Abstract
In patients with gastric cancer (GC), peritoneal recurrence is a common risk and associated with poor prognosis. A novel biomarker for the prediction of high-risk peritoneal recurrence in patients with GC is desirable. The present study investigated the effectiveness of exosome-encapsulated microRNAs (ex-miRNAs) as minimally invasive biomarkers in patients with GC that received curative surgery. Recurrence-specific ex-miRNAs were selected following comparison of miRNA microarray data from patients with TNM stage II GC with peritoneal recurrence (n=3) and without peritoneal recurrence following curative surgery (n=3), and three healthy volunteers. In this analysis, exosome-encapsulated miRNA-21 (ex-miR-21) and exosomal miR-92a (ex-miR-92a) exhibited the greatest alterations in expression patterns. Using plasma exosome samples collected from another 129 patients with stage II and III GC, the present study investigated the potential value of ex-miR-21 and ex-miR-92a as biomarkers. Ex-miRNA levels were measured using TaqMan miRNA assays. Ex-miR-21 levels were significantly higher and ex-miR-92a levels were significantly lower in samples from patients with GC compared with healthy controls. The overall survival (OS) and peritoneal recurrence-free survival (PRFS) were poorer in stage II and III patients with high ex-miR-21 levels than in patients with low miR-21 levels. OS and PRFS of stage II and III patients with low ex-miR92a levels were significantly worse than those with high ex-miR92a levels. Cox multivariate analyses indicated that ex-miR-21 and ex-miR-92a were independent prognostic factors for OS and PRFS in stage II and III GC. A negative correlation was detected between expression levels of miR-21 and programmed cell death protein 4 mRNA, and miR-92a and prostaglandin E receptor 4 mRNA. Therefore, ex-miR-21 and ex-miR-92a may function as effective and minimally invasive biomarkers for the prediction of peritoneal recurrence and the prognosis of patients with stage II/III GC.
Keywords: gastric cancer, liquid biomarker, exosome, miRNA-21, miRNA-92a, prognosis, peritoneal recurrence
Introduction
Gastric cancer (GC) is the third leading cause of cancer-associated death worldwide (1). In particular, the prevalence of GC is high in Japan and other countries in East Asia (2). Although there has been a rise in the overall survival rate due to curative resection (R0) with lymph node dissection plus adjuvant chemotherapy, the survival rate of patients with state II and stage III GC was 30% in 2011 (3). Peritoneal recurrence is the most frequent type of recurrence in patients with GC and is associated with poor prognosis (4). Peritoneal dissemination is detected in 14% of patients with GC at the time of initial diagnosis, and the median survival time for these patients is ~4 months (5). Various treatment regimens have been utilized to treat peritoneal recurrence, including systemic chemotherapy, intraperitoneal chemotherapy, hyperthermia and aggressive surgery; however, none of these treatments have led to a satisfactory clinical outcome (6,7). Among all cases diagnosed as curable without metastasis prior to surgery, 10–20% present peritoneal dissemination during surgery (8). Prophylactic treatment strategies, including extensive intraoperative peritoneal lavage and intraperitoneal chemotherapy are associated with improved survival rates (9), and their effectiveness is associated with the early detection of patients at high risk of developing peritoneal metastasis (10).
Peritoneal fluid lavage cytology (CY) utilizes peritoneal washing as a means to identify patients with a high likelihood of peritoneal recurrence, and CY is a common clinical practice in Japan. However, this procedure is not practiced in Western countries, due to low sensitivity for the detection of recurrence (11). In fact, evidence suggests that patients with negative CY (CY0) occasionally develop peritoneal recurrence post-surgery (12). A prospective randomized study reported peritoneal recurrence in 14.6% of CY0 patients with stage II–III GC that underwent R0 resection (3). This observation supports the need for the discovery of novel biomarkers for the screening of patients with a high risk of peritoneal recurrence.
Circulating microRNAs (miRNAs/miRs) in plasma or serum have attracted attention as minimally invasive biomarkers for the diagnosis and prognosis of various types of cancer, including GC (13,14). miRNAs are small (23–35 nucleotides) non-coding RNAs that negatively regulate the expression of target genes at the post-transcriptional level via RNA interference (13). miRNAs serve an important role in several processes associated with carcinogenesis, including cellular proliferation, apoptosis and differentiation, metastasis (15). Grady and Tewai (16) reported abnormal expression of miRNAs in cancer cells, and the role they serve in the commencement and advancement of cancer in the form of oncogenes or tumor suppressor genes (16). These miRNAs have been identified in an extremely stable form within the exosomes in the plasma and serum, and are protected from endogenous RNase activity (17). Exosomes are vesicles with a small diameter (50–150 nm) derived from the luminal membranes, which are released following fusion with the cell membrane (18). Proteins and selectively packaged RNAs, including miRNAs, are encapsulated in exosomes in a stable and intact form (19). These exosomes may transfer the encapsulated components to other cells (20–22). Exosome-encapsulated miRNAs (ex-miRNAs) have recently attracted attention as promising predictive and prognostic biomarkers in patients with cancer (23–27). However, the potential role of plasma ex-miRNAs in the prediction of peritoneal recurrence in patients with GC, to the best of our knowledge, has not yet been investigated.
The present study aimed to clarify the applicability of circulating plasma ex-miRNAs for the prediction of peritoneal recurrence in patients with stage II and III GC that underwent R0 resection.
Patients and methods
Study design
The inclusion criteria for the healthy controls were: i) Did not have cancer; ii) No abnormalities in the blood test results; iii) normal respiratory function; iv) normal cardiovascular function; and v) normal gastrointestinal examination. The inclusion criteria for the patients with GC were: i) Histopathologically confirmed diagnosis of stage II or III GC; ii) R0 resection (with no tumor cells at the margin); iii) no evidence of hepatic, peritoneal or distant metastasis; iv) no tumor cells in the peritoneal fluid on cytological analysis; v) aged 20–85 years; vi) no previous treatment for cancer except for the initial gastric resection for the primary lesion; and vi) adequate organ function. The present study first profiled peritoneal recurrence-specific plasma ex-miRNAs using a miRNA array. Patients with stage II GC with peritoneal recurrence after surgery (n=3), stage II GC without peritoneal recurrence after surgery (n=3) and healthy control subjects (n=3) were examined. The clinicopathological characteristics of the patients are provided in Table SI. The average age of the 6 patients with GC was 66 years (range, 63–71 years), and there were 4 men and 2 women. The average age of the 3 healthy controls was 65 years old (range, 62–71 years), and there were 2 men and 1 woman. These patients and healthy controls were recruited between January 2006 and October 2006 at Teikyo University Hospital (Tokyo, Japan). Peritoneal recurrence-specific ex-miRNAs were profiled with a miRNA array using plasma exosomes as described below. Subsequently, the potential of selected ex-miRNAs was investigated using other samples collected from 129 patients with GC and 20 healthy controls. The average age of the 129 patients with gastric cancer was 68 years old (range, 36–82 years), and there were 90 men and 39 women. The average age of the 20 healthy controls was 60 years old (range, 53–65 years), and there were 14 men and 6 women. These patients were recruited between November 2006 and December 2015 at Teikyo University Hospital and comprised 49 cases with stage II and 80 cases with stage III GC. The cancer stage was determined according to the tumor-node-metastasis (TNM) classification by the International Union Against Cancer (28). The inclusion criteria of patients were as follows: Japanese patients with GC with TNM stage II or III. Since peritoneal recurrence after surgery was not observed in patients with stage I GC, these patients were excluded from the present study. In addition, recurrent cases without peritoneal dissemination were not included in the present study. The median follow-up period was 3.4 years (range, 0.04–5.5 years). Blood samples were collected prior to treatment, and the primary tumor tissues and matched normal tissues adjacent to the tumor tissues (≥4 cm away) of the same patient were collected during resection. Tissues were frozen in liquid nitrogen (−196°C) and stored at −80°C. until further use. The patients received TS-1 for 1 year as standard adjuvant chemotherapy. The study protocol conformed with the guidelines of the Ethics and Indications Committee of Teikyo University (Tokyo, Japan), and was approved by the Review Board of Teikyo University (approval no. 09-081-3). Written informed consent was obtained from all patients.
Purification of exosomes from plasma samples
Peripheral blood was centrifuged at 1,200 × g for 10 min at 4°C to obtain plasma. Plasma (~1 ml) samples were used for microarray analysis and reverse transcription-quantitative PCR (RT-qPCR). The exosomes were purified from the plasma by ultracentrifugation at 100,000 × g for 70 min at 4°C. The pellets were stored at −80°C for microarray and RT-qPCR analyses.
