Abstract
A subset of cancers rely on telomerase-independent mechanisms to maintain their chromosome ends. The predominant “alternative lengthening of telomeres” pathway appears dependent on homology-directed repair (HDR) to maintain telomeric DNA. However, the molecular changes needed for cells to productively engage in telomeric HDR are poorly understood. To gain new insights into this transition, we monitored the state of telomeres during serial culture of fission yeast (Schizosaccharomyces pombe) lacking the telomerase recruitment factor Ccq1. Rad52 is loaded onto critically short telomeres shortly after germination despite continued telomere erosion, suggesting that recruitment of recombination factors is not sufficient to maintain telomeres in the absence of telomerase function. Instead, survivor formation coincides with the derepression of telomeric repeat-containing RNA (TERRA). In this context, degradation of TERRA associated with the telomere in the form of R-loops drives a severe growth crisis, ultimately leading to a novel type of survivor with linear chromosomes and altered cytological telomere characteristics, including the loss of the shelterin component Rap1 (but not the TRF1/TRF2 ortholog, Taz1) from the telomere. We demonstrate that deletion of Rap1 is protective in this context, preventing the growth crisis that is otherwise triggered by degradation of telomeric R-loops in survivors with linear chromosomes. These findings suggest that upregulation of telomere-engaged TERRA, or altered recruitment of shelterin components, can support telomerase-independent telomere maintenance.
Keywords: telomere, recombination, RNA–DNA hybrid, TERRA, shelterin, ALT pathway
TELOMERES are specialized nucleoprotein structures that protect the ends of linear eukaryotic chromosomes from degradation, aberrant recombination, and end-to-end fusions (de Lange 2009; Dehé and Cooper 2010), while also supporting the formation of subtelomeric heterochromatin (Bühler and Gasser 2009). Telomeric DNA consists of tandem double-stranded G-rich repeats and a single-stranded 3′ overhang (Blackburn 1991). The 3′ ssDNA overhang is sequestered by engaging in higher-order structures, in part due to the binding of sequence-specific telomeric binding proteins (Blackburn 2001). In at least some organisms, the telomere binding protein TRF2 drives formation of a structure called “t-loop”; TRF2 is a component of the shelterin complex that includes the additional factors TRF1, RAP1, TIN2, TPP1, and POT1 in mammalian cells (de Lange 2005). The fission yeast (Schizosaccharomyces pombe) genome encodes several classes of telomere-binding proteins, many of which are orthologous to mammalian shelterin components. For example, Taz1 (related to mammalian TRF1/TRF2) recognizes dsDNA telomere repeats (Cooper et al. 1997), while the conserved Pot1 specifically recognizes the 3′ ssDNA G-rich overhang (Baumann and Cech 2001). These modules are bridged by the shelterin components Rap1 (Chikashige and Hiraoka 2001; Kanoh and Ishikawa 2001), Poz1, and Tpz1 (Miyoshi et al. 2008); Rap1 constitutes the binding interface with Taz1 (Chikashige and Hiraoka 2001; Kanoh and Ishikawa 2001). Cells lacking Taz1 undergo telomere lengthening (Cooper et al. 1997), due at least in part to increased telomerase recruitment (Moser et al. 2011; Dehé et al. 2012). In contrast, loss of Pot1 drives rapid telomere loss (Baumann and Cech 2001), likely due to extensive 5′ end resection (Pitt and Cooper 2010).
In fission yeast, telomeric DNA is composed of ∼300 bp of degenerate GG1-5TTAC[A] repeats (Cooper and Hiraoka 2006). Telomere length is influenced by numerous factors, including mechanisms regulating telomere replication (Martinez and Blasco 2015; Maestroni et al. 2017b), telomerase recruitment and activation (Nandakumar and Cech 2013; Armstrong and Tomita 2017), and the protection of the chromosome end from nucleases (Shore and Bianchi 2009; Longhese et al. 2010). The primary mechanism to counteract telomere loss is the action of the reverse transcriptase telomerase. The fission yeast telomerase is comprised of the conserved catalytic subunit Trt1 (Nakamura et al. 1997), the RNA component TER1 that encodes the template for addition of the degenerate telomeric repeat to support telomere elongation (Leonardi et al. 2008; Webb and Zakian 2008), the conserved subunit Est1 (Beernink et al. 2003), and accessory factors such as the Lsm proteins (Tang et al. 2012) and the La-like protein LARP7/Pof8 (Collopy et al. 2018; Mennie et al. 2018; Páez-Moscoso et al. 2018). In fission yeast, the factor Ccq1 links telomerase recruitment to the 3′ telomeric overhang through Tpz1 and Pot1 to support telomere maintenance (Miyoshi et al. 2008; Tomita and Cooper 2008).
Somatic mammalian cells do not display telomerase activity at sustained levels (Kim et al. 1994; Wright et al. 1996), due primarily to transcriptional silencing of the catalytic subunit TERT (Meyerson et al. 1997; Nakamura et al. 1997), and, therefore, can undergo a limited number of cell divisions before critically short telomeres drive cell cycle arrest and senescence or apoptosis. Thus, cancer cells typically harbor mutations that lead to a gain of telomerase activity (Kim et al. 1994; Shay et al. 2012) likely through reactivation of TERT transcription (Heidenreich and Kumar 2017). However, in ∼15% of cancer cells, the alternative lengthening of telomeres (ALT) pathway—a mechanism first discovered in budding yeast (Lundblad and Blackburn 1993)—can instead support telomere maintenance through homology-directed repair (HDR) pathways (Cesare and Reddel 2010; Apte and Cooper 2017). In fission yeast lacking the telomerase-recruitment factor Ccq1, telomeres gradually shorten with increasing passage number, eventually leading to a growth crisis; the “survivors” that emerge from this crisis can utilize recombination to maintain their telomeres (Miyoshi et al. 2008; Tomita and Cooper 2008). Fission yeast cells can also survive complete telomere loss by circularization of each of their three chromosomes (Nakamura et al. 1998). The generation of these “circular survivors” can occur through multiple pathways, including single-strand annealing (Wang and Baumann 2008).
Despite the heterochromatic nature of chromosome ends, telomeres are transcribed into a group of long noncoding RNA species (Azzalin et al. 2007; Luke et al. 2008; Schoeftner and Blasco 2008; Bah and Azzalin 2012; Bah et al. 2012). The transcription of TElomeric Repeat-containing RNA (TERRA) is evolutionarily conserved, and has been suggested to function in telomere length regulation, heterochromatin establishment, and telomeric HR (Bah and Azzalin 2012; Azzalin and Lingner 2015; Rippe and Luke 2015). TERRA transcription is largely RNA Pol II-dependent, is initiated at subtelomeric region(s) and proceeds toward the 3′ end of the telomeric tract (Bah et al. 2012). The majority of TERRA molecules are <500 bp in fission yeast, whereas in mammalian cells TERRA can be as long as several kilobases (Azzalin et al. 2007; Schoeftner and Blasco 2008; Feuerhahn et al. 2010; Bah and Azzalin 2012; Greenwood and Cooper 2012). The steady-state level of TERRA is negatively regulated by Taz1 and Rap1 in fission yeast, being very low in wild-type cells (Bah et al. 2012; Greenwood and Cooper 2012).
Studies in human cells, and both budding and fission yeast, demonstrate that TERRA molecules localize primarily within the nucleus, and partially colocalize with telomeres (Schoeftner and Blasco 2008; Bah et al. 2012; Cusanelli et al. 2013). A fraction of TERRA molecules is 3′ polyadenylated (Schoeftner and Blasco 2008; Bah et al. 2012; Porro et al. 2014). While the majority of poly(A)-TERRA associates with telomeric DNA, poly(A)+ TERRA is instead largely released from telomeres and resides in the nucleoplasm (Porro et al. 2014; Moravec et al. 2016). We recently reported that telomere shortening in fission yeast leads to derepression of TERRA, similar to observations in budding yeast (Cusanelli et al. 2013; Moravec et al. 2016). This telomere-shortening-induced TERRA is primarily polyadenylated and physically interacts with telomerase, thus promoting its recruitment to the telomere (Moravec et al. 2016). To date, the impact of telomere-associated TERRA on telomere maintenance by recombination in cycling cells has gone largely uninvestigated in fission yeast, although recent evidence suggests that TERRA upregulation in quiescent fission yeast correlates with increased telomere rearrangements driven by recombination (Maestroni et al. 2017a).
Telomere-associated TERRA can form RNA–DNA hybrids at the telomere, resulting in a triple-stranded structure called a “telomeric R-loop” (telR-loop); telR-loops are antagonized by the activity of RNase H, which specifically degrades RNA engaged in such hybrids (Balk et al. 2013, 2014; Arora et al. 2014). Recent evidence indicates that telR-loops may actively engage in HDR in telomerase-negative human and yeast cells. For example, depletion of RNase H1 (alone or in combination with RNase H2) results in accumulated RNA–DNA hybrids at telomeres and greater telomere recombination in cis, while RNase H overexpression leads to decreased telomere recombination in presenescent cells and compromised telomere maintenance in ALT cells (Balk et al. 2013, 2014; Arora et al. 2014).
In order to test the consequences of TERRA upregulation, several studies have taken advantage of engineered, inducible promoters at a TERRA transcriptional start site to construct a transcriptionally inducible telomere (tiTEL) (Maicher et al. 2012; Pfeiffer and Lingner 2012; Arora et al. 2014; Moravec et al. 2016), which can increase telR-loops (Arora et al. 2014). However, the strong transcription driven by a heterologous promoter may also disrupt the heterochromatic state of telomeres and influence the maturation, turnover, and function of natural TERRA molecules. Despite the growing evidence linking TERRA to regulation of telomeric recombination, it is still not clear how endogenous TERRA (and natively formed telR-loops) might be altered to promote telomere recombination and formation of telomerase-deficient survivors in an unperturbed system.
Here, we study the process by which cells adapt to maintain their telomeres by recombination using a fission yeast model in which Ccq1 is deleted. We chose this genetic system as these cells undergo telomere shortening due to defective telomerase recruitment, but are highly proficient at forming linear survivors (Miyoshi et al. 2008; Tomita and Cooper 2008). Given that Ccq1 also supports repression of subtelomeric heterochromatin involving the SHREC (Snf2/HDAC-containing repressor complex) and CLRC (made up of the H3K9 methylase, Clr4, and an E3 ubiquitin ligase complex composed of Cul4, Rik1, Raf1, and Raf2) complexes (Sugiyama et al. 2007; Wang et al. 2016; Armstrong et al. 2018), this ability could be linked to the dual role of Ccq1 in telomerase recruitment and subtelomeric silencing. Consistent with this hypothesis, we find that TERRA levels are higher in ccq1Δ cells at germination, but dramatically rise further during the emergence of linear, recombination-based survivors. TERRA engages in telR-loops at the telomere(s) and promotes a transition to productive telomere maintenance. Degradation of RNA–DNA hybrids by overexpression of RNase H drives these linear survivors back into a growth crisis, from which an alternative type of linear survivor arises with distinct cytological features. Surprisingly, the telomeres in these RNase H-resistant survivors no longer recruit Rap1, despite enhanced Taz1 binding. Importantly, deletion of Rap1 insulates linear survivors from the crisis induced by RNA–DNA hybrid degradation, suggesting that this altered telomere composition functionally promotes telomere maintenance when telomere-associated TERRA is degraded.
