Abstract
Colon cancer is one of the most common malignant tumors in the world; however, the mechanism underlying the progression of colon cancer remains unclear. In the present study, the expression of ubiquitin-specific peptidase 22 (USP22) in paraffin sections of human colon cancer tissues and normal colon tissues were examined using immunohistochemistry. The human colon cancer cell lines HCT116 and HT29 were used for USP22 knockdown experiments, and functional assays were performed. The results demonstrated that compared with normal colon tissues, human colon cancer tissues exhibited upregulated expression of USP22 and this was associated with tumor lymph node metastasis and tumor stage in colon cancer tissues. In addition, upregulated expression of USP22 was significantly correlated with both lower relapse-free survival and lower overall survival rates in patients with colon cancer. When USP22 was silenced in colon cancer cell lines, this resulted in a decrease in cell proliferation and metastatic behaviors. Furthermore, Bmi-1 and Cyclin D2 were found to be positively regulated by USP22, which may have mediated the tumorigenic effects of USP22 in human colon cancer. The results of the present study may have significant implications for examining the underlying mechanisms of cancer development and the potential development of cancer therapeutics.
Keywords: ubiquitin-specific peptidase 22, colon cancer, oncogene, proliferation, metastasis
Introduction
Colon cancer is one of the most common malignancies in the world and one of the leading causes of cancer-associated deaths (1–3). Colon cancer ranks second among the most common types of cancer in China (4,5). The 5-year average survival rate of patients with colon cancer is 50%, and the 5-year survival rate of advanced high-grade colon cancer patients is less than 10% (1–4). At present, the primary therapeutic method for treating patients with colon cancer is surgery combined with adjuvant chemotherapy (5,6). Oxaliplatin, 5-fluorouracil and capecitabine are the primary chemotherapeutic drugs used to treat colon cancer (4,6). Nevertheless, recurrence and multidrug resistance reduce the survival rate of patients with colon cancer (3,7,8). Researchers have demonstrated that multiple mechanisms are involved in the initiation, development, proliferation and metastasis of colon cancer, including the upregulation of oncogenes and downregulation of tumor suppressor genes, and dysregulated expression of miRNAs or other noncoding RNAs (9–11). However, the potential mechanism underlying human colon cancer development remains unclear. Additional studies will facilitate an improved understanding of colon cancer and identify potential targets for new treatments.
USP22 is a ubiquitin-specific protease that belongs to the deubiquitinase family of proteins (12,13). In human tissues, USP22 is primarily expressed in the brain, skeletal muscle and heart tissues, and it is expressed at lower levels in the liver and lung (12,14). USP22 is one subunit of SAGA (Spt-Ada-Gcn5-acetyltransferase), and is involved in the deubiquitination of histones H2A and H2B and the acetylation of histone H4 in order to regulate the transcription and expression of numerous genes (15,16). USP22 participates in a number of physiological processes, including regulating the cell cycle, cell growth, cell differentiation and cell signal transduction (17–19). USP22 has also been demonstrated to be involved in the c-myc and p53 signaling pathways (17). Recently, USP22 has been reported to serve an important role in a number of different types of cancer, including gastric cancer (19), hepatocellular carcinoma (20), non-small-cell lung cancer (21) and breast cancer (22). However, the function of USP22 in human colon cancer has not been demonstrated.
In the present study, the protein expression levels of USP22 in 80 colon cancer tissues and 64 normal colon tissues were determined, and the association between USP22 expression and clinicopathological features was analyzed. Furthermore, the effects of USP22 on colon cancer cell proliferation and metastasis were evaluated through cell functional assays. Finally, the molecular mechanism by which USP22 promotes biological behavior in colon cancer cells was examined.
Materials and methods
Clinical tissue samples
A total of 80 paraffin-embedded colon cancer tissues and 64 normal colon tissues were obtained from 86 male patients and 58 female patients aged 61.2 years (range, 45–78 years) at the First Affiliated Hospital of Anhui Medical University (Hefei, China) between January 2013 and December 2014. These patients with colon cancer were followed up for at least 5 years, and their clinicopathological parameters were collected. Patients with colon cancer and another disease or who had undergone special therapies prior to surgery were excluded. The clinicopathological parameters of these colon cancer patients were determined and confirmed based on WHO systems (23). Every patient provided written informed consent, and the study was approved by the Biomedical Ethics Committee of Anhui Medical University.
Immunohistochemistry (IHC)
USP22 expression in paraffin embedded sections (4 µm thick) of human colon cancer tissues and in normal colon tissues were obtained from the hospital pathology department having been fixed in 10% neutral formalin for 24 h at room temperature and paraffin embedded. The samples were examined by IHC. A two-step histostaining method (Fuzhou Maixin Biotech Co., Ltd.) was used for IHC as described previously (24). Samples were incubated with a USP22 antibody (cat. no. LS-C99567; 1:200; LifeSpan BioSciences, Inc.) for 4 h at room temperature. Subsequently, the samples were incubated with a horseradish peroxidase-conjugated universal detection reagent (Fuzhou Maixin Biotech Co., Ltd.) for 30 min at room temperature. 3,3′-Diaminobenzidine tetrahydrochloride (Fuzhou Maixin Biotech Co., Ltd.) was used for detection. The stained sections were observed and scored under a light microscope (magnification, ×400; Olympus Corporation). Positive signals of USP22 protein in the cells were primarily located in the nucleus and stained dark brown. Sections with ≥10% USP22-positive cells were designated USP22-positive, and sections with <10% USP22-positive cells were designated USP22-negative.
Cell culture
The human colon cancer cell lines SW480, SW620, HCT116, HT29 (colorectal adenocarcinoma cell line), and colon epithelium cell line NCM460 were obtained from American Type Culture Collection and cultured in DMEM with 10% fetal bovine serum (Biological Industries, Inc.), and cells were maintained at 37°C with 5% CO2 in a humidified incubator.
