Anti-Mullerian hormone (Amh) inhibits female reproductive duct development, signals oocyte reserve, and marks polycystic ovarian syndrome. Zebrafish lacks Mullerian ducts and the typical Amh receptor, questioning evolving roles of Amh. Yan et al. made knockout mutations in zebrafish...
Keywords: germ cells, PGC, male fertility, female fertility, gonad development, Genetics of Sex
Abstract
Fetal mammalian testes secrete Anti-Müllerian hormone (Amh), which inhibits female reproductive tract (Müllerian duct) development. Amh also derives from mature mammalian ovarian follicles, which marks oocyte reserve and characterizes polycystic ovarian syndrome. Zebrafish (Danio rerio) lacks Müllerian ducts and the Amh receptor gene amhr2 but, curiously, retains amh. To discover the roles of Amh in the absence of Müllerian ducts and the ancestral receptor gene, we made amh null alleles in zebrafish. Results showed that normal amh prevents female-biased sex ratios. Adult male amh mutants had enormous testes, half of which contained immature oocytes, demonstrating that Amh regulates male germ cell accumulation and inhibits oocyte development or survival. Mutant males formed sperm ducts and some produced a few offspring. Young female mutants laid a few fertile eggs, so they also had functional sex ducts. Older amh mutants accumulated nonvitellogenic follicles in exceedingly large but sterile ovaries, showing that Amh helps control ovarian follicle maturation and proliferation. RNA-sequencing data partitioned juveniles at 21 days postfertilization (dpf) into two groups that each contained mutant and wild-type fish. Group21-1 upregulated ovary genes compared to Group21-2, which were likely developing as males. By 35 dpf, transcriptomes distinguished males from females and, within each sex, mutants from wild types. In adult mutants, ovaries greatly underexpressed granulosa and theca genes, and testes underexpressed Leydig cell genes. These results show that ancestral Amh functions included development of the gonadal soma in ovaries and testes and regulation of gamete proliferation and maturation. A major gap in our understanding is the identity of the gene encoding a zebrafish Amh receptor; we show here that the loss of amhr2 is associated with the breakpoint of a chromosome rearrangement shared among cyprinid fishes.
DEVELOPING mammalian embryos form the rudiments of both male and female sex ducts, the Wolffian and Müllerian ducts, respectively. Over 70 years ago, Alfred Jost conducted remarkable experiments to learn if gonads control sex duct development (Jost 1947). He removed undifferentiated gonads from rabbit fetuses and reimplanted them into the uterus of surrogate rabbit hosts. Gonadectomized kits lost male sex ducts but retained female sex ducts. He concluded that developing testes maintain male ducts (epididymis, seminal vesicles, and vas deferens) but destroy female sex duct anlagen (fallopian tubes and uterus). In contrast, developing ovaries neither maintain male ducts nor destroy female ducts. Subsequent experiments showed that one testis-derived substance (testosterone) maintains male sex duct rudiments and another [anti-Müllerian hormone (AMH), also called Müllerian Inhibiting Substance (MIS)], inhibits female reproductive duct anlagen (Elger 1966; Josso 1972).
Although AMH from testes represses female duct development, AMH from ovaries begins to appear in the third trimester of human fetal development from primary and preantral follicles (Munsterberg and Lovell-Badge 1991). Ovarian AMH expression peaks in juvenile women, declines with age, and disappears at menopause; thus, circulating AMH levels reflect a woman’s ovarian follicle reserve (Visser et al. 2006; Zec et al. 2011). Investigations of Amh mutant mice showed that chromosomal XY males that lack Amh activity develop oviducts, uterus, and vagina in addition to male reproductive ducts (Behringer et al. 1994). Testes in Amh-deficient XY mice attain normal size, but some show Leydig cell hyperplasia (Behringer et al. 1994). Chromosomally female XX Amh mutant juvenile mice have more preantral and small antral follicles and older mutant females have fewer primordial follicles, preantral, and small antral follicles than wild-type siblings (Behringer et al. 1994; Durlinger et al. 1999), suggesting that without AMH, primordial follicles develop more rapidly than normal, which results in larger juvenile ovaries that lose follicles prematurely. This property led to the use of circulating AMH as a marker of polycystic ovarian syndrome (PCOS), the most common problem for couples who visit fertility clinics (Pigny et al. 2003; Diamanti-Kandarakis 2008)].
We next considered the evolution of AMH functions and their relationship to reproductive ducts. Jawless fish lack specialized gamete-transporting sex ducts; lamprey gonads release gametes directly into the body cavity where they are forced out during spawning through genital pores (Applegate 1948; Hardisty 1971). Cartilaginous fish evolved paired Müllerian ducts (or paramesonephric ducts) that condense from intermediate mesoderm parallel to Wolffian ducts (mesonephric ducts), and differentiate into the female reproductive tract, including the fallopian tubes, which collect oocytes released into the coelomic cavity (Wourms 1977). Among bony fish, tetrapods and basally diverging ray-finned fish like spotted gar (Ferrara and Irwin 2001) maintained this ancestral state, but teleosts lost their Müllerian ducts; gonoducts in many teleosts develop from somatic cells posterior to the gonad, and gametes pass from the gonad directly into the ducts rather than into the body cavity (e.g., Suzuki and Shibata 2004; Kossack et al. 2019). We therefore wondered how Amh functions evolved in a teleost given that its eponymous feature of Müllerian duct inhibition is no longer relevant in the absence of a Müllerian duct.
Despite the absence of Müllerian ducts, Amh performs a reproductive function in at least some teleosts because a Y chromosome variant of amh (amhY) plays a role in sex determination in the Patagonian pejerrey (Hattori et al. 2012) and a variant Amh receptor (Amh receptor type II; Amhr2) acts in sex determination in several, but not all, species of pufferfish (Kamiya et al. 2012; Ieda et al. 2018). In addition, amhr2 mutants in medaka show excess germ cell proliferation, premature male meiosis, sex reversal in some chromosomally XY fish, and early-stage follicular arrest in females (Morinaga et al. 2007). We lack, however, full knowledge of the roles these genes play in normal fish development. The situation is even more confusing because zebrafish lacks an amhr2 gene (Rocha et al. 2016), the loss of which we show here to be associated with chromosomal rearrangements that have breakpoints at the expected site of the ancestral amhr2 gene, breakpoints that originated at the base of the cypriniform radiation because we show that this inversion breakpoint is shared by the common carp (Cyprinus carpio).
To help identify ancestral roles, we knocked out amh in the zebrafish Danio rerio. We studied gonad development, reproductive tract function, and transcriptomics to help understand the molecular genetic mechanisms of Amh action. Like mammals, zebrafish expresses amh in Sertoli cells in testes and in granulosa cells in ovaries (Rodríguez-Mari et al. 2005; von Hofsten et al. 2005; Wang and Orban 2007; Chen et al. 2017; Yin et al. 2017). In adult zebrafish organ culture, Amh inhibited the production of Fsh-stimulated androgen, and also inhibited androgen-stimulated proliferation of spermatogonia (Skaar et al. 2011), suggesting a role for Amh in testis function.
Results showed that zebrafish males and females that lack Amh function had enormous gonads due to increased production and/or accumulation of germ cells (Lin et al. 2017). Mutant males developed mature sperm able to fertilize eggs, but at lower rates than wild-type siblings. Young mutant females produced fertile eggs, but older females became sterile as their ovaries accumulated immature follicles that failed to deposit yolk. Reproductive ducts in both males and females were structurally and functionally normal, making unlikely the hypothesis that the inhibition of female sex duct development is a conserved feature of Amh across vertebrates. Juvenile amh mutant zebrafish developing as males retained oocytes longer than their wild-type siblings, which generally develop as hermaphrodites before transitioning to become males or females ∼19–30 days postfertilization (dpf) (Takahashi 1977; Rodríguez-Mari et al. 2005; Wang et al. 2007; Orban et al. 2009). This result suggests that Amh promotes oocyte apoptosis in transitioning juvenile zebrafish. Based on trunk transcriptomes, 21 dpf transitional-stage fish clustered into two groups, one of which expressed more ovary genes, but both groups contained both wild-type and mutant fish, showing that Amh was not playing a sex-specific role at this stage. Transcriptomes of 35 dpf juvenile trunks clustered animals into clearly male and female groups, and within each sex group, wild types separated from mutants, showing that at this stage, Amh action is important for gonad development. Transcriptomic comparisons of wild-type and amh mutant ovaries and testes revealed an ancestral role of Amh in Leydig cell development, oocyte differentiation, and the regulation of germ cell proliferation. We conclude that Amh either was not important for reproductive duct development in the last common ancestor of zebrafish and humans or, more likely, that this role was lost in the zebrafish lineage along with the loss of Müllerian ducts. A shared role of Amh, however, was likely the inhibition of germ cell proliferation both in ovaries and in testes, and that in mammals, the ovary retained this role but the testis apparently lost it. Alternatively, the teleost lineage gained the male germ cell proliferation role of Amh.
Materials and Methods
Animals
CRISPR/Cas9 mutagenesis generated deletions in zebrafish amh (ENSDARG00000014357; http://ensembl.org) using sites identified by ZiFiT Targeter (http://zifit.partners.org/ZiFiT/). Mutagenesis targeted two regions in amh exon 3, GGGATGCTGATAACGAAGGA (site 1) and GGAATGCTTTGGGAACGTGA (site 2), using guide RNAs (gRNAs) synthesized from DNA oligomer templates: aattaatacgactcactataGGGATGCTGATAACGAAGGAgttttagagctagaaatagc and aattaatacgactcactataGGAATGCTTTGGGAACGTGAgttttagagctagaaatagc (IDT, Coralville, IA). MEGAscript T7 Transcription Kit transcribed gRNA and mMESSAGE mMACHINE T3 Transcription Kit (Thermo Fisher Scientific, Waltham, MA) synthesized Cas9 messenger RNA (mRNA). Approximately 2 nl of a solution containing 100 ng/μl Cas9 mRNA and 25 ng/μl of both amh gRNAs was comicroinjected into one-cell embryos of the AB strain. Genomic DNA from injected embryos at 24 hours postfertilization (hpf) provided a template to amplify a 319-bp PCR fragment including both sites (primers: F-AGGGTGTGCATGCTACAGAAGGTAAA and R-TGCCATCTTTTTGCACCATCATTTCCAGCCA). Wild-type alleles have an HpyAV recognition site at site 1 and an HpyCH4IV recognition site at site 2 that are disrupted in amh mutant alleles. Sanger sequencing (GENEWIZ, Plainfield, NJ) verified mutations. We established stable lines for three noncomplementing alleles: deletions of 5 , 10 , or 26 nucleotides designated amh(b1373), amh(b1374), and amh(b1375), respectively (Figure 1, C and D). In addition, we made TALEN-induced deletions in amh (Figure 1E). TALENs targeted the first coding exons of amh and were assembled as previously described (Dranow et al. 2016). TALEN RNAs were synthesized by in vitro transcription using the mMESSAGE mMACHINE kit (Ambion). TALEN pairs were co-injected at the one-cell stage at 50–100 pg for each TALEN. Founders were identified by screening sperm DNA by high-resolution melt analysis (Dahlem et al. 2012), using Light Scanner Master Mix (BioFire Defense), a CFX-96 real-time PCR machine and Precision Melt Analysis software (Bio-Rad, Hercules, CA). Primer sequences for the indicated amplicon used were as follows (wild-type amplicon size in parentheses): F-AGATTTGGGCTGATGCTGAT and R-GTGGGACGAATGACTGACCT (212 bp). After initial identification, subsequent genotyping of offspring was performed by PCR followed by visualization on a 2% agarose gel using the same primers. The mutant allele amh(uc28) was an 11-bp deletion of the bold-faced nucleotides (ACAGTGAGGCACGAAGAGCAGGACAACAACCCGAAGGTCAACCCGCTATC, with TALEN sequences underlined.
Figure 1.
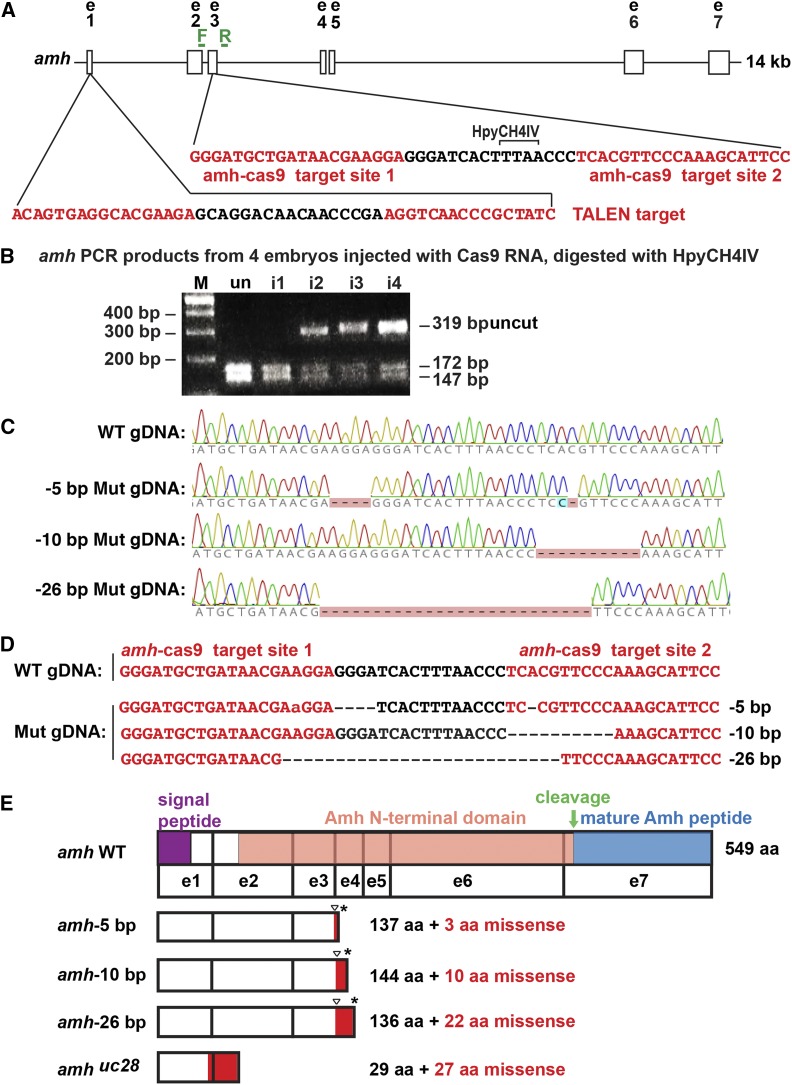
CRISPR/Cas9-induced amh mutants. (A) 14 kb of the amh locus showing two CRISPR target sites (red letters) in exon 3. PCR primers, forward (F) and reverse (R) (green). (B) Assay for injected CRISPR efficacy. PCR analysis of four G0 injected embryos at 1 dpf using genotyping primers F and R shows a 319-bp fragment in wild types that digested with HpyCH4IV to produce fragments of 172 and 147 bp; this site disappeared from amh genes in a large portion of cells in CRISPR-injected embryos. (C) Sequence traces from genomic DNA from a wild-type fish and from three stable mutant lines carrying −5, −10, and −26 bp deletions. (D) Sequences of genomic DNAs from a wild-type fish and three stable mutant lines (Mut). (E) Predicted structure of Amh protein showing the location of the mutation (triangle), the predicted out-of-frame portion (red), and the premature stop codon (*). Protein coding domains: signal peptide, purple; Amh amino-terminal domain, salmon; cleavage site, green arrow; mature Amh peptide, blue. i1-i4, CRISPR-injected 24 hpf embryos; M, length marker; un, uninjected 24 hpf embryos; WT, wild type.
Histology and in situ hybridization
In situ hybridization was performed as described (Rodríguez-Mari et al. 2005) using the probes amh (ENSDARG00000014357), a 375-bp amh fragment including part of exon 7 (primers: F-AGGCTCAGTACCGTTCAGTGTTGC and R-CCAACATCTCCTACAAGACCAACG) (Rodríguez-Mari et al. 2005); bmp15 (ENSDARG00000037491) (Dranow et al. 2016); cyp19a1a (ENSDARG00000041348) (Chiang et al. 2001a); gata4 (ENSDARG00000098952) using a 763-bp fragment including exons 1–6 (primers: F-AGCACCGGGCACCATCATTCTCCG and R-GAGCTGGAGGATCCGCTTGGAGGC); gdf9 (ENSDARG00000003229) using a 979-bp fragment including most of the coding region (primers: F-TGTTGAACCCGACGTGCCCC and R-TGGTGTGCATTGGCGACCCG); gsdf (ENSDARG00000075301) (Yan et al. 2017); bmpr2a (ENSDARG00000011941) using a 914-bp fragment containing a part of the last coding exon and the 3′UTR (primers: bmpr2a +2658 F-GAGAGGGAGGAGAGAACAATGAGAGT and bmpr2a –3572 R-AGGGTACGTATCCACAATAGGTTGGA); bmpr2b (ENSDARG00000020057) giving a 727-bp fragment is in exons 12 and 13 (primers: bmpr2b +2978 F-GGAGTCTTCGTCGTCTCGATTGAAAT and bmpr2b –3705 R-TCACCTCTCCGTCTAGTGTATCAGTG); nr5a1a (ENSDARG00000103176) using a 859-bp fragment including exons 2–6 (primers: F-AAGTGTCCGGTTATCATTACGGCC and R- TGTCTGCAGATGTGATCCAGAAGC); and vasa (ENSDARG00000014373) (Yoon et al. 1997). Histology used paraffin-embedded Bouin’s-fixed tissue sectioned at 10 µm and stained with hematoxylin and eosin (Rodríguez-Mari et al. 2005). The gonadosomatic index was calculated as the weight of the gonad divided by the weight of the fish multiplied by 100.
Transcriptomics
Juvenile wild types and amh-26 homozygous mutants at 21 and 35 dpf were killed in Tricaine followed by isolating the gonad-containing trunk from just posterior of the pectoral fin to just anterior to the anus. Adult wild-type and amh-26 homozygous mutant gonads were dissected from 8 months postfertilization (mpf) adult animals. Trunks or gonads from each fish were individually homogenized in 200 μl TRIzol. Total RNA was extracted following Amores et al. (2011), and enriched for mRNA using Dynabeads Oligo(dt)25 (Thermo Fisher Scientific). We constructed indexed, strand-specific complementary DNA sequencing libraries (NEXTflex qRNA-seq kit, BIOO Scientific), quantified libraries by Qubit fluorometer (Life Technologies), normalized libraries to 2.3 nM, multiplexed and quality-checked libraries (Kapa Library Quantification Kit; Kapa Biosystems), and sequenced them in one lane on an Illumina HiSeq 4000 (paired-end 100 bp).
