Abstract
This article describes several features in the MAFFT online service for multiple sequence alignment (MSA). As a result of recent advances in sequencing technologies, huge numbers of biological sequences are available and the need for MSAs with large numbers of sequences is increasing. To extract biologically relevant information from such data, sophistication of algorithms is necessary but not sufficient. Intuitive and interactive tools for experimental biologists to semiautomatically handle large data are becoming important. We are working on development of MAFFT toward these two directions. Here, we explain (i) the Web interface for recently developed options for large data and (ii) interactive usage to refine sequence data sets and MSAs.
Keywords: multiple sequence alignment, sequence analysis, phylogenetic tree
Multiple sequence alignment (MSA) is an important step in comparative analyses of biological sequences. We provide an online service for computing MSAs on the Web using MAFFT [1, 2]. MAFFT has several different options for computing large MSAs consisting of thousands of sequences. Our service also has some additional functions (interactive sequence selection and phylogenetic inference) for preprocessing and postprocessing MSA. Moreover, these processes can be circularly performed as necessary. Here, we describe usage of these functions, including recently added ones, and several tips for using our online service.
MSA of large data
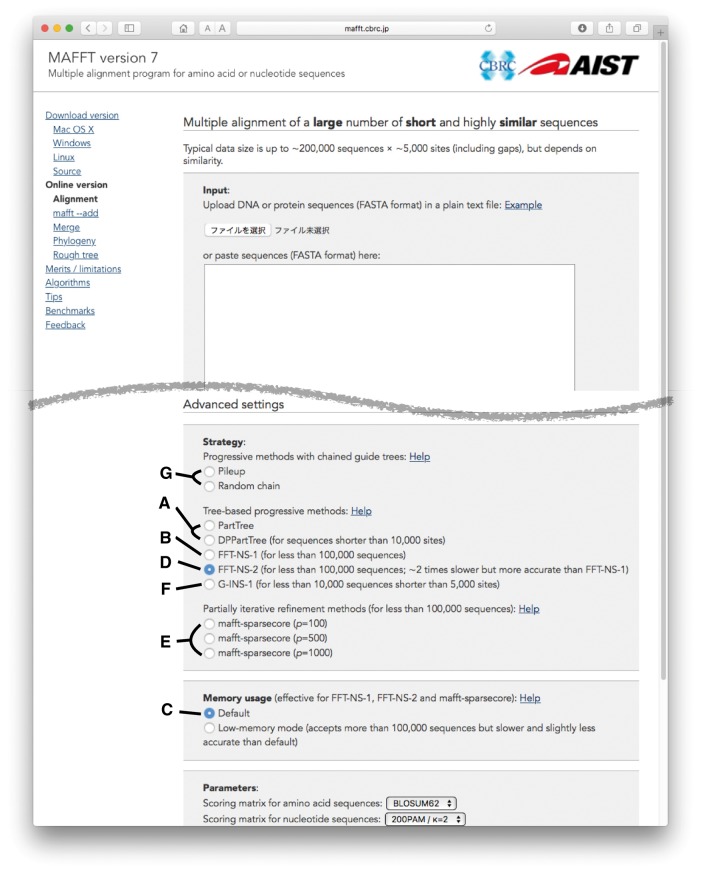
The demand for MSAs with a large number of sequences is increasing along with the advance of sequencing technologies. The default option of MAFFT, FFT-NS-2, is applicable to most cases, but MAFFT has more options for constructing large MSAs. They can be selected in a designated page for large alignment on the MAFFT server: http://mafft.cbrc.jp/alignment/server/large.html. Below, we briefly explain the options available on this page. Headings (A)–(G) correspond to those in Figure 1. Benchmark results of these options are shown in Table 1. Commands for locally running those options are available in the last section.
Figure 1.
Screenshot of input page for large MSAs in MAFFT online service. (A–G) are explained in the main text.
Table 1.