Transmission electron microscopy
The morphology of the isolated exosomes was confirmed by transmission electron microscopy (Hitachi H-7600; Hitachi, Ltd.), as previously described (23). Approximately 5 µl of isolated exosome sample was placed on Parafilm. A carbon coated 400 mesh copper grid was positioned on the top of the drop for 10 sec and washed with a droplet of distilled water. The grid was contrasted by adding a drop of 2% uranyl acetate on Parafilm and placing the grid on top of drop for 10 sec and excess liquid was removed by gently using absorbing paper. After drying, the sample was submitted to transmission electron microscopy. A total of 10 fields of view per sample were analyzed and samples were viewed at a magnification of ×200,000.
Total RNA extraction from exosomes and tissues
Total RNAs (including miRNAs) from exosomes were extracted using the miRNeasy serum/plasma kit (Qiagen, Inc.), and total RNAs (including miRNAs) of the tissues were extracted using the miRNeasy Mini kit (Qiagen, Inc.). The exosomes purified from 1 ml of plasma were diluted with 1 ml of QIAzol Lysis reagent (Qiagen, Inc.). Subsequent extraction and analysis using cartridges (Qiagen China Co., Ltd.) was performed according to the manufacturer's protocol. The quality of the extracted RNA was analyzed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.).
miRNA microarray analysis
Exosomal miRNA expression profiles were investigated with 3D-Gene Human miRNA Oligo chips ver. 20 (Toray Industries, Inc.), according to the manufacturer's protocol. Fluorescence signals were scanned and analyzed using a 3D-gene scanner (Toray Industries, Inc.). A total of 2,578 genes were mounted on the chip. The raw data from each spot were normalized by the subtraction of the background signal mean intensity, as determined by the 95% CI of the signal intensities of all blank spots. Any signal intensity in both duplicate spots at >2 SD of the background signal intensity was considered as a valid measurement.
RT-qPCR for miRNAs from exosomes and tissues
The expression levels of miRNAs from plasma exosomes and tissues were assayed using RT-qPCR. The reverse transcription protocol was: 30 min at 16°C, 30 min at 42°C and 5 min at 85°C. Complementary DNA (cDNA) was synthesized from total RNA using TaqMan MicroRNA primers specific for miRNA-21 (miR-21), miRNA-92a (miR-92a), Caenorhabditis elegans miR-39 (Cel-miR-39) and miRNA-16 (miR-16) (Thermo Fisher Scientific, Inc.) and the TaqMan Micro-RNA Reverse Transcription kit (Thermo Fisher Scientific, Inc.). Cel-miR-39 was selected as the external control, while miR-16 served as an internal control, as previously described (17). In the tissues, cDNA was synthesized from total RNA using TaqMan miRNA primers specific for miR-21 (assay ID 000397), miR-92a (assay ID 000431) and RNA, U6 small nuclear 6, pseudogene (assay ID 001093; Thermo Fisher Scientific, Inc.) and the TaqMan Micro-RNA Reverse Transcription kit (Thermo Fisher Scientific, Inc.). U6 small nuclear 6 was used as an internal control. qPCR was performed using TaqMan Universal PCR Master mix (Thermo Fisher Scientific, Inc.) and the StepOne™ system (Thermo Fisher Scientific, Inc.). PCR reaction mixtures were incubated at 95°C for 10 min for denaturation, followed by 45 amplification cycles of 95°C for 15 sec and 60°C for 1 min, followed by an extension at 40°C for 30 sec. The experiments were repeated three times. Relative quantification of miRNA expression was performed using the 2−ΔΔCq method, as previously described (24,29).
RT-qPCR for prostaglandin E receptor 4 (EP4) and programmed cell death protein 4 (PDCD4) mRNA in tissues
Total RNA was extracted from primary cancer tissues collected from patients with GC using the miRNeasy Mini kit (Qiagen, Inc.). Random hexamer primers and SuperScript II reverse transcriptase (Thermo Fisher Scientific, Inc.) were used to obtain cDNA according to the manufacturer's protocol. RT-qPCR for EP4, PDCD4 and GAPDH (internal control) was performed using the LightCycler (Roche Applied Science). Primers for PDCD4 (cat. no. Hs00377253) and EP4 (cat. no. Hs00168761) were purchased (Thermo Fisher Scientific, Inc.). The sequences for these primers have not been disclosed by the supplier. The amplification of these mRNAs was performed using the TaqMan Universal Master mix II (Thermo Fisher Scientific, Inc.). The thermocycling conditions were as follows: 95°C For 10 min; followed by 45 cycles of 95°C for 15 sec, 60°C for 1 min, and 40°C for 30 sec. The mRNA expression levels of EP4 and PDCD4 were normalized to GAPDH mRNA expression.
Postoperative surveillance
The follow-up program comprised interim history, physical examination, hematology and blood chemistry and was performed every 3 months for the first postoperative year and every 6 months thereafter. Computed tomography or abdominal ultrasonography were examined every 6 months. Evidence of peritoneal recurrence was comprehensively diagnosed using CT, tumor marker (CA125), paracentesis and autopsy Primary tumor tissues from 48 patients with stage II GC (n=24) and stage III GC (n=24) were examined. A significant negative correlation was identified between miR-21 and PDCD4 mRNA expression (P<0.01), as well as between miR-92a and EP4 mRNA expression (P<0.01). These results indicated that PDCD4 expression may be negatively regulated by miR-21, whereas EP4 mRNA expression may be negatively regulated by miR-92a.
Statistical analysis
The data are expressed as the mean ± standard deviation. Experiments were repeated three times. In the clinicopathological study and the survival study, patients were split into two groups, with one group exhibiting high expression levels of ex-miR-21 and ex-miR-92a, and the other group exhibiting low expression levels of these markers. The association between miRNA expression and clinicopathological characteristics was analyzed using Student's t-test, a χ2 test or a one-way ANOVA with a post-hoc Tukey's test. Overall survival (OS) and peritoneal recurrence-free survival (PRFS) were analyzed using the Kaplan-Meier survival curve method, and the resulting data were examined using log-rank and Wilcoxon tests. Cox proportional hazard regression analysis was used to estimate the univariate and multivariate hazard ratios for OS and PRFS. Multivariate analysis was performed for the factors that exhibited significance in the univariate analysis. Correlations were determined using Pearson's rank correlation analysis. Target genes of miR-21 and miR-92a were determineded using miRBase (http://www.mirbase.org/index.shtml) and miRWalk 2.0 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/index.html) databases. All P-values are two-sided, and P<0.05 was considered to indicate a statistically significant difference. Statistical analyses were performed using JMP v9.0 software (SAS Institute, Inc.).
Results
Identification of exosomes in plasma
As presented in Fig. 1, exosomes were identified following the ultracentrifugation of samples from patients with GC. In these samples, round microvesicles with diameters of 50–150 nm were observed.
Figure 1.
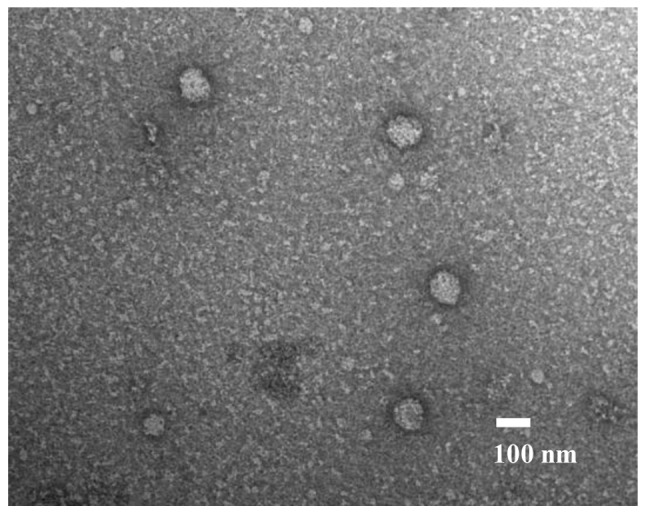
Exosomes isolated from the plasma of patients with gastric cancer captured under a transmission electron microscope. Scale bar, 100 nm.
