Abstract
Lung adenocarcinoma is the most common histologic subtype of lung cancer. The aim of the present study was to assess the expression of hepatoma-derived growth factor (HDGF) and protein kinase Cα (PRKCA) in lung adenocarcinoma (LADC), and to determine the association between the combined expression of these two proteins and clinicopathological characteristics of patients with LADC. The expression of HDGF and PRKCA mRNA was assessed by GEO database analysis, and HDGF and PRKCA protein levels were examined by immunohistochemistry using a tissue microarray. High HDGF and PRKCA expression was observed in LADC tissue compared to normal samples, and increased HDGF and PRKCA expression was associated with AJCC clinical stage, tumor classification, node classification, and lymph node metastasis. GEO database analysis revealed no significant differences between HDGF mRNA and PRKCA mRNA in LADC tissue. However, high PRKCA protein expression was associated with high HDGF protein expression, and patients with high HDGF and PRKCA expression exhibited poorer overall survival rates than patients with low expression levels of the two proteins. The results of the present study suggest that upregulation of both HDGF and PRKCA may be an unfavourable factor for lung adenocarcinoma progression.
Keywords: hepatoma-derived growth factor, protein kinase Cα, lung adenocarcinoma
Introduction
As the most common malignancy in the world, lung cancer arises from the bronchial mucosal epithelium and is the leading cause of cancer mortality worldwide (1). Histologically, lung cancer is generally divided into small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC) (2). NSCLC, including lung squamous cell carcinoma (LSCC) and lung adenocarcinoma (LADC), accounts for ~85% of all lung cancer cases and is the most commonly diagnosed type of lung cancer (2,3). Studies have demonstrated that LADC is overtaking LSCC as the most common histological subtype in various countries, such as the United States and China (4,5). At present, the treatment of LADC mainly includes surgical resection, chemotherapy, radiation therapy, immunotherapy and targeted therapy (6). Although continuous progress has been made, the 5-year survival rate remains low (7,8). Therefore, understanding the pathogenesis based on alterations at the molecular level is essential for identifying useful biomarkers and treating LADC.
Accumulating evidence demonstrates that tumourigenesis is a complex multistep and multistage process involving a number of gene and pathway alterations (9,10). Therefore, the identification of tumour-related genes and signals should be combined with data from clinical studies. Our previous study using miRomics and proteomics revealed a microRNA (miR)-296-3p/protein kinase Cα (PRKCA)/focal adhesion kinase 1 (FAK)/Ras/c-Myc feedback loop modulated by hepatoma-derived growth factor (HDGF) in LADC (11). It was demonstrated that PRKCA was a direct target of miR-296-3p and was regulated by HDGF (11). Together, these factors promoted LADC cell proliferation, migration and invasion, and cisplatin chemotherapy resistance in vitro and in vivo. However, the association of HDGF and PRKCA with clinical characteristics and the prognosis of patients with LADC remain unclear.
As a heparin-binding protein, HDGF was originally purified from the conditioned media of Huh-7 hepatoma cells (12). HDGF overexpression has been reported in various tumour types, including lung cancer (11,13). In addition, increased HDGF expression is a common event in NSCLC and an independent factor for poor prognosis (13). Anti-HDGF antibody treatment enhances the antitumour activities of gemcitabine, bevacizumab and chemotherapy in NSCLC (14). In our previous study, it was demonstrated that HDGF mRNA was highly expressed in LADC tissues and that increased HDGF levels promoted PRKCA expression by inhibiting miR-296-3p in LADC cells (11). PRKCA is a member of the PKC family, and upregulation of PRKCA has been reported in several types of cancer and has been found to regulate various cellular functions, including cell proliferation, survival and metastasis (15–18). Although HDGF and PRKCA overexpression plays a pivotal role in NSCLC progression, the association between HDGF and PRKCA and clinical characteristics, as well as the prognostic effect of combined HDGF and PRKCA expression are not understood in LADC.
Materials and methods
Sample collection
In total, 130 paraffin-embedded primary LADC specimens and 70 normal specimens were included in a tissue array, which was obtained from Shanghai Outdo Biotech Co., Ltd (cat. no. HLug-Ade150Sur). According to the datasheet supplied with the microarray, all patients with LADC underwent surgery or centesis between June 2007 and June 2009 in accordance with medical ethics guidelines in Taizhou Hospital of Zhejiang Province (Zhejiang, China). All patients were followed up, and the last follow up was completed in August 2014. Clinical stage was defined by two or more researchers according to the American Joint Committee on Cancer (AJCC) 7th clinical staging system (19). Patients with a diagnosed relapse and patients who received preoperative radiation, chemotherapy or biotherapy were excluded. Informed consent from the patients and approval from the ethics committees of the Taizhou hospital of Zhejiang province and Kunming Medical University (approval no. KY201726) were obtained prior to the use of these clinical materials for research purposes. Demographic and clinical data were obtained from patients' medical records according to the datasheet supplied with the microarray.
Immunohistochemistry
The tissue array, which included 130 LADC specimens and 70 normal specimens was used for immunohistochemical analysis. The specimens were deparaffinized in 100% xylene and rehydrated in a descending ethanol series (100, 90, 80 and 70% ethanol) and water. Next, the tissue array was incubated in 10 mM citrate buffer for 2 min at 100°C for heat-induced antigen retrieval. Endogenous peroxidase activity and non-specific antigens were blocked with peroxidase-blocking reagent containing 3% hydrogen peroxide and serum (cat. no. SP-9001 kit; Beijing Zhongshan Jinqiao Biotech Co. Ltd. China) for 30 min at room temperature. Samples were then incubated with antibodies against HDGF (1:100; cat. no. 11344-1-AP; Proteintech Group, Inc.) or PRKCA (1:200; cat. no. 21991-1-AP; Proteintech Group, Inc.) overnight at 4°C. After washing, the sections were incubated with undiluted biotin-labelled rabbit anti-goat antibody (cat. no. SP-9001 kit; Beijing Zhongshan Jinqiao Biotech Co. Ltd. China) for 10 min at room temperature and subsequently incubated with undiluted streptavidin-conjugated horseradish peroxidase (cat. no. SP-9001 kit; Beijing Zhongshan Jinqiao Biotech Co. Ltd. China) for 10 min at room temperature. The peroxidase reaction was developed using 3,3′-diaminobenzidine (DAB) chromogen solution in DAB buffer substrate for 2 min at room temperature. Sections were counterstained with haematoxylin for 1 min at room temperature, mounted in neutral gum and imaged using bright-field microscopy (magnification, ×400). Finally, 123 LADC specimens and 64 normal specimens were incorporated into subsequent analysis.
