Abstract
One of the aims of systems biology is to model and discover properties of cells, tissues and organisms functioning as a system. In recent years, studies in the adult Drosophila gut have provided a wealth of information on the cell types and their functions, and the signaling pathways involved in the complex interactions between proliferating and differentiated cells in the context of homeostasis and pathology. Here, we document and discuss how high-resolution ultrastructure studies of organelle morphology have much to contribute to our understanding of how the gut functions as an integrated system.
Keywords: Ultrastructure, Organelle, Electron microscopy, Drosophila midgut
Introduction
The adult Drosophila midgut is a complex tissue with various cell types that interact closely to maintain tissue integrity and perform organ function. The gut consists of a pseudostratified epithelium, a latticework of circular and longitudinal visceral muscles that supports the epithelium, and a tracheal vascular system. The major cell types of the midgut epithelium are the absorptive enterocytes (ECs), characterized by a large nucleus and microvilli-covered luminal surface, the enteroendocrine cells (EEs) that produce various hormones, and the intestinal stem cells (ISCs) that produce ECs and EEs [1,2]. Interactions between these cell types are critical to maintaining tissue integrity and gut function. For example, ISCs proliferation and differentiation are controlled by a complex network integrating autocrine and paracrine signals [3,4]; hormones derived from EEs regulate EC physiology; and EC-derived factors signal to ISCs following gut damage.
Despite the body of knowledge about signaling in the gut, we are still far from understanding how different signals are processed and integrated inside the cell to orchestrate a particular cellular function. As much of the cell is organized in membrane-bound or membrane-free organelles, and that organelles can serve as the venues for signal transduction [5–7], ultrastructure studies examining changes in organelle morphology, size, number, and location can provide cues on how signal integration is coupled with organelle behaviors and inter-organelle communication under healthy or pathological conditions.
In this review, we illustrate, from our ultrastructure analysis of the cell types in the gut, how examining tissue biology at subcellular resolution could lay the groundwork for studying cellular signaling under normal or stressed conditions. We argue that describing organelle phenotypes at the ultrastructure level should play a more prominent part in the phenotypic characterization of tissues, and that such studies are necessary to obtain a systems level understanding of tissue biology and organ function.
Histological and ultrastructure analyses inspired the identification of stem cells in the Drosophila midgut
The concept of a stem cell population in the midgut epithelium dates back to histology analyses in the 1940s. As Albert Miller described in the book “Biology of Drosophila” [8], “the regenerative cells are infrequent, are sometimes wedge-shaped with a tapering apex, and have denser cytoplasm than the active (absorptive) cells; their nuclei are also denser, smaller, and situated near the base”. Later, using electron microscopy (EM), Otto Baumann described that regenerative cells are far smaller in size, have no basal infoldings, fewer mitochondria, and fewer rough endoplasmic reticulums (ERs) [9]. Baumann also noticed that the adherens junction protein Armadillo (Arm) and the non-neuronal isoform of the cell adhesion protein Neuroglian are enriched in regenerative cells.
Definite proof of the existence of stem cells in the adult gut will have to wait experimental evidence provided by two independent studies in 2006 [1,2]. Because escargot (esg) is required to maintain diploidy of imaginal cells [10], Micchelli and Perrimon examined midgut expression of the esgGal4 enhancer trap line, and found that esgGal4 specifically labels the diploid “regenerative” cells. Immunostainings revealed that the mitosis marker phospho-Histone H3 (pH3) is only detectable in esg+cells [1]. In addition, esgGal4-driven cell depletion causes complete and irreversible elimination of mitosis in the midgut [11,12]. Furthermore, using mosaic analysis with a repressible cell marker (MARCM) [13], Micchelli and Perrimon randomly labeled the progenies of mitotic cells and found that both ECs and EEs can arise from the same clones harboring esg+cells [1]. Similarly, Ohlstein and Spradling, inspired by the earlier studies of Miller and Baumann, searched for stem cells in the midgut using a FLP-FRT based lacZ reconstitution system for random lineage labeling and found that only clones arising from regenerative cells have the capacity for long term expansion [2].
Ultrastructure analysis and functional studies of organelles in ISCs
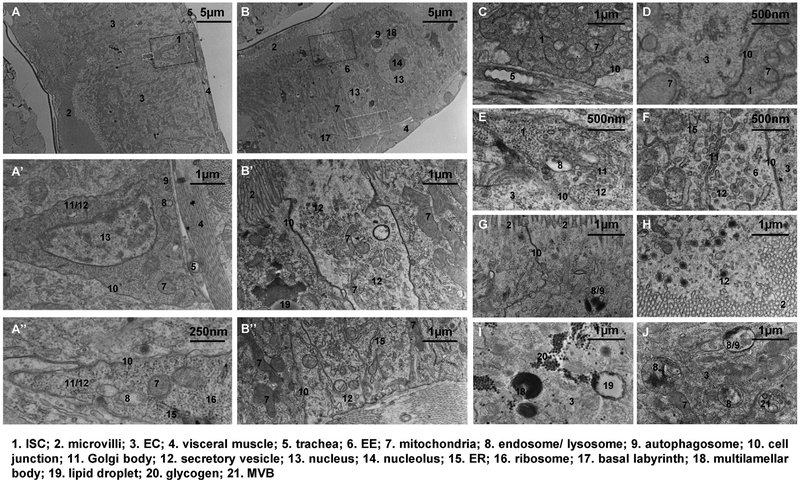
Following on earlier EM findings [9,14,15], we reexamined the ultrastructure of the posterior midgut at higher resolution and in various conditions. ISCs have dense cytoplasm and often appear darker than ECs in electron micrographs [2,8]. Our high-resolution electron micrographs revealed that ISCs have a very high density of free ribosomes (Figure 1A, ribosomes can be easily recognized in the magnified view in 1A″), which might decrease after tissue damage (Figure 2F′). Proteins synthesized on free ribosomes either remain in the cytosol or incorporate into other organelles such as the nucleus and mitochondria [16]. Whether ISCs depend on enriched free ribosomes for their stress response, self-renewal, or differentiation is yet to be determined.