Results of two different benchmarks, ContTest (136 entries, 1467–43 912 sequences) [3] and HomFam (89 entries; 93–93 681 sequences) [4], for some MAFFT options available on our online server
| Method | ContTest |
HomFam |
|||
|---|---|---|---|---|---|
| Accuracy score | CPU time (minutes) | Accuracy score (SP/TC) | CPU time (minutes) | ||
| A | PartTree (partsize = 50) | 0.4103 | 61 | 0.7862/0.5658 | 47 |
| PartTree (partsize = 1000) | 0.4364 | 140 | 0.8258/0.6377 | 94 | |
| DPPartTree (partsize = 50) | 0.4424 | 210 | 0.8413/0.6597 | 160 | |
| DPPartTree (partsize = 1000) | 0.4632 | 1000 | 0.8541/0.6934 | 820 | |
| B | FFT-NS-1 | 0.4856 | 170 | 0.8491/0.6669 | 160 |
| B+C | FFT-NS-1 (memsavetree) | 0.4835 | 280 | 0.8416/0.6667 | 260 |
| D | FFT-NS-2 | 0.4998 | 500 | 0.8759/0.7162 | 460 |
| D+C | FFT-NS-2 (memsavetree) | 0.5099 | 1100 | 0.8611/0.7023 | 990 |
| E | mafft-sparsecore (p = 100) | 0.5153 | 730 | 0.8821/0.7274 | 650 |
| mafft-sparsecore (p = 500) | 0.5361 | 1200 | 0.8970/0.7586 | 1300 | |
| mafft-sparsecore (p = 1000) | 0.5440 | 3400 | 0.9075/0.7810 | 4400 | |
| E+C | mafft-sparsecore (p = 100, memsavetree) | 0.5298 | 1500 | 0.8845/0.7416 | 1300 |
| mafft-sparsecore (p = 500, memsavetree) | 0.5438 | 2000 | 0.8995/0.7638 | 2000 | |
| mafft-sparsecore (p = 1000, memsavetree) | 0.5428 | 4200 | 0.9052/0.7826 | 5000 | |
| F | G-INS-1 | 0.5696 | 55 000 | 0.9306/0.8288 | 49000 |
| G | Randomchain | 0.5425 | 100 | 0.8349/0.6681 | 88 |
Note: The sum-of-pairs (SP) and total-column (TC) scores for HomFam were calculated by the FastSP program [5]. (A–G) correspond to the techniques explained in the main text. Command-line arguments are displayed after performing the calculation on the online service and also listed in the main text. Random numbers are used in (A), (E) and (G). In this test, only one set of random numbers was used for each method. For (E) and (G), seed of random numbers can be specified in the download version (see the last section in the main text) but cannot be specified in the online version. See https://mafft.sb.ecei.tohoku.ac.jp/ for detailed results.
A. PartTree and DPPartTree (Figure 1A) [6] are highly approximate options. These methods recursively cluster sequences and simultaneously compute a distance between the clusters, each of which is represented by a single sequence. The order of the computational time is , where N is the number of sequences. They are fast and applicable to large MSAs, but accuracy is sacrificed because of the approximation of guide tree calculation (Table 1). The PartTree and DPPartTree options share a basic design, but the former uses k-mer-based distance to estimate the similarity between sequences [7], while the latter uses dynamic programming (DP) [8] to estimate the similarity. Accordingly, the latter is slower but more accurate. In the command-line version, the balance between accuracy and speed can also be adjusted by a parameter, partsize, but this parameter is fixed to 1000 in the online service.
B. FFT-NS-1 (Figure 1B): This is another approximate method. Its accuracy is higher than PartTree and DPPartTree in benchmark tests (Table 1). The input sequences are progressively aligned using a guide tree [6, 9, 10]. For constructing the guide tree, pairwise distances are computed based on the number of shared k-mers. The length of k-mer is 6 for both protein and nucleotide data, but 20 amino acids are grouped into six physicochemical groups [11], and an amino acid sequence is converted to a sequence composed of six letters. The current version of MAFFT uses the following formula to compute distance Dij between sequences i and j:
where Sij is alignment score between sequences i and j. f(x, y) adjusts the distance to avoid a case where the distance between unrelated sequences happens to become zero when a long sequence and a short sequence are compared.
where x and y are the lengths of the longer and the shorter sequence i or j, respectively. a, b and c are empirically determined parameters; a = 0.1, b = 10 000 (nucleotide), 2500 (amino acid) and c = 0.01. As Dij is computed for all sequence pairs, the computational time is proportional to N2, where N is the number of sequences. The space complexity is also O(N2) by default.
