Abstract
Helicobacter pylori (H. pylori) infection remains a cause of significant morbidity and mortality worldwide. Comprehensive understanding of the pathogenic mechanism of H. pylori and its interaction with host will contribute to developing novel prophylactical and therapeutical strategies. Here, we first determined microRNA (miRNA) levels in H. pylori-infected patients with gastritis, duodenal ulcer, gastric cancer or mucosa-associated lymphoid tissue lymphoma using miRNA data sets. Thirty-four differentially expressed miRNAs were identified and functional enrichment analysis of those miRNA target genes revealed that H. pylori infection were strongly associated with pathway in cancer and regulation of mRNA synthesis. Using disease connectivity analysis of 28 hub genes, we found that H. pylori may increase the risk of many extragastric diseases (e.g. cardiovascular disease, hemic and lymphatic diseases and nervous system disease). Altogether, our integrated analysis provided a new method to predict pathogen–human disease connectivity based on miRNA-mRNA interaction network and indicated anti-H. pylori therapy as an effective means of human diseases prevention.
Keywords: Helicobacter pylori, miRNA, gastric cancer, cardiovascular disease
Introduction
Helicobacter pylori (H. pylori) is a Gram-negative pathogenic bacterium that can selectively colonize the stomach epithelium for the life of the host without effective eradication. Long-term carriage of H. pylori significantly increases the risk of developing gastritis, duodenal ulcer, gastric adenocarcinoma and mucosa-associated lymphoid tissue (MALT) lymphoma [1–4]. Although several studies have linked H. pylori infection to cardiovascular diseases, neurodegenerative disorders and diabetes mellitus, the association between H. pylori and extragastric diseases remains a matter of debate [5–7].
Despite important progress made in the management, the global emergence of H. pylori antibiotic resistance raises concerns about the efficacy of antibiotic therapy and requires development of new therapeutic strategies [8]. Developing an effective, safe and immunogenic vaccine against H pylori might be a smart strategy. Research on H. pylori vaccine for human use has been ongoing over the past 30 years. To date, the results of all the clinical vaccine trials are disappointing, except for a recent study of a three-dose oral recombinant H pylori vaccine in children [9, 10]. However, there is still a long way to go for this vaccine before being put in the market. Hence, understanding of the molecular pathogenic mechanism of H. pylori and its interaction with host is of crucial significance to the development of novel therapeutic strategies.
Many human diseases are the result of transcriptional misregulation [11]. Previous studies have shown that microRNAs (miRNAs) play regulatory roles in many physiological and pathological processes through downregulating target genes [12]. Various current studies showed the involvement of specific strains of bacteria in different diseases, including prostate cancer, lung cancer and colon cancer using computational approaches [13–16]. Thus, the identified signature miRNAs associated with H. pylori infection will provide novel insights into its carcinogenesis and host mechanisms that are involved in bacterial elimination. Public repository for high-throughput gene expression data, such as the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) project, provides us diverse molecular data and clinical information of many diseases [17]. However, the predictive utility of these data for H. pylori infection analysis has not been systematically explored. In the present study, we aimed at identifying differentially expressed miRNAs associated with H. pylori infection from GEO data sets and investigated the interactions between H. pylori and host, and the possible damage caused by H. pylori infection.
Materials and methods
miRNAs expression data sets
The miRNA expression data sets used in this study were obtained from the GEO data sets, including GSE19769, GSE32174, GSE54397 and GSE23877 [18–21]. The data sets consisted of 31 H. pylori-negative samples and 41 H. pylori-positive samples covering gastritis, duodenal ulcer, gastric cancer and MALT lymphoma caused by H. pylori infection. The information of GEO data sets and patients used in this study was summarized in Table 1.
Table 1.
Descriptions of the GEO data sets and patients used in this study.
| Series accession | Country | Number of controls | Number of H. pylori carriers | Disease associated |
|---|---|---|---|---|
| GSE19769 | Japan | 10 | 9 | Gastritis |
| GSE23877 | Switzerland | 4 | 5 | MALT lymphoma |
| GSE32174 | Spain | 9 | 19 | Duodenal ulcer |
| GSE54397 | South Korea | 8 | 8 | Gastric cancer |
Duodenal ulcer is known as peptic ulcer. Peptic ulcer disease is a break in the lining of the stomach, first part of the small intestine or occasionally the lower esophagus.
Screening for differentially expressed miRNAs
All collected expressed data were analyzed with GEO2R. The adjust P values were used to reduce the false-positive rate using Benjamini and Hochberg (false discovery rate) method by default. The adjusted P value < 0.05 and |logFC| > 1 were set as the cutoff criterion. Hierarchical clustering of differentially expressed miRNAs was carried out and visualized using MeV v4.8.1 [22].
Construction of miRNA-mRNA integrated network
Target genes for differentially expressed miRNAs were retrieved from the miRTarBase [23]. miRNA-mRNA interaction network was generated using Cytoscape 3.4.0 [24].
Protein–protein interaction networks and gene ontology enrichment analysis
The protein–protein interaction network was constructed using STRING 10.0 [25]. Confidence score ≥ 0.9 was set as the cutoff criterion. All of the networks were visualized and analyzed with Cytoscape 3.4.0. The degrees were calculated using Centiscape 2.2 [26]. Hub genes were identified by degree > 40 threshold. Metascape was used for the gene ontology (GO) enrichment analysis [27].
Disease connectivity analysis
Diseases enriched with H. polyri infection-associated genes were identified using the Comparative Toxicogenomic Database (Bonferroni-corrected P value < 0.05) [28].
