Abstract
Two common features of centromeres are their transcription into noncoding centromere RNAs (cen-RNAs) and their assembly into nucleosomes that contain a centromere-specific histone H3 (cenH3). Here, we show that Saccharomyces cerevisiae cen-RNA was present in low amounts in wild-type (WT) cells, and that its appearance was tightly cell cycle-regulated, appearing and disappearing in a narrow window in S phase after centromere replication. In cells lacking Cbf1, a centromere-binding protein, cen-RNA was 5–12 times more abundant throughout the cell cycle. In WT cells, cen-RNA appearance occurred at the same time as loss of Cbf1’s centromere binding, arguing that the physical presence of Cbf1 inhibits cen-RNA production. Binding of the Pif1 DNA helicase, which happens in mid–late S phase, occurred at about the same time as Cbf1 loss from the centromere, suggesting that Pif1 may facilitate this loss by its known ability to displace proteins from DNA. Cen-RNAs were more abundant in rnh1Δ cells but only in mid–late S phase. However, fork pausing at centromeres was not elevated in rnh1Δ cells but rather was due to centromere-binding proteins, including Cbf1. Strains with increased cen-RNA lost centromere plasmids at elevated rates. In cbf1Δ cells, where both the levels and the cell cycle-regulated appearance of cen-RNA were disrupted, the timing and levels of cenH3 centromere binding were perturbed. Thus, cen-RNAs are highly regulated, and disruption of this regulation correlates with changes in centromere structure and function.
Keywords: centromere, centromere RNA, Cbf1, Cse4
CENTROMERES, the segregation elements of eukaryotic chromosomes, are the platform for kinetochore assembly and subsequent microtubule attachment, and hence are absolutely essential for the stability of eukaryotic chromosomes. Centromeres also support sister chromatid cohesion at pericentric DNA, which keeps sisters together until anaphase (Natsume et al. 2013).
Most eukaryotes have so-called regional centromeres, which range in size from 35 to 100 kb in Schizosaccharomyces pombe to 0.1–5 Mb in humans and contain repeated sequences, that are usually assembled into heterochromatin [reviewed in Malik and Henikoff (2002)]. In contrast, the Saccharomyces cerevisiae (hereafter, yeast) centromere, often called a point centromere, is only ∼125-bp long, and is nonrepetitive and nonheterochromatic [see Biggins (2013) for a review of yeast centromeres and their associated proteins].
Yeast centromeres contain three conserved elements (CDEs): CDEI (8 bp), CDEII (78–86 bp), and CDEIII (25 bp) (Figure 1A). CDEI is bound by Cbf1, which—depending on context—activates or represses RNA polymerase II transcription (Cai and Davis 1990; Mellor et al. 1991). Neither CDEI nor CBF1 is essential, but loss of either reduces chromosome stability. CDEII varies somewhat in length and sequence but is always AT-rich. Its size and AT-richness are both essential for centromere function, probably because it is the binding site for a nucleosome containing Cse4, the essential yeast centromere-specific histone H3 variant (cenH3, called CENP-A in humans) (Stoler et al. 1995; Krassovsky et al. 2012). In vivo, centromeric chromatin is thought to wrap around the Cse4 nucleosome, which might be facilitated by Cbf1, whose binding promotes DNA bending (Niedenthal et al. 1993; Kent et al. 2004; Steiner and Henikoff 2015). The size and sequence of CDEIII is essential as it provides a binding site for a four-protein complex called Cbf3, which is required for association of kinetochore proteins (Biggins 2013).
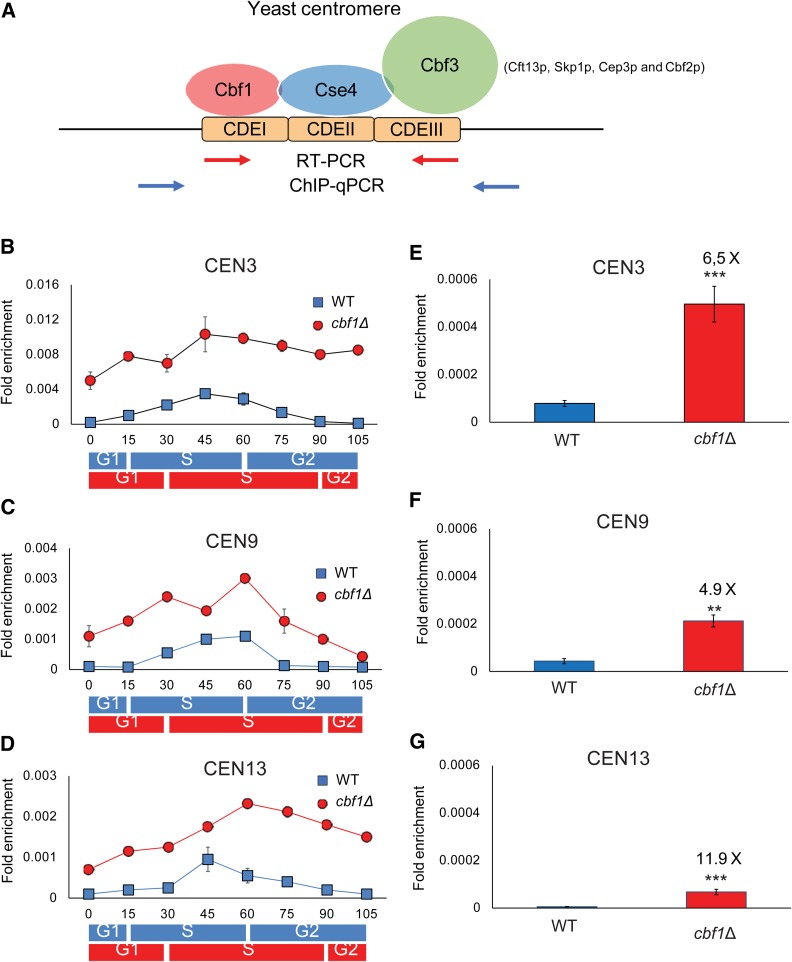
Figure 1.
Cen-RNA is cell cycle-regulated and inhibited by Cbf1. (A) Schematic of centromeric DNA and its binding proteins (Biggins 2013). Cbf1 binds CDEI; CDEII wraps around a nucleosome containing Cse4, a centromere-dedicated histone H3; and CDEIII is bound by a four-protein complex called Cbf3 consisting of Ctf13, Skp1p, Cep3p, and Cbf2p. Blue arrows indicate the position of primers used to amplify centromeric DNA by qPCR and red arrows indicate the position of primers used to amplify cen-RNA by RT-PCR. The primers are specific for each centromere. (B–D) The pattern of cen-RNA abundance at CEN3 (B), CEN9 (C), and CEN13 (D) in 24°-grown WT (blue squares) or cbf1Δ (red circles) cells as they move through a synchronous cell cycle. Samples were collected at the indicated times after release from α factor arrest, total RNA was purified, and cen-RNAs detected by RT-PCR. The approximate timings of cell cycle transitions for WT (blue rectangles) and cbf1Δ cells (red rectangles) are indicated beneath the graphs, as determined from FACS data (Figure S1A). Data are presented as ([RNA]target gene/[ACT1]control). Scales are not the same for the different centromeres. (B–D) The amounts of cen-RNA [(E) CEN3, (F) CEN9, and (G) CEN13] in 30°-grown asynchronous cells determined using RT-PCR. Cen-RNA levels were determined in WT (blue bars) and cbf1Δ (red bars) cells. The levels of cen-RNA at a given centromere in WT cells were defined as one; the fold increases in the levels in cbf1Δ cells are shown above the red bars. Error bars in (E–G) indicate 1 SD from the average value of three independent experiments. P-values were obtained using unpaired two-tailed Student’s t-tests. In all figures, * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, and **** P ≤ 0.0001. CDE, conserved elements; cen-RNA, centromeric RNA; ChIP, chromatin immunoprecipitation; qPCR, quantitative PCR; WT, wild-type.
Regional and point centromeres both require cenH3 nucleosomes to form functional kinetochores (Verdaasdonk and Bloom 2011). While the yeast centromere contains a single cenH3 nucleosome (Furuyama and Biggins 2007; Henikoff and Henikoff 2012; Krassovsky et al. 2012), regional centromeres have multiple cenH3 nucleosomes (Malik and Henikoff 2002). Multiple kinetochore microtubules bind to each regional centromere, while there is a single kinetochore microtubule bound to each yeast centromere (Biggins 2013). Thus, the budding yeast centromere and its kinetochore are scaled-down versions of regional centromeres, yet they share many features with the more complex regional centromeres, including many conserved kinetochore proteins.
Although studied mainly in the context of regional centromeres, centromeric RNA (cen-RNA) transcripts have been described in diverse organisms [reviewed in Caceres-Gutierrez and Herrera (2017) and Talbert and Henikoff (2018)]. At many regional centromeres, cen-RNA acts in concert with RNA interference to form heterochromatin. In addition, there is considerable evidence linking cen-RNAs directly to the segregation function of centromeres. For example, increases and decreases in cen-RNA levels correlate with decreased chromosome stability (Caceres-Gutierrez and Herrera 2017; Talbert and Henikoff 2018). Although there are multiple models for how cen-RNAs affect centromere segregation, a unifying model for their function has not emerged in part because it can be difficult to distinguish the effects of cen-RNAs on heterochromatin formation vs. their effects on chromosome segregation. As S. cerevisiae centromeres are not heterochromatic, analysis of the functions of its cen-RNAs is theoretically less complicated.
Here, we investigate the occurrence and regulation of cen-RNA in budding yeast. The impetus for this work was the finding that the Pif1 DNA helicase binds robustly to all centromeres, and this binding is cell cycle-regulated, occurring in late S/G2 phase, after centromere replication (Chen et al. 2019). Because Pif1 efficiently displaces RNA from RNA–DNA hybrids (Boule and Zakian 2007; Zhou et al. 2014), we considered the possibility that it might remove cen-RNA from centromeres. Here, we report that the appearance of yeast cen-RNA was cell cycle-regulated, occurring in mid–late S phase, about the same time that Pif1 binds centromeres. Cen-RNAs were present in very low amounts in wild-type (WT) cells owing in part to Cbf1 inhibition of cen-RNA production. As Cbf1 binding was transiently lost in late S phase when cen-RNAs were detected, Cbf1 likely acts directly to inhibit cen-RNA synthesis. Cbf1 binding increased in Pif1-depleted cells, which could reflect a role for Pif1 in Cbf1 removal. Cen-RNA levels were higher in the absence of RNaseH1, which removes and degrades RNA in RNA–DNA hybrids, an increase that was limited to mid–late S phase. Likewise, cen-RNAs were more abundant in mid–late S phase in cells deficient for Pif1 and Rrm3, a homolog of Pif1. Together, these data suggest that RNaseH1, Pif1, and Rrm3 contribute to cen-RNA release from centromeres late in the cell cycle. When cen-RNA was made at high levels throughout the cell cycle, Cse4 binding was deregulated.