To build a guide from distances, MAFFT uses a UPGMA-like method with a small modification [12]. When merging clusters L and R into a new cluster P, distance DPC from P to a third cluster C is calculated with:
The resulting tree becomes more imbalanced [13] with smaller values of parameter s (0 ≤ s ≤1). The default s value has been unchanged from 0.1 since the initial release in 2002, but can be specified with the --mixedlinkage flag in the download version.
C. To compute a guide tree with less RAM, a low-memory mode is available but not enabled by default (Figure 1C). If a calculation in the online service requires more RAM than a threshold, then the calculation is terminated and an error message is returned instructing the user to select the low-memory mode. In this mode, instead of storing a full distance matrix in RAM, distances are calculated two times during the tree building step. Accordingly, the calculation time is longer than the normal mode.
D. FFT-NS-2 (Figure 1D): This is the default option of MAFFT. In this method, after performing FFT-NS-1, a new distance matrix and guide tree are recalculated based on the MSA, and then the final MSA is built using the new guide tree. In benchmark tests, the accuracy is generally improved by the recalculation of the guide tree as shown in Table 1. This method is at least two times slower than FFT-NS-1. The low-memory mode (Figure 1C) is also available for FFT-NS-2.
E. mafft-sparsecore (Figure 1E) [12] is a combination of the iterative refinement method [14–16] and the progressive method. It aims to improve the alignment accuracy by partly applying the iterative refinement method, which is known to be more accurate than the progressive method. The procedure was described in Yamada et al. (2016) [12]: (i) the input sequences are sorted by length. From the upper n% of the sorted sequences, p sequences are randomly selected as ‘core’ sequences. The default values of n and p are 50 and 500, respectively. (ii) An MSA of the p core sequences is constructed by an iterative refinement option, G-INS-i. (iii) The remaining sequences are added to the core MSA using the –add option [17], which uses the progressive alignment method. The accuracy and speed are controlled by the parameter p. With larger p, the accuracy is improved, but computational cost becomes higher (Table 1), as more sequences are subjected to the iterative refinement calculation. The memory usage is mainly determined by the progressive alignment stage (iii). The low-memory mode (Figure 1C) is also available for mafft-sparsecore.
F. G-INS-1 (Figure 1F): This gives more accurate MSAs [12, 18] but takes a longer computational time and requires more RAM than other methods. This method uses an accurate guide tree based on all-to-all DP calculation and a scoring function similar to COFFEE [19] in progressive alignment. We are developing a memory-efficient version of G-INS-1, which runs in parallel on distributed memory systems or shared memory systems (manuscript in preparation). This option is experimentally supported at http://mafft.cbrc.jp/alignment/server/large-lsf.html.
G. Pileup (Figure 1G): This is the simplest strategy. The first and the second sequences are first aligned. Then, the other sequences are added to the alignment in the order in the input file. Random chain: This is similar to Pileup, but the order of sequences is randomized. The usefulness of this strategy is controversial [3, 12, 13, 20, 21]. However, because these methods have an advantage in computational simplicity, we have made them available in our service.
Selecting suitable strategies
To select suitable MAFFT options for specific problems, consider the following factors.
For aligning a small number of sequences, the iterative refinement method is known to effectively improve the accuracy, as noted above. However, for large-scale MSAs (the subject of this article), the effect of iterative refinement was recently assessed to be small. More specifically, in Figure 1 in Le et al. [18], the advantage of MAFFT-L-INS-i (an iterative refinement method) over MAFFT-L-INS-1 (a progressive method) was clearly observed for a small number of sequences but not for thousands of sequences. Moreover, a direct application of the iterative refinement method to large sequence data sets is difficult in terms of computational resources.
In benchmarks with ∼1000–100 000 sequences, G-INS-1 outperforms other methods in accuracy as shown in Table 1. The difference is statistically significant in several cases. Thus, this method is first recommended if computational resources allow. We are making an effort to decrease the computational resources required by this method. If it is difficult to apply G-INS-1, then the next candidate would be mafft-sparsecore, which uses the advantage of iterative refinement for small MSAs.