Materials and Methods
S. pombe strain generation and culture conditions
S. pombe strains are described in Supplemental Material, Table S1. Standard manipulations and cell culturing were carried out as described (Moreno et al. 1991). C-terminal GFP or mCherry tagging was performed with the pFa6a-GFP-kanMX6 or pFa6a-mCherry-kanMX6 cassette (Bähler et al. 1998; Snaith et al. 2005). pFa6a-natMX6-nmt41-HA was used to tag Rnh201 at the N-terminus as established (Bähler et al. 1998). In the strain with “transcriptionally inducible” telomere (tiTEL), the thiamine repressible nmt1+ gene promoter (Pnmt1) was inserted 91 bp upstream of the endogenous TERRA transcription start site at Chr I-R (Moravec et al. 2016). A C-terminal GFP cassette was further integrated after the Taz1 ORF in this tiTEL strain (Strain MKSP1722). Fresh presenescent cells for growth assays of ccq1Δ, trt1Δ, rad55Δ, and combinations or variants of these alleles, were obtained by dissection of sporulated heterozygous diploid strains. In the case of h+/h+ diploids, strains were first transformed with plasmid pON177 (Styrkársdottir et al. 1993) to provide the mat-M genes. All strains generated by cassette integration were confirmed by PCR. After genetic crosses, progeny were tested by the segregation of markers, PCR, or presence of the relevant fluorescent protein fusion, as appropriate.
S. pombe were grown at 30°, in yeast extract medium supplemented with amino acids (YE5S) or Edinburgh minimal medium with amino acids (EMM5S). To suppress the nmt1+ or nmt41+ gene promoter (Maundrell 1990), 5 μg/ml thiamine (Sigma-Aldrich) was added to EMM5S medium.
Serial culturing
Single colonies formed from fresh dissections were inoculated in liquid medium. Cell densities were measured 24 hr later using the Moxi-Z mini cell counter (ORFLO). Approximately 28 generations were estimated from a single, germinated cell growing on solid medium for 20 generations to generate a colony, followed by a single overnight culture. Cell cultures were measured for their cell densities and diluted to 5 × 105 cells/ml with fresh medium every 24 hr. At each indicated time point, cells were collected for further analysis or subjected to imaging as described. The increase in generations each day were calculated as log2 [(cell density)/(5 × 105)]. “Survivors” indicate that the culture has achieved a consistent growth rate, typically ∼200 generations (>20 days). Growth curves were plotted using GraphPad Prism 7.01.
Live cell imaging and analysis
Cells for imaging were exponentially grown in YE5S or EMM5S media supplemented with adenine hemi sulfate (0.25 mg/ml). Cells in liquid culture were concentrated and applied to glass slides bearing agarose pads (1.4% agarose in EMM). Samples were sealed under a glass coverslip with VALAP (equal parts by weight: Vaseline, lanolin, and paraffin). Images were acquired on an Applied Precision (now GE Healthcare) Deltavision high performance wide field microscope with solid-state illumination and a Photometrics Evolve 512 EMCCD camera. For imaging of Taz1-GFP in Figure 4A and Figure S3, A–C, each field was captured as a 12 section Z-stack of sample thickness 4.80 μm (optical sectioning every 0.40 μm; this is sufficient to capture the entire nuclear volume to ensure all telomeric foci are visualized); EMCCD Gain = 100; GFP at 10% power, 100 ms exposure. For telomere volume reconstruction (Figure 4D), each field was captured as a 25 section Z-stack of sample thickness 5.0 μm (optical sectioning spacing every 0.2 μm); EMCCD Gain = 100; GFP at 10% power, 50 ms exposure. For cells expressing Rap1-GFP (Figure 5, A and B and Figure S3, C and D) each field was captured as a 24 section Z-stack of sample thickness 4.80 μm (optical sectioning every 0.2 μm; EMCCD Gain = 200; GFP at 10% power, 25 ms exposure. For visualization of Taz1-GFP and Rad52-mCherry (Figure 1, B and C) each field was captured as a 12 section Z-stack of sample thickness 4.8 μm (optical sectioning spacing every 0.4 μm); EMCCD Gain = 100; GFP at 10% power, 50 ms exposure; mCherry at 10% power, 500 ms exposure. Only live, nonmitotic cells that had completed cytokinesis were considered for Taz1-GFP and cell length analysis. In most cases, we display maximum intensity projections, as described in the figure legends.
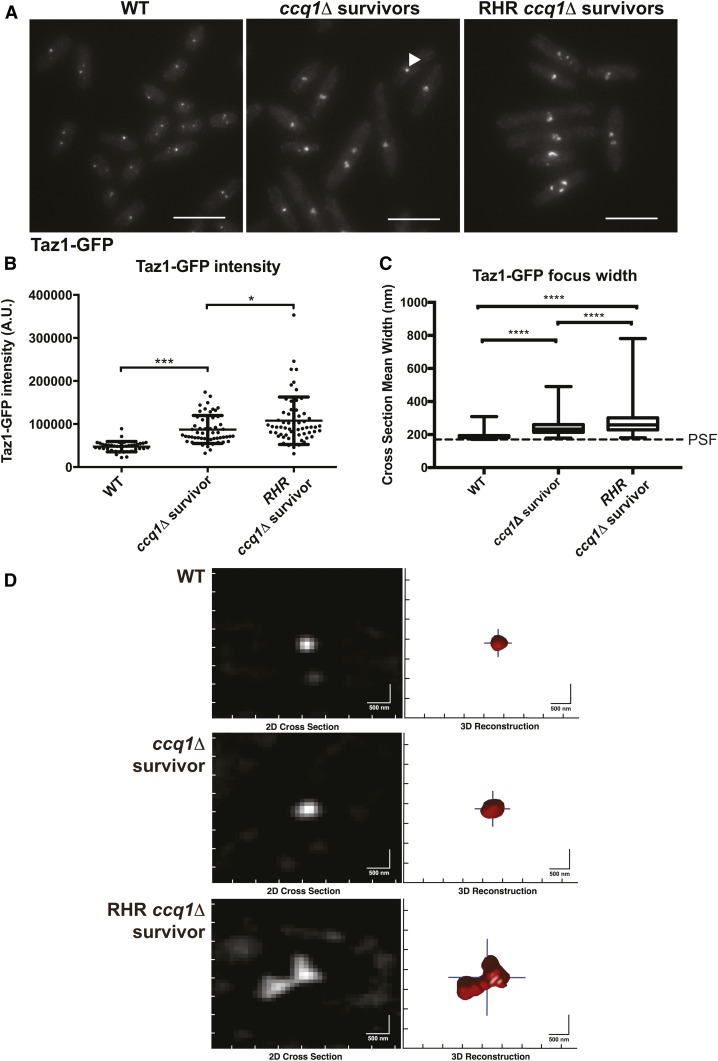
Figure 4.
ccq1Δ survivors that form after constitutive Rnh201 overexpression display increased Taz1-GFP recruitment and have telomeres that appear larger and declustered. (A) Fluorescence imaging of Taz1-GFP reveals changes in telomere appearance in ccq1Δ Taz1-GFP survivors and RHR ccq1Δ Taz1-GFP survivor cells. Maximum intensity projections of fluorescence micrographs of WT Taz1-GFP, ccq1Δ Taz1-GFP survivors or RHR ccq1Δ Taz1-GFP survivor cells obtained as described in the Materials and Methods. Bar, 10 μm. (B) The intensity of focal Taz1-GFP increases in ccq1Δ survivors, and is further enhanced in RHR ccq1Δ survivors. Measurement is the sum intensity of all Taz1-GFP foci. n > 50 cells for each genotype. * P < 0.05, *** P < 0.001 by Student’s t-test corrected for multiple comparisons. (C) Taz1-GFP foci were reconstructed using an algorithm previously developed by our group (see Materials and Methods) on Z-stacks obtained as in (A). The mean cross-section width was determined for each focus; for foci with a cross-section smaller then the point spread function (PSF), the focus will appear with the dimensions of the PSF. Reconstructed WT Taz1-GFP foci (n = 352) are nearly all the size of the PSF (170 nm, dashed line) or smaller. While ccq1Δ survivors display slightly larger Taz1-GFP foci (n = 252), the largest Taz1-GFP foci are seen in the RHR ccq1Δ survivors (n = 132). The box represents the 25th and 75th quartiles, the mean is the black line, and the whiskers include all data. **** P < 0.0001 by Student’s t-test corrected for multiple comparisons. (D) Representative examples of reconstructed Taz1-GFP foci in the three genetic backgrounds. The raw image is on the left, and the telomeric focus is reconstructed on the right. Bar, 500 nm.
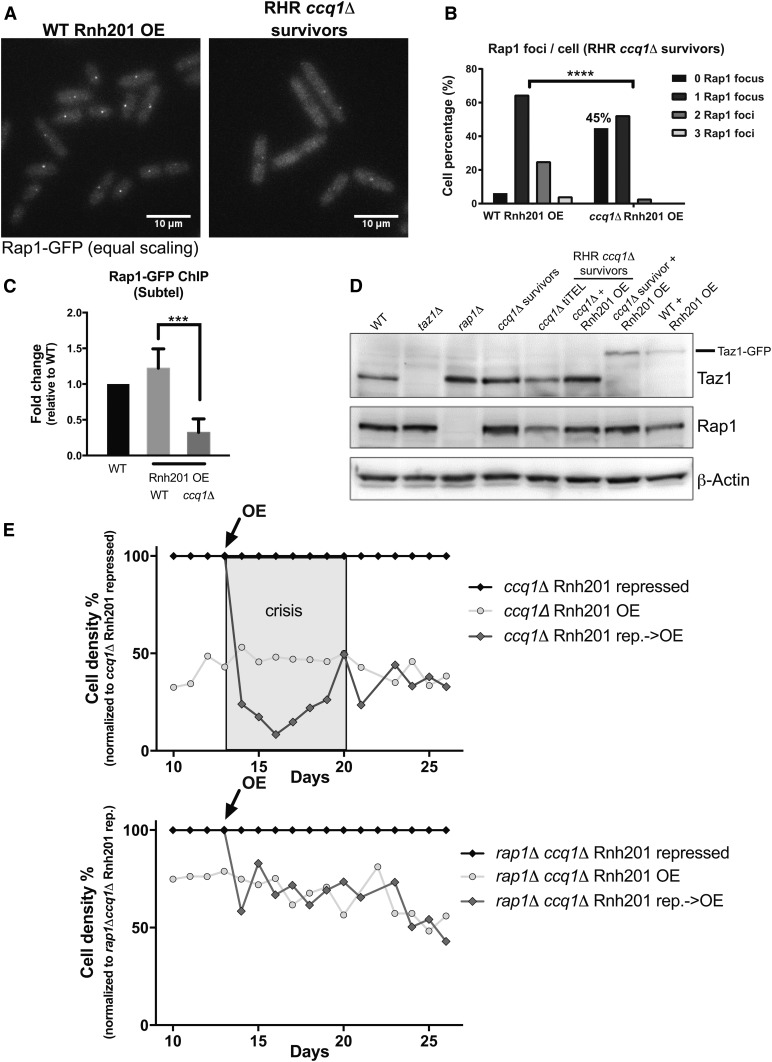
Figure 5.
