Abstract
While novel technologies such as high-throughput screening have advanced together with significant investment by pharmaceutical companies during the past decades, the success rate for drug development has not yet been improved prompting researchers looking for new strategies of drug discovery. Drug repositioning is a potential approach to solve this dilemma. However, experimental identification and validation of potential drug targets encoded by the human genome is both costly and time-consuming. Therefore, effective computational approaches have been proposed to facilitate drug repositioning, which have proved to be successful in drug discovery. Doubtlessly, the availability of open-accessible data from basic chemical biology research and the success of human genome sequencing are crucial to develop effective in silico drug repositioning methods allowing the identification of potential targets for existing drugs. In this work, we review several chemogenomic data-driven computational algorithms with source codes publicly accessible for predicting drug–target interactions (DTIs). We organize these algorithms by model properties and model evolutionary relationships. We re-implemented five representative algorithms in R programming language, and compared these algorithms by means of mean percentile ranking, a new recall-based evaluation metric in the DTI prediction research field. We anticipate that this review will be objective and helpful to researchers who would like to further improve existing algorithms or need to choose appropriate algorithms to infer potential DTIs in the projects. The source codes for DTI predictions are available at: https://github.com/minghao2016/chemogenomicAlg4DTIpred.
Keywords: drug–target interaction, in silico drug repositioning, drug discovery, chemogenomic data, mean percentile ranking, open-source code
Introduction
Drug development is a complex and expensive process. Over the past decades, despite technological advances in drug discovery and increase of investments in pharmaceutical research and development, the number of new drug approvals has remained stagnant [1]. The most significant causes of drug failures are toxicity and a lack of efficacy [2]. Thus, there is an urgent need to develop effective drugs to overcome these limitations [3]. Drug repositioning, the process of finding new uses outside the scope of the original medical indications for existing drugs [4], is considered to be a promising strategy with the benefit of providing a rapid route to clinic than through the traditional drug discovery approaches because of the use of existing knowledge about drugs [5]. The new indication-driven discovery by using repositioning methods has already yielded several successes. For example, HIV protease inhibitors such as nelfinavir can be used as a new class of anticancer drugs [6]. Sunitinib, originally developed for treating renal cell carcinoma, was found to be effective for patients with pancreatic neuroendocrine tumors [7]. Imatinib, developed originally for chronic myeloid leukemia, has shown clinical benefits to the treatment of gastrointestinal stromal tumor [8]. Some other successful repositioning drugs such as thalidomide [9], celecoxib [10] and rapamycin derivatives [11], can be found in the previous reports [12–14].
One of the necessary steps of drug repositioning is to accurately identify the drug–target interactions (DTIs). However, experimental determination of such associations is time-consuming and costly. Thus, computational methods have been proposed alternatively to infer potential DTIs in effective ways. Traditionally, computational methods for DTI predictions include molecular docking simulation, quantitative structure–activity relationship (QSAR) and so forth [15–21]. However, these methods possess inherent limitations. For example, docking simulation requires 3D crystal structure of the drug target, which is difficult to obtain for membrane proteins. Traditional QSAR often handles compound analogs targeting a single molecular target, which is less efficient for processing chemogenomic data with a large library of compounds and many targets.
Unlike QSAR (chemical data-based) and molecular docking (genomic data-based) approaches, chemogenomic data-driven DTI prediction methods simultaneously consider both chemical information and genomic information (often from large-scale screenings of small molecule libraries against a panel of drug target, which may or may not be biologically related). For example, Yamanishi et al. [22] proposed a bipartite graph learning method to infer the relationship between chemical/genomic space and pharmacological space. Kim and coworkers [23] explored the effect of drug–drug interactions (DDIs) on DTI predictions. They used two machine learning algorithms, including support vector machine (SVM) and kernel-based L1-norm regularized logistic regression (KL1LR) to build prediction models. As a result, they concluded that DDI from pharmacological information is a promising feature in predicting DTIs when compared with other data sources such as chemical structures of drugs, and KL1LR is useful for investigating the contributing features. In the work by Wang et al. [24], a two-layer graphical model, called restricted Boltzmann machine, was proposed to predict not only the direct and indirect drug–target relationships but also the drug modes of action, including binding, activation and inhibition, which extended the conventional binary DTI predictions. Most recently, Meng et al. [25] proposed a novel feature-based approach, called predicting drug targets with protein sequence (PDTPS), to infer potential DTIs. In PDTPS, for each protein sequence, position-specific score matrix (PSSM) was first constructed, and the bigram probability feature extraction method was used to represent a given protein sequence based on the calculated PSSM. After this, principal component analysis (PCA) was adopted to reduce the protein sequence feature vector. For each drug compound, the structural features were calculated. As a result, the feature representation of each drug–target pair was obtained by concatenating both protein vector and drug vector. Finally, relevance vector machine was used to predict potential DTIs. Another feature-based approach proposed by Li et al. [26] adopted local binary pattern operator to compute the histogram descriptors for protein sequences. For drug molecules, they calculated the fingerprints, and used PCA to extract the low-dimensional features for both proteins and drugs. Finally, they used the discriminative vector machine classifier to identify DTIs. Some other DTI prediction methods were described in the previous reviews [27–32] and research articles [33–40].
Among these algorithms, many of them are made publicly available. Researchers often compared different algorithms based on the benchmark data set [22], and they adopted two commonly used metrics [i.e. area under the curve (AUC) and area under precision–recall curve (AUPR)] as the evaluation criteria. However, the comparison may be suboptimal and less objective because of differences in program parameter setting and details of cross-validation methods. In this work, we first review chemogenomic data-driven and open-source algorithms published in recent years, and then we compare five representative algorithms based on a new recall-based evaluation metric in the same framework. We hope the reviewed algorithms can be continuously improved to make stronger prediction, and can be optimized to ease reuse and ensure result replication.
Material and methods
Benchmark data set
The benchmark data set used in many DTI prediction studies was originally proposed by Yamanishi and coworkers [22], which has been considered as the golden data set for comparing various DTI prediction algorithms. The data set is composed of drug molecules and protein targets in the KEGG LIGAND and GENES databases [41]. It consists of three matrices: the chemical space matrix, for drugs; the genomics space matrix, for targets; and the drug–target adjacency (interaction/association) matrix Y. denotes the drug similarity matrix, where the chemical similarities between drugs from the KEGG LIGAND database were computed by using the SIMCOMP tool [42]. denotes the target similarity matrix where the sequence similarities between protein targets from the KEGG GENES database [41] were calculated by using normalized Smith–Waterman scores [43]. denotes the interaction matrix with a value if drug interacts with target , 0 otherwise, based on the annotations from the KEGG BRITE [41], BRENDA [44], SuperTarget [45] and DrugBank [46] databases. Here, a 0 value in Y does not necessarily mean that the corresponding target is irrelevant to the drug, but it could be that the DTI is not validated yet by experiments. The benchmark data set is listed in Table 1. The data set includes four subsets grouped by target classification: Enzyme, ion channel (IC), G protein-coupled receptor (GPCR) and nuclear receptor (NR). The largest subset, Enzyme, includes 445 drugs and 664 targets with 2926 known (experimentally validated) DTIs between them, while the smallest subset, NR, consists of only 54 drugs and 26 targets with 90 known interactions. Two subsets of moderate size, IC and GPCR, include 210 and 223 drugs, 204 and 95 targets and 1476 and 635 known interactions, respectively. The sparsity value listed in the last column of Table 1 for each subset is calculated as the ratio of known interactions to all possible interactions between drugs and targets.
