1.89×10−3 |
| MAP1LC3A | 0.736496 | −3.094860 | 1.97×10−3 |
| CTSB | 0.758494 | −3.034600 | 2.41×10−3 |
| PRKDC | 1.470160 | 3.026135 | 2.48×10−3 |
| PARP1 | 1.485479 | 3.024757 | 2.49×10−3 |
| ITGA6 | 0.868587 | −2.995980 | 2.74×10−3 |
| SH3GLB1 | 0.708930 | −2.987360 | 2.81×10−3 |
| DRAM1 | 0.756615 | −2.938770 | 3.30×10−3 |
| FADD | 1.517498 | 2.908387 | 3.63×10−3 |
| ITGA3 | 0.809097 | −2.867680 | 4.14×10−3 |
| APOL1 | 0.843190 | −2.866750 | 4.15×10−3 |
| PTX10N | 1.574460 | 2.833041 | 4.61×10−3 |
| PPP1R15A | 0.842720 | −2.822870 | 4.76×10−3 |
| HSPA5 | 0.698868 | −2.774470 | 5.53×10−3 |
| VAMP7 | 1.347821 | 2.757183 | 5.83×10−3 |
| BNIP3L | 0.734860 | −2.653410 | 7.97×10−3 |
| MAPK1 | 1.497021 | 2.526023 | 1.15×10−2 |
| PINK1 | 0.738836 | −2.496230 | 1.26×10−2 |
| CALCOCO2 | 0.688453 | −2.458060 | 1.40×10−2 |
| HIF1A | 1.101043 | 2.440301 | 1.47×10−2 |
| BCL2L1 | 0.797140 | −2.431050 | 1.51×10−2 |
| DNAJB9 | 0.746170 | −2.406550 | 1.61×10−2 |
| SQSTM1 | 0.705885 | −2.383290 | 1.72×10−2 |
| ATG9A | 0.789362 | −2.358180 | 1.84×10−2 |
| PRKAR1A | 1.414585 | 2.347303 | 1.89×10−2 |
| X10VA1A | 1.174754 | 2.328178 | 1.99×10−2 |
| HSP90AB1 | 1.237892 | 2.277807 | 2.27×10−2 |
| RAB24 | 0.824202 | −2.270130 | 2.32×10−2 |
| VX10GFA | 1.251725 | 2.246293 | 2.47×10−2 |
| ATG5 | 0.659360 | −2.23166 | 2.56×10−2 |
| ATF4 | 0.752327 | −2.221500 | 2.63×10−2 |
| WDR45B | 0.676826 | −2.189410 | 2.86×10−2 |
| XBP1 | 0.778841 | −2.135780 | 3.27×10−2 |
| DNAJB1 | 0.825023 | −2.131930 | 3.30×10−2 |
| ARNT | 1.323780 | 2.109022 | 3.49×10−2 |
| NAF1 | 1.324480 | 2.089132 | 3.67×10−2 |
| MTMR14 | 0.808476 | −2.054530 | 3.99×10−2 |
| CASP8 | 1.271856 | 1.981271 | 4.76×10−2 |
| GAPDH | 1.263585 | 1.973326 | 4.85×10−2 |
Figure 1.
Prognostic autophagy-specific genes in multiple myeloma. The top 20 most significant survival-associated autophagy-related genes. A Z-score >0 indicates that genes are risk factors; otherwise, they are protective factors.
Molecular characteristics of ARGs in MM
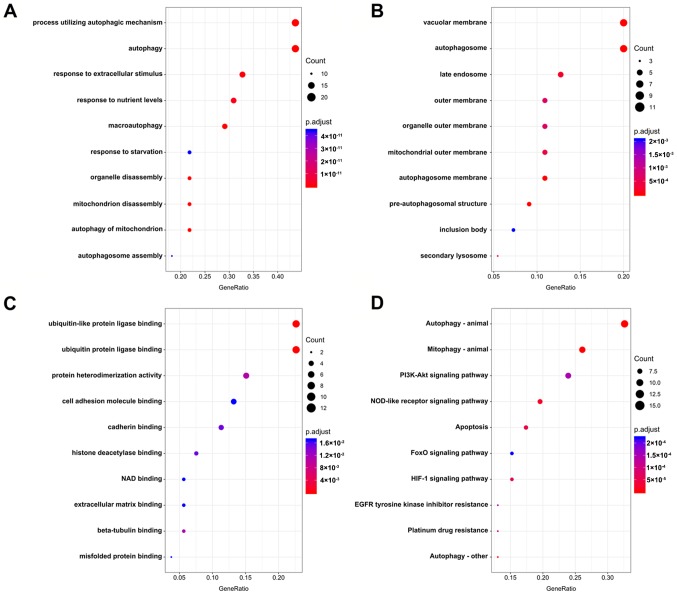
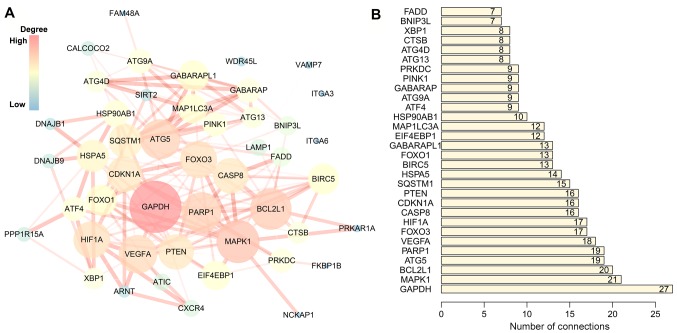
The association of these prognostic ARGs with the GO terms of the biological process (BP), cellular component (CC) and molecular function (MF) categories were analyzed. The top three enriched BP terms were ‘process utilizing autophagic mechanism’, ‘autophagy’ and ‘response to extracellular stimulus’ (Fig. 2A). For CC, the tops three terms were ‘vacuolar membrane’, ‘autophagosome’ and ‘late endosome’ (Fig. 2B). The top three enriched MF terms were ‘ubiquitin-like protein ligase binding’, ‘ubiquitin protein ligase binding’ and ‘protein heterodimerization activity’ (Fig. 2C). Accordingly, genes involved in KEGG pathways were enriched in autophagy-related pathways, including ‘autophagy-animal’, ‘mitophagy-animal’ and ‘PI3K-Akt signaling pathway’ (Fig. 2D; Table II). The PPI network suggested that these genes have important interactions with each other. GAPDH, MAPK1, BCL2L1, ATG5 and PARP1 were at the center of the PPI network, which suggested that these genes had a broader connection to the other genes (Fig. 3).
Figure 2.
Molecular characteristics of prognostic autophagy-specific genes in multiple myeloma. Enriched Gene Ontology (A) biological process, (B) molecular function and (C) cellular component terms, and (D) Kyoto Encyclopedia of Genes and Genomes pathways. NOD, nucleotide-binding oligomerization domain; FoxO, forkhead box class O; HIF-1, hypoxia-inducible factor 1; EGFR, epidermal growth factor receptor.
Table II.
Gene functional enrichment analysis of prognosis-associated autophagy-related genes.
