Abstract
Peptidylprolyl isomerase A (PPIA) has been reported to be correlated with cancer. The present study investigated the prognostic values of PPIA expression levels in cancer by comparing different types of cancer using databases. High expression levels of PPIA were observed in 17 out of 17 cancer types compared with normal adjacent tissues. High expression levels of PPIA were associated with decreased overall survival in low grade glioma, acute myeloid leukemia, lung adenocarcinoma, skin cutaneous melanoma and liver hepatocellular carcinoma (LIHC). The prognostic effect of PPIA expression in LIHC was independent of tumor grade. High expression levels of PPIA were of particular prognostic value in stage 3, American Joint Committee on Cancer Tumor 3, hepatitis B virus negative and sorafenib-administered subgroups in LIHC. The expression level of PPIA was significantly associated with levels of basigin and signal transducer and activator of transcription 3, which may be major effectors of PPIA in the progression of the cancer.
Keywords: liver hepatocellular carcinoma, low grade glioma, acute myeloid leukemia, lung adenocarcinoma, skin cutaneous melanoma, peptidyl-prolyl cis-trans isomerase A, basigin, signal transducer and activator of transcription 3, prognostic factor
Introduction
Primary liver cancer is the sixth most frequent cancer (6%) and the second leading cause of cancer-associated mortality (9%) worldwide (1). Liver hepatocellular carcinoma (LIHC) accounts for 75% of all primary liver cancers (2). The therapy for LIHC has focused primarily on traditional resection and recurrence rates following resection can exceed 70% (3). Sorafenib, a drug that inhibits the activity of tyrosine protein kinases, was recently approved for use in patients and could improve the overall survival (OS) of patients with advanced LIHC from 7.9 to 10.7 months (4). Thus, identifying novel therapeutic targets is urgent for LIHC.
Peptidylprolyl isomerase A (PPIA), also known as cyclophilin A or rotamase A, is an enzyme encoded by the PPIA gene on chromosome 7 (5,6). The PPIA protein catalyzes the cis-trans isomerization of proline imidic peptide bonds, which allows it to regulate a number of biological processes, including intracellular signaling, transcription, inflammation and apoptosis (7–10). PPIA has been reported to interact with molecules that could impact on various pathological processes (11). PPIA could inhibit the activity of interleukin-2 tyrosine kinase (ITK), leading to decreased activation of T cells (12). Basigin (BSG), also known as extracellular matrix metalloproteinase inducer or cluster of differentiation 147 (CD147), which is a member of the immunoglobulin superfamily (13–15), plays fundamental roles in the intercellular recognition involved in various immunological phenomena, including differentiation and development (16). Activation of CD147 receptor with recombinant human PPIA could stimulate the phosphorylation of extracellular signal-regulated kinase 1/2, c-jun NH2-terminal kinase, p38 kinase, protein kinase B and IKB (17–19). PPIA/CD147 signaling has been reported to lead to cell migration, proliferation and differentiation (20–22). PPIA has been reported to interact with signal transducer and activator of transcription 3 (STAT3) in myeloma cells (23). PPIA has been implicated in a broad range of pathological processes, including inflammatory diseases, aging and the progression of cancer metastasis (11). Previous studies have demonstrated that overexpression of PPIA plays key roles in different types of cancer, including hepatocellular carcinoma, lung cancer, pancreatic cancer, breast cancer, colorectal cancer, squamous cell carcinoma and melanoma (24,25). These results were primarily based on laboratory studies; therefore the question remains unanswered as to whether PPIA could serve as a prognostic factor and potential target for therapy in the future. Former studies have suggested that the differences in steady-state between protein levels are largely explained by the variations in transcript concentrations, which means that by examining the different RNA concentrations of the genes, the correlated protein levels of these genes may be identified (26–30). With this hypothesis, the present study analyzed the associations between PPIA expression levels and the survival of different populations with cancer in database platforms such as Oncomine, PrognoScan and UALCAN. In addition, as there have been studies demonstrating the associations between patients' survival and immune cell infiltration in the tumor microenvironment, the present study also investigated the association between immune infiltrates in the tumor microenvironment and PPIA expression levels, with the aim of identifying a link between PPIA expression and immune cell infiltration. The present study aimed to elucidate the association between PPIA expression and cancer prognoses. The associations between the transcription levels of ITK, BSG, STAT3 and PPIA were investigated in order to identify the underlying molecular mechanisms.
Materials and methods
Analysis using the Oncomine database
Oncomine (https://www.oncomine.org/resource/login.html) is a web-based online database containing 715 datasets and 86,733 samples (31,32). This platform provides a powerful set of analysis functions that calculate gene expression signatures, clusters and gene-set modules, automatically extracting biological insights from the data. The expression level of PPIA in different types of cancer was obtained from the Oncomine database. The threshold values were P=0.001, fold-change=1.5.
Analysis in the PrognoScan database
The correlation between the expression level of PPIA and survival in cancer was analyzed using the PrognoScan database (33) (http://www.abren.net/PrognoScan). This database was based on publicly available cancer datasets, which provides Kaplan-Meier plots of gene transcription and survival time in accordance with individual datasets (32). The threshold values were corrected P=0.05 and Cox P=0.05.
Analysis in UALCAN
UALCAN (http://ualcan.path.uab.edu/index.html) provides graphs and plots depicting gene expression and patient survival information for different types of cancer (34). The association between the expression level of PPIA and survival for the different types of cancer that were filtered by PrognoScan was analyzed using UALCAN. In addition, the present study also investigated the types of cancer not included in the PrognoScan, such as liver cancer. The threshold value was P=0.05.
Kaplan-Meier plotter database analysis
A Kaplan-Meier plotter can assess the effect of 54,675 genes on survival using 10,461 cancer samples. The association between PPIA expression and survival in liver cancer was analyzed using the Kaplan-Meier Plotter at (http://kmplot.com/analysis) (35).
Analysis in Tumor IMmune Estimation Resource (TIMER)
TIMER is a resource for the analysis of immune cell infiltrates in the microenvironment of various different types of cancer (36,37) (https://cistrome.shinyapps.io/timer). TIMER applies the deconvolution method to infer levels of tumor-infiltrating immune cells including B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages and dendritic cells from gene expression profiles (32). TIMER also identifies the association between tumor purity and expression level of the input gene. Those genes that are highly expressed in the microenvironment are expected to demonstrate negative associations with tumor purity, while the opposite is expected for genes that are highly expressed in the tumor cells. The present study analyzed the association between PPIA expression with levels of immune infiltrates, as well as the associations between expression levels of PPIA and genes, of which the proteins were reported to interact with PPIA. By default, TIMER currently only provides information regarding the associations between PPIA expression and B cells, CD4+ T cells, CD8+ T cells, macrophages, neutrophils and dendritic cells. In addition to analyzing the associations between PPIA expression and the aforementioned cell types, the present study also investigated the associations between the immunomarkers of other immune cells, including natural killer cells, Treg cells and T cell exhaustion, and the expression levels of PPIA in order to gain an accurate representation of these immune cells and PPIA expression. The biomarkers for natural killer cells were killer cell immunoglobulin-like receptor (KIR)3DL1, KIR2DS4, KIR2DL1, KIR3DL2, KIR2DL3, natural cytotoxicity triggering receptor 2 and CD244 (38–42). The biomarker selected for regulatory T cells was forkhead box protein P3 (43–45). PDCD1 and interleukin (IL)10 were selected as biomarkers for T cell exhaustion (46).