Bioinformatics
The Dupligänger duplicate removal pipeline (Sydes et al. 2019) preprocessed RNA-sequencing (RNA-seq) reads, identified and removed BIOO inline unique molecular identifiers (UMI) from the 5ʹ-end of each read, removed read-through adapters (cutadapt v1.15; Martin 2011; command line options: -n 3 -O 1 -m 30 -a AGATCGGAAGAGC -A AGATCGGAAGAGC–too-short-output–too-short-paired-output), and then removed low-quality sections from both the 5ʹ-ends and 3ʹ-ends (Trimmomatic v0.36; Bolger et al. 2014; command line options: LEADING:10 TRAILING:10 SLIDINGWINDOW:5:10 MINLEN:30). Dupligänger tracked the number of nucleotides removed from the 5ʹ-end and removed reads shorter than 30 nucleotides. We aligned processed paired-end reads to the zebrafish genome (GRCz10, Ensembl version 91) in a splice-aware manner using GSNAP (Wu et al. 2016) (v2017-06-20, command line options:–suboptimal-levels 0–quiet-if-excessive–kmer 15–max-mismatches 0.1–use-splicing–split-output), retaining reads that aligned in a concordant and unique manner. Dupligänger then removed PCR duplicates from the sequence alignment file if both of the following criteria had already been observed in another read pair: the read pair shares 5′ alignment starts for both R1 and R2 after correcting for 5′ trimming, and the read pair shares the same R1 UMI and R2 UMI. We passed deduplicated sequence alignment files to HTSeq count (Anders et al. 2015) (command line options:–mode intersection-strict–type exon–stranded reverse) to obtain per-gene counts for protein-coding genes. DESeq2 provided statistical analysis of fold changes (Love et al. 2015). Analysis of conserved syntenies used the Synteny Database and Genomicus (Catchen et al. 2009; Nguyen et al. 2018).
Data availability
RNA-seq reads are available at the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA512103. Supplemental Material, Table S1 and Table S2 list differentially expressed genes for juvenile trunks or adult gonads for amh mutants and wild-type siblings, respectively. Work was performed under the University of Oregon Institutional Animal Care and Use Committee protocol no. 14-08R. Mutant strains are available on request. Data should be cited associated with this paper. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8184437.
Results
Molecular genetics of induced amh mutations
To identify the roles of Amh in gonad development, we induced frameshift premature stop codon alleles in zebrafish amh (ENSDARG00000014357) using CRISPR/Cas9 and TALEN mutagenesis. CRISPR gRNAs targeted two sites in exon 3 located 16 nucleotides apart (Figure 1A, red). These sites should be translated into the protein’s Amh domain, upstream of the cleavage site that liberates the TGF-β domain that encodes the mature functional Amh protein. To assay CRISPR efficacy, we injected gRNAs and Cas9 RNA into one-cell AB strain embryos, and at 24 hpf, extracted DNA, amplified the target (primer locations in green in Figure 1A), and digested fragments with HpyCH4IV, which cleaves the wild-type but not a mutated site. Three of the four embryos tested had substantially reduced restriction enzyme cleavage (Figure 1B), verifying reagent utility. We raised injected embryos and isolated three mutant lines. Sanger sequencing (Figure 1C) revealed deletions of 5, 10, and 26 nucleotides (Figure 1D, designated below as CRISPR-induced alleles amh-5, amh-10, and amh-26) and a deletion of 11 nucleotides as a TALEN-induced amh(uc28) allele. These frameshift mutations should result in truncated proteins lacking the mature TGF-β domain due to premature stop codons (Figure 1E).
Amh facilitates development of a male phenotype
To learn if amh plays a role in zebrafish sex determination as in some other fish (Hattori et al. 2012; Kamiya et al. 2012; Li et al. 2015), we investigated sex ratios in amh mutant lines. The sex ratio of homozygous wild-type siblings was unbiased (48.4% males; 41 males and 43 females), but homozygous mutants had an average of only 17.8% males (12 males and 54 females; P < 0.05, Wilcoxon rank-sum test), about a third as many as expected, similar to prior results (Lin et al. 2017). We conclude that wild-type amh functions to facilitate the development of males, but is not essential for AB strain zebrafish to develop a male phenotype.
Amh regulates the production of functional gametes
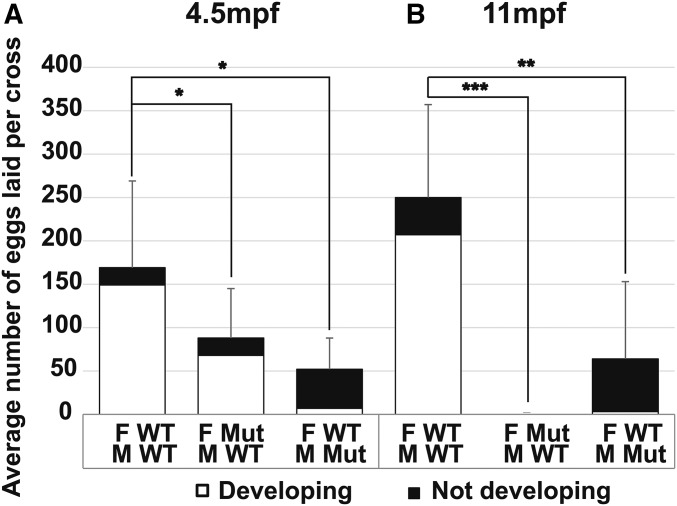
To test female fertility, we mated individual amh mutant females (−26 allele) to AB wild-type males and to test male fertility, we mated individual amh mutant males (−26 allele) to AB wild-type females. For both tests, we counted the number of females that laid eggs, the number of eggs per clutch, and the number of embryos that developed up to 72 hpf. Results showed that homozygous amh mutant females at 4.5 mpf laid about half as many eggs as wild types (87 ± 57 eggs/cross vs. 169 ± 100 eggs/cross), but most eggs from mutant females supported normal embryonic development (744/961 eggs, Figure 2A). Homozygous amh mutant females at 11 mpf failed to lay any eggs at all (Figure 2B). These results show that although young amh mutant females laid fewer eggs than normal, they nevertheless did lay eggs that developed; we conclude that amh mutant females developed functional reproductive ducts and results suggest that Amh is necessary for continued fertility as zebrafish age.
Figure 2.
Fertility tests for adult amh mutants and wild types. (A) Average number of eggs laid per cross from wild-type females crossed to wild-type males (11 crosses), amh-26 mutant females crossed to wild-type males (11 crosses), and wild-type females crossed to amh-26 mutant males (4 crosses) at 4.5 mpf. (B) Average number of eggs laid per cross from wild-type females crossed to wild-type males (8 crosses), amh-26 mutant females crossed to wild-type males (7 crosses), and wild-type females crossed to amh-26 mutant males (6 crosses) at 11 mpf. For each cross, one individual female (either mutant or wild-type sibling) was paired with three nonsibling wild-type males, or for the reciprocal test, one individual male (either mutant or wild-type sibling) was paired with three nonsibling wild-type females. Eggs were collected and counted at 1 dpf and 3 dpf; embryos were scored as developing normally (white bars), or as not developing or improperly developing (black bars). Statistical significance: * 0.05 < P < 0.01, ** 0.01 < P < 0.001, and *** P < 0.001, Wilcoxon rank-sum test. Error bars show SD. F, female; M, male; Mut, mutant; WT, wild type.
Tests of amh mutant male fertility showed that at 4.5 mpf, crosses of single amh-26 homozygous mutant males by three wild-type females resulted in the laying of only ∼27% as many eggs as did wild-type sibling males (45 ± 36 eggs/cross vs. 169 ± 100 eggs/cross, respectively), suggesting that normal Amh activity improves male mating behaviors. Only ∼11% of eggs (5 ± 4 of 45 ± 36) laid by wild-type females mated to mutant males initiated development (Figure 2A), showing that Amh is required for optimal sperm production and/or function. Results for homozygous amh-26 mutant males at 11 mpf showed continuing severe effects on male fertility (Figure 2B). These results indicate that young mutant males make and release mature functional sperm, and thus that their reproductive ducts can transport sperm, at least initially. We conclude that amh function is not required for normal male sex duct development but is necessary for normal rates of functional sperm production. Combined with results from mutant females, we conclude that Amh is not required to construct functional reproductive ducts or to initiate fertility in either sex but is necessary to maintain fertility in both sexes.
Amh promotes juvenile gonad development
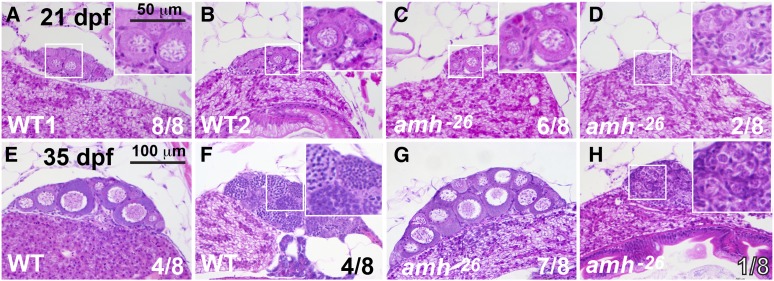
To understand amh mutant gonadal phenotypes, we studied histological sections at several developmental stages. For 21 dpf late-stage larval zebrafish, all eight wild types examined had gonads with stage I oocytes (Selman et al. 1993; Maack and Segner 2003), as expected for zebrafish juvenile hermaphrodites (Takahashi 1977; Rodríguez-Mari et al. 2005, 2010; Wang and Orban 2007). Figure 3, A and B shows two of the eight individuals. Six of eight 21 dpf amh-26 mutants were similar to wild types with stage I oocytes (Figure 3C), but two lacked stage I oocytes and contained only undifferentiated germ cells (Figure 3D). We conclude that most amh mutants develop histologically normal gonads at 21 dpf, although some have gonads with delayed development.
Figure 3.
Gonad histology of 21 dpf and 35 dpf wild-type and amh-mutant fish. (A–D) In histological sections, gonads in all eight 21 dpf wild-type sibling fish contained early oocytes (one gonad shown in each of two individuals in A and B). Gonads in six of eight 21 dpf amh-26 mutants were morphologically like wild-type ovaries (C) and gonads of two of eight 21 dpf amh-26 mutants were undifferentiated (D). At 35 dpf, wild-type fish contained gonads that were clearly either ovaries (4 of 8 fish) (E) or testis (4 of 8 fish) (F). In 35 dpf amh −26 mutants, most fish had ovaries (7 of 8 fish) (G) but one of eight fish had immature testis (H). Smaller boxed regions in several panels are magnified in the larger boxed regions at the right of these panels. Bar in E is 100 µm for all panels; Bar in the higher magnification boxes in A is 50 µm. WT, wild type.
For 35 dpf juveniles, four of the eight wild types examined had stage I–II oocytes (Figure 3E) and four had developing spermatocytes and spermatozoa (Figure 3F). Among the eight amh mutants examined, seven had ovaries with morphologies similar to those in wild types (Figure 3G) and only one fish had gonads that lacked oocytes and possessed developing spermatogonia organized in cysts (Figure 3H) (Maack and Segner 2003). We conclude that most of the 35 dpf mutant juveniles we examined were embarking on a female trajectory, and that the only 35 dpf amh mutant male that we sectioned had gonads that were developmentally delayed with respect to those in wild-type siblings.
In females, Amh inhibits germ cell proliferation and differentiation
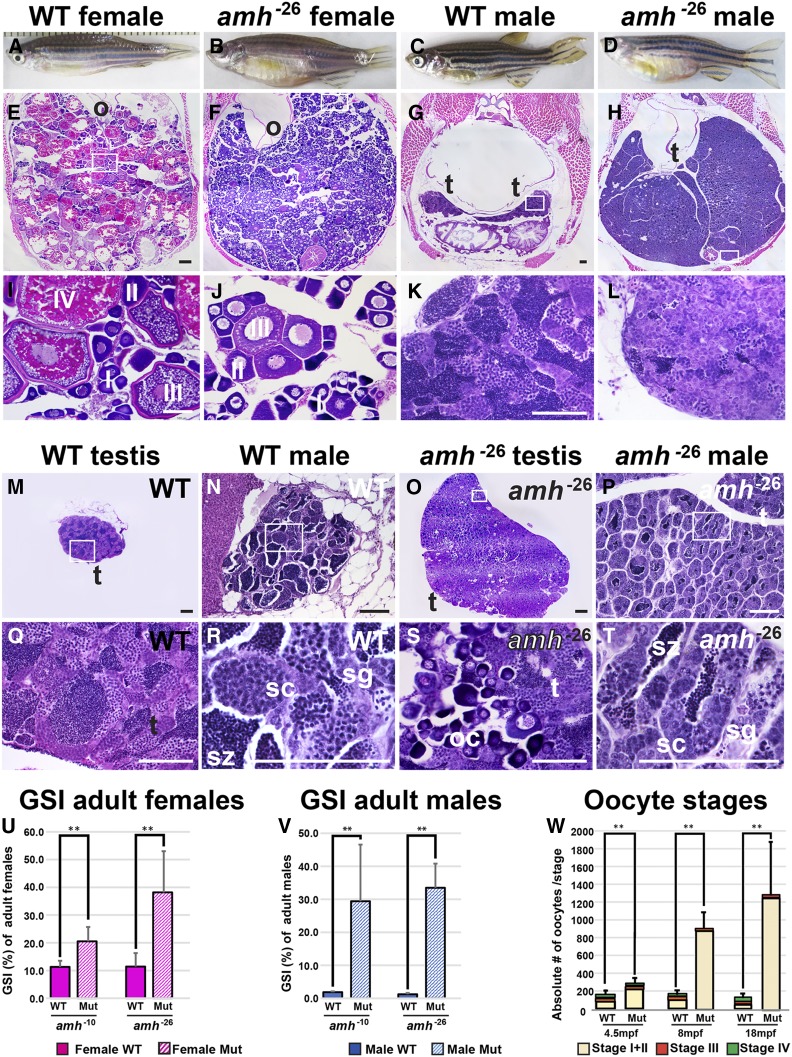
To learn the roles of Amh in adults, we investigated gonad morphology in amh mutants over time. In adult females at 8 mpf, ovaries in wild-type siblings contained oocytes of all stages (Figure 4, A, E, and I). In contrast, amh-26 mutant females had enlarged ovaries that distended the individual’s abdomen (Figure 4, B, F, and J). Averaging results from females homozygous for the amh-10 and amh-26 alleles, the gonadosomatic index [(gonad weight/body weight) × 100] of amh mutants was ∼2.6-fold larger than their respective wild-type siblings, confirming prior results in zebrafish and mouse (Durlinger et al. 1999; Lin et al. 2017). Adult ovaries in 8 mpf amh-26 zebrafish mutants lacked oocytes that had matured beyond stage III (Figure 4, F and J). Young (4.5 mpf) amh-26 mutant ovaries had 2.7 times as many stage I and II oocytes as found in wild-type ovaries (Figure 4W), and by 8 mpf and 18 mpf, the relative proportion of immature oocytes increased to 9- and 35-fold that in wild-type siblings, respectively (Figure 4W). We conclude that Amh activity inhibits oogonia proliferation or maturation. Although young amh-26 mutants had formed stage IV oocytes in the central gonad (average of 23 stage IV oocytes in mutants and 32 in wild types), 8 mpf amh-26 mutant females had few stage IV oocytes in the central gonad (average of 5 oocytes in mutants and 20 in wild types) and 18 mpf amh-26 mutant females had an average of only two stage IV oocytes vs. 17 in wild types (Figure 4W). Homozygotes for the amh-5, amh-10, and amhuc28 alleles displayed similar phenotypes (Figure S3). We conclude that in aging female zebrafish, Amh activity is required to advance ovarian follicles from stage III to more mature stages.
Figure 4.
Amh activity is required for normal gonad morphology in adult zebrafish. (A–D) Eight mpf adult zebrafish: wild types (A, female, six fish sectioned; C, male, seven fish) and amh-26 mutants (B, female, six fish; D, male, seven fish), showing enlarged abdomens in mutants. (E–T) Histological sections of 8 mpf adult gonads: adult female ovaries at low (E and F), and high (I and J) magnification. Cross-sections of an 8 mpf wild-type female sibling (E and I) revealed maturing (stage I and II) and vitellogenic (stage III and IV) follicles. Cross-sections of an 8 mpf amh mutant female (F and J) showed an excess of immature follicles (stage I and II), a few early vitellogenic follicles (stage III), but no late vitellogenic follicles (stage IV) (numbers of oocytes per stage shown in W). M–T illustrate some of the variation in mutant phenotypes. (M and Q) Low and high magnification of dissected wild-type testis. (N and R) Medium and high magnification of a cross section of the abdomen of a different wild-type male. (O and S) Low and high magnification of dissected ovotestis from an amh-26 mutant male showing immature oocytes in the testis. (P and T) Medium and high magnification of the abdomen of a different amh-26 mutant male showing small testis lobules and fewer late stage male gonocytes compared to wild types. Gonadosomatic index (GSI) of adult females (U) and males (V). GSI calculations for females used five wild-type siblings of amh-10 females, five amh-10 mutant females, five wild-type siblings of amh-26 mutant females, and five amh-26 mutant females. GSI calculations for females used five wild-type siblings of amh-10 males, five amh-10 mutant males, eight wild-type siblings of amh-26 mutant males, and five amh-26 mutant males. (W) Number of oocytes per stage at 4.5, 8, and 18 mpf. Oocytes were categorized into three groups: stage I + stage II (beige), stage III (red), and stage IV (green) oocytes in W. The 18 mpf mutant females had mostly stage I + stage II oocytes (W). Statistical significance: ** 0.01 < P < 0.001 and *** P < 0.001, Wilcoxon rank-sum test. Black scale bar in E for E and F; black scale bar in G for G and H; white scale bar in I for I and J; white scale bar in K for K and L. Bar for all, 100 µm. (U–V) Gonadosomatic index (GSI) in percent. In U, Mut refers to amh mutant ovary. In V, Mut refers to amh mutant testis. In U and V, solid boxes, wild types; striped boxes, mutants; red boxes, females; blue boxes, males. I, II, III, IV, ovarian follicle stages 1–4; o, ovary; s, Sertoli cells; sc, spermatocytes; sg, spermatogonia; sz, spermatozoa; t, testis; WT, wild type.
