Cardiovascular disease (CVD) is the leading cause of morbidity and mortality worldwide. Epidemiological and genome-wide association studies have identified environmental and genetic risk factors for CVD. Using human vascular endothelial cells treated with dexamethasone, retinoic acid, caffeine, and selenium...
Keywords: transcription factor, gene regulation, TWAS, gene expression response, genetic variation
Abstract
GWAS and eQTL studies identified thousands of genetic variants associated with complex traits and gene expression. Despite the important role of environmental exposures in complex traits, only a limited number of environmental factors were measured in these studies. Measuring molecular phenotypes in tightly controlled cellular environments provides a more tractable setting to study gene–environment interactions in the absence of other confounding variables. We performed RNA-seq and ATAC-seq in endothelial cells exposed to retinoic acid, dexamethasone, caffeine, and selenium to model genetic and environmental effects on gene regulation in the vascular endothelium—a common site of pathology in cardiovascular disease. We found that genes near regions of differentially accessible chromatin were more likely to be differentially expressed [OR = (3.41, 6.52), ]. Furthermore, we confirmed that environment-specific changes in transcription factor binding are a key mechanism for cellular response to environmental stimuli. Single nucleotide polymorphisms (SNPs) in these transcription response factor footprints for dexamethasone, caffeine, and retinoic acid were enriched in GTEx eQTLs from artery tissues, indicating that these environmental conditions are latently present in GTEx samples. Additionally, SNPs in footprints for response factors in caffeine are enriched in colocalized eQTLs for coronary artery disease (CAD), suggesting a role for caffeine in CAD risk. By combining GWAS, eQTLs, and response genes, we annotated environmental components that can increase or decrease disease risk through changes in gene expression in 43 genes. Interestingly, each treatment may amplify or buffer genetic risk for CAD, depending on the particular SNP or gene considered.
THE vascular endothelium refers to the single layer of cells lining blood vessels that form an interface between the blood and the rest of the body. The endothelium plays an important role in coagulation, thrombosis, leukocyte extravasation, regulation of vascular tone, and angiogenesis (Rajendran et al. 2013). Endothelial dysfunction has been implicated in many pathological processes, including atherosclerosis, hypertension, tumor angiogenesis, wound healing, and preeclampsia (Gimbrone and García-Cardeña 2016). For many of these conditions, genome-wide association studies (GWAS) have been instrumental in identifying a large number of genetic variants associated with disease (e.g., Nikpay et al. 2015). However, the molecular mechanisms linking each variant to the disease phenotype are largely unknown. This is because common risk variants for complex traits tend to fall in noncoding regions and affect regulatory mechanisms that are not yet well characterized (Nica et al. 2010; Gamazon et al. 2015; Gusev et al. 2016; Aguet et al. 2017).
DNase-seq and ATAC-seq (Boyle et al. 2008; Buenrostro et al. 2013) are experimental approaches developed to profile the chromatin landscape and can be used to identify active regulatory elements. Transcription factors bound to chromatin leave characteristic “footprints,” allowing for the identification of many bound transcription factors in a single experiment (Pique-Regi et al. 2011). ATAC-seq offers the advantage of requiring a much smaller number of cells, allowing for increased multiplexing of experiments in small volumes. This enables study of cell lines with differing genetic backgrounds across environmental perturbations, such as the addition of a drug or hormone. Current existing annotations for genetic variants capture regulatory mechanisms in only one arbitrary environment, and thus it is necessary to annotate regulatory regions across multiple conditions. Combining chromatin accessibility data with measures of gene expression, such as RNA-seq, provides insights into the regulatory code governing gene expression. By comparing RNA- and ATAC-seq in the presence/absence of perturbations, it becomes possible to comprehensively study the cellular response and link changes in chromatin accessibility and transcription factor binding to changes in gene expression (Pacis et al. 2015; Nédélec et al. 2016; Alasoo et al. 2018; Gate et al. 2018). The molecular basis of cellular response can be identified by leveraging genetic differences within and across individuals to pinpoint underlying regulatory elements. One such approach that uses genetic variation is quantitative trait loci (QTL) mapping, whereby genetic variants can be linked to differences in gene expression (eQTL) (e.g., Stranger et al. 2007; Aguet et al. 2017) or chromatin accessibility (caQTL) (Degner et al. 2012, among others). Results from these methods, which can be used individually or as complements to one another, have been integrated with GWAS findings to suggest a model in which genetic variants disrupt the chromatin architecture, leading to differences in gene expression that affect complex traits (Gamazon et al. 2015; Gusev et al. 2016; Hormozdiari et al. 2016; Wen et al. 2017).
However, the effect of a genetic variant on a molecular pathway, and, ultimately, on the disease condition, may be modulated by environmental factors (Moyerbrailean et al. 2016b; Knowles et al. 2017, 2018). eQTL studies performed on cells exposed to a stimulus can identify genetic variants whose influence on gene expression response is modulated by the environment, known as response eQTLs (reQTLs) (see for example Maranville et al. 2011; Mangravite et al. 2013; Nédélec et al. 2016; Alasoo et al. 2018; Knowles et al. 2018). Known environmental risk factors for endothelial dysfunction include diabetes, smoking, obesity, high cholesterol, and hypertension (Hadi et al. 2005). Dexamethasone, retinoic acid, caffeine, and selenium are four treatments that have been associated with cardiovascular disease (CVD) and/or endothelial dysfunction (Flores-Mateo et al. 2006; Pan and Baker 2007; Walker 2007; Turnbull et al. 2017), and that produce strong transcriptional responses in endothelial cells (Moyerbrailean et al. 2016b). Dexamethasone is a potent glucocorticoid that serves as a proxy for stress (Sapolsky et al. 2000), and excess may promote calcification within arteriosclerotic lesions (Zhu et al. 2016). Retinoic acid plays a crucial role in the development of the cardiovascular system, and vascular endothelial cells are exposed to high concentrations of retinoic acid and express retinoid receptors (Saito et al. 2007). Caffeine, the most widely consumed stimulant worldwide, promotes vasodilation in the endothelium and has been the subject of numerous studies regarding its association with CVD, with conflicting results (Ding et al. 2014; Turnbull et al. 2017). Selenium is a component of selenoproteins which act as antioxidants, and selenium has been used as a supplement to increase high-density lipoprotein (HDL) cholesterol (Rayman 2012).
Recently, the heterogeneity of the contexts represented in complex datasets has been used as proxy for varying environmental conditions (Knowles et al. 2017; Zhernakova et al. 2017) to identify gene-environment interaction (GxE) effects. The advantage of these approaches resides in the possibility of exploiting pre-existing large-scale eQTL studies that profile gene expression in tissues from a heterogeneous sample of individuals; however, the differences in environmental conditions between individuals is not explicitly ascertained. To characterize genetic and environmental risk factors for cardiovascular disease, it is critical to investigate gene regulation in a relevant cell type under controlled treatment conditions. Here, we developed annotations of genetic variants in key regulatory elements for gene expression response to inform the role of GxE in coronary artery disease.
