Abstract
Thymosin β10 (TMSB10) has been found to be overexpressed and function as an oncogene in several types of cancer. However, there have been limited reports on the role of TMSB10 in bladder cancer (BCa). In the present study, reverse transcription-quantitative PCR was used to quantify the expression of TMSB10 in BCa cell lines, clinical specimens and their corresponding control samples. The protein expression of TMSB10 was also examined in archived tissues from 101 patients with pathologically confirmed BCa by immunohistochemistry. Univariate and multivariate Cox regression models were used to evaluate the prognostic significance of TMSB10 in patients with BCa. The data indicated that the mRNA levels of TMSB10 were significantly overexpressed in BCa cell lines. In addition, the protein levels of TMSB10 were overexpressed in BCa tissues compared with those in adjacent normal tissues. In 55/101 (54.5%) BCa specimens, high expression levels of TMSB10 were noted. Statistical analysis revealed that the high expression of TMSB10 was positively associated with muscular invasion (P<0.05). In addition, a high expression of TMSB10 was associated with shorter overall survival (OS) of patients (P<0.05; log-rank test). The univariate and multivariate analyses suggested that the protein overexpression of TMSB10 was an unfavorable prognostic factor for OS (P<0.05) in patients with BCa. Knockdown of the expression of TMSB10 significantly suppressed cell migration and invasion. In conclusion, TMSB10 can be considered an independent factor for the poor prognosis of patients with BCa. The targeting of TMSB10 can reduce the migration and invasion of BCa cells.
Keywords: thymosin β10, bladder cancer, muscle invasive bladder cancer, prognosis, cell migration
Introduction
Bladder cancer (BCa) is the most common malignancy of the urinary tract (1). In 2017, there were ~79,030 new cases (4.6% of all cancer cases) of BCa in the USA, with a mortality rate of 16,870 (2.8% of all cancer cases) (2). Approximately 20–30% of all newly diagnosed cases of BCa are muscle invasive bladder cancer (MIBCa). Despite extensive cystectomy and adjuvant therapies, including chemotherapy and radiotherapy, the 5-year overall survival (OS) rate for patients with MIBCa is only 47% (3). Cisplatin-containing combination chemotherapy with gemcitabine and cisplatin or methotrexate, vinblastine, adriamycin and cisplatin is the standard treatment used for patients with advanced or metastatic BCa. However, the OS rate of patients with advanced MIBCa remains poor (4). Therefore, the identification of novel biomarkers associated with diagnosis and disease progression is required in order to select high-risk patients who may benefit from combination-treatment strategies.
Thymosin β10 (TMSB10) is an important member of the β-thymosin family, which was first isolated in 1966 by Goldstein et al (5), in addition to other lymphocytopoietic factors. TMSB10 functions as an actin-sequestering protein involved in cell motility (6). In recent years, TMSB10 has been shown to be associated with the occurrence and development of malignant tumors. An accumulation of evidence suggests that the expression of TMSB10 is significantly upregulated in six types of cancer, including human melanoma, gastric, breast and lung cancer, hepatocellular carcinoma and thyroid cancer (7–13), and is involved in a number of cellular processes, including cell proliferation, anti-apoptosis, angiogenesis and metastasis (14,15). However, the expression and role of TMSB10 in BCa have not been examined previously.
The present study reported for the first time, to the best of our knowledge, that TMSB10 is frequently overexpressed in BCa and that it exhibits clinical significance in BCa, as a high expression of TMSB10 was associated with significantly poorer OS. Knockdown of the expression of TMSB10 significantly suppressed BCa cell migration and invasion. Therefore, assessing the tumor expression of TMSB10 may improve disease prognosis and assist in the identification of patients who may benefit from combination treatment.
Materials and methods
Aim, design and setting of the study
The present study aimed to assess the expression of TMSB10 in BCa cell lines and human tissues and to examine the association between TMSB10 and the clinicopathological features and outcomes of patients with BCa. The present study performed in vitro analysis and retrospective analysis of clinical specimens obtained from 101 patients treated at a single institution in China.
Tissue specimens and patient information
Paraffin-embedded BCa tissue samples from a total of 101 cases were clinically and histologically diagnosed at Guangdong Second Provincial General Hospital (Guangzhou, China) between 2002 and 2014. The institutional Ethics Committee approved the study and written informed consent was obtained from each patient.
The age distribution of the patients was between 37 and 82 years, and the mean age was 60.4 years. According to the 2009 World Health Organization (WHO) 7th edition of the TNM classification system (16), 48 patients had primary non-MIBCa and 53 had MIBCa. The pathological grades were classified according to the WHO classification criteria for urothelial cancer. The histological diagnosis was evaluated according to the WHO 2004 guidelines. Grades I, II and III indicate well, moderately, and poorly differentiated tumors, respectively. Grades I and II were classified as low-grade urothelial carcinomas, whereas grade III was classified as high-grade tumors. In the present study, the 101 cases of BCa were divided into 33 cases of high-grade urothelial carcinoma and 68 cases of low-grade urothelial carcinoma. All clinical and pathological features of the patients are shown in Table I.