Small interfering (si)RNA transfection
USP22-siRNA1 (USP22-homo-695), USP22-siRNA2 (USP22-homo-1088) and negative control siRNA were obtained from Shanghai GenePharma Co., Ltd. siRNA (2.5 µg) transfection was performed using Lipofectamine® 2000 (Qiagen China Co., Ltd.) according to the manufacturer's protocol. Western blotting, reverse transcription-quantitative (RT-q)PCR and cell functional assays were performed 48 h after transfection. The sequences of the siRNAs were: siRNA1, 5′-GGAGAAAGAUCACCUCGAATT-3′; siRNA2, 5′-GCAUCAUAGACCAGAUCUUTT-3′; and negative control siRNA, 5′-UUCUCCGAAGGUGUCACGUTT-3′.
Western blotting
Western blot analysis was performed in order to examine the protein levels of USP22 essentially as described in previous studies (24). Protein was extracted from cells using lysis buffer (Beyotime, Inc.) and the protein concentration was determined using the BCA Protein assay kit (Thermo Fisher Scientific, Inc.). Proteins (30 µg) were separated via SDS-PAGE (10% gel) and transferred to polyvinylidene fluoride membranes (EMD Millipore). Membranes were blocked at room temperature with 5% skimmed milk for 45 min and incubated at room temperature for 2 h with the following antibodies: USP22 rabbit polyclonal antibody (LifeSpan BioSciences, Inc.; cat. no. LS-C99567; 1:800), Bmi-1 rabbit polyclonal antibody (Abcam; cat. no. ab126783; 1:500) and Cyclin D2 rabbit polyclonal antibody (OriGene Technologies, Inc.; cat. no. TA323121; 1:500), and subsequently with horseradish peroxidase-conjugated goat anti-rabbit/mouse secondary antibodies (Abcam; cat. no. ab150077; 1:1,000) at room temperature for 1 h. The blots were incubated by WesternBright chemiluminescence (ECL) (Western blotting detection kit; Advansta, Inc.) for 1 min, and developed using ImageQuant™ LAS4000 (GE Healthcare Life Sciences). Relative expression level was normalized to endogenous control β-actin mouse monoclonal antibody (Sigma-Aldrich; Merck KGaA; cat. no. A5316; 1:5,000) using Image J software (version 1.51; National Institutes of Health).
RT-qPCR
RT-quantitative PCR (RT-qPCR) assay was performed in order to investigate the mRNA levels of USP22 in HCT116 and HT29 cell lines following siRNA transfection as previously described (24). Total RNA was isolated from cells and cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and cDNA was synthesized at 42°C for 30 min using the Transcript All-in-One First-Strand cDNA Synthesis Super Mix for qPCR kit (TransGen Biotech, Inc.). The gene expression levels of USP22 were determined using a SYBR Green qPCR kit (TransGen Biotech, Inc.), and calculated using the 2−ΔΔCq method (25). The PCR thermocycling conditions were as follows: 95°C for 40 sec followed by 40 cycles of 95°C for 5 sec, 60°C for 30 sec, and 95°C for 15 sec, 60°C for 57 sec, 95°C for 15 sec. GAPDH was used as the housekeeping gene. The sequences of the primers used were: USP22 forward, 5′-CACTTCTGCGGGACT-3′ and USP22-reverse, 5′-TACGGGATGTGAGGG-3′; and GAPDH-forward 5′-AGCAAGAGCACAAGAGGAAG-3′, and GAPDH-reverse 5′-GGTTGAGCACAGGGTACTTT-3′.
Cell proliferation assays
Cell proliferation was measured using an MTT assay and colony formation assay as previously described (24). For the MTT assay, cells (1,500 per well) were seeded into 96-well plates, and MTT values were detected every day for 5 days. A total of 10 µl of MTT reagent was added into each well. After 2 h, the medium was removed and 100 µl of DMSO detection reagent was added. Light absorption values were detected at 570 nm. For the cell colony formation assay, cells (1,000 per well) were seeded into 6-well plates, when the number of cells in the majority of single colonies were >50, cells were washed three times with PBS and fixed with 90% ethanol for 1 h at room temperature. Subsequently, cells were stained with 0.1% crystal violet for 10 min at room temperature, and the number of visible colonies was counted using the naked eye.
Migration and invasion assays
For the cell migration assay, cells (1×106 per well) were mixed with medium without serum, and plated in upper chambers (Corning Inc.) without Matrigel (BD Biosciences); below the chambers, medium with 10% serum as the chemoattractant was added. After 40 h, chambers were washed three times with PBS and fixed with 4% paraformaldehyde for 1 h at room temperature, and subsequently stained with 0.1% crystal violet for 10 min at room temperature. Finally, the stained cells from four random fields were counted, and the images were captured using a light microscope (magnification, ×100; Olympus Corporation). For the cell invasion assay, cells were plated into 24-well transwell chambers (Corning Inc.) coated with Matrigel (BD Biosciences), the number of invaded cells were counted after 70 h. All other conditions were the same as the migration assay.
Cell cycle assay
For the cell cycle assay, a Cell Cycle and Apoptosis Analysis kit (Beyotime Institute of Biotechnology) was used. Cells were collected, washed with PBS and fixed with 70% ice-cold ethanol >2 h at 4°C. The cells were then washed with 1 ml ice-cold PBS. A total of 0.5 ml PI solution was added into the cells and incubated further for 1 h at 4°C in the dark. Flow cytometry (BD Biosciences FACSVerse) was used to detect the cell cycle stage.