Materials and Methods
Cell culture
Human umbilical vein endothelial cells (HUVECs) were isolated from human umbilical cord tissue collected shortly following birth. Umbilical cord tissue specimens were obtained from 14 healthy, full-term pregnant women, admitted to DMC Hutzel Women’s Hospital (Detroit, MI). All specimens for this study were collected following guidelines approved by the institutional review board (#013213 MP4E) of Wayne State University. Cord specimens, between 10 and 30 cm in length, were first rinsed with warm phosphate-buffered saline (PBS) and a blunt-ended needle was inserted into the umbilical vein at one end of the cord, and subsequently clamped in place. The cord was then purged to remove any excess blood from the vein. The other end of the cord was then sealed, in a manner identical to the first end, and prewarmed 0.25% trypsin-EDTA (Gibco) was then injected into the vein. Following a 20 min incubation at 37°, detached HUVECs were rinsed from the vein, collected by centrifugation, counted, and seeded into an appropriate vessel at 10,000 cells/cm2, in EGM-2 growth medium (Lonza). Expanded cultures were cryopreserved prior to be used in the experiments. For additional three HUVEC lines from a previous study (Moyerbrailean et al. 2016b) we collected ATAC-seq data and used previously published RNA-seq data (dbGaP accession number phs001176.v1.p1).
Treatments
Treatment concentrations were the same as used in Moyerbrailean et al. (2016b) and were derived from the Clinical Guidelines Mayo Clinic Reference Levels (http://www.mayomedicallaboratories.com) and the CDC National Biomonitoring Report Reference Levels (http://www.cdc.gov/biomonitoring/). Concentrations were M for dexamethasone, M for retinoic acid, M for caffeine, and M for selenium. Vehicle controls were included to represent the solvent used to prepare the different treatments, either ethanol (1 μl of ethanol per 10,000 μl of culturing media) or water.
Each individual cell line was treated on a separate day. Culturing conditions were the same as those found in Moyerbrailean et al. (2016b). Specifically, 3 days before treatment, cells were plated (to passage 7) in eight wells in six-well plates at 5000/cm2 in EGM-2. Following a 24 hr recovery period, the medium was changed to a “starvation medium,” composed of phenol-red free EGM-2, without Hydrocortisone and Vitamin C, and supplemented with 2% CS-FBS. Cell starvation was continued for 48 hr prior to treatment. For each individual cell line, control treatments were performed in duplicate. Cells were treated for 6 hr, and were then collected by scraping the plate on ice. For four cell lines, each well was counted in order to remove 50,000 cells to be used for ATAC-seq while the rest were used for RNA sequencing.
RNA library preparation
Treated cells were collected by centrifugation at 2000 rpm and washed twice using ice-cold PBS. Collected pellets were lysed on the plate, using Lysis/Binding Buffer (Ambion), and frozen at −80°. Polyadenylated mRNAs were subsequently isolated from thawed lysates using the Dynabeads mRNA Direct Kit (Ambion) following the manufacturer’s instructions. RNA-seq libraries were prepared using a protocol modified from the NEBNext Ultradirectional (NEB) library preparation protocol to use 96 Barcodes from BIOOScientific added by ligation, as described in Moyerbrailean et al. (2015). The individual libraries were quantified using the KAPA real-time PCR system, following the manufacturer’s instructions and using a custom-made series of standards obtained from serial dilutions of the phi-X DNA (Illumina). Libraries were pooled and sequenced in multiple sequencing runs for an average of 50 M 300 bp PE reads.
ATAC-seq library preparation
We followed the protocol by Buenrostro et al. (2013) to lyse 50,000 cells and prepare ATAC-seq libraries, with the exception that we used the Illumina Nextera Index Kit (Cat #15055290) in the PCR enrichment step. For three of the four individual cell lines (LP-001, LP-002 and H288-L), the cells were not lysed with 0.1% IGEPAL CA-630 before adding the transposase to begin the ATAC-seq protocol. Individual library fragment distributions were assessed on the Agilent Bioanalyzer and pooling proportions were determined using the qPCR Kapa library quantification kit (KAPA Biosystems). Library pools were run on the Illumina NextSeq 500 Desktop sequencer in the Luca/Pique-Regi laboratory. Barcoded libraries of ATAC-seq samples were pooled and sequenced in multiple sequencing runs for an average coverage of 130 M reads.
Alignment of RNA-seq and ATAC-seq
Reads were aligned to the GRCh37 human reference genome using HISAT2 (Kim et al. 2015) (https://ccb.jhu.edu/software/hisat2/index.shtml, version hisat2-2.0.4), and the human reference genome (GRCh37) with the following options:
where <genome> represents the location of the genome file (genome_snp for ATAC-seq alignment, genome_snp_tran for RNA-seq alignment), and <fastqs_R1.gz> and <fastqs_R2.gz> represent that sample’s fastq files.
The multiple sequencing runs were merged for each sample using samtools (version 2.25.0). We removed PCR duplicates and further removed reads with a quality score of (equating to reads mapped to multiple locations) for the RNA-sequencing analysis, only.
Differential gene expression analysis
To identify differentially expressed (DE) genes, we used DESeq2 (Love et al. 2014) (R version 3.2.1, DESeq2 version 1.8.1). Gene annotations from Ensembl version 75 were used and transcripts with <20 reads total were discarded. Bedtools coverage was utilized to count reads with -s to account for strandedness and -split for BED12 input. The counts were then utilized in DESeq2 to determine changes in gene expression under the different treatment conditions. Multiple test correction was performed using the Benjamini-Hochberg procedure (Benjamini and Hochberg 1995) with a significance threshold of 10%. A gene was considered DE if at least one of its transcripts was DE and had an absolute fold change . The model corrected for the library preparation batch and the sample of origin.
Gene ontology analysis
For each treatment, we tested upregulated and downregulated genes separately for enrichment of biological processes gene ontology terms using the Fisher’s test on the AmiGO 2 web browser (http://amigo.geneontology.org/amigo). Multiple test correction was performed using the Benjamini-Hochberg procedure with a significance threshold of 5% (BH-FDR).
Identification of differentially accessible regions following treatment
To identify differentially accessible regions, we used DESeq2 (Love et al. 2014) (R version 3.2.1, DESeq2 version 1.8.1). We separated the genome into 300 bp regions and bedtools coverage was used to count reads in these regions. A total of 508,140 regions with >0.25 reads per million (high accessibility regions) were then utilized in DESeq2 using the following model:
| (1) |
where represents the internal DESeq2 mean of normalized counts of all samples normalized by sequencing depth for region j and experiment n, is the treatment indicator, and parameter is the treatment effect. Differentially accessible regions (DAR) following exposure to dexamethasone, retinoic acid, caffeine, and selenium, were determined at BH FDR %. We annotated the genomic context of these DARs using chromHMM (Ernst and Kellis 2012), which characterizes chromatin states on the basis of histone marks. We downloaded the precomputed HUVEC annotations from ENCODE (ENCODE Project Consortium 2012). Enrichments were performed with the Fisher’s Exact test. DARs were then compared to gene annotations from Ensembl version 75 to identify those that were within 50 kb of a transcription start site, and a Fisher’s exact test was used to test for enrichment of DARs near DE genes.
Overlap of DE genes and DARs across treatments
We used UpSetR (Conway et al. 2017) to visualize DE genes and DARs shared across treatments. To calculate correlations across treatments, we subset genes or regions of chromatin that were differentially expressed or accessible in any treatment and computed Spearman correlations on the log-fold change.
Transcription factor binding footprints
To detect which transcription factors have footprints in each condition we adapted CENTIPEDE (Pique-Regi et al. 2011) to use the fragment length information contained in the ATAC-seq in the footprint model, and to jointly use in parallel the treatment and control conditions in order to ensure that the same footprint shape is used for the same motif in both conditions.
