Abstract
Background and objective
Acinetobacter baumannii (A. baumannii) is a common nosocomial pathogen, which developed multi-drug-resistance to different classes of antibiotics including carbapenems. This study examined ten common carbapenemase genes among 32 carbapenem-resistant A. baumannii clinical isolates recovered from Taif, Saudi Arabia.
Methods
Isolates were phenotypically identified to the genus level by Vitek®2 and API 20NE®. The species level was confirmed by the amplification of blaOXA-51. The susceptibility for 21 different antibiotics was performed by Vitek 2 and modified Kirby-Bauer method. Isolates were genetically screened for 10 carbapenemases. Phylogenetic relatedness between isolates was determined by ERIC-PCR.
Results
Genotypically identified A. baumannii represented 100% of the total phenotypically identified Acinetobacter spp. All the carbapenem-resistant isolates were sensitive to polymyxin B and colistin. Among the other antibiotics, ampicillin/sulbactam and tigecycline were the most effective agents. 90.8% of the isolates were resistant to all ten investigated β–lactams. blaOXA-51, blaIPM, blaNDM and blaOXA-23 were detected in 100%, 87.5%, 62.5% and 59.4% of isolates, respectively. Also, blaVIM and blaOXA-40 were less prevalent and were detected in 9.3% and 3.1% of the isolates, respectively. In addition, blaKPC, blaOXA-48, blaOXA-58, blaOXA-181 were not detected in any isolate. The A. baumannii isolates were categorised into ten genotypes on the basis of the detected carbapenemase genes and ERIC-PCR revealed a remarkable clonal diversity among these isolates.
Conclusion
Class A and class D carbapenemase genes were the most commonly detected among carbapenem resistant A. baumannii (CRAB) clinical isolates.
Keywords: A. baumannii, blaOXA-51, carbapenemases, carbapenems, ERIC-PCR
Introduction
Acinetobacter spp. are recognized as important nosocomial pathogens. They cause a wide range of nosocomial infections including meningitis, urinary tract infection (UTI), bloodstream infection (BSI), wound infection (WI) and ventilator-associated pneumonia (VAP).1 The genus Acinetobacter is strictly aerobic, non-motile, non-fermentative, oxidase-negative, and catalase-positive Gram-negative coccobacilli.2–4
The genus Acinetobacter includes more than 50 species. While some species are pathogenic, the majority are non-pathogenic.5 The most common pathogenic species are Acinetobacter baumannii (A. baumannii) followed by A. calcoaceticus and A. lwoffii. Other occasionally reported pathogenic species include A. haemolyticus and A. johnsonii.6 A. baumannii can be specifically identified on the molecular level by the detection of blaOXA-51, which is absent in other species.7
A. baumannii is considered one of the most troublesome species due to its ability to resist a large number of antibiotics from different classes including carbapenem group.8 In addition to its multi-drug- resistance (MDR), A. baumannii is associated with high morbidity and mortality in different hospital settings, especially the intensive care units (ICUs), where patients are immunocompromised.2,9 According to the classification of Infectious Diseases Society of America (IDSA), A. baumannii is recognized as one of the six most important MDR microorganisms in hospitals worldwide.10
A. baumannii is considered MDR when it resists at least three classes of antimicrobial agents including all penicillins and cephalosporins, fluroquinolones, and aminoglycosides. On the other hand, it is considered an extensive drug resistant (XDR) when the MDR isolate is resistant to carbapenems. Pandrug resistant (PDR) A. baumannii is recognized when the XDR isolate is also resistant to polymyxins and tigecycline.11
Resistance to carbapenems is mediated by different mechanisms including (i) efflux pump, (ii) decreased membrane permeability through the loss of outer membrane porins (OMP) by downregulation of their synthesis and (iii) production of carbapenemases.12
The production of carbapenemases is one of the most important mechanisms responsible for carbapenem resistance among A. baumannii clinical isolates. According to Ambler classification of β-lactamases,13 carbapenemases are related to three different molecular classes: (i) class A carbapenemases eg Klebsiella pneumoniae carbapenemase (KPC) and Imipenem-hydrolysing β-lactamase (IMI),14 (ii) class B carbapenemases or the so-called metallo-β-lactamases eg Verona integrin metallo-β-lactamase (VIM), imipenemase (IMP) and New Delhi metallo-β-lactamases (NDM)14–16 and (iii) class D carbapenemase or the so-called OXA-type carbapenemases eg OXA-23, OXA-40, OXA-48, OXA-51, OXA-58 and OXA-181.17,18
Patients and methods
Bacterial strains
The A. baumannii clinical isolates included in this study were collected from December 2017 to May 2017. They were isolated as a part of the routine hospital laboratory procedures and were further identified and confirmed in the laboratory. The study protocol was approved by Taif University Research Ethics Committee (approval 38-35-0021). Thirty-two non-duplicate non-consecutive clinical isolates of carbapenem-resistant A. baumannii were selected from a total of 45 Acinetobacter spp. clinical isolates. The isolates were recovered from 20 males and 12 females who were admitted to different medical departments at a large tertiary care hospital in Taif, KSA. The patients aged between 7 days to 97 years. The investigated isolates were recovered from different clinical specimens which included blood (n=12), tissue biopsy (n=1), peritoneal fluid (n=1), sputum (n=11), urine (n=2), wound (n=2), and catheter tip (n=3).
Isolation and identification
All strains were primarily isolated on blood agar and, then, purified on MacConkey’s agar (Oxoid, UK). The genus and species level of the recovered Acinetobacter spp. isolates were primarily identified by Vitek®2 and API 20NE®. Molecular confirmation of A. baumannii clinical isolates was achieved through the amplification of the intrinsic blaOXA-51 gene by polymerase chain reaction (PCR).
Antimicrobial susceptibility testing (AST)
All recovered isolates were subjected to AST against 21 different antibiotics. The minimum inhibitory concentration (MIC) was determined for the available 13 antibiotics, in Vitek 2 GN ID card (Biomeriux, France), which are ceftazidime, cefotaxime, cefepime piperacillin/tazobactam, imipenem, meropenem, gentamicin, amikacin, tobramycin, ciprofloxacin, levofloxacin, tetracycline and sulfamethoxazole/trimethoprim. Modified Kirby-Bauer method was used to confirm the results of the Vitek 2 system and to test the susceptibility to ticarcillin, piperacillin, ampicillin/sulbactam, ceftriaxone, netilmicin, polymyxin B, colistin (Biorad, USA) and tigecycline (Wyeth, USA), that are not available in the Vitek® system. Escherichia coli (E. coli) ATCC 25,922 and Klebsiella pneumonia (K. pneumoniae) ATCC 700,603 were used as quality control standard strains. Results were interpreted according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI).19
Preparation of DNA templates
The boiling method was applied for total genomic DNA extraction as previously described.20 Briefly, three to six colonies of the bacterial isolates (depending on colony size) were picked from a tryptic soy agar (Scharlau, Spain) plate and suspended in 100µl of DNase-free water in a sterile 1.5ml microfuge tube to obtain a bacterial suspension equivalent to 1-2×109 CFU/mL. The bacterial suspension was placed in a boiling water bath for 10 min. The lysed suspension was centrifuged at a speed of 13200rpm for 5 mins. The supernatant containing the total genomic DNA extract was transferred to a new sterile DNase free microfuge tube using DNase-free tips. The total genomic DNA extract was stored at −20οC until used.