Materials and Methods
Yeast strains
Yeast strains, plasmids, and primers used in this study are listed in Supplemental Material, Table S1 and S2. All strains were derivatives of YPH499 (Sikorski and Hieter 1989). Deletions of genes eliminated the entire ORF. As expected, the cbf1Δ strain constructed for these experiments was slow growing and benomyl-sensitive, and both phenotypes were suppressed by a centromere plasmid containing CBF1 (Cai and Davis 1990) (Figure S1, C and E). Additionally, MET17 mRNA levels in the same RNA samples used to monitor cen-RNA were ∼50% lower in cbf1Δ vs. WT cells (Thomas et al. 1992; Kuras and Thomas 1995; O’Connell et al. 1995) (Figure S1D). Deletions and tagged genes were confirmed by both Southern and PCR analyses. Because terminally tagged alleles of Cbf1 and Cse4 are not functional (Stoler et al. 1995; Wisniewski et al. 2014), both proteins were internally tagged with nine MYC epitopes as described (Gauss et al. 2005). For CBF1, the epitopes were after codon 9 (Meluh and Koshland 1997) and for Cse4, after codon 81 (Meluh et al. 1998). The structures of the tagged CBF1 genes were confirmed by PCR, Southern analysis (Figure S4A), sequencing, and phenotypic analyses. Cbf1-MYC cells had WT benomyl sensitivity (Figure S4C) and grew about as well as WT cells (Figure S4D). In asynchronous chromatin immunoprecipitations (ChIPs), Cbf1-MYC bound specifically to six centromeres but not to control sites (Figure 2, E–H and Figure S3, C–E). These analyses were important because the size of Cbf1-Myc on a western was 100 kDa rather than the predicted size of 50 kDa (Figure S4B). The difference from the expected size was probably due to the internal tag. Asynchronous cultures were grown at 30°; synchronies were done at 24°.
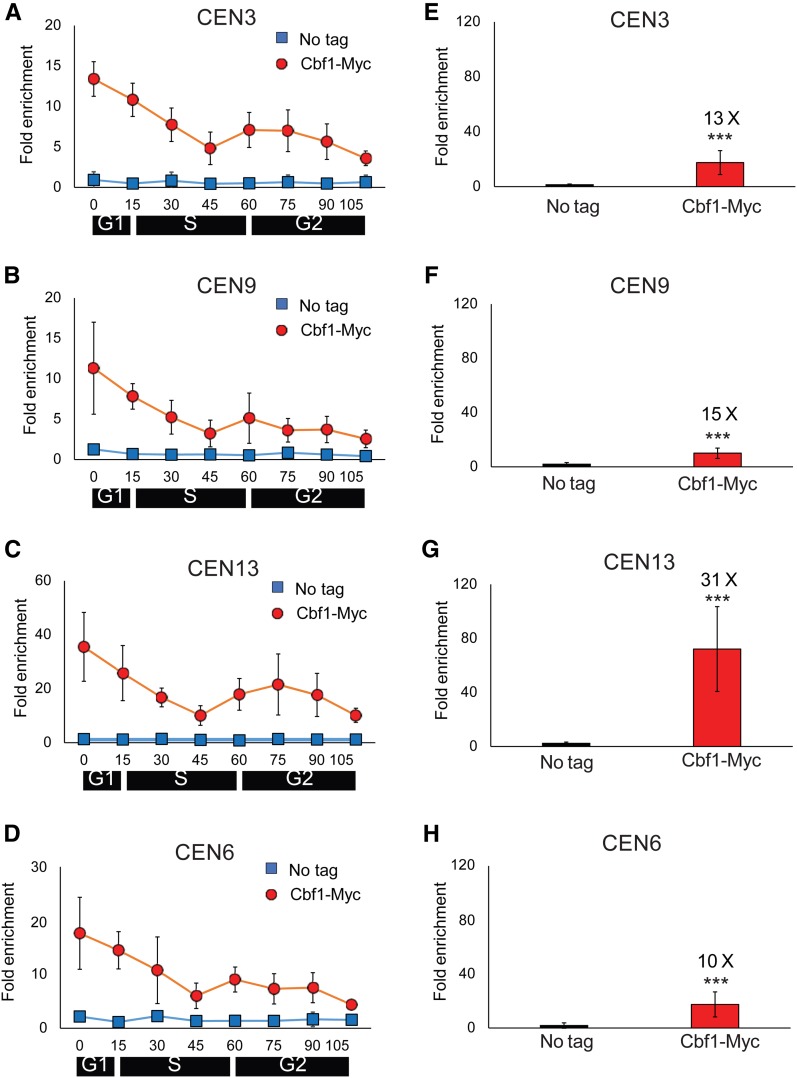
Figure 2.
Cbf1 binding to centromeres is cell cycle-regulated. (A–D) WT cells expressing Cbf1-MYC and growing at 24° were synchronized as described in Figure 1. Samples were removed at the indicated times for ChIP-qPCR analysis of Cbf1-MYC binding (red circles) to (A) CEN3, (B) CEN9, (C) CEN13, and (D) CEN6. Scales are not the same on all panels. ChIP-qPCR was carried out in parallel on a no-tag strain (blue squares). (E–H) show levels of Cbf1-MYC binding in 30°-grown asynchronous cells (red bars) and in a no-tag strain (blue bars). Cbf1-Myc binding was determined at (E) CEN3, (F) CEN9, (G) CEN13, and (H) CEN6. Values are the average of Cbf1-MYC binding in three independent colonies. The fold changes in the levels of binding on a given CEN of Cbf1-MYC relative to WT are shown above the red bars. Error bars and statistical analyses are as in Figure 1. ChIP, chromatin immunoprecipitation; qPCR, quantitative PCR; WT, wild-type.
Cell synchrony
Cells were synchronized and analyzed at 24° as described (Chen et al. 2019). Cells were arrested in α factor (0.015 ng/ml; Princeton University) and collected at 15-min intervals after α factor removal. The quality of each synchrony was monitored by flow cytometry (Chen et al. 2019). For each strain, synchronies were done at least three times on progeny of independent colonies. Samples were analyzed for DNA by ChIP-quantitative PCR (qPCR) and for RNA by reverse transcription polymerase chain reaction (RT-PCR).
RNA purification and RT-PCR
Total RNAs were purified from synchronized and asynchronized cells using a RNeasy Mini Kit (#74106; QIAGEN, Valencia, CA). DNA was eliminated by digestion with a TURBO DNA-free Kit (#AM1907; Invitrogen, Carlsbad, CA). Then, 500 ng of total RNA was analyzed by RT-PCR (SYBR Green One-Step Kit, #172–5151; Bio-Rad, Hercules, CA) as indicated by the manufacturer. The primer sets for RT-PCR are described in Table S2. The process ran for 45 cycles for cen-RNA and for the ACT1 control.
ChIP and qPCR
Asynchronous cells were grown in 50 ml of YPD at 30° and harvested at an OD660 of 0.5. Cells were cross-linked in 1% formaldehyde for 5 min, then quenched with 2.5 ml (2.5 M) glycine for 5 min. Chromatin purification was carried out as described (Chen et al. 2019). Cells were incubated overnight at 4° in 0.02 µg/µl of anti-MYC monoclonal antibody (#631206; Takara) and then coupled to 80 µl of Dynabeads protein G (#10004D; Thermo Fisher Scientific; 4 hr at 4°). Reverse cross-linking of DNA was performed and DNA was purified by QIAquick PCR Purification kit (#28106; QIAGEN). Chromatin immunoprecipitate and input DNA were analyzed by qPCR using iQ SYBR Green Supermix (#170-8882; Bio-Rad) and the CFX96 real-time system (Bio-Rad). Primers are listed in Table S2. At least three biological replicates were conducted for each ChIP. WT cells without a Myc-tagged protein were used as a negative control. ChIP-qPCRs were quantified by ([ChIP/Input] Target site/[ChIP/Input] ARO1). Binding to the telomere of chromosome VI-R was used as a negative control.
Western blotting
Western blotting was carried out as in Tran et al. (2017). Briefly, cell extract was mixed with 1 × SDS-PAGE sample buffer, boiled for 10 min, and pelleted. Samples were analyzed on 15% (37.5:1 polyacrylamide:bis-acrylamide) SDS-PAGE gels and run at 20 V/cm at room temperature. The proteins were transferred to a nitrocellulose membrane at 4° and blocked with 5% nonfat milk in TBST at room temperature using standard protocols. The blot was probed with an anti-MYC monoclonal antibody (#631206; Takara), which was diluted 1:500, and visualized with an HRP-conjugated secondary antibody and ECL detection reagents (GE Healthcare).
Loss rate assay for centromere plasmid
Plasmid loss assays were carried out as described (Chen et al. 2019). The plasmid loss rates are based on three independent experiments for each strain. The plasmid used for loss rates was the CEN6 plasmid pRS316 (Sikorski and Hieter 1989).
Benomyl sensitivity
Cells were grown overnight in YPD at 30°. Indicated strains were spotted in five times serial dilutions from 3 × 107 cells per spot on YPD plates with and without benomyl (10 μg/ml), and grown at 30° for 3 days.
Two-dimensional agarose gel electrophoresis
Replication intermediates were analyzed by standard two-dimensional (2D) agarose gel electrophoresis techniques performed on total genomic DNA isolated from asynchronous cells (Brewer and Fangman 1987, 1991; Huberman et al. 1987). Cells were collected in log phase at an optical density of OD660 of ∼0.6. Collected DNA was digested with MfeI. In the first dimension, DNA was separated in 0.4% agarose at room temperature for 20 hr at 2.0 V/cm. The second dimension was run for 15 hr in 1.1% agarose containing ethidium bromide (0.3 µg/ml) at 4.4 V/cm at 4°. Southern blots were probed using a 32P-labeled probe, whose position is indicated in Figure 5. The extent of pausing was obtained as described (Chen et al. 2019). Quantification of pausing was done in two or more different biological replicates, and was normalized to the WT pause signal to obtain fold increase in the pause in a mutant strain relative to pausing in the otherwise isogenic WT strain.