Figure 1.
Electron micrographs of normal midguts. (A) An ISC localized between two ECs, with the magnified views of the region circled by the red dashed box shown in (A′), the apex region circled by the yellow box shown in (A″). Plastic sections of posterior midguts from wild type (genotype: w1118) young adult flies are used for EM unless noted otherwise. Each image is representative of N>3 sections. (B) An EE localized between two ECs, with the magnified views of the apical region circled by the red dashed box shown in (B′), the basal region circled by the yellow box shown in (B″). (C) An ISC localized next to the trachea, with only the basement membrane between them. (D) Mitochondria in an ISC and its neighbor EC. (E) A Golgi body localized at the apex of an ISC. (F) A Golgi body localized in an EE. (G) Tight junction between two ECs. (H) Secretory vesicles near the microvilli of an EC. (I) Glycogen granules, a lipid droplet, and two multilamellar bodies found within one region of the EC. (J) Endosomes/lysosomes found in an EC. Note that the multivesicular body (MVB) is a type of late endosome and the autophagosome can be considered a type of lysosome that engulfs organelles.
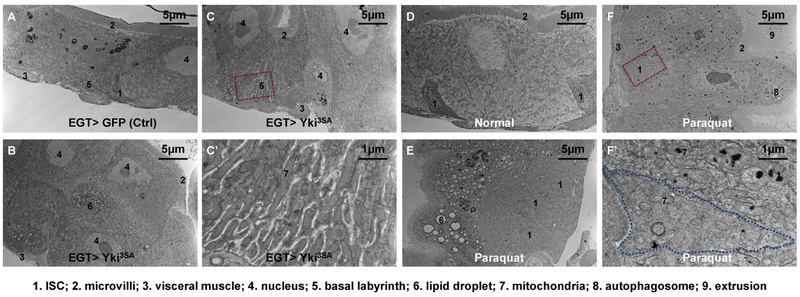
Figure 2.
Electron micrographs of midguts under pathological conditions. (A) The pseudostratified epithelium of midguts expressing GFP in ISCs for 4d with the EGT (esgGal4, UAS-GFP, tubGal80ts) expression driver. (B) The multi-layered epithelium of midguts expressing Yki3SA in ISCs for 4d. (C) A normal EC stretching from the basement membrane to the lumen found in a section of midgut Yki tumor. The magnified view of its extended basal labyrinth is shown in (C′). (D) Midgut epithelium of wild type (genotype: w1118) flies fed on normal food. (E) EC vacuolation and ISC expansion in the midgut epithelium of wild type flies fed on food containing 2 mM paraquat for 2d. Note that under tissue damage conditions the ISCs might no longer be enriched with free ribosomes, but they could be recognized by their cell boundary, small nuclei, and basal localization. (F) Deposition of electron-dense materials in the mitochondria and autophagosomes of ECs, but not in ISCs. Magnified view of an ISC and its neighboring EC is shown in (F′).
ISCs appear to have small mitochondria with much fewer cristae than mitochondria found in ECs (Figure 1A′, A″, C and D). It has been proposed that mitochondria cristae enhance the efficiency of oxidative phosphorylation by acting as proton traps for ATP synthase [17]. Previous studies have connected mitochondria with ISC activity. For example, induction of mitochondria biogenesis by overexpressing dPGC1α/spargel inhibits the levels of reactive oxygen species (ROS) and aging-related ISC overproliferation, leading to an increase in life span [18]. Moreover, knockdown of the mitophagy-related genes Pink1 or Parkin results in increased electron density in the mitochondrial matrix of ISCs, increased the number of swollen mitochondria in ISCs in old flies, and decreased ISC proliferation [19]. However, none of these studies focused on the unique morphology of mitochondria cristae in ISCs. Interestingly, Drosophila germline stem cells also have much fewer cristae in their mitochondria than differentiated germ cells, and knockdown of ATP synthase, but not other members of the oxidative phosphorylation system, inhibits mitochondria cristae formation and germ cell differentiation [20]. The roles of mitochondrial membrane maturation and oxidative phosphorylation in ISCs remain to be elucidated.
