Abstract
Distal-less homeobox 6 antisense RNA 1 (DLX6-AS1) is upregulated in various solid tumors and serves a critical role in the tumorigenesis of cancer. However, to the best of our knowledge, the expression of circulating DLX6-AS1 and its role in the diagnosis of non-small cell lung cancer (NSCLC) have not been previously clarified. The aim of the present study was to investigate the expression and clinical significance of circulating DLX6-AS1 using reverse transcription-quantitative PCR in serum and exosomes derived from patients with NSCLC and healthy donors. The diagnostic value of circulating DLX6-AS1 was identified by receiver operating characteristic curve (ROC) analysis. First, it was revealed that the expression levels of DLX6-AS1 were significantly increased in tumor tissues compared with in adjacent normal tissues. In addition, DLX6-AS1 was highly expressed in NSCLC cell lines compared with in BEAS-2B cells. DLX6-AS1-knockdown inhibited cell proliferation and migration in vitro. It was subsequently demonstrated that the serum DLX6-AS1 level was significantly higher in patients with NSCLC compared with in healthy controls. Additionally, the higher DLX6-AS1 expression was associated with advanced disease stage, positive lymph node metastasis and poor tumor differentiation of NSCLC. ROC analysis demonstrated that the sensitivity and specificity of DLX6-AS1 were higher than those of CYFRA21-1, which is a serum marker for NSCLC. Finally, exosomal DLX6-AS1 expression was increased in patients with NSCLC compared with in healthy controls. The present data implied that circulating DLX6-AS1 was mainly incorporated into exosomes, providing a novel potential diagnostic marker for NSCLC.
Keywords: non-small cell lung cancer, long non-coding RNA, distal-less homeobox 6 antisense RNA 1, exosomes, diagnosis
Introduction
Lung cancer remains a major cause of cancer-associated morbidity and mortality worldwide, particularly in China (1,2). Non-small cell lung cancer (NSCLC) is a major histopathological subtype that accounts for 80–85% of all lung cancer cases (3). Despite significant improvements in clinical strategies for NSCLC, including surgery, targeted therapy, immunotherapy, chemotherapy and radiotherapy, the prognosis of patients with NSCLC remains poor (1). Considerable evidence has demonstrated that the poor prognosis of NSCLC is attributed to tumor metastasis (4), drug resistance (5) and lack of early sensitivity and specificity diagnostic markers (6). Therefore, it is urgent to find novel therapeutic targets for NSCLC.
Long non-coding RNAs (lncRNAs) are a member of the non-coding RNA family, >200 nucleotides in length and without protein translation capacity (7). An increasing number of studies (8,9) have demonstrated that lncRNAs serve an important role in human diseases, including cancer (10). lncRNAs are involved in multiple physiological and pathological processes, including cell proliferation, apoptosis and invasion, as well as tumor metastasis and tumorigenesis (10,11). Notably, lncRNAs are abundantly found in the serum, plasma, saliva and urine, and can serve as potential biomarkers for the early diagnosis and prognosis of NSCLC (12,13). Previous studies (8,13) have indicated that the dysregulation of certain lncRNAs has been identified in NSCLC and serves an important role in the tumorigenesis and progression of NSCLC, providing a novel potential therapeutic target for NSCLC. For example, HOX transcript antisense RNA is overexpressed in NSCLC and promotes tumorigenesis and metastasis by regulating microRNA (miR)-613 (14). Distal-less homebox 6 antisense RNA 1 (DLX6-AS1) is upregulated in various solid tumors, including NSCLC (15–17), renal cell carcinoma (18), hepatocellular carcinoma (19–21), colorectal cancer (22), pancreatic cancer (23,24), glioma (25) and osteosarcoma (26,27), acting as an early diagnostic and prognostic biomarker. However, the expression and function of circulating DLX6-AS1 in NSCLC have not been widely demonstrated.
The present study revealed that DLX6-AS1 was overexpressed not only in tumor tissues compared with in adjacent normal tissues, but also in the serum of patients with NSCLC compared with that of healthy donors. In addition, the circulating DLX6-AS1 expression was significantly associated with advanced disease stage and lymph node metastasis in patients with NSCLC. Furthermore, exosomal DLX6-AS1 was upregulated in patients with NSCLC and had a higher sensitivity and specificity for NSCLC diagnosis than CYFRA21-1, which is a serum diagnostic marker for NSCLC (28), suggesting it could be a potential novel marker for the early diagnosis and metastasis of NSCLC.
Materials and methods
Patient samples and healthy donors
A total of 72 patients who were pathologically confirmed with NSCLC and 64 healthy donors were enrolled at the First Affiliated Hospital of Huzhou University between October 2016 and December 2018. The NSCLC patient group consisted of 39 men and 33 women (age range, 28–81 years; mean age, 61 years). Healthy controls comprised 31 men and 33 women (age range, 29–89 years; mean age, 62 years). Serum samples were obtained from patients on pre-operative day 1 and post-operative day 3, and from healthy controls, and stored at −80°C prior to use. Furthermore, 27 pairs of tumor tissues and matched adjacent normal tissues (>5 cm away from the tumor) were collected immediately after resection and stored in liquid nitrogen until further use. No patients had received adjuvant therapy prior to surgical resection. The pathological diagnosis was confirmed and classified by two certified pathologists based on the 7th Edition of the Union for International Cancer Control (29). Clinical information, including age, sex, smoking history, tumor size, tumor differentiation, lymph node metastasis and TNM stage, was collected. The present study was approved by the Ethics Committee of the First Affiliated Hospital of Huzhou University, and written informed consent was provided by all patients and healthy controls.