Ex-miRNA profile of patients with GC
The clinical characteristics and background information (sex, age, nationality and medical history) of 6 patients with GC and 3 healthy controls whose samples were used in miRNA microarray analyses are described in Table SI. As presented in Table I, the top five upregulated and downregulated ex-miRNAs in the samples collected from all the patients are shown in Table SI were reported. Among the upregulated miRNAs, miR-21-5p (miR-21; MIMAT0000076) expression was markedly altered in the recurrence group compared with the healthy control and non-recurrence groups. Among the downregulated miRNAs, miR-92a-3p (miR-92a; MIMAT0000092) expression exhibited the greatest alterations in the samples from the recurrence group compared with those from the healthy control and non-recurrence groups. Therefore, miR-21 and miR-92a were selected as biomarkers with potential application for the prediction of peritoneal recurrence in patients with GC.
Table I.
Five most up- or downregulated miRNAs in plasma exosomes of patients with stage II GC with peritoneal recurrence according to miRNA array analysis.
| A, Upregulated | |||||
|---|---|---|---|---|---|
| Fold change | |||||
| Ranks | microRNA | MirBase no. | Peritoneal recurrent GC vs. healthy controls | Peritoneal recurrent GC vs. non-recurrent GC | |
| 1 | miR-21-5p | MIMAT 0000076 | 3.28 | 2.71 | |
| 2 | miR-204-3p | MIMAT 0022693 | 3.14 | 2.21 | |
| 3 | miR-6879-5p | MIMAT 0027658 | 3.12 | 2.17 | |
| 4 | miR-3928-3p | MIMAT 0018205 | 3.07 | 2.07 | |
| 5 | miR-4476 | MIMAT 0019003 | 3.03 | 2.14 | |
| B, Downregulated | |||||
| Fold change | |||||
| Ranks | microRNA | MirBase no. | Peritoneal recurrent GC vs. healthy controls | Peritoneal recurrent GC vs. non-recurrent GC | |
| 1 | miR-92a-3p | MIMAT 0000092 | 0.31 | 0.36 | |
| 2 | miR-6850-3p | MIMAT 0027601 | 0.34 | 0.37 | |
| 3 | miR-3944-3p | MIMAT 0018360 | 0.37 | 0.42 | |
| 4 | miR-23b-3p | MIMAT 0000418 | 0.38 | 0.45 | |
| 5 | miR-4686 | MIMAT 0019773 | 0.41 | 0.50 | |
GC, gastric cancer; miR/miRNA, microRNA.
Comparison of ex-miR-21 and ex-miR-92a levels in patients with GC and healthy controls
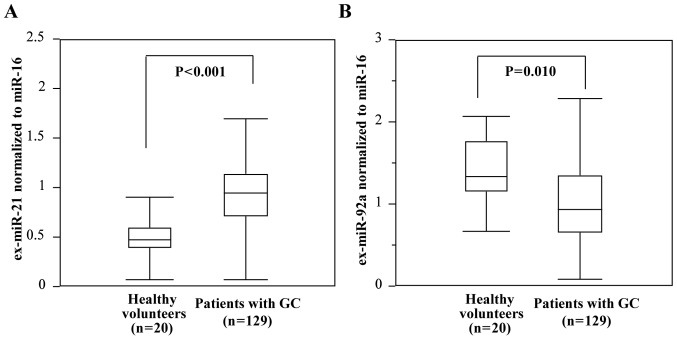
The expression levels of ex-miR-21 were determined to be significantly higher in patients with GC than in healthy controls, whereas those of ex-miR-92a were significantly lower in patients with GC than in healthy controls (Fig. 2).
Figure 2.
Comparison of ex-miR-21 and ex-miR-92a levels between patients with GC and healthy volunteers. (A) Comparison of ex-miR-21 levels between patients with GC (n=129) and healthy volunteers (n=20). (B) Comparison of ex-miR-92a levels between patients with GC (n=129) and healthy volunteers (n=20). ex-, exosome-encapsulated; GC, gastric cancer; miR, microRNA.
Clinicopathological characteristics and ex-miR-21 and ex-miR-92a expression
In Table II, the associations between the expression levels of ex-miR-21 and ex-miR-92a, and clinicopathological characteristics were presented. The patients were split into two groups as follows: One exhibiting high expression levels of ex-miR-21 and ex-miR-92a, and the other with low expression levels of these markers. The cut-off levels were determined as 0.93 for ex-miR-21 and 1.04 for ex-miR-92a, which were the mean levels. A statistically significant association was observed between ex-miR-21 expression and TNM stage. Sex, tumor size, differentiation, lymphatic invasion, venous invasion and lymph node metastasis were not identified to exhibit a significant association with the levels of ex-miR-21. The analysis of ex-miR-92a levels revealed no significant association with any of the clinicopathological characteristics.
Table II.
Association between clinicopathological characteristics and plasma levels of ex-miR-92a and ex-miR-21.
| ex-miR-92a | ex-miR-21 | |||||
|---|---|---|---|---|---|---|
| Variables | High (n=61), n (%) | Low (n=68), n (%) | P-value | High (n=58), n (%) | Low (n=71), n (%) | P-value |
| Sex | 0.33 | 0.57 | ||||
| Male | 40 (65.6) | 50 (73.5) | 42 (72.4) | 48 (67.6) | ||
| Female | 21 (34.4) | 18 (26.5) | 16 (27.6) | 23 (32.4) | ||
| Tumor size, cm | 0.90 | 0.26 | ||||
| <5 | 20 (32.8) | 23 (33.8) | 16 (27.6) | 27 (38.0) | ||
| ≥5 | 41 (67.2) | 45 (66.2) | 42 (72.4) | 44 (62.0) | ||
| Differentiation | 0.80 | 0.80 | ||||
| Well/moderate | 9 (14.8) | 9 (13.2) | 9 (15.5) | 9 (12.7) | ||
| Poorly/other | 52 (85.2) | 59 (86.8) | 49 (84.5) | 62 (87.3) | ||
| Lymphatic invasion | 0.10 | 0.26 | ||||
| Ly(−) | 9 (14.8) | 15 (22.1) | 8 (13.8) | 16 (22.5) | ||
| Ly(+) | 52 (85.2) | 53 (77.9) | 50 (86.2) | 55 (77.5) | ||
| Venous invasion | 0.75 | 0.26 | ||||
| V(−) | 12 (19.7) | 12 (17.6) | 8 (13.8) | 16 (22.5) | ||
| V(+) | 49 (80.3) | 56 (82.4) | 50 (86.2) | 55 (77.5) | ||
| Lymph node metastasis | 0.11 | 0.07 | ||||
| pN(−) | 8 (13.1) | 17 (25.0) | 7 (12.1) | 18 (25.4) | ||
| pN(+) | 53 (86.9) | 51 (75.0) | 51 (87.9) | 53 (74.6) | ||
| TNM Stage | 0.27 | 0.03 | ||||
| II | 22 (36.1) | 27 (39.7) | 16 (27.6) | 33 (46.5) | ||
| III | 39 (63.9) | 41 (60.3) | 42 (72.4) | 38 (53.5) | ||
ex-, exosome-encapsulated; miR, microRNA; Ly, lymphatic invasion; V, venous invasion Poorly, poorly differentiated.
Sensitivity, specificity, and accuracy of ex-miR-21 and ex-miR-92a in the detection of peritoneal recurrence
As presented in Table III, the sensitivity, specificity and accuracy of ex-miR-21 and ex-miR-92a was investigated for the detection of peritoneal recurrence. In this analysis, >60% sensitivity, specificity and accuracy were identified. The specificity and accuracy of ex-miR-21 were markedly higher than those of ex-miR-92a. The levels of ex-miR21 and ex-miR92a between peritoneal recurrence cases and peritoneal recurrence-free cases were determined (Table IV). The results revealed that ex-miR-21 expression was significantly higher in peritoneal recurrence cases than in peritoneal recurrence-free cases. By contrast, ex-miR-92a expression was significantly lower in peritoneal recurrence cases than in peritoneal recurrence-free cases.
Table III.
Sensitivity, specificity and accuracy of ex-miR-21 and ex-miR-92a for predicting peritoneal recurrence.
| Analysis item | ex-miR-21 | ex-miR-92a |
|---|---|---|
| Sensitivity (%) | 45/73 (61.6) | 46/73 (63.0) |
| Specificity (%) | 43/56 (76.8) | 34/56 (60.7) |
| Accuracy (%) | 88/129 (68.2) | 80/129 (62.0) |
ex-, exosome-encapsulated; miR, microRNA.
Table IV.