The Golgi body and the ER are readily detectable in the electron micrographs of ISCs, often localizing at the apex (Figure 1A″and E). These organelles are responsible for the production of a variety of secretory or membrane proteins that are essential for ISCs. The Golgi body produces lipolysis enzymes, which are transported via the coat protein complex I (COPI) to the surface of lipid droplets [21]. Interestingly, depletion of Drosophila COPI or its associated GTPase Arf1 induces necrosis in ISCs but spares differentiated cells [22], suggesting a possible dependency on lipolysis for energy supply in ISCs.
Moreover, the ER-stress responsive transcription factor Xbp1 and the ER-associated degradation pathway component Hrd1 restrict ISC proliferation by preventing ROS production and c-Jun N-terminal kinase (JNK) activation [23]. In response to ER stress or JAK/STAT activation, the PKR-like ER kinase is activated specifically in ISCs to induce proliferation [24]. The ER is also the largest store of releasable Ca2+ in the cell [25]. Calcium channels and transporters located in the ER membrane are crucial for controlling intracellular Ca2+ levels in ISCs [26], which could in turn affect proliferation via Ras/MAPK signaling [12] or affect differentiation via inhibition of Notch activity [27].
A small number of endosomes and autophagosomes can also be recognized in the electron micrographs of ISCs (Figure 1A′, A″ and E). Endocytosis and autophagy play important roles modulating signaling activity in ISCs. For example, endocytosis of the JAK/STAT pathway receptor Domeless is required to prevent excessive JAK/STATactivity [28]. Moreover, dietary lipids can regulate ISC differentiation in newly-eclosed adult flies by modulating the endocytosis of the Notch extracellular domain and Notch pathway ligand Delta [29]. In addition, the asymmetric distribution of endosomes marked by the adaptor protein Smad Anchor for Receptor Activation (Sara) during ISC mitosis induces Notch activity and cell differentiation in the progeny that receives more Sara endosomes [30]. Autophagy mediates cell death and removal of the larval midgut during metamorphosis [31]. In the adult ISCs, ROS-induced auto-phagy is coupled with JNK activation via autophagy-related 9 (Atg9) as part of the stress response machinery [32].
Ultrastructure analysis of other cell types
EEs represent another midgut epithelial cell type that are diploid and thus have much smaller nuclei than the ECs (Figure 1B). Neither ISCs nor EEs have microvilli. However, electron micrographs of EEs differ from ISCs in several ways. First, EEs have long cell bodies spanning across the epithelial layer, whereas ISCs are confined to the basal region (Figure 1A and B) except that some ISCs have very thin apical extension that can reach the luminal surface [2]. Second, in contrast to the loose adherens junctions in ISCs which are thought to allow cell mobility [2] (Figure 1A″ and F), tight junctions are present in both EEs and ECs in the apicolateral regions (Figure 1B′, F and G). Aging-associated deterioration of tight junctions causes impaired intestinal barrier function, raising the levels of gut infection, JNK activity, and ISC proliferation [33]. Finally, consistent with their endocrine functions, EEs are enriched with secretory vesicles, rough ER, and Golgi body (Figure 1B′, B″ and F). Some vesicles are loaded with granules, which likely contain the secretory peptides. Moreover, many secretory vesicles, some presumably ready to fuse with the plasma membrane, can be found at both apical and basal sides of the EEs, suggesting that EE-secretory factors can be delivered to either the luminal fluid or the hemolymph. Previous studies have found that EE-derived factors affect a wide range of local and distant target cell types. For example, EEs produce Slit [34], tachykinin [35], and Activin-β [36] to regulate ISC differentiation, EC lipogenesis, and fat body glucagon signaling, respectively. Based on these EM observations, it will be intriguing to investigate whether the site of secretion determines the target cell type for EE-derived signals, and whether apical and basal secretions are differentially regulated.
ECs are cuboidal or columnar cells with polyploid nuclei at the center and microvilli covering the luminal surface. The differentiation from ISCs to the much larger ECs involves a process of postmitotic cell growth and endoreplication, which is controlled by the Ras/MAPK and InR/PI3K/Target of rapamycin (TOR) pathways [37].
In the lower half of the EC, the plasma membrane is enriched with Na+/K+-ATPase and often forms a basal labyrinth (Figures 1B, 2A and C) by extensive infoldings [9]. Mitochondria are accumulated along the membranes of the basal labyrinth, often in a vertical basal to apical orientation, to provide energy for active ion transport [38]. In mammals, basal labyrinths are prominent in the epithelia of the renal tubules, the renal collecting ducts, and the salivary glands, where they play critical roles in regulating body fluid osmolality [38]. However, the basal labyrinth has not been reported or studied in mammalian ECs.
Our EM analysis identified organelles that are critical for the digestive and metabolic functions of ECs, including secretory vesicles (Figure 1H), lipid droplets (Figure 1B′ and I), and glycogen granules (Figure 1I). In addition, EC endosomes/lysosomes (including MVB and autophagosomes) exhibit diverse morphology (Figure 1G, I and J). Recently it was found that Atg9 loss results in hyperactive TOR signaling and dramatically enlarged ECs [39]. Therefore, autophagy plays an important role in suppressing cell growth in ECs.
