Abstract
Gastric cancer (GC) is the fourth most common tumor and the second most common cause of cancer-associated mortality worldwide. Current tumor biomarkers for GC, such as serum carcinoembryonic antigen and carbohydrate antigen 19-9, are not ideal due to their limited role as prognostic indicators for GC. Proteasome subunit α7 (PSMA7) is a multifunctional protein, which has been revealed to be involved in the development and progression of various types of malignancy. However, little is known about the role of PSMA7 in GC. In the present study, PSMA7 was identified to be overexpressed at the mRNA and protein levels in GC tissues, compared with in non-tumor tissues, using reverse transcription-quantitative PCR and immunohistochemistry. Furthermore, PSMA7 expression is associated with tumor invasion, lymph node metastasis, distant metastasis, and Tumor-Node-Metastasis stage. Univariate and multivariate Cox regression analysis identified that PSMA7 expression is an independent prognostic factor for poor survival. Kaplan-Meier survival curves revealed that high PSMA7 expression is associated with a poor prognosis in patients with GC. Overall, the results of the present study suggested that PSMA7 may be a promising biomarker for the prognosis of GC, and may represent a new diagnostic marker and molecular therapeutic target for GC.
Keywords: proteasome subunit α7, prognosis, gastric cancer
Introduction
Gastric cancer (GC) is one of the most malignant tumors worldwide, and for those diagnosed, the prognosis is still poor (1). GC has recently been ranked globally as the fourth most common malignancy, accounting for >700,000 deaths each year (2), thus making it the second leading cause of cancer-associated mortality worldwide. Due to the non-specific symptoms of early-stage GC, the majority of patients are only diagnosed at an advanced stage (3). Current tumor biomarkers for GC, such as serum carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA199), are not ideal due to their relatively low sensitivity and specificity when regarding diagnosis and prognosis (4,5). Therefore, it is necessary to identify novel sensitive and specific biological markers for GC that can assist in early diagnosis, and subsequently, determine a target therapy for prolonging the life of patients with GC (6,7).
The pathogenesis of GC is complex and only partially known. Previous studies primarily concentrated on the following factors: Likelihood of Helicobacter pylori infection, genetics, lifestyle and diet (8,9). However, major medical advances in molecular technologies over the past two decades have led to a deeper understanding of the pathogenesis of GC. Identifying the biomarkers that are associated with the development and metastasis of GC may assist clinicians in tailoring therapies by identifying those patients who are at a high risk of GC and thus, propose novel molecular targets for its treatment (10). Previous studies have investigated specific genes and proteins, as well as their roles in GC in a search for reliable prognostic markers (8–11). Studying novel molecular prognostic markers may contribute to the elucidation of the molecular mechanisms that underlie GC. Nowadays, tumor targeted therapy is the focus of current research; in addition, the identification of prognostic tumor markers could be used to develop new and effective targeted treatment strategies for GC.
The gene for proteasome subunit α7 (PSMA7), also known as XAPC7 in mammals, is located on the chromosomal anomaly 20q13.33. It has been demonstrated that the 20q region is amplified in numerous types of tumor (11,12). PSMA7 is an α-type subunit of the 20S proteasome core complex and participates in the degradation of proteins through the ubiquitin-proteasome pathway in eukaryotic cells. The protein complex has a molecular mass of ~2,000 kDa and is comprised of an associated 20S proteolytic core and one or two 19S regulatory complexes (13,14). The core particle consists of two α- and two β-rings, which form a cylindrical, four-layered αββα structure (15,16). The α-ring is composed of seven homologous but distinct subunits, α1-7 (17,18), and the β-ring is also composed of seven homologous and distinct β subunits, β1–7 (19–22). Previous studies have demonstrated that PSMA7 serves a role in the retrogradation of a series of proteins essential for the replication of Hepatitis B virus (HBV) (23,24), Hepatitis C virus (25), and human immunodeficiency virus (26). More studies have demonstrated that PSMA7 interacts with a number of proteins implicated in tumorigenesis, such as hypoxia-inducible factor-1α (27), endothelial monocyte-activating polypeptide-II (28), HBV X protein (23,24), c-Abl and Arg tyrosine kinases (29), which are important for the regulation of mammalian genes that are involved in angiogenesis, glucose metabolism and apoptosis.
Previously, downregulation of PSMA7 was detected in K-ras transformed AsPC-1 pancreatic cancer cells compared with untransformed AsPC-1 cells (30), while Richardson et al (31) revealed that PSMA7 expression is elevated in testicular and breast cancer. Romanuik et al (32) observed that high or increased PSMA7 expression is also observed in castration-recurrent prostate cancer. Shi et al (33) detected the overexpression of PSMA7 in liver cancer. Scotto et al (34) and Hu et al (13) reported the overexpression of PSMA7 in cervical cancer and colorectal cancer, respectively. Hu et al (35) revealed that the overexpression of PSMA7 correlated with certain clinical pathological parameters of colorectal cancer and liver metastasis. Honma et al (36) demonstrated that PSMA7 is a potential target for RNA interference-based therapeutics for colorectal cancer. However, little is known about the expression and function of PSMA7 in the development or metastasis of GC. In the present study, the expression profile of PSMA7 was investigated in GC tissue samples, as well as the association between PSMA7 expression and survival rate among patients with GC.
Materials and methods
Human GC tissue samples and clinical information
Tissue microarrays (TMA) were prepared using paraffin-embedded blocks of malignant and benign gastric tissue samples from 735 patients (age range, 19–84 years) and a tissue array sample obtained from The Affiliated Hospital of Nantong University (Nantong, China) between January 2003 and December 2010. The tissue samples included 410 cancer and 212 matched pericarcinomatous tissues: 24 high-grade intraepithelial neoplasia, 27 low-grade intraepithelial neoplasia, 28 intestinal metaplasia and 34 chronic gastritis. The clinical information obtained included sex, age, histological type, differentiation grade, tumor invasion, lymph node metastasis, distant metastasis, Tumor-Node-Metastasis (TNM) stage (37), preoperative serum CA19-9 levels and CEA levels. The 7th edition of the TNM staging in malignant tumors criteria was used to determine tumor stage (37). None of the patients received any form of treatment, including radiotherapy, chemotherapy or immunotherapy, prior to surgical resection. In addition, 60 paired and freshly frozen GC and matching peritumoral tissues were obtained from The Affiliated Xinghua People's Hospital of Yangzhou University Medical College (Xinghua, China). The Ethics Committee of Nanjing Medical University approved the study, and all the following experiments were performed according to the relevant guidelines and regulations. All participants provided written informed consent.