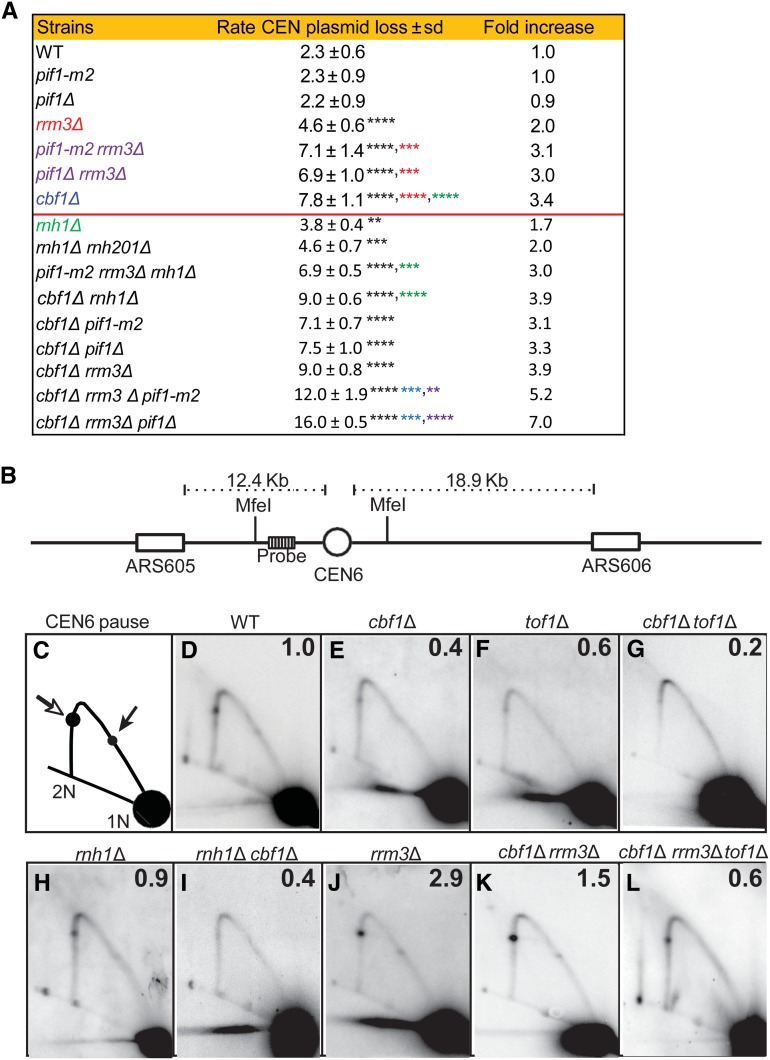
Figure 5.
Stability of a CEN6 plasmid and 2D gel analysis of replication of the chromosomal CEN6 locus, in mutant and WT strains. Methods for determining CEN6 plasmid loss rates and 2D gels are described in the Materials and Methods, and in Chen et al. (2019). (A) CEN plasmid loss rate in WT and mutant strains growing at 30° in YPD. Loss rates for strains above the horizontal red line are from Chen et al. (2019). The second column is the loss rate ± 1 SD for the indicated strain. Asterisks indicate significant differences: black asterisks are significant differences in loss rate of the indicated strain compared to loss rate in WT cells; red asterisks are significant differences between indicated strain and rrm3Δ cells; green asterisks are significant differences between indicated strain and rnh1Δ cells; blue asterisks are significant differences between indicated strain and cbf1Δ cells; and purple asterisks indicate significant differences between indicated strain and pif1Δ (or pif1-m2) rrm3Δ cells. (B) 2D gel electrophoresis and Southern blot hybridization of replication through the chromosomal CEN6 locus in 30°-grown asynchronous WT and mutant cells growing in YPD. DNA was isolated, digested with MfeI restriction enzyme, and analyzed by 2D gels and Southern blotting. (B) Schematic of the chromosomal MfeI fragment that contains CEN6 (open circle) relative to the replication origins (open boxes) on either side. The position of the radiolabeled probe used for Southern blot analysis is indicated. (C) Schematic of 2D gel signal of MfeI-digested DNA using the probe indicated in (B). Arrows mark the pauses at CEN6 along the arc of Y-shaped replication intermediates. The pause produced from replication forks originating from ARS605 are indicated by the open arrow, whereas the pause arising from forks originating from ARS606 are indicated by the solid arrow. 1N indicates nonreplicating linear MfeI fragments. 2N indicates near-fully replicated MfeI fragments. (D–K) Southern blots of 2D gels from cells of the following genotypes: (D) WT, (E) cbf1Δ, (F) tof1Δ, (G) cbf1Δ tof1Δ, (H) rnh1Δ, (I) rnh1Δ cbf1Δ, (J) rrm3Δ, (K) cbf1Δ rrm3Δ, and (L) cbf1Δ rrm3Δ tof1Δ. The signal at the pause was quantified (Tran et al. 2017) and normalized to the pause signal in WT cells to obtain the relative fold change. The average fold difference of mutant over WT from two or more independent biological replicates is shown in the upper right corner of each panel. 2D gel data for tof1Δ and rrm3Δ were published previously (Chen et al. 2019). 2D, two-dimensional; WT, wild-type.
Data availability
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, tables, and Supplemental Materials. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.8939873.
Results
Cen-RNA is cell cycle-regulated, with highest abundance in late S phase
The Pif1 5′–3′ DNA helicase binds centromeres in late S/G2 phase (Chen et al. 2019). To determine if Pif1 has a role in cen-RNA regulation, we first determined the time of cen-RNA appearance. In this paper, we mostly examined three centromeres—CEN3, 9, and 13—which were chosen because all three were studied in an earlier paper on cen-RNAs (Ohkuni and Kitagawa 2011). Because it was used for replication and mini-chromosome loss studies, in some experiments, CEN6 was also examined. Each centromeric DNA and its RNA product was detected by a different set of primers that were unique to a given centromere (Figure 1A). We monitored cen-RNA abundance by RT-PCR as its low abundance and heterogenous size made it difficult to detect with northern analyses (data not shown). Controls indicate that RT-PCR detects cen-RNA; e.g., samples were DNase treated prior to RT-PCR (TURBO DNA; Invitrogen), and no product was obtained when samples were treated with RNase A prior to amplification or if reverse transcriptase was omitted (Figure S1B).
To determine the time of cen-RNA production, a synchronized population of WT cells was obtained by arresting the culture in late G1 phase by incubation in α factor at 24°. Cells were then released from α factor and RT-PCR was used to monitor cen-RNA at three centromeres at 15 min intervals throughout the synchronous cell cycle (Figure 1, B–D, blue squares; primers used to amplify cen-RNA were specific for each centromere). Synchronies were done at 24° because it is easier to detect cell cycle transitions in cells growing more slowly than at the optimal growth temperature of 30°. Progression through the cell cycle was monitored by FACS (see Figure S1 and Figure S2 for FACS of synchronized cells for the WT and mutant strains studied herein). Synchronies were done in triplicate on cultures generated from three independent colonies of a given strain.
At CEN3, cen-RNA was detectable at 15 min after release from α factor (early S phase), peaked in abundance at 45–60 min (late S phase), and decreased to almost background levels by 90 min (post S phase) (Figure 1B). Appearance of cen-RNA was somewhat later for CEN9, first evident at 30 min, peaking at 45–60 min, and no longer detectable by 75 min (Figure 1C). Likewise, at CEN13, cen-RNA was low until 45 min and was back to background levels by 90 min (Figure 1D). Thus, like the binding of Pif1 to centromeres (Chen et al. 2019), the appearance of cen-RNA at all three centromeres was cell cycle-regulated and occurred mainly in mid–late S phase.
We also determined the levels of cen-RNA in asynchronous WT cultures grown at 30° (Figure 1, E–G; blue bars). RNA values for 30°-grown cells cannot be compared directly to values for 24°-grown cells because yeast grows faster at 30° than at 24°(Chen et al. 2019). Levels of cen-RNA varied from centromere to centromere: for example, there was 14 times more cen-RNA at CEN3 (Figure 1E) than at CEN13 (Figure 1G). However, even at CEN3, cen-RNAs levels were very low. In asynchronous cells, cen-RNA was detected only after ∼30 PCR cycles, compared to 16–17 cycles for the abundant ACT1 RNA and ∼26 cycles for the low-abundance telomerase RNA, which is present at ∼20–30 molecules per cell (Mozdy and Cech 2006; P. D. Garcia et al., unpublished results).
While this manuscript was in revision, another group published a paper on yeast cen-RNA whose results partially overlap those reported here (Ling and Yuen 2019b). This group also used RT-qPCR to detect cen-RNA. They focused their analysis on CEN1, 3, and 8. In agreement with our data, they showed that cen-RNAs were present in very low amounts, estimated at considerably lower than one molecule per cell, and were similarly cell cycle-regulated (their cell cycle experiments were done at 28° so the timing of events is not directly comparable to our experiments). They also reported that cen-RNAs are poly-adenylated, and large and heterogenous in size (462–1754 nt). More recently, they reported that cen-RNA levels are elevated in two strains defective in exosome-mediated degradation of nuclear RNA (Ling and Yuen 2019a).
Cbf1 inhibits cen-RNA
Cbf1, a helix-loop-helix sequence-specific DNA-binding protein, binds to the CDEI motif of yeast centromeres (Figure 1A), as well as to the promoters of multiple genes, whose transcription it regulates (Cai and Davis 1990; Mellor et al. 1991). Depending on the gene, Cbf1 has positive or negative effects on transcription.
To determine the effects of Cbf1 on cen-RNA, we monitored the abundance of cen-RNAs at CEN3, 9, and 13 at 30° in asynchronous cbf1Δ cells (Figure 1, E–G; red bars). Cen-RNA levels were ∼5–12× higher in asynchronous cbf1Δ cells than in WT cells (6.5× higher at CEN3, 4.9× at CEN9, and 11.9× at CEN13) (Figure 1, E–G). Of the three centromeres, the fold increase in cen-RNA in cbf1Δ cells was greatest at CEN13. However, because CEN13 cen-RNA was less abundant in WT cells than CEN3 or CEN9 RNA, levels of CEN13 RNA were still lower in cbf1Δ cells than at the other two centromeres. High levels of cen-RNA were suppressed by introducing a centromere plasmid containing a WT copy of CBF1 (Figure S1C). Although the inhibitory effects of Cbf1 on the production of cen-RNA are in agreement with Ling and Yuen (2019b), an earlier paper found that Cbf1 activates cen-RNA transcription (Ohkuni and Kitagawa 2011). The reason for this discrepancy is not clear. In combination with other data (Ling and Yuen 2019b), Cbf1 represses transcription at five of five centromeres. Cen-RNA levels are also higher (1.5–2 times higher than WT) at CEN1, 3, and 8 in htz1Δ cells, where Htz1 is a variant histone H2A that binds near centromeres as well as at many promoters (Ling and Yuen 2019b). Epistasis experiments indicate that Cbf1 and Htz1 affect CEN transcription by different mechanisms.
Cbf1 inhibits cen-RNA production throughout the cell cycle
To determine when Cbf1 represses cen-RNA expression, we synchronized cbf1Δ cells using the α factor protocol described above and determined cen-RNA levels by RT-qPCR throughout the cell cycle at 24° (Figure 1, B–D, red circles). Although cbf1Δ cells grew significantly slower than WT cells at both 24 and 30° (Figure S1F), FACS analysis showed that they spent about the same fraction of the cell cycle in each cell cycle phase (Figure S2). For all three centromeres, there was significantly more cen-RNA at each time point in cbf1Δ (red circles) compared to WT cells (blue squares) (Figure 1, B–D; P < 0.01). Therefore, Cbf1 represses the production of cen-RNA and is also largely responsible for its cell cycle-regulated expression.