It should be noted that the multilamellar body (Figure 1B and I) is a specialized form of lysosome, varying from ~150 nm to ~3 μm in diameter, found in most ECs and less frequently in ISCs or EEs. They could exist alone, in groups, or localize within autophagosomes. Similar multilamellar structures have been studied in type II alveolar cells of the lung [40], in keratinocytes of the skin [41], and in the nervous system of the earthworm [42], where they serve as a reservoir for phospholipids and participate in exocytosis or the biogenesis of other membrane structures. Multilamellar bodies are also found in the mammalian gastrointestinal tract [43], where their function is unknown.
High-resolution electron micrographs could identify non-epithelial cells in the midgut. For example, the trachea cells, characterized by the empty cavity they encircle, can be found on both sides of the visceral muscle layer (Figure 1A and A′). In addition to its physiological function of gas exchange, the trachea can release the TGFβ ligand Dpp to protect the ECs and affect midgut homeostasis [44]. Our electron micrographs indicate that the trachea not only penetrate through the muscle layer, but also make close contact with epithelial cells, especially the ISCs (Figure 1A′ and C). Recent studies suggest that other cell types such as the enteric neurons [45] or hemocytes [46] might also make contacts with the midgut and influence ISC activity via Hh or Dpp signaling, respectively. However, these contacts are infrequent and not easily detectable by EM.
Ultrastructure analysis of the midgut under pathological conditions
With a better understanding of the normal midgut, we wondered whether EM analysis could detect any abnormalities in organelle morphology under pathological conditions. In particular, we examined midguts following oncogene activation or tissue damage.
Consistent with previous studies reporting that Yki activation causes hyperplasia [47], we observed a multilayered midgut epithelium when a constitutively active form of Yki (Yki3SA) is expressed in ISCs (Figure 2A and C). The accumulation of lipid droplets in some cells of Yki tumors (Figure 2B) might indicate apoptosis [48]. Interestingly, in Yki tumors we detected basal labyrinths (Figure 2C) that are more extensive and enriched with more elongated mitochondria (Figure 2C′) than in the normal midgut (Figure 2A). Previously, Yki tumors have been reported to cause excessive body fluid (“bloating syndrome”) in the abdomen [49] — whether or not the bloating syndrome can be attributed to the accelerated water absorption activity of basal labyrinths needs further investigation.
Paraquat is a commonly used herbicide that causes severe lung and gastrointestinal damages when ingested by mammals [50]. In the fly midgut, paraquat feeding induces oxidative stress and ISC proliferation [51]. Interestingly, in addition to the expansion of progenitor cells (Figure 2D and E), three features could be recognized by EM in the midguts of paraquat-fed flies. First, ECs often accumulate large vacuoles of lipid droplets (Figure 2E). A recent report documented similar EC vacuolation in a region of the anterior midgut after pathogenic infection, and speculated that lipid droplets might help alleviate oxidative stress [52]. Second, paraquat causes massive deposition of electron-dense materials (Figure 2E and F), which is also observed in the alveolar cells from dogs intoxicated with paraquat [50]. The electron-dense materials probably correspond to protein aggregates induced by oxidation, and mostly found in mitochondria (Figure 2F′), the major site of ROS production caused by paraquat [53]. Some electron-dense materials also appear in the autophagosomes that have engulfed damaged mitochondria (Figure 2F). Interestingly, the electron-dense materials are found in ECs but not in ISCs (Figure 2F′). The activity of Nrf2/CncC, a major regulator of antioxidant signaling, is more active in the ISCs than in the ECs under homeostatic conditions, but suppressed in the ISCs after paraquat feeding [54]. Although we could not rule out that the pre-existing Nrf2/CncC activity might protect ISCs from oxidative damage, it is also possible that ISC mitochondria properties such as smaller size and fewer cristae contribute to paraquat resistance. Third, following paraquat feeding, we could detect apical cytoplasm extrusion (Figure 2F), which was reported recently as a mechanism for ECs to purge themselves of damaged components and/or bacteria [52].
Conclusion and future perspectives
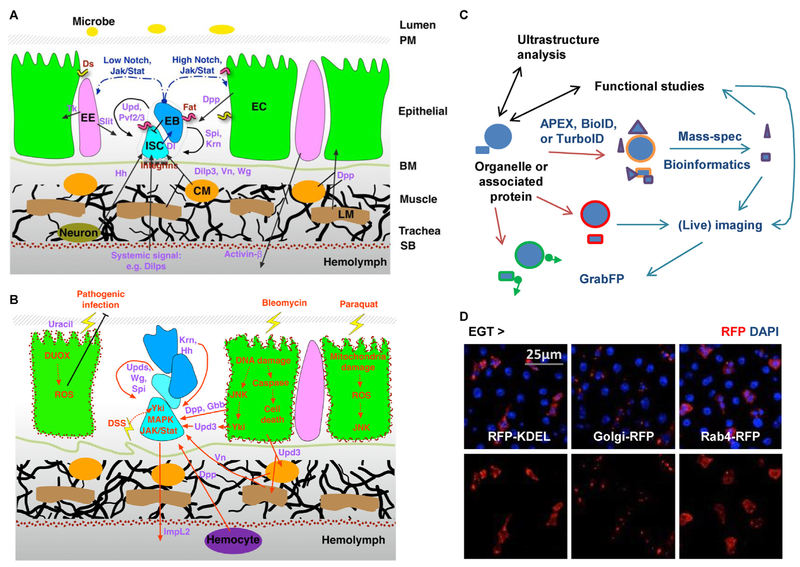
Although previous studies have identified various signaling pathways regulating ISC activity and unveiled a sophisticated network of cell communication in the midgut (Figure 3A and B) [3,4], it is largely unclear when, where, and how these signals are processed inside the cell. Because organelle behaviors are closely related to tissue homeostasis, and tissue pathology is also reflected in organelle defects, further investigation of organelle biology in the midgut should help understand the integration of different signals at a subcellular resolution.