Reverse transcription-quantitative (RT-q) PCR
Total RNA was isolated from freshly frozen GC tissues and matching peritumoral tissues using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol, and cDNA was synthesized using the PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd.). RT-qPCR was performed using the Applied Biosystems StepOne and StepOnePlus Real-Time PCR systems (Thermo Fisher Scientific, Inc.). GAPDH was used for monitoring internal standards. Primers were purchased from GenScript. Relative PSMA7 expression was analyzed using the 2−ΔΔCq method (38). PCR primers were designed with Primer Express Software (version 2.0), and were as follows: PSMA7 forward, 5′-AGTGCGGAAGATCTGTGCTT-3′ and reverse, 5′-TCCGTACGCGTTGTTGTAAT-3′; GAPDH forward, 5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse, 5′-GGCTGTTGTCATACTTCTCATGG-3′. The reaction system (20 µl) contained 2 µl of cDNA template, each primer 20 µM (0.4 ul), 7.2 µl of 2X SYBR®-Green PCR Master Mix (High Rox Plus; Applied Biosystems; Thermo Fisher Scientific, Inc.) and 10 ul aseptic ultra-pure water. The following conditions were used for RT-qPCR: Initial denaturation at 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 10 sec, annealing and extension at 60°C for 30 sec, and dissolution curve at 95°C for 15 sec, 60°C for 1 min and 95°C for 15 sec. The Cq-value for each sample was calculated with the ΔΔCq method and relative results were analyzed using 2−ΔΔCq (38). All results were normalized to GAPDH expression and each experiment was repeated three times.
TMA construction and immunohistochemical (IHC) evaluation
Core tissue biopsies of 2 mm in diameter were obtained from individual samples fixed with 10% formalin overnight at 37°C and paraffin-embedded. IHC analysis of TMA slides was applied to assess the expression of PSMA7, containing 735 gastric tissues. Using the TMA System Quick Ray (UT06; UNITMA Co., Ltd.), manual gastric TMAs were generated at the Affiliated Hospital of Nantong University (Nantong, China).
IHC staining was performed using the Envision technique (39). Briefly, TMA sections (4 µm thick) were dewaxed in xylene and rehydrated in graded alcohol, and then the sections were boiled in citrate solution (0.01 M; pH 6.0) using a microwave for antigen retrieval. Endogenous peroxidase activity was then blocked with 3% H2O2 for 10 min at room temperature, and sections were blocked in 5% bovine serum albumin (Sigma-Aldrich; Merck KGaA) for 10 min at 37°C. Sections were then incubated with a mouse anti-human PSMA7 monoclonal antibody (1:50; catalog no. LS-C114781; LifeSpan Biosciences, Inc.) overnight at 4°C, and with an Envision horseradish peroxidase-conjugated goat anti-mouse-IgG monoclonal antibody at room temperature for 30 min (1:100; catalog no. s35962; Dako; Agilent Technologies, Inc.). Diluted PBS replaced the primary antibody for the negative control. All sections were processed at the same time and under the same conditions. Representative images were captured and analyzed using a light microscope (Carl Zeiss AG). PSMA7 expression was assessed using the semi-quantitative H-score method, which takes both the staining intensity and the percentage of cells with that intensity into account (40). Staining intensity was scored using 4 grades, as follows; i) 0, no staining; ii) 1+, weak staining; iii) 2+, moderate staining; and iv) 3+, intense staining. For each case, the total score was calculated by multiplying the percentage of positive cells by the staining intensity score. The scores were computed, ranging from 0 to 300; the minimum final staining score was 0 (no staining) and the maximum score was 300 (100% of cells with 3+ staining intensity). All scoring was evaluated by two experienced pathologists, who were blinded to the experimental procedure.
Statistical analysis
All data were analyzed using SPSS (version 20.0; IBM Corp.) and STATA (version 12.0; StataCorp) statistical software applications. Continuous PSMA7 expression data from IHC were initially converted into dichotic data (low vs. high) using specific cut-off values. The cut-off values were selected to be significant when taking into account overall survival (OS) using the X-Tile software program (version 3.6.1) for TMA data analysis (Rimm Lab, Yale University) (41,42). A score of 0–130 was considered to indicate no or low expression, while 131–300 was considered to indicate high expression. For subsequent analyses, PSMA7 protein expression levels were considered as either ‘No or Low’ or ‘High’ using these cut-off points prior to analysis. Associations between clinical parameters and the expression of PSMA7 were assessed using χ2 tests. Kaplan-Meier analysis was used to evaluate the associations between the 5-year survival of patients with GC that were exhibiting PSMA7 expression. The significance of the differences between curves was analyzed using a log-rank test. The Cox proportional hazards regression model was used to perform univariate and multivariate analyses. A paired-samples Student's t-test was also used. Each experiment was repeated three times. P<0.05 was considered to indicate a statistically significant difference, and 95% confidence intervals (CI) were used throughout.
Results
Expression of PSMA7 mRNA in GC tissues
The PSMA7 expression at the mRNA level was evaluated using RT-qPCR in 60 paired GC and matching peritumoral tissues. The analysis demonstrated that PSMA7 mRNA expression in gastric carcinoma tissues was significantly increased compared with in the matching peritumoral tissues (P<0.001; Fig. 1).
Figure 1.
