Abstract
Human immunodeficiency virus type 1 (HIV-1) encodes for Tat, a multi-functional regulatory protein involved in transcriptional enhancement and in causing neurotoxicity/central nervous system (CNS) dysfunction. This study examines Sanger sequencing of HIV-1 subtype B Tat from 2006 to 2014 within the Drexel University College of Medicine CNS AIDS Research and Eradication Study (CARES) Cohort to investigate Tat length in patients. The Los Alamos National Laboratory (LANL) database was used as a comparator. Miscoded stop codons were present in the CARES Cohort and LANL and protein variability was highly similar. Tat proteins in CARES and LANL were predominantly 101 residues. There was no observed correlation between Tat length and clinical parameters within the CARES Cohort. Unique Tat lengths found in the CARES Cohort and not in LANL were 31, 36, and 39 residues. When CARES patients were longitudinally examined, sequence lengths of 101 had a low probability of reducing to below 48, and sequences had a high probability of increasing to above 86 residues during their next visit, when below 48 residues in length. This suggests that Tat length is conserved to retain the majority of the proteins function highlighting its importance in viral replication.
Keywords: HIV-1, Subtype B, LANL, Tat, genetic variation, stop codon
1. Introduction
Human immunodeficiency virus type 1 (HIV-1) encodes for the transactivator of transcription (Tat), a small, basic 101 residue protein. Tat has been shown to be primarily responsible for enhancing HIV-1 transcription through interaction with the RNA stem-loop transactivation response (TAR) element, encoded by the viral long terminal repeat (LTR), which has been shown to recruit the host positive transcript elongation factor b (P-TEFb)1,2. While transactivation has been shown to be the primary role of Tat, it has been demonstrated that most of Tat is secreted from the cell3. Tat secretion has been shown to occur through an unconventional pathway; passing through the plasma membrane primarily through oligomerization and pore formation4. Once extracellular, Tat can directly cause neurotoxicity by hyper-activation5 and recruitment of additional immune cells to the central nervous system (CNS), causing low levels of chronic inflammation through activation of bystander cells and release of pro-inflammatory cytokines, such as IL1-β from monocytes and macrophages6–10. Furthermore, Tat produced in the periphery can cross the blood-brain barrier and traffic into the CNS11.
HIV-1-infected patients have been shown to be particularly susceptible to neurocognitive impairment, where the range of symptoms has been collectively referred to as HIV-1-associated neurocognitive disorders (HAND)12. A comprehensive mechanism of HAND has yet to be elucidated, but the secretion of viral proteins, such as gp120, Vpr, and Tat within the CNS has been suggested to be crucial for productive viral infectivity13 and has been demonstrated to impair cognition in vivo14. While antiretroviral therapy (ART) has successfully decreased the mortality associated with HIV-1, the prevalence of HAND has increased in chronically infected patients15.
An additional complication during the course of HIV-1 infection has been its vast genetic variability, which has been shown to occur at an interpretable rate of 1.02 nucleotides per year over the entire genome on average and 0.636 nucleotides per year for the tat gene16. The accumulation of these changes is caused by the error-prone HIV-1 reverse transcriptase in conjunction with selective pressures associated with the immune system and a number of other sources17. This genetic variability within Tat can cause functional alterations, such as a reduction of LTR transactivation ability18,19. As a contributor to the development of HAND, genetic variation within Tat and its subsequent functional alterations have been thought to be critical to understand the etiology of HIV-associated neurocognitive impairment.
The majority of previously reported studies concerning HIV-1 Tat have focused on the functionality of a truncated variant of Tat that is 86 residues long, which has similar levels of transactivation, but differs in a number of other activities20,21. While the frequency of Tat truncations have been examined in isolated cohorts22, it has yet to be evaluated around the world within HIV-1 subtype B sequences. Here, we sought to examine the genetic variation of Tat and the effects of premature truncation of this important viral protein within the Drexel University College of Medicine (Drexel Medicine) CNS AIDS Research and Eradication Study (CARES) cohort through Sanger sequencing performed from 2006 to 2014, as well as the data from the publicly available HIV-1 sequences present in the Los Alamos National Laboratory (LANL) to quantify the frequency of Tat truncations worldwide.
2. Results
2.1. HIV-1 Tat sequences from the CARES Cohort are similar to subtype B sequences in LANL
Analyses were performed on Sanger sequenced PCR amplicons of tat exon I and II amplified from genomic DNA isolated from patient peripheral blood mononuclear cells (PBMC) obtained from patients in the Drexel University College of Medicine (Drexel Medicine) CNS AIDS Research and Eradication Study (CARES) Cohort. Sanger sequencing allows for long, continuous reads of approximately 800 base pairs. The two exonic regions of the tat gene (exon I and II) are 214 and 90 base pairs, respectively. Due to the separation of approximately 2,300 base pairs between the two exons23, they were sequenced separately and reconstructed in silico. The 205 participants in this study were a fair representation of the entire CARES Cohort (Table 1). From these participants, 418 Tat sequences were collected. Of those 418 sequences, 391 contained a stop codon in either their first or second exon (Supplemental Table 1) and were used in further analysis. The other 27 sequences had no stop codon contained in the PCR amplicon that was obtained for Tat exon 1 or 2. Of those 391 Tat sequences, 381 from 201 patients were linked with a patient visit date. These 381 sequences were used for longitudinal analyses. To minimize bias from multiple patient sequences, each CARES Cohort Tat sequence was assigned a weight value. This weight is defined as the reciprocal of the number of Tat sequences generated from a patient; for example, if three Tat sequences arose from a patient, each sequence would have a weight of 1/3. The sum of these weights can be interpreted as the number of patients belonging to a category.
Table 1: Clinical parameters of patients enrolled in the Drexel Medicine CARES Cohort.
