Abstract
The 5-year survival rate of hepatocellular carcinoma (HCC) is <20%; thus, identifying new potential therapeutic targets or novel biomarkers for prognosis prediction is crucial. The present study aimed to screen hub genes by constructing protein-protein interaction (PPI) subnetworks using topological analysis methods, as well as reveal their clinical significance through big data analytics and their association with the clinicopathological features. Firstly, the PPI subnetworks were constructed using four topological analysis methods, including the MCC, DMNC, MNC and degree methods, to obtain 6 hub genes. Subsequently, the hub gene cancer/testis antigen 2 (CTAG2), which affects the prognosis of HCC (overall survival, P=0.035), was acquired by analysing clinical data in The Cancer Genome Atlas database. Meanwhile, CTAG2 expression was significantly associated with the age at diagnosis (P=0.003), T stage (P=0.028), TNM stage (P=0.028) and α-fetoprotein (AFP) expression (P=0.045). Further immunohistochemical analysis of samples collected in our hospital revealed that the expression level of CTAG2 in 46 HCC tissues was significantly higher in comparison with that in paired adjacent tissues. The clinical data indicated that the expression of CTAG2 was significantly correlated with the hepatitis B virus status (P=0.010) and AFP expression (P=0.004). These results were then found to be consistent with the results of big data analytics. Furthermore, Gene Set Enrichment Analysis demonstrated that the function of CTAG2 in HCC may be associated with the cell cycle. Taken together, these findings suggest that CTAG2 may serve as a new potential therapeutic target for HCC patients.
Keywords: hepatocellular carcinoma, topological analysis, cancer/testis antigen 2, therapeutic target, prognosis
Introduction
The 5-year mortality rate of liver cancer ranks third among all types of tumors, and the incidence in female and male patients under the age of 40 years has been increasing annually in recent years (1). Hepatocellular carcinoma (HCC) is the most common primary malignant liver tumor, accounting for approximately 70–85% of all cases of primary liver cancer (2). HCC metastasis and relapse commonly occur, and lack of effective therapeutic strategies is the biggest obstacle to its treatment and favourable prognosis (3). Therefore, it is urgent to identify prognostic biomarkers or potential therapeutic targets to improve the survival rate of HCC patients.
Currently, there are two conventional research strategies of screening for biomarkers or potential therapeutic targets of liver cancer. The first strategy is to perform confirmatory tests on previously reported proteins that influence cell proliferation and prolong cell growth cycle in HCC cell line. For instance, it has been reported that transforming growth factor-α/epidermal growth factor receptor aberrant expression is associated with the progression of liver tumors (4). The second strategy involves screening therapeutic targets by analysing copy number changes and gene expression profiles in limited tumor tissue samples collected at medical institutions. For instance, it has been reported that overexpression of vascular endothelial growth factor A in HCC suggests a non-cell-autonomous mechanism of oncogene activation (5). However, both of these strategies have limitations. The first strategy is the reconfirmation of the reported proteins and studies using the second screening strategy will be influenced by the lack of sample capacity.
The Cancer Genome Atlas (TCGA) database contains a large number of gene microarray data collected from cancer patients. Several previous studies have used bioinformatics analysis of such gene microarray data in order to identify key genes in HCC (6–8). In contrast to these earlier studies, the present study focuses on analysing the protein-protein interaction (PPI) networks using a number of different topological analysis methods to identify the hub genes. The use of different topological analysis methods can provide more biological information due to different prominent topological features or scoring strategies.
In the current study, based on large-scale medical data on HCC extracted from the TCGA database, genome wide data were obtained by bioinformatics analysis, and then hub genes were identified among differentially expressed genes (DEGs) and PPI networks (9). Next, hub genes associated with clinical prognosis and pathological parameters of patients were screened. Furthermore, Gene Set Enrichment Analysis (GSEA) was used to investigate the Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathways associated with the high CTAG2 expression group, which may potentially provide hints for future research into the molecular mechanisms underlying HCC. Overall, the aim of the present study was to establish an effective method for screening potential therapeutic targets.
Materials and methods
Data processing
RNA sequencing (RNA-Seq) and clinical data were downloaded from TCGA database (https://portal.gdc.cancer.gov; accessed on January 18, 2018), including 374 HCC samples and 50 paired paracancerous samples. The present study adheres to TCGA publication guidelines and data access policies. The open source software Bioconductor was used for bioinformatics analysis, and EdgerR package (version 3.4) was downloaded from Bioconductor and applied to screen DEGs. DEGs meeting the criteria of P<0.05 and |log2 fold change|≥2 were considered as statistically significant. Next, a volcano plot was used to present these DEGs. Gene Ontology (GO) analysis of the significant DEGs was also performed based on DAVID tools (https://david.ncifcrf.gov) with a cut-off criteria of P<0.05.
Building the protein-protein interaction (PPI) network and identifying hub genes. Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; http://string.embl.de/) is a database used to search known and predicted interactions between proteins. The interaction between proteins includes direct physical interaction and indirect functional correlation. The present study focused on the first 1,000 upregulated DEGs with the most significant difference, and these were analysed with the STRING online software in order to construct the PPI network. Network visualization and analysis were performed with Cytoscape software (version 3.5.1) (10), using the cytoHubba plug-in to sort biological network nodes. Specifically, four of the most accurate topological analysis methods (11), including the maximal clique centrality (MCC), density of maximum neighbourhood component (DMNC), maximum neighbourhood component (MNC) and degree methods, were applied to construct the subnetworks. The topological characteristics of each topological analysis were different, and therefore the scores are different. Six hub genes were obtained from the intersection of the top 10 genes scored by each of the four topological analyses.