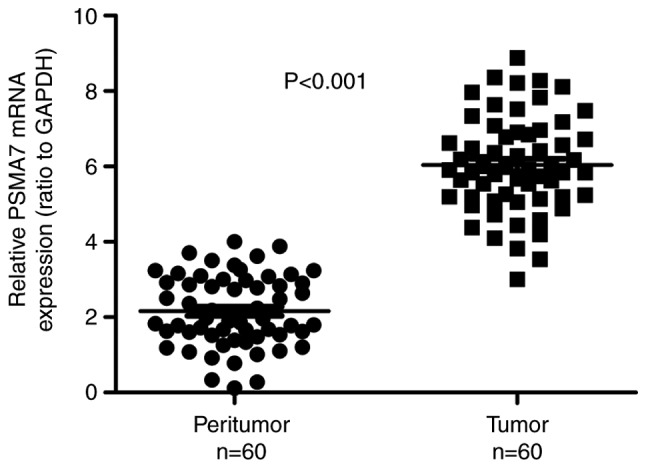
Expression of PSMA7 mRNA in 60 paired gastric tissues. The relative level of PSMA7 mRNA expression in gastric carcinoma tissues was significantly increased compared with that in the matching peritumoral tissues (P<0.001). PSMA7, proteasome subunit α7.
Expression of PSMA7 protein in gastric tissues
IHC was used to examine PSMA7 protein expression in GC tissues, benign gastric disease tissues and matching peritumoral tissues. PSMA7 protein expression was mostly present in the cytoplasm and nucleus (Fig. 2). The PSMA7 IHC data were scored using the semi-quantitative H-score method, taking into account the staining intensity and the percentage of cells at that intensity ranging from 0–300 (40). The cut-off point of PSMA7 expression was ascertained using the X-tiles software program (41,42) for clinical data analysis according to OS among patients with GC. PSMA7 protein expression in GC tissues was higher compared with in chronic gastritis, intestinal metaplasia, low-grade intraepithelial neoplasia, high-grade intraepithelial neoplasia and matching peritumoral tissues (all P<0.05; Table I).
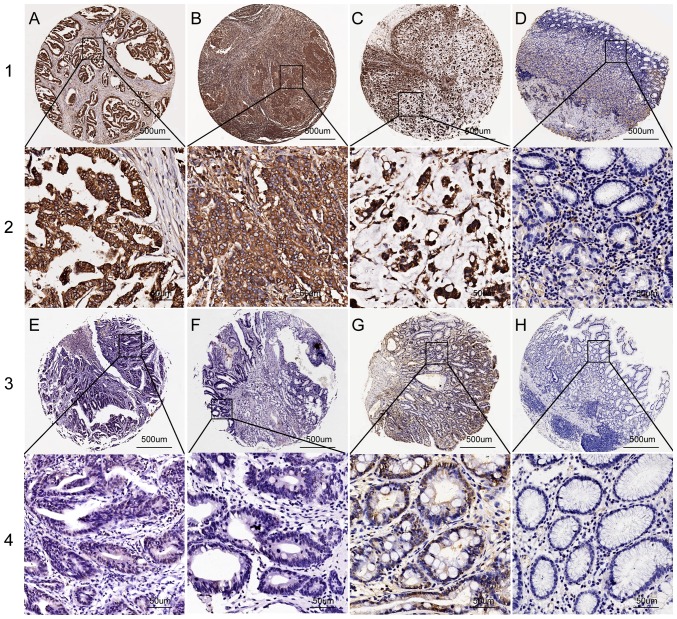
Figure 2.
Representative images of PSMA7 protein expression in gastric tissue TMA sections. Rows 1 and 3 use PSMA7 staining with ×40 magnification (scale bar, 500 µm), and rows 2 and 4 use PSMA7 staining with ×400 magnification (scale bar, 50 µm). (A) Moderately differentiated GC with high PSMA7 expression. (B) Poorly differentiated GC with high PSMA7 expression. (C) Signet ring cell GC with high PSMA7 expression. (D) Peritumoral tissues with low PSMA7 expression. (E) High-grade intraepithelial neoplasia with low PSMA7 expression. (F) Low-grade intraepithelial neoplasia with low PSMA7 expression. (G) Intestinal metaplasia with low PSMA7 expression. (H) Chronic gastritis with no PSMA7 expression. GC, gastric cancer; PSMA7, proteasome subunit α7.
Table I.
PSMA7 expression in gastric tissues.
| PSMA7 expression, n (%) | |||||
|---|---|---|---|---|---|
| Characteristic | Total, n | No or Low | High | χ2 | P-value |
| Gastric cancer | 410 | 159 (38.78) | 251 (61.22) | 75.3782 | <0.001 |
| Peritumoral tissue | 212 | 154 (72.64) | 58 (27.36) | 64.0925 | <0.001 |
| High-grade intraepithelial neoplasia | 24 | 15 (62.50) | 9 (37.50) | 5.3110 | 0.021 |
| Low-grade intraepithelial neoplasia | 27 | 18 (66.67) | 9 (33.33) | 8.1745 | 0.004 |
| Intestinal metaplasia | 28 | 17 (60.71) | 11 (39.29) | 5.2460 | 0.022 |
| Chronic gastritis | 34 | 25 (73.53) | 9 (26.47) | 15.6220 | <0.001 |
PSMA7, proteasome subunit α7.
Association of PSMA7 expression with clinicopathological characteristics in patients with GC
The associations between PSMA7 expression and the clinicopathological parameters of GC are presented as the following (Table II): High PSMA7 positive staining within the cytoplasm and nucleus was significantly associated with tumor invasion (P=0.012), lymph node metastasis (P=0.006), distant metastasis (P=0.006) and TNM stage (P<0.001). PSMA7 expression was not associated with any other clinical parameters, including sex, age, histological type, differentiation and serum levels of CEA and CA199.
Table II.