| Category | ID | Description | GeneRatio | P.adjust | Gene ID (top 10) | Count |
|---|---|---|---|---|---|---|
| Biological process | GO:0061919 | Process utilizing autophagic mechanism | 24/55 | 1.82×10−20 | FOXO1, TM9SF1, ATG4D, GABARAP, SIRT2, GABARAPL1, SUPT20H, ATG13, MAP1LC3A, SH3GLB1 | 24 |
| Biological process | GO:0006914 | Autophagy | 24/55 | 1.82×10−20 | FOXO1, TM9SF1, ATG4D, GABARAP, SIRT2, GABARAPL1, SUPT20H, ATG13, MAP1LC3A, SH3GLB1 | 24 |
| Biological process | GO:0009991 | Response to extracellular stimulus | 18/55 | 5.30×10−13 | FKBP1B, CDKN1A, FOXO1, GABARAP, FOXO3, EIF4EBP1, SIRT2, GABARAPL1, MAP1LC3A, ITGA6 | 18 |
| Biological process | GO:0031667 | Response to nutrient levels | 17/55 | 2.68×10−12 | FKBP1B, CDKN1A, FOXO1, GABARAP, FOXO3 EIF4EBP1, SIRT2, GABARAPL1, MAP1LC3A, SH3GLB1 | 17 |
| Biological process | GO:0016236 | Macro autophagy | 16/55 | 1.38×10−13 | ATG4D, GABARAP, GABARAPL1, ATG13, MAP1LC3A, SH3GLB1, BNIP3L, PINK1, CALCOCO2, HIF1A | |
| Biological process | GO:0042594 | Response to starvation | 12/55 | 4.42×10−11 | CDKN1A, FOXO1, GABARAP, EIF4EBP1, GABARAPL1, MAP1LC3A, SH3GLB1, HSPA5, MAPK1, ATG5 | 12 |
| Biological process | GO:1903008 | Organelle disassembly | 12/55 | 3.16×10−13 | ATG4D, GABARAP, GABARAPL1, ATG13, MAP1LC3A, BNIP3L, PINK1, HIF1A, SQSTM1 ATG9A | 12 |
| Biological process | GO:0061726 | Mitochondrion disassembly | 12/55 | 3.36×10−14 | ATG4D, GABARAP, GABARAPL1, ATG13, MAP1LC3A, BNIP3L, PINK1, HIF1A, SQSTM1, ATG9A | 12 |
| Biological process | GO:0000422 | Autophagy of mitochondrion | 12/55 | 3.36×10−14 | ATG4D GABARAP, GABARAPL1, ATG13, MAP1LC3A, BNIP3L, PINK1, HIF1A, SQSTM1, ATG9A | 12 |
| Biological process | GO:0000045 | Autophagosome assembly | 10/55 | 4.42×10−11 | ATG4D, GABARAP, GABARAPL1, ATG13, MAP1LC3A, SH3GLB1, PINK1, ATG9A, ATG5, WDR45B | 10 |
| Cellular component | GO:0005774 | Vacuolar membrane | 11/55 | 1.65×10−06 | TM9SF1, GABARAP, GABARAPL1, LAMP1, MAP1LC3A, SH3GLB1, DRAM1 VAMP7, CALCOCO2, EVA1A | 11 |
| Cellular component | GO:0005776 | Autophagosome | 11/55 | 3.63×10−13 | TM9SF1, GABARAP, GABARAPL1, LAMP1, MAP1LC3A, SH3GLB1 CALCOCO2, SQSTM1, ATG9A, RAB24 | 11 |
| Cellular component | GO:0005770 | Late endosome | 7/55 | 2.65×10−04 | LAMP1 CXCR4, MAP1LC3A, VAMP7, MAPK1, SQSTM1, ATG9A | 7 |
| Cellular component | GO:0019867 | Outer membrane | 6/55 | 7.08×10−04 | SH3GLB1, PPP1R15A, BNIP3L, PINK1, BCL2L1, CASP8 | 6 |
| Cellular component | GO:0031968 | Organelle outer membrane | 6/55 | 7.08×10−04 | SH3GLB1, PPP1R15A, BNIP3L, PINK1, BCL2L1, CASP8 | 6 |
| Cellular component | GO:0005741 | Mitochondrial outer membrane | 6/55 | 4.38×10−04 | SH3GLB1, PPP1R15A, BNIP3L, PINK1, BCL2L1, CASP8 | 6 |
| Cellular component | GO:0000421 | Autophagosome membrane | 6/55 | 4.42×10−08 | TM9SF1, GABARAP, GABARAPL1, MAP1LC3A, SH3GLB1, CALCOCO2 | 6 |
| Cellular component | GO:0000407 | Pre-autophagosomal structure | 5/55 | 2.23×10−06 | ATG13, SQSTM1, ATG9A, ATG5, WDR45B | 5 |
| Cellular component | GO:0016234 | Inclusion body | 4/55 | 2.07×10−03 | PINK1, SQSTM1, HSP90AB1, ATF4 | 4 |
| Cellular component | GO:0005767 | Secondary lysosome | 3/55 | 3.60×10−04 | LAMP1, MAP1LC3A, SQSTM1 | 3 |
| Molecular function | GO:0044389 | Ubiquitin-like protein ligase binding | 12/53 | 1.81×10−08 | CDKN1A, FOXO1, GABARAP, GABARAPL1, CXCR4, MAP1LC3A, HSPA5, PINK1, HIF1A, SQSTM1 | 12 |
| Molecular function | GO:0031625 | Ubiquitin protein ligase binding | 12/53 | 1.81×10−08 | CDKN1A, FOXO1, GABARAP, GABARAPL1, CXCR4, MAP1LC3A, HSPA5, PINK1, HIF1A, SQSTM1 | 12 |
| Molecular function | GO:0046982 | Protein heterodimerization activity | 8/53 | 1.08×10−02 | ITGA3, BNIP3L, HIF1A, BCL2L1, VEGFA, ATF4, XBP1, ARNT | 8 |
| Molecular function | GO:0050839 | Cell adhesion molecule binding | 7/53 | 1.70×10−02 | ATIC, ITGA6, SH3GLB1, ITGA3, HSPA5, HSP90AB1, DNAJB1 | 7 |
| Molecular function | GO:0045296 | Cadherin binding | 6/53 | 1.49×10−02 | ATIC, ITGA6, SH3GLB1, HSPA5, HSP90AB1, DNAJB1 | 6 |
| Molecular function | GO:0042826 | Histone deacetylase binding | 4/53 | 1.49×10−02 | SIRT2, PARP1, HIF1A, HSP90AB1 | 4 |
| Molecular function | GO:0051287 | NAD binding | 3/53 | 1.70×10−02 | SIRT2, PARP1, GAPDH | 3 |
| Molecular function | GO:0050840 | Extracellular matrix binding | 3/53 | 1.70×10−02 | ITGA6, ITGA3, VEGFA | 3 |
| Molecular function | GO:0048487 | Beta-tubulin binding | 3/53 | 1.12×10−02 | GABARAP, SIRT2, GABARAPL1 | 3 |
| Molecular function | GO:0051787 | Misfolded protein binding | 2/53 | 1.70×10−02 | HSPA5, DNAJB9 | 2 |
| KEGG pathway | hsa04140 | Autophagy-animal | 15/46 | 7.91×10−14 | ATG4D, GABARAP, GABARAPL1, SUPT20H, LAMP1, ATG13, CTSB, SH3GLB1, PTEN, MAPK1 | 15 |
| KEGG pathway | hsa04137 | Mitophagy-animal | 12/46 | 1.73×10−13 | GABARAP, FOXO3, GABARAPL1, BNIP3L, PINK1, CALCOCO2, HIF1A, BCL2L1, SQSTM1, ATG9A | 12 |
| KEGG pathway | hsa04151 | PI3K-Akt signaling pathway | 11/46 | 1.48×10−04 | CDKN1A, FOXO3, EIF4EBP1, ITGA6, ITGA3, PTEN, MAPK1, BCL2L1, HSP90AB1 VEGFA | 11 |
| KEGG pathway | hsa04621 | NOD-like receptor signaling pathway | 9/46 | 2.77×10−05 | GABARAP, GABARAPL1, CTSB FADD, MAPK1, BCL2L1, HSP90AB1, ATG5, CASP8 | 9 |
| KEGG pathway | hsa04210 | Apoptosis | 8/46 | 4.95×10−05 | BIRC5, CTSB, PARP1, FADD, MAPK1, BCL2L1, ATF4, CASP8 | 8 |
| KEGG pathway | hsa04068 | FoxO signaling pathway | 7/46 | 2.22×10−04 | CDKN1A, FOXO1, GABARAP, FOXO3, GABARAPL1, PTEN, MAPK1 | 7 |
| KEGG pathway | hsa04066 | HIF-1 signaling pathway | 7/46 | 6.28×10−05 | CDKN1A, EIF4EBP1, MAPK1, HIF1A, VEGFA, ARNT, GAPDH | 7 |
| KEGG pathway | hsa01521 | EGFR tyrosine kinase inhibitor resistance | 6/46 | 1.48×10−04 | FOXO3, EIF4EBP1, PTEN, MAPK1, BCL2L1, VEGFA | 6 |
| KEGG pathway | hsa01524 | Platinum drug resistance | 6/46 | 1.20×10−04 | BIRC5, CDKN1A, FADD, MAPK1, BCL2L1, CASP8 | 6 |
| KEGG pathway | hsa04136 | Autophagy-other | 6/46 | 1.80×10−06 | ATG4D, GABARAP, GABARAPL1, ATG13, ATG9A, ATG5 | 6 |
P.adjust, adjusted P-value by Benjamini-Hochberg method; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Figure 3.
PPI network of prognostic autophagy-specific genes in multiple myeloma. (A) PPI network of prognosis associated ARGs. The size and brightness of the circle represents the degree of connection. The larger and brighter circles are the hub genes in the network. The thickness of the lines represents the combined score. (B) Top 30 hub genes in the PPI network. ARG, autophagy-related gene; PPI, protein-protein interaction.