Comparison of peritoneal recurrence and ex-miR-21and ex-miR-92a levels.
| miR | Peritoneal recurrence cases | Peritoneal recurrence-free cases | P-value |
|---|---|---|---|
| Ex-miR-21 | 1.04±0.37 | 0.79±0.19 | 0.042a |
| Ex-miR-92a | 0.86±0.41 | 1.28±0.65 | 0.031a |
P<0.05, significant differences between peritoneal recurrence cases and peritoneal recurrence-free cases. ex-, exosome-encapsulated; miR, microRNA.
Association of ex-miR-21 and ex-miR92a levels
The present study examined the combination of ex-miR-21 and ex-miR-92a to clarify the association of these miRNAs (Table V). The numbers of patients in the ex-miR-21high/ex-miR-92alow, ex-miR-21low/ex-miR-92alow, ex-miR-21high/ex-miR92ahigh and ex-miR-21low/ex-miR-92ahigh groups were 28 (21.7%), 40 (31.0%), 30 (23.3%) and 31 (24.0%), respectively.
Table V.
Combination of ex-miR-21and ex-miR-92a levels.
| Combination of ex-miRs | Patients, n (%) |
|---|---|
| ex-miR-21high/ex-miR-92alow | 28 (21.7) |
| ex-miR-21low/ex-miR-92alow | 40 (31.0) |
| ex-miR-21high/ex-miR-92ahigh | 30 (23.3) |
| ex-miR-21low/ex-miR-92ahigh | 31 (24.0) |
ex-, exosome-encapsulated; miR, microRNA.
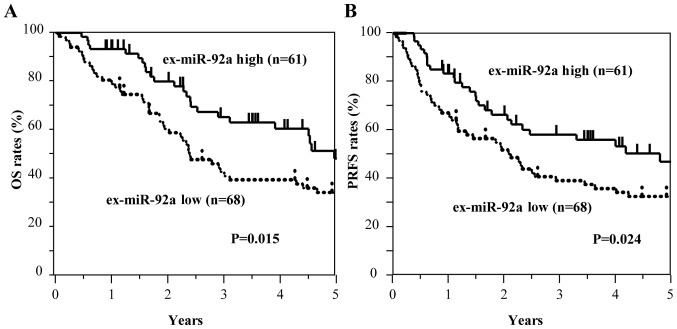
Kaplan-Meier survival curves of OS and PRFS based on ex-miRNA levels
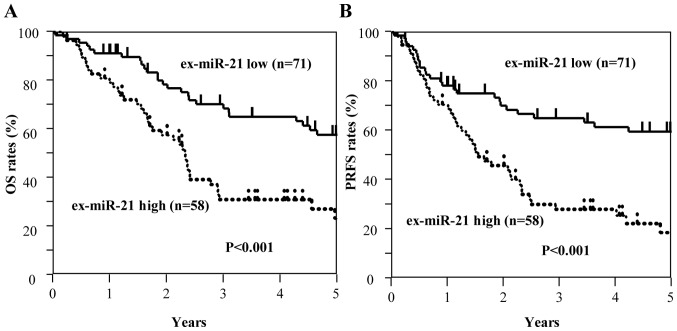
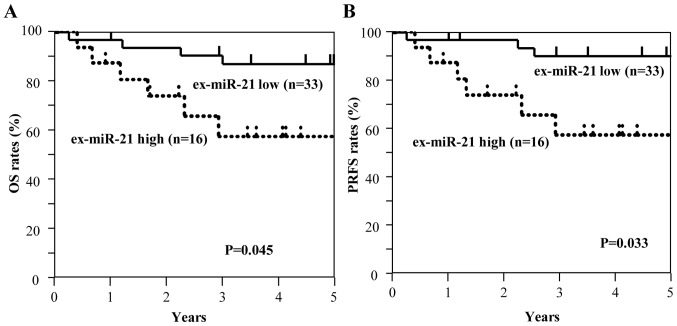
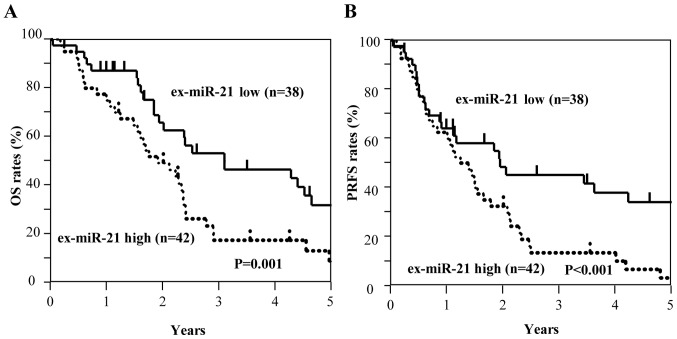
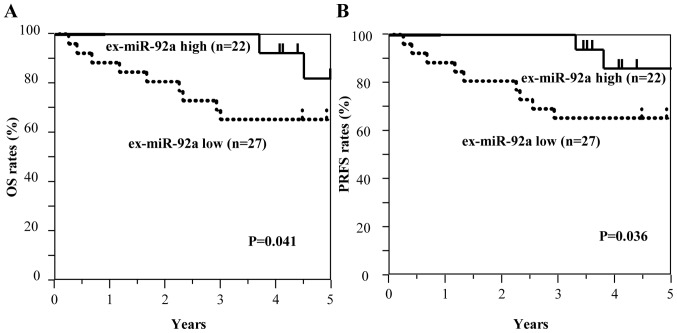
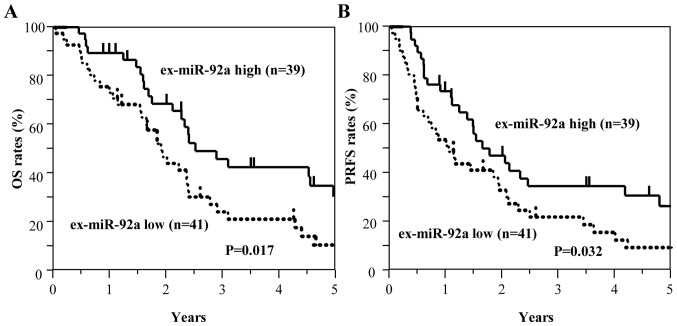
Among all patients with GC, including those with stage II and III GC (n=129), those with high ex-miR-21 expression exhibited significantly worse OS and PRFS compared with those with low ex-miR-21 levels (Fig. 3). Additionally, the present study analyzed the data at each tumor stage. In the analysis of patients with stage II GC (n=49), OS and PRFS rates were significantly lower for patients with high ex-miR-21 levels compared with patients with low ex-miR-21 expression (Fig. 4). In the analysis of patients with stage III GC (n=80), OS and PRFS rates were significantly lower for patients with high ex-miR-21 expression compared with patients with low ex-miR-21 expression (Fig. 5).
Figure 3.
Kaplan-Meier survival curves of OS and PRFS based on ex-miR-21 levels in patients with stage II and III GC. (A) Comparison of OS between patients with stage II and III GC with high and low expression levels of ex-miR-21 (n=129). (B) Comparison of PRFS between patients with stage II and III GC with high and low expression levels of ex-miR-21 (n=129). ex-miR-21, exosome-encapsulated microRNA-21; GC, gastric cancer; OS, overall survival; PRFS, peritoneal recurrence-free survival.
Figure 4.
Kaplan-Meier survival curves of OS and PRFS based on ex-miR-21 levels in patients with stage II GC. (A) Comparison of OS between patients with stage II GC with high and low expression levels of ex-miR-21 (n=49). (B) Comparison of PRFS between patients with stage II GC with high and low expression levels of ex-miR-21 (n=49). ex-miR-21, exosome-encapsulated microRNA-21; GC, gastric cancer; OS, overall survival; PRFS, peritoneal recurrence-free survival.
Figure 5.
Kaplan-Meier survival curves of OS and PRFS based on ex-miR-21 expression in patients with stage III GC. (A) Comparison of OS between patients with stage III GC with high and low expression levels of ex-miR-21 (n=80). (B) Comparison of PRFS between patients with stage III GC with high and low expression levels of ex-miR-21 (n=80). ex-miR-21, exosome-encapsulated microRNA-21; GC, gastric cancer; OS, overall survival; PRFS, peritoneal recurrence-free survival.
By contrast, the low ex-miR-92a expression group exhibited significantly worse OS and PRFS compared with the high ex-miR-92a expression group among all patients with GC (Fig. 6). In the analysis of each tumor stage, the patients with stage II GC and low ex-miR-92a levels exhibited significantly worse OS and PRFS compared with patients with high ex-miR-92a expression (n=49; Fig. 7). Among patients with stage III GC (n=80), the low ex-miR-92a expression group exhibited significantly worse OS and PRFS compared with the high ex-miR-92a expression group (Fig. 8).