Table 1.
Benchmark data set for DTI prediction algorithms
| Data set | Number of drugs | Number of targets | Number of interactions | Sparsity value |
|---|---|---|---|---|
| Enzyme | 445 | 664 | 2926 | 0.010 |
| IC | 210 | 204 | 1476 | 0.034 |
| GPCR | 223 | 95 | 635 | 0.030 |
| NR | 54 | 26 | 90 | 0.064 |
Data set transformation
It should be emphasized that the benchmark data set may be transformed slightly based on the individual prediction algorithms. For algorithms such as bipartite local models (BLMs) [47], the zero components in matrix Y may be transformed to −1, while for algorithms such as dual-network integrated logistic matrix factorization (DNILMF) [48], the original zero elements remain unchanged. Besides the interaction matrix Y, the drug similarity matrix and target similarity matrix may be transformed to corresponding kernel matrices, and , respectively. For example, BLM requires a kernel matrix as input to build a model. A general transformation procedure can be performed in the following way: taking the conversion from to as an example, was first converted to a symmetrical matrix by adding its transposed matrix and then divided by 2. The obtained symmetrical matrix was finally converted to a positive semi-definite matrix by adding an identity matrix with a small value (0.1 in the work) in the main diagonal line for multiple times. A similar procedure was applied to for generating .
Cross-validation and evaluation metric
Stringent cross-validation is important for model evaluation. Different from previous methods [47, 49], which put both positive and negative interaction pairs (considering unknown interactions as negative ones) into the test set. We, in this work, only include the positive interaction pairs in the test set in the process of cross-validation. Specifically, in each split, we removed a random subset of 10% of the known entries in the drug–target adjacency matrix Y as the test set and trained on the remaining 90% of the known DTI. In addition, we ensured each drug has at least one interaction with a target (and vice versa, each target has at least one interaction with a drug as well) in the training matrix as reported by a previous DTI prediction work [50]. Then, we used a ranking-based statistical metric to evaluate different DTI prediction algorithms.
One of the issues with evaluating one-class prediction model is that the data set only encodes positive values (i.e. known DTIs, labeled as 1 in matrix Y). When a 0 value is included in the cell of matrix Y, it does not necessarily mean the drug does not interact with that target but could instead mean that it was not tested against that target. Unlike previous studies [47–49], whereas the authors simply considered zero components in matrix Y as negative samples, we currently only consider positive one as input information (i.e. lack of negative samples) to evaluate the model. Thus, a recall-based evaluation metric, known as mean percentile ranking (MPR) [51, 52], was adopted to evaluate the algorithm performance. Specially, for each drug i in the test set, we generated a ranked list of the targets sorted in descending order by the predicted scores between the current drug with all targets in the data set. Let denote the percentile ranking (PR) of target j with drug i. indicates that drug i is predicted to interact with target j with the highest probability. Similarly, signifies that drug i is predicted to interact with target j with the lowest probability. Herein, the definition of MPR is described as follows:
| (1) |
where denotes the number of drugs in the test set, and can be computed as follows:
| (2) |
where denotes the number of targets in the test set for the current drug . It should be pointed out that lower values of MPR are more desirable, as they indicate higher probability for the experimentally validated DTIs. Conversely, higher values of MPR indicate that drugs are predicted to interact with their targets with lower possibilities. Evidently, randomly produced lists would have an expected MPR of 50%. By using this kind of metric, one can obtain a recommended list of candidate targets for a drug of interest, with top predictions recommended for experimental validation with higher priority.
Open-source chemogenomic data-driven DTI prediction algorithms
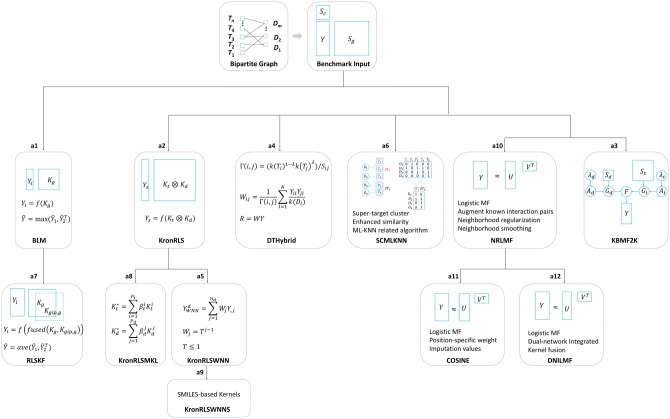
In this section, we review several chemogenomic data-driven and open-source DTI prediction algorithms focusing on model properties and model evolutionary relationships. Table 2 lists the reviewed algorithms with the corresponding Web links.
Table 2.