These two methods can be applied to typical protein sequences with <10 000 sites, but cannot be applied to long DNA sequences. In such a case, FFT-NS-2 or FFT-NS-1 can be useful, as the computational time is proportional to , where L is sequence length, because of the FFT approximation [1]. However, this is only when the input sequences share global homology (from 5′ end to 3′end), and the similarity level is high. MAFFT cannot handle data with genomic rearrangements, such as inversions and translocations. Also note that an MSA can be built only when the sequences are all homologous. It does not make sense to construct an MSA of nonhomologous sequences.
For a data set with much >100 000 sequences, PartTree and DPPartTree, can be applied if the sequences are homologous, as their time complexity is O(NlogN), where N is the number of sequences. However, there are also other popular programs, such as Clustal Omega [4] and UPP [22], for this purpose. The PartTree algorithm contributed to these programs theoretically and/or practically. Clustal Omega uses the mBed algorithm [23] to build a guide tree with a time complexity of . UPP uses PASTA [24] to build a backbone MSA of a small number of sequences and then adds the remaining sequences using hmmalign [25] with the time complexity of O(N). PASTA uses MAFFT-PartTree to generate the initial MSA and MAFFT-L-INS-i (an iterative refinement option for small data) to generate sub-MSAs of closely related sequences. Performance comparison including these methods can be seen on https://mafft.sb.ecei.tohoku.ac.jp/, which also includes detailed benchmark results for subsets with different data sizes.
Similarity level and difference in sequence lengths also should be considered. If the sequences are highly similar to each other and their lengths are also similar, then fast methods, such as FFT-NS-1 or even Pileup should result in a useful MSA. If the input data have fragmentary sequences and full-length sequences, then a two-step strategy sometimes works well. That is (i) align the full-length sequences first and then (ii) add the fragmentary sequences to the full-length MSA using the --addfragments option (see next section).
Use of existing MSA
Each step of the calculation of mafft-sparsecore (Figure 1E) can be separately or manually performed. If a reliable MSA and a set of unaligned sequences are given to http://mafft.cbrc.jp/alignment/server/add_sequences.html, then an MSA of all the sequences is returned, in which the existing MSA is preserved as the original one.
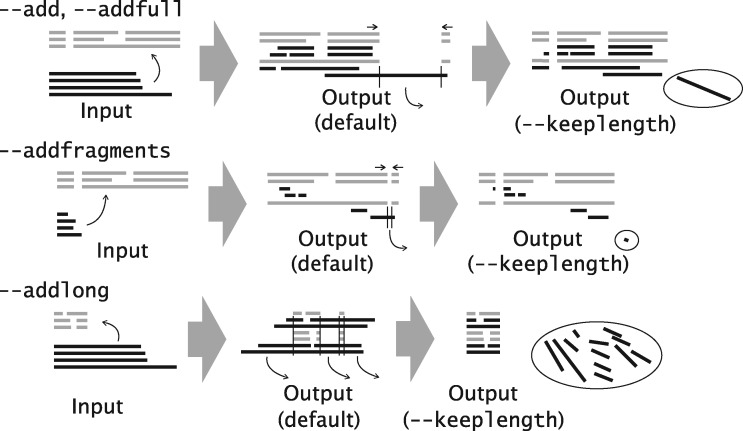
Several variants, --add, --addfull, --addfragments and --addlong, are available. They can be selected according to the relative length of new sequences to the existing MSA as illustrated in Figure 2. The four options work similarly to each other. However, sequences added with the --add option are subjected to distance calculation with time complexity of O(N2), where N is the number of sequences. In the other three options, distances between the sequences in the existing alignment are computed with a time complexity of O(M2), where M is the number of sequences in the existing MSA, to build a tree of the M sequences using the UPGMA-like method (see above). For each of (N−M) sequences to be added, distances to the M sequences are computed to locate the position of the sequence in the tree, followed by the building of an alignment of (M + 1) sequences. Then, a full MSA is built from the (N−M) MSAs. The latter strategy is useful when the new sequences do not overlap with each other (as in the case of fragmentary sequences) and when the phylogenetic relationship between new sequences is not necessary to consider. There are several other tools, such as hmmalign [25], PaPaRa [26] and PAGAN [27], to add sequences to an existing MSA.