Statistical analysis
Survival curves were generated using UALCAN and Kaplan-Meier Plotter. The results generated by Oncomine are presented with P-values, fold-changes and ranks. The threshold values of PrognoScan were corrected P=0.05 and Cox P=0.05. The results of the UALCAN and Kaplan-Meier Plotter analyses are presented with the hazard ratio (HR) and P-value, or Cox P-values from a log-rank test. The correlations in gene expression were assessed using Spearman's correlation tests to determine statistical significance and the strength of the correlation was determined by following a guide for the absolute value: 0.00–0.19, very weak; 0.20–0.39, weak; 0.40–0.59, moderate; 0.60–0.79, strong and 0.80–1.00, very strong. P<0.05 was considered to indicate a statistically significant difference.
Results
Transcription levels of PPIA in cancer
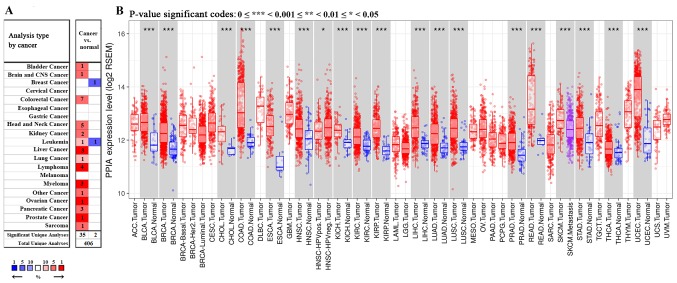
Among the 20 assigned cancer types compared with normal tissues in the present study, under the criteria of a fold-change of 1.5, PPIA transcription levels were upregulated in 35 datasets with 15 different types of cancer, including bladder cancer, brain cancer, colorectal cancer, head and neck cancer, kidney cancer, leukemia, liver cancer, lung cancer, lymphoma, myeloma, ovarian cancer, pancreatic cancer, prostate cancer and sarcoma. The PPIA transcription levels were downregulated in 2 datasets with 2 different types of cancer, including breast cancer and leukemia in the Oncomine database. In particular, PPIA was included in the top 1% of those upregulated in the five cancer types, including liver cancer, lymphoma, myeloma, ovarian cancer and prostate cancer (Fig. 1A). Further consultation in TIMER based on The cancer genome atlas (TCGA) demonstrated that the PPIA transcription levels were all significantly upregulated in the 17 different types of cancer available, including bladder cancer, breast cancer, bile duct carcinoma, colorectal cancer, esophageal carcinoma, head and neck cancer, kidney chromophobe renal cell carcinoma, kidney renal clear cell carcinoma, kidney renal papillary cell carcinoma, liver hepatocellular carcinoma, lung adenocarcinoma, lung squamous cell carcinoma, prostate adenocarcinoma, rectum adenocarcinoma, stomach adenocarcinoma, thyroid carcinoma and uterine corpus endometrial carcinoma, compared with normal adjacent tissue (P<0.001; Fig. 1B).
Figure 1.
PPIA transcription levels in cancer tissues compared with normal tissues. (A) Comparison outcomes from datasets of Oncomine. PPIA was included in the top 1% of those upregulated in the five cancer types, including liver cancer, lymphoma, myeloma, ovarian cancer and prostate cancer. (B) The PPIA transcription levels were all upregulated in the 17 different types of cancer available showed by TIMER. *P<0.05, ***P<0.001. PPIA, peptidyl-prolyl cis-trans isomerase A; TIMER, Tumor IMmune Estimation Resource.
Prognostic potential of PPIA expression levels in cancer
The present study used PrognoScan to identify the association between PPIA expression levels and cancer prognoses. The survival rates of patients with lung cancer, breast cancer, colorectal cancer, brain cancer, ovarian cancer, skin cancer and blood cancer were associated with PPIA transcription level in various datasets (Table I). Overexpression of PPIA was associated with poor relapse free survival of lung adenocarcinoma in datasets GSE32210 and GSE8894 using various primers, and poor OS in datasets GSE32210, GSE13213, jacob-00182-UM and GSE13213. The OS in patients with squamous cell carcinoma of the lung was also associated with PPIA overexpression, as presented in Table I in dataset GSE4573 (HR, 5.57). In breast cancer, there was a certain level of contradiction, although the OS was revealed to be positively correlated with PPIA expression level (HR, 0.83) in dataset GSE9893, the other survival values of breast cancer were negatively correlated with PPIA expression, such as disease free survival in GSE4922-GPL96 (HR, 6.99 and 6.07 with different primers), relapse free survival in GSE1456-GPL96 (HR, 7.41 and 7.12 with different primers), distant metastasis free survival in GSE11121, GSE9195 and GSE2990, and disease specific survival in GSE3494-GPL96 and GSE1456-GPL96. These results indicated that overexpression of PPIA was a risk factor for breast cancer progression and metastasis, and may be harnessed as a therapeutic target. The overexpression of PPIA was associated with the survival rates of patients with brain cancer (HR of OS, 8.47 and 7.34 with different primers), skin cancer (HR of OS, 209.26 and 151.59 with different primers) and multiple myeloma (HR of disease specific survival, 2.29; Table I). The only type of cancer where the survival values were coherently negatively correlated with PPIA expression was ovarian cancer (HR, 0.20 for OS; HR, 0.29 for disease free survival in dataset GSE26712) as presented in Table I. The data of the different primers in colorectal cancer from within the same dataset, GSE17537, were inconsistent, suggesting additional datasets were required.
Table I.
Prognostic value of PPIA mRNA expression in different datasets determined by PrognoScan.