In males, Amh inhibits germ cell proliferation and oocyte development or survival
Males homozygous for each of the four amh mutant alleles displayed several phenotypic differences from wild-type siblings at 8 mpf. First, amh mutant males had much larger abdomens than wild-type siblings (Figure 4, C and D) due to greatly enlarged testes (Figure 4, G, H, K, and L), confirming prior results (Lin et al. 2017). The overgrowth of amh mutant male gonads [∼33.7 times heavier than wild-type sibling gonads; an average of 0.207 ± 0.103 g (SD) for mutant testes (n = 10) vs. 0.006 ± 0.003 g for wild-type testes (n = 13)] was even larger than that of mutant female gonads [2.2-fold, an average of 0.171 ± 0.078 g for mutant ovaries (n = 10) vs. 0.078 ± 0.027 g for wild-type ovaries (n = 10)] (see Figure 4V). We conclude that amh activity is required to inhibit gonad growth both in adult males and in adult females. Adult amh mutant male gonads contained all stages of sperm development, including mature spermatozoa (Figure 4, L, P, and T). Second, the proportion of later stage male gametocytes in amh mutant testes appeared to be greatly reduced compared to wild types and the proportion of immature stages seemed much higher in mutant males than wild type males (Figure 4L vs. Figure 4K). Third, testis tubules were smaller in size but greater in number in amh mutants compared to wild types (Figure 4, G, H, K, L, and M–T). In cross sections, lobules in mutant testes were only 19.3% as large as lobules in wild-type testes (638 ± 272 vs. 3313 ± 611 µm2). Fourth, and most remarkable, more than half of the 8 mpf amh-26 male mutant gonads examined (four out of seven fish) contained early-stage oocytes, but none of the seven 8 mpf wild-type male siblings did (Figure 4, M–T). The finding of ovo-testes in mature adult amh mutants shows that normal amh activity helps to masculinize zebrafish gonad development by inhibiting the production or survival of young oocytes. We conclude that in zebrafish, normal Amh activity is required to regulate the proliferation of spermatogonia, to control the number and size of testis tubules, to govern the rate of maturation of spermatogonia to spermatozoa, and to ensure that immature oocytes disappear from male gonads during the juvenile hermaphrodite stage or to block the formation of oocytes in later development.
Amh and Gsdf appear to act in the same developmental pathway
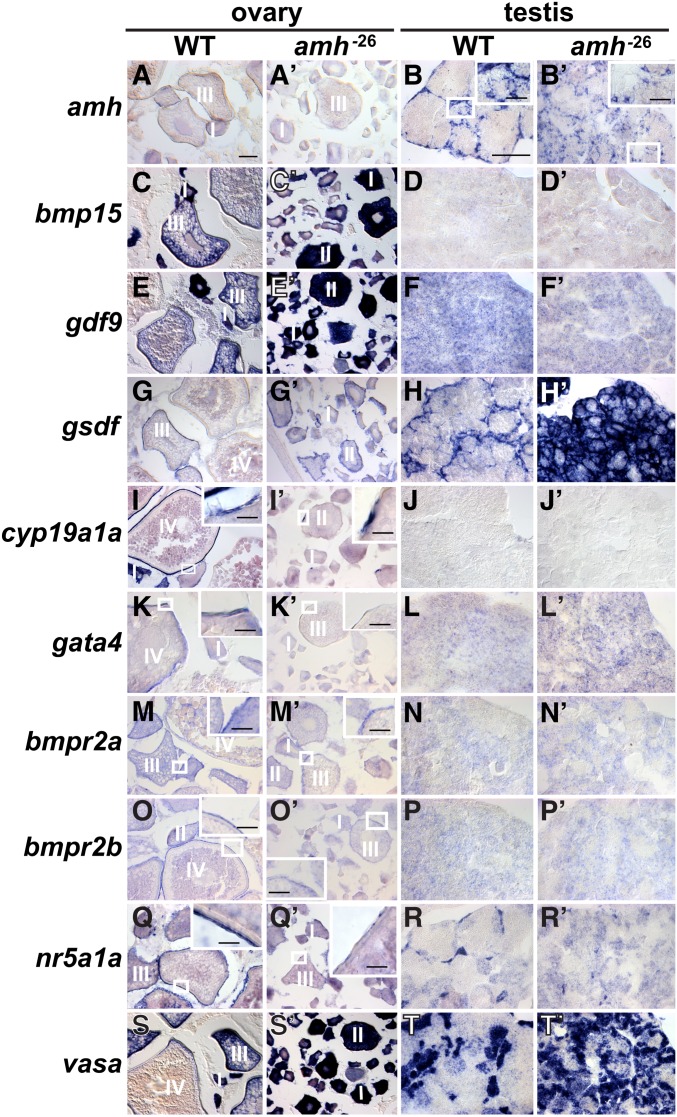
Gsdf, like Amh, is essential to prevent the accumulation of young oocytes as zebrafish females age (Yan et al. 2017). If these two genes act in the same pathway, then double mutant ovaries should have about the same phenotype as each single mutant. Alternatively, if the genes act in parallel pathways, then double mutants should have more severe phenotypes than either single mutant. Analysis of amh;gsdf double mutants revealed female gonad phenotypes that were about the same as in each of the two single mutants: all three genotypes accumulated an enormous number of small oocytes with few stage III oocytes at 8–12 mpf (Figure S1 and data not shown). Males homozygous mutant for either amh or gsdf had enlarged testes compared to wild types, amh mutant males had larger testes even than gsdf mutant males, and amh mutant males became sterile as they aged while gsdf mutant males maintained fertility. Double mutant testes were similar to amh mutants, and not more severe (Figure S1), consistent with the explanation that in males as in females, amh and gsdf act in the same pathway. Furthermore, amh expression was nearly twice as high in gsdf mutant testes as in wild-type testes (Yan et al. 2017), suggesting that Gsdf controls amh. Reciprocally, gsdf expression was 3.4-fold higher in amh mutant adult testes compared to wild-type testes in our RNA-seq results (see Table S2), consistent with the result from in situ hybridization (Figure 5, H and H’), suggesting that Amh controls gsdf. Together the mutant phenotypes and expression data show that the regulation of these two TGF-β family genes are interdependent.
Figure 5.
Gene expression patterns in adult gonads at 8 mpf. Wild-type ovaries (A, C, E, G, I, K, M, O, Q, and S); amh-26 mutant ovaries (A’, C’, E’, G’, I’, K’, M’, O’, Q’, and S’); wild-type testis (B, D, F, H, J, L, N, P, R, and T); amh mutant testis (B’, D’, F’, H’, J’, L’, N’, P’, R’, and T’). In situ hybridization for amh (A, A’, B, and B’), bmp15 (C, C’, D, and D’), gdf9 (E, E’, F, and F’), gsdf (G, G’, H, and H’), cyp19a1a (I, I’, J, and J’), gata4 (K, K’, L, and L’), bmpr2a (M, M’, N, and N’), bmpr2b (O, O’, P, and P’), nr5a1a (Q, Q’, R, and R’), and vasa (S, S’, T, and T’). Small boxed regions in low magnification views are shown in larger boxed regions at higher magnification for B, B’, I, I’ K, K’, M. M’, O, O’, Q, Q’. Bar for main panels represents 100 mm; bar for higher magnification in boxed regions represent 25 mm. I, II, III, IV, ovarian follicle stages 1–4.
Amh activity is required for normal expression of key gonad development genes
To understand in more detail the role of Amh in zebrafish gonad development, we studied the expression of several key regulatory and marker genes in adult wild types and amh mutants by in situ hybridization.
Wild-type adult ovaries at 8 mpf expressed amh mainly in granulosa cells surrounding stage II oocytes (Figure 5A, see also Rodríguez-Mari et al. 2005; von Hofsten et al. 2005). In contrast, amh-26 mutant ovaries at 8 mpf showed little amh expression in somatic cells surrounding oocytes, due either to nonsense-mediated decay or to the failure of amh-expressing cells to form in amh mutants (Figure 5A’). Wild-type males at 8 mpf displayed a well-organized pattern of amh expression in Sertoli cells surrounding testis tubules (Figure 5B, see also Rodríguez-Mari et al. 2005; von Hofsten et al. 2005). Presumptive Sertoli cells also expressed amh in 8 mpf amh mutant males, demonstrating transcript stability, but amh-expressing cells were less organized; testis tubules appeared to be smaller; and amh-expressing cells did not completely surround most testis tubules (Figure 5, B and B’). Homozygous amh-5 and amh-10 mutants showed similar expression patterns (data not shown). We conclude that in adult male zebrafish, amh is required for the organization of Sertoli cells in testis tubules.
Bmp15 is an extracellular signaling protein that 8 mpf wild-type adult zebrafish express mainly in oocytes in early-stage ovarian follicles, and in maturing oocytes in later-stage wild-type follicles (Figure 5C and Clelland et al. 2006; Dranow et al. 2016). Adult amh mutant ovaries appeared to express bmp15 stronger than wild-type ovaries (Figure 5, C and C’) due to the accumulation of younger stages that express high levels of bmp15. Neither wild-type nor mutant testes showed significant bmp15 expression (Figure 5, D and D’). We conclude that Amh function promotes the maturation of ovarian follicles in mature adult ovaries.
Gdf9, like Bmp15, is a TGF-β family member that marks oocytes (Liu and Ge 2007; Dranow et al. 2016). Expression of gdf9 appeared to increase in mature adult amh mutant ovaries compared to wild-type ovaries (Figure 5, E and E’), likely due to accumulating young oocytes in amh mutants. Testes showed negligible gdf9 expression in either amh mutants or in wild-type siblings (Figure 5, F and F’). We conclude that amh function is necessary for the maturation of oocytes to stages in which they appear to downregulate the gdf9 transcript.
Gsdf is an important signaling molecule in fish gonadogenesis (Rondeau et al. 2013; Imai et al. 2015; Zhang et al. 2016). Wild-type ovaries express gsdf in granulosa cells surrounding oocytes (Figure 5G and Gautier et al. 2011a; Yan et al. 2017). Zebrafish amh mutant ovaries also expressed gsdf in epithelial cells surrounding immature oocytes (Figure 5G’). Testes expressed gsdf specifically in Sertoli cells surrounding germ cells (Figure 5H and Gautier et al. 2011a; Yan et al. 2017). Testes lacking amh activity showed substantially greater gsdf expression than normal, suggesting altered Sertoli cell development (Figure 5, H and H’). Expression of the Sertoli cell marker amh in amh mutant testes showed that Sertoli cells were poorly organized with smaller testis tubules (Figure 5, B and B’), which was confirmed by gsdf expression (Figure 5, H and H’) and histology (Figure 4, N and P). Taken together, results from gsdf expression and histology analyses show that amh mutants appeared to have many more testis tubules, but much smaller testis tubules, than normal, consistent with an increase in Sertoli cells or their precursors. We conclude that Amh function in adult male zebrafish is necessary for the organization and number of gsdf-expressing cells and may help regulate gsdf expression.
Aromatase, encoded in zebrafish ovaries by cyp19a1a (and in the brain by cyp19a1b; Chiang et al. 2001a,b), converts testosterone to estrogen (Rouiller-Fabre et al. 1998). As in humans, adult wild-type zebrafish express cyp19a1 in granulosa cells and theca cells in ovarian follicles (Figure 5I and Chiang et al. 2001a,b; Dranow et al. 2016). In contrast, young-stage follicles in adult zebrafish amh mutant ovaries showed fewer cyp19a1a expressing cells in patches that did not completely surround follicles (Figure 5I’). In testes, cyp19a1a expression was not detected in either wild types or amh mutants (Figure 5, J and J’). We conclude that amh activity is required for ovarian follicles to advance to the strongly aromatase-expressing stage and for the organization of granulosa cells around ovarian follicles.
GATA4 in human gonads synergistically activates the AMH promoter by interacting with NR5A1 (SF-1), a process necessary for normal human sex development (Lourenco et al. 2011). In mice, granulosa cells and theca cells express Gata4 (Padua 2014) and in wild-type zebrafish, oocytes express gata4 in early stages and granulosa and theca cells express gata4 in later stages (Figure 5K; Yan et al. 2017). In zebrafish, adult female amh mutants, like wild types, displayed gata4 transcript in young oocytes, but it was patchy in follicular cells due presumably to alterations in follicular maturation (Figure 5K’). Expression of gata4 was low in both wild-type and mutant adult testes (Figure 5, L and L’). These results suggest that amh activity normally helps to upregulate gata4 in granulosa cells of wild-type ovaries.
Bmpr2 is likely the type II receptor for BMP15 (Moore et al. 2003; Pulkki et al. 2012). Zebrafish has two co-orthologs of Bmpr2: bmpr2a is expressed in young oocytes and ovarian follicle cells and bmpr2b is expressed in follicle cells (Li and Ge 2011; Dranow et al. 2016). Our in situ hybridization experiments confirmed the wild-type expression pattern of bmpr2a and showed that in mutant ovaries, bmpr2a expression was reduced in young oocytes but was maintained weakly in stage III follicles (Figure 5, M and M’). For bmpr2b, expression appeared in wild types in follicle cells, but in amh mutants, reduced signal was detected in follicle cells (Figure 5, O and O’). Testes in both wild types and amh mutants appeared to possess little expression of either bmpr2 gene and no difference appeared to distinguish wild types from mutants (Figure 5, N, N’, P, and P’).
NR5A1 (alias steroidogenic factor 1; SF-1) interacts with Gata4 protein in cultured primary rat Sertoli cells to upregulate Amh expression (Tremblay et al. 2001). Zebrafish adult ovaries express nr5a1a (von Hofsten et al. 2005), and our in situ studies showed that this expression is in granulosa cells (Figure 5Q), as it is in mammals. Adult amh mutant females expressed nr5a1a in a much reduced and fragmented, patchy, granulosa cell layer (Figure 5Q’), showing that Amh is important for the organization or development of granulosa cells. Adult wild-type testes expressed nr5a1a in Leydig cells (Figure 5R), but far fewer cells expressed nr5a1a in mutant testes compared to wild-type testes (Figure 5R’), despite the increased number of testis tubules in amh mutants (compare nr5a1a expression in Figure 5, R and R’, to gsdf expression in Figure 5, H and H’). We conclude that in male zebrafish, amh function is required for normal Leydig cell development. These results show that in both male and female adult zebrafish, cells expressing nr5a1a require amh function for normal development, and, because Nr5a1 and Gata4 proteins interact to control Amh expression in mammals (Tremblay et al. 2001; Lourenco et al. 2011), these three genes likely act in a feedback loop.
Vasa, a putative RNA helicase encoded by ddx4, is expressed in germ cells in wild-type zebrafish (Figure 5, S and T; Yoon et al. 1997). Zebrafish amh mutants also expressed ddx4 in germ cells in both males and females (Figure 5, S’ and T’). The intensity of vasa signal in wild-type oocytes diminished as follicles matured (Figure 5S; Yoon et al. 1997), but in adult amh mutants, all oocytes showed high levels of vasa expression, consistent with a failure of oocyte maturation in amh mutant ovaries (Figure 5, S and S’). In adult testes, amh mutants appeared to have more, but smaller, groups of germ cells than did wild types (Figure 5, T and T’). We conclude that differences in ddx4 expression reflect the histological differences between wild-type and amh mutant gonads.
Zebrafish amh mutants help to identify gene regulatory pathways in gonad development
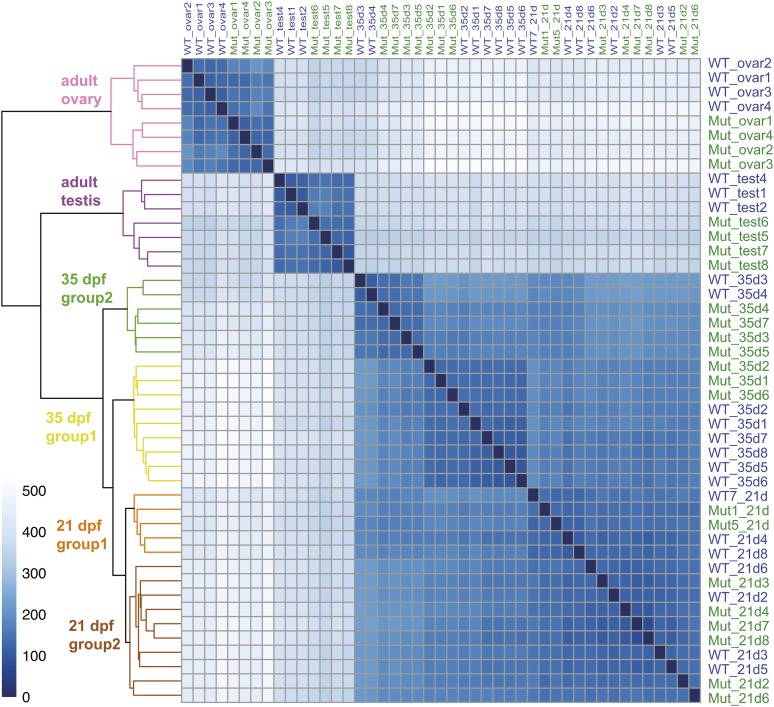
To help understand genetic programs that regulate gonad development, we sequenced 45 strand-specific RNA-seq libraries, each sample derived from a single individual fish at one of three different ages. Fifteen samples comprised the gonad-containing trunks of 21 dpf transitional state juveniles (eight wild types and seven amh-26 mutants). Another 15 trunks were from 35 dpf juveniles (eight wild types and seven amh-26 mutants. The final 15 libraries came from mature adults at 8 mpf, including seven pairs of testes (three individual wild types and four different amh mutants) and eight pairs of ovaries (four wild types and four amh-26 mutants). These 45 RNA-seq libraries produced 396 million paired-end sequence reads, of which 211 million mapped to the Ensembl v91 protein-coding exons of the zebrafish GRCz10 version of the zebrafish reference genome. Two-way similarity clustering (regularized log–transformed Euclidean distances) of all samples produced a clear separation between young juveniles, older juveniles, adult ovaries, and adult testes (Figure 6).
Figure 6.
Heat map and dendrogram of regularized log-transformed Euclidean distances between all 45 RNA-seq samples. Analysis divided samples into six groups: adult ovary, adult testes, two groups of 35 dpf trunks, and two groups of 21 dpf trunks. The intensity of each cell in the panel reflects the number of genes different in the intersecting two samples according to the scale at the left, so the diagonal self-comparisons show no genes differently expressed.
