Abstract
Germplasm collections hold valuable allelic diversity for crop improvement and genetic mapping of complex traits. To gain access to the genetic diversity within the USDA National Small Grain Collection (NSGC), we developed the Barley Recombinant Inbred Diverse Germplasm Population (BRIDG6), a six-row spring barley multiparent population (MPP) with 88 cultivated accessions crossed to a common parent (Rasmusson). The parents were randomly selected from a core subset of the NSGC that represents the genetic diversity of landrace and breeding accessions. In total, we generated 6160 F5 recombinant inbred lines (RILs), with an average of 69 and a range of 37–168 RILs per family, that were genotyped with 7773 SNPs, with an average of 3889 SNPs segregating per family. We detected 23 quantitative trait loci (QTL) associated with flowering time with five QTL found coincident with previously described flowering time genes. A major QTL was detected near the flowering time gene, HvPpd-H1 which affects photoperiod. Haplotype-based analysis of HvPpd-H1 identified private alleles to families of Asian origin conferring both positive and negative effects, providing the first observation of flowering time-related alleles private to Asian accessions. We evaluated several subsampling strategies to determine the effect of sample size on the power of QTL detection, and found that, for flowering time in barley, a sample size >50 families or 3000 individuals results in the highest power for QTL detection. This MPP will be useful for uncovering large and small effect QTL for traits of interest, and identifying and utilizing valuable alleles from the NSGC for barley improvement.
Keywords: barley; multiparent mapping population (MPP); QTL; flowering time; NAM; multiparent advanced generation intercross (MAGIC), multiparental populations, MPP
GERMPLASM collections contain valuable allelic diversity for crop improvement. Germplasm panels assembled from such collections have been used in association mapping studies to characterize the genetic architecture of agronomic traits (e.g., Huang et al. 2010). While there are many examples of the successful application of association panels in plants, they are less useful for characterizing traits that are influenced by alleles at low frequencies or that are difficult to evaluate in germplasm that is not adapted to the evaluation environment (Morrell et al. 2012). To identify the genetic basis of agronomically important genetic variation, and to make use of that variation in plant breeding programs, it is critical to exploit approaches to more efficiently explore and utilize germplasm collections.
Barley is the fourth most important cereal crop in the world, and is used for animal feed, human food, and to produce malt for the brewing and distilling industries. While still used predominantly for animal feed, the proportion of barley used for malt has increased from 10% in the 1960s to 20% since 1980 (Langridge 2018). There are two primary market classes of barley: two-rowed and six-rowed. The name refers to the arrangement of fertile florets forming either two or six “rows” of kernels up and down the length of the rachis. While this floral architecture is controlled primarily by a single gene (vrs1), plant breeders have maintained these populations as separate market classes with relatively few crosses made between two-rowed and six-rowed parents (Komatsuda et al. 2007). For malting and brewing, six-rowed barley has long been preferred in the United States. The six-rowed types have slightly smaller grains and notably higher starch-degrading enzyme activity needed to convert starch to sugars in the brewing process (Weaver 1944). This characteristic is particularly important for brewers in the U.S. that use nonbarley adjuncts as a source of carbohydrate.
The National Small Grains Collection (NSGC) contains 33,176 accessions of barley cultivars, landraces, breeding lines, and genetic stocks (Knüpffer 2009). It is a rich source of phenotypic variation for traits such as disease and insect resistance (Bonman et al. 2005), but screening the entire collection requires significant time and resources, and is limited by the challenge of accurately phenotyping accessions that are adapted to regions much different from where evaluation will take place (Huang et al. 2010). To improve the efficiency and practicality of evaluating diverse germplasm, the NSGC Core (hereafter referred to as “The Core”) was developed to represent the genetic diversity of the NSGC (Bockelman and Valkoun 2010). The Core contains 2417 accessions and has been evaluated for agronomic, morphological, and disease resistance traits, and genotyped with 6913 single nucleotide polymorphisms (SNPs) from the Barley Illumina iSelect 9K SNP (see www.ars-grin.gov/npgs for a list of traits for which data are available) (Muñoz-Amatriaín et al. 2014).
Two common approaches to investigating the genetic basis of the phenotypic variation in a collection include selecting individual accessions to create biparental mapping populations and assembling panels for association mapping. The former approach is limited by the number of allelic variants per locus that can be evaluated. Additionally, small mapping populations have limited statistical power for detection of small effect QTL and tend to inflate estimated effects of loci (Beavis 1998). The major limitations of association mapping are the large number of individuals that are needed for sufficient mapping power, the challenge of accurately accounting for population structure, and the potential inclusion of accessions that are poorly adapted to common evaluation environments, and, thus, difficult to phenotype accurately. While population structure can be modeled appropriately to avoid spurious marker-trait associations (Pritchard and Rosenberg 1999), many of these limitations cannot be addressed within either of these types of experimental populations.
Multi-parent population (MPP) designs can address some of the limitations of biparental and association mapping populations, and improve access to diverse germplasm for investigating trait variation and genetic architecture e.g., (Huang et al. 2015). MPP designs such as nested association mapping (NAM) populations and multiparent advanced generation intercross (MAGIC) populations (Macdonald and Long 2007; Cavanagh et al. 2008; Yu et al. 2008) sample parents from germplasm collections, use controlled crossing schemes, and subsequently advance the population through multiple generations of intermating or single seed descent. The populations can be designed to have a fixed number of founders and family sizes, setting a floor on allele frequency, and improving allelic representation for phenotypic evaluation (Kover et al. 2009). These designs create an expected lower bound on allele frequency that is based on the total population size, number of parents, size of the family created with each parent, and contribution of each parent (i.e., F1, BC1, BC2).
Assuming that the total population size is the limiting factor for phenotyping a population, then increasing the number of parents to capture more diversity will result in the tradeoff of having smaller family sizes to estimate allelic effects. Early NAM designs focused on large family sizes and a smaller number of parents (e.g., Buckler et al. 2009; McMullen et al. 2009; Jordan et al. 2011; Maurer et al. 2015; Bajgain et al. 2016). With sufficient family size, it is possible to use standard analytical approaches in biparental quantitative trait loci (QTL) mapping within a family, but at the expense of sampling less diversity among parents.
Adaptation of barley to production in a wide latitudinal gradient has been possible due to significant changes in flowering time (Jones et al. 2008). Additionally, variation for flowering time can facilitate uniform maturity and harvest, and escape from environmental pressures such as drought and disease (Shavrukov et al. 2017). It is likely that multiple traits are correlated with a response to this latitudinal variation. For example, in barley QTL for the severity of infection by the pathogenic fungus Fusarium graminearum and days-to-flowering (DTF) map to the same regions. It is unclear whether the correlation is due to pleiotropic effects of DTF or a tightly linked resistance gene (de la Pena et al. 1999; Choo et al. 2004; Nduulu et al. 2007). Therefore, a better understanding of the genetic control of flowering time in barley will help illuminate its relation to correlated traits. Two wild barley (Hordeum vulgare spp. spontaneum) NAM populations, the AB-NAM and HEB-25, were used to dissect complex traits, including DTF (Maurer et al. 2015; Nice et al. 2016, 2017). Both populations detect QTL related to flowering time. However, the latitudinal range of wild barley is much narrower than the range over which barley is cultivated (Jones et al. 2008), thus domesticated barley may harbor a greater phenotypic range for flowering time than can be identified in populations involving the wild progenitor.
Here, we describe the development and characterization of a new MPP resource for breeders and geneticists that includes allelic diversity from the primary gene pool of barley. The Barley Recombinant Inbred Diversity Germplasm (BRIDG6) population uses Rasmusson—a six-row malting barley cultivar adapted to the upper Midwest—as a common parent. Rasmusson parentage improves adaptation to the evaluation environment, making it possible to evaluate the effects of genetic variation in lines sampled from The Core. This improves the potential to detect and measure the effect of alleles that contribute to phenotypic diversity. The objectives of this study were to: (1) characterize the allelic diversity from the NSGC captured in the BRIDG6 population; (2) evaluate the effectiveness of the BRIDG6 population to map flowering time; (3) characterize donor parent allele effects for flowering time, and (4) determine the effect of subsampling the BRIDG6 population to inform future MPP design and to identify experimental designs appropriate for mapping additional traits in this MPP.
Materials and Methods
Germplasm selection
The cultivar Rasmusson was chosen as the common parent for its adaptation to the environmental conditions in the upper Midwestern United States and its superior yield (Smith et al. 2010); 92 donor parents were selected from the NSGC Core to cross to Rasmusson. Parents were randomly selected from accessions that had spring growth habit and six-row spike morphology based on USDA NSGC “passport” information. The Core was previously genotyped (Muñoz-Amatriaín et al. 2014) with the Barley Illumina iSelect 9K SNP assay (Comadran et al. 2012). Parents for the MPP were chosen from among lines with <10% missing genotypic data and <10% heterozygosity. After further quality control on SNP genotyping data (described in detail below), one set of three and another set of two parents were found to be genetically identical. A single representative of these parents was retained and redundant families were combined, resulting in 89 families. The GBS and exome capture data for parent CIho15362 were inconsistent, and information for that parent and corresponding progeny in family 619 were removed, resulting in 88 families.
MPP development
Each of the 88 parents was crossed to Rasmusson, which was used as the female parent, to develop the 88 families that comprise the population. The F1 progeny in each family were advanced via single seed descent for four generations to the F5. All generations of self-pollination were carried out in a greenhouse to minimize outcrossing and inadvertent selection. The last generation of inbreeding in the greenhouse was done in winter 2013/2014; however, plants that produced too little seed (n = 352) were sent to a winter nursery in New Zealand in spring of 2014 for seed increase prior to evaluation in the field. All other F5 RILs were stored as remnant seed and grown in the field at the University of Minnesota Agricultural Experiment Station in Saint Paul, MN in the summer of 2014 as a seed increase and to collect phenotypic data. RILs were harvested from the 2014 field experiment and ∼50 g of seed were stored as long-term seeds stocks for future experiments. RILs increased in New Zealand were added to long-term seed stocks and to the 2015 field experiments.
Genotyping and SNP discovery in the BRIDG6 population
Three genotyping approaches were used for the BRIDG6 parents, the progeny RILs, and The Core. The BRIDG6 parents and six-row barley accessions in The Core were genotyped using the barley Infinium 9K iSelect SNP chip (Comadran et al. 2012). The BRIDG6 parents and the progeny RILs were also genotyped using genotyping-by-sequencing (GBS) technology (Elshire et al. 2011; Poland et al. 2012). Furthermore, 78 of the BRIDG6 parents were genotyped using an exome capture design (Mascher et al. 2013), and these data were used for quality control analysis as described below. Analyses of BRIDG6 parents with The Core used the 9K iSelect SNPs, and analyses of the BRIDG6 parents and RILs used the GBS SNPs. The number of intersected SNPs among 9k iSelect chip data, exome capture resequencing data for BRIDG6 parents, and the GBS data for the BRIDG6 progenies were calculated with a Perl script available in the Github repository and displayed as Venn diagram (Supplemental Material, Figure S1).
9k iSelect SNP chip
The Core, which included the BRIDG6 parents, was previously genotyped using the barley 9K iSelect SNP chip (Comadran et al. 2012; Muñoz-Amatriaín et al. 2014). iSelect SNPs were filtered for minor allele frequency (MAF) 0.01 across The Core which requires at least one individual in the BRIDG6 donor parents with the minor allele, markers missing no more than 50% of data, and a line missing no more than 10% of data.
GBS SNP identification and quality control
The BRIDG6 parents and RILs were genotyped using GBS (Elshire et al. 2011). DNA was extracted from leaf tissue collected from a single plant using the Mag-Bind Plant DNA Plus kit from Omega Bio-tek (Norcross, GA), following the manufacturer’s instructions. Genomic DNA was quantified using Quant-iT PicoGreen dsDNA Chip Kit and normalized to 20 ngµl−1. Barcoded GBS libraries were created using Pst1-Msp1 restriction enzymes following the protocol in Poland et al. (2012). The samples were pooled together at 192-plex for progeny and 48-plex for the parents to create pooled libraries and were sequenced on an Illumina Hi-Seq 2500 generating single-end 125 bp sequence reads. Sequencing was performed on an Illumina Hi-Seq 2500 at the North Carolina State University Genome Science Lab, Raleigh, NC to generate paired-end sequence reads.