Figure 2.
Variants of --add option.
Note that the length of the resulting MSA can differ from that of the original MSA. This is because additional gaps are necessary when new sequences have insertions. All-gap sites, if any, in the original MSA are deleted. As such changes in length are not useful in some cases, we have implemented a new option, --keeplength, in which (1) insertions in the new sequences are deleted and (2) all-gap sites in the original MSA are reinserted as shown at the right end in Figure 2. This option is selectable in the online version and sometimes useful for mapping new sequences to a reference MSA.
Interactive sequence choice and visualization
Recently, we have access to huge amounts of sequence data from widely divergent organisms, but the quality of the data is not always high because of the limitations of sequencing technologies. In the case of amino acid sequence data, the difficulty in eukaryotic gene prediction [28–30] also results in errors in data. It might be possible to automatically exclude such problematic data in certain cases, but sometimes, biologically important information is in low-quality sequences, especially when interest is in nonmodel organisms.
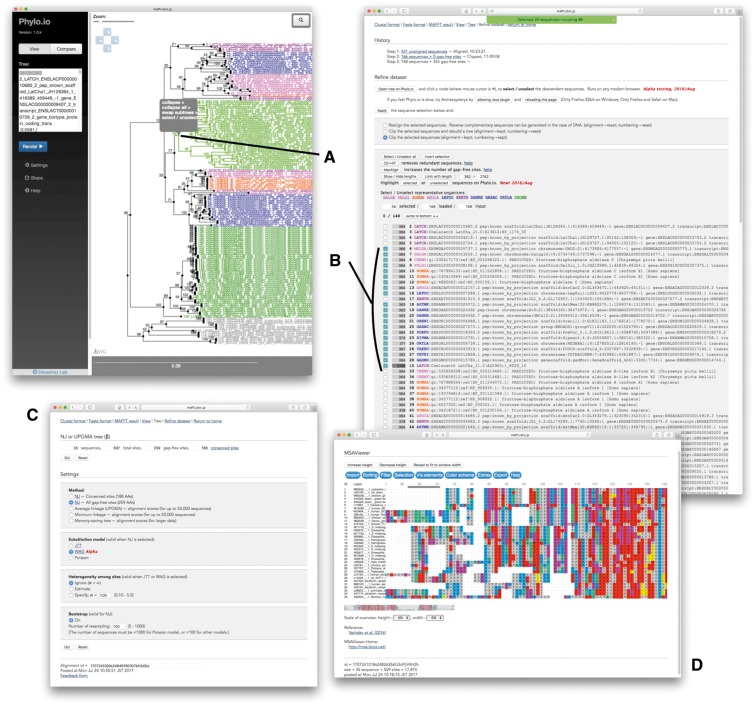
For such cases, it is necessary to manually choose sequences, but this is becoming difficult because of increasing data size. Therefore, an interactive tool to help this process is necessary. Our service has some functions for this purpose as explained in Kuraku et al. [31]. Sequences can be selected/unselected one by one in the sequence selection window (Figure 3B). Moreover, a group of sequences in a single phylogenetic cluster can be selected or unselected in a tree viewer. If you click on a node in a tree (Figure 3A), the descendant sequences under the node are selected or unselected together in the list of sequences (Figure 3B). Automated tools for sequence selection, such as CD-HIT [32] and MaxAlign [33], can also run on our service. The selected sequences are subjected to phylogenetic tree inference using the neighbor-joining method [34] or UPGMA [35] with several options, such as distance measure and the number of bootstrap cycles (Figure 3C). Then, the data set can be further refined using the new tree. The maximum-likelihood method is not supported because of the high computational costs. It must be performed locally or using other online services.
Figure 3.
Interactive sequence selection. A group of sequences in guide tree (A) is selected at a time in sequence selection window (B). Several options for tree estimation can be selected (C). MSA can be visually checked using MSAViewer (D).