Patients and tissue specimens
Tissue samples, including 51 tumor and 51 paired adjacent tissue specimens, were collected from patients at the Department of Hepatobiliary Surgery of the First Affiliated Hospital of China Medical University (Shenyang, China) between March 2013 and December 2015. All patients were diagnosed with HCC and had not received treatment with chemotherapy, radiotherapy or immunotherapy prior to surgery. The study was approved by the Medical Ethics Committee of China Medical University. The Ethics Committee waived the need for the patients to sign a written informed consent due to the retrospective nature of this study. Prior to tissue sample collection, the patients had been informed that their tissue samples would be used for diagnosis and scientific research for clinical treatment at the hospital. Tumor staging was performed by two pathologists, according to the American Joint Committee on Cancer guidelines (version 8, 2017) (12). Primary tumor staging was evaluated in 47 of the HCC patients as follows: T1 (n=6), T2 (n=25), T3 (n=15) and T4 (n=1). A total of 4 medical records did not record the T-stage of the patient. In addition, the present study excluded five patients whose samples had slipped off the microscope slide. So only 46 of 51 patients were in accordance with the inclusion criteria for the patients' tissues to be evaluated via IHC. Clinicopathological data were retrospectively retrieved from the medical records of patients.
Tissue microarray (TMA) and immunohistochemistry (IHC)
All 102 collected tissues were embedded in paraffin blocks, and the representative positions, the location rich in cancer cells rather than a large number of interstitial cells, were marked by checking the slides of the haematoxylin and eosin. The 1.5-mm tissue cores were extracted from the representative position of each sample and carefully transferred to a TMA paraffin receptor block (Pathology Devices, Inc., San Diego, CA, USA). Sections (4 µm) obtained from the TMA blocks were mounted on poly-L-lysine-coated glass slides and prepared for IHC.
For IHC, the slices were deparaffinised and rehydrated, soaked in xylene for 30 min, and then gradually added to ethanol and distilled water for hydration. The slices were immersed in 0.01 M citrate buffer (pH 6.0) at high pressure of 80 kPa for 8 min. Next, 3% H2O2 was used to block endogenous peroxidase activity for 10 min at room temperature, followed by incubation with 10% normal goat serum at 37°C for 30 min to block the binding sites of nonspecific proteins. Subsequently, the slices were incubated overnight at 4°C with diluted primary antibody against CTAG2 (dilution, 1:400; cat. no. bs-6807R; Bioss, Inc., Woburn, MA, USA), and then incubated at 37°C for 30 min with diluted biotinylated secondary antibody (1:200; cat. no. KIT-0105R; MXB Biotechnologies). The slices were further incubated with streptavidin-horseradish peroxidase conjugate at 37°C for 30 min (LSAB kit; Dako, Glostrup, Demark) and dyed with DAB for 25 sec, following which any excess dyestuff was removed by PBS. In the negative control group, the anti-CTAG2 antibody was replaced by PBS, while other steps remained unchanged. For IHC, the slices were conducted under high pressure which made some samples fall off the glass, and so only 46 of 51 paired tissues could be evaluated by IHC.
Evaluation of IHC
The histochemical staining level of TMA blocks was evaluated with blind testing by two experienced clinical pathologists under a light microscope. It is known that CTAG2 is distributed throughout the cell, while a fraction of CTAG2 is highly enriched in a single punctate focus near the nucleus (13). Therefore, evaluation of CTAG2 positive staining was mainly conducted in the cytoplasm. In total, 10 visual fields was selected randomly in each slice under high magnification (×400), and 100 cells were counted in each field of view. Next, the percentage of positive cells was calculated and the mean value was obtained. The immunoreactive score (IRS) was obtained by multiplying the cell staining percentage (0–100%) by the staining intensity score (where a score of 0 indicated no staining, 1 indicated weak staining, 2 indicated moderate staining, and 3 indicated strong staining) (14). The obtained IRS ranged between 0 and 300%.
GSEA and KEGG pathway analysis
GSEA is a method used to analyse genome wide expression profiles at the level of gene sets, which include genes with the same common biological function (http://software.broadinstitute.org/gsea/index.jsp). In the present study, the genome wide expression profiles of 374 HCC patients from TCGA were classified into the high and low CTAG2 expression groups and then analysed with GSEA (version 3.0) (15).
KEGG is a database resource applied to understand high-level functions of biological systems (http://www.genome.jp/kegg). Kegg.v6.1.symbols.gmt that derived from KEGG database is a gene set in the GSEA website's MsigDB database. In the present study, GSEA was conducted by default weighted enrichment statistics, with 1,000 random combinations. The results of GSEA suggested that samples with high CTAG2 expression were significantly enriched in 7 KEGG pathways, according to the calculating criteria of P<0.05, normalized enrichment score (NES) of >1 and false discovery rate (FDR) of <25%.
Statistical analysis
The HCC patient samples from TCGA were divided into the high and low CTAG2 expression groups based on their median value. Survival was analysed by constructing Kaplan-Meier curves (log-rank test), while Cox regression analysis was used to performed univariable and multivariable analysis. Wilcoxon rank-sum test was used to analyse the IHC data. Receiver operating characteristic (ROC) curves were used to select the cut-off value for the expression of CTAG2. Pearson's χ2 test or Fisher's exact test was used to analyse the correlation between the expression of CTAG2 and the clinicopathological characteristics of HCC patients. All the aforementioned statistical tests were completed using SPSS software, version 16.0 (SPSS, Inc., Chicago, IL, USA). In all analyses, P<0.05 was considered as an indicator of a statistically significant difference.
Results
Screening for DEGs
Based on high-throughput RNA-Seq data from TCGA Liver Hepatocellular Carcinoma (TCGA LIHC) project, 5,516 DEGs were obtained, among which 4,750 were upregulated and 766 were downregulated in HCC. In order to visualise the DEGs, a volcano plot was constructed to view their distribution. The red and green points in the plot represent the DEGs with a significantly different expression (Fig. 1). Subsequently, GO analysis of the significant DEGs was performed using DAVID database tools. The results revealed that the significant DEGs are associated with the regulation of transcription and transcription corepressor activity (Table I). A previous study by Wong et al (16) has suggested that a complex of corepressor and proteins promotes cell cycle progression, which in turn promotes tumorigenesis. Therefore, the DEGs associated with transcription corepressor may contribute to tumorigenesis by affecting cell cycle progression.