Quality control on the sequence data used FastQC (Andrews 2010). Reads were aligned to the reference genome assembly (Mascher et al. 2017) using the aln method of Burrows–Wheeler aligner (BWA) version 0.7.10 (Li and Durbin 2009). All other options were default. SNP calling was performed using the TASSEL 5 GBSv2 pipeline (Glaubitz et al. 2014) using 64 base kmer length and minimum kmer count of 5. TASSEL uses a quantitative SNP calling likelihood ratio pHet/pErr >1 approach to score heterozygotes. This approach results in genotype scores in individual samples to be set as homozygous if the ratio of donor to common allele does not pass a cutoff. Homozygous calls that had at least one read supporting the donor allele were set to missing data. VCFtools version 4.2 (Danecek et al. 2011) was used to remove SNPs with MAF <0.3% and a minimum of five reads per site. We genotyped four individuals of the donor parent Rasmusson to obtain a consensus genotype. If a variant had more than one genotype call across the four replicates, the site was set to missing in the entire population. Further quality control on the genotypes involved the removal of SNPs that were missing or heterozygous in Rasmusson, monomorphic SNPs, SNPs segregating for one homozygous and one heterozygous genotype across the RILs. SNPs within 100 bp of another SNP in a family were also removed. Within a family, SNPs with >20% heterozygosity were set to missing. Also within a family, SNPs with significant excess of contribution from one parent based on a χ2 test were set to missing. Samples with >90% missing values were removed from the dataset. Finally, SNPs which had become monomorphic due to the removal of samples with a high degree of missing values subsequently were also removed.
For mixed model association analysis, missing genotype data were imputed using the LD-kNNi method (Money et al. 2015) implemented in TASSEL 5.2.35 (Bradbury et al. 2007). Prior to the imputation, the genotype dataset was filtered to remove SNPs with MAF <0.03 and <10 genotype calls. The parameters used for imputation were as follows: number of high LD sites = 30, number of nearest neighbors = 10, and maximum distance between sites to find LD = 10 Mb. Raw and imputed GBS SNP genotypes are available in The Sequence Read Archive (SRA) at NCBI under BioProject number: PRJNA488050 and The Triticeae Toolbox (T3, https://triticeaetoolbox.org/barley), respectively. To reduce the number of redundant variants, we used LD pruning as implemented in PLINK version 1.9 (Chang et al. 2015) with LD (r2 > 0.8) within windows of 50 kb with a step size of 5 kb.
Exome capture sequencing and SNP identification
A subset of the BRIDG6 parents (n = 78) were also genotyped using the NimbleGen exome capture designed for barley (Mascher et al. 2013) followed by Illumina resequencing. DNA was extracted from leaf tissue collected from a single plant using a standard 2× CTAB isolation protocol (Saghai-Maroof et al. 1984). The barley Roche (Madison, WI) NimbleGen SeqCap EZ developer probe pool was used to construct genomic libraries following the previously described protocol (Jordan et al. 2015). Sequencing was performed on an Illumina Hi-Seq 2500 at the University of Kansas Medical Center Genome Sequencing Facility, Kansas City, KS or the University of Minnesota Genomics Center, Minneapolis, MN to generate 125 bp paired-end sequence reads.
Quality control assessment for sequence reads used FastQC (Andrews 2010). Trimmomatic (Bolger et al. 2014) was used to trim reads based on quality, with a minimum phred score <33. Reads were aligned to the reference genome assembly (Mascher et al. 2017) using the aln option in the BWA version 0.7.12 (Li and Durbin 2009). All other options were default. The verification of mate-pair information (minimum distance used was 200), sorting, conversion to BAM format, and marking of duplicate read pairs was done using PicardTools version 2.300 (http://picard.sourceforge.net). Variant detection and genotype calling were performed using GATK Toolkit version 3.3.0 (commands “HaplotypeCaller”) (McKenna et al. 2010).
VCFtools version 4.2 (Danecek et al. 2011) was used to filter the parental SNP calls. The exome capture data revealed redundant parental lines (≥99% similarity). In these cases, the sample with less missing data was retained (as with the GBS dataset), for a total of 78 unique parental genotypes with exome capture data.
HvPpd-H1 haplotype analysis
We identified the interval containing HvPpd-H1—a locus previously demonstrated to be a major contributor to flowering time variation in barley—using a BLAST search against the barley reference genome (Mascher et al. 2017). Bedtools intersect (Quinlan and Hall 2010) was used to identify SNPs in the exome capture resequencing data that overlapped the locus. SNPs identified in the HvPpd-H1 locus were annotated as intergenic, genic, nonsynonymous, and synonymous using Annovar (Wang et al. 2010).
Nonsynonymous changes at phylogenetically conserved sites are more likely to contribute to phenotypic change (Kono et al. 2016, 2017). SNPs were tested with a likelihood ratio test for sequence conservation (Chun and Fay 2009) implemented in the software BAD_Mutations (Kono et al. 2016, 2017). BAD_Mutations was run with all the 59 publicly available angiosperm genome sequences on Phytozome (https://phytozome.jgi.doe.gov) and Ensembl Plants (http://plants.ensembl.org/) using the default parameters. A SNP was considered deleterious if a logistic regression for masked and unmasked models (Kono et al. 2017) was <0.05. The masked model accounts for reference bias by removing the query species from the comparison, which results in a more conservative estimate of constraint than the unmasked model. The program compute from the libsequence library (Thornton 2003) was used to estimate the minimum number of recombination events (RM) using the four-gamete test (Hudson and Kaplan 1985).
The 78 parental genotypes were used to identify haplotypes using Mesquite v.3.0.4 (Maddison and Maddison 2018). For this analysis, heterozygous SNPs were treated as missing data. Two sequences from wheat, Triticum turgidum subsp. dicoccon (AB691868.1 and AB691869), were used as outgroups to determine the ancestral state of SNPs in the HvPpd-H1 region. To compare the positions of previously reported putative causal variants for photoperiod responsive and insensitive types (Turner et al. 2005), we aligned the HvPpd-H1 resequencing data for four accessions (GenBank IDs AY970701.1, AY970702.1, AY970703.1, AY970704.1) (Turner et al. 2005) to the Morex reference genome in Mesquite v.3.0.4 (Maddison and Maddison 2018).
Genetic characterization of BRIDG6 donor parents and progeny
Using the GBS SNPs, pairwise genetic distance between the donor parents and between each donor parent and Rasmusson was calculated using the dist.gene program from the ape R package (Paradis et al. 2004), ignoring SNPs where either parent in a pair was heterozygous or had a missing genotype. This analysis was done prior to any filtering parameter in the BRIDG6 population in order to describe the maximum amount of diversity represented in each family in the dataset. The degree of population differentiation between the BRIDG6 donor parents and their source population, the six-row NSGC Core accessions, was estimated by calculating the average fixation index using F statistics (FST; Weir and Cockerham 1984). Since there are many more six-row Core accessions (1172) than BRIDG6 donor parents (88), 88 individuals were sampled 1000 times with replacement from the six-row Core accessions to compare to the BRIDG6 donor parents. The estimated variance component for each allele was calculated using the iSelect SNPs for the BRIDG6 donor parents and six-row accessions in the Core using the varcomp function in the R package hierfstat (Goudet 2005). Average FST is reported from the 100 subsamples.
Principal component analysis (PCA) of the BRIDG6 donor parents and the rest of the six-row barley accessions in the Core was conducted with the iSelect SNPs with the program smartPCA from EIGENSOFT version 6.1.4 (Price et al. 2006). Donor parents and NSGC subpopulation assignments are as previously described (Muñoz-Amatriaín et al. 2014; Table 1). PCA of the BRIDG6 donor parents and the population was conducted using the GBS dataset filtered for maximum 10% missing data at any SNP.
Table 1. Parents of the BRIDG6 population.
| Parent GRIN designator | RIL designatora | RILs | Proportion dissimilar SNPs to Rasmusson | Subpopulationb | Cultivation historyc | Accession originc |
|---|---|---|---|---|---|---|
| CIho02205 | 1 | 45 | 0.041 | Asian | Cultivated | Japan |
| CIho02367 | 2 | 72 | 0.032 | Coastal Mediterranean | Cultivated | Khartoum, Sudan |
| CIho02542 | 3 | 38 | 0.031 | Admixed | Cultivar | Apulia, Italy |
| CIho04050 | 4 | 79 | 0.015 | Asian | Landrace | Govi-Altay, Mongolia |
| CIho04184 | 5 | 66 | 0.042 | Asian | Landrace | Takhar, Afghanistan |
| CIho04264 | 6 | 70 | 0.032 | Coastal Mediterranean | Cultivated | Venezuela |
| CIho06020 | 7 | 70 | 0.034 | Coastal Mediterranean | Cultivated | New South Wales, Australia |
| CIho06294 | 8 | 76 | 0.021 | Admixed | Landrace | Malatya, Turkey |
| CIho07247 | 9 | 74 | 0.029 | Admixed | Breeding | Utah, United States |
| CIho10034 | 10 | 75 | 0.022 | Central European | Cultivar | Norway |
| CIho13743 | 11 | 65 | 0.037 | East African | Landrace | Asmara, Eritrea |
| CIho14052 | 12 | 76 | 0.033 | Coastal Mediterranean | Landrace | Biskra, Algeria |
| CIho14216 | 13 | 69 | 0.042 | Asian | Landrace | Mongolia |
| CIho14228 | 14 | 68 | 0.042 | Asian | Landrace | Mongolia |
| CIho14258 | 15 | 80 | 0.044 | Asian | Landrace | Nimruz, Afghanistan |
| CIho14319 | 16 | 87 | 0.020 | Central European | Cultivated | Storstrom, Denmark |
| CIho14881 | 17 | 69 | 0.003 | East African | Landrace | Asmara Provence, Melotti Brewery |
| CIho15349 | 18 | 81 | 0.032 | Coastal Mediterranean | Landrace | Kebili, Tunisia |
| CIho15362 | 19 | 82 | 0.028 | Coastal Mediterranean | Landrace | Bizerte, Tunisia |
| CIho15600 | 20 | 78 | 0.015 | Central European | Breeding | Quebec, Canada |
| PI039590 | 21 | 60 | 0.035 | Coastal Mediterranean | Landrace | Mascara, Algeria |
| PI048133 | 22 | 60 | 0.033 | Coastal Mediterranean | Cultivar | Victoria, Australia |
| PI054915 | 23 | 53 | 0.044 | Asian | Cultivated | Egypt |
| PI057089 | 24 | 68 | 0.041 | Asian | Landrace | Nordland, Norway |
| PI061533 | 25 | 37 | 0.043 | Asian | Landrace | Shanxi, China |
| PI064022 | 26 | 57 | 0.044 | Asian | Breeding | Texas, United States |
| PI069421 | 27 | 59 | 0.017 | Central European | Breeding | Texas, United States |
| PI071075 | 28 | 55 | 0.027 | Unassigned | Landrace | Hebei, China |
| PI087844 | 29 | 72 | 0.026 | Admixed | Breeding | Tashkent, Uzbekistan |
| PI094875 | 30 | 55 | 0.033 | Coastal Mediterranean | Landrace | Hamgyong Puk, North Korea |
| PI119925 | 31 | 56 | 0.018 | Coastal Mediterranean | Cultivated | Asuncion, Paraguay |
| PI129482, PI328052 | 32, 51 | 144 (61, 83) | 0.026 | Unknown | Cultivated | Krakow, Poland |
| PI135758 | 33 | 66 | 0.043 | Asian | Landrace | Sar-e Pol, Afghanistan |
| PI157884 | 34 | 53 | 0.033 | Coastal Mediterranean | Cultivar | Emilia-Romagna, Italy |
| PI163409 | 35 | 62 | 0.030 | Admixed | Cultivated | Buenos Aires, Argentina |
| PI173518 | 36 | 78 | 0.026 | Admixed | Landrace | Samsun, Turkey |
| PI174431 | 37 | 73 | 0.023 | Admixed | Cultivar | France |
| PI178609 | 38 | 51 | 0.024 | Admixed | Landrace | Amasya, Turkey |
| PI181102 | 39 | 37 | 0.040 | Asian | Landrace | Himachal Pradesh, India |
| PI190711 | 40 | 81 | 0.020 | Central European | Landrace | Hokkaido, Japan |
| PI190790 | 41 | 86 | 0.009 | Central European | Landrace | North Korea |
| PI223883 | 42 | 71 | 0.042 | Asian | Landrace | Kondoz, Afghanistan |
| PI270692 | 43 | 71 | 0.033 | Coastal Mediterranean | Landrace | Puno, Peru |