Summary of the clinical parameters of patients with Sanger sequencing data of both exon one and two of HIV-1 subtype B Tat. ART, antiretroviral therapy; MHDS, Modified Hopkins Dementia Score; ≥10 considered normal, <10 considered impaired.
| Clinical data | Tat Sanger patients (mean +/− STD) |
|---|---|
| Number of patients | 205 |
| Race (African American or Black) | 87% |
| Age (years) | 51.42 +/− 8.56 |
| Years Seropositive | 12.78 +/− 6.54 |
| Nadir CD4 (cells/uL) | 256.186 +/− 186.95 |
| Current CD4 (cells/uL) | 605.09 +/− 377.04 |
| Current CD8 (cells/uL) | 1093.05 +/− 710.95 |
| Peak log viral load (copies/mL) | 4.29 +/− 1.29 |
| Current log viral load (copies/mL) | 2.06 +/− 1.12 |
| MHDS | 9.08 +/− 2.85 |
| ART (on) | 89% |
| Gender (male) | 65% |
| Drug use (self-admitted) | 28% |
To compare the CARES Cohort sequences to other subtype B sequences, the Los Alamos National Laboratory (LANL) database was queried. From LANL, only HIV subtype B Tat sequences were retrieved and compared to the CARES Cohort, since the CARES Cohort contains all subtype B sequences. The LANL database contained 5,831 complete HIV-1 subtype B Tat sequences arising from 1,483 patients. These sequences were predominately collected between 2004 to 2008. LANL sequences were assigned weights using the same methodology as the CARES Cohort23. Sequences from LANL with an unknown patient ID were assumed to be individual patients.
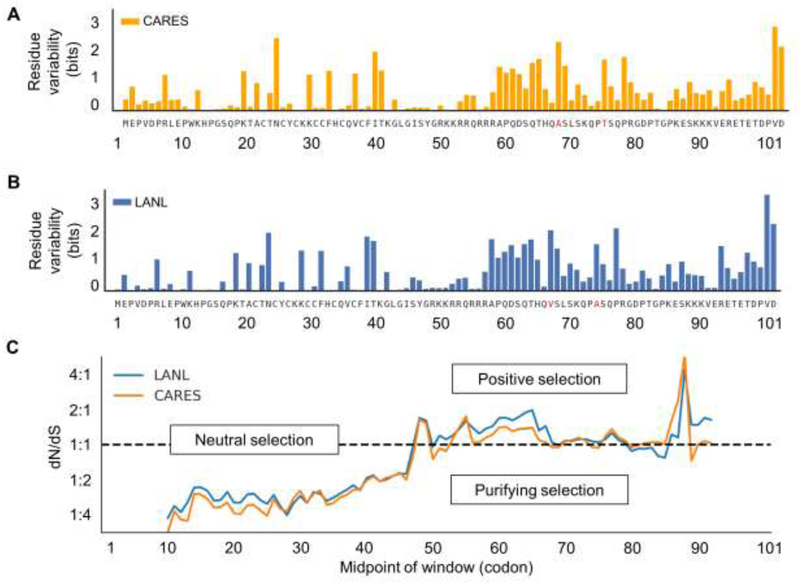
Amino acid variability was assessed between Tat sequences acquired from the CARES cohort or LANL by calculating the Shannon entropy of each residue (Figure 1A and 1B). The three positions with the largest differences in entropy were 92, 24, and 93. The entropy at each position in CARES and LANL were highly similar (r2 = 0.910) (Supplemental Figure 1). The consensus sequence of Tat 101 in the LANL database and CARES Cohort was also almost identical aside from residues 67 and 74, which are an alanine and threonine in the CARES Cohort consensus sequence and a valine and alanine in the LANL consensus sequence, respectively. When position 67 was further examined, valine occurs 19.44% of the time in the CARES Cohort while it occurs 39.93% in LANL. Alanine occurs 32.48% of the time in CARES Cohort while it occurs 31.43% of the time in the LANL database. When position 74 was analyzed, alanine occurred in the CARES Cohort 32.48% of the time while it occurred in the LANL database 40.16% of the time. Threonine occurred in CARES Cohort participants 42.71% of the time while it occurred in the LANL database 40.16% of the time (data not shown). A sequence logo was also generated to compare residue variability between CARES and LANL sequences (Supplemental Figure 2). Overall, the CARES Cohort and LANL database show a similar distribution of amino acid diversity within their Tat sequences.
Figure 1: HIV-1 Tat residue variability between the Drexel Medicine CARES cohort and LANL was highly similar.
A) The Shannon entropy for each residue position within 101-amino acid HIV-1 Tat Sanger sequences (n=391) collected from the Drexel Medicine CARES cohort was calculated. The consensus sequence from all Tats examined was provided for each amino acid position. Residues highlighted in red differ between the consensus sequence of the LANL database. B) The Shannon entropy for each residue position within 101-amino acid HIV-1 Tat Sanger sequences (n=5831) collected from the LANL database was calculated. The consensus sequence from all Tats examined was given for each amino acid position. Residues highlighted in red differ between the consensus sequence of the CARES cohort. C) The average dN/dS ratio found in CARES and LANL across their tat genes using a 20 codon window. The beginning of Tat seems to be preserved and unlikely to undergo any mutations that alter its consensus output. Starting around residue 47, tat seems to maintain its dN/dS ratio around one until the end of the tat gene which seems to have its dN/dS ratio spike upwards around residue 87. This seems to suggest some positive selection around the end of the tat gene that may or may not aid in HIV-1’s survival.
To investigate how evolutionary pressure might influence the tat gene in both the CARES Cohort in Philadelphia and worldwide through the LANL database, the dN/dS ratios of all sequences were calculated using the consensus B tat sequence as a reference (Figure 1C). These results are calculated from observing changes in dN/dS ratio from a 20 codon window that slides one codon at a time. The results indicated that most regions of tat within CARES and LANL show a dN/dS ratio of less than one, suggesting that tat was subjected to minimal evolutionary pressure. The tat dN/dS ratio over the sliding sequence window for sequences derived from the CARES Cohort and LANL database are almost identical, suggesting similar evolutionary pressures applied to both sets of tat sequences. However, there does seem to be a region around residue 89 where the dN/dS ratio spikes upward in both sequences from the LANL database and especially in tat sequences derived from the CARES Cohort. This supports our Shannon entropy calculation as there seems to be an increased entropy for both Tat sequences from patients in the CARES Cohort and LANL database as one approaches the end of the Tat protein.
2.2. Alternative stop codon positions in HIV-1 subtype B Tat
A recent publication examining the Bridging the Evolution and Epidemiology of HIV in Europe (BEEHIVE) study observed various mutated Tat sequences derived from patient RNA, caused by miscoded stop codons22. Inspired by their observations, the Drexel Medicine CARES Cohort was probed for unique patient Tat truncations and assessed for similarities with sequences in the LANL database. The most frequent Tat length observed in patients in the CARES Cohort was 101 residues (observed in 83.0% of patients), followed by 86 (7.5%) and 102 (3.8%) residues, respectively (Supplemental Table 1). To more clearly understand the subtype B Tat length distribution worldwide, sequences present in the LANL database were analyzed by country. Within LANL, the three countries that generated the most subtype B Tat sequences were the United States (573 patients), Brazil (225 patients), and Cyprus (77 patients). The three most frequent Tat lengths found in the LANL database were 101 (1244.4 patients), 102 (69.1 patients), and 86 residues (58.9 patients) (Supplemental Table 2). Tat 101 was the most prevalent Tat length for almost all countries examined (Supplemental Table 3). Interestingly, France appears to have a higher proportion of patients with Tat 86 than other countries (Figure 2A). While all countries were analyzed for geographic distribution, Figure 2A only displays countries that have at least 10 patients and Tat lengths that occurred at least 5 times. The geographical distributions of all Tat lengths can be found in Supplemental Table 3.