Figure 1.
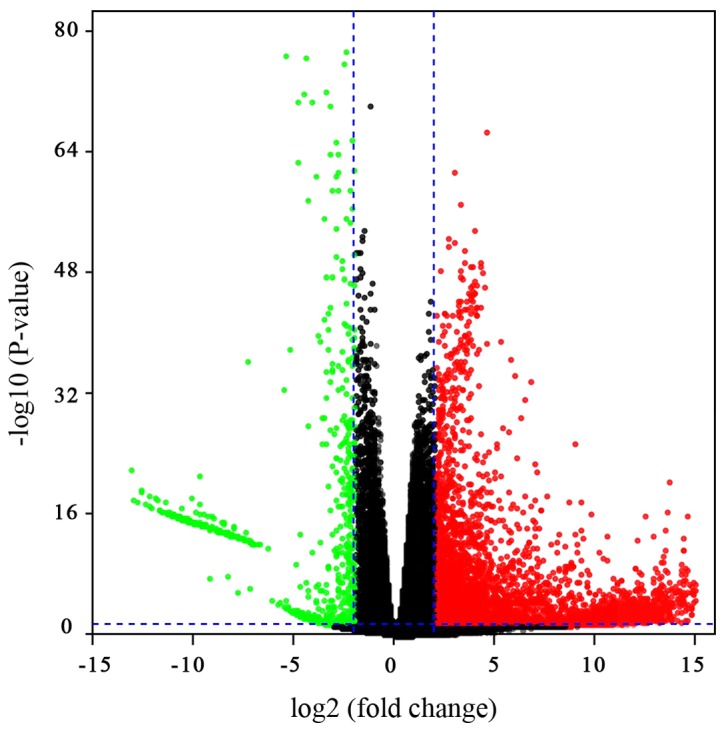
Volcano plot of the differentially expressed genes in patients with hepatocellular carcinoma included in The Cancer Genome Atlas database. The blue vertical lines represent 4-fold upregulation and downregulation, and the horizontal line represents a P-value of 0.05.
Table I.
GO analysis of the significant differentially expressed genes.
| Category | GO term | Count | P-value |
|---|---|---|---|
| BP | Regulation of transcription, DNA-templated | 34 | 0.001676 |
| BP | G-protein coupled receptor signaling pathway | 22 | 0.006075 |
| BP | Multicellular organism development | 16 | 0.003145 |
| BP | Spermatogenesis | 15 | 4.78×10−04 |
| BP | Proteolysis | 15 | 0.005383 |
| CC | Extracellular space | 35 | 2.22×10−04 |
| CC | Transcription factor complex | 9 | 0.004335 |
| CC | Extracellular region | 34 | 0.00815 |
| CC | Intermediate filament | 6 | 0.017465 |
| MF | Sequence-specific DNA binding | 22 | 3.06×10−04 |
| MF | G-protein coupled receptor activity | 20 | 0.001877 |
| MF | Olfactory receptor activity | 14 | 0.003646 |
| MF | Transcription co-repressor activity | 8 | 0.01541 |
| MF | Serine-type endopeptidase activity | 9 | 0.016567 |
Summary of the top 5 GO analysis of BP and MF. GO, Gene Ontology; BP, biological process; CC, cellular component; MF, molecular function.
Establishment of PPI network and hub gene selection
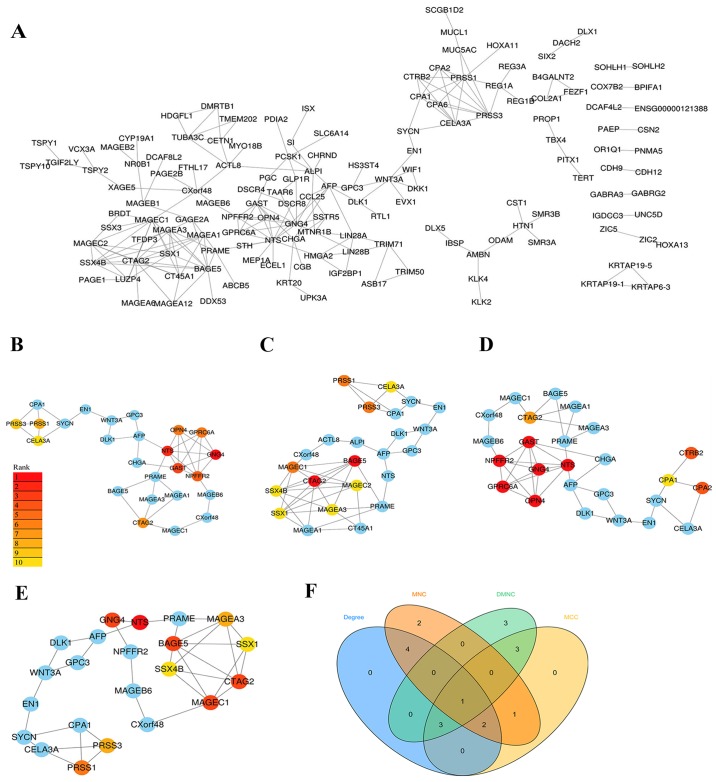
The PPI network was built using the STRING database and visualised with Cytoscape software (Fig. 2A). The PPI enrichment P-value was <1.0×10−16, which indicates that the interaction between the proteins themselves was greater than that of random proteins and that these proteins as a group are at least partially bioconjugated.
Figure 2.
PPI network of DEGs identified in samples from The Cancer Genome Atlas. (A) PPI network of the upregulated DEGs exhibiting statistically significant difference between. Four subnetworks were obtained by different analyses with the (B) MCC, (C) DMNC, (D) MNC and (E) degree methods. Colour labelling from red to yellow represented the top 10 nodes of each subnetwork, while other nodes in the subnetwork appear blue. (F) Venn diagram showing the intersection set of hub genes obtained from the four topological analysis methods. PPI, protein-protein interaction; DEG, differentially expressed gene; MCC, maximal clique centrality; DMNC, density of maximum neighbourhood component; MNC, maximum neighbourhood component.