Genotypic detection of carbapenemases
All carbapenem-resistant isolates were screened for 10 carbapenemase encoding genes; Two class A carbapenemases (blaIPM and blaKPC), two class B carbapenemases (blaNDM, and blaVIM) and six class D carbapenemase (blaOXA-23, blaOXA-40, blaOXA-48, blaOXA-51, blaOXA-58 and blaOXA-181). Target gene amplification was performed using primers (Geumcheon-gu, Seoul, Korea) and the cycling conditions listed in Table 1.
Table 1.
Primers and cycling conditions used for the amplification of carbapenemase genes and the repetitive intergenic consensus
| Primer | Sequence | Gene | Reference | Amplification conditions | Amplicon size (bp) |
|---|---|---|---|---|---|
| ERIC-1R | R:AACCCACGATGTGGGTAGC | - | 50 | Initial denaturation at 95oC for 15 min, then 35 cycles of 95oC for 1 min, 40oC for 1min and 72oC for 5 minutes and one cycle of final elongation at 72oC. | - |
| IPM |
F: GGAATAGRRTGGCTTAAYT R: GGTTTAAYAAARCAMCCACC |
blaIPM | 51 | Initial denaturation at 95oC for 15 min, then 35 cycles of 95oC for 1 min, 40oC for 1min and 72oC for 5 minutes and one cycle of final elongation at 72oC. | 233 |
| KPC |
F: GTATCGCCGTCTAGTTCTGC R: GGTCGTGTTTCCCTTTAGCC |
blaKPC | 52 | Initial denaturation at 95oC for 15 min, then 30 cycles of 95oC for 1 min, 55oC for 1min and 72oC for 5 minutes and one cycle of final elongation at 72oC. | 637 |
| NDM |
F: GGTTTGGCGATCTGGTTTTC R: CGGAATGGCTCATCACGATC |
blaNDM | 53 | Initial denaturation at 95oC for 15 min, then 30 cycles of 95oC for 1 min, 66.7oC for 1min and 72oC for 5 minutes and one cycle of final elongation at 72oC. | 621 |
| VIM |
F: GTTTGGTCGCATATCGCAAC R: GAGCAAKTCYAGACCGCCC |
blaVIM | 51 | Initial denaturation at 95oC for 15 min, then 30 cycles of 95oC for 1 min, 63oC for 1min and 72oC for 5 minutes and one cycle of final elongation at 72oC. | 591 |
| OXA-23 |
F: GATCGGATTGGAGAACCAGA R: ATTTCTGACCGCATTTCCAT |
blaOXA-23 | 54 | Initial denaturation at 95oC for 15 min, then 30 cycles of 95oC for 1 min, 66.7oC for 1min and 72oC for 5 minutes and one cycle of final elongation at 72oC. | 501 |
| OXA-40 |
F: GGTTAGTTGGCCCCCTTAAA R: AGTTGAGCGAAAAGGGGATT |
blaOXA-40 | 55 | Initial denaturation at 95oC for 15 min, then 30 cycles of 95oC for 1 min, 66.6oC for 1min and 72oC for 5 minutes and one cycle of final elongation at 72oC. | 249 |
| OXA-48 |
F: GCTTGATCGCCCTCGATT R: GATTTGCTCCGTGGCCGAAA |
blaOXA-48 | 56 | Initial denaturation at 95oC for 15 min, then 30 cycles of 95oC for 1 min, 60.5oC for 1min and 72oC for 5 minutes and one cycle of final elongation at 72oC. | 281 |
| OXA-51 |
F: TAATGCTTTGATCGGCCTTG R: TGGATTGCACTTCATCTTGG |
blaOXA-51 | 57 | Initial denaturation at 95oC for 15 min, then 30 cycles of 95oC for 1 min, 60.5oC for 1min and 72oC for 5 minutes and one cycle of final elongation at 72 oC. | 353 |
| OXA-58 |
F: AAGTATTGGGGCTTGTGCTG R: CCCCTCTGCGCTCTACATAC |
blaOXA-58 | 54 | Initial denaturation at 95oC for 15 min, then 30 cycles of 95oC for 1 min, 60.5oC for 1min and 72oC for 5 minutes and one cycle of final elongation at 72oC. | 599 |
| OXA-181 |
F: ATGCGTGTATTAGCCTTATCG R: AACTACAAGCGCATCGAGCA |
blaOXA-181 | 22 | Initial denaturation at 95oC for 15 min, then 30 cycles of 95oC for 1 min, 55oC for 1min and 72oC for 5 minutes and one cycle of final elongation at 72oC. | 888 |
Notes: R= (A⁄G), Y= (C⁄ T), K= (G⁄ T) and M= (A⁄C). Boldface in this table clarifies primers and different stages of the PCR cycles.
PCR
The PCR reaction was performed in a final reaction volume of 40µl. The reaction mixture contained 8 µl of the extracted DNA, 8 µl of 5x master mix (HOT FIREPol® Blend Master Mix, Solis BioDyne, Tartu, Estonia), 1.2 µl of the forward primer (10 pmol/μl), 1.2 µl of the reverse primer (10 pmol/μl) and 21.6 µl sterile distilled water. The 0.2 PCR tubes that contained the reaction mixture were placed in the thermal cycler and the reaction was processed for each gene according to cyclic conditions listed in Table 1.
Genotyping of the clinical isolates
Clonal relatedness between A. baumannii clinical isolates was determined by enterobacterial repetitive intergenic consensus-PCR (ERIC-PCR). Amplification of the repeated intergenic consensus regions was performed using Mastercycler® personal (Eppendorf, California, USA), and the primers and cycling conditions described in Table 1. The amplified fragments were run on a 2.5% agarose at 100 volt for 90 mins. The agarose gel was removed from the gel tank and placed on a UV- transilluminator tray in which images were captured.
Fingerprint pattern analysis
The generated ERIC-PCR profiles were analysed by BioNumerics 7.5® software (Applied Maths, Kortrijk, Belgium) as previously described.21 The captured gel images stained with ethidum bromide were uploaded to the software and were analysed to generate the dendrogram.