Open-source chemogenomic data-driven DTI prediction algorithms based on the benchmark data set
| No. | Algorithm | Open-access link | Year | Reference |
|---|---|---|---|---|
| 1 | BLM | http://cbio.mines-paristech.fr/∼yyamanishi/bipartitelocal/ | 2009 | [47] |
| 2 | KronRLS | http://cs.ru.nl/∼tvanlaarhoven/drugtarget2011/ | 2011 | [55] |
| 3 | KBMF2K | http://users.ics.aalto.fi/gonen/bioinfo12.php | 2012 | [59] |
| 4 | DTHybrid | http://alpha.dmi.unict.it/dtweb/dthybrid.php | 2013 | [50] |
| 5 | KronRLSWNN | http://cs.ru.nl/∼tvanlaarhoven/drugtarget2013/ | 2013 | [57] |
| 6 | SCMLKNN | http://web.hku.hk/∼liym1018/projects/drug/drug.html or http://www.bmlnwpu.org/us/tools/PredictingDTI_S2/METHODS.html | 2015 | [63] |
| 7 | RLSKF | https://github.com/minghao2016/RLS-KF | 2016 | [53] |
| 8 | KronRLSMKL | http://www.cin.ufpe.br/∼acan/kronrlsmkl/ | 2016 | [56] |
| 9 | KronRLSWNNS | https://github.com/hkmztrk/SMILESbasedSimilarityKernels | 2016 | [58] |
| 10 | NRLMF | https://github.com/stephenliu0423/PyDTI | 2016 | [49] |
| 11 | COSINE | http://bioinfo.cs.uni.edu/COSINE.html | 2016 | [60] |
| 12 | DNILMF | https://github.com/minghao2016/DNILMF | 2017 | [48] |
Given three matrices, Y (DTI matrix), (kernel matrix of drugs) and (kernel matrix of targets), Bleakley et al. [47] proposed to use multiple BLM to perform DTI predictions (see a1 in Figure 1). BLM essentially transforms the edge-prediction problem of a DTI network into a binary classification problem of points with labels. Specifically, each column of the interaction matrix Y is used as a dependent variable [+1 denotes the positive label (interaction) and −1 denotes the negative label (non-interaction)] in turn, and the target kernel, , is used as the independent variables, and then a kernel-based SVM is used to predict the interactions between current drug i and all targets. The abovementioned process with the predicted scores is based on the target side (). Similarly, the process can also be applied to the drug side (, to generate the prediction scores ), and the final prediction scores are yielded by an aggregated function f, such as the maximum function (), where indicates the transpose of ). Following a similar idea of local models as BLM, Hao et al. [53] adopted a nonlinear kernel fusion (KF) technique [54] combined with the regularized least squares (RLS) algorithm to predict DTIs (RLSKF, see a7 in Figure 1). Two fundamental differences between RLSKF and BLM are (1) RLSKF adopts the RLS algorithm rather than SVM; and (2) RLSKF uses the nonlinear fused kernel (taking target side-based prediction as an example, , where is the Gaussian interaction profile (GIP) kernel calculated from the target interaction profiles of Y, which will be discussed below) instead of the single kernel ( or ) as in BLM. It should also be noted that the fused kernel is derived from an iteration process rather than a simple linear combination such as used in some algorithms such as KronRLS [55].
Figure 1.
Open-source chemogenomic data-driven DTI prediction algorithms clustered by model properties and evolutionary relationships.
We herein summarize and reiterate the key steps of the KF algorithm [54]. First, KF preprocesses the similarity matrices (i.e. , , and in this work. Please note that similarity matrix such as and without kernel properties can also be preprocessed for KF). Taking as an example, its normalized similarity can be calculated by , where D is the diagonal matrix with the diagonal elements of the individual row sums of . From , the local similarity matrix is obtained in the following way:
| (3) |
where represents a set of neighbors of drug , and K is the number of nearest neighbors. Through this operation, the similarities between non-neighboring points set to 0. In a similar way, and are obtained for drug interaction kernel . Then, from two full similarity matrices ( and ) and two local similarity matrices ( and ), the key fusion steps are performed as follows:
| (4) |
| (5) |
where is the status matrix of the drug structural similarity after iterations, is the status matrix of the drug interaction similarity and and are the transpose of and , respectively. Finally, after t steps, the overall similarity is calculated by . Thus, can be used as the input of any DTI prediction algorithm. It is a similar process for the fusion of target-based similarity matrices.
Instead of multiple local models (such as BLM and RLSKF), the following discussed algorithms are all global models indicating that one model can simultaneously predict potential interactions from multiple targets and multiple drugs in the test set. As shown in a2 of Figure 1, van Laarhoven et al. [55] used a simple machine learning method (RLS), combined with a GIP kernel from the DTI network, Y, to infer the potential interaction pairs (named KronRLS here). The GIP kernel can be obtained either from the target side (denoted by ) or from the drug side (denoted by ) based on the DTI matrix Y only. The final combined kernels are obtained by a simple weighted average operation (i.e. for targets, and for drugs). Given and , the Kronecker product of them (denoted by ) is calculated to yield a large pairwise kernel as the independent variables. A long vector stacked by column (denoted by ) of the interaction matrix Y is generated as the dependent variable. As a result, RLS takes the inputs of and to predict the potential interaction scores between drugs and targets globally. It should be emphasized that more efficient implementation based on Eigen decompositions of kernels can be applied to this algorithm [55]. Following KronRLS, two improved algorithms were proposed. The first algorithm is named as KronRLSMKL (see a8 in Figure 1), proposed by Nascimento et al. [56], which extends KronRLS to the multiple kernel learning (MKL) framework. As a result, the kernel weight can be used to indicate the importance of each individual kernel. The second algorithm (KronRLSWNN, see a5 in Figure 1) was proposed by van Laarhoven et al. [57] to extend KronRLS to new drug candidates (i.e. drugs have no interactions with any targets) by inferring the drug profile using the weighted nearest neighbors (WNNs). Having used KronRLSWNN as a basic framework, Öztürk et al. [58] investigated the effects of a series of SMILES-based kernels on the model performance (named KronRLSWNNS, see a9 in Figure 1).
Several of these algorithms are low rank-based models. The first one is KBMF2K (see a3 in Figure 1), which was proposed by Gönen et al. [59]. KBMF2K uses techniques, including dimensionality reduction, matrix factorization and binary classification to perform DTI predictions. Unlike KBMF2K, which transforms and into two low-dimensional matrices ( and , respectively), NRLMF, the second low rank-based model, proposed by Liu et al. [49], transforms the interaction matrix, Y, into two low-dimensional matrices (U and V for target latent variables and drug latent variables, respectively). As a result, the prediction scores of NRLMF are given by (see a10 in Figure 1). NRLMF takes the logistic matrix factorization as the model algorithm, which is especially suitable for binary variables. Furthermore, NRLMF adopts a technique with augmented known interaction pairs to decrease the imbalanced level between positive and negative samples. In the model’s objective function, NRLMF uses a neighborhood regularized approach, and for the postprocessing stage, NRLMF uses a neighborhood smoothing method to generate new drug/target prediction scores instead of the original predicted scores. Based on NRLMF, two improved algorithms were also developed. COSINE (see a11 in Figure 1) was recently proposed by Lim and coworkers [60] to improve prediction performance especially for predicting potential targets for new drugs. Different from NRLMF, COSINE formulates the objective function by using position-specific weight and imputation values, which can overcome the sparseness of the DTI network Y. Later, Hao et al. [48] proposed a DNILMF algorithm to infer potential interactions between drugs and targets (see a12 in Figure 1). Two different points of DNILMF from NRLMF lie in the fact that (1) DNILMF incorporates the ‘trust ensemble’ idea [61] into the logistic function; and (2) DNILMF uses the KF method [54] (same as RLSKF) to enhance the similarity metrics.