Two tree viewers, Phylo.io [36] and Archaeopteryx [37], are used for sequence selection and visualization of phylogenetic trees. Originally, we used Archaeopteryx Java plugin, but modern browsers no longer support Java plugin for security reasons. Thus, we recently adopted Phylo.io, which is written in JavaScript and works with most modern browsers. With the addition of Phylo.io to our service, we have added some new features:
Coloring of sequence title corresponding to the databases in aLeaves [29].
Interactive sequence selection (see above).
Automatic rooting similar to mid-point rooting. This is just for visualization without any biological basis. To estimate the position of root, outgroup or other additional information is necessary.
A JavaScript version of Archaeopteryx is being developed (C. Zmasek, personal communication), and we are planning to use this viewer, too. To visualize MSAs, two tools, Jalview [38] (as Java plugin) and MSAViewer [39] (written in JavaScript; Figure 3C), are available on our service.
Necessity of large MSAs
The relationship between alignment accuracy and data size is still unclear. It is naively expected that the accuracy of an MSA is improved with the number of input sequences. However, highly accurate methods cannot be applied to large data because of computational costs. Useful information related to this issue has recently been reported by Le et al. [18]. In their tests, the accuracy of downstream analysis (protein secondary structure prediction in this case) is improved with the increase of sequences for medium-scale data (<1000 sequences), but with more sequences, the accuracy reaches a sort of plateau. Thus, there may be optimal data size. Their test also suggested that the accuracy of MSA itself hits a maximum point at a smaller number of sequences (around 200) and that the accuracy of MSA decreases with an increase in the number of sequences. This observation is consistent with Sievers et al. [40]. Such optimal data sizes can differ for different problems. For example, in the case of prediction of contact residues based on co-evolution, larger MSAs are generally thought to be necessary [41, 42].
Command-line options
Each method also runs locally. In the current version (7.310; August 2017), the corresponding commands are as follows:
PartTree (Figure 1A)
mafft --parttree --partsize 1000 input > output
DPPartTree (Figure 1A)
mafft --dpparttree --partsize 1000 input > output
FFT-NS-1 (Figure 1B)
mafft --retree 1 input > output
mafft --retree 1 --memsavetree input > output (low-memory mode)
mafft --retree 1 --thread -1 input > output (multithread mode)
With thread -1, the number of physical cores is automatically counted and all cores are used. See http://mafft.cbrc.jp/alignment/software/multithreading.html for detailed information on multithreading.
FFT-NS-2 (Figure 1D)
mafft input > output
mafft --memsavetree input > output (low-memory mode)
mafft --thread -1 input > output (multithread mode)
mafft-sparsecore (Figure 1E)
mafft-sparsecore.rb - p p -n n -s s -i input > output
mafft-sparsecore.rb -p p -n n -s s -A ”--memsavetree” -i input > output (low-memory mode)
mafft-sparsecore.rb -p p -n n -s s -A ”--thread -1” -C ”--thread -1” -i input > output (multithread mode)
p and n are as explained above, and s is seed for random numbers. Flags for the iterative refinement stage and those for the progressive stage can be specified after -C and -A, respectively. See http://mafft.cbrc.jp/alignment/software/sparsecore.html for detailed information.
G-INS-1 (Figure 1F)
mafft --globalpair input > output
mafft --globalpair --thread -1 input > output (multithread mode)
Pileup (Figure 1G)
mafft --pileup input > output
Random chain (Figure 1G)
mafft --randomchain --randomseed s input > output
s is seed for random numbers.
Adding new sequences to an MSA
mafft --add newSequences existingMSA > output
mafft --addfull newSequences existingMSA > output
mafft --addlong newSequences existingMSA > output
mafft --addfragments newSequences existingMSA > output
The --keeplength flag can be added to each command (see above). Add --thread −1 to enable multithreading.
Key Points.
MSA is an important step in phylogeny inference, functional prediction and many other analyses.
The demand for MSAs with a large number of sequences is increasing.
MAFFT has different options for computing large MSAs in both the local and online versions. The online version has additional features for preprocessing and postprocessing MSAs.