Since biological networks are heterogeneous, it is necessary to search for hub genes using different topological analysis methods. There are 11 topological analysis methods in the cytoHubba plug-in of Cytoscape software platform. In the current study, four comparatively accurate methods of topological analysis, including the MCC, MNC, DMNC and degree methods (8), were selected to construct the subnetworks of the PPI network (Fig. 2B-E). Subsequently, based on the hub gene score from the four corresponding models, the intersection was identified with a Venn diagram (Fig. 2F). In total, 6 hub genes appeared in at least three topological analysis methods, including CTAG2, NTS, GNG4, GAST, PRSS1 and PRSS3 (Table II).
Table II.
Hub gene score according to the four topological analysis methods.
| Gene name | MCC | MNC | DMNC | Degree |
|---|---|---|---|---|
| CTAG2 | 63 | 9 | 0.48 | 10 |
| NTS | 127 | – | 0.65 | 12 |
| GNG4 | 128 | – | 0.65 | 10 |
| GAST | 122 | – | 0.65 | 7 |
| PRSS1 | 59 | 8 | – | 9 |
| PRSS3 | 58 | 8 | – | 8 |
MCC, maximal clique centrality; DMNC, density of maximum neighbourhood component; MNC, maximum neighbourhood component; CTAG2, cancer/testis antigen 2.
Association of hub gene expression and prognostic value
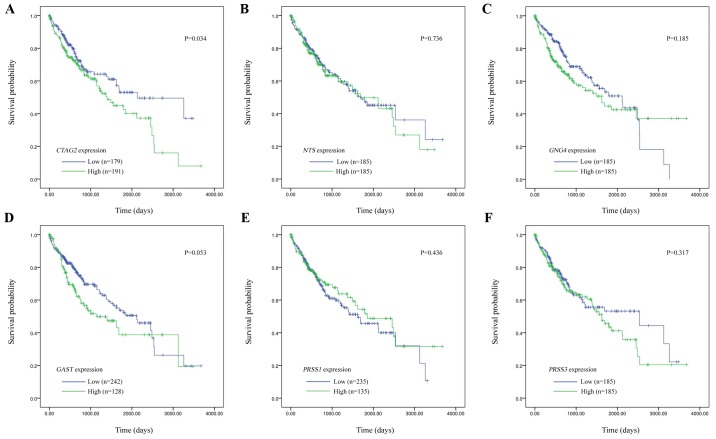
The correlation of hub gene expression with the overall survival of HCC patients from TCGA database was evaluated by Kaplan-Meier survival curves (Fig. 3). According to the RNA-seq expression of tumor tissues of patients in the TCGA LIHC database, the expression levels of CTAG2, NTS, GNG4, GAST, PRSS1 and PRSS3 was grouped into the high-expression and low-expression groups based on the median value. The results revealed that only CTAG2 expression was significantly correlated with prognosis (P=0.034), and the overall survival time of the high-expression group was significantly reduced. By contrast, there was no significant correlation between the expression of other hub genes and patient prognosis (NTS, P=0.736; GNG4, P=0.185; GAST, P=0.053; PRSS1, P=0.436; and PRSS3, P=0.317). Therefore, CTAG2 was regarded as the most important hub gene. Furthermore, based on the univariate Cox regression analysis, TNM stage (P<0.001), α-fetoprotein (AFP) expression (P=0.010) and CTAG2 expression (P=0.035) were found to be significantly correlated with the prognosis of patients. Further multivariate Cox regression analysis (Table III) revealed that the TNM stage, AFP expression and CTAG2 expression were not independent risk factors that influence prognosis.
Figure 3.
Kaplan-Meier curves indicating the correlation of the overall survival of hepatocellular carcinoma patients with different hub genes, including: (A) CTAG2 (P=0.034); (B) NTS (P=0.736); (C) GNG4 (P=0.185); (D) GAST (P=0.053); (E) PRSS1 (P=0.436); and (F) PRSS3 (P=0.317). The patients were divided into two groups according to the expression of hub genes. The blue curve represents the low-expression group, while the green curve represents the high-expression group. CTAG2, cancer/testis antigen 2.
Table III.
Univariate and multivariate Cox regression analysis of clinicopathological data correlated with overall survival in hepatocellular carcinoma patients.
| Univariate analysis | Multivariate analysis (n=264) | ||||
|---|---|---|---|---|---|
| Factor | n | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Age, years (>60/≤60) | 370 | 1.212 (0.854–1.720) | 0.281 | 1.420 (0.875–2.303) | 0.156 |
| Gender (male/female) | 370 | 1.225 (0.859–1.745) | 0.262 | 1.348 (0.838–2.167) | 0.218 |
| TNM stage (1/2-4) | 346 | 1.661 (1.355–2.036) | <0.001 | 1.348 (0.846–2.148) | 0.209 |
| AFP level (high/low) | 277 | 1.855 (1.159–2.970) | 0.010 | 1.528 (0.936–2.495) | 0.090 |
| CTAG2 expression (high/low) | 370 | 1.457 (1.028–2.066) | 0.035 | 1.440 (0.893–2.324) | 0.135 |
HR, hazard ratio; 95% CI, 95% confidence interval; AFP, α-fetoprotein; CTAG2, cancer/testis antigen 2.
Association between CTAG2 expression and the clinicopathological characteristics of HCC patients included in TCGA database
The association of CTAG2 expression with the clinicopathological characteristics of HCC patients included in the TCGA database were investigated. The clinicopathological features of HCC patients from TCGA are displayed in Table SI. According to the RNA-seq expression data of tumor tissue included in TCGA LIHC, the expression of CTAG2 was not associated with the patient gender, but was significantly associated with the patient age (P=0.003), T stage (P=0.028), TNM stage (P=0.028) and AFP expression (P=0.045; Table IV).
Table IV.