Table I.
Correlation between TMSB10 and clinicopathological characteristics of patients with bladder cancer.
| Expression of TMSB10 | ||||
|---|---|---|---|---|
| Clinicopathological characteristics | n | Low, n (%) | High, n (%) | P-value (χ2 test)a |
| Age (years) | ||||
| <65 | 52 | 21 (40.4) | 31 (59.6) | 0.191 |
| ≥65 | 49 | 25 (51.0) | 24 (49.0) | |
| Gender | ||||
| Male | 58 | 28 (48.3) | 30 (51.7) | 0.331 |
| Female | 43 | 18 (41.9) | 25 (58.1) | |
| Histological grade | ||||
| GI | 68 | 31 (45.6) | 37 (54.4) | 0.580 |
| GII–III | 33 | 15 (45.5) | 18 (54.5) | |
| T stage | ||||
| Ta-T1 | 48 | 27 (56.3) | 21 (43.8) | 0.032 |
| T2-T4 | 53 | 19 (35.8) | 34 (64.2) | |
| Distant metastasis | ||||
| Absent | 90 | 41 (45.6) | 49 (54.4) | 0.625 |
| Present | 11 | 5 (45.5) | 6 (54.5) | |
| Multiplicity | ||||
| Single | 71 | 33 (46.5) | 38 (53.5) | 0.473 |
| Multiple | 30 | 13 (43.3) | 17 (56.7) | |
Values in bold are considered statistically significant. TMSB0, thymosin β10.
Microarray data process
The microarray data process and evaluation of microarray datasets [Gene expression Omnibus (GEO) accession nos. GSE3167 and GSE13507] from the BCa and control samples were retrieved from the GEO database (http://www.ncbi.nlm.nih.gov/geo/). Integrative analyses were subsequently performed using the Cancer Genome Atlas (TCGA, http://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) data for bladder urothelial carcinoma (TCGA, Provisional).
Cell lines and transfection
T24, 5637, UM-UC-3, HCV-29 and EJ cells (authenticated via short tandem repeat analysis) were purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences and were grown in RPMI 1640 (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (Corning, Inc.) in 5% CO2 at 37°C. The normal cells comprised primary cells, which were extracted from patients devoid of BCa.
The sequences of oligonucleotides 1 and 2 targeting TMSB10 mRNA, were as follows: RNA i#1: sense 5′-GAAAUCGCCAGCUUCGAUATT-3′ and antisense 5′-UAUCGAAGCUGGCGAUUUCTT-3′. RNAi#2: sense 5′-CGACCAAAGAGACCAUUGATT-3′ and antisense 5′-UCAAUGGUCUCUUUGGUCGTT-3′; These siRNAs were synthesized by Shanghai GenePharma Co., Ltd. Approximately 2×105 cells per well were seeded in a 6-well tissue culture dish on the day before transfection. Transfection with 50 nmol of the small interfering (si)RNAs was performed according to the manufacturer's instructions using Lipofectamineä transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The transfection was performed 48 h prior to subsequent experiments.
Reverse transcription-quantitative PCR (RT-qPCR) analysis
Total RNA samples from the cell lines and freshly frozen tissues were isolated using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's recommendations. The extracted RNA was pretreated with RNase-free DNase and 2.0 µg of total RNA was reverse transcribed into complementary DNA (cDNA) using iScript™ cDNA synthesis Kit (Life Science Research; Bio-Rad, Laboratories, Inc.) according to manufacturer's protocols. To amplify TMSB10 cDNA, the following real-time RT-qPCR cycling conditions were used: Initial denaturation step at 95°C for 10 min, followed by 28 cycles of denaturation at 95°C for 60 sec, primer annealing at 58°C for 30 sec and extension at 72°C for 30 sec. The cycle program was terminated with a final extension step at 72°C for 5 min, and the products were stored at 4°C. The primers were designed using Primer Express software v. 2.0 (Applied Biosystems; Thermo Fisher Scientific, Inc.) with the following sequences: TMSB10, forward 5′-TGGCAGACAAACCAGACATGG-3′ and reverse 5′-CGAAGAGGACGGGGGTAGG-3′; and ACTB, forward 5′-CGTGGACATCCGCAAAGA-3′ and reverse 5′-GAAGGTGG-ACAGCGAGGC-3′. The relative mRNA expression levels were calculated by the 2−ΔΔCq method and normalized to ACTB (17), where Ct represents the quantification cycle for each transcript. Each experiment was performed in triplicate.