Evaluation of staining
Stained tissue sections were reviewed and scored independently by two investigators blinded to the clinical data. For cytoplasmic staining, the score was based on the sum of cytoplasmic staining intensity and the percentage of stained cells. The staining intensity was scored as previously described (scores of 0–3) (20,21), and the percentage of positively stained areas of cells was defined on a scale of 0–3 (0, <10; 1, 10–25; 2, 26–75; and 3, >76%). For nuclear staining, the staining score was defined based on the sum of nuclear staining intensity and the percentage of positive nuclear staining. Nuclear staining intensity were defined as follows: 0, no color; 1, light yellow; 2, light brown; 3, brown (22). The percentage of positive nuclear staining scores were defined as follows: 0, <20; 1, 20–49; 2, 50–79; and 3, >80%. The sum of the staining intensity and staining extent scores (0–6) was used as the final staining score. For statistical analysis, final staining scores of 0–2 and 3–6 in the cytoplasm or 0–3 and 4–6 in the nucleus were considered to represent low and high expression levels, respectively.
Use of databases and bioinformatics analysis
To assess the expression of HDGF mRNA and PRKCA mRNA in NSCLC and LADC tissues, the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/gds/?term) (GSE19188) was searched, and genome-wide gene expression data from the GSE19188 dataset were extracted (23). This dataset includes 91 NSCLC and 65 normal lung tissue samples. Next, gene expression and correlation between HDGF and PRKCA in NSCLC were analysed. In addition, the Kaplan-Meier Plotter database (http://kmplot.com/analysis/index.php?cancer=lung&p=service) was used to investigate the association of HDGF or PRKCA expression and overall survival rates in patients with LADC and the hazard ratio (HR) value was obtained.
Statistical analysis
SPSS 21.0 software (IBM Corp.) was employed to perform all statistical analyses. Data are presented as the mean ± standard deviation. Student's t-test was performed to compare between two groups, and one-way analysis-of-variance with Dunnett's post hoc test was used to perform multiple-comparison tests. The χ2 test was used to determine the differences in HDGF or PRKCA protein expression between lung adenocarcinoma and normal lung tissues, and to determine the association between HDGF and PRKCA protein expression in 123 LADC patients from the tissue array. The associations between clinicopathological characteristics and HDGF and PRKCA expression were also analysed by χ2 test. Kaplan-Meier analysis with the log-rank test was used to assess the prognostic effect of HDGF or PRKCA in 123 patients with LADC. The univariate and multivariate Cox proportional hazards method was used to analyse the association between clinicopathological characteristics and patient survival time. Spearman's correlation analysis was used to perform pairwise comparisons between HDGF and PRKCA mRNA. The range of the correlation coefficient (r) value is −1.0 to 1.0, where when the r value is >0 it is a positive relationship, conversely, when the r value is <0, it's a negative relationship. Chi-square test was used to determine the association between HDGF and PRKCA protein expression in 123 LADC patients. P<0.05 was considered to indicate a statistically significant difference.
Results
HDGF mRNA expression in NSCLC and LADC tissues
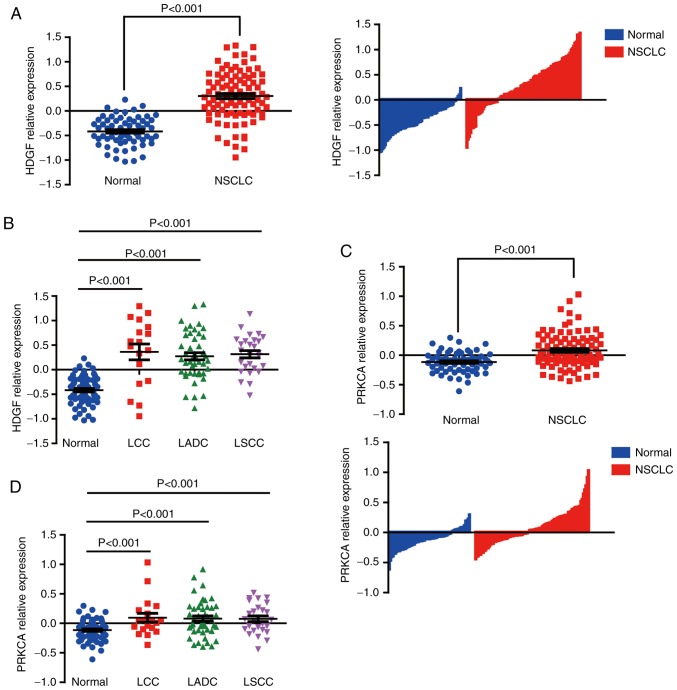
To assess HDGF levels in LADC specimens, genome-wide expression data was extracted from the GEO dataset, GSE19188, in which the genome-wide gene expression analysis was performed by Affymetrix HG-U133_Plus_2 array Platforms (Affymetrix, Inc.) in 91 NSCLC and 65 normal lung tissue samples (23). In this study, the data was normalized by Robust Multi-Array average (RMA) algorithm, the intensities of probe sets <30 were reset to 30 and the intensity values of individual probe sets in each sample were then displayed as log2 of the deviations to the calculated geometric means for that probe sets (23). The expression of HDGF mRNA from GSE19188 was extracted and analyzed, the result showed that HDGF mRNA was highly expressed in NSCLC tissues compared with normal tissues (Fig. 1A). HDGF expression was further analysed in subgroups of NSCLC cases, and HDGF mRNA was revealed to be universally highly expressed in LSCC, large cell lung cancer (LCC) and LADC tissues compared with normal specimens (Fig. 1B).
Figure 1.