| PI282616 | 44 | 83 | 0.020 | Admixed | Landrace | Israel |
| PI296460 | 45 | 47 | 0.037 | East African | Landrace | Senhit Provence, Keren |
| PI298708 | 46 | 62 | 0.037 | East African | Landrace | Kefa, Ethiopia |
| PI320217 | 47 | 56 | 0.032 | Coastal Mediterranean | Cultivated | Western Australia |
| PI327680, PI327716, PI327859 | 48, 59, 50 | 168 (51, 60, 57) | 0.025 | Central European | Landrace | Odesa, Ukraine |
| PI328155 | 52 | 61 | 0.023 | Central European | Landrace | Bulgaria |
| PI328485 | 53 | 69 | 0.034 | Coastal Mediterranean | Landrace | Crete, Greece |
| PI328577 | 54 | 69 | 0.034 | Coastal Mediterranean | Landrace | Peloponnese, Greece |
| PI328632 | 55 | 40 | 0.037 | Coastal Mediterranean | Landrace | Crete, Greece |
| PI329000 | 56 | 82 | 0.038 | East African | Landrace | Unknown Provence |
| PI356719 | 57 | 48 | 0.032 | Coastal Mediterranean | Landrace | Morocco |
| PI362207 | 58 | 51 | 0.028 | Admixed | Cultivar | Yvelines, France |
| PI371817 | 59 | 53 | 0.031 | Admixed | Cultivar | New South Wales, Australia |
| PI382860 | 60 | 56 | 0.036 | East African | Landrace | Gonder Provence, Debark |
| PI386650 | 61 | 63 | 0.037 | East African | Landrace | Gonder Provence, Debark |
| PI387098 | 62 | 87 | 0.037 | Central European | Breeding | Texas, United States |
| PI390281 | 63 | 61 | 0.026 | Central European | Landrace | Macedonia |
| PI392524 | 64 | 66 | 0.034 | Admixed | Breeding | Cape Province, South Africa |
| PI401964 | 65 | 76 | 0.023 | Admixed | Cultivated | Cundinamarca, Colombia |
| PI402037 | 66 | 70 | 0.022 | Admixed | Cultivated | Cundinamarca, Colombia |
| PI402164 | 67 | 64 | 0.015 | Central European | Cultivated | Cundinamarca, Colombia |
| PI410451 | 68 | 64 | 0.042 | Asian | Landrace | Azad Kashmir, Pakistan |
| PI410483 | 69 | 87 | 0.042 | Asian | Landrace | Azad Kashmir, Pakistan |
| PI415348 | 70 | 73 | 0.024 | Central European | Landrace | Macedonia |
| PI428411 | 71 | 51 | 0.020 | Admixed | Cultivar | Federal District Mexico |
| PI434794 | 72 | 82 | 0.016 | Central European | Breeding | Quebec, Canada |
| PI447100 | 73 | 65 | 0.032 | Coastal Mediterranean | Cultivated | Zaragoza, Spain |
| PI449279 | 74 | 74 | 0.029 | Admixed | Breeding | Zaragoza, Spain |
| PI467733 | 75 | 82 | 0.022 | Coastal Mediterranean | Cultivar | Norway |
| PI467758 | 76 | 89 | 0.022 | Central European | Cultivar | Hokkaido, Japan |
| PI531896 | 77 | 73 | 0.038 | Admixed | Cultivated | Victoria, Australia |
| PI531917 | 78 | 64 | 0.038 | Admixed | Cultivated | Egypt |
| PI531986 | 79 | 73 | 0.040 | Admixed | Cultivated | Kafr al-Sheikh, Egypt |
| PI573615 | 80 | 71 | 0.043 | Asian | Landrace | Hubei, China |
| PI573878 | 81 | 62 | 0.042 | Asian | Landrace | Mongolia |
| PI574094 | 82 | 68 | 0.037 | Asian | Landrace | Dhawalagiri, Nepal |
| PI584786 | 83 | 75 | 0.036 | Admixed | Landrace | Mechi, Nepal |
| PI584977 | 84 | 81 | 0.041 | Asian | Landrace | Iraq |
| PI640095 | 85 | 60 | 0.018 | Admixed | Landrace | Mongolia |
| PI640117 | 86 | 66 | 0.034 | Admixed | Breeding | Texas, United States |
| PI640220 | 87 | 74 | 0.021 | Admixed | Landrace | Tashkent, Uzbekistan |
| PI640226 | 88 | 68 | 0.018 | Central European | Breeding | Texas, United States |
| PI640265 | 89 | 89 | 0.018 | East African | Landrace | Shewa Provence, Alem Gena |
| PI640286 | 90 | 63 | 0.020 | Central European | Breeding | Texas, United States |
| PI640376 | 91 | 86 | 0.017 | Central European | Breeding | Texas, United States |
| PI467797 | 92 | 63 | 0.029 | Unassigned | Landrace | Krasnodar, Russia |
RIL designator is used in the naming system for each family of recombinant inbred lines.
Subpopulation assignments are from Muñoz Amatriaín et al. (2014) Table S2. Italicized subpopulation designations were interpolated from principal component analysis clusters.
Cultivation history and accession origin are from passport data in the germplasm resources information network (GRIN).
For analysis of the excess or deficit of contribution from parents to the BRIDG6 individuals, we filtered the original VCF file of GBS genotype calls allowing only 50% missing data across the SNPs using VCFtools (Danecek et al. 2011). Then, accessions were separated by family including the donor parent and common parent (Rasmusson). For each family, markers with MAF <0.05, or heterozygosity ≥0.20 were removed. Genotype calls in the donor parent and Rasmusson were verified or completed (when only one parent was missing and the present parent was not heterozygous) based on the allelic segregation in the RILs. SNPs closer than 100 bp to another marker, within a family, were removed. SNPs with evidence of double crossover were set to missing following a previously described error detection approach (Lincoln and Lander 1992). Segregation distortion in the progeny was estimated for each SNP using the R package qtl (Broman et al. 2003) and the geno.Table function. The excess or deficit of parental contribution was calculated using separate nonindependent chi-square tests at SNPs across families, giving combined statistics indicating deviation from Mendelian inheritance at each SNP.
The folded nucleotide site frequency spectrum (SFS) was calculated using an in-house R script to count the number of times the minor allele was present at each site. The SFS was estimated using the 8101 SNPs that were ultimately used in mapping and the 89 parents (including Rasmusson). Figure S2 shows the percentage of markers that are observed in the data at different minor allele counts (1–44).
Linkage disequilibrium (LD) decay was estimated in the 88 Parents and in the RILs independently as the pairwise correlation (r2) between SNPs. The SNPs were the same used for the GWAS analysis, which included imputed SNPs. The markers were filtered based on MAF <3% in the population. PLINK v1.90b (Chang et. Al. 2015) was used to calculate r2 between all pairs of SNPs using the parameters–ld-window-r2 0–ld-window 999999–ld-window-kb 767855.1. An R script was used to calculate the distance between markers and to plot physical distance vs. r2 to determine LD decay (Figure S3). We estimated the average r2 for nonoverlapping windows of 500 kbp.
Physical mapping of iSelect SNPs
A number of genetic mapping studies have reported genetic map positions for barley SNPs (e.g., Comadran et al. 2012; International Barley Genome Sequencing Consortium et al. 2012; Muñoz-Amatriaín et al. 2014), and the physical locations for a portion of the iSelect SNPs relative to the barley reference genome (Mascher et al. 2017) have been reported previously (Comadran et al. 2012; International Barley Genome Sequencing Consortium et al. 2012; Cantalapiedra et al. 2015; Colmsee et al. 2015). To identify physical positions for the iSelect SNP set, we used the contextual sequences of 7864 SNPs from the iSelect SNPs (typically either 121 or 241 bp long) (Comadran et al. 2012) to perform BLASTn (Altschul et al. 1990) searches against the masked reference genome (Mascher et al. 2017). We configured BLASTn to reject hits with an expected value >0.000001 and identity <90%. For sequences where BLAST did not identify a unique position, we used previously reported genetic map positions (Muñoz-Amatriaín et al. 2014) to infer the most likely chromosome of origin. If two blast hits were inferred to be within 100 kb from each other, we systematically chose the leftmost hit; otherwise, we identified the physical position with the hit closest to the genetic position. The mapping of iSelect SNPs was performed using the Python program SNP_Utils. Approximately 400 SNPs were not aligned due to either no hits above the threshold of e-value ≥0.000001 or identity ≤90% or multiple hits with ≥100 kb without genetic positions. For those 400 SNPs, we used a manual BLAST search of contextual sequence using the IPK web server with default parameters. The BLAST search used no threshold and involved selecting the hits with the highest rank of the identity and scores. If the contextual sequence did not have a unique BLAST hit in the genome, we used SNPMeta (Kono et al. 2014) to identify potential genes where the SNPs reside (see link to Barley_SNP_Annotations in Table S3 for SNPmeta results), then a BLAST search of the gene against the masked reference genome was performed to identify the physical location of the best hit.
Identification of loci controlling flowering time
Based on a literature search, we assembled a list of genes previously reported to be involved in flowering time in barley (Comadran et al. 2012; Alqudah et al. 2014; Russell et al. 2016; Table S5). To estimate the distance between previously reported genes and SNPs, we aligned published versions of the gene sequences to the masked reference genome (Mascher et al. 2017) using a BLAST search, and estimated the genomic interval for each genic region. This search was implemented using the BLAST_to_BED Python program (Hoffman 2016).
Phenotypic evaluation
The BRIDG6 population was evaluated for flowering time in three environments: Crookston, MN in 2014, Saint Paul, MN in 2015, and Fargo, ND in 2015. All experiments were planted in an unreplicated modified augmented design II (MADII) with replicated checks (Lin and Poushinsky 1985). Rasmusson was planted as the primary check in the center of each three row by five column block. Four secondary check varieties, Robust, M61, KLBC4_130i-kk, and Gen2-036, were planted in random blocks throughout the field. DTF was measured as the number of days after planting when 50% of the heads had emerged completely from the boot. Raw phenotypic data are available in T3. Spatial variation was accounted for using a moving average covariate (Technow 2011). Best linear unbiased predictions (BLUPs) were calculated for each RIL across three environments using the R package lme4 (Bates et al. 2015), with family as a fixed effect and line within family and family within environment as random effects. Outlier observations were determined by their studentized residuals using a significance level of α = 0.001, and were removed from further analysis (n = 232). BLUPs were only calculated for lines that had phenotypic observations from all three environments (n = 5189). BLUPs were used for GWAS. Broad sense heritability was calculated from the phenotypic data on a line mean basis across environments. BLUP values for all RILs are available in Table S2.
Mixed-model association analysis
BLUPs and genotypic data were used for association analysis using the gwas2 function in the NAM R package (Xavier et al. 2015). The NAM package makes use of prior information about population structure and relaxes linkage phase assumptions to allow minor allele effects to vary among families. Family was included as a stratification argument and a kinship matrix was calculated among all individuals using the GBS SNP data. For each SNP we calculated the percent of phenotypic variance explained and allele effect relative to Rasmusson. Marker-trait associations were considered significant when −log(P-value) was above a false discovery rate (FDR) threshold of 0.05, which was a −log(P-value) of 5.18 (Benjamini and Hochberg 1995). Since pairwise SNP LD decayed to half the original value at a distance of 5 Mbp, we used that as a window size for QTL (Figure S3). Marker-trait associations were considered to occur at independent loci when significant markers were separated by >5 Mbp. All markers within 5 Mbp of significant SNPs were included in the QTL region defined for subsequent analysis of subsampling strategies (see below). Because some QTL on chromosome 2H were in close proximity, we also determined if SNPs within a QTL were in high LD (r > 0.7) with other significant SNPs on that chromosome. QTL assignments and SNP significance values are available in Table 2. The model permitted the estimation of the allele effect of each allele. Individual SNP allele effects for each family were summarized within a QTL region to compare allele effects across more families.