The generated dendrogram was performed according to the Dice similarity coefficient based on the unweighted pair group method with arithmetic averages (UPGMA) at position tolerance at 0.15.
Results
Isolation and phenotypic identification
According to the identification by Vitek 2 system, all isolates were related to the genus Acinetobacter, in which 56.3% (18/32) were A. baumannii/haemolyticus, 37.5% (12/32) were A. baumannii complex/haemolyticus and 6.3% (2/32) were A. lwoffii. On the other hand, phenotypic identification by API 20NE revealed that all isolates were related to the genus Acinetobacter with different levels of possible identification in which 50% (16/32) of the isolates were A. baumannii/calcoaceticus with excellent identification (98.8%), 40.6% (13/3) were A. baumannii/calcoaceticus with low discrimination (66.5%) and 9.4% (3/32) were A. baumannii/ calcoaceticus with low discrimination (26.5%).
Molecular confirmation of A. baumannii
Molecular investigation of blaOXA-51 revealed that 100% of the phenotypically identified Acinetobacter spp. isolates were positive, so they were considered as A. baumannii.
Antimicrobial susceptibility
Antimicrobial susceptibility testing revealed that polymyxin B and colistin were the most effective agents as all isolates were sensitive to both agents. On the other hand, both tigecycline and ampicillin/sulbactam were ranked as the second effective agents since 75% (24/32) and 62.5% (20/32) of the isolates were sensitive, respectively. It should be mentioned that twelve isolates (37.5%) were intermediately resistant to ampicillin/sulbactam and none of the isolates was resistant to that combination. The isolates had high resistance rates to both 3rd and 4th generation cephalosporins in which at least 90.8% (29/32) of the isolates were resistant.
With regard to the two tested quinolone antibiotics, 100% (32/32) and 78.1% (25/32)of isolates were resistant to ciprofloxacin and levofloxacin, respectively. The resistance rate to aminoglycosides ranged between 56.3 (18/32) and 90.6% (29/32). Lower resistance rates of 56.3% (18/32) and 59.4% (19/32) were observed for gentamicin and tobramycin, respectively as shown in Table 2.
Table 2.
Susceptibility pattern of A. baumannii clinical isolates
| Antibiotic | Susceptibility pattern | |||||
|---|---|---|---|---|---|---|
| Sensitive | Intermediate | Resistant | ||||
| No. | % | No. | % | No. | % | |
| Cefotaxime | 0.00 | 0.00 | 1.00 | 3.12 | 31.00 | 96.88 |
| Ceftazidime | 2.00 | 6.24 | 0.00 | 0.00 | 30.00 | 93.76 |
| Ceftriaxone | 1.00 | 3.12 | 1.00 | 3.12 | 30.00 | 93.76 |
| Cefepime | 0.00 | 0.00 | 3.00 | 9.36 | 29.00 | 90.64 |
| Imipenem | 0.00 | 0.00 | 0.00 | 0.00 | 32.00 | 100.00 |
| Meropenem | 0.00 | 0.00 | 2.00 | 6.24 | 30.00 | 93.76 |
| Ticarcillin | 0.00 | 0.00 | 0.00 | 0.00 | 32.00 | 100.00 |
| Piperacillin | 0.00 | 0.00 | 0.00 | 0.00 | 32.00 | 100.00 |
| Piperacillin/Tazobactam | 0.00 | 0.00 | 0.00 | 0.00 | 32.00 | 100.00 |
| Ampicillin/Sulbactam | 20.00 | 62.50 | 12.00 | 37.50 | 0.00 | 0.00 |
| Gentamicin | 5.00 | 15.63 | 9.00 | 28.13 | 18.00 | 56.25 |
| Tobramycin | 11.00 | 34.38 | 2.00 | 6.24 | 19.00 | 59.38 |
| Amikacin | 3.00 | 9.36 | 0.00 | 0.00 | 29.00 | 90.64 |
| Netilmicin | 8.00 | 25.00 | 0.00 | 0.00 | 25.00 | 75.00 |
| Ciprofloxacin | 0.00 | 0.00 | 0.00 | 0.00 | 32.00 | 100.00 |
| Levofloxacin | 0.00 | 0.00 | 7.00 | 21.88 | 25.00 | 78.12 |
| Tetracycline | 3.00 | 9.36 | 0.00 | 0.00 | 29.00 | 90.64 |
| Tigecycline | 24.00 | 75.00 | 0.00 | 0.00 | 9.00 | 25.00 |
| Sulfamethoxazole/Trimethoprim | 6.00 | 18.75 | 0.00 | 0.00 | 26.00 | 81.25 |
| Polymyxin B | 32.00 | 100.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Colistin | 32.00 | 100.00 | 0.00 | 0.00 | 0.00 | 0.00 |
All A. baumannii isolates had multiple drug-resistance to at least 13 of the 21 tested antibiotics (Table 3). The percentage of isolates resistant to 17–19 antibiotics was 59.4% (19/32). On the other hand, the isolates were resistant to at least eight β-lactams and 12/32 (37.7%) were resistant to the ten tested β-lactams.
Table 3.