| Dataset | Cancer type | Endpoint | Corrected P-value | COX P-value | HR (95% CI) |
|---|---|---|---|---|---|
| GSE31210 | LUAD | Relapse free survival | 1.58×10−10 | 2.98×10−11 | 455.47 (74.93–2768.73) |
| GSE31210 | LUAD | Relapse free survival | 1.36×10−10 | 4.44×10−11 | 395.74 (66.79–2344.72) |
| GSE31210 | LUAD | Overall survival | 2.10×10−06 | 1.94×10−05 | 191.57 (17.18–2136.01) |
| GSE31210 | LUAD | Overall survival | 4.05×10−05 | 3.09×10−05 | 159.62 (14.68–1735.14) |
| GSE9893 | BRCA | Overall survival | 0.001131 | 0.000161 | 0.83 (0.75–0.91) |
| GSE13213 | LUAD | Overall survival | 0.001964 | 0.000172 | 2.55 (1.56–4.15) |
| GSE13213 | LUAD | Overall survival | 0.001602 | 0.000288 | 2.46 (1.51–3.99) |
| GSE4922-GPL96 | BRCA | Disease free survival | 0.000787 | 0.00043 | 6.99 (2.37–20.65) |
| GSE8894 | NSCLC | Relapse free survival | 0.011737 | 0.000637 | 3.50 (1.71–7.19) |
| GSE1456-GPL96 | BRCA | Relapse free survival | 0.013126 | 0.000821 | 7.41 (2.29–23.98) |
| GSE8894 | NSCLC | Relapse free survival | 0.020635 | 0.000857 | 3.36 (1.65–6.85) |
| GSE1456-GPL96 | BRCA | Relapse free survival | 0.006499 | 0.001082 | 7.12 (2.19–23.09) |
| GSE4922-GPL96 | BRCA | Disease Free survival | 0.002932 | 0.002276 | 6.07 (1.91–19.34) |
| jacob-00182-UM | LUAD | Overall survival | 0.041268 | 0.002379 | 0.46 (0.28–0.76) |
| GSE11121 | BRCA | Distant metastasis free survival | 0.01496 | 0.003358 | 17.39 (2.58–117.31) |
| GSE17537 | COADREAD | Disease free survival | 0.008844 | 0.004248 | 0.01 (0.00–0.20) |
| GSE9195 | BRCA | Distant metastasis free survival | 0.008393 | 0.004407 | 29.78 (2.88–307.85) |
| GSE26712 | OV | Overall survival | 0.004054 | 0.004925 | 0.20 (0.06–0.61) |
| GSE4412-GPL96 | LGG | Overall survival | 0.036877 | 0.004997 | 8.47 (1.91–37.66) |
| GSE9195 | BRCA | Distant metastasis free survival | 0.024659 | 0.00592 | 26.26 (2.56–269.20) |
| GSE3494-GPL96 | BRCA | Disease specific survival | 0.047207 | 0.008505 | 6.92 (1.64–29.27) |
| GSE3494-GPL96 | BRCA | Disease specific survival | 0.002599 | 0.009594 | 5.69 (1.53–21.20) |
| GSE4412-GPL96 | LGG | Overall survival | 0.000433 | 0.009663 | 7.34 (1.62–33.25) |
| GSE4573 | LUSD | Overall survival | 0.0207 | 0.010494 | 5.57 (1.49–20.73) |
| GSE1456-GPL96 | BRCA | Disease specific survival | 0.006336 | 0.013111 | 5.75 (1.44–22.92) |
| GSE2658 | Multiple myeloma | Disease specific survival | 0.034132 | 0.013804 | 2.29 (1.18–4.44) |
| GSE13213 | LUAD | Overall survival | 0.012258 | 0.014126 | 2.17 (1.17–4.02) |
| GSE26712 | OV | Disease free survival | 0.008361 | 0.016461 | 0.29 (0.11–0.80) |
| GSE13213 | LUAD | Overall survival | 0.021354 | 0.018883 | 1.94 (1.12–3.37) |
| GSE19234 | SKCM | Overall survival | 0.002576 | 0.019303 | 209.26 (2.38–18399.93) |
| GSE13213 | LUAD | Overall survival | 0.012258 | 0.022418 | 2.07 (1.11–3.86) |
| GSE19234 | SKCM | Overall survival | 0.002576 | 0.023501 | 151.59 (1.97–11682.11) |
| GSE17537 | COADREAD | Disease free survival | 0.006344 | 0.024266 | 20.40 (1.48–281.27) |
| GSE17537 | COADREAD | Disease free survival | 0.009107 | 0.026693 | 18.60 (1.40–246.70) |
| GSE13213 | LUAD | Overall survival | 0.011179 | 0.031871 | 1.83 (1.05–3.17) |
| GSE2990 | BRCA | Distant metastasis free survival | 0.030163 | 0.044086 | 5.10 (1.04–24.91) |
LUAD, lung adenocarcinoma; BRCA, breast invasive carcinoma; NSCLC, non-small cell lung cancer; COADREAD, colorectal cancer; OV, ovarian serous cystadenocarcinoma; LGG, brain lower grade glioma; SKCM, skin cutaneous melanoma; HR, hazard ratio; CI, confidence interval.
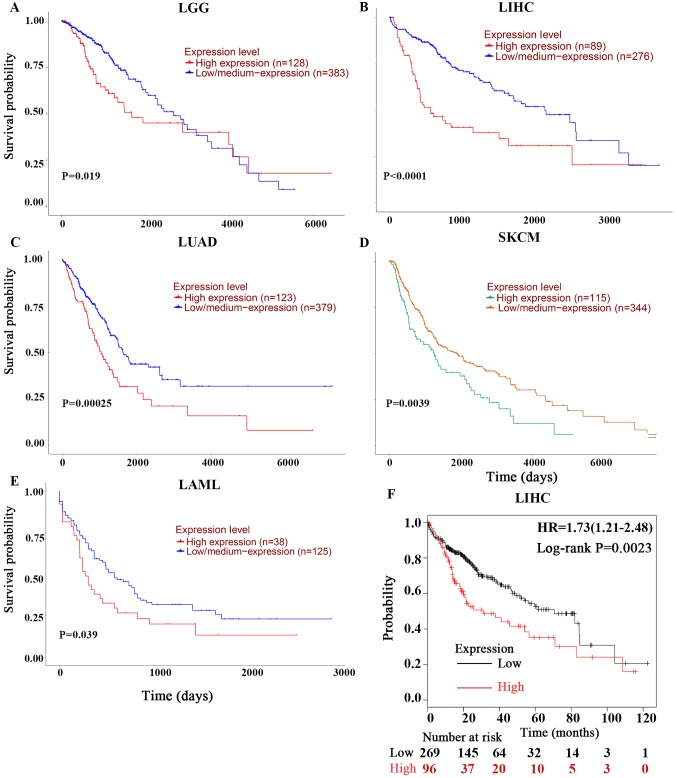
As these outcomes were obtained from individual datasets for each type of cancer, the present study used UALCAN for further investigation into the prognostic potential of PPIA based on the type of cancer. Poor OS of low-grade glioma (LGG), acute myeloid leukemia (LMAL), lung adenocarcinoma (LUAD) and skin cutaneous melanoma (SKCM) were revealed to be associated with PPIA overexpression with P-values of 0.019, 0.00025, 0.0039 and 0.039, respectively (Fig. 2A, C, D and E). Given the lack of LIHC datasets in PrognoScan and the early indication of the PPIA-LIHC association, the present study also investigated the LIHC data in UALCAN and Kaplan-Meier Plotter. It was revealed that overexpression of PPIA was also significantly correlated with poor OS of LIHC (P<0.0001; Fig. 2B and F).
Figure 2.
Kaplan-Meier OS curves of cancer patients comparing PPIA high and low expression levels by UALCAN and Kaplan-Meier Plotter. The OS curves of (A) LGG, (B) LIHC, (C) LUAD, (D) SKCM and (E) LMAL derived from UALCAN. (F) was derived from Kaplan-Meier Plotter. OS, overall survival; PPIA, peptidyl-prolyl cis-trans isomerase A; LGG, brain lower grade glioma; LUAD, lung adenocarcinoma; SKCM, skin cutaneous melanoma; LIHC, liver hepatocellular carcinoma; LMAL, acute myeloid leukaemia.
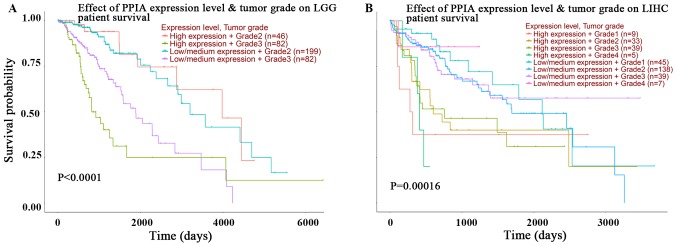
In order to improve the current understanding of the prognostic effects of PPIA transcription, plots assessing tumor grade and PPIA transcription level in LGG and LIHC were investigated using UALCAN. The expression level of PPIA was an independent and more effective predictor of OS than tumor grade in LIHC, but not in LGG (Fig. 3A and B). The prognoses of tumor grade 1, 2, 3 and 4 in those with high PPIA expression levels were all poorer than the prognoses of respective tumor grades with low or median PPIA expression levels in LIHC. The prognosis of low or median PPIA expression in every tumor grade was better than the prognosis of high PPIA expression in every tumor grade, as presented in Fig. 3B. Even the prognosis of grade 4 tumors with low or median expression levels of PPIA was better than the prognosis of grade 1 tumors with high PPIA expression levels, indicating that the PPIA expression level was an independent prognostic factor and that the prognostic value was more precise than the prognostic value of tumor grade in LIHC (Fig. 3B).