Risk score identification
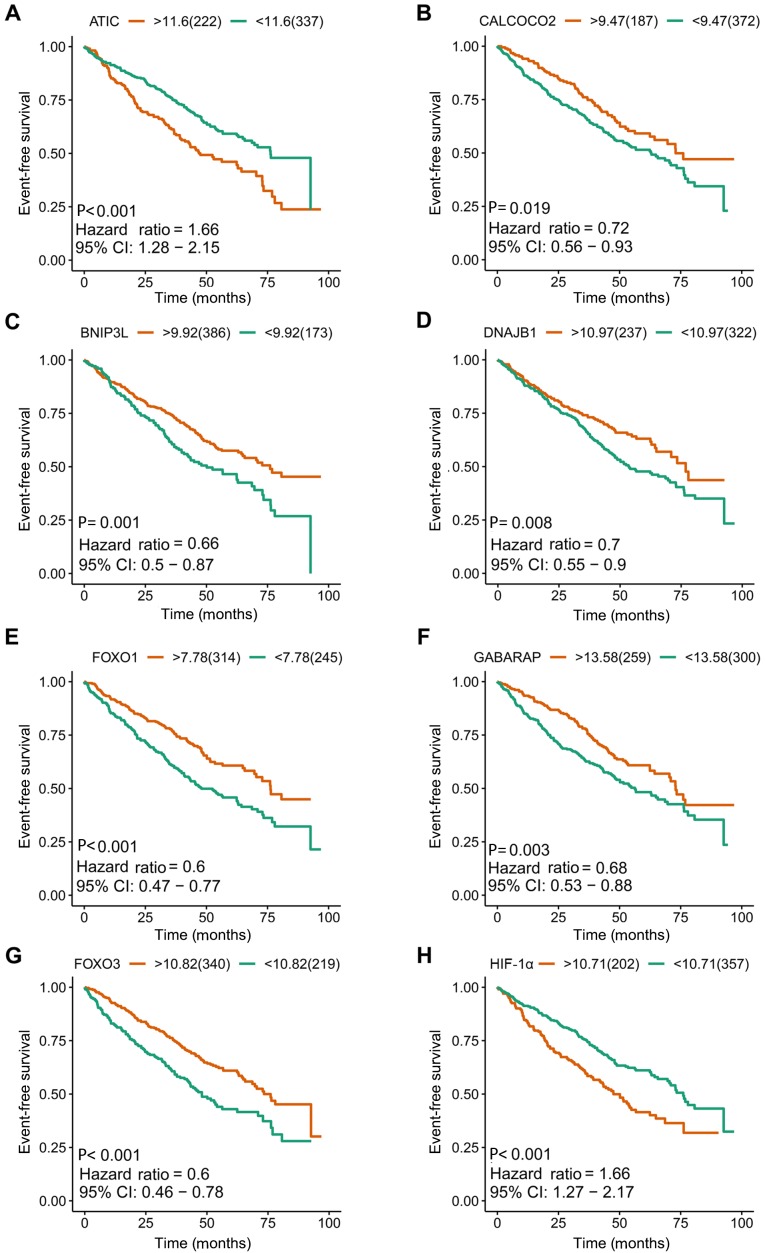
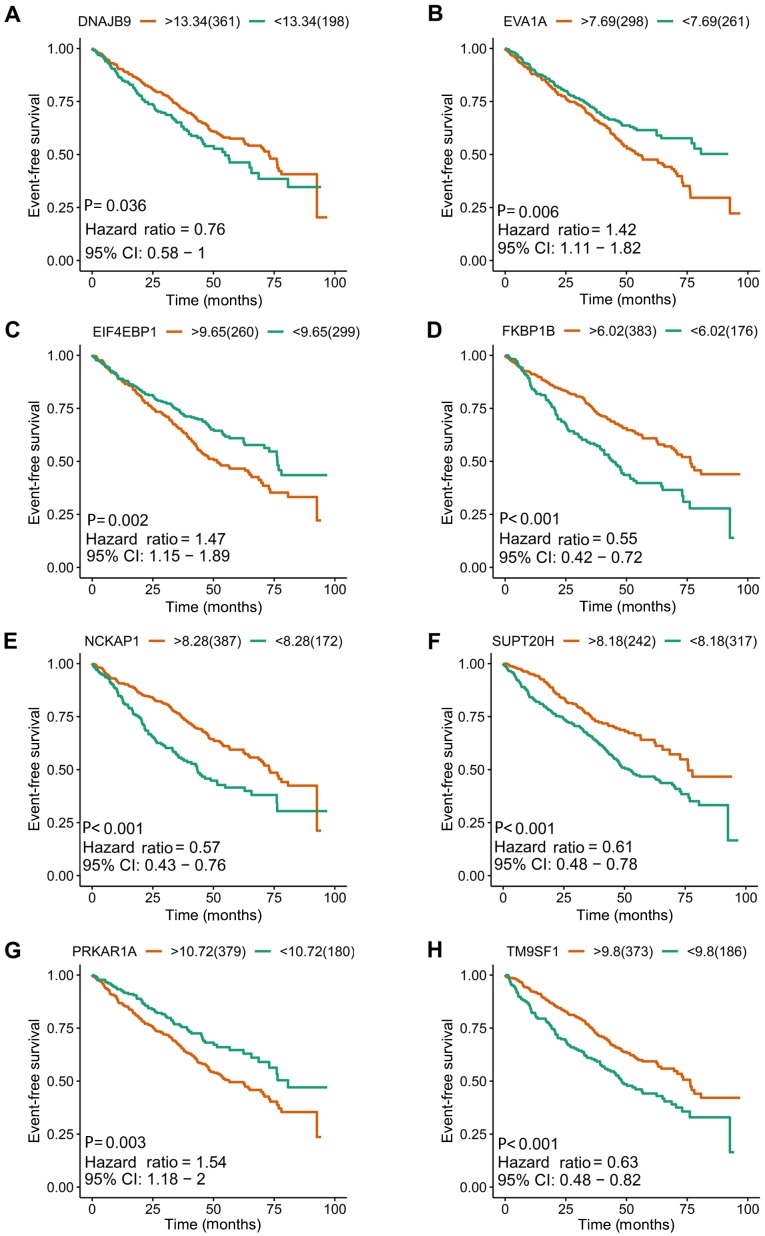
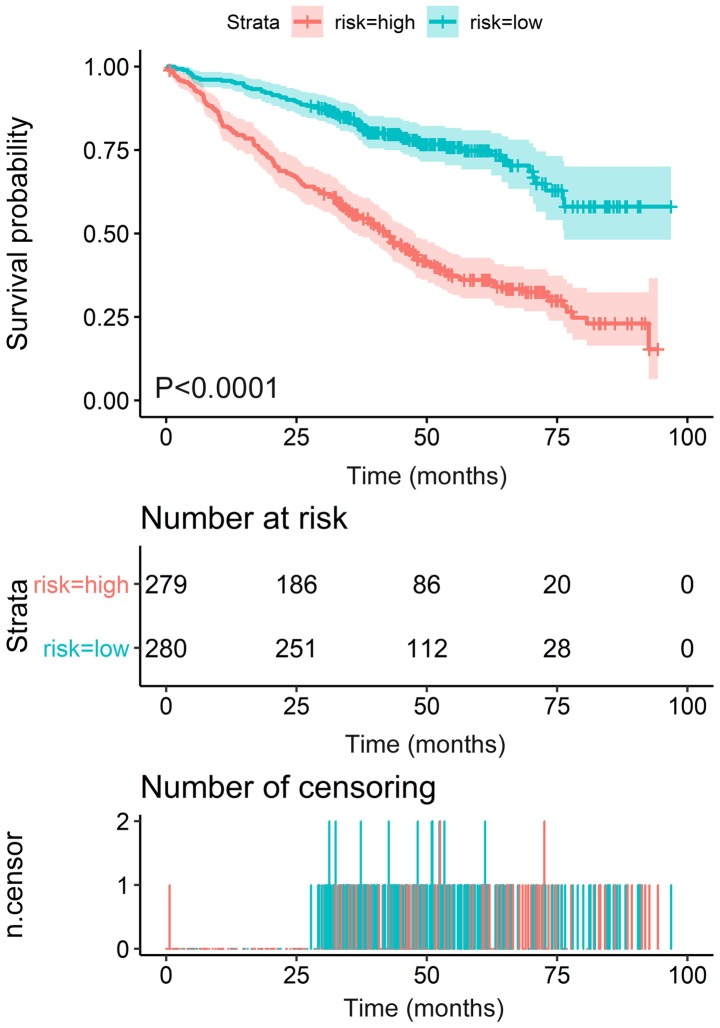
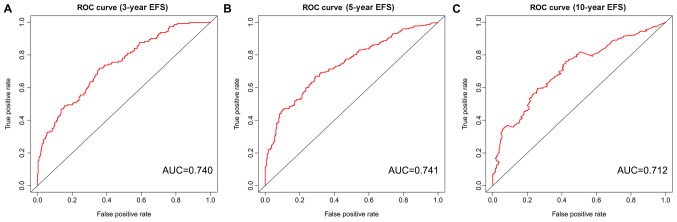
I order to construct novel risk assessment models for the prognosis of patients with MM, prognostic signatures were generated using multivariate Cox regression analysis. Finally, to develop the prognostic signature, 16 prognosis-associated ARGs were included (Figs. 4 and 5). Kaplan-Meier survival plots demonstrated that all 16 genes were significantly associated with the EFS of patients with MM. Furthermore, the risk score was calculated for each patient by multiplying the regression coefficient and expression value for each gene. Risk score=ATIC × 0.3374 + BNIP3L × (−0.2126) + CALCOCO2 × (−0.2682) + DNAJB1 × (−0.3848) + DNAJB9 × (−0.3443) + EIF4EBP1 × 0.1397 + EVA1A × 0.1794 + FKBP1B × (−0.1205) + FOXO1 × (−0.3114) + FOXO3 × (−0.2853) + GABARAP × (−0.3557) + HIF1A × 0.0876 + NCKAP1 × (−0.1487) + PRKAR1A × 0.7314 + SUPT20H × (−0.3261) + TM9SF1 × (−0.1992). Using these prognostic signatures, patients with MM were separated into high- and low-risk groups with distinct clinical outcomes (Fig. 6). The AUC values for the ROC curve were 0.740, 0.741 and 0.712 for 3, 5 and 10 years, respectively (Fig. 7).