Resistance patterns of the A. baumannii clinical isolates
| Isolate No. | Antibiotic resistance profile | Resistance markers |
|---|---|---|
| AC01 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, SAM, CN, TOB, AK, NET, TET, CIP, LEV, SXT | 18 |
| AC02 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, CN, TOB, AK, NET, TET, CIP, LEV, SXT | 16 |
| AC03 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, AK, TET, CIP, LEV, SXT | 13 |
| AC04 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, SAM, CN, TOB, AK, NET, TET, CIP, LEV, TEG, SXT | 19 |
| AC05 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, TET, CIP, LEV, SXT | 13 |
| AC06 | CTX, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, CN, AK, NET, CIP, LEV | 13 |
| AC07 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, CN, TOB, AK, NET, TET, CIP, LEV, SXT | 17 |
| AC08 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, CN, TOB, AK, NET, TET, CIP, LEV, SXT | 17 |
| AC66 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, AK, NET, TET, CIP, LEV, SXT | 15 |
| AC10 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, CN, TOB, CIP, LEV | 13 |
| AC11 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, SAM, CN, TOB, AK, NET, TET, CIP, LEV, TEG, SXT | 19 |
| AC12 | CTX, CAZ, FEP, IPM, MEM, TIC, PIP, TPZ, CN, AK, TET, CIP, LEV, SXT | 14 |
| AC13 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, SAM, CN, TOB, AK, NET, TET, CIP, LEV, SXT | 18 |
| AC14 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, CN, TOB, AK, NET, TET, CIP, LEV, SXT | 17 |
| AC15 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, CN, TOB, AK, NET, TET, CIP, LEV | 16 |
| AC16 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, CN, TOB, AK, NET, TET, CIP, LEV, SXT | 17 |
| AC17 | CTX, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, CN, AK, NET, CIP, LEV | 13 |
| AC18 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, SAM, CN, TOB, AK, TET, CIP, LEV, TEG, SXT | 18 |
| AC19 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, AK, NET, TET, CIP, LEV, SXT | 15 |
| AC20 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, CN, AK, NET, TET, CIP, LEV, SXT | 16 |
| AC21 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, CN, AK, NET, TET, CIP, LEV, SXT | 16 |
| AC22 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, SAM, CN, TOB, AK, NET, TET, CIP, LEV, TEG, SXT | 19 |
| AC23 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, SAM, CN, TOB, AK, NET, TET, CIP, LEV, TEG, SXT | 19 |
| AC24 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, CN, TOB, AK, NET, TET, CIP, LEV, TEG | 17 |
| AC25 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, CN, TOB, AK, NET, TET, CIP, LEV, TEG | 17 |
| AC26 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, CN, TOB, AK, NET, TET, CIP, LEV, SXT | 17 |
| AC27 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, SAM, CN, AK, NET, TET, CIP, LEV, SXT | 17 |
| AC28 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, SAM, CN, TOB, AK, NET, TET, CIP, LEV, SXT | 18 |
| AC29 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, SAM, CN, TOB, AK, NET, TET, CIP, LEV, TEG, SXT | 19 |
| AC30 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, TET, CIP, LEV, SXT | 13 |
| AC31 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, SAM, CN, TOB, AK, NET, TET, CIP, LEV, SXT | 18 |
| AC32 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, SAM, CN, TOB, AK, NET, TET, CIP, LEV, SXT | 18 |
Abbreviations: CTX, cefotaxime; CAZ, ceftazidime; CRO, ceftriaxone; FEP, cefepime; IPM, imipenem; MEM, meropenem; TIC, ticarcillin; PIP, piperacillin; TPZ, piperacillin/tazobactam; SAM, ampicillin/sulbactam; CN, gentamicin; TOB, tobramycin; NET, netilmicin; CIP, ciprofloxacin; LEV, levofloxacin; TET, tetracycline; TEG, tigecycline; SXT, sulfamethoxazole/trimethoprim.
Genotypic detection of carbapenemases
The investigation of class A carbapenemases revealed that 87.5% (28/32) of the isolates were positive for blaIPM gene while none of the isolates harboured the blaKPC gene. With regard to class B carbapenemases (metallo-carbapenemase), it was found that 62.5% (20/32) and 9.4% (3/32) of the isolates harboured blaNDM and blaVIM, respectively (Table 4).
Table 4.
Relationship between genotypic profiles of carbapenemases and phenotypic resistance profiles A. baumannii for β-lactam antibiotics
| No | Genetic Profile | No of isolates | Profiles of detected β -lactamase genes |
Phenotypic resistance profiles to β -lactams | Resistance markers |
|---|---|---|---|---|---|
| 1 | A | 6 | blaOXA-51, blaIPM, blaNDM | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, SAM |
9-10 |
| 2 | B | 8 | blaOXA-51, blaOXA-23, blaIPM | CTX, CRO, FEP, IPM, MEM, TIC, PIP, TPZ CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, SAM |
8-10 |
| 3 | C | 1 | blaOXA-51, blaOXA-23 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, SAM | 10 |
| 4 | D | 6 | blaOXA-51 | CTX, CAZ, FEP, IPM, MEM, TIC, PIP, TPZ CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, SAM |
8-10 |
| 5 | E | 1 | blaOXA-51, blaOXA-40 | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ | 9 |
| 6 | F | 2 | blaOXA-51, blaOXA-23, blaIPM, blaNDM, blaVIM | CTX, CRO, FEP, IPM, MEM, TIC, PIP, TPZ CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ |
8-9 |
| 7 | G | 1 | blaOXA-51, blaOXA-23, blaIPM, blaVIM | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, SAM | 10 |
| 8 | H | 1 | blaOXA-51, blaOXA-23, blaNDM | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ | 9 |
| 9 | I | 6 | blaOXA-51, blaOXA-23, blaIPM, blaNDM | CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ CTX, CAZ, CRO, FEP, IPM, MEM, TIC, PIP, TPZ, SAM |
9-10 |
Class D carbapenemases (OXA-type carbapenemases) were the most prevalent among CRAB isolates where 59.4% (19/32), 3.1% (1/32) and 100% (32/32) of the isolates were positive for blaOXA-23, blaOXA-40 and blaOXA-51 genes, respectively. On the other hand, blaOXA-48 and blaOXA-58 and blaOXA-181 genes were not detected.
Analysis of carbapenemase genetic profiles of the 10 investigated carbapenemase genes revealed nine different genetic profiles (A-I). The most common profile was profile B, which was detected in 25% (8/32) of the isolates followed by the A and J profiles; each was detected in 18.8% (6/32) of the isolates (Table 4). The least detected genetic carbapenemase profiles were C, E, G, H and I that were detected in only one isolate each.