Genome-wide transcriptomics of wild-type adult zebrafish ovaries
Interpretation of gene expression changes in developing mutant gonads requires knowledge of gene expression patterns in adult wild-type gonads (Santos et al. 2007a,b; Sreenivasan et al. 2014; Lee et al. 2017). We sequenced strand-specific RNA-seq libraries from ovaries of four homozygous wild-type adult females at 8 mpf and testes from three homozygous wild-type adult male siblings, all of which were siblings of amh-26 mutants. DESeq2 analysis showed that 16,493 genes were differentially expressed in wild-type adult ovaries vs. testes (Table S2).
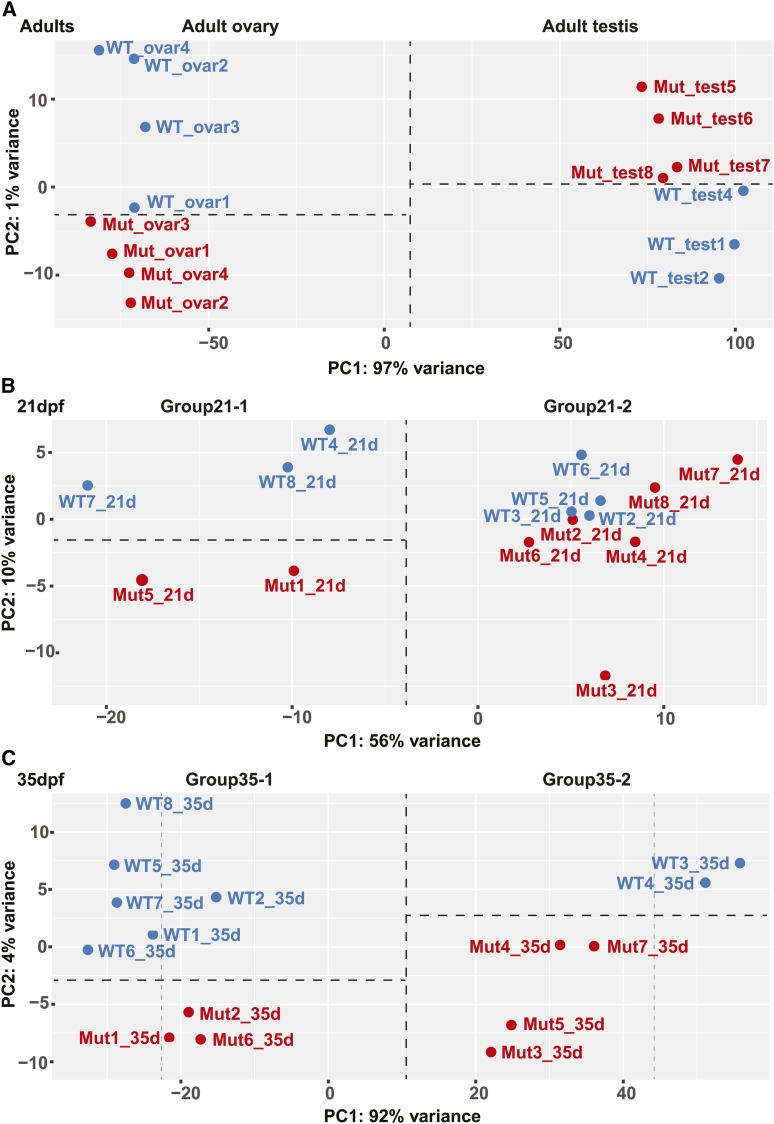
Principal component analysis separated adult testes and ovaries into two distinct groups widely separated in the PC1 axis, which explained 97% of the variance (Figure 7A). Wild-type gonads separated from amh mutant gonads in the PC2 axis, which explained only 1% of the variance. Importantly, amh mutant ovaries tended to occupy the negative portion of the space and wild-type ovaries the positive portion, but the reverse was true for testes (Figure 7A). This result shows that along the PC2 axis, the transcriptomes of mutant ovaries tended to be more like those of wild-type males (i.e., ovaries were masculinized) but the transcriptomes of mutant testes were more like female transcriptomes (i.e., testes were feminized). Masculinization of the ovary transcriptome and feminization of the testis transcriptome reflects the dual roles of amh in males and females.
Figure 7.
Principal component analyses (PCA). DESeq2-generated regularized logs of the 500 most variable genes of (A) adult ovary and testes samples, (B) 21 dpf samples, and (C) 35 dpf samples.
Genes with the highest overexpression in adult zebrafish wild-type ovaries vs. wild-type testes tended to have no human orthologs and no previously assigned functions. For example, three genes were massively upregulated in zebrafish ovaries with respect to testes (zgc:171781, CABZ01059627.2, si:ch211-125e6.12) by 146 million-, 120 million-, and 116 million-fold, respectively. Each of these three genes has several paralogs in zebrafish, but either no orthologs or few orthologs in other species and none have known functions, although ZFIN lists si:ch211-125e6.12 as Pfam:PF00059, a C-type lectin. Of the 100 most upregulated ovary genes, only 21 have gene names that imply function, including 10 zona pellucida genes (zp2.1, zp2.3, zp3.2, zpcx, zp2.5, zp2.6, zp2.2, zp3a.1, zp2l1,and zp3a.2), and only 11 other genes, including the ovary-specific epithelial cell tight junction gene cldnd (1582-fold up) (Clelland and Kelly 2011), the ovary-specific retinol saturase gene retsatl (1393-fold up) (Sreenivasan et al. 2008), the ovary carbonic anhydrase gene ca15b (1337-fold up) (Wang et al. 2013), the primordial germ cell histone gene h1m (1213-fold up) (Müller et al. 2002), two copies of the quinoid dihydropteridine reductase gene qdprb2 (1079- and 490-fold up), the zebrafish ortholog of a gonadal soma nuclear repressor gene required for germ cell development zglp1 (785-fold up) (Li et al. 2007), the oocyte gene cth1 (cysteine three histidine 1, 579-fold up; te Kronnie et al. 1999), the germ plasm aggregation gene birc5b (510-fold up) (Nair et al. 2013), the extracellular matrix protein gene ecm1a (454-fold up), and the immune gene crp2 (C-reactive protein 2, 409-fold up). We hypothesize that the large number of unannotated but highly expressed ovary-specific genes provide essential functions related to eggshells or other species-specific egg functions.
In addition to many genes of unknown function, most known female regulatory genes were also upregulated in wild-type adult zebrafish ovaries compared to testes, including the Wnt-signaling genes axin2 (24-fold up) and rspo1 (2.1-fold up); the Foxl2-related genes foxl2a (ENSDARG00000042180, 49-fold up), foxl2b (ENSDARG00000068417, 7.4-fold up), and foxl3 (ENSDARG00000008010, 5.3-fold up); the zona pellucida gene regulator figla (14-fold up); and other oocyte gene regulators like bmp15 (41-fold up) and gdf9 (26-fold up).
Genome-wide transcriptomics of wild-type adult zebrafish testes
Up-regulated genes in wild-type testes vs. wild-type ovaries included the sperm-specific potassium ion channel gene cngk (9643-fold up) (Fechner et al. 2015). Genes encoding likely sperm components were the next most strongly overexpressed genes in adult wild-type testes vs. ovaries, including ribc1 and ribc2 (6094- and 3730-fold up, respectively), ccdc83 (5898-fold up), and rsph4a and rsph9 (5211- and 3569-fold up). Many genes annotated as being male-specific regulatory genes were also overexpressed in wild-type testis vs. wild-type ovary, including amh (244-fold up), dmrt1 (411-fold up), gsdf (40-fold up), SoxD-related genes (sox9a, 47-fold up; sox8a, 14-fold up; sox8b, 27-fold up; sox10, 4.1-fold up), and dhh and its receptor-encoding genes ptch1 and ptch2 (61-, 3.9-, and 2.9-fold up in testes, respectively). The wt1a and nr0b1 (dax1) genes were only slightly, but significantly, elevated in wild-type testes vs. ovaries (1.7-fold and 3.1-fold, respectively). Although vitellogenin genes appeared to be upregulated in wild-type testes vs. wild-type ovaries, overall counts were so low that fold changes were likely spurious. Vtg peptides have been detected in ovaries (Groh et al. 2013), although we saw no reads from vtg genes in wild-type adult ovaries.
Expression of steroid biosynthetic genes in wild-type gonads
Several steroid biosynthetic genes were differentially expressed comparing adult wild-type ovaries to wild-type testes. A duplication event in the zebrafish lineage after it diverged from Astyanax cavefish produced tandem co-orthologs of the single-copy human gene CYP11A1, which encodes side-chain cleavage enzyme, the first enzyme in steroid biogenesis. The cyp11a1 gene was 25-fold upregulated in zebrafish ovaries but cyp11a2 was 8.1-fold upregulated in testes, suggesting a subfunctionalization event (Force et al. 1999). The gene encoding Hsd17b1, which converts androstenedione to testosterone and estrone (E1) to estradiol (E2), was upregulated in ovaries 175-fold over testes. Females convert testosterone to estrogen by aromatase, and cyp19a1a was upregulated 65-fold in wild-type ovaries compared to wild-type testes. Male mammals and male fish convert testosterone to 11-keto-testosterone, the primary androgen in fish, using Cyp11b1 in mouse (Cyp11c1 in zebrafish) and Hsd11b2 (Wang and Orban 2007; Yazawa et al. 2008; Lee et al. 2017); cyp11c1 was upregulated 1504-fold and hsd11b2 was upregulated 10.1-fold in wild-type testes vs. ovaries. HSD3B1 and HSD3B2 reside in tandem in human but their zebrafish orthologs are on two different chromosomes; we found hsd3b2 upregulated in ovaries (8.1-fold) and hsd3b1 upregulated in testes (6.4-fold).
This data set (Table S2) contributes a substantial resource for understanding the normal functioning of adult zebrafish gonads and a standard for detecting the effects of mutations on gonad development.
Gene expression in 21 dpf zebrafish juveniles
At 21 dpf, zebrafish late-stage larvae are transitioning to become males or females (Takahashi 1977; Maack and Segner 2003; Rodríguez-Mari et al. 2005; Wang et al. 2007). Sequencing the gonad-containing trunks of amh mutants and wild types produced 189 million paired-end reads and after preprocessing (see Table S1), 103 million reads mapped to protein-coding exons. Analysis identified just 24 genes differentially expressed between amh mutants and wild types at 21 dpf. The amh gene itself was underexpressed 6.1-fold in amh mutants, but this change was just outside the limit of significance [adjusted p-value (padj) = 0.106] a result that reflects the relatively small difference between mutant and wild-type gonadal phenotypes as revealed by histology at this stage (see Figure 3, A–D) and the relative stability of transcripts from the mutated amh allele.
Transitional-stage amh mutant fish at 21 dpf expressed a number of gonadal regulatory genes abnormally. The most upregulated gene in 21 dpf amh mutant trunks vs. wild-type trunks was nr0b2a (3.03-fold upregulated). In mammals, Nr0b2(SHP) dimerizes with Nr0b1(DAX1), thereby repressing Nr5a1(SF-1)-mediated activity of the Amh promoter (Tremblay and Viger 2001; Iyer et al. 2006). Furthermore, the loss of nr0b1(dax1) in zebrafish causes female-to-male sex reversal (Chen et al. 2016), in agreement with the reverse situation in which the duplication of NR0B1 in humans causes male-to-female sex reversal (Barbaro et al. 2007). The upregulation of nr0b2a in trunks of 21 dpf amh mutants, as well as in adult mutant ovaries vs. wild-type ovaries (4.8-fold) suggests that amh normally represses nr0b2, and hence female development, in zebrafish. The second most upregulated gene in 21 dpf mutant trunks vs. 21 dpf wild-type trunks, was the Leydig cell marker gene cyp26a1 (2.95-fold up) (Wang et al. 2007), which encodes an enzyme that in zebrafish degrades retinoic acid (Rodríguez-Mari et al. 2013), the signal for entry into meiosis (Koubova et al. 2006; Adolfi et al. 2016). The upregulation of cyp26a1 in amh mutants would likely decrease the level of retinoic acid in mutants, and thus decrease the number of cells entering meiosis, a process that oocytes begin before spermatocytes do, thus suggesting that amh normally depresses cyp26a1 expression at 21 dpf. Other upregulated genes in 21 dpf amh mutants included the proteasome activator psme4a (2.4-fold up in mutants and 10.3-fold up in wild-type testes vs. wild-type ovaries), the lipid metabolism gene trim63a (2.1-fold up), the circadian nuclear receptor gene nr1d1 (1.9-fold up in 21 dpf amh mutants and 6.4-fold up in wild-type testes vs. wild-type ovaries), and the theca cell/Leydig cell marker ptch2 (1.6-fold up in 21 dpf amh mutants and 2.9-fold up in wild-type testes vs. wild-type ovaries) (Yao et al. 2002; Wijgerde et al. 2005; Herpin et al. 2013). Reciprocally, the most downregulated gene in 21 dpf mutant trunks vs. amh wild-type trunks was the complement factor H–related gene cfhl1 (25.1-fold down in 21 dpf mutant trunks and 25-fold down in wild-type ovaries vs. wild-type testes). Only two other genes were significantly downregulated by more than twofold in mutants: an uncharacterized sulfotransferase gene (si:dkey-236e20.3), and a hydroxybutyrate transporter gene slc16a6b (Hugo et al. 2012). We conclude that during the transitional period, the loss of amh function disrupts gonad development but not in a way that appears to strictly downregulate canonical male-related genes as expected by the hypothesis that Amh should upregulate male development.
Unsupervised similarity clustering split the 15 21 dpf animals into two groups (Figure 7B): Group21-1 contained two mutants and three wild types and Group21-2 had six mutants and four wild types. Principal component analysis (Figure 7B) clustered individuals as they had with regularized log–transformed Euclidean distances (Figure 6), bolstering the view that these are biologically meaningful groups. The two groups are separated in the PC1 dimension, which explains 56% of the variance. PC2, which explains 10% of the variance, appeared to further separate Group21-1 into two groups: amh mutants and wild types, but small sample size thwarted statistical analysis of genes differentially expressed between Group21-1 amh mutants and wild types. The finding that Group21-1 and Group21-2 both contain wild-type and mutant individuals shows that at this early stage, amh expression is not the main factor that allocates individual fish into two groups.
To identify biological factors that distinguish the two synthetic 21 dpf groups, we searched for genes differentially expressed between them. Analysis identified 440 genes that met the padj <0.1 criterion for false discovery rate (FDR) (Table S1). Genes upregulated in Group21-1 vs. Group21-2 included several genes encoding components of the chorion, which oocytes begin to produce in stage IB follicles (Selman et al. 1993). These genes included the zona pellucida genes zp2.2 (116-fold upregulated in Group21-1 vs. Group21-2 and upregulated 938-fold in wild-type ovary vs. wild-type testis), zp2.5 (98-fold up and 958-fold up in wild-type ovary vs. wild-type testis), and 13 other zp genes. Zona pellucida genes in mouse and likely in zebrafish are controlled by the germ-cell transcription factor gene figla (factor in germline-alpha) (Liang et al. 1997; Onichtchouk et al. 2003; Mold et al. 2009); consistent with this role, figla was upregulated in Group21-1 vs. Group21-2 (32-fold; 14-fold up in wild-type ovary vs. wild-type testis), Group21-1 also upregulated the follicle stage I and II tight junction gene cldnd (96-fold; 5.6-fold up in wild-type ovary vs. wild-type testis), and other oocyte genes like the oocyte carbonic anhydrase gene ca15b (72-fold up; 1337-fold up in wild-type ovary vs. wild-type testis) (Wang et al. 2013), zar1 (66-fold up; 148-fold up in wild-type ovary vs. wild-type testis) (Miao et al. 2017), gdf9 (18-fold up; 26-fold up in wild-type ovary vs. wild-type testis), and dazl (11-fold up; not differentially expressed in wild-type ovary vs. wild-type testis) (Howley and Ho 2000; Clelland and Kelly 2011; Dranow et al. 2016). Germ cells in Group21-1 gonads were apparently entering meiosis because they upregulated the synaptonemal complex gene sycp2l relative to Group21-2 (14.6-fold up; 14-fold up in wild-type ovary vs. wild-type testis). The strong expression of many oocyte genes shows that Group21-1 juveniles had substantially more developing oocytes than Group21-2. Vitellogenin genes were also upregulated in Group21-1 trunks relative to Group21-2 trunks, including vtg4 (32-fold up) and vtg2 (31-fold). Vitellogenin genes were most likely expressed in liver, which was present in trunk preparations, but might also have been expressed in adipose cells in the ovary (Wang et al. 2005). This result suggests that Group21-1 gonads were already secreting estrogen that upregulated vtg expression, but the only granulosa or theca cell marker that was upregulated in Group21-1 compared to Group21-2 was cyp11a1 (27-fold up), which encodes the enzyme catalyzing the first and rate-limiting step in steroid biogenesis. We conclude that genes overexpressed in Group21-1 vs. Group21-2 characterize developing oocytes.
Reciprocally, Group21-2 upregulated 18 genes relative to Group21-1 fish (padj < 0.1). Of these 18 genes, 14 were also upregulated in wild-type testes relative to wild-type ovaries (cap2, stard13a, si:ch211-133n4.4, col15a1b, adamts12, mmp13b, elf3, ift74, mhc1uka, b3gat1b, cyp27b1, BX004785.2, si:ch211-286b4.4, gstm.2) an average of 49-fold; none were downregulated in wild-type testis relative to wild-type ovaries; and four were not differentially expressed in wild-type gonads (pomk, si:ch211-226h7.5, BX005421.3, zgc:162154). We conclude that Group21-2, which was not expressing female genes, were expressing male genes, although few of these genes had previously been recognized as testis-related genes. Note, however, that the three most upregulated genes in Group21-2 relative to Group 21-1 (si:ch211-226h7.5, BX005421.3, zgc:162154) were not differentially expressed in our ovary-vs.-testis comparison, suggesting that they may be transiently expressed in zebrafish transitioning to stable male development. We conclude that Group21-2 fish were embarking on a male pathway or were developmentally delayed with respect to Group21-1 fish.
We assessed the functional significance of the 440 genes differentially expressed between Group21-1 and Group21-2 samples using gene ontology (GO) analysis of biological processes (Mi et al. 2013). GO analysis identified 18 gene clusters at an FDR < 0.05. The top three clusters were strongly influenced by germ cell development. The highest loading enrichment cluster was “piRNA metabolic process” (FDR = 8.70E−03), and contained three genes (henmt1, pld6, and asz1) that were upregulated in Group21-1 (putative females). The next highest loading enrichment cluster was “positive regulation of acrosome reaction,” (FDR = 1.7E−12), with 11 of 12 genes annotated as zona pellucida genes or containing a zona pellucida domain. All were upregulated in Group21-1 samples. These same 12 genes were also the basis for the third (“egg coat formation”) and fourth (“binding of sperm to zona pellucida”) enrichment clusters. Together, the examination of individual dysregulated genes and the unbiased GO analysis agree that that among the 21 dpf fish, Group21-1 juveniles are embarking on a female development and Group21-2 are becoming males. This is the first demonstration of a difference between developing males and females at this early age by whole-genome transcriptomic analysis.