Two global algorithms in the separate cluster are DTHybrid and SCMLKNN (see a4 and a6 in Figure 1, respectively). DTHybrid was proposed by Alaimo et al. [50], which was inspired by the work from Zhou and coworkers [62] by plugging the drug and target similarity information into the resource allocation equation. SCMLKNN designed by Shi et al. [63] is a multi-label K-nearest neighbors-related algorithm, which incorporates the super-target clustering idea for handling missing interactions and combines additional information for enhancing the similarity measures. The final confidence score of a DTI pair is obtained by a product of two kinds of probabilistic scores generated from the models. It should be pointed out, in this work, that the different algorithms were only described briefly with the key components highlighted. For more details, one can refer to the original studies and the open-source codes.
Algorithm comparison procedure
We performed the following evaluation procedures for a more rigorous comparison of the reviewed algorithms based on the recall-based statistical metric. Step 1: The adjacency matrix, Y, was first split for 10-fold cross-validation in the abovementioned approach. Briefly, only positive pairs were used to perform the subset splitting to compare the recall-based evaluation metric. In each fold, at least one link (known interaction) was kept in each row and each column of Y, respectively. Five trials of 10-fold cross-validation processes were performed to yield 50 fold matrices with test set data points included in each matrix. Each fold matrix was used to build the model and predict the data points in the test set. Step 2: Optionally, the similarity matrices were converted to the corresponding kernel matrices as shown in the data set transformation section for the kernel-based algorithms such as BLM. Step 3: Multiple models were built based on similarity matrix either from targets or from drugs (or based on matrices from both targets and drugs), as well as each fold matrix (generated from Step 1). Step 4: The recall-based evaluation metric, MPR, was calculated from the test set data points in each fold matrix. The source codes for comparison of the discussed algorithms can be found at: https://github.com/minghao2016/chemogenomicAlg4DTIpred.
Results and discussion
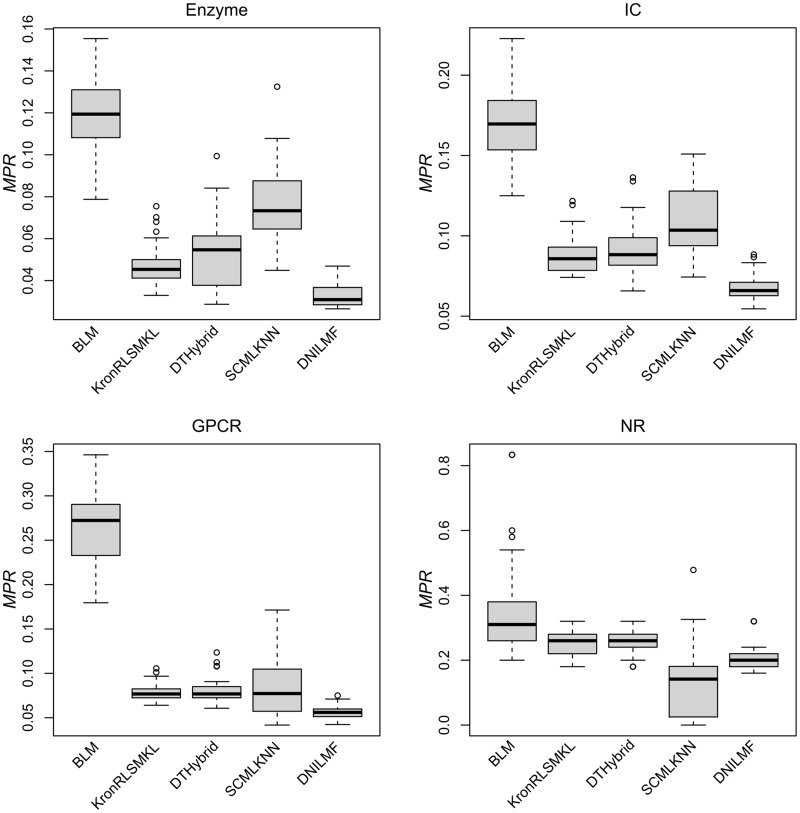
In this work, we reviewed several chemogenomic data-driven and open-source algorithms based on model properties and model evolutionary relationships. We re-implemented five algorithms selected based on the clusters (Figure 1) in R programming language [64]. DTI prediction algorithms were validated by using the MPR evaluation metric. Additionally, two traditional metrics, AUC and AUPR, were also reported. Table 3 lists the results of MPR, AUC and AUPR for the representative algorithms based on five trials of 10-fold cross-validation. For MPR, the smaller the value, the better of the algorithm's performance is. For AUC and AUPR, a larger value indicates better performance. For calculating AUC and AUPR, the whole data set was randomly divided into 10 parts with approximately equal size for the positive and ‘negative’ interaction pairs, respectively. Each part was selected in turn as the test set, while the remaining nine parts were served as the training set. It is evident that the data set possesses the imbalanced property. Therefore, AUPR is more suitable to be used to evaluate such imbalanced data set compared with AUC. As shown in Table 3, for all of the four subsets (Enzyme, IC, GPCR and NR), KronRLSMKL consistently outperforms (sometimes slightly though) other algorithms in terms of AUPR, followed by DNILMF. BLM exhibits the lowest AUPR value. From these results, it can be noticed that the representative algorithms performed with a similar trend when evaluated using MPR, except that DNILMF has the lowest MPR value (indicating best performance), followed by KronRLSMKL in three larger data sets (i.e. Enzyme, IC and GPCR), which will be discussed in the following. Figure 2 shows the boxplots of MPR from five trials of 10-fold cross-validation for the five algorithms based on the benchmark data set.
Table 3.