Figure 3.
Future perspectives of organelle studies in the midgut. (A) The organization of different cell types and the cell communication network in the midgut under homeostatic conditions. Ligands for different signaling pathways are highlighted in purple. CM: circular visceral muscle; LM: longitudinal visceral muscle; PM: peritrophic membrane; SB: serosal barrier. (B) Activation of multiple signals in the midgut under stressed conditions. Red arrows indicate signals that are positively affected by tissue damage. DUOX: dual oxidase; DSS: dextran sulfate sodium. (C) Development of new tools will establish a versatile interdisciplinary platform and transform future studies of organelle biology in the midgut. (D) Examples of cell type specific organelle labeling. Midguts expressing RFP-labeled ER (-KDEL), Golgi body, or endosome (Rab4) in ISCs for 4d were co-stained with DAPI.
Ultrastructure analyses and functional studies could be used to corroborate and inspire each other (Figure 3C). For example, an EM observation may help formulate a hypothesis on the function of a particular type of organelle or organelle-associated protein. Conversely, one could use EM, with immunogold labeling, to examine the organelle association of a candidate protein identified from a functional screen or bioinformatics analysis. Importantly, because very limited areas can be imaged with EM, researchers could use optical microscopy to corroborate EM findings in larger areas of the midgut. A number of UAS-fluorescent reporter lines labeling major types of organelles are available from Bloomington Drosophila Stock Center and are valuable resources for optical microscopy (examples in Figure 3D). In addition, CRISPR/Cas9-mediated genome editing makes it easy to generate knock-in flies labeling organelle-associated proteins [55,56]. Furthermore, expansion microscopy allows us to physically magnify biological specimens fixed on swellable polymer and perform scalable super-resolution imaging using ordinary confocal microscopes [57].
In addition to CRISPR, the advent of several new techniques will likely transform future studies of organelle biology (Figure 3C). For example, tagging organelles with proximity labeling enzymes (APEX, BioID, or TurboID) can help characterize the proteome of organelles in specific cell types and under different conditions [58–60], which may help for example to identify candidates that might integrate signaling pathways. Organelles, or their associated proteins, once marked with a fluorescent reporter, could be examined for real-time analysis using a recently established long-term midgut live imaging platform, which tracks cell behaviors in a living fly for 12–16 h [61]. Furthermore, GFP-labeled organelles could be manipulated with the GrabFP (grab Green Fluorescent Protein) toolbox for controlled localization [62], which might help address whether subcellular localization matters for organelle function; and whether the distribution of specific organelles during asymmetric stem cell division affects cell fate. Ultimately, an integration of new techniques with conventional ultrastructure analysis and functional studies will make the fly midgut an even more powerful system to tackle fundamental cell biology questions in the context of a complex tissue.
EM method
Samples were fixed in the routine fixative [2.5% Glutaraldehyde 1.25% Paraformaldehyde and 0.03% picric acid in 0.1 M sodium cacodylate buffer (pH 7.4)] for at least 2 h at room temperature, washed in 0.1M cacodylate buffer and postfixed with 1% Osmiumtetroxide (OsO4)/1.5% Potassiumferrocyanide (KFeCN6) for 1 h, washed 2 × in water, 1 × Maleate buffer (MB) 1x and incubated in 1% uranyl acetate in MB for 1hr followed by 2 washes in water and subsequent dehydration in grades of alcohol (10 min each; 50%, 70%, 90%, 2 × 10 min 100%). The samples were then put in propyleneoxide for 30 min and infiltrated ON in a 1:1 mixture of propyleneoxide and TAAB Epon (Marivac Canada Inc. St. Laurent, Canada). The following day the samples were embedded in TAAB Epon and polymerized at 60°°C for 48 h. Ultrathin sections (about 60 nm) were cut on a Reichert Ultracut-S microtome, picked up on to copper grids stained with lead citrate and examined in a JEOL 1200EX Transmission electron microscope and images were recorded with an AMT 2k CCD camera.
Acknowledgement
We thank Afroditi Petsakou and Ruei-jiun Hung for comments on the manuscript; Harvard Medical School EM Facility for electron microscopy support; Lucy O’Brien for discussion of midgut live imaging. This work is funded by National Institute of General Medical Sciences (GM067761). N.P. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Conflict of interest
Nothing declared.