As in CENTIPEDE, we need to start from candidate binding sites for a given motif model. For each transcription factor we scan the entire human genome (hg19) for matches to its DNA recognition motif using position weight matrix (PWM) models from TRANSFAC and JASPAR as previously described (Pique-Regi et al. 2011). Then for each candidate location l we collect all the ATAC-seq fragments which are partitioned into four bins depending on the fragment length: (1) [39–99], (2) [100–139], (3) [140–179], (4) [180–250]. For each fragment, the two Tn5 insertion sites were calculated as the position 4 bp after the 5′-end in the 5′ to 3′ direction. Then, for each candidate motif, a matrix X was constructed to count Tn5 insertion events: each row represented a sequence match to motif in the genome (motif instance), and each column a specific cleavage site at a relative base pair and orientation with respect to the motif instance. We built a matrix for each fragment length bin, each using a window half-size S = 150 bp resulting in columns, where W is the length of the motif in base pair.
Finally, we fit the CENTIPEDE model in a subset of ∼5000 instances to learn footprint shapes for each factor as in Moyerbrailean et al. (2016a). Then we used CENTIPEDE to analyze the remaining motif instances in the genome and kept those with a CENTIPEDE posterior probability higher than 0.99 to denote locations where the transcription factors are bound, also referred to as “footprints.” We further subset these bound transcription factor sites by including only transcription factors that were active genome-wide in any condition, defined by a significant enrichment (Fisher’s exact test, FDR < 10%) of binding sites in high accessibility regions (see above) compared to a randomly selected set of 500,000 chromatin regions genome-wide (not high accessible regions). There were 882 active factors across all treatments.
Identification of response factors and genetic variants in their binding sites
In order to identify transcription factors that are important for response to treatments, we calculated enrichment scores for each active motif in regions of differentially accessible chromatin using a Fisher’s exact test. We defined motifs that were significantly enriched or depleted in opening or closing chromatin (BH FDR %; Table S3 and Table S4).
For the enrichment analysis with TORUS, we relaxed the threshold for response factors to nominal significance (P-value ) to get better estimates for the confidence intervals for the enrichment parameters. For each tissue and in each treatment, we then annotated each single nucleotide polymorphism (SNP) from the 1000 Genomes project (1000 Genomes Project Consortium 2015) as (1) not in a footprint for a treatment response factor; (2) in a footprint for a treatment response factor (Table S2). SNPs in footprints are generally enriched in eQTLs (Wen et al. 2016). Thus, to account for this treatment-independent effect, we also generated an annotation for SNPs in active footprints in either control condition, which should represent a baseline annotation of candidate regulatory variants. We note that the controls are largely redundant, sharing >93% similarity.
Enrichment of GTEx eQTL and CAD variants in footprints for response factors
To identify treatments for which GTEx eQTLs (Aguet et al. 2017) are enriched in footprints for response factors, we focused on the three artery tissues present in GTEx: tibial, coronary, and aorta. GTEx identified 11,945, 4378, and 9203 genes with an eQTL (eGenes) in the tibial artery, coronary artery, and aorta tissues, respectively. We then used the Torus package (Wen et al. 2016) with the “-est” option to calculate the enrichment of eQTLs in response factors for each treatment. Torus quantitatively assesses the enrichment of molecular annotations (e.g., SNPs in response factors) in GWAS or eQTL datasets. TORUS is based on a hierarchical model, where the prior probability of association for each SNP in a locus is modeled with a logistic function that has one hyper-parameter for each category (i.e., nonresponse factor control footprint, response factor footprint, and distance to the transcription start site as overall baseline). The confidence interval for each enrichment parameter is estimated using a profile likelihood procedure. In this analysis, the summary statistics for all SNPs that can be imputed and tested in the eQTL or GWA study are used, regardless of SNP P-value. Here we used the annotations generated above for SNPs in footprints for response factors and control. As additional annotation, we included distance of the SNP to the transcriptional start site of the gene.
We meta-analyzed the enrichments for each treatment across tissues using a fixed effects model with inverse variance weights. For the CAD enrichment, we used the same annotations as above on the summary statistics of the CAD GWAS, minus the distance of the SNP to the transcriptional start site of a gene. GWAS SNPs were partitioned into blocks based on linkage disequilibrium (LD) pattern.
Genotyping and colocalization
We performed colocalization as previously described using enloc (Wen et al. 2017). Briefly, enloc integrates QTL annotations into GWAS analysis by calculating the enrichment of QTL in complex trait-associated genetic variants, which is then used to identify colocalized SNPs between the two annotations. We performed the colocalization between the CARDIoGRAM CAD summary statistics and eQTLs from all GTEx tissues independently. We considered the lead SNP from a locus with a cumulative posterior inclusion probability (PIP) of >0.5 to be colocalized; 168 SNPs colocalize, corresponding to 139 genes.
To identify polymorphic colocalized SNPs in our dataset, DNA was isolated from umbilical cords and sent to be genotyped using ultralow sequencing performed by Gencove. In our dataset, 21 of the colocalized SNPs (corresponding to 14 eGenes) are polymorphic, and, therefore, can be tested for reQTL.
reQTL mapping
For reQTL mapping of the colocalized GTEx and CAD GWAS SNPs, we tested all 21 SNPs for reQTLs, corresponding to 33 gene-SNP pairs. We used a linear model to identify reQTLs, testing for the effect of genotype dosage on quantile-normalized gene expression fold change between treatment and control. We generated an empirical null distribution by permuting sample labels, and corrected the treatment P-values based on the ranks observed in the permutation. We performed 2820 permutations, corresponding to 3x the number of tests in the treated samples.
TWAS
We used GAMBIT (https://github.com/corbinq/GAMBIT) to conduct a TWAS on the CARDIoGRAM GWAS using the three artery tissues from GTEx as reference gene expression sets. Using a threshold of , there were 44, 17, and 49 genes significantly associated with CAD for the tibial artery, coronary artery, and aorta, respectively, for a total of 74 unique genes in at least one of the three arteries.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Raw sequencing data, including RNA-seq on all 17 individuals, ATAC-seq on 4 individuals, and genotypes for all 17 individuals, have been deposited in the NCBI dbGaP (https://www.ncbi.nlm.nih.gov/gap/) under study ID phs001176.v3.p1.Supplemental text, Supplemental Material, Figure S1, and Tables S1–S8 are available at http://genome.grid.wayne.edu/FUNGEI/data_tables/.
Results
Exposure of endothelial cells to environmental perturbations leads to changes in gene expression and chromatin accessibility
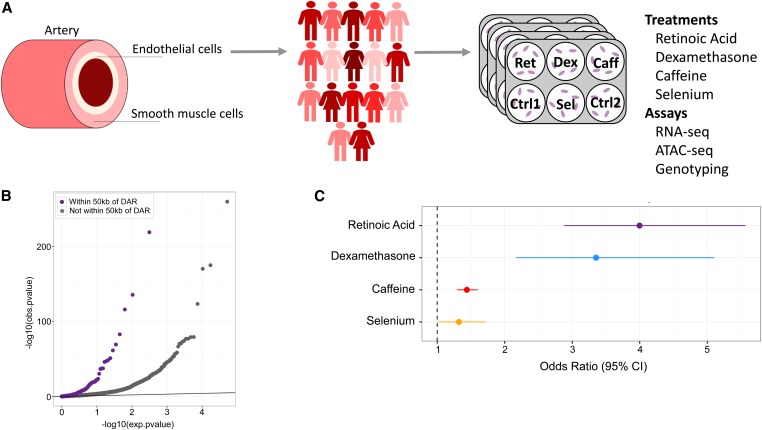
To determine the impact of genetic variation on endothelial cell response to environmental perturbations, we treated human umbilical vein endothelial cells (HUVECs) from 17 unrelated, healthy donors with four compounds (dexamethasone, retinoic acid, caffeine, and selenium) and two vehicle controls. These treatments have been associated with cardiovascular disease and/or endothelial dysfunction (Flores-Mateo et al. 2006; Pan and Baker 2007; Walker 2007; Turnbull et al. 2017) and have been shown in a previous study (Moyerbrailean et al. 2016b) to produce strong transcriptional responses in HUVECs. We then assessed changes in gene expression with RNA-seq in all samples and chromatin accessibility with ATAC-seq in four samples at 6 hr following treatment (Figure 1A).