Loss of Rap1 promotes formation of RHR ccq1Δ survivors in the context of constitutive Rnh201 overexpression. (A) Fluorescence imaging of Rap1-GFP reveals decreased recruitment to the telomere upon long-term induction of Rnh201 in ccq1Δ survivor cells. Fluorescence micrographs of WT or ccq1Δ Rap1-GFP survivor cells after long-term Rnh201 overexpression (RHR ccq1Δ survivors). Maximum intensity projections obtained as described in the Materials and Methods. Bar, 10 μm (B) Quantification of the number of Rap1-GFP foci after long-term induction of Rnh201 in the WT and ccq1Δ backgrounds. At least 200 cells were analyzed. P < 0.0001 by K-S test. (C) The loss of Rap1 association with telomeres is also observed upon Rnh201 overexpression by chromatin immunoprecipitation-qPCR. Normalized to WT cells and plotted as the mean with SD from three biological replicates. *** P = 0.0002 by Student’s t-test. (D) Taz1 levels increase in the absence of Rap1 and in all varieties of ccq1Δ survivors while Rap1 levels are unaffected. Whole cell extracts were separated on SDS-PAGE, blotted, and probed with antibodies to Taz1, Rap1, or β-actin (as a loading control). (E) Deletion of Rap1 prevents the growth crisis upon Rnh201 overexpression in ccq1Δ survivors. Serial culturing of either ccq1Δ (top) or rap1Δccq1Δ (bottom) Pnmt41-rnh201+ cells under constitutive repression (black), overexpression (light gray circles), or induced induction of overexpression (OE at day 13, black arrow; dark gray diamonds). Cell density is normalized to the Rnh201-repressed condition. The gray box indicates the period of telomere crisis upon overexpression of Rnh201 in ccq1Δ cells that is absent in rap1Δccq1Δ cells.
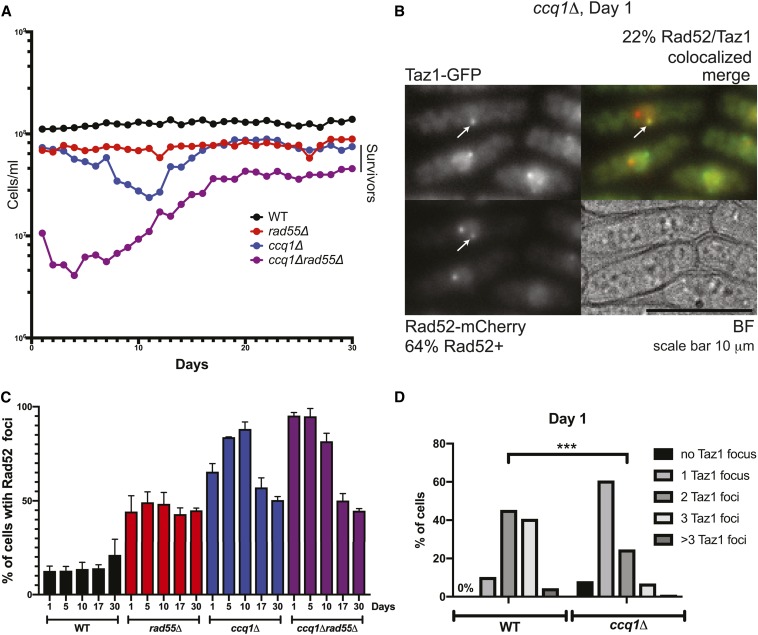
Figure 1.
Recombination factors are loaded at telomeres shortly after germination. (A) Serial culturing of WT, ccq1Δ, rad55Δ, and ccq1Δrad55Δ cells. Plot of cell density vs. number of days in culture since germination, diluted once every 24 hr. (B) Telomeres are recognized as DNA damage shortly after germination in ccq1Δ cultures. Representative micrographs of ccq1Δ cells expressing Taz1-GFP and Rad52-mCherry immediately after germination. The percent of cells with the indicated phenotype are noted. BF, brightfield; Bar, 10 μm (C) The percent of cells with Rad52-GFP foci mirrors growth rate, with ccq1Δrad55Δ cells displaying high levels of Rad52 foci at germination. WT, ccq1Δ, rad55Δ, and ccq1Δrad55Δ cells expressing Rad52-GFP were imaged on the indicated days of the growth assay in (A). n > 100 cells for each strain and timepoint from at least three biological replicates. Images displayed are maximum intensity projections of Z-stacks obtained as described in the Materials and Methods. (D) The number of Taz1-GFP foci is less in ccq1Δ cells than WT cells on Day 1. P < 0.001 by K-S test.
Images were analyzed in ImageJ (Fiji; Schindelin et al. 2012). To calculate the number of Taz1-GFP foci per cell, or to examine the colocalization of Taz1-GFP and Rad52-mCherry, maximum Z-projections were generated. For quantitative measurement of Taz1-GFP recruitment to telomeres, the intensity of each Taz1-GFP focus was measured from mean intensity projections as the integrated pixel density of the visibly occupied area. The background GFP signal was removed from measurement of each focus by subtracting the integrated density of an empty area of the same size as the focus within the nucleoplasm of the same cell. The intensities of all foci were then summed to give a single measurement per cell.
Real-time PCR to measure TERRA levels
RT-PCR protocols were adapted from our previous work (Bah et al. 2012). Total RNA was extracted using MasterPure yeast RNA purification kit (Epicentre). To completely eliminate DNA contamination, RNA was treated three times with RNase-free DNase I (provided with the kit). cDNA was generated using SuperScript III (Invitrogen). Each 20 μl reverse transcription reaction contained 5 μg total RNA, 1 μl 10 μM oligo for total TERRA (Sp C-rich telo: 5′-TGTAACCGTGTAACCACGTAACCTTGTAACCC-3′) and 1 μl 1 μM reverse control oligo for actin [Sp ACT1(2): 5′-CATCTGTTGGAAAGTAGAAAGAGA AGCAAG-3′], or 2 μl 40 μM oligo(dT) [in the case of poly(A)+ TERRA], 1 μl 10 mM dNTP, 4 μl 5× First-Strand Buffer, 2 μl 0.1 M DTT, and 1 μl SuperScript III RT enzyme. The reaction was incubated at 50° for 1 hr and the remaining RNA was degraded by RNase H (NEB) and RNase A (Sigma-Aldrich) by incubating at 37° for 20 min. Real-time PCR was performed on C1000 Touch Thermal Cycler (Bio-Rad), using FastStart Universal SYBR Green Master (Rox) (Roche). For TERRA PCR amplification, each 20 μl reaction contained 2 μl of cDNA, 1 μl of each 10 μM primer (SpRACE290-320_4: 5′-GGGCCCAATAGTGGGGGCATTGTATTTGTG-3′; Sp-subtelo_425: 5′-GAAGTTCACTCAGTCATAATTAATTGGGTAACGGAG-3′). For actin PCR amplification, each 20 μl reaction contained 2 μl of 1:10 diluted cDNA, 1 μl of each 10 μM primer (SpACT1FOR2: 5′-CCGGTATTCATGAGGCTACT-3′; SpACT1REV2: 5′-GGAGGAGCAACAATCTTGAC-3′).
qPCR steps: one cycle of 95° for 5 min; 45 cycles of 95° for 10 sec, and 60° for 30 sec, followed by a melting curve. Relative TERRA levels were calculated for each sample after normalization against actin using the ΔΔCt method. Ct values for each reaction were determined using the software Bio-Rad CFX Manager 2.1. Duplicate Ct values were averaged in each run. Each RNA sample was reverse-transcribed into cDNA twice, and each cDNA sample was analyzed by RT-PCR at least three times. Ct values of the same sample were averaged and plotted in GraphPad Prism 6. No RT and no template reactions were always included to control for DNA contamination.
Southern blots
To measure telomere length, experiments were performed as described (Bennett et al. 2016). Briefly, genomic DNA (25 μg) was digested with EcoRI-HF (NEB) at 37° overnight, electrophoresed on a 1% agarose gel, denatured and transferred to Zeta-Probe GT membrane (Bio-Rad), then hybridized with a telomere-DIG probe in ExpressHyb Solution (Clontech). After hybridization, the membrane was washed and incubated with anti-DIG-AP conjugate (the same kit, 1:1000 dilution). The blot was then treated with SuperSignal West Femto Maximum Sensitivity Kit (ThermoFisher/Pierce), and imaged on VerSaDoc imaging system (Bio-Rad) using the software Quantity One 4.6.9.
Pulsed‐field gel electrophoresis
Experiments were performed as in Nakamura et al. (1998), Ferreira and Cooper (2001) with minor modifications, as described in Lorenzi et al. (2015). Briefly, cells were washed in TSE (10 mM Tris–HCl (pH 7.5), 0.9 M d‐Sorbitol, 45 mM EDTA) and resuspended at 5 × 107 cells with 10 mg/ml zymolyase‐20T (Seikagaku Biobusiness) and 50 mg/ml lysing enzyme (Sigma). One volume of 2% low melt agarose (Bio‐Rad) in TSE was added and the suspension dispensed into plug molds. Plugs were incubated in TSE containing 5 mg/ml zymolyase‐20T and 25 mg/ml lysing enzyme at 37° for 1 hr. Plugs were incubated for 90 min at 50° in 0.25 M EDTA, 50 mM Tris–HCl (pH 7.5), 1% SDS and then for 3 days at 50° in 0.5 M EDTA, 10 mM Tris–HCl (pH 9.5), 1% lauroyl sarcosine, 1 mg/ml proteinase K. Plugs were washed in 10 mM Tris–HCl (pH 7.5), 10 mM EDTA and incubated with 0.04 mg/ml PMSF for 1 hr at 50°. Plugs were washed again in 10 mM Tris–HCl (pH 7.5), 10 mM EDTA, and then digested with 100 U of NotI (New England Biolabs) for 24 hr at 37°. Plugs were equilibrated in 0.5 × TAE, loaded on a 1% agarose gel, and ran in a CHEF‐DR III system (Bio‐Rad) for 16 hr at 14° using the following program: 60–120 sec switch time, 120° angle, 6 V/cm electric field. After running, gels were processed as for Southern blot and hybridized with a mix of L, I, M, and C probes recognizing unique sequences from the most terminal NotI fragments of the left arm of chromosome I, right arm of chromosome I, left arm of chromosome II, and right arm of chromosome II, respectively. For Southern blotting, DNA was denatured in gel and transferred to a positively charged nylon membrane (GE Osmonics), and processed as described above.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) of Rap1-GFP-expressing cells was carried out as described previously (Lorenzi et al. 2015) with the following modifications: 75 OD of yeast cells at an OD600 of 0.6–1.0 were cross-linked in 1% formaldehyde for 30 min prior to quenching with glycine. Cross-linked material was washed in ice-cold PBS and resuspended in 1 ml lysis buffer [50 mM HEPES-KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, protease inhibitor cocktail (Roche)]. Cells were transferred to 1.5-ml screw-cap tubes, and subjected to mechanical lysis with silica beads (0.5 mm diameter, BioSpec) using the Mini-BeadBeater-16 instrument (BioSpec), shaking 1 min 30 sec, six to eight times at 3-min intervals. Lysates were centrifuged for 30 min at 16,000 × g, and pellets were resuspended in 500 μl lysis buffer and subjected to sonication using the S-4000 sonicator (Misonix) for 15 min (Amplitude 4, 5 sec pulse with 15 sec cool-down intervals). After centrifugation at 10,000 × g for 15 min, the supernatant containing DNA fragments was collected. Lysis buffer with protease inhibitor cocktail (Roche) was added to adjust the volume to ∼1.4 ml, with 120 μl taken as the input sample. Immunoprecipitations were performed on a rotating wheel at 4° overnight with GFP-Trap beads (Chromotek). The beads were then washed four times with lysis buffer, lysis buffer plus 500 mM NaCl, wash buffer (10 mM Tris-HCl pH 7.5, 0.25 M LiCl, 0.5% NP40, 0.5% sodium deoxycholate), and lysis buffer, respectively. Immunoprecipitated chromatin was eluted by adding 120 μl elution buffer (1% SDS, 100 mM NaHCO3, 40 μg/ml RNase A) and incubated at 37° for 1 hr. The input sample was treated in parallel by adding 6 μl of 20% SDS, 11 μl of 1 M NaHCO3, and RNase A. Cross-links were reversed at 65° overnight (16 hr) and DNA was purified using a PCR purification kit (Qiagen). DNA content was quantified by RT-PCR using iTaq Universal SYBR Green Supermix (Bio-Rad) on a C1000 Thermal Cycler instrument (Bio-Rad) with primers: subtel-for (5′-TATTTCTTTATTCAACTTACCGCACTTC-3′) and subtel-rev (5′-CAGTAGTGCAGTGTATTATGATAATTAAAATGG-3′).