Gene Expression Profiling Interactive Analysis (GEPIA)
GEPIA (http://gepia.cancer-pku.cn/) is a public web server used for evaluation of cancer and normal gene expression profiling and interactive analysis according to The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) database (30). DLX6-AS1 expression levels in lung adenocarcinoma (LUAD, n=483) and lung squamous cell carcinoma (LUSC, n=486) were compared with their normal cohorts (n=347; n=338). Association between DLX6-AS1 expression and TNM stage was investigated in LUAD and LUSC cohort. Additionally, patients with LUAD and LUSC were divided into two groups according to DLX6-AS1 expression. Overall survival and disease-free survival were performed using the Kaplan-Meier method.
Cell lines and culture
The human NSCLC A549, H1299 and 95D, and normal human epithelial BEAS-2B cell lines were obtained from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences, and cultured in DMEM (HyClone; GE Healthcare Life Sciences) containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin (Sigma-Aldrich; Merck KGaA) at 37°C in a humidified incubator with 5% CO2.
Cell transfection
The two specific small interfering RNAs (siRNAs) of lncRNA DLX6-AS1 (20 µM) and scrambled siRNA (20 µM) were designed and synthesized by Guangzhou RiboBio Co., Ltd.. Once they reached 50–70% confluence, the A549 and H1299 cells were transfected with Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. At 2 days after transfection, cells were harvested and the DLX6-AS1 expression was measured by reverse transcription-quantitative PCR (RT-qPCR) to confirm the transfection efficiency. The siRNA sequences used in the experiment were as follows: si-DLX6-AS1#1 forward, 5′-GGCUAACACAUCCAUGGAAdTdT-3′ and reverse, 5′-UUCCAUGGAUGUGUUAGCCdTdT-3′; si-DLX6-AS1#2 forward, 5′-GCCGCUUGUCUUACUUAAAdTdT-3′ and reverse, 5′-UUUAAGUAAGACAAGCGGCdTdT-3′; and scrambled siRNA forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse, 5′-ACGUGACACGUUCGGAGAATT-3′.
RNA extraction and RT-qPCR
Total RNA from tissues, cells and exosomes was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), as described previously (31). Complementary DNA was reverse synthesized using the PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd.), according to the manufacturer's protocol. RNA (500 ng), 2 µl PrimeScript™ RT Master Mix (Perfect Real Time) and RNase free water (up to 10 µl), were mixed together and incubated at 37°C for 15 min and 85°C for 5 sec. qPCR was performed with SYBR Green PCR Master Mix using the 7500 system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The qPCR reaction system (10 µl) included the following: 5 µl 2X SYBR Green PCR Master Mix, 0.3 µl forward primer (10 µM), 0.3 µl reverse primer (10 µM), 50 ng cDNA and RNase free water. The thermocycling conditions used for the qPCR were as follows 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. Gene expression was normalized to 18S ribosomal RNA (18sRNA), and the relative expression level was calculated using the 2−ΔΔCq method (32). The primer sequences used in the present study were as follows: DLX6-AS1 forward, 5′-AGTTTCTCTCTAGATTGCCTT-3′ and reverse, 5′-ATTGACATGTTAGTGCCCTT-3′; 18sRNA forward, 5′-GTAACCCGTTGAACCCCATT-3′ and reverse, 5′-CCATCCAATCGGTAGTAGCG-3′.
Western blotting
Exosomes were lysed in RIPA buffer (cat. no. P0013B; Beyotime Institute of Biotechnology) containing protease and phosphatase inhibitor (Beyotime Institute of Biotechnology) and the total protein concentration was measured using a bicinchoninic acid assay (Beyotime Institute of Biotechnology). An equal amount of protein (30 µg) was loaded onto 10–15% SDS-PAGE gels. The proteins were then transferred onto 0.45-µm PVDF membranes (EMD Millipore). The membranes were blocked with 5% non-fat milk at room temperature for 1 h, and incubated with specific primary antibodies at 4°C overnight, followed by incubation with horseradish peroxidase-conjugated goat-anti-mouse secondary antibody (cat. no. A0286; dilution, 1:1,000; Beyotime Institute of Biotechnology) at room temperature for 1 h. Bands were visualized using enhanced chemiluminescence (Beyotime Institute of Biotechnology). Relative expression level of proteins was normalized to β-actin using ImageJ v1.6.0 software (National Institutes of Health). The primary antibodies used in the present study were as follows: Anti-CD81 (cat. no. sc-166029; dilution, 1:200; Santa Cruz Biotechnology, Inc.); anti-CD9 (cat. no. sc-13118; dilution, 1:200; Santa Cruz Biotechnology, Inc.); anti-Alix (cat. no. sc-53540; dilution, 1:200; Santa Cruz Biotechnology, Inc.); and anti-β-actin (cat. no. 4967S; dilution, 1:2,000; Cell Signaling Technology, Inc.).
Exosomes isolation
Plasma (5 ml) was collected and divided into two parts, and exosomes were isolated using ExoQuick Exosome Precipitation solution (System Biosciences, LLC) according to the manufacturer's protocol. Exosomes were resuspended in PBS for transmission electron microscopy and nanoparticle tracking analysis, in RIPA buffer for western blotting or TRIzol® for RNA extraction.