Figure 2: HIV-1 Tat length was highly similar between the Drexel Medicine CARES Cohort and the LANL database.
A) The geographic distribution of Tat Sanger sequences submitted to the LANL database was plotted based on the frequency of the Tat lengths present. Countries with less than 10 patients and Tat lengths that occurred less than 5 times were omitted. The number of sequences per country is listed above each column. All data can be found in Supplemental Table 3. Country codes presented in this figure are as follows: Argentina (AR), Australia (AU), Brazil (BR), Canada (CA), Switzerland (CH), China (CN), Cyprus (CY), Germany (DE), Denmark (DK), Spain (ES), France (FR), United Kingdom (GB), Japan (JP), The Republic of Korea (KR), Peru (PE), Sweden (SE), Thailand (TH), and the United States (US). B) The proportion of cumulative Tat length was plotted for both the LANL database (yellow) and the CARES Cohort (blue). All data can be found in Supplemental Tables 1 and 2. C) The distribution of patient Tat sequences between the LANL database and the CARES Cohort were grouped into sets of 10 residues. The number represents the number of patients (CARES=205; LANL=1483) that had a stop codon in the corresponding 10 residue window.
The distribution of Tat lengths in the CARES Cohort was highly similar to the distribution Tat lengths in the LANL database (Figure 2B). Large increases occurred in both the CARES Cohort and LANL database in the cumulative density graph around position 101 with a smaller increase around position 86. This has indicated that most sequences in LANL and CARES are 101 or 86 residues long. In agreement with previously reported results22, full-length 101 residue Tat was the most prevalent out of the 205 patients analyzed in the Drexel Medicine CARES Cohort (83.0%). A comparison between the various lengths of HIV-1 Tat within the Drexel Medicine CARES Cohort and LANL subtype B Sanger sequences revealed truncations unique to the Drexel Medicine CARES Cohort. Premature stop codons were present that caused Tat variants 31, 36, and 39 residues in length, which were not present in the LANL analysis. The LANL dataset also contained Tat variants not present in the Drexel Medicine CARES Cohort, which were 11, 16, 34, 38, 42, 43, 52, 54, 55, 59, 61, 63, 70, 71, 72, 75, 77, 89, 93, 96, 98, 105, 106, 107, 113, and 124 residues in length. Despite these differences, the relative frequencies of Tat lengths 101, 86, and 102 in CARES and LANL were shown to be similar. When we examined the distribution of different stop codon types between LANL and CARES, we found that they were predominantly amber (TAG; UAG stop codon; Supplemental Tables 1 and 2).
2.3. Longitudinal analyses of CARES Cohort patient Tat sequences
To investigate the distribution of the Tat lengths within the CARES Cohort over time, a frequency table was generated between Tat length and year the patient was sampled (Supplementary Table 4). The predominant lengths across all years were 101 and 86 residues, with some minor deviation from the consensus length. Rare Tat length variants were observed in the analysis, including lengths: 10, 31, 36, 39, 95, and 99. However, Tat length 95 was only observed in a single patient in 2013. Likewise, Tat length 36 was only observed in one of two samples examined that year and Tat 39 as observed in 1 patient in 2008. Tat length 99 was observed in 4.11 patients across 2007, 2009, and 2014. Tat lengths 10 and 31 were observed in multiple patients in most years of the CARES Cohort. These results were subsequently converted into a bubble plot, where larger bubbles corresponded to a higher frequency of a specific Tat length (Figure 3A). Tat length 101 was the most prevalent within the patients assessed.
Figure 3: HIV-1 Tat length was predominately 101 residues within the Drexel Medicine CARES Cohort.
A) Tat length was plotted per year for Sanger sequences within the CARES Cohort. The size of the bubble corresponds to the frequency of a specific Tat length. The color of each Tat length can be found within the key. The numbers on top of the graph represent the total sequence weight for that year. All data can be found in Supplemental Table 4. B) Selected patients, that had at least three visits and were recorded with a Tat length other than 101 residues, were subjected to longitudinal analysis.
This dataset was further analyzed by selecting patients that had at least three visits and were noted to have a different length of Tat other than 101 residue version. A potential concern with such a limited input is an increase in sequencing errors. We have previously optimized our Tat amplification and sequencing protocol to minimize this from occurring2,24. It is still important to note that these sequences may have come from integrated defective provirus. To date, we have been unable to determine whether these were derived from defective proviruses, since only tat exons I and II were sequenced, however, a full-length replication competent provirus may still be unable to transcribe these truncated Tats depending on the chromatin architecture surrounding the integrated provirus. To determine whether a potential reduction in Sanger sequencing efficiency occurred in patients with reduced viral load, sequencing studies have indicated that there was no difference in efficiency (data not shown). Of the 205 patients, 35 patients had at least three visits and could be subjected to longitudinal analysis. Of those 35 patients, 20 of them consistently showed 101 residue Tat sequences while 15 had exhibited a variable Tat length throughout the visits. When Tat length was longitudinally examined, most sequences were either 101 or 86 residues long (Figure 3B). There appears to be a high probability that when a patient yields a Tat sequence of less than 48 residues, the Tat sequence reverts to a length greater than 48 residues during the next time point examined (likelihood = 83.33%). The only exception to this trend was patient A0013, which had a sequence length of 31 residues followed by a length of 10 residues during the subsequent time point. Conversely, when a patient yields predominantly a 101 residue Tat sequence, the likelihood that this sequence shortens below 86 residues during their subsequent visit was only 2.5%.