Statistical analysis
Each experiment was repeated three times. An unpaired two-tailed t-tests was used to analyze the mean values between two groups, and a one-way ANOVA was used to compare the mean values among multiple groups with a Bonferroni's multiple comparison post-hoc test. A Pearson's χ2 test was used for analysis of data from tissue immunohistochemistry and clinicopathological parameter association analysis. Patient relapse-free survival (RFS) and overall survival (OS) analysis was performed using Kaplan-Meier curves, and the log-rank test was used for statistical analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of USP22 is upregulated in human colon cancer tissues
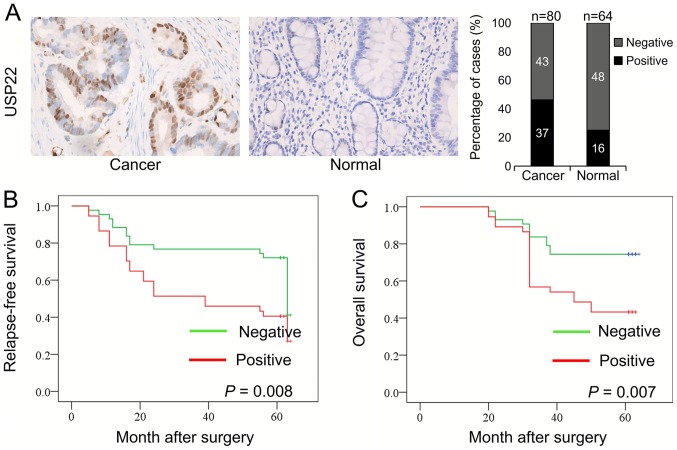
A total of 80 colon cancer tissues and 64 normal colon tissues were collected, and the expression of USP22 in these tissues was determined using IHC. Positive signals for USP22 protein in the cells were primarily located in the nucleus (Fig. 1A). The expression of USP22 was significantly higher in colon cancer tissues compared with normal tissues. The percentages of USP22-positive and USP22-negative cells in these tissues were calculated. Among the 80 colon cancer tissues, 37 (46.25%) were USP22-positive, and 43 (53.75%) were USP22-negative. Among the 64 normal colon tissues, 16 (25%) were USP22-positive, and 48 (75%) were USP22-negative. There was a significant difference in USP22 expression between colon cancer tissues and normal colon tissues (P=0.009; Table I). Therefore, the expression of USP22 was significantly higher in human colon cancer tissues compared with the normal colon tissues.
Figure 1.
Expression of USP22 in human colon cancer tissues and normal colon tissues. (A) Representative images of USP22 expression in colon cancer and normal colon tissues were examined using immunohistochemistry. Percentage of cases which stained positive or negative for USB-22 expression. Kaplan-Meier curves were constructed of (B) RFS and (C) OS rates in the USP22-negative and USP22-positive groups of patients with colon cancer. USP22, ubiquitin-specific peptidase 22.
Table I.
Expression of USP22 in colon cancer and normal tissues.
| USP22 expression | |||
|---|---|---|---|
| Group | n | Negative, n (%) | Positive, n (%) |
| Colon cancer | 80 | 43 (53.75) | 37 (46.25)a |
| Normal | 64 | 48 (75.0) | 16 (25.0) |
P=0.009. USP22, ubiquitin-specific peptidase 22.
Association between USP22 expression and clinicopathological parameters and survival rates in patients with colon cancer
The clinicopathological parameters of the 80 colon cancer patients were collected to analyze the association between USP22 expression and clinicopathological parameters. Patient age, tumor size, lymph node metastasis, tumor grade and the Tumor-Node-Metastasis staging system were included in this analysis (23). Table II shows that the percentage of tissues with high USP22 expression in colon cancer patients with lymph node metastasis (20 out of 31; 64.5%) was significantly higher compared with patients with colon cancer without lymph node metastasis (17 out of 49; 34.7%; P=0.009). The percentage of tissues with high USP22 expression in late-stage colon cancer (stage III–IV; 19 out of 30, 63.3%) was significantly higher compared with patients with early-stage colon cancer (stage I–II; 18 out of 50, 36.0%; P=0.018). However, there were no significant associations between USP22 expression and patient age, tumor size or tumor grade (all P>0.05).
Table II.
Association between USP22 expression and clinicopathological parameters in patients with colon cancer.
| Parameter | n | USP22 positive expression, n (%) | P-value |
|---|---|---|---|
| Age, years | 0.698 | ||
| ≤55 | 20 | 10 (50.0) | |
| >55 | 60 | 27 (45.5) | |
| Tumor size, cm | 0.779 | ||
| ≤5 | 66 | 31 (47.0) | |
| >5 | 14 | 6 (42.9) | |
| Lymph node metastasis | 0.009b | ||
| Absent | 49 | 17 (34.7) | |
| Present | 31 | 20 (64.5) | |
| Grade | 0.210 | ||
| Well | 10 | 3 (30.0) | |
| Moderate | 58 | 26 (44.8) | |
| Poor | 12 | 8 (66.7) | |
| Stage | 0.018a | ||
| I + II | 50 | 18 (36.0) | |
| III+ IV | 30 | 19 (63.3) |
P<0.05
P<0.01. USP22, ubiquitin-specific peptidase 22.
Kaplan-Meier curves were generated to analyze the association between USP22 expression and RFS and OS in the 80 colon cancer samples. All patients were followed-up for ≥5 years. Compared with patients in the USP22-negative group, patients in the USP22-positive group exhibited significantly lower RFS (P=0.008) and OS rates (P=0.007; Fig. 1B and C). Therefore, high expression of USP22 was associated with a poor prognosis.