Figure 3.
Kaplan-Meier OS curves of different grade cancer patients comparing PPIA high and low expression levels by UALCAN. The OS curves of different grades and PPIA expression levels of (A) LIHC and (B) LGG respectively. OS, overall survival; LGG, brain lower grade glioma; LIHC, liver hepatocellular carcinoma; PPIA, peptidyl-prolyl cis-trans isomerase A.
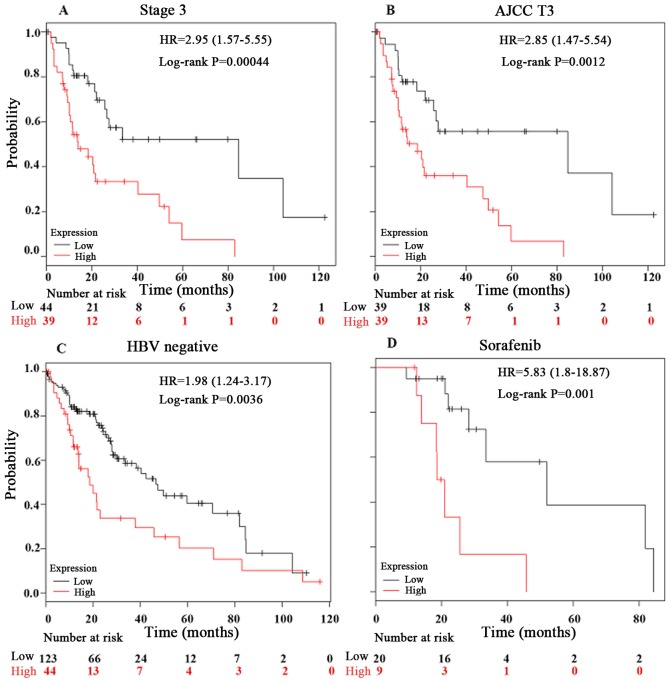
Furthermore, the present study investigated the prognostic effect of PPIA expression in LIHC in detail using a Kaplan-Meier Plotter. In the hepatitis B virus (HBV)-free, AJCC T3, sorafenib-administered and stage 3 tumor groups, the PPIA expression level was a more robust method of predicting prognosis, with log rank P-values of 0.0036, 0.0012, 0.001 and 0.00044, respectively (Fig. 4). The prognostic value of PPIA overexpression in stage 3 and AJCC T3 groups implied that PPIA played a specific role in advanced LIHC, and may possess therapeutic value in advanced LIHC. The prognostic value of PPIA in the sorafenib-administered group also indicated that PPIA served a specific role in LIHC progression in the poorer OS subgroup of the sorafenib-administered group, which meant that PPIA could serve as a therapeutic target specifically for sorafenib-administered patients with LIHC.
Figure 4.
Kaplan-Meier OS curves of LIHC patients in different conditions comparing peptidyl-prolyl cis-trans isomerase A high and low expression levels by Kaplan-Meier Plotter. The liver hepatocellular carcinoma OS curves of (A) stage 3, (B) AJCC T3, (C) HBV negative and (D) sorafenib-administered subgroups respectively. OS, overall survival; HBV, hepatitis B virus; HR, hazard ratio.
Association between PPIA expression levels and the immune cell infiltration levels in cancer
The association between PPIA expression levels and immune cell infiltration levels in the tumor microenvironment was investigated using TIMER in the present study. The purity of each solid tumor tested was not associated with PPIA expression level, suggesting that PPIA was not highly expressed by immune cells in the tumor microenvironment, but was more likely to be overexpressed in cancer cells (Table II). However, certain associations were observed between PPIA expression level and immune cell infiltration levels, including CD4+ T cells and macrophages, in four solid tumors, particularly in LUAD and SKCM (Table II). By default, TIMER can currently only provide the information regarding correlations between PPIA expression and B cells, CD4+ T cells, CD8+ T cells, macrophages, neutrophils and dendritic cells. In addition to identifying the correlations between PPIA expression and the aforementioned cell types, the present study also investigated the immunomarkers of other immune cells, including natural killer cells, Treg cells, T cell exhaustion and the expression level of PPIA in order to gain an accurate representation of the associations between these immune cells and PPIA expression. The associations between NK cells, Treg cells, T cell exhaustion and PPIA expression were weak, with no biomarker observed as uniformly associated with PPIA expression level (Table III).
Table II.
Correlations between immune infiltrating cells and peptidyl-prolyl cis-trans isomerase A transcription level.
| Cancer | Cell types | Correlation coefficient | P-value | Statistic difference | Correlation strength |
|---|---|---|---|---|---|
| LGG | Purity | −0.00865 | 0.850256 | No | Very weak |
| B Cell | −0.08926 | 0.051135 | No | Very weak | |
| CD8+ T cell | −0.03738 | 0.414905 | No | Very weak | |
| CD4+ T cell | −0.16085 | 0.000426 | Yes | Very weak | |
| Macrophage | −0.19021 | 3.13×10−05 | Yes | Very weak | |
| Neutrophil | −0.09131 | 0.04671 | Yes | Very weak | |
| Dendritic cell | −0.05931 | 0.196445 | No | Very weak | |
| LIHC | Purity | −0.02851 | 0.59712 | No | Very weak |
| B Cell | −0.04497 | 0.405739 | No | Very weak | |
| CD8+ T cell | −0.02699 | 0.618908 | No | Very weak | |
| CD4+ T cell | −0.21739 | 4.78×10−05 | Yes | Weak | |
| Macrophage | −0.06422 | 0.236874 | No | Very weak | |
| Neutrophil | −0.18394 | 0.000596 | Yes | Very weak | |
| Dendritic cell | −0.1004 | 0.064432 | No | Very weak | |
| LUAD | Purity | 0.090959 | 0.043308 | Yes | Very weak |
| B Cell | −0.27619 | 6.36×10−10 | Yes | Weak | |
| CD8+ T cell | −0.12476 | 0.005834 | Yes | Very weak | |
| CD4+ T cell | −0.38104 | 3.58×10−18 | Yes | Weak | |
| Macrophage | −0.25887 | 7.24×10−09 | Yes | Weak | |
| Neutrophil | −0.24369 | 5.84×10−08 | Yes | Weak | |
| Dendritic cell | −0.21245 | 2.19×10−06 | Yes | Weak | |
| SKCM | Purity | 0.038591 | 0.409982 | No | Very weak |
| B Cell | −0.13361 | 0.004615 | Yes | Very weak | |
| CD8+ T cell | −0.3377 | 3.82×10−13 | Yes | Weak | |
| CD4+ T cell | −0.33597 | 3.14×10−13 | Yes | Weak | |
| Macrophage | −0.36791 | 5.75×10−16 | Yes | Weak | |
| Neutrophil | −0.46714 | 7.02×10−26 | Yes | Moderate | |
| Dendritic cell | −0.34575 | 5.73×10−14 | Yes | Weak |
CD, cluster of differentiation; LGG, brain lower grade glioma; LUAD, lung adenocarcinoma; SKCM, skin cutaneous melanoma; LIHC, liver hepatocellular carcinoma.