Western blots
Experiments were carried out as described (Lorenzi et al. 2015). Cells were collected at exponential phase and samples were prepared by TCA precipitation. Antibodies were as follows: a rabbit polyclonal anti-Rap1 and a rabbit polyclonal anti-Taz1 antibody (kind gifts from J. Kanoh and J. P. Cooper, respectively); and a mouse monoclonal anti-beta actin antibody (mAbcam 8224) used as loading control.
DNA immunoprecipitation
Log phase cells were harvested, washed with water, and flash frozen in liquid nitrogen. The frozen pellet was resuspended in 1 ml of RA1 buffer (Macherey-Nagel) supplemented with 1% β-mercaptoethanol. The cell suspension was mixed with 300 μl of phenol:chloroform:isoamyl alcohol (25:24:1 saturated with 10 mM Tris-Cl pH 8.0, 1 mM EDTA), and 100 μl of acid-washed glass beads. Cells were lysed by mechanical shaking using a cell disruptor (Disruptor Gene) for 3 min using the 30 sec on/off mode at power 4. Cell extracts were centrifuged (13,000 × g, 20 min, 4°). Pellets were washed with 70% ethanol, air-dried, resuspended in 200 μl of Tris-EDTA, and sonicated with a Bioruptor (Diagenode) to obtain 100–500 bp long fragments. Sheared nucleic acids (10 μg) were diluted in 1 ml of IP buffer (0.1% SDS, 1% Triton X‐100, 10 mM HEPES pH 7.7, 0.1% sodium deoxycholate, 275 mM NaCl) and incubated overnight on a rotating wheel at 4° in presence of 1 μg of S9.6 antibody (Kerafast) and 20 μl of protein G sepharose beads (GE Healthcare) blocked with Escherichia coli DNA and bovine serum albumin. Beads were washed four times with IP buffer and bound nucleic acids were isolated using the Wizard SV gel and PCR clean-up kit (Promega). Collected DNA fragments were quantified using RT-PCR as described above with the following primers: oF1 (5′-GAAGTTCACTCAGTCATAATTAATTGGGTAACGGAG-3′) and oR1 (5′-GGGCCCAATAGTGGGGGCATTGTATTTGTG-3′).
Data availability statement
Table S1 describes all fission yeast strains used in this work, and are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.9342314.
Results
Telomere recombination likely precedes the growth crisis in ccq1Δ cells
Immediately following germination, ccq1Δ cells propagated in liquid culture grow slightly slower than WT cells (Figure 1A and Figure S1A) but already have substantially shorter telomeres as measured by Southern blot (Figure S1B). After 10 days of serial culturing, ccq1Δ cells are in the depth of the growth crisis, which is followed by the emergence of survivors that have growth rates similar to the starting population, consistent with earlier studies (Flory et al. 2004; Miyoshi et al. 2008; Tomita and Cooper 2008; Harland et al. 2014) (Figure 1A and Figure S1A). These observations suggest that presenescent ccq1Δ cells have already undergone substantial telomere loss prior to entry into the growth crisis (Figure 1A and Figure S1, A and B), consistent with previous studies in budding yeast (Abdallah et al. 2009; Khadaroo et al. 2009) and with the activation of the ATR-dependent Chk1 checkpoint (Tomita and Cooper 2008) and formation of Crb2 foci (Carneiro et al. 2010) in early generation ccq1Δ cells. In light of these results, we considered whether telomere recombination was occurring shortly after germination, but was insufficient to maintain telomere length. To assess this, we serially cultured cells lacking both Ccq1 and Rad55/Rhp55, which ultimately survive primarily through chromosome circularization (Miyoshi et al. 2008). Consistent with our hypothesis [and the observed role for the recombination factor Rad52 in antagonizing senescence in budding yeast models of telomere dysfunction (Lundblad and Blackburn 1993; Abdallah et al. 2009)], ccq1Δ/rad55Δ cells grow much more poorly than ccq1Δ cells immediately following germination (Figure 1A and Figure S1A). This suggests that the activation of recombination at critically short or deprotected telomeres occurs early after germination of ccq1Δ cells but is insufficient to maintain telomere length.
Shortly after germination, we also observe Rad52(Rad22)-mCherry foci in ∼65% of ccq1Δ cells (n = 233) compared to only 2% of WT cells (n = 239) (Figure 1, B and C). While only a subset of these foci colocalize with Taz1-GFP (22% of ccq1Δ cells; Figure 1B), it is likely that the remaining Rad52-mCherry foci correspond to critically short telomeres that cannot recruit sufficient Taz1 to be visualized. Indeed, the percentage of cells with 2–3 Taz1-GFP foci decreases from ∼85% for WT cells to only 30% of ccq1Δ cells (Figure 1D). Nearly all cells lacking both Ccq1 and Rad55 have Rad52-GFP foci at this initial time point (Figure 1C), further suggesting that Rad52 foci are tied to telomere dysfunction. Although cells lacking Rad55 alone grow more slowly than WT (Figure 1A) and have consistent, persistent Rad52-GFP foci (Figure 1C) over the course of the serial culturing assay, they lack the dynamic increase and subsequent decrease in Rad52-mCherry foci seen during telomere crisis and escape. Taken together, these results suggest that recombination is active at ccq1Δ telomeres shortly after germination but that additional factors must underlie the ability of cells to productively use recombination to maintain their telomeres, as seen in survivors.
TERRA levels increase in recombination-based survivors, but driving TERRA production cannot prevent telomere crisis in presenescent ccq1Δ cells
Studies in both budding yeast and human cells suggest that TERRA upregulation underlies productive recombination to maintain telomeres in the absence of telomerase, which correlates with elevated levels of telR-loops (Arora et al. 2012; Balk et al. 2013; Yu et al. 2014; Graf et al. 2017). Consistent with the role for Ccq1 in maintaining subtelomeric heterochromatin (Sugiyama et al. 2007; Wang et al. 2016; Armstrong et al. 2018), we find that TERRA levels, which are already ∼10-fold higher in ccq1Δ cells shortly after germination than WT cells, are dramatically increased in ccq1Δ survivors, irrespective of whether Taz1-GFP—a convenient cytological marker for telomeres—was expressed during serial culturing (Figure 2A). Indeed, we observe a progressive increase in TERRA during the emergence of ccq1Δ survivors as detected by quantitative RT-PCR (Figure 2B). Moreover, we detect an increase in telR-loops by DNA:RNA-IP/RT-PCR in ccq1Δ survivors (Figure 2C). We were also curious whether TERRA levels increase in survivors derived from other genetic backgrounds. Consistent with the ability of TERRA to support ongoing telomeric recombination in budding yeast (Balk et al. 2013), we observe mild TERRA induction in recombination-competent trt1Δ survivors, which, when serially cultured in liquid media, likely contain both linear and circular types of survivors (Nakamura et al. 1998) (Figure S1C). By contrast, although ccq1Δ/rad55Δ cells show very high levels of TERRA shortly after germination, ccq1Δ/rad55Δ survivors have nearly undetectable TERRA levels, far below what is seen in WT cells, likely due to complete loss of telomeric repeats (Figure 2D). Taken together, these results show that TERRA production is quickly stimulated in cells lacking Ccq1 and is either further increased during formation of survivors (in ccq1Δ recombination-competent strains) or is lost (in recombination-deficient genetic backgrounds that undergo chromosome circularization such as ccq1Δ/rad55Δ).
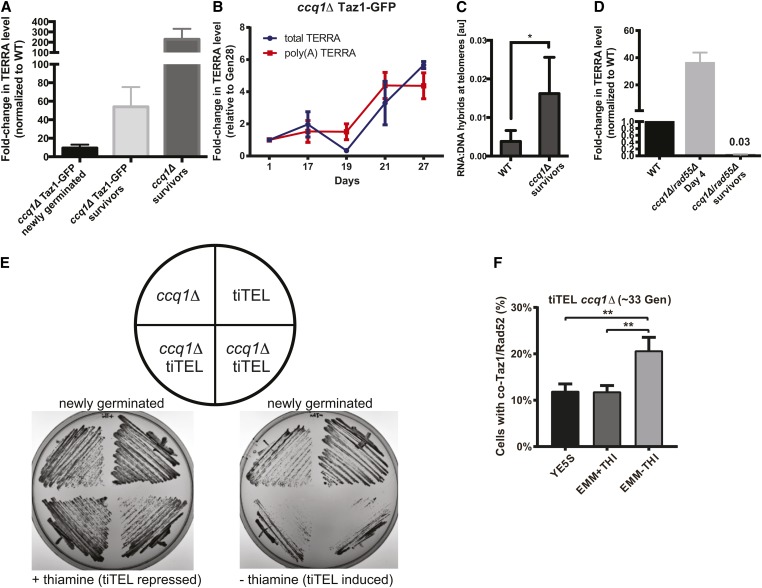
Figure 2.