Gonadal gene expression in 35 dpf juveniles
Sequencing the individual trunks of 15 juveniles at 35 dpf (seven amh-28 mutants and eight wild-type siblings) produced 93 million paired-end reads and after preprocessing (see Table S2), 48 million reads mapped to protein-coding exons. Analysis of differential expression between wild-type and amh mutant samples identified 75 differentially expressed genes (Table S1). Unlike the 21 dpf late-stage larvae, 35 dpf mutant juveniles showed significant downregulation of amh expression, the fourth most downregulated gene in mutants (14.1-fold down).
The most differentially expressed upregulated gene in 35 dpf amh mutants was the butyrophilin subfamily immunoregulator gene si:dkey-208m12.2 (85-fold up in mutants). Other strongly upregulated genes in amh mutants were also immune related, including the novel fish interferon-stimulated gene gig2l (22-fold up) (Zhang et al. 2013) and interferon-stimulated gene-15 (isg15; 4.8-fold up), and the interferon-induced genes mxe (4.1-fold up) and mxb (5.0-fold up) (Novel et al. 2013). Interferon regulatory factor-7 (irf7; 4.5-fold up) is positively correlated with male-related genes in turbot (Ribas et al. 2016) and is a paralog of the trout master sex-determining gene sdY, a duplicated, truncated copy of irf9 (Yano et al. 2012). Downregulated genes in 35 dpf amh mutants vs. wild-type siblings included the complement factor genes cfhl2 (18-fold down, in amh 35 dpf mutant trunks and 5.1-fold up in wild-type testes vs. wild-type ovaries) and cfhl1 (18.7-fold down in amh 35 dpf mutant trunks and 25-fold up in wild-type mature testes vs. ovaries); cfhl1 was also the most strongly downregulated gene in 21 dpf amh mutant trunks vs. wild-type trunks (25.1-fold down). These results suggest that at 35 dpf, gonads developing in amh mutants may experience cell damage that evokes an inflammatory response.
Similarity clustering based on globally correlated gene expression patterns resolved 35 dpf samples into two distinct groups, and within those two major groups, wild types separated from amh mutants but with short branches in the tree (Figure 6). Principal component analysis sorted 35 dpf animals into the same two groups (Group35-1 and Group35-2), primarily along PC1, which explained 92% of the variance (Figure 7C). Because each synthetic group included both amh mutants and wild types, differences other than genotype at the amh locus were important for distinguishing between major groups at 35dp. Within each of the two groups separated along PC1, mutants tended to occupy the lower portion of the plot and wild types the upper portion along the PC2 axis (Figure 7C), even though this axis explained only 4% of the overall variance. Separation along PC2 may have resulted from expression changes in genes downstream of Amh function.
Analysis of genes differentially expressed between these Group35-1 and Group35-2 yielded 8728 differentially expressed genes (Table S1). The most differentially expressed genes between the trunks of Group35-1 and Group35-2 juveniles encode the egg yolk protein Vitellogenin-1 (vtg1, 929-fold upregulated in Group35-1), with other vtg genes also highly upregulated (e.g., vtg2, 445-fold up and vtg4, 319-fold up in Group35-1). The strong upregulation of vtg gene expression in Group35-1 animals suggests first, that they are developing as females and second, that their livers had activated vtg genes due to secretion of higher levels of estrogen than Group35-2 fish, and thus, third, that their granulosa and theca cells were already functioning. Group35-1 animals also expressed differentially the female-enriched cell-cycle gene btg4 (Small et al. 2009) (431-fold up), as well as several zona pellucida-encoding genes including zp2l1 (247-fold up), zpcx (265-fold up), and zp2.2 (214-fold up) along with their putative regulator figla (165-fold up). Group35-1 animals also expressed the meiosis gene sycp2l (120-fold up). These results show that Group35-1 animals had initiated a female pattern of developmental gene expression.
Reciprocally, the most upregulated gene in Group35-2 relative to Group35-1 was transglutaminase-1-like-2 (tgm1l2; 65-fold up), which has not previously been documented as sex-specific and has an unclear human ortholog, but was greatly overexpressed in wild-type testes vs. wild-type ovaries (72-fold up). This finding suggests the hypothesis that Group35-2 fish were embarking on a male developmental pathway. Group35-2 had increased expression of a number of other male-specific genes relative to Group35-1, including amh (46-fold up); the sperm-specific potassium ion channel gene cngk (18-fold up) (Fechner et al. 2015); an acyl-CoA thioesterase gene acot17 (17-fold up) that was also overexpressed by adult wild-type testes vs. wild type ovaries (6.3-fold overexpressed in testes), but whose expression is otherwise unstudied; ankar, which human testes overexpress compared to any other organ (Fagerberg et al. 2014) (14-fold up in Group35-2); the male factor dmrt1 (13-fold up) (Webster et al. 2017); heat shock transcription factor 5 (hsf5; 11-fold up), whose human ortholog is expressed almost exclusively in testis (Fagerberg et al. 2014); fank1, the mammalian ortholog of which is exclusively expressed in pachytene spermatocytes and spermatids (Zheng et al. 2007) (11-fold up); the sperm-motility gene t-complex-associated-testis-expressed-1 (tcte1; 9.8-fold up); and the testosterone-synthesizing enzyme gene cyp11c1 (8.2-fold up in Group35-2). These results show that Group35-2 individuals were becoming males. Group35-2 individuals also had increased expression of the synaptonemal complex encoding genes sycp3 (16-fold up in Group35-2) and sycp2 (8.7-fold up), and the DNA meiotic recombinase-1 gene (dmc1; 11-fold up), likely reflecting a large number of spermatogonia preparing to undergo meiosis in Group35-2 fish compared to fewer meiotic cells in Group35-1 individuals. We conclude that Group35-2 fish were beginning to mature their testes, as judged by their stronger expression of male-related genes compared to Group35-1 (putative females).
Within Group35-2, all of the amh mutant samples were substantially shifted in the PC1 dimension toward Group35-1 with respect to the wild-type samples. Because Group35-1 were expressing female genes and Group35-2 were expressing male genes, this finding shows that 35 dpf fish lacking amh activity tend to be feminized in terms of their gene expression. Likewise, within Group35-1, all three of the amh mutants were closer to Group35-2 than five of the six wild types in the PC1 axis. This result suggests that zebrafish juveniles developing as females tend to be somewhat masculinized in the absence of amh activity. These observations confirm the utility of Amh in both male and female development.
Expression patterns of the 35 dpf putatively male (Group35-2) fish were strongly correlated to expression patterns of the 21 dpf Group21-2 (not obviously female) fish. Of the 18 genes that were significantly differentially upregulated in putative nonfemale Group21-2 fish (Figure 7C), 11 (zgc:162154, BX005421.3, BX004785.2, mhc1uka, elf3, adamts12, mmp13b, cap2, stard13a, si:ch211-133n4.4, col15a1b) were also significantly differentially upregulated in the male gene-expressing 35 dpf cohort (Group35-2, Figure 7C) at an average of 2.9-fold, with the amount of upregulation highly correlated between the 21 dpf and 35 dpf data sets (correlation coefficient of 0.97). The seven other genes significantly upregulated in the nonfemale Group21-2 (pomk, ift74, b3gat1b, cyp27b1, si:ch211-286b4.4, gstm.2, si:ch211-226h7.5) were not differentially expressed between the two 35 dpf synthetic groups. Of the 25 most upregulated differentially expressed genes in the male-like Group35-2 relative to Group35-1, all but one were also upregulated in wild-type testis relative to wild-type ovary an average of 1922-fold (tgm1l2, zgc:158427, amh, cngk, acot17, sycp3, gstk4, ankar, dmrt1, si:ch211-242f23.3, pimr214, hormad1, si:dkeyp-50b9.1, ifit16, hsf5, dmc1, fank1, si:dkeyp-80c12.8, ttc29, spag16, tcte1, dnah6, hbaa2, tekt1). Only one gene upregulated in Group35-2 (CABZ01076758.1) was not differentially expressed in wild-type testis compared to wild-type ovary. We conclude that, despite the fact that upregulated genes in the nonfemale 21 dpf group were mostly not previously known to be male-related genes, their continued upregulation in the group of 35 dpf fish that were expressing many clearly male genes shows that fish in Group21-2 were also developing male characteristics. These experiments thus identify a previously unknown cohort of sex-specific genes expressed early in gonadogenesis.
GO analysis of differentially expressed genes comparing the two 35 dpf groups yielded 44 enrichment clusters (FDR P < 0.05). The most significantly enriched cluster contained 22 genes enriched for “negative regulation of mitotic cell cycle phase transition” (FDR = 3.0E−02). Among these genes were mitotic checkpoint genes (bub1bb, bub1, hus1, mad1l1, and rad17), and a variety of DNA repair genes (oraov1, orc1, mre11a, blm, msh6). All were upregulated in female-like Group35-1. The second cluster included 30 genes enriched for “mitotic cell cycle checkpoint” (FDR = 5.93E−03), with an expanded list of checkpoint and DNA repair genes similar to the first cluster, including rad9a, rad9b, eme1, and msh2. All were upregulated in Group35-1 except rad9b, suggesting a negative correlation of the co-orthologs rad9a and rad9b and their possible subfunctionalization. The third enrichment cluster comprised 41 genes enriched for “DNA-dependent DNA replication” (FDR = 3.60E−03). These included a variety of DNA polymerases (polg, poln, pola2, pole2, pold2) and associated DNA binding proteins (orc1, orc3, orc6, rpa1, msh2, cdc45, wdhd1). All but poln were upregulated in Group35-1 female-like fish. This cluster also included the upregulated early-onset breast cancer and Fanconi anemia gene brca2(fancd1). Nine other Fanconi anemia genes were also significantly upregulated in the DESeq2 analysis for Group35-1 vs. Group35-2 (fanca, fancb, fancc, fancd2, fance, fancf, fancg, fanci, fancm). These GO enrichment terms for the two 35 dpf juvenile groups differed markedly from the GO terms discovered for the two 21 dpf late-stage larval groups. At 21 dpf, germ cell functions (Piwi-interacting RNAs and eggshell genes) dominated GO terms differentially expressed between the two groups, but at 35 dpf, cell cycle and DNA-repair genes were most differentially expressed between the two groups.
Amh activities regulate adult testis gene expression patterns
To help understand the molecular genetic basis for abnormal testis morphologies caused by loss of amh function, we sequenced seven libraries of 8 mpf adult testes, one each for three wild types and four amh-26 mutants. Sequencing produced 54 million paired-end reads, of which 24 million passed quality filters and mapped to the zebrafish genome assembly. DESeq2 identified 3902 differentially expressed genes (Table S2). Similarity clustering (Figure 6) and PCA based on correlated gene expression (Figure 7A) placed all seven testis samples together, with clear separation between wild types and amh mutants. As predicted, amh was significantly downregulated in mutant testes (amh 6.7-fold down in mutants).
The first three most upregulated genes in mutant testes vs. wild-type testes were the same as the first three most upregulated genes in wild-type ovaries vs. wild-type testes (CABZ01059627.2, si:ch211-125e6.12, zgc:171781, upregulated ∼239,000-, 116,000-, and 90,000-fold respectively). This result shows that amh mutant testes greatly upregulated ovary-specific genes, suggesting a partial feminization of adult amh mutant testes, which could happen by the retention of the early oocytes that were detected by histology (see Figure 4, O and S).
Leydig cell markers were mostly downregulated in amh mutant testes compared to wild-type testes, consistent with our previous analysis of nr5a1a expression, which labels Leydig cells (Figure 5, R and R’). Zebrafish orthologs of 16 of 50 Leydig cell marker genes (Uhlén et al. 2015) were differentially expressed in zebrafish amh mutant testes vs. wild-type testes. The zebrafish orthologs of 14 of these 16 human Leydig cell marker genes (DHH, APOE, AMH, FDX1, CYP17A1, AK1, CNTRL, SGPL1, EPHX1, CCS, ELOA2, ACLY, TMF1, CLEC16A) were downregulated in amh-mutant testes an average of 3.6-fold, and orthologs of only two (CACNA1H and PRSS12) were upregulated (average of 2.e8-fold). The downregulated genes included dhh (down 13-fold), which triggers Leydig cell differentiation (Yao et al. 2002), and cyp17a1 (down 4.3-fold), which encodes the second enzyme in the testosterone biosynthesis pathway. We conclude that loss of amh function disrupts the specification, proliferation, or functioning of Leydig cells in adult zebrafish testis, supporting conclusions from our in situ hybridization analyses (see Figure 5, R and R’).
Sertoli cell marker genes responded in various ways to the loss of amh activity. Zebrafish has orthologs of 38 of 50 human Sertoli cell marker genes (Uhlén et al. 2015). Several Sertoli cell genes were upregulated in mutant testis relative to wild-type testis (gsdf, 3.4-fold up; fndc7a, 2.40-fold; and ddx3a, 1.30-fold) but seven were downregulated in mutants, including amh itself (2.8-fold down, as well as arid4a, alg11, fndc3a, lrig1, abhd2b, sox8b, and cst3, an average of 2.1-fold down (Table S2). Many Sertoli cell regulatory genes, including acvr2aa, acvr2ab, dmrt1, fshb, fshr, hsd17b4, inha, nr0b1, sox9a, and sox9b, were not differentially expressed between mutant and wild-type testes. These results suggest that amh tends to inhibit some Sertoli cell functions, but not strongly. Recall that our in situ hybridization data showed mainly a change in the spatial distribution of Sertoli cells (see Figure 5, B, B’, H, and H’).
Germ cells in amh mutant testes seemed to develop rather normally from a histological perspective, although mutant testes accumulated many more germ cells than normal (see Figure 5, U and U’). The Human Protein Atlas lists 50 genes strongly expressed in spermatogonia, but only 15 of these have orthologs or closely related paralogs in zebrafish. None of these 15 germ cell genes were upregulated in amh mutant testes vs. wild-type testes and only three [brd2a, CU929144.1 (alias cfap46), and meiob] were downregulated (an average of 2.3-fold), while 12 (acvr2ba, acvr2b, brip1, cbl, dazl, dmrt1, dync1h1, hmga2, mgea5, mgea5l, mtmr3, nanos3, plk4, uchl1) were not differentially expressed. For 51 human spermatocyte marker genes in the Human Protein Atlas, only 19 have zebrafish orthologs. One gene (BCL6) has two zebrafish co-orthologs, and although one co-ortholog (bcl6a) was upregulated (2.5-fold) in amh mutant testes, its co-ortholog (bcl6b) was downregulated (5.5-fold). The zebrafish orthologs of seven human spermatocyte genes were downregulated an average of 3.1-fold (bcl6b, clgn, cremb, ccdc65, crema, rnf32, tekt1) consistent with a role for amh in spermatogenesis.
GO analysis of amh mutant testes vs. wild-type testes identified 10 GO clusters (FDR < 0.05). The highest loading cluster included 22 genes enriched for “interciliary transport,” including intraflagellar transport proteins (e.g., ift27, ift57, ift74, and ift81, all downregulated in mutants two- to threefold) and tetratricopeptide repeat domains (ttc21b and ttc26; 1.9- and 2.1-fold downregulated). The second cluster contained 38 genes enriched for “axoneme assembly,” including dynein-related genes downregulated in mutants (e.g., dnah3, dnah5, dnah12, dnai1.2), and genes encoding coiled-coil domain proteins (ccdc39, ccdc103, ccdc114, and ccdc151). Downregulation of genes involved in microtubule assembly is consistent with defects in the ability of amh mutants to produce mature sperm with fully developed tails. We conclude that sperm maturation was suppressed in amh mutant testes, which likely contributed to observed loss of fertility as animals aged.
Amh activities regulate adult ovary gene expression patterns
To understand the genetic effects of Amh on zebrafish ovary development, we investigated gene expression patterns of ovaries from four amh mutants and four of their wild-type siblings in eight individual libraries. Sequencing produced 114 million paired-end reads, 60 million of which mapped to the Ensembl v91 protein coding exons in GRCz10. Similarity clustering using the entire 45-sample data set placed all ovary samples together on a long branch, indicating a unique transcriptional profile (Figure 6). Within the eight ovary samples, amh mutant ovaries occupied a different branch from wild-type ovaries (Figure 6); this result contrasts to the 21 dpf and 35 dpf samples where amh mutants and wild types intermixed within each age group and within subgroups (Figure 6). We conclude that developmental processes that depend on Amh are stronger in adult ovaries than in 21 dpf or 35 dpf animals.
DESeq2 identified 7426 genes differentially expressed in mutant vs. wild-type ovaries (Table S2). Although amh was downregulated in mutant ovaries compared to wild-type ovaries (2.4-fold down), this difference was just short of reaching statistical significance (padj = 0.102). A comparison of adult amh mutant ovaries to adult wild-type ovaries showed that the top six most differentially expressed genes in terms of fold change encode vitellogenins. Zebrafish express vitellogenin genes not only in their livers in response to estrogen as do egg-laying tetrapods, but also in adipocytes in their ovaries (Wang et al. 2005). Overexpression of vtg genes in amh mutant ovaries is not due to contamination from liver in our samples by dissection errors because zebrafish liver marker genes, such as fga, fgb, fabp10a, hmgcra, and hmgcrb, as well as zebrafish orthologs of human liver marker genes, including apoa2, a1bg, ahsg, f2, cfhr2, hpx, and f9 (Uhlén et al. 2015), were not differentially expressed between mutant and wild-type ovaries in our samples. This result is consistent with our morphological studies, which showed that the mutant ovary accumulates enormous quantities of follicles stalled in a previtellogenic state (Figure 4, E and F). If a negative feedback mechanism were in place that senses yolky oocytes and inhibits the transcription of vtg genes in ovarian cell types (Wang et al. 2005), then the absence of yolky oocytes would result in continuous upregulation of vtg gene expression.
