Abstract
Small RNAs (sRNAs) are important short-length molecules with regulatory functions essential for plant development and plasticity. High-throughput sequencing of total sRNA populations has revealed that the largest share of sRNA remains uncategorized. To better understand the role of sRNA-mediated cellular regulation, it is necessary to create accurate and comprehensive catalogues of sRNA and their sequence features, a task that currently relies on nontrivial bioinformatic approaches. Although a large number of computational tools have been developed to predict features of sRNA sequences, these tools are mostly dedicated to microRNAs and none integrates the functionalities necessary to describe units from all sRNA pathways thus far discovered in plants. Here, we review the different classes of sRNA found in plants and describe available bioinformatics tools that can help in their detection and categorization.
Keywords: small RNA categorization, sRNA structural features, sRNA function prediction, sRNA sequencing
Introduction
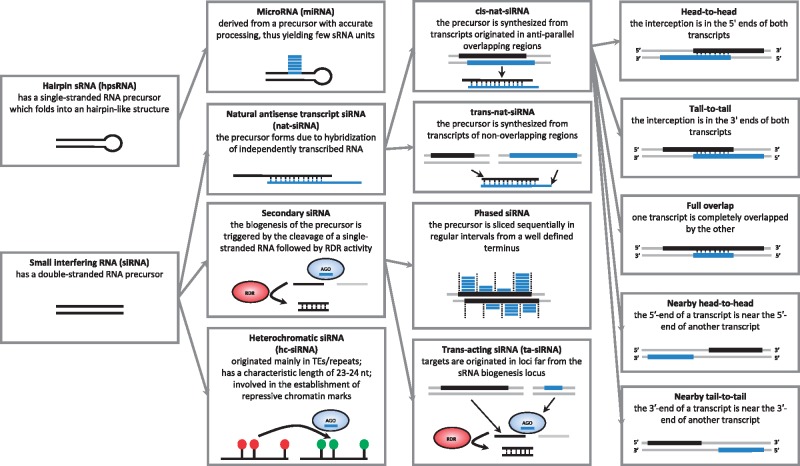
Over the past few years, the scientific community has centered efforts to unravel the complex world of RNA molecules that are not translated into a protein, but that rather have a regulatory function in the cell [1]. Such regulatory RNAs are involved in the control of the concentration of messenger RNA (mRNA) and comprise, among other subclasses, the small RNAs (sRNAs). sRNAs have been shown to have key regulatory functions in development, response to biotic and abiotic stressors, genome stability and transposon control [2]. With the advent of next-generation sequencing of small RNA (sRNA-seq), it has become feasible to survey entire sRNA populations from diverse plant species, cell types, developmental time-points or from different experimental treatments. The identification and classification of sRNA from such high-throughput data is a nontrivial computation task, as plants can produce millions of sRNA from diverse pathways, which are collectively captured in a single sequencing experiment. Accurate sorting of sRNA by class requires categorizing sRNA according to their precursors, structural properties of the mature molecules, as well as functional aspects, such as their potential target sites (Figure 1). A number of computational tools have been developed to detect known sRNA in newly synthesized sequencing libraries, and to help in the identification of novel candidates. For many biologists, a key bottleneck to in silico sRNA analysis is to find software that is tailored to their specific research question and to the data type at hand. In this manuscript, we provide an inventory of various computational tools for the identification and categorization of plant sRNA. We start by describing a simplified classification scheme based on structural and functional sRNA properties, which is adopted in subsequent sections to organize currently available computational tools.
Figure 1.
A stratified classification scheme for sRNA in plants.
sRNAs in plants
Plant sRNA biology has been extensively reviewed elsewhere [3, 4]. Briefly, in plants, sRNAs are mostly 21–24 nucleotides (nt) in length, and result from cleavage of double-stranded RNA substrates by dicer-like (DCL) enzymes. The RNA substrates, themselves, can originate either from a single-stranded RNA precursor with a stem-loop conformation, or from a double helix. If the sRNA originates from a hairpin structure, they are referred to as hairpin-derived sRNA (hpsRNA), and if they originate from a double helix, they are referred to as small interfering RNA (siRNA). The hpsRNA class can be further considered a microRNA (miRNA) if the hairpin is processed in such a way that it produces only one or few functional units. siRNAs comprise all other classes of known sRNA: secondary siRNA such as trans-acting (ta)-siRNA, natural antisense transcript (nat)-siRNA and heterochromatic (hc)-siRNA.
In the case of secondary siRNA, two nonmutually exclusive groups can be defined: phased siRNA, which is originated from a precursor that is processed in a precise and sequential manner; and ta-siRNA, which is a plant specific sRNA type with targets originated in trans.
In the case of nat-siRNA, the precursor double helix is derived from overlapping RNA segments produced independently of each other, while secondary siRNA and hc-siRNA precursors are preceded by the action of a RNA-dependent RNA polymerase (RDR) over single-stranded RNA. Considering the physical distance between NAT producing loci, two main categories emerge: cis-NAT and trans-NAT. cis-NATs are transcribed from the same genomic loci but typically from opposite DNA strands and thus form perfect pairs, while trans-NAT are transcribed from distant genomic locations. Cis-NAT overlapping regions do not have a characteristic length and can occur in five orientations [5]:
Head-to-head: Consists in the interception in the 5′ ends of both transcripts
Tail-to-tail: Comprises the interception in the 3′ ends of both transcripts
Completely overlapping: A transcript on one strand of the genome is overlapped by the entire length of the other transcript on the opposite strand
Nearby head-to-head: Nearby transcripts in a head-to-head manner where the 5′-end of a transcript is near the 5′-end of another transcript in the genome
Nearby tail-to-tail: Nearby transcripts in a tail-to-tail manner where the 3′-end of a transcript is near the 3′-end of another transcript in the genome
To become active in plants, sRNAs must load into Argonaute (AGO) proteins, which guide silencing complexes to their targets according to sequence pairing principles. When associated with AGO, sRNA can regulate genomes at the transcriptional (TS) or posttranscriptional (PTS) level depending on the specific AGO to which the sRNA binds. Both modes of action have been intensively studied, but PTS mechanisms such as mRNA cleavage and translation inhibition are better understood. PTS is typically observed for miRNA, secondary siRNA and nat-siRNA, while TS is often associated with the action of hc-siRNA. Functional siRNA characterization is key to identify hc-siRNA, as no clear structural features to discriminate between hc-siRNA and other siRNA have been defined to date.
Computational approaches for sRNA detection and categorization
Software for sRNA categorization is typically designed to deal with sequences as input. Some software tools identify precursors in segments with a length of dozens or even hundreds of nucleotides, while others focus on short matured fragments of about 20 nt, and there are also platforms that combine information from both forms. A comprehensive overview of tools is given in the following sections, and further detailed in Table 1. Depending on the application, sRNA analysis can be complex, often involving preliminary steps such as data preprocessing and quality controls, as well as downstream analysis such as gene ontology annotation and pathway discovery. A number of recent computational platforms have tried to integrate various software modules by stringing together existing tools into single analysis pipelines [6–12]. The description of modules that do not directly deal with sRNA identification and categorization is outside of the scope of this document and will not be discussed here.
Table 1.
