Errors during meiosis are the leading cause of birth defects and miscarriages in humans. Thus, the coordinated control of meiotic events is critical for the faithful inheritance of the genome with each generation...
Keywords: meiosis, recombination, crossovers, synaptonemal complex, C. elegans, chromosome axis, cohesin
Abstract
Crossovers (COs) between homologous chromosomes are critical for meiotic chromosome segregation and form in the context of the synaptonemal complex (SC), a meiosis-specific structure that assembles between aligned homologs. During Caenorhabditis elegans meiosis, central region components of the SC (SYP proteins) are essential to repair double-strand DNA breaks (DSBs) as COs. Here, we investigate the relationships between the SYP proteins and conserved pro-CO factors by examining the immunolocalization of these proteins in meiotic mutants where SYP proteins are absent, reduced, or mislocalized. Although COs do not form in syp null mutants, pro-CO factors COSA-1, MSH-5, and ZHP-3 nevertheless colocalize at DSB-dependent sites during late prophase, reflecting an inherent affinity of these factors for DSB repair sites. In contrast, in mutants where SYP proteins are present but form aggregates or display abnormal synapsis, pro-CO factors consistently track with SYP-1 localization. Further, pro-CO factors usually localize to a single site per SYP-1 structure, even in SYP aggregates or in mutants where the SC forms between sister chromatids, suggesting that CO regulation occurs within these aberrant SC structures. Moreover, we find that the meiotic cohesin REC-8 is required to ensure that SC formation occurs between homologs and not sister chromatids. Taken together, our findings support a model in which SYP proteins promote CO formation by promoting the localization of pro-CO factors to recombination events within an SC compartment, thereby ensuring that pro-CO factors identify a recombination event within an SC structure and that CO maturation occurs only between properly aligned homologous chromosomes.
DURING sexual reproduction, the generation of haploid gametes from diploid germ cells involves substantial reorganization of chromosomes within the nucleus and the formation of specialized meiosis-specific chromosome structures. In preparation for segregating to opposite poles at the meiosis I division, homologous chromosome pairs align along their full lengths and assemble a structure known as the synaptonemal complex (SC) between them. The SC structure is composed of axial elements that assemble along the lengths of conjoined pairs of sister chromatids (known as lateral elements in the context of an assembled SC) and a set of proteins that comprise the central region of the SC that link the parallel-aligned homolog axes (Cahoon and Hawley 2016; Cahoon and Libuda 2019). Multiple SC central region proteins function together to span the distance between the lateral elements and are required for normal meiosis in all organisms that assemble the SC. How the SC central region and its constituent proteins contribute to a successful outcome of meiosis remains a subject of active investigation.
Four different components of the SC central region have been identified in Caenorhabditis elegans, termed SYP-1, SYP-2, SYP-3, and SYP-4. These SYP proteins localize between the lateral elements of the SC, and are interdependent for localization and stability (Colaiacovo et al. 2003; Schild-Prüfert et al. 2011). Analysis of syp mutants has demonstrated that the SYP proteins are required both to stabilize homolog pairing between homologous chromosomes and to promote the formation of crossover (CO) recombination events, which are required for proper chromosome segregation during meiosis I (MacQueen et al. 2002; Colaiacovo et al. 2003; Smolikov et al. 2007b, 2009). In addition to this role in promoting CO formation, the SYP proteins also play a role in limiting the number of COs that form during meiosis. Partial depletion of SYP proteins by RNA interference (RNAi) causes in an increase in the number of chromosomes with more than one CO and an attenuation of CO interference (Libuda et al. 2013). Further, recent studies have suggested a liquid crystalline-like behavior of the SC central region proteins and revealed dynamic properties of the SYPs that change during the course of meiotic prophase progression (Rog and Dernburg 2015; Mlynarczyk-Evans and Villeneuve 2017; Pattabiraman et al. 2017; Rog et al. 2017). Moreover, studies have found that CO recombination events are linked to post-translational modifications of the SYP proteins (Kim et al. 2015; Nadarajan et al. 2016, 2017; Pattabiraman et al. 2017). Despite these advances in our understanding, how the SYP proteins promote the formation of COs between homologs during meiosis remains poorly understood.
In the context of an assembled SC, a set of pro-CO factors (MSH-5, COSA-1, ZHP-1, ZHP-2, ZHP-3, and ZHP-4) are loaded on chromosomes during C. elegans meiosis to promote and license the repair of a subset of programmed DNA double-strand breaks (DSBs) as COs between homologs (Kelly et al. 2000; Jantsch et al. 2004; Bhalla et al. 2008; Yokoo et al. 2012; Nguyen et al. 2018; Zhang et al. 2018). Following the formation of DSBs by the conserved endonuclease SPO-11, the pro-CO factor MSH-5 (a component of the meiosis-specific MutSγ complex) and COSA-1 (a cyclin-related protein specific to metazoan meiosis) form multiple DSB-dependent foci in early pachytene prior to reducing down in number in late pachytene, marking the six CO sites (one CO per chromosome) in C . elegans meiosis (Kelly et al. 2000; Yokoo et al. 2012; Woglar and Villeneuve 2018). In contrast, ZHP-1, ZHP-2, ZHP-3, and ZHP-4 (RING domain-containing proteins) coat the SC in early pachytene, before reducing and retracting down in a DSB-dependent manner to distinct foci that colocalize with the six CO sites marked by MSH-5 and COSA-1 in late pachytene (Jantsch et al. 2004; Bhalla et al. 2008; Yokoo et al. 2012; Nguyen et al. 2018; Zhang et al. 2018). Although analysis of null mutants of the pro-CO factors demonstrates that these proteins are interdependent for their localization and are required for CO formation (Yokoo et al. 2012; Rog et al. 2017), the mechanism of how the pro-CO factors function with one another and the SC to establish a CO is unknown. Recent evidence in C. elegans indicates that the SC proteins envelop CO-designated sites marked by the pro-CO factors (Woglar and Villeneuve 2018); however, the relationship between the pro-CO factors and the SC is still largely unclear.
Here, we address how the SC central region proteins promote the formation of interhomolog COs by investigating how the SYPs contribute to localization of conserved pro-CO factors that normally localize at CO sites. Our findings indicate that meiotic chromosome structures collaborate together with recombination events to control the localization of pro-CO factors, such that: (1) SYP proteins dictate the context in which pro-CO factors attempt to locate recombination intermediates and (2) correctly assembled chromosome axes restrict SYP proteins to load only between paired homologs. These features likely promote the formation of interhomolog COs by ensuring that CO maturation occurs only in a productive manner, between properly aligned and synapsed homologous chromosomes.
Materials and Methods
C. elegans strains, genetics, and culture conditions
All strains are from the Bristol N2 background, and were maintained and crossed at 20° under standard conditions. Temperatures used for specific experiments are indicated below. For all experiments with meiotic mutants, homozygous mutant worms were derived from balanced heterozygous parents by selecting progeny lacking a dominant marker [Uncoordinated (Unc) and/or GFP markers] associated with the balancer.
The following strains were used in this study:
N2: Bristol wild-type strain.
AV198: spo-11(ok79) IV; syp-1(me17) V / nT1[unc-?(n754) let-? qIs50] (IV;V).
AV276: syp-2(ok307) V / nT1[unc-?(n754) let-?(m435)] (IV;V).
AV278: spo-11 IV; syp-2(ok307) V / nT1[unc-?(n754) let-? qIs50] (IV;V).
AV307: syp-1(me17) V / nT1[unc-?(n754) let-? qIs50] (IV;V).
AV596: cosa-1(tm3298)/ qC1[qIs26] (III).
AV630: meIs8[unc-119(+) Ppie-1::gfp::cosa-1] II.
AV647: meIs8[unc-119(+) Ppie-1::gfp::cosa-1] II; spo-11(me44) IV / nT1[unc-?(n754) let-? qIs50] (IV;V).
AV671: meIs8[unc-119(+) Ppie-1::gfp::cosa-1] II; him-3(e1256) IV.
AV686: meIs8[unc-119(+) Ppie-1::gfp::cosa-1] II; rec-8(ok978) IV / nT1[qIs51] (IV;V).
AV687: syp-3(ok758) I / hT2[bli-4(e937) let-?(q758) qIs48] (I;III); meIs8[unc-119(+) Ppie-1::gfp::cosa-1] II.
AV688: meIs8[unc-119(+) Ppie-1::gfp::cosa-1] II; syp-2(ok307) V / nT1[unc-?(n754) let-?(m435)] (IV;V).
AV689: meIs8[unc-119(+) Ppie-1::gfp::cosa-1] II; him-3(gk149) IV / nT1[qIs51] (IV;V).
AV695: meIs8[unc-119(+) Ppie-1::gfp::cosa-1] II; mnT12 (X;IV).
AV697: meIs8[unc-119(+) Ppie-1::gfp::cosa-1] II; htp-3(y428) ccIs4251 I / hT2[bli-4(e937) let-?(q782) qIs48] (I,III).
AV699: meIs8[unc-119(+) Ppie-1::gfp::cosa-1] II; syp-1(me17) V / nT1[unc-?(n754) let-? qIs50] (IV;V).
AV700: him-3(gk149) IV / nT1[qIs51] (IV;V); syp-2(ok307) V / nT1[qIs51] (IV;V).
CB1256: him-3(e1256) IV.
CV2: syp-3(ok758) I / hT2[bli-4(e937) let-?(q758) qIs48] (I;III).
TY4986: htp-3(y428) ccIs4251 I / hT2[bli-4(e937) let-?(q782) qIs48] (I,III).
VC418: him-3(gk149) IV / nT1[qIs51] (IV;V).
VC666: rec-8(ok978) IV / nT1[qIs51] (IV;V).
DLW1: cosa-1(tm3298)/ qC1[qIs26] III; rec-8(ok978) IV/nT1 [qIs51] (IV;V).
DLW12: GFP::COSA-1 II; rec-8 (ok978)/ nT1 [qIs51] (IV;V)IV; syp-2(ok307) V/nT1
(IV;V).