Western blotting
The samples were prepared for immunoblotting as previously described. The cells cultured at 70–80% confluence were washed twice with ice-cold phosphate-buffered saline (PBS) and lysed on ice in radio-immunoprecipitation assay buffer (Cell Signaling Technology, Inc.) with complete protease inhibitor cocktail (Roche Applied Science). The concentration of total protein was determined using Bicinchoninic Acid Protein Assay kit (Pierce; Thermo Fisher Scientific, Inc). The samples were subsequently heated for 10 min at 98°C. The freshly-frozen tissue samples were homogenized in liquid nitrogen and lysed using SDS-PAGE sample buffer. The protein concentrations were determined with the Bradford assay reagent (Bio-Rad Laboratories, Inc.). Equal quantities of protein (30 µg) were separated on a 10.5% SDS polyacrylamide gel and subjected to electrophoresis. The gels were transferred onto PVDF membranes (Immobilon P; Millipore Corp.; Merck KGaA) and the membranes were blocked using 5% fat-free milk in Tris-buffered saline containing 0.1% Tween-20 for 1 h at room temperature. The membranes then were probed with anti-TMSB10 (cat. no. ab14338; 1:1,000 dilution; Abcam) or anti-GAPDH (cat. no. ab181602; 1:10,000 dilution; Abcam) at 4°C overnight. Membranes were washed for three times with Tris-buffered saline and Polysorbate 20 and incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (cat. no. 2030230; 1:3,000 dilution; Invitrogen; Thermo Fisher Scientific, Inc.) for 1 h at room temperature. The bands were detected using enhanced chemiluminescence reagent (Amersham; GE Healthcare Life Sciences) following the manufacturer's protocols and quantified via densitometry using Image J software (V1.8.0; National Institutes of Health).
Transwell invasion assay
The T24 and EJ cells were initially transfected with TMSB10 siRNA or control siRNA. A total of 5×104 transfected cells were seeded onto a thick layer of Matrigel in Transwell inserts (BD Biosciences) and cultured for an additional 24 h. The invasive cells that adhered to the lower surface of the filter were washed with PBS, fixed with 4% paraformaldehyde and stained with 0.05% crystal violet. The invasive cells were counted under a light microscope (magnification, ×100; Carl Zeiss, Inc.).
Wound healing assay
A total of 1×106 transfected cells were seeded and allowed grow at 70–80% confluence. The cells were subsequently starved for 24 h. The cell monolayers were wounded with a sterile plastic tip and cultured in serum-free medium. Cell migration was monitored every 12 h using light microscopy (magnification, ×100; Nikon Corporation). The Image J software (V1.8.0; National Institutes of Health) was used to quantify the percentage of cell wound closure, in a blinded manner.
Immunohistochemical (IHC) analysis
The protein expression of TMSB10 was examined in the 101 human BCa tissue specimens via immunohistochemistry. Samples were fixed in 4% paraformaldehyde buffer at 25°C for 24 h. After embedding in paraffin sections (4 µm thick) were incubated at 60°C for 1 h. The sections were deparaffinized with xylene twice for 15 min, rehydrated in a descending alcohol series (100, 95 and 70% ethanol; each rinse, 5 min), placed in EDTA antigen retrieval buffer and microwaved. Endogenous peroxidase activity was inhibited using a solution of 3% hydrogen peroxide in methanol and non-specific binding was blocked using 1% bovine serum albumin. Samples were subsequently incubated with anti-TMSB10 rabbit polyclonal antibodies (cat. no. ab14338; 1:50; Abcam) at 4°C overnight. Normal goat serum was used as a negative control in place of primary antibodies. After washing with (PBS) 4 times (each wash, 4 min), sections were incubated at room temperature for 30 min with biotinylated anti-rabbit secondary antibodies (1:50; cat. no. ab6720; Abcam) and stained for 0.5–1 min with The EnVision™ Detection System (cat. no. K5007, Dako; Agilent Technologies, Inc.). Samples were then treated with hematoxylin for 30 sec at 25°C. Following dehydration and soaking in xylene, the stained slices were sealed and observed under a light microscope (magnification, ×200 and ×400).
The degree of immunostaining was scored by two independent observers. The proportion of TMSB10-expressing cells was scored as 1 (<25% positive tumor cells), 2 (25–50%), 3 (50–75%) or 4 (>75%). Staining intensity was scored as 0 (no staining), 1 (weak, light yellow), 2 (moderate, yellowish-brown), and 3 (strong, brown). Staining intensity and the proportion of positive cells in each section were multiplied (to obtain values of 0, 1, 2, 3, 4, 6, 8, 9 or 12). Tumor samples with scores >6 were considered to exhibit a high expression and samples with scores ≤6 were determined to have a low TMSB10 expression. The AxioVision Rel.4.6 computerized image analysis system, with assistance from the automatic measurement program (Carl Zeiss), was used to analyze inconsistencies in IHC stain intensities in tumors and normal tissues.
