Abstract
As technologies change, MG-RAST is adapting. Newly available software is being included to improve accuracy and performance. As a computational service constantly running large volume scientific workflows, MG-RAST is the right location to perform benchmarking and implement algorithmic or platform improvements, in many cases involving trade-offs between specificity, sensitivity and run-time cost. The work in [Glass EM, Dribinsky Y, Yilmaz P, et al. ISME J 2014;8:1–3] is an example; we use existing well-studied data sets as gold standards representing different environments and different technologies to evaluate any changes to the pipeline. Currently, we use well-understood data sets in MG-RAST as platform for benchmarking. The use of artificial data sets for pipeline performance optimization has not added value, as these data sets are not presenting the same challenges as real-world data sets. In addition, the MG-RAST team welcomes suggestions for improvements of the workflow. We are currently working on versions 4.02 and 4.1, both of which contain significant input from the community and our partners that will enable double barcoding, stronger inferences supported by longer-read technologies, and will increase throughput while maintaining sensitivity by using Diamond and SortMeRNA. On the technical platform side, the MG-RAST team intends to support the Common Workflow Language as a standard to specify bioinformatics workflows, both to facilitate development and efficient high-performance implementation of the community’s data analysis tasks.
Keywords: metagenome analysis, cloud, distributed workflows
Introduction
The ever-increasing amount of DNA sequence data [1] has motivated significant developments in biomedical research. Currently, however, many researchers continue to struggle with large-scale computing and data management requirements. Numerous approaches have been proposed and are being pursued to alleviate this burden on application scientists. The approaches include focusing on the user-interface layer while relying primarily on legacy technology [2]; reimplementing significant chunks of code in new languages [3]; and developing clean-slate designs [4]. Breakthroughs that appreciably reduce computational burden, such as Diamond [5], are the exception. While important, few if any of the solutions contribute to solving the central problem: data analysis is becoming increasingly expensive in terms of both time and cost, with reference databases growing rapidly and data volumes rising. In essence, more and more data are being produced without sufficient resources to analyze the data. All indicators show that this trend will continue in the foreseeable future [1].
We strongly believe that a change in how the research community handles routine data analytics is required. While we cannot predict the outcome of this evolutionary process, scalable, flexible and—most important—efficient platforms will, in our opinion, be part of any ‘new computational ecosystem’. MG-RAST [6] is one such platform that handles hundreds of submissions daily, often aggregating >0.5 terabytes in a 24 h period.
MG-RAST is a hosted, open-source, open-submission platform (‘Software as a Service’) that provides robust analysis of environmental DNA data sets (where environment is broadly defined). The system has three main components: a workflow, a data warehouse and an API (with a Web frontend). The workflow combines automated quality control, automated analysis and user-driven parameter- and database-flexible analysis. The data warehouse supports data archiving, discovery and integration. The platform is accessible via a Web interface [7], as well as a RESTful API [8].
Analysis of environmental DNA (i.e. metagenomics) presents a number of challenges, including feature extraction (e.g. gene calling) from (mainly unassembled) often lower-quality sequence data; data warehousing; movement of often large data sets to be compared against many equally large data sets; and data discovery. A key insight (see Lessons learned, L1) is that the challenges faced here are distinct from the challenges facing groups that render services for individual genomes or even sets of genomes [9].
Several hosted systems currently provide services in this field: JGI IMG/M [10], EBI MG-Portal [11] and MG-RAST [6]. Myriad stand-alone tools exist, including integrative user-friendly interfaces [12]; feature prediction tools [5, 13]; tools that ‘bin’ individual reads using codon frequencies, read abundance and cross-sample abundance [14–17]; and sets of marker genes reducing the search space for analysis with associated visualization tools [18]. MG-RAST seeks to select the best-in-class implementations and provide a hosted resource-efficient service that implements a balance between custom analysis and one-size-fits-all recipes. The approach taken in MG-RAST to achieve this goal is by defining parameters late—during download or analysis—not a priori before running a set of analyses tools. The analysis workflow in MG-RAST is identical across all data sets, except for data set-specific operations such as host DNA removal and variations in filtering to accommodate different user-submitted data types.
While many approaches to metagenome analysis exist, we chose an approach that allows large-scale analysis and massive comparisons. The core principle for the design of MG-RAST was to provide consistent analyses as deep and unbiased as possible at affordable computational cost. Other approaches, such as comprehensive genome and protein binning, adopted by IMG/M, or the profile hidden Markov model-based approaches using MG-Portal, do add value and provide valuable alternative analyses. These portals complement each other’s capabilities, and we routinely share best practices with them. MG-RAST’s strong suit is handling raw reads directly from a sequencing service. It has been extended to handle assembled metagenomes and metatranscriptomes as well. In its current form, however, it does not support metagenomics assembly or a genome-centric approach to metagenomics (i.e. binning).