Main features of computational tools for sRNA characterization
| # | Tool | Type |
Focus | Input | Analysis |
Year | Refe- rence | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Local | Web server | Precursor |
Mature |
Target |
||||||||||||||||
| Conser- vation | Structure |
Dicing |
Machine learning | Conser- vation | Isomir detection | Machine learning | PTS |
|||||||||||||
| Hairpin | NAT | Precise excision | Phasing | Compleme- ntarity | Degra- dome | Machine learning | Expre- ssion | |||||||||||||
| 1 | SeqBuster | x | miRNA | SL + TG | x | x | x | x | 2010 | [13] | ||||||||||
| 2 | QuickMIRSeq | x | miRNA | SL + TG | x | x | x | x | 2017 | [14] | ||||||||||
| 3 | IsomiRage | x | miRNA | SL + TG | x | x | x | x | 2014 | [15] | ||||||||||
| 4 | sRNAbench | x | miRNA | SL + TG | x | x | x | x | x | 2014 | [16] | |||||||||
| 5 | isomiRex | x | miRNA | SL + TG | x | x | x | x | 2013 | [17] | ||||||||||
| 6 | isomiRID | x | miRNA | SL + TG | x | x | x | x | 2013 | [18] | ||||||||||
| 7 | RNAFold | x | x | hpsRNA | TG | x | 2003 | [19] | ||||||||||||
| 8 | UNAFold | x | hpsRNA | TG | x | 2003 | [20] | |||||||||||||
| 9 | Mirinho | x | miRNA | SL + TG | x | x | x | 2015 | [21] | |||||||||||
| 10 | miRNAFold | x | x | hpsRNA | TG | x | 2016 | [22] | ||||||||||||
| 11 | MIRFINDER | x | miRNA | SS + TG | x | x | x | 2004 | [23] | |||||||||||
| 12 | MIRcheck | x | miRNA | SS + TG | x | x | x | 2004 | [24] | |||||||||||
| 13 | microHarvester | x | x | miRNA | SS + TG | x | x | x | 2006 | [25] | ||||||||||
| 14 | MiMatcher | x | miRNA | TS + TG | x | x | 2005 | [26] | ||||||||||||
| 15 | mirTour | x | miRNA | SS + TG | x | x | x | 2011 | [27] | |||||||||||
| 16 | C-mii | x | miRNA | SS + TG | x | x | x | x | 2012 | [28] | ||||||||||
| 17 | mirDeepFinder | x | miRNA | SL + TG + DF | x | x | x | x | x | 2012 | [29] | |||||||||
| 18 | PlantMiRNAPred | x | miRNA | TG | x | x | 2011 | [30] | ||||||||||||
| 19 | plantMirP | x | miRNA | TG | x | x | 2016 | [31] | ||||||||||||
| 20 | NOVOMIR | x | miRNA | TG | x | x | 2010 | [32] | ||||||||||||
| 21 | HuntMi | x | miRNA | TG | x | x | 2013 | [33] | ||||||||||||
| 22 | miRNAPrediction | x | miRNA | SS + TG | + | x | 2012 | [34] | ||||||||||||
| 23 | SplamiR | x | miRNA | TG | x | + | x | 2011 | [35] | |||||||||||
| 24 | miPlantPreMat | x | mirRNA | TG | x | x | x | 2014 | [36] | |||||||||||
| 25 | MiRPara | x | miRNA | TG | x | x | 2011 | [37] | ||||||||||||
| 26 | miRDup | x | miRNA | SS + TG | x | x | 2013 | [38] | ||||||||||||
| 27 | miRduplexSVM | x | miRNA | TG | x | x | 2015 | [39] | ||||||||||||
| 28 | MaturePred | x | miRNA | TG | x | x | 2011 | [40] | ||||||||||||
| 29 | miRLocator | x | miRNA | TG | x | x | 2015 | [41] | ||||||||||||
| 30 | ShortStack | x | hpsRNA/ miRNA/ ta-siRNA | SA + TG | x | x | x | x | x | 2013 | [42] | |||||||||
| 31 | mirDeep-P | x | miRNA | SL + TG | x | x | x | 2011 | [43] | |||||||||||
| 32 | miRPlant | x | miRNA | SL + TG | x | x | x | 2014 | [44] | |||||||||||
| 33 | miRA | x | miRNA | SA + TG | x | x | 2015 | [45] | ||||||||||||
| 34 | PIPmiR | x | miRNA | SL + TG | x | x | x | 2011 | [46] | |||||||||||
| 35 | Mir-PREFeR | x | miRNA | SA + TG | x | x | 2014 | [47] | ||||||||||||
| 36 | miRCat2 | x | miRNA | SL + TG | x | x | 2017 | [48] | ||||||||||||
| 37 | miReader | x | miRNA | SL | x | x | 2013 | [49] | ||||||||||||
| 38 | UEA sRNA Workbench | x | miRNA/ta-siRNA | SL + TG + D | x | x | x | x | x | x | x | 2012 | [7] | |||||||
| 39 | pssRNAMiner | x | ta-siRNA | SL + TG + PI | x | x | 2008 | [50] | ||||||||||||
| 40 | Shortran | x | miRNA/ta-siRNA | SL + TG | x | x | x | x | x | 2012 | [51] | |||||||||
| 41 | TasExpAnalysis | x | ta-siRNA | SL + TG + D | x | x | x | x | 2014 | [52] | ||||||||||
| 42 | PhaseTank | x | Secondary/ phased siRNA | SL + TG | x | x | 2015 | [53] | ||||||||||||
| 43 | NASTI-seq/R | x | nat-siRNA | SL | x | 2013 | [54] | |||||||||||||
| 44 | NATpipe | x | nat-siRNA | SL + TG + D | x | x | x | 2016 | [55] | |||||||||||
| 45 | Cleaveland | x | PTS | SS + TG + DF | x | x | 2014 | [56] | ||||||||||||
| 46 | PAREsnip | x | PTS | SS + TG + DF | x | x | 2012 | [57] | ||||||||||||
| 47 | SoMART | x | miRNA/ta-siRNA | SL + TG + DF | x | x | x | x | x | x | x | 2012 | [58] | |||||||
| 48 | SeqTar | x | PTS | SL + TG + DF | x | 2012 | [59] | |||||||||||||
| 49 | miRNA Digger | x | miRNA | SL + TG + DF | x | x | x | x | x | x | 2016 | [60] | ||||||||
| 50 | psRNATarget | x | PTS | SS + TG | x | 2011 | [61] | |||||||||||||
| 51 | TAPIR | x | x | PTS | SS + TG | x | 2010 | [62] | ||||||||||||
| 52 | Targetfinder | x | PTS | SS + TG | x | 2010 | [63] | |||||||||||||
| 53 | Target-align | x | x | PTS | SS + TG | x | 2010 | [64] | ||||||||||||
| 54 | PsRobot | x | x | hpsRNA/miRNA | SS + TG | x | x | x | x | 2012 | [65] | |||||||||
| 55 | p-TAREF | x | x | miRNA | SS + TG | x | x | 2011 | [66] | |||||||||||
| 56 | microRNA- Target | x | miRNA | SS + TG | x | x | 2014 | [67] | ||||||||||||
| 57 | PlantMirnaT | x | miRNA | SS + RL + TG + DF | x | x | x | x | 2015 | [68] | ||||||||||
| 58 | MTide | x | miRNA | SL + TG + DF | x | x | x | x | x | x | 2015 | [69] | ||||||||
| 59 | sPARTA | x | PTS | SS + TS + DF | x | x | 2014 | [70] | ||||||||||||
| 60 | imiRTP | x | miRNA | SS + TS + DF | x | x | 2011 | [71] | ||||||||||||
Notes: Modules for downstream analysis or that are not applicable to plants are not mentioned. DF: degradation fragments; RL: RNA sequencing; SA: sRNA-seq alignments; SL: sRNA-seq library; SS: sRNA sequence; PI: phase initiator sequence; PS: precursor sequence; TR: transcript; TG: transcript or genomic sequence.
Detecting known sRNA
An obvious choice when trying to identify new sRNA candidates is to search for known sequences that have been experimentally confirmed. Owing to their popularity, a large number of databases of experimentally validated miRNA have been built up, comprising several species and kingdoms. miRBase [72] is the most famous miRNA repository, and provides extensive information on precursors, mature sequences and their targets. Unfortunately, similarly detailed databases for other sRNA categories do not currently exist. To our knowledge, there is only one public database (tasiRNAdb) for secondary sRNA in plants, which is strictly dedicated to ta-siRNA [52]. tasiRNAdb provides information not only about mature ta-siRNA but also their precursors and targets in 18 plant species. We are not aware of repositories of mature nat-siRNA or hc-siRNA. Still, there is one databases of NATs in plants: PlantNATsDB [73]. This resource contains a large inventory of precomputed NATs for 70 plant species, but it focuses on genes ignoring extensive intergenic regions.
