Abstract
Abundant evidence indicates that long noncoding RNAs (lncRNAs) and circular RNAs (circRNAs) serve important roles in tumorigenesis and tumour progression. However, their diagnostic and treatment value for esophageal squamous carcinoma (ESCC) remains unknown. A microarray (SBC human ceRNA array V1.0) was performed to assess the expression profiles and biological functions of lncRNAs, circRNAs and mRNAs in ESCC and para-cancerous tissues from three patients. Microarray data were validated by reverse transcription-quantitative polymerase chain reaction for a group of genes. A number of lncRNA-microRNAs (miRNA) and circRNA-miRNA-mRNA networks were constructed. Bioinformatics tools, including gene ontology and Kyoto Encyclopedia of Genes and Genomes biological pathway analyses, were used to predict the functions of differentially expressed lncRNAs, circRNAs and potentially co-expressed target genes. The results revealed that compared with the expression levels of para-cancerous tissues, 1,384 lncRNAs, 2,046 circRNAs and 936 mRNAs were frequently altered in ESCC tissues. Co-expression networks of lncRNAs-miRNAs-circRNAs-mRNAs were constructed based on the correlation analyses among the differentially expressed RNAs. Furthermore, using bioinformatics methods, correlation expression networks were constructed that included cis- and trans-regulatory elements. Therefore, these results suggest that lncRNAs and circRNAs may be involved in the pathogenesis and development of ESCC. These findings provide a novel and systematic perspective on the potential function of noncoding RNAs in ESCC.
Keywords: esophageal squamous carcinoma, long noncoding RNAs, circularRNAs, mRNAs
Introduction
Esophageal carcinomas (ECs) are classified as either squamous cell carcinomas or adenocarcinomas. Esophageal squamous cell carcinomas (ESCCs) comprise over 90% of ECs (1). ESCC is one of the most common malignancies and has a poor outcome in China; there were an estimated 298,500 deaths in 2012. Furthermore, among the patients aged 0–74 years, the mortality rate was 9.65%. In males, the mortality rate was 10.80%, while in female the mortality rate was 7.68% (2). China had a 20-fold higher incidence rate than low-risk western Africa in 2002 (3). The 5-year survival rate of patients with esophageal cancer with localized disease is less than 20% (4). Therefore, to improve patient outcome, the molecular mechanism of ESCC requires further investigation.
Approximately 5, 70 and 80% of the genomic regions of bacteria, unicellular eukaryotes and invertebrates, respectively, are annotated as noncoding (5). Noncoding RNAs include long noncoding RNAs (lncRNAs), circular RNAs (circRNAs) and microRNAs (miRNAs). An increasing number of studies reported that lncRNAs, circRNAs and miRNAs may serve crucial roles in tumorigenesis and tumour progression (6–11).
LncRNAs are a class of longer transcripts that range from 200 nt to 100 kb and have little protein-coding potential (7). Previous studies have reported that lncRNAs can regulate multiple cellular and disease processes, including cell differentiation, stem cell pluripotency, cell proliferation and apoptosis, and cancer metastasis (8–11). To the best of our knowledge, the majority of the lncRNAs functions remain unclear. Regulation via lncRNAs does not occur independently; rather, regulation occurs through a large and complex network that involves mRNAs, miRNAs and proteins (6). LncRNAs may regulate gene expression by both cis- and trans-regulation (12). Based on this knowledge, a systematic study was performed, in order to understand the function of lncRNAs and mRNAs.
CircRNAs are another novel type of endogenous non-coding RNA, widespread in mammalian cells (13). Unlike linear RNA, circRNA consists of RNA molecules with covalently joined 3′- and 5′- ends formed by a back-splice event; this type of RNA therefore presents as a covalently closed continuous loop structure and is considered to be a major subtype of gene transcription (14). The characteristics of circRNA include the following: Exists in nearly all types of species, is expressed in a tissue- and disease-dependent manner; and is more stable in tissue and circulation compared with linear RNA, since it resists RNase (15). Recently, progress has been made on the study of the formation and biogenesis of circRNAs, adding to the evidence and possibilities for its biological value (16). Similar to lncRNAs, abundant evidence indicates that circRNAs may serve important roles in non-alcoholic steatohepatitis, bladder cancer, hepatocellular carcinoma, Hirschsprung's disease, colorectal cancer and lung cancer (17–22). A number of studies have revealed that circRNAs are differentially expressed in various cancerous tissues or cell lines (such as HEK293 and HeLa) and can function as ‘miRNA sponges’, regulators of splicing and transcription, and modifiers of parental gene expression (23–25). CircRNAs may negatively regulate the activity of miRNAs by competing endogenous RNA (ceRNA) network that is composed of lncRNAs, circRNAs and mRNAs (26). If the dynamic balance of a given ceRNA network is disturbed, tumorigenesis may occur (27).
To the best of our knowledge, the role of noncoding RNAs in tumorigenesis and progression of ESCC remain unclear. In the present study, using a microarray, the differential expression patterns of lncRNAs, circRNAs and mRNAs were investigated in ESCC. Subsequently, bioinformatics methods were used to identify the associated pathways and gene ontology items, based on the association between the different RNA species.
Furthermore, the lncRNA and circRNA profiles were investigated and a number of lncRNA-mRNA and circRNA-miRNA-mRNA networks were constructed. Finally, the functions of the lncRNAs and circRNAs were predicted by constructing a co-expression network. These findings may provide novel data for the diagnosis and pathogenesis of ESCC, in addition to novel targets for ESCC treatment.
Materials and methods
Patients and samples
A total of 6 ESCC tissue samples and para-carcinoma tissues were obtained from 3 male patients (mean, 45 years), who underwent surgical treatment without preoperative chemotherapy or radiotherapy at the Affiliated Cancer Hospital of Xinjiang Medical University (Urumqi, China) between July 2014 and April 2017 (Table I). The Union for International Cancer Control TNM classification (7th edition, 2009) was used to assess the anatomic extent of ESCC (28). The cancerous and paracancerous tissues were matched samples. The samples were stored at the Regional Bank of Tumour Resources of the Xinjiang Uygur Autonomous Region at −80°C until use.
Table I.
Details of ESCC and para-carcinoma specimen used for microarray analysis.
| Sample ID | Sex | Age, years | Tumor size, cm | TNM |
|---|---|---|---|---|
| ESCC-1 | Male | 70 | 5.5 | T3N0M0 |
| ESCC-2 | Male | 55 | 5 | T3N0M0 |
| ESCC-3 | Male | 44 | 4.5 | T3N0M0 |
TNM, Tumor-Node-Metastasis; ESCC, esophageal squamous carcinoma.