Figure 4.
Kaplan-Meier survival plots of eight included prognostic predictors for multiple myeloma. (A) ATIC, (B) CALCOCO2, (C) BNIP3L, (D) DNAJB1, (E) FOXO1, (F) GABARAP, (G) FOXO3 and (H) HIF-1α. ATIC, 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase; CI, confidence interval; CALCOCO2, calcium binding and coiled-coil domain 2; BNIP3L, BCL2 interacting protein 3 like; DNAJB1, DnaJ heat shock protein family (Hsp 40) member B1; FOXO1, forkhead box O1; GABARAP, GABA type A receptor-associated protein; FOXO3, forkhead box O3; HIF-1α, hypoxia-inducible factor 1 subunit α.
Figure 5.
Kaplan-Meier survival plots of the other eight included prognostic predictors for multiple myeloma. (A) DNAJB9, (B) EVA1A, (C) EIF4EBP1, (D) FKBP1B, (E) NCKAP1, (F) SUPT20H, (G) PRKAR1A and (H) TM9SF1. DNAJB9, DnaJ heat shock protein family (Hsp 40) member B9; CI, confidence interval; EIF4EBP1, eukaryotic translation initiation factor 4E binding protein 1; EVA1A, eva-1 homolog A; FKBP1B, FKBP prolyl isomerase 1B; NCKAP1, NCK associated protein 1; SUPT20H, SPT20 homolog, SAGA complex component; PRKAR1A, protein kinase cAMP-dependent type I regulatory subunit α; TM9SF1, transmembrane 9 superfamily member 1.
Figure 6.
Kaplan-Meier survival analysis of the risk score in multiple myeloma. The Kaplan-Meier survival plot indicated that the risk score of event-free survival could separate patients into two groups with distinct clinical outcomes.
Figure 7.
ROC curves risk score of EFS in multiple myeloma. Analysis at (A) 3, (B) 5 and (C) 10 years. ROC, receiver operating characteristic; AUC, area under the curve; EFS, event-free survival.
Hallmarks of high- and low-risk groups
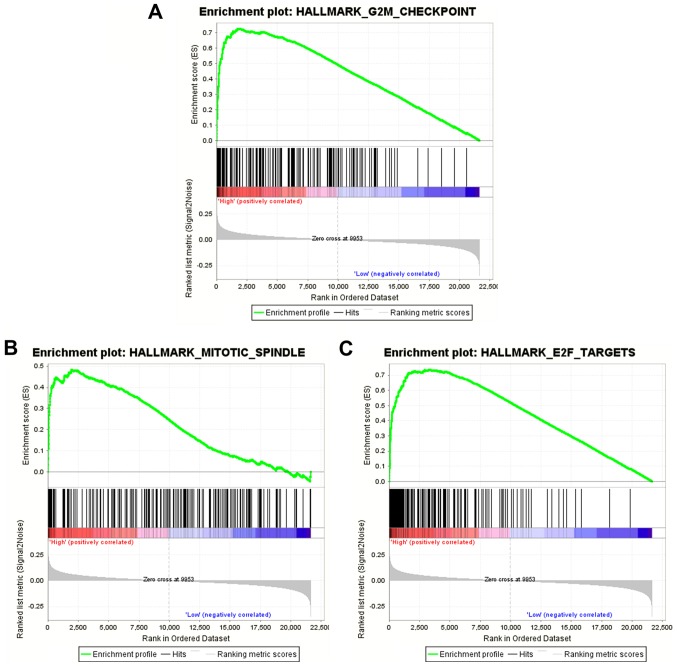
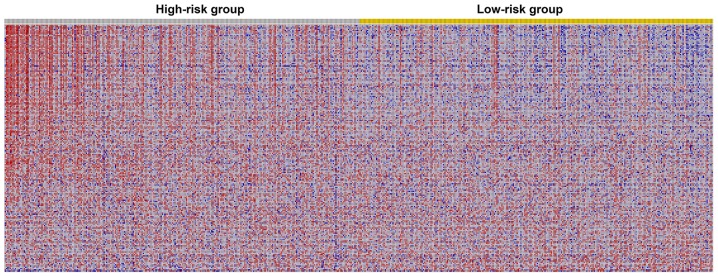
The results of the GSEA analysis indicated that five hallmarks were significantly associated with high-risk patients, including ‘G2M_CHECKPOINT’, ‘MITOTIC_SPINDLE’, ‘E2F_TARGETS’, ‘MYC_TARGETS_V1’ and ‘MYC_TARGETS_V2’. Fig. 8 shows the three most significant hallmarks, and heatmaps of the G2M checkpoint revealed that genes in the high-risk group differed greatly from genes in the low-risk group (Fig. 9).
Figure 8.
Gene set enrichment analysis of prognostic autophagy-related genes in multiple myeloma. (A) G2M_CHECKPOINT, (B) MITOTIC_SPINDLE and (C) E2F_TARGETS.
Figure 9.
Heatmap of prognostic autophagy-related genes for multiple myeloma. Blue represents low expression value while red represent high expression value. The heatmap revealed that most genes enriched in hallmark ‘G2M checkpoint’ were activated in the high-risk group.
Validation of the autophagy-associated risk score in other independent cohorts
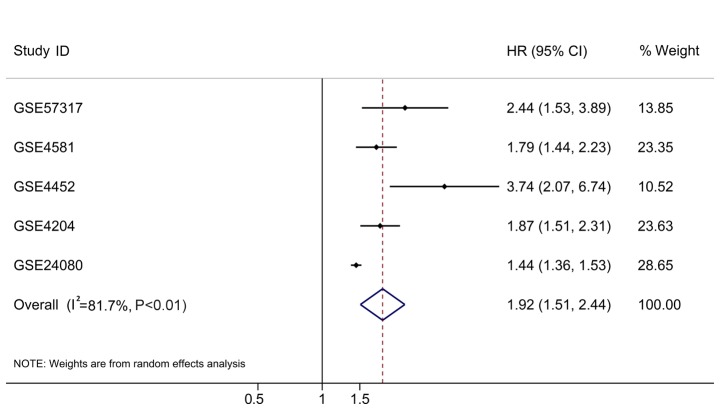
In addition to the original testing cohort (GSE24080), four independent cohorts met the inclusion criteria, including GSE57317, GSE4581, GSE4452 and GSE4204. The expression data of the 16 ARGs were extracted from these microarray datasets and the prognostic signatures were calculated based on the aforementioned formula. The HRs were all >1 (Table III). To have a comprehensive view of the clinical role of the risk score based on all the rational cases, the overall HR was 1.92 (95% CI, 1.51–2.44; P<0.01; Table III; Fig. 10) with 1,631 cases, which supported the findings from the testing cohort.
Table III.