Sequence aligners such as BLAST [74] are commonly used to query such databases. In fact, the platforms supporting tasiRNAdb and PlantNATsDB implement their own online BLAST modules. Searches for long precursors can be performed using standard BLAST parameters; however, mature sequences pose additional challenges because of their small size. When searching for mature sequences, perfect matches reduce the odds of getting hits by chance when compared with the use of mismatches and gaps. On the other hand, allowing for mismatches and gaps is often necessary when studying close inter-species homologues. While single-nucleotide polymorphisms are a well-known source of genomic variation, mature sRNA can be subject to further modifications. For example, miRNA variants (i.e. ‘isomiRs’) have been identified as a result of inaccurate DCL cleavage, sequence editing events and even nucleotide additions to the mature sRNA [75, 76]. To deal with isomiRs, several tools rely on alignment algorithms and a preprocessing scheme that consists on sequence terminal trimming and nucleotide additions to simulate known sequence modifications. This is the case in applications such as seqBuster [13], QuickMIRSeq [14], IsomiRage [14], sRNAbench [16], isomiRex [17] and isomiRID [18]. As ‘template isomiRs’ are a result of dicing shifts, they can be detected if perfect complementarity between the sRNA candidate, and a known miRNA precursor (or pre-miRNA) is observed. On the other hand, simulating ‘non-template isomiRs’ by creating all possible combinations with 1–3 nt extensions in the 5′ and 3′ ends of known miRNA, and by trimming canonical mature sequences, has been central for their identification. To reduce false positives, some of these tools perform additional processing steps typically exploring features of sRNA-seq libraries. The simplest procedure consists in using read abundance cutoffs as done in isomiRID. seqBuster uses several filters to eliminate sequences with low read abundance and computes z-scores to distinguish true isoforms from sequencing errors. QuickMIRSeq uses multiple samples simultaneously with the rationale that noisy background reads are not captured consistently in multiple samples unlike true miRNA, even if they show low expression levels.
Computer-aided de novo sRNA categorization
Once known mature sRNAs are identified in sRNA-seq data, the remaining sequences (which usually comprise the large majority of the initial sRNA-seq sets) are typically mapped to a genomic reference (if available) to eliminate sequencing chimeras and artifacts. Mature sequences mapping to previously characterized ribosomal RNA (rRNA), transfer RNA (tRNA), small nucleolar RNA (snoRNA) and small nuclear RNA (from databases like for example Rfam [77]) are filtered out by some computational frameworks [29, 57], as these are thought to be mostly non-DCL fragmentation products that have a low chance of entering functional sRNA pathways [57, 78]. Still, doing so can eliminate true sRNA, as tRNA-, rRNA- and snoRNA-derived sRNAs have been identified in multiple plant species [79]. Removing low complexity and low copy reads is also a common practice to reduce noisy data [29, 42], but again, care must be taken in doing so because important sequences can be missed (e.g. hc-siRNAs are typically derived from repetitive regions). If BLAST is a popular mapping tool suitable to work with long precursor sequences, aligners like BWA [80] and bowtie [81] are primary choices when it comes to mapping libraries of short reads to a reference. An accurate mapping is of importance in the sense that meaningful clues for sRNA categorization can be obtained from the sequence and chromatin context of the mapped loci.
The identification and classification of putatively functional sRNA is a challenging computational task. While the majority of computational tools have been tailored to animal data, several of these tools can also be applied to other species, including plants. However, sRNA biology differs considerably between plants and animals, and several plant-specific computational tools have been developed. Existing computational methods can be broadly divided in five main groups: (i) those that explore conservation principles; (ii) those that rely on structural features such as the spatial conformation of the precursor(s); (iii) inspired by machine learning; (iv) rule-based and (v) target-centered. In practice, this distinction can be difficult as most modern tools consist of pipelines involving a mixture of these methods.
Below we discuss computational tools specific to each plant sRNA class. Because sRNA biogenesis and function can be treated separately, emphasis is given to each of these facets in distinct sections.
Identification of hairpin structures and miRNA classification
The root concept underlying hpsRNA and miRNA categorization is the biogenesis from a RNA transcript with capacity to fold into a hairpin-shaped precursor. As hpsRNA are not well understood and given the popularity of miRNA, most hairpin detectors were developed having in mind pre-miRNA. In truth, a large number of tools branded as miRNA detectors are no more than hairpin or pre-miRNA predictors, as they do not even provide a location for the putative mature miRNA inside the pre-miRNA. These tools must therefore be examined carefully to avoid erroneous conclusions [38].
The inference of RNA secondary structure is central to many computational methods designed to detect hairpin structures. Algorithms such as RNAfold [19] and UNAFold [20] explore thermodynamic principles applied to RNA runners and under the premise that the minimal folding free energy index for miRNA precursors is significantly lower than for other products frequently captured during sequencing such as tRNA, rRNA or mRNA [82]. It is important to recall that plant miRNA stem-loops are more heterogeneous when compared with animals (usually they are larger and can contain big bugles); hence, the parameters of the algorithms need to be adjusted according to whether the input data are derived from plant or animal species [83, 84]. Once calibrated, these ab initio methods can predict hairpin structures without additional knowledge.
Traditional RNA folders are computationally intensive, characterized by a cubic time complexity, which is suboptimal for large inputs. Mirinho [21] and miRNAFold [22] are folders with a square time complexity, recently introduced to tackle this problem.
Because homology search is inherently simpler than folding estimation, most software combines conservation principles with RNA secondary structure predictions to decrease processing time, but also to increase accuracy. In the case of MIRFINDER [23], a miRNA reference set from Arabidopsis is used to tune a pipeline similar to the one described above. This tool performs a search for new miRNA by comparing queries against a reference, and explores three principles: (i) the reference miRNA sequence is conserved between the query and the reference species, independently of the rest of the precursor sequence having diverged; (ii) the precursor sequence must be able to form a stem-loop secondary structure; and (iii) for two miRNA orthologs the location on the arm of the stem-loop secondary structures is the same in both species. To fulfill the first condition, a search for mature miRNA is made with BLAST, and the second condition is verified with RNAfold. Other tools that use similar comparative genomics principles include MIRcheck [24], microHarvester [25], MiMatcher [26], miRTour [27], C-mii [28] and miRDeepFinder [29]. For example, miRDeepFinder uses a set of miRNA candidates as queries and searches in a reference for segments with potential to form pre-miRNA. The hit sites are extended (700 nt by default) upstream and downstream to capture and examine precursor candidates with the miRNA located in one arm of the stem at either the 5′ or 3′ end. After a miRNA candidate is identified, miRDeepFinder extracts the complementary miRNA* sequence considering a 3′ overhang of 2 nt characteristic of the miRNA-miRNA* duplex.
In a slightly different approach, machine learning methods have been used to train classifiers capable of distinguishing plant pre-miRNA from other RNA sequences. One argument in favor of these latter approaches is that comparative methods have limited capacity to detect miRNA sequences and precursors with low similarity to the reference set, while machine learning models can capture more general features that overcome this weakness. PlantMiRNAPred [30] and plantMirP [31] are part of this list, both of which were developed using support vector machines (SVMs). PlantMiRNAPred uses a classifier trained with data from several plant species. A set of 68 features extracted from pre-miRNA and optimized using information gain and feature similarity criteria was considered for training the final classifier, including information about the sequence composition, k-mers, secondary structure, energy and thermodynamics-related parameters. Interestingly, the authors compared PlantMiRNAPred with triplet-SVM [85] and microPred [86], two tools following a similar philosophy but developed using human data. Indeed, these two methods show discriminative capacity when applied to plants, but the accuracy of PlantMiRNAPred is considerably higher, illustrating the need for kingdom-specific tools. Other machine learning algorithms have been applied to pre-miRNA detection, including Markov models in NOVOMIR [32], random forest in HuntMi [33] and C5.0 decision trees in miRNAprediction [34]. In a less usual scheme, SplamiR [35] combines software for detecting primary transcripts that undergo splicing events with a machine learning classification system to identify candidate pre-miRNA among generated putative pre-miRNA.