Table 2. Twenty-three QTL associated with flowering time detected in the BRIDG6 population.
| QTLa | LD groupb | Maximum -log(p)c | Location of most significant SNP (cM) | Location of most significant SNP (bp) | Number of significant SNPd | Total number of SNP in QTL region (5 Mbp)e | Max. Families segregatingf | Min. Families Segregatingg |
|---|---|---|---|---|---|---|---|---|
| 1_01 | A | 9.2 | 50.22 | 427,905,991 | 1 | 13 | 26 | 26 |
| 1_02 | B | 13.42 | 88.84 | 524,189,956 | 1 | 50 | 22 | 22 |
| 2_01 | C | 142.43 | 20.01 | 27,204,805 | 102 | 268 | 47 | 11 |
| 2_02 | D | 6.03 | 43.17 | 60,578,410 | 1 | 21 | 44 | 44 |
| 2_03 | E | 7.98 | 57.86 | 181,205,852 | 2 | 6 | 36 | 17 |
| 2_04 | F | 7.75 | 58.01 | 187,898,571 | 1 | 1 | 24 | 24 |
| 2_05 | E | 5.88 | 58.01 | 197,287,292 | 2 | 11 | 42 | 29 |
| 2_06 | E | 14.27 | 58.8 | 241,566,109 | 4 | 9 | 31 | 23 |
| 2_07 | EG | 10.36 | 58.8 | 257,828,128 | 2 | 4 | 37 | 13 |
| 2_08 | EH | 6.47 | 58.8 | 403,342,684 | 1 | 7 | 38 | 38 |
| 2_09 | I | 7.9 | 58.8 | 437,926,040 | 1 | 6 | 22 | 22 |
| 2_10 | J | 10.18 | 58.8 | 486,598,181 | 1 | 1 | 18 | 18 |
| 2_11 | K | 17.17 | 59.61 | 493,756,332 | 1 | 2 | 14 | 14 |
| 2_12 | K | 10.16 | 60.2 | 521,775,169 | 2 | 6 | 38 | 28 |
| 2_13 | K | 20.95 | 60.2 | 549,097,349 | 10 | 14 | 30 | 17 |
| 2_14 | K | 8.17 | 60.2 | 564,309,170 | 2 | 2 | 33 | 29 |
| 2_15 | L | 8.7 | 62.04 | 582,332,455 | 1 | 4 | 15 | 15 |
| 2_16 | M | 6.66 | 58.01 | 763,099,837 | 1 | 76 | 37 | 37 |
| 3_01 | N | 23.19 | 51.71 | 123,315,601 | 3 | 9 | 40 | 16 |
| 5_01 | O | 5.49 | 50.04 | 403,946,354 | 1 | 10 | 30 | 30 |
| 5_02 | P | 7.31 | 144.97 | 630,784,439 | 1 | 41 | 47 | 47 |
| 7_01 | Q | 32.44 | 34.85 | 39,192,808 | 41 | 197 | 41 | 13 |
| 7_02 | R | 44.56 | 131.1 | 646,266,549 | 2 | 99 | 42 | 13 |
Maximum–log(P-value) for any SNP in the QTL.
Flowering time QTL. Association determined by FDR threshold >.0.05.
QTL with the same letter include SNPs in high LD (r > 0.7)
Number of significant SNP in the QTL.
Total number of SNPs within 5 Mbp of QTL.
Maximum number of families for which a significant SNP in the QTL segregates.
Minimum number of families for which a significant SNP in the QTL segregates.
Isolating the effect of individual QTL
To isolate the effect of the most significant QTL for flowering time (QTL 2.1, see Results), which includes the linkage group 2H region around HvPpd-H1, we ran the association analysis using the significant SNPs in the next most significant QTL (36 SNPs in a 5 Mbp window located on linkage group 7H) as covariates. This analysis was performed using the R NAM package (Xavier et al. 2015). The allele effects at each SNP were identified, comparing lines according to the haplotypes identified in the parents.
Mapping population subsampling strategy
To evaluate the relationship between association panel size and marker trait associations, we compared two main sampling strategies: subsampling by individuals and subsampling by families. For the subsampling based on individuals, we tested four different population sizes: 264, 528, 968, and 2024. We compared random selection conditioned on sampling the same number of individuals from each family (3, 6, 11, and 23 individuals per family respectively) vs. random selection across the entire BRIDG6 regardless of the representation of families. Random selection based on families evaluated the use of a fixed number of individuals (25) sampled from 10, 25, 50, and 75 families. Each of the 12 subsampling strategies was run 100 iterations. The same phenotypic and genotypic data used for mapping in the total population was filtered for MAF of 0.05 in each subsample. Mixed-model association analysis was conducted using the methods described above. A marker significance threshold of FDR 0.05 in each subsample was calculated by taking the logarithm of α = 0.05 divided by the number of markers found significant in each subsample, therefore the significance threshold varied by bootstrap. The power of each subsampling strategy to detect QTL was determined by the number of times a QTL, that was identified using the total data set, was identified in the subsample. A QTL was counted if at least one of the significant SNPs in the subsample was present in the physical region where the QTL was identified in the total BRIDG6 sample.
Data and code availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. The computer code used for all analyses (unless otherwise specified) and Figures are available at https://github.com/UMN-BarleyOatSilphium/BRIDG6. Raw GBS SNP genotypes are available in The Sequence Read Archive (SRA) at NCBI under BioProject number PRJNA488050. Imputed GBS SNP genotype and DTF phenotype data are available at The Triticeae Toolbox (T3, https://triticeaetoolbox.org/barley). We have made supplemental files available through the GSA Figshare portal. The computer code used for all analyses is available at https://github.com/UMN-BarleyOatSilphium/BRIDG6. Accessory files not appropriate as supplementary materials are available through a public archive system from our university library (https://doi.org/10.13020/c5kj-af95). Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7757252.
Results
Development of a spring, six-row barley, MPP
The BRIDG6 population was developed by crossing 92 spring six-row barley landrace and breeding accessions from the Core to the modern cultivar, Rasmusson. Comparative analysis of the Core parents showed that two (PI129482 and PI328052) and three (PI327680, PI327716, PI327859) of the parents were duplicates of the same accession. RILs from each of these five pseudoreplicated parents were collapsed into two families, resulting in a total of 89 families. Family HR619 was removed from GWAS analysis due to unknown parentage identified through inconsistencies between donor parent and population genotype. The resulting 88 families had a range of 37–168 F5 RILs, with an average family size of 69 RILs. In total, the BRIDG6 population contains 6336 F5 RILs (Table 1).
Genetic characterization
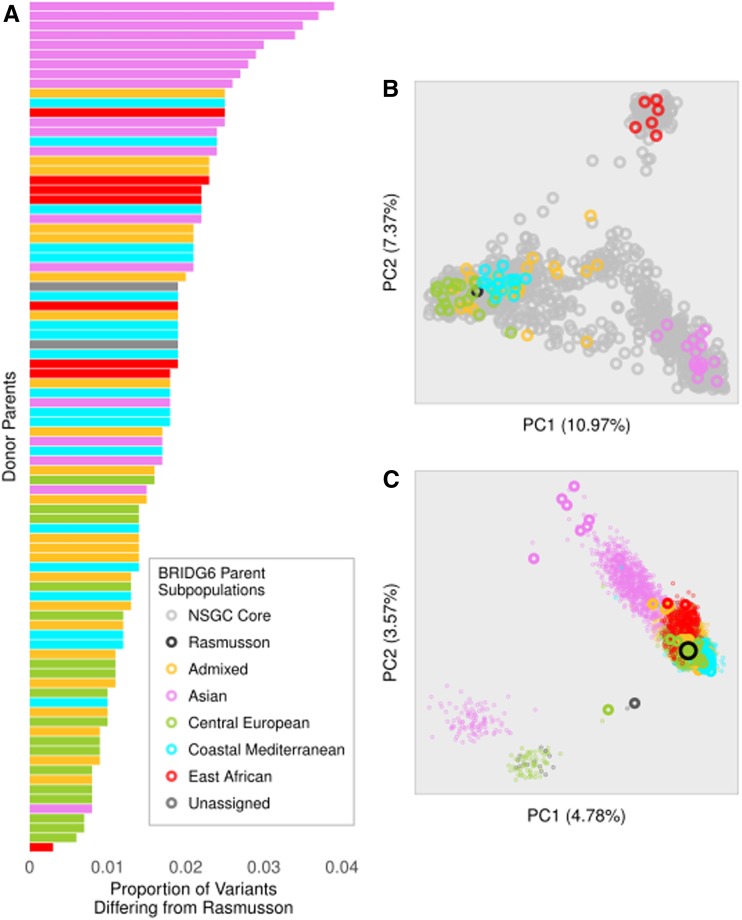
To examine the degree to which the BRIDG6 donor parents represent genetic diversity in six-row spring barley in the Core, we calculated FST. We used an existing dataset containing 6648 iSelect SNPs available for 1172 spring six-row barley accessions in The Core. We obtained the physical positions of 7757 of the 7864 SNPs on the iSelect chip (Table S1). Clho14881 was not included because it was missing >10% of the iSelect SNP data. The average FST between the BRIDG6 donor parents and the six-row Core accessions was −0.0011, and the maximum FST value for any SNP was 0.04, indicating little change in allele frequency between the BRIDG6 parents and their source population (Figure S1). Furthermore, PCA demonstrated that the BRIDG6 parents were evenly sampled from the four major subpopulations in The Core (Figure 1B). PCA of the donor parents and the BRIDG6 population revealed residual population structure that largely corresponds to the geographic origin of parents of individual families (Figure 1C). Lines designated “Unassigned” did not clearly cluster with one subpopulation.
Figure 1.
Genetic diversity in the BRIDG6 parents and six-row spring barley in the National Small Grains Collection Core. (A) The proportion of variants differing between Rasmusson (the common parent) and each BRIDG6 donor parent. Donor parents are sorted from the greatest (top) to least (bottom) genetic distance from Rasmusson. Analysis was conducted using 588,482 GBS SNPs prior to filtering steps required for subsequent analyses. (B) Principal component analysis (PCA) of the BRIDG6 parents and 1172 six-row National Small Grains Collection Core accessions, which are plotted as small gray open circles. Rasmusson, is plotted as a large black open circle, and BRIDG6 parents are open circles colored by subpopulation assignments in orange (Admixed), purple (Asian), green (Central European), blue (Coastal Mediterranean), red (East African), and dark gray (Unassigned). Analysis was conducted using 6648 iSelect 9K SNPs. (C) PCA of the BRIDG6 donor parents and 6059 recombinant inbred lines. BRIDG6 parents and the population are colored as described in (B), except large points represent parents and small points represent the population. Analysis was conducted using 5332 GBS SNPs with maximum 10% missing data.
To characterize the genetics of the BRIDG6 population, we genotyped Rasmusson, the 88 donor parents, and the 6079 RILs using GBS, obtaining an initial dataset of 593,645 SNPs. After removing SNPs with >80% missing data, there were 180,529 SNPs. SNPs present as heterozygous in Rasmusson were removed, leaving 178,841 SNPs. After removing monomorphic markers in the population, there were 37,783 SNPs. SNPs closer than 100 bp in any given family were filtered, resulting in 31,076 SNPs. Then, 49 individuals that have >90% missing values were removed. This was followed by the removal of monomorphic SNPs, which resulted in 29,741 SNPs in the data. Further filtering for MAF 0.03 left 14,657 SNPs. Then, 27 SNPs were removed due to being missing in Rasmusson. All SNPs mapped to the unordered portion of the genome reference were removed; there were 14,050 SNPs remaining in the data. The 14,050 SNPs were used for imputation purposes. SNPs in high LD were removed after imputation, leaving 11,046 SNPs in the data set. Before the GWAS analysis, a filter for MAF 0.05 was applied, leaving 8101 SNPs for downstream analysis.
The proportion of variants differing between each donor parent and Rasmusson, determined by percent polymorphic loci in the GBS genotyping, ranged from 0.3 to 4.4%, with the average of 3.3% (Figure 1A). The final dataset used for association analysis, after quality control to remove highly missing samples or markers, monomorphic, or low MAF markers included 5141 RIL accessions and Rasmusson genotyped for 8021 SNPs. The minimum number of SNPs segregating in a family was 2099, and the maximum was 4740, with an average of 3511 SNPs per family. Across all chromosomes and all families, the mean and median distance between GBS SNPs were 566 and 127 kbp, respectively.