Results
Identification of differentially expressed miRNAs associated with H. pylori infection
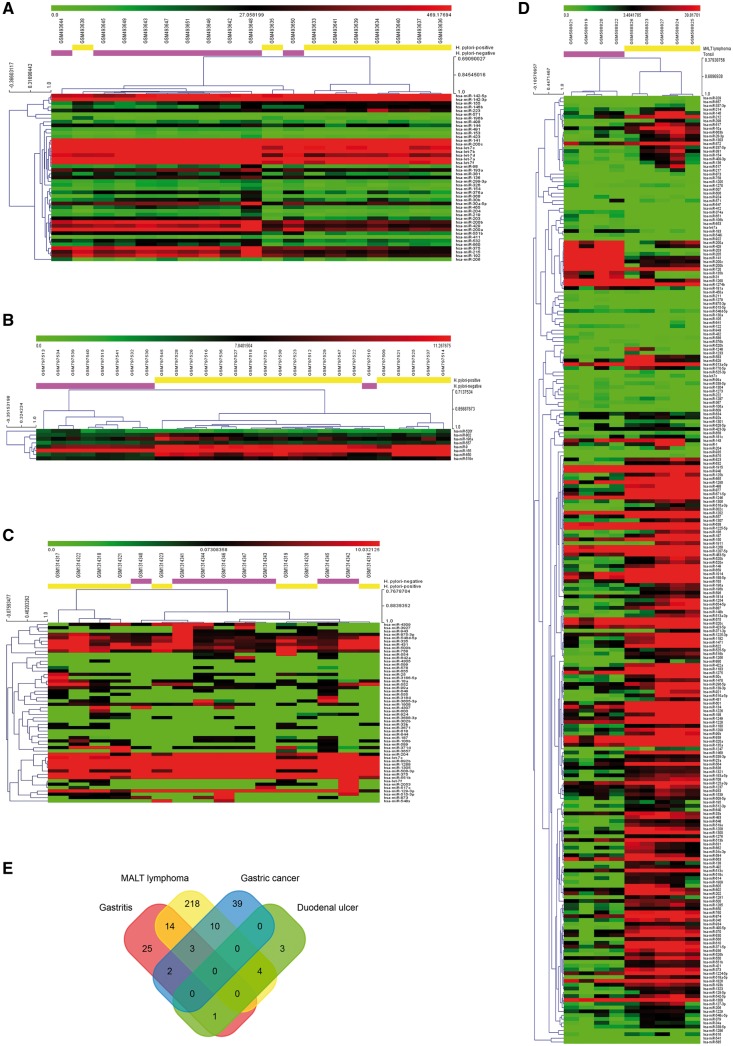
To identify aberrant miRNAs associated with H. pylori infection, we analyzed four public GEO data sets (GSE19769, GSE23877, GSE32174 and GSE54397). The derived data sets consisted of 31 H. pylori-negative samples and 41 H. pylori-positive samples from Asia and Europe (Supplementary Table S1). Four common H. pylori infection-related diseases (gastritis, duodenal ulcer, gastric cancer and MALT lymphoma) were included in this study. In total, 45, 249, 8 and 54 differentially expressed miRNAs were identified from GSE19769, GSE23877, GSE32174 and GSE54397, respectively (Figure 1A–D). As shown in the Venn diagram (Figure 1E), we filtered out 34 differentially expressed miRNAs that at least occurred in two data sets. The expression of these 34 miRNAs in the data sets was listed in Supplementary Table S2. miRNAs with the smallest P values will be the most reliable. Among these 34 miRNAs, 3 miRNAs (hsa-let-7c, hsa-miR-204 and hsa-miR-551 b) were overlapped in three data sets. These 34 miRNAs were considerate to be H. pylori infection related miRNAs.
Figure 1.
Identification of differentially expressed miRNAs associated with H. pylori infection. (A-D) Heat maps of miRNA microarray analysis. Each column contains the data from a specific gene and each row contains data from single sample. Upregulated expression levels are in red and downregulated in green. The fold-changes (log2 transformed) of miRNA expression in H. pylori-negative samples versus H. pylori-positive samples from GSE19769 (A; gastritis), GSE32174 (B; duodenal ulcer), GSE54397 (C; gastric cancer), GSE23877 (D; MALT lymphoma). In total, 45 249 8 and 54 differentially expressed miRNAs were identified from GSE19769, GSE23877, GSE32174 and GSE54397, respectively. (E) Venn diagram of overlap of differentially expressed miRNAs. We filtered out 34 differentially expressed miRNAs that at least occurred in two datasets. Among these 34 miRNAs, hsa-let-7c, hsa-miR-204 and hsa-miR-551 b were overlapped in three data sets.
Targets prediction and miRNAs-targets interaction
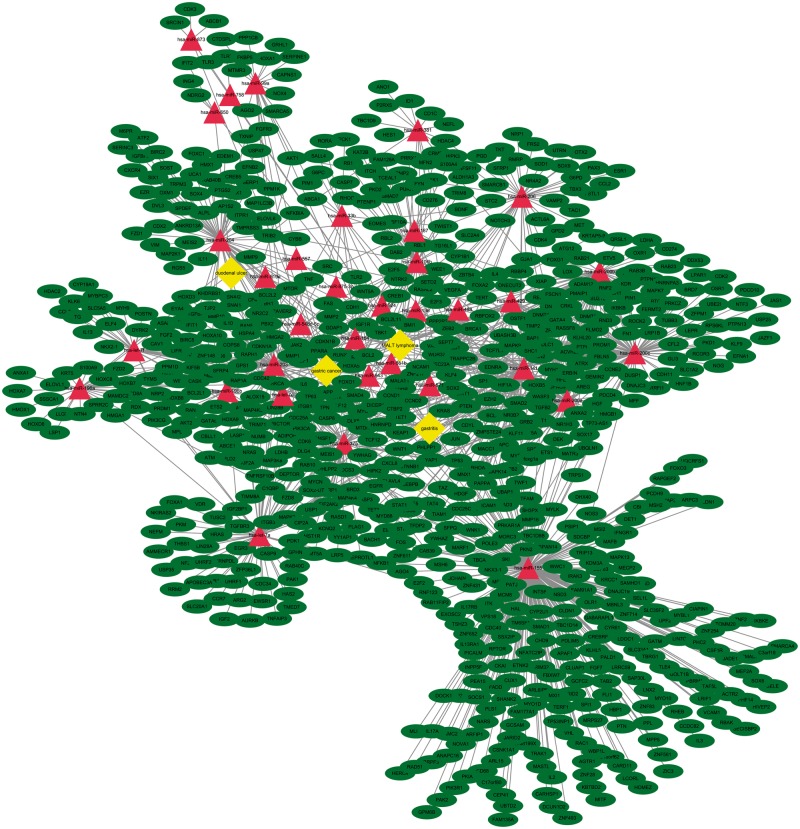
As an miRNA exerts its biological regulatory function through all of its target genes at the posttranscriptional level, to understand the potential role of these 34 miRNAs in the pathogenesis of H. pylori, we obtained 765 highly reliable miRNA target genes from miRTarBase data set. These target genes are validated by at least one of strongly experimental methods (reporter assay, Western blot or quantitative real-time poymerase chain reaction). Six miRNAs (hsa-miR-551 b, hsa-miR-557, hsa-miR-571, hsa-miR-875-3p, hsa-miR-924 and hsa-miR-943) have no strong experimental evidences supported. We found that miR-155 regulates the most target genes (247 genes) in the miRNA-mRNA regulatory network (Figure 2). Among these target genes, 25.9% genes have 2 or more miRNA predictive binding sites, especially BCL2 can be targeted by up to 9 of the 34 miRNAs, and 32 genes have 5 or more predictive miRNA binding sites (Table 2). These 765 miRNA target genes may play important roles during H. pylori colonization and persistent infection.