Figure 1.
Changes in gene expression and chromatin accessibility in HUVECs treated with dexamethasone, retinoic acid, caffeine, and selenium. (A) Diagram of artery layers and study design. HUVECs were treated separately with four compounds and vehicle controls prior to RNA- and ATAC-seq (Materials and Methods). (B) QQ-plot of P-values from DESeq2 analysis of gene expression in response to retinoic acid. The purple dots represent genes within 50 kb of a DAR, while the gray dots represent genes not within 50 kb of a DAR. (C) Enrichment of DE genes within 50 kb of a DAR. Odds ratio was estimated using Fisher’s exact test, and for all treatments P.
Each treatment caused significant transcriptional responses, with >2500 differentially expressed genes in each environment (Benjamini-Hochberg (BH) , ) (Table 1). By gene ontology analysis, we identified an enrichment of differentially expressed genes implicated in biological processes relevant for endothelial function. For example, response to wounding is enriched in genes downregulated in response to dexamethasone, and regulation of blood vessel diameter is enriched in genes downregulated in response to retinoic acid (Table S1).
Table 1. Number of differentially expressed genes and differentially accessible regions.
| Treatment | DE genes | DARs |
|---|---|---|
| Retinoic acid | 4,874 | 222 |
| Dexamethasone | 2,879 | 133 |
| Caffeine | 5,790 | 1038 |
| Selenium | 10,329 | 196 |
Across the four samples for which ATAC-seq was obtained, we identified between 133 and 1038 DARs in a given treatment condition (Table 1). The smaller number of DARs compared with DE genes could be due to the timescale at which we measured response, the increased stochasticity of chromatin opening and closing relative to gene expression, and/or insufficient statistical power. We used chromHMM (Ernst and Kellis 2012) chromatin states derived from ENCODE HUVECs histone marks (Dunham et al. 2012) to annotate DARs. We found an enrichment of DARs in strong enhancer regions in dexamethasone, retinoic acid, and selenium (OR = 5.31, 3.89, and 3.51, respectively. P for all). However, caffeine DARs are only slightly enriched in strong enhancers (OR , P ). Instead, DARs in caffeine are most enriched for active promoters (OR , P ). This difference between treatments could indicate that different types of factors are involved in each response.
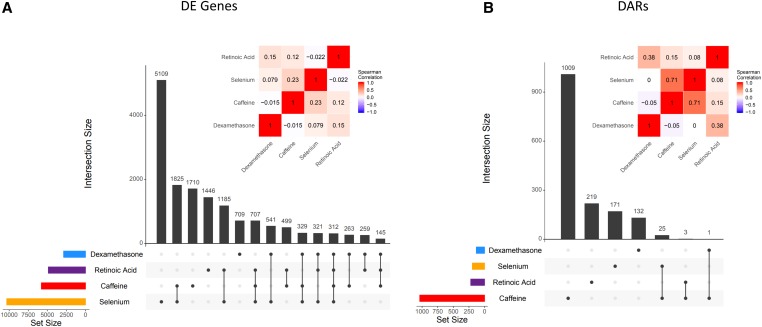
We compared the degree to which transcriptional and chromatin changes were shared across treatments by overlapping DE genes and DARs, respectively, across treatments (Figure 2). Thousands of DE genes are shared by at least two treatments, with 312 genes being differentially expressed across all four. Due to the reduced number of DARs, we have less power to detect sharing across treatments (Figure 2B). As a complementary approach, we calculated the correlation of the log-fold change across all treatments for genes differentially expressed (Figure 2A) and regions differentially accessible (Figure 2B) in any condition.
Figure 2.
Shared DE genes and DARs across treatments. (A) DE gene sharing. Vertical bars indicate the number of genes differentially expressed in each group of treatments. Inset heatmap depicts the Spearman correlation of the logFC of all DE genes between treatments. (B) DAR sharing. Vertical bars indicate the number of DARs in each group of treatments. Inset heatmap depicts the Spearman correlation of the logFC of all DARs between treatments.
To investigate the relationship between differential chromatin accessibility and transcriptional response, we calculated the enrichment of DE genes within 50 kb of a DAR. DE genes in all treatments were significantly enriched near DARs identified in the same treatment. The greatest enrichments were found for dexamethasone and retinoic acid with odds ratios of 3.35 and 3.99, respectively (Figure 1, B and C and Figure S1; Fisher’s exact test; P ). The significant enrichment for DE genes among DARs in dexamethasone and retinoic acid could be due to their mechanism of action, specifically that both treatments are known to work largely through specific nuclear receptors that bind the DNA and influence gene expression directly. Overall these results suggest that changes in chromatin accessibility are an important mechanism through which cells regulate gene expression in response to environmental perturbation.
Specific transcription factors regulate response to treatment
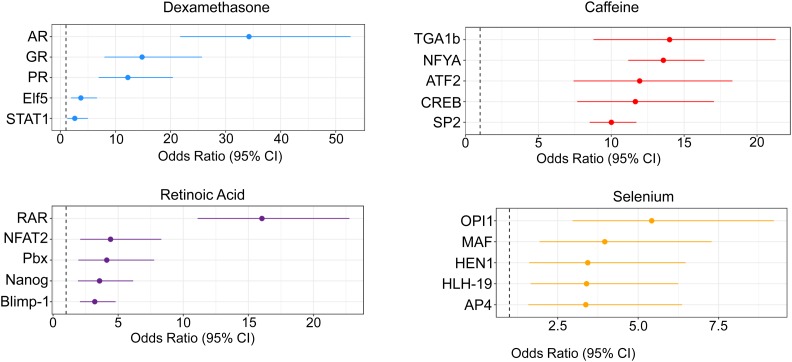
In addition to informing on open chromatin, ATAC-seq data can be analyzed to characterize the transcription factor landscape governing response to treatment. This is particularly important given that the vast majority of genetic variants found to influence complex traits act through gene regulatory regions. In order to identify transcription factors controlling response to each treatment, we used CENTIPEDE (Pique-Regi et al. 2011) to perform footprinting analysis on the ATAC-seq data. Across all conditions, we identified a total of 882 active transcription factors, defined as those with motifs that have CENTIPEDE footprints and are associated with open chromatin (Materials and Methods, Table S7). We hypothesized that transcription factors that are important in the regulation of a particular treatment will be more likely to be found in DARs. The factors with the greatest enrichments in DARs are shown in Figure 3. We refer to these enriched factors as response factors. We identified 293 factors with footprints in regions that become more accessible after treatment, and 38 factors in regions that are less accessible after treatment (Table 2). For well-studied treatments, like retinoic acid and dexamethasone, our approach identified transcription factors known to be central to the regulation of gene expression in response to those treatments. For example, the top retinoic acid response factor was the retinoic acid receptor (Figure 3B). For dexamethasone, the androgen receptor (AR), glucocorticoid receptor (GR), and progesterone receptor (PR) have similar binding sites, and likely all represent sites of GR binding (Schoenmakers et al. 2000; Severson et al. 2018). While for these treatments, we have prior information on the major mechanisms that regulate the cellular response, for caffeine and selenium less is known, and our data can be used to identify the relevant factors (Table S3 and Table S4). Caffeine acts as a phosphodiesterase inhibitor, which increases intracellular cyclic AMP (cAMP) (Essayan 2001). One of the top caffeine response factors in opening chromatin is CREB (cAMP response element-binding protein). In response to selenium treatment the top enriched factors are in regions that become less accessible after treatment (Figure 3).