2.4. Correlation between HIV-1 Tat length and clinical parameters
Previous studies by us2,25–30 and others31–33 have identified alterations in the genetic architecture the HIV-1 genome that have been correlated to changes in HIV disease severity. Based on these studies, we have performed additional studies to determine whether any correlations between the clinical parameters discussed in Table 1 and the Tat lengths observed within the CARES Cohort could be identified. These clinical parameters have included age, race, years seropositive, nadir CD4 count, latest CD4 count, latest CD8 count, peak log viral load, latest log viral load, MHDS, current ART status, gender, and current drug use. The CARES Cohort contained samples predominantly from Tat 101 and relatively little of other Tat lengths (Figure 4A). As a result, the lack of significant numbers of shorter Tat sequences made it difficult to obtain definitive clinical correlations with respect to shorter or longer versions of Tat. However, when the analysis was performed, there was no significant correlation between Tat length and any given clinical parameter. The parameter with the highest positive and negative correlation with Tat length was the Modified Hopkins Dementia Score (MHDS) (r2 = 0.022) and age (r2 = 0.021), respectively (Figure 4B). The lack of information from smaller Tat lengths necessitated caution when using these values to associate clinical parameters with Tat length. Relatively little information was available on the clinical parameters of the sequences deposited in LANL for HIV-1 subtype B, which did not permit similar analyses to be performed with the LANL HIV-1 Tat sequence dataset. None of the metadata variables were shown to significantly correlate with Tat length (data not shown) given a Bonferroni corrected significance threshold (threshold = 0.05 / 12 = 0.00417). Additionally, a post-hoc power analysis was conducted to determine whether there were enough samples to determine whether there was a true relationship between Tat length and other clinical parameters. The only clinical parameter that showed a definitive power to determine a true power was age (power = 0.818). Collectively, these results have suggested that there was no correlation between Tat length and the tested clinical parameters.
Figure 4: HIV-1 Tat length does not correlate to any clinical parameter assessed in the Drexel Medicine CARES Cohort.
A) Tat lengths from the Drexel Medicine CARES Cohort were plotted with the clinical parameters assessed in Table 1. A post-hoc power analysis was also calculated to determine if there was appropriate power (>0.8), however, only age had the appropriate power. B) The coefficient of determination (r2) of the clinical parameters from Panel A. ART, antiretroviral therapy; MHDS, Modified Hopkins Dementia Score. There were no significant results after adjusting the significance threshold (p < 4.16e-3) using Bonferroni correction (data not shown).
2.5. Smaller Tat truncations transactivate the HIV-1 LTR potentially due to stop codon read through
To examine functional alterations caused by these reported premature stop codon mutations, Tat truncation variants were constructed in a pcDNA3.1/Hygro(+) backbone (Primers listed in Supplemental Table 5). In order to transactivate the HIV-1 LTR, Tat requires residues 1–57, residues 1–48 interacting with Cyclin T1, a subunit of P-TEFb, and residues 49–57 interacting with the UCU bulge on the HIV-1 TAR RNA stem-loop (Figure 5A). The full-length Tat 101 sequence was used for the mutagenesis, meaning that after each stop codon the remainder of Tat was encoded.
Figure 5: Tat truncations were not capable of transactivating the HIV-1 LTR.
A) A schematic of the functional domains of Tat. The arrows under the figure (yellow) denote the locations of the stop codons present in the CARES cohort. The residues required for transactivation of the LTR (1–57) was also denoted below the figure. B) β-galactosidase expression was assessed after transfection with the listed Tat constructs. Experiments were performed as quadruplicates on three separate occasions (n=3). Error bars are the mean with 99% confidence intervals. Cells are mock transfected cells. EV corresponds to an empty vector control transfection. All Tat constructs correspond to a nucleotide sequence to encode a 101 amino acid protein with a stop codon at the indicated position. Student’s t-test was performed comparing the β-galactosidase expression of each truncated Tat with Tat 101’s expression. **** p<1.46e-6. C) β-galactosidase expression was assessed after transfection with the listed Tat constructs. 10_stop encodes only the first 10 residues of Tat, while 10 contains the nucleotide sequence for the full-length protein with a stop codon mutation at position 11. Experiments were performed as quadruplicates on three separate occasions (n=3). Error bars are the mean with 99% confidence intervals. Student’s t-tests was performed comparing Tat 10 and 10_stop and 36 and 36_stop, separately. **** p<1.00e-6.
Interestingly, Tat 10 was capable of increasing transactivation approximately 20% in comparison to the empty vector control (Figure 5B), which led to further examination of this extreme truncation. This proposed read through did not seem to occur in Tat 31 and Tat 39 but appeared to have occurred to a lesser degree in Tat 36. One of the first controls was to determine if the proximity of the promoter may be reading through the first stop codon, allowing for leaky expression of full-length Tat 101. In order to examine this, an additional construct was generated that encoded only the first 10 residues of Tat. This Tat10_stop construct was unable to transactivate the LTR, supporting that the production of low levels of full-length Tat 101 were probably responsible for the increase in LTR transactivation (Figure 5C). This was also repeated with a Tat 36 construct, which also resulted in a similar trend. Overall, these truncated Tat variants, which did not encode for the entire transactivation domain, were unable to transactivate the LTR.
3. Discussion
The transactivation domain has been shown to be essential for Tat function and spans from position 1–48. The domain responsible for the interaction with TAR, a secondary RNA stem loop encoded by the HIV-1 LTR, has been shown to be residues 48–58. Investigations commonly utilize Tat 72 or 86 and those lengths transactivate to similar levels compared to 101 residues in most cells. However, the predominant length of Tat from patients was 101 residues in length, and other studies22,34 have observed this trend. Based on this information, the miscoded stop codons causing Tat lengths of 86, 95, 99, and 102 residues were predicted not to alter LTR transactivation (Figure 5B). Conversely, the truncated Tat lengths of 10, 31, 36, and 39 residues in length were rationalized to reduce transactivation by acting as less efficient transactivator proteins or by possibly acting as dominant negative inhibitor. The first four domains of Tat are responsible for transactivation, the first three domains (residues 1–48) bind to Cyclin-T1, a component of P-TEFb, and the fourth domain (residues 49–57) binds to TAR. These smaller truncations do not contain the domain that interacts with TAR, but have portions of the first three domains, which may allow it to bind to P-TEFb. In this scenario, the Tat truncations would bind to P-TEFb and prevent the co-localization of the host transcription factor to the HIV-1 LTR, thus reducing transcription of the virus. This may explain why short Tat lengths were likely selected against with a high probability of reverting back to longer Tat variant lengths in subsequent visits and why long Tat lengths were selected for, as opposed to being selected against, and have a low probability of maintaining any short forms of Tat in subsequent visits; at least this was what was observed in the CARES cohort. However, this was not observed in patient A0013. Although it should be noted that in early 2008 the patient had a 31-residue Tat and the next visit was not until late 2011. During this three-year duration, it would not be surprising if this patient returned to a predominantly longer Tat isoform before returning to a 10-residue form of Tat. In a separate study, 58 different patients from the Drexel Medicine CARES Cohort had Illumina next generation sequencing performed on Tat exon 1 and 2. When these samples were examined to determine the frequency of this extreme truncation, it was found to be present in 34 patients, however, it accounted for a relatively low proportion of the Tat lengths sequenced. In 82.4% (28/34) of the patients examined, Tat 10 was less than 20% of the total Tat sequenced (Supplemental Figure 3).