TERRA is upregulated during ccq1Δ survivor formation but a tiTEL cannot prevent a growth crisis. (A) TERRA levels are higher in ccq1Δ cells shortly after germination, and are dramatically increased in survivors. Quantitative RT-PCR analysis for G-rich TERRA normalized to WT cells and plotted as the mean with SD from three replicates. (B) TERRA levels increase concomitant with the emergence of ccq1Δ survivors between days 19 and 27. Quantitative RT-PCR analysis for both total and poly(A+) TERRA normalized to Day1 ccq1Δ cells and plotted as the mean with SD from three replicates. (C) Telomeric RNA/DNA hybrids increase in ccq1Δ survivor cells as assessed by DNA-IP using the S9.6 antibody (see Materials and Methods). Mean of three replicates plotted with the SD. * P < 0.05. (D) Newly germinated ccq1Δrad55Δ cells contain high levels of total TERRA shortly after germination but TERRA is depleted in ccq1Δrad55Δ survivors as determined by RT-PCR as in (A). (E and F) tiTEL induction leads to a more rapid and pronounced growth crisis in newly germinated ccq1Δ cells. (E) Culturing of two ccq1Δ tiTEL clones on either EMM + thiamine (tiTEL repressed) or – thiamine (tiTEL induced) solid media. tiTEL induction leads to poorer growth than seen for ccq1Δ cells alone. (F) Increased loading of recombination factors on telomeres in newly germinated ccq1Δ tiTEL cells upon induction. Fluorescence micrographs of newly germinated (33 generations) ccq1Δ tiTEL cells expressing Taz1-GFP and Rad52-mCherry were quantified for colocalization. ** P < 0.01 by Student’s t-test.
Our data suggest that TERRA upregulation during the emergence of survivors from the growth crisis in ccq1Δ cells supports recombination-based telomere maintenance. However, we were curious whether driving TERRA production from the time of germination would be sufficient to avoid the growth crisis in ccq1Δ cells altogether. To examine this, we took advantage of the tiTEL system in which the thiamine-regulated nmt1+ promoter drives TERRA overexpression from a single telomere (Moravec et al. 2016). Surprisingly, ccq1Δ tiTEL cells serially restruck on plates under conditions of TERRA induction grow much more slowly than ccq1Δ cells or tiTEL cells grown under the same conditions (Figure 2E). Interestingly, we find that Rad52-mCherry is enriched on telomeres 1 day after tiTEL induction (but not in cells in which the tiTEL was repressed), consistent with signals that slow cell growth (Figure 2F). Thus, engineered TERRA overexpression cannot overcome the passage of cells through a period of catastrophic growth crisis.
Degradation of RNA–DNA hybrids drives ccq1Δ survivors to enter a second growth crisis from which new survivors emerge
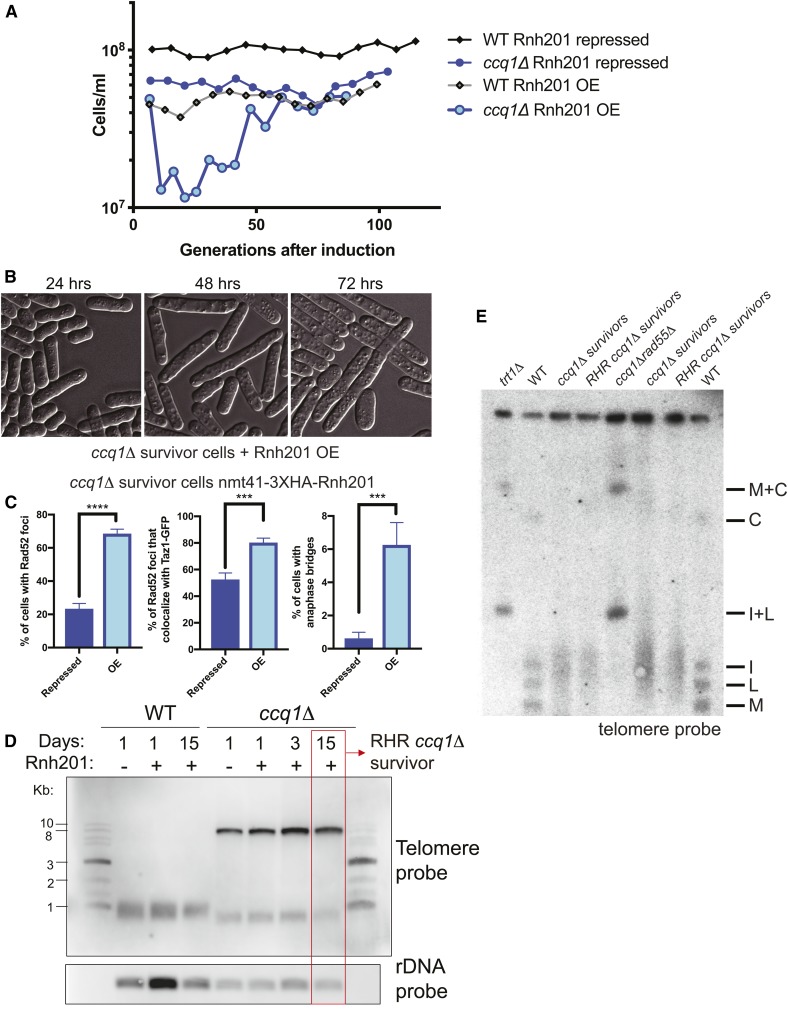
To test whether TERRA upregulation (and specifically TERRA engaged in telR-loops) contributes to telomere maintenance by recombination in ccq1Δ cells, we sought to develop an approach to induce degradation of telR-loops. To this end, we integrated the inducible nmt41+ promoter in front of the Rnh201 gene, which encodes an RNase H2 homolog capable of digesting RNA specifically engaged in RNA–DNA hybrids including TERRA in budding yeast when overexpressed (Huertas and Aguilera 2003; El Hage et al. 2010; Balk et al. 2013). We found that Rnh201 overexpression (Figure S2, A and B) is sufficient to suppress telR-loops in S. pombe (Figure S2C), although we note that, even in the repressed state (with thiamine), Rnh201 is overexpressed relative to WT cells (Figure S2B). Strikingly, high overexpression of Rnh201 caused ccq1Δ survivors to rapidly reenter a growth crisis (Figure 3A), characterized by cell cycle arrest as indicated by the accumulation of very long cells (Figure 3B). We also observed hallmarks of telomere crisis 24 hr after induction of Rnh201 overexpression in ccq1Δ survivors, including a greater than threefold increase in the percent of cells with Rad52 foci (to >60% of all cells), with 80% of these Rad52 foci occurring at the telomere as assessed by colocalization with Taz1-GFP (Figure 3C and Figure S2D). In addition, Rnh201 overexpression in ccq1Δ survivors leads to an increase in cells with anaphase bridges decorated with Rad52, suggestive of fused or entangled telomeres due to the ongoing crisis (Figure 3C and Figure S2D). Ultimately, “new” survivors emerged ∼50 generations after Rnh201 induction (Figure 3A) despite persistent expression of Rnh201 (Figure S2E). These “new” survivor cells, which we term RNase H2-resistant (RHR) ccq1Δ survivors, maintain their telomere length as assessed by Southern blot (Figure 3D). Moreover, pulsed-field gel electrophoresis demonstrates that, like the initial ccq1Δ survivors, RHR ccq1Δ survivors maintain linear chromosomes (Figure 3E) without any apparent telomere-telomere fusions (M+C and I+L bands) that appear in circular trt1Δ and ccq1Δrad55Δ survivors.
Figure 3.
Degradation of RNA–DNA hybrids induces a second growth crisis in ccq1Δ survivors, ultimately leading to a new type of RNase H-resistant ccq1Δ survivor. (A) Rnh201 induction returns ccq1Δ survivors into a growth crisis. WT or ccq1Δ Taz1-GFP survivor cells (MKSP1213 and MKSP1214) were modified to insert the nmt41+ promoter upstream of the rnh201+ coding sequence. Strains were then cultured in a repressed state (with thiamine) or induced state (without thiamine), as indicated and diluted every 24 hr (B) DIC imaging shows that Rnh201 overexpression leads to a new growth crisis in ccq1Δ survivors. The first 3 days of liquid culturing corresponding to (A) are shown. Note the increase in cell length, indicating checkpoint arrest. (C) Rnh201 overexpression (OE) leads to increased Rad52-mCherry loading at telomeres and anaphase bridges. (D and E) The telomeres of RHR ccq1Δ survivors selected after constitutive Rnh201 overexpression are similar in length and structure to the initial ccq1Δ survivors. (C) Southern blot of EcoRI-digested genomic DNA as in Figure S1B. The rDNA probe serves as a loading control. (E) The RHR ccq1Δ survivors, like ccq1Δ survivors, retain linear chromosomes as assessed by pulsed-field gel electrophoresis followed by Southern blotting. Genomic DNA was digested with NotI and hybridized to C, I, L, and M probes, which detect the terminal fragments of chromosomes I and II. Bands corresponding to chromosome end fusions are indicated by M+C and I+L bands, observed in the trt1Δ and ccq1Δrad55Δ circular survivors.
RHR ccq1Δ survivors display altered recruitment of telomere binding proteins and compartmentalization of telomeres
To gain further insight into the mechanisms that RHR ccq1Δ survivors employ to maintain linear chromosomes, we visualized telomeres by expression of Taz1-GFP. Taz1-GFP intensity in the initial ccq1Δ survivors is increased compared to WT cells (Figure 4, A and B), with some cells displaying a single, very bright Taz1-GFP focus (Figure 4A, center panel, arrowhead). The generation of RHR ccq1Δ survivors led to an even higher Taz1-GFP intensity and more declustered, expanded telomere foci (Figure 4, A and B). To further quantitate this defect, we took advantage of an image analysis pipeline we initially developed to render and measure the size of heterochromatin foci in fission yeast (Schreiner et al. 2015). In WT cells, the true size of telomere clusters is nearly always smaller than the diffraction limit of our microscope, and thus they appear equivalent to the point spread function (PSF; Figure 4, C and D). The mean size of the Taz1-associated telomere clusters in the initial ccq1Δ survivors is greater than the PSF (Figure 4, C and D). However, the telomere foci in the RHR ccq1Δ survivors are highly expanded (Figure 4, A–C) and irregularly shaped (Figure 4D). These effects were not influenced by Taz1-GFP expression during the serial culturing, as integration of Taz1-GFP into ccq1Δ and RHR ccq1Δ survivors led to cytologically indistinguishable telomere appearance (Figure S3, A and B).
In budding yeast, telomere clustering is promoted by the shelterin component, Rap1, acting in concert with Sir3 (Gotta et al. 1996; Ruault et al. 2011; Hoze et al. 2013). Loss of Rap1 alone is insufficient to cause telomere declustering in fission yeast (Figure S3C) and Rap1 foci appear unaltered in ccq1Δ survivors (Figure S3, D and E). However, we observe a profound loss of Rap1-GFP from RHR ccq1Δ survivor telomeres (Figure 5A) despite normal (or higher) levels of Taz1-GFP recruitment (Figure 4). Indeed, in 45% of cells we could not detect any Rap1-GFP at the telomere, while two or more Rap1-GFP-associated telomere clusters were seen only very rarely (Figure 5B). ChIP followed by qPCR further supported this observation, as we find a significant decrease in the association of Rap1-GFP with the subtelomere upon long-term Rnh201 overexpression in ccq1Δ survivors (Figure 5C), although we acknowledge that loss of Rap1 binding could be overestimated due to telomere rearrangements in these cells (Figure 3C). To gain further insight into the mechanisms at play, we measured the expression of both Taz1 and Rap1. At the protein level, we find that Taz1 is increased in ccq1Δ survivors (whether derived in the absence or presence of the GFP tag) and is maintained in RHR ccq1Δ survivors (Figure 5D), suggesting that enhanced expression may support the elevated levels of Taz1-GFP at the telomere in both types of survivors. By contrast, Rap1 expression levels remain nearly constant in all conditions (Figure 5D), arguing that altered recruitment, not less expression, underlies the loss of Rap1 from the telomere.