Downregulation of USP22 expression
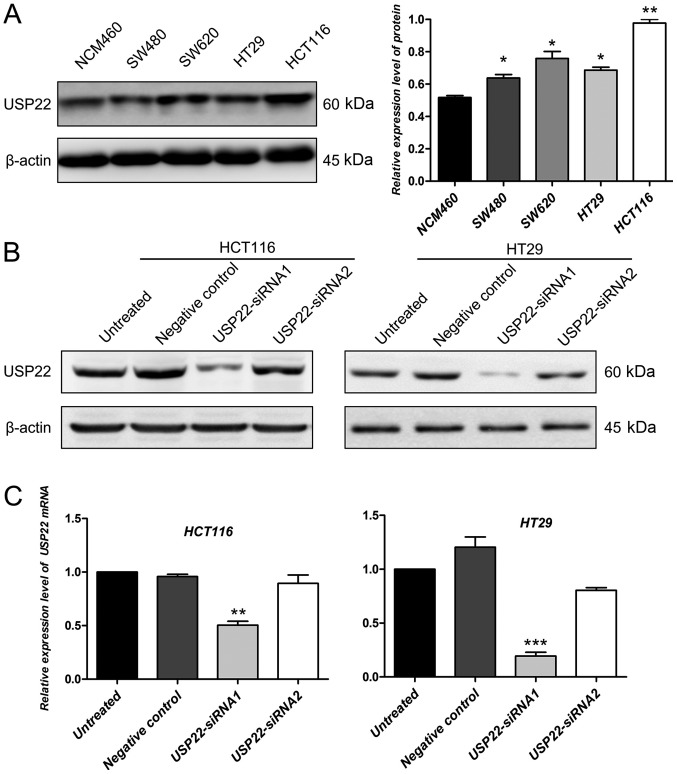
The protein expression levels of USP22 in colon cancer cell lines (included HT29, colorectal adenocarcinoma cell line) and a colon epithelium cell line were determined by western blotting. Compared with the colon epithelium cell line, the expression of USP22 was significantly higher in colon cancer cell lines (P<0.05; Fig. 2A). The human colon cancer cell line HCT116 and colorectal adenocarcinoma cell line HT29 were selected to perform the cell functional experiments using siRNA transfection for USP22 knockdown. Compared with untreated cells and the negative control, in both cell lines, USP22-siRNA1 decreased the protein expression level of USP22 (Fig. 2B). However, the decrease in USP22 protein levels in cells transfected with siRNA2 was not significant compared with that in control cells (Fig. 2B). To confirm this result, the mRNA expression levels of USP22 were also examined in untreated, negative control, and USP22-siRNA1 and USP22-siRNA2 transfected cancer cells. Compared with the control, the mRNA expression levels of USP22 decreased significantly in cells transfected with USP22-siRNA1 in HCT116 and HT29 cells (P<0.01; Fig. 2C). Therefore, cells transfected with USP22- siRNA1 were used for all subsequent experiments.
Figure 2.
Expression of USP22 in cells transfected with siRNA. (A) Protein expression levels of USP22 were examined in colon cancer cell lines and a colon epithelium cell line. *P<0.05, **P<0.01 vs. NCM460. (B) Protein expression levels of USP22 were determined following transfection of HCT116 and HT29 cell lines with USP22-siRNA1, USP22-siRNA2, negative control siRNA or no vector. β-Actin was used as the loading control. (C) mRNA expression levels of USP22 were examined following transfection with USP22-siRNA1, USP22-siRNA2, negative control siRNA or no vector. **P<0.01, ***P<0.001 vs. NCM460. USP22, ubiquitin-specific peptidase 22; siRNA, small interfering RNA.
Knockdown of USP22 reduces proliferation in human colon cancer cells
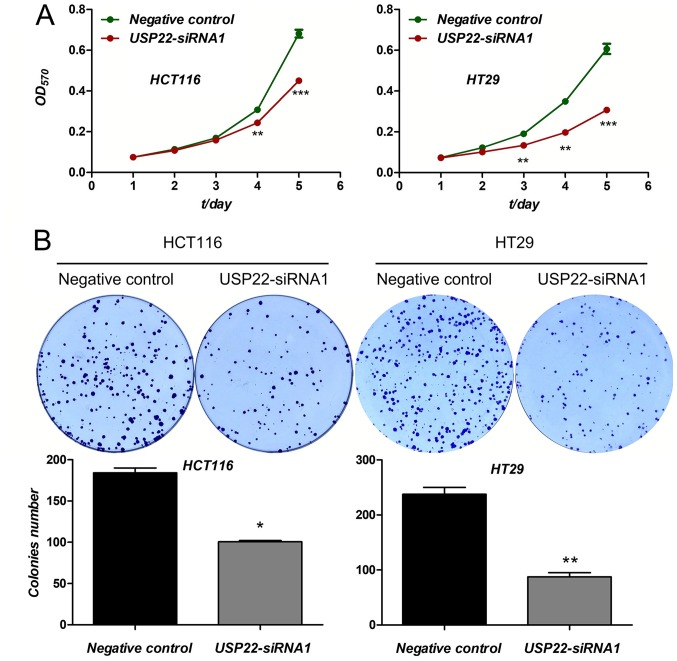
The effect of USP22 knockdown on proliferation as determined. Compared with the negative control, cell viability, evaluated using an MTT assay, was significantly decreased in cells transfected with USP22-siRNA1 in both cancer cell lines after 3 (HCT116 cells) and 4 days (HT29 cells) (P<0.01; Fig. 3A). Furthermore, compared with the negative control, cell colony formation was also reduced in both cancer cell lines transfected with USP22-siRNA1 (P<0.05; Fig. 3B). Therefore, knockdown of USP22 significantly reduced cell proliferation in human colon cancer cells. These results suggest that USP22 acts as a promoter of tumor cell proliferation in human colon cancer cells.