Figure 3.
Identification of response factors. Topmost enriched transcription factor footprints in DARs in each condition. For selenium, the top factors are enriched in regions of closing chromatin. For all other treatments, the top factors are enriched in regions of opening chromatin.
Table 2. Number of response factors enriched in opening and closing chromatin and SNPs in response factors footprints.
| Treatment | Opening chromatin | Closing chromatin | Annotated SNPs |
|---|---|---|---|
| Retinoic acid | 17 | 0 | 438,507 |
| Dexamethasone | 15 | 0 | 218,793 |
| Caffeine | 261 | 16 | 1,887,282 |
| Selenium | 0 | 22 | 710,611 |
Identifying latent environmental factors in large-scale eQTL datasets from vascular tissues
Large-scale efforts to identify expression eQTLs, such as GTEx (Aguet et al. 2017), have discovered genetic regulators of gene expression in many different tissues, including three types of artery. However, GTEx samples likely represent a composite of different environmental exposures. Here, we propose that latent, unobserved environmental factors contribute to eQTL effects detected in GTEx, and that some of these eQTLs may represent cases of significant interaction effects in specific environmental contexts. To indirectly test this hypothesis, we considered that interactions with latent environments represented by our four in vitro treatments (retinoic acid, caffeine, selenium, and dexamethasone) would be enriched in response factor binding sites. We used centiSNPs (Moyerbrailean et al. 2016b), an extension of the CENTIPEDE framework, to annotate SNPs in response factor footprints, as these SNPs are most likely to disrupt transcription factor binding and modify the transcriptional response. We found that out of 4,104,579 SNPs in footprints for active factors, 56% are in footprints for response factors for at least one of the treatments. We then estimated enrichments for our treatment-specific annotations in eQTLs from the artery samples in GTEx. Using the EM-DAP1 algorithm implemented in the software package TORUS (Wen et al. 2016), we found enrichment of eQTLs in footprints for dexamethasone, retinoic acid, and caffeine response factors, but not for selenium (Figure 4A). These results suggest that exposure to retinoic acid, caffeine, and dexamethasone (or a closely related factor; e.g., cortisol elevated by stress) are represented as latent environments in the GTEx samples and can modulate eQTL effects.
Figure 4.
Latent environments in GTEx and CAD risk. (A) Enrichment of GTEx eQTLs in SNPs within response factor footprints for each treatment. (B) Enrichment of coronary artery disease risk loci in SNPs within response factor footprints for each treatment. (C) For the genes shown in (D), number of concordant (treatment changes gene expression in the same direction as TWAS risk) and discordant (vice versa) gene/treatment pairs. (D) Comparison of logFC of gene expression vs. CAD TWAS z-score. A positive logFC indicates that the treatment increases gene expression. A positive TWAS value indicates that increased expression of the gene is associated with CAD risk.
Latent environments in coronary artery disease
To identify which conditions may be enriched for putative latent GxE in coronary artery disease (CAD), we performed an enrichment analysis for SNPs in footprints for response factors using CAD GWAS data (Nikpay et al. 2015) (Figure 4B). Among the treatment conditions we explored, we found a strikingly high enrichment of 14.5-fold for CAD-associated variants that are found in caffeine response factor footprints. This enrichment is also significantly higher than that found for footprints that are identified in the control condition, which represent regulatory variants that do not interact with the treatments considered here. This suggests a large number of latent GxE effects in cardiovascular disease risk.
To further investigate the role of gene expression changes induced by environmental perturbations in CAD risk, we used Transcriptome Wide Association Study (TWAS). TWAS (Gamazon et al. 2015; Barbeira et al. 2018; Gusev et al. 2018) is an approach based on Mendelian randomization that was developed to infer the causal relationship between gene expression and complex trait variation, using genetic information (genotypes) as instrumental variables. Importantly, TWAS assigns a direction of effect of gene expression on the trait (i.e., whether increased or decreased expression is associated with trait risk). We have performed a TWAS on the CARDIoGRAM GWAS using the three artery tissues from GTEx as reference gene expression sets using the GAMBIT software (https://github.com/corbinq/GAMBIT). There were 44, 17, and 49 genes significantly associated with CAD () for the tibial artery, coronary artery, and aorta, respectively, for a total of 74 unique genes in at least one of the three arteries; 43 of these genes were differentially expressed in at least one condition in our data. For these genes, we compared the TWAS z-score to the change in gene expression in each condition for which the gene is differentially expressed (Figure 4D). Positive TWAS z-scores indicate that increased expression of the gene is associated with increased risk of CAD. Building on the conceptual framework established by Marigorta et al. (2017) for coherent and incoherent genetic effects, we consider instances where the TWAS effect and gene expression response act in the same direction to be concordant, indicating that the treatment effect on that gene amplifies the genetic CAD risk (i.e., treatment increases gene expression, and increased gene expression is associated with increased CAD risk). Neither concordant nor discordant effects predominate for any treatment, indicating that the effect of these treatments on CAD risk is not broadly protective or harmful. Rather, it is a composite of effects at many genes, which can be additive to the genetic effect, or, as we show below, can be further influenced by interactions with genetic factors (GxE).
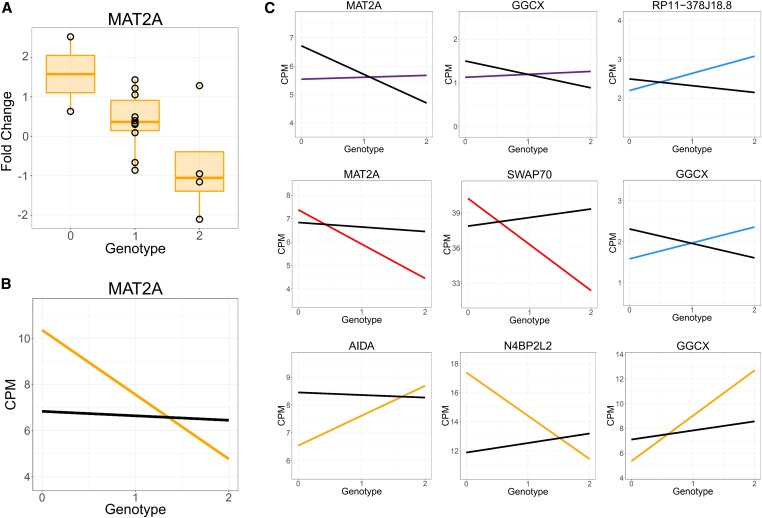
To further dissect the molecular mechanism for GxE in CAD risk, we integrated GTEx artery eQTLs with the GWAS summary statistics and GxE effects in gene expression in our data. We first performed colocalization of GTEx eQTLs with CAD GWAS hits using enloc (Wen et al. 2017) (Materials and Methods) and identified 168 SNP-gene pairs; 40 GTEx tissues had at least one colocalized eQTL. The most represented tissues for colocalized SNPs were the aorta and tibial artery, with 56 and 28 colocalized SNPs, respectively (Table S5). Of the colocalized genes, 14 can be tested for reQTL in our data and 6 have a reQTL in at least one of the treatment conditions considered (permutation P < 0.05, see Table S6). Although with our sample size we do not have the power to detect genome-wide significant signals, we provide examples of our reQTLs (Figure 5). The reQTL for MAT2A acts in different directions depending on the treatment. In Figure 5, A and B, we show MAT2A has a negative reQTL effect following treatment with selenium. However, the reQTL effect is positive following treatment with retinoic acid (first panel of Figure 5C).