Association of CTAG2 expression with the clinicopathological features of hepatocellular carcinoma patients from The Cancer Genome Atlas database.
| CTAG2 expression, n (%) | |||||
|---|---|---|---|---|---|
| Characteristic | Categories | n | Low | High | P-value |
| Sex | Female | 121 | 56 (46.3) | 65 (53.7) | 0.598 |
| Male | 250 | 123 (49.2) | 127 (50.8) | ||
| Age, years | <60 | 169 | 96 (56.8) | 73 (43.2) | 0.003 |
| ≥60 | 201 | 83 (41.3) | 118 (58.7) | ||
| T stage | 1 | 181 | 96 (53.0) | 85 (47.0) | 0.028 |
| 2 | 94 | 39 (41.5) | 55 (58.5) | ||
| 3 | 80 | 40 (50) | 40 (50) | ||
| 4 | 13 | 2 (15.4) | 11 (84.6) | ||
| TNM stage | 1 | 171 | 93 (54.4) | 78 (45.6) | 0.028 |
| 2–4 | 176 | 75 (42.6) | 101 (57.4) | ||
| AFP level | Low (<upper limit) | 120 | 67 (55.8) | 53 (44.2) | 0.045 |
| High (≥upper limit) | 158 | 69 (43.7) | 89 (56.3) | ||
P-value was obtained from Pearson's χ2 test or fisher's exact test. CTAG2, cancer/testis antigen 2; AFP, α-fetoprotein.
Immunostaining of CTAG2 in HCC tissue samples from our hospital
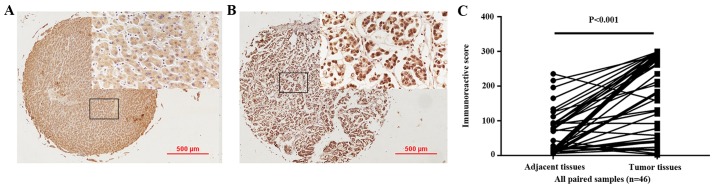
In the present study, immunohistochemical staining was conducted to detect CTAG2 expression in 46 hepatoma and 46 paired adjacent tissues collected at the First Affiliated Hospital of China Medical University (Fig. 4). Among the 46 paired tissues included in the TMA, the median IRS in the cancer tissues was 224.25% (interquartile range, 73.00–290.25%), whereas the median IRS in the cancer adjacent tissues was 22.00% (interquartile range, 13–90.25%). These results suggested that the positive expression of CTAG2 was significantly higher in carcinoma tissue compared with that in adjacent tissues (P<0.001; Fig. 4C).
Figure 4.
CTAG2 exhibited increased expression in hepatocellular carcinoma tissues obtained from patients at our hospital, as compared with that in adjacent tissues. Representative micrographs show immunohistochemical staining of CTAG2 in (A) adjacent and (B) tumor tissues (magnification, ×40). The square frame indicate the region that is magnified in the insert (magnification, ×400; scale bar, 500 µm). (C) Immunoreactive scores for CTAG2 staining in tumor tissues were significantly higher as compared with those in adjacent tissues. The P-value was obtained using the Wilcoxon rank-sum test (all paired samples, n=46, P<0.001). CTAG2, cancer/testis antigen 2.
Association between CTAG2 expression and clinicopathological characteristics of HCC patients from our hospital
The clinicopathological parameters of HCC patients from the First Affiliated Hospital of China Medical University are displayed in Table SII. In order to determine the correlation between the CTAG2 expression and clinicopathological factors, the IRS cut-off value of the high and low protein expression was defined by the ROC curves. The ROC curve analysis with respect to AFP expression level was used to determine the cut-point (208.5%; P=0.011) of the IRS (Fig. S1). Next, the association of CTAG2 expression with the clinicopathological data of HCC patients admitted to our hospital was examined. As shown in Table V, the results demonstrated that the expression of CTAG2 was statistically irrelevant to the patient gender, age at diagnosis, carcinoembryonic antigen expression, glypican-3 expression and differentiation level. By contrast, CTAG2 expression was found to be significantly associated with the hepatitis B virus (HBV) status (P=0.010) and AFP expression (P=0.004) of patients, and marginally significantly with the T stage (P=0.056).
Table V.
Association of CTAG2 expression with the clinicopathological features of hepatocellular carcinoma patients from the First Affiliated Hospital of China Medical University.
| CTAG2 expression, n (%) | |||||
|---|---|---|---|---|---|
| Characteristic | Category | n | Low | High | P-value |
| Sex | Female | 8 | 5 (62.5) | 3 (37.5) | 0.361 |
| Male | 38 | 17 (44.7) | 21 (55.3) | ||
| Age, years | <60 | 34 | 14 (41.2) | 20 (58.8) | 0.129 |
| ≥60 | 12 | 8 (66.7) | 4 (33.3) | ||
| T stage | 1–2 | 28 | 16 (57.1) | 12 (42.9) | 0.056 |
| 3–4 | 15 | 4 (26.7) | 11 (73.3) | ||
| HBV status | Negative | 11 | 9 (81.8) | 2 (18.2) | 0.010 |
| Positive | 35 | 13 (37.1) | 22 (76.1) | ||
| AFP level | Low (<upper limit) | 18 | 13 (72.2) | 5 (27.8) | 0.004 |
| High (≥upper limit) | 25 | 7 (28.0) | 18 (72.0) | ||
| Differentiation | High | 17 | 9 (52.9) | 8 (47.1) | 0.494 |
| Low | 26 | 11 (42.3) | 15 (57.7) | ||
| CEA | Negative | 8 | 5 (62.5) | 3 (37.5) | 0.199 |
| Positive | 12 | 4 (33.3) | 8 (66.7) | ||
| GPC-3 | Negative | 7 | 3 (42.9) | 4 (57.1) | 0.606 |
| Positive | 26 | 14 (53.8) | 12 (46.2) | ||
P-value was obtained from Pearson's χ2 test or Fisher's exact test. CTAG2, cancer/testis antigen 2; HBV, hepatitis B virus; AFP, α-fetoprotein; CEA, carcinoembryonic antigen; GPC-3, glypican-3.