Many ovarian regulatory genes were greatly underexpressed in adult mutant ovaries compared to wild-type ovaries. Granulosa cell marker genes (Hatzirodos et al. 2015) tended to be downregulated in amh mutant ovaries vs. wild-type ovaries, including aromatase (cyp19a1a, 18.7-fold down in mutant ovaries), nr5a2 (12.4-fold down), luteinizing hormone receptor (lhcgr; 12.6-fold down), gata4 (25.8-fold down), the estrogen receptors esr1 (4.0-fold down) and esr2b (6.9-fold down), foxl2a (ENSDARG00000042180, 5.7-fold down), foxl2b (ENSDARG00000068417, 4.4-fold down), and slc35g1 (7.3-fold down). Theca cell marker genes (Hatzirodos et al. 2015) were also downregulated: insl3 (23.6-fold down), nid1b (10.3-fold down), nr5a1a (16.1-fold down), nr5a1b (2.9-fold down), star (3.8-fold down), cyp11a2 (3.4-fold down), and hsd3b1 (3.4-fold down). These results show that expression of marker genes for both granulosa cells and theca cells are downregulated and confirm in situ hybridization results (Figure 5) that showed significant disruption of follicle cell morphologies in amh mutants.
Given the great enlargement of amh mutant gonads (Figure 4, A, B, E, and F), it was unexpected to find that many key oocyte marker genes were not expressed differentially between amh mutants and wild types, including vasa, dnd1, piwi paralogs, bmp15, gdf9, nanos paralogs, sycp synaptonemal complex genes, and zona pellucida genes (Liu et al. 2006). This finding was further surprising given that most of these genes were misregulated in the putative female vs. putative male synthetic groups for the 21 dpf and 35 dpf time points. Some markers of meiosis were upregulated in mutant ovaries compared to wild-type ovaries [spo11 (2.1-fold up), rad51d (3.1-fold up)], but others, like dmc1 and synaptonemal complex genes, were not differentially expressed. These results may suggest that the large number of mutant oocytes accumulated by mutant ovaries had stalled at an early stage of meiosis.
GO analysis (PANTHER; Mi et al. 2013) identified 49 enrichment clusters comparing genes differentially expressed between adult amh mutant ovaries and adult wild-type ovaries (FDR < 0.05). The highest loading cluster was “ribosomal large subunit assembly” (FDR = 2.59E−02), and comprised 17 genes, including various ribosomal protein genes (rpl3, rpl5a, rpl5b, rpl6, rpl11, rpl12, and rpl23a) that were upregulated in amh mutant ovaries compared to wild-type ovaries. The second cluster was “cytoplasmic translation” (FDR = 1.49E−03), and comprised 28 genes, including additional upregulated ribosomal protein genes (e.g., rpl7, rpl9, rpl22l1, rpl26, rpl29, and rpl31) and translation initiation factors (etf1b, eif3a, eif3m, eif4h, and eif4bb). The third cluster was “maturation of large subunit-rRNA” (FDR = 4.81E−02) and contained 18 upregulated ribosomal biogenesis protein genes (e.g., wdr12, nsa2, las1l, and rpf2), as well as additional ribosomal proteins (e.g. nhp2, rpl7, rpl10a, and rpl35). These clusters reflect the massive accumulation of ribosomes that maturing eggs normally store. Other GO enrichments indicated coordination of replication, transcription, and translation. These results are expected from the morphological studies that showed massive changes in oocyte accumulation and defects in follicle development (Figure 4, E, F, I, and J).
The lack of amhr2 in the genomes of zebrafish and other cyprinids is associated with a chromosome rearrangement breakpoint
AMH in mammals binds to the receptor AMHR2. Humans lacking function of either AMH or AMHR2 have persistent Müllerian ducts and Amh;Amhr2 double mutant mice have the same phenotype as either single mutant, showing that the ligand and receptor act in the same pathway (Imbeaud et al. 1996; Mishina et al. 1996). Amhr2 makes a dimer with one of the Bmpr1 proteins, and the zebrafish mutant phenotype for bmpr1bb mimics the amh mutant phenotype reported here for the enlarged testes and accumulation of immature oocytes, but the bmpr1bb mutant males did not retain oocytes like the amh mutant testes did (Neumann et al. 2011). In addition, the bmpr1bb mutants did not appear to alter the sex ratio as did the amh mutants (Neumann et al. 2011). Percomorph fish genomes generally contain an ortholog of Amhr2 (e.g., stickleback, Gasterosteus aculeatus, ENSGACG00000006672). In contrast to percomorphs, zebrafish is an otophysan teleost, and at least two suborders of otophysans also possess an amhr2 gene, the characiform suborder, including both red-bellied piranha (Pygocentrus nattereri, ENSPNAG00000001197) and cavefish (Astyanax mexicanus, ENSAMXG00000024722), and the siluriform suborder, including channel catfish (Ictalurus punctatus, ENSIPUG00000006414). In contrast, zebrafish in the cypriniform suborder of otophysans appears to lack amhr2 (Ribas et al. 2016).
To understand whether this apparent loss of amhr2 is unique to zebrafish (which could then either be a zebrafish-specific loss or a genome assembly error) or whether it might represent an event shared among cypriniforms, we studied conserved syntenies. Results showed that the amhr2-containing region of cavefish (Figure S2A) corresponds to three widely scattered portions of the zebrafish genome (Figure S2, B and C), with a break in conserved syntenies occurring at the predicted location of amhr2 and its nearest neighbor (ENSAMXG00000024723, cell division cycle associated 7), which is also present in most percomorphs but is missing from zebrafish. Duplicates of the zebrafish orthologs of genes flanking amhr2 in cavefish that originated in the teleost genome duplication are on two different zebrafish chromosomes, Dre2 and Dre22 (Figure S2). These data are consistent with the hypothesis that both amhr2 and its neighbor ENSAMXG00000024723 disappeared in the zebrafish lineage after it diverged from other otophysans associated with a chromosome inversion breakpoint at the ancestral site of amhr2.
To determine whether the rearrangement breakpoint at the expected position of amhr2 is zebrafish specific, we examined the genomes of two other cypriniform fish: common carp (C. carpio) and goldfish (Carassius auratus). BLASTP searches using cavefish Amhr2 against common carp and goldfish genomes did not identify an Amhr2 ortholog, but brought back Tgfbr2b as the most similar protein, suggesting that these cyprinids, like zebrafish, have no ortholog of amhr2. (Note that BLASTP searches of cavefish Amhr2 vs. zebrafish brought back as the two top hits Bmpr2a and then Bmpr2b.) Conserved synteny analysis showed that zebrafish and common carp share gene orders at the location predicted for amhr2 (Figure S2). This result is predicted by the hypothesis that a chromosome rearrangement with a breakpoint in or near amhr2 destroyed this gene and that the event occurred after cyprinid otophysans diverged from characiform and siluriform otophysan teleosts, but before the divergence of the zebrafish and carp lineages.
Discussion
In human male fetuses, Sertoli cells secrete AMH, which causes developing Müllerian ducts to disappear, while in adult human females, AMH suppresses the initiation of primary follicle growth, serves as a marker for ovarian reserve, and provides an assay for conditions like PCOS (Carlsson et al. 2006; Diamanti-Kandarakis 2008). Teleost fish do not have Müllerian ducts but nevertheless maintain an amh gene (Adolfi et al. 2019). To investigate conserved roles of Amh in vertebrates that lack a Müllerian duct, we made zebrafish amh null activity alleles.
Results showed that amh activity promotes, but is not essential for, male development in zebrafish because homozygous amh mutants were only ∼20% as likely to develop into males as their wild-type siblings (see also Lin et al. (2017). In addition, most mature adult male mutant gonads we examined contained a few early-stage oocytes, while at the same age, wild-type siblings did not. This finding shows that Amh normally masculinizes zebrafish by inhibiting the development or survival of young oocytes. Several fish species expand the male-biasing role of amh, having evolved a modified gene duplicate that has become the major sex determinant. For example, Patagonian pejerrey and Nile tilapia possess amh gene duplicates that independently became the primary sex determinant (Hattori et al. 2012; Li et al. 2015); ling cod also possesses a male-specific amh duplication (Rondeau et al. 2016). A further demonstration of the role of the amh pathway is the finding that variants of the Amh receptor gene amhr2 provide the primary sex determinant in several species of pufferfish (Kamiya et al. 2012; Ieda et al. 2018). In tetrapods and most teleosts, Amhr2 is expressed in Leydig and Sertoli cells (di Clemente et al. 1994; Racine et al. 1998) and mediates Amh signaling. Zebrafish, however, has no ortholog of Amhr2, which we show here is associated with an inversion breakpoint. Because we show that Amh is critical for zebrafish gonad development, the function of Amhr2 is likely performed by another Bmpr2 paralog. Zebrafish has two bmpr2 ohnologs; bmpr2a is expressed in young oocytes and in ovarian follicle cells and bmpr2b is expressed in follicle cells (Li and Ge 2011; Dranow et al. 2016). We found that bmpr2a was overexpressed in adult wild-type testis vs. wild-type ovary (4.7-fold), and both bmpr2a and bmpr2b were underexpressed in amh mutant ovary vs. wild-type ovary (4.7-fold and 6.2-fold, respectively), but we did not detect differential expression of any bmpr2 genes comparing putative female and putative male groups of 21 dpf or 35 dpf juveniles.
Young zebrafish amh mutant females are fertile, showing that they have functional reproductive ducts. As female amh mutants age, however, they become sterile, showing that Amh supports continued female fertility. Zebrafish males lacking amh activity are less effective than their wild-type male siblings at stimulating wild-type females to lay eggs, showing that Amh action improves male mating behavior. It is as yet unknown whether this difference is related to changes in brain organization that depend on the developmental availability of Amh. Likewise, mouse mutants lacking either Amh or Amhr2 show feminized spinal motor neurons and some feminized behaviors (Wang et al. 2009). Some wild-type eggs that were fertilized by mutant males develop to hatching, showing that functional male reproductive ducts form without the benefit of Amh. Nevertheless, eggs laid by wild-type females in the presence of amh mutant males are less likely to develop than those fertilized by wild-type males, showing that Amh helps optimize sperm production, function, or release. We conclude that in zebrafish, Amh is not necessary for the development of reproductive ducts or for the initiation of functional gamete formation, but is necessary for continued fertility in both sexes.
Wild-type siblings of our amh mutants had gonads with stage I oocytes at 21 dpf as expected (Takahashi 1977; Selman et al. 1993; Rodríguez-Mari et al. 2005, 2010; Wang and Orban 2007), but some 21 dpf amh mutants had gonads that contained only undifferentiated germ cells, suggesting that amh activity helps to accelerate gonad development. By 35 dpf, about half of wild-type siblings were continuing to develop oocytes and the other half were forming spermatocytes, but most amh mutants were developing normal-looking ovaries and few were developing spermatogonia. We conclude that amh mutants are slow to adapt a male phenotype, and many never do, leading to a female-biased sex ratio among amh mutants.
Ovaries in mature adult amh mutant females were swollen by immature oocytes to nearly three times the size of ovaries in wild-type siblings, confirming previous results (Lin et al. 2017). The ovarian phenotype of zebrafish amh mutants mimics the phenotype of gsdf mutants in zebrafish (Yan et al. 2017), of Amh receptor mutants in female medaka (Hattori et al. 2012), and of PCOS in humans (Diamanti-Kandarakis 2008). We conclude that Amh represses primordial oocyte proliferation but stimulates oocyte maturation in zebrafish. Suppression of oocyte maturation appears to be a role of Amh shared by fish and mammals because mice lacking Amh show premature depletion of the primordial follicle pool (Durlinger et al. 1999), likely because Amh slows follicle growth in zebrafish and mammals as it does in humans (Carlsson et al. 2006). Zebrafish males lacking amh activity also had greatly enlarged gonads (Lin et al. 2017), showing that in zebrafish, Amh helps slow gonad growth both in males and in females. In contrast to females, however, males even at 18 mpf appeared to contain mature gametes, although these males were sterile, demonstrating (somewhat paradoxically) that, although Amh normally nudges juvenile zebrafish toward a male pathway, its activity in older fish is required for oocyte maturation (no amh mutant female produced mature eggs at 11 mpf) but appears only to accelerate spermatocyte maturation (3% of eggs laid by wild-type females developed after mating with mutant males at 11 mpf).
The finding of the similarity of the phenotypes of zebrafish amh and gsdf mutants raised the question of whether these genes regulate germ cell proliferation and differentiation by acting in the same or different pathways. Because results showed that amh;gsdf double mutant gonads were no more compromised than either single mutant, we conclude that amh and gsdf act in the same pathway. Sertoli cells and granulosa cells both express both amh and gsdf (Rodríguez-Mari et al. 2005; von Hofsten et al. 2005; Gautier et al. 2011a,b; Yan et al. 2017). Zebrafish lacks an ortholog of amhr2, which encodes the Amh receptor found in other vertebrates, and the receptor for Gsdf is unknown, so the relative positions of Amh and Gsdf in a shared developmental pathway are currently ripe for further investigation.
Differences in gene expression patterns in mutants compared to wild types give clues to how genes exert their effects. We studied altered gene expression patterns in amh mutants in two ways: by in situ hybridization and by whole genome transcriptome analyses. In situ hybridization experiments revealed reduced amh transcript accumulation in granulosa cells and disrupted organization of amh-expressing Sertoli cells. The lack of Amh activity appeared to result in an upregulation of gsdf expression in Sertoli cells both in our in situ hybridization data and in our RNA-seq data (3.39-fold upregulated in mutant testes; Table S2), suggesting that if Amh and Gsdf act in the same pathway, Amh may repress gsdf activity. Reciprocally, amh expression was upregulated 1.8-fold in our gsdf mutants (Yan et al. 2017), showing interdependent regulation of amh and gsdf genes. The downregulation of the aromatase gene in zebrafish amh mutants and the failure of aromatase-expressing cells to completely envelop oocytes suggests that Amh is required for proper development of granulosa cells to the stage appropriate for maximal aromatase expression. With depressed aromatase activity, levels of estrogen should diminish, thereby inhibiting oocyte maturation, which we observed. An additional contributor to endocrine disruption would be our finding of the greatly reduced expression of nr5a1a, which encodes a nuclear receptor transcription factor that regulates steroidogenesis genes (Val et al. 2003). Our in situ hybridization results compared to histological phenotypes paint a picture of disrupted development stemming from amh-expressing granulosa and Sertoli in zebrafish amh mutants that leads to disordered organization of these helper cells, their failure to support sex steroid output of theca and Leydig cells, followed by failure of proper germ cell maturation and inhibition of germ cell proliferation.
Genome-wide transcriptional analyses further provided an unbiased probe of the mechanisms of normal gonad development and the roles of Amh. A comparison of wild-type ovary to wild-type testis identified hundreds of genes with previously unknown functions, many of which are lineage-specific, with strong differential expression. These genes likely encode eggshell and sperm components. These data provide, for the first time, information related to function for many genes known previously only by sequence.
Principal component analysis showed that the transcriptomes of adult amh mutant ovaries were shifted in the direction of wild-type testis transcriptomes and mutant adult testis transcriptomes were shifted in the direction of wild-type ovary transcriptomes. This result likely reflects the depressed ability of amh mutant ovaries to convert testosterone to estrogen and the observed retention of immature oocytes in adult amh mutant testes.
Analysis of the transcriptomes of gonad-containing trunks of juveniles provided important insights into both normal zebrafish sex determination and the roles of Amh in gonad development and physiology. At 21 dpf, laboratory strain zebrafish have morphologically undifferentiated gonads (Takahashi 1977; Maack and Segner 2003; Rodríguez-Mari et al. 2005; Wang et al. 2007), so it was a surprise to find that unsupervised transcriptome similarity clustering divided late-stage larvae into two distinct groups. Each of these two groups contained both amh mutants and wild types, showing that factors other than amh function distinguish these two groups. Differential expression analysis showed that one group overexpressed ovary genes and the other overexpressed genes that were also overexpressed in wild-type testes vs. wild-type ovaries, although few of these genes had previously been recognized as testis marker genes. We conclude that, despite little morphological differentiation in 21 dpf gonads, they had already embarked on a female or male developmental program. This is the first report of the genome-wide transcriptional differentiation of zebrafish gonads at such an early age.
The small number of genes (16) that were differentially upregulated in amh mutant trunks vs. wild-type trunks at 21 dpf included regulators of steroidogenesis and meiosis. Two upregulated genes in mutants affect development or function of steroid-producing Leydig cells: nr0b2, which inhibits steroidogenic gene expression in mouse Leydig cells (Volle et al. 2007), and ptch2, part of a receptor complex for desert hedgehog signaling in Leydig cells (Yao et al. 2002; Wijgerde et al. 2005; Herpin et al. 2013). Amh also likely helps regulate the initiation of meiosis. Nr0b2 in mouse reduces the level of retinoic acid, thereby reducing the expression of genes essential for mitotic germ cells to enter meiosis (Volle et al. 2007) and we found that nr0b2 was the most upregulated gene in 21 dpf amh mutants. Late-stage larval zebrafish amh mutants also upregulated the Leydig cell marker gene cyp26a1, which encodes an enzyme that degrades retinoic acid, the regulator of entry into meiosis. We conclude that Amh from Sertoli cells is required for normal Leydig cell development and function and likely helps regulate the timing of meiosis as early as 21 dpf.
Transcriptomes of 35 dpf trunks also separated samples into two groups, each of which contained both mutants and wild types. Within each group, mutants separated from wild types, showing that between 21 dpf and 35 dpf, amh had begun to exert a significant effect on gonad development. One group evidently had significant levels of estrogen because they were strongly expressing estrogen-induced vitellogenin genes, and in addition they strongly expressed zona pellucida eggshell genes. The other group overexpressed a number of testis-specific genes, including amh and dmrt1, as well as genes not previously recognized as testis genes but overexpressed in wild-type testis vs. wild-type ovary, thus providing novel insight into potential functions of these genes previously known only by sequence. Expression levels of genes in putatively male 35 dpf animals were strongly correlated to their levels in the nonfemale 21 dpf group, confirming that some late-stage larval fish had already begun to initiate a male developmental pathway. GO enrichment terms between putative males and putative females in 35 dpf juveniles changed from a focus on germ cell functions like Piwi-interacting RNAs and eggshell genes at 21 dpf to an emphasis on cell cycle and DNA-repair genes at 35 dpf, consistent with more cells undergoing meiosis.
A comparison of amh mutant juveniles to wild-type juveniles showed that numerous immune-related genes were upregulated in mutants. These genes included irf7, which is a paralog of the trout master sex-determining gene sdY that has been shown to be a duplicated, truncated interferon regulatory factor 9 (irf9) (Yano et al. 2012). The upregulation of immune-related genes might reflect an inflammatory response to cell damage that accompanies the disruption of the development of Leydig cells and granulosa cells observed in histological sections and reflected in our in situ hybridization and transcriptome studies.