Figure 6.
Kaplan-Meier survival curves of OS and PRFS based on ex-miR-92a levels in patients with stage II and III GC. (A) Comparison of OS in patients with stage II and III GC with high and low expression levels of ex-miR-92a (n=129). (B) Comparison of PRFS in patients with stage II and III GC with high and low expression levels of ex-miR-92a (n=129). ex-miR-92a, exosome-encapsulated microRNA-92a; GC, gastric cancer; OS, overall survival; PRFS, peritoneal recurrence-free survival.
Figure 7.
Kaplan-Meier survival curves of OS and PRFS based on ex-miR-92a levels in patients with stage II GC. (A) Comparison of OS between patients with stage II GC with high and low expression levels of ex-miR-92a (n=49). (B) Comparison of PRFS between patients with stage II GC with high and low expression levels of ex-miR-92a (n=49). ex-miR-92a, exosome-encapsulated microRNA-92a; GC, gastric cancer; OS, overall survival; PRFS, peritoneal recurrence-free survival.
Figure 8.
Kaplan-Meier survival curves of OS and PRFS based on ex-miR-92a levels in patients with stage III GC. (A) Comparison of OS between patients with stage III GC with high and low expression levels of ex-miR-92a (n=80). (B) Comparison of PRFS between patients with stage III GC with high and low expression levels of ex-miR-92a (n=80). ex-miR-92a, exosome-encapsulated microRNA-92a; GC, gastric cancer; OS, overall survival; PRFS, peritoneal recurrence-free survival.
These results suggest that high ex-miR-21 expression and low ex-miR-92a expression were associated with peritoneal recurrence and poor prognosis in patients with stage II and III GC.
Univariate and multivariate Cox analyses for OS and PRFS
For univariate analysis, sex, tumor size, lymph node metastasis, lymphatic invasion, venous invasion, differentiation, stage, ex-miR-21 expression and ex-miR-92a expression were examined. Multivariate analysis was performed for the variables that exhibited significance in univariate analysis. The results of the univariate and multivariate Cox analyses for OS and PRFS in all patients (n=129) were presented in Table VI. In the univariate analysis for OS and PRFS, tumor size, lymph node metastasis, stage, and ex-miR-21 and ex-miR-92a levels were significant, whereas stage, and ex-miR-21 and ex-miR-92a levels were determined to be significant for OS and PRFS in the multivariate analysis.
Table VI.
Univariate and multivariate Cox analyses for OS in patients with stage II and III gastric cancer.
| A, OS | ||||||
|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | |||||
| Variables | Regression coefficient | Hazard ratio (95% CI) | P-value | Regression coefficient | Hazard ratio (95% CI) | P-value |
| Sex | 0.10 | 1.10 (0.66–1.94) | 0.720 | |||
| Tumor size | 0.65 | 1.92 (1.13–3.44) | 0.015 | 0.19 | 1.21 (0.68–2.26) | 0.518 |
| Lymph node metastasis | 0.80 | 2.23 (1.09–5.38) | 0.026 | −0.39 | 0.68 (0.23–2.01) | 0.474 |
| Lymphatic invasion | 0.40 | 1.50 (0.80–3.13) | 0.219 | |||
| Venous invasion | 0.57 | 1.78 (0.92–3.85) | 0.087 | |||
| Differentiation | 0.13 | 1.14 (0.61–2.38) | 0.691 | |||
| Stage | 1.47 | 4.34 (2.43–8.35) | 0.001 | 1.67 | 5.30 (2.45–13.41) | 0.001 |
| ex-miR-21 | 1.16 | 3.20 (1.95–5.34) | 0.011 | 1.02 | 2.77 (1.66–4.71) | 0.027 |
| ex-miR-92a | −0.30 | 0.41 (0.21–0.68) | 0.014 | −0.78 | 0.46 (0.27–0.75) | 0.022 |
| B, PRFS | ||||||
| Univariate analysis | Multivariate analysis | |||||
| Variables | Regression coefficient | Hazard ratio (95% CI) | P-value | Regression coefficient | Hazard ratio (95% CI) | P-value |
| Sex | 0.68 | 1.97 (1.18–3.46) | 0.009 | 0.15 | 1.16 (0.66–2.11) | 0.616 |
| Tumor size | 0.94 | 2.56 (1.26–6.15) | 0.008 | −0.31 | 0.73 (0.26–2.12) | 0.552 |
| Lymph node metastasis | 0.49 | 1.63 (0.87–3.39) | 0.130 | |||
| Lymphatic invasion | 0.52 | 1.69 (0.90–3.50) | 0.104 | |||
| Venous invasion | 0.19 | 1.21 (0.65–2.51) | 0.567 | |||
| Differentiation | 1.65 | 5.19 (2.92–9.95) | 0.001 | 1.80 | 6.07 (2.87–14.73) | 0.001 |
| Stage | 1.11 | 3.03 (1.88–4.78) | 0.012 | 1.02 | 2.76 (1.36–5.27) | 0.025 |
| ex-miR-21 | −0.54 | 0.58 (0.31–0.83) | 0.023 | −0.69 | 0.67 (0.36–0.91) | 0.032 |
| ex-miR-92a | ||||||
ex-, exosome-encapsulated; miR, microRNA; OS, overall survival; PRFS, peritoneal recurrence-free survival.
Subsequently, the present study conducted analysis at each tumor stage. In the univariate analysis of patients with stage II GC, tumor size, ex-miR-21 levels and ex-miR-92a levels exhibited significance for OS and PRFS (Table VII). In the multivariate analysis of patients with stage II GC, ex-miR-21 and ex-miR-92a levels were significantly associated with OS and PRFS. In the univariate analysis of patients with stage III GC, lymph node metastasis, and ex-miR-21 and ex-miR-92a levels were significantly associated with OS and PRFS (Table VIII), whereas ex-miR-21 and ex-miR-92a levels exhibited significance for OS and PRFS in the multivariate analysis.
Table VII.
Univariate and multivariate Cox analyses for OS and PRFS in patients with stage II gastric cancer.
| A, OS | ||||||
|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | |||||
| Variables | Regression coefficient | Hazard ratio (95% CI) | P-value | Regression coefficient | Hazard ratio (95% CI) | P-value |
| Sex | 1.63 | 3.03 (0.98–11.27) | 0.065 | |||
| Tumor size | 1.11 | 5.09 (1.00–12.72) | 0.041 | 1.20 | 3.03 (0.98–11.27) | 0.055 |
| Lymph node metastasis | −0.19 | 0.83 (0.28–2.59) | 0.740 | |||
| Lymphatic invasion | −0.24 | 0.78 (0.25–2.90) | 0.691 | |||
| Venous invasion | 1.25 | 3.48 (0.69–63.39) | 0.153 | |||
| Differentiation | −0.66 | 0.52 (0.17–1.72) | 0.265 | |||
| ex-miR-21 | 1.15 | 3.17 (1.28–10.93) | 0.024 | 1.66 | 5.25 (1.49–18.65) | 0.035 |
| ex-miR-92a | −1.44 | 0.24 (0.04–0.88) | 0.031 | −1.40 | 0.25 (0.04–0.93) | 0.037 |
| B, PRFS | ||||||
| Univariate analysis | Multivariate analysis | |||||
| Variables | Regression coefficient | Hazard ratio (95% CI) | P-value | Regression coefficient | Hazard ratio (95% CI) | P-value |
| Sex | 1.23 | 3.43 (1.08–13.08) | 0.036 | 1.28 | 2.94 (0.95–10.93) | 0.061 |
| Tumor size | −0.19 | 0.83 (0.27–2.58) | 0.737 | |||
| Lymph node metastasis | 0.14 | 1.53 (0.33–7.07) | 0.580 | |||
| Lymphatic invasion | 1.28 | 3.59 (0.71–65.45) | 0.141 | |||
| Venous invasion | −0.67 | 0.51 (0.17–1.70) | 0.257 | |||
| Differentiation | 1.06 | 2.88 (1.80–9.00) | 0.022 | 1.54 | 3.36 (1.49–10.65) | 0.031 |
| ex-miR-21 | −1.48 | 0.23 (0.04–0.85) | 0.026 | −1.43 | 0.24 (0.04–0.91) | 0.035 |
| ex-miR-92a | ||||||
ex-, exosome-encapsulated; miR, microRNA; OS, overall survival; PRFS, peritoneal recurrence-free survival.
Table VIII.