Written informed consent was obtained from all subjects. This experimental study was approved and supervised by the Ethics Committee of The Third Affiliated Teaching Hospital of Xinjiang Medical University, Affiliated Cancer Hospital.
RNA extraction
Total RNA was extracted from ESCC and para-carcinoma tissues using Takara RNAiso (Takara Bio), according to the manufacturer's protocols and was evaluated for a RIN number to inspect the RNA integrity by an Agilent Bioanalyzer 2100 (Agilent Technologies). Characterized total RNA was further purified using an RNeasy mini kit (Qiagen GmBH) and an RNase-Free DNase Set, according to the manufacturer's protocols. RNA quantity and quality were measured by NanoDrop ND-2000 (Thermo Fisher Scientific, Inc.), and RNA integrity was assessed using standard denaturing 10% agarose gel electrophoresis following RNA extraction and prior to sample labelling.
Microarray assay
Six tissue samples, including 3 ESCC samples and 3 control samples, were sent to Shanghai BioChip Co., Ltd. for ceRNA microarray analysis to identify differentially expressed lncRNAs, circRNAs and mRNAs.
Ribosomal RNA was removed from total RNA using a Ribo-Zero rRNA Removal kit (Qiagen GmBH), and the RNA(-rRNA) was subsequently amplified and labelled using a Low Input Quick Amp WT Labeling kit (Agilent Technologies), according to the manufacturer's protocols. Labelled cRNA was purified using an RNeasy mini kit (Qiagen GmBH). Each slide was hybridized with 1.65 µg Cy3-labelled cRNA using a Gene Expression Hybridization kit (Agilent Technologies) in a hybridization oven (Agilent Technologies), according to the manufacturer's instructions, at 65°C for 17 h. Following which the slides were washed in staining dishes (Thermo Fisher Scientific, Inc.) with a Gene Expression Wash Buffer kit (Agilent Technologies), according to the manufacturer's protocols.
Slides were scanned by Agilent Microarray Scanner (cat. no. G2565CA, Agilent Technologies) with default settings, dye channel: Green, scan resolution=3 µm, and photomultiplier tube 100%, 20bit. Data were extracted with Feature Extraction software 10.7 (Agilent Technologies). Raw data were normalized by Quantile algorithm, using the limma packages of the R software (version 3.5.1) (18). Following quantile normalization of the raw data, low intensity filtering was performed.
The lncRNAs, circRNAs and mRNAs with 3 samples flagged as ‘P’ or ‘A’ (all target values) were retained for further analysis. When the profile differences were compared between two groups, including the disease group vs. the control group, the fold-change between the groups for each lncRNA, circRNA and mRNA was computed. A scatter plot and volcano plots were prepared to visualize the association between the fold-change and the statistical significance of lncRNA, circRNA and mRNA expression patterns among samples. The statistical significance of differentially regulated lncRNAs, circRNAs and mRNAs between the ESCC group and the para-carcinoma control group was evaluated based on the P-value. Significantly differentially expressed transcripts were calculated by screening for a fold-change ≥2.0 and P<0.05.
Hierarchical clustering analysis
To generate an overview of lncRNA, circRNA and mRNA expression profiles between the two groups, hierarchical clustering analysis was performed based on the expression values of all expressed transcripts and significantly differentially expressed transcripts using the Cluster and Tree View program (R software) (18).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RT-qPCR was used to confirm the lncRNA and circRNA expression profiles obtained from the microarray data. All the samples were normalized to the signal generated by GAPDH (Sangon Biotech, Co., Ltd.) (25). Data are shown as the fold-change (2−ΔΔCq) (29). Student's t-tests were used and P<0.05 was considered to indicate a statistically significant difference. The cDNA was used as the template in an SYBR-Green Real-Time PCR Master Mixes (Invitrogen; Thermo Fisher Scientific, Inc.) and in triplicate subjected to denaturation at 94°C for 4 min and 35 cycles of 94°C for 30 sec, and 60°C for 30 sec, followed by extension at 72°C for 10 min using the specific primers. The primer sequences were as follows: Circ-0025580: F, 5′-CACGAGGGGCTTGTAGGATA-3′; R, 5′-AGGAAACCAAGCCACCAAG-3′. Circ-0024108: F, 5′-AGGCAAGGGATAACTCTTCTAACAC-3′; R, 5′-TTGGCAAATCTGGCGTGTAA-3′. Circ-0025933: F, 5′-GGAATGGAACGACATGCAAA-3′; R, 5′-GACACACATTGTATTTTCACGACAGT-3′. lnc-KLHDC7A-6:2: F, 5′-GGGCGTGAGGTGTGTGTTTA-3′; R, 5′-CGCTTACAAGCAGCAGGTAG-3′. LOC440173: F, 5′-GAGGTACCAAGAGAAGTGCTGATG-3′; R, 5′-GTTAATGCTTTCGGCCAAGATC-3′. EPB41L4A-AS1: F, 5′-GTCATCTATGGAGAGGAAAGGTACAAA-3′; R, 5′-TGTCACCCCAAACCTCAAATG-3′. SMAD5-AS1_3: F, 5′-GTTCTGGTGGTGATGGCATTG-3′; R, 5′-CATCTGGCTCAGGGTGTTCA-3′. GAPDH: F, 5′-TGACTTCAACAGCGACACCCA-3′; R, 5′-CACCCTGTTGCTGTAGCCAAA-3′.
Correlation and co-expression analysis
The co-expression analysis was performed by calculating the Pearson's correlation coefficient (PCC) between coding genes and noncoding transcripts, according to their expression levels. The absolute value of parameters PCC ≥0.90, P<0.05 and false discovery rate <0.01 was recommended and retained for further analysis.