Like many hosted applications (not just in bioinformatics), MG-RAST started out as a traditional database-oriented system using largely traditional design patterns. While expanding the number of machines able to execute MG-RAST workflows, we learned that data access input and output (I/O) is as limiting a factor as the processing power or memory (see Lessons learned, L2 and L8). MG-RAST has rapidly adapted [19–21] to meet the needs of a growing user community, as well as the changing technology landscape. We have run MG-RAST workflows on several computational platforms, including OpenStack [22], Amazon’s AWS [23], Microsoft’s Azure [24], several local clusters and even laptops on occasion. In many ways, MG-RAST has evolved to be the counterpoint to the now-abundant one-offs that are routinely implemented in many laboratories for sequence analysis. It offers reproducibility and was designed for efficient execution [9] (see Lessons learned, L9).
To date, MG-RAST has processed >295 000 data sets from 23 000 researchers. As of June 2017, over 1 trillion individual sequences totaling >40 terabase pairs have been processed, and the total volume of data generated is well over half a petabyte of data. A fair assessment is that we do a lot of the heavy lifting of high-volume automated analysis of amplicon and shotgun metagenomes as well as metatranscriptomes for a large user community. Currently, only 20% (44 000) of the data sets in MG-RAST are publicly available. Data are frequently shared by researchers with only their collaborators. In future releases, we will introduce a series of features to incentivize data publication.
The vast majority of the data sets in MG-RAST represent user submissions; <3000 are data sets extracted from SRA by the developers. Determining identity between any two data sets is far from trivial if available metadata does not provide sufficient evidence—one more reason to incentivize metadata (see Lessons learned, L7). However, the developers are working jointly with researchers at EBI to synchronize the contents of EBI’s ENA with the contents of MG-RAST. Currently, to the best of our combined knowledge, there is little overlap between the data sets in SRA/ENA and those in MG-RAST.
The analysis shown in Figure 1 is typical for one class of user queries. We note that in addition to requesting SEED annotations, the user might also request annotations from the M5NR sub-databases (i.e. namespaces) such as KEGG pathways [25], KEGG orthologues [26], COG [27] and RefSeq [28]. Providing a smart data product that can be projected with no computation onto other namespaces (read annotation databases) saves a significant amount of computational resources (see Lessons learned, L3).
Figure 1.
MG-RAST data and analysis results can be reused for other purposes. Here, we show a muscle [29] alignment of (the prodigal translations) of filtered sequences from the following unauthenticated API call: http://api.metagenomics.anl.gov//annotation/sequence/mgm4662210.3?evalue=10&type=function&source=Subsystems&filter=Inosine-5.
Compared with previous versions of MG-RAST, the latest version has increased throughput dramatically while using the same amount of resources: ∼22 million core-hours annually are used to run the MG-RAST workflow for user-based submissions. In addition, the RESTful API has allowed a rethinking and restructuring of the user interface (different model–view–controller pattern) and, most important, the reproducibility of results using containers.
Implementation
While MG-RAST started as an application built on a traditional LAMP stack [30], we quickly realized that a single database could not provide sufficient flexibility to support various underlying components at the scale required. Instead, we chose to rely on an open API [8] that provides the ability to change underlying components as required by scale.
We note that MG-RAST is not a comprehensive analysis tool offering every conceivable kind of boutique analysis. By making some data-filtering parameters user adjustable at analysis or download time, MG-RAST provides flexibility. Via the API, users and developers can pipe MG-RAST data and results into their in-house analysis procedures. Figure 1 shows an MG-RAST API query for sequences with similarities to proteins with the SEED Subsystem namespace annotation inosine-5′ phosphate dehydrogenase from the soil metagenome mgm4662210.3 that is streamed into a filtering and alignment procedure. A key feature of MG-RAST (see Lessons learned, L6) is its ability to adjust database match parameter at query time—a function frequently not recognized by researchers and in some cases missed even by studies comparing systems [31].
MG-RAST has been designed to treat every data set with the same pipeline. Given the expected volume and variety of datasets, per-data set optimization of parameters has not been a design goal. The system is optimized for robust handling of a wide variety of input types, and users can perform optimizations within sets of parameters that filter the pipeline results. The automatic setting of, for instance, detection thresholds for dramatically different data types and research questions is not the role of a data analysis platform. While this one-size-fits-all nature of the processing might somewhat limit sensitivity and potentially limit downstream scientific inquiry, these limitations are counterbalanced by the vast scope of the consistently analyzed data universe that the uniformly applied workflows and data management and discovery systems enable researchers to access. We believe that relying on smart data products that enable adjustment of parameters after processing and using custom downstream analysis scripts more than compensate for any reduction in sensitivity (see Lessons learned, L3 and L6).
Backend components
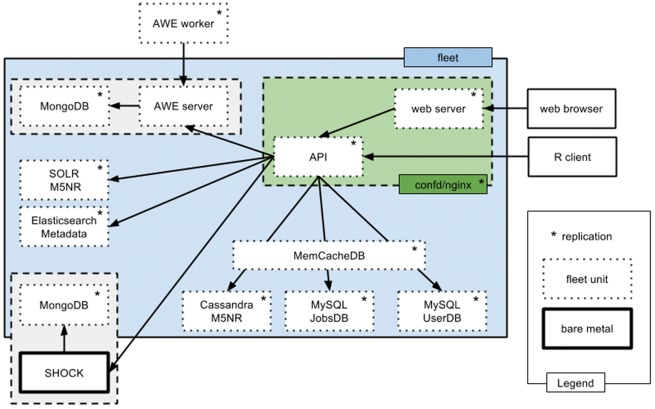
Figure 2 shows the current design of the MG-RAST backend components, using various databases and caching systems [32–35] as appropriate to support the API with the performance needed.