Association of PSMA7 expression with clinicopathological characteristics of patients with gastric cancer.
| PSMA7 expression | |||||
|---|---|---|---|---|---|
| Characteristic | Total, n | No or low (%) | High (%) | χ2 | P-value |
| Total | 410 | 159 (38.78) | 251 (61.22) | ||
| Sex | 2.0689 | 0.150 | |||
| Male | 301 | 123 (40.86) | 178 (59.14) | ||
| Female | 109 | 36 (33.03) | 73 (66.97) | ||
| Age, years | 0.0070 | 0.934 | |||
| ≤60 | 148 | 57 (38.51) | 91 (61.49) | ||
| >60 | 262 | 102 (38.93) | 160 (61.07) | ||
| Histological type | 5.7666 | 0.217 | |||
| Tubular | 322 | 116 (36.02) | 206 (63.98) | ||
| Mucinous | 29 | 13 (44.83) | 16 (55.17) | ||
| Mixed (tubular and mucinous) | 10 | 6 (60.00) | 4 (40.00) | ||
| Signet ring cell | 31 | 16 (51.61) | 15 (48.39) | ||
| Othersa | 18 | 8 (44.44) | 10 (55.56) | ||
| Differentiation | 6.6376 | 0.084 | |||
| Well | 21 | 11 (52.38) | 10 (47.62) | ||
| Moderate | 98 | 38 (38.78) | 60 (61.22) | ||
| Poor | 252 | 89 (35.32) | 163 (64.68) | ||
| Othersb | 39 | 21 (53.85) | 18 (46.15) | ||
| Tumor invasion | 12.7703 | 0.012 | |||
| Tis | 30 | 18 (60.00) | 12 (40.00) | ||
| T1 | 53 | 27 (50.94) | 26 (49.06) | ||
| T2 | 70 | 29 (41.43) | 41 (58.57) | ||
| T3 | 222 | 74 (33.33) | 148 (66.67) | ||
| T4 | 35 | 11 (31.43) | 24 (68.57) | ||
| Lymph node metastasis | 12.4718 | 0.006 | |||
| N0 | 163 | 80 (49.08) | 83 (50.92) | ||
| N1 | 81 | 28 (34.57) | 53 (65.43) | ||
| N2 | 89 | 28 (31.46) | 61 (68.54) | ||
| N3 | 77 | 23 (29.87) | 54 (70.13) | ||
| Distant metastasis | 7.4160 | 0.006 | |||
| M0 | 386 | 156 (40.41) | 230 (59.59) | ||
| M1 | 24 | 3 (12.50) | 21 (87.50) | ||
| TNM stage | 24.1716 | <0.001 | |||
| 0 | 30 | 19 (63.33) | 11 (36.67) | ||
| I | 73 | 40 (54.79) | 33 (45.21) | ||
| II | 141 | 54 (38.30) | 87 (61.70) | ||
| III | 142 | 40 (28.17) | 102 (71.83) | ||
| IV | 24 | 6 (25.00) | 18 (75.00) | ||
| Preoperative CEA, ng/ml | 5.1842 | 0.075 | |||
| ≤5 | 177 | 62 (35.03) | 115 (64.97) | ||
| >5 | 44 | 13 (29.55) | 31 (70.45) | ||
| Unknown | 189 | 84 (44.44) | 105 (55.56) | ||
| Preoperative CA199, ng/ml | 1.0034 | 0.605 | |||
| ≤37 | 191 | 79 (41.36) | 112 (58.64) | ||
| >37 | 30 | 11 (36.67) | 19 (63.33) | ||
| Unknown | 189 | 69 (36.51) | 120 (63.49) | ||
Papillary adenocarcinoma, 9 cases; adeno squamous carcinoma, 3 cases; squamous cell carcinoma, 4 cases; neuroendocrine carcinoma, 2 cases.
Including undifferentiated and unknown. CA199, carbohydrate antigen 19-9; CEA, carcinoembryonic antigen; M0, metastasis-negative; M1, metastasis-positive; N0, node-negative; N1, 1–2 nodes; N2, 3–6 nodes; N3, >6 nodes; PSMA7, proteasome subunit α7; T1, lamina propria; T2, muscularis propria; T3, subserosa; T4, adjacent structures; Tis, carcinoma in situ; TNM, Tumor-Node-Metastasis.
Upregulation of PSMA7 protein is associated with poor prognosis of GC
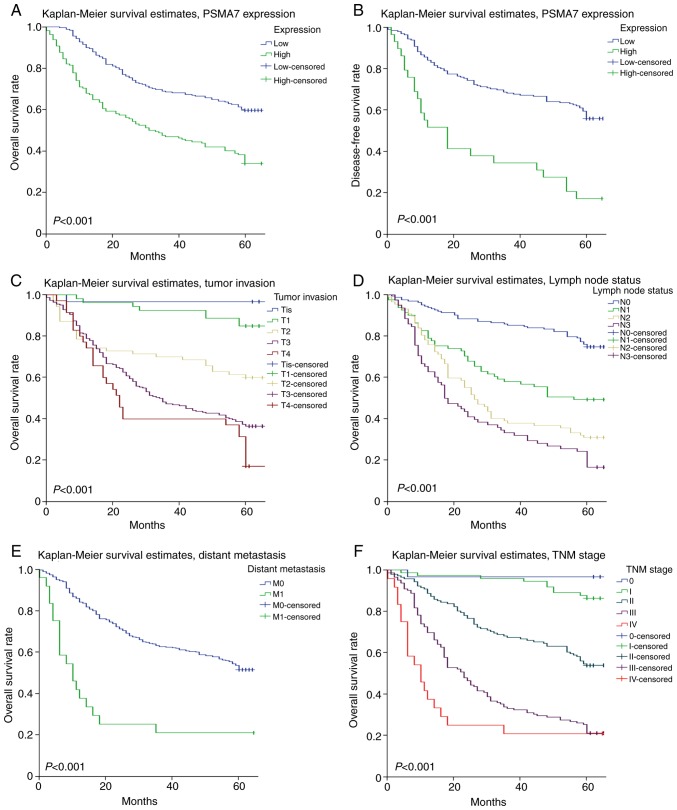
Univariate and multivariate analyses were used to investigate the prognostic value of PSMA7 expression in GC. Univariate Cox regression analysis suggested that high PSMA7 expression [hazard ratio (HR), 1.005; 95% CI, 1.003–1.007; P<0.001], age (HR, 3.135; 95% CI, 2.209–4.450; P<0.001), histological differentiation (HR, 1.393; 95% CI, 1.131–1.715; P=0.002), tumor invasion (HR, 2.059; 95% CI, 1.714–2.411; P<0.001), lymph node metastasis (HR, 1.773; 95% CI, 1.576–1.995; P<0.001), distant metastasis (HR, 3.316; 95% CI, 2.063–5.330; P<0.001) and TNM stage (HR, 2.059; 95% CI, 1.764–2.402; P<0.001) are positive prognostic factors (Table III). Multivariate Cox regression analysis confirmed PSMA7 expression (HR, 0.995; 95% CI, 0.993–0.998; P<0.001), lymph node metastasis (HR, 1.592; 95% CI, 1.305–1.941; P<0.001) and TNM stage (HR, 1.592; 95% CI, 1.219–2.079; P<0.001) to be independent prognostic factors (Table III). Kaplan-Meier survival analysis revealed that patients with higher PSMA7 expression levels showed poor OS and disease-free survival rates compared with those with lower PSMA7 expression levels (P<0.001; Fig. 3A and B). In addition, patients with deep tumor invasion, lymph node metastasis, distant metastasis and poor TNM stage had significantly lower OS compared with patients with shallow tumor invasion, no lymph node metastasis or distant metastasis and good TNM stage (P<0.001; Fig. 3C-F).