Table III.
Correlation between transcription levels of immune markers and peptidyl-prolyl cis-trans isomerase A.
| Cancer type | Cell type | Genes | Correlation | P-value | Correlation strength |
|---|---|---|---|---|---|
| LGG | NK | KIR3DL1 | 0.0015 | 0.97385 | Very weak |
| KIR2DS4 | 0.0112 | 0.80683 | Very weak | ||
| KIR2DL1 | 0.0820 | 0.07294 | Very weak | ||
| KIR3DL2 | 0.0208 | 0.64963 | Very weak | ||
| KIR2DL3 | 0.0412 | 0.36846 | Very weak | ||
| NCR2 | −0.0379 | 0.40798 | Very weak | ||
| CD244 | 0.1050 | 0.02149 | Very weak | ||
| Treg | FOXP3 | −0.0679 | 0.13784 | Very weak | |
| T cell exhaustion | PDCD1 | 0.0772 | 0.09136 | Very weak | |
| IL10 | −0.0735 | 0.10823 | Very weak | ||
| LIHC | NK | KIR3DL1 | −0.1743 | 0.00113 | Very weak |
| KIR2DS4 | −0.0291 | 0.58918 | Very weak | ||
| KIR2DL1 | 0.0016 | 0.97592 | Very weak | ||
| KIR3DL2 | −0.0103 | 0.84931 | Very weak | ||
| KIR2DL3 | −0.0264 | 0.62414 | Very weak | ||
| NCR2 | 0.0622 | 0.24882 | Very weak | ||
| CD244 | 0.0603 | 0.26366 | Very weak | ||
| Treg | FOXP3 | −0.2146 | 5.72×10−05 | Weak | |
| T cell exhaustion | PDCD1 | 0.0626 | 0.24587 | Very weak | |
| IL10 | 0.0189 | 0.72558 | Very weak | ||
| LUAD | NK | KIR3DL1 | −0.0965 | 0.03209 | Very weak |
| KIR2DS4 | −0.0593 | 0.18822 | Very weak | ||
| KIR2DL1 | −0.0846 | 0.06032 | Very weak | ||
| KIR3DL2 | −0.0348 | 0.44042 | Very weak | ||
| KIR2DL3 | −0.0415 | 0.35798 | Very weak | ||
| NCR2 | −0.0189 | 0.67590 | Very weak | ||
| CD244 | −0.0532 | 0.23789 | Very weak | ||
| Treg | FOXP3 | −0.0824 | 0.06729 | Very weak | |
| T cell exhaustion | PDCD1 | −0.0166 | 0.71271 | Very weak | |
| IL10 | −0.0876 | 0.05174 | Very weak | ||
| SKCM | NK | KIR3DL1 | −0.0635 | 0.17510 | Very weak |
| KIR2DS4 | −0.0132 | 0.77886 | Very weak | ||
| KIR2DL1 | −0.0452 | 0.33463 | Very weak | ||
| KIR3DL2 | −0.1534 | 0.00099 | Very weak | ||
| KIR2DL3 | −0.1012 | 0.03030 | Very weak | ||
| NCR2 | −0.0941 | 0.04414 | Very weak | ||
| CD244 | −0.1413 | 0.00244 | Very weak | ||
| Treg | FOXP3 | −0.0960 | 0.04015 | Very weak | |
| T cell exhaustion | PDCD1 | −0.1698 | 0.00027 | Very weak | |
| IL10 | −0.3034 | 3.33×10−11 | Weak |
LGG, brain lower grade glioma; LUAD, lung adenocarcinoma; SKCM, skin cutaneous melanoma; LIHC, liver hepatocellular carcinoma; NK, natural killer; IL, interleukin; FOXP3, forkhead box protein P3; CD, cluster of differentiation; NCR, natural cytotoxicity triggering receptor 2; KIR, killer cell immunoglobulin-like receptor; Treg, regulatory T cell. The bold print is used to highlight stronger than ‘very weak’ correlations (absolute correlation value, 0.00–0.19) in the ‘Correlation’ row and statistically significant values (P<0.05) in the ‘P-value’ row.
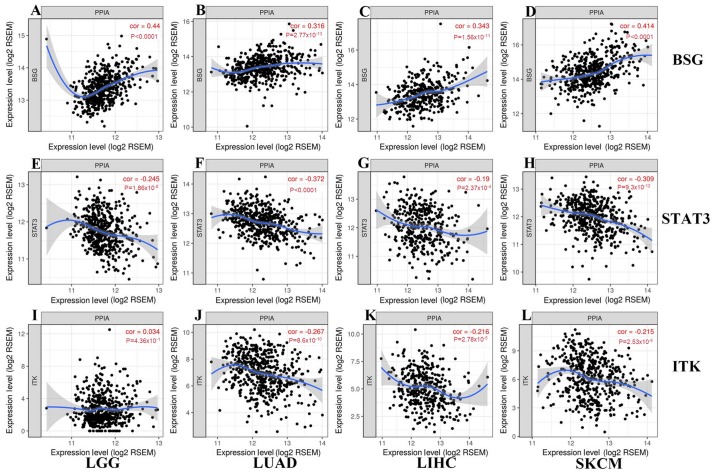
PPIA expression level is correlated with BSG and STAT3 expression levels
BSG, STAT3 and ITK were reported to interact with PPIA on the protein level, and so the present study further investigated the associations between the expression levels of PPIA and these genes. BSG and STAT3 were correlated with PPIA in each cancer type, with a positive correlation for BSG, and a negative correlation for STAT3 (Fig. 5). The expression levels of ITK were negatively correlated with PPIA in LIHC, LUAD and SKCM, but not LGG (Fig. 5).
Figure 5.
Correlations between transcription levels of BSG, STAT3, ITK and PPIA in each cancer type generated from TIMER. As shown in (A-D) and (E-H), BSG and STAT3 correlated with PPIA in each cancer type, with a (A-D) positive correlation for BSG, and a (E-H) negative correlation for STAT3. As shown in (J-L) and (I), the expression levels of ITK were negatively correlated with PPIA in (K) LIHC, (J) LUAD and (L) SKCM, but not (I) LGG. LGG, brain lower grade glioma; LIHC, liver hepatocellular carcinoma; STAT3, signal transducer and activator of transcription 3; TIMER, Tumor IMmune Estimation Resource; BSG, basigin; PPIA, peptidyl-prolyl cis-trans isomerase A; ITK, interleukin-2 tyrosine kinase; LUAD, lung adenocarcinoma; SKCM, skin cutaneous melanoma.
Discussion
In the present study, the expression level of PPIA was observed to be associated with the survival of patients with different types of cancer by narrowing down cancer types in various databases. The OS of patients with LGG, LMAL, LUAD, SKCM and LIHC were associated with PPIA expression level. The OS of LIHC was revealed to be highly associated with PPIA expression and the prognostic effect was independent of tumor grade. The disease free survival, relapse free survival, distant metastasis free survival and disease specific survival of breast cancer were also negatively associated with PPIA expression level.
Sorafenib was reported to improve the OS of inoperable patients with LIHC (HR, 0.69), although PFS was not improved (4). The OS of patients with high PPIA expression levels was significantly decreased compared with patients with low PPIA expression levels in the sorafenib-administered group. This indicated that PPIA played a specific role in the progression of LIHC in the poorer OS subgroup of the sorafenib-administered group and that inhibition of PPIA expression or PPIA inhibitor ciclosporin A may be beneficial for sorafenib-administered patients. The association between PPIA transcription level and OS of LIHC groups of sorafenib-administered, HBV-free, AJCC T3 and stage 3 tumors was more profound, suggesting that the high value of PPIA expression level could act as a prognostic factor and demonstrates the potential of PPIA as a new therapeutic target for advanced LIHC in these settings.