Fingerprint pattern analysis
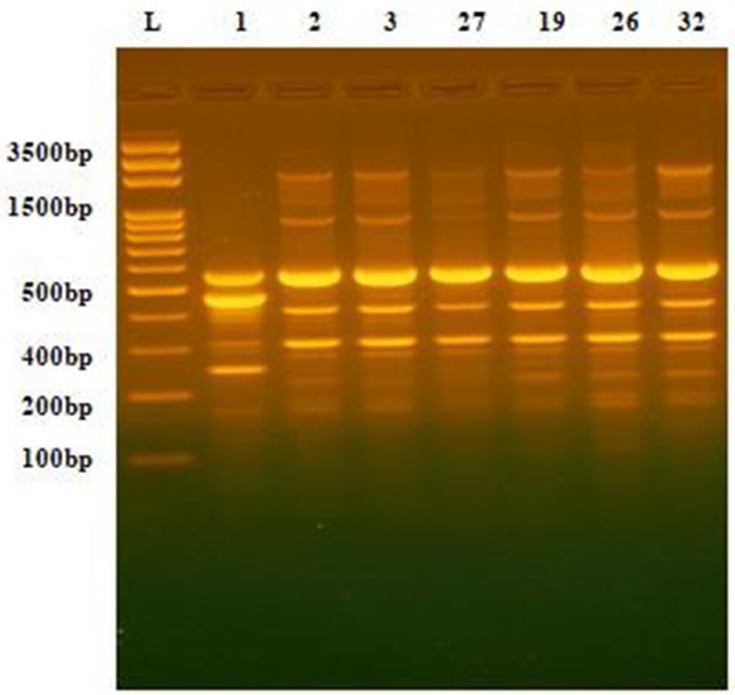
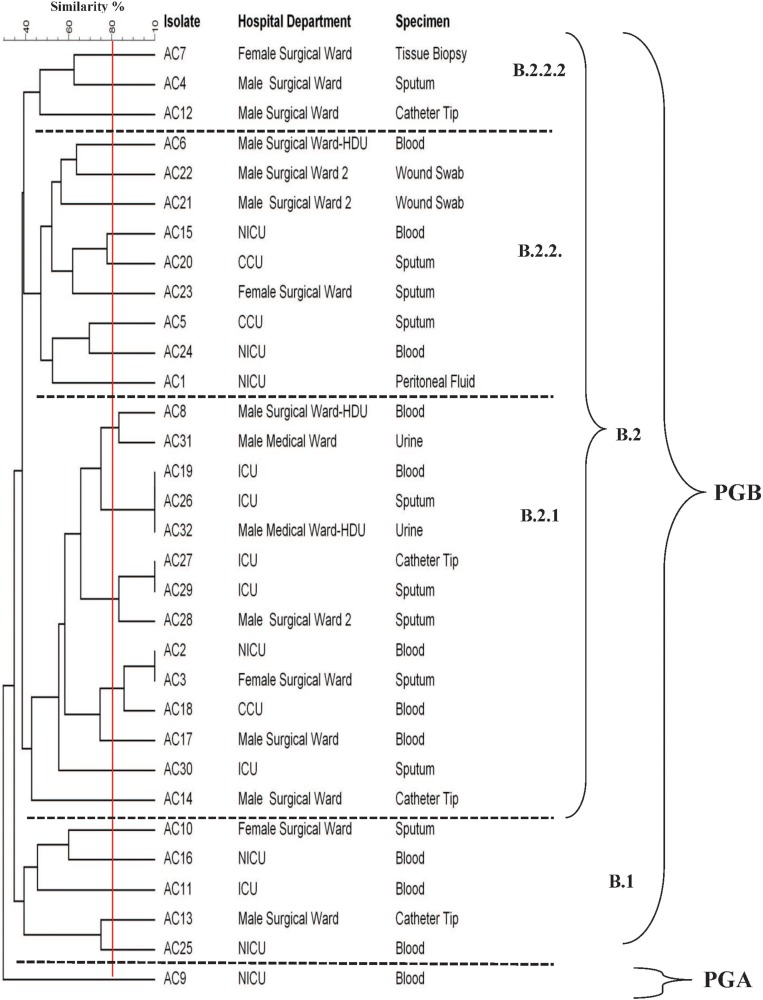
The DNA fingerprint patterns of A. baumannii isolates were generated by ERIC-PCR as shown in Figure 1. The generated dendrogram at 80% similarity demonstrated 28 different fingerprint profiles with high clonal variability. Twenty-five isolates had 25 different profiles and 7 isolates had the remaining three profiles. The three profiles included one single profile for AC19, AC26 and AC32, one single profile for AC27 and AC29 isolates and one single profile for AC2 and AC3 isolates (Figure 2). The generated UPGMA dendrogram categorized the 32 isolates into two main phylogenetic groups (A and B). While phylogenetic group A (PGA) included only one isolate (AC9), phylogenetic group B (PGB) hosted 31 isolates (96.9%). PGB was further subdivided into two main sub-phylogenetic groups (PGB.1 and PGB.2). As shown in Figure 2, PGB 2 was further sub-classified into three main clades B.2.1, B.2.2.1 and B.2.2.2. Subgroup B2.2.1 represented 43.8% (14/32) of the investigated isolates.
Figure 1.
DNA fingerprint pattern generated by ERIC-PCR for Acinetobacter clinical isolates. L, 100bp DNA ladder; lanes 1, 2, 3, 27,19, 26 and 32 represent the code No. of the isolates.
Figure 2.
Clonal relationship between A. baumannii clinical isolates generated by UPGMA dendrogram.
Discussion
A. baumannii is one of the most important opportunistic pathogens that cause hospital-acquired epidemics.2,10 Patients with complicated infections by A. baumannii have been treated with carbapenems as drugs of choice to save their lives. However, the number of CRAB has been increasing in the KSA and worldwide.22 Fortunately, recently approved antibiotics and antibiotic/inhibitor combinations such as ceftazidime/avibactam, meropenem/vaborbactam, ceftolozane/tazobactam, plazomicin, and eravacycline are effective in the treatment of infections caused by CRAB.23
In the present study, 32 molecularly confirmed CRAB clinical isolates were chosen out of 45 phenotypically identified Acinetobacter spp. isolates to investigate (i) antimicrobial susceptibility to 21 different antibiotics; (ii) the rate of harbouring 10 carbapenemase genes and (iii) the clonal relationship between the investigated resistant strains. To our knowledge from the available literature, this is the first report from Taif area.
All the investigated CRAB isolates were MDR. The isolates were resistant to 13–19 of the 21 investigated antibiotics. Apart from ampicillin/sulbactam combination, for which none of the isolates was resistant, at least 90.8% of the isolates were resistant to all the ten investigated β–lactams.