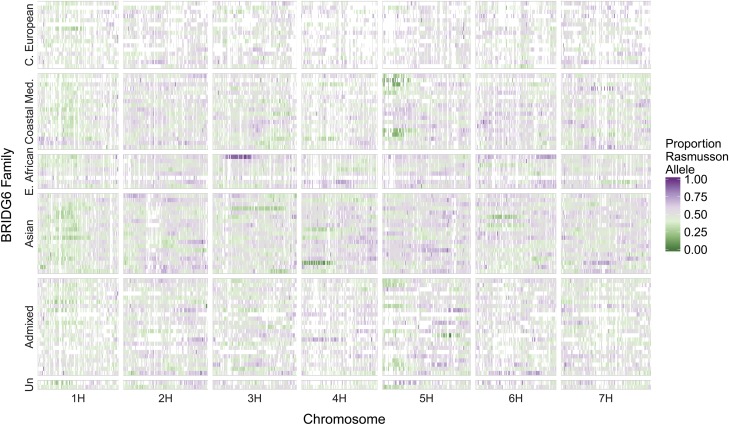
SNPs that differ between Rasmusson and donor parents have an expected mean frequency in progeny of 0.50. We tested for deviations from the expected mean frequency of SNPs in each of the BRIDG6 families. In general, there were no distinct patterns of excess or deficit of donor parent alleles apparent by chromosome or subpopulation with the exception of slight excess of the donor parent allele across chromosome 1H (Figure 2). Several regions that had extreme excess and deficit of Rasmusson alleles were observed at the family level. Nine families with parents from the Coastal Mediterranean subpopulation exhibited an overrepresentation of the donor parent allele on the short arm of chromosome 5H, starting at base position 395,158, and extending for 525.7 Mbp (796 SNPs). In contrast, the family derived from PI296460 of East African origin exhibited an excess contribution from Rasmusson on chromosome 3H, starting at base position 27,030,833 and extending for 165.5 Mbp (697 SNPs).
Figure 2.
Segregation distortion across 88 BRIDG6 families. Families (Y-axis) are sorted by donor parent subpopulation and then from earliest days to flowering (top) to latest days to flowering (bottom) of the donor parent within each subpopulation. The X-axis shows the seven barley chromosomes. Average frequency of the Rasmusson allele per family is calculated for each of 200 windows per chromosome. The estimation of the proportion of alleles from each parent was achieved by dividing the number of markers with a donor allele present in each RIL by the total number of SNPs in each window. Purple indicates the Rasmusson allele, green indicates the donor parent allele, and gray represents equilibrium, or allele frequency of 0.50. Markers not segregating within a family are represented in white. Analysis used an average of 2097 segregating SNPs per family.
Phenotypic variation for flowering time
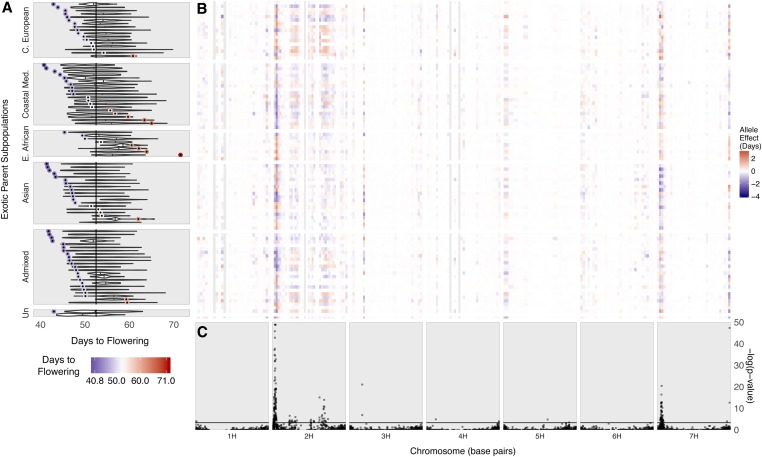
Flowering time was evaluated on the BRIDG6 population because of its importance in local adaptation, and to characterize the utility of the population for mapping. The broad sense heritability on a line mean basis for flowering time in the BRIDG6 population was 0.92. The donor parents of the BRIDG6 population flowered 40–78 days after planting, with a mean DTF of 55.7. Rasmusson DTF was ∼53. The BLUP values of the BRIDG6 progeny for DTF ranged from 43.2 to 69.4, with a mean value of 53.1. The mean DTF for each family ranged from 49.0 to 58.4. The mean DTF of families in the East African subpopulation was significantly greater than the population mean (56.4, P < 0.001). The range in DTF within a family was between 6.3 and 24.1 days, with a mean range of 14.1 days (Figure 3A). The mean variance for DTF of families in the Coastal Mediterranean subpopulation (mean 17.2, P < 0.01) and families in the East African subpopulation (mean 7.7, P < 0.01) were significantly different than the mean family variance. There was a significant correlation between the donor parent flowering time and corresponding family mean with a correlation coefficient of 0.34 (P < 0.01). The correlation between the difference in DTF between the donor parents and Rasmusson and the family variance was −0.01 and was not significant (P > 0.10).
Figure 3.
BRIDG6 population flowering time and allele effects across the genome. (A) Phenotypic distribution of days to flowering (DTF) in the BRIDG6 parents and population. The vertical line is the average DTF for the common parent, Rasmusson. Each donor parent DTF is plotted colored from early (blue) to late (red). Corresponding family DTF BLUPs are plotted as a violin plot. Families (Y-axis) are sorted by donor parent subpopulation and then from earliest DTF (top) to latest DTF (bottom) of the donor parent. (B) Additive effect estimates of alleles contributed by each donor parent for flowering time. Allele effect estimates for each family (Y-axis) are in days relative to the common parent, Rasmusson. For clarity, allele effect estimates are binned over 20 Mbp on each chromosome (X-axis) and the largest allele effect within each bin is plotted. The legend increments are colored from earlier (blue) to later (red). Effect color is centered around zero (white) for the reference allele. Gray regions across families indicate gaps between SNPs >20 Mbp. (C) Marker-trait associations for flowering time. Nested association mapping was conducted across 88 families using 7773 GBS SNPs. All QTL are represented by at least 1 significant marker, but 15 markers with –log(P-value) >50 on chromosome 2H not shown. The horizontal line indicates the 0.05 false discovery rate significance threshold = 3.308.
Mapping flowering time
We detected 23 QTL regions associated with flowering time that were distributed on every chromosome except chromosome 6H (Figure 3C). We observed some inflated P-values in Q-Q plots of each chromosome, indicating that our model may not have completely accounted for population structure related to subpopulation and/or NAM design (data not shown). The most significant QTL was located on chromosome 2H and contained HvPpd-H1 (Table 3), the major determinant of barley photoperiod response (Laurie et al. 1995; Decousset et al. 2000), with 120 significant SNPs that each segregated in 11–47 of the families. Each of the 23 QTL regions includes between 1 and 120 significant SNPs (Table 2). Seven of the significant QTL harbor eight previously identified flowering-time-related genes (Figure S4 and Table 3).
Table 3. Nearest gene to flowering time QTL.
| QTL –log(p) | Nearest candidate gene | Allele effect | Segregating Families (n) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| QTL | SNP | SNP –log(p) | Distance to gene | Linkage Group | Minimum | Maximum | |||
| 1_02 | 1H2_201180558 | 0 | 13.42 | Ppd-H2/HvFT3 | 0.082 | 1H | −0.061 | 0.047 | 83 |
| 2_01 | 2H1_29202906 | 15.67 | 142.43 | PpdH1 | 0.077 | 2H | −0.181 | 0.376 | 37 |
| 2_02 | 2H1_68788788 | 0 | 6.03 | HvFT4 | 0.046 | 2H | −0.047 | 0.043 | 38 |
| 2_12 | 2H2_131280495 | 5.06 | 10.16 | eps2/HvCEN | 1.435 | 2H | −0.212 | 0.217 | 67 |
| 3_01 | 3H1_119234365 | 0.46 | 23.19 | HvFT2 | 0.02 | 3H | −0.017 | 0.027 | 25 |
| 5_01 | 5H2_19078474 | 0 | 5.49 | CO3 | 0.057 | 5H | −0.035 | 0.032 | 51 |
| 7_01 | 7H1_39482671 | 11.58 | 32.44 | Vrn-H3/HvFT1 | 0.198 | 7H | −0.656 | 1.744 | 48 |
| 7_01 | 7H1_49195592 | 6.44 | 32.44 | CO8 | 0.021 | 7H | −1.044 | 0.879 | 71 |
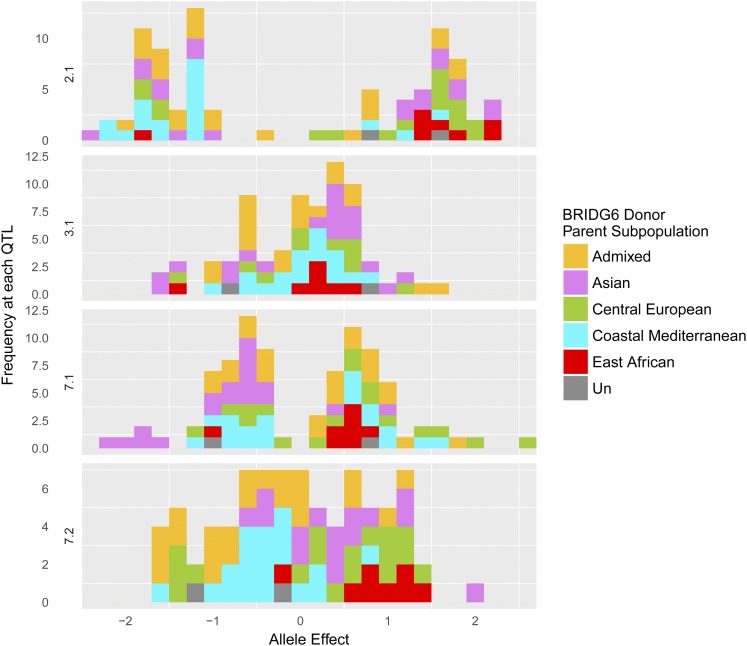
At significant SNPs, both positive and negative allelic effects relative to Rasmusson were detected in various families, indicating an allelic series. For example, for QTL 2.1 near HvPpd-H1, the marker with the largest allele effect (2H1_27204805) had a –log(P-value) of 142.4, and had an effect of −2.3 days in family PI094875 of Coastal Mediterranean origin, and an effect of +1.9 days in the family created from donor parent PI223883, a cultivar from the Asian subpopulation (Figure 3B). Across the 88 families, 45 families had an allelic effect that decreased DTF relative to Rasmusson, and 43 families had an allele effect that increased DTF at QTL 2.1. The distribution of allele effects of two of the largest effect QTL (2.1 and 7.1; Figure 4) are bimodal, and allele effects are roughly normally distributed for all other QTL (Figure S2).
Figure 4.
Distribution of allele effect estimates at the four top QTL for flowering time (2.1, 3.1, 7.1, 7.2). The frequency of observations (Y-axis) are plotted at each allele effect relative to Rasmusson (X-axis). Estimates are the maximum allele effect per family for any significant SNP in the QTL. Bars are colored by the subpopulation assignment of the donor parent and are stacked. Orange (Admixed), purple (Asian), green (Central European), blue (Coastal Mediterranean), red (East African), and dark gray (Unassigned).