There was no negative correlation observed between tumor purity levels and the expression levels of PPIA in the different types of cancer, suggesting that PPIA was not highly expressed in immune cells in the tumor microenvironment, but was more likely to be highly expressed in cancer cells. Notably, in the cancer types with more positive correlations between tumor purity and PPIA expression, the negative correlations between immune cells and PPIA expression levels tended to be more visible, which also implied that the PPIA expression upregulation occurred in cancer cells rather than background immune cells. PPIA has been reported to be ubiquitously expressed in all 27 normal tissues tested (47). Furthermore, PPIA could be secreted from cancer cells, such as head and neck squamous cell carcinoma and HBV-positive hepatoma cell spontaneously, or from epithelial breast cancer cells by irradiation (48–50).
PPIA was reported to interact with BSG, which could lead to decreased activity and proliferation of lymphocytes (19,51,52), causing immune inhibition of cancer cells. Studies by Lu et al (53) and de la Iglesia et al (54) demonstrated that BSG overexpression was highly associated with poor prognosis of lung adenocarcinoma and breast cancer by stimulating the production of matrix metalloproteinases. Zhao et al (55) demonstrated that downregulation of BSG induced malignant melanoma cell apoptosis via the regulation of IGFBP2 expression. Lu et al (53) revealed that basolateral CD147 induced hepatocyte polarity loss by E-cadherin ubiquitination and degradation in the progression of hepatocellular carcinoma. Consistent with previous studies (19,51,52), the present study demonstrated that high PPIA expression levels in different types of cancer were positively correlated with high BSG expression and negatively correlated with high immune cell infiltrates, which could be a reason for the lower OS compared with patients with low PPIA expression levels.
Previous studies indicated that STAT3 interacts with PPIA (23), suppressing PTEN loss-induced malignant cell transformation in astrocytes (54) and impairing invasiveness of intestinal tumors (56,57). The results of the present study revealed an inferior OS rate in the high PPIA and low STAT3 expression groups coincided with the results of the aforementioned studies.
ITK is a protein tyrosine kinase that participates in the intracellular signaling events leading to T cell activation and it is highly expressed in T cells (58). The activity of ITK could be silenced by PPIA (12). The present study demonstrated that the PPIA expression levels was negatively correlated with CD4+ T cell counts in three types of cancer tissue, with LGG excluded, which, according to previous studies (12,58), means that the expression levels of PPIA and ITK should be negatively correlated. ITK expression was negatively correlated with PPIA expression in three different types of cancer, with the exception of LGG. Further investigations into the literature revealed that glioblastoma and other types of cancer introduced into the intracranial compartment could harbor AIDS-level CD4 counts, which may be due to the sequestration of T cells caused by S1P1 loss from the T cell surface (59). A similar phenomenon occurred in immune cell infiltration, with CD4+ T cells being negatively correlated with PPIA expression at a weak level in three other types of cancer and the association with LGG was demonstrated to be very weak, with a correlation coefficient of −0.16085. Previous studies demonstrated that in inflammation, extracellular PPIA could recruit leukocytes by interacting with CD147. However, in the cancer-infiltrating environment, this aspect of PPIA has not yet been studied and according to the results of the present study, this leukocyte chemoattractant effect of PPIA in the tumor infiltrating environment was not as strong as in the inflammation environment demonstrated by former studies (19,60–62), suggesting that there may be other stronger effectors in the tumor environment. CD4+ T cells expressing more CD147 migrated more readily to PPIA (60). The association between PPIA and neutrophils remains to be elucidated. The neutrophils in SKCM were moderately correlated with PPIA expression in the present study, which to the best of our knowledge, has not yet been reported, despite the abundance of studies indicating that neutrophils regulated by CXCL5 played an important role in the progression of melanoma (63–65). Table III presents the associations between PPIA expression and biomarkers of natural killer cells, Treg cells and T cell exhaustion in the four different types of cancer, which were very weak, with only two exceptions of Treg cells in LIHC and IL10 in SKCM, which were both weakly correlated with PPIA expression.
To conclude, the present study demonstrated that PPIA expression was upregulated in all 17 types of cancer assessed when compared with normal tissues. The upregulation of PPIA was associated with poor OS of patients with LGG, LMAL, LUAD, SKCM and LIHC. In LIHC, the upregulation was strongly associated with poor overall of patients in the HBV-free, AJCC T3, sorafenib-administered and tumor stage 3 groups, and the prognostic effect was independent of tumor grade. The expression of PPIA in these types of cancer may have a negative effect by interacting with BSG and STAT3.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- LIHC
liver hepatocellular carcinoma
- LGG
low grade glioma
- LMAL
acute myeloid leukemia
- LUAD
lung adenocarcinoma
- SKCM
skin cutaneous melanoma
- PPIA
peptidyl-prolyl cis-trans isomerase A
- BSG (CD147)
basigin
- STAT3
signal transducer and activator of transcription 3
- ITK
interleukin-2 tyrosine kinase
- NK
natural killer
- Treg
regulatory T cell
- TCGA
the cancer genome atlas
Funding
The present study was supported by the National Key Research and Development Program of China (grant no. 2018YFC1313200) and the Key Research and Development Program of Shandong Province (grant no. 2016CYJS01A03).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) and The Cancer Genome Atlas (www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) repositories.
Authors' contributions
JY and SW designed the study. SW, ML and LX performed the data analyses and drafted the manuscript. ML, LX, SW and JY revised the paper. All the authors approved the final version of the manuscript and agreed to be accountable for all aspects of the work.