Additional information on strains:
qIs48 contains [Pmyo-2::gfp; Ppes-10::gfp; Pges-1::gfp].
qIs50 contains [Pmyo-2::gfp; Ppes-10::gfp; PF22B7.9::gfp].
qIs51 contains [Pmyo-2::gfp; Ppes-10::gfp; PF22B7.9::gfp].
syp-1 partial depletion by RNAi
Partial depletion of syp-1 by RNAi was performed as in Libuda et al. (2013). Notably, partial depletion of syp-2 and syp-3 has been shown to also function similarly to partial depletion of syp-1 with regards to affecting CO numbers in C. elegans (Libuda et al. 2013). Worms were synchronized at the L1 phase by bleaching adults and allowing the resultant eggs to hatch on unseeded NGM plates at 20° for 20–24 hr. Synchronized L1s were then washed off the unseeded NGM plates with M9 and placed on NGM + IPTG + Amp plates that were poured within 30 days of use, and that had been freshly seeded 1 day before use with Escherichia coli HT115 cells containing either a fragment of the syp-1/F26D2.2 gene in the L4440 vector (Ahringer laboratory RNAi library) or the empty vector (referred to as “control RNAi” in figures and text). The RNAi plates with L1s were then placed at 25° for 40–48 hr and then their gonads were dissected for immunofluorescence.
Immunofluorescence
Immunofluorescence was performed as in Libuda et al. (2013). Gonads from adult worms at 18–24 hr post-L4 stage were dissected in 1× egg buffer with 0.1% Tween on VWR Superfrost Plus slides, fixed for 5 min in 1% paraformaldehyde, flash frozen with liquid nitrogen, and then fixed for 1 min in 100% methanol at −20°. Slides were washed 3 × 5 min in 1× PBS Tween (PBST) and blocked for 1 hr in 0.7% BSA in 1× PBST. Primary antibody dilutions were made in 1× PBST and added to slides. Slides were covered with a parafilm coverslip and incubated in a humid chamber overnight (14–18 hr). Slides were washed 3 × 10 min in 1× PBST. Secondary antibody dilutions were made at 1:200 in 1× PBST using Invitrogen goat or donkey Alexa Fluor-labeled antibodies, and added to slides. Slides were covered with a parafilm coverslip and placed in a humid chamber in the dark for 2 hr. Slides were washed 3 × 10 min in 1× PBST in the dark. All washes and incubations were performed at room temperature, unless otherwise noted. Next, 2 μg/ml DAPI was added to slides and slides were subsequently incubated in the dark with a parafilm coverslip in a humid chamber. Slides were washed once for 5 min in 1× PBST prior to mounting with Vectashield and a 20 × 40 mm coverslip with a 170 ± 5 μm thickness. Slides were sealed with nail polish immediately following mounting and then stored at 4° prior to imaging. For structured illumination microscopy (SIM) imaging, slides were made as described above with the following modification. All SIM slides were mounted in Prolong Gold (P36930; Thermo Fisher Scientific) and left to harden at room temperature for 2–3 days prior to imaging. All slides were imaged (as described below) within 2 weeks of preparation. The following primary antibody dilutions were used: rabbit anti-GFP (1:1000) (Yokoo et al. 2012), chicken anti-GFP (1:1000) (13970; Abcam), guinea pig anti-ZHP-3 (1:500) (Bhalla et al. 2008), rabbit anti-MSH-5 (1:10000) (#3875.00.02; Novus), guinea pig anti-SYP-1 (1:200) (MacQueen et al. 2002), goat anti-SYP-1 (1:1500) (Harper et al. 2011), guinea pig anti-HIM-8 (1:250) (Phillips et al. 2009), and chicken anti-HTP-3 (1:500) (MacQueen et al. 2005).
Imaging
Immunofluorescence slides were imaged at 512 × 512 pixel dimensions on an Applied Precision DeltaVision microscope with a 63× lens and a 1.5× optivar. Images were acquired as Z-stacks at 0.2 μm intervals and deconvolved with Applied Precision softWoRx deconvolution software. For quantification of GFP::COSA-1 foci, nuclei that were in the last four-to-five rows of late pachytene and were completely contained within the image stack were analyzed. Foci were quantified manually from deconvolved three-dimensional (3D) stacks. For quantification of chiasmata and visualization of chiasmata, individual chromosomes from a single diakinesis nucleus were cropped and rotated in three dimensions using Volocity 3D rendering software. Images shown are projections through 3D data stacks encompassing whole nuclei, generated with a maximum-intensity algorithm with the softWoRx software. For quantification of HIM-8, early-to-midpachytene regions of the gonad were visualized in IMARIS (Bitplane) and the distance between each HIM-8 focus was determined using the “Measurements” tool in IMARIS. HIM-8 foci were considered paired if the distance between the foci was ≤ 0.7 µm and unpaired if the distance was > 0.7 µm. To determine SC association with the HIM-8 foci, the nuclei were visualized and rotated in 3D using IMARIS to track the SC traces in and out of each HIM-8 focus. A single SC trace was determined by following a single SC track in and out of the HIM-8 focus. For two SC traces, the HIM-8 focus displayed two SC tracks coming in and two SC tracks coming out of the HIM-8 focus.
For SIM, slides were imaged at 2430 × 2430 pixel dimensions on a Zeiss ELYRA S.1/LSM 880 microscope with a Plan Apochromat 63× (1.4 NA) oil lens. Images were acquired as a Z-stack at 0.110 µm intervals with three rotations, and were processed using the Zeiss ZEN software for both SIM reconstruction and channel alignment (alignment calibration based off 100 nm TetraSpeck beads from Thermo Fisher Scientific). Maximum-intensity projections were generated using FIJI (National Institutes of Health). Images were adjusted for brightness and contrast. For quantification of SC trace lengths, individual nuclei were 3D cropped from the midpachytene region using IMARIS. The “Filament tracer” tool in IMARIS was used to trace the SC in each cropped nucleus.
Statistics
Most of the P-values reported are two-tailed and calculated from Mann–Whitney U-tests, which are robust nonparametric statistical tests appropriate for the relevant data sets. A Wilcoxon test was used for the rec-8 mutant SC trace comparisons. Each test used is indicated next to the reported P-value in the Results section.
Data availability
All strains are available upon request. Supplemental Material, Figure S1A shows the localization of COSA-1, ZHP-3, and MSH-5 in syp-1 and syp-3 mutants. Figure S1B has unadjusted images of GFP::COSA-1 localization in wild-type, syp-2, and syp-3 mutants. Figure S2A shows the localization of COSA-1 and MSH-5 in wild-type, him-3, htp-3, and rec-8 mutants. Figure S2B shows the localization of COSA-1 and ZHP-3 in wild-type worms and the rec-8 mutant. Figure S3 shows localization of COSA-1 and SYP-1 in syp-1 partial depletions combined with wild-type, him-3, and rec-8 mutants. Figure S4 shows representative images and the quantification of DAPI-staining bodies at diakinesis from wild-type, rec-8, cosa-1, rec-8; cosa-1, and rec-8; syp-2 mutants. Figure S5 shows the localization of RAD-51 in wild-type, syp-2 (ok307), and syp-2; rec-8 mutants at both late pachytene (A) and diakinesis (B). Table S1 has the number of GFP::COSA-1 foci per nucleus in late pachytene from wild-type, syp-1, syp-2, syp-3, rec-8, rec-8;syp-2, and him-3 mutants. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.9600878.
Results
Pro-CO factors colocalize to DSB-dependent events in late pachytene of mutants lacking SC central region proteins
Many of the pro-CO factors display distinct localization patterns that often colocalize with the SYP proteins during pachytene (Figure 1). ZHP-3 (an E3 ligase) localizes along the lengths of the chromosomes in nuclei at the midpachytene stage of meiotic prophase, and MSH-5 forms foci in excess of the number of COs at this stage (Jantsch et al. 2004; Bhalla et al. 2008; Zhang et al. 2018). Upon transition to the late pachytene stage, COSA-1 is detected as six bright foci at nascent CO sites that colocalize with MSH-5 and correspond to the six CO sites (one per chromosome) in C. elegans [(Yokoo et al. 2012); Table S1]. Additionally, at late pachytene, the ZHP-3 tracks gradually retract in a DSB-dependent manner to form distinct foci that colocalize with COSA-1 and MSH-5 (Yokoo et al. 2012).
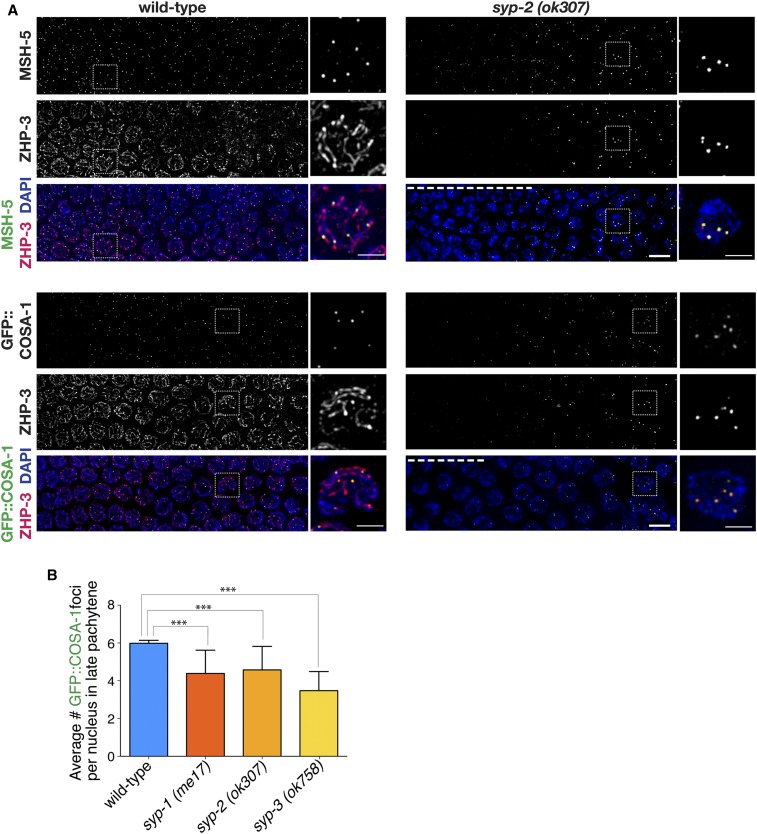
Figure 1.