Figure 3.
Knockdown of USP22 reduces proliferation in human colon cancer cells. (A) MTT assays were performed for 5 days in colon cancer cells following transfection with USP22-siRNA1 or negative control siRNA. **P<0.01, ***P<0.001. (B) Colony formation assays were performed in colon cancer cells transfected with USP22-siRNA1 or negative control siRNA. *P<0.05, **P<0.01. USP22, ubiquitin-specific peptidase 22; siRNA, small interfering RNA; OD, optical density.
Knockdown of USP22 reduces metastasis in human colon cancer cells
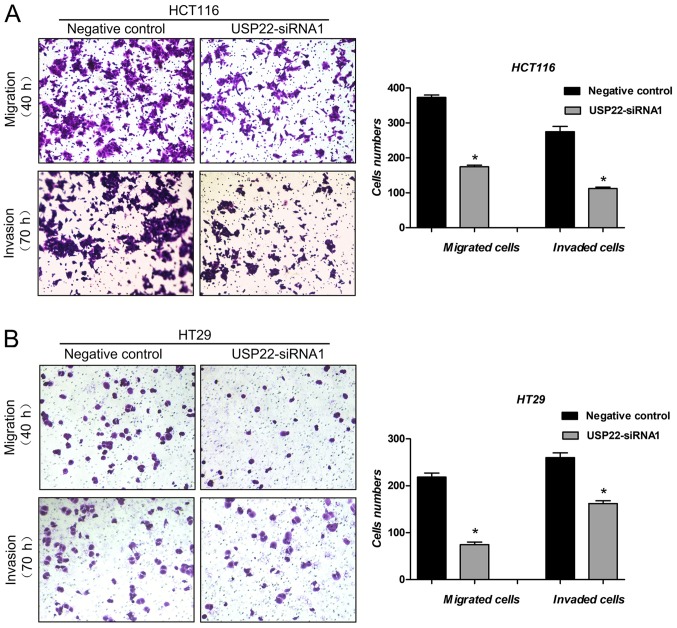
Cell migration and invasion assays were used to examine the effect of USP22 on metastasis of human colon cancer cells. Compared with the negative control, in cells transfected with USP22-siRNA1, both cell migration (P<0.05) and cell invasion (P<0.05) were significantly decreased in both colon cancer cell lines (Fig. 4A and B). These results suggest that USP22 promotes metastasis in human colon cancer cells.
Figure 4.
Knockdown of USP22 reduces metastasis in human colon cancer cells. (A) Migration and (B) invasion assays were performed following transfection with USP22-siRNA1 or negative control siRNA. *P<0.05 vs. respective negative control. Magnification, ×100. USP22, ubiquitin-specific peptidase 22; siRNA, small interfering RNA.
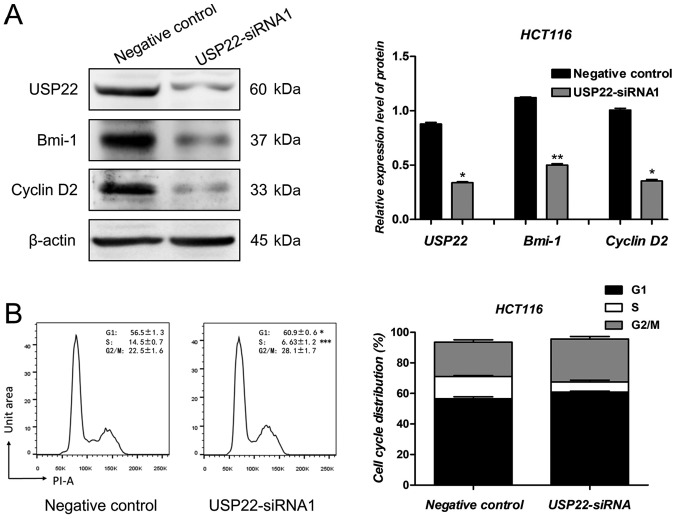
USP22 regulates the expression of Bmi-1 and Cyclin D2
Several candidate genes were selected to identify the downstream mechanism by which USP22 exerted its effects on human colon cancer cells (data not shown). Similar to previous results, the protein expression levels of USP22 decreased significantly following transfection with USP22-siRNA1 in HCT116 cells. Compared with the negative control cells, Bmi-1 and Cyclin D2 protein expression levels were decreased significantly in cells transfected with USP22-siRNA1 (P<0.05; Fig. 5A). As reported previously, Bmi-1 and Cyclin D2 are oncogenes in human colon cancer cells (26–28). To determine whether USP22 knockdown affected cell cycle regulation, flow cytometry analysis was performed following transfection with USP22-siRNA1 and negative control siRNA. As shown in Fig. 5B, the number of USP22-siRNA1 cells in the G1 phase increased significantly compared with the negative control (P<0.05; Fig. 5B). Therefore, Bmi-1 and Cyclin D2 may be involved in the promotion of proliferation and metastasis of colon cancer cells by USP22.
Figure 5.
USP22 regulates the expression of Bmi-1 and Cyclin D2. (A) Protein expression levels of USP22, Bmi-1 and Cyclin D2 were examined following transfection of HCT116 cells with USP22-siRNA1 or negative control siRNA. β-Actin was used as a loading control. *P<0.05, **P<0.01. (B) Cell cycle distribution was determined by flow cytometry analysis. The percentage of HCT116 cells in each cell cycle phase is shown. *P<0.05. ***P<0.001. USP22, ubiquitin-specific peptidase 22; siRNA, small interfering RNA; PI, propidium iodide.