Table III.
Univariate and multivariate analysis of prognostic factors for overall survival in gastric cancer.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| HR | P-value | 95% CI | HR | P-value | 95% CI | |
| PSMA7 expression | ||||||
| High vs. low or no | 1.005 | <0.001 | 1.003–1.007 | 0.995 | 0.001 | 0.993–0.998 |
| Age, years | ||||||
| ≤60 vs. >60 | 3.135 | <0.001 | 2.209–4.450 | |||
| Sex | ||||||
| Male vs. female | 1.109 | 0.903 | 0.747–1.390 | |||
| Histological type, tubular vs. mucinous vs. mixed (tubular and mucinous) vs. signet ring cell vs. othersa | 1.015 | 0.802 | 0.901–1.145 | |||
| Differentiation, well vs. moderate vs. poor vs. othersb | 1.393 | 0.002 | 1.131–1.715 | |||
| Tumor invasion, Tis vs. T1 vs. T2 vs. T3 vs. T4 | 2.059 | <0.001 | 1.714–2.411 | |||
| Lymph node metastasis, N0 vs. N1 vs. N2 vs. N3 | 1.773 | <0.001 | 1.576–1.995 | 1.592 | <0.001 | 1.305–1.941 |
| Distant metastasis, M0 vs. M1 | 3.316 | <0.001 | 2.063–5.330 | |||
| TNM stage, 0 vs. I vs. II vs. III vs. IV | 2.059 | <0.001 | 1.764–2.402 | 1.592 | <0.001 | 1.219–2.709 |
| CEA, ≤5 vs. >5 | 1.151 | 0.053 | 0.998–1.328 | |||
| CA199, ≤37 vs. >37 | 1.133 | 0.081 | 0.985–1.303 | |||
Papillary adenocarcinoma, 9 cases; adeno squamous carcinoma, 3 cases; squamous cell carcinoma, 4 cases; neuroendocrine carcinoma, 2 cases.
Including undifferentiated and unknown. CA199, carbohydrate antigen 19-9; CEA, carcinoembrynoc antigen; CI, confidence interval; HR, hazard ratio; M0, metastasis-negative; M1, metastasis-positive; N0, node-negative; N1, 1–2 nodes; N2, 3–6 nodes; N3, >6 nodes; PMSA7, proteomic subunit α7; T1, lamina propria; T2, muscularis propria; T3, subserosa; T4, adjacent structures; Tis, carcinoma in situ; TNM, Tumor-Node-Metastasis.
Figure 3.
Survival curves of gastric carcinoma generated by the Kaplan-Meier method and the log-rank test. (A) OS curves comparing PSMA7 expression levels. (B) Disease-free survival curves comparing PSMA7 expression levels. (C) OS curves comparing levels of tumor invasion. (D) OS curves comparing lymph node metastasis status. (E) OS curves comparing distant metastasis status. (F) OS curves comparing TNM stage. M0, metastasis-negative; M1, metastasis-positive; N0, node-negative; N1, 1–2 nodes; N2, 3–6 nodes; N3, >6 nodes; OS, overall survival; PSMA7, proteasome subunit α7; T1, lamina propria, submucosa; T2, muscularis propria; T3, subserosa; T4, adjacent structures; Tis, carcinoma in situ; TNM, Tumor-Node-Metastasis.
Discussion
The majority of GC cases are characterized by late clinical presentation, rapid progression and poor survival. Considering the high morbidity and mortality rates of GC, a number of studies have been dedicated to identifying available and new prognostic markers. Several studies have demonstrated that PSMA7 serves a key role in the development of tumors (12,13). However, other studies have indicated that PSMA7 expression is a poor prognostic factor (14,35). Hu et al (13) highlighted the associations between PSMA7 expression and the clinicopathological characteristics of colorectal cancer and suggested that PSMA7 may be a molecular target for colorectal cancer therapy. PSMA7 protein and mRNA levels in GC tissues were measured by TMA, IHC and RT-qPCR in the present study. The results of the present study indicated that the expression of PSMA7 protein and mRNA is higher in GC tissues compared with in benign tissues. These experimental results preliminarily demonstrate that PSMA7 is a tumor-associated antigen, which coincides with the findings of Shi et al (33). D'Errico et al (43) revealed that PSMA7 expression was upregulated in gastric intestinal type adenocarcinoma and gastric mixed adenocarcinoma compared with gastric mucosa samples. Li et al (44) revealed that PSMA7 was overexpressed in GC compared with normal tissues; however, only the PSMA7 expression at the mRNA level was investigated. The results of the present study confirmed that overexpression of PSMA7 predicted poor prognosis at the mRNA and the protein level.