Transmission electron microscopy
Exosomes were fixed with 4% paraformaldehyde for 4 h at 22°C, followed by 1% glutaraldehyde for 30 min at 22°C. The samples were dropped onto formvar carbon-coated grids and allowed to absorb for 10 min, followed by fixation with 4% paraformaldehyde at room temperature for 10 min. Following PBS washing, exosomes were stained with 2% uranyl acetate solution for 1 min at room temperature. Images were observed under a Tecnai G2 spititi electron microscopy.
Nanoparticle tracking analysis (NTA)
The size distribution and concentration of exosomes were analyzed by NTA, according to the manufacturer's instructions. Briefly, exosomes were resuspended and diluted at 1:200 in PBS, and the particle concentration of exosomes was assessed using a NanoSight NS500 Instrument (Malvern Instruments, Ltd.).
Real-time cell analysis (RTCA) assays
Cell proliferation and migration were assessed using the RTCA xCell Ligence system, as previously described (33). For cell proliferation assays, A549 or H1299 cells were seeded at 6,000 or 10,000 cells/well density and incubated for 30 min at room temperature. The plate was loaded and the data were collected every 15 min for 60 h. For cell migration assays, 165 µl DMEM containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) was added into the lower chamber and 30 µl serum-free medium was added into upper chamber. A549 or H1299 cells were seeded at 40,000 or 60,000 cells/well density in the upper chamber. The plate was loaded and the data were collected every 15 min for 48 h.
Statistical analysis
All data are presented as the mean ± standard deviation of three independent experiments. Comparisons between two groups were performed by Student's t-test, and comparisons between more than two groups were performed using one-way ANOVA followed by a Bonferroni post hoc test. The diagnostic values of circulating DLX6-AS1 and CYFRA21-1 for NSCLC were analyzed using receiver operating characteristic curve (ROC) analysis. Survival curves were analyzed using the Kaplan-Meier method with the log-rank test. Data were analyzed using SPSS (version 21.0; IBM Corp.). P<0.05 was considered to indicate a statistically significant difference.
Results
DLX6-AS1 is upregulated in NSCLC
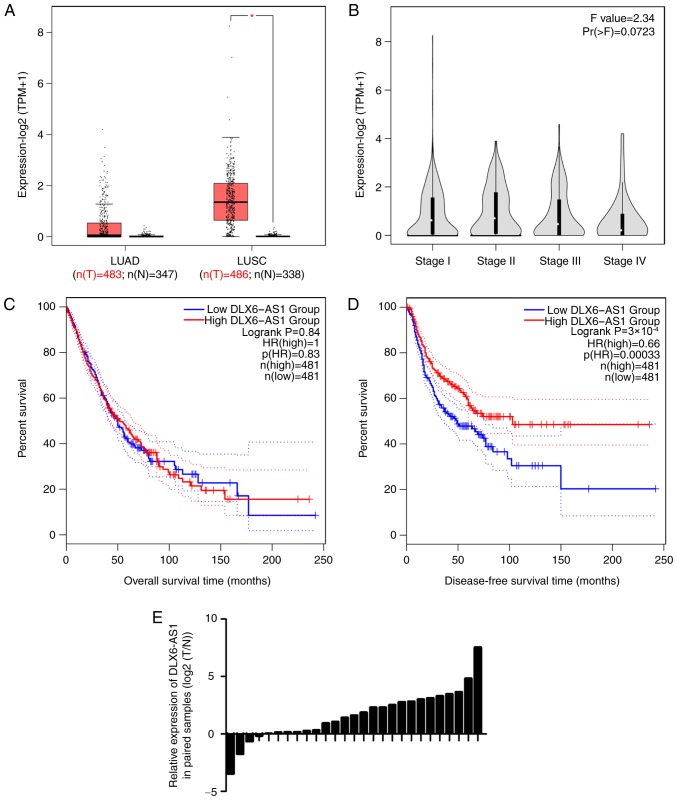
The present study first investigated DLX6-AS1 expression by using GEPIA tool (30), which provides single gene, cancer type and multiple gene analysis. The results demonstrated that DLX6-AS1 expression was significantly increased in lung squamous cell carcinoma (Fig. 1A), but was not associated with TNM stage (Fig. 1B). In addition, patients with low DLX6-AS1 expression had a poor prognosis with a shorter disease-free survival (Fig. 1C and D). Additionally, it was found that DLX6-AS1 was overexpressed in 23 pairs (accounting for 85% of total samples) of tumor tissues, compared with in adjacent normal tissues from patients with NSCLC (Fig. 1E).
Figure 1.
DLX6-AS1 is overexpressed in NSCLC. (A) DLX6-AS1 expression data were obtained from the Gene Expression Profiling Interactive Analysis database. (B) Associations between DLX6-AS1 expression and TNM stage of patients with NSCLC were analyzed. (C) Kaplan-Meier survival analysis of overall survival of patients with NSCLC with low and high DLX6-AS1 expression. (D) Kaplan-Meier survival analysis of disease-free survival of patients with NSCLC with low and high DLX6-AS1 expression. (E) DLX6-AS1 expression was detected in 27 pairs of tumor and adjacent normal tissues. Data were analyzed using Student's t test. *P<0.05. DLX6-AS1, distal-less homeobox 6 antisense RNA 1; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; NSCLC, non-small cell lung cancer; TPM, transcripts per million; n(T), number (Tumor); n(N), number (Normal).
DLX6-AS1 expression was studied in NSCLC cell lines, and it was identified that DLX6-AS1 exhibited significantly higher expression levels in A549, H1299 and 95D cells, compared with in the normal human epithelial BEAS-2B cells (Fig. S1A). Subsequently, the effect of DLX6-AS1 on cell proliferation and migration in A549 and H1299 cells transfected with two DLX6-AS1 siRNAs or scrambled siRNA was investigated. The RT-qPCR results demonstrated that DLX6-AS1 expression was significantly reduced following transfection (Fig. S1B). DLX6-AS1-knockdown suppressed A549 and H1299 proliferation and migration in vitro (Fig. S1C-F).