Figure 2.
Backend of MG-RAST version 4 using several database systems to enable efficient querying via the API.
A major criterion for success of the workflow is the ability to scale to the throughput levels required. Algorithmic changes (e.g. adoption of Diamond [5]) can help, but the design of the execution environment—most specifically its portability—is the single key to scaling (see Lessons learned, L4 and L5).
Access to data
In computational biology, shared filesystems traditionally are used to serve data to the computational resources. Sharing data between multiple computers is necessary because the data typically require more computational resources than a single machine can provide. Shared filesystems can render data accessible on several computers. This approach, however, limits the range of available platforms or requires significant time for configuring access or moving data into the platform. In addition, many shared filesystems exhibit poor scaling performance for science applications. Slow or inadequate shared filesystems have been observed by almost every practitioner of bioinformatics (see Lessons learned, L2). This situation has forced the use of complex I/O middleware to transform science I/O workloads into patterns that can scale in various science domains, including quantum chromodynamics and astrophysics [36], molecular dynamics [37], fusion science [38] and climate [39].
Rather than adopting this approach, we conducted a detailed analysis of our workloads, which revealed that individual computational units (e.g. cluster nodes) typically use a small fraction of the data and do not require access to the entire data set. Consequently, we chose to centralize data into a single point and access it in a RESTful way, thus providing efficient access while requiring no configuration for the vast majority of computing systems. A single object store can support distributed streaming analysis of data across many computers (see Lessons learned, L8).
The SHOCK object store [40] provides secure access to data and, most important, to subsets of the data. A computational client node can request a number of sequence records or sets of records meeting specific criteria. Data are typically streamed at significant fractions of line speed, and as results are frequently returned as indices that are much smaller than the original data files, writing is extremely efficient. Furthermore, the data are primarily write-once, which significantly simplifies the design of the object store with respect to data consistency.
Data in SHOCK is available to third parties via a RESTful API, and thus, SHOCK supports the reuse of both data and results.
Execution format
Executing workflows across a number of systems requires that the code be made available in suitable binary form on those platforms. Among the emerging challenges, reproducibility is a key problem for scientific disciplines that rely on the results of sequence analysis at scale without the ability to validate every single computational step in depth.
Virtual machines have been used to provide stable and portable execution environments [41] for a number of years. However, because of many technical details (e.g. significant number of binary formats required to cover all platforms) and significant overhead [42] in execution, containers provide a more suitable platform for most scientific computations.
In particular, the relatively recent advent of binary Linux containers (notably, Docker) in computing affords a novel way to distribute execution environments. Containers reduce the set of requirements for any given software package to one: a container. We have devised a scalable system [43] to execute scientific workflows across a number of containers connected only via a RESTful interface to an object store. With increasing numbers of systems supporting containerized execution [44] and with compatibility mechanisms [45] emerging to support legacy installations, Linux containers are quickly becoming the lingua franca of binary execution environments (see Lessons learned, L5). As with all of MG-RAST, the recipes for building the containers (‘Dockerfiles’) are available as open source on github, and the binary containers are available on DockerHub. The resulting containers are not specific to the MG-RAST systems, and the binary containers and the recipes are available to third parties for their adoption.
Current MG-RAST workflow
MG-RAST has been used for tens of thousands of data sets. This extensive use has led to a level of stability and robustness that few sequence analysis workflows can match.
The workflow (version 4.01) consists of the following logical steps:
Data hygiene Providing quality control and normalization steps that also include mate pair merging with ea-utils fastq-join [46–48]. The focus, however, is on artifact removal and host DNA removal [48, 49].
Feature extraction Using a predictor that has been shown to be robust against sequence noise (FragGeneScan [50]) to predict potentially protein-coding features, and using a purposefully simple similarity-based approach to predict ribosomal RNAs using VSEARCH [51]. The similarity-based predictions use a version of M5RNA [52] that was clustered at 70% identity to find candidate ribosomal RNA sequences.
Data reduction Clustering of predicted features at 90% identity (protein coding) and 97% (ribosomal RNA). Features overlapping with predicted ribosomal RNA (rRNA) sequences are removed. For each cluster, the longest representative is used.
Feature annotation Using similarity-based mapping of cluster representatives using super nonredundant M5NR [52] with a parallelized version of BLAT [53] for candidate proteins and ribosomal RNAs. This creates ‘annotations’ with M5NR database identifiers only.
Profile creation Mapping the M5NR identifiers to several functional namespaces (e.g. RefSeq or SEED), hierarchical namespaces (e.g. COG and Subsystems), pivoting into functional and taxonomic categories, and thus creating a reduced fingerprint (‘profile’) for each namespace and hierarchy.
Database load Uploading profiles to the various MG-RAST backend databases that support the API.
We note that the approach taken to sequence analysis is different from the state of the art for more or less complete microbial genomes [54].