Figure 5.
reQTLs. (A) Boxplot depicting logFC for selenium reQTL for MAT2A (B) Same reQTL as in (A), but with trendlines representing gene expression in the treatment and control conditions. (C) All reQTLs. Colors indicate gene expression in the treatment condition (blue, dexamethasone; red, caffeine; orange, selenium; purple, caffeine; black, control).
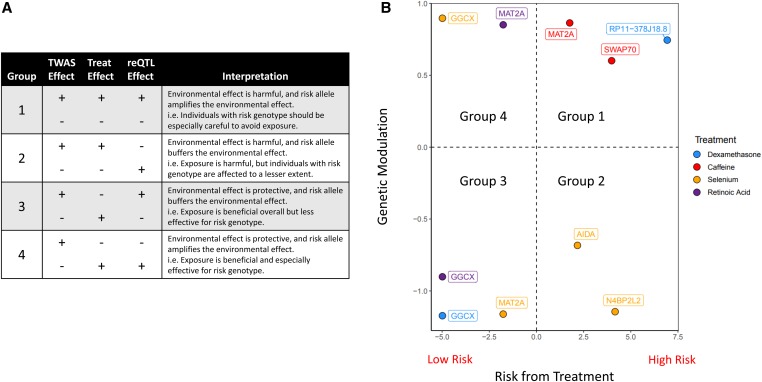
We now develop a conceptual framework that integrates TWAS, gene expression changes, and reQTLs to illustrate how genetic variation modulates the effect of an environmental exposure on a complex trait (Figure 6). We describe eight possible scenarios, which can be divided into four groups (Figure 6A) based on the relationships between TWAS and treatment effect direction, and treatment and reQTL effect direction. A positive TWAS value indicates that increased expression of the gene is associated with increased risk. A positive gene expression change indicates that the treatment increases gene expression. The treatment effect can act in the same direction as the TWAS effect, indicating that the treatment increases risk (Groups 1 and 2), or they can act in opposite directions, where the treatment buffers risk (Groups 3 and 4).
Figure 6.
reQTLs modulate environmental effects on complex traits. (A) Table relating TWAS, gene expression, and reQTL effects. (B) Risk associated with treatment vs. genetic modulation by reQTLs. The risk associated with treatment represents the TWAS effect multiplied by the sign of the logFC of the treatment. Positive values indicate that the treatment amplifies CAD risk. Genetic modulation represents the reQTL effect multiplied by the sign of the logFC of the treatment. Positive values indicate that the risk allele amplifies the treatment effect.
Because our reQTLs originated from GWAS-eQTL colocalization, we have polarized the reQTL effect to the CAD risk allele. Therefore, a positive reQTL effect indicates that the risk allele causes an increased change in gene expression relative to the nonrisk allele following exposure to a treatment. The reQTL and treatment effect can act in the same direction, indicating that the risk allele amplifies the environmental effect (Groups 1 and 4), or opposite direction, buffering the environmental effect (Groups 2 and 3).
Of the 10 reQTL we identified, six are in Groups 1 and 3, where the reQTL increases disease risk by amplifying harmful environmental effects (Group 1) or buffering protective environmental effects (Group 3) (Figure 6B). The remaining four are in Groups 2 and 4, where the reQTL decreases disease risk by buffering harmful environmental effects (Group 2) or enhancing protective environmental effects (Group 4). These results show that environmental effects on disease risk are contingent on the individual’s genotype, with certain exposures amplifying a specific genetic risk but buffering others.
Discussion
Vascular endothelial dysfunction has been implicated in a variety of cardiovascular and noncardiovascular diseases. Here we have characterized the transcriptional and chromatin landscape in endothelial cells exposed to dexamethasone, retinoic acid, caffeine, and selenium.
While all of these treatments cause significant changes in gene expression and chromatin accessibility, they act through varied pathways. Dexamethasone and retinoic acid are steroid hormones, whose action at the molecular and transcriptional level has been well-studied (Oakley and Cidlowski 2013; di Masi et al. 2015). Caffeine and selenium have been investigated at the molecular level, particularly with regard to caffeine’s role in inhibiting the breakdown of cAMP and selenium’s role as a cofactor (Echeverri et al. 2010; Roman et al. 2014). However, the regulators of gene expression response to these compounds are less characterized, especially in endothelial cells. We were able to confirm with retinoic acid and dexamethasone that our approach in analyzing ATAC-seq data identifies biologically relevant response factors, as we found enrichment of known intracellular steroid receptors like the retinoic acid receptor and glucocorticoid receptor. Upon extension of this framework to caffeine and selenium, we identified key response factors for these treatments that were previously unknown (e.g., Sp2 for caffeine).
Combining the ATAC-seq data with known genetic variation in human populations, we have generated a set of annotations for genetic variants in the footprints of treatment response factors (1300 motifs genome-wide) that may affect binding, and, therefore, gene expression response in the different conditions considered. Using this annotation in combination with large-scale genetic studies of gene expression (GTEx) and CAD risk (Cardiogram), we detected potential latent environmental effects and assigned a putative mechanism of action for variants associated with these molecular and organismal traits, through disruption of transcription factor binding. We found that dexamethasone, retinoic acid, and caffeine were enriched in GTEx eQTLs from three different arteries in our response factor annotations. This is not surprising given the widespread use of caffeine in the general population and the expression of the glucocorticoid receptor and retinoic acid receptor in most GTEx tissues (Aguet et al. 2017). Interestingly, we did not find an enrichment for the selenium annotation, which is expected considering that we are exposed to only limited amounts of selenium on a daily basis. Our observation that genetic loci associated with risk of coronary artery disease are highly enriched in the binding sites of transcription factors that regulate response to caffeine, suggests that caffeine, or a compound that activates a similar regulatory response, modulates coronary artery disease risk. This is an intriguing finding given the extensive number of studies that have explored the link between caffeine, coffee consumption, and heart disease (Ding et al. 2014; Grosso et al. 2017; Turnbull et al. 2017).
Here, we developed a novel conceptual framework that combines information on specific environmental perturbations collected in vitro with large-scale heterogeneous datasets interrogating genetic effects on both molecular and organismal phenotypes. While our study demonstrates that there is a significant enrichment for genetic associations in context specific regulatory annotations, our small sample size had limited power to directly validate these latent GxE effects. Nevertheless, this approach can be used to guide the design of future studies to directly test for GxE. Increasing the sample size would likely have resulted in a greater number of genome-wide significant reQTLs, but there is a trade-off between sample size and the number of environmental conditions which can be assayed in one experiment. For example, one of the first studies of reQTLs identified 26 reQTLs in 120 LCLs in response to dexamethasone (Maranville et al. 2011). More recently, 503 and 244 reQTLs were identified for Salmonella and Listeria in 175 macrophages (Nédélec et al. 2016). In a panel of 45 iPSC-derived cardiomyocyte lines treated with varying concentrations of doxorubicin, 447 reQTL were discovered (Knowles et al. 2018), while 387 reQTL were discovered in 86 iPSC-derived macrophage lines treated with interferon gamma and Salmonella infection (Alasoo et al. 2018). Based on these and other studies, it is difficult to determine an ideal sample size, as power to identify reQTLs depends on the stimulus considered and the method used.
