Abstract
Purpose
The oncogenic roles of lncRNA LINC01116 have been reported in several types of cancer, while its involvement in gastric cancer is unknown. This study aimed to investigate the involvement of LINC01116 in gastric cancer.
Methods
Gene expression was detected by qPCR. Correlations were analyzed by linear regression. Overexpression and siRNA silencing techniques were used to analyze gene functions. Cell invasion and migration were analyzed by Transwell assays.
Results
LINC01116 and lncRNA CASC11 were both upregulated in cancer tissues compared to cancer-adjacent tissues. Expression levels of LINC01116 and CASC11 were increased with the increase in clinical stages. Expression levels of LINC01116 and CASC11 were positively correlated. Overexpression of LINC01116 mediated the upregulated CASC11 in gastric cancer cells, and CASC11 overexpression also led to overexpressed LINC01116. Overexpression of LINC01116 and CASC11 led to promoted invasion and migration of gastric cancer cells. Rescue experiments showed that CASC11 knockdown attenuated the effects of LINC01116 overexpression. Overexpression of LINC01116 failed to significantly affect cancer cell proliferation.
Conclusion
LINC01116 promoted cancer cell invasion and migration in gastric cancer by positively interacting with CASC11.
Keywords: gastric cancer, lncRNA LINC01116, lncRNA CASC11
Introduction
Gastric cancer is the 4th most frequently diagnosed malignancy and one of the leading causes of cancer-related deaths.1 Development of surgical techniques has improved the overall survival of gastric cancer patients at early stages.2,3 However, the early diagnosis rate of gastric cancer is low and most patients are diagnosed at advanced stages with tumor metastasis to local or even distant regions, leading to unacceptable high mortality rate.4 The development of gastric cancer requires the aberrant expression of tumor suppressors or oncogenes.5 However, pathological pathways involved in gastric cancer still haven’t been fully elucidated.
The human genome annotation ENCODE project has revealed that more than 80% of the human genome can be transcribed but more than 98% transcripts were not related to protein-coding,6 indicating the existence of millions of non-coding genes that only transcribe non-coding RNAs (ncRNAs).7 Long (> 200 nt) ncRNAs are emerging classes of functional ncRNAs involved in diverse pathological and pathological processes.8,9 A growing body of literature has shown that lncRNAs are critical determinants in cancer biology,10 and regulation of lncRNA expression provides new insights to cancer treatment and diagnosis.11 However, function of most lncRNAs in cancer remains unclear. It has been reported that lncRNA LINC01116 (hereafter lncRNA was omitted) plays oncogenic roles in several types of cancer,12,13 while its involvement in gastric cancer is unknown. We therefore investigated the function of LINC01116 in gastric cancer.
Materials And Methods
Patients
From January 2016 to January 2018, 76 patients with gastric cancer were enrolled in Cancer Hospital of China Medical University to serve as research subjects. Inclusion criteria: 1) gastric cancer patients confirmed by histopathological examinations; 2) no history of malignancies. Exclusion criteria: 1) patients with other medical conditions besides gastric cancer; 2) patients who were transferred from other hospital or had been treated within 3 months before admission. The 76 patients included 39 males and 37 females, and age ranged from 32 to 69 years, and the mean age was 51.2±8.1 years. According to AJCC staging, there were 18, 18, 20 and 20 cases at stage 1-IV, respectively. This study was approved by Ethics Committee of Cancer Hospital of China Medical University. All procedures performed in studies involving human participants were conducted in accordance with the Declaration of Helsinki. All patients provided written form informed consent.
Specimens And Cell Lines
All patients were diagnosed by histopathological biopsy, and tumor and adjacent normal tissues (about 0.1 g) were collected during this procedure. All tissue specimens were confirmed by 3 pathologists.
SNU-1 and Hs 746T gastric cancer cell lines were used to perform all in vitro cell experiments. Cells of both cell lines were bought from ATCC (Manassas, VA, USA). A mixture of 90% RPMI-1640 Medium supplemented and 10% FBS was used to cultivate the cells in an incubator (37°C, 5% CO2).