Acknowledgement
The authors thank Daron M. Standley, RIMD, Osaka University, for inspiring discussion.
Biographies
Kazutaka Katoh is an associate professor in the Department of Genome Informatics, Research Institute for Microbial Diseases, Osaka University. His research interests are in bioinformatics and molecular evolution.
John Rozewicki is a researcher in the Department of Genome Informatics, Research Institute for Microbial Diseases, Osaka University. His research focus is designing systems to conduct bioinformatics research using high-performance computing and networked server infrastructure.
Kazunori D. Yamada is an assistant professor in graduate school of information sciences, Tohoku University. His research interests include development of amino acid sequence alignment methods and machine learning.
Research Institute for Microbial Diseases, Osaka University, conducts basic research in the areas of infectious disease, immunology and oncology.
Funding
Japan Society for the Promotion of Science (JSPS) KAKENHI (grant number JP16K07464) and the Platform Project for Supporting Drug Discovery and Life Science Research from Japan Agency for Medical Research and Development (AMED).
References
- 1. Katoh K, Misawa K, Kuma K, et al. . MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 2002;30:3059–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Katoh K, Standley DM.. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 2013;30(4):772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fox G, Sievers F, Higgins DG.. Using de novo protein structure predictions to measure the quality of very large multiple sequence alignments. Bioinformatics 2016;32(6):814–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sievers F, Wilm A, Dineen D, et al. . Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 2011;7:539.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mirarab S, Warnow T.. FastSP: linear time calculation of alignment accuracy. Bioinformatics 2011;27:3250–8. [DOI] [PubMed] [Google Scholar]
- 6. Katoh K, Toh H.. PartTree: an algorithm to build an approximate tree from a large number of unaligned sequences. Bioinformatics 2007;23:372–4. [DOI] [PubMed] [Google Scholar]
- 7. Higgins DG, Sharp PM.. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 1988;73:237–44. [DOI] [PubMed] [Google Scholar]
- 8. Needleman SB, Wunsch CD.. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol 1970;48:443–53. [DOI] [PubMed] [Google Scholar]
- 9. Hogeweg P, Hesper B.. The alignment of sets of sequences and the construction of phyletic trees: an integrated method. J Mol Evol 1984;20:175–86. [DOI] [PubMed] [Google Scholar]
- 10. Feng DF, Doolittle RF.. Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J Mol Evol 1987;25:351–60. [DOI] [PubMed] [Google Scholar]
- 11. Dayhoff MO, Schwartz RM, Orcutt BC.. A model of evolutionary change in proteins In Dayhoff MO, Ech RV (eds), Atlas of Protein Sequence and Structure. Maryland: National Biomedical Research Foundation; 1978, 345–52. [Google Scholar]
- 12. Yamada KD, Tomii K, Katoh K.. Application of the mafft sequence alignment program to large data-reexamination of the usefulness of chained guide trees. Bioinformatics 2016;32(21):3246–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Boyce K, Sievers F, Higgins DG.. Simple chained guide trees give high-quality protein multiple sequence alignments. Proc Natl Acad Sci USA 2014;111:10556–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barton GJ, Sternberg MJ.. A strategy for the rapid multiple alignment of protein sequences. confidence levels from tertiary structure comparisons. J Mol Biol 1987;198:327–37. [DOI] [PubMed] [Google Scholar]
- 15. Berger MP, Munson PJ.. A novel randomized iterative strategy for aligning multiple protein sequences. Comput Appl Biosci 1991;7:479–84. [DOI] [PubMed] [Google Scholar]
- 16. Gotoh O. Optimal alignment between groups of sequences and its application to multiple sequence alignment. Comput Appl Biosci 1993;9:361–70. [DOI] [PubMed] [Google Scholar]
- 17. Katoh K, Frith MC.. Adding unaligned sequences into an existing alignment using MAFFT and LAST. Bioinformatics 2012;28:3144–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Le Q, Sievers F, Higgins DG.. Protein multiple sequence alignment benchmarking through secondary structure prediction. Bioinformatics 2017;33(9):1331–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Notredame C, Holm L, Higgins DG.. COFFEE: an objective function for multiple sequence alignments. Bioinformatics 1998;14:407–22. [DOI] [PubMed] [Google Scholar]