Gene Ontology (GO) and Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway enrichment analysis
The DAVID 6.7 functional annotation clustering tool was used to analyse the potential functions of lncRNAs, circRNAs and co-expressed genes (18). The permutated P-value cut-off was set at <0.05. GO is used to describe biological genes and perform expression analyses of product attributes and includes the following three subgroups: Biological process (BP), cellular component (CC) and molecular function (MF) (30). GO classifications are calculated for selected genes using a particular branch of the hypergeometric distribution relationship for each gene to obtain a P-value <0.05 for the enrichment items. GO suggests a role based on the differences in the gene analysis. It can also indicate enriched genes that are differentially categorized entries based on changes in different samples (18). GO enrichment analysis of significantly differentially expressed mRNAs can reveal the role of obviously differentially regulated lncRNAs and circRNAs. Therefore, lncRNAs and circRNAs have functions that may be embodied in associated mRNA genes. The top 10 enriched GO terms of the two groups, ranked according to fold enrichment and enrichment score, are presented. The KEGG pathway analysis was performed to determine the involvement of co-expressed genes in different biological pathways. In addition, the -log10 (P-value) denotes the enrichment score, indicating the significance of the pathway correlations (31).
LncRNA target gene prediction and KEGG analysis
To identify the neighbouring target genes of differentially expressed lncRNAs that may interact via cis- or trans-regulatory effects, differentially expressed lncRNAs (fold-change >5) were selected for potential target gene prediction. LncRNAs have been found to regulate gene expression in both cis- and trans-manners using cis- and trans-regulatory mechanism-based algorithms (32). The co-expressed protein-coding genes were defined as cis-regulated genes when one differentially expressed lncRNA was indicated within 10 kb on the same chromosome or as trans-regulated genes if the aforementioned criterion was not met.
Using gene annotations from UCSC (http://genome.ucsc.edu/), lncRNAs and potential target genes were paired and visualized using the UCSC Genome Browser. The genes transcribed within a 10-kb region upstream or downstream of lncRNAs were considered cis target genes. The RNA plex software was used to choose trans-acting target genes as described previously (33). Biological pathways were defined by KEGG.
Correlation analysis of circRNAs and mRNAs in ESCC
An Agilent circRNA and mRNA expression profile microarray was used to screen the differentially expressed circRNA and mRNA gene expression. The regulation of the mRNA target expression of circRNAs was evaluated to investigate whether circRNAs could act as ‘miRNA sponges’. CircRNA-miRNA interaction analysis was conducted by Cytoscape 3.2.1 software (Cytoscape Consortium). The size of each node represents the number of putative miRNAs that were functionally connected to each circRNA.
LncRNA-miRNA-circRNA-mRNA ceRNA networks
LncRNA-mRNA and circRNA-miRNA co-expression networks were constructed based on the correlation analysis between the differentially expressed lncRNA and mRNA, and circRNAs and miRNAs. The expression of differentially expressed circRNA-miRNA, lncRNA-mRNA and miRNA-mRNA pairs was analysed by Pearson's correlation coefficient.
Statistical analysis
The results are reported as the mean ± standard deviation for triplicate measurements. Statistically significant differences between groups were estimated by two-tailed Student's t-test using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA). Pearson's coefficient was used to compare the microarray data and qPCR results. P<0.05 was considered to indicate a statistically significant difference.
Results
Evaluation of extracted RNA
Three clear bands were observed, namely, 28S, 18S and 5S, and RNA integrity of the 28S/18S band was ~2. The inclusion criteria for RNA were set at an optical density (OD) A260/280 ratio between 1.8 and 2.1 and an OD A260/230 ratio >1.8. All quality criteria that were predefined by the manufacturer for successful microarray analysis were fulfilled for each array.
Differentially expressed lncRNA, mRNA and circRNA profiles by microarray
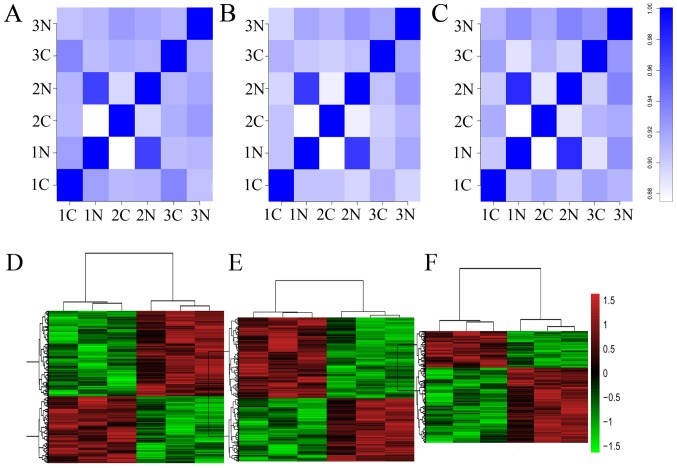
As indicated in the top row of Fig. 1, a correlation plot was used to detect the correlation between samples and to verify the homogeneity between biological replicates. In addition, as presented in the hierarchical clustering in Fig. 1, 1,384 lncRNAs, 2,046 circRNAs and 936 mRNAs were differentially expressed, with fold-changes ≥2.0 and P<0.05 in ESCC and para-carcinoma tissues. Among them, 608 and 776 lncRNAs were upregulated and downregulated, respectively; 1,148 and 898 circRNAs were upregulated and downregulated, respectively; and 306 and 630 mRNAs were upregulated and downregulated (fold-change ≥2.0 and P<0.05) in three ESCC tissue samples compared with those of controls.
Figure 1.
Correlation plot and heat map of lncRNAs, circRNAs and mRNAs in ESCC and para-carcinoma tissues. (A) The correlation among the six samples based on expression values of significantly altered lncRNAs. (B) The correlation among the six samples based on expression values of significantly altered circRNAs. (C) The correlation among the six samples based on expression values of significantly altered mRNAs. Heat map based on the expression values of significantly altered (D) lncRNAs, (E) cirRNAs and (F) mRNAs with fold changes ≥2.0 and P<0.05. lncRNAs, long noncoding RNAs; circRNAs, circularRNAs; ESCC, esophageal squamous carcinoma; C, ESCC tissues; N, control tissues.