Based on the association between the PSMA7 protein expression levels and the clinicopathological values in patients with GC, data analysis indicated that high expression of PSMA7 in GC was associated with tumor invasion, lymph node metastasis, distant metastasis and TNM stage in the present study. These results indicated that PSMA7 may be a potential biomarker for prognosis of GC, which coincides with previous studies (13,35). However, Tan et al (12) verified that PSMA7 inhibits the proliferation, tumorigenicity and invasion of A549 cells in vitro and in vivo. This may be associated with the tissue specificity of PSMA7 expression in different types of tumor cell (45). For patients with GC, CEA and CA199 are the most commonly used serum tumor markers. However, in the present study, high PSMA7 expression was not significantly associated with the serum levels of CEA and CA199; this may be due to its relatively low sensitivity and specificity for diagnosis and prognosis (4,5).
Univariate analyses indicated that PSMA7 expression and six other factors (age, differentiation, tumor invasion, lymph node metastasis, distant metastasis and TNM stage) have statistically significant associations with OS. All seven factors were included in the multivariate Cox proportional hazards model to adjust for the effects of the covariates. Multivariate analysis demonstrated that PSMA7 expression, lymph node metastasis and TNM stage were independent risk factors in the prognosis of patients with GC. Kaplan-Meier survival analysis indicated that elevated expression of PSMA7 was closely associated with reduced OS and DFS in patients with GCs as opposed to in those with low expression levels. Hu et al (35) revealed that the knockdown of PSMA7 by short hairpin RNA in the RKO cell line inhibited their anchorage-independent growth, as well as migration and invasion. Furthermore, PSMA7 depletion was able to strongly suppress the tumorigenic ability of the RKO cells in vivo. These results indicated that PSMA7 may serve as a novel predictor of prognosis in patients with GC (35). However, Li et al (44) found that high mRNA expression of PSMA7 was associated with improved OS, which is different from the results of the present study. The sample size for biostatistics should be expanded in future studies. There were some limitations to the present study; although the expression and prognostic significance of PSMA7 in GC have each been evaluated, the specific functions and molecular mechanisms of PSMA7 in GC remain to be further investigated in vivo and in vitro. In subsequent studies the authors hope to identify the possible signaling mechanisms involved in PSMA7 expression-triggered proliferation, differentiation, invasion and metastasis in GC.
It is well known that a single gene cannot be responsible for or directly connected with each step in the occurrence and progression of GC. It has been reported previously that certain factors are associated with PSMA7 in the colon carcinoma cell line HCT116 and murine models, including nucleotide-binding oligomerization domain-containing protein 1 (NOD1) (46), nuclear factor-kappa B (NF-κB) (46), CD44 (13) and c-Abl (27,47), among others. Yang et al (46) verified that PSMA7 regulates the tumorigenesis of colorectal cancer through the inhibition of NOD1-mediated apoptosis and NF-κB activation. Hu et al (13) revealed that CD44 may be a downstream element via PSMA7 depletion in colorectal cancer cell tumorigenicity and metastasis. Future studies are required in order to investigate these factors and their associations with PSMA7 in GC, and to improve the current understanding of the molecular mechanism underlying the role of PSMA7 in GC. In conclusion, to the best of our knowledge, the present study proves for the first time that PSMA7 is overexpressed at the mRNA and the protein level in GC and that it is an independent prognostic factor for GC. PSMA7 may, therefore, be a potential candidate for the diagnosis and targeted therapy of patients with GC.
Acknowledgements
Not applicable.
Funding
This project was supported by the National Natural Science Foundation of China (grant nos. 81201596, 81773100) and Special Fund of Clinical Medicine in Jiangsu Province (grant no. BL2013038).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding authors upon reasonable request.
Authors' contributions
SX and QT were responsible for RNA extraction and reverse transcription reaction. LZ and LJ performed polymerase chain reaction. DW and PX prepared tissue sections. XW and XZ conducted immunohistochemistry. LZ, LJ, GT and TY contributed to the follow-up and analysis of patient's characteristics. SX drafted the manuscript. ZF and LL made substantial contributions to article design and manuscript revision, and gave final approval of the version to be published. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
The present study was reviewed and approved by the Medical Ethics Committee of Xinghua People's Hospital (Xinghua, China), and all the patients or their families provided written informed consent
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Shimizu D, Kanda M, Kodera Y. Review of recent molecular landscape knowledge ofgastric cancer. Histol Histopathol. 2017;33:11–26. doi: 10.14670/HH-11-898. [DOI] [PubMed] [Google Scholar]
- 2.Daniyal M, Ahmad S, Ahmad M, Asif HM, Akram M, Ur Rehman S, Sultana S. Risk factors and epidemiology of gastric cancer in Pakistan. Asian Pac J Cancer Prev. 2015;16:4821–4824. doi: 10.7314/APJCP.2015.16.12.4821. [DOI] [PubMed] [Google Scholar]
