Abstract
The diversity in sperm shape and size represents a powerful paradigm to understand how selection drives the evolutionary diversification of cell morphology. Experimental work on the sperm biology of the male-hermaphrodite nematode Caenorhabditis elegans has elucidated diverse factors important for sperm fertilization success, including the competitive superiority of larger sperm. Yet despite extensive research, the molecular mechanisms regulating C. elegans sperm size and the genetic basis underlying natural variation in sperm size remain unknown. To address these questions, we quantified male sperm size variation of a worldwide panel of 97 genetically distinct C. elegans strains, allowing us to uncover significant genetic variation in male sperm size. Aiming to characterize the molecular genetic basis of C. elegans male sperm size variation using a genome-wide association study, we did not detect any significant quantitative trait loci. We therefore focused on the genetic analysis of pronounced sperm size differences observed between recently diverged laboratory strains (N2 vs. LSJ1/2). Using mutants and quantitative complementation tests, we demonstrate that variation in the gene nurf-1 underlies the evolution of small sperm in the LSJ lineage. Given the previous discovery that this same nurf-1 variation was central for hermaphrodite laboratory adaptation, the evolution of reduced male sperm size in LSJ strains likely reflects a pleiotropic consequence. Together, our results provide a comprehensive quantification of natural variation in C. elegans sperm size and first insights into the genetic determinants of Caenorhabditis sperm size, pointing at an involvement of the NURF chromatin remodeling complex.
Keywords: Androdioecy, Caenorhabditis, male function, nurf-1, NURF chromatin remodeling complex, sperm competition, sperm dimorphism
SPERM morphology can show extreme variation and is often associated with variation in competitive ability and thus male reproductive success (Smith 1984; Birkhead and Moller 1998; Snook 2005; Birkhead et al. 2009; Pitnick et al. 2009; Ramm et al. 2014) . Furthermore, disparities of specific sperm traits, such as cell size or flagellum length, are not only common among species, but also within species (Pitnick et al. 2009). Numerous studies focusing on intraspecific variation, through comparison of sperm traits across populations or by using artificial selection on sperm traits, have uncovered extensive levels of heritable variation in diverse sperm characteristics (Ward 1998; Morrow and Gage 2001a; Joly et al. 2004; Pitnick et al. 2009; Simmons and Moore 2009). Despite such studies on diverse invertebrate and vertebrate taxa, the quantitative and molecular genetic architecture of sperm traits associated with competitive ability remain largely undescribed. Therefore, although the developmental genetics of spermatogenesis has been elucidated in great detail from model organisms, such as the fly Drosophila melanogaster (Demarco et al. 2014) or the nematode Caenorhabditis elegans (Ellis and Stanfield 2014), it is largely unknown whether uncovered genes are also a substrate for evolution to affect intraspecific variation in sperm characteristics relevant for competitive ability.
Here we aimed to quantify and characterize intraspecific genetic variation of a well-defined sperm trait, cell size, known to modulate sperm competitive ability in C. elegans. In this androdioecious (male-hermaphrodite) species, both males and hermaphrodites produce sperm, so that hermaphrodites can either self-fertilize or outcross with males. C. elegans shows a pronounced sperm size dimorphism: male sperm are larger and consistently outcompete smaller hermaphrodite sperm when both types of sperm are present in the hermaphrodite reproductive tract (Ward and Carrel 1979; LaMunyon and Ward 1995). Male sperm size is also critical when multiple males compete for fertilization, with larger sperm often outcompeting smaller sperm (LaMunyon and Ward 1998; Murray et al. 2011). In addition, increased levels of male-male competition in experimental contexts can lead to the evolution of larger male sperm size (LaMunyon and Ward 2002; Palopoli et al. 2015), consistent with the relevance of sperm size for male competitive ability. Although self-fertilization is the predominant mode of C. elegans reproduction, with rare occurrence of males and outcrossing events in natural populations (Jovelin et al. 2003; Barrière and Félix 2005; Sivasundar and Hey 2005), ample natural variation in diverse male traits exists (Hodgkin and Doniach 1997; Teotónio et al. 2006; Palopoli et al. 2008, 2015; Morran et al. 2009; Anderson et al. 2010; Noble et al. 2015; Alcorn et al. 2016), including male sperm size (Ward and Carrel 1979; LaMunyon and Ward 1995, 1998, 1999, 2002; Murray et al. 2011; Palopoli et al. 2015). Moreover, gonochoristic (male-female) Caenorhabditis species exhibit, on average, much larger male sperm than the three androdioecious species, C. briggsae, C. elegans, and C. tropicalis, in which male-male competition is much weaker (LaMunyon and Ward 1999; Vielle et al. 2016).
While the size of amoeboid Caenorhabditis sperm can be a critical factor for sperm competitive ability, recent work has uncovered crucial roles of genetic factors in Caenorhabditis sperm competition that act independently of sperm size (Thomas et al. 2012; Ting et al. 2014, 2018; Fierst et al. 2015; Hansen et al. 2015; Yin et al. 2018; Yin and Haag 2019). Most prominently, genome shrinkage observed in the three androdioecious Caenorhabditis species involves a strong bias in the loss of male-expressed genes, including the parallel loss of the male secreted short (mss) gene family, which is critical for sperm competitive ability in gonochoristic species (Thomas et al. 2012; Fierst et al. 2015; Yin et al. 2018). Hence, not only sperm size, but also diverse cellular and genetic components jointly determine sperm competitive ability in C. elegans and other Caenorhabditis species. One question emerging from this updated view on C. elegans sperm competition is thus whether natural variation in C. elegans sperm size is indeed closely linked to variation in male fertilization success and male-male competitive ability.
Another large gap in our understanding of C. elegans sperm competition is the absence of information on the molecular genetic determinants of sperm size and its natural variation. Although the genetic regulation of spermatogenesis has been elucidated in great detail (L’Hernault 2006; Geldziler et al. 2011; Chu and Shakes 2013; Ellis and Stanfield 2014), no specific genes regulating C. elegans sperm size have so far been identified. Spermatogenesis of C. elegans hermaphrodites and males seem essentially identical: meiosis is initiated by the formation of primary spermatocytes, followed by two rapid, mostly symmetrical divisions resulting in four haploid spermatids and an anucleate residual body (Ward et al. 1981; Shakes et al. 2009; Vielle et al. 2016). The cell size of the primary spermatocyte is a key determinant of final spermatid size (Vielle et al. 2016). Therefore, sperm size seems to be mostly determined prior to or at the time of primary spermatocyte formation. However, it remains unknown how genetic factors contribute to this process. In addition, it is unclear to what extent the differential presence or activity of such potential genes explain reported differences in sperm size across sexes, genotypes, or species.
In this study, we therefore focused on the characterization of natural variation and its genetic basis in C. elegans male sperm size, using a worldwide collection of nearly 100 wild isolates (Andersen et al. 2012; Cook et al. 2017). First, we tested how well observed natural variation in male sperm size correlates with variation in male reproductive performance, as well as with morphological traits of both males and hermaphrodites, such as body size and hermaphrodite sperm size. Second, we aimed to characterize the molecular genetic basis of intraspecific variation in C. elegans male sperm, by performing a genome-wide association study and an in-depth genetic analysis of recently diverged laboratory strains that display strong sperm size differences.