Searches for genomic sequences with the potential to form fold-back stem-loop structures do not yield high-confidence putatively functional miRNA, as many more inverted repeats can be found than the number of miRNA expected for a given organism. For example, in Arabidopsisthaliana, 138 864 inverted repeat structures have been identified [24] but <1000 miRNA confirmed [72]. To increase miRNA detection accuracy, miPlantPreMat [36] and miRPara [37] feed properties of mature miRNA sequences and their precursors to machine learning models. Both combine SVM classifiers in a hierarchical architecture. miPlantPreMat works with classifiers individually trained to recognize mature and precursor sequences, while miRPara explores inter-kingdom differences.
Although the determinants for miRNA location inside a precursor remain poorly understood, efforts have been made to develop computational procedures for their detection. For example, miRDup [38] was designed to infer the precise positions and length of mature miRNA within a candidate pre-miRNA through random forest classifiers that use sequence and structural features. In addition, tools such as MiRduplexSVM [39], MaturePred [40] and miRLocator [41] use classifiers to extract the position of miRNA duplexes from hairpins.
High-confidence miRNA classification requires additional criteria [87]. For example, the precursor must be diced at specific loci producing only one or a reduced number of mature miRNA. When that happens, piles of sRNA accumulate at these genomic positions. To inspect this feature, it is necessary to access the layout of sRNAs along the precursor. This can be directly assessed using the short-read alignment patterns from sRNA-seq data. Such a functionality can be found in most modern tools, including Shortstack [42], mirDeep-P [43], mirPlant [44], miRA [45], PIPmiR [46], miR-PREFeR [47] and miRCat2 [48]. MirDeep-P and mirPlant are extensions of the popular tool mirDeep [88]. While mirDeep was developed for animal applications, mirDeep-P and mirPlant were specifically designed for plant-based sRNA analysis. Following the mapping of sRNA reads to a reference genome with bowtie, mirDeep-P extracts RNA segments to further determine secondary structure and checks if sRNA spatial distribution patterns are compatible with dicer activity. The mature candidates and the respective pre-miRNA are then filtered according to plant-specific criteria based on known properties of plant miRNA genes. A significant difference between mirPlant and mirDeep-P is that in the latter case, the precursor region is determined based on the genomic region overlapping reads, while in miRPlant the precursor region is determined based on the highest expressed read, which is presumably the mature miRNA. The authors of miRPlant argue that this strategy reduces the number of false negatives, as it guarantees that the mature miRNA is located at the end of one arm of the hairpin. In the case of Shortstack, giving the mapped reads and a reference as input, a de novo sRNA cluster discovery is performed by analyzing local patterns of read coverage. Each genomic region overlapping an sRNA cluster is then subjected to a hairpin-folding analysis with RNAfold. Afterward, hairpins are annotated either as hpsRNA or miRNA loci, depending on how strong is the evidence for precise excision given by local sRNA patterns. miRA tries to maximize the flexibility in parameter settings to enable a conservation-independent miRNA analysis; the authors argue that the use of standard parameters for all plant species is suboptimal because of the complex and nonhomogeneous nature of miRNA precursors in plants. In miR-PREFeR, expression information from multiple sRNA-seq libraries can, in addition, be used to decrease false positives and improve the reliability of the predictions.
Another less common solution is miReader [49], which aims at identifying mature miRNA directly from sRNA-seq data, thanks to an embedded algorithm for de novo contig assembly using short reads.
Detection of secondary siRNA and ta-siRNA
In contrast with miRNA-encoding MIR genes, secondary siRNA precursors such as those encoded by ta-siRNA loci or TAS genes lack a specific secondary structure, and thus require alternative computational prediction strategies. The computational identification of new secondary siRNA is strongly focused on the detection of phasing patterns. This kind of analysis requires sRNA-seq data and a genomic reference, and can be executed with tools such as UEA sRNA Workbench [7], ShortStack [42], pssRNAMiner [50] and shortran [51], which implement variants of the method described in [89]. In this approach, sRNA clusters are determined from the mapped reads, and the occurrence of significant phasing patterns inside these regions (Figure 1) is calculated considering a hypergeometric distribution. The sRNA thought to be phase-initiators can also be mapped to the reference to help identify the start and stop coordinates of the precursor, and restrict the inspection of secondary siRNA candidates to clusters inside that region [50]. In the ‘one hit’ initiator case, functional siRNA must be searched in both the 5′ and 3′ direction of the initial cleavage coordinate, as phasing is a bidirectional process. To mimic patterns of DCL slicing, both UEA sRNA Workbench and ShortStack introduce a shift that pushes the start position of the segment located in the opposite strand 2 nt downstream.
The TasExpAnalysis module, available online through the tasiRNAdb [52] platform, combines phasing detection with a search for known TAS and ta-siRNA in user-provided sRNA and sequencing reads from endonucleolytic mRNA cleavage products, also known as degradome. This tool follows a target-centered approach, where after mapping sRNA-seq and degradome reads to a TAS candidate, it checks the consistency between the TAS cleavage position and the 5′ end of the degradome fragments. Next, it searches for an sRNA from the provided library that can fit the role of phase initiator. Statistical tests to detect phasing are then performed using the mapped sRNA and assuming a hypergeometric distribution.
PhaseTank [53] implements a slightly different methodology. After defining phased clusters that contain at least four-phased sRNA in 84 nt regions, a nonstatistical phased score is computed to express the chance of a region to be a producer of phased siRNA. This score depends on patterns of sRNA distribution and abundance in the region. The triggering sRNA is then determined following sequence complementarity principles along with the fact that the cleavage site must occur at positions 9–11 nt of the sRNA from its 5′ terminal.
Some tools like UEA sRNA Workbench do not provide an indication of the phase initiator(s). Using standard tools for PTS target prediction to test diverse sRNA candidates that fall around the initial cleavage site(s) can be a solution. On the other hand, ignoring positional information about the initial cleavage allows a more liberal approach, not restricted to known sRNA but considering potentially unidentified sRNA to be starters of the process [89, 90]. Distinguishing ta-siRNA from other secondary siRNA is done simply by comparing the mapping locations of the siRNA and its target transcript; if in trans, the siRNA is incorporated in the ta-siRNA group.
Finding NAT pairs and nat-siRNA
Genome-wide identification of NAT from multiple organisms is nowadays possible using the large collections of sequencing data freely available online. Annotated genomes have been used in combination with other highly abundant expressed sequences. In silico methods for detecting NAT suffer from several shortcomings depending on the source of sequence information [91]. For example, the use of mRNA can come with information about the orientations of the transcripts, but the amount of mRNA sequence information available can be limited, reflecting specific tissues or development stages [92]. Either way, computational resources and databases dedicated to NATs are scarce.
Current methods for the detection of new NATs are simplistic and based on two main pillars: the sequence complementarity between candidate pairs and the potential for transcripts to hybridize. Although the main criterion for the recognition of NAT pairs is the presence of overlapping transcript clusters, the length of the overlay is a parameter artificially defined and variable from study to study [5]. Other parameters to define NATs have a heuristic basis and lack clear standardization. As an example, in [91], cis-NAT pairs from Arabidopsis were studied using annotated and anchored full-length complementary DNA (cDNA), by applying the following criteria: (i) cDNAs of both transcripts can be uniquely mapped to the genome with at least 96% sequence identity; (ii) the two transcripts are derived from overlapping loci but opposite strands; (iii) the size of the overlaid fragment must be longer than 50 nucleotides; and (iv) the sense and antisense transcripts must have distinct splicing patterns. Other studies implemented comparable but slightly altered approaches for rice [93, 94] and Arabidopsis [95].