GSEA of HCC samples in TCGA database
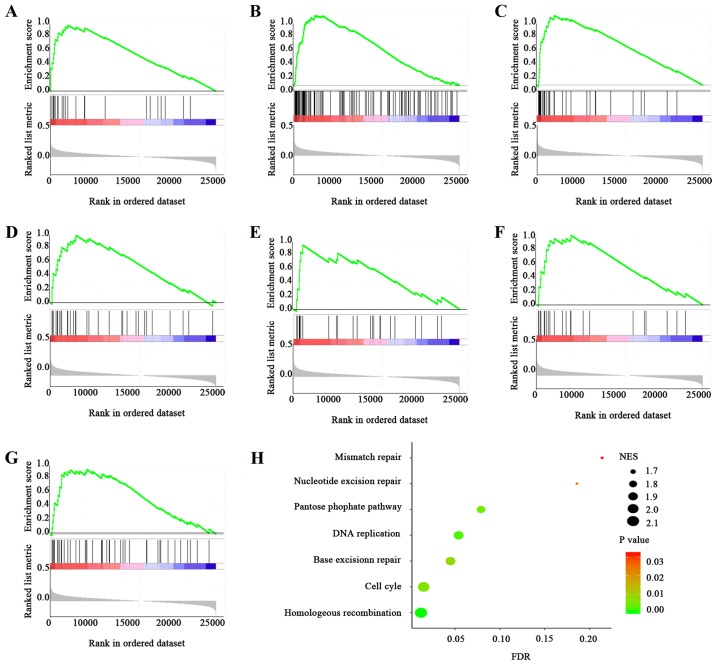
In order to obtain functional level information of CTAG2, the GSEA method was used based on KEGG database. The samples of 374 HCC patients included in TCGA database were divided into two groups according to CTAG2 expression, and GSEA was performed. The results suggested that samples with high CTAG2 expression were associated with enrichment in 7 pathways (Fig. 5), including homologous recombination (P<0.001), cell cycle (P=0.006), DNA replication (P=0.002), base resection and repair (P=0.008), pentose phosphate pathway (P=0.004), mismatch repair (P=0.032) and nucleotide excision repair (P=0.022). Among these, the genes involved in cell cycle pathway were significantly enriched in the high CTAG2 expression group (NES=2.05, P=0.006, FDR=0.016). The GSEA thus indicated that the function of CTAG2 in HCC may be correlated to cell cycle and DNA replication signalling pathways.
Figure 5.
GSEA of hepatocellular carcinoma data from The Cancer Genome Atlas database. Enrichment plots for significant pathways identified by GSEA include (A) homologous recombination, (B) cell cycle, (C) DNA replication, (D) base excision repair, (E) pentose phosphate pathway, (F) mismatch repair and (G) nucleotide excision repair. (H) Graph indicating the enrichment of each pathway, with the size of each point representing the NES value and the colour representing the P-value. GSEA, gene set enrichment analysis.
Discussion
At present, the main treatments for HCC include surgical resection, tumor ablation and liver transplantation; however, the therapeutic effect of these methods is limited for numerous patients, and the recurrence rate of HCC within 5 years after surgical resection has been reported to be 70% (17). According to the survival data available in the SEER database, the 5-year survival rate of HCC patients is <20% (18). The poor prognosis is associated with the inaccuracy of the existing HCC staging system, which results in patients not receiving adequate personalised treatment and monitoring. For instance, the TNM system of the Union for International Cancer Control is a tumor staging system commonly used to evaluate tumor progression in patients (19). However, there are great differences between the recurrence risk and survival conditions among patients with similar staging (20). Therefore, it is urgent and essential to identify suitable biomarkers and classify molecular subtypes to supplement the TNM staging system. Meanwhile, the biomarkers screened may also be developed into potential therapeutic targets, providing support for the development of new drugs for the treatment of HCC.
In the present study, 374 liver cancer and 50 paired paracancerous samples from the TCGA database were analysed, and the DEGs were detected. A PPI network was built using STRING database, while the subnetworks were constructed with four different topological analysis methods (21). It has been reported that the four methods used in the current study, including the MCC, DMNC, MNC and degree methods, are more accurate when analysing PPIs in the biological network (11). For instance, using MCC, a greater number of required proteins can be selected from the top ranked list, from high to low degree scores (11). In addition, different basic proteins are identified by DMNC, which scores the biological network with different methods. Therefore, these methods of topological analysis were used in the present study to construct the protein subnetworks. Subsequently, the intersection was determined according to the Venn graph, and six pivotal genes were identified in each subnetwork, which were as follows: CTAG2, NTS, GNG4, GAST, PRSS1 and PRSS3. The correlation between the expression levels of the six hub genes and the prognosis of HCC was then analysed, and CTAG2 was finally selected as the key research object. The aforementioned analytical methods may provide a new perspective for screening hub genes.
The CTA family includes tumor-associated antigens with a specific expression pattern (22,23). CTA proteins recognised by immune cells are encoded in tumor cells by certain special genes. These genes are normally silent in cells other than the human normal testicular and malignant cells. CTAG2 is a member of the CTA family that is mainly used for tumor diagnosis, metastasis monitoring and tumor treatment (24–26). It is highly expressed in numerous tumors, such as melanoma (27), multiple myeloma (28), urothelial carcinoma (29), prostate cancer (30), colorectal cancer (31) and breast cancer (9). Previous research was only conducted in a small number of samples and reported that the mRNA expression of CTAG2 in HCC tissues was higher than that in normal tissues (32). In the current study, a large number of samples were used to confirm the high expression of CTAG2 in HCC. According to our study, examination of the association of CTAG2 expression in 374 HCC patients included in the TCGA database with various clinicopathological indexes indicated that the expression of CTAG2 was significantly correlated with patient age (P=0.003), T stage (P=0.028), TNM stage (P=0.028) and AFP level (P=0.045). Furthermore, according to univariate Cox analysis, CTAG2 expression (overall survival, P=0.035) was correlated with the prognosis of the patients with HCC. By contrast, a previous study reported that the level of CTAG2 mRNA expression was not significantly correlated with clinical pathological parameters in patients (28). The discrepancy in the results obtained in the present study and those of the previous report may be due to the difference in the sample size and source. In addition, bioinformatics analysis of different racial classifications could not be contacted, since the majority of TCGA data involve Caucasian patients. Therefore, tissues from Chinese patients were further analysed by IHC in the present study. The results revealed that the positive expression of CTAG2 in carcinoma tissues was significantly higher when compared with that in adjacent tissues. Thus, it is possible that CTAG2 is highly expressed in tumor tissues for different ethnic groups.
