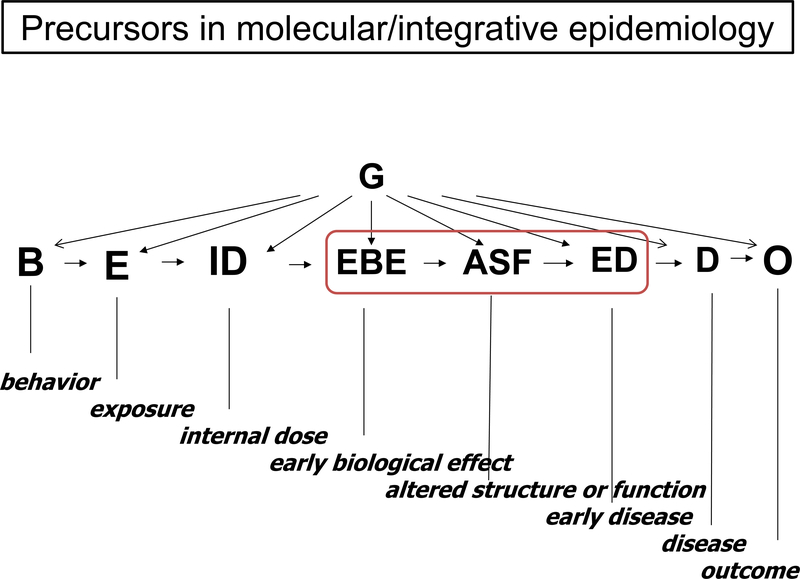
Cancer precursors are the tissue antecedents of cancer. They are distinguished from biomarkers in that they reflect a morphologic as opposed to a merely biochemical or genetic association with a specific cancer. Precursors to cancers are increasingly recognized as universal, relevant to carcinogenesis, and providing a unique potential for primary and secondary prevention. As the molecular, imaging, and genomic tools to investigate them evolve, it is becoming increasingly evident that virtually all malignancies are preceded by these clinically silent states where the molecular lesions that characterize the specific cancer emerge. In terms of both time and antecedent molecular events, cancer is the visible iceberg; the 90% submerged is the precursor. We also take it as axiomatic that all morphologic changes must be accompanied by an extensive and proximal scaffolding of molecular alterations. Biologically, molecular and morphologic lesions must correspond, but our understanding of their relationship is still limited. Vogelstein’s classic paradigm [1] for the molecular steps in progression to colon cancer and similar schemas for other tumors remain incomplete. A key theme of this review is that the most direct path to understanding the molecular taxonomy of tumors and what determines their progression (precursor>tumor>metastasis) should emphasize investigating precursor lesions in population studies (Figure 1). Here, detailed information on exposure and outcome combined with biomarker studies (serial where possible) can characterize molecular steps in progression and refine their relation to morphology.
Figure 1.
The Figure is adapted from previous integrative[13, 14] and molecular[15, 16] epidemiology schema. Precursors loosely reside in the region between ‘early biological effects’ and ‘early disease’. ‘Early biological effects’ imply something more than dose but still proximal on the pathway to disease. ‘Altered structure and function’, implies biologically manifest changes with a morphology component that must be accompanied by molecular events. This comes closed to what we understand as a precursor, with the caution that such events may be evanescent (reversible) and lack specificity for a neoplastic end point. ‘Early disease’ progresses further along the pathway but may reflect ‘in situ’ cancer, typically undetectable clinical states. All of these will be fruitful for study, and their investigation in the context of population study designs will be required to refine our knowledge of molecular and morphology correlates.
New technologies and the rapidly expanding database of molecular and genomic lesions in tumors are expected to reveal the underpinnings of malignancy with attendant benefits: new insight into etiology, reinvigoration of screening and prevention, and potential to enhance the therapeutic armamentarium. Large scale advanced technology initiatives such as the Tumor Cancer Genome Atlas (TCGA) focuses on tumor tissue characterization and ENCODE on functional elements of the genome[2]. These efforts and others that aim to elucidate the somatic underpinnings (as well as expression, methylation, and other manifestations of a dynamic genome) of neoplasia have appropriately focused on those with frank malignancy. Regarding the environmental exposures that account for most cancer, metabolomic, microbiome, and related approaches[3, 4] will offer insight into chemical and infectious exposures while mobile technologies will allow more quantitative, detailed and accurate assessment of sleep, physical activity, light exposure, diet and circadian variation in unprecedented detail. A priority is to extend this exciting body of work to the earlier steps on the pathway to malignancy[5], i.e. cancer precursors. These approaches will not only to assess clues to their origin, but will investigate what determines progression.