Using data from MG-RAST
A key problem of current big data bioinformatics is the barrier to reuse of data and results. Comparing results of an expensive computational procedure with results from another laboratory can be problematic if the procedures used are not identical (potentially compromising integrity of the study). Another common approach is to not reuse existing results but to do an expensive reanalysis of both data sets, thus duplicating the work originally performed. One key problem with this approach is that data-driven science is no longer reviewable, as no reviewer can be expected to retrace the steps of the investigators while duplicating their computational work. If the data and results (also intermediate results) were available as reproducible entities, the problem of data uncertainty and costly recomputations would disappear.
This exceptional waste of computer time is acceptable default behavior in a discipline that is rich in computation and poor in data. In a data-rich ecosystem, however, either the terms of engagement have to change or the percentage of the research budgets allocated to computational resources has to dramatically increase. One of the key goals of MG-RAST is to provide a wealth of data sets and the underlying analysis results. Both the Web-based user interface and the RESTful API make these results accessible. To get closer to our goal of transparent and reproducible MG-RAST data analysis, we already execute all workflow steps in containers. The missing building block—which we are currently working on and which will enable every interested party to easily to execute, compare or modify our analysis pipeline—is support for the Common Workflow Language (CWL) [55] in our workflow engine. We think that producing data with a CWL workflow adds more value because it adds executable provenance (see Lessons learned, L4). Executable provenance is critical, as it allows recreation of the results on a wide variety of computational platforms.
Using profiles generated by MG-RAST
Profiles are the primary data product generated by MG-RAST, and they feed into the Web user interface and the various other tools. They encode the abundance of entities in a given sample combining information from several databases. Most important, profiles include information on the quality of the underlying observation (e.g. results of sequence similarity search) (Figure 3). Profiles are a compressed representation of the environmental samples, allowing large-scale comparisons.
Figure 3.
MG-RAST profile encoding abundance and matching parameter information as well as information on the observed entities.
Another critical feature is the ability to adjust matching parameters (e.g. minimal alignment length required for inclusion) at analysis time, allowing data reuse without the need for recomputing the profile with different cutoffs. With this ‘smart data product’, data consumers can switch between reference databases and parameter sets without recomputing the underlying sequence similarity searches (see Lessons learned, L3).
Metadata—making data discoverable
A key component of data reuse is the much-discussed ‘metadata’ (or ‘data describing data’). With tens of thousands of data sets available, the ability to identify the relevance of data sets has become critical. Approaches include ‘simple’ machine-readable encoding of data items such as pH, temperature and location and the use of controlled vocabularies to allow unambiguous encoding of, for example, anatomical organs via [56] or geographical features using the ENVO ontology [57].
Machine-readable metadata, such as the concepts championed by the Genomic Standards Consortium (GSC) [58], is key. GSC metadata is intentionally kept as simple and lightweight as possible while trying to meet the needs of the data producers and data consumers. Despite its simplicity, however, for the occasional user (e.g. a scientist depositing data), it is still cumbersome. Tools such as Metazen [59] help bridge the gap between data scientists and occasional users. MG-RAST implements the core MIxS [60] checklist, as well as all available environmental packages [61].
GSC-compliant, machine-readable markup of data sets at the time of upload to or deposition in online resources offers a unique opportunity. Data become discoverable, and analysis is made easier. MG-RAST incentivizes the addition of metadata by offering priority access to the compute resources to data sets with valid GSC metadata (see Lessons learned, L7).
Web user interface
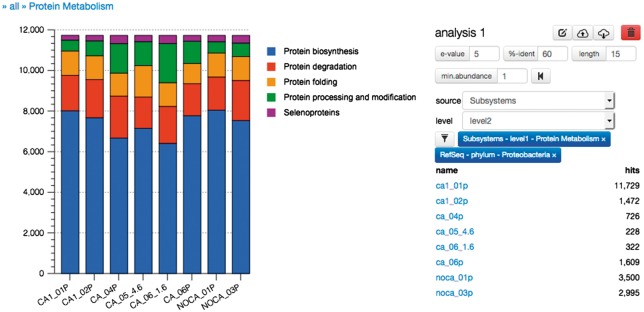
Not all scientists spend a significant fraction of their time on the command line or enjoy using the command line to solve their bioinformatics questions. Extracting and displaying the relative abundance of proteins from proteins classified as part of the subsystem class ‘Protein Metabolism’from the phylum Proteobacteria are simple via the Web interface (Figure 4) but require many command line invocations.
Figure 4.
Relative abundance of protein functional classes (‘Subsystems’) in Proteobacteria (‘RefSeq Phylum’) displayed as a waterfall diagram for data sets in study mgp128 as displayed by the version 4.0 MG-RAST graphical user interface.