Materials and Methods
Strains and culture conditions
All strains were maintained at 20° on 2.5% agar Nematode Growth Medium (NGM) plates seeded with the Escherichia coli strain OP50 (Stiernagle 2006). The following strains/genotypes were used in this study: 95 C. elegans wild isolates (Supplemental Material, Table S1) (Andersen et al. 2012; Cook et al. 2017) and laboratory strains N2, LSJ1, LSJ2, CX12311(kyIR1 V, CB4856 > N2; qqIR1 X, CB4856 > N2), CX13248 (kyIR84 II, LSJ2 > N2), nurf-1(n4295) (MT13649), isw-1(n3294) (MT17795), isw-1(n3297) (MT16012), pyp-1(n4599) IV/nT1 [qIs51] (MT14910), PD4790, and C. plicata (SB355). Males homozygous for pyp-1 die during larval stages and were thus scored as heterozygotes. Hermaphrodites of the mutant fog-2(q71) (strain CB4108) do not produce any self-sperm, i.e., they are effectively females (Schedl and Kimble 1988), which were used for certain mating assays. The strain PD4790 contains an integrated transgene [mls12 (myo-2::GFP, pes-10::GFP, F22B7.9::GFP)] in the N2 reference genetic background, expressing green fluorescent protein (GFP) in the pharynx. Additional information on wild isolates is available from C. elegans Natural Diversity Resource (CeNDR; http://elegansvariation.org) (Cook et al. 2017)
Measurements of male and hermaphrodite sperm size
Males were collected from strain cultures at the L4 stage to be maintained on NGM plates containing only males to measure their spermatid size at stage L4 + 24 hr from synchronized and unmated males. Hermaphrodite spermatids were dissected from young virgin adults (at around mid-L4 + 24 hr), at which stage most individuals contained both spermatids and activated sperm (spermatozoa), the latter of which were excluded from analysis. To measure sperm size, male or hermaphrodite spermatids were released into sperm medium (50 mM HEPES, pH 7.8, 50 mM NaCl, 25 mM KCl, 5 mM CaCl2, 1 mM MgSO4, 1 mg/ml BSA) by needle dissection (Nelson and Ward 1980). Images of the spermatids were captured using Nomarski optics (×60 or ×63 objectives). ImageJ software (Rasband, 1997–2014) was used to calculate length and width of each spermatid to obtain measures of cross-sectional area assuming an ellipse shape: π × (length/2) × (width/2) (Vielle et al. 2016).
Male mating ability
All mating assays (Figure 3 and Figure 4) were performed on mating plates (35 mm diameter NGM plates seeded with a spot of 20 µl OP50) using unmated males, fog-2 females, or hermaphrodites that had been isolated at the L4 larval stage 24 hr (hermaphrodites and females) or 36 hr (males) prior to the assay. Following established protocols (Wegewitz et al. 2008; Murray et al. 2011), 10 virgin fog-2 females were transferred onto a mating plate and allowed to roam for 30 min on the bacterial lawn. Next, a single male was added to the plate and allowed to mate for 8 hr, after which it was discarded. Females were left on the mating plate for an additional hour and were then picked as single animals onto fresh NGM plates. Females were scored as not fertilized if they did not contain any embryos in the uterus 24 hr later. Offspring production of fertilized females was followed over four consecutive days.
Figure 3.
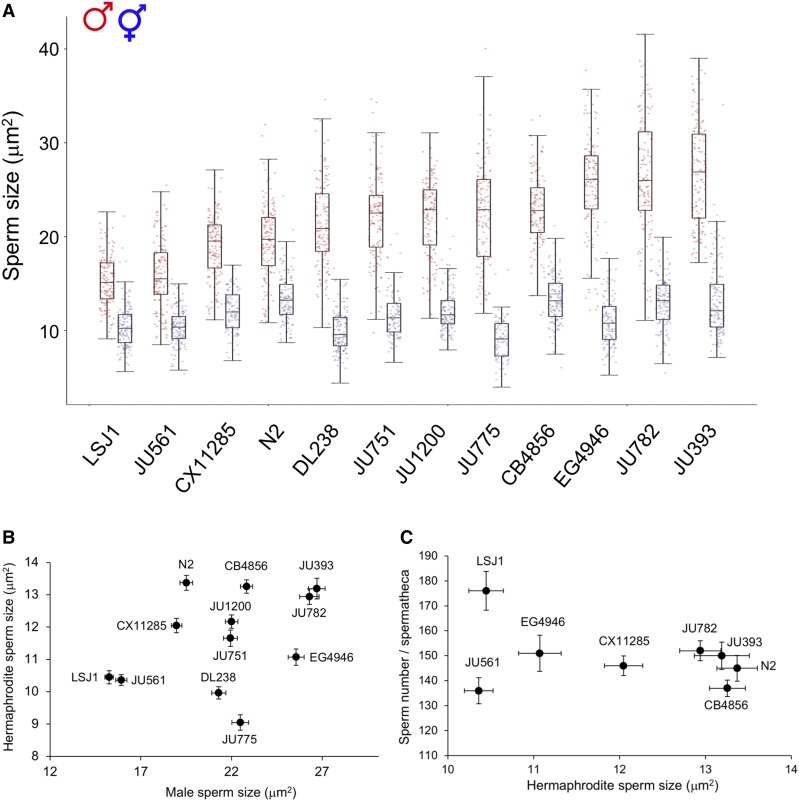
No correlation of C. elegans male and hermaphrodite sperm size across strains. (A) Hermaphrodite sperm size across 12 strains. Strains with smallest (LSJ1) to largest (JU393) male sperm are arranged from left to right. Effect of strain genotype and sex on C. elegans sperm size (ANOVA, effect sex: F1,3207 = 6267.55, P < 0.0001; effect strain: F11, 3207 = 107.14, P < 0.0001; interaction sex × strain: F11, 3207 = 50.90, P < 0.0001). For each strain, 87–152 hermaphrodite spermatids from 7 to 13 individuals were measured (male sperm size data are the same as shown in Figure 1). (B) Absence of significant correlation between male and hermaphrodite sperm size across 12 strains, inferred from least-squares regression of strain mean values (F1, 11 = 1.38, R2 = 0.11, P = 0.27). (C) No correlation between average hermaphrodite sperm size and sperm number across eight strains (F1, 7 = 0.59, R2 = 0.10, P = 0.43). Hermaphrodite self-sperm number were established by measuring all sperm contained within a single spermatheca of 12–31 individuals per strain.
Figure 4.
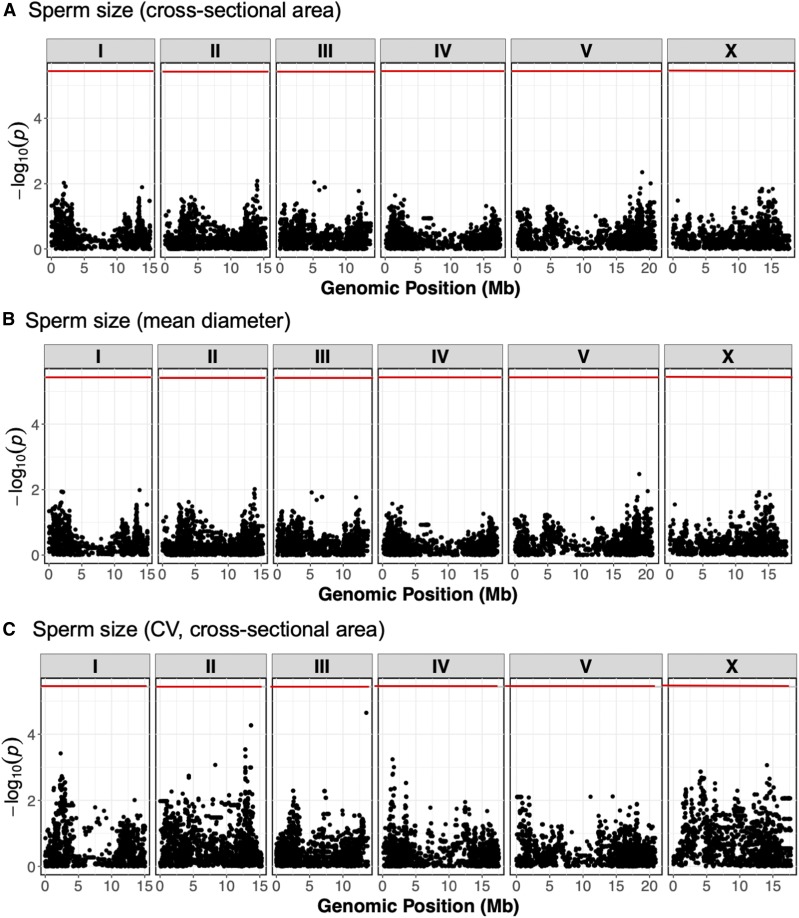
Genome-wide association mapping for C. elegans male sperm size. Manhattan plots of single-marker based GWA mappings show no significant genomic regions for least-squares mean estimates (LSM) of (A) sperm cross-sectional area, (B) sperm mean diameter and (C) coefficient of variation (CV) (sperm cross-sectional area). Each dot represents an SNV that is present in at least 5% of the assayed population. The genomic location of each SNV is plotted on the x-axis, and the statistical significance is plotted on the y-axis. The Bonferroni-corrected significance threshold is shown as a red horizontal line.