Univariate and multivariate Cox analyses for OS and PRFS in patients with stage III gastric cancer.
| A, OS | ||||||
|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | |||||
| Variables | Regression coefficient | Hazard ratio (95% CI) | P-value | Regression coefficient | Hazard ratio (95% CI) | P-value |
| Sex | −0.28 | 0.76 (0.43–1.38) | 0.352 | |||
| Tumor size | −0.05 | 0.95 (0.52–1.84) | 0.864 | |||
| Lymph node metastasis | 1.54 | 4.69 (1.03–82.91) | 0.045 | 1.26 | 3.54 (0.74–63.31) | 0.131 |
| Lymphatic invasion | 0.48 | 1.61 (0.74–4.23) | 0.243 | |||
| Venous invasion | 0.44 | 1.72 (0.71–3.85) | 0.234 | |||
| Differentiation | −0.10 | 0.90 (0.40–2.60) | 0.833 | |||
| ex-miR-21 | 0.89 | 2.42 (1.40–4.31) | 0.014 | 1.26 | 3.54 (1.74–3.31) | 0.025 |
| ex-miR-92a | −0.65 | 0.52 (0.30–0.89) | 0.017 | −0.55 | 0.58 (0.33–0.99) | 0.047 |
| B, PRFS | ||||||
| Univariate analysis | Multivariate analysis | |||||
| Variables | Regression coefficient | Hazard ratio (95% CI) | P-value | Regression coefficient | Hazard ratio (95% CI) | P-value |
| Sex | −0.01 | 0.99 (0.56–1.86) | 0.962 | |||
| Tumor size | 1.79 | 5.99 (1.32–5.94) | 0.041 | 1.41 | 4.11 (0.87–7.36) | 0.080 |
| Lymph node metastasis | 0.58 | 1.78 (0.83–4.65) | 0.150 | |||
| Lymphatic invasion | 0.38 | 1.47 (0.75–3.21) | 0.273 | |||
| Venous invasion | −0.24 | 0.79 (0.35–2.27) | 0.625 | |||
| Differentiation | 0.81 | 2.26 (1.33–3.94) | 0.024 | 0.87 | 2.07 (1.41–3.98) | 0.028 |
| ex-miR-21 | −0.56 | 0.57 (0.34–0.96) | 0.033 | −0.43 | 0.51 (0.28–0.91) | 0.044 |
| ex-miR-92a | ||||||
ex-, exosome-encapsulated; miR, microRNA; OS, overall survival; PRFS, peritoneal recurrence-free survival.
These observations indicated that ex-miR-21 and ex-miR-92a levels were independent predictive biomarkers for peritoneal recurrence and prognosis in patients with stage II and III GC.
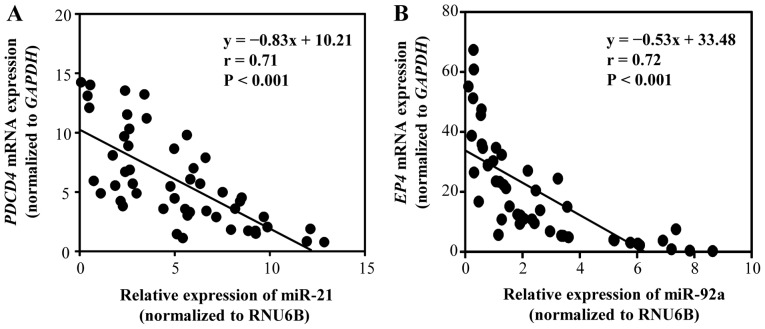
Correlation between target genes, and miR-21 and miR-92a levels in GC tissues
We investigated the correlation between miR-21 and PDCD4 mRNA expression, and miR-92a and EP4 mRNA expression in GC tissues (Fig. 9). PDCD4 for miR-21 and EP4 for miR-92a were selected as potential target genes, based on the results of searches using miRBase and miRWalk databases (Table SII). Primary tumor tissues from 48 patients with stage II GC (n=24) and stage III GC (n=24) were examined. A significant negative correlation was identified between miR-21 and PDCD4 mRNA expression (P<0.01), as well as between miR-92a and EP4 mRNA expression (P<0.01). These results indicated that PDCD4 expression may be negatively regulated by miR-21, whereas EP4 mRNA expression may be negatively regulated by miR-92a.
Figure 9.
Correlation between target gene expression levels and miR-21 or miR-92a expression in GC tissues. (A) Correlation between miR-21 and PDCD4 mRNA expression in GC tissues (n=48). (B) Correlation between miR-92a and EP4 mRNA expression in GC tissues (n=48). EP4, prostaglandin E receptor 4; miR, microRNA; PDCD4, programmed cell death protein 4; RNU6B, RNA, U6 small nuclear 6, pseudogene.
Discussion
The present study demonstrated the potential role of ex-miR-21 and ex-miR-92a as non-invasive biomarkers for the prediction of peritoneal recurrence and prognosis in patients with stage II/III GC that underwent R0 resection.
Several previous studies have revealed the potential of ex-miRNAs as stable biomarkers for patients with cancer (30,31). Additionally, our previous studies demonstrated the usefulness of ex-miR-451a as a biomarker to predict recurrence and prognosis in patients with non-small cell lung cancer and pancreatic cancer (25,26). Furthermore, ex-miR-21 serves as an independent predictive biomarker for recurrence and prognosis in patients with colorectal cancer (24). However, to the best of our knowledge, no studies have examined the clinical significance of ex-miRNAs in the prediction of peritoneal recurrence in patients with GC.
The present study evaluated the ex-miRNAs specific for peritoneal recurrence using a miRNA microarray. In this analysis, miR-21 exhibited significant upregulation, whereas miR-92a was the most downregulated miRNA in samples from patients with stage II GC that exhibited peritoneal recurrence after R0 resection compared with in samples from patients with stage II GC that did not show any recurrence after R0 resection and healthy controls. Based on these results, miR-21 and miR-92a were selected as biomarkers for the prediction of peritoneal recurrence. Furthermore, the results revealed that ex-miR-21 expression was significantly higher and ex-miR-92a expression was significantly lower in patients with GC compared with in healthy controls. Our previous studies demonstrated that ex-miR-21 levels were significantly higher in patients with colorectal cancer and non-small cell lung cancer than in healthy controls (24,25). Additionally, Huang et al (32) reported significantly lower levels of serum miR-92a in patients with GC than in healthy controls. By contrast, Zhu et al (33) reported that plasma miR-92a, but not ex-miR-92a, is elevated in patients with GC compared with in healthy controls. While the reason underlying this discrepancy remains unknown, it may be associated with the instability of miRNAs in plasma samples (33). Furthermore, the present study examined the association between ex-miR-21 and ex-miR-92a levels and clinicopathological characteristics, and revealed that high ex-miR-21 expression was significantly associated with tumor stage. Zhao et al (34) reported a significant association between plasma miR-21 levels and tumor size, lymph node metastasis and progression of tumor stage. The present study failed to observe any association between ex-miR-92a expression and clinicopathological characteristics.
While studies have examined the diagnostic and prognostic values of circulating plasma/serum free-miRNAs in patients with GC, few reports have focused on the plasma/serum ex-miRNAs (32–35). The present study demonstrated that high ex-miR-21 levels and low ex-miR-92a levels were significantly associated with poor OS and PRFS in patients with stage II and III GC that underwent R0 resection. Cox multivariate analysis revealed that ex-miR-21 and ex-miR-92a were independent prognostic factors. Although the prognostic value of plasma/serum ex-miR-21 in patients with GC has been reported previously (36,37), the value of ex-miR-21 as a predictive biomarker for peritoneal recurrence, to the best of our knowledge, has not been examined. Only a few studies have reported plasma/serum miR-92a as a biomarker for diagnosis in patients with GC (38). To the best of our knowledge, the present study is the first report to clarify the predictive value of ex-miR-21 and ex-miR-92a for peritoneal recurrence and prognosis in patients with GC.
