Abstract
Parasitoid wasps are among the most speciose animals, yet have relatively few available genomic resources. We report a draft genome assembly of the wasp Diachasma alloeum (Hymenoptera: Braconidae), a host-specific parasitoid of the apple maggot fly Rhagoletis pomonella (Diptera: Tephritidae), and a developing model for understanding how ecological speciation can “cascade” across trophic levels. Identification of gene content confirmed the overall quality of the draft genome, and we manually annotated ∼400 genes as part of this study, including those involved in oxidative phosphorylation, chemosensation, and reproduction. Through comparisons to model hymenopterans such as the European honeybee Apis mellifera and parasitoid wasp Nasonia vitripennis, as well as a more closely related braconid parasitoid Microplitis demolitor, we identified a proliferation of transposable elements in the genome, an expansion of chemosensory genes in parasitoid wasps, and the maintenance of several key genes with known roles in sexual reproduction and sex determination. The D. alloeum genome will provide a valuable resource for comparative genomics studies in Hymenoptera as well as specific investigations into the genomic changes associated with ecological speciation and transitions to asexuality.
Keywords: Hymenoptera, sequential speciation, de novo genome assembly, genome evolution, chemosensory genes
Introduction
The Hymenoptera may be the largest order of insects due to the immense diversity of parasitic wasps (i.e., “parasitoids”) that lay their eggs into or on other insect species (LaSalle and Gauld 1993; Austin and Dowton 2000; Whitfield 2003; Forbes et al. 2018). The great diversity of parasitoid wasps may be a consequence of their close relationship with their insect hosts. When a specialist parasitoid shifts to a new host, this change can propel the evolution of reproductive isolating barriers between wasp populations using the new and ancestral hosts (Feder and Forbes 2010). The evolution of reproductive isolating barriers following a host shift is a well-documented phenomenon in host specialist insects (Forbes et al. 2017), but the study of genomic changes that accompany such phenomena is still in its early stages.
Diachasma alloeum (Hymenoptera: Braconidae) is a specialist parasitoid of the fruit fly Rhagoletis pomonella (Diptera: Tephritidae). After the introduction of domesticated apples to the United States from Europe, R. pomonella infesting native hawthorn fruits experienced a host shift and subsequently evolved reproductive isolating barriers in what has become a well-known example of incipient ecological speciation (Walsh 1867; Bush 1966, 1994; Nosil 2012). This new “apple maggot fly” was sequentially colonized by D. alloeum, which appears to have shifted from its ancestral host, the blueberry maggot Rhagoletis mendax (Forbes et al. 2009). Two reproductive isolating barriers (i.e., diapause emergence and host fruit volatile discrimination) have evolved in parallel in R. pomonella and D. alloeum, and in both fly and wasp, these traits appear to have a genetic basis (Dambroski et al. 2005; Forbes and Feder 2006; Forbes et al. 2009). This phenomenon of “sequential” or “cascading” speciation may be an important driver of new biodiversity (Stireman et al. 2006; Abrahamson and Blair 2007; Hood et al. 2015).
Reproductive isolation in genus Diachasma has also arisen as a consequence of the loss of sexual reproduction, a general pattern observed in many hymenopteran insects (van der Kooi et al. 2017; Tvedte et al. 2019). Asexual Diachasmamuliebre appears to have split from its sexual sister Diachasmaferrugineum between 0.5 and 1 Ma (Wharton and Marsh 1978; Forbes et al. 2013). Although the decay of genes involved in sexual traits has been observed in multiple asexual parasitoid wasps (Ma et al. 2014; Kraaijeveld et al. 2016), there is a lack of comparative assessments of genomic molecular evolution between sexual and asexual Hymenoptera.
Here, we report the de novo genome assembly of the parasitoid wasp D. alloeum, adding to the genomic resources for parasitoid wasps, which are underrepresented among available hymenopteran genomes (Branstetter et al. 2018). We performed a series of descriptive analyses to assess the overall quality and content of the D. alloeum genome, and then focused on annotation and evolutionary analyses of gene families with potential relevance to speciation and sex determination in Diachasma.