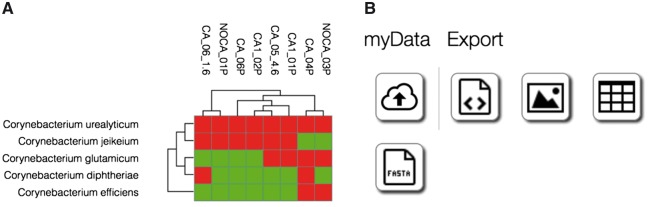
For these users, MG-RAST provides a graphical user interface (GUI) implemented in JavaScript/HTML5. The GUI provides guidance for nontrivial procedures such as data upload and validation, data sharing and data discovery, as well as data analysis Figure 5A. Data export in various formats is also supported Figure 5B.
Figure 5.
(A) Heatmap and clustering of the occurrence of Corynebacteria in study mgp128 as displayed by the MG-RAST web frontend. (B) Data export options available for the data and visualization, including sequences and abundance in tabular and JSON format.
User’s view of MG-RAST
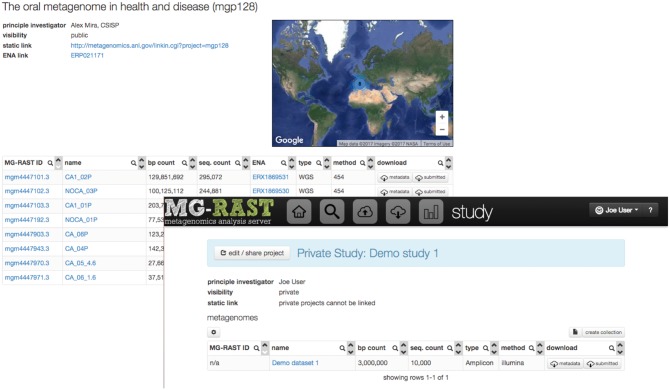
Every user has a different view of the data in MG-RAST. All users have access to the public metagenomics data, but shared or private data available to the user are linked to the user’s login information. Each data set has a unique identifier and information on visibility; until the data are made publicly available, temporary identifiers are used to minimize the number of data sets mentioned in the literature without being publicly available. Figure 6 provides a comparison of public and private data sets and highlights the sharing and data organization capabilities of the platform.
Figure 6.
Public study (with permanent unique identifier mgp128) and private study set with temporary identifier. A study groups multiple data sets, provides a single identifier and allows sharing via simply providing an email address for the person the data are to be shared with.
A key design feature of MG-RAST is to allow private data sets; users are in charge of uploading, sharing and releasing the data. Once submitted, data are private to the submitting user. The submitting user is reminded to share their data at their earliest convenience.
In addition to data, the processing pipeline and the data warehousing, MG-RAST provides an analytical tool set. It is implemented as a user-friendly Web application and consuming the profiles generated by the MG-RAST pipeline.
Future work
As technologies change, MG-RAST is adapting. Newly available software is being included to improve accuracy and performance. As a computational service constantly running large-volume scientific workflows, MG-RAST is the right location to perform benchmarking and implement algorithmic or platform improvements, in many cases involving trade-offs between specificity, sensitivity and run-time cost. The work in [62] is an example. We use existing well-studied data sets as gold standards representing different environments and different technologies to evaluate any changes to the pipeline. Currently, we use well-understood data sets in MG-RAST as a platform for benchmarking. The use of artificial data sets for pipeline performance optimization has not added value because these data sets do not present the same challenges as real-world data sets do.
The MG-RAST team welcomes suggestions for improvements of the workflow. We are currently working on versions 4.02 and 4.1, both of which contain significant input from the community and our partners that will enable double barcoding and stronger inferences supported by longer-read technologies and will increase throughput while maintaining sensitivity by using Diamond and SortMeRNA.
On the technical platform side, the MG-RAST team intends to support the CWL as a standard to specify bioinformatics workflows, to facilitate both development and efficient high-performance implementation of the community’s data analysis tasks.
Lessons learned
L1. Analyzing large-scale environmental DNA is different from genomics.
Because of the absence of high-quality assembled data (in most projects) and the lack of good models for removing contaminations upstream, a metagenomics portal site has to take over quality control and normalization and become good at it.
L2. Data I/O is as limiting as CPU and RAM.
A bad tradition in bioinformatics is ignoring the cost of I/O. Large-scale distributed systems need to model the I/O cost explicitly and design their solution to include I/O cost as well as CPU cost.
L3. Using smart data products helps avoid costly recomputations and empowers downstream tool builders.
The bad tradition of downloading raw data and creating spreadsheets with results is not sustainable. While bioinformatics is not yet able to fully rely on disseminating data as research objects [63], we need to move toward them.
L4. The use of reproducible workflows such as CWL [55, 64] is a crucial requirement for any service generating data meant for reuse.
Providing a detailed, portable, executable recipe for how the data were generated is important to data consumers. In addition, making the recipes available supports improvement to the workflows by third parties.
L5. Containers should be used to capture the execution environment.
Containers (e.g. Linux containers) capture the environment in a reproducible format.
Workflows without their environment are less than useful.
L6. Data reuse is critical for saving computational cost.
While the reproducibility resulting from reproducible execution environments is great, providing intermediate results adds significantly more value to reviewers and fosters reuse of computational results for a variety of purposes such as building software to improve existing components (e.g. feature predictors) or use the data for scientific projects.
L7. Metadata is invaluable and should be required.