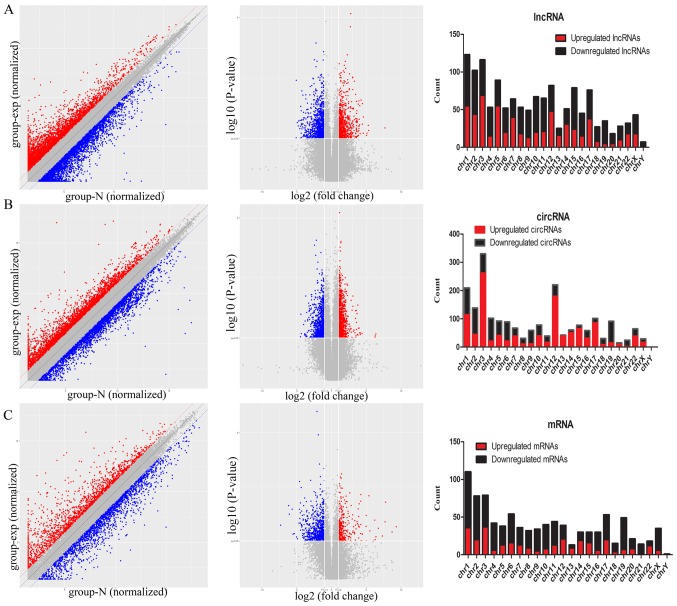
The scatter plots and volcano plots data suggested that the expression of lncRNAs, circRNAs and mRNAs in ESCC tissues differed from that of matched para-carcinoma tissues (Fig. 2).
Figure 2.
Detected results for all lncRNAs, circRNAs and mRNAs. Scatter plot and volcano plots based on the expression values of significantly altered (A) lncRNAs, (B) circRNAs and (C) mRNAs in ESCC. The upregulated and downregulated lncRNAs, circRNAs and mRNAs are marked by red or blue bars, respectively. The right vertical line corresponds to 2-fold upregulation, the left vertical line corresponds to 2-fold downregulation, and the horizontal line indicates a P=0.05. The numbers of dysregulated lncRNAs, circRNAs and mRNAs identified in ESCC tissues and matched para-carcinoma tissues are presented. lncRNAs, long noncoding RNAs; circRNAs, circularRNAs; ESCC, esophageal squamous carcinoma.
The differentially regulated lncRNAs, circRNAs and mRNAs were widely distributed among all chromosomes (Fig. 2). The dysregulated lncRNAs were classified into six categories according to their association with protein-coding genes: 34% were intergenic, 29% were exonic sense, 13% were exonic antisense, 11% were intronic sense, 7% were bidirectional and 6% were intronic antisense. A total of 120 lncRNAs had a fold-change ≥10, including 46 upregulated lncRNAs and 74 downregulated lncRNAs. Lnc-SNX10-1:1 (fold-change: Approximately 208) was the most upregulated lncRNA. An overview of the coding gene profile indicated that 40 circRNAs had a fold-change ≥10 (up: 20; down: 20) and 118 mRNAs had a fold-change ≥10 (up: 36; down: 82).
Validation of deregulated lncRNAs and circRNAs
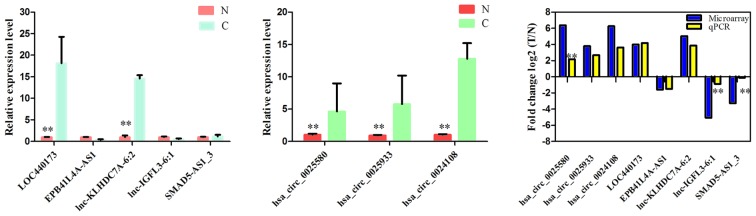
To validate the microarray profiling expression data, RT-qPCR was performed and revealed four deregulated expressed lncRNAs and three circRNAs. RT-qPCR assays indicated that the expression of lncRNAs lnc-KLHDC7A-6:2 and LOC440173 was upregulated, whereas the expression of EPB41L4A-AS1 and SMAD5-AS1_3 was downregulated (Fig. 3A). In addition, circRNAs hsa_circ_0025580, hsa_circ_0024108 and hsa_circ_0025933 were upregulated in ESCC compared with those of the control (Fig. 3B). The expression levels that were detected by the two methods were consistent with each other, demonstrating the high reliability of the microarray expression results (Fig. 3C). Therefore, the RT-qPCR data verified the validity of the microarray results.
Figure 3.
Validation of the expression of significant transcripts. (A) The relative expression levels of five lncRNAs in ESCC tissues and control tissues. (B) The relative expression levels of three circRNAs in ESCC tissues and control tissues. (C) The comparison between microarray data and qPCR results of each lncRNA and circRNA. The relative expression levels of four lncRNAs and three circRNAs. Data are presented as the mean ± standard deviation (n=20). **P<0.01. lncRNAs, long noncoding RNAs; circRNAs, circularRNAs; ESCC, esophageal squamous carcinoma; C, ESCC tissues; N, control tissues; qPCR, quantitative polymerase chain reaction.
Delineation of GO and KEGG pathway analysis
LncRNAs can regulate the expression of neighboring and overlapping coding genes (10). In addition, circRNAs can regulate their parental gene transcription (14). Therefore, GO analysis of the genes that produced differently expressed lncRNAs and circRNAs was performed. The top 10 generally changed GO terms in all comparison groups were classified as BP, CC, and MF and were ranked by fold enrichment or enrichment score as listed.
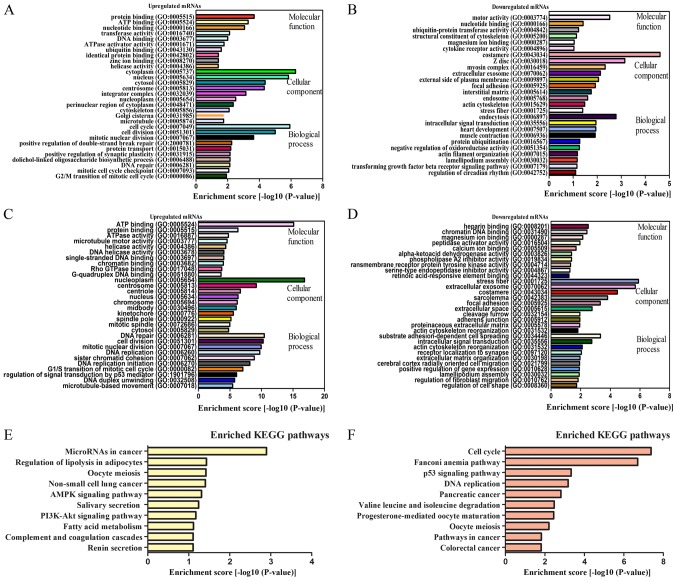
Compared with those of para-carcinoma tissues, data of the present study indicated that the mRNAs that were upregulated by lncRNAs and were associated with biological processes were associated to cell cycle and cell division (Fig. 4A). By contrast, the downregulated transcripts were most relevant for endocytosis and intracellular signal transduction (Fig. 4B). The mRNAs that were upregulated by circRNAs and were associated with biological processes were also associated with DNA repair and cell division (Fig. 4C), whereas the downregulated transcripts were most relevant for substrate adhesion-dependent cell spreading and intracellular signal transduction. The cell cycle and intracellular signal transduction GO terms were correlated with cancer and served an important role in the control of cell proliferation and gene expression (Fig. 4D).