DSB-dependent colocalization of pro-CO factors to late pachytene foci in syp mutants. (A) Immunofluorescence images of mid-to-late pachytene region of germ lines from wild-type and syp-2(ok307) mutant worms, with meiotic prophase progressing from left to right. In wild-type nuclei at the midpachytene stage (left sides of wild-type panels), ZHP-3 is localized along the lengths of the chromosomes, MSH-5 is detected as foci in excess of the eventual number of COs, and COSA-1 foci are not detected. Upon transition to late pachytene, COSA-1 foci are detected at nascent CO sites, colocalized with MSH-5, and ZHP-3 tracks gradually reduce and retract toward the COSA-1 foci. In the syp-2 mutant panels, nuclei at the left sides of the images exhibit characteristic DAPI signals reflecting prolonged persistence of chromosome clustering and chromosome movement (dashed white line). However, upon eventual release from chromosome clustering and transition to a late pachytene-like dispersed chromosome organization, ZHP-3 is detected as foci, most of which colocalize with MSH-5 (top) or GFP::COSA-1 (bottom). Both Jantsch et al. (2004) and Zhang et al. (2018) have also published ZHP-3 localization in syp null mutants. As the ZHP-3, MSH-5, and COSA-1 foci in the syp mutants are of weaker intensity than in wild-type, signal intensities in the syp-2 images were boosted relative to controls to enable visualization of the foci (see Figure S1 for unadjusted images). Dashed box indicates the nucleus that is enlarged in the adjacent image and scale bars on the enlarged images represent 2 µm. All other bars, 5 μm. (B) Quantitation of GFP::COSA-1 foci in late pachytene nuclei for syp null mutants (Table S1). Number of asterisks represents degree of statistical significance from a Mann–Whitney U-test (*** = P < 0.0001). Error bars represent SD. Number of nuclei scored for GFP::COSA-1: wild-type, n = 505; syp-1(me17), n = 223; syp-2(ok307), n = 101; syp-3(ok758), and n = 99. CO, crossover; DSB, DNA double-strand break.
In syp null mutants, meiotic recombination is initiated by the formation of DSBs, but repair of these breaks does not yield interhomolog COs and the germ lines exhibit a prolonged clustering stage (MacQueen et al. 2002; Colaiacovo et al. 2003; Smolikov et al. 2007b, 2009). Despite the lack of interhomolog COs, syp null mutants display a few foci of COSA-1 and MSH-5, specifically during late pachytene when the prolonged clustering stage ends (Figure 1A and Figure S1). As was also shown in previous studies (Jantsch et al. 2004; Zhang et al. 2018), localization of ZHP-3 as tracks along the chromosomes in early-to-midpachytene requires the SYP proteins, and upon eventual release from the prolonged clustering in syp mutants, ZHP-3 localizes as foci during late pachytene. Further, we found that most of these ZHP-3 foci in the syp mutants colocalize with MSH-5 or COSA-1 (Figure 1A and Figure S1). Notably, the ZHP-3, MSH-5, and COSA-1 foci observed in the syp mutants are of weaker intensity than those observed in wild-type or other mutant situations where SYP proteins are present (Figure S1B). In contrast to the highly reproducible number of COSA-1 foci (six per nucleus) detected in wild-type worms, the number of COSA-1 foci observed in syp mutants are reduced and significantly different from wild-type (Figure 1B and Table S1; P < 0.0001, Mann–Whitney U-test). Thus, in the absence of the SYPs, pro-CO factors are able to associate into foci, even though these foci are incapable of forming COs.
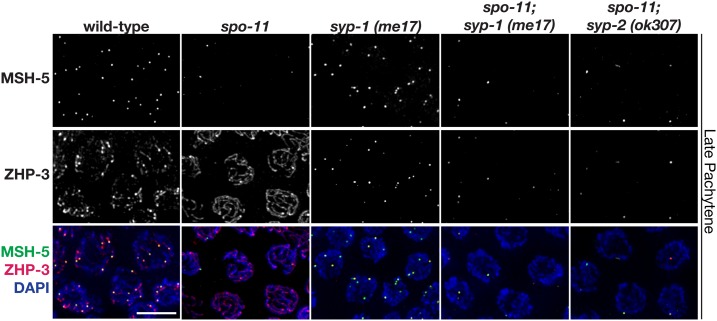
To determine if these pro-CO factor foci in syp mutants are forming at DSB sites, we assessed pro-CO factor localization in syp mutants that lacked SPO-11, the conserved endonuclease that forms programmed DSBs during meiosis (Dernburg et al. 1998). Similar to the spo-11 single mutant (which lacks endogenous DSBs) (Dernburg et al. 1998), late pachytene nuclei in spo-11; syp-1 and spo-11; syp-2 double mutants typically have only an occasional MSH-5 focus (zero-to-one focus per nucleus; Figure 2). This result demonstrates that in the absence of synapsis, the localization of pro-CO factors is dependent on programmed DSBs. Further, this result reflects the proclivity of these factors to associate both with each other and with abnormal recombination intermediates present on the chromosomes in this context (Pattabiraman et al. 2017).
Figure 2.
Colocalization of pro-CO factors in syp mutants is DSB-dependent. Immunolocalization of MSH-5 and ZHP-3 in representative late pachytene nuclei from wild-type, spo-11, syp-1(me17), spo-11; syp-1(me17), and spo-11; syp-2(ok307). Similar to the spo-11 single mutant, late pachytene nuclei in spo-11; syp-1 and spo-11; syp-2 double mutants typically have only an occasional MSH-5 focus (zero-to-one), indicating that the presence of multiple foci in the syp single mutants is DSB-dependent. Representative images of the syp-2 single mutant are in Figure 1. Bar, 5 μm. CO, crossover; DSB, DNA double-strand break.
Pro-CO factors specifically associate with SYP stretches along chromosomes in mutants with limited synapsis
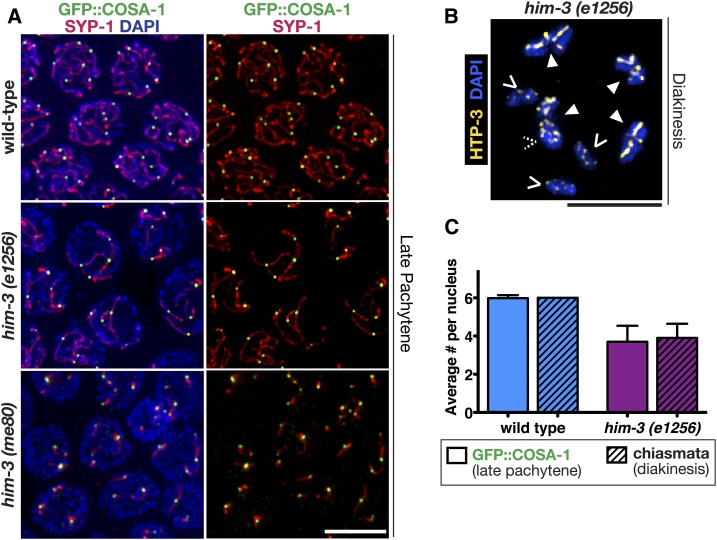
To further understand the relationship between the SC and the loading of pro-CO factors, we examined the localization of pro-CO factors and SYP-1 in mutants with abnormal SC formation. Specifically, we assessed worms homozygous for partial loss-of-function mutations affecting the chromosome axis protein HIM-3 (Figure 3 and Figure S2). In him-3(e1256) mutants, SYP-1 loads only on a subset of chromosome pairs (usually four chromosomes) (Zetka et al. 1999). In these him-3(e1256) mutants, we found that COSA-1 and MSH-5 foci only associated with the synapsed chromosomes displaying extensive SYP-1 stretches during late pachytene (Figure 3A and Figure S2). Further, for him-3(e1256) mutants, the average number of COSA-1 foci detected in late pachytene nuclei corresponded to the eventual number of chiasmata present in diakinesis-stage oocytes (Figure 3B and Table S1; P < 0.0001, Mann–Whitney U-test), indicating that the COSA-1 foci detected in this mutant represent bona fide CO events. In comparison, the him-3(me80) mutants exhibit a more severe synapsis defect where only short discontinuous stretches of synapsis occur on a subset of autosomes (Figure 3A; Couteau et al. 2004; Nabeshima et al. 2005). Additionally, him-3(me80) mutants also display increases in double COs determined by both genetic assays and diakinesis bivalents containing two chiasmata (Couteau et al. 2004; Nabeshima et al. 2004). Similar to him-3(e1256), COSA-1 foci in him-3(me80) were also invariably associated with the limited SYP-1 stretches (Figure 3A). Similar to prior studies in wild-type worms and in strains partially depleted for syp-1 (Yokoo et al. 2012; Libuda et al. 2013), we found that all COSA-1 foci were associated with a SYP-1 stretch in either him-3 partial loss-of-function mutant [90/90 nuclei contained COSA-1 associated with the SC in him-3(e1256) and 90/90 nuclei contained COSA-1 associated with the SC in him-3(me80)].
Figure 3.
GFP::COSA-1 specifically associates with synapsed chromosome segments in mutants with limited synapsis. (A) Immunofluorescence images of representative nuclei in the late pachytene regions of germ lines from worms of the indicated genotypes, in which the synaptonemal complex central region protein SYP-1 (red) localizes: along the lengths of paired homologs (wild-type), along the lengths of a subset of chromosomes [him-3(e1256) mutant], or in several short stretches [him-3(me80) mutant]. All GFP::COSA-1 (green) are associated with the chromosomes or chromosome segments where SYP-1 localizes. (B) Immunofluorescence image of a representative diakinesis nucleus from the him-3(e1256) mutant, shown with DAPI (blue) and chromosome axis component HTP-3 (yellow) to visualize the chiasmata. Chiasmata were visualized and counted using three-dimensional rotations; solid arrowheads indicate bivalents connected by chiasmata, while carets indicate achiasmate chromosomes (univalent) (dashed caret indicates a univalent hidden in this projection). (C) Bar graph depicting quantitation of GFP::COSA-1 foci in late pachytene nuclei (bars without a pattern), and chiasmata (bars with diagonal lines), in diakinesis nuclei for wild-type (blue bars) and the him-3(e1256) (purple bars) partial loss-of-function chromosome axis mutant (Table S1); error bars indicate SD (P < 0.0001, Mann–Whitney U-test). Number of late pachytene nuclei scored for COSA-1 foci: wild-type, n = 505 and him-3(e1256), n = 161. Number of nuclei scored for chiasmata: wild-type, n = 28 and him-3(e1256), n = 40. Bars, 5 μm.