Statistical analysis
All statistical analyses were conducted using SPSS (version 19.0) software (IBM Corp.). Data were expressed as the mean ± standard deviation and compared using a paired t-test between two groups. One-way ANOVA followed by an LSD post-hoc test was used for comparisons among groups. The cutoff value for TMSB10 was selected using receiver operating characteristic (ROC) curve analysis. The associations between the expression of TMSB10 and clinicopathological features were examined using Pearson's χ2 test or Fisher's exact test. OS was measured from the initiation of treatment until patient mortality. The survival curves were plotted using the Kaplan-Meier method and compared using the log-rank test. Univariate and multivariate Cox regression models were used to evaluate prognostic significance. P<0.05 was considered to indicate a statistically significant difference.
Results
TMSB10 is overexpressed in human BCa tissues, notably in MIBCa
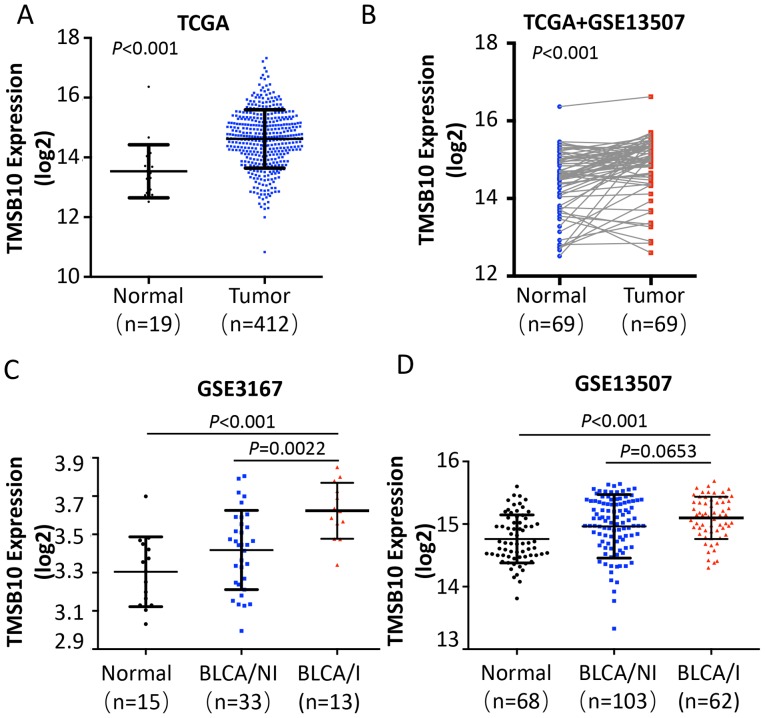
To determine the clinical significance of TMSB10 in BCa, publicly available microarray data (TCGA data, and GSE3167 and GSE13507 data for BCa) were initially analyzed. The analysis revealed that the expression of TMSB10 was upregulated in BCa tissues compared with that noted in adjacent normal tissues (ANT) (Fig. 1A and B). Of note, the expression of TMSB10 was considerably higher with regard to MIBCa (Fig. 1C and D).
Figure 1.
Microarray data reveals TMSB10 is upregulated in BCa. (A) Expression levels of TMSB10 were markedly elevated in BCa tissues compared with those in adjacent normal tissues as assessed by analyzing the TCGA BCa sequencing dataset (normal, n=19; BCa, n=412). (B) Reverse transcription-quantitative PCR analysis of the expression of TMSB10 in 69 paired BCa tissues compared with that in matched adjacent normal tissues as assessed by analyzing the TCGA data and GSE13507 data. Expression levels of TMSB10 were markedly elevated in BCa/I tissues compared with those in BCa/NI tissues as assessed by analyzing the (C) GSE3167 and (D) GSE13507 sequencing datasets. BCa, bladder cancer; TMSB10, thymosin β10; TCGS, The Cancer Genome Atlas; BCa/I, BCA with muscle invasion; BCa/NI, BCa with no muscle invasion.