The NASTI-seq R package [54] is one of the few computational tools currently available for NAT discovery. This software is specialized in cis-NAT detection using strand-specific RNA sequencing data. It models the probability of finding read enrichment in each strand using a binomial model and identifies cis-NAT conditional on additional spatial criteria such as the location in opposite strands and the proximity in the genome.
To our knowledge, the only tool implementing an engine for generic NAT search (including trans-NAT) is NATpipe [55]. This is a pipeline for NAT prediction for organisms without a reference genome. Using transcriptomic data, it performs a BLAST-based search to preselect NAT pair candidates. Then, the annealing potential of these candidates is explored by RNAplex [96]. The secondary structure is analyzed and instances containing bubbles in the annealed region comprising >10% of its length are rejected. If sRNA-seq data are available, NATpipe can perform a search for prospective nat-siRNA by looking to phasing patterns in the annealed region in a similar way to what is done by tools for ta-siRNA detection. To distinguish trans-nat-siRNA from cis-nat-siRNA, it is necessary to keep in mind that the concept of trans implies transcripts not sharing a common genomic location.
Detecting hc-siRNA and the respective generating loci
In plants, hc-siRNAs are typically 24 nt long and mostly derive from transposons, repeats and heterochromatic regions. Their biogenesis is primarily connected with the activity of PolIV-RDR2-DCL3 [99, 101]. hc-siRNAs are central for RNA-directed DNA methylation (RdDM), which is the pathway responsible for de novo DNA methylation. A hallmark of RdDM is the presence of cytosine methylation in all DNA sequence contexts (CG, CHG and CHH, where H can be C, A or T) [99, 103]. Some transposon families can switch the production of siRNA from 24 to 21–22 nt when methylation is lost [97, 98]. This transition starts with the synthesis of transcripts by Pol II that are afterward degraded into 21–22 nt siRNA. Some of these siRNA can enter a noncanonical RdDM pathway dependent on PolII, RDR6 and DCL2/4 [98].
To date, there are no public tools specifically developed to detect hc-siRNA. This limitation is in part because of the fact that hc-siRNA biology remains unclear, and experimental tests to functionally validate hc-siRNA are difficult to establish. Although certain families of TEs have been described to produce hc-siRNAs when epigenetic marks are changed [97, 98], the reason for their involvement in hc-siRNA biogenesis remains poorly understood. So far, the identification of hc-siRNA has focused mostly on the abundance of 24 nt sRNA mapping to differentially methylated regions that arise in RdDM mutants [97–103], the correlation with the presence or absence of histone marks [104] and variations in gene expression. However, numerous epigenetically activated sRNAs have been identified, which seem to lack the functional properties to be included in the hc-siRNA category. Rather they show PTS activity [97, 105], but the structural features that separate these from true hc-siRNA are unclear. Although the length of mature sRNA sequences is somewhat predictive, and has been taken as a way to discriminate putative hc-siRNA from other types of siRNA (hc-siRNAs are typically 24 nt in length), this feature—by itself—can be misleading. For example, miR163 is an example of a 24 nt long miRNA, which has an exceptionally long length for a DCL1-dependent sequence, and that despite its length primarily binds to AGO1 exerting posttranscriptional regulation [100].
Function prediction in plants
Established nomenclature for miRNA annotation [78] does not require the identification of a functional target sequence, as target prediction can be notoriously difficult. Moreover, target sequences are not steady entities, but can arise de novo, or be lost through mutational events over evolutionary time.
However, the structural features that distinguish other (non-miRNA) sRNA classes remain obscure and can often only be clearly delineated based on knowledge of their target sequences. For example, sorting sRNA by length and matching them to heterochromatic regions, TEs or repeats are naive approaches often used to identify hc-siRNA, but these approaches are insufficient to discriminate hc-siRNA from epigenetically activated siRNA involved in PTS [98, 105]. Hence, knowing the mode of action of a given sRNA sequence would appear to be a fundamental aspect of sRNA classification.
Methods for PTS target prediction
Posttranscriptional regulation in plants can occur in two ways: target cleavage [106] and translational repression [107]. A negative correlation between sRNA expression levels and those of the target transcript is usually taken as evidence for target cleavage. Alternatively, translational repression can happen after binding of the sRNA-AGO complex to the 5′ untranslated region or the open reading frame of target RNA, which inhibits the recruitment or movement of ribosomes through the mRNA [108].
Targeting of plant mRNA follows rules that are significantly different from those in animals and therefore, tools developed for the animal kingdom are suboptimal for plants. Studies with miRNA have revealed a number of key differences: in animals, a seed region of around 8 nt demands near-perfect sRNA/mRNA complementarity, while in plants this complementarity must be preserved throughout the complete miRNA; in animals, miRNAs have a positional preference for the 3′-UTR of the target, while in plants this is not observed [83, 84].
Target cleavage can be identified through the analysis of degradation fragments captured by sequencing (i.e. the degradome). The underlying idea is to use experimental evidence given by the degradome to discriminate between random degradation products and RNA segments precisely targeted by AGO proteins. Methods such as Cleaveland [56], PAREsnip [57] SoMART [58], SeqTar [59] and miRNA Digger [60], jointly explore degradome data, sRNA and other transcripts to detect PTS-sRNA and their targets [57, 56]. Taking miRNA Digger as an illustrative example: miRNA Digger starts by scanning the degradome for potential cleavage sites after mapping the degradation segments to a genomic reference. The mapping loci are then tested for the presence of RNA with hairpin-folding capacity. With sRNA-seq data available, it then looks for marks of miRNA-miRNA* duplexes, plus AGO-enriched miRNA(*)s, in case such the libraries are provided.
Other prediction-based algorithms such as psRNATarget [61] and TAPIR [62] only require candidate sRNA-target pair as input. The analysis is typically performed in two main steps: (i) search for the best sRNA/mRNA complementarity location in the target candidate and (ii) measure target accessibility. The strength of these parameters is in some cases used to discriminate between translational and posttranscriptional inhibition. For example, in psRNATarget, a modified version of the Smith–Waterman algorithm [109] is used to look for optimal sRNA/mRNA alignments, and the UPE score (which is the energy required to ‘open’ secondary structure around target site on mRNA) is determined with RNAup [110]. In cases where mismatches are detected in the central complementary region of the sRNA sequence, the software assumes that the sRNA is likely involved in protein translational inhibition rather than in mRNA cleavage, as cleavage activity is known to be reduced when sRNA-mRNA complementarity is poor. psRNATarget is available via a Web portal, working with an efficient computing back-end pipeline that parallelizes processing on a Linux cluster. TAPIR is another popular tool that follows similar principles used in psRNATarget. It allows a fast search using FASTA and for more precise results uses RNAhybrid [111]. Targetfinder [63] and Target-align [64] are counterparts that fall in the same category. PsRobot [65] is an interesting example on how to take advantage of the large amount of deep sequencing data currently available in a meta-analysis. Its core includes a modified Smith–Waterman algorithm and a simple scoring methodology to search for candidate targets. The user is offered extra information about the predicted targets, such as their conservation across species, degradome profiles and target expression in diverse sRNA-related mutants, something that can help to judge the reliability of the predictions.
Machine learning principles have also been used to predict PTS targets: p-TAREF [66] explores dinucleotide variation around the sRNA-target sites using support vector regression, and microRNA-Target [67] implements a PCA-SVM classifier that uses multiple sequence, structure and thermodynamic features to characterize miRNA–target interaction.
More recently, a new breed of tools that include PlantMirnaT [68] explore deep sequencing miRNA and mRNA expression profiles to identify condition-specific miRNA-mRNA target pairs.
Unfortunately, the methods developed to date for PTS target prediction still suffer from relatively high false-positive rates [64, 112] and inconsistent results across platforms are common. This has spurned the development of pipelines that integrate several of these algorithms to obtain consensus predictions. For example, Mtide [69] combines degradome analysis by Cleveland, target prediction by TAPIR and miRNA prediction using a plant-adapted version of miRDeep2 [113] with a set of rules to determine miRNA–target interactions. Other platforms that combine multiple software packages to perform a target-centered analysis include sPARTA [70] and imiRTP [71].