To determine if CO regulation is still occurring within the SC structures of him-3(e1256) worms, we decided to test whether we could experimentally perturb CO interference in this mutant. In previous work, we showed that partial depletion of any of the SYP proteins (SYP-1, SYP-2, and SYP-3) by 60–70% results in an increased number of COSA-1 foci and attenuates CO interference (Libuda et al. 2013). Combining the partial depletion of SYP-1 with the him-3(e1256) mutant resulted in an increased occurrence of SYP-1 stretches with >more than two COSA-1 foci (Figure S3). In 14% of control RNAi-treated him-3(e1256) nuclei, we observe two COSA-1 foci along a SYP-1 stretch. Upon syp-1 partial RNAi treatment, the number of SYP-1 stretches containing more than two COSA-1 foci significantly increased to 41%. This result suggests that CO interference is likely still acting along the SYP-1 stretches that form on the chromosomes in him-3(e1256) partial loss-of-function mutants.
Pro-CO factors associate with SYP protein aggregates formed in null mutants lacking meiotic chromosome axis components
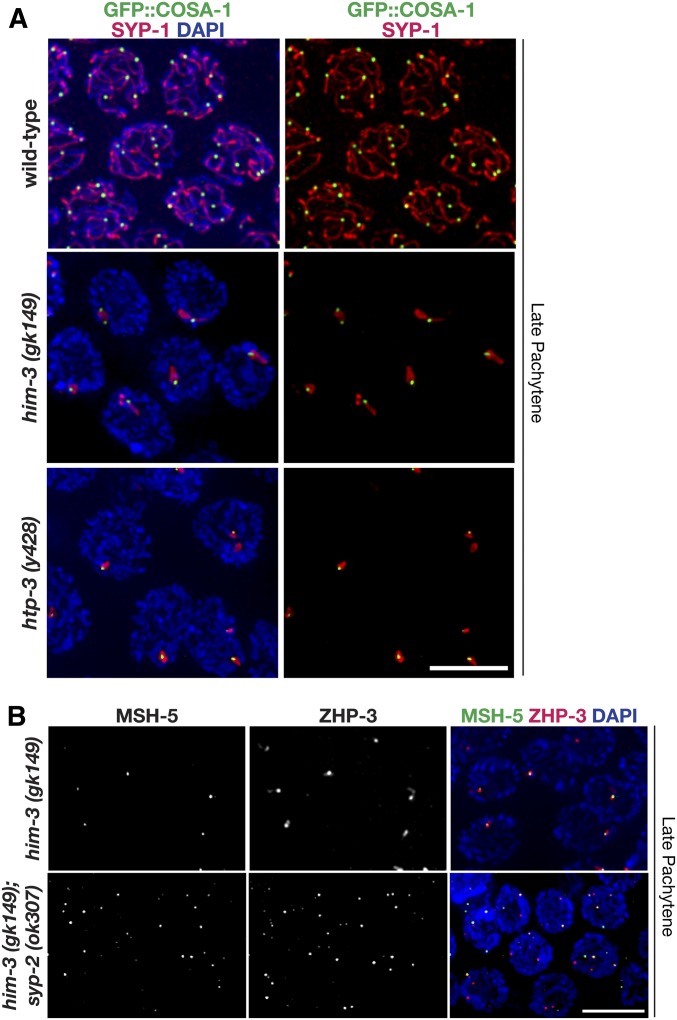
To determine if the localization of pro-CO factors is directed by SYP proteins, we examined pro-CO factor localization in mutants where SYPs form aggregates in the nucleoplasm (Figure 4 and Figure S2). Mutants null for the lateral elements HTP-3 and HIM-3 are both unable to form COs and are unable to load SYP proteins onto chromosomes, instead forming a SYP protein aggregate within the nucleoplasm (Couteau et al. 2004; Goodyer et al. 2008). Specifically, him-3(gk149) null mutants typically contain a single elongated SYP-1 aggregate in late pachytene nuclei (Figure 4A). Likewise, htp-3(y428) null mutants usually contain one or sometimes two aggregates per nucleus in late pachytene (Figure 4A). Both him-3 and htp-3 mutants displayed COSA-1 and MSH-5 foci associated with the SYP aggregate (Figure 4 and Figure S2). Notably, this similar result for pro-CO factor localization in him-3 and htp-3 null mutants occurs despite the fact DSBs are formed in him-3(gk149), but not in htp-3(y428) worms (Couteau et al. 2004; Goodyer et al. 2008). Further, in most cases, we find that only a single COSA-1 focus was associated with a given SYP-1 aggregate (Figure 4A). Similarly, Rog et al. (2017) also showed that the pro-CO factors ZHP-3 and COSA-1 also localize to SC aggregates as a single focus in htp-3(tm3655) null mutants. Collectively, these data suggest that COSA-1 has a strong tendency to associate with SYP-1, and that the ability to limit COSA-1 foci to a single site on a given SYP-1 structure is retained even when the SYP proteins are concentrated in a nucleoplasmic aggregate.
Figure 4.
Pro-CO factors associate with SYP-1 aggregates in mutants lacking meiotic chromosome axis components. (A) Immunolocalization of SYP-1 (red) and GFP::COSA-1 (green) in nuclei from the late pachytene regions of null mutants lacking chromosome axis components HIM-3 or HTP-3. SC assembly is severely impaired in both the him-3(gk149) and htp-3(y428) null mutants, and SYP proteins instead assemble into abnormal aggregates known as polycomplexes. GFP::COSA-1 localization is consistently associated with these abnormal SYP-1 structures in both mutants. The representative image of late pachytene in wild-type is repeated from Figure 3. Rog et al. (2017) also showed that ZHP-3 and COSA-1 also localize to SC aggregates in htp-3(tm3655) null mutants. (B) Immunolocalization of MSH-5 (green) and ZHP-3 (red) in nuclei from late pachytene from him-3(gk149) and the him-3(gk149); syp-2(ok307) double mutant. Whereas MSH-5 and ZHP-3 are usually detected together at a single site per nucleus in the him-3(gk149) mutant, multiple foci are detected in nuclei in the him-3(gk149); syp-2(ok307) double mutant (as in the syp-2 single mutant, see Figure 1). Bars, 5 μm. CO, crossover; SC, synaptonemal complex.
To test whether the pro-CO factors are held in a SYP-dependent manner within the aggregates formed in the him-3(gk149) null mutant, we assessed MSH-5 and ZHP-3 localization in an him-3(gk149); syp-2(ok307) double mutant. In contrast to the him-3(gk149) null mutant, we found that the him-3(gk149); syp-2(ok307) double mutant (Figure 4) looked similar to the syp-2(ok307) single mutant (Figure 1), where MSH-5 and ZHP-3 localize to multiple DSB-dependent chromosomal sites in late pachytene nuclei. This finding suggests that aggregation of SYP proteins to a single site preferentially stabilizes the association of pro-CO factors and directs them to colocalize together with SYP-1 in a single compartment. When this constraint is released, the pro-CO factors are free to associate together at DSB-dependent chromosomal sites.
rec-8 mutants inappropriately assemble SCs between sister chromatid pairs
The proclivity of pro-CO factors to be targeted to synapsed regions raised the possibility that inappropriate synapsis between nonhomologous chromosomes and/or sister chromatids might also direct the localization of pro-CO factors to these incorrectly synapsed regions. Multiple studies have suggested that worms mutant for a meiosis-specific cohesin protein REC-8 may undergo nonhomologous and/or sister chromatid synapsis due to extensive homolog pairing defects (Pasierbek et al. 2001; Severson et al. 2009). Further, electron microscopy in rec-8 mutant mouse spermatocytes has indicated that SC formation may occur between sister chromatids in this context (Xu et al. 2005).
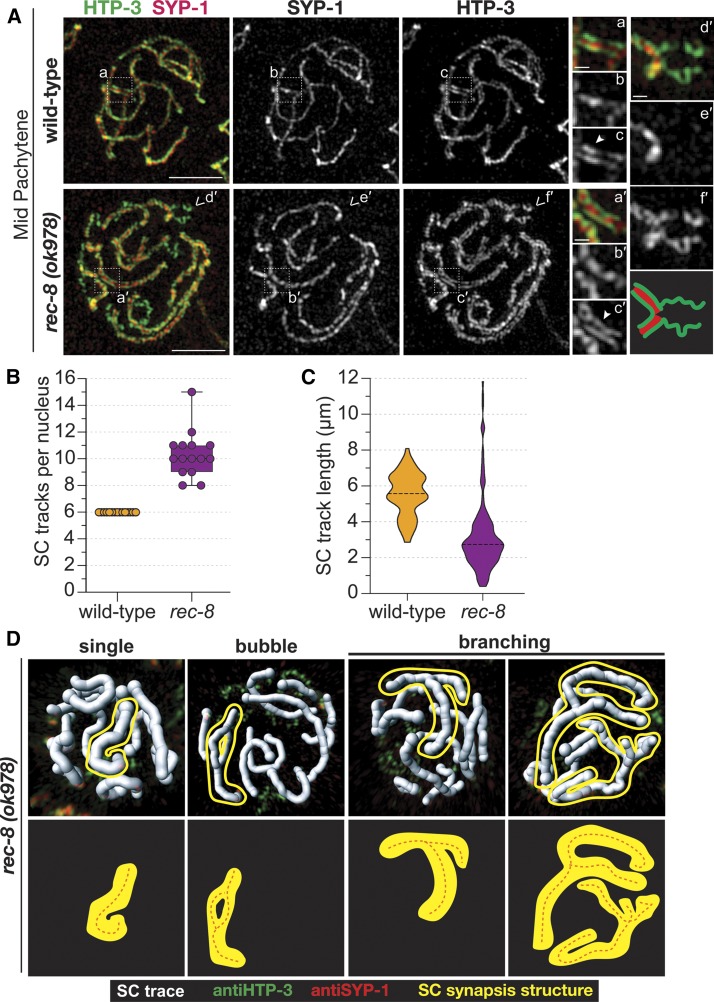
To characterize chromosome synapsis along the 12 sets of sister chromatids in a C. elegans rec-8(ok978) null mutant, we used super resolution microscopy. We found that both wild-type and rec-8 null mutants displayed tripartite SCs, with SYP-1 localizing between the two HTP-3 tracks suggesting that the overall structure of the SC is unaltered in rec-8 null mutants (Figure 5A). Notably, after tracing the SC segments in midpachytene nuclei, we observed that the number of SC segments in rec-8(ok978) null mutants is significantly higher than in wild-type worms. While wild-type worms always displayed 6 SC traces per nucleus representing the six homolog pairs, rec-8 null mutants showed on average 10 SC traces per nucleus (Figure 5B), suggesting that inappropriate synapsis is occurring among the 12 pairs of sister chromatids (P < 0.0001, Wilcoxon test). Further, the lengths of the SC traces in rec-8 mutants were on average shorter than the average lengths of the wild-type traces (3.13 and 5.53 µm, respectively; P < 0.0001, Mann–Whitney U-test; Figure 5C). Some of these shorter traces in the rec-8 mutants had unsynapsed HTP-3 segments extending from the synapsed region (Figure 5, A and D), likely representing regions of partial synapsis along the chromosome. However, it is also possible that some of these short-synapsed regions are the result the chromosome self-synapsing.