Comparison of open-source algorithms based on MPR, AUC and AUPR for the benchmark data set
| Data | Method | MPR (mean ± SE) | AUC (mean ± SE) | AUPR (mean ± SE) |
|---|---|---|---|---|
| Enzyme | BLM | 0.119 ± 0.002 | 0.923 ± 0.003 | 0.750 ± 0.003 |
| KronRLSMKL | 0.047 ± 0.001 | 0.993 ± 0.000 | 0.963 ± 0.001 | |
| DTHybrid | 0.053 ± 0.002 | 0.986 ± 0.001 | 0.939 ± 0.001 | |
| SCMLKNN | 0.076 ± 0.002 | 0.986 ± 0.000 | 0.839 ± 0.002 | |
| DNILMF | 0.033 ± 0.001 | 0.996 ± 0.000 | 0.951 ± 0.001 | |
| IC | BLM | 0.169 ± 0.003 | 0.899 ± 0.002 | 0.684 ± 0.009 |
| KronRLSMKL | 0.088 ± 0.002 | 0.990 ± 0.001 | 0.953 ± 0.003 | |
| DTHybrid | 0.090 ± 0.002 | 0.989 ± 0.002 | 0.918 ± 0.002 | |
| SCMLKNN | 0.109 ± 0.003 | 0.975 ± 0.000 | 0.823 ± 0.004 | |
| DNILMF | 0.068 ± 0.001 | 0.996 ± 0.000 | 0.947 ± 0.002 | |
| GPCR | BLM | 0.266 ± 0.006 | 0.752 ± 0.010 | 0.326 ± 0.009 |
| KronRLSMKL | 0.079 ± 0.001 | 0.987 ± 0.003 | 0.833 ± 0.005 | |
| DTHybrid | 0.080 ± 0.002 | 0.969 ± 0.002 | 0.768 ± 0.014 | |
| SCMLKNN | 0.086 ± 0.005 | 0.968 ± 0.003 | 0.650 ± 0.011 | |
| DNILMF | 0.056 ± 0.001 | 0.987 ± 0.001 | 0.826 ± 0.008 | |
| NR | BLM | 0.349 ± 0.020 | 0.777 ± 0.050 | 0.211 ± 0.091 |
| KronRLSMKL | 0.254 ± 0.006 | 0.979 ± 0.001 | 0.613 ± 0.060 | |
| DTHybrid | 0.257 ± 0.005 | 0.917 ± 0.003 | 0.566 ± 0.087 | |
| SCMLKNN | 0.135 ± 0.016 | 0.951 ± 0.002 | 0.342 ± 0.009 | |
| DNILMF | 0.205 ± 0.005 | 0.952 ± 0.011 | 0.605 ± 0.063 | |
| kd | BLM | 0.320 ± 0.003 | 0.755 ± 0.009 | 0.233 ± 0.015 |
| KronRLSMKL | 0.166 ± 0.003 | 0.817 ± 0.004 | 0.200 ± 0.002 | |
| DTHybrid | 0.126 ± 0.002 | 0.957 ± 0.001 | 0.686 ± 0.004 | |
| SCMLKNN | 0.181 ± 0.005 | 0.908 ± 0.002 | 0.526 ± 0.008 | |
| DNILMF | 0.122 ± 0.002 | 0.966 ± 0.001 | 0.721 ± 0.004 |
Figure 2.
MPR of five representative DTI prediction algorithms based on the benchmark data set. For Enzyme, IC and NR, all results differ significantly except KronRLSMKL VS. DTHybrid (P < 0.01, t-test). For GPCR, all results differ significantly except KronRLSMKL versus DTHybrid, KronRLSMKL versus SCMLKNN and DTHybrid versus SCMLKNN (P < 0.01, t-test).
For three relatively larger data sets (i.e. Enzyme, IC and GPCR), all of the five representative algorithms keep the same trend of prediction performance, where DNILMF outperforms the other four algorithms consistently, and BLM shows large MPR values compared with others. Both KronRLSMKL and DTHybrid show comparable results, which are both (slightly) better than the ones from SCMLKNN. Interestingly, for NR, which is the smallest data set, SCMLKNN exhibits the best MPR value. However, for all four data sets, these algorithms already gave a large improvement over a purely random model with MPR expected as of 50% (especially for the larger data sets). We emphasize that the current results of MPR were calculated based on the original parameter settings from the reported algorithms with slight differences, and the parameters were fixed during the cross-validations. Therefore, it is anticipated that the performance might be improved by fully exploiting the parameter space. As reported in the previous work [65], the nested cross-validation can be used to perform parameter tuning from the inner loop, and the outer loop is used to evaluate the model. In this work, we took the DTHybrid algorithm as an example to perform nested cross-validation. As a result, the model with optimal parameters (i.e. lambda and alpha used by DTHybrid) derived from the nested cross-validation exhibits similar or slightly better results than the one using fixed parameters.
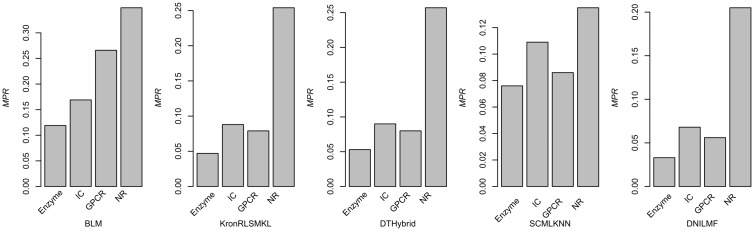
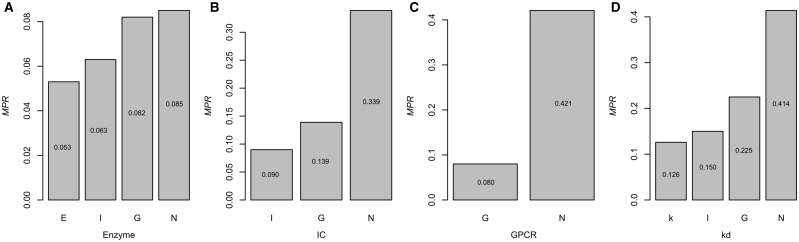
From these results, we notice that the size of samples in the data set can have an impact on the model performance (Figure 3). Most models would show improved performance with the increase of samples. Interestingly, though IC includes more data points compared with GPCR, algorithms from KronRLSMKL, DTHybrid, SCMLKNN and DNILMF consistently give relative better results for GPCR. One of the possible reasons for this result may be that the ratio of the number of targets to the number of drugs in Y for GPCR is much less than the ratio in the IC group. To further investigate the influence of data size, we subsampled three larger data sets (i.e. Enzyme, IC and GPCR in descending order of size) to the sizes approximate to that of the smaller data sets and calculated the MPR values. For computational efficiency, we took DTHybrid as the tested algorithm. To subsample a larger data set, for example, for the Enzyme data set (i.e. 664 targets and 445 drugs), three subsamples were generated with the approximated size of IC (i.e. 204 targets and 221 drugs), GPCR (i.e. 95 targets and 125 drugs) and NR (i.e. 30 targets and 55 drugs). Similarly, two subsamples for the IC data set and one subsample for the GPCR data sets were generated. It showed that when the size of a larger data set decreased, the corresponding performance also reduced (Figure 4). This indicates that the data set size is indeed a factor to influence the model performance.
Figure 3.
Performance of five representative algorithms on four benchmark subsets with different data sizes.
Figure 4.
Influence of data size on model performance of DTHybrid. (A) subsample the original Enzyme (E) data set into approximated size of IC (I), GPCR (G) and NR (N) data sets, respectively; (B) subsample the original IC (I) data set into approximated size of GPCR (G), NR (N) data sets, respectively; (C) subsample the original GPCR (G) data set into approximated size of NR (N) data set; and (D) subsample the original kd (k) data set into approximated size of IC (I), GPCR (G) and NR (N) data sets, respectively.