Male sperm number transferred during single insemination (ejaculate size)
We quantified the number of sperm transferred during single insemination events (Figure 2H and Figure S1, E–H). For each strain to be tested (N2, CB4856, LSJ1, JU561, CX11285, EG4946, JU393, and JU782), unmated fog-2 females were individually mated with an excess of 20–30 males, aged 36 hr post-L4 larval stage, to increase the chance of mating. Mating was monitored every few minutes by observation through the stereoscope. When a male was engaged in mating, it was kept under constant surveillance for spicule insertion and visualization of sperm flow from the male vas deferens to the female uterus, until mating was completed. Immediately after the end of mating, the inseminated female was isolated and fixed in ice-cold methanol and the mated male was removed from the male pool. A new virgin female was then mated with the males. Next, fixed females were washed twice in M9 and mounted in DAPI-containing Vectashield (Vector Laboratories, Burlingame, CA). Sperm number was counted on images taken at ×40 magnification as Z-stacks covering the entire thickness of the gonad using an Olympus BX61 microscope with a CoolSnap HQ2 camera (Poullet et al. 2015, 2016).
Figure 2.
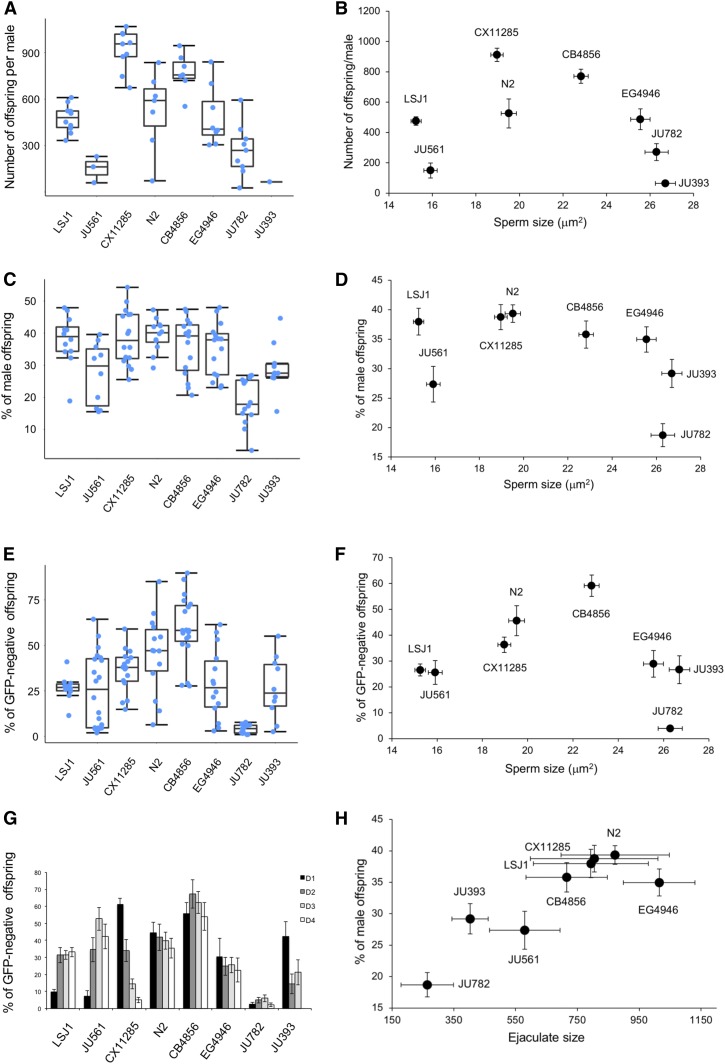
Covariation of male sperm size with male reproductive performance. Male reproductive performance in eight C. elegans strains with different average male sperm size. Strains with smallest (LSJ1) to largest (JU393) male sperm are arranged from left to right. (A) Significant strain variation in the number of offspring sired by a single male during 8 hr of mating with up to 10 fog-2 females (Kruskal–Wallis, χ2 = 37.78, df = 7, P < 0.0001) and (B) absence of correlation with average male sperm size (ρSpearman = −0.29, P = 0.49). (C) Significant strain variation in male fertilization success when competing with hermaphrodite self-sperm (strain CB4856) (ANOVA, F7,103 = 11.20, P < 0.0001) and (D) absence of correlation between male fertilization success and male sperm size (ρSpearman = −0.45, P = 0.45). (E) Significant strain variation in male-male competitive ability (in fertilization success) sperm (ANOVA, F7,113 = 14,66, P < 0.0001). Male competitive ability of a given strain (vs. the GFP-positive strain PD4790) quantified by the proportion of GFP-negative offspring produced over 4 days after mating. (F) No correlation between competitive ability and male sperm size (ρSpearman = 0.00, P = 1). (G) Details of time course of progeny production across the 4 days after mating event (same data as in E). (H) Ejaculate size, as measured by the number of sperm deposited by one male in a single mating (same data as in Figure S1E), and male fertilization success (C) show a significant correlation (ρSpearman = 0.74, P = 0.036). Sample sizes: n = 10–20 per strain per experiment. Box and whiskers plots: boxes are delimited by the data’s first and third quartiles, broken by a band at the median, and flanked by whiskers of which length is equal to 1.5 × the interquartile range.
Hermaphrodite-male sperm competition
We measured variation in competition between hermaphrodite and male sperm by measuring male fertilization success of the eight strains when mated to hermaphrodites of a tester strain (the wild isolate CB4856) (Figure 4A). L4 hermaphrodites of the CB4856 strain were isolated 24 hr prior to mating, and L4 males of the eight strains were isolated 36 hr prior to mating. On the next day, one single CB4856 adult hermaphrodite was mated with an excess of 20–30 males for each strain to be tested and kept under surveillance for single mating as described above. As soon as mating was completed, the male was discarded from the male pool and the hermaphrodite was isolated onto a fresh NGM plate. Offspring (and male) production was scored for 3 days after the mating assay (i.e., until completion of the reproductive span).
Male-male sperm competition
We measured second-male sperm precedence of the eight C. elegans strains using a tester strain expressing GFP in the pharynx (PD4790), following previously used protocols (Murray et al. 2011) (Figure 2, E–G). fog-2 females were first mated with PD4970 males, then with males of the eight strains. Males and fog-2 females were isolated at the L4 stage and maintained in isolation for 36 hr prior to mating assays. Each mating plate (N = 20) was established by adding 10 fog-2 females and 20 PD4790 males, which were allowed to mate for 15 hr, so that all females were fertilized (confirmed by the presence of embryos in the uterus). Ten fertilized fog-2 females were then randomly allocated to each new mating plate and allowed to mate with 20 males of each of the eight strains examined. Plates were kept under surveillance for single mating as described above. Upon completion of a mating event, both male and female were removed, and offspring production of the female was observed for the next 4 days. Total offspring were counted using a regular stereoscope and GFP-expressing offspring were counted with a fluorescence stereoscope.
Quantification of hermaphrodite self-sperm number
We quantified the number of self-sperm in synchronized young adult hermaphrodites, i.e., adults containing one or two embryos in their uterus (Figure 5C). Animals were fixed overnight in ice-cold methanol (−20°), washed three times in 1× PBS containing 0.05% Tween and mounted in Vectashield (Vector Laboratories) supplemented with DAPI. Sperm images were acquired from adults containing oocytes to ensure that the sperm to oocyte transition had occurred. Imaging of the anterior spermatheca was performed with an Olympus BX61 microscope using a ×63 objective with epifluorescence. Z-sections (1 µm) of the entire spermatheca were taken and sperm number counted (cell counter plugin in ImageJ) (Poullet et al. 2015, 2016)
Figure 5.