TMSB10 is overexpressed in BCa cell lines and its knockdown suppresses BCa cell migration and invasion
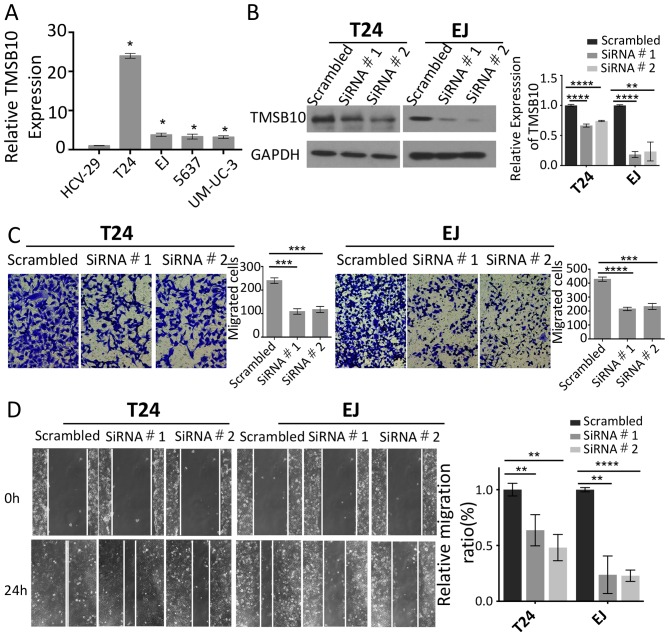
To investigate the expression levels of TMSB10 and its effects on the metastatic ability of BCa cells, the mRNA expression of TMSB10 was measured in four BCa cell lines (EJ, T24, 5637 and UM-UC-3) and one normal bladder epithelial cell line (HCV-29). The results indicated that TMSB10 was overexpressed in all four BCa cell lines compared with the corresponding expression of this gene in the HCV-29 cell line (Fig. 2A). The expression of TMSB10 was elevated in the T24 and EJ cells. Therefore, TMSB10 was silenced in these cells in order to assess cell motility. Efficient knockdown of the expression of TMSB10 was confirmed by western blotting (Fig. 2B). Wound healing and migration assays were performed, and it was shown that the knockdown of TMSB10 reduced the number of invasive cells and significantly suppressed mobility compared with the corresponding parameters noted in the vector control cells (Fig. 2C and D).
Figure 2.
Effects of TMSB10 silencing on cancer cell migration and invasion. (A) Reverse transcription-quantitative PCR analysis of the mRNA expression of TMSB10 in HCV-29 normal bladder cells and five cultured bladder cancer cell lines. GAPDH was used as a loading control. (B) Knockdown efficiency of endogenous expression of TMSB10 on protein levels was determined by western blotting. (C) T24 and EJ cells were transfected with two individual TMSB10 siRNAs or a scrambled control followed by Transwell invasion assays (magnification, ×100). (D) Effect on migration was assessed using wound healing assays (magnification, ×100). *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001. TMSB10, thymosin β10; siRNA, small interfering RNA.
Expression of TMSB10 correlates with the clinicopathological features of BCa
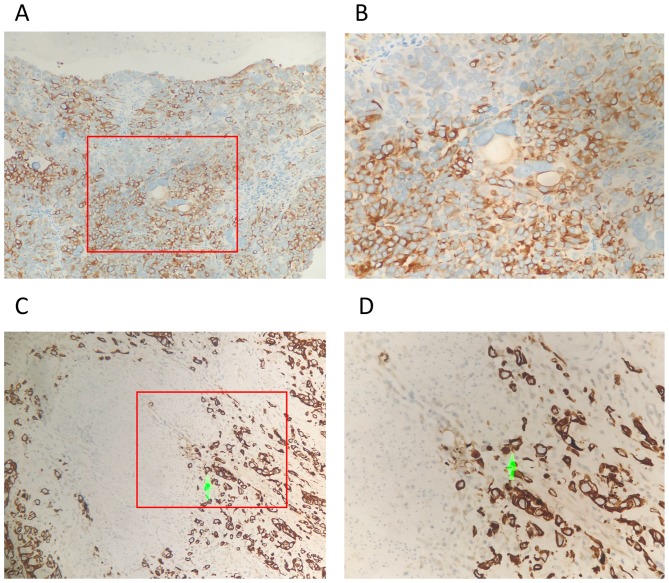
The association between the protein expression of TMSB10 and the clinicopathological features of BCa were further assessed using 101 archived paraffin-embedded human BCa specimens. The percentage of TMSB10 protein overexpression in patients with BCa was 54.5% (55/101; Fig. 3A-D). By contrast, 30.0% (6/20) of the adjacent normal tissues exhibited overexpression of TMSB10 protein, which was significantly lower than that noted in the BCa tissues (P<0.05, Table II). A positive association between clinicopathological features and the overexpression of TMSB10 protein was noted for muscular invasion (absent vs. present, P<0.05). A previous study suggested that the incidence of UCB markedly increases after the age of 65 years (18). Therefore, the age of 65 was used as a cut-off threshold in the present study to monitor the correlation between TMSB10 and age. However, based on the analysis, no significant associations were found between the protein expression of TMSB10 and age (≥65 vs. <65 years, P>0.05), gender (male vs. female, P>0.05), histological grade (GI vs. GII–III, P>0.05), distant metastases (absent vs. present, P>0.05) or multiplicity (single vs. multiple, P>0.05) (Table I).
Figure 3.