HDGF and PRKCA are upregulated in NSCLC samples in a dataset from the GEO database. (A) HDGF mRNA expression in NSCLC and normal tissues. (B) HDGF mRNA expression in different subtypes of NSCLC. (C) PRKCA mRNA expression in NSCLC and normal tissues. (D) PRKCA mRNA expression in different subtypes of NSCLC. HDGF, hepatoma-derived growth factor; PRKCA, protein kinase Cα; NSCLC, non-small cell lung cancer; LCC, large cell lung cancer; LADC, lung adenocarcinoma; LSCC, lung squamous cell carcinoma.
PRKCA mRNA expression in NSCLC and LADC tissues
PRKCA expression was also analysed in NSCLC tissues from the GEO GSE19188 dataset. Results similar to those noted for HDGF expression were observed. Upregulated PRKCA expression was observed in NSCLC specimens, and also in NSCLC subgroups (Fig. 1C and D). No differences were observed in PRKCA mRNA between LCC, LSCC and LADC samples.
HDGF protein expression in LADC tissues
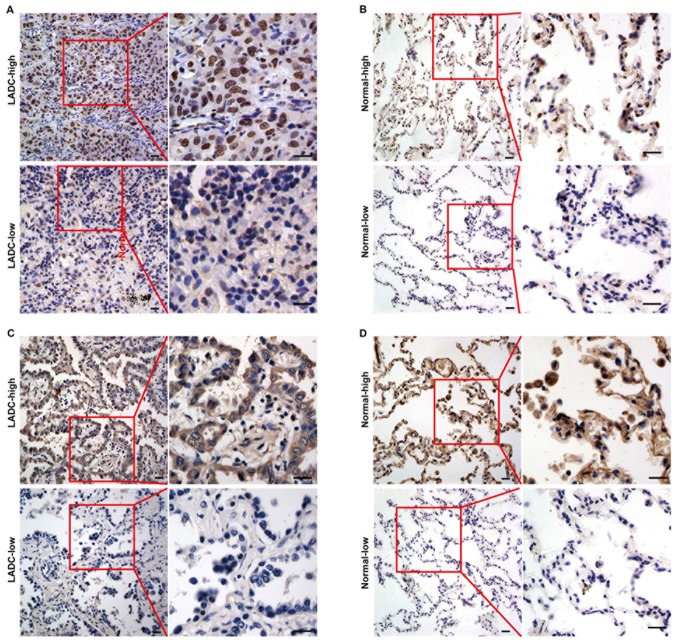
HDGF protein levels and subcellular localization was determined in 123 LADC samples and 64 normal lung tissues via immunohistochemical staining using a tissue array. High and low expression of HDGF protein in LADC and normal lung tissues are presented in Fig. 2A and B, respectively. As shown in Fig. 2A, HDGF protein was mainly expressed in the nuclei of LADC cells. However, HDGF protein was observed in the cytoplasm and nuclei of normal epithelial cells (Fig. 2B). It was observed that 32.8% (21/64) of normal lung samples and 56.9% (70/123) of LADC tissues exhibited high HDGF protein expression, and the expression levels were significantly different between the tissue types (P=0.002) (Table I).
Figure 2.
HDGF and PRKCA expression in LADC tissues by immunohistochemistry. (A) HDGF expression in LADC tissues. (B) HDGF expression in normal lung tissues. (C) PRKCA expression in LADC tissues. (D) PRKCA expression in normal lung tissues. HDGF, hepatoma-derived growth factor; PRKCA, protein kinase Cα; LADC, lung adenocarcinoma. Magnification, ×400.
Table I.
Expression of HDGF and PRKCA in lung adenocarcinoma tissue compared with that in normal lung tissue.
| Expression level | |||||
|---|---|---|---|---|---|
| Protein | Group | Cases, n | Low, n (%) | High, n (%) | P-value |
| HDGF | Cancer | 123 | 53 (43.1) | 70 (56.9) | 0.002 |
| Normal | 64 | 43 (67.2) | 21 (32.8) | ||
| PRKCA | Cancer | 123 | 55 (44.7) | 68 (55.3) | 0.007 |
| Normal | 64 | 42 (65.6) | 22 (34.4) | ||
HDGF, hepatoma-derived growth factor; PRKCA, protein kinase Cα.
PRKCA protein expression in LADC tissues
The expression and subcellular localization of PRKCA protein were also examined in 123 LADC samples and 64 normal lung tissues via immunohistochemical staining. High and low expression of PRKCA protein in LADC and normal lung tissue are presented in Fig. 2C and D, respectively. PRKCA protein was predominantly localized in the cytoplasm of LADC cells (Fig. 2C) and normal lung epithelial cells (Fig. 2D). Furthermore, statistical analysis showed that high PRKCA expression protein was significantly more common in LADC samples (55.3%, 68/123) compared with that in normal lung samples (34.4%, 22/64) (P=0.007) (Table I).
Association between clinicopathological characteristics and HDGF in LADC patients
The association between HDGF expression and clinical features was investigated. As presented in Table II, expression of HDGF was associated with node (N) classification (P<0.001), lymph node metastasis (P<0.001) and AJCC clinical stage (P<0.001). However, a significant association was not observed between HDGF expression and patient sex (P=0.436), age P=0.294) and tumour (T) classification (P=0.074).
Table II.
Association of clinicopathological factors and expression of HDGF in patients with lung adenocarcinoma.
| HDGF expression | ||||
|---|---|---|---|---|
| Factors | n | Low, n (%) | High, n (%) | P-value |
| Sex | ||||
| Male | 67 | 31 (46.3) | 36 (53.7) | 0.436 |
| Female | 56 | 22 (39.3) | 34 (60.7) | |
| Age, years | ||||
| ≤60 | 67 | 26 (38.8) | 41 (61.2) | 0.294 |
| >60 | 56 | 27 (48.2) | 29 (51.8) | |
| T classification | ||||
| T1-T2 | 103 | 48 (46.6) | 55 (53.4) | 0.074 |
| T3-T4 | 20 | 5 (25.0) | 15 (75.0) | |
| N classification | ||||
| N0-N1 | 80 | 48 (60.0) | 32 (40.0) | <0.001 |
| N2-N3 | 43 | 5 (11.6) | 38 (88.4) | |
| Lymph node metastasis | ||||
| Negative | 61 | 38 (62.3) | 23 (37.7) | <0.001 |
| Positive | 62 | 15 (24.2) | 47 (75.8) | |
| Clinical stage | ||||
| I–II | 76 | 47 (61.8) | 29 (38.2) | <0.001 |
| III–IV | 47 | 6 (12.8) | 41 (87.2) | |
HDGF, hepatoma-derived growth factor; T, tumour; N, node.