Users require encouragement to provide metadata. We aim to make users submit metadata as early as possible, and to incentivize users, we provide high-quality tools that make metadata collection easy.
L8. The complexity of shared filesystems should be avoided whenever possible.
Relying on RESTful interfaces instead of shared filesystems provides cross-cloud execution capabilities, allowing us to run on almost any computational platform including the cheapest computational platform available.
L9. Portals are the right place for performance engineering.
While many biomedical informatics groups are computationally proficient, the convergence of large-scale processing and domain expertise makes portal sites an ideal location for optimization. Running many workflows thousands of times and providing services to many other groups is a good platform for accumulating expertise.
Discussion
As more environmental DNA sequence data become available to the research community, a new set of challenges emerges. These challenges require a change in approach to computing at the community level. We describe a domain-specific portal that, like its European companion system [11], acts as an integrator of data and efficiently implements domain-specific workflows. The lessons learned about building scale-out infrastructure dedicated to executing bioinformatics workflows and the resulting middleware systems [19, 20, 40, 59, 65] will benefit both the community of users and researchers attempting to build efficient sequence analytics workflows.
Reproducible efficient execution of domain-specific workflows is a central contribution of the MG-RAST system. Provisioning of data and results via a Web interface and a RESTful API is another key aspect. Encouraging data reuse by provisioning both data and results (as well as intermediate files) via a stable API is a key function that serves the community of bioinformatics developers, who can use precomputed data that are well described by a workflow, rather than implementing their own (frequently subpar preprocessing steps), and thus can focus on their key mission.
By providing preanalyzed data (using an open recipe that is available to the community for discussion and improvement), MG-RAST can help reduce the current ‘method uncertainty’, where individual data sets analyzed with different analysis strategies can lead to dramatically different interpretations.
The role of MG-RAST is not one-size-fits-all. Rather than being the one and only analysis mechanism, MG-RAST is a well-designed high-performance system on top of an efficient scale-out platform [66] that can take some of the heavy lifting off the shoulders of individual researchers. Researchers can add their own custom boutique analyses at a fraction of the computational and development cost, allowing them to focus on their specific problem and thus maximizing overall productivity.
With the state of the art of sequencing technology shifting, MG-RAST will adapt to extract maximum value by, for example, explicitly supporting value-added information from longer sequences with multiple features, for example for taxonomy calling. We also anticipate that the currently used alignment-based methods will be supplemented by profile-based methods for performance reasons within a few years.
Key Points
Analyzing the growing volume of biomedical environmental sequence data requires cost-effective, reproducible and flexible analysis platforms and data reuse and is significantly different from analyzing (almost) complete genomes.
The hosted MG-RAST service provides a Linux container-based workflow system and a RESTful API that allow data and analysis reuse.
Community portals are the right location for performance engineering, as they operate at the required scale.
Acknowledgements
The authors thank Dion Antonopoulos, Gail Pieper and Robert Ross for their input and help.
Biographies
Folker Meyer is a Senior Computational Biologist at Argonne National Laboratory; a Professor at the Department of Medicine, University of Chicago; and a Senior Fellow at the Computation Institute at the University of Chicago. He is also deputy division director of the Biology Division at Argonne National Laboratory and a senior fellow at the Institute of Genomics and Systems Biology (a joint Argonne National Laboratory and University of Chicago Institute).
Saurabh Bagchi is a Professor in the School of Electrical and Computer Engineering and the Department of Computer Science (by courtesy) at Purdue University. He is the founding Director of CRISP, a university-wide resiliency center at Purdue.
Somali Chaterji is a biomedical engineer and medical data analyst. She is a Research Faculty at Purdue University, specializing in high-performance computing infrastructures and algorithms for synthetic biology and epigenomics.
Wolfgang Gerlach, PhD is a Bioinformatics Senior Software Engineer at the University of Chicago with a joint appointment at Argonne National Laboratory.
Ananth Grama is a Professor of Computer Science at Purdue University. He also serves as the Associate Director of the Center for Science of Information, a Science and Technology Center of the National Science Foundation.
Travis Harrison is a Bioinformatics Senior Software Engineer at the University of Chicago with a joint appointment at Argonne National Laboratory.
Tobias Paczian is a Senior Developer at the University of Chicago with a joint appointment at Argonne National Laboratory. He has more than a decade of experience building User Interfaces for bioinformatics applications.
William L. Trimble, PhD is a postdoctoral researcher at Argonne National Laboratory with a background in physics and data science.
Andreas Wilke is a Principal Bioinformatics Specialist Argonne National Laboratory with a joint appointment at the University of Chicago. He has more than a decade of experience building bioinformatics applications.
Funding
The work reported in this article was supported in part by a grant from the National Institutes of Health (NIH) grant 1R01AI123037-01. Work on this article was also supported by NSF award 1645609. This work was supported in part by the NIH award U01HG006537 ‘OSDF: Support infrastructure for NextGen sequence storage, analysis, and management’, by the Gordon and Betty Moore Foundation with the grant ‘6-34881, METAZen-Going the Last Mile for Solving the Metadata Crisis)’. This material was based on work supported by the US Department of Energy, Office of Science, under contract DE-AC02-06CH11357.