Validation of the autophagy-related risk score in other independent cohorts.
| GEO datasets | HR | LCI | UCI | P-value | Number of samples | Platform |
|---|---|---|---|---|---|---|
| GSE57317 | 2.442 | 1.533 | 3.892 | <0.001 | 55 | GPL570 |
| GSE4581 | 1.793 | 1.438 | 2.235 | <0.001 | 414 | GPL570 |
| GSE4452 | 3.737 | 2.072 | 6.740 | <0.001 | 65 | GPL570 |
| GSE4204 | 1.867 | 1.508 | 2.312 | <0.001 | 538 | GPL570 |
| GSE24080 | 1.442 | 1.363 | 1.526 | <0.001 | 559 | GPL570 |
| Combination | 1.920 | 1.510 | 2.440 | <0.001 | 1,631 | NA |
GEO, Gene Expression Omnibus; HR, hazard ratio; LCI, lower 95% confidence interval; UCI, upper 95% confidence interval; NA, not applicable.
Figure 10.
Individual and comprehensive HRs in five independent multiple myeloma cohorts. HR, hazard ratio; CI, confidence interval.
Discussion
The role of autophagy in tumors is well known, and the function of autophagy in the development and treatment of MM has been previously reported (29–30). However, the clinical significance of ARGs, particularly their prognostic effect in MM has not been extensively studied. Furthermore, there is no comprehensive analysis of the prognostic significance of all ARGs. Therefore, in the present study, the expression levels of ARGs were analyzed, and the prognostic value of ARGs was subsequently examined. Finally, a prognostic model was constituted using the prognostic ARGs. Furthermore, this model could predict the prognosis of patients well and provided novel ideas for clinical applications in MM. Given the clinical significance of these prognostic ARGs in MM, if drugs could be used to intervene in their expression, they may provide novel directions for clinical MM treatment.
With the assistance of GEO GSE24080 from the MAQC-II Project, the prognostic value of all 234 ARGs in MM was evaluated. By performing univariate Cox analysis, 55 prognostic ARGs were identified, which indicated that autophagy served an essential role in the development of MM and could influence the outcome of patients with MM. As, in addition to autophagy, the ARGs could have multiple functions, GO and KEGG pathway analyses were also conducted. Indeed, most pathways that were enriched were autophagy-related pathways. Interestingly, certain other annotations were identified, including ‘response to extracellular stimulus’, ‘vacuolar membrane’ and ‘late endosome’. For KEGG pathways, the ‘PI3K-Akt signaling pathway’ was identified. It has been documented that there was a close association between PI3K-Akt signaling and autophagy (53–55). In general, activation of the PI3K-Akt signaling pathway may induce autophagy in numerous types of cancer (53–58).
To assess the prognosis of patients with MM more accurately, multivariate Cox regression analysis was used to analyze prognostic significance of ARGs. The results narrowed the scope of ARGs to 16. Due to the limited predictive power of individual prognostic indicators, these ARGs were combined into a prognostic assessment model. The prognostic model exhibited AUCs of >0.7, suggesting that it had a moderate power to evaluate the prognosis of patients with MM. The establishment of such a prognostic index confirmed the role of autophagy for the development and patient prognosis of MM; however, it also provided novel biomarkers for clinical applications in MM. Notably, another four independent cohorts were used to validate the results based on the 559 patients from GSE24080, and the current ARG-based risk score succeeded in yielding concordant prognostic values in individual cohorts and when all cohorts were combined (1,631 cases).
To identify why these ARGs may hold any value for the prognosis of MM, the mechanisms involved in the development of MM were explored. However, among these 16 prognostic ARGs, 11 (ATIC, CALCOCO2, DNAJB1, DNAJB9, EVA1A, FKBP1B, GABARAP, NCKAP1, PRKAR1A, SUPT20H and TM9SF1) have not been reported to be associated with MM, to the best of our knowledge. Their roles in MM are yet to be determined; however, several well-established ARGs for MM, including HIF1A, EIF4EBP1, FOXO1, FOXO3 and BNIP3L were identified as prognostic ARGs in the present study.
Among the five previously identified ARGs associated with MM, HIF1A is the most widely studied (59,60). Hypoxia, a central characteristic for cancer incidence and progression, occurs when most types of cancer are evolving (59–64). HIF1A is a hypoxia-inducible factor, and constitutive expression of HIF1A in MM indicates that suppression of HIF1A-mediated transcription could become a favorable target for MM (65–68). For instance, chetomin, an inhibitor of the HIF1A/p300 interaction, can inhibit tumor cell growth of MM (69). Due to the function of HIF1A in inducing autophagy, the suppressive effects of inhibitors of HIF1A could exert their effect by modulating the autophagy of MM cells (70–73). The expression levels of EIF4EBP1, which is a target of mTOR, have been reported to be upregulated for MM cases (74). As a master regulator of protein synthesis control, phosphorylation of EIF4EBP1 has a close functional association with Myc and mTOR (75). In Myc-dependent tumor initiation and maintenance of MM, the mTOR-dependent phosphorylation of EIF4EBP1 is required for tumor cell survival (75). This may explain the prognostic role of EIF4EBP1 in MM, which, to the best of our knowledge, has not been reported previously. FOXO1 has been reported to act as a tumor suppressor for MM (76). The activation of FOXO1 could subsequently inhibit the tumor growth and induce cell autophagy and cell death (77–80). FOXO3, another family member of the FOXO family, has been studied in MM (81). In primary MM cells, FOXO transcription factors are highly phosphorylated (81). The activation of FOXO3 has been observed in response to thiadiazolidinone, a non-competitive inhibitor of glycogen synthase kinase-3 (82). An increase in FOXO3a expression was observed following treatment of MM cells with 4-chlorobenzoyl berbamine, a novel berbamine derivative (83). The prognostic roles of FOXO1 and FOXO3 have not yet been investigated, to the best of our knowledge, and the present study demonstrated their prognostic value for MM. As for BNIP3L, an increase in BNIP3L occurs when MM cells are treated with panobinostat, a pan-histone deacetylase inhibitor used to treat MM (84). However, its prognostic value, to the best of our knowledge, has not been documented. The activation of BNIP3L under drug treatment may improve the understanding of its potential mechanism in the development of MM. However, the molecular mechanism of these 16 prognostic ARGs in MM requires further exploration.
There were several limitations of the present study. Firstly, of the 16 identified prognostic ARGs, 11 have not been previously reported to be associated with MM. Their prognostic value needs to be confirmed by other cohorts. Additionally, the potential mechanism of these 11 ARGs in MM progression remains unclear, which requires additional research. Secondly, the efficacy of the prognostic model based on 16 ARGs identified in the present study has to be validated using other independent samples. Furthermore, multiple detection methods are also required for validation of the results of the present study using clinical samples. For instance, RNA-sequencing, microarray or reverse transcription-quantitative PCR, are also promising starting points for future research investigating ARGs in MM.
In conclusion, the present study focused on autophagy, an important phenomenon for tumors, and extracted ARGs with prognostic value for MM. Furthermore, an ARG-based MM prognostic evaluation model, with moderate performance in predicting the clinical outcome patients with MM, was constructed. However, the results require validation, and the working principles and molecular mechanisms of the ARGs in MM require additional research.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Program of Scientific and Technology Project (Guilin Science Research and Technology Development; grant no. 2016012706-2), Sanming Project of Medicine in Shenzhen (grant no. SZSM201602087), National Natural Science Foundation of China (grant no. 81460038), Guangxi Natural Science Foundation of China (grant no. 2017GXNSFAA198178) and Shenzhen Futian Public Welfare Scientific Research Project (grant nos. FTWS2017020 and FTWS2018005).
Availability of data and materials
All data generated or analyzed during the present study are included in this published article.