Association between clinicopathological characteristics and PRKCA in LADC patients
The association of PRKCA expression and clinical features of LADC patients was investigated. Statistical analysis of PRKCA immunohistochemical staining demonstrated that PRKCA expression was associated with T classification (P=0.015), N classification (P<0.001), lymph node metastasis (P=0.002) and AJCC clinical stage (P<0.001), but no significant association was observed between PRKCA expression and sex (P=0.988) or age (P=0.476) (Table III).
Table III.
Association of clinicopathological factors and expression of PRKCA in patients with lung adenocarcinoma.
| PRKCA expression | ||||
|---|---|---|---|---|
| Factors | n | Low, n (%) | High, n (%) | P-value |
| Sex | ||||
| Male | 67 | 30 (44.8) | 37 (55.2) | 0.988 |
| Female | 56 | 25 (44.6) | 31 (55.4) | |
| Age, years | ||||
| ≤60 | 67 | 28 (41.8) | 39 (58.2) | 0.476 |
| >60 | 56 | 27 (48.2) | 29 (51.8) | |
| T classification | ||||
| T1-T2 | 103 | 51 (49.5) | 52 (50.5) | 0.015 |
| T3-T4 | 20 | 4 (20.0) | 16 (80.0) | |
| N classification | ||||
| N0-N1 | 80 | 47 (58.8) | 33 (41.3) | <0.001 |
| N2-N3 | 43 | 8 (18.6) | 35 (81.4) | |
| Lymph node metastasis | ||||
| Negative | 61 | 36 (59) | 25 (41) | 0.002 |
| Positive | 62 | 19 (30.6) | 43 (69.4) | |
| Clinical stage | ||||
| I–II | 76 | 46 (60.5) | 30 (39.5) | <0.001 |
| III–IV | 47 | 9 (19.1) | 38 (80.9) | |
PRKCA, protein kinase Cα; T, tumour; N, node.
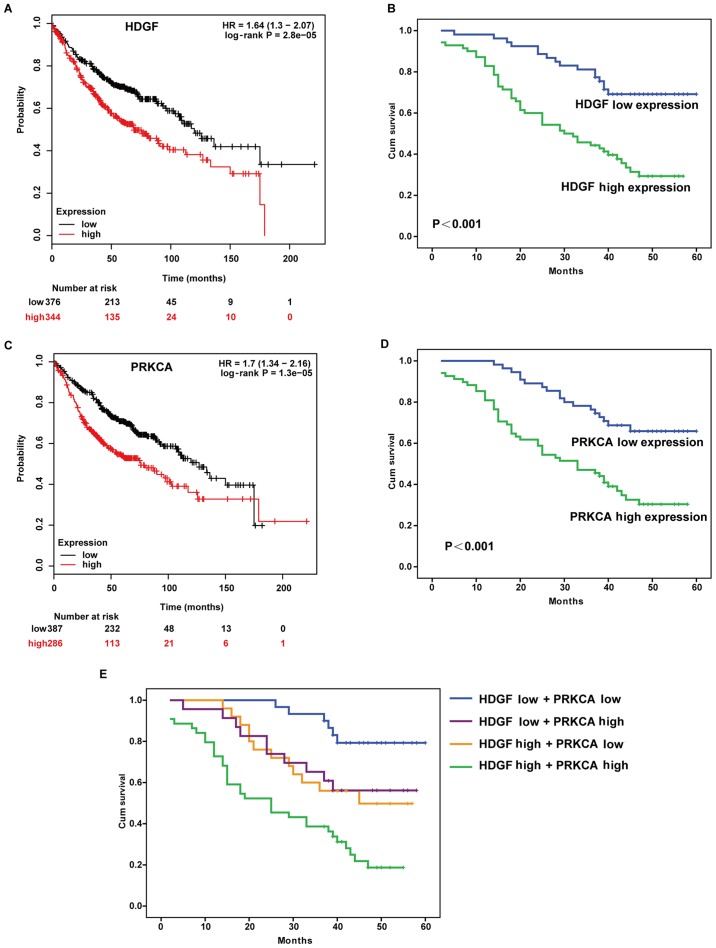
High HDGF expression is associated with a lower cumulative overall survival rate in LADC patients
To investigate the prognostic value of HDGF expression in LADC patients, the Kaplan-Meier Plotter database was used to analyse the association between HDGF expression and patient survival. The results demonstrated that patients with high HDGF expression exhibited a poorer overall survival rate and higher risk (HR=1.64) compared with patients with low HDGF expression (Fig. 3A). The prognostic effect of HDGF in 123 LADC patients from the tissue array was assessed using the log-rank test, and significantly lower overall survival rate was observed in patients with high HDGF expression (P<0.001; Fig. 3B).
Figure 3.
High expression of HDGF and PRKCA is associated with poor prognosis in LADC. (A) Overall survival rate in LADC patients based on HDGF expression was predicted using Kaplan-Meier Plotter database. (B) The cumulative overall survival rate was analysed in 123 LADC tissues from the tissue array. (C) Overall survival rate in LADC patients based on PRKCA expression was predicted using Kaplan-Meier Plotter database. (D) The cumulative overall survival rate was analysed in 123 LADC tissues from the tissue array. (E) The cumulative overall survival rates were analysed by Kaplan-Meier survival analysis in 123 LADC patients based on combined HDGF and PRKCA expression. HDGF, hepatoma-derived growth factor; PRKCA, protein kinase Cα; LADC, lung adenocarcinoma; HR, hazard ratio; Cum, cumulative.