Low sample sizes appeared to have caused misleading results in Figures 2A and 3A. During the later years of the CARES cohort, NGS has replaced Sanger sequencing for acquiring HIV-1 genomic information. For comparison, 18.56 patients underwent Sanger sequencing in 2014. In Figure 3A, it appeared that France yielded a higher proportion of Tat 86 than other country. However, this was likely because the France patient sample size was only slightly over the threshold of 10 patients used in the analysis with 10.86 patients after removing low frequency Tat lengths. With a low sample size, the small number of Tat 86 patients appeared to comprise a higher proportion of the French population than probably exists.
Previous investigations concerning Tat truncations, such as the BEEHIVE study22, did not report any Tat truncations that occurred within Tat exon I. This could have been due to the lack of their presence, sequencing bias, or they simply that they may not have been reported at the time the analysis was performed. Our analysis extended into the first exon, addressing whether function altering truncations were present within patient samples. Additionally, these analyses have demonstrated that full-length Tat 101 was the most prevalent Tat length in Sanger sequences from HIV-1 subtype B patient samples worldwide. Examining the open reading frame of the tat gene at positions that prematurely truncate Tat at positions 31, 36, 39 also coincide with the 3’ end of the vpr gene. Similarly, the miscoded stop codons in the second exon of the tat gene, 86, 95, 99, and 102, are within the env gene and the second exon of the rev gene. To our knowledge, an analysis on premature truncations within the Vpr, Rev, or Env (gp41) proteins has yet to be performed.
A shortcoming of this study has been that the defective status of the sequenced proviral genomes was not assessed. To address this deficiency, one would have required the use of single genome amplification and this procedure was not within the scope of this study at the time these studies were initiated. Even though these Tat truncations were readily detectable via Sanger sequencing, it remains to be determined whether these viruses were capable of producing these Tat proteins and whether they might impact HAND progression. While the data collected here do not include full-length sequences of other genes, future studies will attempt to use longer PCR amplicons and 3rd generation NGS to determine the impact of Tat truncations on those genes. For instance, when there is a premature stop in Tat does that correlate with a virus that has a defective envelope gene? This could be either at the same spot in the genome or in a different part of the reading frame. Rather, is Tat the only predicted defective protein? In the future we intend to examine the production of these Tat truncations within a functional HIV-1 genome and functional alterations other than transactivation.
The apparent lack of correlation of the differing Tat lengths to the clinical parameters in the Drexel Medicine CARES cohort was an expected result due to the predominant number of Tat sequences being 101 residues in length (Figure 4A). A post-hoc power analysis was performed to determine if there was appropriate power (>0.8). Only age had the appropriate power to support statistical analyses. Further assessment was performed and the parameters with the highest positive and negative correlations with Tat length were MHDS (r2 = 0.022) and age (r2 = 0.021) (Figure 4B). The implication of these results demonstrated that it is unlikely that Tat length alone can account for increases or decreases in viral load or CD4+ T-cell count. However, it would be interesting to determine the potential function of the truncated Tat. Given the lack of the TAR binding domain, or in some cases portions of the transactivation domain, it would be assumed that the majority would not be functional in transactivation of the LTR. However, it would be interesting to see if they are secreted from the cell at a different rate and therefore pathogenic. This may be especially important in the CNS. The CARES cohort is currently collecting NGS data, utilizing the Illumina platform, of the HIV-1 quasispecies within patients. From this NGS data, the Tat sequences will be isolated and assessed for additional stop codon mutations. They will also be assessed for the proportion of the quasispecies that the truncated Tat is found in. was it the predominant sequence? The study here was performed as Sanger sequencing and we know from previous work on the LTR that the Sanger sequence provides the predominant sequence about 80% of the time, when compared to next generation results29. If it was not the predominant sequence, what proportion of the sequence does it make up? These types of data could potentially then be used to examine associations with neurocognitive impairment.
4. Methods
4.1. HIV-1-infected patient enrollment and PBMC collection
Patients in the Drexel CNS AIDS Research and Eradication Study (CARES) Cohort were recruited from the Partnership Comprehensive Care Practice of the Division of Infectious Diseases and HIV Medicine at Drexel University College of Medicine (Drexel Medicine), located in Philadelphia, Pennsylvania. Patients in the Drexel Medicine CARES cohort were recruited under protocol 1201000748 (Brian Wigdahl, PI), which adheres to the ethical standards of the Helsinki Declaration (1964, amended most recently in 2008)2. All patients provided written consent upon enrollment.
Following enrollment, patient clinical data was collected and entered into individual case report forms with anonymous identifiers. Clinical information collected directly from the patient included: age, self-identified gender, race, illicit drug use history, and approximate year of seroconversion. Additional clinical information was gathered directly from the patient chart, including: initial, nadir, and current CD4+ T-cell counts; initial, nadir, and current CD8+ T-cell counts; peak and latest viral loads; disease status; current ART status; current ART regimen; past ART treatments; AIDS-defining illnesses; admission of illicit drug, alcohol, and tobacco; any notations on mental health history including: inpatient stays for mental health reasons, any history of depression, schizophrenia, and other neurological diseases; and any notations on other complicating conditions related to HIV-1 infection. After clinical information was collected and the neurological status was assessed, blood samples (~50 mL) were drawn from each patient enrolled in the study and blood samples were used for drug screening, serum analysis, and PBMC isolation as previously described2.
4.2. Computational methodologies
Tat exon I and exon II were amplified from genomic DNA isolated from patient-derived PBMCs as previously described2,24. Direct Sanger sequencing of a PCR product was the predominant means of generating the sequences. Additionally, a 4.4 kb fragment read, which consisted of amplifying a 4.4 kb fragment from the 3’-end of the viral genome followed by isolation of individual fragments by subcloning and Sanger sequencing was used. With respect to the potential for sequencing errors, a stringent PHRED score cutoff of 40 was used to ensure that only quality nucleotide calls are analyzed. A PHRED score of 40 corresponds to a 99.99% accuracy. Overall, 205 patients had a sequence for both Tat exon 1 and exon 2.