Precursors provide a substrate for early intervention and prevention. Frank malignancy represents the endpoint of a series of molecular steps and is difficult to treat or reverse. Precursors represent a more malleable and potentially reversible state where less aggressive interventions may avert or slow progression. Screening may offer the greatest advantage when detection of an asymptomatic cancer/precursor is possible and a curative intervention (in the so-called Detectable Preclinical Phase) exists, as it does for cervix[6] and colorectal[7] and selected skin precursors (i.e. actinic keratosis). In other cancers such as breast[8], prostate, lung and melanoma, screening-based early detection has a more complex risk-benefit and further research is needed to understand how to best avoid adverse outcomes and identify the most critical determinants of progression. Some of the clearest gains in avoiding cancer deaths have been made from cervical and colon cancer screening. Although they are not explicit precursors as earlier defined, infectious agents associated with specific malignancies offer opportunities for cancer prevention where vaccination is established, i.e., HBV (liver), HPV (cervix, anus, oral) and for identification of individuals at elevated risk for follow-up based on infection with HCV (liver), EBV (Burkitt’s), and HTLV1 (ATL).
The molecular dissection of cancer promises to redefine the taxonomy of cancer, eventually superseding the traditional morphologic definition. Among the diverse molecular abnormalities observed in a given tissue sample, it is not always easy to identify the ‘drivers’ that specify the malignant phenotype. A comparison of tissue obtained in precursors, early and advanced cancer can provide an unparalleled opportunity to dissect out the critical elements.
Biomarkers are increasingly proposed as plausible surrogates to follow cancer and provide diagnostic, prognostic or therapeutic guidance. Complementary markers (that fill ‘gaps’ in screening by other modalities like imaging or genetics) are urgently needed to improve screening, detect early recurrence, identify individuals at highest risk or with poor prognosis. Early identification of the molecular features of individuals fated to progress could allow early and potentially more effective treatment to those at greatest risk and spare others.
Study design impacts the ability to conduct informative research on precursors. The ideal is a defined population setting with rich exposure data, outcome information, and repeated access to tissue for biomarker study. Cohort studies which obtain information and biospecimens prior to disease are similarly free of recall bias, but extensive tissue and in depth questionnaire information are lacking. Case-control studies obtain tissue and questionnaire data that focuses on the disease of interest, but the impact of clinically manifest cancer in the host can alter laboratory assays or cause recall bias in retrospective study designs. In contrast to studies focusing on cancer, individuals with precursor conditions are free of the clinical consequences of cancer. Subjects are undisturbed by late clinical or treatment sequelae of frank cancer and therefore effect-cause bias is absent.
Studies in precursors can a) focus heavily on the associated malignancy, b) be mostly free of clinical effects of cancer in the host, c) obtain targeted biospecimens of the tissue of interest. Precursors are much more common than the underlying cancers they precede. Therefore, they can sometimes offer better statistical power to test selected hypotheses (in comparison with a general population group). A limitation is that morphologic manifestations of preneoplastic changes may be evanescent or reversible, of example, dysplasia in the respiratory epithelium on those who quit smoking. Progression of a precursor to the malignancy of interest will be substantially higher in an identified group with the precursor than in a similar age-matched comparison group, although precursors develop in younger individuals than do the corresponding cancers. So, studies focused on environmental, genetic, laboratory or clinical features of progression will enjoy improved power.
The factors that propel or delay a precursor along the path to malignancy are mechanistically and clinically important, as well as potential targets for early interventions. Studies focused on treatment of precursors or early cancers with chemopreventive agents have had limited success. With an understanding of the key molecular lesions that drive progression, this effort could be specifically targeted and rationally based. Improved outcomes and more efficient trails would result since an intermediate endpoint might allow us to track response without waiting for morbid events.
Exposures accumulate through life and damage target tissues. Yet even for the best established and quantified exposures like cigarette smoking, no ‘molecular dosimeter’ can be quantitated. For malignancies like lung cancer where credible risk models exist based on measured exposure, a tissue dosimeter of damage related to development of lung cancer or of a premalignant state (i.e., for squamous cell carcinoma, field defects) would be valuable since it could improve risk models, help evaluate ameliorative interventions, improve models for association with inherited genes, and enhance screening. Morphologic changes do not track overall damage well, so identification of the underlying molecular alterations could be a key advance. In some cases, exposures are unknown or weak predictors of malignancy, but genetic risk as defined by family history is a strong marker of risk. There are several examples of gene mutations responsible for cancer predisposition including retinoblastoma, colorectal cancer (familial adenomatous polyposis) and early onset breast cancer. For some conditions such as monoclonal B-Cell lymphocytosis, a precursor to CLL, despite increased occurrence in high risk families, neither an associated set of exposures nor a specific gene that confers high risk is known. In general, SNPs identified in GWAS as associated with various malignancies have yet to neither show a strong impact in risk models nor find utility in screening[9].
Precursors must represent some of the earliest events on the path to malignancy. Cancer genomes have 10’s to 100’s of somatic alterations per tumor, accompanied by intertumor heterogeneity[10]. With progression, further molecular alterations may supervene, accelerated by genomic instability. Characterizing the earliest alterations present in the precursor state may prove efficient in defining the necessary and sufficient conditions, and in identifying which events are drivers and which are epiphenomenon.