References
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Micchelli C, Perrimon N: Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 2006, 439: 475–479. [DOI] [PubMed] [Google Scholar]
- 2.Ohlstein B, Spradling A: The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 2006, 439: 470–474. [DOI] [PubMed] [Google Scholar]
- 3.Biteau B, Hochmuth CE, Jasper H: Maintaining tissue homeostasis: dynamic control of somatic stem cell activity. Cell Stem Cell 2011, 9:402–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang H, Edgar BA: Intestinal stem cells in the adult Drosophila midgut. Exp Cell Res 2011, 317:2780–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miaczynska M, Pelkmans L, Zerial M: Not just a sink: endosomes in control of signal transduction. Curr Opin Cell Biol 2004, 16:400–406. [DOI] [PubMed] [Google Scholar]
- 6.Brookes PS, Levonen AL, Shiva S, Sarti P, Darley-Usmar VM: Mitochondria: regulators of signal transduction by reactive oxygen and nitrogen species. Free Radic Biol Med 2002, 33: 755–764. [DOI] [PubMed] [Google Scholar]
- 7.Asare A, Levorse J, Fuchs E: Coupling organelle inheritance with mitosis to balance growth and differentiation. Science 2017, 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller A: The internal anatomy and histology of the imago of Drosophila melanogaster In Demerec M. Biology of Drosophila, vol. 435 Hafner; 1950. [Google Scholar]
- 9.Baumann O: Posterior midgut epithelial cells differ in their organization of the membrane skeleton from other drosophila epithelia. Exp Cell Res 2001, 270:176–187. [DOI] [PubMed] [Google Scholar]
- 10.Fuse N, Hirose S, Hayashi S: Diploidy of Drosophila imaginal cells is maintained by a transcriptional repressor encoded by escargot. Genes Dev 1994, 8:2270–2281. [DOI] [PubMed] [Google Scholar]
- 11.Jin Y, Patel PH, Kohlmaier A, Pavlovic B, Zhang C, Edgar BA: Intestinal stem cell pool regulation in Drosophila. Stem Cell Rep 2017, 8:1479–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • 12.Xu C, Luo J, He L, Montell C, Perrimon N: Oxidative stress induces stem cell proliferation via TRPA1/RyR-mediated Ca(2+) signaling in the Drosophila midgut. Elife 2017, 6.The ER regulates ISC proliferation via cytosolic Ca2+. High levels of Ca2+ in ISCs, induced by knockdown of sarco/endoplasmic reticulum Ca2+-ATPase (SERCA), can activate Ras/MAPK signaling to drive overproliferation. In contrast, reduction of Ca2+ levels in ISCs by knockdown of the ER Ca2+ channel Ryanodine receptor (RyR) can inhibit Ras/MAPK activity and proliferation.
- 13.Lee T, Luo L: Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 1999, 22:451–461. [DOI] [PubMed] [Google Scholar]
- 14.Marianes A, Spradling AC: Physiological and stem cell compartmentalization within the Drosophila midgut. Elife 2013, 2:e00886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shanbhag S, Tripathi S: Epithelial ultrastructure and cellular mechanisms of acid and base transport in the Drosophila midgut. J Exp Biol 2009, 212:1731–1744. [DOI] [PubMed] [Google Scholar]
- 16.Cooper G: The endoplasmic reticulum In The cell: a molecular approach. 2nd ed. Sinauer Associates; 2000. [Google Scholar]
- 17.Strauss M, Hofhaus G, Schroder RR, Kuhlbrandt W: Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J 2008, 27:1154–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rera M, Bahadorani S, Cho J, Koehler CL, Ulgherait M, Hur JH, Ansari WS, Lo T Jr, Jones DL, Walker DW: Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 homolog. Cell Metab 2011, 14:623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •• 19.Koehler CL, Perkins GA, Ellisman MH, Jones DL: Pink1 and Parkin regulate Drosophila intestinal stem cell proliferation during stress and aging. J Cell Biol 2017, 216:2315–2327.Inhibition of the mitochondrial quality control system in ISCs by knockdown of Pink1 or Parkin results in mitochondria abnormality, inducing senescence and suppression of proliferation.
- • 20.Teixeira FK, Sanchez CG, Hurd TR, Seifert JR, Czech B, Preall JB, Hannon GJ, Lehmann R: ATP synthase promotes germ cell differentiation independent of oxidative phosphorylation. Nat Cell Biol 2015, 17:689–696.Drosophila germline stem cell mitochondria have fewer cristae, compared to differentiated germ cells. Inhibition of cristae formation by knockdown of ATP synthase can prevent germline stem cells from differentiating. Whereas the mechanism connecting mitochondria cristae formation with germline differentiation is still unclear, it does not seem to depend on the oxidative phosphorylation activity of mitochondria.
- 21.Beller M, Sztalryd C, Southall N, Bell M, Jackle H, Auld DS, Oliver B: COPI complex is a regulator of lipid homeostasis. PLoS Biol 2008, 6:e292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • 22.Singh SR, Zeng X, Zhao J, Liu Y, Hou G, Liu H, Hou SX: The lipolysis pathway sustains normal and transformed stem cells in adult Drosophila. Nature 2016, 538:109–113.Suppression of the lipolysis pathway by knocking down the Golgi membrane trafficking proteins COPI or Arf1 causes ISC death and engulfment by ECs.