CARES participants who did not undergo Sanger sequencing for Tat exons I and II were excluded from the subsequent analyses. Otherwise, there were no inclusion/exclusion criteria. CARES participant information was summarized for the following characteristics: age, years seropositive, nadir CD4 count, current CD4 count, current CD8 count, peak log viral load, current log viral load, modified Hopkins dementia scale (MHDS)35, current HAART adherence, gender, and self-admitted drug use. Once the necessary categories were coded, the mean and standard deviation of each category was calculated. Missing values in the columns were excluded in mean and standard deviation calculation.
CARES Tat exon sequences were trimmed and combined into one DNA strand before translation using purpose built scripts. All HIV-1 Tat protein sequences were acquired from translation of the combined tat CDS sequences. Once translated, the Tat sequences were linked to their corresponding patient metadata and sequence length was calculated for further analysis. Afterwards, the correlation between Tat length and all previously mentioned categories were calculated to measure the potential influence of Tat length on these other traits. Twenty seven Tat sequences did not contain a stop codon and were excluded from the analysis because of their unknown length.
To investigate the distribution of Tat length in the CARES Cohort over time, a frequency table was generated between Tat length and year the patient was sampled. This allows the visualization of Tat sequence length frequency collected in a year. Afterwards, 15 patients of the original 205 patients, that had at least three visits and yielded an observed Tat length other than 101, were longitudinally examined for changes in Tat length.
The LANL HIV-1 sequence database was then screened for subtype B sequences containing full Tat coding sequence (CDS) on May 26th, 2018. All other search parameters were unmodified. These sequences were downloaded without alignment, in a Fasta format, without gap handling, including HXB2 Reference Sequence (K03455), named as Subtype Country Year Name Accession, and separated into their exonic sequences. The exons were then joined and translated for comparison to CARES distribution of Tat lengths using purpose built scripts. To minimize bias from multiple patient sequences, each CARES Tat sequence was assigned a weight value. This weight was defined as the reciprocal of the number of Tat sequences generated from a patient; for example, if three Tat sequences arose from a patient, each sequence would have a weight of 1/3. The sum of these weights was interpreted as the number of patients belonging to a category. LANL sequences were assigned weights using the same methodology with CARES sequences23. Sequences with an unknown patient ID were assigned a weight of 1. Afterwards, the LANL data was analyzed by country to investigate the geographical distribution of Tat length23.
The nucleotide sequences obtained from the CARES Cohort and LANL database were converted into amino acids before performing the analyses presented. The residue specific variability for CARES and LANL Tat sequences were then analyzed by calculating their Shannon entropy. The standard HIV-1 reference (HXB2) Tat is 86 residues long, which would only allow entropy calculation of the first 86 residues of Tat if used to assign positional values. To circumvent this, CARES and LANL Tat sequences were aligned with a consensus subtype B Tat reference sequence from the 2000 HIV Sequence Compendium36 using the default settings of Clustal Omega v. 1.2.437. Afterwards, all positions where gaps existed in the reference sequence were removed in all sequences. The Shannon entropy for LANL and CARES Tat residues were then calculated using a modified method to determine amino acid occurrence proportion at each position. This modified method takes into account the proportion of amino acids that was found in each patient at a position and all proportions were divided by the number of patients found. These values were then summed to determine the amino acid occurrence proportion found in each residue position and then used for Shannon entropy calculation. The CARES and LANL alignments were also inputted into WebLogo38 to generate the sequence logos found in Supplemental Figure 2.
On June 11th, 2019 the LANL database was queried for a consensus Tat gene that translated into Tat 101 to use as a reference gene for dN/dS analysis. This was found on the LANL curated alignment” page where the selected Alignment Type was “Consensus/Ancestral” and the pre-selected region of the HIV-1 genome was the “TAT” option. All other options were left as default and “Get Alignment” was selected. From the resulting fasta file, the “CONSENSUS_B” sequence was acquired, ungapped, and aligned with CARES and LANL sequences using Clustal Omega as previously described in the Methods. Sequences were then ungapped in all positions where gaps existed in the reference. Sequences from the CARES Cohort and the LANL database that still contained gap characters and/or ambiguous DNA characters (e.g. ‘M’,’N’,’Y’) were disregarded for this analysis. A sliding window of 20 codons was used for calculation of dN, dS, and dN/dS for each position for all sequences from the LANL database and CARES Cohort compared to the consensus reference. All dN and dS values were averaged for all sequences at each position and then dN/dS was calculated. The dN/d ratio was then plotted using the average codon position for each given window position.
As an example of the modified calculation, assume a cohort has three patients. Patient one has 3 Tat sequences, and patients two and three both have 1 Tat sequence. To calculate the modified Shannon entropy of residue 1 of this cohort, the proportion of each amino acid occurrence at residue 1 for each patient needs to be calculated. In this example, the proportion of residue 1 for patient 1 was 0.6667 for Methionine and 0.3333 for Isoleucine. In patient two, it was 1.0 for Methionine and for patient three, 1.0 for Isoleucine. These proportions were then divided by the total number of patients in the cohort, which was 3 in this case. Afterwards, these are summed together by unique amino acid. In this example, Methionine would have a value of 0.5556 and Isoleucine would have a value of 0.4444. These represent the weighted probabilities of a particular amino acid occurring at residue 1. Afterwards, Shannon entropy is calculated using those probabilities.
Bioinformatic analysis was performed using Python v. 3.6.539 along with the Biopython v. 1.6840, Pandas v. 0.23.041, NumPy v. 1.14.342, SciPy v. 1.1.043, Matplotlib v. 2.2.244, Seaborn v.0.9.045, dnds v.2.146, pysam v.0.15.247, and rpy2 v.2.9.548 external libraries. The patient longitudinal figure creation and power calculations were performed using R v.3.4.449 along with the ggplot2 v.2.2.150, scales v.0.5.051, and pwr v.1.1.252 external libraries.
4.3. Tat truncation variants
Tat101Flag, inserted into a pcDNA3.1/Hygro (+), vector was previous gifted to us by Dr. Zachary Klase (University of the Sciences, PA). Site-directed mutagenesis (ThermoFisher) was used to initially introduce a stop codon before the C-terminal Flag tag before substituting specific residues into stop codons (see Supplemental Table 5 for oligonucleotide sequences for each variant). Site-directed mutagenesis was performed as previously described by the manufacturer. The PCR product was transformed into One Shot MAX Efficiency DH5α-T1R chemically competent E. coli. Sequencing analysis confirmed cloning accuracy.