Transcriptomes of adult amh mutant testes differed substantially from those of adult wild-type testes. First, mutant testes upregulated several genes that were greatly overexpressed in wild-type ovaries, likely reflecting the oocytes that histological sections revealed in amh mutant testes. These results show that Amh acts to block oocyte development in zebrafish testes. Second, adult amh mutant testes underexpressed most Leydig cell marker genes, consistent with our histology results and the finding that mouse Amh mutant males have disrupted Leydig cell development (Behringer et al. 1994). In contrast to the strongly altered Leydig cell markers in amh mutant transcriptomes, Sertoli cell marker genes were not strongly altered in amh mutants. We conclude that, although Amh is produced by and secreted from Sertoli cells, the lack of Amh function alters the developmental activities of Leydig cells, a conserved feature of Amh function. Thus, Leydig cells must express a receptor for Amh even though zebrafish lacks an ortholog of Amhr2, which helps form the Amh receptor in mammals and most fish. Third, amh mutant testes underexpressed testis-biased genes, consistent with the finding of ovotestes in amh mutants. Fourth, Despite amh mutant males accumulating substantial quantities of testis lobules, they greatly underexpressed genes involved in the production of mature sperm, verifying histology. The enormous testes in zebrafish amh mutants are consistent with zebrafish organ culture experiments that showed that Amh inhibits androgen-stimulated proliferation of spermatogonia (Skaar et al. 2011); thus, with less Amh, the inhibition should lessen, resulting in the accumulation of spermatogonia we observed. Together, our findings show that Amh signaling is required for normal development of Leydig cells, for the disappearance of ovotestes, for accelerating sperm maturation, and for the inhibition of spermatocyte proliferation.
Transcriptome analyses and in situ hybridization studies showed differences between adult ovaries in amh mutants and wild types, including the underexpression of marker genes for both granulosa cells and theca cells. In contrast, many oocyte marker genes were not differentially expressed, which is a bit surprising given the misexpression of many of these genes in the putatively female groups (Group21-1 and Group35-1) in late-stage larval and juvenile zebrafish. On the other hand, amh adult mutant ovaries overexpressed many genes involved in translation, as expected from the massive accumulation of young oocytes in mutant ovaries, and they underexpressed some ovary regulatory genes like foxl2a, cyp19a1a, and gata4, reflecting the loss of control of oocyte proliferation.
In tetrapods and most teleosts, Amhr2 is expressed in both Leydig and Sertoli cells (di Clemente et al. 1994; Racine et al. 1998) and it mediates Amh signaling. Reference genomes of zebrafish and as we show, other cyprinids, however, have no ortholog of Amhr2, and gene loss is associated with a chromosome break at the ancestral site of the gene, thus, Amhr2 function is likely performed by another Bmpr2 paralog. In zebrafish, bmpr2a is overexpressed in adult wild-type testis vs. wild-type ovary and both bmpr2a and bmpr2b are underexpressed in amh mutant ovary vs. wild-type ovary, making these genes candidates for the elusive Amh receptor.
Acknowledgments
We thank the University of Oregon Genomics and Cell Characterization Core Facility (Doug Turnbull and Maggie Weitzman) for their wonderful expertise, Clay Small for comments on the manuscript, and the National Institutes of Health for support by grants R01 GM-085318 (J.H.P.), and IOS-1456737 (B.D.).
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8184437.
Communicating editor: D. Greenstein
Literature Cited
- Adolfi M. C., Herpin A., Regensburger M., Sacquegno J., Waxman J. S. et al. , 2016. Retinoic acid and meiosis induction in adult vs. embryonic gonads of medaka. Sci. Rep. 6: 34281 10.1038/srep34281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adolfi M. C., Nakajima R. T., Nóbrega R. H., and Schartl M., 2019. Intersex, hermaphroditism, and gonadal plasticity in vertebrates: evolution of the mullerian duct and Amh/Amhr2 signaling. Annu Rev Anim Biosci. 7: 149–172. 10.1146/annurev-animal-020518-114955 [DOI] [PubMed] [Google Scholar]
- Amores A., Catchen J., Ferrara A., Fontenot Q., and Postlethwait J. H., 2011. Genome evolution and meiotic maps by massively parallel DNA sequencing: spotted gar, an outgroup for the teleost genome duplication. Genetics 188: 799–808. 10.1534/genetics.111.127324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., and Huber W., 2015. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applegate, V. C., 1948 Sea lamprey investigations. 2. Egg development, maturity, egg production, and percentage of unspawned eggs of sea lampreys, Petromyzon marinus, captured in several Lake Huron tributaries, Fisheries Division Reports 1161. Michigan Department of Natural Resources, Lansing, MI, pp. 1–28. [Google Scholar]
- Barbaro M., Oscarson M., Schoumans J., Staaf J., Ivarsson S. A. et al. , 2007. Isolated 46,XY gonadal dysgenesis in two sisters caused by a Xp21.2 interstitial duplication containing the DAX1 gene. J. Clin. Endocrinol. Metab. 92: 3305–3313. 10.1210/jc.2007-0505 [DOI] [PubMed] [Google Scholar]
- Behringer R. R., Finegold M. J., and Cate R. L., 1994. Mullerian-inhibiting substance function during mammalian sexual development. Cel 79: 415–425. 10.1016/0092-8674(94)90251-8 [DOI] [PubMed] [Google Scholar]
- Bolger A., Lohse M., and Usadel B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson I. B., Scott J. E., Visser J. A., Ritvos O., Themmen A. P. et al. , 2006. Anti-Mullerian hormone inhibits initiation of growth of human primordial ovarian follicles in vitro. Hum. Reprod. 21: 2223–2227. 10.1093/humrep/del165 [DOI] [PubMed] [Google Scholar]
- Catchen J. M., Conery J. S., and Postlethwait J. H., 2009. Automated identification of conserved synteny after whole-genome duplication. Genome Res. 19: 1497–1505. 10.1101/gr.090480.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhang H., Wang F., Zhang W., and Peng G., 2016. nr0b1 (DAX1) mutation in zebrafish causes female-to-male sex reversal through abnormal gonadal proliferation and differentiation. Mol. Cell. Endocrinol. 433: 105–116. 10.1016/j.mce.2016.06.005 [DOI] [PubMed] [Google Scholar]
- Chen W., Liu L., and Ge W., 2017. Expression analysis of growth differentiation factor 9 (Gdf9/gdf9), anti-mullerian hormone (Amh/amh) and aromatase (Cyp19a1a/cyp19a1a) during gonadal differentiation of the zebrafish, Danio rerio. Biol. Reprod. 96: 401–413. 10.1095/biolreprod.116.144964 [DOI] [PubMed] [Google Scholar]
- Chiang E. F., Yan Y. L., Guiguen Y., Postlethwait J., and Chung B., 2001a Two Cyp19 (P450 aromatase) genes on duplicated zebrafish chromosomes are expressed in ovary or brain. Mol. Biol. Evol. 18: 542–550. 10.1093/oxfordjournals.molbev.a003833 [DOI] [PubMed] [Google Scholar]
- Chiang E. F., Yan Y. L., Tong S. K., Hsiao P. H., Guiguen Y. et al. , 2001b Characterization of duplicated zebrafish cyp19 genes. J. Exp. Zool. 290: 709–714. 10.1002/jez.1121 [DOI] [PubMed] [Google Scholar]
- Clelland E., Kohli G., Campbell R. K., Sharma S., Shimasaki S. et al. , 2006. Bone morphogenetic protein-15 in the zebrafish ovary: complementary deoxyribonucleic acid cloning, genomic organization, tissue distribution, and role in oocyte maturation. Endocrinology 147: 201–209. 10.1210/en.2005-1017 [DOI] [PubMed] [Google Scholar]
- Clelland E. S., and Kelly S. P., 2011. Exogenous GDF9 but not Activin A, BMP15 or TGFbeta alters tight junction protein transcript abundance in zebrafish ovarian follicles. Gen. Comp. Endocrinol. 171: 211–217. 10.1016/j.ygcen.2011.01.009 [DOI] [PubMed] [Google Scholar]
- Dahlem T. J., Hoshijima K., Jurynec M., Gunther D., Starker C. et al. , 2012. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 8: e1002861 10.1371/journal.pgen.1002861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamanti-Kandarakis E., 2008. Polycystic ovarian syndrome: pathophysiology, molecular aspects and clinical implications. Expert Rev. Mol. Med. 10: e3 10.1017/S1462399408000598 [DOI] [PubMed] [Google Scholar]
- di Clemente N., Wilson C., Faure E., Boussin L., Carmillo P. et al. , 1994. Cloning, expression, and alternative splicing of the receptor for anti-Müllerian hormone. Mol. Endocrinol. 8: 1006–1020. [DOI] [PubMed] [Google Scholar]
- Dranow D. B., Hu K., Bird A. M., Lawry S. T., Adams M. T. et al. , 2016. Bmp15 is an oocyte-produced signal required for maintenance of the adult female sexual phenotype in zebrafish. PLoS Genet. 12: e1006323 10.1371/journal.pgen.1006323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durlinger A. L., Kramer P., Karels B., de Jong F. H., Uilenbroek J. T. et al. , 1999. Control of primordial follicle recruitment by anti-Mullerian hormone in the mouse ovary. Endocrinology 140: 5789–5796. 10.1210/endo.140.12.7204 [DOI] [PubMed] [Google Scholar]
- Elger W., 1966. Die rolle der fetalen Androgene in der Sexualdifferenzierung des Kaninchens und ihre Abgrenzung gene andere Hormonale und somatische Faktoren durch Anwendung eines starken Antiandrogens. Arch. Anat. Micr. Morph. Exp. 55: 657–743. [PubMed] [Google Scholar]
- Fagerberg L., Hallström B. M., Oksvold P., Kampf C., Djureinovic D. et al. , 2014. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics 13: 397–406. 10.1074/mcp.M113.035600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fechner S., Alvarez L., Bönigk W., Müller A., Berger T. K. et al. , 2015. A K(+)-selective CNG channel orchestrates Ca(2+) signalling in zebrafish sperm. eLife 4: e07624. 10.7554/eLife.07624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara A. M., and Irwin E. R., 2001. A standardized procedure for internal sex identification in lepisosteidae. N. Am. J. Fish. Manage. 21: 956–961. [DOI] [Google Scholar]
- Force A., Lynch M., Pickett F. B., Amores A., Yan Y. L. et al. , 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier A., Le Gac F., and Lareyre J. J., 2011a The gsdf gene locus harbors evolutionary conserved and clustered genes preferentially expressed in fish previtellogenic oocytes. Gene 472: 7–17. 10.1016/j.gene.2010.10.014 [DOI] [PubMed] [Google Scholar]
- Gautier A., Sohm F., Joly J. S., Le Gac F., and Lareyre J. J., 2011b The proximal promoter region of the zebrafish gsdf gene is sufficient to mimic the spatio-temporal expression pattern of the endogenous gene in Sertoli and granulosa cells. Biol. Reprod. 85: 1240–1251. 10.1095/biolreprod.111.091892 [DOI] [PubMed] [Google Scholar]
- Groh K. J., Schonenberger R., Eggen R. I., Segner H., and Suter M. J., 2013. Analysis of protein expression in zebrafish during gonad differentiation by targeted proteomics. Gen. Comp. Endocrinol. 193: 210–220. 10.1016/j.ygcen.2013.07.020 [DOI] [PubMed] [Google Scholar]
- Hardisty M. W., 1971. Gonadogenesis, sex differentiation and gametogenesis, pp. 295–359 in The Biology of Lampreys, edited by Hardisty M. W. and Potter I. C.. Academic Press, London. [Google Scholar]
- Hattori R. S., Murai Y., Oura M., Masuda S., Majhi S. K. et al. , 2012. A Y-linked anti-Mullerian hormone duplication takes over a critical role in sex determination. Proc. Natl. Acad. Sci. USA 109: 2955–2959. 10.1073/pnas.1018392109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzirodos N., Hummitzsch K., Irving-Rodgers H. F., and Rodgers R. J., 2015. Transcriptome comparisons identify new cell markers for theca interna and granulosa cells from small and large antral ovarian follicles. PLoS One 10: e0119800 10.1371/journal.pone.0119800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herpin A., Adolfi M. C., Nicol B., Hinzmann M., Schmidt C. et al. , 2013. Divergent expression regulation of gonad development genes in medaka shows incomplete conservation of the downstream regulatory network of vertebrate sex determination. Mol. Biol. Evol. 30: 2328–2346. 10.1093/molbev/mst130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howley C., and Ho R. K., 2000. mRNA localization patterns in zebrafish oocytes. Mech. Dev. 92: 305–309. 10.1016/S0925-4773(00)00247-1 [DOI] [PubMed] [Google Scholar]
- Hugo S. E., Cruz-Garcia L., Karanth S., Anderson R. M., Stainier D. Y. et al. , 2012. A monocarboxylate transporter required for hepatocyte secretion of ketone bodies during fasting. Genes Dev. 26: 282–293. 10.1101/gad.180968.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda R., Hosoya S., Tajima S., Atsumi K., Kamiya T. et al. , 2018. Identification of the sex-determining locus in grass puffer (Takifugu niphobles) provides evidence for sex-chromosome turnover in a subset of Takifugu species. PLoS One 13: e0190635 10.1371/journal.pone.0190635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T., Saino K., and Matsuda M., 2015. Mutation of Gonadal soma-derived factor induces medaka XY gonads to undergo ovarian development. Biochem. Biophys. Res. Commun. 467: 109–114. 10.1016/j.bbrc.2015.09.112 [DOI] [PubMed] [Google Scholar]
- Imbeaud S., Belville C., Messika-Zeitoun L., Rey R., di Clemente N. et al. , 1996. A 27 base-pair deletion of the anti-mullerian type II receptor gene is the most common cause of the persistent mullerian duct syndrome. Hum. Mol. Genet. 5: 1269–1277. 10.1093/hmg/5.9.1269 [DOI] [PubMed] [Google Scholar]
- Iyer A. K., Zhang Y. H., and McCabe E. R., 2006. Dosage-sensitive sex reversal adrenal hypoplasia congenita critical region on the X chromosome, gene 1 (DAX1) (NR0B1) and small heterodimer partner (SHP) (NR0B2) form homodimers individually, as well as DAX1-SHP heterodimers. Mol. Endocrinol. 20: 2326–2342. 10.1210/me.2005-0383 [DOI] [PubMed] [Google Scholar]
- Josso N., 1972. Permeability of membranes to the Mullerian-inhibiting substance synthesized by the human fetal testis in vitro: a clue to its biochemical nature. J. Clin. Endocrinol. Metab. 34: 265–270. 10.1210/jcem-34-2-265 [DOI] [PubMed] [Google Scholar]
- Jost A., 1947. Recherches sur la différenciation sexuelle de l’embryon de lapin. Arch. Anat. Microsc. Morphol. Exp. 36: 271–315. [Google Scholar]
- Kamiya T., Kai W., Tasumi S., Oka A., Matsunaga T. et al. , 2012. A trans-species missense SNP in Amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (fugu). PLoS Genet. 8: e1002798 10.1371/journal.pgen.1002798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kossack M. E., High S. K., Hopton R. E., Yan Y. L., Postlethwait J. H. et al. , 2019. Female sex development and reproductive duct formation depend on Wnt4a in zebrafish. Genetics 211: 219–233. 10.1534/genetics.118.301620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koubova J., Menke D. B., Zhou Q., Capel B., Griswold M. D. et al. , 2006. Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proc. Natl. Acad. Sci. USA 103: 2474–2479. 10.1073/pnas.0510813103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. L. J., Horsfield J. A., Black M. A., Rutherford K., Fisher A. et al. , 2017. Histological and transcriptomic effects of 17alpha-methyltestosterone on zebrafish gonad development. BMC Genomics 18: 557 10.1186/s12864-017-3915-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. W., and Ge W., 2011. Spatiotemporal expression of bone morphogenetic protein family ligands and receptors in the zebrafish ovary: a potential paracrine-signaling mechanism for oocyte-follicle cell communication. Biol. Reprod. 85: 977–986. 10.1095/biolreprod.111.092239 [DOI] [PubMed] [Google Scholar]
- Li M., Sun Y., Zhao J., Shi H., Zeng S. et al. , 2015. A tandem duplicate of anti-mullerian hormone with a missense SNP on the Y chromosome is essential for male sex determination in nile Tilapia, Oreochromis niloticus. PLoS Genet. 11: e1005678 10.1371/journal.pgen.1005678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Lu M. M., Zhou D., Hammes S. R., and Morrisey E. E., 2007. GLP-1: a novel zinc finger protein required in somatic cells of the gonad for germ cell development. Dev. Biol. 301: 106–116. 10.1016/j.ydbio.2006.07.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L., Soyal S. M., and Dean J., 1997. FIGalpha, a germ cell specific transcription factor involved in the coordinate expression of the zona pellucida genes. Development 124: 4939–4947. [DOI] [PubMed] [Google Scholar]
- Lin Q., Mei J., Li Z., Zhang X., Zhou L. et al. , 2017. Distinct and cooperative roles of amh and dmrt1 in self-renewal and differentiation of male germ cells in zebrafish. Genetics 207: 1007–1022. 10.1534/genetics.117.300274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., and Ge W., 2007. Growth differentiation factor 9 and its spatiotemporal expression and regulation in the zebrafish ovary. Biol. Reprod. 76: 294–302. 10.1095/biolreprod.106.054668 [DOI] [PubMed] [Google Scholar]
- Liu X., Wang H., and Gong Z., 2006. Tandem-repeated Zebrafish zp3 genes possess oocyte-specific promoters and are insensitive to estrogen induction. Biol. Reprod. 74: 1016–1025. 10.1095/biolreprod.105.049403 [DOI] [PubMed] [Google Scholar]
- Lourenco D., Brauner R., Rybczynska M., Nihoul-Fekete C., McElreavey K. et al. , 2011. Loss-of-function mutation in GATA4 causes anomalies of human testicular development. Proc. Natl. Acad. Sci. USA 108: 1597–1602. 10.1073/pnas.1010257108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Anders S., Kim V., and Huber W., 2015. RNA-Seq workflow: gene-level exploratory analysis and differential expression. F1000Res. 4: 1070 10.12688/f1000research.7035.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maack G. S., and Segner H., 2003. Morphological development of the gonads in zebrafish. J. Fish Biol. 62: 895–906. 10.1046/j.1095-8649.2003.00074.x [DOI] [Google Scholar]