Figure 2.
Regulatory network between miRNA and their target genes. Target genes for differentially expressed miRNAs were retrieved from the miRTarBase. These target genes were validated by at least one of strongly experimental methods (reporter assay, Western blot or quantitative real-time polymerase chain reaction). We also linked these differentially expressed miRNAs to the diseases that were diagnosed in the patients. This network was generated using Cytoscape 3.4.0 and consisted of 4 H. pylori infection-related diseases, 28 miRNAs and 765 genes. Rectangle (yellow) represents diseases, ellipses (green) represents genes and triangle represents miRNAs.
Table 2.
Target genes with five or more predictive miRNA-binding sites
| Target genes | miRNA counts | miRNA |
|---|---|---|
| BCL2 | 9 | hsa-let-7a, hsa-miR-33b, hsa-miR-136, hsa-miR-200b, hsa-miR-200c, hsa-miR-204, hsa-miR-206, hsa-miR-375, hsa-miR-429 |
| MYC | 7 | hsa-let-7a, hsa-let-7c, hsa-let-7f, hsa-miR-33b, hsa-miR-106b, hsa-miR-155, hsa-miR-429 |
| ZEB2 | 7 | hsa-miR-141, hsa-miR-154, hsa-miR-200a, hsa-miR-200b, hsa-miR-200c, hsa-miR-203, hsa-miR-429 |
| MALAT1 | 7 | hsa-miR-141, hsa-miR-200a, hsa-miR-200b, hsa-miR-200c, hsa-miR-204, hsa-miR-375, hsa-miR-429 |
| HMGA2 | 6 | hsa-let-7a, hsa-let-7c, hsa-miR-33b, hsa-miR-154, hsa-miR-196a, hsa-miR-204, |
| CCND2 | 6 | hsa-let-7a, hsa-miR-106b, hsa-miR-154, hsa-miR-155, hsa-miR-203, hsa-miR-206, |
| CDKN1A | 6 | hsa-let-7a, hsa-let-7f, hsa-miR-106b, hsa-miR-196a, hsa-miR-203, hsa-miR-519e |
| VEGFA | 6 | hsa-miR-106b, hsa-miR-200b, hsa-miR-200c, hsa-miR-203, hsa-miR-206, hsa-miR-429 |
| PTEN | 6 | hsa-miR-106b, hsa-miR-141, hsa-miR-155, hsa-miR-200a, hsa-miR-200c, hsa-miR-429 |
| ZEB1 | 6 | hsa-miR-33b, hsa-miR-141, hsa-miR-200a, hsa-miR-200b, hsa-miR-200c, hsa-miR-429 |
Functional enrichment analysis of miRNA target genes
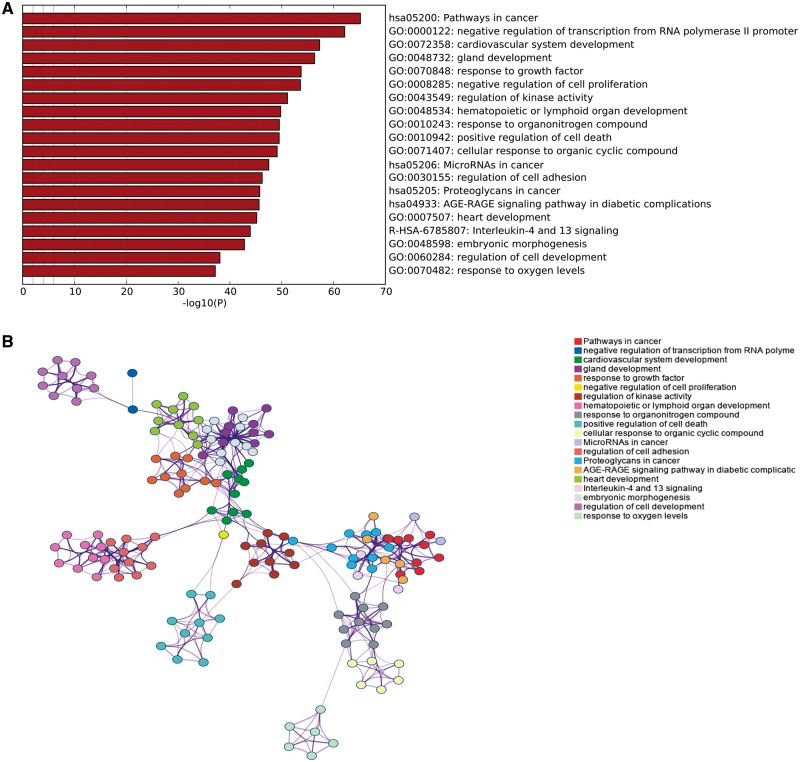
Next, we performed GO and KEGG pathway enrichment analysis using Metascape for 765 H. pylori infection-associated genes, and the most significantly enriched gene sets were pathways in cancer (hsa05200) and negative regulation of transcription from RNA polymerase II promoter (GO: 0000122) (Figure 3A). This analysis also revealed that H. pylori infection was largely related to cardiovascular system development (GO: 0072358), gland development (GO: 0048732), hematopoietic or lymphoid organ development (GO: 0048534), proteoglycans in cancer (hsa05205) and heart development (GO: 0007507). Interestingly, all these biological processes identified with a significant P value were closely interrelated except for response to oxygen levels (GO: 0070482) (Figure 3B). These results indicate that H. pylori infection could negatively regulate the synthesis of mRNA, many of which may influence the development of cancer and other human diseases.