Interestingly, Chen et al (5) reported the importance of the exosome-dependent molecular transfer or signaling pathway activation in the four stages (exfoliation, survival, adhesion and invasion and angiogenesis) of peritoneal dissemination in GC. The present study demonstrated that ex-miR-21 and ex-miR-92a levels differ significantly between patients with peritoneal recurrence and patients without peritoneal recurrence. The mechanism underlying the peritoneal metastasis may be mediated by alterations in the levels of ex-miR-21 and ex-miR-92a. The results of our preliminary experiments confirmed that the alterations in ex-miRNA levels in the peritoneal cavity affect the ex-miRNA levels in the peripheral blood (data not shown). A previous study reported that the tumor-derived exosomes containing miRNAs can initiate pre-metastatic niche formation by inducing metastasis of host cells (39). In particular, ex-miR-21 is known to affect the growth and metastasis of tumor cells via the activation of Toll-like receptors on the surrounding immune cells (40). These results indicated that ex-miR-21 may contribute to the formation of a pre-metastatic niche in the peritoneum. In addition, the phenotype of cancer stem cells is enhanced following overexpression of miR-21, leading to the promotion of invasion, migration and tumorigenesis (41). miR-92a, a member of the miR-17-92 cluster, may be closely linked to the functions of the E2F family of transcription factors, which are important regulators of the cell cycle and apoptosis (42). ex-miR-21 and ex-miR-92a may support peritoneal tumor invasion and recurrence following curative resection of GC. Further studies are required to elucidate these mechanisms.
miRNAs specifically target protein-coding mRNAs either by direct cleavage of the target mRNA or through the inhibition of protein synthesis (43). miRNAs regulate the gene expression at the post-transcriptional level by binding to the 3′-untranslated regions of specific mRNAs (44). miR-21 drives tumorigenesis via the inhibition of negative regulations of the RAS/mitogen-activated protein kinase/extracellular signal-regulated kinase signaling pathway, and miR-21 overexpression downregulates the expression of PDCD4, PTEN and tropomyosin 1, thereby promoting cell proliferation and cancer progression (45,46). miR-92a suppresses cell proliferation and invasion via the EP4/Notch 1 signaling pathway, and the restoration of miR-92a expression may result in the suppression of cell proliferation and the induction of apoptosis through the downregulation of EP4 receptor in GC (42). Furthermore, PDCD4 has been reported as a target gene of miR-21, whereas EP4 is a known target of miR-92a (24). The present study observed a significant inverse correlation between miR-21 and PDCD4 mRNA levels, and miR-92a and EP4 mRNA levels, in GC tissues. These results suggest that PDCD4 mRNA expression is regulated by miR-21, whereas miR-92a regulates EP4 expression.
In conclusion, the present study demonstrated the applications of plasma ex-miR-21 and ex-miR-92a for the prediction of peritoneal recurrence in patients with GC. However, the small sample size of the retrospective marker analyses was a limitation of the present study. Therefore, a larger scale prospective study is required to confirm the usefulness of ex-miR-21 and ex-miR-92a as biomarkers. Furthermore, in our preliminary study, the ex-miR-21 and ex-miR-92a levels of some patients normalized following surgery. These points will be considered in future studies.
Supplementary Material
Acknowledgements
The authors would like to thank Miss J. Tamura (Department of Surgery, Teikyo University School of Medicine) for her technical assistance.
Glossary
Abbreviations
- GC
gastric cancer
- miRNA
microRNA
- RT-qPCR
reverse transcription-quantitative PCR
Funding
This study is partly supported by Grants-in-Aid for Scientific Research from the Japanese Society for the Promotion of Science (grant nos. 15K10150, 17K10655 and 18K08716).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
HI conceived and designed the study. NS and HI wrote the manuscript. NS and HI performed the experiments. RF, TF, YS, DT, HM, YI, YK, MH and TK collected the clinical data. HI, TF and RF reviewed and edited the manuscript. All authors read and approved the manuscript, and agree to be held accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work is appropriate investigated and resolved.
Ethics approval and consent to participate
The study protocol conformed to the guidelines of the Teikyo University Ethics Committee, and was approved by the review board of Teikyo University (approval no. 09-081-3). Written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Hundahl SA, Menck HR, Mansour EG, Winchester DP. The National Cancer Data Base report on gastric carcinoma. Cancer. 1997;80:2333–2341. doi: 10.1002/(SICI)1097-0142(19971215)80:12<2333::AID-CNCR15>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 2.Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, Taniyama K, Sasaki N, Schlemper RJ. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345:784–789. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- 3.Sasako M, Sakuramoto S, Katai H, Kinoshita T, Furukawa H, Yamaguchi T, Nashimoto A, Fujii M, Nakajima T, Ohashi Y. Five-year outcomes of a randomized phase III trial comparing adjuvant chemotherapy with S-1 versus surgery alone in stage II or III gastric cancer. J Clin Oncol. 2011;29:4387–4393. doi: 10.1200/JCO.2011.36.5908. [DOI] [PubMed] [Google Scholar]
- 4.Bando E, Yonemura Y, Takeshita Y, Taniguchi K, Yasui T, Yoshimitsu Y, Fushida S, Fujimura T, Nishimura G, Miwa K. Intraoperative lavage for cytological examination in 1,297 patients with gastric carcinoma. Am J Surg. 1999;178:256–262. doi: 10.1016/S0002-9610(99)00162-2. [DOI] [PubMed] [Google Scholar]
- 5.Chen KB, Chen J, Jin XL, Huang Y, Su QM, Chen L. Exosome-mediated peritoneal dissemination in gastric cancer and its clinical applications. Biomed Rep. 2018;8:503–509. doi: 10.3892/br.2018.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sugarbaker PH, Yonemura Y. Clinical pathway for the management of resectable gastric cancer with peritoneal seeding: Best palliation with a ray of hope for cure. Oncology. 2000;58:96–107. doi: 10.1159/000012086. [DOI] [PubMed] [Google Scholar]
- 7.Ishigami H, Fujiwara Y, Fukushima R, Nashimoto A, Yabusaki H, Imano M, Imamoto H, Kodera Y, Uenosono Y, Amagai K, et al. Phase III trial comparing intraperitoneal and intravenous paclitaxel plus S-1 versus cisplatin plus S-1 in patients with gastric cancer with peritoneal metastasis: PHOENIX-GC trial. J Clin Oncol. 2018;36:1922–1929. doi: 10.1200/JCO.2018.77.8613. [DOI] [PubMed] [Google Scholar]
- 8.Jeung HC, Rha SY, Jang WI, Noh SH, Chung HC. Treatment of advanced gastric cancer by palliative gastrectomy, cytoreductive therapy and postoperative intraperitoneal chemotherapy. Br J Surg. 2002;89:460–466. doi: 10.1046/j.0007-1323.2001.02048.x. [DOI] [PubMed] [Google Scholar]
- 9.Kuramoto M, Shimada S, Ikeshima S, Matsuo A, Yagi Y, Matsuda M, Yonemura Y, Baba H. Extensive intraoperative peritoneal lavage as a standard prophylactic strategy for peritoneal recurrence in patients with gastric carcinoma. Ann Surg. 2009;250:242–246. doi: 10.1097/SLA.0b013e3181b0c80e. [DOI] [PubMed] [Google Scholar]
- 10.Liang P, Hu X. Strategies of diagnosis and treatment for peritoneal metastasis of gastric cancer. Zhonghua Wei Chang Wai Ke Za Zhi. 2017;20:500–503. (In Chinese) [PubMed] [Google Scholar]
- 11.La Torre M, Rossi Del Monte S, Ferri M, Cosenza G, Mercantini P, Ziparo V. Peritoneal washing cytology in gastric cancer. How, when and who will get a benefit? A review. Minerva Gastroenterol Dietol. 2011;57:43–51. [PubMed] [Google Scholar]
- 12.Abe S, Yoshiwara H, Tabata H, Tachibana M, Monden N, Nakamura T, Nagaoka S. Curative resection of gastric cancer: Limitation of peritoneal lavage cytolology in predicating the outcome. J Surg Oncol. 1995;59:226–229. doi: 10.1002/jso.2930590405. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, Wang L, Wu Z, Sun R, Jin H, Ma J, Liu L, Ling R, Yi J, Wang L, et al. Three dysregulated microRNAs in serum as novel biomarkers for gastric cancer screening. Med Oncol. 2014;31:298. doi: 10.1007/s12032-014-0298-8. [DOI] [PubMed] [Google Scholar]