Authors' contributions
FZ analyzed the data, generated figures and wrote the manuscript. XW, HZ and ZHY performed bioinformatics analysis and wrote the manuscript. ZZY made substantial contributions to the design of the study, conducted data analysis and figure generation, and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Boga JA, Caballero B, Potes Y, Perez-Martinez Z, Reiter RJ, Vega-Naredo I, Coto-Montes A. Therapeutic potential of melatonin related to its role as an autophagy regulator: A review. J Pineal Res. 2018;66:e12534. doi: 10.1111/jpi.12534. [DOI] [PubMed] [Google Scholar]
- 2.Guillaume JD, Celano SL, Martin KR, MacKeigan JP. Determining the impact of metabolic nutrients on autophagy. Methods Mol Biol. 2019;1862:151–162. doi: 10.1007/978-1-4939-8769-6_11. [DOI] [PubMed] [Google Scholar]
- 3.Liang ZG, Lin GX, Yu BB, Su F, Li L, Qu S, Zhu XD. The role of autophagy in the radiosensitivity of the radioresistant human nasopharyngeal carcinoma cell line CNE-2R. Cancer Manag Res. 2018;10:4125–4134. doi: 10.2147/CMAR.S176536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao Y, Onda K, Sugiyama K, Yuan B, Tanaka S, Takagi N, Hirano T. Antitumor effects of arsenic disulfide on the viability, migratory ability, apoptosis and autophagy of breast cancer cells. Oncol Rep. 2019;41:27–42. doi: 10.3892/or.2018.6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiao YN, Wu LN, Xue D, Liu XJ, Tian ZH, Jiang ST, Han SY, Li PP. Marsdenia tenacissima extract induces apoptosis and suppresses autophagy through ERK activation in lung cancer cells. Cancer Cell Int. 2018;18:149. doi: 10.1186/s12935-018-0646-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun T, Liu H, Ming L. Multiple roles of autophagy in the sorafenib resistance of hepatocellular carcinoma. Cell Physiol Biochem. 2017;44:716–727. doi: 10.1159/000485285. [DOI] [PubMed] [Google Scholar]
- 7.Wu JS, Li L, Wang SS, Pang X, Wu JB, Sheng SR, Tang YJ, Tang YL, Zheng M, Liang XH. Autophagy is positively associated with the accumulation of myeloid-derived suppressor cells in 4-nitroquinoline-1-oxide-induced oral cancer. Oncol Rep. 2018;40:3381–3391. doi: 10.3892/or.2018.6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Y, Liu X, Qin Z, Hu L, Wang X. Low-frequency ultrasound enhances chemotherapy sensitivity and induces autophagy in PTX-resistant PC-3 cells via the endoplasmic reticulum stress-mediated PI3K/Akt/mTOR signaling pathway. Onco Targets Ther. 2018;11:5621–5630. doi: 10.2147/OTT.S176744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu J, Zhao B, Xiong P, Wang C, Zhang J, Tian X, Huang Y. Curcumin induces autophagy via inhibition of yes-associated protein (YAP) in human colon cancer cells. Med Sci Monit. 2018;24:7035–7042. doi: 10.12659/MSM.910650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duan X, Chen B, Cui Y, Zhou L, Wu C, Yang Z, Wen Y, Miao X, Li Q, Xiong L, He J. Ready player one? Autophagy shapes resistance to photodynamic therapy in cancers. Apoptosis. 2018;23:587–606. doi: 10.1007/s10495-018-1489-0. [DOI] [PubMed] [Google Scholar]
- 11.Han Y, Fan S, Qin T, Yang J, Sun Y, Lu Y, Mao J, Li L. Role of autophagy in breast cancer and breast cancer stem cells (Review) Int J Oncol. 2018;52:1057–1070. doi: 10.3892/ijo.2018.4270. [DOI] [PubMed] [Google Scholar]
- 12.Ianniciello A, Rattigan KM, Helgason GV. The Ins and outs of autophagy and metabolism in hematopoietic and leukemic stem cells: Food for thought. Front Cell Dev Biol. 2018;6:120. doi: 10.3389/fcell.2018.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacomin AC, Taillebourg E, Fauvarque MO. Deubiquitinating enzymes related to autophagy: New therapeutic opportunities? Cells. 2018;7(pii):E112. doi: 10.3390/cells7080112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin S, Wei J, You L, Liu H, Qian W. Autophagy regulation and its dual role in blood cancers: A novel target for therapeutic development (Review) Oncol Rep. 2018;39:2473–2481. doi: 10.3892/or.2018.6370. [DOI] [PubMed] [Google Scholar]
- 15.Feldmann A, Bekbulat F, Huesmann H, Ulbrich S, Tatzelt J, Behl C, Kern A. The RAB GTPase RAB18 modulates macroautophagy and proteostasis. Biochem Biophys Res Commun. 2017;486:738–743. doi: 10.1016/j.bbrc.2017.03.112. [DOI] [PubMed] [Google Scholar]
- 16.Han Q, Deng Y, Chen S, Chen R, Yang M, Zhang Z, Sun X, Wang W, He Y, Wang F, et al. Downregulation of ATG5-dependent macroautophagy by chaperone-mediated autophagy promotes breast cancer cell metastasis. Sci Rep. 2017;7:4759. doi: 10.1038/s41598-017-04994-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pajares M, Jimenez-Moreno N, Garcia-Yague AJ, Escoll M, de Ceballos ML, Van Leuven F, Rábano A, Yamamoto M, Rojo AI, Cuadrado A. Transcription factor NFE2L2/NRF2 is a regulator of macroautophagy genes. Autophagy. 2016;12:1902–1916. doi: 10.1080/15548627.2016.1208889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang C, Wang H, Zhang D, Luo W, Liu R, Xu D, Diao L, Liao L, Liu Z. Phosphorylation of ULK1 affects autophagosome fusion and links chaperone-mediated autophagy to macroautophagy. Nat Commun. 2018;9:3492. doi: 10.1038/s41467-018-05449-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bednarczyk M, Muc-Wierzgon M, Waniczek D, Fatyga E, Klakla K, Mazurek U, Wierzgoń J. Autophagy-related gene expression in colorectal cancer patients. J Biol Regul Homeost Agents. 2017;31:923–927. [PubMed] [Google Scholar]
- 20.Cao QH, Liu F, Yang ZL, Fu XH, Yang ZH, Liu Q, Wang L, Wan XB, Fan XJ. Prognostic value of autophagy related proteins ULK1, Beclin 1, ATG3, ATG5, ATG7, ATG9, ATG10, ATG12, LC3B and p62/SQSTM1 in gastric cancer. Am J Transl Res. 2016;8:3831–3847. [PMC free article] [PubMed] [Google Scholar]
- 21.Chen D, Chen J, Guo Y, Li Y. Cinobufacini promotes apoptosis of bladder cancer cells by influencing the expression of autophagy-related genes. Oncol Lett. 2018;15:7104–7110. doi: 10.3892/ol.2018.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li WL, Xiong LX, Shi XY, Xiao L, Qi GY, Meng C. IKKβ/NFκBp65 activated by interleukin-13 targets the autophagy-related genes LC3B and beclin 1 in fibroblasts co-cultured with breast cancer cells. Exp Ther Med. 2016;11:1259–1264. doi: 10.3892/etm.2016.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin P, He RQ, Dang YW, Wen DY, Ma J, He Y, Chen G, Yang H. An autophagy-related gene expression signature for survival prediction in multiple cohorts of hepatocellular carcinoma patients. Oncotarget. 2018;9:17368–17395. doi: 10.18632/oncotarget.24089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin P, He Y, Wen DY, Li XJ, Zeng JJ, Mo WJ, Li Q, Peng JB, Wu YQ, Pan DH, et al. Comprehensive analysis of the clinical significance and prospective molecular mechanisms of differentially expressed autophagy-related genes in thyroid cancer. Int J Oncol. 2018;53:603–619. doi: 10.3892/ijo.2018.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma Y, Zhang Y, Zhao Y, Wang X, Lin Y, Ma A. Expression of autophagy-related genes in cerebrospinal fluid of patients with tuberculous meningitis. Exp Ther Med. 2018;15:4671–4676. doi: 10.3892/etm.2018.5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng LQ, Li SY, Li CX. Expression profiling analysis of autophagy-related genes in perineural invasion of cutaneous squamous cell carcinoma. Oncol Lett. 2018;15:4837–4848. doi: 10.3892/ol.2018.7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moussay E, Kaoma T, Baginska J, Muller A, Van Moer K, Nicot N, Nazarov PV, Vallar L, Chouaib S, Berchem G, Janji B. The acquisition of resistance to TNFα in breast cancer cells is associated with constitutive activation of autophagy as revealed by a transcriptome analysis using a custom microarray. Autophagy. 2011;7:760–770. doi: 10.4161/auto.7.7.15454. [DOI] [PubMed] [Google Scholar]