All the isolates were sensitive to polymyxin B and colistin. Therefore, these antibiotics remain the best empirical therapeutic choices for the treatment of infection caused by CRAB in high-risk patients in Taif. The previous findings are in line with a recent report that suggests that polymyxins are often the last line of treatment for recalcitrant infections by CRAB isolates.24 Nonetheless, it should be mentioned that isolates resistant to both carbapenems and colistin have been reported elsewhere in the Kingdome of Saudi Arabia.25–28
While, the resistance rates to aminoglycosides, quinolones and tetracycline ranged between 56.3 to 100%, only 25% of the isolates were resistant to tigecycline. Higher rates of resistance to tigecycline have been reported in other areas of KSA.29–31
In spite of the fact that carbapenemases can’t be inhibited by serine class A β–lactamases inhibitors,32,33 it was found that 62.5% of the isolates were sensitive to ampicillin/sulbactam and 37.5% were intermediately resistant. The antibacterial effect of ampicillin/sulbactam against the carbapenem-resistant A. baumannii is probably due to the antibacterial activity exerted by sulbactam against A. baumannii, rather than the inhibition of their carbapenemases. In a previous study, sulbactam was used successfully to treat 14 patients with VAP caused by MDR Acinetobacter spp.34
The significant prevalence of MDR in nosocomial infections caused by A. baumannii along with the rapid development of XDR and PDR limits the therapeutic options for the treatment of serious infections caused by A. baumannii.35,36 Herein, the rate of XDR among our isolates was 71.1% (32/45). Fortunately, none of the isolates exhibited resistance to both tigecycline and polymyxin. Therefore, according to the criteria proposed by Manchanda, et al, none of the isolates was categorised as PDR.11
In this study, all isolates harboured blaOXA51 either alone or in combination with either blaOXA-40, or blaOXA-23. In this regard, blaOXA-51 is located on the chromosome of A. baumannii7 and therefore, such a gene has been utilized for molecular confirmation of A. baumannii. blaOXA-40 was detected in only one isolate (3.1%), which is lower than its incidence reported by others across other regions in KSA like Aseer and the Eastern region, in which the incidence of blaOXA-40 in such regions ranged between 13 to 30%.25,37,38 On the other hand, blaOXA-23 was detected in 19 isolates (59.4%). This resistance gene has been reported as one of the most detected carbapenemases in Saudi Arabia and the Gulf area.9,39 The incidence of blaOXA-23 in KSA is controversial. For instance, similar26,28,38 and higher rates (16–18) have been reported in the Eastern region and Riyadh. The blaOXA-23 gene is plasmid-encoded.33 Therefore, the horizontal dissemination of harbouring plasmids may explain the widespread of blaOXA-23 in Taif and other parts of the KSA. One of the isolates harboured blaOXA-40 while none of the isolates harboured the blaOXA-48, blaOXA-58 and blaOXA-181 genes. blaOXA-58 was previously reported in Aseer and Riyadh at low incidences of 3.6%26 and 1.6%,25 respectively. On the other hand, while blaOXA-40 was prevalent in 30% of A. baumannii isolated from diabetic patients,37 blaOXA-181 and blaOXA-48 have not been reported in KSA.40
The current study revealed that 20 (62.5%) of CRAB isolates harboured two blaOXA genes. One isolate (3.1%) harboured blaOXA-40 and 19 isolates (59.4%) harboured both blaOXA51 and blaOXA-23. The presence of more than one type of blaOXA genes is common in the A. baumannii in KSA.40
The investigated isolates were screened for three metallo-carbapenemase genes, namely, NDM, VIM and IMP. Metallo-β-lactamases are not inhibited by β-lactamase inhibitors, which are active against serine-based, class A β–lactamases like clavulanate, sulbactam, or tazobactam.33,41 No clinically available inhibitors are currently available to block metallo-β-lactamases. Out of the 32 investigated A. baumannii isolates, 24 (75%) harboured one to three metalloenzymes. The genes of blaNDM, blaIPM, blaVIM were incident in 46.9%, 71.9%, and 9.4%, of the isolates, respectively.
To our knowledge from the literature, blaNDM was first detected in 2008 in K. pneumonia and E. coli in a patient returning to Sweden from India.42 In this study, blaNDM was detected in 62.5% of the isolates. Based on the previous finding, we conclude that blaNDM was probably introduced to KSA through the large number of Indian labours working in the Kingdom. blaNDM has been found to be located on several types of plasmids.43 The location of blaNDM on the plasmids contributes to its rapid horizontal spreading among Gram-negative bacteria by conjugation.44 In this study, blaNDM was detected in 9.4% of the isolates. Lower rates (2.4%) and higher rates (30%), for blaNDM, were reported among A. baumannii collected across the KSA38 and in the Eastern District of the KSA, respectively.31
The incidences of blaIMP and blaVIM in this study were 71.9% and 9.4%, respectively. Both blaIMP and blaVIM reside with other resistance genes on integrons associated with transposons. This facilitates their translocation between the chromosome and plasmids33 and their rapid horizontal dissemination.45
The present study revealed that two to five class B and D carbapenemases co-existed in 81.3% of the investigated CRAB clinical isolates. The presence of multiple genes responsible for resistance to carbapenems has been well documented in the KSA.28,39,40,46
The data on the clonal relationship between the A. baumannii isolates reflects their diversity in society and helps in choosing and designing some preventive methods to limit their spreading. ERIC-PCR was performed in this study because it is a rapid and reliable method for studying the phylogenetic relationship between the isolates.47 The ERIC-PCR data in this study shed the light on the remarkable clonal diversity among CRAB clinical isolates (28 clones among 32 strains) collected from a tertiary care hospital in Taif, KSA rather than the predominance of certain epidemic clones. This suggests the circulation of different A. baumannii clones in the hospital. The strains possessing the same ERIC-PCR pattern did not necessarily exhibit the same antibiotic resistance pattern or harbour the same resistance genes. This may be attributed to the process of horizontal resistance gene transfer between bacteria under the high selection pressure in the hospital.
Annually, more than 1.5 million Muslims from across the globe gather in Makkah to perform the rites of pilgrimage and Umrah. The mass gatherings in the Holy City provide a good environment for exchanging microorganisms and the dissemination of various antibiotic resistance genes between the visitors and the Makkans.42 French Muslim pilgrims were screened for the acquired bacteria during Hajj. One of the acquired isolates of A. baumannii harboured a blaOXA-72,36 which is rarely detected in KSA.39,40 The location of Taif near Makkah and the continuous mixing of the population of both cities might be one of the factors involved in the high rate of MDR observed in A. baumannii isolates collected from Taif.
In conclusion, this study exposes the problem of carbapenem-resistance in Taif area. Data obtained herein emphasize the necessity of the application of strict infection control measures.48,49 Generally, there should be protocols for screening high-risk patients for carbapenem-resistant pathogens before their admission to healthcare facilities. Also, stewardship guidelines are recommended to restrict the irrational use of antibiotics in healthcare facilities, community pharmacies and agricultural settings.