Figure 5.
Synapsis occurs between sister chromatid pairs in rec-8 mutants. (A) Structured illumination microscopy images of SYP-1 (red) and chromosome axis component HTP-3 (green) in representative midpachytene nuclei, showing that SYP-1 localizes between pairs of HTP-3 tracks in both wild-type and rec-8(ok978); this indicates that synapsis occurs between sister chromatid pairs in the rec-8 mutant. White dashed box indicates the enlarged region of the SC depicted in the smaller images on the right. The solid arrowhead identifies a region where both lateral elements of the SC are visible, indicated by the two tracks of HTP-3. The carets indicate a region of unsynapsed HTP-3, which is enlarged in (d′–f′) with a cartoon diagram of the unsynapsed region below (f′) (red, SYP-1 and green, HTP-3). Bars for whole-nucleus images represent 2 µm and scale bars for smaller enlarged SC segments represent 250 nm. (B) Box plot depicting the number of SC tracks per nucleus showing that rec-8(ok978) (purple) mutants display on average 10 SC tracks per nucleus, while wild-type (yellow) only has 6 SC tracks per nucleus. (C) Violin plots showing the distribution of the SC track length in micrometers from wild-type (yellow) and rec-8(ok978) (purple). Number of midpachytene nuclei traced for the SC: wild-type, n = 15 (three total gonads) and rec-8(ok978), n = 15 (three total gonads). (D) 3D surfaces, generated in IMARIS, showing the SC traces (white) in each nucleus from rec-8(ok978). Each SC trace contains both HTP-3 (green) and SYP-1 (red). Multiple SC synapsis structures were observed in rec-8(ok978) mutants: single SC track, bubble SC track, Y-shaped branching SC track, and multi-branching SC track. A representative example of each SC synapsis structure is outlined in yellow and depicted as a diagram below each 3D surface image with the orange dashed line representing the SC trace. 3D, three-dimensional; SC, synaptonemal complex.
Some of the SC traces in the rec-8 null mutants displayed lengths longer than those of the wild-type traces, with traces ranging from 8 to nearly 12 µm long. We determined that these long SC traces were created by multi-chromosome and/or chromatid synapsis events (Figure 5D). Among these events, we observed three different classes of multi-chromosome synapsis: (1) a single branching Y-shaped structure, (2) a bubble-shaped structure, and (3) very large multi-branching structures (as if four chromatids are synapsing along different regions) (Figure 5D). Taken together, the formation of these large aberrant synapsis events and the average 10 SC traces observed in rec-8 mutants suggests that these mutants are assembling SCs between the sister chromatids, and possibly with nonhomologous chromosomes.
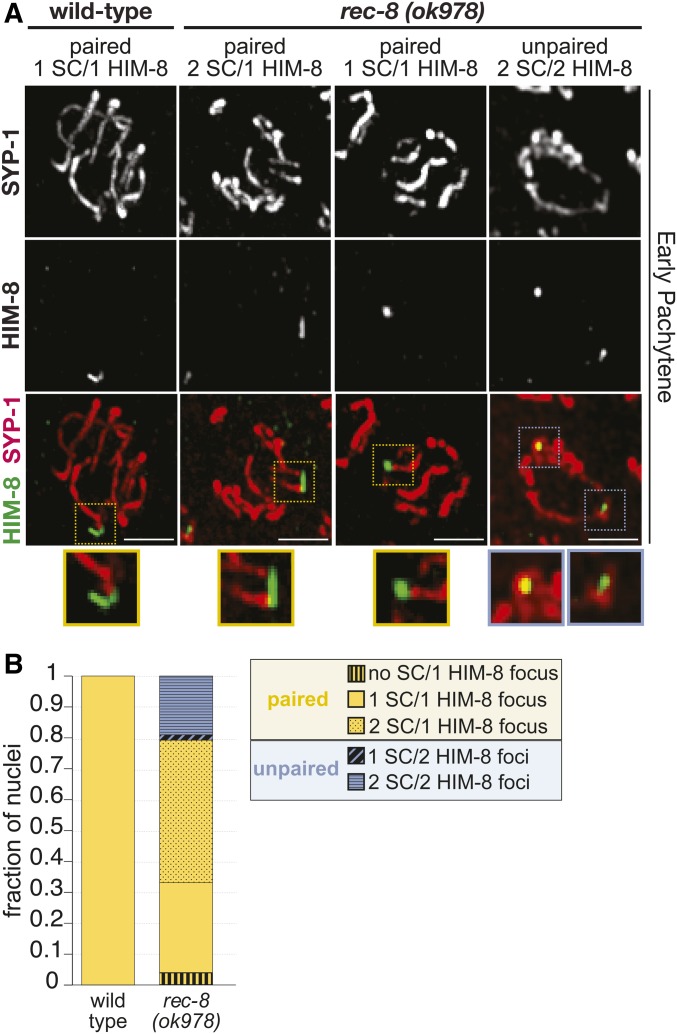
To further determine if rec-8 null mutants are assembling SCs between sister chromatids, we also assayed homolog pairing on the X chromosome using the pairing center protein HIM-8, which binds to the pairing center region on one end of the X chromosome and is required for X chromosome pairing (MacQueen et al. 2005; Phillips et al. 2009). In wild-type worms, 100% of HIM-8 foci were paired (≤ 0.7 µm apart) and only had a single SC track extending from the paired HIM-8 focus, indicating the SC assembled between the homologs (Figure 6). While previous work using FISH has shown that rec-8 RNAi nuclei display extensive homolog pairing defects along the autosomes (Pasierbek et al. 2001), we found that rec-8 null mutants displayed only a slight X chromosome pairing defect with ∼80% of the HIM-8 foci being paired. Strikingly, nearly 60% of these paired HIM-8 foci in rec-8 null mutants displayed two SC tracks extending from the HIM-8 focus (Figure 6). Additionally, of the 20% unpaired HIM-8 foci, 92% of these foci had an SC associated with each unpaired HIM-8 focus. The occurrence of two sets of SC tracks extending from paired HIM-8 foci and SCs associating with both unpaired HIM-8 foci strongly suggests that rec-8 null mutants are assembling SCs between sister chromatids.
Figure 6.
Paired X chromosomes in rec-8 mutants exhibit two stretches of SC. (A) Immunolocalization of HIM-8 (green) and SYP-1 (red) in early pachytene nuclei from rec-8(ok978) and wild-type. As some SCs from the top and bottom halves of the nuclei are superimposed in the full projections encompassing whole nuclei, partial projections showing half nuclei are shown. Colored dashed boxes indicate the enlarged regions of the HIM-8 focus and SC depicted in the smaller images below, with the color indicating paired (yellow) or unpaired (blue) HIM-8 foci. Bars, 5 µm. (B) Stacked bar plot showing the fraction of nuclei displaying paired (yellow) or unpaired (blue) HIM-8 foci. HIM-8 foci were considered paired if the distance between the foci was ≤ 0.7 µm (see Materials and Methods). All HIM-8 foci in wild-type are paired, and in rec-8(ok978) mutants 80% of the HIM-8 foci are paired. In wild-type, all of the paired HIM-8 foci are associated with one SC track (solid bar) indicating SCs between homologous chromosomes. However, within rec-8 mutant nuclei, we observed differences in the number of SC tracks associating with either the paired or unpaired HIM-8 focus/foci. Among the paired HIM-8 foci (yellow), the majority of the rec-8 mutant nuclei displayed HIM-8 foci associated with two SC tracks (dotted bar) suggesting SC assembly between sister chromatids. Paired HIM-8 foci were also observed to not be associated with SC tracks (vertical striped bar) or associated with one SC track (solid bar). Among the unpaired (blue) HIM-8 foci in rec-8 mutants, the majority of the nuclei displayed HIM-8 foci where each was associated with an SC track (horizontal striped bar), also suggesting that the SC is assembling between sister chromatids in this context. Unpaired HIM-8 foci were also observed where only one focus was associated with an SC track (diagonal striped bar). Number of early pachytene nuclei scored for HIM-8: wild-type, n = 90 and rec-8(ok798), n = 116. SC, synaptonemal complex.
Localization of pro-CO factors tracks with SYP stretches when synapsis occurs incorrectly between sister chromatid pairs
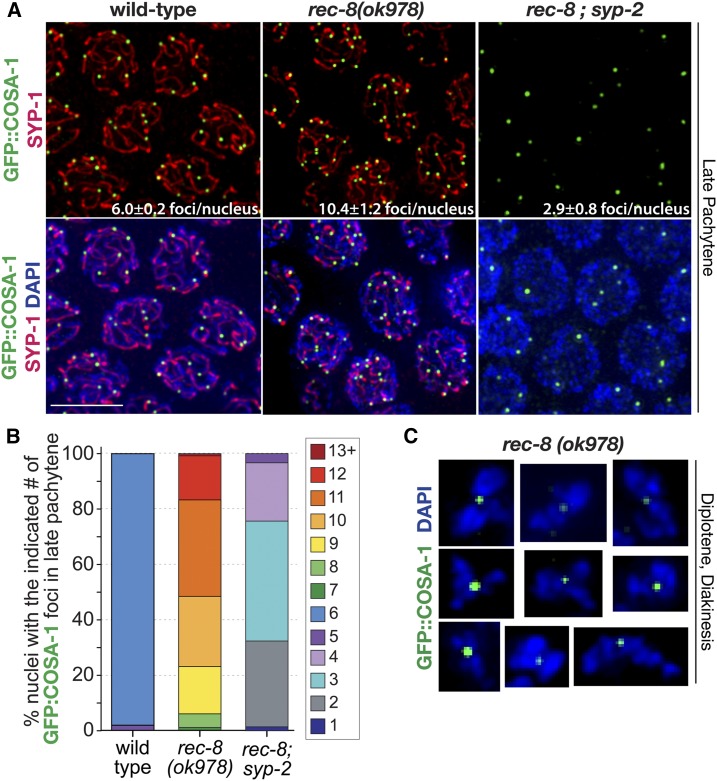
To test whether the localization of pro-CO factors could be mistargeted to events along incorrectly synapsed regions, we investigated pro-CO factor localization in the rec-8(ok978) null mutant. We found that the inappropriately synapsed sister chromatids in rec-8(ok978) mutants still enable the localization of pro-CO factors to sites along the synapsed sisters. Specifically, rec-8 null mutants displayed COSA-1 foci along the SYP-1 stretches in late pachytene nuclei (Figure 7A), and between condensed pairs of sister chromatids in diplotene- and diakinesis-phase nuclei (Figure 7C). Interestingly, 99% of rec-8 mutants localized COSA-1 to ≤ 12 sites in each late pachytene nucleus (Figure 7B and Table S1). Further, ZHP-3 and MSH-5 similarly associate with COSA-1 foci, strongly suggesting that recombination may be occurring between sister chromatids in this mutant (Figure S2).