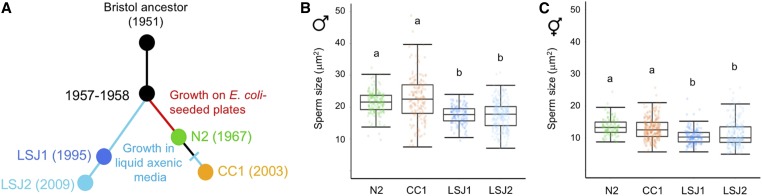
LSJ1 and LSJ2 strains exhibit strongly reduced male and hermaphrodite sperm size. (A) Laboratory evolution of LSJ and N2 lineages in the laboratory, after isolation of the common ancestral strain “Bristol,” derived from a single hermaphrodite individual, in 1951. LSJ1 and LSJ2 were cultivated in axenic liquid medium and N2 was cultivated on agar plates [after McGrath et al. (2011)]. (B) Male sperm size: LSJ1 and LSJ2 exhibit significantly reduced male sperm size compared to N2 and CC1 (ANOVA, effect strain: F3, 658 = 65.62, P < 0.0001). (C) Hermaphrodite sperm size: LSJ1 and LSJ2 exhibit significantly reduced hermaphrodite sperm size compared to N2 and CC1 (ANOVA, effect strain: F3, 666 = 40.95, P < 0.0001). For male sperm measurements, 135–225 sperm were analyzed from 9 to 15 individuals of each strain. For hermaphrodite sperm measurements, 123–248 sperm were analyzed from 9 to 19 individuals of each strain. Values with the same letter are not significantly different from each other (Tukey’s honestly significant difference, P < 0.05). Box and whiskers plots: boxes are delimited by the data’s first and third quartiles, broken by a band at the median, and flanked by whiskers of which length is equal to 1.5 × the interquartile range.
Body size measurements
Synchronized populations were used to isolate unmated males in the mid-L4 stage and were scored 24 hr later. Hermaphrodites were scored as early adults when they contained between one and two embryos in the uterus. Animals were then anesthetized in sodium azide on an agar pad and whole-animal images were captured immediately after under Nomarski optics (×20). Body length and width were measured with the ImageJ software and body volume was calculated as that of a cylinder (π × (width/2)2 × length).
Primary spermatocyte measurements
Extruded gonads from unmated males at 24 hr post-L4 were obtained by dissection in levamisole-containing M9. Gonads were fixed in 4% paraformaldehyde for 10 min and permeabilized for 5 min in 1× PBS with 0.1% Triton X-100. Gonads were next stained for actin with phalloidin (1:500 dilution; Sigma-Aldrich, St. Louis, MO) overnight at 4° in a humidified chamber. Slides were mounted in Vectashield supplemented with DAPI (Vector Laboratories) and observed under an epifluorescence microscope. Primary spermatocyte area was measured by outlining cell boundaries using ImageJ software (Vielle et al. 2016).
RNA interference experiments
RNA interference (RNAi) by bacterial feeding for C. elegans (N2) and C. plicata (SB355) was performed as previously described (Timmons and Fire 1998; Kamath et al. 2003). Briefly, control RNAi (HT115) and nurf-1 clone (provided by the Ahringer laboratory) were seeded on standard NGM with 50 μg/ml of ampicillin and 1 mM of IPTG and grown at room temperature for at least 24 hr before experiment. Worms were fed RNAi and control bacteria from the L1 stage and spermatid size was measured in the early adult stage (L4 + 24 hr).
Genome-wide association mapping
Genome-wide association mapping was performed using phenotype data from 97 C. elegans isotypes (Table S2). We used the cegwas R package for association mapping (Cook et al. 2017). This package uses the EMMA algorithm for performing association mapping and correcting for population structure (Kang et al. 2008), which is implemented by the GWAS function in the rrBLUP package (Endelman 2011). Specifically, the GWAS function in the rrBLUP package was called with the following command: rrBLUP::GWAS(pheno = ph, geno = y, K = kin, min.MAF = 0.05, n.core = 1, P3D = FALSE, plot = FALSE). The kinship matrix used for association mapping was generated using a whole-genome high-quality single-nucleotide variant (SNV) set from CeNDR release 20160408 (Cook et al. 2016; Evans et al. 2017; Zdraljevic et al. 2017) and the A.mat function from the rrBLUP package. SNVs previously identified using Restriction site–associated DNA sequencing (Andersen et al. 2012) that had at least 5% minor allele frequency in this strain set were used for performing genome-wide association mappings. Burden test analyses were performed using RVtests (Zhan et al. 2016) and the variable-threshold method (Price et al. 2010). We called SNVs using bcftools (Li 2011) with settings previously described (Cook et al. 2016, 2017; Zhan et al. 2016). We next performed imputation using BEAGLE v4.1 (Cook et al. 2017) with window set to 8000, overlap set to 3000, and ne set to 17,500. Within RVtests, we set the minor allele frequency range from 0.003 to 0.05 for burden testing.
Statistical analyses
Statistical tests were performed using R, JMP, or SPSS. Data for parametric tests were transformed where necessary to meet the assumptions of ANOVA procedures (homogeneity of variances and normal distributions of residuals); all size data were log-transformed. For post hoc comparisons, Tukey’s honestly significant difference procedure was used. For data, where ANOVA assumptions could not be met, we used nonparametric tests (e.g., Kruskal–Wallis).
Broad-sense heritability (H2) was estimated using the lmer function in the lme4 package (Bates et al. 2015) with the linear mixed model (phenotype ∼1 + (1|strain). H2 was then calculated as the fraction of the total variance explained by the random component (strain) of the mixed model.
Data availability
All raw data are provided in Additional File 1. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.9389255.
Results
Natural variation in C. elegans male sperm size
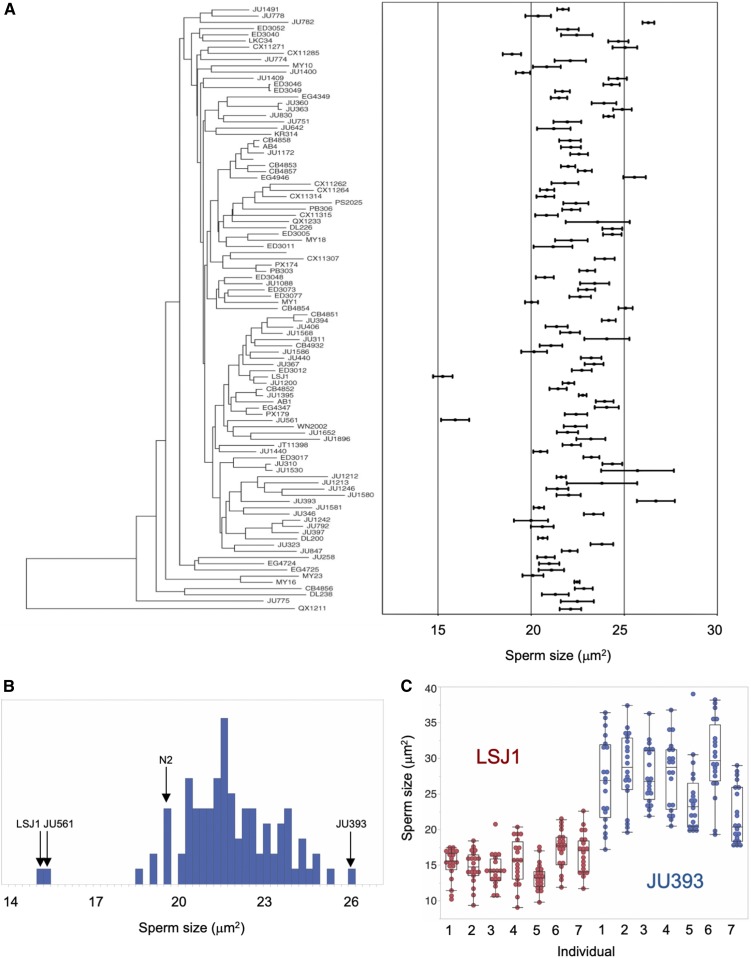
We quantified male sperm size variation of a worldwide collection of 97 C. elegans strains (Andersen et al. 2012), including two related laboratory strains (N2 and LSJ1), using measures of spermatid cross-sectional area. Average male sperm size, ranging from 15 to 27 µm2, varied significantly across strains (Figure 1 and Table S2). 90% of strains exhibited a male sperm size between 20 and 25 µm2, and we detected only two significant outliers: the wild strain JU561 (France) and the laboratory-adapted strain LSJ1 (McGrath et al. 2011) with the smallest male sperm size (Figure 1, A and B). As found previously for C. elegans and other Caenorhabditis species (Vielle et al. 2016), we also detected high levels of interindividual and intraindividual variation in male sperm size for most strains (Figure 1, A and C).