High PRKCA expression is associated with a lower cumulative overall survival rate in LADC patients
The association between PRKCA expression and patient survival was investigated using the Kaplan-Meier Plotter database. Patients with high PRKCA expression exhibited a poorer overall survival rate and higher risk (HR=1.7) compared with patients with low PRKCA expression (Fig. 3C). For 123 patients with LADC with prognostic information, log-rank analysis was performed, and it was found that patients with high PRKCA expression exhibited worse prognosis than patients with low PRKCA expression (P<0.001; Fig. 3D).
High expression of HDGF or PRKCA is not an independent prognostic factor for LADC patients
Kaplan-Meier analysis revealed that high HDGF expression and high PRKCA expression were associated with a lower survival rate (Fig. 3A-D). Patients with high expression of both HDGF and PRKCA displayed a significantly decreased survival rate compared with patients with high expression of only one of the proteins, or with low expression of both (HDGF high and PRKCA low; HDGF low and PRKCA high; HDGF low and PRKCA low) (Fig. 3E). Notably, low expression of HDGF and PRKCA was associated with a higher survival rate (Fig. 3E). Univariate Cox regression analysis of overall survival duration revealed that T classification (P=0.004), N classification (P<0.001), lymph node metastasis (P<0.001), AJCC clinical stage (P<0.001), HDGF expression (P<0.001) and PRKCA expression (P<0.001) were significantly associated with patient survival (Table IV). To determine whether HDGF or PRKCA is an independent prognostic factor for LADC, multivariate analysis was performed. HDGF and PRKCA protein expression levels in LADC patients were adjusted for T classification, N classification, lymph node metastasis, AJCC clinical stage, HDGF expression and PRKCA expression. HDGF and PRKCA expression levels were not independent prognostic factors for LADC (P=0.103 and P=0.094, respectively) (Table IV).
Table IV.
Univariate and multivariate Cox regression analysis of overall survival rate.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Factors | P-value | HR | 95% CI | P-value | HR | 95% CI |
| Sex, male vs. female | 0.080 | 0.635 | 0.382–1.056 | – | – | – |
| Age, ≤60 vs. >60 years | 0.862 | 0.957 | 0.583–1.571 | – | – | – |
| T classification, T1-T2 vs. T3-T4 | 0.004 | 1.663 | 1.172–1.665 | 0.793 | 0.949 | 0.639–1.408 |
| N classification, N0-N1 vs. N2-N3 | 0.000 | 2.358 | 1.834–3.031 | 0.592 | 1.178 | 0.647–2.147 |
| Lymph node metastasis, negative vs. positive | 0.000 | 4.964 | 2.769–8.899 | 0.758 | 1.205 | 0.368–3.949 |
| Clinical stage, I–II vs. III–IV | 0.000 | 3.033 | 2.142–4.295 | 0.131 | 1.854 | 0.832–4.130 |
| HDGF expression, low vs. high | 0.000 | 3.170 | 1.794–5.601 | 0.103 | 1.675 | 0.901–3.115 |
| PRKCA expression, low vs. high | 0.000 | 2.909 | 1.681–5.034 | 0.094 | 1.677 | 0.915–3.076 |
HDGF, hepatoma-derived growth factor; PRKCA, protein kinase Cα; T, tumour; N, node; HR, hazard ratio; CI, confidence interval.
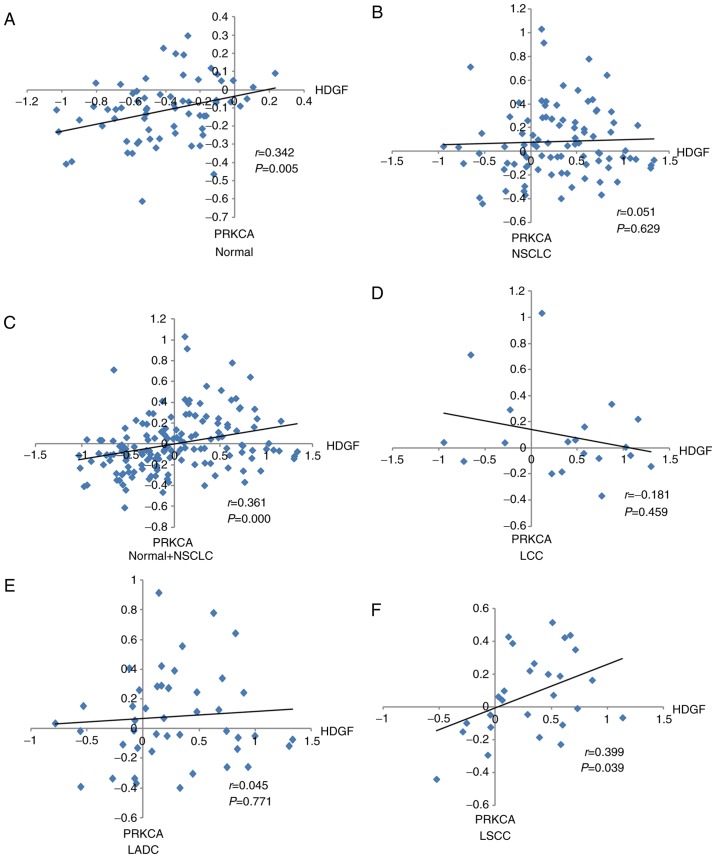
Association between HDGF expression and PRKCA expression in LADC patients
Our previous study demonstrated that PRKCA was a downstream factor of HDGF and that HDGF regulated PRKCA expression via miR-296-3p (6). In the present study, χ2 testing demonstrated that PRKCA protein expression was associated with HDGF protein expression in LADC patients (P=0.021) (Table V). Correlation analysis of HDGF and PRKCA mRNA levels in different tissue types was performed. As presented in Fig. 4, a positive correlation between HDGF mRNA and PRKCA mRNA was demonstrated in normal lung tissues (r=0.342), normal and NSCLC (r=0.361), and LSCC (r=0.399) tissues (Fig. 4A, C and F); however, no significant correlations were observed between HDGF mRNA and PRKCA mRNA in NSCLC (r=0.051), LCC (r=−0.181) and LADC (r=0.045) samples (Fig. 4B, D and E). In addition, combined normal and NSCLC groups used to perform correlation analysis showed that no correlation between HDGF mRNA and PRKCA mRNA in NSCLC tissues was reversed by normal lung tissues (Fig. 4A, B and C). The aforementioned suggested that combined normal tissues used to perform the correlation analysis may have a different effect on tumour tissues.