- 3.Kanda M, Kodera Y. Recent advances in the molecular diagnostics of gastric cancer. World J Gastroenterol. 2015;21:9838–9852. doi: 10.3748/wjg.v21.i34.9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mu JS, Huang WD, Tan ZJ, Li ML, Zhang L, Ding QC, Wu XS, Lu JH, Liu YB, Dong Q, Xu HN. Dopamine receptor D2 is correlated with gastric cancer prognosis. Oncol Lett. 2017;13:1223–1227. doi: 10.3892/ol.2017.5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emoto S, Ishigami H, Yamashita H, Yamaguchi H, Kaisaki S, Kitayama J. Clinical significance of CA125 and CA72-4 in gastric cancer with peritoneal dissemination. Gastric Cancer. 2012;15:154–161. doi: 10.1007/s10120-011-0091-8. [DOI] [PubMed] [Google Scholar]
- 6.Kanda M, Fujii T, Takami H, Suenaga M, Inokawa Y, Yamada S, Nakayama G, Sugimoto H, Koike M, Nomoto S, Kodera Y. Combination of the serum carbohydrate antigen 19-9 and carcinoembryonic antigen is a simple and accurate predictor of mortality in pancreatic cancer patients. Surg Today. 2014;44:1692–1701. doi: 10.1007/s00595-013-0752-9. [DOI] [PubMed] [Google Scholar]
- 7.Yong H, Zhu H, Zhang S, Zhao W, Wang W, Chen C, Ding G, Zhu L, Zhu Z, Liu H, et al. Prognostic value of decreased expression of RBM4 in human gastric cancer. Sci Rep. 2016;6:28222. doi: 10.1038/srep28222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng ZQ. The present situation and future of personalized medicine of precise medicine. Acta Universitatis Medicinalis Nanjing. 2016;11:1283–1284. [Google Scholar]
- 9.Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118:3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 10.Tsugane S, Sasazuki S. Diet and the risk of gastric cancer: Review of epidemiologica evidence. Gastric Cancer. 2007;10:75–83. doi: 10.1007/s10120-007-0420-0. [DOI] [PubMed] [Google Scholar]
- 11.Kanda M, Shimizu D, Sueoka S, Nomoto S, Oya H, Takami H, Ezaka K, Hashimoto R, Tanaka Y, Kobayashi D, et al. Prognostic relevance of SAMSN1 expression in gastric cancer. Oncol Lett. 2016;12:4708–4716. doi: 10.3892/ol.2016.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan JY, Huang X, Luo YL. PSMA7 inhibits the tumorigenicity of A549 human lung adenocarcinoma cells. Mol Cell Biochem. 2012;366:131–137. doi: 10.1007/s11010-012-1290-2. [DOI] [PubMed] [Google Scholar]
- 13.Hu XT, Chen W, Wang D, Shi QL, Zhang FB, Liao YQ, Jin M, He C. The proteasome subunit PSMA7 located on the 20q13 amplicon is overexpressed and associated with liver metastasis in colorectal cancer. Oncol Rep. 2008;19:441–446. [PubMed] [Google Scholar]
- 14.Du H, Huang X, Wang S, Wu Y, Xu W, Li M. PSMA7, a potential biomarker of diseases. Protein Pept Lett. 2009;16:486–489. doi: 10.2174/092986609788167824. [DOI] [PubMed] [Google Scholar]
- 15.Pickart CM. Ubiquitin enters the new millennium. Mol Cell. 2001;8:499–504. doi: 10.1016/S1097-2765(01)00347-1. [DOI] [PubMed] [Google Scholar]
- 16.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 17.Groll M, Ditzel L, Löwe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 18.Unno M, Mizushima T, Morimoto Y, Tomisugi Y, Tanaka K, Yasuoka N, Tsukihara T. The structure of the mammalian 20S proteasome at 2.75 A resolution. Structure. 2002;10:609–618. doi: 10.1016/S0969-2126(02)00748-7. [DOI] [PubMed] [Google Scholar]
- 19.Takagi K, Saeki Y, Yashiroda H, Yagi H, Kaiho A, Murata S, Yamane T, Tanaka K, Mizushima T, Kato K. Pba3-Pba4 heterodimer acts as a molecular matchmaker in proteasome α-ring formation. Biochem Biophys Res Commun. 2014;450:1110–1114. doi: 10.1016/j.bbrc.2014.06.119. [DOI] [PubMed] [Google Scholar]
- 20.Tomko RJ, Jr, Hochstrasser M. Molecular architecture and assembly of the eukaryotic proteasome. Annu Rev Biochem. 2013;82:415–445. doi: 10.1146/annurev-biochem-060410-150257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirano Y, Kaneko T, Okamoto K, Bai M, Yashiroda H, Furuyama K, Kato K, Tanaka K, Murata S. Dissecting beta-ring assembly pathway of the mammalian 20S proteasome. EMBO J. 2008;27:2204–2213. doi: 10.1038/emboj.2008.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishii K, Noda M, Yagi H, Thammaporn R, Seetaha S, Satoh T, Kato K, Uchiyama S. Disassembly of the self-assembled, double-ring structure of proteasome α7 homo-tetradecamer by α6. Sci Rep. 2015;5:18167. doi: 10.1038/srep18167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang J, Kwong J, Sun EC, Liang TJ. Proteasome complex as a potential cellular target of hepatitis B virus X protein. J Virol. 1996;70:5582–5591. doi: 10.1128/jvi.70.8.5582-5591.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z, Torii N, Furusaka A, Malayaman N, Hu Z, Liang TJ. Structural and functional characterization of interaction between hepatitis B virus X protein and the proteasome complex. J Biol Chem. 2000;275:15157–15165. doi: 10.1074/jbc.M910378199. [DOI] [PubMed] [Google Scholar]
- 25.Krüger M, Beger C, Welch PJ, Barber JR, Manns MP, Wong-Staal F. Involvement of proteasome alpha-subunit PSMA7 in hepatitis C virus internal ribosome entry site-mediated translation. Mol Cell Biol. 2001;21:8357–8364. doi: 10.1128/MCB.21.24.8357-8364.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Apcher GS, Heink S, Zantopf D, Kloetzel PM, Schmid HP, Mayer RJ, Kruger E. Human immunodeficiency virus-1 Tat protein interacts with distinct proteasomal alpha and beta subunits. FEBS Lett. 2003;553:200–204. doi: 10.1016/S0014-5793(03)01025-1. [DOI] [PubMed] [Google Scholar]