Figure 1.
Natural variation in C. elegans male sperm size. (A) Quantification of male spermatid cross-sectional area (mean ± SEM) in 97 C. elegans strains, arranged with respect to the neighbor-joining tree of Andersen et al. (2012) based on genome-wide SNP data. There is significant genetic (and interindividual) variation in sperm size (ANOVA, effect strain: F96, 13539 = 24.75, P < 0.0001; effect individual(strain): F580, 13539 = 3.08, P < 0.0001). Twenty spermatids from each of seven individuals were measured per strain (N = 140) with the exception of strains JU397 and KR413 for which 20 spermatids from each of six individuals (N = 120) were measured (Table S2). (B) Histogram of sperm size across strains (least-squares mean estimates) shows the significant outlier trait values for the two strains (JU561 and LSJ1) with smallest male sperm. (C) Illustration of inter- and intraindividual variation in male sperm size for strains with smallest (LSJ1) vs. largest male sperm size (JU393) (N = 20 sperm per individual).
Coefficients of variation (CV), i.e., the ratio of the SD to the mean, in sperm characters have been predicted and shown to be lower in species or genotypes experiencing higher levels of sperm competition (Gomendio et al. 2006; Calhim et al. 2007; Immler et al. 2008; Kleven et al. 2008; Fitzpatrick and Baer 2011). Therefore, we tested whether C. elegans strains with larger male sperm showed reduced variability. However, we did not detect a negative correlation between mean and CV of within-individual (ρPearson = 0.14, P = 0.17, N = 97) or between-individual (ρPearson = −0.09, P = 0.36, N = 97) male sperm size (Table S2), as expected under such a scenario.
Natural variation in C. elegans male sperm size: testing for covariation with male reproductive performance, hermaphrodite sperm size, and body size
Several studies have shown that sperm size may explain differences in male reproductive success and competitive ability among C. elegans wild isolates (LaMunyon and Ward 1998; Wegewitz et al. 2008; Murray et al. 2011). Using assays similar to those described in these previous studies, we tested whether the observed natural variation in C. elegans male sperm size correlates with variation in male mating ability, fertilization success, and male competitive ability, using eight strains with divergent male sperm size (Figure 2, A–G and Figure S1). Although we found significant heritable variation in all measured phenotypes, C. elegans male sperm size did not significantly correlate with any of them. In addition, the polymorphic plugging phenotype of males, i.e., the deposition of a gelatinous plug on the vulva after copulation, likely representing a trait of male competitive ability as it affects mating ability of subsequent males (Barker 1994; Hodgkin and Doniach 1997; Palopoli et al. 2008), did not depend on sperm size as the average male sperm size did not differ between plugging vs. nonplugging strains (ANOVA, F1,96 = 0.09, P = 0.77) (Table S3).
These results imply that additional sperm characteristics or other morphological and behavioral traits need to be considered to account for natural variation in male competitive ability and overall male reproductive performance. Consistent with this idea, we found that sperm transfer during a single mating (ejaculate size) (Figure S1, E–H) rather than sperm size shows a strong positive correlation with male fertility when mated to hermaphrodites (Figure 2H).
An unresolved question is whether genetic mechanisms regulating C. elegans sperm size are shared between the sexes. Previously, a weak positive correlation between the average sperm size of hermaphrodites and males was only found in C. tropicalis but not in C. elegans or C. briggsae (Vielle et al. 2016). This analysis was based on a small set of strains (n = 5), so we revisited this question using 12 strains differing in male sperm size. In agreement with previous reports (Baldi et al. 2011; Vielle et al. 2016), hermaphrodite sperm showed significant genetic variation and were substantially smaller than male sperm in all strains (Figure 3A and Table S4). Again, as in Vielle et al. (2016), there was no significant cross-sexual correlation in sperm size (Figure 3B). Given the presence of significant natural variation in C. elegans hermaphrodite sperm size, we tested whether this variation correlates with differential sperm production, which could be indicative of a potential trade-off between hermaphrodite sperm size and sperm number. Across eight strains hermaphrodite sperm production differed significantly (ANOVA, F7,184 = 5.37, P < 0.0001) (Table S5), but we found no correlation between hermaphrodite sperm size and number (Figure 3C).
Finally, we tested whether natural variation in C. elegans sperm size may reflect fixed allometric relationships between body and cell size. Such positive correlations between sperm size (length or cell size) and animal body size or mass have been frequently observed in diverse invertebrate taxa (Pitnick et al. 2009). Size of amoeboid sperm of nematodes, including Caenorhabditis, partly correlates with male body size across species (LaMunyon and Ward 1999; Vielle et al. 2016). In contrast, whether intraspecific variation in body size is linked to variation in sperm size in C. elegans had so far not been evaluated. Measuring early adult body size of hermaphrodites and males in a subset of strains, we found significant variation across strains and sex (Table S6); however, we did not detect any positive correlation between average sperm size and body size (length) in either sex (Figure S2, A and B). In males only, sperm size was correlated with body width (F1,10 = 16.86, R2 = 0.65, P = 0.0027) (Figure S2, C and D). A larger male body width could potentially allow for larger gonad width, thus allowing production of larger spermatocytes. On the other hand, male body width may also simply increase as consequence of storing of larger sperm. Further experiments are thus required to consolidate the evidence for a positive scaling relationship between male body size and sperm size.
Genome-wide association mapping of male sperm size
The significant variation in male sperm size enabled the mapping of genomic regions that could underlie this variation using genome-wide association studies, as has been performed successfully for a variety of traits using this C. elegans isolate panel (Andersen et al. 2012; Ghosh et al. 2012; Ashe et al. 2013). Broad-sense heritability was low (∼14%) for both sperm size cross-sectional area and diameter, and we found no significant genomic regions for these two traits and additional sperm size traits, including CV measurements (Figure 4). This result suggests that many loci could regulate differences in sperm size. Additionally, we used rare-variant based burden testing (Price et al. 2010; Bates et al. 2015; Zhan et al. 2016) to look for association of genes affected by deleterious rare variants with sperm area and diameter. As with marker-based association testing, we did not identify any significant genomic regions (data not shown).
Because specific natural niches could drive mating preferences, we investigated any effect of geography (e.g., latitude/longitude of strain origin) (Table S1, CeNDR: https://www.elegansvariation.org) on average male sperm size but found no such relationships (Spearman rank correlations, all P > 0.05).