Figure 7.
Pro-CO factors associate with SYP-1 stretches and between sister chromatid pairs in rec-8 mutants. (A) Immunolocalization of SYP-1 and GFP::COSA-1 in fields of nuclei from the late pachytene regions of wild-type, rec-8(ok978), and rec-8; syp-2 germ lines. Average number of COSA-1 foci per nucleus and SD is labeled on the image for each genotype. Bar, 5 μm. (B) Stacked bar graph showing percentages of nuclei with indicated numbers of GFP::COSA-1 foci in late pachytene for wild-type, rec-8(ok978), and rec-8; syp-2 (Table S1). Number of late pachytene nuclei scored for COSA-1 foci: wild-type, n = 505; rec-8(ok978), n = 245; and rec-8; syp-2, n = 204. (C) Immunolocalization of GFP::COSA-1 in DAPI-stained diplotene and diakinesis bivalents from rec-8(ok978) germ lines. CO, crossover.
The average number of COSA-1 foci formed in rec-8(ok978) null mutants was consistent with the average number of SC traces per nucleus (10.4 ± 1.2 COSA-1 foci per nucleus and 10 SC traces per nucleus; Figure 5A, Figure 7B, and Table S1). This observation in rec-8(ok978) null mutants may reflect an imposed limitation of COSA-1 foci by CO interference occurring along synapsed sister chromatids and multi-chromatid synapsis events. In support of this suggestion, partial depletion of SYP-1 in the rec-8(ok978) null mutant background resulted in the frequent occurrence of SYP-1 stretches harboring two COSA-1 foci, even while reducing the fraction of chromosomes associated with SYP-1 stretches (Figure S3). Together, our data reinforce the suggestion that the SC central region may regulate CO numbers and the distribution along the length of a chromosome (or chromatid) in which the SC has assembled.
Regions of desynapsis in rec-8 null mutants fail to repair DSBs
Previous studies have shown that rec-8 null mutants will frequently equationally separate the sister chromatids at the first meiotic division, and that the sister chromatids are held together in a DSB-dependent manner prior to the first meiotic division (Severson et al. 2009; Severson and Meyer 2014). As COSA-1 localization is DSB-dependent (Yokoo et al. 2012), our finding of COSA-1 foci between condensed sister chromatid pairs during diakinesis in rec-8 null mutants suggests that sister chromatids are held together in a COSA-1-dependent manner at a DSB site (Figure 7C). Previous studies found that in rec-8 mutants, the absence of COSA-1 causes the separation of the sister chromatids at diakinesis (Crawley et al. 2016) and that the absence of the SC results in severe chromosome fragmentation (Colaiacovo et al. 2003) (Figure S4). Further, we found that the number of COSA-1 foci are significantly reduced in the rec-8; syp-2 double mutant (Figure 7B and Table S1). Thus, the SC central region between sister chromatids in rec-8 mutants is important for the efficient loading of COSA-1. Additionally, the loading of COSA-1 in rec-8 mutants is likely marking a DSB-dependent event occurring between sister chromatids that may be used to equationally separate the sister chromatids at meiosis I.
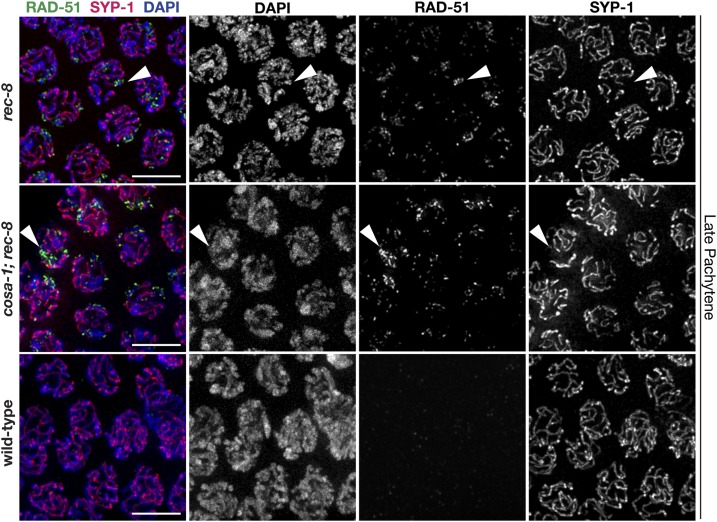
The previously reported striking chromosome fragmentation defect in the rec-8; syp-2 double mutant [Colaiacovo et al. 2003; Figure S4] suggests that the intersister associations created by the SC central region proteins in rec-8 mutants are required to complete DSB repair. Therefore, in the absence of the SC,rec-8; syp-2 double mutants likely lose both intersister and interhomolog associations, and accumulate unrepaired DSBs, which leads to the extensive chromosome fragments, DNA bridges, and DNA aggregates observed in the majority of the diakinesis nuclei of rec-8; syp-2 mutants (Colaiacovo et al. 2003; Figure S4). In accordance with this chromosome fragmentation phenotype during diakinesis in rec-8; syp-2 double mutants, the recombinase RAD-51 [a marker of DSBs, (Colaiacovo et al. 2003)] is accumulated extensively along chromosomes through diakinesis in this context (Figure S5), unlike in syp-2 mutants that only accumulate RAD-51 through late pachytene (Colaiacovo et al. 2003). Thus, if synapsis during pachytene is required to enable DNA repair in rec-8 single mutants, then the unsynapsed regions observed in rec-8 mutants should accumulate DNA damage. In support of this hypothesis, RAD-51 does indeed accumulate on late pachytene chromosome stretches where SYP-1 is absent in rec-8 single mutants (Figure 8). Similar to rec-8 single mutants, rec-8; cosa-1 double mutants also accumulate RAD-51 on unsynapsed chromosome regions at late pachytene (Figure 8). Thus, the maintenance of sister chromatid interactions by the SC in rec-8 mutants is indeed critical for DSB repair during meiosis. Overall, these data and previous published results suggest that meiotic DSB repair requires the SC central region proteins to promote partner associations critical for accessing DNA repair templates.
Figure 8.
Regions of desynapsis in rec-8 null mutants fail to repair DSBs. Immunolocalization of RAD-51 and SYP-1 in the late pachytene regions of wild-type, rec-8(ok978), and cosa-1; rec-8(ok978) germ lines. Arrowheads indicate chromatids that failed to load SYP-1 and have accumulated RAD-51 foci. Bars, 5 µm. DSB, DNA double-strand break.
Discussion
Relationship between the SC and pro-CO factors
Many studies have implicated a connection between the SC proteins and crossing over in C. elegans (Colaiacovo et al. 2003; Nabeshima et al. 2004, 2005; Smolikov et al. 2007a; Martinez-Perez et al. 2008; Libuda et al. 2013; Pattabiraman et al. 2017; Woglar and Villeneuve 2018; Zhang et al. 2018). To regulate where COs can form, our data indicate that the SC central region proteins in C. elegans have the capacity to promote the localization of the pro-CO factors COSA-1, MSH-5, and ZHP-3 to recombination events. Previous data in C. elegans demonstrate that the pro-CO factors, which are interdependent for localization, normally load in the context of an assembled SC (Yokoo et al. 2012; Woglar and Villeneuve 2018). Even in the context of an SC aggregate, the pro-CO factors are still interdependent for localization (Rog et al. 2017). Moreover, recent studies in C. elegans have shown that the SC central region proteins are preferentially stabilized on chromosomes containing CO or CO-like events (Machovina et al. 2016; Nadarajan et al. 2016; Pattabiraman et al. 2017). Further, our previous work demonstrated that meiotic chromosome structures both limit and respond to CO formation (Libuda et al. 2013). Collectively, these previous results paired with our current findings suggest a close reciprocal relationship between the SYP proteins and COs. Notably, a recent study in Saccharomyces cerevisiae identified a site on Zip1 (an SC central region protein that is an analog of SYP-1) that is required for normal Zip3/ZHP-3 localization in meiosis (Voelkel-Meiman et al. 2019). Interestingly, this site on Zip1 is directly adjacent to a site required for synapsis. Thus, the C. elegans SC central region proteins may directly interact with the pro-CO factors to restrict them to DSB events that occur within the context of the SC.
The localization of pro-CO factors to DSB-dependent sites in syp mutants (in which COs fail to form) suggests that pro-CO factors have an inherent capacity to associate with a DSB repair intermediate prior to CO formation. Previous studies have suggested that the pro-CO factors associate with a DSB repair intermediate after RAD-51 unloading, but before double Holliday junction resolution (Schvarzstein et al. 2014). Since the CO fate of a DSB is thought to occur very early in DSB repair [reviewed in Lake and Hawley (2016)], if not at the formation of the DSB, it is possible that in syp mutants the pro-CO factors could be associating with an early CO-competent repair intermediate that ultimately fails to establish a CO and is resolved through a different repair pathway. Alternatively, the pro-CO factors could be associated with a repair intermediate that they normally do not localize to in a wild-type situation. However, it is possible that there are other models that could explain this relationship between the SC and pro-CO factors. Thus, future studies assessing how DSBs are repaired in syp mutants may provide insight into the types of repair intermediates that the pro-CO factors have an inherent affinity for in the absence of SYP proteins.