A central mystery of cancer is the contrast between the characteristic features that fundamentally define neoplasia of a specific organ (unchecked growth, inevitable progression towards metastasis) and the differences in speed of progression, outcome and response to treatment in individual tumors. How is this clinical unity maintained when the total set of somatic mutations documented in sets of tumors appears variable (along with the specific environmental and genetic factors)? The minimal modular (one from column A, one from Column B, etc.) set of mutations and their shared characteristics remains poorly understood. While there is broad agreement that an integrated ‘system’ of molecular alterations best characterizes this situation, understanding the features of the earliest altered tissue states can provide insight into the puzzle. Cross sectional comparisons will be useful (that is, conducting these studies on different individuals with early lesions and cancer) but having serial studies of individual tumors derived from interdisciplinary studies (Figure 1) will best demonstrate the specific changes that drive progression.[11–13]
Formidable barriers remain. Existing populations-based studies often fail to identify, characterize or focus on precursors. The longitudinal investigations best suited to following their evolution are time consuming, expensive and may lack provision to collect biospecimens. Tissue resources to study precursors tend to be difficult to obtain and smaller in quantity (compared to frank tumors). For some malignancies, the precise precursor lesion is controversial. In general, the exposure and genetic factors related to precursors and their progression are less well understood than those for the associated malignancy.
Notwithstanding these formidable challenges, we strongly advocate systematic study of early lesions to leverage the application of advanced technologies in cancer. Contrasting the findings in precursor lesions with those in cancer will shed light on pathways of progression, enhance fundamental understanding of the molecular roots of cancer, provide new avenues of prevention and perhaps reveal how the ‘system’ itself evolves to acquire the malignant phenotype. To provide the maximum impact and render interpretable the results of refined laboratory approaches on the relevant tissues, these studies need to be conducted in populations that are well characterized with regard to study design, exposure assessment, biomarker and tissue availability, and include clinical outcomes such as survival.
REFERNECES
- 1.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nature Medicine 2004;10:789–99. [DOI] [PubMed] [Google Scholar]
- 2.Dunham I, Kundaje A, Aldred SF, et al. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rappaport SM. Biomarkers intersect with the exposome. Biomarkers : biochemical indicators of exposure, response, and susceptibility to chemicals 2012;17:483–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi Q, Qin L, Wei W, Geng F, Fan R, Shin YS et al. Single-cell proteomic chip for profiling intracellular signaling pathways in single tumor cells. Proceedings of the National Academy of Sciences of the United States of America 2012;109:419–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Umar A, Dunn BK, Greenwald P. Future directions in cancer prevention. Nature Reviews Cancer 2012;12:835–48. [DOI] [PubMed] [Google Scholar]
- 6.Pierce Campbell CM, Menezes LJ, Paskett ED, Giuliano AR. Prevention of invasive cervical cancer in the United States: past, present, and future. Cancer Epidemiology, Biomarkers Prevention 2012;21:1402–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson CM, Cassells AN, Greene MA, Beach ML, Tobin JN, Dietrich AJ. Barriers to colorectal cancer screening among publicly insured urban women: no knowledge of tests and no clinician recommendation. Journal of the National Medical Association 2011;103:746–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Assi V, Warwick J, Cuzick J, Duffy SW. Clinical and epidemiological issues in mammographic density. Nature Reviews Clinical oncology 2012;9:33–40. [DOI] [PubMed] [Google Scholar]
- 9.Wacholder S, Hartge P, Prentice R, Garcia-Closas M, Feigelson HS, Diver WR, et al. Performance of common genetic variants in breast-cancer risk models. The New England Journal of Medicine 2010;362:986–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet 2010;11:685–96. [DOI] [PubMed] [Google Scholar]
- 11.Ogino S, Chan AT, Fuchs CS, Giovannucci E. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut 2011;60:397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perera FP, Weinstein IB. Molecular epidemiology and carcinogen-DNA adduct detection: new approaches to studies of human cancer causation. Journal of Chronic Diseases 1982;35:581–600. [DOI] [PubMed] [Google Scholar]
- 13.Caporaso NE. Integrative study designs--next step in the evolution of molecular epidemiology? Cancer Epidemiology, Biomarkers & Prevention 2007;16:365–6. [DOI] [PubMed] [Google Scholar]
- 14.Spitz MR, Caporaso NE, Sellers TA. Integrative cancer epidemiology--the next generation. Cancer Discovery 2012;2:1087–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perera FP. Molecular cancer epidemiology: a new tool in cancer prevention. J Natl Cancer Inst 1987;78:887–98. [PubMed] [Google Scholar]
- 16.Rothman N, Stewart WF, Schulte PA. Incorporating biomarkers into cancer epidemiology: a matrix of biomarker and study design categories. Cancer Epidemiology, Biomarkers & Prevention 1995; 4:301–11. [PubMed] [Google Scholar]