Besides data set size, data quality also has an impact. Thus, we also evaluated the five algorithms based on the kd data set used in the previous work [65]. Originally, the kd data set contains drug–target pairs with regression-format kd data from the study by Davis and coworkers [66]. We converted it to a data set of binary data as done by Pahikkala and coworkers [65]. We also performed several preprocessing steps for generating proper data format for the current algorithms, which means that the generated data set should guarantee each row and each column have at least one known interaction, respectively. Finally, we obtained 373 targets and 65 drugs and the corresponding similarity matrices. Having kept the default parameters, we obtained the MPR values for BLM, KronRLSMKL, DTHybrid, SCMLKNN and DNILMF as of 0.320, 0.166, 0.126, 0.181 and 0.122, respectively. The results for the kd data set show a similar trend of algorithms performance with those from the Enzyme, IC and GPCR data sets, except that DTHybrid outperforms KronRLSMKL in the kd data set as indicated by MPR values. In terms of AUPR, the DNILMF algorithm also presents the best performance for the kd data set as compared with other algorithms. We also investigated the influence of data size on the performance based on the kd data set. As shown in Figure 4, when we subsampled the original kd data (i.e. 373 targets and 65 drugs) to the approximate size of the smaller data sets in the well-used benchmark (i.e. IC: 204 targets and 57 drugs; GPCR: 95 targets and 44 drugs; NR: 26 targets and 31 drugs), the performance also decreased.
It is evident that DNILMF shows better performance for all the subsets except NR. However, we notice that the enhanced performance of DNILMF is not only derived from the proposed algorithm itself but also from the KF method [48, 53, 54], which is an important but understudied approach in the DTI prediction field. In fact, the KF method can be applied to any algorithm, as it is independent of the model itself. In this work, we combined two kinds of kernels (in the DNILMF algorithm) including the drug kernel from structural information (or target kernel from sequence information) and the drug GIP kernel from the interaction matrix Y (or target GIP kernel of Y). However, multiple kernels are allowed to KF.
Besides the KF technology, many other methods were also proposed to improve the data set itself, which are independent of algorithms. For example, in the SCMLKNN [63] algorithm, the authors proposed the super-target technique, which first clusters the targets into protein families on the basis of sequence similarity. By performing such operation, the data sparsity problem can be solved to some extent. In the KronRLSWNN algorithm [57], the authors proposed to use a WNN to infer the interaction profiles for new drugs, which have no interaction data with any targets. In both RLSKF [53] and DNILMF [48], a similar process was also used to infer those profiles for both new drugs and new targets. Technologies such as KF, super-target clustering and WNN, which are unsupervised methods, are more straightforward and flexible to combine with other algorithms, as they are obtained before the model building step. Therefore, such unsupervised technologies are often adopted by researchers who do not have statistical/mathematical background because of the simplicity and easy implementation compared with supervised algorithms, which often require optimization process with complex mathematical knowledge.
Besides the mentioned algorithms above, in fact, there are many algorithms from other scientific disciplines such as implicit feedback [51, 52], which can be smoothly transformed and applied to tackle DTI predictions. Indeed, progress for one scientific field may be accelerated by ‘borrowing’ ideas, concepts or theories from a different discipline. For example, NRLMF borrows the logistic matrix factorization technique used by collaborative filtering [51] with enhanced objective function, and DNILMF borrows the ‘trust ensemble’ idea from the recommender systems field [61] by adding similarity fusion technique and extending the original one to dual integration. Thus, with the development of algorithms in various research fields, it is beneficial to transfer across-discipline methods into the DTI prediction field.
In this work, we assessed five open-access DTI prediction algorithms based on the experimental setting where both training and test sets share common drugs/targets, and proposed to use a recall-based metric to evaluate the models. Algorithms, which can handle new drug/target scenarios, will be studied in the future, and additional and multiple evaluation metrics for estimating one-class classification problems may be taken into consideration. Despite of the many applications in previous work, the benchmark data sets used are rather limited. In fact, as more DTI data becomes available in the public domain, it will be beneficial to apply the DTI algorithms to diverse data sets for comparison. In summary, the current work compares and analyzes the performance on DTI predictions using the commonly used benchmark data set and DTI data in the kd data set. Such review and comparative work may provide insights for advancing the state of art for DTI predictions by developing new methods to improve scalability and gain stronger generalization abilities, as well as to effectively incorporate negative samples and better handle regression-format data.
Conclusion
A set of open-source and chemogenomic data-driven DTI prediction algorithms was reviewed based on the model properties and model evolutionary relationships. Five representative algorithms were compared primarily based on recall-based evaluation metric, MPR. The selected algorithms were re-implemented in R programming language for straightforward comparison, ease of reuse and further improvement. Algorithms such as DNILMF combined with the KF technology exhibited better performance. We believe this review will be helpful to researchers by facilitating decision making for choosing DTI algorithms, as well as by providing a common ground to understand the state of art for future enhancement of DTI algorithms.
Key Points.
Open-source chemogenomic data-driven computational algorithms for predicting DTIs were reviewed.
Five representative algorithms were re-implemented in R programming language.
Recall-based metric, MPR, was used to evaluate different algorithms.
Ming Hao is a Postdoctoral Researcher at National Center for Biotechnology Information, National Institutes of Health. His research focuses on bioinformatics and chemoinformatics related to drug design.
Stephen H. Bryant is a Senior Investigator at National Center for Biotechnology Information, National Institutes of Health. His structure group conducts basic research in bioinformatics and cheminformatics.
Yanli Wang is a Lead Staff Scientist at National Center for Biotechnology Information, National Institutes of Health. Her research interests include computational methods for drug discovery and data mining chemogenomic data.
Funding
This research was supported by the Intramural Research Program of the NIH, National Library of Medicine.