Cbf1 centromere binding is cell cycle-regulated
To gain insight into the mechanism by which Cbf1 inhibits cen-RNA transcription, we determined if it was centromere-associated at the time when cen-RNA appears. Cbf1-MYC cells were synchronized, and Cbf1-MYC binding to CEN3, 6, 9, and 13 was determined by ChIP-qPCR at 24° (Figure 2, A–D and Figure S3, A and B). The pattern of Cbf1-MYC binding was very similar at the four centromeres (red circles). Binding was highest in late G1 phase (0 time point), decreased as cells moved through S phase, and was lowest at 45 min. The time of lowest Cbf1-MYC binding corresponded to the time of peak appearance of cen-RNAs in WT cells (Figure 1, B–D, blue squares). Levels of Cbf1 binding were also examined at six centromeres by ChIP-qPCR in asynchronous cells grown at 30° (Figure 2, E–H and Figure S3, C and D). Levels of Cbf1 binding varied somewhat from centromere to centromere with particularly high levels at CEN13 (31× the no-tag control; Figure 2G). Thus, CEN13 produced less cen-RNA and bound more Cbf1 than the other centromeres examined. We conclude that the repressive effects of Cbf1 on centromere transcription are relieved by removal of Cbf1.
Cen-RNA levels are suppressed in mid–late S phase by RNaseH1
Rnh1 removes and degrades RNA in RNA–DNA hybrids (R-loops) (Cerritelli and Crouch 2009; Zimmer and Koshland 2016). Because the R-loop-specific S9.6 antibody stains human centromeres, and this staining is RNaseH1-sensitive, human cen-RNA is thought to be in R-loops (Kabeche et al. 2018). To determine if S. cerevisiae cen-RNA was RNaseH1-sensitive, cen-RNA levels were determined in rnh1Δ cells. In 30°-grown asynchronous rnh1Δ cells, cen-RNA abundance was significantly higher than in WT cells at CEN3, 9 and 13 (Figure 3, A–C; 1.7–2.4× higher; P < 0.05). In contrast, levels of the Cbf1 regulated MET17 mRNA were not affected by deleting RNH1 (Figure S1D; P > 0.05). Deletion of RNH201, the catalytic subunit of RNaseH2, had little or no effect on cen-RNA levels (Figure 3, A–C).
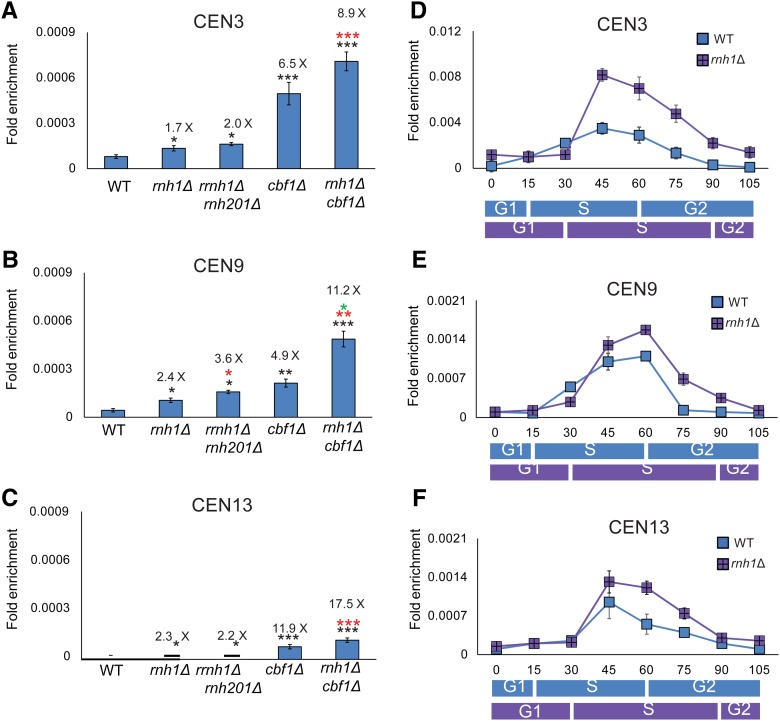
Figure 3.
Cen-RNA abundance is increased in rnh1Δ cells in mid–late S phase. Cen-RNA levels were determined by RT-PCR in asynchronous cultures grown in YPD at 30° (A) CEN3, (B) CEN9, and (C) CEN13 in WT and mutant strains (names of strains are under bars). The numbers above the bars indicate the fold difference in cen-RNA in mutants compared to WT cells. Black asterisks indicate P-values relative to WT. Red asterisks indicate P-values relative to rnh1Δ. Green asterisks indicate P-values relative to cbf1Δ. (D–F) show cen-RNA levels in 24°-grown cells synchronized as described in the legend for Figure 1 at (D) CEN3, (E) CEN9, and (F) CEN13. RNA levels in WT cells (blue square) and rnh1Δ cells (purple square) are shown at each time point. The scale for (D) (CEN3) is different from those of (E and F). The approximate timings of the cell cycle transitions for WT (blue rectangle) and rnh1Δ (purple rectangle) are indicated beneath the graphs, as determined by FACs analysis (Figure S1A and Figure S2F). cen-RNA, centromeric RNA; WT, wild-type.
We also examined the impact of RNaseH1 on cen-RNA throughout a synchronous cell cycle at 24° (Figure 3, D–F; purple squares). At all three centromeres, cen-RNA was significantly higher (P < 0.001) in rnh1Δ vs. WT cells (blue squares). In contrast to the cell cycle-wide increase seen in cbf1Δ, the increase in cen-RNA in rnh1Δ cells was limited to late S phase (45–75 min for CEN3; Figure 3D; 60 and 75 min for CEN9 and 13; Figure 3, E and F).
To determine if Cbf1 and RNase H1 increased cen-RNA by the same mechanism, we determined cen-RNA levels in 30°-grown asynchronous rnh1Δ cbf1Δ cells (Figure 3, A–C). At all three centromeres, levels of cen-RNA were significantly elevated in the double mutant compared to either single mutant (for direct comparison, the values for cen-RNA in cbf1Δ cells from Figure 1, E–G are reshown in Figure 3, A–C). Thus, Cbf1 and RNaseH1 affect cen-RNA levels by different mechanisms.
The increase in cen-RNA in rnh1Δ cells suggests a model where yeast cen-RNA is in R-loops. However, in WT yeast cells, cen-RNAs are not precipitated by the S9.6 antibody (Gómez-González et al. 2011; Stirling et al. 2012; Wahba et al. 2016) (our unpublished results). Thus, it is possible that the effects of RNaseH1 deletion on cen-RNA levels are indirect.
Cen-RNA levels are suppressed in mid–late S phase by Pif1 and Rrm3
Pif1 and Rrm3 are members of the Pif1 family of 5′–3′ DNA helicases that are present in almost all eukaryotes, and some bacteria [reviewed in Geronimo and Zakian (2016) and Pohl and Zakian (2019)]. The multifunctional Pif1 has numerous functions, including maintenance of mitochondrial DNA, inhibition of telomerase, promoting replication past G-quadruplex motifs, and facilitating break-induced replication. Rrm3 is best known for promoting fork progression past nonnucleosomal proteins complexes, including those at centromeres and tRNA genes (Ivessa et al. 2003; Chen et al. 2019). Pif1 also promotes replication at centromeres (Chen et al. 2019) and tRNA genes (Osmundson et al. 2017; Tran et al. 2017), but only in rrm3Δ cells. Because Pif1 binds robustly to all centromeres late in the cell cycle (Chen et al. 2019) and actively unwinds RNA–DNA hybrids (Boule and Zakian 2007; Zhou et al. 2014), we tested the effects of Pif1 (and Rrm3) on cen-RNA (Figure 4). Because pif1-m2 cells grow much better than pif1Δ cells, which are respiratory-deficient, we used pif1-m2 cells, which have reduced nuclear but normal mitochondrial Pif1 functions (Schulz and Zakian 1994; Myung et al. 2001; Ribeyre et al. 2009; Wilson et al. 2013).
Figure 4.
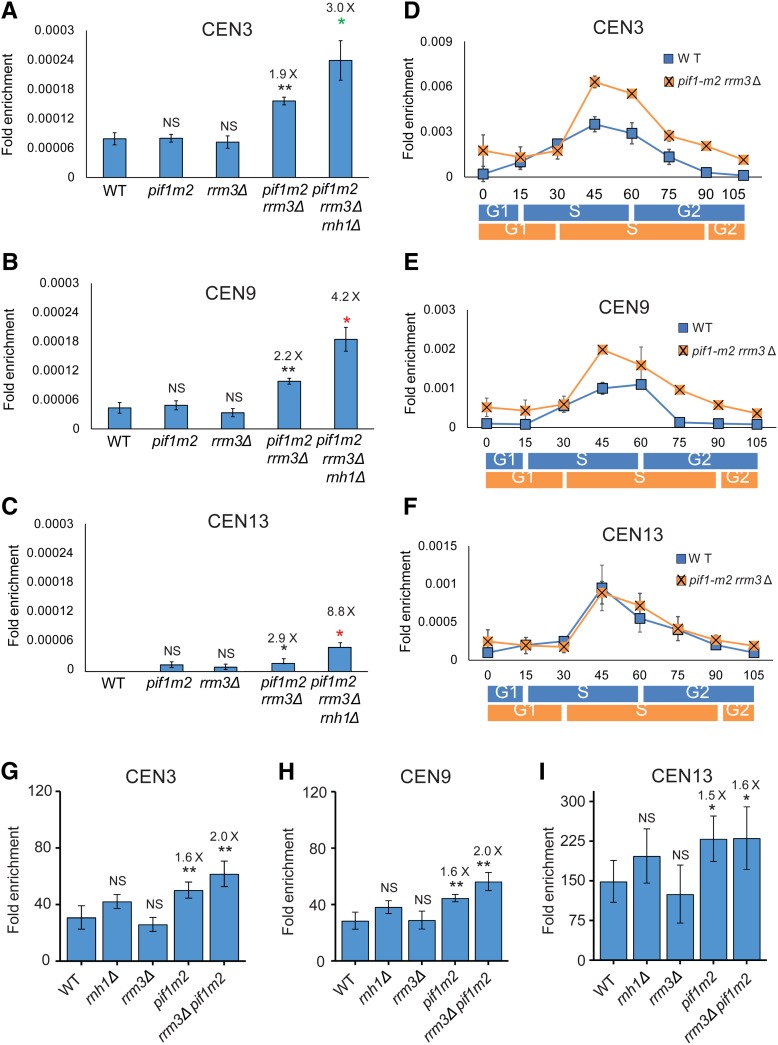
Cen-RNA abundance is increased in late S phase in cells lacking Pif1 and Rrm3. Cen-RNA levels were determined by RT-PCR in asynchronous cultures grown in YPD at 30° at (A) CEN3, (B) CEN9, and (C) CEN13 in WT and mutant strains (names of strains are under bars). The numbers above the bars indicate the fold differences in cen-RNA in mutants compared to WT cells. Black asterisks indicate P-values relative to WT. Red asterisks indicate P-values relative to pif1-m2 rrm3Δ. Green asterisks indicate P-values relative to rnh1Δ. (D–F) show cen-RNA levels in 24°-grown cells synchronized as described in Figure 1 legend at (D) CEN3, (E) CEN9, and (F) CEN13. The scales for the three panels are different. RNA levels in WT cells (blue square) and pif1-m2 rrm3Δ cells (orange square) are shown. The approximate timings of the cell cycle transitions for WT (blue rectangle) and pif1-m2 rrm3Δ (orange square) are indicated below the graphs as determined by FACs data (Figure S1A and Figure S2G). (G–I) show Cbf1-Myc binding in WT and mutant strains (names of strains are under bars). The numbers above the bars indicate the fold differences in Cbf1-Myc binding to mutant strains. cen-RNA, centromeric RNA; NS, not significant; WT, wild-type.