RT-qPCR
Tissue specimens were ground in liquid nitrogen to make fine powers, followed by the addition of RNAzol reagent (Sigma-Aldrich) to extract total RNAs. In vitro cultivated cells were directly mixed with RNAzol reagent to extract total RNAs. SSRT III was used to synthesize cDNA through reverse transcription. To detect the expression of LINC01116 and CASC11, Applied Biosystems™ PowerUp™ SYBR™ Green Master Mix was used to prepare PCR reaction systems with 18S rRNA as endogenous control. Expression of LINC01116 and CASC11 was normalized to 18S rRNA using 2−ΔΔCq method.
Vectors, siRNAs And Cell Transfection
Full length LINC01116 and CASC11 genomic DNAs were inserted into pcDNA3.1 vectors to establish LINC01116 and CASC11 expression vectors. Vector construction was performed by Sangon (Shanghai, China). LINC01116 and CASC11 siRNA and siRNA negative control (NC) were also designed and synthesized by Sangon. Cells of SNU-1 and Hs 746T cell lines were harvested at 70–80% confluence and all cell transfections were performed by lipofectamine 2000 reagent (Thermo Fisher Scientific, Inc.,) to transfect 10 nM vectors (empty vector as NC group) or 40 nM siRNA (NC siRNA as NC group) into 106 cells. Cells without transfections were control (C) cells. Cells were harvested at 24h after transfection to perform subsequent experiments.
Transwell Migration And Invasion Assay
Cells were harvested at 24h after transfection to test cell migration and invasion. Briefly, 1ml serum-free RPMI-1640 Medium was used to resuspend cell pellets containing 3×104 cells. The upper chamber was filled with cell suspension with 0.1 mL in each well, while the lower chamber as filled with a mixture of 80% RPMI-1640 Medium and 20% FBS. Cell migration and invasion were allowed for 2h. After that, with the lower surface of the membrane was stained with 0.5% crystal violet (Sigma-Aldrich, USA). Both assays were performed in the same way except that the upper chambed was coated with Matrigel (356234, Millipore, USA) before use.
Statistical Analysis
There biological replicates were included in each experiment. Comparisons between tumor and tumor adjacent normal tissues were performed by paired t test. Comparison among different clinical stages or among different cell treatment groups were performed by ANOVA (one-way) and Tukey’s test. Linear regression was performed to analyze the correlation between LINC01116 and CASC11. Patients were divided into high and low LINC01116/CASC11 group with the median expression level in cancer tissues as the cutoff values. Correlations between patients’ age, gender, smoking habit, drinking habit, lymph node metastasis and distant metastasis were analyzed by Chi-squared t test. p<0.05 was the level of statistical significant.
Results
LINC01116 And CASC11 Were Both Upregulated In Gastric Cancer Tissues Than In Cancer-Adjacent Tissues
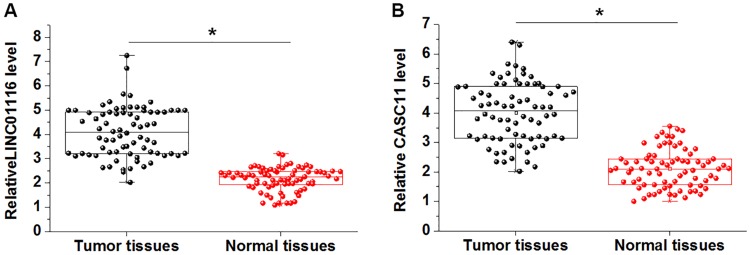
The differential expression of LINC01116 and CASC11 in cancer tissues and cancer-adjacent tissues from the 76 gastric cancer patients was detected by RT-qPCR. The normalized expression levels of LINC01116 in cancer tissues and cancer-adjacent tissues were 4.12±0.96 and 2.37±0.33, respectively. It was observed that LINC01116 was significantly upregulated in tumor tissues (1.74-fold, Figure 1A, p<0.05). The normalized expression levels of CASC11 in cancer tissues and cancer-adjacent tissues were 4.03±0.90 and 2.24±0.38, respectively. In addition, CASC11 was also significantly upregulated in tumor (1.80-fold, Figure 1B, p<0.05).