The growth crisis induced by RNA–DNA hybrid degradation can be ameliorated by deletion of Rap1
The loss of Rap1 from the telomere of RHR ccq1Δ survivors suggests that Rap1 may negatively influence TERRA-independent telomere maintenance by recombination. To test this hypothesis, we compared the growth response of ccq1Δ survivors to critical Rnh201 overexpression in the presence and absence of Rap1 (Figure 5E). We carried out serial culturing of ccq1Δ Pnmt41-rnh201+ and rap1Δccq1Δ Pnmt41-rnh201+ cells under three conditions: constitutive Rnh201 repression (black, in the presence of thiamine), constitutive Rnh201 overexpression (OE; light gray circles) and switching from repression (at 13 days) to overexpression (arrow, dark gray diamonds). Consistent with Figure 3A, critical induction of Rnh201 in ccq1Δ cells at day 13 leads to a growth crisis (days 14–19; see shaded gray box), followed by recovery at day 20 to achieve a growth rate similar to cells cultured under conditions of constitutive Rnh201 overexpression. By contrast, although critical Rnh201 overexpression in cells lacking Rap1 at day 13 mildly suppresses cell growth (as does Rnh201 overexpression in WT cells, Figure 3A), there is no growth crisis (Figure 5E and Figure S4A). We note that ccq1 and rap1 show a mild synthetic growth defect under conditions of Rnh201 repression (Figure S4B); to account for this, the plots in Figure 5, D–E represent the relative effect of RNA–DNA hybrid degradation. Raw growth plots reveal the same qualitative trend (Figure S4B). Loss of Rap1 is known to induce telomere elongation in telomerase-positive fission yeast cells (Kanoh and Ishikawa 2001), which is tied to enhanced recruitment of telomerase to the telomere (Moser et al. 2011; Dehe et al. 2012). We therefore addressed the possibility that loss of Rap1 reconstitutes telomerase-dependent telomere elongation in these cells. However, deletion of the essential telomerase component Est1 has no effect on the growth of RHR ccq1Δ survivors (Figure S4C), consistent with previous work demonstrating that increased telomere length and telomerase recruitment in the absence of Rap1 is Ccq1-dependent (Tomita and Cooper 2008; Moser et al. 2011). Thus, Rap1 appears to attenuate productive telomere recombination under conditions where RNA–DNA hybrids are degraded. How RHR ccq1Δ survivors displace Rap1 from the telomere despite its continued expression (Figure 5D) and the persistence of telomeric Taz1 (Figure 4), will require further study.
Discussion
Here, we find that loss of the telomerase recruitment factor Ccq1 leads to progressive telomere shortening despite early loading of Rad52 onto telomeres, suggesting that additional factors are required for productive telomere lengthening by recombination. We provide evidence that the spontaneous increase of telR-loop-engaged TERRA promotes productive telomere maintenance by recombination in fission yeast. In presenescent cells, increased telomere-associated TERRA is detrimental and telomere recombination is inefficient. By contrast, during survivor emergence, TERRA levels increase spontaneously (and/or are selected for during competition in liquid culture), form telR-loops, and promotes efficient telomere maintenance. Long-term degradation of telR-loops compromises the growth of ccq1Δ survivors, ultimately leading to a new survivor type with altered biochemical and cytological characteristics, including loss of telomeric Rap1. Because deletion of Rap1 is sufficient to avoid the growth crisis induced upon RNA–DNA hybrid degradation in ccq1Δ survivors, we propose that TERRA-independent recombination pathways might be engaged to maintain telomeres by recombination.
A switch from telomerase to recombination
As we place increased accumulation of telomere-associated TERRA as a key step in the transition to productive recombination, we favor a model in which changes to chromatin state within the subtelomere occur upon telomere shortening, supporting derepression of TERRA (Rippe and Luke 2015; Moravec et al. 2016). Indeed, induction of TERRA has been suggested to occur as part of the more global loss of the telomere position effect upon telomere shortening (Mandell et al. 2005; Arnoult et al. 2012; Cusanelli et al. 2013). Further, depletion of the histone chaperone ASF1 can induce a switch to the ALT pathway, suggesting that changes in the chromatin landscape are sufficient to activate HDR-dependent telomere maintenance (O’Sullivan et al. 2014). Ccq1 also contributes to recruitment of the SHREC complex (Sugiyama et al. 2007) and the CLRC complex (Wang et al. 2016; Armstrong et al. 2018) to telomeres to promote heterochromatization. Disrupting the phosphorylation of Ccq1 by the DNA damage checkpoint kinases ATM and ATR, which enhances its interaction with Est1, abrogates telomerase recruitment without adversely affecting recruitment of SHREC (Moser et al. 2011); in this context survivors undergo chromosome circularization (Moser et al. 2011), suggesting that derepression of the subtelomere is critical for the maintenance of linear chromosomes by recombination. Combined with the observation that TERRA levels begin to increase after germination in cells lacking Ccq1 (Figure 2B), we propose that the combination of a more transcriptionally permissive chromatin state combined with shortened telomeres (Graf et al. 2017) together drives high levels of TERRA, consistent with the ability of this genetic background to maintain linear chromosomes via recombination (Miyoshi et al. 2008; Tomita and Cooper 2008). The slow emergence of survivors over many generations (and our failure to circumvent the growth crisis by TERRA expression from the tiTEL; Figure 2, E and F) could reflect transmissibility of heterochromatic marks at the subtelomere and/or the requirement for additional changes, for example to checkpoint signaling, that remain to be uncovered.
Support of recombination by telomere-associated TERRA
When telomere-associated TERRA is degraded, postsenescent ccq1Δ survivors reenter a growth crisis (Figure 3, A and B). This finding is in line with the observation that TERRA is necessary for recombination at telomeres in budding yeast (Balk et al. 2013). Further, elevated levels of TERRA are characteristic of human ALT cell lines established from tumors or immortalized in vitro (Lovejoy et al. 2012; Arora et al. 2014). TERRA could act to promote HDR-dependent telomere maintenance in several ways: (1) by stabilizing D-loop structures to facilitate recombination between telomeres (Balk et al. 2013, 2014); (2) by promoting an increase in telomere mobility to support telomere–telomere interactions necessary for recombination (Arora et al. 2012); and/or (3) by promoting break-induced replication (Dilley et al. 2016). While discriminating between these explicit molecular mechanisms will require further study, our data clearly support a model in which spontaneous derepression of TERRA plays a critical role in telomere maintenance in ccq1Δ survivors.
Distinct roles of TERRA in the presence or absence of telomerase
The effect that TERRA has on telomere length regulation varies, in part depending on the presence or absence of telomerase (Azzalin and Lingner 2008; Schoeftner and Blasco 2008; Redon et al. 2010, 2013; Pfeiffer and Lingner 2012; Balk et al. 2013; Cusanelli et al. 2013). In addition, our recent work revealed that TERRA molecules in fission yeast can be further grouped into at least two subsets: one is poly(A)+ TERRA, which loses all telomeric tracts and is soluble in the nucleoplasm; the other is poly(A)– TERRA, which has G-rich telomeric sequences, and is bound at telomeric DNA (Moravec et al. 2016). While poly(A)+ TERRA stimulates telomerase recruitment in telomerase-positive cells (Moravec et al. 2016), here we suggest that poly(A)– TERRA (or G-rich TERRA) engaged in telR-loops promotes telomere recombination in cells with insufficient telomerase function. Our observation that TERRA produced from a tiTEL cannot prevent ccq1Δ cell senescence but also precipitates a more rapid growth crisis in presenescent cells (Figure 2, E and F) is consistent with the suggestion that increased TERRA production can be deleterious to telomere maintenance in some contexts (Balk et al. 2013). We note, however, that the tiTEL predominantly produces poly(A)+ TERRA and that nascent TERRA is limited to a single telomere; thus, the tiTEL system may model the role for TERRA in telomerase-dependent (Moravec et al. 2016), but not recombination-dependent, telomere maintenance.
TERRA dependent and independent recombination-based telomere maintenance
The “addiction” of ccq1Δ survivors to telomere-associated TERRA (suggested by the ability of Rnh201 overexpression to induce a second growth crisis) can be overcome, as evidenced by the formation of RNase H2-resistant ccq1Δ survivors. This mechanism is necessarily distinct from so-called HAATI survivors, which replace the telomeric repeats with “generic” heterochromatin, as this pathway requires the presence of Ccq1 (Jain et al. 2010). What changes might support a productive, TERRA-independent telomere recombination mechanism that can maintain telomeres in the absence of Ccq1? Our data suggest that these RHR ccq1Δ survivors display altered recruitment of telomere binding proteins, including loss of Rap1 (but not Taz1) from the telomere (Figure 4 and Figure 5). This result is somewhat surprising, as Taz1 binds directly to Rap1 and promotes its recruitment to the telomere in fission yeast, thereby bridging the double-stranded telomeric repeat region to Poz1 and the shelterin components that associate with the single-stranded telomeric overhang (Chikashige and Hiraoka 2001; Kanoh and Ishikawa 2001; Pan et al. 2015). Moreover, loss of Taz1 and Rap1 phenocopy one another with respect to increased telomere length and derepression of TERRA (Miller et al. 2005; Bah et al. 2012; Greenwood and Cooper 2012), although the mechanisms at play may be distinct (Miller et al. 2005; Dehe et al. 2012). We note, however, that recent evidence supports allosteric regulation of Rap1 binding to Tpz1-Poz1 to regulate telomere length (Kim et al. 2017), suggesting that the Rap1-Taz1 interface may also be subject to modulation. Further studies will be required to test if changes in the composition and/or conformation of shelterin are tied to the mechanisms allowing for telomere maintenance in the absence of telR-loops.
Importantly, loss of Rap1 is not simply a consequence of the mechanisms that support telomere maintenance in RHR ccq1Δ survivors, because deletion of Rap1 ameliorates the growth crisis apparent upon degradation of RNA–DNA hybrids in ccq1Δ survivors (Figure 5E). How Rap1 eviction from the telomeres promotes the maintenance of linear chromosomes in this context is not yet clear, although we have ruled out the participation of telomerase (Figure S4C). Given that Rap1 is a known repressor of TERRA (Bah et al. 2012), one possibility is that there is a further stimulation of TERRA production that restores telomere-engaged TERRA. Alternatively, decreased telomeric Rap1 could weaken the association of telomeres with the nuclear envelope, which has been suggested to repress telomere recombination in budding yeast (Schober et al. 2009). Lastly, Rap1 and Taz1 could have negative and positive influences, respectively, on HDR mechanisms such as BIR. Ultimately, testing how changes in shelterin composition may act independently of TERRA to promote telomere maintenance by ALT will require further study.