Available research has reported the mRNA expression of CTAG2 in HCC, without investigating the protein level. In the current study, IHC was conducted and confirmed that CTAG2 was highly expressed in HCC tissues at the protein level. Subsequently, the clinical data revealed that the expression of CTAG2 was correlated with the HBV status (P=0.010) and AFP level (P=0.004). These results are basically consistent with the results of the TCGA data analytics in the present study.
Analysing the association of CTAG2 with KEGG pathways may be beneficial for revealing the molecular mechanism of its influence on HCC. It has been reported that GSEA has significant advantages over traditional gene expression analysis (33). This method aims to detect biological processes in the whole gene network by focusing on gene sets. GSEA was conducted in the present study and demonstrated that CTAG2 expression was associated with several signalling pathways, including the cell cycle pathway. CTAG2 is known to affect the cell cycle due to being involved in mitosis by interacting with the centrosomal protein pericentrin (13). Meanwhile, according to the analysis performed in the present study, the expression of CTAG2 was associated with the T stage (primary tumor topography). Therefore, it is suggested that CTAG2 may influence the tumor size by affecting the cell cycle. Numerous antineoplastic drugs work by influencing the cell cycle, while CTAG2 promotes tumor growth by influencing the cell cycle; therefore, CTAG2 may serve as a cell cycle-specific therapeutic target.
In conclusion, the current research aimed to analyse the PPI networks in HCC by using different topological analysis methods to identify the hub genes. The study revealed that data and immunohistochemical analyses are effective methods for screening potential therapeutic targets. Based on the results, it can be inferred that CTAG2 can be used as a potential HCC therapeutic target or prognostic biomarker, and thus patients with high expression of CTAG2 should be monitored and treated appropriately. However, further research is needed in the future to reveal the mechanism underlying the effect of CTAG2 on the development of HCC.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was supported in part by grants from the National Natural Science Foundation of China (grant no. 81601370) and Shenyang S&T Projects (grant nos. F16-094-1-00 and Z18-4-020).
Availability of data and materials
RNA-Seq data analysed during the present study are available in The Cancer Genome Atlas (https://portal.gdc.cancer.gov).
Authors' contributions
HG was the major contributor in designing this study and provided guidance for bioinformatic analysis. MW participated in designing this work and revised the manuscript for important intellectual content, and provided approval for the final version to be published. JL drafted the manuscript and performed the experiments. JL performed the bioinformatic analysis. ZY participated in drafting of the manuscript for important intellectual content and evaluated the histochemical staining level of TMA blocks. QL assisted in samples collection and evaluated the histochemical staining level of TMA blocks. MS participated in the bioinformatic analysis and conducted the statistical analyses on the data. All authors agreed to be accountable for all aspects of the present study in ensuring that questions related to the accuracy or integrity of the work are appropriately resolved.
Ethics approval and consent to participate
The study was approved by the Medical Ethics Committee of China Medical University. The Ethics Committee waived the need for the patient to sign a written informed consent due to the retrospective nature of the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol. 2006;45:529–538. doi: 10.1016/j.jhep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 3.Hung AK, Guy J. Hepatocellular carcinoma in the elderly: Meta-analysis and systematic literature review. World J Gastroenterol. 2015;21:12197–12210. doi: 10.3748/wjg.v21.i42.12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kiss A, Wang NJ, Xie JP, Thorgeirsson SS. Analysis of transforming growth factor (TGF)-alpha/epidermal growth factor receptor, hepatocyte growth Factor/c-met, TGF-beta receptor type II, and p53 expression in human hepatocellular carcinomas. Clin Cancer Res. 1997;3:1059–1066. [PubMed] [Google Scholar]
- 5.Chiang DY, Villanueva A, Hoshida Y, Peix J, Newell P, Minguez B, LeBlanc AC, Donovan DJ, Thung SN, Solé M, et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68:6779–6788. doi: 10.1158/0008-5472.CAN-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liao H, Liao M, Xu L, Yan X, Ren B, Zhu Z, Yuan K, Zeng Y. Integrative analysis of h-prune as a potential therapeutic target for hepatocellular carcinoma. EBioMedicine. 2019;41:310–319. doi: 10.1016/j.ebiom.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin B, Wang W, Du G, Huang GZ, Han LT, Tang ZY, Fan DG, Li J, Zhang SZ. Identifying hub genes and dysregulated pathways in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2015;19:592–601. [PubMed] [Google Scholar]
- 8.Zheng Y, Long J, Wu L, Zhang H, Li L, Zheng Y, Wang A, Lin J, Yang X, Sang X, et al. Identification of hub genes involved in the development of hepatocellular carcinoma by transcriptome sequencing. Oncotarget. 2017;8:60358–60367. doi: 10.18632/oncotarget.19483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiong Y, You W, Wang R, Peng L, Fu Z. prediction and validation of hub genes associated with colorectal cancer by integrating PPI network and gene expression data. BioMed Res Int. 2017;2017:2421459. doi: 10.1155/2017/2421459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR, Winchester DP. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more ‘personalized’ approach to cancer staging. CA Cancer J Clin. 2017;67:93–99. doi: 10.3322/caac.21388. [DOI] [PubMed] [Google Scholar]