- 23.Wang L, Zeng X, Ryoo HD, Jasper H: Integration of UPRER and oxidative stress signaling in the control of intestinal stem cell proliferation. PLoS Genet 2014, 10:e1004568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Ryoo HD, Qi Y, Jasper H: PERK limits Drosophila lifespan by promoting intestinal stem cell proliferation in response to ER stress. PLoS Genet 2015, 11:e1005220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashby MC, Tepikin AV: ER calcium and the functions of intracellular organelles. Semin Cell Dev Biol 2001, 12:11–17. [DOI] [PubMed] [Google Scholar]
- 26.Deng H, Gerencser AA, Jasper H: Signal integration by Ca(2+) regulates intestinal stem-cell activity. Nature 2015, 528: 212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • 27.He L, Si G, Huang J, Samuel ADT, Perrimon N: Mechanical regulation of stem-cell differentiation by the stretch-activated Piezo channel. Nature 2018, 555:103–106.High levels of Ca2+, induced by SERCA knockdown in Piezo+ISCs (a subpopulation of ISCs that are destined to become EEs), can stimulate EE production by inhibiting Notch signaling.
- •• 28.Ren W, Zhang Y, Li M, Wu L, Wang G, Baeg GH, You J, Li Z, Lin X: Windpipe controls Drosophila intestinal homeostasis by regulating JAK/STAT pathway via promoting receptor endocytosis and lysosomal degradation. PLoS Genet 2015, 11:e1005180.Endosomes regulate ISC proliferation and differentiation in the midgut. The single transmembrane protein Windpipe (Wdp) acts as a negative feedback regulator of JAK/STAT signaling in ISCs by promoting endocytic degradation of the JAK/STAT pathway receptor Domeless.
- 29.Obniski R, Sieber M, Spradling AC: Dietary lipids modulate notch signaling and influence adult intestinal development and metabolism in Drosophila. bioRxiv 2018, 273813 10.1101/273813; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montagne C, Gonzalez-Gaitan M: Sara endosomes and the asymmetric division of intestinal stem cells. Development 2014, 141:2014–2023. [DOI] [PubMed] [Google Scholar]
- 31.Denton D, Shravage B, Simin R, Mills K, Berry DL, Baehrecke EH, Kumar S: Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr Biol 2009, 19:1741–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •• 32.Tang HW, Liao HM, Peng WH, Lin HR, Chen CH, Chen GC: Atg9 interacts with dTRAF2/TRAF6 to regulate oxidative stress-induced JNK activation and autophagy induction. Dev Cell 2013, 27:489–503.The autophagy core component Atg9 interacts with Drosophila tumor necrosis factor receptor-associated factor 2 (dTRAF2) to mediate ROS-associated JNK signaling, including JNK-induced autophagosome formation and ISC proliferation.
- •• 33.Resnik-Docampo M, Koehler CL, Clark RI, Schinaman JM, Sauer V, Wong DM, Lewis S, D’Alterio C, Walker DW, Jones DL: Tricellular junctions regulate intestinal stem cell behaviour to maintain homeostasis. Nat Cell Biol 2017, 19:52–59.Midguts of old flies exhibit dramatic mislocalization of tight junction proteins. Knockdown of the tricellular junction component Gli (which is presumably involved in tight junction assembly) in ECs impairs intestinal barrier function in aging flies, and induces ISC proliferation via JNK activation.
- 34.Biteau B, Jasper H: Slit/Robo signaling regulates cell fate decisions in the intestinal stem cell lineage of Drosophila. Cell Rep 2014, 7:1867–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song W, Veenstra JA, Perrimon N: Control of lipid metabolism by tachykinin in Drosophila. Cell Rep 2014, 9:40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • 36.Song W, Cheng D, Hong S, Sappe B, Hu Y, Wei N, Zhu C, O’Connor MB, Pissios P, Perrimon N: Midgut-derived Activin regulates glucagon-like action in the fat body and glycemic control. Cell Metab 2017, 25:386–399.EE-derived signals can modulate the function of a different organ and affect systemic physiology. In response to chronic high-sugar diet, Activin-β production in the EEs is up-regulated to enhance AKH/glucagon signaling in the fat body, causing hyperglycemia.
- • 37.Xiang J, Bandura J, Zhang P, Jin Y, Reuter H, Edgar BA: EGFR-dependent TOR-independent endocycles support Drosophila gut epithelial regeneration. Nat Commun 2017, 8:15125.EGFR/ Ras/MAPK, EGFR/ Ras/MAPK and InR/PI3K/TOR signaling pathways control EC growth. Ras/Raf signaling upregulates E2f1 levels post-transcriptionally to promote EC endoreplication.
- •• 38.Pavelka M, Roth J: Basal labyrinth In Functional ultrastructure. Edited by Pavelka M, Roth J, Springer; 2010:178–179.Autophagy is coupled with cell growth in ECs. Atg9 interacts with and stablizes TSC2, a major suppressor of TOR signaling. Atg9 depletion in ECs activates TOR and thus causes a dramatic increase in cell size.