Materials and Methods
We isolated genomic DNA from wasps collected in Fennville, MI. Illumina paired-end, mate pair, and TruSeq Synthetic Long Read (TSLR) libraries were sequenced on an Illumina HiSeq2000. The library from a single haploid male enabled the initial contig assembly, and pooled samples were required to achieve the minimum DNA mass needed for other library preparations. Paired-end and mate pair reads were de novo assembled using SOAPdenovo2 v2.04 (Luo et al. 2012) and TSLR “reads” were added using PBJelly v2 (English et al. 2012). We removed putative microbial contaminant sequences from the assembly that were identified by both BlobTools (Laetsch and Blaxter 2017) and a separate custom pipeline developed by Wheeler et al. (2013) and modified as described in Poynton et al. (2018). We separately assembled the mitochondrial genome de novo using NOVOplasty v2.6.3 (Dierckxsens et al. 2017).
We used ten wasps of each sex to generate two (pooled male and pooled female) paired-end RNASeq libraries and sequenced read libraries using an Illumina HiSeq2500. The input DNA required for library preparation precluded the use of the same biological samples for genome and transcriptome sequencing runs. We combined read data sets and assembled a transcriptome de novo with Trinity (Release April 13, 2014) (http://trinityrnaseq.github.io/; last accessed May 2015) (Grabherr et al. 2011; Haas et al. 2013). Annotation of the D. alloeum genome assembly was performed by the NCBI using their Eukaryotic Genome Annotation Pipeline (https://www.ncbi.nlm.nih.gov/genome/annotation_euk/process/; last accessed July 2019), with experimental support from the RNAseq and transcriptome. Manual annotations were added to a D. alloeum project on the i5k workspace (https://apollo.nal.usda.gov/diaall/jbrowse/; last accessed May 2018; Poelchau et al. 2015). See Supplementary Material online for additional information on genome sequencing, assembly, and annotation.
Results and Discussion
Quality Assessment of Genome Assembly
Libraries from a combination of single and pooled wasp samples contained 182.88 Gb total sequence data. The de novo genome assembly Dall1.0 (GenBank accession: GCA_001412515.1) had 3,968 scaffolds with a total scaffold length of 388.8 Mb and a scaffold N50 of 645,583 bp (supplementary table S1, Supplementary Material online). The presence of prokaryotic-like sequences in eukaryotic genome projects may reflect contamination in sequencing libraries or an actual association between microorganisms and hosts. Of the D. alloeum scaffolds, we annotated 656 as likely bacterial contaminants and an additional scaffold (Dall2.0 RefSeq accession: NW_021680771.1) as an apparent lateral gene transfer event from a Rickettsia species (see Supplementary Material online). The likely bacterial contaminating scaffolds were removed from the D. alloeum assembly, and the assembly containing the remaining 3,313 scaffolds is available as Dall2.0 (GCA_001412515.3).
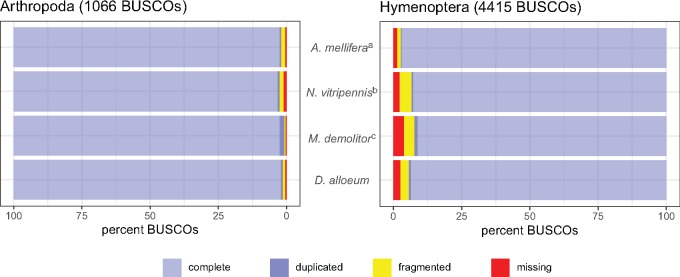
A common metric used to assess the relative completeness of a genome assembly is the identification of conserved single-copy genes, performed here using BUSCO v3 (Simão et al. 2015). We found 1,059/1,066 (99%) Arthropoda BUSCOs and 4,300/4,415 (97%) Hymenoptera BUSCOs in the D. alloeum genome, most of which were complete and single-copy (fig. 1). These values are similar to BUSCO gene content in other published hymenopteran genomes, including Apis mellifera, Nasonia vitripennis, and Microplitis demolitor (fig. 1 and see Supplementary Material online). Our de novo assembly of the D. alloeum mitochondrial sequence using NOVOplasty (Dierckxsens et al. 2017) produced a 15,936 bp sequence with a complete set of 13 protein coding genes, two rRNA sequences, and 20 tRNA sequences (GenBank accession NW_021683654.1). In addition, our annotation of 65/68 (96%) of the canonical suite of nuclear-encoded mitochondrial genes provided additional evidence for a high-quality genome assembly (see Supplementary Material online).