Serum DLX6-AS1 expression is increased in NSCLC
The present study investigated the serum expression levels of DLX6-AS1 in patients with NSCLC and healthy controls. The results demonstrated that serum DLX6-AS1 expression was significantly increased in patients with NSCLC compared with healthy controls (Fig. 2A). The association between the serum expression levels of DLX6-AS1 and clinicalpathological features of patients with NSCLC was also investigated. The data indicated that the serum expression levels of DLX6-AS1 were significantly higher in patients with advanced disease stage, positive lymph node metastasis and poor tumor differentiation compared with in patients with earlier disease stage, negative lymph node metastasis and well/moderate tumor differentiation, respectively (Fig. 2B-D). However, DXL6-AS1 expression was not associated with sex, age, smoking history and tumor size (Table I).
Figure 2.
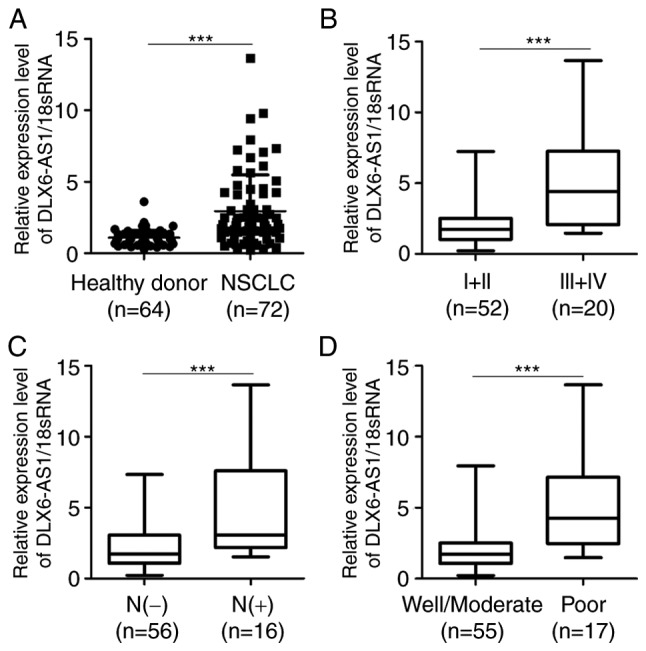
Serum DLX6-AS1 is highly expressed in samples from patients with NSCLC and associated with clinicopathological characteristics. (A) Serum level of DLX6-AS1 was measured by reverse transcription-quantitative PCR in patients with NSCLC and healthy donors. (B) Associations between the serum DLX6-AS1 expression and TNM stage of patients with NSCLC. (C) Associations between the serum DLX6-AS1 expression and lymph node metastasis of patients with NSCLC. (D) Associations between the serum DLX6-AS1 expression and tumor differentiation of patients with NSCLC. Data were analyzed using Student's t-test. *P<0.05 and ***P<0.001. 18sRNA, 18S ribosomal RNA; DLX6-AS1, distal-less homeobox 6 antisense RNA 1; NSCLC, non-small cell lung cancer.
Table I.
Association between DLX6-AS1 expression and clinicopathological characteristics of patients with non-small cell lung cancer.
| Features | n | DLX6-AS1 (mean ± SD) | t | P-value |
|---|---|---|---|---|
| Sex | 1.902 | 0.065 | ||
| Male | 39 | 3.15±3.00 | ||
| Female | 33 | 2.00±1.31 | ||
| Age, years | 0.066 | 0.197 | ||
| >65 | 35 | 2.92±2.95 | ||
| ≤65 | 37 | 2.96±2.12 | ||
| Smoking history | 1.347 | 0.075 | ||
| Yes | 37 | 3.33±2.88 | ||
| No | 35 | 2.52±2.08 | ||
| Tumor size, cm | 0.981 | 0.102 | ||
| >3 | 30 | 3.71±3.11 | ||
| ≤3 | 42 | 2.69±1.90 | ||
| Tumor differentiation | 5.294 | <0.001 | ||
| Well/moderate | 55 | 2.08±1.46 | ||
| Poor | 17 | 5.37±3.40 | ||
| TNM | 2.334 | 0.022 | ||
| I–II | 52 | 2.52±2.26 | ||
| III–IV | 20 | 4.03±2.95 | ||
| Lymphovascular invasion | 6.005 | <0.001 | ||
| Present | 16 | 5.69±2.40 | ||
| Absent | 56 | 2.15±1.98 |
DLX6-AS1, distal-less homeobox 6 antisense RNA 1.
Serum DLX6-AS1 expression is reduced in patients post-operatively
To investigate the effect of surgery on the serum DLX6-AS1 expression, alterations in DLX6-AS1 expression were compared between pre- and post-operative serum samples from a set of 20 matched patients, who were selected to our follow-up study. The data demonstrated that the serum DLX6-AS1 expression was significantly lower in post-operative samples than in pre-operative samples (Fig. 3). These results suggest that circulating DLX6-AS1 is derived from tumor tissues, providing a potential marker for the early diagnosis of NSCLC.
Figure 3.