Figure 1.
LINC01116 and lncRNA CASC11 were both upregulated in gastric cancer tissues than in cancer-adjacent tissues. RT-qPCR results showed that LINC01116 (A) and CASC11 (B) were both upregulated in gastric cancer tissues than in tumor adjacent normal tissues (*p<0.05).
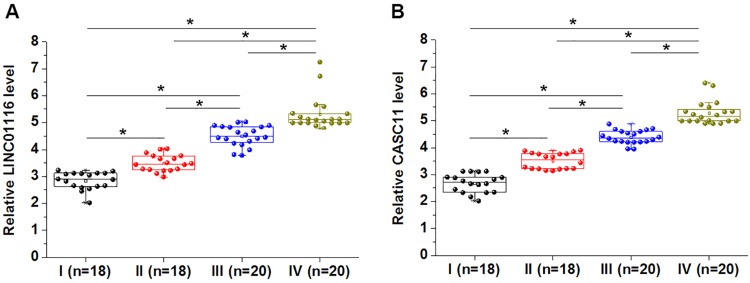
LINC01116 And CASC11 Expression Levels Increased With Increase In Clinical Stages
Comparisons of expression levels of LINC01116 and CASC11 among 4 clinical stages revealed that LINC01116 (Figure 2A) and CASC11 (Figure 2B) expression in tumor tissues were significantly upregulated with the increase of clinical stages (p<0.05). Chi square test showed that expression levels of LINC01116 and CASC11 were not significantly correlated with patients’ age, gender, and smoking and drinking habits, however, there are closely correlated with lymph node metastasis (both p<0.05) and distant metastasis (both p<0.01).
Figure 2.
LINC01116 and CASC11 expression levels were increased with the increase in clinical stages. Expression levels of LINC01116 (A) and CASC11 (B) in tumor tissues were significantly increased with the increase in clinical stages (*p<0.05).
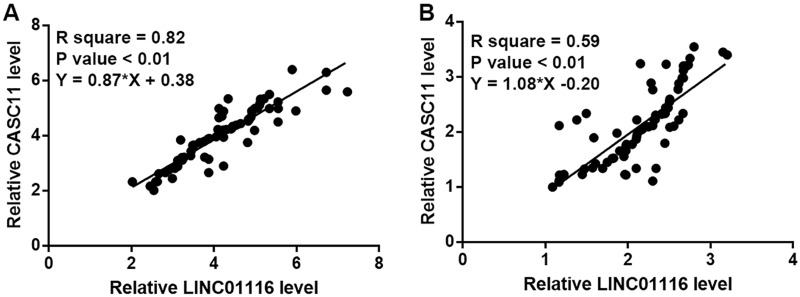
Expression levels of LINC01116 and CASC11 were positively correlated. Linear regression was performed to analyze the correlation between expression levels of LINC01116 and CASC11. It was observed that LINC01116 and CASC11 were positively and significantly correlated in tumor tissues of gastric cancer patients (Figure 3A). In addition, a significant and positive correlation between LINC01116 and CASC11 was also observed in adjacent normal tissues (Figure 3B).
Figure 3.
Expression levels of LINC01116 and CASC11 were positively correlated in tumor tissues and adjacent normal tissues. Linear regression showed that expression levels of LINC01116 and CASC11 were positively correlated in both tumor tissues (A) and adjacent normal tissues (B).