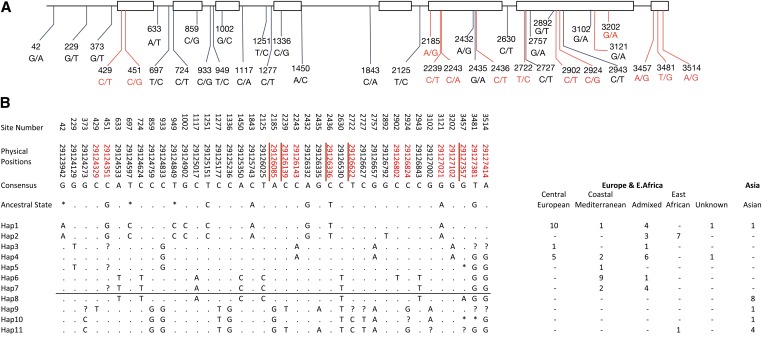
HvPpd-H1haplotype analysis
Exome capture sequencing allowed for a more in-depth view of the haplotypes that contributed to the increase and decrease in DTF. Based on exome capture sequencing, there were 39 SNPs in the HvPpd-H1 locus among the 78 BRIDG6 parents sampled. Of these, 14 were nonsynonymous variants, 11 were silent variants, and 14 were in noncoding regions (see Figure 5 and Table S3). The 39 SNPs contributed to 11 distinct haplotypes, with direct evidence of five recombination events based on the four-gamete test (RM). Three haplotypes were found in only one accession each (haplotypes 5, 9 and 10). Haplotype 1 (Figure 5B) is present primarily in Central European parents, haplotype 2 is present primarily in East African parents, haplotypes 3 and 4 are found mostly in Central European and Admixed parents, haplotype 6 is present in Coastal Mediterranean parents, and haplotype 7 is found in parents from admixture ancestry and two Coastal Mediterranean parents. There are 13 SNPs that are private to parents with Asian origin, five of which result in an amino acid change relative to Rasmusson (Figure 5). One of the haplotypes at high frequency in Asia (haplotype 8) differs by one nonsynonymous SNP from haplotype 7 observed in Coastal Mediterranean and Admixed ancestry parents. Despite the similarity between these two haplotypes, they have distinctive allelic effects. While haplotype 7 (Figure 5B) results in a mixture of positive and negative effects, Asian haplotype 8 is associated with increased DTF among all families carrying the derived variant. Alignments of sequences from four accessions used by Turner et al. (2005) indicated that the putative causative variant was located in linkage group 2H at position 291,273,881 bp. It was previously suggested that this variant was a G to T mutation (Turner et al. 2005). However, our analysis using wheat as an outgroup indicates that the mutation was a T to G change. This SNP is found in parents of diverse origins, but it is not segregating in Asian parents despite considerable variation in the phenotype among these parents. This suggests that the putative causative variant detected in a panel of European origin may not be the primary causative variant for flowering time variation among Asian accessions. Among European, Coastal Mediterranean, and East African families where the SNP is segregating, the mean flowering time for lines carrying the ancestral or derived states did not show significant difference (t-test P-value = 0.077) as one would expect based on previous predictions (Mamanova et al. 2010). However, families derived from parents carrying the ancestral state (T) at this SNP in Haplotype 2 (but not Haplotype 1) flower 9 days later than those carrying the derived mutation (G). These observations suggest that although this variant is not solely responsible for flowering time variation, it might have a larger contribution to this trait congruent with previous findings (Turner et al. 2005).
Figure 5.
Structure of HvPpd-H1 (A) The eight exons in HvPpd-H1 are shown as white rectangles, and the 39 polymorphisms identified in exome capture sequencing for 78 donor parents and Rasmusson. Mutations resulting in a nonsynonymous amino acid change are depicted in red. (B) Haplotype alignment of HvPpd-H1 showing haplotypes private to Asia. Nucleotide positions for each segregating site are shown in the first row. The physical position of each of the SNPs in HvPpd-H1 which is located in linkage group 2H are shown in the second row. The third row has the consensus nucleotides for the alignment. Ancestral state for each SNP derived from Wheat are shown in the fourth row. Nucleotide state similar to consensus is shown as a point, missing data are represented with an asterisk and heterozygous genotypes set to missing are represented with a question mark. The black horizontal line separates haplotypes private to Asia (bottom) for those private to Europe and East Africa (top). The number in the right table indicate how many times the haplotype appears in each of the subpopulations. Mutations resulting in a nonsynonymous amino acid change are represented with the physical positions colored in red font.
Grouping the estimated allelic effects per family of QTL 2.1 (estimated in isolation of other significant QTL), by the HvPpd-H1 haplotypes identified in the parents showed that two haplotypes confer positive effects and two haplotypes negative effects among the European and East African material, while three other haplotypes private to Asia confer positive effects and one haplotype confers a negative effect among Asian families (Figure S2).
Effect of subsampling the BRIDG6 population on flowering time QTL detection
Development of the BRIDG6 population intentionally included many parents to capture as much variation in The Core as possible. However, the total size of the BRIDG6 population is potentially prohibitive for evaluating some phenotypic traits. Therefore, to examine the balance between the need for genetic variation and sufficient family size to maintain power to detect QTL in this population, we tested various subsampling methods varying in population size and composition.
To determine the effect of sample size and sample composition to detect QTL, we conducted mixed-model association in bootstrapped samples from the BRIDG6 population using three strategies: randomly sampling different numbers of families and holding the family size constant, randomly sampling different population sizes while representing all families, and randomly sampling different population sizes without regard to families. Based on 69 cases (3 strategies × 23 QTL), there was not a significant difference between the strategies using similar population sizes either by random selection of families or by random selection of individuals (with or without considering families) (Table S4). The two QTL most significantly associated with flowering time (QTL 2.1 and 7.2) were detected in >70% of bootstrapped samples in all scenarios tested except for a sample size of 264 (Table S4). There was a significant difference between the number and frequency of QTL detected at sample sizes >500 individuals in each strategy. Randomly selecting families detected more QTL at higher frequency than randomly selecting individuals. The most noteworthy case was for samples of 625 individuals where subsampling based on families resulted in four of the largest QTL detected >50% of the time, compared to two QTL in the sampling of 528 random individuals.
Discussion
MPPs, such as BRIDG6, provide a comprehensive and potentially enduring resource for gene discovery and crop improvement. Their large size and the time and resources needed to create immortal lines (RILs, doubled haploids) necessitate careful design and genotyping to insure long-lasting utility. The major objective of the BRIDG6 population was to provide access to novel alleles and an unobstructed path from genetic discovery to breeding. We chose to use a large number of donor parents for the BRIDG6 population compared to previous MPP, with the goal of capturing more of the genetic diversity available in domesticated barley. This provides a better resource for dissecting the genetic architecture of complex traits and mining allelic diversity for crop improvement.
The SNPs genotyped in this population were mapped to physical positions on the barley reference genome, allowing for straightforward comparison with other SNP mapping experiments and identification of potential candidate genes. We examined the genetic composition of the population, and showed that it captures much of the diversity of the NSGC collection with relatively even representation of donor and common parent alleles. We used BRIDG6 to successfully identify major and minor QTL for flowering time. Moreover, we identify new haplotypes for HvPpd-H1 found primarily in barley lines from Asia, suggesting that the broad sampling in the BRIDG6 population has the potential to better explore and characterize genetic variation in barley.
BRIDG6 experimental design reduces the loss of founder alleles
An important consideration when developing a MPP is whether to impose selection to homogenize the population for a key trait or traits to facilitate QTL identification and subsequent integration in breeding. Cultivated barley can be distinguished by spike row type (six-row vs. two-row) and vernalization requirement (spring vs. winter growth habit) (cf. Hamblin et al. 2010; Poets et al. 2015b). Segregation of either of these two characters can complicate phenotyping of other traits, and requires subsequent selection to fix these traits to fit the appropriate market class (e.g., six-row or spring habit), which slows breeding progress. On the other hand, selection during RIL family development to fix a character can result in “founder dropout,” the loss of the allelic contribution of a parent at a locus that impedes the comparison of parental allelic contributions in MPPs (Macdonald and Long 2007; Figure 2). Founder dropout or segregation distortion can also result from purifying selection, unintentional selection during population development, or genetic drift (Macdonald and Long 2007). The AB-NAM used two-row wild barley accessions as donor parents, but selection for spring growth habit and six-row heads resulted in a significant decrease in the frequency of wild barley alleles in several regions including the HvVrs1 locus, which controls spike morphology (Nice et al. 2016). In BRIDG6, the population was homogenized for six-row spike through the choice of only six-row donor parents, thus minimizing selection during population development. As a result, the donor parent alleles are well represented across the progeny with the exception of a few families that favored one of the parents along a single chromosomal region. Depending on expected uses of an MPP, designs may benefit from selecting parents homogeneous for a major trait but with the potential to retain all donor alleles represented in the population for other traits. Studies designed to identify novel QTL for yield or other quantitative agronomic traits might benefit from a diverse set of founders that captures a variety of alleles at a locus in a population design where structure can be accounted for, as occurs in the BRIDG6 population. A more diverse population like the AB-NAM (Nice et al. 2016, 2017) is more suited to identifying novel large effect loci (e.g., disease resistance).
Flowering time variation
The BRIDG6 donor parents exhibit large variation for DTF, but the population as a whole moved closer to Rasmusson’s flowering time (Figure 3A) making this population a more uniform panel to study trait variation. Similar to BRIDG6, previous MPP studies in barley have identified flowering time QTL mapping to regions containing genes known to be associated with flowering time variation. Of the 14 genes considered jointly by these studies, eight were identified in BRIDG6 (Table 3), seven in HEB-25 (Maurer et al. 2015), seven in the German MAGIC population (Sannemann et al. 2015), six in the AB-NAM (Nice et al. 2017), and two in the Core (Muñoz-Amatriaín et al. 2014). The flowering time gene, Ppd-1H, was identified in all five studies, Vrn-H3 in four of the studies, HvCEN, HvFT4 and Vrn-1H in three of the studies, and the rest in one or two of the five studies. Interestingly, only Ppd-1H and HvFT4 were detected in the Core, while these two and six others were identified in BRIDG6. Even when a subsample of BRIDG6 (n = 2024) with a size similar to the Core (2417) was used, five flowering time genes were identified (Table S4). These five MPP represent a wide range of genetic diversity in terms of donor parents from German varieties (MAGIC) to wild barley (HEB-25 and AB-NAM), yet identify similar numbers of QTL and many of the same flowering time genes.
Since the AB-NAM and BRIDG6 share the common recipient parent Rasmusson, it is possible to more directly compare QTL in cultivated and wild barley. For the five loci detected in both the AB-NAM and the BRIDG6 populations, the order of the estimated effect size for each QTL was significantly different (Spearman rank correlation test ρ (4) = 0.3, P-value = 0.6833), but in both cases HvPpd-H1 was the largest effect QTL detected. This locus was also the major QTL identified in the HEB-25 population (Maurer et al. 2015). As demonstrated by our bootstrapping comparisons, a population size of 25 families (∼700 individuals) comparable to the AB-NAM has the power to detect most of the larger effect QTL, but not many of the smaller effect QTL. Despite the parents of the AB-NAM being more diverse, the increased population size of the BRIDG6 allowed for detection of more flowering time QTL.
Allelic variation at HvPpd-H1
The genetic diversity captured in the BRIDG6 population allowed us to identify multiple loci with opposite effects and four novel haplotypes at a single locus with the largest effect on flowering time variation, indicating an allelic series. The detection of such alleles in the BRIDG6 population demonstrates the power of this population as a tool to identify novel sources of genetic diversity. Asian barleys have a largely independent origin and distinct genetic composition relative to lines currently cultivated in Europe and the New World (Morrell and Clegg 2007; Poets et al. 2015a). The haplotypes identified explain newly discovered flowering time differences in Asian lines and among families from more Western origins. The variation at HvPpd-H1 is consistent with an allelic series at the locus. The presence of multiple alleles that alter phenotype is consistent with the contribution of HvPpd-H1 to flowering time variation in the majority of BRIDG6 families. Our ability to identify haplotypes private to Asia that were not reported previously can be attributed to the use of parental accessions that better represent the diversity of a larger barley collection. These two characteristics bode well for the utility of the BRIDG6 population to be employed successfully to study the architecture of other agronomically important traits, and the discovery of new alleles useful for breeding.
Interactions among multiple loci is remarkable in a family from a Coastal Mediterranean donor parent (Figure 3B bottom row in the Coastal Mediterranean block) were HvPpd-H1 in linkage group 2H has an early allele for flowering time, and a late allele at HvFT1 in linkage group 7H. Although the family carries an early flowering allele at HvPpd-H1, on average the individuals in this family flower later than Rasmusson as it would be expected when considering the HvFT1 allele in combination with the HvPpd-H1 allele. The opposite situation is observed in a family from an East African donor parent (Figure 3B third family from the bottom row in the East African block), which has a late flowering allele at HvPpd-H1 and an early allele at HvFT1, but in this case, on average, the family flowers later than Rasmusson, indicating the effect of HvPpd-H1 over HvFT1. This is yet another example where the phenotype of the donor parent hides the effect of alleles that contribute in a direction that is contrary to the overall phenotype of the parent.