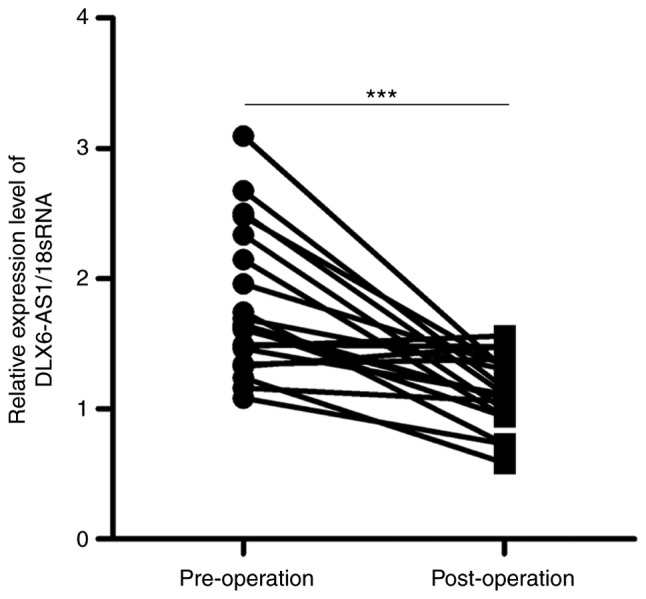
DLX6-AS1 expression is altered in patients pre- and post-operatively. Serum DLX6-AS1 expression was detected by reverse transcription-quantitative PCR in patients pre- and post-operatively. The data were analyzed using Student's t-test. ***P<0.001. 18sRNA, 18S ribosomal RNA; DLX6-AS1, distal-less homeobox 6 antisense RNA 1.
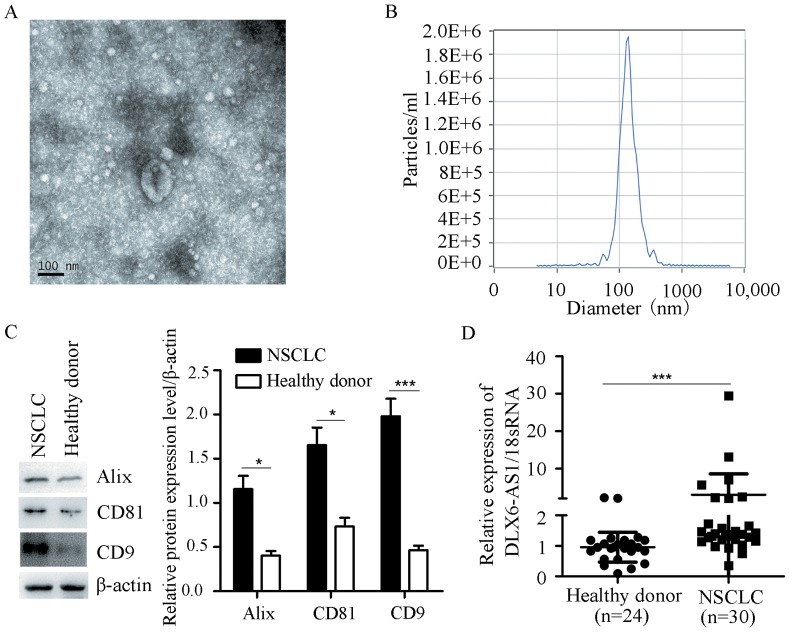
Exosomal DLX6-AS1 expression in patients with NSCLC and healthy donors
Previous studies have demonstrated that exosomes are abundantly present in the serum, and exosomal lncRNAs are more stable than free lncRNAs (34,35). Therefore, the exosomal DLX6-AS1 levels were examined in patients with NSCLC and healthy donors. The results revealed that exosomes were abundant in the serum of patients with NSCLC and healthy controls (Fig. 4A) according to electron microscopy. The concentration and size of particles were detected by NTA, and the majority of particles were distributed between 39.8 and 136 nm (Fig. 4B). Exosomal markers (CD9, CD81 and Alix) were measured by western blot analysis (Fig. 4C). In addition, the exosomal DLX6-AS1 expression was significantly increased in patients with NSCLC compared with in healthy controls (Fig. 4D).
Figure 4.
Exosomal DLX6-AS1 expression in patients with NSCLC and healthy controls. (A) Exosomes were identified by electron microscopy. Scale bar, 100 nm. (B) Concentration and sizes were measured by nanoparticle tracking analysis. (C) CD9, CD81, Alix and β-actin expression was detected by western blotting. (D) Exosomal DLX6-AS1 was measured by reverse transcription-quantitative PCR. The data were analyzed using Student's t-test. *P<0.05 and ***P<0.001. 18sRNA, 18S ribosomal RNA; DLX6-AS1, distal-less homeobox 6 antisense RNA 1; NSCLC, non-small cell lung cancer.
Diagnostic value of circulating DLX6-AS1 in patients with NSCLC
To further explore the diagnostic value of DLX6-AS1 in NSCLC, ROC curve analysis was performed (Fig. 5). The results indicated that the area under curve of DLX6-AS1 was 0.806 and the sensitivity and specificity were 0.775 and 0.859, respectively. However, the area under curve of CYFRA21-1 was 0.600. The sensitivity reached 0.606 and specificity was 0.547. These results suggested that circulating DLX6-AS1 could serve as a novel biomarker for the early diagnosis of NSCLC.
Figure 5.