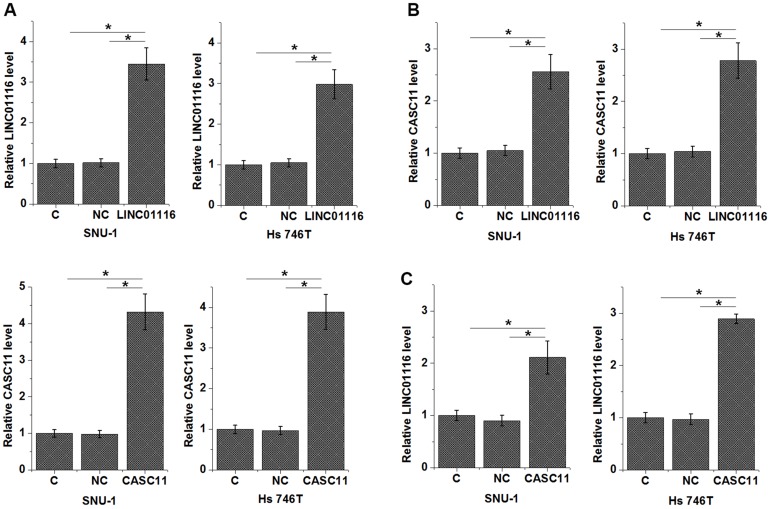
LINC01116 And CASC11 Upregulated Each Other In Cells Of Gastric Cancer Cell Lines
LINC01116 and CASC11 expression vectors were transfected into cells of both SNU-1 and Hs 746T cells. Overexpression of LINC01116 and CASC11 was observed at 24h after transfection (Figure 4A, p<0.05). Comparing to control (C) and negative control (NC) groups, cells with LINC01116 overexpression showed significantly upregulated CASC11 expression (Figure 4B, p<0.05). In addition, CASC11 overexpression also led to overexpressed LINC01116 (Figure 4C, p<0.05).
Figure 4.
LINC01116 and CASC11 upregulated each other in cells of gastric cancer cell lines. Overexpression of LINC01116 and CASC11 was observed at 24h after transfection (A). Overexpression of LINC01116 mediated the upregulated CASC11 in gastric cancer cells (B), and CASC11 overexpression also led to overexpressed LINC01116 (C) (*p<0.05).
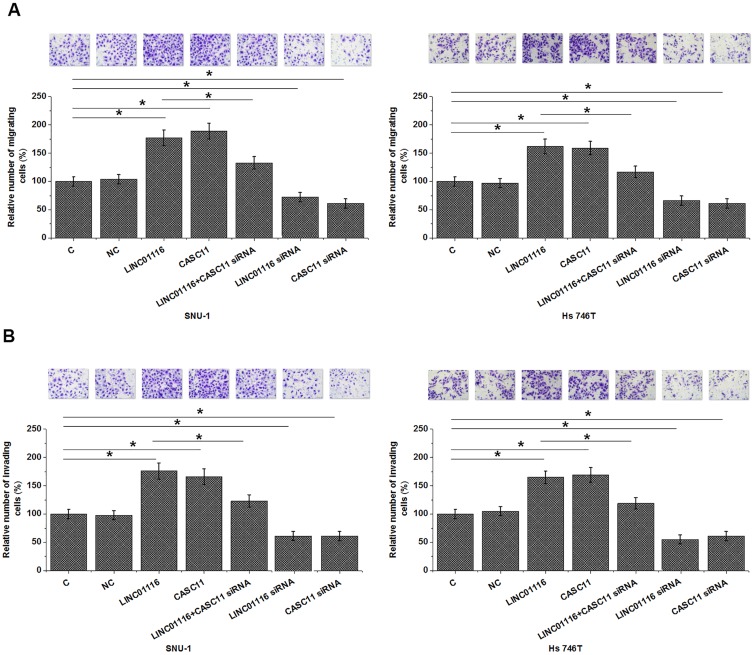
Interaction Between LINC01116 And CASC11 Participate In The Migration And Invasion Of Gastric Cancer Cells
Comparing to control (C) and negative control (NC) groups, cells with overexpression of LINC01116 and CASC11 showed significantly promoted migration (Figure 5A, p<0.05) and invasion (Figure 5B, p<0.05). However, cells with LINC01116 and CASC11 silencing showed significantly inhibited migration (Figure 5A, p<0.05) and invasion (Figure 5B, p<0.05). Rescue experiments showed that CASC11 knockdown attenuated the effects of LINC01116 overexpression. Overexpression of LINC01116 failed to significantly affect cancer cell proliferation (data not shown).