Figure 3.
Functional enrichment analysis of miRNA target genes. (A) Top 20 clusters from Metascape pathway enrichment analysis of the 765 H. pylori infection-associated genes. Length of bars represent log10 (P value) based on the best-scoring term within each cluster. (B) Relationships among these top 20 clusters enrichment terms displayed as a network analyzed by Metascape. Nodes of the same color belonged to the same cluster. Terms with a similarity score > 0.3 were linked by an edge. The network was visualized with Cytoscape with ‘force-directed’ layout and with edge bundled for clarity.
Disease connectivity analysis
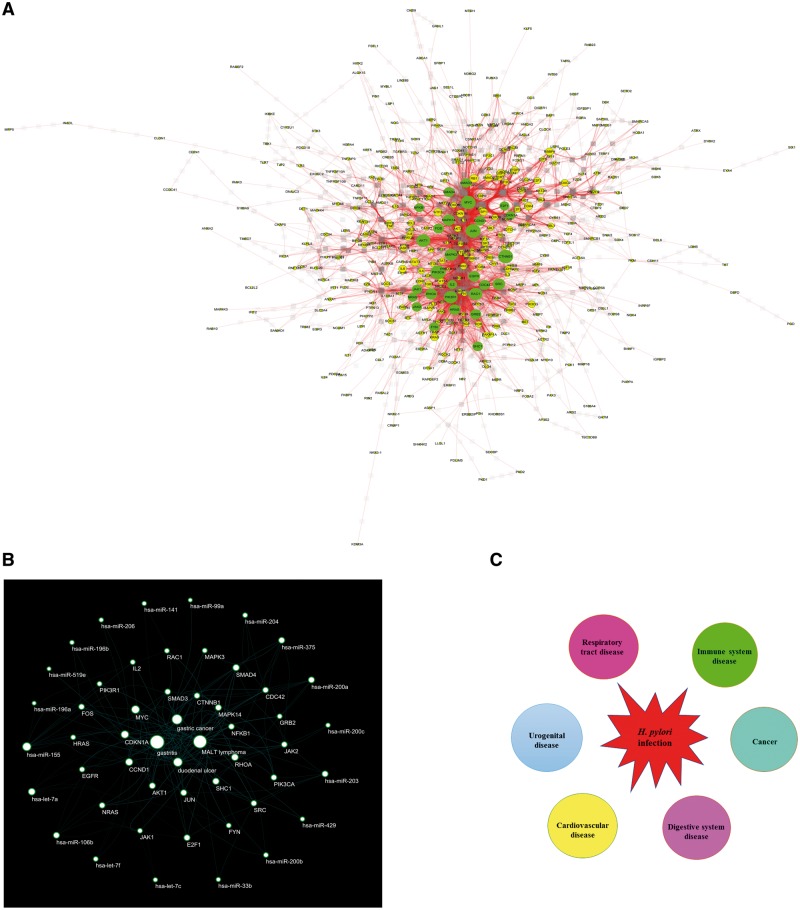
To reduce interference of unrelated genes and the complexity of the list of H. pylori infection associated genes, 28 genes were identified as hub genes (Figure 4A and Table 3). We found that these hub genes could interact with each other and are target of 21 of 28 H. pylori infection associated miRNAs (Figure 4B). Disease connectivity analysis of these 28 hub genes showed H. pylori infection significantly associated with cancer, digestive system disease, urogenital disease, cardiovascular disease, respiratory tract disease and immune system disease (Figure 4C and Table 4). H. pylori infection is also strongly associated with blood disease, lymphatic disease, endocrine system disease, nervous system disease and skin disease (Supplementary Table S3). Taken together, these results demonstrate that H. pylori colonizes the gastric epithelium, but beyond gastric diseases.
Figure 4.
Hub genes and disease connectivity analysis. (A) The protein–protein interaction network of 765 genes was generated using STRING (confidence score ≥ 0.9) and visualized with Cytoscape 3.4.0. The node properties, such as degree, were calculated using Centiscape 2.2. In total, 28 hub genes were identified according to degree > 40 threshold. (B) STRING-generated interaction network among the 28 hub genes and miRNA-mRNA interaction predicted with miRTarBase were described in ‘Materials and methods’ section. Then miRNA–hub gene–disease network was visualized with Cytoscape 3.4.0. This network consists of 4 H. pylori infection-related diseases, 21 miRNAs and 28 genes. (C) Diseases or conditions enriched with H. pylori infection associated genes were identified using the ‘set analyzer’ tool of the Comparative Toxicogenomic Database with Bonferoni-corrected P value < 0.05. The corrected P value is calculated by the hypergeometric distribution and adjusted for multiple testing using the Bonferroni method. Diseases with the smallest P values mean the most reliable connectivity between a disease and genes analyzed.
Table 3.