Table V.
Association between HDGF and PRKCA protein expression in patients with lung adenocarcinoma.
| HDGF | ||||
|---|---|---|---|---|
| PRKCA | High, n | Low, n | χ2 | P-value |
| High | 45 | 23 | 5.324 | 0.021 |
| Low | 25 | 30 | ||
HDGF, hepatoma-derived growth factor; PRKCA, protein kinase Cα; LADC, lung adenocarcinoma.
Figure 4.
Correlation between HDGF and PRKCA expression levels in NSCLC samples from the Gene Expression Ontology database. Correlation between HDGF and PRKCA expression in (A) normal lung tissues, (B) NSCLC samples, (C) NSCLC and normal tissues, (D) LCC tissues, (E) LADC tissues and (F) LSCC tissues. HDGF, hepatoma-derived growth factor; PRKCA, protein kinase Cα; NSCLC, non-small cell lung cancer; LCC, large cell lung cancer; LADC, lung adenocarcinoma; LSCC, lung squamous cell carcinoma.
Discussion
Tumourigenesis is a multifactorial and multistep process involving the accumulation of alterations in genetic, epigenetic and environmental factors (24). Among them, the activation of oncogenes, inactivation of suppressor genes and changes in cell signalling pathways represent key changes (9,10). As the most common histological subtype of lung cancer, LADC is associated with EGFR mutations and abnormal expression of microRNAs and genes (25–29). Our previous study demonstrated that HDGF modulated PRKCA expression via the transcriptional regulation of miR-296-3p in LADC (11). However, HDGF and PRKCA expression levels in normal lung and LADC tissues were examined, and correlations between their expression and clinicopathological parameters were unclear (11).
HDGF is a unique multifunctional protein that is involved in growth-promoting effects, vascular growth and formation, and antiapoptotic effects in various malignancies (30,31). Studies have demonstrated that high HDGF expression is a novel prognostic factor in several types of tumour, including gall bladder cancer (32), endometrial carcinoma (33) and cholangiocarcinoma (34). In the present study, HDGF was found to be highly expressed in LADC tissues by immunohistochemical staining, and these levels were associated with N classification, lymph node metastasis and AJCC clinical stage. Furthermore, the patient prognosis was reduced in patients with high HDGF expression compared to the patients with low HDGF expression. These findings are similar to a previous study in which the researcher analyzed the HDGF expression and the prognosis of patients in human endometrial carcinoma (33).
PRKCA is a serine/threonine protein kinase and a member of the PKC family (17). The PKC family has been implicated in various cellular functions, including cell proliferation, survival and metastasis via the regulation of the ERK-MAPK, NF-κB and PI3K/AKT pathways (11,35,36). PRKCA is upregulated in several human cancer types, including breast cancer (37), colon carcinoma (38), NSCLC cell lines (39) and hematological malignancies (40). A study by Lahn et al (41) revealed that PRKCA is highly expressed in ≤20% of patients with NSCLC by analyzing the PRKCA protein and mRNA levels in NSCLC specimens from an independent tumor tissue bank and a publicly available gene expression array data. Significantly increased PRKCA mRNA expression was also observed in lung cancer tissues in another study, and PRKCA upregulation was found to be modulated by miR-203 (39). Most studies were performed in cell lines, and few studies have examined PRKCA expression in lung cancer biopsies. In the present study, PRKCA was revealed to be highly expressed in LADC tissues, and high PRKCA expression was associated with T classification, N classification, lymph node metastasis and AJCC clinical stage, and negatively associated with patient prognosis.
In a previous study, HDGF was found to modulate PRKCA expression via miR-296-3p in LADC (11); however, the associations between HDGF and PRKCA expression and clinicopathological parameters were unclear. In the present study, PRKCA protein expression was found to be positively associated with HDGF protein expression in LADC tissues, but no correlations were observed between PRKCA and HDGF mRNA levels in patients with LADC. These findings are consistent with the previous study that demonstrated that PRKCA expression is post-transcriptionally regulated by miR-296-3p, which is modulated by HDGF (11). In addition, high HDGF and PRKCA expression was associated with a significantly reduced survival rate, whereas patients with low HDGF and PRKCA expression had a higher survival rate. However, HDGF and PRKCA expression levels were not independent prognostic factors for LADC.