Stabilized pro-CO factor localization may require both SYPs and a recombination intermediate
Based on our data and previous data from other groups, we suggest that pro-CO factors have a set of conditions that need to be met for strong localization to a recombination intermediate (Colaiacovo et al. 2003; Nabeshima et al. 2004, 2005; Smolikov et al. 2007a; Martinez-Perez et al. 2008; Libuda et al. 2013; Crawley et al. 2016; Pattabiraman et al. 2017; Woglar and Villeneuve 2018; Zhang et al. 2018). First, pro-CO factors are drawn to an SC compartment. Second, once inside the SC compartment, the pro-CO factors locate a recombination intermediate, at which point the pro-CO factors may be stabilized by the SC and/or trigger a reorganization of the SC compartment locally around that repair intermediate, the latter of which has been shown to occur in C. elegans (Woglar and Villeneuve 2018). Alternatively, this reorganization of SC proteins might promote a structural change in the recombination intermediate that has a higher affinity for pro-CO factors influencing their stabilization at the repair intermediate. Intriguingly, pro-CO factors fail to form a focus along chromosomes with SCs in spo-11 mutant nuclei that do not form any DNA lesions (Yokoo et al. 2012; Pattabiraman et al. 2017); however, pro-CO factors can form a focus within an SC aggregate that lacks recombination events in htp-3 mutants (Rog et al. 2017). This difference in pro-CO factor localization between spo-11 and htp-3 suggests that a chromosome-associated SC may be inherently different from a nucleoplasmic SC aggregate. Thus, the formation of an SC compartment in conjunction with recombination intermediates may be dictating the ability of pro-CO factors to stably colocalize with one another.
REC-8 and chromosome synapsis
During C. elegans meiosis, the meiotic-specific cohesin protein REC-8 functions in both sister chromatid cohesion and homolog pairing (Pasierbek et al. 2001; Severson et al. 2009; Severson and Meyer 2014). For both of these functions, REC-8 works together with two other meiotic-specific cohesins (COH-3 and COH-4), but genetic mutant analysis of all three of the cohesin proteins suggests that REC-8 may have additional roles separate from the sister chromatid cohesion and pairing role with COH-3/COH-4 (Severson et al. 2009; Severson and Meyer 2014; Crawley et al. 2016). We have identified a possible role for REC-8 in establishing synapsis between homologs, thereby enabling the recruitment of pro-CO factors to recombination events between homologs.
In multiple organisms it has been shown that the SC is two vertically stacked layers, with each layer connecting one sister chromatid of each homolog (Cahoon et al. 2017; Köhler et al. 2017). However, it is unclear what is ensuring that the SC is assembled between the homologs and not between sister chromatids, since each sister contains a chromosome axis that has the lateral element proteins. Our data in the rec-8 null mutant indicate that at the pairing center in wild-type situations, the initiation of SC assembly is regulated to ensure the SC assembles between the homologs and not the sisters. Further, we find that occasionally the SC assembles between multiple chromatids in rec-8 null mutants. Given that the SC assembles between pairs of sister chromatids in this mutant, it is compelling to suggest that these multi-chromatid synapsis events may represent the four sister chromatids for the same chromosome engaging with one another along different corresponding chromosomal regions. Alternatively, these events could represent nonhomologous synapsis. While it is clear that REC-8 is required to assemble the SC between the homologs, it is unknown if REC-8 performs this SC assembly function by: (1) promoting SC assembly between the homologs or (2) by preventing SC assembly between the sisters. Future studies are needed to elucidate the relationship between SC assembly and REC-8.
Meiotic DSB repair and sister chromatid associations in rec-8 mutants
Several studies indicate that the vast majority, if not all, of DSB repair during meiotic prophase I is by recombination-based mechanisms that require access to a repair template [reviewed in Hunter (2015)]. Previous studies have found that unlike other meiotic chromosome structure mutants [such as syp mutants and him-3 null mutants, in which all DSBs are repaired by late pachytene and midpachytene, respectively (Colaiacovo et al. 2003; Couteau et al. 2004)], rec-8 null mutants are unable to efficiently repair DSBs by the end of late pachytene, thereby resulting in both the persistence (or continued formation) of DSBs through diplotene and the eventual fragmentation of chromosomes in diakinesis (Alpi et al. 2003; Hayashi et al. 2007). While homologs are unpaired in syp and him-3 null mutants, sisters are still held together by cohesion, and therefore DSBs are able to repair, albeit not as COs (Colaiacovo et al. 2003; Couteau et al. 2004). In the absence of both the SC central region and cohesion, as occurs in rec-8; syp-2 double mutants, DSBs are unable to repair, thereby resulting in prevalent chromosome fragmentation. Further, in rec-8 single mutants, we also found the persistence (or continued formation) of DSBs along single chromatids that were not associated with their sister via SC formation. Taken together, these results reinforce the notion that: (1) interactions between chromatids or homologs are required to access DNA templates during recombination-based repair, and (2) recombination is required for the repair of most DSBs during meiotic prophase I.
CO events between homologs are known to be required in most organisms to maintain connections between homologs during diakinesis [reviewed in Hunter (2015)]. In C. elegans, an extreme form of CO interference exists such that only one CO is formed between homologs, therefore the pro-CO factors are localized to a single CO event per pair of homologs during late pachytene. In rec-8 null mutants, we find that the pro-CO factors are largely recruited to single events along the SC formed between sister chromatids in late pachytene. Further, sister chromatid pairs are held together in a COSA-1-dependent manner at diakinesis in rec-8 null mutants. Given these results, it is compelling to hypothesize that the pro-CO factors are marking and enabling CO formation between sisters, thereby joining sister chromatids together as pairs during diakinesis in rec-8 null mutants. Future experiments investigating the specific DSB repair outcomes that can occur in rec-8 null mutants may elucidate the nature of these events marked by the pro-CO factors.
Meiotic chromosome structures and limiting COs
The SC central region proteins are required for both the promotion and inhibition of crossing over during meiosis (MacQueen et al. 2002; Colaiacovo et al. 2003; Hayashi et al. 2010; Libuda et al. 2013; Pattabiraman et al. 2017). Even in the context of compromised or aberrant SC between either sister chromatids or homologs, we find that an assembled SC between DNA molecules is still capable of regulating the amount of crossing over. For example, the numbers we observed for COSA-1 foci formation in rec-8(ok978) null mutants is consistent with interference occurring along synapsed sister chromatid pairs. As there are 12 pairs of sister chromatids in the rec-8(ok978) null mutants and we only very rarely observe > 12 COSA-1 foci (0.8% of all nuclei), this result may reflect an imposed limitation of COSA-1 foci by the number of pairs of sister chromatids. Further, we see an increase in COSA-1 foci along stretches of SYP-1 if CO interference is perturbed in wild-type, him-3(e1256) partial loss-of-function, and rec-8(ok978) null strains. Given our previous study demonstrating a role for the SC in promoting CO interference (Libuda et al. 2013), in these cases of sister chromatid synapsis within the rec-8(ok978) null mutant, it is possible that these partially synapsed sister chromatid pairs are being recognized as a signal “module” or chromosome in which interference can act. Hence, interference, which is occurring along one set of sisters, may be transmitted along the other set of sister chromatids to which the pair is partially synapsed. Overall, these results further support the hypothesis that a fully assembled SC may serve as the scaffolding along which a signal may be propagated in C. elegans.
Acknowledgments
We thank A. Dernburg, A. Villeneuve, and M. Zetka for antibodies; the Caenorhabditis Genetics Center [funded by National Institutes of Health grant (NIH) P40 OD-010440], B. Meyer, and A. Villeneuve for strains; and A. Villeneuve, K. Hillers, and members of the Libuda laboratory, especially N. Kurhanewicz and E. Toraason, for comments on the manuscript. This work was supported by the NIH (grants R00 HD-076165 and R35 GM-128890 to D.E.L. and a Jane Coffin Childs Postdoctoral Fellowship to C.K.C. D.E.L. is also a Searle Scholar and recipient of a March of Dimes Basil O’Connor Starter Scholar award.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.9600878.