Expression of TMSB10 in BCa. Representative immunostaining of TMSB10 in BCa samples without muscle invasion at (A) ×200 magnification and (B) ×400 magnification. BCa samples with muscle invasion at (C) ×200 magnification and (D) ×400 magnification. Green arrows indicate sites of muscle invasion. Expression of TMSB10 was low in BCa samples without muscle invasion and high in BCa samples with muscle invasion. BCa, bladder cancer; TMSB10, thymosin β10.
Table II.
Protein expression of TMSB10 is increased in bladder cancer.
| Protein expression of TMSB10 | ||||
|---|---|---|---|---|
| Group | n | Low, n (%) | High, n (%) | P-value (χ2 test)a |
| Tumor tissues | 101 | 46 (45.5) | 55 (54.5) | 0.039 |
| Normal tissues | 20 | 14 (70.0) | 6 (30.0) | |
Values in bold are considered statistically significant. TMSB10, thymosin β10.
High expression of TMSB10 is associated with poor survival and prognosis in BCa
The prognostic value of the protein expression of TMSB10 in patients with BCa was further examined in 101 BCa tissue samples using immunohistochemical techniques. Kaplan-Meier survival analysis indicated that patients with high protein expression levels of TMSB10 exhibited lower OS compared with patients with low protein expression levels of TMSB10 (P<0.05, Fig. 4). In addition, univariate analysis was performed and identified the following seven prognostic parameters: Age, gender, histological grade, distant metastasis, multiplicity, muscular invasion and protein expression of TMSB10. It was found that the overexpression of TMSB10 protein was an independent risk factor for poor disease prognosis (P<0.05; Table III). However, no significant associations were found between the protein expression of TMSB10 and age, histological grade or distant metastases when all data were separated according to gender (Tables SI–IV).
Figure 4.
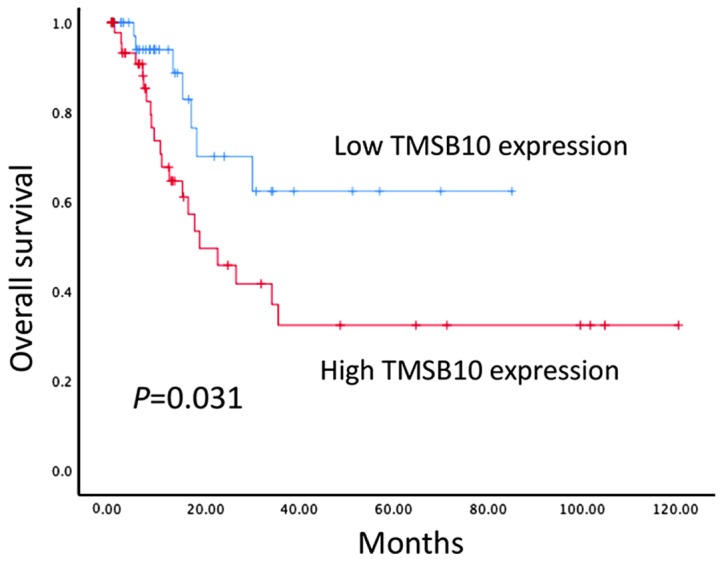
Protein expression of TMSB10 is associated with overall survival in BCa. Kaplan-Meier analysis of overall survival in relation to the expression of TMSB10 in patients with BCa. Patients were divided into two groups: high TMSB10 expression (n=55) and low TMSB10 expression (n=46). Kaplan-Meier analysis of overall survival in relation to the expression of TMSB10 in 101 bladder cancer patients revealed significant separation between curves. BCa, bladder cancer; TMSB10, thymosin β10.
Table III.
Summary of univariate and multivariate Cox regression analyses of overall survival duration in patients with bladder cancer.
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Characteristics | HR (95% CI) | P-valuea | HR (95% CI) | P-valuea |
| Age (≥65/<65 years) | 0.859 (0.400–1.842) | 0.696 | ||
| Gender (female/male) | 0.766 (0.362–1.620) | 0.484 | ||
| Histological grade (GI/GII–III) | 1.359 (0.596–3.103) | 0.466 | ||
| T stage (T2-T4/Ta-T1) | 2.351 (1.081–5.113) | 0.031 | 2.338 (1.062–5.146) | 0.035 |
| Distant metastasis (present/absent) | 3.015 (1.216–7.478) | 0.017 | 3.402 (1.341–8.632) | 0.010 |
| Multiplicity (multiple/single) | 1.568 (0.722–3.405) | 0.225 | ||
| TMSB10 (high/low) | 0.402 (0.171–0.946) | 0.037 | 0.418 (0.177–0.988) | 0.047 |
Values in bold are considered statistically significant. TMSB10, thymosin β10; HR, hazard ratio; CI, confidence interval.