References
- 1.NHGRI. DNA sequencing costs. https://www.genome.gov/sequencingcosts/.[TQ1]
- 2. Afgan E, Baker D, van den Beek M, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res 2016;44:W3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Doring A, Weese D, Rausch T, et al. SeqAn an efficient, generic C ++ library for sequence analysis. BMC Bioinformatics 2008;9:11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xia F, Dou Y, Xu J. Families of FPGA-based accelerators for BLAST algorithm with multi-seeds detection and parallel extension. In: Elloumi M, Küng J, Linial M, et al. (eds), Bioinformatics Research and Development: Second International Conference, BIRD 2008 Vienna, Austria, July 7-9, 2008 Proceedings Berlin, Heidelberg: Springer Berlin Heidelberg, 2008, 43–57.
- 5. Buchfink B, Xie C, Huson DH.. Fast and sensitive protein alignment using DIAMOND. Nat Methods 2015;12:59–60. [DOI] [PubMed] [Google Scholar]
- 6. Meyer F, Paarmann D, D'Souza M, et al. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 2008;9:386.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wilke A, Bischof J, Gerlach W, et al. The MG-RAST metagenomics database and portal in 2015. Nucleic Acids Res 2016;44:D590–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wilke A, Bischof J, Harrison T, et al. A RESTful API for accessing microbial community data for MG-RAST. PLoS Comput Biol 2015;11:e1004008.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Desai N, Antonopoulos D, Gilbert JA, et al. From genomics to metagenomics. Curr Opin Biotechnol 2012;23:72–6. [DOI] [PubMed] [Google Scholar]
- 10. Chen IA, Markowitz VM, Chu K, et al. IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Res 2017;45:D507–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mitchell A, Bucchini F, Cochrane G, et al. EBI metagenomics in 2016–an expanding and evolving resource for the analysis and archiving of metagenomic data. Nucleic Acids Res 2016;44:D595–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huson DH, Weber N.. Microbial community analysis using MEGAN. Methods Enzymol 2013;531:465–85. [DOI] [PubMed] [Google Scholar]
- 13. Kopylova E, Noe L, Touzet H.. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012;28:3211–7. [DOI] [PubMed] [Google Scholar]
- 14. Kang DD, Froula J, Egan R, et al. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 2015;3:e1165.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eren AM, Esen OC, Quince C, et al. Anvi'o: an advanced analysis and visualization platform for 'omics data. PeerJ 2015;3:e1319.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Imelfort M, Parks D, Woodcroft BJ, et al. GroopM: an automated tool for the recovery of population genomes from related metagenomes. PeerJ 2014;2:e603.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alneberg J, Bjarnason BS, de Bruijn I, et al. Binning metagenomic contigs by coverage and composition. Nat Methods 2014;11:1144–6. [DOI] [PubMed] [Google Scholar]
- 18. Segata N, Waldron L, Ballarini A, et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods 2012;9:811–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tang W, Bischof J, Desai N, et al. Workload characterization for MG-RAST metagenomic data analytics service in the cloud. In: Proceedings of IEEE International Conference on Big Data, Washington, DC, USA, 2014. IEEE Press, Piscataway, NJ, USA.
- 20. Tang W, Wilkening J, Bischof J, et al. Building scalable data management and analysis infrastructure for metagenomics. In: 5th International Workshop on Data-Intensive Computing in the Clouds, Poster at Supercomputing2013.
- 21. Wilke A, Wilkening J, Glass EM, et al. An experience report: porting the MG-RAST rapid metagenomics analysis pipeline to the cloud. Concurr Comput 2011;23:2250–7. [Google Scholar]
- 22.openstack.org. OpenStack. https://www.openstack.org/.
- 23. Amazon Inc. Amazon Web Services. https://aws.amazon.com/.
- 24. Microsoft Inc. Azure. https://azure.microsoft.com/.
- 25. Kanehisa M, Goto S, Sato Y, et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 2014;42:D199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. KEGG. KAAS - KEGG automatic annotation server. http://www.genome.jp/kegg/kaas/.
- 27. Tatusov RL, Fedorova ND, Jackson JD, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 2003;4:41.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. O'Leary NA, Wright MW, Brister JR, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 2016;44:D733–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004;32:1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wikipedia. https://en.wikipedia.org/wiki/LAMP_(software_bundle).
- 31. Plummer E, Twin J, Bulach DM, et al. A comparison of three bioinformatics pipelines for the analysis of preterm gut microbiota using 16S rRNA gene sequencing data. J Proteom Bioinform 2016;283–91. [Google Scholar]
- 32. Alexandre R. Instant Apache Solr for Indexing Data How-to. Packt Publishing Limited, 2013. [Google Scholar]
- 33. Elasticsearch BV, Elastic search. https://www.elastic.co/products/elasticsearch.
- 34. Cassandra. http://cassandra.apache.org/.
- 35. Inc. O. MySQL. https://www.mysql.com/.