Acknowledgments
We thank J. Kanoh and J. P. Cooper for sharing antibodies. We are also indebted to the Yeast Genomic Resource Center (YGRC) at Osaka University for providing access to strains, as well as the many researchers who have deposited strains at this resource. This work was supported by the New Innovator Award (National Institutes of Health, Office of the Director) - DP2OD008429 (to M.C.K.), the Yale Institute for Biological, Physical, and Engineering Sciences (to M.C.K. and H.W.B.) and the Yale Science, Technology and Research Scholars Program (STARS II) (to H.W.B.). Research in the Azzalin laboratory was supported by the European Molecular Biology Organization (IG3576) and the Fundação para a Ciência e a Tecnologia (IF/01269/2015; PTDC/MED-ONC/28282/2017; PTDC/BIA-MOL/29352/2017).
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25386/genetics.9342314.
Communicating editor: J. Surtees
Literature Cited
- Abdallah P., Luciano P., Runge K. W., Lisby M., Geli V. et al. , 2009. A two-step model for senescence triggered by a single critically short telomere. Nat. Cell Biol. 11: 988–993. [corrigenda: Nat. Cell Biol. 12: 520 (2010)] 10.1038/ncb1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apte M. S., and Cooper J. P., 2017. Life and cancer without telomerase: ALT and other strategies for making sure ends (don’t) meet. Crit. Rev. Biochem. Mol. Biol. 52: 57–73. 10.1080/10409238.2016.1260090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong C. A., and Tomita K., 2017. Fundamental mechanisms of telomerase action in yeasts and mammals: understanding telomeres and telomerase in cancer cells. Open Biol. 7: pii: 160338. 10.1098/rsob.160338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong C. A., Moiseeva V., Collopy L. C., Pearson S. R., Ullah T. R. et al. , 2018. Fission yeast Ccq1 is a modulator of telomerase activity. Nucleic Acids Res. 46: 704–716. 10.1093/nar/gkx1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoult N., Van Beneden A., and Decottignies A., 2012. Telomere length regulates TERRA levels through increased trimethylation of telomeric H3K9 and HP1α. Nat. Struct. Mol. Biol. 19: 948–956. [corrigenda: Nat. Struct. Mol. Biol. 20: 244 (2013)] 10.1038/nsmb.2364 [DOI] [PubMed] [Google Scholar]
- Arora R., Brun C. M., and Azzalin C. M., 2012. Transcription regulates telomere dynamics in human cancer cells. RNA 18: 684–693. 10.1261/rna.029587.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R., Lee Y., Wischnewski H., Brun C. M., Schwarz T. et al. , 2014. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat. Commun. 5: 5220 10.1038/ncomms6220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin C. M., and Lingner J., 2008. Telomeres: the silence is broken. Cell Cycle 7: 1161–1165. 10.4161/cc.7.9.5836 [DOI] [PubMed] [Google Scholar]
- Azzalin C. M., and Lingner J., 2015. Telomere functions grounding on TERRA firma. Trends Cell Biol. 25: 29–36. 10.1016/j.tcb.2014.08.007 [DOI] [PubMed] [Google Scholar]
- Azzalin C. M., Reichenbach P., Khoriauli L., Giulotto E., and Lingner J., 2007. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318: 798–801. 10.1126/science.1147182 [DOI] [PubMed] [Google Scholar]
- Bah A., and Azzalin C. M., 2012. The telomeric transcriptome: from fission yeast to mammals. Int. J. Biochem. Cell Biol. 44: 1055–1059. 10.1016/j.biocel.2012.03.021 [DOI] [PubMed] [Google Scholar]
- Bah A., Wischnewski H., Shchepachev V., and Azzalin C. M., 2012. The telomeric transcriptome of Schizosaccharomyces pombe. Nucleic Acids Res. 40: 2995–3005. 10.1093/nar/gkr1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A. et al. , 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951. [DOI] [PubMed] [Google Scholar]
- Balk B., Maicher A., Dees M., Klermund J., Luke-Glaser S. et al. , 2013. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat. Struct. Mol. Biol. 20: 1199–1205. 10.1038/nsmb.2662 [DOI] [PubMed] [Google Scholar]
- Balk B., Dees M., Bender K., and Luke B., 2014. The differential processing of telomeres in response to increased telomeric transcription and RNA-DNA hybrid accumulation. RNA Biol. 11: 95–100. 10.4161/rna.27798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P., and Cech T. R., 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292: 1171–1175. 10.1126/science.1060036 [DOI] [PubMed] [Google Scholar]
- Beernink H. T., Miller K., Deshpande A., Bucher P., and Cooper J. P., 2003. Telomere maintenance in fission yeast requires an Est1 ortholog. Curr. Biol. 13: 575–580. 10.1016/S0960-9822(03)00169-6 [DOI] [PubMed] [Google Scholar]
- Bennett H. W., Liu N., Hu Y., and King M. C., 2016. TeloPCR-seq: a high-throughput sequencing approach for telomeres. FEBS Lett. 590: 4159–4170. 10.1002/1873-3468.12444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn E. H., 1991. Structure and function of telomeres. Nature 350: 569–573. 10.1038/350569a0 [DOI] [PubMed] [Google Scholar]
- Blackburn E. H., 2001. Switching and signaling at the telomere. Cell 106: 661–673. 10.1016/S0092-8674(01)00492-5 [DOI] [PubMed] [Google Scholar]
- Bühler M., and Gasser S. M., 2009. Silent chromatin at the middle and ends: lessons from yeasts. EMBO J. 28: 2149–2161. 10.1038/emboj.2009.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro T., Khair L., Reis C. C., Borges V., Moser B. A. et al. , 2010. Telomeres avoid end detection by severing the checkpoint signal transduction pathway. Nature 467: 228–232. 10.1038/nature09353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare A. J., and Reddel R. R., 2010. Alternative lengthening of telomeres: models, mechanisms and implications. Nat. Rev. Genet. 11: 319–330. 10.1038/nrg2763 [DOI] [PubMed] [Google Scholar]
- Chikashige Y., and Hiraoka Y., 2001. Telomere binding of the Rap1 protein is required for meiosis in fission yeast. Curr. Biol. 11: 1618–1623. 10.1016/S0960-9822(01)00457-2 [DOI] [PubMed] [Google Scholar]
- Collopy L. C., Ware T. L., Goncalves T., í Kongsstovu S., Yang Q., et al. , 2018. LARP7 family proteins have conserved function in telomerase assembly. Nat. Commun. 9: 557 10.1038/s41467-017-02296-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. P., and Hiraoka Y., 2006. Fission yeast telomeres, in Telomeres, edited by de Lange T. Lundblad V., and Blackburn E. H.. Cold Spring Harbor Laboratory Press, New York. [Google Scholar]
- Cooper J. P., Nimmo E. R., Allshire R. C., and Cech T. R., 1997. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385: 744–747. 10.1038/385744a0 [DOI] [PubMed] [Google Scholar]
- Cusanelli E., Romero C. A., and Chartrand P., 2013. Telomeric noncoding RNA TERRA is induced by telomere shortening to nucleate telomerase molecules at short telomeres. Mol. Cell 51: 780–791. 10.1016/j.molcel.2013.08.029 [DOI] [PubMed] [Google Scholar]
- Dehé P. M., and Cooper J. P., 2010. Fission yeast telomeres forecast the end of the crisis. FEBS Lett. 584: 3725–3733. 10.1016/j.febslet.2010.07.045 [DOI] [PubMed] [Google Scholar]
- Dehé P. M., Rog O., Ferreira M. G., Greenwood J., and Cooper J. P., 2012. Taz1 enforces cell-cycle regulation of telomere synthesis. Mol. Cell 46: 797–808. 10.1016/j.molcel.2012.04.022 [DOI] [PubMed] [Google Scholar]
- de Lange T., 2005. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19: 2100–2110. 10.1101/gad.1346005 [DOI] [PubMed] [Google Scholar]
- de Lange T., 2009. How telomeres solve the end-protection problem. Science 326: 948–952. 10.1126/science.1170633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilley R. L., Verma P., Cho N. W., Winters H. D., Wondisford A. R. et al. , 2016. Break-induced telomere synthesis underlies alternative telomere maintenance. Nature 539: 54–58. 10.1038/nature20099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hage A., French S. L., Beyer A. L., and Tollervey D., 2010. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 24: 1546–1558. 10.1101/gad.573310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira M. G., and Cooper J. P., 2001. The fission yeast Taz1 protein protects chromosomes from Ku-dependent end-to-end fusions. Mol. Cell 7: 55–63. 10.1016/S1097-2765(01)00154-X [DOI] [PubMed] [Google Scholar]
- Feuerhahn S., Iglesias N., Panza A., Porro A., and Lingner J., 2010. TERRA biogenesis, turnover and implications for function. FEBS Lett. 584: 3812–3818. 10.1016/j.febslet.2010.07.032 [DOI] [PubMed] [Google Scholar]
- Flory M. R., Carson A. R., Muller E. G., and Aebersold R., 2004. An SMC-domain protein in fission yeast links telomeres to the meiotic centrosome. Mol. Cell 16: 619–630. 10.1016/j.molcel.2004.10.027 [DOI] [PubMed] [Google Scholar]
- Gotta M., Laroche T., Formenton A., Maillet L., Scherthan H. et al. , 1996. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol. 134: 1349–1363. 10.1083/jcb.134.6.1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf M., Bonetti D., Lockhart A., Serhal K., Kellner V. et al. , 2017. Telomere length determines TERRA and R-loop regulation through the cell cycle. Cell 170: 72–85.e14. 10.1016/j.cell.2017.06.006 [DOI] [PubMed] [Google Scholar]
- Greenwood J., and Cooper J. P., 2012. Non-coding telomeric and subtelomeric transcripts are differentially regulated by telomeric and heterochromatin assembly factors in fission yeast. Nucleic Acids Res. 40: 2956–2963. 10.1093/nar/gkr1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland J. L., Chang Y. T., Moser B. A., and Nakamura T. M., 2014. Tpz1-Ccq1 and Tpz1-Poz1 interactions within fission yeast shelterin modulate Ccq1 Thr93 phosphorylation and telomerase recruitment. PLoS Genet. 10: e1004708 10.1371/journal.pgen.1004708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidenreich B., and Kumar R., 2017. TERT promoter mutations in telomere biology. Mutat. Res. 771: 15–31. 10.1016/j.mrrev.2016.11.002 [DOI] [PubMed] [Google Scholar]
- Hozé N., Ruault M., Amoruso C., Taddei A. and Holcman D., 2013. Spatial telomere organization and clustering in yeast Saccharomyces cerevisiae nucleus is generated by a random dynamics of aggregation-dissociation. Mol. Biol. Cell 24: 1791–1800, s1–10. 10.1091/mbc.e13-01-0031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P., and Aguilera A., 2003. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell 12: 711–721. 10.1016/j.molcel.2003.08.010 [DOI] [PubMed] [Google Scholar]
- Jain D., Hebden A. K., Nakamura T. M., Miller K. M., and Cooper J. P., 2010. HAATI survivors replace canonical telomeres with blocks of generic heterochromatin. Nature 467: 223–227. 10.1038/nature09374 [DOI] [PubMed] [Google Scholar]
- Kanoh J., and Ishikawa F., 2001. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr. Biol. 11: 1624–1630. 10.1016/S0960-9822(01)00503-6 [DOI] [PubMed] [Google Scholar]
- Khadaroo B., Teixeira M. T., Luciano P., Eckert-Boulet N., Germann S. M. et al. , 2009. The DNA damage response at eroded telomeres and tethering to the nuclear pore complex. Nat. Cell Biol. 11: 980–987. 10.1038/ncb1910 [DOI] [PubMed] [Google Scholar]