Fig. 1.
—BUSCO analysis of Diachasma alloeum and additional hymenopteran genome assemblies. aApis mellifera assembly reported in Weinstock et al. (2006). bNasonia vitripennis assembly reported in Werren et al. (2010). cMicroplitis demolitor assembly reported in Burke et al. (2018).
We used RepeatModeler (Smit et al. 2015), PASTEClassifier (Hoede et al. 2014, version 1.0), and RepeatMasker (Smit et al. 2010) for de novo repeat identification, repeat reclassification, and repeat quantification, respectively (see Supplementary Material online). Remarkably, nearly half (49%) of the D. alloeum genome consisted of repetitive sequences, although a substantial contributor (30%) was from unclassified repetitive sequences.
Chemosensory Gene Repertoire in D. alloeum
Chemoreception in arthropods is mediated by three major families of receptors: odorant receptors (ORs), gustatory receptors (GRs), and ionotropic receptors (IRs) (Clyne et al. 1999, 2000; Benton et al. 2009). In addition, two major families of water-soluble proteins are responsible for transport and/or quenching of ligands to chemosensory receptors: odorant binding proteins (OBPs) and chemosensory proteins (CSPs) (Vieira and Rozas 2011; Pelosi et al. 2014; Larter et al. 2016). Chemosensory discrimination of fruit volatiles is an important axis of divergence among host fly-associated populations of D. alloeum, initiating reproductive isolating barriers between these wasps (Forbes et al. 2009).
Previous characterizations of chemosensory genes in hymenopteran insects, in particular the gene-rich receptor families, demonstrate that automated gene prediction pipelines are generally poor at accurately predicting these gene models (Robertson and Wanner 2006; Croset et al. 2010; Robertson et al. 2010, 2018; Zhou et al. 2015). We therefore manually annotated a total of 321 gene models that represents the full inventory of five chemosensory gene families in D. alloeum (table 1 and see Supplementary Material online). The OR, GR, and IR gene families were larger in D. alloeum and other parasitoid wasps relative to A. mellifera. We found D. alloeum OR lineages in addition to clusters of GRs present in the braconid wasps D. alloeum and M. demolitor but absent in the well-studied hymenopterans N. vitripennis or A. mellifera (see Supplementary Material online). We also observed an increased number of IRs in D. alloeum relative to another Microplitis species, M. mediator (see Supplementary Material online). Although we identified chemosensory gene clusters specific to D. alloeum, the extensive gene duplication, gene loss, and sequence divergence in these families resulted in poor phylogenetic resolution and indeterminate orthology between gene family members. The difficulty in attributing gene expansions to D. alloeum is compounded by the relative lack of genome resources for parasitoid wasps.
Table 1.
Chemosensory Gene Content of Selected Hymenopteran Insects
| Organism | ORs | GRs | IRs | OBPs | CSPs | Citations |
|---|---|---|---|---|---|---|
| Diachasma alloeum | 187 (14) | 39(1) | 51 (5) | 15 (0) | 9 (0) | This study |
| Apis mellifera | 163 (11) | 10(0) | 10 (0a) | 21 (0a) | 6 (0a) | Robertson and Wanner (2006); Forêt and Maleszka (2006); Forêt et al. (2007); Croset et al. (2010); Elsik et al. (2014) |
| Nasonia vitripennis | 225 (76) | 47(11) | 99(54) | 90 (8) | 9 (0) | Robertson et al. (2010); Robertson et al. (2018); Werren et al. (2010); Vieira et al. (2012) |
| Microplitis demolitor b | 218 (4) | 85(1) | Zhou et al. (2015) | |||
| Microplitis mediator | 17 (0a) | 20 (0a) | 3 (0a) | Zhang et al. (2009); Wang et al. (2016); Peng et al. (2017) |
Note.—Intact gene counts are outside parentheses and pseudogene counts are inside parentheses.