We show that, in principle, we can leverage molecular phenotyping data in response to tightly controlled treatments to characterize the complex interplay between genotypes and environment and their role in complex traits, highlighting the importance of latent environmental exposures in large-scale datasets. When considering CAD risk, both concordant and discordant GxE effects on gene expression are observed across multiple genes without either trend predominating. This would indicate that the effect of these treatments on CAD risk is not broadly protective or harmful when all genes are considered. Environmental effects therefore may explain the discrepancies between transcriptional risk scores and genetic risk scores (Marigorta et al. 2017), and the limited portability of genetic risk scores even within populations (Mostafavi et al. 2019). In the context of precision medicine, these latent environmental exposures need to be accounted for when predicting genetic risk for diseases, as genetic effects measured in large datasets may indeed represent GxE, and, consequently, accurate prediction of genetic risk can be achieved only by accounting for these interactions.
Acknowledgments
We thank Anna Di Rienzo, members of the Luca and Pique-Regi laboratories, and the anonymous reviewers for helpful discussions and comments. This work was supported by the American Heart Association (14SDG20450118 to F.L.) and the National Institute of General Medical Sciences of the National Institutes of Health (R01GM109215 to F.L. and R.P.-R. and F30GM131580 to A.S.F.). The authors declare that they have no competing interests.
Footnotes
Communicating editor: E. Hauser
Literature Cited
- 1000 Genomes Project Consortium, Auton, A., L. D. Brooks, R. M. Durbin, E. P. Garrison, et al., 2015. A global reference for human genetic variation. Nature 526: 68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguet, F., K. G. Ardlie, B. B. Cummings, E. T. Gelfand, G. Getz et al., 2017. Genetic effects on gene expression across human tissues. Nature 550: 204–213 [corrigenda: Nature 553: 530 (2018)]. 10.1038/nature24277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alasoo, K., J. Rodrigues, S. Mukhopadhyay, A. J. Knights, A. L. Mann et al., 2018. Shared genetic effects on chromatin and gene expression indicate a role for enhancer priming in immune response. Nat. Genet. 50: 424–431. 10.1038/s41588-018-0046-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbeira, A. N., S. P. Dickinson, R. Bonazzola, J. Zheng, H. E. Wheeler et al., 2018. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 9: 1825 10.1038/s41467-018-03621-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y., and Y. Hochberg, 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Boyle, A. P., S. Davis, H. P. Shulha, P. Meltzer, E. H. Margulies et al., 2008. High-resolution mapping and characterization of open chromatin across the genome. Cell 132: 311–322. 10.1016/j.cell.2007.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro, J. D., P. G. Giresi, L. C. Zaba, H. Y. Chang, and W. J. Greenleaf, 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10: 1213–1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway, J. R., A. Lex, and N. Gehlenborg, 2017. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33: 2938–2940. 10.1093/bioinformatics/btx364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner, J. F., A. Pai, R. Pique-Regi, J.-B. B. Veyrieras, D. J. Gaffney et al., 2012. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature 482: 390–394. 10.1038/nature10808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Masi, A., L. Leboffe, E. De Marinis, F. Pagano, L. Cicconi et al., 2015. Retinoic acid receptors: from molecular mechanisms to cancer therapy. Mol. Aspects Med. 41: 1–115. 10.1016/j.mam.2014.12.003 [DOI] [PubMed] [Google Scholar]
- Ding, M., S. N. Bhupathiraju, A. Satija, R. M. van Dam, and F. B. Hu, 2014. Long-term coffee consumption and risk of cardiovascular disease: a systematic review and a dose-response meta-analysis of prospective cohort studies. Circulation 129: 643–659. 10.1161/CIRCULATIONAHA.113.005925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encode Project Consortium, 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri, D., F. R. Montes, M. Cabrera, A. Galán, and A. Prieto, 2010. Caffeine’s vascular mechanisms of action. Int. J. Vasc. Med. 2010: 834060 10.1155/2010/834060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst, J., and M. Kellis, 2012. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods 9: 215–216. 10.1038/nmeth.1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essayan, D. M., 2001. Cyclic nucleotide phosphodiesterases. J. Allergy Clin. Immunol. 108: 671–680. 10.1067/mai.2001.119555 [DOI] [PubMed] [Google Scholar]
- Flores-Mateo, G., A. Navas-Acien, R. Pastor-Barriuso, and E. Guallar, 2006. Selenium and coronary heart disease: a meta-analysis. Am. J. Clin. Nutr. 84: 762–773. 10.1093/ajcn/84.4.762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamazon, E. R., H. E. Wheeler, K. P. Shah, S. V. Mozaffari, K. Aquino-Michaels et al., 2015. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 47: 1091–1098. 10.1038/ng.3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gate, R. E., C. S. Cheng, A. P. Aiden, A. Siba, M. Tabaka et al., 2018. Genetic determinants of co-accessible chromatin regions in activated T cells across humans. Nat. Genet. 50: 1140–1150. 10.1038/s41588-018-0156-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimbrone, M. A., and G. García-Cardeña, 2016. Endothelial cell dysfunction and the pathobiology of atherosclerosis. Circ. Res. 118: 620–636. 10.1161/CIRCRESAHA.115.306301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosso, G., J. Godos, F. Galvano, and E. L. Giovannucci, 2017. Coffee, caffeine, and health outcomes: an umbrella review. Annu. Rev. Nutr. 37: 131–156. 10.1146/annurev-nutr-071816-064941 [DOI] [PubMed] [Google Scholar]
- Gusev, A., A. Ko, H. Shi, G. Bhatia, W. Chung et al., 2016. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 48: 245–252. 10.1038/ng.3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev, A., N. Mancuso, H. Won, M. Kousi, H. K. Finucane et al., 2018. Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat. Genet. 50: 538–548. 10.1038/s41588-018-0092-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadi, H. A., C. S. Carr, and J. Al Suwaidi, 2005. Endothelial dysfunction: cardiovascular risk factors, therapy, and outcome. Vasc. Health Risk Manag. 1: 183–198. [PMC free article] [PubMed] [Google Scholar]