- 36. Bent J, Gibson G, Grider G, et al. A Checkpoint Filesystem for Parallel Applications. 2009. [Google Scholar]
- 37. Jens Freche WF, Sutmann G.. High-Throughput Parallel-I/O using SIONlib for Mesoscopic Particle Dynamics Simulations on Massively Parallel Computers, Advances in Parallel Computing; Volume 19: Parallel Computing: From Multicores and GPU's to Petascale, 371–78, DOI: 10.3233/978-1-60750-530-3-371. IOS Press, Amsterdam.
- 38. Jay FL, Scott K, Karsten S, et al. Flexible IO and integration for scientific codes through the adaptable IO system (ADIOS). In: Proceedings of the 6th International Workshop on Challenges of Large Applications in Distributed Environments Boston, MA: ACM, 2008, 15–24.
- 39. Dennis JM, Edwards J, Loy R, et al. An application level parallel I/O library for earth system models. Int J High Perform Comput Appl 2012;26:43–53. [Google Scholar]
- 40. Bischof J, Wilke A, Gerlach W, et al. Shock: active storage for multicloud streaming data analysis. In: 2nd IEEE/ACM International Symposium on Big Data Computing Limassol, Cyprus, 2015.
- 41. Wilkening J, Wilke A, Desai N, et al. Using Clouds for Metagenomics: A Case Study CLUSTER. New Orleans, LA: IEEE Computer Society, 2009, 1–6. [Google Scholar]
- 42. Felter W, Ferriera A, Rajamony R, et al. An Updated Performance Comparison of Virtual Machines and Linux Containers.http://domino.research.ibm.com/library/cyberdig.nsf/papers/0929052195DD819C85257D2300681E7B/$File/rc25482.pdf.
- 43. Gerlach W, Tang W, Keegan K, et al. Skyport: container-based execution environment management for multi-cloud scientific workflows. In: Proceedings of the 5th International Workshop on Data-Intensive Computing in the Clouds 2014, 25–32. IEEE Press, Piscataway, NJ, USA.
- 44. Kurtzer G, Sochat V, Bauer M.. Singularity: Scientific containers for mobility of compute. PLoS ONE 2017;12(5):e0177459.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. udocker. https://github.com/indigo-dc/udocker.
- 46. Keegan KP, Trimble WL, Wilkening J, et al. A platform-independent method for detecting errors in metagenomic sequencing data: DRISEE. PLoS Comput Biol 2012;8:e1002541.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Marcais G, Kingsford C.. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 2011;27:764–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Aronesty E. Comparison of sequencing utility programs. Open Bioinform J 2013;7:1–8. [Google Scholar]
- 49. Langmead B, Salzberg SL.. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rho M, Tang H, Ye Y.. FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res 2010;38:e191.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rognes T, Flouri T, Nichols B, et al. VSEARCH: a versatile open source tool for metagenomics. PeerJ 2016;4:e2584.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wilke A, Harrison T, Wilkening J, et al. The M5nr: a novel non-redundant database containing protein sequences and annotations from multiple sources and associated tools. BMC Bioinformatics 2012;13:141.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res 2002;12:656–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Overbeek R, Bartels D, Vonstein V, et al. Annotation of bacterial and archaeal genomes: improving accuracy and consistency. Chem Rev 2007;107:3431–47. [DOI] [PubMed] [Google Scholar]
- 55. Amstutz P, Crusoe MR, Tijanić N, et al. Common Workflow Language, v1.0. 10.6084/m9.figshare.3115156.v2. [DOI]
- 56. Mungall CJ, Torniai C, Gkoutos GV, et al. Uberon, an integrative multi-species anatomy ontology. Genome Biol 2012;13:R5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Buttigieg PL, Morrison N, Smith B, et al. The environment ontology: contextualising biological and biomedical entities. J Biomed Semantics 2013;4:43.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Field D, Sterk P, Kottmann R, et al. Genomic standards consortium projects. Stand Genomic Sci 2014;9:599–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bischof J, Harrison T, Paczian T, et al. Metazen - metadata capture for metagenomes. Stand Genomic Sci 2014;9:18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yilmaz P, Kottmann R, Field D, et al. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat Biotechnol 2011;29:415–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Glass EM, Dribinsky Y, Yilmaz P, et al. MIxS-BE: a MIxS extension defining a minimum information standard for sequence data from the built environment. ISME J 2014;8:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Trimble WL, Keegan KP, D'Souza M, et al. Short-read reading-frame predictors are not created equal: sequence error causes loss of signal. BMC Bioinformatics 2012;13:183.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sean B, Buchan I, De Roure D, et al. Why linked data is not enough for scientists. Fut Gener Comput Syst 2013;29(2): 599–611. [Google Scholar]
- 64. Crusoe MR, Brown CT.. Walking the talk: adopting and adapting sustainable scientific software development processes in a small biology lab. J Open Res Softw 2016;4:e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tang W, Wilkening J, Desai N, et al. A scalable data analysis platform for metagenomics. In: 2013 IEEE International Conference on Big Data, Silicon Valley, CA, USA, 2013. IEEE Press, Piscataway, NJ, USA.
- 66. Michael M, Moreira JE, Shiloach D, et al. Scale-up x scale-out: a case study using Nutch/Lucene. In: 2007 IEEE International Parallel and Distributed Processing Symposium 2007, 1.