- 28.Zhang H, Lu X, Wang N, Wang J, Cao Y, Wang T, Zhou X, Jiao Y, Yang L, Wang X, et al. Autophagy-related gene expression is an independent prognostic indicator of glioma. Oncotarget. 2017;8:60987–61000. doi: 10.18632/oncotarget.17719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gandolfi S, Prada CP, Richardson PG. How I treat the young patient with multiple myeloma. Blood. 2018;132:1114–1124. doi: 10.1182/blood-2017-05-693606. [DOI] [PubMed] [Google Scholar]
- 30.Raje NS, Bhatta S, Terpos E. Role of the RANK/RANKL pathway in multiple myeloma. Clin Cancer Res. 2019;25:12–20. doi: 10.1158/1078-0432.CCR-18-1537. [DOI] [PubMed] [Google Scholar]
- 31.Zhu B, Ju S, Chu H, Shen X, Zhang Y, Luo X, Cong H. The potential function of microRNAs as biomarkers and therapeutic targets in multiple myeloma. Oncol Lett. 2018;15:6094–6106. doi: 10.3892/ol.2018.8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu D, Yang C, Zhang Z, Cong Y, Xiao M. Knockdown of Linc00515 inhibits multiple myeloma autophagy and chemoresistance by upregulating miR-140-5p and downregulating ATG14. Cell Physiol Biochem. 2018;48:2517–2527. doi: 10.1159/000492690. [DOI] [PubMed] [Google Scholar]
- 33.Ma R, Zhang Y, Wang W, Wu J, Yang Q, Xu W, Jiang S, Han Y, Yu K, Zhang S. Inhibition of autophagy enhances the antitumour activity of tigecycline in multiple myeloma. J Cell Mol Med. 2018;22:5955–5963. doi: 10.1111/jcmm.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mei H, Xiang Y, Mei H, Fang B, Wang Q, Cao D, Hu Y, Guo T. Pterostilbene inhibits nutrient metabolism and induces apoptosis through AMPK activation in multiple myeloma cells. Int J Mol Med. 2018;42:2676–2688. doi: 10.3892/ijmm.2018.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su N, Wang P, Li Y. Role of Wnt/β-catenin pathway in inducing autophagy and apoptosis in multiple myeloma cells. Oncol Lett. 2016;12:4623–4629. doi: 10.3892/ol.2016.5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng Z, Liu T, Zheng J, Hu J. Clarifying the molecular mechanism associated with carfilzomib resistance in human multiple myeloma using microarray gene expression profile and genetic interaction network. Onco Targets Ther. 2017;10:1327–1334. doi: 10.2147/OTT.S130742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Desantis V, Saltarella I, Lamanuzzi A, Mariggiò MA, Racanelli V, Vacca A, Frassanito MA. Autophagy: A new mechanism of prosurvival and drug resistance in multiple myeloma. Transl Oncol. 2018;11:1350–1357. doi: 10.1016/j.tranon.2018.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yun Z, Zhichao J, Hao Y, Ou J, Ran Y, Wen D, Qun S. Targeting autophagy in multiple myeloma. Leuk Res. 2017;59:97–104. doi: 10.1016/j.leukres.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 39.Shi L, Campbell G, Jones WD, Campagne F, Wen Z, Walker SJ, Su Z, Chu TM, Goodsaid FM, Pusztai L, et al. The MicroArray quality control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nat Biotechnol. 2010;28:827–838. doi: 10.1038/nbt.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ioannidis JPA. The proposal to lower P value thresholds to .005. JAMA. 2018;319:1429–1430. doi: 10.1001/jama.2018.1536. [DOI] [PubMed] [Google Scholar]
- 41.Ostaszewski M, Kieffer E, Danoy G, Schneider R, Bouvry P. Clustering approaches for visual knowledge exploration in molecular interaction networks. BMC Bioinformatics. 2018;19:308. doi: 10.1186/s12859-018-2314-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He RQ, Zhou XG, Yi QY, Deng CW, Gao JM, Chen G, Wang QY. Prognostic signature of alternative splicing events in bladder urothelial carcinoma based on spliceseq data from 317 cases. Cell Physiol Biochem. 2018;48:1355–1368. doi: 10.1159/000492094. [DOI] [PubMed] [Google Scholar]
- 44.Liang L, Zeng JH, Qin XG, Chen JQ, Luo DZ, Chen G. Distinguishable prognostic signatures of left- and right-sided colon cancer: A study based on sequencing data. Cell Physiol Biochem. 2018;48:475–490. doi: 10.1159/000491778. [DOI] [PubMed] [Google Scholar]
- 45.Lin P, He RQ, Ma FC, Liang L, He Y, Yang H, Dang YW, Chen G. Systematic analysis of survival-associated alternative splicing signatures in gastrointestinal pan-adenocarcinomas. EBioMedicine. 2018;34:46–60. doi: 10.1016/j.ebiom.2018.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin P, Wen DY, Li Q, He Y, Yang H, Chen G. Genome-wide analysis of prognostic lncRNAs, miRNAs, and mRNAs forming a competing endogenous RNA network in hepatocellular carcinoma. Cell Physiol Biochem. 2018;48:1953–1967. doi: 10.1159/000492519. [DOI] [PubMed] [Google Scholar]
- 47.Yang H, Lin P, Wu HY, Li HY, He Y, Dang YW, Chen G. Genomic analysis of small nucleolar RNAs identifies distinct molecular and prognostic signature in hepatocellular carcinoma. Oncol Rep. 2018;40:3346–3358. doi: 10.3892/or.2018.6715. [DOI] [PubMed] [Google Scholar]
- 48.Zhang R, Lin P, Yang X, He RQ, Wu HY, Dang YW, Gu YY, Peng ZG, Feng ZB, Chen G. Survival associated alternative splicing events in diffuse large B-cell lymphoma. Am J Transl Res. 2018;10:2636–2647. [PMC free article] [PubMed] [Google Scholar]
- 49.Lu X, Sun W, Tang Y, Zhu L, Li Y, Ou C, Yang C, Su J, Luo C, Hu Y, Cao J. Identification of key genes in hepatocellular carcinoma and validation of the candidate gene, cdc25a, using gene set enrichment analysis, meta-analysis and cross-species comparison. Mol Med Rep. 2016;13:1172–1178. doi: 10.3892/mmr.2015.4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ni Y, Song C, Jin S, Chen Z, Ni M, Han L, Wu J, Jin Y. Gene set enrichment analysis: A genome-wide expression profile-based strategy for discovering functional microRNA-disease relationships. J Int Med Res. 2018;46:596–611. doi: 10.1177/0300060517693719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J, Vasaikar S, Shi Z, Greer M, Zhang B. WebGestalt 2017: A more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017;45:W130–W137. doi: 10.1093/nar/gkx356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zyla J, Marczyk M, Weiner J, Polanska J. Ranking metrics in gene set enrichment analysis: Do they matter? BMC Bioinformatics. 2017;18:256. doi: 10.1186/s12859-017-1674-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han X, Zhong Z, Kou J, Zheng Y, Liu Z, Jiang Y, Zhang Z, Gao Z, Cong L, Tian Y, Yang L. ROS Generated by upconversion nanoparticle-mediated photodynamic therapy induces autophagy via PI3K/AKT/mTOR signaling pathway in M1 peritoneal macrophage. Cell Physiol Biochem. 2018;48:1616–1627. doi: 10.1159/000492283. [DOI] [PubMed] [Google Scholar]
- 54.Li X, Huang Q, Wang M, Yan X, Song X, Ma R, Jiang R, Zhao D, Sun L. Compound K inhibits autophagy-mediated apoptosis through activation of the PI3K-Akt signaling pathway thus protecting against Ischemia/reperfusion injury. Cell Physiol Biochem. 2018;47:2589–2601. doi: 10.1159/000491655. [DOI] [PubMed] [Google Scholar]
- 55.Liu M, Zhao G, Zhang D, An W, Lai H, Li X, Cao S, Lin X. Active fraction of clove induces apoptosis via PI3K/Akt/mTOR-mediated autophagy in human colorectal cancer HCT-116 cells. Int J Oncol. 2018;53:1363–1373. doi: 10.3892/ijo.2018.4465. [DOI] [PubMed] [Google Scholar]
- 56.Luo X, Ye S, Jiang Q, Gong Y, Yuan Y, Hu X, Su X, Zhu W. Wnt inhibitory factor-1-mediated autophagy inhibits Wnt/β-catenin signaling by downregulating dishevelled-2 expression in non-small cell lung cancer cells. Int J Oncol. 2018;53:904–914. doi: 10.3892/ijo.2018.4442. [DOI] [PubMed] [Google Scholar]