Ethics approval and consent to participate
The present study was reviewed and approved by the Institutional Review Board of Shandong Cancer Hospital affiliated to Shandong University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Moore LE, Fung ET, McGuire M, Rabkin CC, Molinaro A, Wang Z, Zhang F, Wang J, Yip C, Meng XY, Pfeiffer RM. Evaluation of apolipoprotein A1 and posttranslationally modified forms of transthyretin as biomarkers for ovarian cancer detection in an independent study population. Cancer Epidemiol Biomarkers Prev. 2006;15:1641–1646. doi: 10.1158/1055-9965.EPI-05-0980. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed I, Lobo DN. Malignant tumours of the liver. Surgery. 2009;25:34–41. [Google Scholar]
- 3.Bruix J, Sherman M, American Association for the Study of Liver Diseases Management of hepatocellular carcinoma: An update. Hepatology. 2011;53:1020–1022. doi: 10.1002/hep.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 5.Haendler B, Hofer E. Characterization of the human cyclophilin gene and of related processed pseudogenes. Eur J Biochem. 1990;190:477–482. doi: 10.1111/j.1432-1033.1990.tb15598.x. [DOI] [PubMed] [Google Scholar]
- 6.Holzman TF, Egan DA, Edalji R, Simmer RL, Helfrich R, Taylor A, Burres NS. Preliminary characterization of a cloned neutral isoelectric form of the human peptidyl prolyl isomerase cyclophilin. J Biol Chem. 1991;266:2474–2479. [PubMed] [Google Scholar]
- 7.Hoffmann H, Schiene-Fischer C. Functional aspects of extracellular cyclophilins. Biol Chem. 2014;395:721–735. doi: 10.1515/hsz-2014-0125. [DOI] [PubMed] [Google Scholar]
- 8.Ramachandran S, Venugopal A, Kutty VR, A V, G D, Chitrasree V, Mullassari A, Pratapchandran NS, Santosh KR, Pillai MR, Kartha CC. Plasma level of cyclophilin A is increased in patients with type 2 diabetes mellitus and suggests presence of vascular disease. Cardiovasc Diabetol. 2014;13:38. doi: 10.1186/1475-2840-13-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei Y, Jinchuan Y, Yi L, Jun W, Zhongqun W, Cuiping W. Antiapoptotic and proapoptotic signaling of cyclophilin A in endothelial cells. Inflammation. 2013;36:567–572. doi: 10.1007/s10753-012-9578-7. [DOI] [PubMed] [Google Scholar]
- 10.Kazui T, Inoue N, Yamada O, Komatsu S. Selective cerebral perfusion during operation for aneurysms of the aortic arch: A reassessment. Ann Thorac Surg. 1992;53:109–114. doi: 10.1016/0003-4975(92)90767-X. [DOI] [PubMed] [Google Scholar]
- 11.Nigro P, Pompilio G, Capogrossi MC. Cyclophilin A: A key player for human disease. Cell Death Dis. 2013;4:e888. doi: 10.1038/cddis.2013.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brazin KN, Mallis RJ, Fulton DB, Andreotti AH. Regulation of the tyrosine kinase Itk by the peptidyl-prolyl isomerase cyclophilin A. Proc Natl Acad Sci USA. 2002;99:1899–1904. doi: 10.1073/pnas.042529199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kasinrerk W, Fiebiger E, Stefanová I, Baumruker T, Knapp W, Stockinger H. Human leukocyte activation antigen M6, a member of the Ig superfamily, is the species homologue of rat OX-47, mouse basigin, and chicken HT7 molecule. J Immunol. 1992;149:847–854. [PubMed] [Google Scholar]
- 14.Yurchenko V, Constant S, Bukrinsky M. Dealing with the family: CD147 interactions with cyclophilins. Immunology. 2006;117:301–309. doi: 10.1111/j.1365-2567.2005.02316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyauchi T, Masuzawa Y, Muramatsu T. The basigin group of the immunoglobulin superfamily: Complete conservation of a segment in and around transmembrane domains of human and mouse basigin and chicken HT7 antigen. J Biochem. 1991;110:770–774. doi: 10.1093/oxfordjournals.jbchem.a123657. [DOI] [PubMed] [Google Scholar]
- 16.Kanekura T, Chen X, Kanzaki T. Basigin (CD147) is expressed on melanoma cells and induces tumor cell invasion by stimulating production of matrix metalloproteinases by fibroblasts. Int J Cancer. 2002;99:520–528. doi: 10.1002/ijc.10390. [DOI] [PubMed] [Google Scholar]
- 17.Jin ZG, Lungu AO, Xie L, Wang M, Wong C, Berk BC. Cyclophilin A is a proinflammatory cytokine that activates endothelial cells. Arterioscler Thromb Vasc Biol. 2004;24:1186–1191. doi: 10.1161/01.ATV.0000130664.51010.28. [DOI] [PubMed] [Google Scholar]
- 18.Seko Y, Fujimura T, Taka H, Mineki R, Murayama K, Nagai R. Hypoxia followed by reoxygenation induces secretion of cyclophilin A from cultured rat cardiac myocytes. Biochem Biophys Res Commun. 2004;317:162–168. doi: 10.1016/j.bbrc.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 19.Yurchenko V, Zybarth G, O'Connor M, Dai WW, Franchin G, Hao T, Guo H, Hung HC, Toole B, Gallay P, et al. Active site residues of cyclophilin A are crucial for its signaling activity via CD147. J Biol Chem. 2002;277:22959–22965. doi: 10.1074/jbc.M201593200. [DOI] [PubMed] [Google Scholar]
- 20.Kim SH, Lessner SM, Sakurai Y, Galis ZS. Cyclophilin A as a novel biphasic mediator of endothelial activation and dysfunction. Am J Pathol. 2004;164:1567–1574. doi: 10.1016/S0002-9440(10)63715-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang H, Li M, Chai H, Yan S, Lin P, Lumsden AB, Yao Q, Chen C. Effects of cyclophilin A on cell proliferation and gene expressions in human vascular smooth muscle cells and endothelial cells. J Surg Res. 2005;123:312–319. doi: 10.1016/j.jss.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 22.Obchoei S, Sawanyawisuth K, Wongkham C, Kasinrerk W, Yao Q, Chen C, Wongkham S. Secreted cyclophilin A mediates G1/S phase transition of cholangiocarcinoma cells via CD147/ERK1/2 pathway. Tumour Biol. 2015;36:849–859. doi: 10.1007/s13277-014-2691-5. [DOI] [PubMed] [Google Scholar]
- 23.Bauer K, Kretzschmar AK, Cvijic H, Blumert C, Löffler D, Brocke-Heidrich K, Schiene-Fischer C, Fischer G, Sinz A, Clevenger CV, Horn F. Cyclophilins contribute to Stat3 signaling and survival of multiple myeloma cells. Oncogene. 2009;28:2784–2795. doi: 10.1038/onc.2009.142. [DOI] [PubMed] [Google Scholar]
- 24.Lee J, Kim SS. Current implications of cyclophilins in human cancers. J Exp Clin Cancer Res. 2010;29:97. doi: 10.1186/1756-9966-29-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye Y, Huang A, Huang C, Liu J, Wang B, Lin K, Chen Q, Zeng Y, Chen H, Tao X, et al. Comparative mitochondrial proteomic analysis of hepatocellular carcinoma from patients. Proteomics Clin Appl. 2013;7:403–415. doi: 10.1002/prca.201100103. [DOI] [PubMed] [Google Scholar]
- 26.Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 27.Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Corrigendum: Global quantification of mammalian gene expression control. Nature. 2013;495:126–127. doi: 10.1038/nature11848. [DOI] [PubMed] [Google Scholar]
- 28.Li JJ, Biggin MD. Gene expression. Statistics requantitates the central dogma. Science. 2015;347:1066–1067. doi: 10.1126/science.aaa8332. [DOI] [PubMed] [Google Scholar]