In asynchronous cells growing at 30°, the amount of cen-RNA at CEN3, 9, and 13 was not significantly different in pif1-m2 or rrm3Δ compared to WT cells (P > 0.05; Figure 4, A–C). However, the amount of cen-RNA was about two-to-three times higher in pif1-m2 rrm3Δ vs. WT cells, and these differences were significant at all three centromeres (Figure 4, A–C; P < 0.01 at CEN3 and 9, and P < 0.05 at CEN13). The amount of cen-RNA was even higher in asynchronous pif1-m2 rrm3Δ rnh1Δ cells, and again this difference was significant compared to either pif1-m2 rrm3Δ or rnh1Δ cells (Figure 4, A–C; P < 0.05; the data for rnh1Δ cells are from Figure 3, A–C; P < 0.05).
Similar results were seen for 24°-grown synchronized cells (Figure 4, D–F). There was no significant difference in cen-RNA levels in pif1-m2 or rrm3Δ single mutants in late S/G2 phase (Figure S5). However, for CEN3 and 9, cen-RNA was significantly higher at 45–70 min in doubly mutant pif1-m2 rrm3Δ (orange squares) compared to WT cells (blue squares) (P < 0.05). Effects on cen-RNA levels were not detected at CEN13 in synchronized doubly mutant cells (Figure 4F), perhaps because the lower temperature provides more time for RNaseH1 to act at this centromere. However, cen-RNA levels were almost three times higher at CEN13 in 30°-grown asynchronous pif1-m2 rrm3Δ cells (P < 0.05; Figure 4C).
These data suggest that Pif1 and Rrm3 have redundant functions in removal of cen-RNA. Given that R-loops are preferred substrates for both Pif1 (Boule and Zakian 2007; Zhou et al. 2014) and Pfh1, the S. pombe Pif1 family helicase (Mohammad et al. 2018), it is possible that Pif1/Rrm3 act in parallel with RNaseH1 to displace cen-RNA from centromeric DNA. Unlike RNaseH1, DNA helicases cannot degrade the RNA they release. Thus, helicase-released cen-RNA is probably degraded by the exosome (Ling and Yuen 2019a).
Cbf1 binding to centromeres is reduced in Pif1-depleted cells
Pif1 centromere binding (Chen et al. 2019) and Cbf1 removal both occur in late S phase (Figure 2, A–D). Because Pif1 displaces Rap1 from DNA in vitro (Koc et al. 2016), and Rrm3 promotes replication fork progression past stable protein complexes (Ivessa et al. 2003), we tested if one or both helicases affect Cbf1 removal from centromeres. In asynchronous cells, Cbf1 binding at all three centromeres was not significantly different from its binding in WT cells in either rrm3Δ or rnh1Δ cells (Figure 4, G–I). However, Cbf1 binding was significantly higher in pif1-m2 than in WT cells (Figure 4, G–I; P < 0.01 at CEN3 and 9, and P < 0.05 at CEN13). Levels of Cbf1 binding in rrm3Δ pif1-m2 cells were similar to levels in pif1-m2 cells. These data suggest that Pif1, but not Rrm3, promotes removal of Cbf1 in late S phase. Because Pif1 binds centromeres after centromere replication (Chen et al. 2019), Cbf1 removal likely occurs after centromere replication (discussed in more detail below). Also, the increased abundance of cen-RNA in rnh1Δ cells did not affect Cbf1 binding (Figure 4, G–I).
Mutations that increase cen-RNA are associated with increased loss of a CEN6 mini-chromosome
Our studies identified three conditions where cen-RNA levels were elevated: cbf1Δ, rnh1Δ, and pif1-m2 rrm3Δ cells (Figure 1, Figure 3, and Figure 4). Levels of cen-RNA were also higher in htz1Δ cells (Ling and Yuen 2019b). To assess if increased cen-RNA is associated with changes in chromosome stability, we determined the loss rate of a CEN6 mini-chromosome at 30° in WT and mutant cells (Figure 5A).
We reported recently that the loss rate of this CEN6 mini-chromosome was 3.4× higher in cbf1Δ (7.8% loss per cell division) and 3.0× higher in pif1Δ rrm3Δ (6.9%) compared to WT cells (2.3% loss rate) (P < 0.01 for both strains) (Chen et al. 2019). For convenience, these previously published values are shown in Figure 5A. Likewise, mini-chromosome loss was significantly higher in rnh1Δ (3.8%) and rnh1Δ rnh201Δ (4.6%) compared to WT cells (1.7× and 2× higher than WT; Figure 5A; the rate in rnh1Δ rnh201Δ was not significantly different from the rate in rnh1Δ cells; P > 0.05).
We also determined the loss rates of the CEN6 mini-chromosome in double and triple mutants. The loss rate in cbf1Δ rnh1Δ cells (9.0%) was significantly higher than in rnh1Δ cells (3.8%, P < 0.0001) or cbf1Δ cells (7.8%, P < 0.0001), indicating that Rnh1 and Cbf1 affected mini-chromosome stability by different mechanisms, just as they affected cen-RNA levels by different mechanisms (Figure 3, A–C). Likewise, CEN6 mini-chromosome loss was higher in cbf1Δ rrm3Δ pif1Δ cells (16%) than in either cbf1Δ (7.8%) or pif1Δ rrm3Δ cells (6.9%) (P < 0.001) (Figure 5A). CEN6 mini-chromosome loss rates were similar in pif1-m2 rrm3Δ rnh1Δ (6.9%) vs. pif1-m2 rrm3Δ (7.1%) (P > 0.05) cells, but higher than in rnh1Δ (3.8%) cells (P < 0.05). Pif1 and, especially, Rrm3 promote replication through centromeres (Chen et al. 2019), which probably contributes to their role in centromere function. This interpretation is supported by the two times higher loss rate of the CEN6 mini-chromosome in rrm3Δ cells (Figure 5A), which had WT levels of cen-RNA (Figure 4, A–C). A CEN8 mini-chromosome is also lost at elevated rates in cbf1Δ (around four times higher than WT), htz1Δ (∼2.8×), and cbf1Δ htz1Δ (13.5×) cells (Ling and Yuen 2019b).
Cbf1, but not RNA–DNA hybrids, slows fork progression during centromere replication
Replication forks pause as they move through centromeres, due at least in part to the Cbf3 protein complex that is centromere-associated during most of the cell cycle (Greenfeder and Newlon 1992). From analogy with Cbf3, Cbf1 might also impede fork progression. In this model, fork progression through centromeres should be improved in cbf1Δ cells owing to the removal of a protein impediment to fork progression. Another possibility is that Cbf1 affects pausing by suppressing the production of cen-RNA. In this model, fork pausing should increase in cbf1Δ compared to WT cells, as cen-RNA levels were high throughout the cell cycle in this strain (Figure 1, B–D). In other contexts, R-loops cause fork stalling. If yeast cen-RNA is in R-loops at the time of centromere replication, R-loops might impede fork progression at centromeres. This model predicts that stalling should be stronger in rnh1Δ compared to WT cells.
To test these possibilities, we used 2D gel electrophoresis to study replication through chromosomal CEN6 in WT and mutant cells (Figure 5, B–L). The 2D gel analysis showed that pausing was reduced at CEN6 in cbf1Δ cells (∼0.4× WT; Figure 5E; the numbers in the upper right-hand corner of the 2D gel panels indicate the levels of pausing at the centromere relative to pausing in WT cells). These data suggest that the physical presence of Cbf1 is responsible for about one-half of the replication pausing at the chromosome VI centromere. This interpretation is supported by 2D gel analysis in cbf1Δ tof1Δ cells (Figure 5G). Tof1 stabilizes nonnucleosomal protein complexes, making them greater obstacles for fork progression (Hodgson et al. 2007). As fork stalling at CEN6 was almost undetectable in the double mutant (Figure 5G), the negative impact of Cbf1 on fork progression is due to its physical presence. An inference from these data is that Cbf1 is centromere-associated at the time of centromere replication. This finding supports the conclusion that centromere transcription occurs after DNA replication, as inferred from the timing of its appearance (Figure 1) and the timing of Cbf1 removal (Figure 2). Additionally, fork pausing at CEN6 was similar in rnh1Δ and WT cells (0.9× WT; Figure 5H), and in rnh1Δ cbf1Δ and cbf1Δ cells (0.4× WT; Figure 5I). Thus, R-loops are not responsible for fork pausing at centromeres.
Rrm3 promotes fork progression past protein complexes at many sites, including centromeres. Thus, pausing at centromeres is around three times higher in its absence than in WT cells (Ivessa et al. 2003; Chen et al. 2019) (see also Figure 5J). About one-half of this pausing remained in cbf1Δ rrm3Δ cells (1.5× WT; Figure 5K). These data indicate that Rrm3 promotes replication past CEN-bound Cbf1, but Cbf1 is not the only Rrm3-sensitive replication obstacle at centromeres. Pausing was reduced even more in cbf1Δ rrm3Δ tof1Δ cells (0.6× WT; Figure 5L). Together, the 2D gels show that proteins, not R-loops, cause pausing at centromeres. These data also establish that defects in fork progression do not explain increased mini-chromosome loss in either cbf1Δ or rnh1Δ cells (but they could in rrm3Δ cells) (Figure 5A).
Cbf1 is required for proper levels of Cse4 binding, especially in G1 and S phase
Although CENP-A interacts with cen-RNA in flies, mouse, humans, and corn (Caceres-Gutierrez and Herrera 2017; Talbert and Henikoff 2018), we did not detect cen-RNA association with immunoprecipitated epitope-tagged Cse4 in WT or cbf1Δ cells (A. Chan, data not shown). As an alternative approach to detect an impact of cen-RNAs on Cse4, we asked if elevated levels of cen-RNA altered Cse4 centromere binding. We used ChIP to determine Cse4 binding to six different centromeres in WT and mutant cells, where cen-RNA was more abundant throughout the cell cycle (cbf1Δ) or only in late S/G2 phase (rnh1Δ and pif1-m2 rrm3Δ cells; Figure 6). The tested mutations increased cen-RNA levels to different extents: 5–12× (cbf1Δ), ∼2× (rnh1Δ), or ∼2–3× (pif1-m2 rrm3Δ) (Figure 1, Figure 3, and Figure 4). As a negative control, we examined Cse4 binding to the right telomere of chromosome VI (Figure 6G).
Figure 6.