References
- 1. Booth B, Zemmel R.. Prospects for productivity. Nat Rev Drug Discov 2004;3(5):451–6. 10.1038/nrd1384 [DOI] [PubMed] [Google Scholar]
- 2. Kola I, Landis J.. Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov 2004;3(8):711–16. [DOI] [PubMed] [Google Scholar]
- 3. Iorio F, Rittman T, Ge H, et al. . Transcriptional data: a new gateway to drug repositioning? Drug Discov Today 2013;18(7–8):350–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chong CR, Sullivan DJ.. New uses for old drugs. Nature 2007;448(7154):645–6. 10.1038/448645a [DOI] [PubMed] [Google Scholar]
- 5. Ashburn TT, Thor KB.. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov 2004;3(8):673–83. 10.1038/nrd1468 [DOI] [PubMed] [Google Scholar]
- 6. Chow WA, Jiang C, Guan M.. Anti-HIV drugs for cancer therapeutics: back to the future?. Lancet Oncol 2009;10(1):61–71. 10.1016/S1470-2045(08)70334-6 [DOI] [PubMed] [Google Scholar]
- 7. Delbaldo C, Faivre S, Dreyer C, et al. . Sunitinib in advanced pancreatic neuroendocrine tumors: latest evidence and clinical potential. Ther Adv Med Oncol 2012;4(1):9–18. 10.1177/1758834011428147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Druker BJ. Imatinib as a paradigm of targeted therapies. Adv Cancer Res 2004;91:1–30. [DOI] [PubMed] [Google Scholar]
- 9. Bartlett JB, Dredge K, Dalgleish AG.. The evolution of thalidomide and its IMiD derivatives as anticancer agents. Nat Rev Cancer 2004;4(4):314–22. 10.1038/nrc1323 [DOI] [PubMed] [Google Scholar]
- 10. Steinbach G, Lynch PM, Phillips RK, et al. . The effect of celecoxib, a cyclooxygenase-2 inhibitor, in familial adenomatous polyposis. N Engl J Med 2000;342(26):1946–52. 10.1056/NEJM200006293422603 [DOI] [PubMed] [Google Scholar]
- 11. Koehl GE, Schlitt HJ, Geissler EK.. Rapamycin and tumor growth: mechanisms behind its anticancer activity. Transplant Rev 2005;19(1):20–31. 10.1016/j.trre.2005.01.001 [DOI] [Google Scholar]
- 12. Cappelli C, Castellano M, Pirola I, et al. . Reduced thyroid volume and nodularity in dyslipidaemic patients on statin treatment. Clin Endocrinol 2008;68(1):16–21. 10.1111/j.1365-2265.2007.02982.x [DOI] [PubMed] [Google Scholar]
- 13. Gu S, Tian Y, Chlenski A, et al. . Valproic acid shows potent antitumor effect with alteration of DNA methylation in neuroblastoma. Anti-Cancer Drugs 2012;23(10):1054. 10.1097/CAD.0b013e32835739dd [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li YY, Jones SJ.. Drug repositioning for personalized medicine. Genome Med 2012;4(3):27.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hao M, Li Y, Wang Y, et al. . Combined 3D-QSAR, molecular docking, and molecular dynamics study on piperazinyl-glutamate-pyridines/pyrimidines as potent P2Y12 antagonists for inhibition of platelet aggregation. J Chem Inf Model 2011;51(10):2560–72. 10.1021/ci2002878 [DOI] [PubMed] [Google Scholar]
- 16. Hao M, Zhang X, Ren H, et al. . In silico identification of structure requirement for novel thiazole and oxazole derivatives as potent fructose 1, 6-bisphosphatase inhibitors. Int J Mol Sci 2011;12(12):8161–80. 10.3390/ijms12118161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li Y, Hao M, Ren H, et al. . Exploring the structure requirement for PKCθ inhibitory activity of pyridinecarbonitrile derivatives: an in silico analysis. J Mol Graph Model 2012;34:76–88. [DOI] [PubMed] [Google Scholar]
- 18. Hao M, Li Y, Zhang S-W, et al. . Investigation on the binding mode of benzothiophene analogues as potent factor IXa (FIXa) inhibitors in thrombosis by CoMFA, docking and molecular dynamic studies. J Enzyme Inhib Med Chem 2011;26(6):792–804. 10.3109/14756366.2011.554414 [DOI] [PubMed] [Google Scholar]
- 19. Cai J, Li C, Liu Z, et al. . Predicting DPP-IV inhibitors with machine learning approaches. J Comput Aided Mol Des 2017;31(4):393–402. 10.1007/s10822-017-0009-6 [DOI] [PubMed] [Google Scholar]
- 20. Myint K-Z, Wang L, Tong Q, et al. . Molecular fingerprint-based artificial neural networks QSAR for ligand biological activity predictions. Mol Pharm 2012;9(10):2912–23. 10.1021/mp300237z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hou X, Li K, Yu X, et al. . Protein flexibility in docking-based virtual screening: discovery of novel lymphoid-specific tyrosine phosphatase inhibitors using multiple crystal structures. J Chem Inf Model 2015;55(9):1973–83. 10.1021/acs.jcim.5b00344 [DOI] [PubMed] [Google Scholar]
- 22. Yamanishi Y, Araki M, Gutteridge A, et al. . Prediction of drug-target interaction networks from the integration of chemical and genomic spaces. Bioinformatics 2008;24(13):i232–i40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim S, Jin D, Lee H.. Predicting drug-target interactions using drug-drug interactions. PLoS One 2013;8(11):e80129.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang Y, Zeng J.. Predicting drug-target interactions using restricted Boltzmann machines. Bioinformatics 2013;29(13):i126–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Meng FR, You ZH, Chen X, et al. . Prediction of drug-target interaction networks from the integration of protein sequences and drug chemical structures. Molecules 2017;22(7):1119. 10.3390/molecules22071119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li Z, Han P, You Z, et al. . In silico prediction of drug-target interaction networks based on drug chemical structure and protein sequences. Sci Rep 2017;7(1):11174. 10.1038/s41598-017-10724-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen X, Yan CC, Zhang X, et al. . Drug-target interaction prediction: databases, web servers and computational models. Brief Bioinform 2016;17(4):696–712. 10.1093/bib/bbv066 [DOI] [PubMed] [Google Scholar]
- 28. Mousavian Z, Masoudi-Nejad A.. Drug-target interaction prediction via chemogenomic space: learning-based methods. Expert Opin Drug Metab Toxicol 2014;10(9):1273–87. 10.1517/17425255.2014.950222 [DOI] [PubMed] [Google Scholar]
- 29. Ding H, Takigawa I, Mamitsuka H, et al. . Similarity-based machine learning methods for predicting drug-target interactions: a brief review. Brief Bioinform 2014;15(5):734–47. 10.1093/bib/bbt056 [DOI] [PubMed] [Google Scholar]
- 30. Cheng T, Hao M, Takeda T, et al. . Large-scale prediction of drug-target interaction: a data-centric review. AAPS J 2017;19(5):1264–75. 10.1208/s12248-017-0092-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang L, You ZH, Chen X, et al. . RFDT: a rotation forest-based predictor for predicting drug-target interactions using drug structure and protein sequence information. Curr Protein Pept Sci 2016;18(999):1. 10.2174/1389203718666161114111656 [DOI] [PubMed] [Google Scholar]
- 32. Huang YA, You ZH, Chen X.. A systematic prediction of drug-target interactions using molecular fingerprints and protein sequences. Curr Protein Pept Sci 2017;18:1–11. [DOI] [PubMed] [Google Scholar]