We argue that PTS target prediction could be further improved by considering additional biological criteria, such as the capacity of sRNA to load into specific classes of AGO proteins that are known to be required for PTS. Sequence features of sRNA that predict AGO loading have been recently obtained from machine learning approaches applied to AGO-IP sRNA-seq libraries [114]. This information could compliment the abovementioned computational tools for PTS target prediction.
Methods for TS target prediction
There is currently no computational tool in the public domain to predict transcriptional silencing targets from genomic data. This kind of inference is still in an early stage of development and is typically done based on indirect observations and assumptions about DNA/chromatin properties. For example, the presence/absence of methylation in CHH sequence context, the correlation with the abundance of 24 nt sRNA mapping in the vicinity of these marks and variations in the concentration of mRNA from the candidate target are used as proxies to predict RdDM targets [97, 101, 102, 105].
Future perspectives
The biology of sRNA is complex and poses numerous computational challenges. The computational categorization of sRNA is far from being solved. sRNA prediction based on sequencing data is either inaccurate or lacks dedicated tools altogether. Although less attention has been paid to plants than to animals, algorithms for predicting various aspects of sRNA biogenesis and function in plants can be found dispersed over the internet. These are mostly individual modules, making sRNA cataloging a hard assignment for nonspecialists. Future work should focus on incorporating existing tools into a unifying framework. This would aid in the automation of sRNA analysis, and shift focus away from the assembly of pipelines to their applications.
Currently, the majority of tools focus on miRNA, although hc-siRNAs are by far the most numerous. This bias is most likely because of the fact that miRNA have well-defined structural features in comparison with other sRNA categories. In addition, miRNAs are easily validated experimentally, which helps in calibrating computational algorithms for miRNA detection and prediction. The investment in proper software should coevolve with experimental procedure for acquiring sRNA data. This development is necessary to be able to maximize the knowledge that can be extracted from such data.
Key Points.
Characterizing sRNA in terms of their biogenesis and function is essential for understanding regulatory mechanisms underlying plant development and adaptation.
Deep sequencing data of total sRNA indicate that a large fraction of sRNA sequences remains to be catalogued.
Numerous computational algorithms have been developed to facilitate the detection and categorization of plant sRNA, but existing software is mostly dedicated to miRNA.
By adapting existing software in combination with public data sources, it is possible to craft more accurate and automated in silico tools applicable to a wider spectrum of sRNA classes.
Biographies
Lionel Morgado is a PhD candidate at the Groningen Bioinformatics Centre (University of Groningen, The Netherlands). The focus of his research is the development of high-throughput computational methods to study small RNAs in plants.
Frank Johannes is an assistant professor for population epigenetics and epigenomics at the Technical University of Munich. He combines bioinformatic and statistical genetic approaches with high-throughput molecular data to characterize patterns of epigenetic variation in populations of plants. The goal is to understand how this variation arises, how stable it is across generations and to what extent it determines agriculturally and evolutionarily relevant plant traits. www.johanneslab.org.
Funding
Lionel Morgado acknowledges support from the University of Groningen. Frank Johannes acknowledges support from the Technical University of Munich-Institute for Advanced Study funded by the German Excellence Initiative and the European Union Seventh Framework Programme under grant agreement #291763.
References
- 1. Bernstein BE, Birney E, Dunham I, et al. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mirouze M. The small RNA-based odyssey of epigenetic information in plants: from cells to species. DNA Cell Biol 2012;12:1650–6. doi:10.1089/dna.2012.1681. [DOI] [PubMed] [Google Scholar]
- 3. Axtell MJ. Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 2013;64:137–59. [DOI] [PubMed] [Google Scholar]
- 4. Borges F, Martienssen RA.. The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol 2015;16:727–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Osato N, Suzuki Y, Ikeo K, et al. Transcriptional interferences in cis natural antisense transcripts of humans and mice. Genetics 2007;176(2):1299–306. doi:10.1534/genetics.106.069484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rueda A, Barturen G, Lebrón R, et al. sRNAtoolbox: an integrated collection of small RNA research tools. Nucl Acids Res 2015;43:W467–73. doi:10.1093/nar/gkv555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stocks MB, Moxon S, Mapleson D, et al. The UEA sRNA workbench: a suite of tools for analysing and visualizing next generation sequencing microRNA and small RNA datasets. Bioinformatics 2012;28:2059–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Müller S, Rycak L, Winter P, et al. omiRas: a Web server for differential expression analysis of miRNAs derived from small RNA-Seq data. Bioinformatics 2013;29(20):2651–2. [DOI] [PubMed] [Google Scholar]
- 9. Patra D, Fasold M, Lagenberger D, et al. plantDARIO: web based quantitative and qualitative analysis of small RNA-seq data in plants. Front Plant Sci 2014;5:708.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen CJ, Servant N, Toedling J, et al. ncPRO-seq: a tool for annotation and profiling of ncRNAs in sRNA-seq data. Bioinformatics 2012;28:3147–9. [DOI] [PubMed] [Google Scholar]
- 11. Icay K, Chen P, Cervera A, et al. SePIA: RNA and small RNA sequence processing, integration, and analysis. BioData Min 2016;9(1):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wan C, Gao J, Ban R, et al. CPSS 2.0: a computational platform update for the analysis of small RNA sequencing data. Bioinformatics 2017, in press. doi:10.1093/bioinformatics/btx066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pantano L, Estivill X, Marti E.. SeqBuster, a bioinformatic tool for the processing and analysis of small RNAs datasets, reveals ubiquitous miRNA modifications in human embryonic cells. Nucl Acids Res 2010;38:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhao S, Gordon W, Du S, et al. QuickMIRSeq: a pipeline for quick and accurate quantification of both known miRNAs and isomiRs by jointly processing multiple samples from microRNA sequencing. BMC Bioinformatics 2017;18:180.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Muller H, Marzi MJ, Nicassio F.. IsomiRage: from functional classification to differential expression of miRNA isoforms. Front Bioeng Biotechnol 2014;2:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barturen G, Rueda A, Hamberg M, et al. sRNAbench: profiling of small RNAs and its sequence variants in single or multi-species high-throughput experiments. Methods Next Gen Seq 2014;43:W467–73. [Google Scholar]
- 17. Sablok G, Milev I, Minkov G, et al. isomiRex: web-based identification of microRNAs, isomiR variations and differential expression using next-generation sequencing datasets. FEBS Lett 2013;587:2629–34. [DOI] [PubMed] [Google Scholar]
- 18. De Oliveira LF, Christoff AP, Margis R.. isomiRID: a framework to identify microRNA isoforms. Bioinformatics 2013;29:2521–3. [DOI] [PubMed] [Google Scholar]
- 19. Hofacker IL. Vienna RNA secondary structure server. Nucl Acids Res 2003;31:3429–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Markham NR, Zuker M.. UNAFold: software for nucleic acid folding and hybridization. Methods Mol Biol 2008;453:3–31. [DOI] [PubMed] [Google Scholar]
- 21. Higashi S, Fournier C, Gautier C, et al. Mirinho: an efficient and general plant and animal pre-miRNA predictor for genomic and deep sequencing data. BMC Bioinformatics 2015;16:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tav C, Tempel S, Poligny L, et al. miRNAFold: a Web server for fast miRNA precursor prediction in genomes. Nucleic Acids Res 2016;44(W1):W181–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bonnet E, Wuyts J, Rouze P, et al. Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc Natl Acad Sci USA 2004;101:11511–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jones-Rhoades MW, Bartel DP.. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 2004;14(6):787–99. [DOI] [PubMed] [Google Scholar]
- 25. Dezulian T, Remmert M, Palatnik JF, et al. Identification of plant microRNA homologs. Bioinformatics 2006;22:359–60. [DOI] [PubMed] [Google Scholar]