Discussion
The role of TMSB10 in tumor metastasis is controversial. It has been shown that TMSB10 is downregulated in ovarian cancer compared with its expression in normal ovarian tissues (19), whereas others have shown that this protein acts as a tumor suppressor by disrupting the actin structure and inhibiting the Ras signaling pathway (20). However, the majority of studies indicate that TMSB10 serves an oncogenic role. The overexpression of TMSB10 is correlated with distant metastases and lymph node metastases in lung cancer. In addition, the expression of TMSB10 has been significantly associated with other factors, including VEGF, VEGF-C, and microvascular and lymphatic vessel density in lung cancer (21). Califano et al (13) found that the overexpression of TMSB10 correlated with the anaplastic histotype of thyroid tumors, which is considered the most aggressive thyroid histotype. The same study showed that the gene expression of TMSB10 was associated with the oncogenic transformation of highly malignant rat thyroid epithelial cells caused by several viruses (13). High expression levels of TMSB10 have also been associated with lymph node metastases in papillary thyroid carcinoma (22). Recent studies by Zhang et al reported that the expression of TMSB10 correlated with clinicopathological features, poor prognosis and distant metastases in patients with breast cancer. In addition, they found that TMSB10 promoted the proliferation, invasion and migration of breast cancer cells in vitro and in vivo via the AKT/FOXO signaling pathway (9). To the best of our knowledge, the present study is the first to assess the expression and association of TMSB10 with the clinicopathological features of BCa.
In the present study, following the analysis of publicly available microarray data, it was found that the mRNA expression levels of TMSB10 in BCa were significantly higher than those in adjacent noncancerous tissues. It is noteworthy that the overexpression of TMSB10 was associated with MIBCa. Similarly, the mRNA levels of TMSB10 were overexpressed in all four of the BCa cell lines assessed compared with those noted in the HCV-29 normal bladder epithelial cell line. In addition, knocking down the expression of TMSB10 caused a marked reduction in the migration and invasion of BCa cell lines.
High expression levels of TMSB10 were observed in 54.5% (55/101) of BCa tissues in the present study by immunohistochemical detection. In addition, the overexpression of TMSB10 protein was positively associated with cancer cell invasion to the muscularis mucosae. These results support the hypothesis that TMSB10 may be an oncogene in BCa and further suggested that it may serve an important role in the metastasis of BCa. The results also revealed that the expression of TMSB10 correlated with OS and that it was an independent prognostic factor for low OS rates in patients with BCa. The present study suggested that TMSB10 may serve a crucial role in muscle invasion of bladder tissues and that high expression levels may represent a novel indicator for muscle invasion in BCa and poor prognosis of the disease.
The present study has limitations, such as the retrospective study design and the lack of experimental evidence to support a specific regulatory mechanism between TMSB10 and the activation of BCa cell metastasis. Furthermore, the processes of invasion and apoptosis were not thoroughly addressed. Therefore, the examination of further subjects is required in order to determine the exact role of TMSB10 in a carcinogenic environment. In addition, the cohort size was not sufficiently large to support the use of TMSB10 as a prognostic marker. However, similar conclusions may be deduced following the investigation of TMSB10 in larger sample sizes of BCa specimens, based on the survival curve shown in the present study. In the future, follow up of the patients is planned to examine differences in recurrence, kidney function and infection in returning patients with disease progression.
In conclusion, the present study is the first, to the best of our knowledge, to report that TMSB10 is frequently overexpressed and has considerable clinical applications in BCa. A high expression of TMSB10 was associated with significantly lower OS. In addition, the assessment of tumor expression of TMSB10 may improve prognosis and assist in the identification of patients who may benefit from combination treatment. The in vitro analyses demonstrated that the migration and invasion of cancer cells were significantly reduced by silencing TMSB10.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from Guangdong Medical Research Foundation of China (grant no. A2018326).
Availability of data and materials
All data generated or analyzed during the present study are included in this published article.