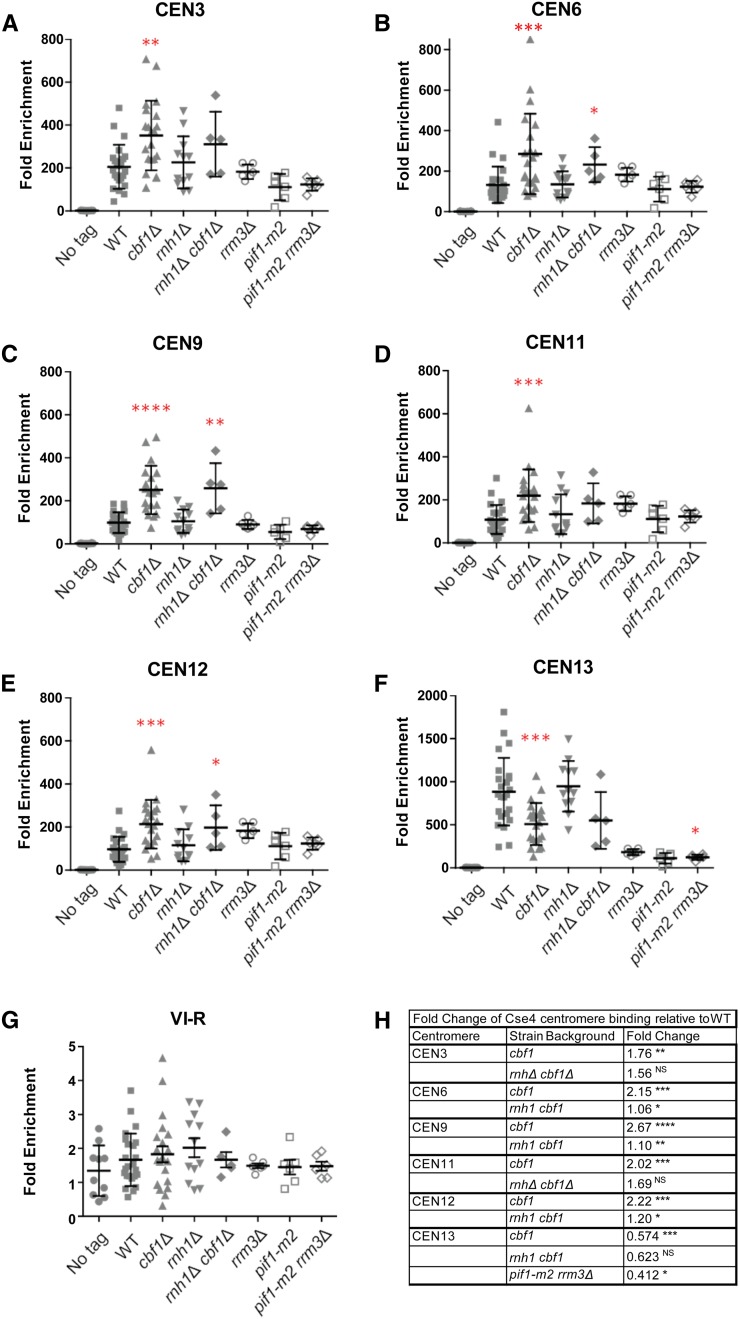
Altered levels of cen-RNA correlate with changes in Cse4 centromere binding. Scatter plots of Cse4-Myc9 binding to centromeres in asynchronous WT and mutant cells growing in YPD at 30°, as determined by ChIP. (A) CEN3, (B) CEN6, (C) CEN9, (D) CEN11, (E) CEN12, (F) CEN13, and (G) VI-R telomere (control). Quantified as ([ChIP/Input] Target site/[ChIP/Input] ARO1). Cross bars indicate means and SD. The paired two sample for means Student’s t-test, with the hypothesized mean difference set at 0, was used to calculate significance. In all figures, * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, and **** P ≤ 0.0001. Graphs show data points from several ChIP experiments consisting of three biological replicates per experiment. (H) Table showing centromeres and strains where there were significant changes in Cse4 binding compared to binding at the same CEN in WT cells. The third column shows fold change in Cse4 binding in the indicated mutants relative to WT. The scales for CEN13 and telomere VI-R are different from those of other panels in the figure. cen-RNA, centromeric RNA; ChIP, chromatin immunoprecipitation; NS, no significance; WT, wild-type.
Compared to WT, at five of the six centromeres, Cse4 binding was significantly higher in cbf1Δ cells (1.8–2.7× higher; P-values 0.01–0.0001; Figure 6, A–E), while at CEN13, Cse4 binding was significantly lower in cbf1Δ cells (0.6× WT; Figure 6F; P = 0.001) (see Figure 6H for fold differences in binding and individual P-values.) However, the level of Cse4 binding to CEN13 in WT cells was ∼5–10× higher than at the other five centromeres. Therefore, even though Cse4 binding to CEN13 was lower in cbf1Δ compared to WT cells, this level of binding was still higher than the increased Cse4 binding to the other five centromeres in cbf1Δ cells. There were no significant effects of rnh1Δ, rrm3Δ, or pif1-m2 (Figure 6) on Cse4 binding at any of the six centromeres (P > 0.05). In pif1-m2 rrm3Δ cells, Cse4 binding was significantly changed (P < 0.05) only at CEN13, where Cse4 binding was 0.4× lower than in WT cells (Figure 6, F and H).
There are at least two explanations for why Cse4 binding was perturbed in cbf1Δ but not rnh1Δ or pif1-m2 rrm3Δ cells. First, cen-RNA levels were higher in cbf1Δ cells than in the other two mutant strains, and higher levels might be necessary to impact Cse4 binding. Second, in contrast to rnh1Δ and pif1-m2 rrm3Δ cells where the increase in cen-RNA was limited to late S/G2 phase (Figure 3 and Figure 4, D–F), cen-RNA was more abundant throughout the cell cycle in cbf1Δ cells (Figure 1, B–D). Thus, a second hypothesis to explain the effects of cbf1Δ on Cse4 binding is that it is due to the unscheduled production of cen-RNA.
To distinguish between the two possibilities, we examined Cse4 binding in rnh1Δ cbf1Δ cells, which had even higher cen-RNA levels than cbf1Δ cells (Figure 3, A–C). Thus, if more cen-RNA was the cause of altered Cse4 binding, Cse4 binding should be affected more in rnh1Δ cbf1Δ cells than in cbf1Δ cells. However, Cse4 binding was not significantly different (P > 0.05) in rnh1Δ cbf1Δ vs. cbf1Δ cells at any of the six centromeres. Together, these results suggest that it is not high cen-RNA levels per se that impact Cse4 binding, but rather its unscheduled production.
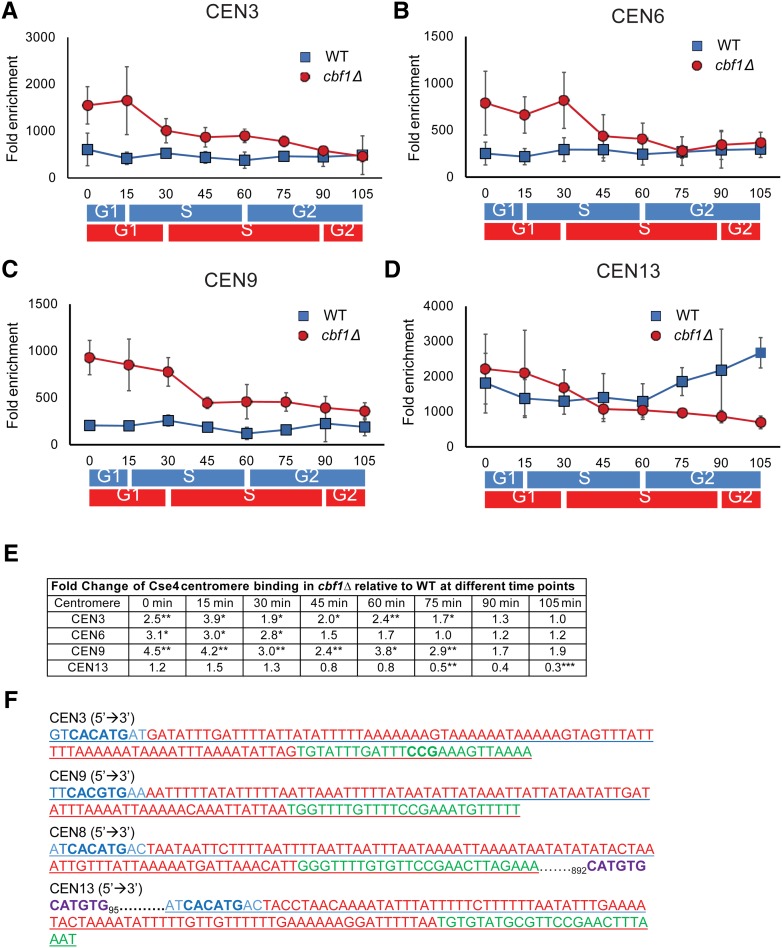
We also examined Cse4 binding to four centromeres in synchronous cultures of WT and cbf1Δ cells (Figure 7). At CEN3, 6, and 9, Cse4 binding was constant throughout the cell cycle in WT cells (Figure 7, A–C; blue squares). Again, CEN13 was different from the other centromeres: (1) in WT cells, Cse4 binding in G1 phase was ∼3–9× higher than at the other centromeres, and (2) Cse4 binding increased as cells moved through S phase to the end of the cell cycle.
Figure 7.
The timing of Cse4 binding is altered in synchronized cbf1Δ cells. Myc9-tagged Cse4 binding to different centromeres was determined by ChIP in synchronized cells grown at 24° in YPD in WT and cbf1Δ cells. Data are presented as ([ChIP/Input]Target site/[ChIP/Input]ARO). The binding of Cse4-Myc9 in WT (blue squares) and cbf1Δ (red circles) cells at (A) CEN3, (B) CEN6, (C) CEN9, and (D) CEN13. Blue and red bars indicate approximate positions in the cell cycle as determined by FACS in, respectively, WT and cbf1Δ cells (see Figure S1A). For CEN3 (A) and CEN9 (C), binding was significantly higher in cbf1Δ cells than in WT cells from 0 to 75 min (P < 0.05). For CEN6 (B), binding was significantly higher in cbf1Δ cells than in WT cells from 0 to 30 or 45 min (P < 0.05). At CEN 13 (D), Cse4 binding was significantly lower in cbf1Δ cells than in WT cells only at 75 and 105 min (P < 0.01). However, binding of Cse4 to CEN13 in WT cells was significantly higher than to any of the three other centromeres throughout the cell cycle (P < 0.05). (E) Table showing time points for each centromere where Cse4 binding was significantly different from that in WT cells. For each time point, fold change is the difference at that time point in cbf1Δ cells compared to WT. (F) Analysis of sequences for CEN3 (chosen because it has a nonpalindromic CDEI site), CEN9 (chosen because it has a palindromic CDEI site), CEN8, and CEN13. Blue letters indicate CDEI site. Red color indicates CDEII site. Green color indicates CDEIII site. Purple color indicates a second potential Cbf1-binding site. Other than CEN13, none of the other 15 centromeres has an additional CDEI consensus site < 1000 bp from the core centromere. CDE, conserved elements; ChIP, chromatin immunoprecipitation; WT, wild-type.