Future mapping with BRIDG6 and translating to barley improvement
A challenge of utilizing large MPPs is accurately evaluating traits that are more difficult or time-consuming to measure than flowering time. New methods of high-throughput phenotyping using aerial imaging could be used to collect data on other traits for the entire population. Alternatively, strategies for subsampling from the population that maintain the benefits of the design and balance the practical constraints of collecting high quality phenotypic data will facilitate the use of the BRIDG6 in the future. We used DTF to explore how subsampling the BRIDG6 population influences the number of significant marker trait associations detected. The same QTL that were detected across all 88 BRIDG6 families were detected with low confidence in samples of as few as five families, but sample size had to be as large as 50 families for 70% of QTL to be detected in >50% of bootstrapped samples. This trend indicates that within our study design, samples of at least 3000 individuals or 50 families should be phenotyped to detect all important QTL for highly quantitative traits controlled by small effect QTL. However, if the goal is to detect only large effect QTL, these numbers could be substantially reduced.
Perhaps the simplest use of the BRIDG6 population is to screen the parents for polymorphism and then evaluate the appropriate families for the trait of interest. Recently, Carter et al. (2019) used three families from the BRIDG6 population to map the location of a gene encoding a host nucleotide-binding leucine-rich repeat (NLR) resistance protein, designated Pbr1 (for AvrPphB Response 1). AvrPphB encodes an effector protease from the plant pathogen, Pseudomonas syringae pv. phaseolicola, which activates specific NLR genes by cleaving a second host protein, PBS1 (DeYoung et al. 2012). After identifying a 23 Mb region based on GWAS of infiltration assays with AvrPphB, they searched the entire BRIDG6 population and found 18 recombinants in the region and were able to narrow the interval to 3 Mb containing the Pbr1gene. Thus, the selective use of a subsample, and the power to examine a large number of recombinants, hastened the discovery a gene controlling a complicated trait.
Another strategy to efficiently utilize BRIDG6 would be to evaluate the donor parents for the trait of interest, and choose a subset of families based on parental phenotypes. This has been observed in other studies where simulation tools based on parental genotype and phenotype data provided more accurate predictions of progeny variance (Bernardo 2015; Mohammadi et al. 2015). However, this does not guarantee segregating progeny for quantitative traits, as parental phenotype and progeny variance were not correlated for DTF in this study. This strategy could be informed by what is already known about the genetic architecture of the trait of interest, such as choosing families based on haplotype diversity at an important QTL.
Flowering time influences many other important agronomic traits, and, therefore, variation for flowering time in a mapping population can affect the ability to detect QTL for those traits. In particular, disease severity is often associated with flowering time or maturity. For example, DTF and level of Fusarium head blight (FHB) disease are highly correlated, whereby late flowering is associated with lower disease (de la Pena et al. 1999; Choo et al. 2004; Nduulu et al. 2007). Coincident QTL for DTF and FHB could be due to a pleiotropic effect of late flowering contributing to the host escaping disease, or to tight linkage of genes controlling DTF and FHB resistance. Experimental means to mitigate the effect of flowering time include matching the timing of inoculation with the pathogen, or the timing of disease assessment to DTF; however, differences in weather conditions during disease development can obscure the genetic signal for resistance. Analyses controlling for known QTL associated with flowering time could aid the discovery of QTL for disease resistance. To avoid this problem, we can consider sampling the BRIDG6 for homogenous allele effects at a confounding QTL, such as HvPpd-H1. Preliminary analyses of the genetic architecture of bacterial leaf streak infection shows that including photoperiod sensitivity as a fixed effect when calculating BLUPs partially accounts for variation that is not due to heritable response to infection (data not shown).
Another subsampling approach could involve creating smaller families that still represent both alleles at most loci rather than selecting individuals from a family at random. Optimizing the number of individuals from each family needed to represent allelic diversity may show that trait genetic architecture can be understood in a smaller BRIDG6 Core. However, development of a Core will inevitably reduce the number of lower frequency variants and reduce the power to map QTL and identify potential allelic series. The development of a Core should balance contributions of allelic richness and divergence (Brown and Schoen 1994). This same method was used to develop the NSGC Core and other core germplasm collections (Muñoz-Amatriaín et al. 2014). Additional studies are needed to explore these approaches to utilizing BRIDG6 for other important traits that are more expensive or difficult to measure.
Acknowledgments
The authors thank Ed Schiefelbein, Guillermo Velasquez, and Karen Beaubien for technical support during population development, field trial management (Minnesota), and genotyping. They also thank Rich Horsley for field data collection in Fargo, ND. This research was supported with funding from the United States Department of Agriculture-National Institute of Food and Agriculture Triticeae Coordinated Agricultural Project (USDA-NIFA TCAP), No. 2011-68002-30029 and US National Science Foundation Plant Genome Program grant DBI-1339393.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7757252.
Communicating editor: E. King
Literature Cited
- Alqudah A. M., Sharma R., Pasam R. K., Graner A., Kilian B. et al. , 2014. Genetic dissection of photoperiod response based on GWAS of pre-anthesis phase duration in spring barley. PLoS One 9: e113120 [corrigenda: PLoS One 10: e0123748 (2015)]. 10.1371/journal.pone.0113120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Andrews, S., 2010 A quality control tool for high throughput sequence data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Bajgain P., Rouse M. N., Tsilo T. J., Macharia G. K., Bhavani S. et al. , 2016. Nested association mapping of stem rust resistance in wheat using genotyping by sequencing. PLoS One 11: e0155760 10.1371/journal.pone.0155760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D., Maechler M., Bolker B., and Walker S., 2015. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67: 1–48. 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- Beavis W. D., 1998. QTL analyses: power, precision, and accuracy, pp. 145–162 in Molecular Dissection of Complex Traits, edited by Paterson A. H. CRC Press, New York. [Google Scholar]
- Benjamini Y., and Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57: 289–300. [Google Scholar]
- Bernardo R., 2015. Genomewide selection of parental inbreds: classes of loci and virtual biparental populations. Crop Sci. 55: 2586–2595. [Google Scholar]
- Bockelman H. E., and Valkoun J., 2010. Barley germplasm conservation and resources, pp. 144–159 in Barley: Improvement, Production, and Uses, edited by Ullrich S. E. Wiley-Blackwell, Oxford. [Google Scholar]
- Bolger A. M., Lohse M., and Usadel B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonman J. M., Bockelman H. E., Jackson L. F., and Steffenson B. J., 2005. Disease and insect resistance in cultivated barley accessions from the USDA national small grains collection. Crop Sci. 45: 1271–1280. 10.2135/cropsci2004.0546 [DOI] [Google Scholar]
- Bradbury P. J., Zhang Z., Kroon D. E., Casstevens T. M., Ramdoss Y. et al. , 2007. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635. 10.1093/bioinformatics/btm308 [DOI] [PubMed] [Google Scholar]
- Broman K. W., Wu H., Sen S., and Churchill G. A., 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. 10.1093/bioinformatics/btg112 [DOI] [PubMed] [Google Scholar]
- Brown A. H. D., and Schoen D. J., 1994. A revised measure of association of gene diversity values. Hereditas 120: 77–79. 10.1111/j.1601-5223.1994.00077.x [DOI] [Google Scholar]
- Buckler E. S., Holland J. B., Bradbury P. J., Acharya C. B., Brown P. J. et al. , 2009. The genetic architecture of maize flowering time. Science 325: 714–718. 10.1126/science.1174276 [DOI] [PubMed] [Google Scholar]
- Cantalapiedra C. P., Boudiar R., Casas A. M., Igartua E., and Contreras-Moreira B., 2015. BARLEYMAP: physical and genetic mapping of nucleotide sequences and annotation of surrounding loci in barley. Mol. Breed. 35: 13 10.1007/s11032-015-0253-1 [DOI] [Google Scholar]
- Carter M. E., Helm M., Chapman A. V. E., Wan E., Restrepo Sierra A. M. et al. , 2019. Convergent evolution of effector protease recognition by Arabidopsis and barley. Mol. Plant-Microbe Interact. 32: 550–555. 10.1094/MPMI-07-18-0202-FI [DOI] [PubMed] [Google Scholar]
- Cavanagh C., Morell M., Mackay I., and Powell W., 2008. From mutations to MAGIC: resources for gene discovery, validation and delivery in crop plants. Curr. Opin. Plant Biol. 11: 215–221. 10.1016/j.pbi.2008.01.002 [DOI] [PubMed] [Google Scholar]
- Chang C. C., Chow C. C., Tellier L. C., Vattikuti S., Purcell S. M. et al. , 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4: 7 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo T. M., Vigier B., Shen Q. Q., Martin R. A., Ho K. M. et al. , 2004. Barley traits associated with resistance to fusarium head blight and deoxynivalenol accumulation. Phytopathology 94: 1145–1150. 10.1094/PHYTO.2004.94.10.1145 [DOI] [PubMed] [Google Scholar]
- Chun S., and Fay J. C., 2009. Identification of deleterious mutations within three human genomes. Genome Res. 19: 1553–1561. 10.1101/gr.092619.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmsee C., Beier S., Himmelbach A., Schmutzer T., Stein N. et al. , 2015. BARLEX - the barley draft genome explorer. Mol. Plant 8: 964–966. 10.1016/j.molp.2015.03.009 [DOI] [PubMed] [Google Scholar]
- Comadran J., Kilian B., Russell J., Ramsay L., Stein N. et al. , 2012. Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nat. Genet. 44: 1388–1392. 10.1038/ng.2447 [DOI] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C. A., Banks E. et al. ; 1000 Genomes Project Analysis Group, 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decousset L., Griffiths S., Dunford R. P., Pratchett N., and Laurie D. A., 2000. Development of STS markers closely linked to the Ppd-H1 photoperiod response gene of barley (Hordeum vulgare L.). Theor. Appl. Genet. 101: 1202–1206. 10.1007/s001220051598 [DOI] [Google Scholar]
- de la Pena R. C., Smith K. P., Capettini F., Muehlbauer G. J., Gallo-Meagher M. et al. , 1999. Quantitative trait loci associated with resistance to Fusarium head blight and kernel discoloration in barley. Theor. Appl. Genet. 99: 561–569. 10.1007/s001220051269 [DOI] [PubMed] [Google Scholar]
- DeYoung B. J., Qi D., Kim S., Burke T. P., and Innes R. W., 2012. Activation of a plant nucleotide binding-leucine rich repeat disease resistance protein by a modified self protein. Cell. Microb. 14: 1071–1084. 10.1111/j.1462-5822.2012.01779.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshire R. J., Glaubitz J. C., Sun Q., Poland J. A., Kawamoto K. et al. , 2011. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6: e19379 10.1371/journal.pone.0019379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaubitz J. C., Casstevens T. M., Lu F., Harriman J., Elshire R. J. et al. , 2014. TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS One 9: e90346 10.1371/journal.pone.0090346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J., 2005. Hierfstat, a package for R to compute and test hierarchical F statistics. Mol. Ecol. Notes 5: 184–186. 10.1111/j.1471-8286.2004.00828.x [DOI] [Google Scholar]
- Hamblin M. T., Close T. J., Bhat P. R., Chao S., Kling J. G. et al. , 2010. Population structure and linkage disequilibrium in US barley germplasm: implications for association mapping. Crop Sci. 50: 556–566. 10.2135/cropsci2009.04.0198 [DOI] [Google Scholar]
- Hoffman, P. J., 2016 BLAST_to_BED: convert BLAST XML alignments to BED format. Available at: https://github.com/mojaveazure/BLAST_to_BED