Pseudogene counts were not addressed explicitly in the study.
Zhou et al. (2015) provided counts of truncated models and pseudogenes for ORs and GRs, however, these sequences were not published and therefore were not used in building phylogenies.
In summary, this gene set is an important resource for future studies of the evolutionary history of Diachasma chemosensory genes. It will be critical to ascertain the members of the D. alloeum chemosensory repertoire that operate specifically in chemosensory behavior. Although the families are generally well conserved across insects, the challenge of orthology assessment and the limited functional study of these genes make it difficult to estimate the precise chemosensory inventory of D. alloeum. ORs operate specifically in odorant recognition, and the expansion of OR genes in insects may have been adaptive during the transition to terrestrial life (Robertson et al. 2003, but see Missbach et al. 2014). Although relatively understudied, the IR family has a likely protostome origin, and conservation of multiple orthologs initially identified in Drosophilamelanogaster suggest an important function of IR genes in olfaction across insects (Rytz et al. 2013). Conversely, the origin of GRs dates back to the Placozoa, and GR-like genes in basal animals function in development, not chemosensation (Robertson 2015; Saina et al. 2015). The OBP and CSP transporter families have roles in chemical ligand delivery to chemosensory receptors but also function in release of pheromones, reproductive processes, and embryonic development (Pelosi et al. 2018). Transcriptome data sets used for D. alloeum gene predictions were taken from pooled whole male and female wasps, so we cannot exclude the possibility that some genes have nonchemosensory roles. Future studies should incorporate tissue-specific RNA data sets to provide stronger support for genetic components of chemosensation in D. alloeum.
Chemosensory genes are promising candidates for differential selective regimes in apple and hawthorn populations of D. alloeum. Rhagoletis pomonella host flies use olfactory cues from ripening fruit to identify suitable sites for mating and oviposition (Linn et al. 2003). Like R. pomonella, D. alloeum parasitoids have demonstrated odor preferences for their host fruits, representing a potential prezygotic reproductive barrier preventing mating between wasp populations utilizing different hosts (Forbes et al. 2009). Evolutionary rate and differential expression analyses of chemosensory genes in D. alloeum populations could be potential areas of inquiry.
Chemosensory gene evolution could also be influenced by transitions in reproductive strategies in Diachasma. Wasp courtship is mediated by the male perception of sex pheromones produced by females (Boush and Baerwald 1967). Across arthropods, chemosensory genes demonstrate differential expression in males and females (Zhou et al. 2012; Shiao et al. 2013; Eyun et al. 2017). Chemosensory genes showing strong sex bias may be candidates for degradation in an asexual genome, such as those involved in female signaling or male recognition of mate signals (Normark et al. 2003; Tabata et al. 2017). Future studies could assess sex-specific expression of chemosensory genes in D. alloeum and corresponding evolutionary patterns in its asexual relative D. muliebre.
Diachasma a lloeum Contains Canonical Genes Involved in Reproduction and Sex Determination
Hymenoptera is an insect order characterized by haplodiploid sex determination, providing an opportunity for studying the evolution of reproductive modes, including transitions from sexual to asexual systems. Meiosis is essential to obligate sexual reproduction, such that loss of sex may be accompanied by the subsequent degradation of meiotic genetic machinery (Schurko and Logsdon 2008). However, identical sets of meiosis genes in D. alloeum (sexual) and D. muliebre (asexual) (Tvedte et al. 2017) and population genetic data implying that the asexual D. muliebre undergoes recombination (Forbes et al. 2013) together suggests that asexual wasps retain meiotic production of gametes despite the loss of sexual reproduction. Given the apparent lack of male production in D. muliebre, a noncanonical form of meiosis could facilitate the maintenance of genetic variation and promote the persistence of this asexual lineage.