- 26. Lindow M, Krogh A.. Computational evidence for hundreds of non-conserved plant microRNAs. BMC Genomics 2005;6:119–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Milev I, Yahubyan G, Minkov I, et al. miRTour: plant miRNA and target prediction tool. Bioinformation 2011;6:248–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Numnark S, Mhuantong W, Ingsriswang S, et al. C-mii: a tool for plant miRNA and target identification. BMC Genomics 2012;13:S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xie F, Xiao P, Chen D, et al. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol Biol 2012;80:75–84. [DOI] [PubMed] [Google Scholar]
- 30. Xuan P, Guo M, Liu X, et al. PlantMiRNAPred: efficient classification of real and pseudo plant pre-miRNAs. Bioinformatics 2011;27:1368–76. [DOI] [PubMed] [Google Scholar]
- 31. Yao Y, Ma C, Deng H, et al. plantMirP: an efficient computational program for the prediction of plant pre-miRNA by incorporating knowledge-based energy features. Mol Biosyst 2016;12:3124–31. [DOI] [PubMed] [Google Scholar]
- 32. Teune JH, Steger G.. NOVOMIR: de novo prediction of MicroRNA-coding regions in a single plant-genome. J Nucleic Acids 2010;2010:1–10. doi:10.4061/2010/495904 pmid:20871826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gudys A, Szczesniak M, Sikora M, et al. HuntMi: an efficient and taxon-specific approach in pre-miRNA identification. BMC Bioinformatics 2013;14:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Williams PH, Eyles R, Weiller G.. Plant microRNA prediction by supervised machine learning using C5.0 decision trees. J Nucleic Acids 2012;2012:652979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thieme CJ, Gramzow L, Lobbes D, et al. SplamiR–prediction of spliced miRNAs in plants. Bioinformatics 2011;27:1215–23. [DOI] [PubMed] [Google Scholar]
- 36. Meng J, Liu D, Sun C, et al. Prediction of plant pre-microRNAs and their microRNAs in genome-scale sequences using structure-sequence features and support vector machine. BMC Bioinformatics 2014;15:6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu Y, Wei B, Liu H, et al. MiRPara: a SVM-based software tool for prediction of most probable microRNA coding regions in genome scale sequences. BMC Bioinformatics 2011;12:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Leclercq M, Diallo AB, Blanchette M.. Computational prediction of the localization of microRNAs within their pre-miRNA. Nucleic Acids Res 2013;41:7200–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Karathanasis N, Tsamardinos I, Poirazi P.. MiRduplexSVM: a high-performing miRNA-duplex prediction and evaluation methodology. PLoS One 2015;10:e0126151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xuan P, Guo M, Huang Y, et al. MaturePred: efficient identification of MicroRNAs within novel plant pre-miRNAs. PLoS One 2011;6:e27422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cui H, Zhai J, Ma C.. MiRLocator: machine learning-based prediction of mature MicroRNAs within plant pre-miRNA sequences. PLoS One 2015;10:e0142753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Axtell MJ. ShortStack: comprehensive annotation and quantification of small RNA genes. RNA 2013;19:740–51. doi: 10.1261/rna.035279.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang X, Li L.. miRDeep-P: a computational tool for analyzing the microRNA transcriptome in plants. Bioinformatics 2011;27(18):2614–15. doi:10.1093/bioinformatics/btr430. [DOI] [PubMed] [Google Scholar]
- 44. An J, Lai J, Sajjanhar A, et al. miRPlant: an integrated tool for identification of plant miRNA from RNA sequencing data. BMC Bioinformatics 2014;15:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Evers M, Huttner M, Dueck A, et al. miRA: adaptable novel miRNA identification in plants using small RNA sequencing data. BMC Bioinformatics 2015;16:370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Breakfield NW, Corcoran DL, Petricka JJ, et al. High-resolution experimental and computational profiling of tissue-specific known and novel miRNAs in Arabidopsis. Genome Res 2012;22(1):163–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lei J, Sun Y.. miR-PREFeR: an accurate, fast and easy-to-use plant miRNA prediction tool using small RNA-Seq data. Bioinformatics 2014;30:2837–9. [DOI] [PubMed] [Google Scholar]
- 48. Paicu C, Mohorianu I, Stocks MB, et al. miRCat2: accurate prediction of plant and animal microRNAs from next-generation sequencing datasets. Bioinformatics 2017;33:2446–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ashwani J, Shankar R.. miReader: discovering novel miRNAs in species without sequenced genome. PLoS One 2013;8(6):e66857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dai X, Zhao PX.. pssRNAMiner: a plant short small RNA regulatory cascade analysis server. Nucl Acids Res 2008;36(2):W114–18. doi:10.1093/nar/gkn297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gupta V, Markmann K, Pedersen CN, et al. Shortran: a pipeline for small RNA-seq data analysis. Bioinformatics 2012;28:2698–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang C, Li G, Zhu S, et al. tasiRNAdb: a database of ta-siRNA regulatory pathways. Bioinformatics 2014;30(7):1045–6. doi: 10.1093/bioinformatics/btt746. [DOI] [PubMed] [Google Scholar]
- 53. Guo Q, Qu X, Weibo J.. PhaseTank: genome-wide computational identification of phasiRNAs and their regulatory cascades. Bioinformatics 2015;31:284–6. doi:10.1093/bioinformatics/btu628. pmid:25246430. [DOI] [PubMed] [Google Scholar]
- 54. Li S, Liberman L, Mukherjee N, et al. Integrated detection of natural antisense transcripts using strand-specific RNA sequencing data. Genome Res 2013;23:1730–9. doi:10.1101/gr.149310.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yu D, Meng Y, Zuo Z, et al. NATpipe: an integrative pipeline for systematical discovery of natural antisense transcripts (NATs) and phase-distributed nat-siRNAs from de novo assembled transcriptomes. Sci Rep 2016;6:21666 10.1038/srep21666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Brousse C, Liu Q, Beuclair L, et al. A non-canonical plant microRNA target site. Nucleic Acids Rese 2014;42(8):5270–9. doi:10.1093/nar/gku157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Folkes L, Moxon S, Woolfenden HC, et al. PAREsnip: a tool for rapid genome-wide discovery of small RNA/target interactions evidenced through degradome sequencing. Nucl Acids Res 2012;40(13):e103. doi:10.1093/nar/gks277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li F, Orban R, Baker B.. SoMART: a webserver for plant miRNA, tasiRNA and target gene analysis. Plant J 2012;70:891–901. [DOI] [PubMed] [Google Scholar]
- 59. Zheng Y, Li YF, Sunkar R, et al. SeqTar: an effective method for identifying microRNA guided cleavage sites from degradome of polyadenylated transcripts in plants. Nucl Acids Res 2012;40:e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yu L, Shao C, Ye X, et al. miRNA Digger: a comprehensive pipeline for genome-wide novel miRNA mining. Sci Rep 2016;6:18901. doi:10.1038/srep18901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dai X, Zhao PX.. psRNATarget: a plant small RNA target analysis server. Nucl Acids Res 2011;39:W155–9. doi:10.1093/nar/GKR319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bonnet E, He Y, Billiau K, et al. TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics 2010;26:1566–8. [DOI] [PubMed] [Google Scholar]
- 63. Fahlgren N, Carrington JC.. miRNA target prediction in plants. Methods Mol Biol 2010;592:51–7. [DOI] [PubMed] [Google Scholar]
- 64. Xie F, Zhang B.. Target-align: a tool for plant microRNA target identification. Bioinformatics 2010;26:3002–3. [DOI] [PubMed] [Google Scholar]
- 65. Wu HJ, Ma YK, Chen T, et al. PsRobot: a web-based plant small RNA meta-analysis toolbox. Nucl Acids Res 2012;40:W22–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jha A, Shankar R.. Employing machine learning for reliable miRNA target identification in plants. BMC Genomics 2011;12(1):636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Meng J, Shi L, Luan Y.. Plant microRNA-target interaction identification model based on the integration of prediction tools and support vector machine. PLoS One 2014;9(7):e103181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rhee S, Chae H, Kim S.. PlantMirnaT: miRNA and mRNA integrated analysis fully utilizing characteristics of plant sequencing data. Methods 2015;83:80–7. [DOI] [PubMed] [Google Scholar]
- 69. Zhang Z, Jiang L, Wang J, et al. MTide: an integrated tool for the identification of miRNA–target interaction in plants. Bioinformatics 2015;31(2):290–1. doi:10.1093/bioinformatics/btu633. [DOI] [PubMed] [Google Scholar]
- 70. Kakrana A, Hammond R, Patel P, et al. sPARTA: a parallelized pipeline for integrated analysis of plant miRNA and cleaved mRNA data sets, including new miRNA target-identification software. Nucleic Acids Res 2015;42:e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ding J, Yu S, Ohler U, et al. imiRTP: an integrated method to identifying miRNA-target interactions in Arabidopsis thaliana In: IEEE International Conference on Bioinformatics and Biomedicine. 2011, Atlanta, GA, USA: IEEE, pp. 100–4.