- 13.Maine EA, Westcott JM, Prechtl AM, Dang TT, Whitehurst AW, Pearson GW. The cancer-testis antigens SPANX-A/C/D and CTAG2 promote breast cancer invasion. Oncotarget. 2016;7:14708–14726. doi: 10.18632/oncotarget.7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han Q, Sun ML, Liu WS, Zhao HS, Jiang LY, Yu ZJ, Wei MJ. Upregulated expression of ACTL8 contributes to invasion and metastasis and indicates poor prognosis in colorectal cancer. Onco Targets Ther. 2019;12:1749–1763. doi: 10.2147/OTT.S185858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Chen W, Wei W, Lou J. Oncogene TUBA1C promotes migration and proliferation in hepatocellular carcinoma and predicts a poor prognosis. Oncotarget. 2017;8:96215–96224. doi: 10.18632/oncotarget.21894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong PP, Miranda F, Chan KV, Berlato C, Hurst HC, Scibetta AG. Histone demethylase KDM5B collaborates with TFAP2C and Myc to repress the cell cycle inhibitor p21(cip) (CDKN1A) Mol Cell Biol. 2012;32:1633–1644. doi: 10.1128/MCB.06373-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 18.Wang S, Sun H, Xie Z, Li J, Hong G, Li D, Mallampati S, Zhou X, Zhou C, Zhang H, et al. Improved survival of patients with hepatocellular carcinoma and disparities by age, race and socioeconomic status by decade, 1983–2012. Oncotarget. 2016;7:59820–59833. doi: 10.18632/oncotarget.10930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan AC, Fan ST, Poon RT, Cheung TT, Chok KS, Chan SC, Lo CM. Evaluation of the seventh edition of the American Joint Committee on Cancer tumor-node-metastasis (TNM) staging system for patients undergoing curative resection of hepatocellular carcinoma: Implications for the development of a refined staging system. HPB (Oxford) 2013;15:439–448. doi: 10.1111/j.1477-2574.2012.00617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greene FL, Sobin LH. The staging of cancer: A retrospective and prospective appraisal. CA Cancer J Clin. 2008;58:180–190. doi: 10.3322/CA.2008.0001. [DOI] [PubMed] [Google Scholar]
- 21.Gao H, Wang H, Yang W. Identification of key genes and construction of microRNA-mRNA regulatory networks in multiple myeloma by integrated multiple GEO datasets using bioinformatics analysis. Int J Hematol. 2017;106:99–107. doi: 10.1007/s12185-017-2216-2. [DOI] [PubMed] [Google Scholar]
- 22.Almeida LG, Sakabe NJ, deOliveira AR, Silva MC, Mundstein AS, Cohen T, Chen YT, Chua R, Gurung S, Gnjatic S, et al. CTdatabase: A knowledge-base of high-throughput and curated data on cancer-testis antigens. Nucleic Acids Res 37 (Database Issue) 2009:D816–D819. doi: 10.1093/nar/gkn673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen YT, Scanlan MJ, Venditti CA, Chua R, Theiler G, Stevenson BJ, Iseli C, Gure AO, Vasicek T, Strausberg RL, et al. Identification of cancer/testis-antigen genes by massively parallel signature sequencing. Proc Natl Acad Sci USA. 2005;102:7940–7945. doi: 10.1073/pnas.0502583102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caballero OL, Chen YT. Cancer/testis (CT) antigens: Potential targets for immunotherapy. Cancer Sci. 2009;100:2014–2021. doi: 10.1111/j.1349-7006.2009.01303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao J, Caballero OL, Yung WK, Weinstein JN, Riggins GJ, Strausberg RL, Zhao Q. Tumor subtype-specific cancer-testis antigens as potential biomarkers and immunotherapeutic targets for cancers. Cancer Immunol Res. 2014;2:371–379. doi: 10.1158/2326-6066.CIR-13-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hofmann O, Caballero OL, Stevenson BJ, Chen YT, Cohen T, Chua R, Maher CA, Panji S, Schaefer U, Kruger A, et al. Genome-wide analysis of cancer/testis gene expression. Proc Natl Acad Sci USA. 2008;105:20422–20427. doi: 10.1073/pnas.0810777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lotem M, Merims S, Frank S, Hamburger T, Nissan A, Kadouri L, Cohen J, Straussman R, Eisenberg G, Frankenburg S, et al. Adjuvant autologous melanoma vaccine for macroscopic stage III disease: Survival, biomarkers, and improved response to CTLA-4 blockade. J Immunol Res. 2016;2016:8121985. doi: 10.1155/2016/8121985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Duin M, Broyl A, de Knegt Y, Goldschmidt H, Richardson PG, Hop WC, van der Holt B, Joseph-Pietras D, Mulligan G, Neuwirth R, et al. Cancer testis antigens in newly diagnosed and relapse multiple myeloma: Prognostic markers and potential targets for immunotherapy. Haematologica. 2011;96:1662–1669. doi: 10.3324/haematol.2010.037978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dyrskjot L, Zieger K, Kissow Lildal T, Reinert T, Gruselle O, Coche T, Borre M, Ørntoft TF. Expression of MAGE-A3, NY-ESO-1, LAGE-1 and PRAME in urothelial carcinoma. Br J Cancer. 2012;107:116–122. doi: 10.1038/bjc.2012.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulkarni P, Shiraishi T, Rajagopalan K, Kim R, Mooney SM, Getzenberg RH. Cancer/testis antigens and urological malignancies. Nat Rev Urol. 2012;9:386–396. doi: 10.1038/nrurol.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shantha Kumara HM, Grieco MJ, Caballero OL, Su T, Ahmed A, Ritter E, Gnjatic S, Cekic V, Old LJ, Simpson AJ, et al. MAGE-A3 is highly expressed in a subset of colorectal cancer patients. Cancer Immu. 2012;12:16. [PMC free article] [PubMed] [Google Scholar]
- 32.Wang XY, Chen HS, Luo S, Zhang HH, Fei R, Cai J. Comparisons for detecting NY-ESO-1 mRNA expression levels in hepatocellular carcinoma tissues. Oncol Rep. 2009;21:713–719. [PubMed] [Google Scholar]
- 33.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq data analysed during the present study are available in The Cancer Genome Atlas (https://portal.gdc.cancer.gov).