Laboratory-derived strains LSJ1 and LSJ2 exhibit strongly reduced male and hermaphrodite sperm size
Across all 97 C. elegans strains measured, LSJ1 exhibited the smallest male sperm size (Figure 1A). LSJ1 is a laboratory-derived strain and shares a common ancestor with the reference strain N2 (Sterken et al. 2015) (Figure 5A), which shows a much larger male sperm size (Figure 1A and Figure 5B). The two lineages diverged between 1957 and 1958. N2 was then maintained on agar plates seeded with E. coli for ∼15 years, while LSJ1 was maintained in axenic liquid culture for close to 40 years before cultures were cryopreserved. LSJ2 is a derivative of LSJ1 that was kept in liquid culture for another 14 years prior to freezing (McGrath et al. 2009, 2011; Sterken et al. 2015; Large et al. 2017). We therefore also measured LSJ2, which displayed a similarly small sperm size as LSJ1 (Figure 5B). In contrast, the N2-derived strain CC1, grown in liquid axenic medium for only 4 years, did not differ from N2 in male sperm size (Figure 5B). Given the common, inbred, and likely isogenic “Bristol ancestor” of LSJ and N2-CC1 lineages, these results suggest that the evolution of reduced male sperm size in the LSJ lineage occurred due to de novo mutations before 1995. Of note, the LSJ1 and LSJ2 strains also showed significantly reduced hermaphrodite sperm size relative to N2 (and CC1) (Figure 5C).
Reduced male sperm size of LSJ strains is caused by genetic variation in nurf-1
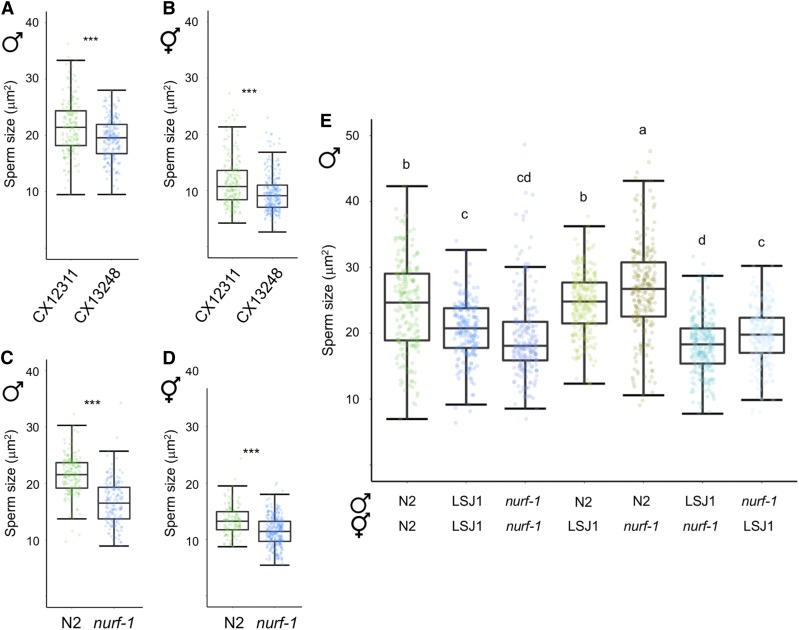
Whole-genome short-read sequencing identified 188 and 94 new mutations fixed in the LSJ2 and N2 lineages, respectively (McGrath et al. 2011). These mutations include a 60 bp deletion in the 3′ end of nurf-1, which encodes the ortholog of the BPTF subunit of the NURF chromatin remodeling complex (Andersen et al. 2006; McGrath et al. 2011; Large et al. 2016). This variant is predicted to replace the last 16 amino acids of the protein with 11 novel residues and is known to underlie multiple life history differences between N2 and LSJ2, including reproductive timing, progeny production, growth rate, life span, and Dauer formation (Large et al. 2016, 2017). Moreover, the NURF complex had previously been shown to function in the C. briggsae sperm-oocyte decision (Chen et al. 2014) as well as Drosophila spermatogenesis (Kwon et al. 2009). We thus reasoned that the nurf-1 deletion specific to the LSJ lineage provides a good candidate explaining reduced male (and hermaphrodite) sperm size. This hypothesis was supported by the observation that RNAi knockdown of nurf-1 resulted in a significant reduction of male sperm size in the N2 strain (Figure S3). To further test our hypothesis, we first examined the introgression line CX13248 (kyIR87) containing the LSJ2 region surrounding nurf-1 in an N2-like background (CX12311, which is of N2 genotype, except for introgressed npr-1 and glb-5 alleles from the strain CB4856) (McGrath et al. 2011). The kyIR87 introgression contains the 60 bp deletion along with LSJ2 alleles of eight additional variants, including an SNV in the intron of nurf-1 that was fixed in the N2 lineage (Large et al. 2016). Consistent with our hypothesis, sperm size of the CX13248 strain containing the nurf-1 deletion was significantly smaller compared to CX12311, both in males (Figure 6A) and hermaphrodites (Figure 6B). In addition, sperm size of the nurf-1(n4295) deletion mutant (N2 background) (Andersen et al. 2006) was also strongly reduced in both sexes (Figure 6, C and D); therefore, strains containing two different deletions in the 3′ coding region of nurf-1 result in reduced C. elegans sperm size. Specifically, the 60 bp nurf-1 deletion of LSJ strains covers the 3′ coding region (plus stop codon and 8 bp of the 3′ UTR region), and the nurf-1(n4295) deletion spans 1078 bp of the 3′ coding region (Large et al. 2016). Interestingly, the 60 bp and n4295 deletions differentially affect NURF-1.B and NURF-1.D, two of the four main isoforms of NURF-1, described to play antagonistic roles in the regulation of gametogenesis: while NURF-1.B is involved in sperm production, NURF-1.D is necessary for the switch to oogenesis (Xu et al. 2019). The nurf-1(n4295) mutation affects three exons found only in the 3′-terminal region of the nurf-1.d transcript. In contrast, the 60 bp deletion covers not only the very end of the 3′ coding region, but also the STOP codon and 8 bp of the 3′ UTR, present in both nurf-1.b and nurf-1.d transcripts (Xu et al. 2019). Our finding that sperm size is reduced in both n4295 mutant and LSJ strains thus suggests that NURF-D or both NURF-D and NURF-B affect sperm size determination.
Figure 6.
Reduced male sperm size of LSJ strains is caused by variation in nurf-1. (A and B) A near-isogenic line (CX13248) with the LS2 genomic region containing the nurf-1 deletion exhibits reduced sperm size relative to the N2-like parent CX12311 in (A) males (ANOVA, F1, 336 = 23.11, P < 0.0001) and (B) hermaphrodites (ANOVA, F1, 420 = 30.35, P < 0.0001). (C and D) Sperm size of the deletion mutant nurf-1(n4295) is reduced in (C) males (ANOVA, F1, 322 = 184.30, P < 0.0001) and (D) hermaphrodites (ANOVA, F1, 383 = 61.64, P < 0.0001). (E) Quantitative complementation tests using the strains N2, LSJ1, and nurf-1(n4295) (ANOVA, effect strain: F6, 1502 = 97.09, P < 0.0001). Values with the same letter are not significantly different from each other (Tukey’s honestly significant difference, P < 0.05). Box and whiskers plots: boxes are delimited by the data’s first and third quartiles, broken by a band at the median, and flanked by whiskers of which length is equal to 1.5 × the interquartile range (* P < 0.05, ** P < 0.01, *** P < 0.001).
To test whether nurf-1 is the causal gene underlying reduced sperm size in the LSJ lineage, we performed a quantitative complementation test (Long et al. 1996) taking advantage of the two nurf-1 deletion alleles present in LSJ1 and nurf-1(n4295) (Figure 6E). The recessive phenotype caused by either nurf-1 deletion was confirmed by the large, N2-like sperm size in F1 males derived from crosses between N2 and LSJ1 and between N2 and nurf-1(n4295) (Figure 6E). However, F1 males derived from bidirectional crosses between LSJ1 and nurf-1(n4295) exhibited small sperm size, comparable to parental strains (Figure 6E). We conclude that variation in the gene nurf-1 underlies the evolution of reduced male sperm size in LSJ strains.
Collectively, our experiments suggest that the 60 bp deletion in nurf-1 is the causal variant responsible for the decreased sperm size in males and hermaphrodites in the LSJ lineage. First, quantitative complementation indicates nurf-1 to be the causal gene. Second, the LSJ1/LSJ2 strains are outliers with regards to sperm size, suggesting that a mutation occurred in this lineage, like the 60 bp deletion, to reduce sperm size. Finally, the n4295 allele, which phenocopies the LSJ strains, is genetically similar to the 60 bp deletion, affecting the C terminus of the protein. However, the intron SNV in nurf-1, derived in the N2 lineage, cannot be completely ruled out to explain observed sperm size differences.