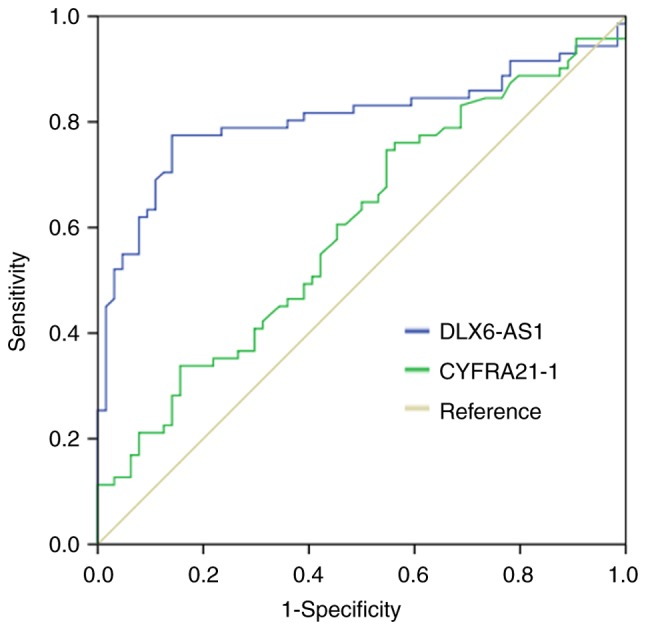
Diagnostic value of serum DLX6-AS1 in NSCLC. The diagnostic value of circulating DLX6-AS1 and CYFRA21-1 was examined by receiver operating curve analysis in NSCLC. DLX6-AS1, distal-less homeobox 6 antisense RNA 1; NSCLC, non-small cell lung cancer.
Discussion
In recent years, an increasing number of functional lncRNAs have been identified through next generation sequencing technology or gene microarrays. Accumulating evidence has demonstrated the effect of lncRNAs on cell proliferation, migration, invasion and metastasis, which can be regarded as an effective biomarker for the early diagnosis and prognosis of NSCLC (36,37). Notably, lncRNAs are abundantly present in bodily fluids, including serum, plasma, urine and saliva (35). Circulating lncRNAs have great potential for diagnosis in liquid biopsy due to being easily and non-invasively obtained from patients (35). For instance, three circulating lncRNA signatures are significantly elevated in patients with NSCLC compared with in healthy donors, demonstrating their critical role in the tumorigenesis of NSCLC (38). The present study investigated the expression of circulating DLX6-AS1 and its role in the clinical diagnosis of NSCLC.
DLX6-AS1 has been identified as an oncogene in various solid tumors, including NSCLC (15–17), renal cell carcinoma (18), hepatocellular carcinoma (19–21), colorectal cancer (22), pancreatic cancer (23,24), glioma (25) and osteosarcoma (26,27). Previous studies have demonstrated that DLX6-AS1 expression is upregulated in NSCLC (15,16). DLX6-AS1-knockdown significantly suppresses cell proliferation, migration and invasion, and promotes cell apoptosis via the miR-144/proline rich 11 signaling pathway (15), suggesting that DLX6-AS1 can be regarded as a promising target for NSCLC therapy.
The present study revealed that DLX6-AS1 was highly expressed in NSCLC, particularly in squamous cell lung carcinoma (data from GEPIA database and study cohort), which was consistent with previous studies (15–17). Patients with lower DLX6-AS1 expression exhibited a poorer prognosis than those with higher expression, since the relative expression of DLX6-AS1 in stage III+IV disease was lower than in stage I+II disease. Recent studies have demonstrated that lncRNAs are easily detected in the serum or plasma of patients and provide a non-invasive target for NSCLC (36,37). Therefore, the present study investigated the circulating DLX6-AS1 expression in NSCLC and revealed that the serum DLX6-AS1 expression was significantly higher in patients with NSCLC compared with in healthy controls. The higher expression levels of DLX6-AS1 were associated with advanced disease stage, positive lymph node metastasis and poor tumor differentiation, suggesting that DLX6-AS1 may be a promising therapeutic target for NSCLC. In addition, the serum expression of DLX6-AS1 was reduced in patients post-operatively.
An increasing number of studies have indicated that exosomes containing lncRNAs stably exist in various conditions and serve a critical role in metastasis, drug resistance and immunosuppression, providing a novel therapeutic target for NSCLC (36,39–42). Therefore, exosomal DLX6-AS1 expression in NSCLC and healthy control samples was investigated. The results indicated that exosomal DLX6-AS1 levels were significantly increased in NSCLC samples compared with in samples from healthy controls. In addition, ROC analysis results demonstrated that circulating DLX6-AS1 had a higher sensitivity and specificity for NSCLC diagnosis than CYFRA21-1. These results suggest that exosomal DLX6-AS1 may serve as a potential diagnostic marker for NSCLC. However, further investigation is required to determine the underlying mechanisms of circulating DLX6-AS1 on NSCLC progression.
Supplementary Material
Acknowledgements
The authors would like to thank Professor Qilin Shi (Department of Pathology, the First People's Hospital of Huzhou, Huzhou, China) and Professor Hui Xia (Department of Pathology, the First People's Hospital of Huzhou, Huzhou, China) for their help in pathological diagnosis.
Funding
This work was supported by grants from Zhejiang Province Science and Technology Department of Public Welfare Project (grant no. LGF18H160019), the Scientific Technology Projects of Health and Medicine of Zhejiang Province (grant nos. WKJ-ZJ-1830, 2017KY642 and 2019KY207), and Huzhou Science and Technology Fund (grant nos. 2017GY33, 2017GY32 and 2018GY04).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
DX and XW designed and conceived the study. XZ, HG and HY performed the experiments. XZ, YB and XW analyzed the data. XZ, DX and XW wrote the manuscript. All authors have read the manuscript.