- Huang B. E., Verbyla K. L., Verbyla A. P., Raghavan C., Singh V. K. et al. , 2015. MAGIC populations in crops: current status and future prospects. Theor. Appl. Genet. 128: 999–1017. 10.1007/s00122-015-2506-0 [DOI] [PubMed] [Google Scholar]
- Huang X., Wei X., Sang T., Zhao Q., Feng Q. et al. , 2010. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 42: 961–967. 10.1038/ng.695 [DOI] [PubMed] [Google Scholar]
- Hudson R. R., and Kaplan N. L., 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111: 147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Barley Genome Sequencing Consortium; Mayer K. F., Waugh R., Langridge P., Close T. J., Wise R. P. et al. , 2012. A physical, genetic and functional sequence assembly of the barley genome. Nature 491: 711–716. 10.1038/nature11543 [DOI] [PubMed] [Google Scholar]
- Jones H., Leigh F. J., Mackay I., Bower M. A., Smith L. M. J. et al. , 2008. Population-based resequencing reveals that the flowering time adaptation of cultivated barley originated east of the Fertile Crescent. Mol. Biol. Evol. 25: 2211–2219. 10.1093/molbev/msn167 [DOI] [PubMed] [Google Scholar]
- Jordan D. R., Mace E. S., Cruickshank A. W., Hunt C. H., and Henzell R. G., 2011. Exploring and exploiting genetic variation from unadapted sorghum germplasm in a breeding program. Crop Sci. 51: 1444–1457. 10.2135/cropsci2010.06.0326 [DOI] [Google Scholar]
- Jordan K. W., Wang S., Lun Y., Gardiner L. J., Maclachlan R. et al. , 2015. A haplotype map of allohexaploid wheat reveals distinct patterns of selection on homoeologous genomes. Genome Biol. 16: 48 10.1186/s13059-015-0606-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knüpffer H., 2009. Triticeae genetic resources in ex situ genebank collections, pp. 31–79 in Genetics and Genomics of the Triticeae. Plant Genetics and Genomics: Crops and Models, edited by Muehlbauer G. and Feuillet C.. Springer, New York: 10.1007/978-0-387-77489-3_2 [DOI] [Google Scholar]
- Komatsuda T., Pourkheirandish M., He C., Azhaguvel P., Kanamori H. et al. , 2007. Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc. Natl. Acad. Sci. USA 104: 1424–1429. 10.1073/pnas.0608580104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono T. J., Seth K., Poland J. A., and Morrell P. L., 2014. SNPMeta: SNP annotation and SNP Metadata collection without a reference genome. Mol. Ecol. Resour. 14: 419–425. 10.1111/1755-0998.12183 [DOI] [PubMed] [Google Scholar]
- Kono T. J., Fu F., Mohammadi M., Hoffman P. J., Liu C. et al. , 2016. The role of deleterious substitutions in crop genomes. Mol. Biol. Evol. 33: 2307–2317. 10.1093/molbev/msw102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono T. J. Y., Lei L., Shih C. H., Hoffman P. J., and Morrell P. L., 2017. Comparative genomics approaches accurately predict deleterious variants in plants. G3 (Bethesda) 8: 3321–3329. 10.1534/g3.118.200563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kover P. X., Valdar W., Trakalo J., Scarcelli N., Ehrenreich I. M. et al. , 2009. A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 5: e1000551 10.1371/journal.pgen.1000551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langridge P., 2018. Economic and academic importance of barley, pp. 1–10 in The Barley Genome, edited by Stein N. and Muehlbauer G.. Springer International Publishing AG, Basel, Switzerland [Google Scholar]
- Laurie D. A., Pratchett N., Snape J. W., and Bezant J. H., 1995. RFLP mapping of five major genes and eight quantitative trait loci controlling flowering time in a winter× spring barley (Hordeum vulgare L.) cross. Genome 38: 575–585. 10.1139/g95-074 [DOI] [PubMed] [Google Scholar]
- Li H., and Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., and Poushinsky G., 1985. A modified augmented design (type 2) for rectangular plots. Can. J. Plant Sci. 65: 743–749. 10.4141/cjps85-094 [DOI] [Google Scholar]
- Lincoln S. E., and Lander E. S., 1992. Systematic detection of errors in genetic linkage data. Genomics 14: 604–610. 10.1016/S0888-7543(05)80158-2 [DOI] [PubMed] [Google Scholar]
- Macdonald S. J., and Long A. D., 2007. Joint estimates of quantitative trait locus effect and frequency using synthetic recombinant populations of Drosophila melanogaster. Genetics 176: 1261–1281. 10.1534/genetics.106.069641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison, W. P., and D. R. Maddison, 2018 Mesquite: a modular system for evolutionary analysis. Version 3.0.4. Available at: http://mesquiteproject.org
- Mamanova L., Coffey A. J., Scott C. E., Kozarewa I., Turner E. H. et al. , 2010. Target-enrichment strategies for next-generation sequencing. Nat. Methods 7: 111–118. 10.1038/nmeth.1419 [DOI] [PubMed] [Google Scholar]
- Mascher M., Richmond T. A., Gerhardt D. J., Himmelbach A., Clissold L. et al. , 2013. Barley whole exome capture: a tool for genomic research in the genus Hordeum and beyond. Plant J. 76: 494–505. 10.1111/tpj.12294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher M., Gundlach H., Himmelbach A., Beier S., Twardziok S. O. et al. , 2017. A chromosome conformation capture ordered sequence of the barley genome. Nature 544: 427–433. 10.1038/nature22043 [DOI] [PubMed] [Google Scholar]
- Maurer A., Draba V., Jiang Y., Schnaithmann F., Sharma R. et al. , 2015. Modelling the genetic architecture of flowering time control in barley through nested association mapping. BMC Genomics 16: 290 10.1186/s12864-015-1459-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K. et al. , 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen M. D., Kresovich S., Villeda H. S., Bradbury P., Li H. et al. , 2009. Genetic properties of the maize nested association mapping population. Science 325: 737–740. 10.1126/science.1174320 [DOI] [PubMed] [Google Scholar]
- Mohammadi M., Tiede T., and Smith K. P., 2015. PopVar: a genome-wide procedure for predicting genetic variance and correlated response in biparental breeding populations. Crop Sci. 55: 2068–2077. 10.2135/cropsci2015.01.0030 [DOI] [Google Scholar]
- Money D., Gardner K., Migicovsky Z., Schwaninger H., Zhong G. Y. et al. , 2015. LinkImpute: fast and accurate genotype imputation for nonmodel organisms. G3 (Bethesda) 5: 2383–2390. 10.1534/g3.115.021667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell P. L., and Clegg M. T., 2007. Genetic evidence for a second domestication of barley (Hordeum vulgare) east of the Fertile Crescent. Proc. Natl. Acad. Sci. USA 104: 3289–3294. 10.1073/pnas.0611377104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell P. L., Buckler E. S., and Ross-Ibarra J., 2012. Crop genomics: advances and applications. Nat. Rev. Genet. 13: 85–96. 10.1038/nrg3097 [DOI] [PubMed] [Google Scholar]
- Muñoz-Amatriaín M., Cuesta-Marcos A., Endelman J. B., Comadran J., Bonman J. M. et al. , 2014. The USDA barley core collection: genetic diversity, population structure, and potential for genome-wide association studies. PLoS One 9: e94688 10.1371/journal.pone.0094688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nduulu L. M., Mesfin A., Muehlbauer G. J., and Smith K. P., 2007. Analysis of the chromosome 2(2H) region of barley associated with the correlated traits Fusarium head blight resistance and heading date. Theor. Appl. Genet. 115: 561–570. 10.1007/s00122-007-0590-5 [DOI] [PubMed] [Google Scholar]
- Nice L. M., Steffenson B. J., Brown-Guedira G. L. Akhunov E. D., Liu C. et al. , 2016. Development and genetic characterization of an advanced backcross–nested association mapping (AB-NAM) population of wild× cultivated barley. Genetics 203: 1453–1467. 10.1534/genetics.116.190736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nice L. M., Steffenson B. J., Blake T. K., Horsley R. D., Smith K. P. et al. , 2017. Mapping agronomic traits in a wild barley advanced backcross–nested association mapping population. Crop Sci. 57: 1199–1210. 10.2135/cropsci2016.10.0850 [DOI] [Google Scholar]
- Paradis E., Claude J., and Strimmer K., 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290. 10.1093/bioinformatics/btg412 [DOI] [PubMed] [Google Scholar]
- Poets A. M., Fang Z., Clegg M. T., and Morrell P., 2015a Barley landraces are characterized by geographically heterogeneous genomic origins. Genome Biol. 16: 173 10.1186/s13059-015-0712-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poets A. M., Mohammadi M., Seth K., Wang H., Kono T. J. et al. , 2015b The effects of both recent and long-term selection and genetic drift are readily evident in North American barley breeding populations. G3 (Bethesda) 6: 609–622. 10.1534/g3.115.024349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland J. A., Brown P. J., Sorrells M. E., and Jannink J. L., 2012. Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS One 7: e32253 10.1371/journal.pone.0032253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A. L., Patterson N. J., Plenge R. M., Weinblatt M. E., Shadick N. A. et al. , 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38: 904–909. 10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- Pritchard J. K., and Rosenberg N. A., 1999. Use of unlinked genetic markers to detect population stratification in association studies. Am. J. Hum. Genet. 65: 220–228. 10.1086/302449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R., and Hall I. M., 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J., Mascher M., Dawson I. K., Kyriakidis S., Calixto C. et al. , 2016. Exome sequencing of geographically diverse barley landraces and wild relatives gives insights into environmental adaptation. Nat. Genet. 48: 1024–1030. 10.1038/ng.3612 [DOI] [PubMed] [Google Scholar]
- Saghai-Maroof M. A., Soliman K. M., Jorgensen R. A., and Allard R. W., 1984. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. USA 81: 8014–8018. 10.1073/pnas.81.24.8014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sannemann W., Huang E. B., Mathew B., and Léon J., 2015. Multi-parent advanced generation inter-cross in barley: high-resolution quantitative trait locus mapping for flowering time as a proof of concept. Mol. Breed. 35: 86 10.1007/s11032-015-0284-7 [DOI] [Google Scholar]
- Shavrukov Y., Kurishbayev A., Jatayev S., Shvidchenko V., Zotova L. et al. , 2017. Early flowering as a drought escape mechanism in plants: how can it aid wheat production. Front. Plant Sci. 8: 1950 10.3389/fpls.2017.01950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K. P., Rasmusson D. C., Schiefelbein E., Wiersma J. J., Wiersma J. V. et al. , 2010. Registration of ‘Rasmusson’ barley. J. Plant Regist. 4: 167–169. 10.3198/jpr2009.10.0622crc [DOI] [Google Scholar]
- Technow, F., 2011 R package mvngGrAd: moving grid adjustment in plant breeding field trials. R package version 0.1.5. https://www.uni-hohenheim.de/fileadmin/einrichtungen/plant-breeding/software/mvngGrAd_vignettes.pdf.
- Thornton K., 2003. libsequence: a C++ class library for evolutionary genetic analysis. Bioinformatics 19: 2325–2327. 10.1093/bioinformatics/btg316 [DOI] [PubMed] [Google Scholar]
- Turner A., Beales J., Faure S., Dunford R. P., and Laurie D. A., 2005. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310: 1031–1034. 10.1126/science.1117619 [DOI] [PubMed] [Google Scholar]
- Wang K., Li M., and Hakonarson H., 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38: e164 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver J. C., 1944. United States malting barley production. Ann. Assoc. Am. Geogr. 34: 97–131 10.1080/00045604409357256 [DOI] [Google Scholar]
- Weir B. S., and Cockerham C. C., 1984. Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- Xavier A., Xu S., Muir W. M., and Rainey K. M., 2015. NAM: association studies in multiple populations. Bioinformatics 31: 3862–3864. 10.1093/bioinformatics/btv448 [DOI] [PubMed] [Google Scholar]
- Yu J., Holland J. B., McMullen M. D., and Buckler E. S., 2008. Genetic design and statistical power of nested association mapping in maize. Genetics 178: 539–551. 10.1534/genetics.107.074245 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. The computer code used for all analyses (unless otherwise specified) and Figures are available at https://github.com/UMN-BarleyOatSilphium/BRIDG6. Raw GBS SNP genotypes are available in The Sequence Read Archive (SRA) at NCBI under BioProject number PRJNA488050. Imputed GBS SNP genotype and DTF phenotype data are available at The Triticeae Toolbox (T3, https://triticeaetoolbox.org/barley). We have made supplemental files available through the GSA Figshare portal. The computer code used for all analyses is available at https://github.com/UMN-BarleyOatSilphium/BRIDG6. Accessory files not appropriate as supplementary materials are available through a public archive system from our university library (https://doi.org/10.13020/c5kj-af95). Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7757252.