- 57.Wang J, Sun P, Chen Y, Yao H, Wang S. Novel 2-phenyloxypyrimidine derivative induces apoptosis and autophagy via inhibiting PI3K pathway and activating MAPK/ERK signaling in hepatocellular carcinoma cells. Sci Rep. 2018;8:10923. doi: 10.1038/s41598-018-29199-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yin S, Yang S, Pan X, Ma A, Ma J, Pei H, Dong Y, Li S, Li W, Bi X. MicroRNA155 promotes ox-LDL-induced autophagy in human umbilical vein endothelial cells by targeting the PI3K/Akt/mTOR pathway. Mol Med Rep. 2018;18:2798–2806. doi: 10.3892/mmr.2018.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Daskalaki I, Gkikas I, Tavernarakis N. Hypoxia and selective autophagy in cancer development and therapy. Front Cell Dev Biol. 2018;6:104. doi: 10.3389/fcell.2018.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Du L, Shen T, Liu B, Zhang Y, Zhao C, Jia N, Wang Q, He Q. Shock wave therapy promotes cardiomyocyte autophagy and survival during hypoxia. Cell Physiol Biochem. 2017;42:673–684. doi: 10.1159/000477885. [DOI] [PubMed] [Google Scholar]
- 61.Liang Y, Chen X, Liang Z. MicroRNA-320 regulates autophagy in retinoblastoma by targeting hypoxia inducible factor-1alpha. Exp Ther Med. 2017;14:2367–2372. doi: 10.3892/etm.2017.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Niu G, Zhu D, Zhang X, Wang J, Zhao Y, Wang X. Role of hypoxia-inducible factors 1a (HIF1a) in SH-SY5Y cell autophagy induced by oxygen-glucose deprivation. Med Sci Monit. 2018;24:2758–2766. doi: 10.12659/MSM.905140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang H, Zhang D, Jia S, Huang S, Xiao L, Ma L, Liu G, Gong K, Xu L. Effect of sustained hypoxia on autophagy of genioglossus Muscle-derived stem cells. Med Sci Monit. 2018;24:2218–2224. doi: 10.12659/MSM.906195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Z, Deng M, Liu Z, Wu S. Hypoxia-induced miR-210 promoter demethylation enhances proliferation, autophagy and angiogenesis of schwannoma cells. Oncol Rep. 2017;37:3010–3018. doi: 10.3892/or.2017.5511. [DOI] [PubMed] [Google Scholar]
- 65.Bosseler M, Marani V, Broukou A, Lequeux A, Kaoma T, Schlesser V, François JH, Palissot V, Berchem GJ, Aouali N, Janji B. Inhibition of HIF1a-dependent upregulation of Phospho-l-Plastin resensitizes multiple myeloma cells to frontline therapy. Int J Mol Sci. 2018;19(pii):E1551. doi: 10.3390/ijms19061551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Coudre C, Alani J, Ritchie W, Marsaud V, Sola B, Cahu J. HIF-1a and rapamycin act as gerosuppressant in multiple myeloma cells upon genotoxic stress. Cell Cycle. 2016;15:2174–2182. doi: 10.1080/15384101.2016.1196302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Filippi I, Saltarella I, Aldinucci C, Carraro F, Ria R, Vacca A, Naldini A. Different adaptive responses to hypoxia in normal and multiple myeloma endothelial cells. Cell Physiol Biochem. 2018;46:203–212. doi: 10.1159/000488423. [DOI] [PubMed] [Google Scholar]
- 68.Muz B, Kusdono HD, Azab F, de la Puente P, Federico C, Fiala M, Vij R, Salama NN, Azab AK. Tariquidar sensitizes multiple myeloma cells to proteasome inhibitors via reduction of hypoxia-induced P-gp-mediated drug resistance. Leuk Lymphoma. 2017;58:2916–2925. doi: 10.1080/10428194.2017.1319052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Viziteu E, Grandmougin C, Goldschmidt H, Seckinger A, Hose D, Klein B, Moreaux J. Chetomin, targeting HIF-1a/p300 complex, exhibits antitumour activity in multiple myeloma. Br J Cancer. 2016;114:519–523. doi: 10.1038/bjc.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang P, Long M, Zhang S, Cheng Z, Zhao X, He F, Liu H, Ming L. Hypoxia inducible factor-1a regulates autophagy via the p27-E2F1 signaling pathway. Mol Med Rep. 2017;16:2107–2112. doi: 10.3892/mmr.2017.6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang X, Wu TT, Jiang L, Rong D, Zhu YQ. Deferoxamine-induced migration and odontoblast differentiation via ROS-dependent autophagy in dental pulp stem cells. Cell Physiol Biochem. 2017;43:2535–2547. doi: 10.1159/000484506. [DOI] [PubMed] [Google Scholar]
- 72.Zhang W, Zhang J. Dexmedetomidine preconditioning protects against lung injury induced by ischemia-reperfusion through inhibition of autophagy. Exp Ther Med. 2017;14:973–980. doi: 10.3892/etm.2017.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu SM, Rao T, Yang X, Ning JZ, Yu WM, Ruan Y, Yuan R, Li CL, Jiang K, Hu W, et al. Autophagy may play an important role in varicocele. Mol Med Rep. 2017;16:5471–5479. doi: 10.3892/mmr.2017.7253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Seegmiller AC, Wang HY, Hladik C, Chen W. Uniform expression of Notch1, suppressor of B-cell-specific gene expression, in plasmablastic lymphoma. Arch Pathol Lab Med. 2011;135:770–775. doi: 10.5858/2009-0691-OA.1. [DOI] [PubMed] [Google Scholar]
- 75.Pourdehnad M, Truitt ML, Siddiqi IN, Ducker GS, Shokat KM, Ruggero D. Myc and mTOR converge on a common node in protein synthesis control that confers synthetic lethality in Myc-driven cancers. Proc Natl Acad Sci USA. 2013;110:11988–11993. doi: 10.1073/pnas.1310230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu X, Zhang Y, Wang Z, Wang X, Zhu G, Han G, Chen G, Hou C, Wang T, Shen B, et al. Metabotropic glutamate receptor 3 is involved in B-cell-related tumor apoptosis. Int J Oncol. 2016;49:1469–1478. doi: 10.3892/ijo.2016.3623. [DOI] [PubMed] [Google Scholar]
- 77.Kinoshita S, Ri M, Kanamori T, Aoki S, Yoshida T, Narita T, Totani H, Ito A, Kusumoto S, Ishida T, et al. Potent antitumor effect of combination therapy with sub-optimal doses of Akt inhibitors and pomalidomide plus dexamethasone in multiple myeloma. Oncol Lett. 2018;15:9450–9456. doi: 10.3892/ol.2018.8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kishino A, Hayashi K, Hidai C, Masuda T, Nomura Y, Oshima T. XBP1-FoxO1 interaction regulates ER stress-induced autophagy in auditory cells. Sci Rep. 2017;7:4442. doi: 10.1038/s41598-017-02960-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Miki Y, Tanji K, Mori F, Utsumi J, Sasaki H, Kakita A, Takahashi H, Wakabayashi K. Autophagy mediators (FOXO1, SESN3 and TSC2) in Lewy body disease and aging. Neurosci Lett. 2018;684:35–41. doi: 10.1016/j.neulet.2018.06.052. [DOI] [PubMed] [Google Scholar]
- 80.Shen M, Cao Y, Jiang Y, Wei Y, Liu H. Melatonin protects mouse granulosa cells against oxidative damage by inhibiting FOXO1-mediated autophagy: Implication of an antioxidation-independent mechanism. Redox Biol. 2018;18:138–157. doi: 10.1016/j.redox.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.De Bruyne E, Bos TJ, Schuit F, Van Valckenborgh E, Menu E, Thorrez L, Atadja P, Jernberg-Wiklund H, Vanderkerken K. IGF-1 suppresses Bim expression in multiple myeloma via epigenetic and posttranslational mechanisms. Blood. 2010;115:2430–2440. doi: 10.1182/blood-2009-07-232801. [DOI] [PubMed] [Google Scholar]
- 82.Zhou Y, Uddin S, Zimmerman T, Kang JA, Ulaszek J, Wickrema A. Growth control of multiple myeloma cells through inhibition of glycogen synthase kinase-3. Leuk Lymphoma. 2008;49:1945–1953. doi: 10.1080/10428190802304966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shen JK, Du HP, Ma Q, Yang M, Wang YG, Jin J. 4-Chlorobenzoyl berbamine, a novel berbamine derivative, induces apoptosis in multiple myeloma cells through the IL-6 signal transduction pathway and increases FOXO3a-Bim expression. Oncol Rep. 2013;30:425–432. doi: 10.3892/or.2013.2431. [DOI] [PubMed] [Google Scholar]
- 84.Liu XF, Zhou Q, Hassan R, Pastan I. Panbinostat decreases cFLIP and enhances killing of cancer cells by immunotoxin LMB-100 by stimulating the extrinsic apoptotic pathway. Oncotarget. 2017;8:87307–87316. doi: 10.18632/oncotarget.20263. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the present study are included in this published article.