Figure 5.
Interaction between LINC01116 and CASC11 participate in the migration and invasion of gastric cancer cells. Overexpression of LINC01116 and CASC11 led to promoted, while siRNA silencing led to suppressed migration (A) and invasion (B) of gastric cancer cells. Rescue experiments showed that CASC11 knockdown attenuated the effects of LINC01116 overexpression (*p<0.05).
Discussion
LINC01116 plays oncogenic roles in in several types of cancer,12,13 while no study has reported its involvement in gastric cancer. This paper is the first time to report the oncogenic function of LINC01116 in gastric cancer and showed that the actions of LINC01116 in gastric cancer are likely achieved through its positive interactions with oncogenic CASC11.14
The oncogenic function of CASC11 has been investigated in several types of cancers.14–16 In the development of hepatocellular carcinoma, CASC11 inactivates PTEN signaling and activates PI3K/AKT signaling through epigenetic pathways to mediate the cancer cell migration and invasion as well as epithelial-mesenchymal transition.15 CASC11 also promotes colorectal cancer growth and metastasis through its interactions with WNT/β-catenin pathway.16 In a recent study Zhang et al also showed that the overexpression of CASC11 is at least partially responsible for the accelerated cancer cell proliferation, migration and invasion in gastric cancer.16 Our study also observed the upregulated expression of CASC11 in tumor tissues than in adjacent healthy tissues of gastric cancer patients, and overexpression of CASC11 also resulted in promoted cancer cell migration and invasion. Therefore, our data confirmed the oncogenic role of CASC11 in gastric cancer.
Our study first reported the oncogenic role of LINC01116 in gastric cancer. It is known that overexpression of LINC01116 promotes cancer cell proliferation in ovarian cancer, osteosarcoma and breast cancer.12,13,17 However, our study did not observed significantly altered cell proliferation abilities of two gastric cancer cell lines after LINC01116 overexpression. Therefore, although LINC01116 has oncogenic functions in different types of gastric cancer, but its regulatory effects on cancer cell behaviors may be different in different types of cancer.
LINC01116 and CASC11 participate in cancer biology through the interactions with multiple tumor suppression and oncogenic pathways.12–17 Our present study proved that LINC01116 and CASC11 can form a positive feedback regulation loop to participate in the regulation of cancer cell migration and invasion in gastric cancer. Therefore, the interaction between LINC01116 and CASC11 may serve as bridge to connect multiple signaling pathways to promote gastric cancer. LINC01116 has interactions with miR-145,17 which has interactions with wnt/β-catenin.18 It is known that CASC11 can interact with wnt/β-catenin to participate in cancer biology.16 Therefore, miR-145 may be a mediator between LINC01116 and CASC11.
It is worth noting that analysis of TCGA dataset revealed no such strong correlation between LINC01116 and CASC11. All patients included in this study are Han Chinese. Therefore, the gene expression regulation network in GC may be different in different populations. Our future studies will include patients from different races to further expression the interactions between LINC01116 and CASC11 in GC. We also failed to analyze the prognostic values of LINC01197 and CASC11. Our future studies will carry out a 5-year follow-up study to analyze their prognostic values.
Conclusion
In conclusion, LINC01116 plays an oncogenic role in gastric cancer possibly by forming a positive feedback regulation loop with CASC11.
Informed Consent
Informed consent was obtained from all individual participants included in the study.
Ethics Approval
This study was approved by the Ethics Committee of Cancer Hospital of China Medical University. All procedures performed in studies involving human participants were conducted in accordance with the Declaration of Helsinki.
Funding
We received financial support from the Key Projects of Liaoning Natural Science Foundation Program (20170540566).