Figure 4.
GO and KEGG pathway analyses. GO annotation of (A) upregulated lncRNAs and co-expressed mRNAs, (B) downregulated lncRNAs and co-expressed mRNAs, (C) upregulated circRNAs and co-expressed mRNAs, (D) downregulated circRNAs and co-expressed mRNAs with top enrichment scores that fell in the domains of BP, CC and MF. KEGG pathway enrichment analysis of (E) dysregulated lncRNAs and co-expressed mRNAs and (F) dysregulated circRNAs and co-expressed mRNAs with the top enrichment scores. lncRNAs, long noncoding RNAs; circRNAs, circularRNAs; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; BP, biological processes; CC, cellular components; MF, molecular functions.
The KEGG pathway enrichment analysis was designed to identify pathways and molecular interactions associated with mRNAs. For lncRNAs, our data indicated that 7 pathways were associated with upregulated mRNAs and that 10 were associated with downregulated mRNAs. For all lncRNA studies, the microRNAs in cancer were the most common pathway of dysregulated protein-coding genes (Fig. 4E).
For genes that were co-expressed with circRNAs, the top enriched KEGG pathway for upregulated mRNAs was associated with the cell cycle, and for downregulated mRNAs, the top enriched pathway was associated with valine, leucine and isoleucine degradation (Fig. 4F). Cell cycle signaling was the top pathway in dysregulated protein-coding genes. Based on these results, these pathways may contribute significantly to the pathogenesis and development of ESCC.
Cis- and trans-regulation function prediction of lncRNAs
In order to understand the underlying function of lncRNAs, cis- or trans-regulatory role of dysregulated lncRNAs in nearby coding genes was investigated.
The networks were constructed to determine the association of the differentially expressed lncRNAs and adjacent coding genes. Each lncRNA had a different number of neighbouring coding genes. For example, lnc-HOXD11-1:1 had a maximum number of 4 adjacent coding genes, whereas ENST00000529769 was indicated to have only 1 nearby coding gene (Fig. 5A).
Figure 5.
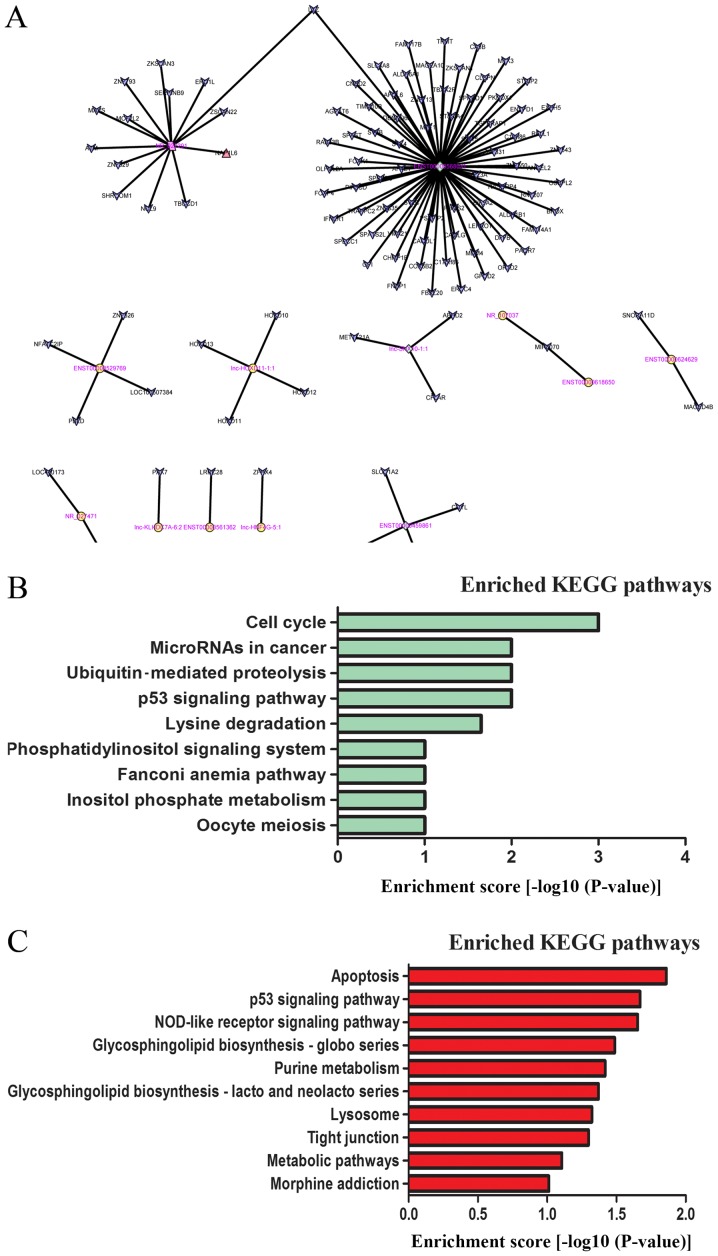
The network of lncRNAs and target genes. (A) LncRNAs and the potential cis- and trans-regulated target genes located nearby are presented in the network. The square nodes represent the lncRNAs that have a cis function, and the circle nodes represent the lncRNAs that have a trans function. The rhombus nodes represent the lncRNAs with both cis and trans functions. The tiny blue arrow nodes indicate the target genes. The size of the node represents the fold-change of lncRNA levels, with a larger size indicating a lower P-value. (B) KEGG pathway enrichment analysis of potential target genes regulated by lncRNAs in a (B) cis manner and (C) trans manner with the top enrichment scores. lncRNAs, long noncoding RNAs; KEGG, Kyoto Encyclopedia of Genes and Genomes.
To identify lncRNA target genes and their corresponding pathways, the DAVID 6.7 software was used to analyse the potential functions of specific lncRNAs. KEGG pathway enrichment analysis of 1,945 mRNAs was used, the genes for which lncRNAs (fold-change >5) may serve a cis-regulatory role. The results indicated that these mRNAs were implicated in a number of biological processes, including the cell cycle, microRNA expression in cancer, ubiquitin-mediated proteolysis and the p53 signalling pathway (Fig. 5B).