In many hymenopterans, development into male versus female forms is based on allelic states at a single locus, a mechanism known as complementary sex determination (CSD) (van Wilgenburg et al. 2006). In A. mellifera specifically, sex determination depends on the csd gene (Hasselmann et al. 2008). We found no evidence of the csd locus in D. alloeum, however our inability to consistently rear wasps in the laboratory at the current time precludes our ability to definitively rule out CSD as a sex determination mechanism. In CSD and non-CSD hymenopterans, a well-conserved sex determination regulatory cascade includes transformer and doublesex, both displaying sex-specific splicing (Geuverink and Beukeboom 2014). We annotated male and female isoforms of transformer and doublesex genes in D. alloeum (GenBank accessions THK33055.1, THK33056.1, THK32977.1, THK32978.1).
Sex determination genes may be targets of selection in asexual Hymenoptera. Across insects, male production occurs due to alternative splicing of transformer rendering the protein nonfunctional, leading to male-splicing of doublesex. Conversely, translation of full-length transformer into functional protein mediates the splicing of female-specific doublesex isoforms (Verhulst et al. 2010). RNA-seq read mapping patterns supported sex-specific transformer isoforms in D. alloeum (see Supplementary Material online). In all-female Diachasma species, we would expect selection to preserve the full-length transformer gene. In doublesex, the female isoform in D. alloeum is shorter (see Supplementary Material online), similar to splicing patterns in other insects (Cho et al. 2007; Oliveira et al. 2009). The single exon specific to males may be subject to future degradation following sex loss in asexual Diachasma species.
Additional genes contributing to sex-specific traits (e.g., sperm production, pheromones, pigmentation) may be candidates for degradation in asexual wasps (van der Kooi and Schwander 2014; Kraaijeveld et al. 2016). The high quality of D. alloeum assembly provides a suitable framework for future studies of the effects of sexual and asexual reproductive modes on patterns of molecular evolution across the wasp genome.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank the W.M. Keck Center for Comparative and Functional Genomics at the University of Illinois at Urbana-Champaign for genomic and RNA library construction and sequencing; Chris Fields at the High Performance Computing for Biology Center at the Roy J. Carver Biotechnology Center at the University of Illinois at Urbana-Champaign for performing the PBJelly scaffolding with TSLR reads; Austin Ward at the Biology Department at the University of Iowa for generating custom scripts for generating nonredundant protein data sets; Samuel Cummings at the Biology Department at the University of Iowa for contributing to annotation of chemosensory genes; and Monica Poelchau at the United States Department of Agriculture-Agricultural Research Service for generating the official gene set for D. alloeum. This work was supported by the United States Department of Agriculture/National Institute of Food and Agriculture as a grant to H.M.R. (A2008-35302-18819) and J.L.F. (2015-67013-23289) and the National Science Foundation as a grant to A.A.F. (DEB-1145355), J.L.F. (DEB-1638997), and J.H.W. (DEB1257053, IOS1456233).
Data deposition: This project has been deposited at GenBank under the accessions PRJNA284396 (whole genome) and PRJNA283787 (transcriptome).
Literature Cited
- Abrahamson WG, Blair CP.. 2007. Sequential radiation through host-race formation: herbivore diversity leads to diversity in natural enemies In: Tilmon KJ, editor. Specialization, speciation, and radiation: the evolutionary biology of herbivorous insects. Berkeley (CA: ): University of California Press; p. 188–200. [Google Scholar]
- Austin A, Dowton M.. 2000. The Hymenoptera: an introduction In: Austin A, Dowton M editors. Hymenoptera: evolution, biodiversity and biological control. Clayton (AU: ): CSIRO Publishing; p. 3–16. [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB.. 2009. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136(1):149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boush GM, Baerwald RJ.. 1967. Courtship behavior and evidence for a sex pheromone in the apple maggot parasite, Opius alloeus (Hymenoptera: Braconidae). Ann Entomol Soc Am. 60(4):865–866. [Google Scholar]