4.4. Cell culture
P4 MAGI CCR5+ (P4R5)53 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% FBS, 0.4% penicillin and streptomycin, 0.8% Kanamycin, 0.75% sodium bicarbonate, and puromycin (1 μg/mL) at 37°C in humidified air with 5% CO2. These cells represent a HeLa-based cell line with a single integrated HIV-1 LTR driving expression of a β-galactosidase reporter. These cells have also been engineered to express CD4, CXCR4, and CCR5 on their surface through expression from non-integrated episome, with a puromycin-positive selection marker.
4.5. Transient transfection
P4R5 cells were transfected using Lipofectamine 2000 (ThermoFisher) with the various Tat truncations. Empty vector (EV; 100 ng) or each of the Tat variants was added to each well in a 96-well plate. Experiments were performed in quadruplicate on three separate occasions (n=3). Cells were harvested at 24 hours after transfection and processed using a Tropix Galacto-Star reporter procedure (Applied Biosystems) to quantify β-galactosidase expression, plates were read using a GLOMAX luminometer (Promega).
4.6. Statistical analyses
Significance was assessed using an unpaired two tailed Student’s t-test (α = 0.05) between β-galactosidase expression of truncated Tats with Tat 101’s expression (Figure 5A). Student’s two-tailed t-test was also used to calculate the significance of the different Tat lengths compared to their appropriate length stop construct (Figure 5B). To account for multiple comparisons, the threshold for significance was subjected to a Bonferroni correction. Student’s t-tests were performed using Python’s SciPy library.
Supplementary Material
Highlights.
Accumulation of HIV-1 Tat truncations was observed in both CARES and LANL
Most Tat sequences in the CARES Cohort and LANL database were 101 residues long
No correlation was observed between Tat length and clinical parameters within CARES
Longitudinal analysis of CARES patients showed a high likelihood of Tat 86 or 101 occurrence after visits where Tat length was below 86 residues
Specific Tat truncations appear to transactivate the HIV-1 LTR potentially due to stop codon read through
Acknowledgements
The authors were funded in part by the Public Health Service, National Institutes of Health, through grants from the National Institute of Neurological Disorders and Stroke (NINDS) R01 NS089435 (PI, Michael R. Nonnemacher), the NIMH Comprehensive NeuroAIDS Center (CNAC) P30 MH092177 (Kamel Khalili, PI; Brian Wigdahl, PI of the Drexel subcontract involving the Clinical and Translational Research Support Core) and under the Ruth L. Kirschstein National Research Service Award T32 MH079785 (PI, Jay Rappaport; with Brian Wigdahl serving as the PI of the Drexel University College of Medicine component and Olimpia Meucci as Co-Director). The contents of the paper are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Publisher's Disclaimer: This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Availability
The datasets generated during and/or analyzed during the current study are available in the Tat length analysis repository, https://github.com/DamLabResources/Tat-length-analysis. All Tat sequences have been deposited under BioProject ID XXX (Number will be provided upon publication).
Additional information
The corresponding author is responsible for submitting a competing financial interests statement on behalf of all authors of the paper. This statement must be included in the submitted article file.
Competing interests
The authors declare no competing interests.
References
- 1.Dingwall C et al. Human immunodeficiency virus 1 tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc Natl Acad Sci U S A 86, 6925–6929 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li L et al. Development of co-selected single nucleotide polymorphisms in the viral promoter precedes the onset of human immunodeficiency virus type 1-associated neurocognitive impairment. J Neurovirol 17, 92–109, 10.1007/s13365-010-0014-1 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rayne F et al. Phosphatidylinositol-(4,5)-bisphosphate enables efficient secretion of HIV-1 Tat by infected T-cells. EMBO J 29, 1348–1362, 10.1038/emboj.2010.32 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mele AR et al. Defining the molecular mechanisms of HIV-1 Tat secretion: PtdIns(4,5)P2 at the epicenter. Traffic, 10.1111/tra.12578 (2018). [DOI] [PMC free article] [PubMed]
- 5.Fields JA et al. Mechanisms of HIV-1 Tat neurotoxicity via CDK5 translocation and hyper-activation: role in HIV-associated neurocognitive disorders. Curr HIV Res 13, 43–54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Albini A et al. HIV-1 Tat protein mimicry of chemokines. Proc Natl Acad Sci U S A 95, 13153–13158 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bachani M, Sacktor N, McArthur JC, Nath A & Rumbaugh J Detection of anti-tat antibodies in CSF of individuals with HIV-associated neurocognitive disorders. J Neurovirol 19, 82–88, 10.1007/s13365-012-0144-8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofman FM, Dohadwala MM, Wright AD, Hinton DR & Walker SM Exogenous tat protein activates central nervous system-derived endothelial cells. J Neuroimmunol 54, 19–28 (1994). [DOI] [PubMed] [Google Scholar]
- 9.Hudson L et al. Detection of the human immunodeficiency virus regulatory protein tat in CNS tissues. J Neurovirol 6, 145–155 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Rayne F, Debaisieux S, Bonhoure A & Beaumelle B HIV-1 Tat is unconventionally secreted through the plasma membrane. Cell Biol Int 34, 409–413, 10.1042/CBI20090376 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Banks WA, Robinson SM & Nath A Permeability of the blood-brain barrier to HIV-1 Tat. Exp Neurol 193, 218–227, 10.1016/j.expneurol.2004.11.019 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Dahiya S, Irish BP, Nonnemacher MR & Wigdahl B Genetic variation and HIV-associated neurologic disease. Adv Virus Res 87, 183–240, 10.1016/B978-0-12-407698-3.00006-5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agostini S et al. Inhibition of Non Canonical HIV-1 Tat Secretion Through the Cellular Na(+),K(+)-ATPase Blocks HIV-1 Infection. EBioMedicine 21, 170–181, 10.1016/j.ebiom.2017.06.011 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carey AN, Sypek EI, Singh HD, Kaufman MJ & McLaughlin JP Expression of HIV-Tat protein is associated with learning and memory deficits in the mouse. Behav Brain Res 229, 48–56, 10.1016/j.bbr.2011.12.019 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gannon P, Khan MZ & Kolson DL Current understanding of HIV-associated neurocognitive disorders pathogenesis. Curr Opin Neurol 24, 