Mutants of different NURF-complex components exhibit reduced sperm and spermatocyte size
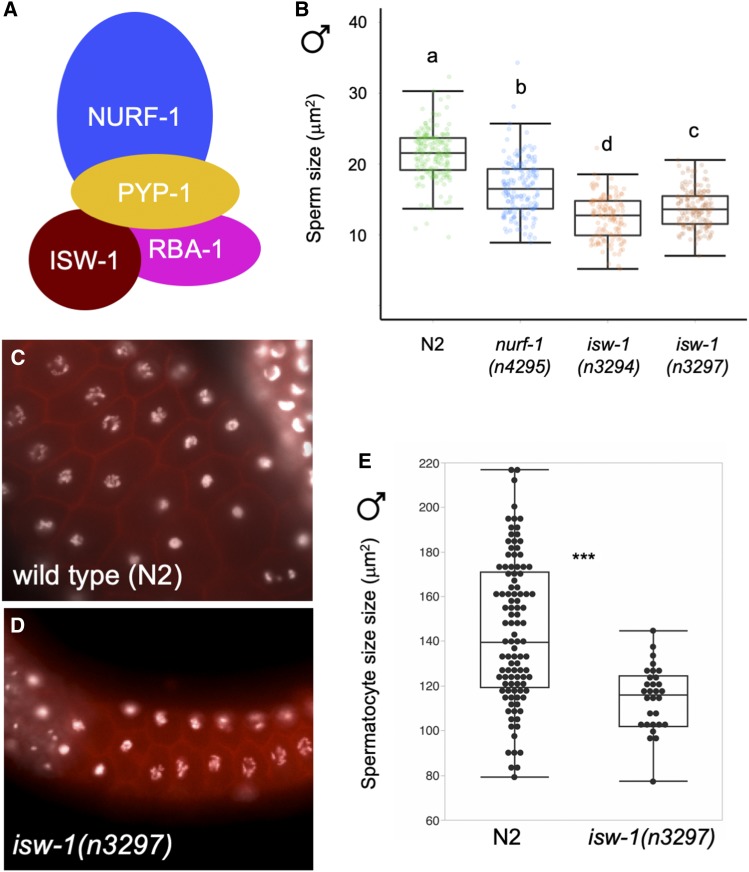
Observed sperm size reduction caused by the two independent nurf-1 deletions implies a potential role of the NURF chromatin remodeling complex (Figure 7A) in C. elegans sperm size determination. We therefore tested whether mutants of another complex member, the ATPase component ISW-1 (Figure 7B) (Tsukiyama et al. 1995; Andersen et al. 2006) show altered sperm size. Indeed, males of two independent deletion mutants of isw-1 (Andersen et al. 2006) exhibited strongly reduced male sperm size, and even smaller than in nurf-1(n4295) (Figure 7B). (In addition, males heterozygous for pyp-1(n4599), another member of the NURF complex, also made smaller sperm; data not shown.) Using the isw-1(n3297) allele (Andersen et al. 2006), we also found that hermaphrodite sperm size was significantly reduced compared to the N2 wild-type (Figure S4). Overall germline structure and organization of strains with small sperm size (including LSJ1 and LSJ2) appeared intact, except for a fraction of isw-1 mutant individuals that displayed severe errors, such as displacement of spermatids into the distal region. Given that Caenorhabditis sperm size differences (between species, genotypes within species, and sexes within species) are developmentally established at the primary spermatocyte stage (Vielle et al. 2016), we measured primary male spermatocyte size in isw-1(n3297) animals with intact germline structure: primary spermatocyte size was on average significantly smaller than in the wild-type N2 strain (Figure 7, C–E). Size variation of C. elegans sperm observed in NURF-complex mutants was thus introduced prior to, or at, the primary spermatocyte stage, as observed for sperm size variation occurring within and between different Caenorhabditis species (Vielle et al. 2016). Taken together, these data suggest that the NURF chromatin remodeling complex likely acts, directly or indirectly, in C. elegans sperm size determination. Furthermore, the role of nurf-1 in sperm size determination seems to be evolutionarily conserved because nurf-1 RNAi also reduced sperm size of a gonochoristic Caenorhabditis species, C. plicata, which normally exhibits substantially larger sperm (Vielle et al. 2016) (Figure S5).
Figure 7.
Mutants of the NURF complex exhibit reduced size of both male sperm and primary spermatocytes. (A) Illustration of NURF-complex components. Adapted from Alkhatib and Landry (2011). (B) nurf-1 and isw-1 mutants display reduced male sperm size (ANOVA, effect strain: F4, 728 = 192.23, P < 0.0001). (C and D) Microscopy images of primary spermatocytes in (C) wild type (N2) and (D) isw-1(n3297) (DAPI: white, Phalloidin: red). (E) Primary spermatocytes (area measurements) of the mutant isw-1(n3297) are significantly smaller than in the wild type (N2) (ANOVA, effect strain: F1, 119 = 83.47, P < 0.0001). Values with the same letter are not significantly different from each other (Tukey’s honestly significant difference, P < 0.05). Box and whiskers plots: boxes are delimited by the data’s first and third quartiles, broken by a band at the median, and flanked by whiskers of which length is equal to 1.5 × the interquartile range. (* P < 0.05, ** P < 0.01, *** P < 0.001)
Male sperm size is reduced independently of body size in NURF-complex mutants
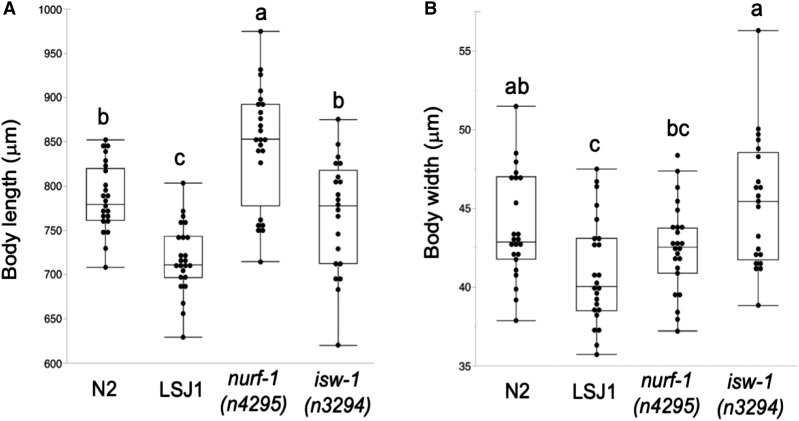
An earlier study has shown that the 60 bp deletion in nurf-1 is responsible for reduced hermaphrodite body length in LSJ2, and similarly, the deletion allele nurf-1(n4295) was shown to exhibit a significantly reduced hermaphrodite body length relative to N2 (Large et al. 2016). We therefore hypothesized that perturbing activity of the NURF chromatin complex may cause systemic size reduction of diverse tissues and organs, including spermatids. Inconsistent with this hypothesis, we found male body size of nurf-1(n4295) with small sperm size to be significantly larger, rather than smaller than in the wild-type N2 strain (Figure 8); in addition, the isw-1(n3294) mutant with very small sperm had the same male body size as N2 (Figure 8). Male sperm size reduction in NURF mutants thus occurs independently of body size, suggesting that reduced male sperm size of LSJ strains is not necessarily a pleiotropic consequence of reduction in male body size.
Figure 8.
Male sperm size reduction in NURF mutants occurs independently of body size. Significant differences in body size of strains N2, LSJ1, nurf-1(n4295), and isw-1(n3294). (A) Body length (ANOVA, F3, 91 = 23.36, P < 0.0001). (B) Body width (ANOVA, F3, 91 = 6.83, P = 0.0003). Values with the same letter are not significantly different from each other (Tukey’s honestly significant difference, P < 0.05).