In the absence of Cbf1, Cse4 binding was significantly higher in G1 and through much of S phase at CEN3, 6, and 9 (1.7–4.5× increase; Figure 7, A–C; red circles). However, this increase was most dramatic in G1/early S phase. Again, the pattern was different at CEN13: Cse4 binding was similar to binding in WT cells except at two time points (75 and 105 min, P < 0.01) when binding was significantly lower than in WT cells (Figure 7D). Thus, both the timing and amount of Cse4 binding are disturbed in the absence of Cbf1.
In contrast to our results, a recent paper concluded that Cse4 chromatin binding is reduced in cbf1Δ cells (Ling and Yuen 2019b). The authors used chromosome spreads to monitor Cse4 binding and found that ∼100% of WT cells had a single cluster of Cse4 binding, while only ∼85% did in cbf1Δ cells. However, yeast centromeres are clustered in WT but to a lesser extent in cbf1Δ cells (E. Yeh and K. Bloom, personal communication). The most likely explanation for the different results by the two groups is that Cse4 binding to centromeres is difficult to detect by chromosome spreads when centromeres are not clustered. Moreover, ChIP is a much more quantitative assay than chromosome spreads as it determines binding at individual centromeres rather than on a per-cell basis.
Atypical behavior of CEN13 can be explained by its having two Cbf1-binding sites
Despite the relative uniformity of the 16 yeast centromeres, CEN13 often had different properties than the other centromeres. Like CEN3 and 9, CEN13 RNA levels were RNaseH1-sensitive in mid–late S phase (Figure 1, B–D; blue squares and Figure 3). However, the amount of cen-RNA at CEN13 was much lower than at CEN3 or CEN9 (Figure 1, E–G). The cell cycle pattern of Cbf1 binding to CEN13 was the same as at other centromeres (Figure 2, A–D), but the level of Cbf1 binding to CEN13 was two-to-three times higher than at the other centromeres (Figure 2, E–H), and deletion of CBF1 had an almost two times greater effect on the level of CEN13 cen-RNA than on other cen-RNAs (Figure 1, E–G). Compared to five other centromeres, Cse4 binding was 5–10× higher at CEN13 in WT cells (Figure 6, A–F). Thus, at CEN13, higher Cbf1 binding was associated with lower cen-RNA and higher Cse4.
An examination of the sequence of the 16 yeast centromeres suggests an explanation for the atypical behavior of CEN13 (Figure 7F). There are two Cbf1 consensus binding sites near CEN13, one at CDEI and another 97 bp upstream of CDEI (Figure 7F). We speculate that occupancy of this second site explains why there was less cen-RNA and more Cbf1 at CEN13. The only other centromere with a second nearby Cbf1 consensus binding site is CEN8, but this site is 1000 bp downstream of CDEI (Figure 7F). Because single CBF1-binding sites are only occupied near centromeres (Cai and Davis 1990; Kent et al. 2004), the second Cbf1 site at CEN8 is probably not occupied. Perhaps, the doubling of Cbf1-binding sites at CEN13 is a precursor of the centromeric heterochromatin seen at regional centromeres.
Discussion
In agreement with Ling and Yuen (2019b), we show that cen-RNA was present in very low amounts in WT yeast. Low abundance of cen-RNA is conserved, as in multicellular eukaryotes, it is estimated to be present in about a one-to-one ratio with the DNA from which it is transcribed (Talbert and Henikoff 2018). In addition, yeast cen-RNA was detected during a narrow window of the cell cycle that corresponded to mid–late S phase, which reduced its level on a per-cell basis even more (Figure 1, B–D).
Levels of cen-RNA increased 5–12×, and cell cycle-limited expression of cen-RNA was lost in cbf1Δ cells (Figure 1, B–G) (Ling and Yuen 2019b). Consistent with Cbf1 being a negative regulator of cen-RNA, Cbf1 binding to centromeres was low when cen-RNA levels were high (Figure 2, A–D). Thus, Cbf1-mediated repression of cen-RNA likely involves centromere binding. However, Cbf1 is unlikely to bind to the cen-RNA promoters, as cen-RNAs are much larger (462–1754 nt) than the core centromere (Ling and Yuen 2019b). In WT cells, Pif1 binds to all centromeres (Chen et al. 2019) at about the time that cen-RNA appeared (Figure 1). As Cbf1 binding was higher in pif1-m2 cells (Figure 4, G–I), Pif1 may use its protein eviction activity (Koc et al. 2016) to displace Cbf1 from centromeres.
Cen-RNA was also elevated in rnh1Δ (Figure 3) and pif1-m2 rrm3Δ cells (Figure 4). However, in contrast to the increased cen-RNA in cbf1Δ cells, the extent of the increase in these strains was more modest (two-to-threefold) and was limited to mid–late S phase. As the only known activity of RNAseH1 is the removal and degradation of RNA in RNA–DNA hybrids, the cen-RNA increase in rnh1Δ cells (Figure 3) suggests a model where some or all of the cen-RNA in WT cells in mid–late S phase is in R-loops. This conclusion is consistent with the Pif1/Rrm3 sensitivity of cen-RNA (Figure 4), as Pif1 family helicases are very efficient at releasing RNA from RNA–DNA hybrids (Boule and Zakian 2007; Zhou et al. 2014; Mohammad et al. 2018). An R-loop structure for cen-RNA, combined with its transient occurrence, can explain why cen-RNA is present in very low amounts, not only in S. cerevisiae but also in multicellular organisms. Thus, R-loops may be a conserved feature of cen-RNAs (Kabeche et al. 2018).
However, other data argue against an R-loop model for S. cerevisiae cen-RNA. Specifically, several groups do not detect cen-RNA in WT cells by immunoprecipitation with the S9.6 antibody that detects R-loops (Gómez-González et al. 2011; Stirling et al. 2012; Wahba et al. 2016) (our unpublished results). At this point, it is not clear how to reconcile these conflicting data. One possibility is that difficulties detecting yeast cen-RNAs with the S9.6 antibody are due to their transient occurrence, as S9.6 detects centromeric R-loops at all S. cerevisiae centromeres in hpr1Δ cells, a strain where R-loops throughout the genome have longer half-lives (Castellano-Pozo et al. 2013). Nonetheless, more data are needed to establish that S. cerevisiae cen-RNAs are in R-loops.
In addition to affecting cen-RNA, loss of Cbf1 also correlated with changes in the timing and levels of Cse4 binding (Figure 6 and Figure 7, A–D). Both high levels of cen-RNA and its unscheduled appearance were needed to affect Cse4 binding. The budding yeast centromere is associated with a single Cse4 nucleosome (Furuyama and Biggins 2007; Krassovsky et al. 2012), but there is some Cse4 in centromere-proximal regions that is not involved in kinetochore assembly (Coffman et al. 2011; Lawrimore et al. 2011; Haase et al. 2013). One explanation for the increased centromere-associated Cse4 in cbf1Δ cells is that the pool of centromere-peripheral Cse4 is higher in this background. If so, based on a shear size of ∼300 bp and the primers used to amplify centromeric DNA (Figure 1A), the additional Cse4 must be bound within ∼300 bp from the core centromere. An increase in peripheral Cse4 binding might be facilitated by the absence of Cbf1 and perhaps lead to competition for kinetochore assembly between the two closely spaced Cse4 sites, thereby impairing segregation fidelity. Another possibility is that the composition of the single Cse4 nucleosome is changed in cbf1Δ cells, as it is not clear if there are one or two Cse4 molecules per centromeric nucleosome (Biggins 2013).
In combination, the data in this paper suggest the following working model for centromere transcription. In this model, loss of Cbf1 binding in mid–late S phase allows centromere transcription. As Cbf1 was centromere-associated at the time of centromere replication, centromere transcription must occur after centromere replication (Figure 5G). The low levels of cen-RNA shown here and in Ling and Yuen (2019b) suggest that in WT cells, each centromere is transcribed no more than once (or a very few times) in a given cell cycle. If S. cerevisiae cen-RNAs remain base-paired to centromeric DNA, as they do in human cells (Kabeche et al. 2018), it would explain why there are so few rounds of transcription. Our model also proposes that the absence of cbf1 allows the occurrence of multiple rounds of centromere transcription throughout a given cell cycle, and this free cen-RNA might interfere with kinetochore assembly.
Given the ubiquitous nature of cen-RNAs, it seems reasonable that they affect one or more aspects of centromere function rather than being low-level noise in the system. Consistent with this idea, in diverse organisms, positive and negative changes in cen-RNA abundance affect chromosome stability. However, establishing direct roles for cen-RNAs is challenging. Part of the difficulty is that many of the genes whose mutation affect cen-RNA have other functions. For example, while deletion of CBF1, PIF1/RRM3, RNH1, and HTZ1 increased both cen-RNA and centromere dysfunction (Figure 5A) (Ling and Yuen 2019b), each of these genes is pleiotropic, so the increased centromere instability associated with their loss may be linked to a phenotype other than increased cen-RNA. Nonetheless, it is striking that although each of the four mutations increase cen-RNA by different mechanisms, mini-chromosome loss was elevated in each strain.
Acknowledgments
We thank A. Aguilera, K. Cimprich, D. Koshland, and B. Luke for answering questions about R-loops and providing protocols to detect them; D. Rubinstein for help with statistical analysis of the data in Figure 6; M. Basrai, S. Biggins, K. Bloom, and T. Kunkel for discussions on cen-RNAs and sharing data prior to publication; C. J. Decoste and the Princeton Molecular Biology FACS facility for help with flow cytometry; and Carly Geronimo for comments and suggestions on the manuscript. This work was funded by grant 1R35 GM-118279 to V.A.Z. from the National Institutes of Health. C.-F. C. was funded in part by a grant from the New Jersey Commission on Cancer Research (NJCCR). T.J.P. was funded in part by grants from the FORD Foundation, the Burroughs Wellcome Fund Postdoctoral Enrichment Program, and the NJCCR.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.8939873.