The degree, betweenness and closeness of 28 hub genes analyzed with Centiscape 2.2
| Name | Degree | Betweenness | Closeness |
|---|---|---|---|
| PIK3CA | 74 | 0.03170252 | 0.46114519 |
| CTNNB1 | 72 | 0.08612305 | 0.46590909 |
| JUN | 72 | 0.06560389 | 0.46784232 |
| MYC | 71 | 0.10257151 | 0.47978723 |
| AKT1 | 66 | 0.06789642 | 0.47423764 |
| PIK3R1 | 65 | 0.0222343 | 0.45145145 |
| RAC1 | 64 | 0.05136969 | 0.4500998 |
| MAPK3 | 61 | 0.04631864 | 0.45694022 |
| SRC | 58 | 0.02860764 | 0.44302554 |
| MAPK14 | 57 | 0.04698078 | 0.46399177 |
| RHOA | 56 | 0.03652856 | 0.44521224 |
| EGFR | 53 | 0.04216526 | 0.44042969 |
| HRAS | 53 | 0.02578029 | 0.42993327 |
| GRB2 | 51 | 0.01993267 | 0.4187558 |
| FOS | 49 | 0.0236865 | 0.44831014 |
| CDC42 | 49 | 0.03134559 | 0.41759259 |
| SMAD4 | 49 | 0.02615774 | 0.42110177 |
| CCND1 | 48 | 0.02644287 | 0.44565217 |
| SHC1 | 47 | 0.01052397 | 0.41605166 |
| FYN | 47 | 0.01772007 | 0.42427093 |
| JAK2 | 46 | 0.01814914 | 0.40851449 |
| JAK1 | 45 | 0.0171368 | 0.42467043 |
| SMAD3 | 45 | 0.02195681 | 0.42627599 |
| IL2 | 44 | 0.01386536 | 0.42993327 |
| E2F1 | 43 | 0.01451441 | 0.40231936 |
| NFKB1 | 42 | 0.03770201 | 0.43574879 |
| CDKN1A | 42 | 0.01303738 | 0.4254717 |
| NRAS | 42 | 0.00935047 | 0.40412186 |
Table 4.
Top 25 enriched diseases of 28 hub genes analyzed with the Comparative Toxicogenomics Database
| Disease name | Disease categories | P value | Corrected P value | Annotated genes quantity | Annotated genes | Genome frequency |
|---|---|---|---|---|---|---|
| Digestive system neoplasms | Cancer digestive system disease | 4.16E-30 | 2.55E-27 | 22 | AKT1 CCND1 CDKN1A CTNNB1 E2F1 EGFR FOS FYN HRAS IL2 JAK2 JUN MAPK14 MAPK3 MYC NFKB1 NRAS PIK3CA RAC1 RHOA SMAD4 SRC | 1118/42 703 genes: 2.62% |
| Neoplasms by histologic type | Cancer | 6.77E-30 | 4.16E-27 | 24 | AKT1 CCND1 CDC42 CDKN1A CTNNB1 E2F1 EGFR FOS FYN HRAS IL2 JAK2 JUN MAPK14 MAPK3 MYC NFKB1 NRAS PIK3CA PIK3R1 RAC1 RHOA SMAD3 SMAD4 | 1742/42 703 genes: 4.08% |
| Neoplasms by site | Cancer | 6.19E-29 | 3.80E-26 | 25 | AKT1 CCND1 CDC42 CDKN1A CTNNB1 E2F1 EGFR FOS FYN HRAS IL2 JAK2 JUN MAPK14 MAPK3 MYC NFKB1 NRAS PIK3CA PIK3R1 RAC1 RHOA SMAD3 SMAD4 SRC | 2325/42 703 genes: 5.44% |
| Cardiovascular diseases | Cardiovascular disease | 6.85E-27 | 4.21E-24 | 21 | AKT1 CCND1 CDKN1A CTNNB1EGFR FOS HRAS IL2 JAK2 JUN MAPK14 MAPK3 MYC NFKB1 NRAS PIK3CA RAC1 SHC1 SMAD3 SMAD4 SRC | 1267/42 703 genes: 2.97% |
| Pathologic processes | Pathology (process) | 3.98E-26 | 2.44E-23 | 21 | AKT1 CCND1 CDC42 CDKN1A CTNNB1 EGFR FOS HRAS IL2 JAK2 JUN MAPK3 MYC NFKB1 NRAS PIK3CA PIK3R1 RAC1 RHOA SMAD4 SRC | 1378/42 703 genes: 3.23% |
| Pathological conditions, signs and symptoms | 5.74E-26 | 3.52E-23 | 24 | AKT1 CCND1 CDC42 CDKN1A CTNNB1 EGFR FOS FYN HRAS IL2 JAK2 JUN MAPK3 MYC | 2542/42 703 genes: 5.95% | |
| NFKB1 NRAS PIK3CA PIK3R1 RAC1 RHOA SHC1 SMAD3 SMAD4 SRC | ||||||
| Carcinoma | Cancer | 6.12E-26 | 3.76E-23 | 19 | AKT1 CCND1 CDC42 CDKN1A CTNNB1 E2F1 EGFR FOS HRAS JUN MAPK3 MYC NFKB1 NRAS PIK3CA PIK3R1 RAC1 RHOA SMAD4 | 895/42 703 genes: 2.10% |
| Neoplasms, glandular and epithelial | Cancer | 7.82E-26 | 4.80E-23 | 20 | AKT1 CCND1 CDC42 CDKN1A CTNNB1 E2F1 EGFR FOS HRAS IL2 JUN MAPK3 MYC NFKB1 NRAS PIK3CA PIK3R1 RAC1 RHOA SMAD4 | 1144/42 703 genes: 2.68% |
| Neoplasms | Cancer | 1.11E-25 | 6.85E-23 | 25 | AKT1 CCND1 CDC42 CDKN1A CTNNB1 E2F1 EGFR FOS FYN HRAS IL2 JAK2 JUN MAPK14 MAPK3 MYC NFKB1 NRAS PIK3CA PIK3R1 RAC1 RHOA SMAD3 SMAD4 SRC | 3141/42 703 genes: 7.36% |
| Digestive system diseases | Digestive system disease | 1.42E-24 | 8.72E-22 | 23 | AKT1 CCND1 CDKN1A CTNNB1 E2F1 EGFR FOS FYN HRAS IL2 JAK2 JUN MAPK14 MAPK3 MYC NFKB1 NRAS PIK3CA RAC1 RHOA SMAD3 SMAD4 SRC | 2419/42 703 genes: 5.66% |