- 33. Keiser MJ, Roth BL, Armbruster BN, et al. . Relating protein pharmacology by ligand chemistry. Nat Biotechnol 2007;25(2):197–206. 10.1038/nbt1284 [DOI] [PubMed] [Google Scholar]
- 34. Cheng T, Li Q, Wang Y, et al. . Identifying compound-target associations by combining bioactivity profile similarity search and public databases mining. J Chem Inf Model 2011;51(9):2440–8. 10.1021/ci200192v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mizutani S, Pauwels E, Stoven V, et al. . Relating drug-protein interaction network with drug side effects. Bioinformatics 2012;28(18):i522–i8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cheng F, Li W, Wu Z, et al. . Prediction of polypharmacological profiles of drugs by the integration of chemical, side effect, and therapeutic space. J Chem Inf Model 2013;53(4):753–62. 10.1021/ci400010x [DOI] [PubMed] [Google Scholar]
- 37. Jaeger S, Min J, Nigsch F, et al. . Causal network models for predicting compound targets and driving pathways in cancer. J Biomol Screen 2014;19(5):791–802. 10.1177/1087057114522690 [DOI] [PubMed] [Google Scholar]
- 38. Meslamani J, Rognan D.. Enhancing the accuracy of chemogenomic models with a three-dimensional binding site kernel. J Chem Inf Model 2011;51(7):1593–603. 10.1021/ci200166t [DOI] [PubMed] [Google Scholar]
- 39. Chen B, Ding Y, Wild DJ, Tropsha A.. Assessing drug target association using semantic linked data. PLoS Comput Biol 2012;8(7):e1002574.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen X, Liu MX, Yan GY.. Drug-target interaction prediction by random walk on the heterogeneous network. Mol Biosyst 2012;8(7):1970–8. 10.1039/c2mb00002d [DOI] [PubMed] [Google Scholar]
- 41. Kanehisa M, Goto S, Hattori M, et al. . From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res 2006;34:D354–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hattori M, Okuno Y, Goto S, et al. . Development of a chemical structure comparison method for integrated analysis of chemical and genomic information in the metabolic pathways. J Am Chem Soc 2003;125(39):11853–65. 10.1021/ja036030u [DOI] [PubMed] [Google Scholar]
- 43. Smith TF, Waterman MS.. Identification of common molecular subsequences. J Mol Biol 1981;147(1):195–7. 10.1016/0022-2836(81)90087-5 [DOI] [PubMed] [Google Scholar]
- 44. Schomburg I, Chang A, Ebeling C, et al. . BRENDA, the enzyme database: updates and major new developments. Nucleic Acids Res 2004;32(90001):431D–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Günther S, Kuhn M, Dunkel M, et al. . SuperTarget and Matador: resources for exploring drug-target relationships. Nucleic Acids Res 2007;36:D919–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wishart DS, Knox C, Guo AC, et al. . DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res 2008;36:D901–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bleakley K, Yamanishi Y.. Supervised prediction of drug-target interactions using bipartite local models. Bioinformatics 2009;25(18):2397–403. 10.1093/bioinformatics/btp433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hao M, Bryant SH, Wang Y.. Predicting drug-target interactions by dual-network integrated logistic matrix factorization. Sci Rep 2017;7:40376.. 10.1038/srep40376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu Y, Wu M, Miao C, et al. . Neighborhood regularized logistic matrix factorization for drug-target interaction prediction. PLoS Comput Biol 2016;12(2):e1004760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Alaimo S, Pulvirenti A, Giugno R, et al. . Drug-target interaction prediction through domain-tuned network-based inference. Bioinformatics 2013;29(16):2004–8. 10.1093/bioinformatics/btt307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Johnson CC. Logistic matrix factorization for implicit feedback data. Adv Neural Inf Process Syst 2014;27. [Google Scholar]
- 52. Hu Y, Koren Y, Volinsky C. Collaborative filtering for implicit feedback datasets. In: IEEE International Conference on Data Mining Pisa, Italy, 2008.
- 53. Hao M, Wang Y, Bryant SH.. Improved prediction of drug-target interactions using regularized least squares integrating with kernel fusion technique. Anal Chim Acta 2016;909:41–50. 10.1016/j.aca.2016.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang B, Mezlini AM, Demir F, et al. . Similarity network fusion for aggregating data types on a genomic scale. Nat Methods 2014;11(3):333–7. 10.1038/nmeth.2810 [DOI] [PubMed] [Google Scholar]
- 55. van Laarhoven T, Nabuurs SB, Marchiori E.. Gaussian interaction profile kernels for predicting drug-target interaction. Bioinformatics 2011;27(21):3036–43. 10.1093/bioinformatics/btr500 [DOI] [PubMed] [Google Scholar]
- 56. Nascimento AC, Prudêncio RB, Costa IG.. A multiple kernel learning algorithm for drug-target interaction prediction. BMC Bioinformatics 2016;17(1):46.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. van Laarhoven T, Marchiori E.. Predicting drug-target interactions for new drug compounds using a weighted nearest neighbor profile. PLoS One 2013;8(6):e66952.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Öztürk H, Ozkirimli E, Özgür A.. A comparative study of SMILES-based compound similarity functions for drug-target interaction prediction. BMC Bioinformatics 2016;17(1):128.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gönen M. Predicting drug-target interactions from chemical and genomic kernels using Bayesian matrix factorization. Bioinformatics 2012;28(18):2304–10. [DOI] [PubMed] [Google Scholar]
- 60. Lim H, Gray P, Xie L, et al. . Improved genome-scale multi-target virtual screening via a novel collaborative filtering approach to cold-start problem. Sci Rep 2016;6(1):388860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ma H, King I, Lyu MR. Learning to recommend with social trust ensemble. In: Proceedings of SIGIR Boston, USA, 2009.
- 62. Zhou T, Kuscsik Z, Liu J-G, et al. . Solving the apparent diversity-accuracy dilemma of recommender systems. Proc Natl Acad Sci USA 2010;107(10):4511–5. 10.1073/pnas.1000488107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shi J-Y, Yiu S-M, Li Y, et al. . Predicting drug-target interaction for new drugs using enhanced similarity measures and super-target clustering. Methods 2015;83:98–104. 10.1016/j.ymeth.2015.04.036 [DOI] [PubMed] [Google Scholar]
- 64. R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/2017.
- 65. Pahikkala T, Airola A, Pietila S, et al. . Toward more realistic drug-target interaction predictions. Brief Bioinform 2015;16(2):325–37. 10.1093/bib/bbu010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Davis MI, Hunt JP, Herrgard S, et al. . Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol 2011;29(11):1046–51. 10.1038/nbt.1990 [DOI] [PubMed] [Google Scholar]