- Kim J. K., Liu J., Hu X., Yu C., Roskamp K. et al. , 2017. Structural basis for shelterin bridge assembly. Mol. Cell 68: 698–714.e5. 10.1016/j.molcel.2017.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N. W., Piatyszek M. A., Prowse K. R., Harley C. B., West M. D. et al. , 1994. Specific association of human telomerase activity with immortal cells and cancer. Science 266: 2011–2015. 10.1126/science.7605428 [DOI] [PubMed] [Google Scholar]
- Leonardi J., Box J. A., Bunch J. T., and Baumann P., 2008. TER1, the RNA subunit of fission yeast telomerase. Nat. Struct. Mol. Biol. 15: 26–33. 10.1038/nsmb1343 [DOI] [PubMed] [Google Scholar]
- Longhese M. P., Bonetti D., Manfrini N., and Clerici M., 2010. Mechanisms and regulation of DNA end resection. EMBO J. 29: 2864–2874. 10.1038/emboj.2010.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzi L. E., Bah A., Wischnewski H., Shchepachev V., Soneson C. et al. , 2015. Fission yeast Cactin restricts telomere transcription and elongation by controlling Rap1 levels. EMBO J. 34: 115–129. 10.15252/embj.201489559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy C. A., Li W., Reisenweber S., Thongthip S., Bruno J. et al. , 2012. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 8: e1002772 10.1371/journal.pgen.1002772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B., Panza A., Redon S., Iglesias N., Li Z. et al. , 2008. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol. Cell 32: 465–477. 10.1016/j.molcel.2008.10.019 [DOI] [PubMed] [Google Scholar]
- Lundblad V., and Blackburn E. H., 1993. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell 73: 347–360. 10.1016/0092-8674(93)90234-H [DOI] [PubMed] [Google Scholar]
- Maestroni L., Audry J., Matmati S., Arcangioli B., Geli V. et al. , 2017a Eroded telomeres are rearranged in quiescent fission yeast cells through duplications of subtelomeric sequences. Nat. Commun. 8: 1684 10.1038/s41467-017-01894-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestroni L., Matmati S., and Coulon S., 2017b Solving the telomere replication problem. Genes (Basel) 8: pii: E55. 10.3390/genes8020055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maicher A., Kastner L., Dees M., and Luke B., 2012. Deregulated telomere transcription causes replication-dependent telomere shortening and promotes cellular senescence. Nucleic Acids Res. 40: 6649–6659. 10.1093/nar/gks358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell J. G., Bahler J., Volpe T. A., Martienssen R. A., and Cech T. R., 2005. Global expression changes resulting from loss of telomeric DNA in fission yeast. Genome Biol. 6: R1 10.1186/gb-2004-6-1-r1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez P., and Blasco M. A., 2015. Replicating through telomeres: a means to an end. Trends Biochem. Sci. 40: 504–515. 10.1016/j.tibs.2015.06.003 [DOI] [PubMed] [Google Scholar]
- Maundrell K., 1990. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J. Biol. Chem. 265: 10857–10864. [PubMed] [Google Scholar]
- Mennie A. K., Moser B. A., and Nakamura T. M., 2018. LARP7-like protein Pof8 regulates telomerase assembly and poly(A)+TERRA expression in fission yeast. Nat. Commun. 9: 586 10.1038/s41467-018-02874-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson M., Counter C. M., Eaton E. N., Ellisen L. W., Steiner P. et al. , 1997. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell 90: 785–795. 10.1016/S0092-8674(00)80538-3 [DOI] [PubMed] [Google Scholar]
- Miller K. M., Ferreira M. G., and Cooper J. P., 2005. Taz1, Rap1 and Rif1 act both interdependently and independently to maintain telomeres. EMBO J. 24: 3128–3135. 10.1038/sj.emboj.7600779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T., Kanoh J., Saito M., and Ishikawa F., 2008. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science 320: 1341–1344. 10.1126/science.1154819 [DOI] [PubMed] [Google Scholar]
- Moravec M., Wischnewski H., Bah A., Hu Y., Liu N. et al. , 2016. TERRA promotes telomerase-mediated telomere elongation in Schizosaccharomyces pombe. EMBO Rep. 17: 999–1012. 10.15252/embr.201541708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar A., and Nurse P., 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194: 795–823. 10.1016/0076-6879(91)94059-L [DOI] [PubMed] [Google Scholar]
- Moser B. A., Chang Y. T., Kosti J., and Nakamura T. M., 2011. Tel1ATM and Rad3ATR kinases promote Ccq1-Est1 interaction to maintain telomeres in fission yeast. Nat. Struct. Mol. Biol. 18: 1408–1413. 10.1038/nsmb.2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T. M., Morin G. B., Chapman K. B., Weinrich S. L., Andrews W. H. et al. , 1997. Telomerase catalytic subunit homologs from fission yeast and human. Science 277: 955–959. 10.1126/science.277.5328.955 [DOI] [PubMed] [Google Scholar]
- Nakamura T. M., Cooper J. P., and Cech T. R., 1998. Two modes of survival of fission yeast without telomerase. Science 282: 493–496. 10.1126/science.282.5388.493 [DOI] [PubMed] [Google Scholar]
- Nandakumar J., and Cech T. R., 2013. Finding the end: recruitment of telomerase to telomeres. Nat. Rev. Mol. Cell Biol. 14: 69–82. 10.1038/nrm3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan R. J., Arnoult N., Lackner D. H., Oganesian L., Haggblom C. et al. , 2014. Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nat. Struct. Mol. Biol. 21: 167–174. 10.1038/nsmb.2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Páez-Moscoso D. J., Pan L., Sigauke R. F., Schroeder M. R., Tang W. et al. , 2018. Pof8 is a La-related protein and a constitutive component of telomerase in fission yeast. Nat. Commun. 9: 587 10.1038/s41467-017-02284-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L., Hildebrand K., Stutz C., Thoma N., and Baumann P., 2015. Minishelterins separate telomere length regulation and end protection in fission yeast. Genes Dev. 29: 1164–1174. 10.1101/gad.261123.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer V., and Lingner J., 2012. TERRA promotes telomere shortening through exonuclease 1-mediated resection of chromosome ends. PLoS Genet. 8: e1002747 10.1371/journal.pgen.1002747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt C. W., and Cooper J. P., 2010. Pot1 inactivation leads to rampant telomere resection and loss in one cell cycle. Nucleic Acids Res. 38: 6968–6975. 10.1093/nar/gkq580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porro A., Feuerhahn S., Delafontaine J., Riethman H., Rougemont J. et al. , 2014. Functional characterization of the TERRA transcriptome at damaged telomeres. Nat. Commun. 5: 5379 10.1038/ncomms6379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon S., Reichenbach P., and Lingner J., 2010. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 38: 5797–5806. 10.1093/nar/gkq296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon S., Zemp I., and Lingner J., 2013. A three-state model for the regulation of telomerase by TERRA and hnRNPA1. Nucleic Acids Res. 41: 9117–9128. 10.1093/nar/gkt695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippe K., and Luke B., 2015. TERRA and the state of the telomere. Nat. Struct. Mol. Biol. 22: 853–858. 10.1038/nsmb.3078 [DOI] [PubMed] [Google Scholar]
- Ruault M., De Meyer A., Loiodice I., and Taddei A., 2011. Clustering heterochromatin: Sir3 promotes telomere clustering independently of silencing in yeast. J. Cell Biol. 192: 417–431. 10.1083/jcb.201008007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M. et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Meth. 9: 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober H., Ferreira H., Kalck V., Gehlen L. R., and Gasser S. M., 2009. Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev. 23: 928–938. 10.1101/gad.1787509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S., and Blasco M. A., 2008. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 10: 228–236. 10.1038/ncb1685 [DOI] [PubMed] [Google Scholar]
- Schreiner S. M., Koo P. K., Zhao Y., Mochrie S. G., and King M. C., 2015. The tethering of chromatin to the nuclear envelope supports nuclear mechanics. Nat. Commun. 6: 7159 10.1038/ncomms8159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay J. W., Reddel R. R., and Wright W. E., 2012. Cancer. Cancer and telomeres–an ALTernative to telomerase. Science 336: 1388–1390. 10.1126/science.1222394 [DOI] [PubMed] [Google Scholar]
- Shore D., and Bianchi A., 2009. Telomere length regulation: coupling DNA end processing to feedback regulation of telomerase. EMBO J. 28: 2309–2322. 10.1038/emboj.2009.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snaith H. A., Samejima I., and Sawin K. E., 2005. Multistep and multimode cortical anchoring of tea1p at cell tips in fission yeast. EMBO J. 24: 3690–3699. 10.1038/sj.emboj.7600838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styrkársdottir U., Egel R., and Nielsen O., 1993. The smt-0 mutation which abolishes mating-type switching in fission yeast is a deletion. Curr. Genet. 23: 184–186. 10.1007/BF00352020 [DOI] [PubMed] [Google Scholar]
- Sugiyama T., Cam H. P., Sugiyama R., Noma K., Zofall M. et al. , 2007. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell 128: 491–504. 10.1016/j.cell.2006.12.035 [DOI] [PubMed] [Google Scholar]
- Tang W., Kannan R., Blanchette M., and Baumann P., 2012. Telomerase RNA biogenesis involves sequential binding by Sm and Lsm complexes. Nature 484: 260–264. 10.1038/nature10924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K., and Cooper J. P., 2008. Fission yeast Ccq1 is telomerase recruiter and local checkpoint controller. Genes Dev. 22: 3461–3474. 10.1101/gad.498608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Cohen A. L., Letian A., Tadeo X., Moresco J. J. et al. , 2016. The proper connection between shelterin components is required for telomeric heterochromatin assembly. Genes Dev. 30: 827–839. 10.1101/gad.266718.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., and Baumann P., 2008. Chromosome fusions following telomere loss are mediated by single-strand annealing. Mol. Cell 31: 463–473. 10.1016/j.molcel.2008.05.028 [DOI] [PubMed] [Google Scholar]
- Webb C. J., and Zakian V. A., 2008. Identification and characterization of the Schizosaccharomyces pombe TER1 telomerase RNA. Nat. Struct. Mol. Biol. 15: 34–42. 10.1038/nsmb1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright W. E., Piatyszek M. A., Rainey W. E., Byrd W., and Shay J. W., 1996. Telomerase activity in human germline and embryonic tissues and cells. Dev. Genet. 18: 173–179. [DOI] [PubMed] [Google Scholar]
- Yu T. Y., Kao Y. W., and Lin J. J., 2014. Telomeric transcripts stimulate telomere recombination to suppress senescence in cells lacking telomerase. Proc. Natl. Acad. Sci. USA 111: 3377–3382. 10.1073/pnas.1307415111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Table S1 describes all fission yeast strains used in this work, and are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.9342314.