Discussion
In the present study, the role of USP22 in human colon cancer was determined through clinical analyses and cell functional assays. IHC analysis and clinicopathological parameter association analysis showed that the expression of USP22 was higher in colon cancer tissues compared with normal colon tissues, and that overexpression of USP22 was positively correlated with tumor lymph node metastasis and tumor stage. Kaplan-Meier curves showed that USP22 was negatively associated with patient RFS and OS rates in the 80 patients with colon cancer. Cell functional assays were performed. Knockdown of USP22 by siRNA transfection significantly decreased cell viability, cell colony formation, cell migration and cell invasion. Therefore, USP22 promoted both cell proliferation and cell metastasis in colon cancer cells and upregulation of USP22 predicted poor prognosis for patients with colon cancer.
USP22 belongs to the deubiquitinase family of proteins and strictly regulates gene expression through histone deubiquitination or acetylation procession (12,13,15,16). It has been reported that USP22 serves an important role in driving transcription and cell cycle progression (17–19). USP22 has been demonstrated to be a vital biomarker of cancer stem cells (16,29). In the present study, the oncogenic role of USP22 in human colon cancer was determined. As reported previously, Wang et al (30) showed that USP22 was upregulated in malignant colon carcinoma, and expression was associated with the degree of differentiation, invasion, lymph node metastasis and tumor stage in patients with colon carcinoma (30). The results of the present study are consistent with the results observed by Wang et al (30). However, Ao et al (31) reported that USP22 promoted cell proliferation but inhibited cell invasion in SW480 colon cancer through the STAT3/MMP9 pathway (31). In these experiments, USP22 increased the proliferation in HCT116 and SW480 colon cancer cell lines, whereas the metastasis promoting effects on colon cancer cells were cell specific. Furthermore, it has been reported that USP22 positively regulates c-Myc and promotes tumorigenic activity in human breast cancer (22). Tang et al (32) reported that elevated expression of USP22 was associated with poor prognosis in breast cancer patients. Ma et al (19) demonstrated that USP22 maintained gastric cancer cell stemness and promoted gastric cancer progression by stabilizing the Bmi-1 protein (19). USP22 was also reported to serve as an oncogene in human hepatocellular carcinoma; overexpression of USP22 indicated poor prognosis for patients with hepatocellular carcinoma and USP22 mediated multidrug resistance in hepatocellular carcinoma (20,32). USP22 also served as an oncogene in a number of different types of cancer, including non-small-cell lung cancer (21), papillary thyroid carcinoma (29) and glioma (33). Therefore, USP22 has been shown to be oncogenic in a large number of different types of cancer including colon cancer, and specific inhibitors of USP22 may serve as potential therapeutic options for treating patients with cancer where upregulation of USP22 is observed.
It was determined that Bmi-1 and Cyclin D2 were positively regulated by USP22. As reported previously, Bmi-1 is oncogenic in human colon cancer cells; the expression level of Bmi-1 is associated with tumor progression and prognosis of colon cancer, and Bmi-1 promotes migration and invasion of colon cancer stem cells by regulating E-cadherin (26,27). Furthermore, Bmi-1 has also been reported to serve as an oncogene in human hepatocellular carcinoma (34), oral cancer (35), breast cancer (36), gastric cancer (37) and lung cancer (38) amongst others. Cyclin D2 is an important protein involved in cell cycle regulation. Cyclin D2 promotes both proliferation and metastasis of human colon cancer cells (28,39). Furthermore, Cyclin D2 also serves as an oncogene in a number of different types of cancer, including breast cancer (40), prostate cancer (41), oral squamous carcinoma (42) and non-small-cell lung cancer (43). Therefore, Bmi-1 and Cyclin D2 are important oncogenes in cancer and may mediate the promoting role of USP22 in human colon cancer.
In conclusion, the present study systematically examined the oncogenic role of USP22 in human colon cancer. Overexpression of USP22 is associated with enhanced malignant properties in colon cancer cells and a poorer prognosis in patients with colon cancer. The present study highlights the role of USP22 in cancer progression, and USP22 may serve as a potential therapeutic target for treating patients with colon cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Scientific Research Foundation of Anhui Medical University, National Natural Science Foundation of China (grant nos. 81572350 and 81500373), Natural Science Foundation of Anhui Province (grant no. 1608085MH193) and the Excellent Talents Supporting Program of the University of Anhui Education Department (grant no. gxyq2018011).
Availability of data and material
The datasets used and/or analyzed during the present study are available from the author upon reasonable request.