Acknowledgment
This study was funded by the Deanship of Scientific Research, Taif University, KSA (Research Project Number 1-437-5211).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Rahman A, Iza N, Ismail S, et al. Acinetobacter spp. infections in Malaysia: a review of antimicrobial resistance trends, mechanisms and epidemiology. Front Microbiol. 2017;8:2479. doi: 10.3389/fmicb.2017.02479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim B, Bae W, Kim K, Lee H, Yoon J. Nosocomial infection due to Acinetobacter baumannii in korean ICUs a multicenter study. Crit Care Med. 2018;46(1):343. doi: 10.1097/01.ccm.0000528728.54507.7e29337802 [DOI] [Google Scholar]
- 3.Almaghrabi MK, Joseph MR, Assiry MM, Hamid ME. Multidrug-resistant Acinetobacter baumannii: an emerging health threat in Aseer Region, Kingdom of Saudi Arabia. Can J Infect Dis Med Microbiol. 2018;2018. doi: 10.1155/2018/9182747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Majid Bouzari D, Abedini F, Yadegari S. Identification of genomic species of Acinetobacter isolated from burns of ICU patients. Arch Iran Med. 2015;18(10):638–642. [PubMed] [Google Scholar]
- 5.Al Atrouni A, Joly-Guillou M-L, Hamze M, Kempf M. Reservoirs of non-baumannii Acinetobacter species. Front Microbiol. 2016;7:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong D, Nielsen TB, Bonomo RA, Pantapalangkoor P, Luna B, Spellberg B. Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin Microbiol Rev. 2017;30(1):409–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turton JF, Woodford N, Glover J, Yarde S, Kaufmann ME, Pitt TL. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J Clin Microbiol. 2006;44(8):2974–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pakharukova N, Tuittila M, Paavilainen S, et al. Structural basis for Acinetobacter baumannii biofilm formation. Proc Natl Acad Sci. 2018;115(21):5558–5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehlers M, Hughes J, Kock M. Prevalence of carbapenemases in Acinetobacter baumannii. In: antibiotic resistant bacteria-a continuous challenge in the new millennium. InTech. 2012:213–246. [Google Scholar]
- 10.Antunes L, Visca P, Towner KJ. Acinetobacter baumannii: evolution of a global pathogen. Pathog Dis. 2014;71(3):292–301. doi: 10.1111/2049-632X.12125 [DOI] [PubMed] [Google Scholar]
- 11.Manchanda V, Sanchaita S, Singh N. Multidrug resistant Acinetobacter. J Glob Infect Dis. 2010;;2(3):291. doi: 10.4103/0974-777X.68538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye Y, Xu L, Han Y, Chen Z, Liu C, Ming L. Mechanism for carbapenem resistance of clinical Enterobacteriaceae isolates. Exp Ther Med. 2018;15(1):1143–1149. doi: 10.3892/etm.2017.5485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quale J, Spelman D, Hooper D, Bloom A. Overview of carbapenemase producing GRam-negative bacilli. UpToDate: Waltham, MA, USA, Accessed On. 2016;21. [Google Scholar]
- 14.Queenan AM, Bush K. Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev. 2007;20(3):440–458. doi: 10.1128/CMR.00001-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meini MR, Llarrull L, Vila A. Evolution of metallo-β-lactamases: trends revealed by natural diversity and in vitro evolution. Antibiotics. 2014;3(3):285–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berrazeg M, Diene S, Medjahed L, et al. New Delhi Metallo-beta-lactamase around the world: an eReview using google maps. Euro Surveill. 2014;19(20):20809. doi: 10.2807/1560-7917.ES2014.19.20.20809 [DOI] [PubMed] [Google Scholar]
- 17.Opazo A, Domínguez M, Bello H, Amyes SG, González-Rocha G. OXA-type carbapenemases in Acinetobacter baumannii in South America. J Infect Dev Ctries. 2012;6(04):311–316. [DOI] [PubMed] [Google Scholar]
- 18.Antunes NT, Lamoureaux TL, Toth M, Stewart NK, Frase H, Vakulenko SB. Class D β-lactamases: are they all carbapenemases? Antimicrob Agents Chemother. 2014;2119–2125. doi: 10.1128/AAC.02522-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cockerill FR. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-first Informational Supplement. Wayne, PA: CLSI; 2011. [Google Scholar]
- 20.Englen M, Kelley L. A rapid DNA isolation procedure for the identification of Campylobacter jejuni by the polymerase chain reaction. Lett Appl Microbiol. 2000;31(6):421–426. [DOI] [PubMed] [Google Scholar]
- 21.Ocampo AM, Chen L, Cienfuegos AV, et al. High frequency of non-CG258 clones of carbapenem-resistant Klebsiella pneumoniae with distinct clinical characteristics: A two-year surveillance in five Colombian tertiary care hospitals. Antimicrob Agents Chemother. 2015;60(1):332–342. doi: 10.1128/AAC.01775-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khatun R, Shamsuzzaman S. Detection of OXA-181/OXA-48 carbapenemase producing Enterobacteriaceae in Bangladesh. IMC J Med Sci. 2015;9(2):45–51. [Google Scholar]
- 23.Bassetti M, Peghin M, Vena A, Giacobbe DR. Treatment of infections due to MDR Gram-negative bacteria. Front Med. 2019;16. doi: 10.3389/fmed.2019.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheah S-E, Li J, Tsuji BT, Forrest A, Bulitta JB, Nation RL. Colistin and polymyxin B dosage regimens against Acinetobacter baumannii: differences in activity and the emergence of resistance. Antimicrob Agents Chemother. 2016;60(7):3921–3933. doi: 10.1128/AAC.02927-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elabd FM, Al-Ayed MS, Asaad AM, Alsareii SA, Qureshi MA, Musa H-A-A. Molecular characterization of oxacillinases among carbapenem-resistant Acinetobacter baumannii nosocomial isolates in a Saudi hospital. J Infect Public Health. 2015;8(3):242–247. doi: 10.1016/j.jiph.2014.10.002 [DOI] [PubMed] [Google Scholar]
- 26.Al-Agamy MH, Jeannot K, El-Mahdy TS, et al. First detection of GES-5 carbapenemase-producing Acinetobacter baumannii isolate. Microb Drug Resist. 2017;23(5):556–562. doi: 10.1089/mdr.2016.0152 [DOI] [PubMed] [Google Scholar]
- 27.AlMasoudi SB, Aly M, AlHumidi NQ, Halwani MA. Incidence and prevalence of Acinetobacter baumannii in king fahd general hospital, Saudi Arabia. Life Sci J. 2013;10(4):1702–1710. [Google Scholar]
- 28.Al-Agamy MH, Shibl AM, Ali MS, Khubnani H, Radwan HH, Livermore DM. Distribution of β-lactamases in carbapenem-non-susceptible Acinetobacter baumannii in Riyadh, Saudi Arabia. J Glob Antimicrob Resist. 2014;2(1):17–21. doi: 10.1016/j.jgar.2013.08.004 [DOI] [PubMed] [Google Scholar]
- 29.Abdalhamid B, Hassan H, Itbaileh A, Shorman M. Characterization of carbapenem-resistant Acinetobacter baumannii clinical isolates in a tertiary care hospital in Saudi Arabia. New Microbiol. 2014;37(1):65–73. [PubMed] [Google Scholar]
- 30.Aljindan R, Bukharie H, Alomar A, Abdalhamid B. Prevalence of digestive tract colonization of carbapenem-resistant Acinetobacter baumannii in hospitals in Saudi Arabia. J Med Microbiol. 2015;64(4):400–406. doi: 10.1099/jmm.0.000033 [DOI] [PubMed] [Google Scholar]