Overall, the current study revealed that expression levels of HDGF and PRKCA are significantly associated in LADC, and the inhibition of HDGF and PRKCA expression may represent an effective approach for the treatment of the disease. The present study provides the molecular foundation for the application of HDGF and PRKCA inhibitors as a therapeutic strategy in the future.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural Science Foundation of China (grant nos. 81702295, 81660389 and 81602029), the China Postdoctoral Science Foundation (grant no. 2017M613008), the Yunnan Province Applied Foundation Project (grant nos. 2017FB127 and 2017FB126), the Yunnan Province-Kunming Medical University Joint Foundation for Applied Basic Research (grant no. 2015FB074) and the Integrative Therapy Innovation Team for Yunnan Regional Cancer (grant no. 2017HC006).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
HHJ, QFF, CLG, RLL, ZL and BZZ performed the research. RCL, WYF and XS designed the study. CYL, YW and YBX performed the statistical analysis, and HHJ, QFF and XS wrote the paper. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Informed consent from the patients and approval from the Ethics Committee of the Taizhou Hospital of Zhejiang Province and Kunming Medical University (approval no. KY201726) were obtained prior to the use of clinical materials for research purposes.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.McIntyre A, Ganti AK. Lung cancer-a global perspective. J Surg Oncol. 2017;115:550–554. doi: 10.1002/jso.24532. [DOI] [PubMed] [Google Scholar]
- 3.Glatzer M, Elicin O, Ramella S, Nestle U, Putora PM. Radio(chemo)therapy in locally advanced nonsmall cell lung cancer. Eur Respir Rev. 2016;25:65–70. doi: 10.1183/16000617.0053-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel MI, Cheng I, Gomez SL. US lung cancer trends by histologic type. Cancer. 2015;121:1150–1152. doi: 10.1002/cncr.29180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang L, Li M, Wu N, Chen Y. Time trends in epidemiologic characteristics and imaging features of lung adenocarcinoma: A population study of 21,113 cases in China. PLoS One. 2015;10:e0136727. doi: 10.1371/journal.pone.0136727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ettinger DS, Wood DE, Aisner DL, Akerley W, Bauman J, Chirieac LR, D'Amico TA, DeCamp MM, Dilling TJ, Dobelbower M, et al. Non-small cell lung cancer, version 5.2017, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2017;15:504–535. doi: 10.6004/jnccn.2017.0050. [DOI] [PubMed] [Google Scholar]
- 7.Jung CY, Antonia SJ. Tumor immunology and immune checkpoint inhibitors in non-small cell lung cancer. Tuberc Respir Dis (Seoul) 2018;81:29–41. doi: 10.4046/trd.2017.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rabbani M, Kanevsky J, Kafi K, Chandelier F, Giles FJ. Role of artificial intelligence in the care of patients with nonsmall cell lung cancer. Eur J Clin Invest. 2018:48. doi: 10.1111/eci.12901. doi: 10.1111/eci.12901. [DOI] [PubMed] [Google Scholar]
- 9.Walter AO, Sjin RT, Haringsma HJ, Ohashi K, Sun J, Lee K, Dubrovskiy A, Labenski M, Zhu Z, Wang Z, et al. Discovery of a mutant-selective covalent inhibitor of EGFR that overcomes T790M-mediated resistance in NSCLC. Cancer Discov. 2013;3:1404–1415. doi: 10.1158/2159-8290.CD-13-0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Passiglia F, Listí A, Castiglia M, Perez A, Rizzo S, Bazan V, Russo A. EGFR inhibition in NSCLC: New findings.. and opened questions? Crit Rev Oncol Hematol. 2017;112:126–135. doi: 10.1016/j.critrevonc.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 11.Fu Q, Song X, Liu Z, Deng X, Luo R, Ge C, Li R, Li Z, Zhao M, Chen Y, et al. miRomics and proteomics reveal a miR-296-3p/PRKCA/FAK/Ras/c-Myc feedback loop modulated by HDGF/DDX5/β-catenin complex in lung adenocarcinoma. Clin Cancer Res. 2017;23:6336–6350. doi: 10.1158/1078-0432.CCR-16-2813. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura H, Kambe H, Egawa T, Kimura Y, Ito H, Hayashi E, Yamamoto H, Sato J, Kishimoto S. Partial purification and characterization of human hepatoma-derived growth factor. Clin Chim Acta. 1989;183:273–284. doi: 10.1016/0009-8981(89)90361-6. [DOI] [PubMed] [Google Scholar]
- 13.Bao CH, Liu K, Wang XT, Ma W, Wang JB, Wang C, Jia YB, Wang NN, Tan BX, Song QX, Cheng YF. Prognostic role of hepatoma-derived growth factor in solid tumors of Eastern Asia: A systematic review and meta-analysis. Asian Pac J Cancer Prev. 2015;16:1803–1811. doi: 10.7314/APJCP.2015.16.5.1803. [DOI] [PubMed] [Google Scholar]
- 14.Zhao J, Ma MZ, Ren H, Liu Z, Edelman MJ, Pan H, Mao L. Anti-HDGF targets cancer and cancer stromal stem cells resistant to chemotherapy. Clin Cancer Res. 2013;19:3567–3576. doi: 10.1158/1078-0432.CCR-12-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baier G. The PKC gene module: Molecular biosystematics to resolve its T cell functions. Immunol Rev. 2003;192:64–79. doi: 10.1034/j.1600-065X.2003.00018.x. [DOI] [PubMed] [Google Scholar]
- 16.Guo Y, Bao Y, Ma M, Zhang S, Zhang Y, Yuan M, Liu B, Yang Y, Cui W, Ansong E, et al. Clinical significance of the correlation between PLCE 1 and PRKCA in esophageal inflammation and esophageal carcinoma. Oncotarget. 2017;8:33285–33299. doi: 10.18632/oncotarget.16635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paraboschi EM, Rimoldi V, Soldá G, Tabaglio T, Dall'Osso C, Saba E, Vigliano M, Salviati A, Leone M, Benedetti MD, et al. Functional variations modulating PRKCA expression and alternative splicing predispose to multiple sclerosis. Hum Mol Genet. 2014;23:6746–6761. doi: 10.1093/hmg/ddu392. [DOI] [PubMed] [Google Scholar]
- 18.Rosenberg S, Simeonova I, Bielle F, Verreault M, Bance B, Le Roux I, Daniau M, Nadaradjane A, Gleize V, Paris S, et al. A recurrent point mutation in PRKCA is a hallmark of chordoid gliomas. Nat Commun. 2018;9:2371. doi: 10.1038/s41467-018-04622-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edge SB, Byrd DR, Compton CC, et al. AJCC 7th Edition Cancer Staging Manual. 2010. https://cancerstaging.org/references-tools/deskreferences/Pages/default.aspx. 2015 Oct 22;