- Branstetter M, et al. 2018. Genomes of the Hymenoptera. Curr Opin Insect Sci. 25:65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke GR, Walden KK, Whitfield JB, Robertson HM, Strand MR.. 2018. Whole genome sequence of the parasitoid wasp Microplitis demolitor that harbors an endogenous virus mutualist. G3 (Bethesda) 8:2875–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush GL. 1966. The taxonomy, cytology, and evolution of the genus Rhagoletis in North America (Diptera, Tephritidae). B Mus Compar Zool. 134:431–562. [Google Scholar]
- Bush GL. 1994. Sympatric speciation in animals: new wine in old bottles. Trends Ecol Evol. 8:285–288. [DOI] [PubMed] [Google Scholar]
- Cho S, Huang ZY, Zhang J.. 2007. Sex‐specific splicing of the honeybee doublesex gene reveals 300 million years of evolution at the bottom of the insect sex‐determination pathway. Genetics 177(3):1733–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne PJ, et al. 1999. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron 22(2):327–338. [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Carlson JR. 2000. Candidate taste receptors in Drosophila. Science 287:1830–1834. [DOI] [PubMed] [Google Scholar]
- Croset V, et al. 2010. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 6(8):e1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambroski HR, et al. 2005. The genetic basis for fruit odor discrimination in Rhagoletis flies and its significance for sympatric host shifts. Evolution 59(9):1953–1964. [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G.. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsik CG, et al. 2014. Finding the missing honey bee genes: lessons learned from a genome upgrade. BMC Genomics 15(1):86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English AC, et al. 2012. Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS One 7(11):e47768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyun S, et al. 2017. Evolutionary history of chemosensory-related gene families across the Arthropoda. Mol Biol Evol. 34(8):1838–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder JL, Forbes AA.. 2010. Sequential speciation and the diversity of parasitic insects. Ecol Entomol. 35:67–76. [Google Scholar]
- Forbes AA, Bagley RK, Beer MA, Hippee AC, Widmayer HA.. 2018. Quantifying the unquantifiable: why Hymenoptera—not Coleoptera—is the most speciose animal order. BMC Ecol. 18(1):21.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes AA, et al. 2017. Revisiting the particular role of host shifts in initiating insect speciation. Evolution 71(5):1126–1137. [DOI] [PubMed] [Google Scholar]
- Forbes AA, Feder JL.. 2006. Divergent preferences of Rhagoletis pomonella host races for olfactory and visual fruit cues. Entomol Exp Appl. 119(2):121–127. [Google Scholar]
- Forbes AA, Powell THQ, Stelinski LL, Smith JJ, Feder JL.. 2009. Sequential sympatric speciation across trophic levels. Science 323(5915):776–779. [DOI] [PubMed] [Google Scholar]
- Forbes AA, Rice LA, Stewart NB, Yee WL, Neiman M.. 2013. Niche differentiation and colonization of a novel environment by an asexual parasitic wasp. J Evol Biol. 26(6):1330–1340. [DOI] [PubMed] [Google Scholar]
- Forêt S, Maleszka R.. 2006. Function and evolution of a gene family encoding odorant binding-like proteins in a social insect, the honey bee (Apis mellifera). Genome Res. 16:1404–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forêt S, Wanner KW, Maleszka R.. 2007. Chemosensory proteins in the honey bee: insights from the annotated genome, comparative analyses and expressional profiling. Insect Biochem Mol. 37(1):19–28. [DOI] [PubMed] [Google Scholar]
- Geuverink E, Beukeboom LW.. 2014. Phylogenetic distribution and evolutionary dynamics of the sex determination genes doublesex and transformer in insects. Sex Dev. 8(1–3):38–49. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29(7):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 8(8):1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselmann M, et al. 2008. Evidence for the evolutionary nascence of a novel sex determination pathway in honeybees. Nature 454(7203):519–522. [DOI] [PubMed] [Google Scholar]