- 39.Wen JK, Wang YT, Chan CC, Hsieh CW, Liao HM, Hung CC, Chen GC: Atg9 antagonizes TOR signaling to regulate intestinal cell growth and epithelial homeostasis in Drosophila. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balis JU, Conen PE: The role of alveolar inclusion bodies in the developing lung. Lab Invest 1964, 13:1215–1229. [PubMed] [Google Scholar]
- 41.Suzuki H, Kurosumi K: Lamellar granules and keratohyalin granules in the epidermal keratinocytes, with special reference to their origin, fate and function. J Electron Microsc (Tokyo) 1972, 21:285–292. [PubMed] [Google Scholar]
- 42.Al-Yousuf Sa: Multilamellar bodies In An atlas of cells-ultra-structure. Edited by Al-Yousuf Sa, Doha Modern Printing Press; 1992:161–166. [Google Scholar]
- 43.Schmitz G, Muller G: Structure and function of lamellar bodies, lipid-protein complexes involved in storage and secretion of cellular lipids. J Lipid Res 1991, 32:1539–1570. [PubMed] [Google Scholar]
- 44.Li Z, Zhang Y, Han L, Shi L, Lin X: Trachea-derived dpp controls adult midgut homeostasis in Drosophila. Dev Cell 2013, 24:133–143. [DOI] [PubMed] [Google Scholar]
- 45.Han H, Pan C, Liu C, Lv X, Yang X, Xiong Y, Lu Y, Wu W, Han J, Zhou Z, et al. : Gut-neuron interaction via Hh signaling regulates intestinal progenitor cell differentiation in Drosophila. Cell Discov 2015, 1:15006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ayyaz A, Li H, Jasper H: Haemocytes control stem cell activity in the Drosophila intestine. Nat Cell Biol 2015, 17:736–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karpowicz P, Perez J, Perrimon N: The Hippo tumor suppressor pathway regulates intestinal stem cell regeneration. Development 2010, 137:4135–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boren J, Brindle KM: Apoptosis-induced mitochondrial dysfunction causes cytoplasmic lipid droplet formation. Cell Death Differ 2012, 19:1561–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • 49.Kwon Y, Song W, Droujinine IA, Hu Y, Asara JM, Perrimon N: Systemic organ wasting induced by localized expression of the secreted insulin/IGF antagonist ImpL2. Dev Cell 2015, 33:36–46.Yki-induced, Yki-induced ISC tumor could cause abdomen bloating and the degeneration of multiple organs including the ovary, fat body, and muscle. The organ-wasting phenotypes could be partially attributed to the induction of secreted insulin/IGF antagonist ImpL2 in Yki tumor.
- 50.Williams JH, Whitehead Z, Van Wilpe E: Paraquat intoxication and associated pathological findings in three dogs in South Africa. J S Afr Vet Assoc 2016, 87:e1–e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi NH, Kim JG, Yang DJ, Kim YS, Yoo MA: Age-related changes in Drosophila midgut are associated with PVF2, a PDGF/VEGF-like growth factor. Aging Cell 2008, 7: 318–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •• 52.Lee KZ, Lestradet M, Socha C, Schirmeier S, Schmitz A, Spenle C, Lefebvre O, Keime C, Yamba WM, Bou Aoun R, et al. : Enterocyte purge and rapid recovery is a resilience reaction of the gut epithelium to pore-forming toxin attack. Cell Host Microbe 2016, 20:716–730.When exposed to hemolysin, a pore-forming toxin secreted by the pathogenic bacteria Serratia marcescens, Drosophila ECs could extrude most of their apical cytoplasm as a protective mechanism to purge damaged organelles such as mitochondria.
- 53.Cocheme HM, Murphy MP: Complex I is the major site of mitochondrial superoxide production by paraquat. J Biol Chem 2008, 283:1786–1798. [DOI] [PubMed] [Google Scholar]
- 54.Hochmuth CE, Biteau B, Bohmann D, Jasper H: Redox regulation by Keap1 and Nrf2 controls intestinal stem cell proliferation in Drosophila. Cell Stem Cell 2011, 8:188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nagarkar-Jaiswal S, DeLuca SZ, Lee PT, Lin WW, Pan H, Zuo Z, Lv J, Spradling AC, Bellen HJ: A genetic toolkit for tagging intronic MiMIC containing genes. Elife 2015, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, O’Connor-Giles KM: Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 2014, 196:961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karagiannis ED, Boyden ES: Expansion microscopy: development and neuroscience applications. Curr Opin Neurobiol 2018, 50:56–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen CL, Hu Y, Udeshi ND, Lau TY, Wirtz-Peitz F, He L, Ting AY, Carr SA, Perrimon N: Proteomic mapping in live Drosophila tissues using an engineered ascorbate peroxidase. Proc Natl Acad Sci U S A 2015, 112:12093–12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han S, Li J, Ting AY: Proximity labeling: spatially resolved proteomic mapping for neurobiology. Curr Opin Neurobiol 2017, 50:17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Branon TC, Bosch JA, Sanchez AD, Udeshi ND, Svinkina T, Steven A Carr, Feldman JL, Perrimon N, Ting AY: Directed evolution of TurboID for efficient proximity labeling in living cells and organisms. bioRxiv 2017, 196980 10.1101/196980; 2017. [DOI] [Google Scholar]
- 61.Martin J, Sanders EN, Moreno-Roman P, Balachandra S, Du X, Koyama LAJ, O’Brien LE: Long-term live imaging of the Drosophila adult midgut reveals real-time dynamics of cell division, differentiation, and loss. bioRxiv 2018, 271742 10.1101/271742; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harmansa S, Alborelli I, Bieli D, Caussinus E, Affolter M: A nanobody-based toolset to investigate the role of protein localization and dispersal in Drosophila. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]