- Hormozdiari, F., M. van de Bunt, A. V. Segre, X. Li, J. W. J. Joo et al., 2016. Colocalization of GWAS and eQTL signals detects target genes. Am. J. Hum. Genet. 99: 1245–1260. 10.1016/j.ajhg.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D., B. Langmead, and S. L. Salzberg, 2015. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12: 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles, D. A., J. R. Davis, H. Edgington, A. Raj, M.-J. Favé et al., 2017. Allele-specific expression reveals interactions between genetic variation and environment. Nat. Methods 14: 699–702. 10.1038/nmeth.4298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles, D. A., C. K. Burrows, J. D. Blischak, K. M. Patterson, D. J. Serie et al., 2018. Determining the genetic basis of anthracycline-cardiotoxicity by molecular response QTL mapping in induced cardiomyocytes. eLife 7: pii: e33480. 10.7554/eLife.33480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M. I., W. Huber, and S. Anders, 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangravite, L. M., B. E. Engelhardt, M. W. Medina, J. D. Smith, C. D. Brown et al., 2013. A statin-dependent QTL for GATM expression is associated with statin-induced myopathy. Nature 502: 377–380. 10.1038/nature12508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maranville, J. C., F. Luca, A. L. Richards, X. Wen, D. B. Witonsky et al., 2011. Interactions between glucocorticoid treatment and cis-regulatory polymorphisms contribute to cellular response phenotypes. PLoS Genet. 7: e1002162 10.1371/journal.pgen.1002162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marigorta, U. M., L. A. Denson, J. S. Hyams, K. Mondal, J. Prince et al., 2017. Transcriptional risk scores link GWAS to eQTLs and predict complications in Crohn’s disease. Nat. Genet. 49: 1517–1521. 10.1038/ng.3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafavi, H., A. Harpak, D. Conley, J. K. Pritchard, and M. Przeworski, 2019. Variable prediction accuracy of polygenic scores within an ancestry group. bioRxiv. doi: 10.1101/629949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyerbrailean, G. A., G. O. Davis, C. T. Harvey, D. Watza, X. Wen et al., 2015. A high-throughput RNA-seq approach to profile transcriptional responses. Sci. Rep. 5: 14976 10.1038/srep14976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyerbrailean, G. A., C. A. Kalita, C. T. Harvey, X. Wen, F. Luca et al., 2016a Which genetics variants in dnase-seq footprints are more likely to alter binding? PLoS Genet. 12: e1005875 10.1371/journal.pgen.1005875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyerbrailean, G. A., A. L. Richards, D. Kurtz, C. A. Kalita, G. O. Davis et al., 2016b High-throughput allele-specific expression across 250 environmental conditions. Genome Res. 26: 1627–1638. 10.1101/gr.209759.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nédélec, Y., J. Sanz, G. Baharian, Z. A. Szpiech, A. Pacis et al., 2016. Genetic ancestry and natural selection drive population differences in immune responses to pathogens. Cell 167: 657–669.e21. 10.1016/j.cell.2016.09.025 [DOI] [PubMed] [Google Scholar]
- Nica, A. C., S. B. Montgomery, A. S. Dimas, B. E. Stranger, C. Beazley et al., 2010. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 6: e1000895 10.1371/journal.pgen.1000895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikpay, M., A. Goel, H.-H. Won, L. M. Hall, C. Willenborg et al., 2015. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 47: 1121–1130. 10.1038/ng.3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley, R. H., and J. A. Cidlowski, 2013. The biology of the glucocorticoid receptor: new signaling mechanisms in health and disease. J. Allergy Clin. Immunol. 132: 1033–1044. 10.1016/j.jaci.2013.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacis, A., L. Tailleux, A. M. Morin, J. Lambourne, J. L. MacIsaac et al., 2015. Bacterial infection remodels the DNA methylation landscape of human dendritic cells. Genome Res. 25: 1801–1811. 10.1101/gr.192005.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, J., and K. M. Baker, 2007. Retinoic acid and the heart. Vitam. Horm. 75: 257–283. 10.1016/S0083-6729(06)75010-5 [DOI] [PubMed] [Google Scholar]
- Pique-Regi, R., J. F. Degner, A. A. Pai, D. J. Gaffney, Y. Gilad et al., 2011. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 21: 447–455. 10.1101/gr.112623.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran, P., T. Rengarajan, J. Thangavel, Y. Nishigaki, D. Sakthisekaran et al., 2013. The vascular endothelium and human diseases. Int. J. Biol. Sci. 9: 1057–1069. 10.7150/ijbs.7502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayman, M. P., 2012. Selenium and human health. Lancet 379: 1256–1268. 10.1016/S0140-6736(11)61452-9 [DOI] [PubMed] [Google Scholar]
- Roman, M., P. Jitaru, and C. Barbante, 2014. Selenium biochemistry and its role for human health. Metallomics. 6: 25–54. 10.1039/C3MT00185G [DOI] [PubMed] [Google Scholar]
- Saito, A., A. Sugawara, A. Uruno, M. Kudo, H. Kagechika et al., 2007. All-trans retinoic acid induces in vitro angiogenesis via retinoic acid receptor: possible involvement of paracrine effects of endogenous vascular endothelial growth factor signaling. Endocrinology 148: 1412–1423. 10.1210/en.2006-0900 [DOI] [PubMed] [Google Scholar]
- Sapolsky, R. M., L. M. Romero, and A. U. Munck, 2000. How do glucocorticoids influence stress responses? Integrating permissive, suppressive, stimulatory, and preparative actions. Endocr. Rev. 21: 55–89. 10.1210/edrv.21.1.0389 [DOI] [PubMed] [Google Scholar]
- Schoenmakers, E., G. Verrijdt, B. Peeters, G. Verhoeven, W. Rombauts et al., 2000. Differences in DNA binding characteristics of the androgen and glucocorticoid receptors can determine hormone-specific responses. J. Biol. Chem. 275: 12290–12297. 10.1074/jbc.275.16.12290 [DOI] [PubMed] [Google Scholar]
- Severson, T. M., Y. Kim, S. E. P. Joosten, K. Schuurman, P. van der Groep et al., 2018. Characterizing steroid hormone receptor chromatin binding landscapes in male and female breast cancer. Nat. Commun. 9: 482 10.1038/s41467-018-02856-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger, B. E., A. C. Nica, M. S. Forrest, A. Dimas, C. P. Bird et al., 2007. Population genomics of human gene expression. Nat. Genet. 39: 1217–1224. 10.1038/ng2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull, D., J. V. Rodricks, G. F. Mariano, and F. Chowdhury, 2017. Caffeine and cardiovascular health. Regul. Toxicol. Pharmacol. 89: 165–185. 10.1016/j.yrtph.2017.07.025 [DOI] [PubMed] [Google Scholar]
- Walker, B. R., 2007. Glucocorticoids and cardiovascular disease. Eur. J. Endocrinol. 157: 545–559. 10.1530/EJE-07-0455 [DOI] [PubMed] [Google Scholar]
- Wen, X., Y. Lee, F. Luca, and R. Pique-Regi, 2016. Efficient integrative multi-SNP association analysis via deterministic approximation of posteriors. Am. J. Hum. Genet. 98: 1114–1129. 10.1016/j.ajhg.2016.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, X., R. Pique-Regi, and F. Luca, 2017. Integrating molecular QTL data into genome-wide genetic association analysis: probabilistic assessment of enrichment and colocalization. PLoS Genet. 13: e1006646 10.1371/journal.pgen.1006646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhernakova, D. V., P. Deelen, M. Vermaat, M. van Iterson, M. van Galen et al., 2017. Identification of context-dependent expression quantitative trait loci in whole blood. Nat. Genet. 49: 139–145. 10.1038/ng.3737 [DOI] [PubMed] [Google Scholar]
- Zhu, D., N. A. Rashdan, K. E. Chapman, P. W. Hadoke, and V. E. MacRae, 2016. A novel role for the mineralocorticoid receptor in glucocorticoid driven vascular calcification. Vascul. Pharmacol. 86: 87–93. 10.1016/j.vph.2016.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Raw sequencing data, including RNA-seq on all 17 individuals, ATAC-seq on 4 individuals, and genotypes for all 17 individuals, have been deposited in the NCBI dbGaP (https://www.ncbi.nlm.nih.gov/gap/) under study ID phs001176.v3.p1.Supplemental text, Supplemental Material, Figure S1, and Tables S1–S8 are available at http://genome.grid.wayne.edu/FUNGEI/data_tables/.