275–283, 10.1097/WCO.0b013e32834695fb (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dampier W et al. HIV-1 Genetic Variation Resulting in the Development of New Quasispecies Continues to Be Encountered in the Peripheral Blood of Well-Suppressed Patients. PLoS One 11, e0155382, 10.1371/journal.pone.0155382 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L et al. Impact of Tat Genetic Variation on HIV-1 Disease. Adv Virol 2012, 123605, 10.1155/2012/123605 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boven LA et al. Brain-derived human immunodeficiency virus-1 Tat exerts differential effects on LTR transactivation and neuroimmune activation. J Neurovirol 13, 173–184, 10.1080/13550280701258399 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Ronsard L et al. Impact of Genetic Variations in HIV-1 Tat on LTR-Mediated Transcription via TAR RNA Interaction. Front Microbiol 8, 706, 10.3389/fmicb.2017.00706 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kukkonen S, Martinez-Viedma Mdel P, Kim N, Manrique M & Aldovini A HIV-1 Tat second exon limits the extent of Tat-mediated modulation of interferon-stimulated genes in antigen presenting cells. Retrovirology 11, 30, 10.1186/1742-4690-11-30(2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez-Huertas MR et al. The presence of HIV-1 Tat protein second exon delays fas protein-mediated apoptosis in CD4+ T lymphocytes: a potential mechanism for persistent viral production. J Biol Chem 288, 7626–7644, 10.1074/jbc.M112.408294 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Kuyl AC et al. The evolution of subtype B HIV-1 tat in the Netherlands during 1985–2012. Virus Res 250, 51–64, 10.1016/j.virusres.2018.04.008 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Foley B et al. HIV Sequence Compendium 2018 (Theoretical Biology and Biophysics Group, 2018). [Google Scholar]
- 24.Aiamkitsumrit B et al. Defining differential genetic signatures in CXCR4- and the CCR5-utilizing HIV-1 co-linear sequences. PLoS One 9, e107389, 10.1371/journal.pone.0107389 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antell GC et al. Utilization of HIV-1 envelope V3 to identify X4- and R5-specific Tat and LTR sequence signatures. Retrovirology 13, 32, 10.1186/s12977-016-0266-9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antell GC et al. Evidence of Divergent Amino Acid Usage in Comparative Analyses of R5- and X4-Associated HIV-1 Vpr Sequences. Int J Genomics 2017, 4081585, 10.1155/2017/4081585 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shah S et al. Functional properties of the HIV-1 long terminal repeat containing single-nucleotide polymorphisms in Sp site III and CCAAT/enhancer binding protein site I. Virol J 11, 92, 10.1186/1743-422X-11-92 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Link RW, Nonnemacher MR, Wigdahl B & Dampier W Prediction of Human Immunodeficiency Virus Type 1 Subtype-Specific Off-Target Effects Arising from CRISPR-Cas9 Gene Editing Therapy. CRISPR J 1, 294–302, 10.1089/crispr.2018.0020 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nonnemacher MR et al. HIV-1 Promoter Single Nucleotide Polymorphisms Are Associated with Clinical Disease Severity. PLoS One 11, e0150835, 10.1371/journal.pone.0150835 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spector C, Mele AR, Wigdahl B & Nonnemacher MR Genetic variation and function of the HIV-1 Tat protein. Med Microbiol Immunol 208, 131–169, 10.1007/s00430-019-00583-z (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darcis G et al. The Impact of HIV-1 Genetic Diversity on CRISPR-Cas9 Antiviral Activity and Viral Escape. Viruses 11, 10.3390/v11030255 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hemelaar J Implications of HIV diversity for the HIV-1 pandemic. J Infect 66, 391–400, 10.1016/j.jinf.2012.10.026 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Wertheim JO et al. Maintenance and reappearance of extremely divergent intra-host HIV-1 variants. Virus Evol 4, 10.1093/ve/vey030 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopez-Huertas MR et al. Modifications in host cell cytoskeleton structure and function mediated by intracellular HIV-1 Tat protein are greatly dependent on the second coding exon. Nucleic Acids Res 38, 3287–3307, 10.1093/nar/gkq037 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Tang XP, McArthur JC, Scott J & Gartner S Analysis of human immunodeficiency virus type 1 gp160 sequences from a patient with HIV dementia: evidence for monocyte trafficking into brain. J Neurovirol 6 Suppl 1, S70–81 (2000). [PubMed] [Google Scholar]
- 36.Kuiken CL et al. HIV Sequence Compendium 2000 (Theoretical Biology and Biophysics Group, 2000). [Google Scholar]
- 37.Sievers F et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular systems biology 7, 539, 10.1038/msb.2011.75 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crooks GE, Hon G, Chandonia JM & Brenner SE WebLogo: a sequence logo generator. Genome Res 14, 1188–1190, 10.1101/gr.849004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Rossum G Python tutorial (Centrum voor Wiskunde en Informatica (CWI)). [Google Scholar]
- 40.Cock PJA et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423, 10.1093/bioinformatics/btp163 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKinney W Data Structures for Statistical Computing in Python. Proceedings of the 9th Python in Science Conference, 51–56 (2010). [Google Scholar]
- 42.van der Walt S, Colbert SC & Varoquaux G The NumPy Array: A Structure for Efficient Numerical Computation. Computing in Science \& Engineering 13, 22–30, 10.1109/MCSE.2011.37 (2011). [DOI] [Google Scholar]
- 43.Jones E, O. E., Peterson P, et al. SciPy: Open Source Scientific Tools for Python doi: http://www.scipy.org/ (2001).
- 44.Hunter JD Matplotlib: A 2D Graphics Environment. Computing in Science \& Engineering 9, 90–95, 10.1109/MCSE.2007.55 (2007). [DOI] [Google Scholar]
- 45.Waskom M et al. (2014).
- 46.Qualieh A Dnds (April 30th, 2018).
- 47.Heger A PySam (April 30th, 2018).
- 48.Gautier L rpy2 (2017).
- 49.R Core Team (2014). R: A language and environment for statistical computing R Foundation for Statistical Computing, Vienna, Austria: URL http://www.R-project.org/.(2018). [Google Scholar]
- 50.Wickham H ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag, 2009). [Google Scholar]
- 51.Wickham H scales: Scale Functions for Visualization (2017).
- 52.Champely S pwr: Basic Functions for Power Analysis (2018).
- 53.Naldini L, Blomer U, Gage FH, Trono D & Verma IM Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci U S A 93, 11382–11388, 10.1073/pnas.93.21.11382 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.