Authors' contributions
WW designed the study. XY collected the data and HW performed the experiments. AX, XZ and YZ performed the statistical analyses. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Each patient signed informed consent for the present study. The present study was approved by Biomedical Ethics Committee of Anhui Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Meng X, Fu R. miR-206 regulates 5-FU resistance by targeting Bcl-2 in colon cancer cells. Onco Targets Ther. 2018;11:1757–1765. doi: 10.2147/OTT.S159093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evert J, Pathak S, Sun XF, Zhang H. A study on effect of oxaliplatin in MicroRNA expression in human colon cancer. J Cancer. 2018;9:2046–2053. doi: 10.7150/jca.24474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y, Gong P, Hou JX, Huang W, Ma XP, Wang YL, Li J, Cui XB, Li N. miR-34a regulates multidrug resistance via positively modulating OAZ2 signaling in colon cancer cells. J Immunol Res. 2018;2018:7498514. doi: 10.1155/2018/7498514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dou H, Shen R, Tao J, Huang L, Shi H, Chen H, Wang Y, Wang T. Curcumin Suppresses the Colon Cancer Proliferation by Inhibiting Wnt/β-catenin pathways via miR-130a. Front Pharmacol. 2017;8:877. doi: 10.3389/fphar.2017.00877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li L, Ma BB. Colorectal cancer in Chinese patients: Current and emerging treatment options. Onco Targets Ther. 2014;7:1817–1828. doi: 10.2147/OTT.S48409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mishra J, Drummond J, Quazi SH, Karanki SS, Shaw JJ, Chen B, Kumar N. Prospective of colon cancer treatments and scope for combinatorial approach to enhanced cancer cell apoptosis. Crit Rev Oncol Hematol. 2013;86:232–250. doi: 10.1016/j.critrevonc.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu YR, Liang L, Zhao JM, Zhang Y, Zhang M, Zhong WL, Zhang Q, Wei JJ, Li M, Yuan J, et al. Twist1 confers multidrug resistance in colon cancer through upregulation of ATP-binding cassette transporters. Oncotarget. 2017;8:52901–52912. doi: 10.18632/oncotarget.17548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He K, Chen D, Ruan H, Li X, Tong J, Xu X, Zhang L, Yu J. BRAFV600E-dependent Mcl-1 stabilization leads to everolimus resistance in colon cancer cells. Oncotarget. 2016;7:47699–47710. doi: 10.18632/oncotarget.10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Segditsas S, Tomlinson I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene. 2006;25:7531–7537. doi: 10.1038/sj.onc.1210059. [DOI] [PubMed] [Google Scholar]
- 10.Hollis M, Nair K, Vyas A, Chaturvedi LS, Gambhir S, Vyas D. MicroRNAs potential utility in colon cancer: Early detection, prognosis, and chemosensitivity. World J Gastroenterol. 2015;21:8284–8292. doi: 10.3748/wjg.v21.i27.8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu YJ, Nangia-Makker P, Farhana L, Majumdar APN. A novel mechanism of lncRNA and miRNA interaction: CCAT2 regulates miR-145 expression by suppressing its maturation process in colon cancer cells. Mol Cancer. 2017;16:155. doi: 10.1186/s12943-017-0725-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gong Z, Liu J, Xie X, Xu X, Wu P, Li H, Wang Y, Li W, Xiong J. Identification of potential target genes of USP22 via ChIP-seq and RNA-seq analysis in HeLa cells. Genet Mol Biol. 2018;41:488–495. doi: 10.1590/1678-4685-gmb-2017-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang A, Ning Z, Lu C, Gao W, Liang J, Yan Q, Tan G, Liu J. USP22 induces cisplatin resistance in lung adenocarcinoma by regulating γH2AX-Mediated DNA damage repair and Ku70/Bax-mediated apoptosis. Front Pharmacol. 2017;8:274. doi: 10.3389/fphar.2017.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee HJ, Kim MS, Shin JM, Park TJ, Chung HM, Baek KH. The expression patterns of deubiquitinating enzymes, USP22 and Usp22. Gene Expr Patterns. 2006;6:277–284. doi: 10.1016/j.modgep.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Armour SM, Bennett EJ, Braun CR, Zhang XY, McMahon SB, Gygi SP, Harper JW, Sinclair DA. A high-confidence interaction map identifies SIRT1 as a mediator of acetylation of USP22 and the SAGA coactivator complex. Mol Cell Biol. 2013;33:1487–1502. doi: 10.1128/MCB.00971-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell. 2008;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X, Zang H, Luo Y, Wu J, Fang Z, Zhu W, Li Y. High expression of USP22 predicts poor prognosis and advanced clinicopathological features in solid tumors: A meta-analysis. Onco Targets Ther. 2018;11:3035–3046. doi: 10.2147/OTT.S148662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458:438–444. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]
- 19.Ma Y, Fu HL, Wang Z, Huang H, Ni J, Song J, Xia Y, Jin WL, Cui DX. USP22 maintains gastric cancer stem cell stemness and promotes gastric cancer progression by stabilizing BMI1 protein. Oncotarget. 2017;8:33329–33342. doi: 10.18632/oncotarget.16445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ling S, Li J, Shan Q, Dai H, Lu D, Wen X, Song P, Xie H, Zhou L, Liu J, et al. USP22 mediates the multidrug resistance of hepatocellular carcinoma via the SIRT1/AKT/MRP1 signaling pathway. Mol Oncol. 2017;11:682–695. doi: 10.1002/1878-0261.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu G, Cai J, Wang L, Jiang LY, Huang JB, Hu R, Ding F. MicroRNA-30e-5p suppresses non-small cell lung cancer tumorigenesis by regulating USP22-mediated Sirt1/JAK/STAT3 signaling. Exp Cell Res. 2018;362:268–278. doi: 10.1016/j.yexcr.2017.11.027. [DOI] [PubMed] [Google Scholar]