Ethics approval and consent to participate
The study was undertaken on agreement of the Ethics Committee of the First Affiliated Hospital of Huzhou University (approval no. 2017014), and written informed consent was provided by all patients or healthy donors.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4.Hsu PC, Tian B, Yang YL, Wang YC, Liu S, Urisman A, Yang CT, Xu Z, Jablons DM, You L. Cucurbitacin E inhibits the Yes-associated protein signaling pathway and suppresses brain metastasis of human nonsmall cell lung cancer in a murine model. Oncol Rep. 2019;42:697–707. doi: 10.3892/or.2019.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imakita T, Matsumoto H, Hirano K, Morisawa T, Sakurai A, Kataoka Y. Impact on prognosis of rebiopsy in advanced non-small cell lung cancer patients after epidermal growth factor receptor-tyrosine kinase inhibitor treatment: A systematic review. BMC Cancer. 2019;19:105. doi: 10.1186/s12885-019-5309-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamard C, Mignard X, Pecuchet N, Mathiot N, Blons H, Laurent-Puig P, Leroy K, Lupo A, Chapron J, Giraud F, et al. IHC, FISH, CISH, NGS in non-small cell lung cancer: What changes in the biomarker era? Rev Pneumol Clin. 2018;74:327–338. doi: 10.1016/j.pneumo.2018.09.013. (In French) [DOI] [PubMed] [Google Scholar]
- 7.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang L, Li Z, Wang R. Long noncoding RNAs in lung cancer: Regulation patterns, biologic function and diagnosis implications (Review) Int J Oncol. 2019 Jul 29; doi: 10.3892/ijo.2019.4850. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim LJ, Wong SYS, Huang F, Lim S, Chong SS, Ooi LL, Kon OL, Lee CG. Roles and regulation of long non-coding RNAs in hepatocellular carcinoma. Cancer Res. 2019 Jul 23; doi: 10.1158/0008-5472.CAN-19-0255. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 10.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 11.Fatica A, Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 12.Dai SP, Jin J, Li WM. Diagnostic efficacy of long non-coding RNA in lung cancer: A systematic review and meta-analysis. Postgrad Med J. 2018;94:578–587. doi: 10.1136/postgradmedj-2018-135862. [DOI] [PubMed] [Google Scholar]
- 13.Lu T, Wang Y, Chen D, Liu J, Jiao W. Potential clinical application of lncRNAs in non-small cell lung cancer. Onco Targets Ther. 2018;11:8045–8052. doi: 10.2147/OTT.S178431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang C, Yang Y, Yang Y, Guo L, Huang J, Liu X, Wu C, Zou J. Long noncoding RNA (lncRNA) HOTAIR affects tumorigenesis and metastasis of non-small cell lung cancer by upregulating miR-613. Oncol Res. 2018;26:725–734. doi: 10.3727/096504017X15119467381615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Y, Ni R, Wang J, Liu Y. Knockdown of lncRNA DLX6-AS1 inhibits cell proliferation, migration and invasion while promotes apoptosis by downregulating PRR11 expression and upregulating miR-144 in non-small cell lung cancer. Biomed Pharmacother. 2019;109:1851–1859. doi: 10.1016/j.biopha.2018.09.151. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Li P, Zhao W, Yang R, Chen S, Bai Y, Dun S, Chen X, Du Y, Wang Y, et al. Expression of long non-coding RNA DLX6-AS1 in lung adenocarcinoma. Cancer Cell Int. 2015;15:48. doi: 10.1186/s12935-015-0201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun W, Zhang L, Yan R, Yang Y, Meng X. LncRNA DLX6-AS1 promotes the proliferation, invasion, and migration of non-small cell lung cancer cells by targeting the miR-27b-3p/GSPT1 axis. Onco Targets Ther. 2019;12:3945–3954. doi: 10.2147/OTT.S196865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng X, Hu Z, Ke X, Tang H, Wu B, Wei X, Liu Z. Long noncoding RNA DLX6-AS1 promotes renal cell carcinoma progression via miR-26a/PTEN axis. Cell Cycle. 2017;16:2212–2219. doi: 10.1080/15384101.2017.1361072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L, He X, Jin T, Gang L, Jin Z. Long non-coding RNA DLX6-AS1 aggravates hepatocellular carcinoma carcinogenesis by modulating miR-203a/MMP-2 pathway. Biomed Pharmacother. 2017;96:884–891. doi: 10.1016/j.biopha.2017.10.056. [DOI] [PubMed] [Google Scholar]
- 20.Li D, Tang X, Li M, Zheng Y. Long noncoding RNA DLX6-AS1 promotes liver cancer by increasing the expression of WEE1 via targeting miR-424-5p. J Cell Biochem. 2019;120:12290–12299. doi: 10.1002/jcb.28493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu DM, Zheng ZH, Zhang YB, Fan SH, Zhang ZF, Wang YJ, Zheng YL, Lu J. Down-regulated lncRNA DLX6-AS1 inhibits tumorigenesis through STAT3 signaling pathway by suppressing CADM1 promoter methylation in liver cancer stem cells. J Exp Clin Cancer Res. 2019;38:237. doi: 10.1186/s13046-019-1239-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Zhou FR, Pan ZP, Shen F, Huang LQ, Cui JH, Cai K, Guo XL. Long noncoding RNA DLX6-AS1 functions as a competing endogenous RNA for miR-577 to promote malignant development of colorectal cancer. Eur Rev Med Pharmacol Sci. 2019;23:3742–3748. doi: 10.26355/eurrev_201905_17800. [DOI] [PubMed] [Google Scholar]