The potential target genes of the 462 differentially expressed lncRNAs (fold-change >5), which may serve a trans-regulatory role, were predicted. In all, 2,893 target genes were indicated. In addition, 282 differentially expressed genes that had been identified by the mRNA arrays (fold-change >1.5) were matched to these target genes. A number of well-known tumour-associated pathways, including those that involve apoptosis and the p53 signalling pathway, were indicated to be associated with tumorigenesis (Fig. 5C).
Construction of the circRNA-miRNA co-expression network
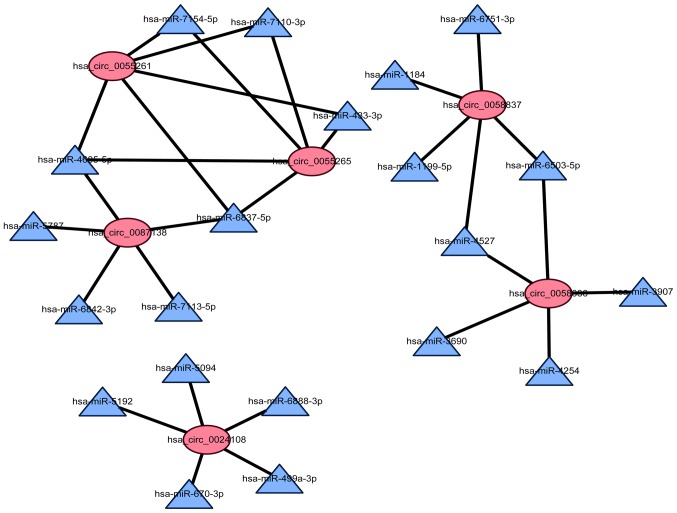
To determine the function of circRNAs, a circRNA-miRNA co-expression network was subsequently constructed, based on bioinformatics analysis. All of the differentially expressed circRNA were predicted according to the complementary miRNA sequence. An entire network of circRNA/miRNA interactions was delineated using Cytoscape. Furthermore, the graph was enlarged to display the top 6 up- and downregulated circRNAs and their corresponding miRNAs (Fig. 6). In the network, the ellipses represent circRNAs, and the triangles represent miRNAs.
Figure 6.
The network of circRNAs-miRNAs. The network consisted of 6 circRNAs and 31 miRNAs. In the circRNA-miRNA network, the ellipse represents circRNA and the triangle represents miRNA, and their association is represented by one edge. miRNAs, microRNAs.
Construction of ceRNA networks
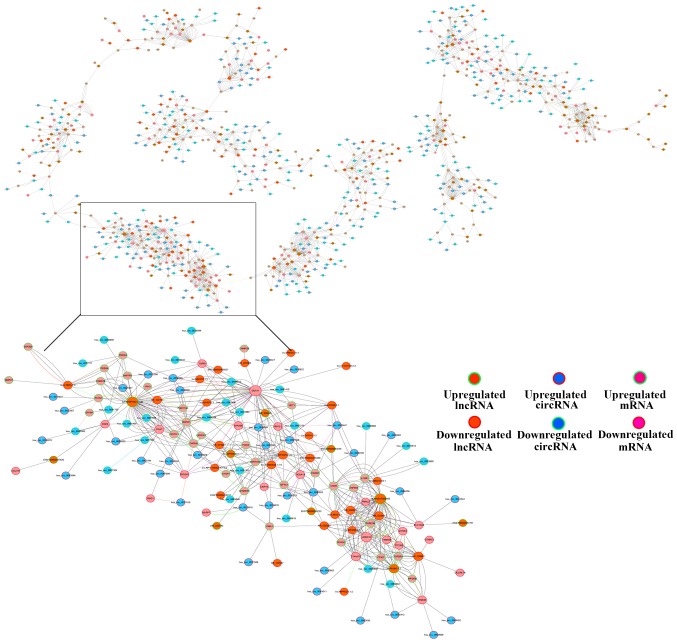
Based on the ceRNA hypothesis, ceRNAs can compete for the same miRNA response elements (MREs) to regulate each other (26). Bioinformatics methods to establish a ceRNA network in ESCC were used according to our microarray data, which included the circRNA, lncRNA and mRNA data (Fig. 7). The association among circRNA, lncRNA, mRNA and miRNA were established using the expression values. The size of the circle represents the ability of a given type of RNA to interact with other genes.
Figure 7.
Competing endogenous RNA network in ESCC. The competing endogenous RNA network was based on lncRNA/miRNA, circRNA/miRNA and miRNA/mRNA interactions. In this network, each point represents a gene, and a straight line between the genes represents an miRNA. ESCC, esophageal squamous carcinoma; lncRNAs, long noncoding RNAs; circRNAs, circularRNAs; miRNAs, microRNAs.
In the present study, a total of 185 lncRNAs, 332 circRNAs and 318 mRNAs were selected to generate a network map with Cytoscape. The red colour and green colour represent up- and downregulation, respectively. The size of the circle represents the size of the P-value, with a larger size resulting from a smaller fold-change. The local graphic was enlarged to display the ceRNA network. For instance, circ-0078344, circ-0026599, circ-0036698 and NR-102741 are ceRNAs of different miRNAs that target transforming grown factor β induced (TGFBI) (27). TGFBI is a type of protein that is induced by the transforming growth factor beta secretory protein. The analysis revealed that TGFBI is associated with cancer in the general biological process subgroup, and may be a biological marker of cancer. These RNA interactions provided a novel perspective on the tumorigenic mechanisms of ESCC.
Discussion
Previous studies have focused on protein-coding genes until the discovery of numerous non-coding RNAs, including lncRNAs and circRNAs (12,14,16). This study, to the best of our knowledge, presents the first simultaneous investigation of lncRNAs, circRNAs and mRNAs in clinical ESCC tissues.
A growing number of studies have confirmed that lncRNAs may be one of the most important factors for the control of gene expression (8–11,34). LncRNA has been widely reported to participate in a wide range of biological processes, and its dysregulated expression affects many human disease phenotypes, including those of cancers (35–37). Similar to lncRNAs, circRNAs are a large class of noncoding RNAs (ncRNAs). CircRNAs were recently described as pivotal gene regulators in humans, due to the different ways that they control transcription and translation (38). As increased attention is paid to the roles of circRNAs as oncogenes and tumour suppressors in cancer, circRNAs have exhibited great potential as diagnostic and prognostic biomarkers (19,39).