Communicating editor: F. Cole
Literature Cited
- Alpi A., Pasierbek P., Gartner A., and Loidl J., 2003. Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma 112: 6–16. 10.1007/s00412-003-0237-5 [DOI] [PubMed] [Google Scholar]
- Bhalla N., Wynne D. J., Jantsch V., and Dernburg A. F., 2008. ZHP-3 acts at crossovers to couple meiotic recombination with synaptonemal complex disassembly and bivalent formation in C. elegans. PLoS Genet. 4: e1000235 10.1371/journal.pgen.1000235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon C. K., and Hawley R. S., 2016. Regulating the construction and demolition of the synaptonemal complex. Nat. Struct. Mol. Biol. 23: 369–377. 10.1038/nsmb.3208 [DOI] [PubMed] [Google Scholar]
- Cahoon C. K., and Libuda D. E., 2019. Leagues of their own: sexually dimorphic features of meiotic prophase I. Chromosoma. DOI: 10.1007/s00412-019-00692-x. 10.1007/s00412-019-00692-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon C. K., Yu Z., Wang Y., Guo F., Unruh J. R. et al. , 2017. Superresolution expansion microscopy reveals the three-dimensional organization of the Drosophila synaptonemal complex. Proc. Natl. Acad. Sci. USA 114: E6857–E6866. 10.1073/pnas.1705623114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- M. P. Colaiacovo, MacQueen A. J., Martinez-Perez E., McDonald K., Adamo A. et al. , 2003. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell 5: 463–474. 10.1016/S1534-5807(03)00232-6 [DOI] [PubMed] [Google Scholar]
- Couteau F., Nabeshima K., Villeneuve A., and Zetka M., 2004. A component of C. elegans meiotic chromosome axes at the interface of homolog alignment, synapsis, nuclear reorganization, and recombination. Curr. Biol. 14: 585–592. 10.1016/j.cub.2004.03.033 [DOI] [PubMed] [Google Scholar]
- O. Crawley, Barroso C., Testori S., Ferrandiz N., Silva N. et al. , 2016. Cohesin-interacting protein WAPL-1 regulates meiotic chromosome structure and cohesion by antagonizing specific cohesin complexes. Elife 5: e10851 10.7554/eLife.10851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg A. F., McDonald K., Moulder G., Barstead R., Dresser M. et al. , 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94: 387–398. 10.1016/S0092-8674(00)81481-6 [DOI] [PubMed] [Google Scholar]
- Goodyer W., Kaitna S., Couteau F., Ward J. D., Boulton S. J. et al. , 2008. HTP-3 links DSB formation with homolog pairing and crossing over during C. elegans meiosis. Dev. Cell 14: 263–274. 10.1016/j.devcel.2007.11.016 [DOI] [PubMed] [Google Scholar]
- Harper N. C., Rillo R., Jover-Gil S., Assaf Z. J., Bhalla N. et al. , 2011. Pairing centers recruit a Polo-like kinase to orchestrate meiotic chromosome dynamics in C. elegans. Dev. Cell 21: 934–947. 10.1016/j.devcel.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M., Chin G. M., and Villeneuve A. M., 2007. C. elegans germ cells switch between distinct modes of double-strand break repair during meiotic prophase progression. PLoS Genet. 3: e191 10.1371/journal.pgen.0030191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M., Mlynarczyk-Evans S., and Villeneuve A. M., 2010. The synaptonemal complex shapes the crossover landscape through cooperative assembly, crossover promotion and crossover inhibition during Caenorhabditis elegans meiosis. Genetics 186: 45–58. 10.1534/genetics.110.115501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N., 2015. Meiotic recombination: the essence of heredity. Cold Spring Harb. Perspect. Biol. 7: a016618. 10.1101/cshperspect.a016618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- V. Jantsch, Pasierbek P., Mueller M. M., Schweizer D., Jantsch M. et al. , 2004. Targeted gene knockout reveals a role in meiotic recombination for ZHP-3, a Zip3-related protein in Caenorhabditis elegans. Mol. Cell Biol. 24: 7998–8006. 10.1128/MCB.24.18.7998-8006.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly K. O., Dernburg A. F., Stanfield G. M., and Villeneuve A. M., 2000. Caenorhabditis elegans MSH-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics 156: 617–630. https://www.genetics.org/content/156/2/617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Kostow N., and Dernburg A. F., 2015. The chromosome axis mediates feedback control of CHK-2 to ensure crossover formation in C. elegans. Dev. Cell 35: 247–261. 10.1016/j.devcel.2015.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler S., Wojcik M., Xu K., and Dernburg A. F., 2017. Superresolution microscopy reveals the three-dimensional organization of meiotic chromosome axes in intact Caenorhabditis elegans tissue. Proc. Natl. Acad. Sci. USA 114: E4734–E4743. 10.1073/pnas.1702312114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake C. M., and Hawley R. S., 2016. Becoming a crossover-competent DSB. Semin. Cell Dev. Biol. 54: 117–125. 10.1016/j.semcdb.2016.01.008 [DOI] [PubMed] [Google Scholar]
- D. E. Libuda, Uzawa S., Meyer B. J., and Villeneuve A. M., 2013. Meiotic chromosome structures constrain and respond to designation of crossover sites. Nature 502: 703–706. 10.1038/nature12577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machovina T. S., Mainpal R., Daryabeigi A., McGovern O., Paouneskou D. et al. , 2016. A surveillance system ensures crossover formation in C. elegans. Curr. Biol. 26: 2873–2884. 10.1016/j.cub.2016.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A. J., Colaiacovo M. P., McDonald K., and Villeneuve A. M., 2002. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 16: 2428–2442. 10.1101/gad.1011602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A. J., Phillips C. M., Bhalla N., Weiser P., Villeneuve A. M. et al. , 2005. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell 123: 1037–1050. 10.1016/j.cell.2005.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez E., Schvarzstein M., Barroso C., Lightfoot J., Dernburg A. F. et al. , 2008. Crossovers trigger a remodeling of meiotic chromosome axis composition that is linked to two-step loss of sister chromatid cohesion. Genes Dev. 22: 2886–2901. 10.1101/gad.1694108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlynarczyk-Evans S., and Villeneuve A. M., 2017. Time-course analysis of early meiotic prophase events informs mechanisms of homolog pairing and synapsis in Caenorhabditis elegans. Genetics 207: 103–114. 10.1534/genetics.117.204172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K., Villeneuve A. M., and Hillers K. J., 2004. Chromosome-wide regulation of meiotic crossover formation in Caenorhabditis elegans requires properly assembled chromosome axes. Genetics 168: 1275–1292. 10.1534/genetics.104.030700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K., Villeneuve A. M., and Colaiacovo M. P., 2005. Crossing over is coupled to late meiotic prophase bivalent differentiation through asymmetric disassembly of the SC. J. Cell Biol. 168: 683–689. 10.1083/jcb.200410144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadarajan S., Mohideen F., Tzur Y. B., Ferrandiz N., Crawley O. et al. , 2016. The MAP kinase pathway coordinates crossover designation with disassembly of synaptonemal complex proteins during meiosis. Elife 5: e12039 10.7554/eLife.12039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadarajan S., Lambert T. J., Altendorfer E., Gao J., Blower M. D. et al. , 2017. Polo-like kinase-dependent phosphorylation of the synaptonemal complex protein SYP-4 regulates double-strand break formation through a negative feedback loop. Elife 6: e23437. 10.7554/eLife.23437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H., Labella S., Silva N., Jantsch V., and Zetka M., 2018. C. elegans ZHP-4 is required at multiple distinct steps in the formation of crossovers and their transition to segregation competent chiasmata. PLoS Genet. 14: e1007776 10.1371/journal.pgen.1007776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasierbek P., Jantsch M., Melcher M., Schleiffer A., Schweizer D. et al. , 2001. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 15: 1349–1360. 10.1101/gad.192701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattabiraman D., Roelens B., Woglar A., and Villeneuve A. M., 2017. Meiotic recombination modulates the structure and dynamics of the synaptonemal complex during C. elegans meiosis. PLoS Genet. 13: e1006670 10.1371/journal.pgen.1006670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C. M., Meng X., Zhang L., Chretien J. H., Urnov F. D. et al. , 2009. Identification of chromosome sequence motifs that mediate meiotic pairing and synapsis in C. elegans. Nat. Cell Biol. 11: 934–942. 10.1038/ncb1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rog O., and Dernburg A. F., 2015. Direct visualization reveals kinetics of meiotic chromosome synapsis. Cell Rep. 10: 1639–1645. 10.1016/j.celrep.2015.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rog O., Köhler S., and Dernburg A. F., 2017. The synaptonemal complex has liquid crystalline properties and spatially regulates meiotic recombination factors. Elife 6: e21455. 10.7554/eLife.21455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild-Prüfert K., Saito T. T., Smolikov S., Gu Y., Hincapie M. et al. , 2011. Organization of the synaptonemal complex during meiosis in Caenorhabditis elegans. Genetics 189: 411–421. 10.1534/genetics.111.132431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvarzstein M., Pattabiraman D., Libuda D. E., Ramadugu A., Tam A. et al. , 2014. DNA helicase HIM-6/BLM both promotes MutSγ-dependent crossovers and antagonizes MutSγ-independent interhomolog associations during Caenorhabditis elegans meiosis. Genetics 198: 193–207. 10.1534/genetics.114.161513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson A. F., and Meyer B. J., 2014. Divergent kleisin subunits of cohesin specify mechanisms to tether and release meiotic chromosomes. Elife 3: e03467 10.7554/eLife.03467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson A. F., Ling L., van Zuylen V., and Meyer B. J., 2009. The axial element protein HTP-3 promotes cohesin loading and meiotic axis assembly in C. elegans to implement the meiotic program of chromosome segregation. Genes Dev. 23: 1763–1778. 10.1101/gad.1808809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolikov S., Eizinger A., Hurlburt A., Rogers E., Villeneuve A. M. et al. , 2007a Synapsis-defective mutants reveal a correlation between chromosome conformation and the mode of double-strand break repair during Caenorhabditis elegans meiosis. Genetics 176: 2027–2033. 10.1534/genetics.107.076968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolikov S., Eizinger A., Schild-Prufert K., Hurlburt A., McDonald K. et al. , 2007b SYP-3 restricts synaptonemal complex assembly to bridge paired chromosome axes during meiosis in Caenorhabditis elegans. Genetics 176: 2015–2025. 10.1534/genetics.107.072413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolikov S., Schild-Prüfert K., and Colaiacovo M. P., 2009. A yeast two-hybrid screen for SYP-3 interactors identifies SYP-4, a component required for synaptonemal complex assembly and chiasma formation in Caenorhabditis elegans meiosis. PLoS Genet. 5: e1000669 10.1371/journal.pgen.1000669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkel-Meiman K., Cheng S. Y., Parziale M., Morehouse S. J., Feil A. et al. , 2019. Crossover recombination and synapsis are linked by adjacent regions within the N terminus of the Zip1 synaptonemal complex protein. PLoS Genet. 15: e1008201 10.1371/journal.pgen.1008201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woglar A., and Villeneuve A. M., 2018. Dynamic architecture of DNA repair complexes and the synaptonemal complex at sites of meiotic recombination. Cell 173: 1678–1691.e16. 10.1016/j.cell.2018.03.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Beasley M. D., Warren W. D., van der Horst G. T., and McKay M. J., 2005. Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev. Cell 8: 949–961. 10.1016/j.devcel.2005.03.018 [DOI] [PubMed] [Google Scholar]
- Yokoo R., Zawadzki K. A., Nabeshima K., Drake M., Arur S. et al. , 2012. COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell 149: 75–87. 10.1016/j.cell.2012.01.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetka M. C., Kawasaki I., Strome S., and Muller F., 1999. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev. 13: 2258–2270. 10.1101/gad.13.17.2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- L. Zhang, Kohler S., Rillo-Bohn R., and Dernburg A. F., 2018. A compartmentalized signaling network mediates crossover control in meiosis. Elife 7: e30789 10.7554/eLife.30789.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All strains are available upon request. Supplemental Material, Figure S1A shows the localization of COSA-1, ZHP-3, and MSH-5 in syp-1 and syp-3 mutants. Figure S1B has unadjusted images of GFP::COSA-1 localization in wild-type, syp-2, and syp-3 mutants. Figure S2A shows the localization of COSA-1 and MSH-5 in wild-type, him-3, htp-3, and rec-8 mutants. Figure S2B shows the localization of COSA-1 and ZHP-3 in wild-type worms and the rec-8 mutant. Figure S3 shows localization of COSA-1 and SYP-1 in syp-1 partial depletions combined with wild-type, him-3, and rec-8 mutants. Figure S4 shows representative images and the quantification of DAPI-staining bodies at diakinesis from wild-type, rec-8, cosa-1, rec-8; cosa-1, and rec-8; syp-2 mutants. Figure S5 shows the localization of RAD-51 in wild-type, syp-2 (ok307), and syp-2; rec-8 mutants at both late pachytene (A) and diakinesis (B). Table S1 has the number of GFP::COSA-1 foci per nucleus in late pachytene from wild-type, syp-1, syp-2, syp-3, rec-8, rec-8;syp-2, and him-3 mutants. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.9600878.