- 22.Kim D, Hong A, Park HI, Shin WH, Yoo L, Jeon SJ, Chung KC. Deubiquitinating enzyme USP22 positively regulates c-Myc stability and tumorigenic activity in mammalian and breast cancer cells. J Cell Physiol. 2017;232:3664–3676. doi: 10.1002/jcp.25841. [DOI] [PubMed] [Google Scholar]
- 23.Bosman FT, Carneiro F, Hruban RH, Theise ND, editors. 4th. Vol. 3. International Agency for Research on Cancer; Lyon: 2010. WHO classification of tumours of the digestive system. [Google Scholar]
- 24.Wang B, Wu ZS, Wu Q. CMIP promotes proliferation and metastasis in human glioma. Biomed Res Int. 2017;2017:5340160. doi: 10.1155/2017/5340160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Li DW, Tang HM, Fan JW, Yan DW, Zhou CZ, Li SX, Wang XL, Peng ZH. Expression level of Bmi-1 oncoprotein is associated with progression and prognosis in colon cancer. J Cancer Res Clin Oncol. 2010;136:997–1006. doi: 10.1007/s00432-009-0745-7. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Z, Bu X, Chen H, Wang Q, Sha W. Bmi-1 promotes the invasion and migration of colon cancer stem cells through the downregulation of E-cadherin. Int J Mol Med. 2016;38:1199–1207. doi: 10.3892/ijmm.2016.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlormann W, Naumann S, Renner C, Glei M. Influence of miRNA-106b and miRNA-135a on butyrate-regulated expression of p21 and Cyclin D2 in human colon adenoma cells. Genes Nutr. 2015;10:50. doi: 10.1007/s12263-015-0500-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li ZH, Yu Y, Du C, Fu H, Wang J, Tian Y. RNA interference-mediated USP22 gene silencing promotes human brain glioma apoptosis and induces cell cycle arrest. Oncol Lett. 2013;5:1290–1294. doi: 10.3892/ol.2013.1188. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Wang Z, Zhu L, Guo T, Wang Y, Yang J. Decreased H2B monoubiquitination and overexpression of ubiquitin-specific protease enzyme 22 in malignant colon carcinoma. Hum Pathol. 2015;46:1006–1014. doi: 10.1016/j.humpath.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 31.Ao N, Liu Y, Bian X, Feng H, Liu Y. Ubiquitin-specific peptidase 22 inhibits colon cancer cell invasion by suppressing the signal transducer and activator of transcription 3/matrix metalloproteinase 9 pathway. Mol Med Rep. 2015;12:2107–2113. doi: 10.3892/mmr.2015.3661. [DOI] [PubMed] [Google Scholar]
- 32.Tang B, Tang F, Li B, Yuan SG, Xu Q, Tomlinson S, Jin JF, Hu W, He SQ. High USP22 expression indicates poor prognosis in hepatocellular carcinoma. Oncotarget. 2015;6:12654–12667. doi: 10.18632/oncotarget.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao H, Tang H, Huang Q, Qiu B, Liu X, Fan D, Gong L, Guo H, Chen C, Lei S, et al. MiR-101 targets USP22 to inhibit the tumorigenesis of papillary thyroid carcinoma. Am J Cancer Res. 2016;6:2575–2586. [PMC free article] [PubMed] [Google Scholar]
- 34.Shao Y, Zhang D, Li X, Yang J, Chen L, Ning Z, Xu Y, Deng G, Tao M, Zhu Y, Jiang J. MicroRNA-203 increases cell radiosensitivity via directly targeting Bmi-1 in hepatocellular carcinoma. Mol Pharm. 2018;15:3205–3215. doi: 10.1021/acs.molpharmaceut.8b00302. [DOI] [PubMed] [Google Scholar]
- 35.Kim JS, Choi DW, Kim CS, Yu SK, Kim HJ, Go DS, Lee SA, Moon SM, Kim SG, Chun HS, et al. MicroRNA-203 induces apoptosis by targeting Bmi-1 in YD-38 oral cancer cells. Anticancer Res. 2018;38:3477–3485. doi: 10.21873/anticanres.12618. [DOI] [PubMed] [Google Scholar]
- 36.Yan Y, Wang Y, Zhao P, Ma W, Hu Z, Zhang K. BMI-1 promotes self-renewal of radio- and temozolomide (TMZ)-resistant breast cancer cells. Reprod Sci. 2017;24:1620–1629. doi: 10.1177/1933719117697255. [DOI] [PubMed] [Google Scholar]
- 37.Li N, Jiang K, Fang LP, Yao LL, Yu Z. Knockdown of long noncoding RNA CCAT1 inhibits cell growth, invasion and peritoneal metastasis via downregulation of Bmi-1 in gastric cancer. Neoplasma. 2018;65:736–744. doi: 10.4149/neo_2018_171206N801. [DOI] [PubMed] [Google Scholar]
- 38.Koh H, Park H, Chandimali N, Huynh DL, Zhang JJ, Ghosh M, Gera M, Kim N, Bak Y, Yoon DY, et al. MicroRNA-128 suppresses paclitaxel-resistant lung cancer by inhibiting MUC1-C and BMI-1 in cancer stem cells. Oncotarget. 2017;8:110540–110551. doi: 10.18632/oncotarget.22818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Xue J, Kuang H, Zhou X, Liao L, Yin F. microRNA-1297 inhibits the growth and metastasis of colorectal cancer by suppressing Cyclin D2 expression. DNA Cell Biol. 2017;36:991–999. doi: 10.1089/dna.2017.3829. [DOI] [PubMed] [Google Scholar]
- 40.Zhong H, Yang J, Zhang B, Wang X, Pei L, Zhang L, Lin Z, Wang Y, Wang C. LncRNA GACAT3 predicts poor prognosis and promotes cell proliferation in breast cancer through regulation of miR-497/CCND2. Cancer Biomark. 2018;22:787–797. doi: 10.3233/CBM-181354. [DOI] [PubMed] [Google Scholar]
- 41.Huang F, Zhao H, Du Z, Jiang H. MiR-615 inhibits prostate cancer cell proliferation and invasion by directly targeting Cyclin D2. Oncol Res. 2019;27:293–299. doi: 10.3727/096504018X15190399381143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeng Q, Tao X, Huang F, Wu T, Wang J, Jiang X, Kuang Z, Cheng B. Overexpression of miR-155 promotes the proliferation and invasion of oral squamous carcinoma cells by regulating BCL6/Cyclin D2. Int J Mol Med. 2016;37:1274–1280. doi: 10.3892/ijmm.2016.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li YL, Wang J, Zhang CY, Shen YQ, Wang HM, Ding L, Gu YC, Lou JT, Zhao XT, Ma ZL, Jin YX. MiR-146a-5p inhibits cell proliferation and cell cycle progression in NSCLC cell lines by targeting CCND1 and CCND2. Oncotarget. 2016;7:59287–59298. doi: 10.18632/oncotarget.11040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the author upon reasonable request.