- 14.Ma GJ, Gu RM, Zhu M, Wen X, Li JT, Zhang YY, Zhang XM, Chen SQ. Plasma post-operative miR-21 expression in the prognosis of gastric cancers. Asian Pac J Cancer Prev. 2013;14:7551–7554. doi: 10.7314/APJCP.2013.14.12.7551. [DOI] [PubMed] [Google Scholar]
- 15.Deng D, Liu Z, Du Y. Epigenetic alterations as cancer diagnostic, prognostic, and predictive biomarkers. Adv Genet. 2010;71:125–176. doi: 10.1016/B978-0-12-380864-6.00005-5. [DOI] [PubMed] [Google Scholar]
- 16.Grady WM, Tewai M. The next thing in prognostic molecular markers: MicroRNA signatures of cancer. Gut. 2010;59:706–708. doi: 10.1136/gut.2009.200022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kowal J, Tkach M, Thery C. Biogenesis and secretion of exosome. Curr Opin Cell Biol. 2014;29:116–125. doi: 10.1016/j.ceb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Palma J, Yaddannapudi SC, Pigati L, Havens MA, Jeong S, Weiner GA, Weimer KM, Stern B, Hastings ML, Duelli DM. MicroRNAs are exported from malignant cells in customized particles. Nucleic Acids Res. 2012;40:9125–9138. doi: 10.1093/nar/gks656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silva M, Melo SA. Non-coding RNAs in exosomes: New players in cancer biology. Curr Genomics. 2015;16:295–303. doi: 10.2174/1389202916666150707154719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simons M, Raposo G. Exosomes-vesicular carriers for intercelluar communication. Curr Opin Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 21.Ge Q, Zhou Y, Lu J, Bai Y, Xie X, Lu Z. MiRNA in plasma exosome is stable under different storage conditions. Molecules. 2014;19:1567–1575. doi: 10.3390/molecules19021568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hannafon BN, Ding WQ. Intercellular communication by exosome-derived microRNAs in cancer. Int J Mol Sci. 2013;14:14240–14269. doi: 10.3390/ijms140714240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dejima H, Iinuma H, Kanaoka R, Matsutani N, Kawamura K. Exosomal microRNA in plasma as a non-invasive biomarker for the recurrence of non-small cell lung cancer. Oncol Lett. 2017;13:1256–1263. doi: 10.3892/ol.2017.5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsukamoto M, Iinuma H, Yagi T, Matsuda K, Hashiguchi Y. Circulating exosomal microRNA-21 as a biomarker in each tumor stage of colorectal cancer. Oncology. 2017;92:360–370. doi: 10.1159/000463387. [DOI] [PubMed] [Google Scholar]
- 25.Kanaoka R, Iinuma H, Dejima H, Sakai T, Uehara H, Matsutani N, Kawamura M. Usefulness of plasma exosomal microRNA-451a as a noninvasive biomarker for early prediction of recurrence and prognosis of non-small cell lung cancer. Oncology. 2018;94:311–323. doi: 10.1159/000487006. [DOI] [PubMed] [Google Scholar]
- 26.Takahasi K, Iinuma H, Wada K, Minezaki S, Kawamura S, Kainuma M, Ikeda Y, Shibuya M, Miura F, Sano K. Usefulness of exosome-encapsulated microRNA-451a as a minimally invasive biomarker for prediction of recurrence and prognosis in pancreatic ductal adenocarcinoma. J Hepatobiliary Pancreat Sci. 2018;25:155–161. doi: 10.1002/jhbp.524. [DOI] [PubMed] [Google Scholar]
- 27.Kumata Y, Iinuma H, Suzuki Y, Tsukahara D, Midorikawa H, Igarashi Y, Soeda N, Kiyokawa T, Horikawa M, Fukushima R. Exosome-encapsulated microRNA-23b as a minimally invasive liquid biomarker for the prediction of recurrence and prognosis of gastric cancer patients in each tumor stage. Oncol Rep. 2018;40:319–330. doi: 10.3892/or.2018.6418. [DOI] [PubMed] [Google Scholar]
- 28.Bertero L, Massa F, Metovic J, Zanetti R, Castellano I, Ricardi U, Papotti M, Cassoni P. Eighth edition of the UICC classification of malignant tumours: An overview of the changes in the pathological TNM classification criteria-what has changed and why? Virchows Arch. 2018;472:519–531. doi: 10.1007/s00428-017-2276-y. [DOI] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Cheng L, Sharples RA, Scicluna BJ, Hill AF. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J Extracell Vesicles. 2014;3:23743. doi: 10.3402/jev.v3.23743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun Z, Chen C, Su Y, Wang W, Yang S, Zhou Q, Wang G, Li Z, Song J, Zhang Z, et al. Regulatory mechanisms and clinical perspectives of circRNA in digestive system neoplasms. J Cancer. 2019;10:2885–2891. doi: 10.7150/jca.31167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang S, Wang J, Li J, Luo Q, Zhao M, Zheng L, Dong X, Chen C, Che Y, Liu P, et al. Serum microRNA expression profile as a diagnostic panel for gastric cancer. Jpn J Clin Oncol. 2016;46:811–818. doi: 10.1093/jjco/hyw085. [DOI] [PubMed] [Google Scholar]
- 33.Zhu C, Ren C, Han J, Ding Y, Du J, Dai N, Dai J, Ma H, Hu Z, Shen H, et al. A five-microRNA panel in plasma was identified as a potential biomarker for early detection of gastric cancer. Br J Cancer. 2014;110:2291–2299. doi: 10.1038/bjc.2014.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao G, Jiang T, Liu Y, Huai G, Lan C, Li G, Jia G, Wang K, Yang M. Droplet digital PCR-based circulating microRNA detection serves as a promising diagnostic method for gastric cancer. BMC Cancer. 2018;18:676. doi: 10.1186/s12885-018-4601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang Z, Zhu D, Wu L, He M, Zhou X, Zhang L, Zhang H, Wang W, Zhu J, Cheng W, et al. Six serum-based miRNAs as potential diagnostic biomarkers for gastric cancer. Cancer Epidemiol Biomarkers Prev. 2017;26:188–196. doi: 10.1158/1055-9965.EPI-16-0607. [DOI] [PubMed] [Google Scholar]
- 36.Komatsu S, Ichikawa D, Tsujiura M, Konishi H, Takeshita H, Nagata H, Kawaguchi T, Hirajima S, Arita T, Shiozaki A, et al. Prognostic impact of circulating miR-21 in the plasma of patients with gastric carcinoma. Anticancer Res. 2013;33:271–276. [PubMed] [Google Scholar]
- 37.Zheng Y, Cui L, Sun W, Zhou H, Yuan X, Huo M, Chen J, Lou Y, Guo J. MicroRNA-21 is a new marker of circulating tumor cells in gastric cancer patients. Cancer Biomark. 2011-2012;10:71–77. doi: 10.3233/CBM-2011-0231. [DOI] [PubMed] [Google Scholar]
- 38.Liu HN, Wu H, Tseng YJ, Chen YJ, Zhang DY, Zhu L, Dong L, Shen XZ. Serum microRNA signatures and metabolomics have high diagnostic value in gastric cancer. BMC Cancer. 2018;18:415. doi: 10.1186/s12885-018-4343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rana S, Malinowska K, Zöller M. Exosomal tumor microRNA modulates premetastatic organ cells. Neoplasia. 2013;15:281–295. doi: 10.1593/neo.122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Y, Luo F, Liu Y, Shi L, Lu X, Xu W, Liu Q. Exosomal miR-21 derived from arsenite-transformed human bronchial epithelial cells promotes cell proliferation associated with arsenite carcinogenesis. Arch Toxicol. 2015;89:1071–1082. doi: 10.1007/s00204-014-1291-x. [DOI] [PubMed] [Google Scholar]
- 41.Jiang J, Yang P, Guo Z, Yang R, Yang H, Yang F, Li L, Xiang B. Overexpression of microRNA-21 strengthens stem cell-like characteristics in a hepatocellular carcinoma cell line. World J Surg Oncol. 2016;14:278. doi: 10.1186/s12957-016-1028-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shin VY, Siu MT, Liu X, Ng EKO, Kwong A, Chu KM. MiR-92 suppresses proliferation and induces apoptosis by targeting EP4/Notch1 axis in gastric cancer. Oncotarget. 2018;9:24209–24220. doi: 10.18632/oncotarget.24819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 44.Szafranska AE, Davison TS, John J, Cannon T, Sipos B, Maghnouj A, Labourier E, Hahn SA. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442–4452. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 45.Motoyama K, Inoue H, Mimori K, Tanaka F, Kojima K, Uetake H, Sugihara K, Mori M. Clinicopathological and prognostic significance of PDCD4 and microRNA-21 in human gastric cancer. Int J Oncol. 2010;36:1089–1095. doi: 10.3892/ijo_00000590. [DOI] [PubMed] [Google Scholar]
- 46.Cao Z, Yoon JH, Nam SW, Lee JY, Park WS. PDCD4 expression inversely correlated with miR-21 levels in gastric cancers. J Cancer Res Clin Oncol. 2012;138:611–619. doi: 10.1007/s00432-011-1140-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.