- Hoede C, et al. 2014. PASTEC: an automatic transposable element classification tool. PLoS One 9(5):e91929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood GR, et al. 2015. Sequential divergence and the multiplicative origin of community diversity. Proc Natl Acad Sci U S A. 112(44):E5980–E5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraaijeveld K, et al. 2016. Decay of sexual trait genes in an asexual parasitoid wasp. Genome Biol Evol. 8(12):3685–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laetsch DR, Blaxter ML.. 2017. BlobTools: interrogation of genome assemblies. F1000Res. 6:1287. [Google Scholar]
- Larter NK, Sun JS, Carlson JR.. 2016. Organization and function of Drosophila odorant binding proteins. Elife 5:e20242.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaSalle J, Gauld ID.. 1993. Hymenoptera: their diversity, and their impact on the diversity of other organisms In: LaSalle J, Gauld ID, editors. Hymenoptera and biodiversity. Wallingford (United Kingdom): CAB International; p. 1–26. [Google Scholar]
- Linn C, et al. 2003. Fruit odor discrimination and sympatric host race formation in Rhagoletis. Proc Nat Acad Sci U S A. 100(20):11490–11493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience 1(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma WJ, Pannebakker BA, Beukeboom LW, Schwander T, Van de Zande L.. 2014. Genetics of decayed sexual traits in a parasitoid wasp with endosymbiont-induced asexuality. Heredity 113(5):424–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missbach C, et al. 2014. Evolution of insect olfactory receptors. Elife 3:e02115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normark BB, Judson OP, Moran NA.. 2003. Genomic signatures of ancient asexual lineages. Biol J Linn Soc. 79(1):69–84. [Google Scholar]
- Nosil P. 2012. Ecological speciation. New York: Oxford University Press. [Google Scholar]
- Oliveira D, et al. 2009. Identification and characterization of the doublesex gene of Nasonia. Insect Mol Biol. 18(3):315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi P, Iovinella I, Felicioli A, Dani FR.. 2014. Soluble proteins of chemical communication: an overview across arthropods. Front Physiol. 5:320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi P, Iovinella I, Zhu J, Wang G, Dani FR.. 2018. Beyond chemoreception: diverse tasks of soluble olfactory proteins in insects. Biol Rev. 93(1):184–200. [DOI] [PubMed] [Google Scholar]
- Peng Y, et al. 2017. Identification of odorant binding proteins and chemosensory proteins in Microplitis mediator as well as functional characterization of chemosensory protein 3. PLoS One 12(7):e0180775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poelchau M, et al. 2015. The i5k Workspace@ NAL—enabling genomic data access, visualization and curation of arthropod genomes. Nucleic Acids Res. 43(D1):D714–D719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poynton HC, et al. 2018. The toxicogenome of Hyalella azteca: a model for sediment ecotoxicology and evolutionary toxicology. Environ Sci Technol. 52(10):6009–6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM. 2015. The insect chemoreceptor superfamily is ancient in animals. Chem Senses. 40(9):609–614. [DOI] [PubMed] [Google Scholar]
- Robertson HM, et al. 2018. Genome sequence of the wheat stem sawfly, Cephus cinctus, representing an early-branching lineage of the Hymenoptera, illuminates evolution of hymenopteran chemoreceptors. Genome Biol Evol. 10:2997–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM, Gadau J, Wanner KW.. 2010. The insect chemoreceptor superfamily of the parasitoid jewel wasp Nasonia vitripennis. Insect Mol Biol. 19:121–136. [DOI] [PubMed] [Google Scholar]
- Robertson HM, Wanner KW.. 2006. The chemoreceptor superfamily in the honey bee, Apis mellifera: expansion of the odorant, but not gustatory, receptor family. Genome Res. 16(11):1395–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM, Warr CG, Carlson JR.. 2003. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc Nat Acad Sci U S A. 100(Suppl 2):14537–14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rytz R, Croset V, Benton R.. 2013. Ionotropic receptors (IRs): chemosensory ionotropic glutamate receptors in Drosophila and beyond. Insect Biochem Mol. 43(9):888–897. [DOI] [PubMed] [Google Scholar]
- Saina M, et al. 2015. A cnidarian homologue of an insect gustatory receptor functions in developmental body patterning. Nat Commun. 6:6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurko AM, Logsdon JM Jr.. 2008. Using a meiosis detection toolkit to investigate ancient asexual “scandals” and the evolution of sex. Bioessays 6:579–589. [DOI] [PubMed] [Google Scholar]