- 20. Sievers F, Hughes GM, Higgins DG.. Systematic exploration of guide-tree topology effects for small protein alignments. BMC Bioinformatics 2014;15:338.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tan G, Gil M, Löytynoja AP, et al. . Simple chained guide trees give poorer multiple sequence alignments than inferred trees in simulation and phylogenetic benchmarks. Proc Natl Acad Sci USA 2015;112:E99–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nguyen NPD, Mirarab S, Kumar K, et al. . Ultra-large alignments using phylogeny-aware profiles. Genome Biol 2015;16:124.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Blackshields G, Sievers F, Shi W, et al. . Sequence embedding for fast construction of guide trees for multiple sequence alignment. Algorithms Mol Biol 2010;5:21.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mirarab S, Nguyen N, Guo S, et al. . PASTA: ultra-large multiple sequence alignment for nucleotide and amino-acid sequences. J Comput Biol 2015;22(5):377–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Finn RD, Clements J, Eddy SR.. Hmmer web server: interactive sequence similarity searching. Nucleic Acids Res 2011;39:W29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Berger SA, Stamatakis A.. Aligning short reads to reference alignments and trees. Bioinformatics 2011;27:2068–75. [DOI] [PubMed] [Google Scholar]
- 27. Löytynoja A, Vilella AJ, Goldman N.. Accurate extension of multiple sequence alignments using a phylogeny-aware graph algorithm. Bioinformatics 2012;28:1684–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gotoh O, Morita M, Nelson DR.. Assessment and refinement of eukaryotic gene structure prediction with gene-structure-aware multiple protein sequence alignment. BMC Bioinformatics 2014;15:189.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nagy A, Patthy L.. MisPred: a resource for identification of erroneous protein sequences in public databases. Database 2013;2013:bat053.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yandell M, Ence D.. A beginner’s guide to eukaryotic genome annotation. Nat Rev Genet 2012;13:329–42. [DOI] [PubMed] [Google Scholar]
- 31. Kuraku S, Zmasek CM, Nishimura O, et al. . aLeaves facilitates on-demand exploration of metazoan gene family trees on mafft sequence alignment server with enhanced interactivity. Nucleic Acids Res 2013;41:W22–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li W, Jaroszewski L, Godzik A.. Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics 2001;17(3):282–3. [DOI] [PubMed] [Google Scholar]
- 33. Gouveia-Oliveira R, Sackett PW, Pedersen AG.. MaxAlign: maximizing usable data in an alignment. BMC Bioinformatics 2007;8:312.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Saitou N, Nei M.. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987;4(4):406–25. [DOI] [PubMed] [Google Scholar]
- 35. Sokal RR, Michener CD.. A statistical method for evaluating systematic relationships. University of Kansas Scientific Bulletin 1958;28:1409–38. [Google Scholar]
- 36. Robinson O, Dylus D, Dessimoz C.. Phylo.io: interactive viewing and comparison of large phylogenetic trees on the web. Mol Biol Evol 2016;33(8):2163–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Han MV, Zmasek CM.. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics 2009;10:356.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Waterhouse AM, Procter JB, Martin DM, et al. . Jalview version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009;25(9):1189–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yachdav G, Wilzbach S, Rauscher B, et al. . MSAViewer: interactive JavaScript visualization of multiple sequence alignments. Bioinformatics 2016;32(22):3501–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sievers F, Dineen D, Wilm A, et al. . Making automated multiple alignments of very large numbers of protein sequences. Bioinformatics 2013;29(8):989–95. [DOI] [PubMed] [Google Scholar]
- 41. Kamisetty H, Ovchinnikov S, Baker D.. Assessing the utility of coevolution-based residue-residue contact predictions in a sequence- and structure-rich era. Proc Natl Acad Sci USA 2013;110:15674–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Marks DS, Hopf TA, Sander C.. Protein structure prediction from sequence variation. Nat Biotechnol 2012;30(11):1072–80. [DOI] [PMC free article] [PubMed] [Google Scholar]