- 20.Liu Z, Li L, Yang Z, Luo W, Li X, Yang H, Yao K, Wu B, Fang W. Increased expression of MMP9 is correlated with poor prognosis of nasopharyngeal carcinoma. BMC Cancer. 2010;10:270. doi: 10.1186/1471-2407-10-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tu L, Liu Z, He X, He Y, Yang H, Jiang Q, Xie S, Xiao G, Li X, Yao K, Fang W. Over-expression of eukaryotic translation initiation factor 4 gamma 1 correlates with tumor progression and poor prognosis in nasopharyngeal carcinoma. Mol Cancer. 2010;9:78. doi: 10.1186/1476-4598-9-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu H, Jin X, Yuan Y, Deng P, Jiang L, Zeng X, Li XS, Wang ZY, Chen QM. Prognostic value rom integrative analysis of transcription factors c-Jun and Fra-1 in oral squamous cell carcinoma: A multicenter cohort study. Sci Rep. 2017;7:7522. doi: 10.1038/s41598-017-05106-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Philipsen S. Expression data for early stage NSCLC. 2010. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE19188. 2017 Jun 10;
- 24.Cai S, Cai J, Jiang WG, Ye L. Kidins220 and tumour development: Insights into a complexity of cross-talk among signalling pathways (Review) Int J Mol Med. 2017;40:965–971. doi: 10.3892/ijmm.2017.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bica-Pop C, Cojocneanu-Petric R, Magdo L, Raduly L, Gulei D, Berindan-Neagoe I. Overview upon miR-21 in lung cancer: Focus on NSCLC. Cell Mol Life Sci. 2018;75:3539–3551. doi: 10.1007/s00018-018-2877-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jing P, Zhao N, Xie N, Ye M, Zhang Y, Zhang Z, Li M, Lai X, Zhang J, Gu Z. miR-24-3p/FGFR3 Signaling as a novel axis is involved in epithelial-mesenchymal transition and regulates lung adenocarcinoma progression. J Immunol Res. 2018;2018:2834109. doi: 10.1155/2018/2834109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li W, Qiu T, Guo L, Ling Y, Gao Y, Ying J, He J. Primary and acquired EGFR T790M-mutant NSCLC patients identified by routine mutation testing show different characteristics but may both respond to osimertinib treatment. Cancer Lett. 2018;423:9–15. doi: 10.1016/j.canlet.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 28.Lin L, Zhao J, Hu J, Zou G, Huang F, Han J, He Y, Cao X. Current smoking has a detrimental effect on survival for epidermal growth factor receptor (EGFR) and anaplastic lymphoma kinase (ALK) negative advanced non-squamous Non-small cell lung cancer (NSCLC) patients treated with pemetrexed continuation maintenance. J Cancer. 2018;9:2140–2146. doi: 10.7150/jca.22378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su C, Cheng X, Li Y, Han Y, Song X, Yu D, Cao X, Liu Z. MiR-21 improves invasion and migration of drug-resistant lung adenocarcinoma cancer cell and transformation of EMT through targeting HBP1. Cancer Med. 2018;7:2485–2503. doi: 10.1002/cam4.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamura H, Izumoto Y, Kambe H, Kuroda T, Mori T, Kawamura K, Yamamoto H, Kishimoto T. Molecular cloning of complementary DNA for a novel human hepatoma-derived growth factor. Its homology with high mobility group-1 protein. J Biol Chem. 1994;269:25143–25149. [PubMed] [Google Scholar]
- 31.Enomoto H, Nakamura H, Liu W, Nishiguchi S. Hepatoma-derived growth factor: Its possible involvement in the progression of hepatocellular carcinoma. Int J Mol Sci. 2015;16:14086–14097. doi: 10.3390/ijms160614086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li M, Shen J, Wu X, Zhang B, Zhang R, Weng H, Ding Q, Tan Z, Gao G, Mu J, et al. Downregulated expression of hepatoma- derived growth factor (HDGF) reduces gallbladder cancer cell proliferation and invasion. Med Oncol. 2013;30:587. doi: 10.1007/s12032-013-0587-7. [DOI] [PubMed] [Google Scholar]
- 33.Wang L, Jiang Q, Hua S, Zhao M, Wu Q, Fu Q, Fang W, Guo S. High nuclear expression of HDGF correlates with disease progression and poor prognosis in human endometrial carcinoma. Dis Markers. 2014;2014:298795. doi: 10.1155/2014/298795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Sun J, Yang G, Liu Z, Guo S, Zhao R, Xu K, Wu X, Zhang Z. Downregulation of the expression of HDGF attenuates malignant biological behaviors of hilar cholangiocarcinoma cells. Mol Med Rep. 2015;12:4713–4719. doi: 10.3892/mmr.2015.3922. [DOI] [PubMed] [Google Scholar]
- 35.Kim YD, Jeon JY, Woo HJ, Lee JC, Chung JH, Song SY, Yoon SK, Baek SH. Interleukin-1beta induces MUC2 gene expression and mucin secretion via activation of PKC-MEK/ERK, and PI3K in human airway epithelial cells. J Korean Med Sci. 2002;17:765–771. doi: 10.3346/jkms.2002.17.6.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee HY, Crawley S, Hokari R, Kwon S, Kim YS. Bile acid regulates MUC2 transcription in colon cancer cells via positive EGFR/PKC/Ras/ERK/CREB, PI3K/Akt/IkappaB/NF-kappaB and p38/MSK1/CREB pathways and negative JNK/c-Jun/AP-1 pathway. Int J Oncol. 2010;36:941–953. doi: 10.3892/ijo_00000573. [DOI] [PubMed] [Google Scholar]
- 37.O'Brian C, Vogel VG, Singletary SE, Ward NE. Elevated protein kinase C expression in human breast tumor biopsies relative to normal breast tissue. Cancer Res. 1989;49:3215–3217. [PubMed] [Google Scholar]
- 38.Kopp R, Noelke B, Sauter G, Schildberg FW, Paumgartner G, Pfeiffer A. Altered protein kinase C activity in biopsies of human colonic adenomas and carcinomas. Cancer Res. 1991;51:205–210. [PubMed] [Google Scholar]
- 39.Wang C, Wang X, Liang H, Wang T, Yan X, Cao M, Wang N, Zhang S, Zen K, Zhang C, Chen X. miR-203 inhibits cell proliferation and migration of lung cancer cells by targeting PKCα. PLoS One. 2013;8:e73985. doi: 10.1371/journal.pone.0073985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lahn M, Sundell K, Köhler G. The role of protein kinase C-alpha in hematologic malignancies. Acta Haematol. 2006;115:1–8. doi: 10.1159/000089458. [DOI] [PubMed] [Google Scholar]
- 41.Lahn M, Su C, Li S, Chedid M, Hanna KR, Graff JR, Sandusky GE, Ma D, Niyikiza C, Sundell KL, et al. Expression levels of protein kinase C-alpha in non-small-cell lung cancer. Clin Lung Cancer. 2004;6:184–189. doi: 10.3816/CLC.2004.n.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.