- 31.El-Mahdy TS, Al-Agamy MH, Al-Qahtani AA, Shibl AM. Detection of bla OXA-23-like and bla NDM-1 in Acinetobacter baumannii from the Eastern Region, Saudi Arabia. Microb Drug Resist. 2017;23(1):115–121. doi: 10.1089/mdr.2015.0304 [DOI] [PubMed] [Google Scholar]
- 32.Bennett P. Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. Brit J Pharmacol. 2008;153(S1):S347–S357. doi: 10.1038/sj.bjp.0707607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perez-Llarena FJ, Bou G. β-Lactamase inhibitors: the story so far. Curr Med Chem. 2009;16(28):3740–3765. doi: 10.2174/092986709789104957 [DOI] [PubMed] [Google Scholar]
- 34.Wood GC, Scott DH, Martin AC, Timothy CF, Bradley AB. Comparison of ampicillin-sulbactam and imipenem-cilastatin for the treatment of Acinetobacter ventilator-associated pneumonia. Clin Infec Dis. 2002;34(11):1425–1430. doi: 10.1086/340055 [DOI] [PubMed] [Google Scholar]
- 35.Nowak J, Zander E, Stefanik D, et al. High incidence of pandrug-resistant Acinetobacter baumannii isolates collected from patients with ventilator-associated pneumonia in Greece, Italy and Spain as part of the MagicBullet clinical trial. J Antimicrob Chemother. 2017;72(12):3277–3282. doi: 10.1093/jac/dkx322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leangapichart T, Gautret P, Griffiths K, et al. Acquisition of a high diversity of bacteria during the Hajj pilgrimage, including Acinetobacter baumannii with blaOXA-72 and Escherichia coli with blaNDM-5 carbapenemase genes. Antimicrob Agents Chemother. 2016;60(10):5942–5948. doi: 10.1128/AAC.00669-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alsultan A, Evans B, Elsayed E, et al. High frequency of carbapenem-resistant Acinetobacter baumannii in patients with diabetes mellitus in Saudi Arabia. J Med Microbiol. 2013;62(6):885–888. doi: 10.1099/jmm.0.057216-0 [DOI] [PubMed] [Google Scholar]
- 38.Al-Sultan AA, Evans BA, Aboulmagd E, et al. Dissemination of multiple carbapenem-resistant clones of Acinetobacter baumannii in the Eastern District of Saudi Arabia. Fronts Microbiol. 2015;6. doi: 10.3389/fmicb.2015.00634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Memish ZA, Assiri A, Almasri M, et al. Molecular characterization of carbapenemase production among Gram-negative bacteria in Saudi Arabia. Microb Drug Resist. 2015;21(3):307–314. [DOI] [PubMed] [Google Scholar]
- 40.Ibrahim ME. Prevalence of Acinetobacter baumannii in Saudi Arabia: risk factors, antimicrobial resistance patterns and mechanisms of carbapenem resistance. Ann Clin Microbiol Antimicrob. 2019;18(1). doi: 10.1186/s12941-12018-10301-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drawz SM, Bonomo RA. Three decades of β-lactamase inhibitors. Clin Microbiol Rev. 2010;23(1):160–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haseeb A, Faidah HS, Bakhsh AR, et al. Antimicrobial resistance among pilgrims: a retrospective study from two hospitals in Makkah, Saudi Arabia. Int J Infect Dis. 2016;47(2016):92–94. [DOI] [PubMed] [Google Scholar]
- 43.Jaidane N, Naas T, Oueslati S, et al. Whole-genome sequencing of NDM-1-producing ST85 Acinetobacter baumannii isolates from Tunisia. Inter J Antimicrob Agents. 2018;52(6):916–921. [DOI] [PubMed] [Google Scholar]
- 44.Nordmann P, Poirel L, Walsh TR, Livermore DM. The emerging NDM carbapenemases. Trends Microbiol. 2011;19(12):588–595. [DOI] [PubMed] [Google Scholar]
- 45.Palzkill T. Metallo-β-lactamase structure and function. Ann N Y Acad Sci. 2013;1277(1):91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alyamani EJ, Khiyami MA, Booq RY, Alnafjan BM, Altammami MA, Bahwerth FS. Molecular characterization of extended-spectrum beta-lactamases (ESBLs) produced by clinical isolates of Acinetobacter baumannii in Saudi Arabia. Ann Clin Microbiol Antimicrob. 2015;14(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bertrand X, Dowzicky MJ. Antimicrobial susceptibility among gram-negative isolates collected from intensive care units in North America, Europe, the Asia-Pacific Rim, Latin America, the Middle East, and Africa between 2004 and 2009 as part of the tigecycline evaluation and surveillance trial. Clin Therap. 2012;34(1):124–137. [DOI] [PubMed] [Google Scholar]
- 48.Zowawi HM. Antimicrobial resistance in Saudi Arabia: an urgent call for an immediate action. Sau Med J. 2016;37(9):935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perez F, Van Duin D. Carbapenem-resistant Enterobacteriaceae: a menace to our most vulnerable patients. Clev Clin J Med. 2013;80(4):225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.El-Badawy MF, Tawakol WM, El-Far SW, et al. Molecular identification of aminoglycoside-modifying enzymes and plasmid-mediated quinolone resistance genes among Klebsiella pneumoniae clinical isolates recovered from Egyptian patients. Int J Microbio. 2017;2017. doi: 10.1155/2017/8050432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balsalobre LC, Dropa M, Lincopan N, Mamizuka EM, Matté GR, Matté MH. Detection of metallo-β-lactamases encoding genes in environmental isolates of Aeromonas hydrophila and Aeromonas jandaei. Lett Appl Microbiol. 2009;49(1):142–145. [DOI] [PubMed] [Google Scholar]
- 52.Novovic K, Mihajlovic S, Vasiljevic Z, Filipic B, Begovic J, Jovcic B. Carbapenem-resistant Acinetobacter baumannii from Serbia: revision of CarO classification. PLoS One. 2015;10(3):e0122793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tarashi S, Goudarzi H, Erfanimanesh S, Pormohammad A, Hashemi A. Phenotypic and molecular detection of Metallo-beta-lactamase genes among imipenem resistant Pseudomonas aeruginosa and Acinetobacter baumannii strains isolated from patients with burn injuries. Arch Clin Infect Dis. 2016;11(4). doi: 10.5812/archcid.39036 [DOI] [Google Scholar]
- 54.Qi C, Malczynski M, Parker M, Scheetz MH. Characterization of genetic diversity of carbapenem-resistant Acinetobacter baumannii clinical strains collected from 2004 to 2007. J Clin Microbiol. 2008;46(3):1106–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Woodford N, Ellington MJ, Coelho JM, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006;27(4):351–353. [DOI] [PubMed] [Google Scholar]
- 56.van der Zwaluw K, de Haan A, Pluister GN, Bootsma HJ, de Neeling AJ, Schouls LM. The carbapenem inactivation method (CIM), a simple and low-cost alternative for the Carba NP test to assess phenotypic carbapenemase activity in Gram-negative rods. PLoS One. 2015;10(3):e0123690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shamsizadeh Z, Nikaeen M, Esfahani BN, Mirhoseini SH, Hatamzadeh M, Hassanzadeh A. Detection of antibiotic resistant Acinetobacter baumannii in various hospital environments: potential sources for transmission of Acinetobacter infections. Environ Health Prev Med. 2017;22(1):22–44. [DOI] [PMC free article] [PubMed] [Google Scholar]