Communicating editor: S. Lacefield
Literature Cited
- Biggins S., 2013. The composition, functions, and regulation of the budding yeast kinetochore. Genetics 194: 817–846. 10.1534/genetics.112.145276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boule J. B., and Zakian V. A., 2007. The yeast Pif1p DNA helicase preferentially unwinds RNA DNA substrates. Nucleic Acids Res. 35: 5809–5818. 10.1093/nar/gkm613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer B. J., and Fangman W. L., 1987. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51: 463–471. 10.1016/0092-8674(87)90642-8 [DOI] [PubMed] [Google Scholar]
- Brewer B. J., and Fangman W. L., 1991. Mapping replication origins in yeast chromosomes. Bioessays 13: 317–322. 10.1002/bies.950130702 [DOI] [PubMed] [Google Scholar]
- Caceres-Gutierrez R., and Herrera L. A., 2017. Centromeric non-coding transcription: opening the black box of chromosomal instability? Curr. Genomics 18: 227–235. 10.2174/1389202917666161102095508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M., and Davis R. W., 1990. Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell 61: 437–446. 10.1016/0092-8674(90)90525-J [DOI] [PubMed] [Google Scholar]
- Castellano-Pozo M., Santos-Pereira J. M., Rondon A. G., Barroso S., Andujar E. et al. , 2013. R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Mol. Cell 52: 583–590. 10.1016/j.molcel.2013.10.006 [DOI] [PubMed] [Google Scholar]
- Cerritelli S. M., and Crouch R. J., 2009. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 276: 1494–1505. 10.1111/j.1742-4658.2009.06908.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. F., Pohl T. J., Pott S., and Zakian V. A., 2019. Two pif1-family DNA helicases cooperate in centromere replication and segregation in Saccharomyces cerevisiae. Genetics 211: 105–119. 10.1534/genetics.118.301710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman V. C., Wu P., Parthun M. R., and Wu J. Q., 2011. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J. Cell Biol. 195: 563–572. 10.1083/jcb.201106078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama S., and Biggins S., 2007. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc. Natl. Acad. Sci. USA 104: 14706–14711. 10.1073/pnas.0706985104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss R., Trautwein M., Sommer T., and Spang A., 2005. New modules for the repeated internal and N-terminal epitope tagging of genes in Saccharomyces cerevisiae. Yeast 22: 1–12. 10.1002/yea.1187 [DOI] [PubMed] [Google Scholar]
- Geronimo C. L., and Zakian V. A., 2016. Getting it done at the ends: Pif1 family DNA helicases and telomeres. DNA Repair (Amst.) 44: 151–158. 10.1016/j.dnarep.2016.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-González B., Garcia-Rubio M., Bermejo R., Gaillard H., Shirahige K. et al. , 2011. Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. EMBO J. 30: 3106–3119. 10.1038/emboj.2011.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfeder S. A., and Newlon C. S., 1992. Replication forks pause at yeast centromeres. Mol. Cell. Biol. 12: 4056–4066. 10.1128/MCB.12.9.4056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase J., Mishra P. K., Stephens A., Haggerty R., Quammen C. et al. , 2013. A 3D map of the yeast kinetochore reveals the presence of core and accessory centromere-specific histone. Curr. Biol. 23: 1939–1944. 10.1016/j.cub.2013.07.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., and Henikoff J. G., 2012. “Point” centromeres of Saccharomyces harbor single centromere-specific nucleosomes. Genetics 190: 1575–1577. 10.1534/genetics.111.137711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson B., Calzada A., and Labib K., 2007. Mrc1 and Tof1 regulate DNA replication forks in different ways during normal S phase. Mol. Biol. Cell 18: 3894–3902. 10.1091/mbc.e07-05-0500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman J. A., Spotila L. D., Nawotka K. A., el-Assouli S. M., and Davis L. R., 1987. The in vivo replication origin of the yeast 2 microns plasmid. Cell 51: 473–481. 10.1016/0092-8674(87)90643-X [DOI] [PubMed] [Google Scholar]
- Ivessa A. S., Lenzmeier B. A., Bessler J. B., Goudsouzian L. K., Schnakenberg S. L. et al. , 2003. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol. Cell 12: 1525–1536. 10.1016/S1097-2765(03)00456-8 [DOI] [PubMed] [Google Scholar]
- Kabeche L., Nguyen H. D., Buisson R., and Zou L., 2018. A mitosis-specific and R loop-driven ATR pathway promotes faithful chromosome segregation. Science 359: 108–114. 10.1126/science.aan6490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent N. A., Eibert S. M., and Mellor J., 2004. Cbf1p is required for chromatin remodeling at promoter-proximal CACGTG motifs in yeast. J. Biol. Chem. 279: 27116–27123. 10.1074/jbc.M403818200 [DOI] [PubMed] [Google Scholar]
- Koc K. N., Singh S. P., Stodola J. L., Burgers P. M., and Galletto R., 2016. Pif1 removes a Rap1-dependent barrier to the strand displacement activity of DNA polymerase delta. Nucleic Acids Res. 44: 3811–3819. 10.1093/nar/gkw181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krassovsky K., Henikoff J. G., and Henikoff S., 2012. Tripartite organization of centromeric chromatin in budding yeast. Proc. Natl. Acad. Sci. USA 109: 243–248. 10.1073/pnas.1118898109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L., and Thomas D., 1995. Identification of the yeast methionine biosynthetic genes that require the centromere binding factor 1 for their transcriptional activation. FEBS Lett. 367: 15–18. 10.1016/0014-5793(95)00528-H [DOI] [PubMed] [Google Scholar]
- Lawrimore J., Bloom K. S., and Salmon E. D., 2011. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J. Cell Biol. 195: 573–582. 10.1083/jcb.201106036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Y. H., and Yuen K. W. Y., 2019a Centromeric non-coding RNA as a hidden epigenetic factor of the point centromere. Curr. Genet. DOI: 10.1007/s00294-019-00988-6. 10.1007/s00294-019-00988-6 [DOI] [PubMed] [Google Scholar]
- Ling Y. H., and Yuen K. W. Y., 2019b Point centromere activity requires an optimal level of centromeric noncoding RNA. Proc. Natl. Acad. Sci. USA 116: 6270–6279. 10.1073/pnas.1821384116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik H. S., and Henikoff S., 2002. Conflict begets complexity: the evolution of centromeres. Curr. Opin. Genet. Dev. 12: 711–718. 10.1016/S0959-437X(02)00351-9 [DOI] [PubMed] [Google Scholar]
- Mellor J., Rathjen J., Jiang W., Barnes C. A., and Dowell S. J., 1991. DNA binding of CPF1 is required for optimal centromere function but not for maintaining methionine prototrophy in yeast. Nucleic Acids Res. 19: 2961–2969. 10.1093/nar/19.11.2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh P. B., and Koshland D., 1997. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 11: 3401–3412. 10.1101/gad.11.24.3401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh P. B., Yang P., Glowczewski L., Koshland D., and Smith M. M., 1998. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94: 607–613. 10.1016/S0092-8674(00)81602-5 [DOI] [PubMed] [Google Scholar]
- Mohammad J. B., Wallgren M., and Sabouri N., 2018. The Pif1 signature motif of Pfh1 is necessary for both protein displacement and helicase unwinding activities, but is dispensable for strand-annealing activity. Nucleic Acids Res. 46: 8516–8531. 10.1093/nar/gky654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy A. D., and Cech T. R., 2006. Low abundance of telomerase in yeast: implications for telomerase haploinsufficiency. RNA 12: 1721–1737. 10.1261/rna.134706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K., Chen C., and Kolodner R. D., 2001. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411: 1073–1076. 10.1038/35082608 [DOI] [PubMed] [Google Scholar]
- Natsume T., Muller C. A., Katou Y., Retkute R., Gierlinski M. et al. , 2013. Kinetochores coordinate pericentromeric cohesion and early DNA replication by Cdc7-Dbf4 kinase recruitment. Mol. Cell 50: 661–674. 10.1016/j.molcel.2013.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedenthal R. K., Sen-Gupta M., Wilmen A., and Hegemann J. H., 1993. Cpf1 protein induced bending of yeast centromere DNA element I. Nucleic Acids Res. 21: 4726–4733. 10.1093/nar/21.20.4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell K. F., Surdin-Kerjan Y., and Baker R. E., 1995. Role of the Saccharomyces cerevisiae general regulatory factor CP1 in methionine biosynthetic gene transcription. Mol. Cell. Biol. 15: 1879–1888. 10.1128/MCB.15.4.1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkuni K., and Kitagawa K., 2011. Endogenous transcription at the centromere facilitates centromere activity in budding yeast. Curr. Biol. 21: 1695–1703. 10.1016/j.cub.2011.08.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmundson J. S., Kumar J., Yeung R., and Smith D. J., 2017. Pif1-family helicases cooperatively suppress widespread replication-fork arrest at tRNA genes. Nat. Struct. Mol. Biol. 24: 162–170. 10.1038/nsmb.3342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl T. J., and Zakian V. A., 2019. Pif1 family DNA helicases: a helpmate to RNase H? DNA Repair (Amst.). DOI: 10.1016/j.dnarep.2019.06.004. 10.1016/j.dnarep.2019.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeyre C., Lopes J., Boule J. B., Piazza A., Guedin A. et al. , 2009. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 5: e1000475 10.1371/journal.pgen.1000475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz V. P., and Zakian V. A., 1994. The saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell 76: 145–155. 10.1016/0092-8674(94)90179-1 [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., and Hieter P., 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner F. A., and Henikoff S., 2015. Diversity in the organization of centromeric chromatin. Curr. Opin. Genet. Dev. 31: 28–35. 10.1016/j.gde.2015.03.010 [DOI] [PubMed] [Google Scholar]
- Stirling P. C., Chan Y. A., Minaker S. W., Aristizabal M. J., Barrett I. et al. , 2012. R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev. 26: 163–175. 10.1101/gad.179721.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S., Keith K. C., Curnick K. E., and Fitzgerald-Hayes M., 1995. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 9: 573–586. 10.1101/gad.9.5.573 [DOI] [PubMed] [Google Scholar]
- Talbert P. B., and Henikoff S., 2018. Transcribing centromeres: noncoding RNAs and kinetochore assembly. Trends Genet. 34: 587–599. 10.1016/j.tig.2018.05.001 [DOI] [PubMed] [Google Scholar]
- Thomas D., Jacquemin I., and Surdin-Kerjan Y., 1992. MET4, a leucine zipper protein, and centromere-binding factor 1 are both required for transcriptional activation of sulfur metabolism in Saccharomyces cerevisiae. Mol. Cell. Biol. 12: 1719–1727. 10.1128/MCB.12.4.1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran P. L. T., Pohl T. J., Chen C. F., Chan A., Pott S. et al. , 2017. PIF1 family DNA helicases suppress R-loop mediated genome instability at tRNA genes. Nat. Commun. 8: 15025 10.1038/ncomms15025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdaasdonk J. S., and Bloom K., 2011. Centromeres: unique chromatin structures that drive chromosome segregation. Nat. Rev. Mol. Cell Biol. 12: 320–332. 10.1038/nrm3107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba L., Costantino L., Tan F. J., Zimmer A., and Koshland D., 2016. S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev. 30: 1327–1338. 10.1101/gad.280834.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. A., Kwon Y., Xu Y., Chung W. H., Chi P. et al. , 2013. Pif1 helicase and Poldelta promote recombination-coupled DNA synthesis via bubble migration. Nature 502: 393–396. 10.1038/nature12585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski J., Hajj B., Chen J., Mizuguchi G., Xiao H. et al. , 2014. Imaging the fate of histone Cse4 reveals de novo replacement in S phase and subsequent stable residence at centromeres. Elife 3: e02203 10.7554/eLife.02203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R., Zhang J., Bochman M. L., Zakian V. A., and Ha T., 2014. Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA. Elife 3: e02190 10.7554/eLife.02190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer A. D., and Koshland D., 2016. Differential roles of the RNases H in preventing chromosome instability. Proc. Natl. Acad. Sci. USA 113: 12220–12225. 10.1073/pnas.1613448113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, tables, and Supplemental Materials. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.8939873.