Author Contributions
All authors contributed towards data analysis, drafting and critically revising the paper, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107 [DOI] [PubMed] [Google Scholar]
- 2.Ono H, Kondo H, Gotoda T, et al. Endoscopic mucosal resection for treatment of early gastric cancer. Gut. 2001;48(2):225–229. doi: 10.1136/gut.48.2.225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan YK, Fielding JW. Early diagnosis of early gastric cancer. Eur J Gastroenterol Hepatol. 2006;18(8):821–829. doi: 10.1097/00042737-200608000-00004 [DOI] [PubMed] [Google Scholar]
- 4.Kagawa S, Shigeyasu K, Ishida M, et al. Molecular diagnosis and therapy for occult peritoneal metastasis in gastric cancer patients. World J Gastroenterol. 2014;20(47):17796–17803. doi: 10.3748/wjg.v20.i47.17796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23(5):700–713. doi: 10.1158/1055-9965.EPI-13-1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrow J, Frankish A, Gonzalez JM, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22(9):1760–1774. doi: 10.1101/gr.135350.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mattick JS. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2001;2(11):986–991. doi: 10.1093/embo-reports/kve230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. doi: 10.1038/nrg3606 [DOI] [PubMed] [Google Scholar]
- 9.Chen X, Yan CC, Zhang X, You ZH. Long non-coding RNAs and complex diseases: from experimental results to computational models. Brief Bioinform. 2017;18(4):558–576. doi: 10.1093/bib/bbw060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–719. doi: 10.4161/rna.20481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol. 2013;26(2):155–165. doi: 10.1038/modpathol.2012.160 [DOI] [PubMed] [Google Scholar]
- 12.Fang YN, Huang ZL, Li H, et al. LINC01116 promotes the progression of epithelial ovarian cancer via regulating cell apoptosis. Eur Rev Med Pharmacol Sci. 2018;22(16):5127–5133. doi: 10.26355/eurrev_201808_15707 [DOI] [PubMed] [Google Scholar]
- 13.Zhang B, Yu L, Han N, et al. LINC01116 targets miR-520a-3p and affects IL6R to promote the proliferation and migration of osteosarcoma cells through the Jak-stat signaling pathway. Biomed Pharmacother. 2018;107:270–282. doi: 10.1016/j.biopha.2018.07.119 [DOI] [PubMed] [Google Scholar]
- 14.Zhang L, Kang W, Lu X, Ma S, Dong L, Zou B. LncRNA CASC11 promoted gastric cancer cell proliferation, migration and invasion in vitro by regulating cell cycle pathway. Cell Cycle. 2018;17(15):1886–1900. doi: 10.1080/15384101.2018.1502574 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Han Y, Chen M, Wang A, Fan X. STAT3-induced upregulation of lncRNA CASC11 promotes the cell migration, invasion and epithelial-mesenchymal transition in hepatocellular carcinoma by epigenetically silencing PTEN and activating PI3K/AKT signaling pathway. Biochem Biophys Res Commun. 2019;508(2):472–479. doi: 10.1016/j.bbrc.2018.11.092 [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z, Zhou C, Chang Y, et al. Long non-coding RNA CASC11 interacts with hnRNP-K and activates the WNT/beta-catenin pathway to promote growth and metastasis in colorectal cancer. Cancer Lett. 2016;376(1):62–73. doi: 10.1016/j.canlet.2016.03.022 [DOI] [PubMed] [Google Scholar]
- 17.Hu HB, Chen Q, Ding SQ. LncRNA LINC01116 competes with miR-145 for the regulation of ESR1 expression in breast cancer. Eur Rev Med Pharmacol Sci. 2018;22(7):1987–1993. doi: 10.26355/eurrev_201804_14726 [DOI] [PubMed] [Google Scholar]
- 18.Mayorga ME, Penn MS. miR-145 is differentially regulated by TGF-beta1 and ischaemia and targets Disabled-2 expression and wnt/beta-catenin activity. J Cell Mol Med. 2012;16(5):1106–1113. doi: 10.1111/j.1582-4934.2011.01385.x [DOI] [PMC free article] [PubMed] [Google Scholar]