| Gastrointestinal diseases | Digestive system disease | 2.73E-23 | 1.67E-20 | 18 | AKT1 CCND1 CDKN1A CTNNB1 EGFR FYN HRAS JAK2 JUN MAPK3 MYC NFKB1 NRAS PIK3CA RHOA SMAD3 SMAD4 SRC | 978/42703 genes: 2.29% |
| Gastrointestinal neoplasms | Cancer digestive system disease | 1.68E-21 | 1.03E-18 | 16 | AKT1 CCND1 CDKN1A CTNNB1 EGFR FYN HRAS JUN MAPK3 MYC NFKB1 NRAS PIK3CA RHOA SMAD4 SRC | 748/42 703 genes: 1.75% |
| Hemic and lymphatic diseases | 1.68E-21 | 1.03E-18 | 16 | AKT1 CCND1 CDC42 CTNNB1 FYN HRAS IL2 JAK2 MAPK14 MYC NRAS PIK3CA PIK3R1 RHOA SMAD4 SRC | 748/42 703 genes: 1.75% | |
| Liver neoplasms | Cancer digestive system disease | 2.04E-21 | 1.25E-18 | 14 | CCND1 CTNNB1 E2F1 EGFR FOS HRAS IL2 JAK2 JUN MAPK14 MAPK3 MYC PIK3CA RAC1 | 417/42 703 genes: 0.98% |
| Neoplastic processes | Cancer pathology (process) | 2.58E-21 | 1.59E-18 | 14 | CCND1 CTNNB1 EGFR FOS HRAS IL2 JUN MAPK3 MYC NFKB1 PIK3CA RAC1 RHOA SRC | 424/42 703 genes: 0.99% |
| Liver diseases | Digestive system disease | 4.74E-21 | 2.91E-18 | 19 | CCND1 CDKN1A CTNNB1 E2F1 EGFR FOS FYN HRAS IL2 JAK2 JUN MAPK14 MAPK3 MYC NFKB1 PIK3CA RAC1 SMAD3 SMAD4 | 1624/42 703 genes: 3.80% |
| Intestinal diseases | Digestive system disease | 1.42E-19 | 8.70E-17 | 14 | AKT1 CCND1 CDKN1A CTNNB1 EGFR JAK2 JUN MYC NFKB1 NRAS PIK3CA SMAD3 SMAD4 SRC | 564/42 703 genes: 1.32% |
| Female urogenital diseases | Urogenital disease (female) | 2.03E-19 | 1.25E-16 | 17 | AKT1 CCND1 CDKN1A CTNNB1 EGFR FOS HRAS IL2 JAK2 MAPK3 MYC NFKB1 PIK3CA PIK3R1 RHOA SMAD3 SRC | 1289/42 703 genes: 3.02% |
| Colonic diseases | Digestive system disease | 4.14E-19 | 2.54E-16 | 13 | AKT1 CCND1 CDKN1A CTNNB1 EGFR JAK2 JUN MYC NFKB1 NRAS PIK3CA SMAD4 SRC | 441/42 703 genes: 1.03% |
| Lung neoplasms | Cancer respiratory tract disease | 5.55E-19 | 3.41E-16 | 13 | AKT1 CCND1 CDKN1A CTNNB1 EGFR FOS HRAS IL2 JUN MAPK14 MAPK3 MYC PIK3CA | 451/42 703 genes: 1.06% |
| Respiratory tract neoplasms | Cancer respiratory tract disease | 6.41E-19 | 3.93E-16 | 13 | AKT1 CCND1 CDKN1A CTNNB1 EGFR FOS HRAS IL2 JUN MAPK14 MAPK3 MYC PIK3CA | 456/42 703 genes: 1.07% |
| Thoracic neoplasms | Cancer | 6.59E-19 | 4.05E-16 | 13 | AKT1 CCND1 CDKN1A CTNNB1 EGFR FOS HRAS IL2 JUN MAPK14 MAPK3 MYC PIK3CA | 457/42 703 genes: 1.07% |
| Female urogenital diseases and pregnancy complications | 1.16E-18 | 7.10E-16 | 17 | AKT1 CCND1 CDKN1A CTNNB1 EGFR FOS HRAS IL2 JAK2 MAPK3 MYC NFKB1 PIK3CA PIK3R1 RHOA SMAD3 SRC | 1430/42 703 genes: 3.35% | |
| Immune system diseases | Immune system disease | 1.73E-18 | 1.06E-15 | 16 | AKT1 CCND1 CDC42 CDKN1A CTNNB1 FOS FYN IL2 JUN MYC NFKB1 NRAS PIK3CA PIK3R1 RHOA SMAD3 | 1158/42 703 genes: 2.71% |
| Colorectal neoplasms | Cancer digestive system disease | 3.89E-18 | 2.39E-15 | 12 | AKT1 CCND1 CDKN1A CTNNB1 EGFR JUN MYC NFKB1 NRAS PIK3CA SMAD4 SRC | 369/42 703 genes: 0.86% |
Discussion
This integrated study of 31 H. pylori-negative samples and 41 H. pylori-positive samples provides numerous novel insights into H. pylori infection associated diseases and delineates multiple potential opportunities for therapeutic intervention. Several of the differentially expressed miRNAs identified in this study, particularly hsa-let-7c, hsa-miR-204 and hsa-miR-551 b, are amenable in principle to diagnostic biomarkers and therapeutic targets. Previous studies demonstrated that downregulation of has-miR-204 promoted epithelial–mesenchymal transition in H.pylori-induced gastric cancer by targeting SOX4 [29]. Decreased hsa-let-7c significantly associated with the increasing severity of the histological lesions in H. pylori-related carcinogenesis, but the molecular mechanisms are still largely unclear [30]. So far, no evidence has shown that hsa-miR-551 b was clearly associated with H. pylori infection and may serve as a novel H. pylori infection-related miRNA biomarker in clinical use.