- 27.Semenza GL. O2-regulated gene expression: Transcriptional control of cardiorespiratory physiology by HIF-1. J Appl Physiol (1985) 2004;96:1170–1172. doi: 10.1152/japplphysiol.00770.2003. [DOI] [PubMed] [Google Scholar]
- 28.Tandle AT, Calvani M, Uranchimeg B, Zahavi D, Melillo G, Libutti SK. Endothelial monocyte activating polypeptide-II modulates endothelial cell responses by degrading hypoxia- inducible factor-1alpha through interaction with PSMA7, a component of the proteasome. Exp Cell Res. 2009;315:1850–1859. doi: 10.1016/j.yexcr.2009.03.021. [DOI] [PubMed] [Google Scholar]
- 29.Li D, Dong Q, Tao Q, Gu J, Cui Y, Jiang X, Yuan J, Li W, Xu R, Jin Y, et al. c-Abl regulates proteasome abundance by controlling the ubiquitin-proteasomal degradation of PSMA7 subunit. Cell Rep. 2015;10:484–496. doi: 10.1016/j.celrep.2014.12.044. [DOI] [PubMed] [Google Scholar]
- 30.Ohnami S, Matsumoto N, Nakano M, Aoki K, Nagasaki K, Sugimura T, Terada M, Yoshida T. Identification of genes showing differential expression in antisense K-ras-transduced pancreatic cancer cells with suppressed tumorigenicity. Cancer Res. 1999;59:5565–5571. [PubMed] [Google Scholar]
- 31.Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, Liao X, Iglehart JD, Livingston DM, Ganesan S. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 32.Romanuik TL, Wang G, Morozova O, Delaney A, Marra MA, Sadar MD. LNCaP Atlas: Gene expression associated with in vivo progression to castration-recurrent prostate cancer. BMC Med Genomics. 2010;3:43. doi: 10.1186/1755-8794-3-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi YY, Wang HC, Yin YH, Sun WS, Li Y, Zhang CQ, Wang Y, Wang S, Chen WF. Identification and analysis of tumour-associated antigens in hepatocellular carcinoma. Br J Cancer. 2005;92:929–934. doi: 10.1038/sj.bjc.6602460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scotto L, Narayan G, Nandula SV, Arias-Pulido H, Subramaniyam S, Schneider A, Kaufmann AM, Wright JD, Pothuri B, Mansukhani M, Murty VV. Identification of copy number gain and overexpressed genes on chromosome arm 20q by an integrative genomic approach in cervical cancer: Potential role in progression. Genes Chromosomes Cancer. 2008;47:755–765. doi: 10.1002/gcc.20577. [DOI] [PubMed] [Google Scholar]
- 35.Hu XT, Chen W, Zhang FB, Shi QL, Hu JB, Geng SM, He C. Depletion of the proteasome subunit PSMA7 inhibits colorectal cancer cell tumorigenicity and migration. Oncol Rep. 2009;22:1247–1252. doi: 10.3892/or_00000561. [DOI] [PubMed] [Google Scholar]
- 36.Honma K, Takemasa I, Matoba R, Yamamoto Y, Takeshita F, Mori M, Monden M, Matsubara K, Ochiya T. Screening of potential molecular targets for colorectal cancer therapy. Int J Gen Med. 2009;2:243–257. doi: 10.2147/ijgm.s5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edge SB, Carolyn CC. The American Joint Committee on cancer: The 7th edition of the AJCC cancer staging manualand the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Bai J, Yong HM, Chen FF, Mei PJ, Liu H, Li C, Pan ZQ, Wu YP, Zheng JN. Cullin1 is a novel marker of poor prognosis and a potential therapeutic target in human breast cancer. Ann Oncol. 2013;24:2016–2022. doi: 10.1093/annonc/mdt147. [DOI] [PubMed] [Google Scholar]
- 40.Tang Q, Liu YF, Zhu XJ, Li YH, Zhu J, Zhang JP, Feng ZQ, Guan XH. Expression and prognostic significance of the alpha B-crystallin gene in human hepatocellular carcinoma. Hum Pathol. 2009;40:300–305. doi: 10.1016/j.humpath.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 41.Zhao W, Zhu H, Zhang S, Yong H, Wang W, Zhou Y, Wang B, Wen J, Qiu Z, Ding G, et al. Trop2 is overexpressed in gastric cancer and predicts poor prognosis. Oncotarget. 2016;7:6136–6145. doi: 10.18632/oncotarget.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen C, Wang X, Huang X, Yong H, Shen J, Tang Q, Zhu J, Ni J, Feng Z. Nicotinamide N-methyltransferase: A potential biomarker for worse prognosis in gastric carcinoma. Am J Cancer Res. 2016;6:649–663. [PMC free article] [PubMed] [Google Scholar]
- 43.D'Errico M, de Rinaldis E, Blasi MF, Viti V, Falchetti M, Calcagnile A, Sera F, Saieva C, Ottini L, Palli D, et al. Genome-wide expression profile of sporadic gastric cancers with microsatellite instability. Eur J Cancer. 2009;45:461–469. doi: 10.1016/j.ejca.2008.10.032. [DOI] [PubMed] [Google Scholar]
- 44.Li Y, Huang J, Sun J, Xiang S, Yang D, Ying D, Lu M, Li H, Ren G. The transcription levels and prognostic values of seven proteasome alpha subunits in human cancers. Oncotarget. 2017;8:4501–4519. doi: 10.18632/oncotarget.13885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holmila R, Sklias A, Muller DC, Degli Esposti D, Guilloreau P, McKay J, Sangrajrang S, Srivatanakul P, Hainaut P, Merle P, et al. Targeted deep sequencing of plasma circulating cell-free DNA reveals Vimentin and Fibulin 1 as potential epigenetic biomarkers for hepatocellular carcinoma. PLoS One. 2017;12:e0174265. doi: 10.1371/journal.pone.0174265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang L, Tang Z, Zhang H, Kou W, Lu Z, Li X, Li Q, Miao Z. PSMA7 directly interacts with NOD1 and regulates its function. Cell Physiol Biochem. 2013;31:952–959. doi: 10.1159/000350113. [DOI] [PubMed] [Google Scholar]
- 47.Liu X, Huang W, Li C, Li P, Yuan J, Li X, Qiu XB, Ma Q, Cao C. Interaction between c-Abl and Arg tyrosine kinases and proteasome subunit PSMA7 regulates proteasome degradation. Mol Cell. 2006;22:317–327. doi: 10.1016/j.molcel.2006.04.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding authors upon reasonable request.