- 23.An Y, Chen XM, Yang Y, Mo F, Jiang Y, Sun DL, Cai HH. LncRNA DLX6-AS1 promoted cancer cell proliferation and invasion by attenuating the endogenous function of miR-181b in pancreatic cancer. Cancer Cell Int. 2018;18:143. doi: 10.1186/s12935-018-0643-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J, Ye Z, Mei D, Gu H, Zhang J. Long noncoding RNA DLX6-AS1 promotes tumorigenesis by modulating miR-497-5p/FZD4/FZD6/Wnt/β-catenin pathway in pancreatic cancer. Cancer Manag Res. 2019;11:4209–4221. doi: 10.2147/CMAR.S194453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Zhang H, Wu X. Long noncoding RNA DLX6-AS1 accelerates the glioma carcinogenesis by competing endogenous sponging miR-197-5p to relieve E2F1. Gene. 2019;686:1–7. doi: 10.1016/j.gene.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 26.Zhang RM, Tang T, Yu HM, Yao XD. LncRNA DLX6-AS1/miR-129-5p/DLK1 axis aggravates stemness of osteosarcoma through Wnt signaling. Biochem Biophys Res Commun. 2018;507:260–266. doi: 10.1016/j.bbrc.2018.11.019. [DOI] [PubMed] [Google Scholar]
- 27.Zhang N, Meng X, Mei L, Zhao C, Chen W. LncRNA DLX6-AS1 promotes tumor proliferation and metastasis in osteosarcoma through modulating miR-641/HOXA9 signaling pathway. J Cell Biochem. 2019 Mar 6; doi: 10.1002/jcb.28426. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 28.Wei L, Wu W, Han L, Yu W, Du Y. A quantitative analysis of the potential biomarkers of non-small cell lung cancer by circulating cell-free DNA. Oncol Lett. 2018;16:4353–4360. doi: 10.3892/ol.2018.9198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jhun BW, Lee KJ, Jeon K, Suh GY, Chung MP, Kim H, Kwon OJ, Sun JM, Ahn JS, Ahn MJ, et al. Clinical applicability of staging small cell lung cancer according to the seventh edition of the TNM staging system. Lung Cancer. 2013;81:65–70. doi: 10.1016/j.lungcan.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 30.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X, Hu J, Zhong L, Wang N, Yang L, Liu CC, Li H, Wang X, Zhou Y, Zhang Y, et al. Quercetin stabilizes apolipoprotein E and reduces brain Aβ levels in amyloid model mice. Neuropharmacology. 2016;108:179–192. doi: 10.1016/j.neuropharm.2016.04.032. [DOI] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Yan G, Du Q, Wei X, Miozzi J, Kang C, Wang J, Han X, Pan J, Xie H, Chen J, Zhang W. Application of real-time cell electronic analysis system in modern pharmaceutical evaluation and analysis. Molecules. 2018;23:E3280. doi: 10.3390/molecules23123280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jakobsen KR, Paulsen BS, Baek R, Varming K, Sorensen BS, Jorgensen MM. Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma. J Extracell Vesicles. 2015;4:26659. doi: 10.3402/jev.v4.26659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L, Duan W, Yan S, Xie Y, Wang C. Circulating long non-coding RNA colon cancer-associated transcript 2 protected by exosome as a potential biomarker for colorectal cancer. Biomed Pharmacother. 2019;113:108758. doi: 10.1016/j.biopha.2019.108758. [DOI] [PubMed] [Google Scholar]
- 36.Li C, Lv Y, Shao C, Chen C, Zhang T, Wei Y, Fan H, Lv T, Liu H, Song Y. Tumor-derived exosomal lncRNA GAS5 as a biomarker for early-stage non-small-cell lung cancer diagnosis. J Cell Physiol. 2019;234:20721–20727. doi: 10.1002/jcp.28678. [DOI] [PubMed] [Google Scholar]
- 37.Cheng Y, Dai X, Yang T, Zhang N, Liu Z, Jiang Y. Low long noncoding RNA growth arrest-specific transcript 5 expression in the exosomes of lung cancer cells promotes tumor angiogenesis. J Oncol. 2019;2019:2476175. doi: 10.1155/2019/2476175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang Q, Ni Z, Cheng Z, Xu J, Yu H, Yin P. Three circulating long non-coding RNAs act as biomarkers for predicting NSCLC. Cell Physiol Biochem. 2015;37:1002–1009. doi: 10.1159/000430226. [DOI] [PubMed] [Google Scholar]
- 39.Zhang R, Xia Y, Wang Z, Zheng J, Chen Y, Li X, Wang Y, Ming H. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem Biophys Res Commun. 2017;490:406–414. doi: 10.1016/j.bbrc.2017.06.055. [DOI] [PubMed] [Google Scholar]
- 40.Zhang W, Cai X, Yu J, Lu X, Qian Q, Qian W. Exosome-mediated transfer of lncRNA RP11838N2.4 promotes erlotinib resistance in non-small cell lung cancer. Int J Oncol. 2018;53:527–538. doi: 10.3892/ijo.2018.4412. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Lei Y, Guo W, Chen B, Chen L, Gong J, Li W. Tumor-released lncRNA H19 promotes gefitinib resistance via packaging into exosomes in nonsmall cell lung cancer. Oncol Rep. 2018;40:3438–3446. doi: 10.3892/or.2018.6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mathieu M, Martin-Jaular L, Lavieu G, Théry C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. 2019;21:9–17. doi: 10.1038/s41556-018-0250-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.