In this study, we also constructed miRNA-target mRNA network based on mirTarbase database. The result showed that miR-155, miR-203, miR-204 and miR-200 family (miR-200a, miR-200 b, miR-200c, miR-141 and miR-429) were central miRNAs. Eleven target genes (BCL2, MYC, ZEB2, MALAT1, HMGA2, CCND2, CDKN1A, VEGFA, PTEN and ZEB1) most connected with differentially expressed miRNAs in this network. The data here suggest that these miRNAs and target genes may also exert important biological functions in the progression of H. pylori infection. As expected, GO enrichment analysis of miRNA target genes showed that the most significantly enriched gene sets were pathways in cancer and negative regulation of transcription from RNA polymerase II promoter. We also identified a number of previously unappreciated biological processes and pathways closely related to H. pylori infection, such as hematopoietic or lymphoid organ development, heart development.
A total of 28 target genes were identified as hubs that occupy central positions in protein–protein interaction network. Several hubs have shown important regulatory functions during H. pylori infection. For example, H. pyloriCagA promotes the expression of the proto-oncogenes c-JUN and regulates FasL-dependent T-cell apoptosis [31, 32]. c-Myc acts as an important mediator of macrophage activation and contributes to the mucosal inflammatory response to H. pylori infection by its transactivation of the ornithine decarboxylase promoter [33]. However, we are still largely unknown about the functional role of these hub genes identified in this study, such as CTNNB1 and RHOA, providing a new direction in the future research.
Disease connectivity analysis of 28 hub genes predicted that 228 items of human diseases are related to H. pylori infection, including cancer, cardiovascular disease, digestive system disease, urogenital disease and immune system disease, many of which are overlapped with previous reports [34–37]. H. pylori infection is also significantly associated with hemic and lymphatic diseases, respiratory tract disease, endocrine system disease, nervous system disease and skin disease. The possible mechanism includes molecular mimicry, epithelial injury and chronic inflammation [38–40]. However, evidence now supports the tenet that bacterial dysbiosis plays an important role in human disease [41]. H. pylori infection not only directly injures host cells but also changes the gastric environment and mucosal barrier, initiating the inflammatory cascades and altering the microbiota [42]. These findings demonstrate the importance of examining H. pylori infection in the treatment of other extra-gastric diseases.
Conclusions
Our current prediction work demonstrated a new method to predict pathogen–human disease connectivity based on miRNA-mRNA interaction network. Furthermore, this integrated network analysis revealed a profound impact of H. pylori infection on various human extra-gastric diseases. The curated omics data may also provide an important resource for future studies.
Key Points.
Comprehensive analysis of H. pylori infection-associated miRNAs.
Integrated analysis of H. pylori infection-associated human extra-gastric diseases.
A new method to predict pathogen–human disease connectivity based on miRNA-mRNA interaction network.
Supplementary Material
Jue Yang is a PhD in the State Key Laboratory of Functions and Applications of Medicinal Plants, Guizhou Medical University, Guiyang, China.
Hui Song is a technician in the Key Laboratory of Endemic and Ethnic Diseases (Guizhou Medical University), Ministry of Education, Guiyang, China, the Key Laboratory of Medical Molecular Biology (Guizhou Medical University), Guizhou Province, Guiyang, China.
Kun Cao is a PhD in the Department of General Surgery, Affiliated Hospital of Guizhou Medical University, Guiyang, China.
Jialei Song is a PhD in the Laboratory of Cell Biochemistry and Topogenic Regulation, College of Bioengineering and Faculty of Sciences, Chongqing University, Chongqing, China.
Jianjiang Zhou is a Professor in the Key Laboratory of Endemic and Ethnic Diseases (Guizhou Medical University), Ministry of Education, Guiyang, China, the Key Laboratory of Medical Molecular Biology (Guizhou Medical University), Guizhou Province, Guiyang, China.
Funding
This study was supported by the National Natural Science Foundation of China (grant numbers 31560326, 31760328), and the Science and Technology Foundation of Guizhou Province (grant number Qiankehepintairencai [2017] 5652).
References
- 1. The EUROGAST Study Group. An international association between Helicobacter pylori infection and gastric cancer. Lancet 1993;341:1668. [PubMed] [Google Scholar]
- 2. Tjandra JJ. Helicobacter pylori in peptic ulcer disease. Del Med J 1994;66:557–8. [PubMed] [Google Scholar]
- 3. Parsonnet J, Friedman GD, Vandersteen DP, et al. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med 1991;325(16):1127–31. [DOI] [PubMed] [Google Scholar]
- 4. Warren JR, Marshall B.. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet 1983;1:1273–5. [PubMed] [Google Scholar]
- 5. Lai CY, Yang TY, Lin CL, et al. Helicobacter pylori infection and the risk of acute coronary syndrome: a nationwide retrospective cohort study. Eur J Clin Microbiol Infect Dis 2015;34(1):69–74. [DOI] [PubMed] [Google Scholar]
- 6. Wang XL, Zeng J, Feng J, et al. Helicobacter pylori filtrate impairs spatial learning and memory in rats and increases bamyloid by enhancing expression of presenilin-2. Front Aging Neurosci 2014;6:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang GH, Wu JS, Yang YC, et al. Gastric Helicobacter pylori infection associated with risk of diabetes mellitus, but not prediabetes. J Gastroenterol Hepatol 2014;29(10):1794–9. [DOI] [PubMed] [Google Scholar]
- 8. Megraud F, Coenen S, Versporten A, et al. Helicobacter pylori resistance to antibiotics in Europe and its relationship to antibiotic consumption. Gut 2013;62(1):34–42. [DOI] [PubMed] [Google Scholar]
- 9. Czinn SJ, Blanchard T.. Vaccinating against Helicobacter pylori infection. Nat Rev Gastroenterol Hepatol 2011;8(3):133–40. [DOI] [PubMed] [Google Scholar]