However, comprehensive analyses of differentially expressed profiles of lncRNAs and circRNAs in ESCC, to the best of our knowledge, have yet to be reported. To probe the functions of lncRNAs and circRNAs in ESCC tumours, the genome-wide expression profiles of lncRNAs and circRNAs were examined in the present study in three ESCC specimens and matched adjacent tissues using a microarray assay.
From the results of the present study, it was determined that hundreds of lncRNAs, circRNAs and mRNAs were differentially expressed in tumour tissues compared with those of the control group. Overall, 1,384 lncRNAs, 2,046 circRNAs and 936 mRNAs were observed to be significantly differentially expressed in ESCC. Four dysregulated lncRNAs and three circRNAs were further verified by RT-qPCR. Therefore, the RT-qPCR data verified the microarray data, demonstrating the reliability of the microarray results. These differentially expressed lncRNAs, circRNAs and mRNAs were subsequently integrated into hierarchical categories, according to heat maps and hierarchical clustering.
To further study the function of these differentially expressed lncRNAs and circRNAs, GO and KEGG pathway analyses were performed. The annotation results of the most significant GO terms of lncRNAs were cell cycle, cell division, endocytosis and intracellular signal transduction. The notably changed GO terms of circRNAs were DNA repair, cell division, substrate adhesion-dependent cell spreading and intracellular signal transduction. These biological processes identified coding genes that were associated with the development of ESCC (40,41).
KEGG pathway analysis for the differentially expressed lncRNAs and circRNAs revealed 10 pathways. These pathways, including miRNAs in cancer, the 5′ AMP-activated protein kinase signalling pathway, PI3K-Akt signalling pathway, p53 signalling pathway and cell cycle, may serve pivotal roles in the tumorigenic mechanisms of ESCC, as they were more likely to be identified in the ESCC group compared with in the normal control group. This suggests that dysregulated lncRNAs and circRNAs may affect these targets by regulating the associated pathways in ESCC.
Increasing evidence indicates that lncRNAs and circRNAs are extensively targeted by miRNAs and that they function as ceRNAs (20,42). CeRNAs include lncRNAs, circRNAs and mRNAs, as these transcripts can compete for the same MREs and mutually regulate each other (23). ceRNAs have been implicated in both physiological conditions and cancer development. If the balance in the intricate ceRNA network is disturbed, carcinogenesis may occur. To date, to the best of our knowledge, no information on ceRNAs in ESCC has been reported. A lncRNA-miRNA-circRNA-mRNA ceRNA network in ESCC was constructed based on our microarray data. As indicated in Fig. 7, the association among these types of RNA is extremely complex. Compared with those of the control group, the lncRNAs and circRNAs that were dysregulated in the ESCC group were identified. In the ceRNA network, 185 lncRNAs and 332 circRNAs were included. In order to illustrate the ceRNA network, TGFBI was selected, as it serves a role in the activation of morphogenesis, cell proliferation, adhesion, migration, differentiation, chemoresistance and inflammation (43). The RNAs circ-0078344, circ-0026599, circ-0036698 and NR-102741 are ceRNAs of different miRNAs that target TGFBI. Theoretically, lncRNAs and circRNAs may regulate the tumorigenesis of ESCC by targeting TGFBI. However, the functions of lncRNAs, circRNAs and their associated ceRNAs in ESCC remain unclear. Further studies on these RNA interactions are required, in order to provide a novel perspective of the tumorigenic mechanisms of ESCC.
In conclusion, the present study presented a profile of dysregulated lncRNAs, circRNAs and mRNAs in ESCC, as determined by microarray analysis. GO and KEGG pathway analyses were performed to annotate the potential functions of differentially expressed lncRNAs and circRNAs. Co-expression networks were constructed for lncRNA-miRNA-circRNA-mRNA. Our data may establish a foundation for further functional research into lncRNAs and circRNAs in ESCC. Therefore, these results suggest that ncRNAs may serve an important diagnostic and therapeutic role in ESCC.
Acknowledgements
The authors would like to thank The Xinjiang Cancer Biobank of Tumor Affiliated Hospital of Xinjiang Medical University (Urumuqi, China) for providing samples of esophageal cancer.
Glossary
Abbreviations
- BP
biological process
- CC
cellular component
- ceRNA
competing endogenous RNA
- circRNAs
circularRNAs
- EC
esophageal carcinomas
- ESCC
esophageal squamous carcinoma
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- lncRNAs
long noncoding RNAs
- miRNAs
microRNAs
- MF
molecular function
- MREs
miRNA response elements
- ncRNAs
non-coding RNAs
- PCC
Pearson's correlation coefficient
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
Funding
The present study was supported by the National Natural Science Foundation of China (grant nos. 81760498).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JS and YL conceived the study and JZ designed the study. JS, YL and MH performed the experiments. WS, XD and YZ collected clinical samples and analyzed the data. JS and YRL wrote the paper. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Third Affiliated Teaching Hospital of Xinjiang Medical University, Affiliated Cancer Hospital's Protection of Human Subjects Committee. Written informed consent was obtained from all subjects.