- Martin M., 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet 17: 1–10. [Google Scholar]
- Mi H., Muruganujan A., and Thomas P. D., 2013. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41: D377–D386. 10.1093/nar/gks1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao L., Yuan Y., Cheng F., Fang J., Zhou F. et al. , 2017. Translation repression by maternal RNA binding protein Zar1 is essential for early oogenesis in zebrafish. Development 144: 128–138. 10.1242/dev.144642 [DOI] [PubMed] [Google Scholar]
- Mishina Y., Rey R., Finegold M. J., Matzuk M. M., Josso N. et al. , 1996. Genetic analysis of the Mullerian-inhibiting substance signal transduction pathway in mammalian sexual differentiation. Genes Dev. 10: 2577–2587. 10.1101/gad.10.20.2577 [DOI] [PubMed] [Google Scholar]
- Mold D. E., Dinitz A. E., and Sambandan D. R., 2009. Regulation of zebrafish zona pellucida gene activity in developing oocytes. Biol. Reprod. 81: 101–110. 10.1095/biolreprod.108.071720 [DOI] [PubMed] [Google Scholar]
- Moore R. K., Otsuka F., and Shimasaki S., 2003. Molecular basis of bone morphogenetic protein-15 signaling in granulosa cells. J. Biol. Chem. 278: 304–310. 10.1074/jbc.M207362200 [DOI] [PubMed] [Google Scholar]
- Morinaga C., Saito D., Nakamura S., Sasaki T., Asakawa S. et al. , 2007. The hotei mutation of medaka in the anti-Mullerian hormone receptor causes the dysregulation of germ cell and sexual development. Proc. Natl. Acad. Sci. USA 104: 9691–9696. 10.1073/pnas.0611379104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K., Thisse C., Thisse B., and Raz E., 2002. Expression of a linker histone-like gene in the primordial germ cells in zebrafish. Mech. Dev. 117: 253–257. 10.1016/S0925-4773(02)00174-0 [DOI] [PubMed] [Google Scholar]
- Munsterberg A., and Lovell-Badge R., 1991. Expression of the mouse anti-mullerian hormone gene suggests a role in both male and female sexual differentiation. Development 113: 613–624. [DOI] [PubMed] [Google Scholar]
- Nair S., Marlow F., Abrams E., Kapp L., Mullins M. C. et al. , 2013. The chromosomal passenger protein birc5b organizes microfilaments and germ plasm in the zebrafish embryo. PLoS Genet. 9: e1003448 10.1371/journal.pgen.1003448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann J. C., Chandler G. L., Damoulis V. A., Fustino N. J., Lillard K. et al. , 2011. Mutation in the type IB bone morphogenetic protein receptor Alk6b impairs germ-cell differentiation and causes germ-cell tumors in zebrafish. Proc. Natl. Acad. Sci. USA 108: 13153–13158. 10.1073/pnas.1102311108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen N. T. T., Vincens P., Roest Crollius H., and Louis A., 2018. Genomicus 2018: karyotype evolutionary trees and on-the-fly synteny computing. Nucleic Acids Res. 46: D816–D822. 10.1093/nar/gkx1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novel P., Fernandez-Trujillo M. A., Gallardo-Galvez J. B., Cano I., Manchado M. et al. , 2013. Two Mx genes identified in European sea bass (Dicentrarchus labrax) respond differently to VNNV infection. Vet. Immunol. Immunopathol. 153: 240–248. 10.1016/j.vetimm.2013.03.003 [DOI] [PubMed] [Google Scholar]
- Onichtchouk D., Aduroja K., Belting H. G., Gnugge L., and Driever W., 2003. Transgene driving GFP expression from the promoter of the zona pellucida gene zpc is expressed in oocytes and provides an early marker for gonad differentiation in zebrafish. Dev. Dyn. 228: 393–404. 10.1002/dvdy.10392 [DOI] [PubMed] [Google Scholar]
- Orban L., Sreenivasan R., and Olsson P. E., 2009. Long and winding roads: testis differentiation in zebrafish. Mol. Cell. Endocrinol. 312: 35–41. 10.1016/j.mce.2009.04.014 [DOI] [PubMed] [Google Scholar]
- Padua M. B. Fox S. C., Jiang T., Morse D. A., and Tevosian S. G., 2014. Simultaneous gene deletion of gata4 and gata6 leads to early disruption of follicular development and germ cell loss in the murine ovary. Biol. Reprod. 91: 24 10.1095/biolreprod.113.117002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigny P., Merlen E., Robert Y., Cortet-Rudelli C., Decanter C. et al. , 2003. Elevated serum level of anti-mullerian hormone in patients with polycystic ovary syndrome: relationship to the ovarian follicle excess and to the follicular arrest. J. Clin. Endocrinol. Metab. 88: 5957–5962. 10.1210/jc.2003-030727 [DOI] [PubMed] [Google Scholar]
- Pulkki M. M., Mottershead D. G., Pasternack A. H., Muggalla P., Ludlow H. et al. , 2012. A covalently dimerized recombinant human bone morphogenetic protein-15 variant identifies bone morphogenetic protein receptor type 1B as a key cell surface receptor on ovarian granulosa cells. Endocrinology 153: 1509–1518. 10.1210/en.2010-1390 [DOI] [PubMed] [Google Scholar]
- Racine C., Rey R., Forest M. G., Louis F., Ferré A. et al. , 1998. Receptors for anti-müllerian hormone on Leydig cells are responsible for its effects on steroidogenesis and cell differentiation. Proc. Natl. Acad. Sci. USA 95: 594–599. 10.1073/pnas.95.2.594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas L., Robledo D., Gomez-Tato A., Vinas A., Martinez P. et al. , 2016. Comprehensive transcriptomic analysis of the process of gonadal sex differentiation in the turbot (Scophthalmus maximus). Mol. Cell. Endocrinol. 422: 132–149. 10.1016/j.mce.2015.11.006 [DOI] [PubMed] [Google Scholar]
- Rocha A., Zanuy S., and Gomez A., 2016. Conserved anti-mullerian hormone: anti-mullerian hormone type-2 receptor specific interaction and intracellular signaling in teleosts. Biol. Reprod. 94: 141 10.1095/biolreprod.115.137547 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Mari A., Yan Y. L., Bremiller R. A., Wilson C., Canestro C. et al. , 2005. Characterization and expression pattern of zebrafish Anti-Mullerian hormone (Amh) relative to sox9a, sox9b, and cyp19a1a, during gonad development. Gene Expr. Patterns 5: 655–667. 10.1016/j.modgep.2005.02.008 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Mari A., Canestro C., Bremiller R. A., Nguyen-Johnson A., Asakawa K. et al. , 2010. Sex reversal in zebrafish fancl mutants is caused by Tp53-mediated germ cell apoptosis. PLoS Genet. 6: e1001034 10.1371/journal.pgen.1001034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Mari A., Cañestro C., BreMiller R. A., Catchen J. M., Yan Y. L. et al. , 2013. Retinoic acid metabolic genes, meiosis, and gonadal sex differentiation in zebrafish. PLoS One 8: e73951 10.1371/journal.pone.0073951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondeau E. B., Messmer A. M., Sanderson D. S., Jantzen S. G., von Schalburg K. R. et al. , 2013. Genomics of sablefish (Anoplopoma fimbria): expressed genes, mitochondrial phylogeny, linkage map and identification of a putative sex gene. BMC Genomics 14: 452 10.1186/1471-2164-14-452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondeau E. B., Laurie C. V., Johnson S. C., and Koop B. F., 2016. A PCR assay detects a male-specific duplicated copy of Anti-Mullerian hormone (amh) in the lingcod (Ophiodon elongatus). BMC Res. Notes 9: 230 10.1186/s13104-016-2030-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouiller-Fabre V., Carmona S., Merhi R. A., Cate R., Habert R. et al. , 1998. Effect of anti-Mullerian hormone on Sertoli and Leydig cell functions in fetal and immature rats. Endocrinology 139: 1213–1220. 10.1210/endo.139.3.5785 [DOI] [PubMed] [Google Scholar]
- Santos E. M., Paull G. C., Van Look K. J., Workman V. L., Holt W. V. et al. , 2007a Gonadal transcriptome responses and physiological consequences of exposure to oestrogen in breeding zebrafish (Danio rerio). Aquat. Toxicol. 83: 134–142. 10.1016/j.aquatox.2007.03.019 [DOI] [PubMed] [Google Scholar]
- Santos E. M., Workman V. L., Paull G. C., Filby A. L., Van Look K. J. et al. , 2007b Molecular basis of sex and reproductive status in breeding zebrafish. Physiol. Genomics 30: 111–122. 10.1152/physiolgenomics.00284.2006 [DOI] [PubMed] [Google Scholar]
- Selman K., Wallace R. A., Sarka A., and Qi X., 1993. Stages of Oocyte Development in the Zebrafish, Brachydanio rerio. J. Morphol. 218: 203–224. 10.1002/jmor.1052180209 [DOI] [PubMed] [Google Scholar]
- Skaar K. S., Nobrega R. H., Magaraki A., Olsen L. C., Schulz R. W. et al. , 2011. Proteolytically activated, recombinant anti-mullerian hormone inhibits androgen secretion, proliferation, and differentiation of spermatogonia in adult zebrafish testis organ cultures. Endocrinology 152: 3527–3540. 10.1210/en.2010-1469 [DOI] [PubMed] [Google Scholar]
- Small C. M., Carney G. E., Mo Q., Vannucci M., and Jones A. G., 2009. A microarray analysis of sex- and gonad-biased gene expression in the zebrafish: evidence for masculinization of the transcriptome. BMC Genomics 10: 579 10.1186/1471-2164-10-579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasan R., Cai M., Bartfai R., Wang X., Christoffels A. et al. , 2008. Transcriptomic analyses reveal novel genes with sexually dimorphic expression in the zebrafish gonad and brain. PLoS One 3: e1791 10.1371/journal.pone.0001791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasan R., Jiang J., Wang X., Bartfai R., Kwan H. Y. et al. , 2014. Gonad differentiation in zebrafish is regulated by the canonical Wnt signaling pathway. Biol. Reprod. 90: 45 10.1095/biolreprod.113.110874 [DOI] [PubMed] [Google Scholar]
- Suzuki A., and Shibata N., 2004. Developmental process of genital ducts in the medaka, Oryzias latipes. Zool. Sci. 21: 397–406. 10.2108/zsj.21.397 [DOI] [PubMed] [Google Scholar]
- Sydes, J., P. Batzel, T. A. Titus, and J. H. Postlethwait, 2019 Dupligänger: a reference genome-based, UMI-cognizant, 5’-trimming-aware PCR duplicate removal pipeline, version 0.97. Zenodo. 10.5281/zenodo.2530972 [DOI]
- Takahashi H., 1977. Juvenile hermaphroditism in the zebrafish, Brachydanio rerio. Bull. Fac. Fish. Hokkaido Univ. 28: 57–65. [Google Scholar]
- te Kronnie G., Stroband H., Schipper H., and Samallo J., 1999. Zebrafish CTH1, a C3H zinc finger protein, is expressed in ovarian oocytes and embryos. Dev. Genes Evol. 209: 443–446. 10.1007/s004270050276 [DOI] [PubMed] [Google Scholar]
- Tremblay J. J., and Viger R. S., 2001. Nuclear receptor Dax-1 represses the transcriptional cooperation between GATA-4 and SF-1 in Sertoli cells. Biol. Reprod. 64: 1191–1199. 10.1095/biolreprod64.4.1191 [DOI] [PubMed] [Google Scholar]
- Tremblay J. J., Robert N. M., Robert S., and Viger V., 2001. Modulation of endogenous GATA-4 activity reveals its dual contribution to Müllerian Inhibiting Substance gene transcription in Sertoli Cells. Mol. Endocrinol. 15: 1636–1650. [DOI] [PubMed] [Google Scholar]
- Uhlén M., Fagerberg L., Hallström B. M., Lindskog C., Oksvold P. et al. , 2015. Proteomics. Tissue-based map of the human proteome. Science 347: 1260419 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- Val P., Lefrancois-Martinez A. M., Veyssiere G., and Martinez A., 2003. SF-1 a key player in the development and differentiation of steroidogenic tissues. Nucl. Recept. 1: 8 10.1186/1478-1336-1-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser J. A., de Jong F. H., Laven J. S., and Themmen A. P., 2006. Anti-Mullerian hormone: a new marker for ovarian function. Reproduction 131: 1–9. 10.1530/rep.1.00529 [DOI] [PubMed] [Google Scholar]
- Volle D. H., Duggavathi R., Magnier B. C., Houten S. M., Cummins C. L. et al. , 2007. The small heterodimer partner is a gonadal gatekeeper of sexual maturation in male mice. Genes Dev. 21: 303–315. 10.1101/gad.409307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hofsten J., Larsson A., and Olsson P. E., 2005. Novel steroidogenic factor-1 homolog (ff1d) is coexpressed with anti-Mullerian hormone (AMH) in zebrafish. Dev. Dyn. 233: 595–604. 10.1002/dvdy.20335 [DOI] [PubMed] [Google Scholar]
- Wang H., Tan J. T., Emelyanov A., Korzh V., and Gong Z., 2005. Hepatic and extrahepatic expression of vitellogenin genes in the zebrafish, Danio rerio. Gene 356: 91–100. 10.1016/j.gene.2005.03.041 [DOI] [PubMed] [Google Scholar]
- Wang H., Teng Y., Xie Y., Wang B., Leng Y. et al. , 2013. Characterization of the carbonic anhydrases 15b expressed in PGCs during early zebrafish development. Theriogenology 79: 443–452. 10.1016/j.theriogenology.2012.10.016 [DOI] [PubMed] [Google Scholar]
- Wang P. Y., Protheroe A., Clarkson A. N., Imhoff F., Koishi K. et al. , 2009. Mullerian inhibiting substance contributes to sex-linked biases in the brain and behavior. Proc. Natl. Acad. Sci. USA 106: 7203–7208. 10.1073/pnas.0902253106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. G., and Orban L., 2007. Anti-Mullerian hormone and 11 beta-hydroxylase show reciprocal expression to that of aromatase in the transforming gonad of zebrafish males. Dev. Dyn. 236: 1329–1338. 10.1002/dvdy.21129 [DOI] [PubMed] [Google Scholar]
- Wang X. G., Bartfai R., Sleptsova-Freidrich I., and Orban L., 2007. The timing and extent of ‘juvenile ovary’ phase are highly variable during zebrafish testis differentiation. J. Fish Biol. 70: 33–44. 10.1111/j.1095-8649.2007.01363.x [DOI] [Google Scholar]
- Webster K. A., Schach U., Ordaz A., Steinfeld J. S., Draper B. W. et al. , 2017. Dmrt1 is necessary for male sexual development in zebrafish. Dev. Biol. 422: 33–46. 10.1016/j.ydbio.2016.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijgerde M., Ooms M., Hoogerbrugge J. W., and Grootegoed J. A., 2005. Hedgehog signaling in mouse ovary: Indian hedgehog and desert hedgehog from granulosa cells induce target gene expression in developing theca cells. Endocrinology 146: 3558–3566. 10.1210/en.2005-0311 [DOI] [PubMed] [Google Scholar]
- Wourms J. P., 1977. Reproduction and development in chondrichthyan fishes. AMER. ZOOL 17: 379–410. [Google Scholar]
- Wu T. D., Reeder J., Lawrence M., Becker G., and Brauer M. J., 2016. GMAP and GSNAP for genomic sequence alignment: enhancements to speed, accuracy, and functionality. Methods Mol. Biol. 1418: 283–334. 10.1007/978-1-4939-3578-9_15 [DOI] [PubMed] [Google Scholar]
- Yan Y. L., Desvignes T., Bremiller R., Wilson C., Dillon D. et al. , 2017. Gonadal soma controls ovarian follicle proliferation through Gsdf in zebrafish. Dev. Dyn. 246: 925–945. 10.1002/dvdy.24579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano A., Guyomard R., Nicol B., Jouanno E., Quillet E. et al. , 2012. An immune-related gene evolved into the master sex-determining gene in rainbow trout, Oncorhynchus mykiss. Curr. Biol. 22: 1423–1428. 10.1016/j.cub.2012.05.045 [DOI] [PubMed] [Google Scholar]
- Yao H. H., Whoriskey W., and Capel B., 2002. Desert hedgehog/patched 1 signaling specifies fetal Leydig cell fate in testis organogenesis. Genes Dev. 16: 1433–1440. 10.1101/gad.981202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazawa T., Uesaka M., Inaoka Y., Mizutani T., Sekiguchi T. et al. , 2008. Cyp11b1 is induced in the murine gonad by luteinizing hormone/human chorionic gonadotropin and involved in the production of 11-ketotestosterone, a major fish androgen: conservation and evolution of the androgen metabolic pathway. Endocrinology 149: 1786–1792. 10.1210/en.2007-1015 [DOI] [PubMed] [Google Scholar]
- Yin Y., Tang H., Liu Y., Chen Y., Li G. et al. , 2017. Targeted disruption of aromatase reveals dual functions of cyp19a1a during sex differentiation in zebrafish. Endocrinology 158: 3030–3041. 10.1210/en.2016-1865 [DOI] [PubMed] [Google Scholar]
- Yoon C., Kawakami K., and Hopkins N., 1997. Zebrafish vasa homologue RNA is localized to the cleavage planes of 2- and 4-cell-stage embryos and is expressed in the primordial germ cells. Development 124: 3157–3165. [DOI] [PubMed] [Google Scholar]
- Zec I., Tislaric-Medenjak D., Megla Z. B., and Kucak I., 2011. Anti-Mullerian hormone: a unique biochemical marker of gonadal development and fertility in humans. Biochem. Med. (Zagreb) 21: 219–230. 10.11613/BM.2011.031 [DOI] [PubMed] [Google Scholar]
- Zhang X., Guan G., Li M., Zhu F., Liu Q. et al. , 2016. Autosomal gsdf acts as a male sex initiator in the fish medaka. Sci. Rep. 6: 19738 10.1038/srep19738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. B., Liu T. K., Jiang J., Shi J., Liu Y. et al. , 2013. Identification of a novel Gig2 gene family specific to non-amniote vertebrates. PLoS One 8: e60588 10.1371/journal.pone.0060588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Zheng H., and Yan W., 2007. Fank1 is a testis-specific gene encoding a nuclear protein exclusively expressed during the transition from the meiotic to the haploid phase of spermatogenesis. Gene Expr. Patterns 7: 777–783. 10.1016/j.modgep.2007.05.005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RNA-seq reads are available at the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA512103. Supplemental Material, Table S1 and Table S2 list differentially expressed genes for juvenile trunks or adult gonads for amh mutants and wild-type siblings, respectively. Work was performed under the University of Oregon Institutional Animal Care and Use Committee protocol no. 14-08R. Mutant strains are available on request. Data should be cited associated with this paper. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8184437.