- 72. Kozomara A, Griffiths-Jones S.. miRBase: integrating microRNA annotation and deep-sequencing data. Nucl Acids Res 2001;39(1):D152–7. doi:10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chen D, Yuan C, Zhang J, et al. PlantNATsDB: a comprehensive database of plant natural antisense transcripts. Nucl Acids Res 2012;40(1):D1187–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Altschul S, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol 1990;215(3):403–10. doi:10.1016/S0022- 2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 75. Iida K, Jin H, Zhu JK.. Bioinformatics analysis suggests base modification of tRNA and miRNA in Arabidopsis thaliana. BMC Genomics 2009;10:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ebhardt HA, Tsang HH, Dai DC, et al. Meta-analysis of small RNA-sequencing errors reveals ubiquitous post-transcriptional RNA modifications. Nucl Acids Res 2009;37:2461–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gardner PP, Daub J, Tate JG, et al. Rfam: updates to the RNA families database. Nucl Acids Res 2009;37:D136–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Meyers BC, Axtell MJ, Bartel B, et al. Criteria for annotation of plant microRNAs. Plant Cell 2008;20:3186–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang Y, Li H, Sun Q, et al. Characterization of small RNAs derived from tRNAs, rRNAs and snoRNAs and their response to heat stress in wheat seedlings. PLoS One 2016;11:e0150933.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Li H, Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Langmead B, Trapnell C, Pop M, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009;10(3):R25. doi:10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zhang B, Pan X, Cannon CH, et al. Conservation and divergence of plant microRNA genes. Plant J 2006;46(2):243–59. [DOI] [PubMed] [Google Scholar]
- 83. Mendes ND, Freitas AT, Sagot MF.. Current tools for the identification of miRNA genes and their targets. Nucl Acids Res 2009;37:2419–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Gomes CPC, Cho JH, Hood L, et al. A review of computational tools in microRNA discovery. Front Genet 2013;4:81. doi: 10.3389/fgene.2013.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Xue C, Li F, He T, et al. Classification of real and pseudo microRNA precursors using local structure-sequence features and support vector machine. BMC Bioinformatics 2005;6:310–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Batuwita R, Palade V.. MicroPred: effective classification of pre-miRNAs for human miRNA gene prediction. Bioinformatics 2009;25:989–95. [DOI] [PubMed] [Google Scholar]
- 87. Kozomara A, Griffiths-Jones S.. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 2014;42:D68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Friedlander MR, Chen W, Adamidi C, et al. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol 2008;26:407–15. [DOI] [PubMed] [Google Scholar]
- 89. Chen HM, Li YH, Wu SH.. Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc Natl Acad Sci USA 2007;104(9):3318–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Moxon S, Schwach F, Dalmay T, et al. A toolkit for analyzing large-scale plant small RNA datasets. Bioinformatics 2008;24(19):2252–3. doi:10.1093/bioinformatics/btn428. [DOI] [PubMed] [Google Scholar]
- 91. Wang XJ, Gaasterland T, Chua NH.. Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol 2005;6:R30. doi:10.1186/gb-2005-6-4-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lavorgna G, Dahary D, Lehner B, et al. In search of antisense. Trends Biochem Sci 2004;29(2):88–94. [DOI] [PubMed] [Google Scholar]
- 93. Osato N, Yamada H, Satoh K, et al. Antisense transcripts with rice full-length cDNAs. Genome Biol 2003;5:R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zhou X, Sunkar R, Jin H, et al. Genome-wide identification and analysis of small RNAs originated from natural antisense transcripts in Oryza sativa. Genome Res 2009;19(1):70–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Jen CH, Michalopoulos I, Westhead DR, et al. Natural antisense transcripts with coding capacity in Arabidopsis may have a regulatory role that is not linked to double-stranded RNA degradation. Genome Biol 2005;6:R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Tafer H, Hofacker IL.. RNAplex: a fast tool for RNA-RNA interaction search. Bioinformatics 2008;24(22):2657–63. doi: 10.1093/bioinformatics/btn193. [DOI] [PubMed] [Google Scholar]
- 97. McCue A, Nuthikattu S, Reeder SH, et al. Gene expression and stress response mediated by the epigenetic regulation of a transposable element small RNA. PLoS Genet 2012;8:e1002474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Nuthikattu S, McCue A, Panda K, et al. The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21-22 nucleotide small interfering RNAs. Plant Physiol 2013;162:116–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Stroud H, Greenberg MVC, Feng S, et al. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 2013;152:352–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Wu L, Zhou H, Zhang Q, et al. DNA methylation mediated by a microRNA pathway. Mol Cell 2010;38:465–75. [DOI] [PubMed] [Google Scholar]
- 101. Mari-Ordonez A, Marchais A, Etcheverry M, et al. Reconstructing de novo silencing of an active plant retrotransposon. Nat Genet 2013;45:1029–39. [DOI] [PubMed] [Google Scholar]
- 102. Zhang Q, Wang D, Lang Z, et al. Methylation interactions in Arabidopsis hybrids require RNA-directed DNA methylation and are influenced by genetic variation. Proc Natl Acad Sci USA 2016;113(29):E4248–56. doi:10.1073/pnas.1607851113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Lister R, O’Malley RC, Tonti-Filippini J, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 2008;133:523–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Li X, Wang X, He K, et al. High-resolution mapping of epigenetic modifications of the rice genome uncovers interplay between DNA methylation, histone methylation, and gene expression. Plant Cell 2008;20:259–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Creasey KM, Zhai J, Borges F, et al. MiRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature 2014;508:411–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Llave C, Kasschau KD, Rector MA, et al. Endogenous and silencing-associated small RNAs in plants. Plant Cell 2002;14:1605–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, et al. Widespread translational inhibition by plant miRNAs and siRNAs. Science 2008;320(5880):1185–90. [DOI] [PubMed] [Google Scholar]
- 108. Iwakawa HO, Tomari Y.. Molecular insights into microRNA-mediated translational repression in plants. Mol Cell 2013;52:591–601. [DOI] [PubMed] [Google Scholar]
- 109. Smith TF, Waterman MS.. Identification of common molecular subsequences. J Mol Biol 1981;147:195–7. [DOI] [PubMed] [Google Scholar]
- 110. Lorenz R, Bernhart SH, Hoener zu Siederdissen C, et al. ViennaRNA package 2.0. Algorithms Mol Biol 2011;6:26. doi:10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Kruger J, Rehmsmeier M.. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucl Acids Res 2006;34:W451–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Srivastava PK, Moturu TR, Pandey P, et al. A comparison of performance of plant miRNA target prediction tools and the characterization of features for genome-wide target prediction. BMC Genomics 2014;15:348. doi:10.1186/1471-2164-15-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Friedlander MR, Mackowiak SD, Li N, et al. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucl Acids Res 2012;40:37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Morgado L, Jansen RC, Johannes F.. Learning sequence patterns of AGO-sRNA affinity from high-throughput sequencing libraries to improve in silico functional small RNA detection and classification in plants. bioRxiv 2107:173575. doi: 10.1101/173575 [DOI] [Google Scholar]