- 10. Zeng M, Mao XH, Li JX, et al. Efficacy, safety, and immunogenicity of an oral recombinant Helicobacter pylori vaccine in children in China: a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2015;386(10002):1457–64. [DOI] [PubMed] [Google Scholar]
- 11. Lee TI, Young RA.. Transcriptional regulation and its misregulation in disease. Cell 2013;152(6):1237–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136(2):215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Khan S, Zakariah M, Rolfo C, et al. Prediction of Mycoplasma hominis proteins targeting in mitochondria and cytoplasm of host cells and their implication in prostate cancer etiology. Oncotarget 2017;8(19):30830–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Khan S, Zakariah M, Palaniappan S.. Computational prediction of Mycoplasma hominis proteins targeting in nucleus of host cell and their implication in prostate cancer etiology. Tumour Biol 2016;37(8):10805–13. [DOI] [PubMed] [Google Scholar]
- 15. Khan S, Imran A, Khan AA, et al. Systems biology approaches for the prediction of possible role of chlamydia pneumoniae proteins in the etiology of lung cancer. PLoS One 2016;11(2):1:e0148530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Khan S. Potential role of Escherichia coli DNA mismatch repair proteins in colon cancer. Crit Rev Oncol Hematol 2015;96(3):475–82. [DOI] [PubMed] [Google Scholar]
- 17. Edgar R, Domrachev M, Lash AE.. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30(1):207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Matsushima K, Isomoto H, Inoue N, et al. MicroRNA signatures in Helicobacter pylori-infected gastric mucosa. Int J Cancer 2011;128(2):361–70. [DOI] [PubMed] [Google Scholar]
- 19. Lario S, Ramírez-Lázaro MJ, Aransay AM, et al. microRNA profiling in duodenal ulcer disease caused by Helicobacter pylori infection in a Western population. Clin Microbiol Infect 2012;18(8):E273–82. [DOI] [PubMed] [Google Scholar]
- 20. Chang H, Kim N, Park JH, et al. Different microRNA expression levels in gastric cancer depending on Helicobacter pylori infection. Gut Liver 2015;9(2):188–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Craig VJ, Cogliatti SB, Rehrauer H, et al. Epigenetic silencing of MicroRNA-203 dysregulates ABL1 expression and drives helicobacter-associated gastric lymphomagenesis. Cancer Res 2011;71(10):3616–24. [DOI] [PubMed] [Google Scholar]
- 22. Saeed AI, Sharov V, White J, et al. TM4: a free, open- source system for microarray data management and analysis. Biotechniques 2003;34:374–8. [DOI] [PubMed] [Google Scholar]
- 23. Chou CH, Shrestha S, Yang CD, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res 2018;46(D1):D296–302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13(11):2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 2013;41:D808–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Scardoni G, Petterlini M, Laudanna C.. Analyzing biological network parameters with CentiScaPe. Bioinformatics 2009;25(21):2857–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tripathi S, Pohl MO, Zhou Y, et al. Meta- and orthogonal integration of influenza “OMICs” data defines a role for UBR4 in virus budding. Cell Host Microbe 2015;18(6):723–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Davis AP, Grondin CJ, Johnson RJ, et al. The comparative toxicogenomics database: update 2017. Nucleic Acids Res 2017;45(D1):D972–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou X, Li L, Su J, et al. Decreased miR-204 in H. pylori-associated gastric cancer promotes cancer cell proliferation and invasion by targeting SOX4. PLoS One 2014;9(7):e101457. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30. Fassan M, Saraggi D, Balsamo L, et al. Let-7c down-regulation in Helicobacter pylori-related gastric carcinogenesis. Oncotarget 2016;7(4):4915–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Meyer-ter-Vehn T, Covacci A, Kist M, et al. Helicobacter pylori activates mitogen-activated protein kinase cascades and induces expression of the proto-oncogenes c-fos and c-jun. J Biol Chem 2000;275(21):16064–72. [DOI] [PubMed] [Google Scholar]
- 32. Sharma S, Wang J, Denning TL, et al. Reactive oxygen species stimulated by Helicobacter pylori trigger c-Jun/AP-1 activation that regulates Fas ligand (FasL)-dependent T-cell apoptosis. Gastroenterology 2002;122:A10. [Google Scholar]
- 33. Cheng Y, Chaturvedi R, Asim M, et al. Helicobacter pylori-induced macrophage apoptosis requires activation of ornithine decarboxylase by c-Myc. J Biol Chem 2005;280(23):22492–6. [DOI] [PubMed] [Google Scholar]
- 34. Uemura N. Helicobacter pylori infection and the development of gastric cancer in Japan. Nihon Rinsho 2003;61:25–9. [PubMed] [Google Scholar]
- 35. Patel P, Mendall MA, Carrington D, et al. Association of Helicobacter pylori and Chlamydia pneumoniae infections with coronary heart disease and cardiovascular risk factors. BMJ 1995;311(7007):711–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lai LH, Sung JJ.. Helicobacter pylori and benign upper digestive disease. Best Pract Res Clin Gastroenterol 2007;21(2):261–79. [DOI] [PubMed] [Google Scholar]
- 37. Al-Marhoon MS. Is there a role for Helicobacter pylori infection in urological diseases? Urol J 2008;5(3):139–43. [PubMed] [Google Scholar]
- 38. Lamb DJ, El-Sankary W, Ferns GA.. Molecular mimicry in atherosclerosis: a role for heat shock proteins in immunisation. Atherosclerosis 2003;167(2):177–85. [DOI] [PubMed] [Google Scholar]
- 39. Buzás GM. Metabolic consequences of Helicobacter pylori infection and eradication. World J Gastroenterol 2014;20(18):5226–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Graham DY. Helicobacter pylori update: gastric cancer, reliable therapy, and possible benefits. Gastroenterology 2015;148(4):719–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schwabe RF, Jobin C.. The microbiome and cancer. Nat Rev Cancer 2013;13(11):800–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fox JG, Wang TC.. Inflammation, atrophy, and gastric cancer. J Clin Invest 2007;117(1):60–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.