- 29.Morimoto S, Yahara K. Identification of stress responsive genes by studying specific relationships between mRNA and protein abundance. Heliyon. 2018;4:e00558. doi: 10.1016/j.heliyon.2018.e00558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Beyer A, Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell. 2016;165:535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 31.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan JH, Zhou H, Cooper L, Huang JL, Zhu SB, Zhao XX, Ding H, Pan YL, Rong L. LAYN is a prognostic biomarker and correlated with immune infiltrates in gastric and colon cancers. Front Immunol. 2019;10:6. doi: 10.3389/fimmu.2019.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: A new database for meta-analysis of the prognostic value of genes. BMC Med Genomics. 2009;2:18. doi: 10.1186/1755-8794-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menyhárt O, Nagy Á, Győrffy B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R Soc Open Sci. 2018;5:181006. doi: 10.1098/rsos.181006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, Jiang P, Shen H, Aster JC, Rodig S, et al. Comprehensive analyses of tumor immunity: Implications for cancer immunotherapy. Genome Biol. 2016;17:174. doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Graef T, Moesta AK, Norman PJ, Abi-Rached L, Vago L, Older Aguilar AM, Gleimer M, Hammond JA, Guethlein LA, Bushnell DA, et al. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. J Exp Med. 2009;206:2557–2572. doi: 10.1084/jem.20091010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vivian JP, Duncan RC, Berry R, O'Connor GM, Reid HH, Beddoe T, Gras S, Saunders PM, Olshina MA, Widjaja JM, et al. Killer cell immunoglobulin-like receptor 3DL1-mediated recognition of human leukocyte antigen B. Nature. 2011;479:401–405. doi: 10.1038/nature10517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goodridge JP, Burian A, Lee N, Geraghty DE. HLA-F and MHC class I open conformers are ligands for NK cell Ig-like receptors. J Immunol. 2013;191:3553–3562. doi: 10.4049/jimmunol.1300081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cantoni C, Bottino C, Vitale M, Pessino A, Augugliaro R, Malaspina A, Parolini S, Moretta L, Moretta A, Biassoni R. NKp44, a triggering receptor involved in tumor cell lysis by activated human natural killer cells, is a novel member of the immunoglobulin superfamily. J Exp Med. 1999;189:787–796. doi: 10.1084/jem.189.5.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eissmann P, Beauchamp L, Wooters J, Tilton JC, Long EO, Watzl C. Molecular basis for positive and negative signaling by the natural killer cell receptor 2B4 (CD244) Blood. 2005;105:4722–4729. doi: 10.1182/blood-2004-09-3796. [DOI] [PubMed] [Google Scholar]
- 43.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 44.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 45.Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, Rudensky AY. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 46.Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12:492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 47.Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chevalier F, Depagne J, Hem S, Chevillard S, Bensimon J, Bertrand P, Lebeau J. Accumulation of cyclophilin A isoforms in conditioned medium of irradiated breast cancer cells. Proteomics. 2012;12:1756–1766. doi: 10.1002/pmic.201100319. [DOI] [PubMed] [Google Scholar]
- 49.Ralhan R, Masui O, Desouza LV, Matta A, Macha M, Siu KW. Identification of proteins secreted by head and neck cancer cell lines using LC-MS/MS: Strategy for discovery of candidate serological biomarkers. Proteomics. 2011;11:2363–2376. doi: 10.1002/pmic.201000186. [DOI] [PubMed] [Google Scholar]
- 50.Tian X, Zhao C, Zhu H, She W, Zhang J, Liu J, Li L, Zheng S, Wen YM, Xie Y. Hepatitis B virus (HBV) surface antigen interacts with and promotes cyclophilin a secretion: Possible link to pathogenesis of HBV infection. J Virol. 2010;84:3373–3381. doi: 10.1128/JVI.02555-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Igakura T, Kadomatsu K, Taguchi O, Muramatsu H, Kaname T, Miyauchi T, Yamamura K, Arimura K, Muramatsu T. Roles of basigin, a member of the immunoglobulin superfamily, in behavior as to an irritating odor, lymphocyte response, and blood-brain barrier. Biochem Biophys Res Commun. 1996;224:33–36. doi: 10.1006/bbrc.1996.0980. [DOI] [PubMed] [Google Scholar]
- 52.Staffler G, Szekeres A, Schütz GJ, Säemann MD, Prager E, Zeyda M, Drbal K, Zlabinger GJ, Stulnig TM, Stockinger H. Selective inhibition of T cell activation via CD147 through novel modulation of lipid rafts. J Immunol. 2003;171:1707–1714. doi: 10.4049/jimmunol.171.4.1707. [DOI] [PubMed] [Google Scholar]
- 53.Lu M, Wu J, Hao ZW, Shang YK, Xu J, Nan G, Li X, Chen ZN, Bian H. Basolateral CD147 induces hepatocyte polarity loss by E-cadherin ubiquitination and degradation in hepatocellular carcinoma progress. Hepatology. 2018;68:317–332. doi: 10.1002/hep.29798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de la Iglesia N, Konopka G, Puram SV, Chan JA, Bachoo RM, You MJ, Levy DE, Depinho RA, Bonni A. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev. 2008;22:449–462. doi: 10.1101/gad.1606508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao S, Wu L, Kuang Y, Su J, Luo Z, Wang Y, Li J, Zhang J, Chen W, Li F, et al. Downregulation of CD147 induces malignant melanoma cell apoptosis via the regulation of IGFBP2 expression. Int J Oncol. 2018;53:2397–2408. doi: 10.3892/ijo.2018.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee J, Kim JC, Lee SE, Quinley C, Kim H, Herdman S, Corr M, Raz E. Signal transducer and activator of transcription 3 (STAT3) protein suppresses adenoma-to-carcinoma transition in Apcmin/+ mice via regulation of Snail-1 (SNAI) protein stability. J Biol Chem. 2012;287:18182–18189. doi: 10.1074/jbc.M111.328831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Musteanu M, Blaas L, Mair M, Schlederer M, Bilban M, Tauber S, Esterbauer H, Mueller M, Casanova E, Kenner L, et al. Stat3 is a negative regulator of intestinal tumor progression in Apc(Min) mice. Gastroenterology. 2010;138:1003–1011.e1-e5. doi: 10.1053/j.gastro.2009.11.049. [DOI] [PubMed] [Google Scholar]
- 58.Gomez-Rodriguez J, Kraus ZJ, Schwartzberg PL. Tec family kinases Itk and Rlk/Txk in T lymphocytes: Cross-regulation of cytokine production and T-cell fates. FEBS J. 2011;278:1980–1989. doi: 10.1111/j.1742-4658.2011.08072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chongsathidkiet P, Jackson C, Koyama S, Loebel F, Cui X, Farber SH, Woroniecka K, Elsamadicy AA, Dechant CA, Kemeny HR, et al. Sequestration of T cells in bone marrow in the setting of glioblastoma and other intracranial tumors. Nat Med. 2018;24:1459–1468. doi: 10.1038/s41591-018-0135-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Damsker JM, Bukrinsky MI, Constant SL. Preferential chemotaxis of activated human CD4+ T cells by extracellular cyclophilin A. J Leukoc Biol. 2007;82:613–618. doi: 10.1189/jlb.0506317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sherry B, Yarlett N, Strupp A, Cerami A. Identification of cyclophilin as a proinflammatory secretory product of lipopolysaccharide-activated macrophages. Proc Natl Acad Sci USA. 1992;89:3511–3515. doi: 10.1073/pnas.89.8.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu Q, Leiva MC, Fischkoff SA, Handschumacher RE, Lyttle CR. Leukocyte chemotactic activity of cyclophilin. J Biol Chem. 1992;267:11968–11971. [PubMed] [Google Scholar]
- 63.Forsthuber A, Lipp K, Andersen L, Ebersberger S, Graña-Castro ', Ellmeier W, Petzelbauer P, Lichtenberger BM, Loewe R. CXCL5 as regulator of neutrophil function in cutaneous melanoma. J Invest Dermatol. 2019;139:186–194. doi: 10.1016/j.jid.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 64.Drewes CC, Alves AC, Hebeda CB, Copetti I, Sandri S, Uchiyama MK, Araki K, Guterres SS, Pohlmann AR, Farsky SH. Role of poly(ε-caprolactone) lipid-core nanocapsules on melanoma-neutrophil crosstalk. Int J Nanomedicine. 2017;12:7153–7163. doi: 10.2147/IJN.S140557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hölzel M, Tüting T. Inflammation-induced plasticity in melanoma therapy and metastasis. Trends Immunol. 2016;37:364–374. doi: 10.1016/j.it.2016.03.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) and The Cancer Genome Atlas (www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) repositories.