Authors' contributions
BW and GY conceived and designed the study, and analyzed and interpreted the results. BW, ZW and TZ performed experiments and wrote this manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The institutional Ethics Committee of Guangdong Second Provincial General Hospital (Guangdong, China) approved the study and written informed consent was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Antoni S, Ferlay J, Soerjomataram I, Znaor A, Jemal A, Bray F. Bladder cancer incidence and mortality: A global overview and recent trends. Eur Urol. 2017;71:96–108. doi: 10.1016/j.eururo.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 4.Bellmunt J, Petrylak DP. New therapeutic challenges in advanced bladder cancer. Semin Oncol. 2012;39:598–607. doi: 10.1053/j.seminoncol.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Goldstein AL, Slater FD, White A. Preparation, assay, and partial purification of a thymic lymphocytopoietic factor (thymosin) Proc Natl Acad Sci USA. 1966;56:1010–1017. doi: 10.1073/pnas.56.3.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erickson-Viitanen S, Ruggieri S, Natalini P, Horecker BL. Thymosin beta 10, a new analog of thymosin beta 4 in mammalian tissues. Arch Biochem Biophys. 1983;225:407–413. doi: 10.1016/0003-9861(83)90047-4. [DOI] [PubMed] [Google Scholar]
- 7.Weterman MA, van Muijen GN, Ruiter DJ, Bloemers HP. Thymosin beta-10 expression in melanoma cell lines and melanocytic lesions: A new progression marker for human cutaneous melanoma. Int J Cancer. 1993;53:278–284. doi: 10.1002/ijc.2910530218. [DOI] [PubMed] [Google Scholar]
- 8.Oien KA, Vass JK, Downie I, Fullarton G, Keith WN. Profiling, comparison and validation of gene expression in gastric carcinoma and normal stomach. Oncogene. 2003;22:4287–4300. doi: 10.1038/sj.onc.1206615. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X, Ren D, Guo L, Wang L, Wu S, Lin C, Ye L, Zhu J, Li J, Song L, et al. Thymosin beta 10 is a key regulator of tumorigenesis and metastasis and a novel serum marker in breast cancer. Breast Cancer Res. 2017;19:15. doi: 10.1186/s13058-016-0785-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu CR, Ma CS, Ning JY, You JF, Liao SL, Zheng J. Differential thymosin beta 10 expression levels and actin filament organization in tumor cell lines with different metastatic potential. Chin Med J (Engl) 2004;117:213–218. [PubMed] [Google Scholar]
- 11.Theunissen W, Fanni D, Nemolato S, Di Felice E, Cabras T, Gerosa C, Van Eyken P, Messana I, Castagnola M, Faa G. Thymosin beta 4 and thymosin beta 10 expression in hepatocellular carcinoma. Eur J Histochem. 2014;58:2242. doi: 10.4081/ejh.2014.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takano T, Hasegawa Y, Miyauchi A, Matsuzuka F, Yoshida H, Kuma K, Amino N. Quantitative analysis of thymosin beta-10 messenger RNA in thyroid carcinomas. Jpn J Clin Oncol. 2002;32:229–232. doi: 10.1093/jjco/hyf054. [DOI] [PubMed] [Google Scholar]
- 13.Califano D, Monaco C, Santelli G, Giuliano A, Veronese ML, Berlingieri MT, de Franciscis V, Berger N, Trapasso F, Santoro M, et al. Thymosin beta-10 gene overexpression correlated with the highly malignant neoplastic phenotype of transformed thyroid cells in vivo and in vitro. Cancer Res. 1998;58:823–828. [PubMed] [Google Scholar]
- 14.Sribenja S, Li M, Wongkham S, Wongkham C, Yao Q, Chen C. Advances in thymosin beta10 research: Differential expression, molecular mechanisms, and clinical implications in cancer and other conditions. Cancer Invest. 2009;27:1016–1022. doi: 10.3109/07357900902849640. [DOI] [PubMed] [Google Scholar]
- 15.Sribenja S, Wongkham S, Wongkham C, Yao Q, Chen C. Roles and mechanisms of β-thymosins in cell migration and cancer metastasis: An update. Cancer Invest. 2013;31:103–110. doi: 10.3109/07357907.2012.756111. [DOI] [PubMed] [Google Scholar]
- 16.Sobin LH, Gospodarowicz MK, Wittekind C. International against cancer (UICC). TNM classification of malignant tumours. 7th edn. New York. 2009:Wiley. [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 19.Lee SH, Zhang W, Choi JJ, Cho YS, Oh SH, Kim JW, Hu L, Xu J, Liu J, Lee JH. Overexpression of the thymosin beta-10 gene in human ovarian cancer cells disrupts F-actin stress fiber and leads to apoptosis. Oncogene. 2001;20:6700–6706. doi: 10.1038/sj.onc.1204683. [DOI] [PubMed] [Google Scholar]
- 20.Lee SH, Son MJ, Oh SH, Rho SB, Park K, Kim YJ, Park MS, Lee JH. Thymosin {beta}(10) inhibits angiogenesis and tumor growth by interfering with Ras function. Cancer Res. 2005;65:137–148. [PubMed] [Google Scholar]
- 21.Gu Y, Wang C, Wang Y, Qiu X, Wang E. Expression of thymosin beta10 and its role in non-small cell lung cancer. Hum Pathol. 2009;40:117–124. doi: 10.1016/j.humpath.2008.06.023. [DOI] [PubMed] [Google Scholar]
- 22.Zhang XJ, Su YR, Liu D, Xu DB, Zeng MS, Chen WK. Thymosin beta 10 correlates with lymph node metastases of papillary thyroid carcinoma. J Surg Res. 2014;192:487–493. doi: 10.1016/j.jss.2014.05.066. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during the present study are included in this published article.