- Shiao M, et al. 2013. Transcriptional profiling of adult Drosophila antennae by high-throughput sequencing. Zool Stud. 52(1):42. [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. RepeatMasker Open-3.0. 1996-2010. http://www.repeatmasker.org, last accessed October 2018. [Google Scholar]
- Smit AFA, Hubley R. RepeatModeler Open-1.0. 2008-2015. http://www.repeatmasker.org, last accessed October 2018. [Google Scholar]
- Stireman JO, Nason JD, Heard SB, Seehawer JM.. 2006. Cascading host-associated genetic differentiation in parasitoids of phytophagous insects. Proc R Soc B. 273(1586):523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata J, Ichiki RT, Moromizato C, Mori K.. 2017. Sex pheromone of a coccoid insect with sexual and asexual lineages: fate of an ancestrally essential sexual signal in parthenogenetic females. J R Soc Interface. 14(128):20170027.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tvedte ES, Forbes AA, Logsdon JJ.. 2017. Retention of core meiotic genes across diverse Hymenoptera. J Hered. 108(7):791–806. [DOI] [PubMed] [Google Scholar]
- Tvedte ES, Logsdon JM Jr, Forbes AA.. 2019. Sex loss in insects: causes of asexuality and consequences for genomes. Curr Opin Insect Sci. 31:77–83. [DOI] [PubMed] [Google Scholar]
- van der Kooi CJ, Matthey-Doret C, Schwander T.. 2017. Evolution and comparative ecology of parthenogenesis in haplodiploid arthropods. Evol Lett. 1(6):304–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kooi CJ, Schwander T.. 2014. On the fate of sexual traits under asexuality. Biol Rev. 89(4):805–819. [DOI] [PubMed] [Google Scholar]
- van Wilgenburg E, Driessen G, Beukeboom LW.. 2006. Single locus complementary sex determination in Hymenoptera: an “unintelligent” design? Front Zool. 3(1):1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhulst EC, van de Zande L, Beukeboom LW.. 2010. Insect sex determination: it all evolves around transformer. Curr Opin Genet Dev. 20(4):376–383. [DOI] [PubMed] [Google Scholar]
- Vieira FG, et al. 2012. Unique features of odorant-binding proteins of the parasitoid wasp Nasonia vitripennis revealed by genome annotation and comparative analyses. PLoS One 7(8):e43034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira FG, Rozas J.. 2011. Comparative genomics of the odorant-binding and chemosensory protein gene families across the Arthropoda: origin and evolutionary history of the chemosensory system. Genome Biol Evol. 3:476–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh BD. 1867. The apple‐worm and the apple‐maggot. J Hort. 2:338–343. [Google Scholar]
- Wang S, et al. 2016. Cloning and expression profile of ionotropic receptors in the parasitoid wasp Microplitis mediator (Hymenoptera: Braconidae). J Insect Physiol. 90:27–35. [DOI] [PubMed] [Google Scholar]
- Weinstock GM, et al. 2006. Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443:931–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, et al. 2010. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science 327(5963):343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton RA, Marsh PM.. 1978. New world Opiinae (Hymenoptera: Braconidae) parasitic on Tephritidae (Diptera). J Wash Acad Sci. 147–167. [Google Scholar]
- Wheeler D, Redding AJ, Werren JH.. 2013. Characterization of an ancient lepidopteran lateral gene transfer. PLoS One 8(3):e59262.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield JB. 2003. Phylogenetic insights into the evolution of parasitism in Hymenoptera. Adv Parasitol. 54:69–100. [DOI] [PubMed] [Google Scholar]
- Zhang S, Zhang Y, Su H, Gao X, Guo Y.. 2009. Identification and expression pattern of putative odorant-binding proteins and chemosensory proteins in antennae of the Microplitis mediator (Hymenoptera: Braconidae). Chem Senses. 34(6):503–512. [DOI] [PubMed] [Google Scholar]
- Zhou X, et al. 2012. Phylogenetic and transcriptomic analysis of chemosensory receptors in a pair of divergent ant species reveals sex-specific signatures of odor coding. PLoS Genet. 8(8):e1002930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, et al. 2015. Chemoreceptor evolution in hymenoptera and its implications for the evolution of eusociality. Genome Biol Evol. 7(8):2407–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.