Patient consent for publication
All patients signed written informed consent for the publication.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Forghanifard MM, Taleb Sh, Abbaszadegan MR. Notch signaling target genes are directly correlated to esophageal squamous cell carcinoma tumorigenesis. Pathol Oncol Res. 2015;21:463–467. doi: 10.1007/s12253-014-9849-8. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Zuo T, Zeng H, Zhang S, He J. National cancer incidence and mortality in China 2012. Chin J Cancer Res. 2016;28:1–11. doi: 10.1007/978-3-319-42740-9_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y. Epidemiology of esophageal cancer. World J Gastroenterol. 2013;19:5598–5606. doi: 10.3748/wjg.v19.i34.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 6.Rong D, Sun H, Li Z, Liu S, Dong C, Fu K, Tang W, Cao H. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8:73271–73281. doi: 10.18632/oncotarget.19154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han X, Wang L, Ning Y, Li S, Wang Z. Long non-coding RNA AFAP1-AS1 facilitates tumor growth and promotes metastasis in colorectal cancer. Biol Res. 2016;49:36. doi: 10.1186/s40659-016-0094-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou M, Hou Y, Yang G, Zhang H, Tu G, Du YE, Wen S, Xu L, Tang X, Tang S, et al. LncRNA-Hh strengthen cancer stem cells generationin Twist-positive breast cancer via activation of Hedgehog signaling pathway. Stem Cells. 2016;34:55–66. doi: 10.1002/stem.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu S, Wang P, You Z, Meng H, Mu G, Bai X, Zhang G, Zhang J, Pang D. The longnon-coding RNAEPB41L4A-AS2 inhibits tumor proliferation and is associated with favorable prognoses in breast cancer and other solid tumors. Oncotarget. 2016;7:20704–20717. doi: 10.18632/oncotarget.8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee S, Kopp F, Chang TC, Sataluri A, Chen B, Sivakumar S, Yu H, Xie Y, Mendell JT. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell. 2016;164:69–80. doi: 10.1016/j.cell.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in cancer: Opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 16.Geng Y, Jiang J, Wu C. Function and clinical significance of circRNAs in solid tumors. J Hematol Oncol. 2018;11:98. doi: 10.1186/s13045-018-0643-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin X, Feng CY, Xiang Z, Chen YP, Li YM. CircRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of nonalcoholic steatohepatitis. Oncotarget. 2016;7:66455–66467. doi: 10.18632/oncotarget.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang M, Zhong Z, Lv M, Shu J, Tian Q, Chen J. Comprehensive analysis of differentially expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in bladder carcinoma. Oncotarget. 2016;7:47186–47200. doi: 10.18632/oncotarget.9706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z, Yang J, Fan J, Liu L, Qin W. Hsa_circ_0001649: A circularRNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16:161–169. doi: 10.3233/CBM-150552. [DOI] [PubMed] [Google Scholar]
- 20.Peng L, Chen G, Zhu Z, Shen Z, Du C, Zang R, Su Y, Xie H, Li H, Xu X, et al. CircularRNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in Hirschsprung's disease. Oncotarget. 2017;8:808–818. doi: 10.18632/oncotarget.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y, Yang S, Zeng Z, Liao W, Ding YQ, Liang L. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7:26680–26691. doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao JT, Zhao SH, Liu QP, Lv MQ, Zhou DX, Liao ZJ, Nan KJ. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract. 2017;213:453–456. doi: 10.1016/j.prp.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 23.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 24.Han D, Li J, Wang H, Su X, Hou J, Gu Y, Qian C, Lin Y, Liu X, Huang M, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 25.Chen L, Zhang S, Wu J, Cui J, Zhong L, Zeng L, Ge S. CircRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene. 2017;36:4551–4561. doi: 10.1038/onc.2017.89. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Salmena L, Poliseno L, Tay Y, Kats L, Pandolf PP. A ceRNA hypothesis: The rosetta stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou B, Yu JW. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-β1. Biochem Biophys Res Commun. 2017;487:769–775. doi: 10.1016/j.bbrc.2017.04.044. [DOI] [PubMed] [Google Scholar]
- 28.Sobin LH, Gospodarowicz MK, Wittekind C. TNM classification of malignant tumours. 7th. Oxford: Wiley-Blackwell; 2009. Oesophagus including oesophagogastric junction; pp. 66–72. [Google Scholar]
- 29.Coates DE, Zafar S, Milne TJ. Quantitative real-time gene profiling of human alveolar osteoblasts. Methods Mol Biol. 2017;1537:447–459. doi: 10.1007/978-1-4939-6685-1_27. [DOI] [PubMed] [Google Scholar]
- 30.The Gene Ontology Consortium, corp-author. Gene ontology consortium: Going forward. Nucleic Acids Res. 2015;43((Database Issue)):D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petruk S, Sedkov Y, Riley KM, Hodgson J, Schweisguth F, Hirose S, Jaynes JB, Brock HW, Mazo A. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell. 2006;127:1209–1221. doi: 10.1016/j.cell.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tafer H, Hofacker IL. RNAplex: A fast tool for RNA-RNA interaction search. Bioinformatics. 2008;24:2657–2663. doi: 10.1093/bioinformatics/btn193. [DOI] [PubMed] [Google Scholar]
- 34.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou M, Wang X, Li J, Hao D, Wang Z, Shi H, Han L, Zhou H, Sun J. Prioritizing candidate disease-related long non-coding RNAs by walking on the heterogeneous lncRNA and disease network. Mol Biosyst. 2015;11:760–7569. doi: 10.1039/C4MB00511B. [DOI] [PubMed] [Google Scholar]
- 36.Sun J, Shi H, Wang Z, Zhang C, Liu L, Wang L, He W, Hao D, Liu S, Zhou M. Inferring novel lncRNA-disease associations based on a random walk model of a lncRNA functional similarity network. Mol Biosyst. 2014;10:2074–2081. doi: 10.1039/C3MB70608G. [DOI] [PubMed] [Google Scholar]
- 37.Sahu A, Singhal U, Chinnaiyan AM. Long noncoding RNAs in cancer: From function to translation. Trends Cancer. 2015;1:93–109. doi: 10.1016/j.trecan.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Yang J, Zhou P, Le Y, Zhou C, Wang S, Xu D, Lin HK, Gong Z. Circular RNAs in cancer: Novel insights into origins, properties, functions and implications. Am J Cancer Res. 2015;5:472–480. [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Zhang Y, Huang L, Zhang J, Pan F, Li B, Yan Y, Jia B, Liu H, Li S, Zheng W. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol. 2015;8:16020–16025. [PMC free article] [PubMed] [Google Scholar]
- 40.Cai X, Hu X, Tan X, Cheng W, Wang Q, Chen X, Guan Y, Chen C, Jing X. Metformin induced AMPK activation, G0/G1 phase cell cycle arrest and the inhibition of growth of esophageal squamous cell carcinomas in vitro and in vivo. PLoS One. 2015;10:e0133349. doi: 10.1371/journal.pone.0133349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng HH, Zhang X, Cao PG. MMP-1/PAR-1 signal transduction axis and its prognostic impact in esophageal squamous cell carcinoma. Braz J Med Biol Res. 2012;45:86–92. doi: 10.1590/S0100-879X2011007500152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou M, Diao Z, Yue X, Chen Y, Zhao H, Cheng L, Sun J. Construction and analysis of dysregulated lncRNA-associated ceRNA network identified novel lncRNA biomarkers for early diagnosis of human pancreatic cancer. Oncotarget. 2016;7:56383–56394. doi: 10.18632/oncotarget.10891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu J, Chen X, Liao Z, He C, Hu X. TGFBI protein high expression predicts poor prognosis in colorectal cancer patients. Int J Clin Exp Pathol. 2015;8:702–710. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.