Discussion
Our survey of intraspecific variation in C. elegans male sperm size uncovered significant heritable variation for this trait, generally thought to associate with sperm competitive ability. Yet, examining a subset of strains with divergent male sperm size, we did not detect a strong effect of male sperm size on competitive ability and reproductive performance. The second part of our study focused on the genetic basis underlying the evolutionary reduction in sperm size of laboratory-adapted strains, LSJ1 and LSJ2, which exhibit the smallest sperm size among all strains examined. This analysis, by means of mutants and quantitative complementation tests, shows that genetic changes in the nucleosome remodeling factor nurf-1 underlie the evolution of small male sperm size in the LSJ lineage. These and additional experimental results suggest that the NURF chromatin remodeling complex acts in C. elegans sperm size regulation.
Natural variation in C. elegans sperm size
Although we observed significant natural genetic variation in C. elegans male sperm size, the vast majority of isolates show a relatively narrow average sperm size range, from 20 to 25 µm2, and the only two significant outliers were the strains JU561 and LSJ1, with reduced male sperm size (∼15 µm2) (Figure 1). Performing a genome-wide association study to detect potential genetic loci explaining variation in male sperm size did not yield any quantitative trait loci, likely due to low statistical power and/or complex genetic trait architecture. As previously observed for some C. elegans isolates and other Caenorhabditis species (Vielle et al. 2016), male sperm size showed extreme variability within single individuals, and to a lesser extent, among populations of genetically identical individuals (Figure 1). Such within-genotype variability is often observed for sperm size traits, such as sperm length in taxa with flagellate sperm (Ward 1998; Morrow and Gage 2001b; Miller et al. 2003; Joly et al. 2004). Possibly, such high sperm size variance could reflect a means to maximize both average size and number of sperm produced, or developmentally decreasing sperm size variance may come at the cost of reduced sperm production speed (Parker and Begon 1993; Gomendio et al. 2006; Vielle et al. 2016).
Unlike a previous study (Murray et al. 2011), we did not find any correlations between male sperm size and multiple traits related to male reproductive performance and competitive ability. This mixed evidence suggests that male sperm size is not a reliable predictor and sole key determinant of male reproductive success, in line with studies reporting diverse genetic factors affecting Caenorhabditis sperm competitive ability independently of sperm size (Thomas et al. 2012; Ting et al. 2014, 2018; Fierst et al. 2015; Hansen et al. 2015; Yin et al. 2018; Yin and Haag 2019). Moreover, many other traits that we did not measure here, including male mating behavior or hermaphrodite receptivity, will contribute to overall male reproductive success. We also found that a rarely measured trait—the number of sperm transferred per mating (ejaculate size)—was strongly correlated with the number of male offspring sired across different strains (Figure 2H). Although we did not detect a potential trade-off between male sperm size and ejaculate size (Figure S1F), an earlier study (Murray et al. 2011) did report that the rate of sperm production was reduced in C. elegans strains with larger male sperm. Trade-offs between male sperm size and sperm number could thus potentially shape and limit the extent of sperm size evolution. Importantly, the observed absence of a tight relationship between male sperm size and competitive ability might also represent a signature of reduced male conflict and sperm competition, in line with reports of low rates of outcrossing in C. elegans wild populations (Barrière and Félix 2005, 2007; Félix and Braendle 2010) and reduced maintenance of (male) mating function (Chasnov and Chow 2002; Teotónio et al. 2006; Thomas et al. 2012; Chasnov 2013; Yin et al. 2018). Nevertheless, C. elegans male sperm seem to maintain the evolutionary potential to respond to changes in the extent of male mating competition: several experimental evolution studies have shown that male sperm size may rapidly evolve toward increased size in response to increased male-male competition (LaMunyon and Ward 2002; Palopoli et al. 2015; Poullet et al. 2016).
We also aimed to assess to what extent C. elegans sperm size differs between sexes, given that little is known about natural genetic variation in hermaphrodite sperm size and whether male and hermaphrodite sperm size may correlate. Specifically, a significant correlation between male and hermaphrodite sperm size could be indicative of shared genetic regulation of sperm size across sexes. Our analyses of intraspecific variation in C. elegans hermaphrodite sperm size confirmed previous reports (Ward and Carrel 1979; LaMunyon and Ward 1995, 1998; Vielle et al. 2016) that for any given strain, hermaphrodite sperm are always significantly smaller than male sperm (Figure 3A). As for male sperm size, we detected substantial levels of heritable variation in hermaphrodite sperm size. Although the two strains with smallest male sperm (LSJ1 and JU561) also had the smallest hermaphrodite sperm, male and hermaphrodite sperm size did not correlate across examined strains (Figure 3B). The C. elegans sperm size dimorphism is thought to reflect differential selection on conflicting sex-specific size optima: larger male sperm to increase sperm competitive ability vs. smaller hermaphrodite sperm to accelerate sperm production to allow for a rapid switch to oogenesis, which is critical for early maturity and reproduction (Hodgkin and Barnes 1991; Cutter 2004); C. elegans hermaphrodites are protandrous with initial production of sperm, stored in the spermathecae, before switching to oocyte production. Therefore, the sequential spermatogenesis-oogenesis switch in C. elegans causes a trade-off between maximal sperm number (i.e., potential number of self-progeny) and minimal age at maturity (i.e., generation time). Consequently, evolution of small hermaphrodite sperm size results from selection for rapid self-sperm production, consistent with the fact that hermaphrodite sperm are always drastically smaller than male sperm (Figure 3A). In the same fashion, evolution of increased hermaphrodite sperm production may lead to smaller self-sperm. However, we did not find evidence for a trade-off between hermaphrodite sperm size and number across a set of C. elegans strains differing in sperm size (Figure 3C). Importantly, the evolution of small hermaphrodite sperm in androdioecious species seems not only to result from selection for improved self-fertilization but also from direct developmental effects, when spermatogenesis takes place in a female soma, that reduce hermaphrodite sperm size (Baldi et al. 2011). Disentangling these different evolutionary and developmental mechanisms in shaping C. elegans sperm size dimorphism therefore remains a major challenge.
Genetic variation in nurf-1 explains the evolution of small sperm and suggests a role for the NURF chromatin remodeling complex in C. elegans sperm size determination
We uncovered strong differences in male sperm size between two very closely related C. elegans laboratory lineages, N2 vs. LSJ1/LSJ2. Given their recent evolutionary divergence, and thus close genetic similarity (McGrath et al. 2011), we successfully applied a candidate approach to demonstrate that variation in the gene nurf-1, encoding a subunit of the NURF chromatin remodeling complex, explains reduced LSJ sperm size. The observation that evolutionary reduction of C. elegans male sperm size is caused by variation in nurf-1 suggests that this gene acts in C. elegans sperm size determination. Our analysis of multiple mutants in NURF-complex genes, all of which displayed reduced sperm size, not only in males but also in hermaphrodites, confirms this idea. As a subunit of the NURF chromatin remodeling complex, NURF-1 is a BPTF ortholog, involved in histone modification on nucleosomes and remodeling of nucleosomes through recruitment of ISWI, to modulate transcription (Badenhorst et al. 2002; Andersen et al. 2006; Wysocka et al. 2006; Ruthenburg et al. 2011). Members of the C. elegans NURF chromatin remodeling complex are expressed in diverse tissues and organs, including the developing germline (Reece-Hoyes et al. 2007; Feng et al. 2012; Craig et al. 2013). Consistent with these expression patterns, nurf-1 mutations affect diverse phenotypes in C. elegans, ranging from vulval development (Andersen et al. 2006) to multiple life history traits (Large et al. 2016), and most relevantly in the context of our findings, a very recent study showed that nurf-1 plays specific roles in spermatogenesis and the sperm-oocyte decision (Xu et al. 2019). Together with our findings that nurf-1 variants of LSJ strains and mutants of NURF-complex genes (nurf-1, isw-1, pyp-1) reduce sperm size, this experimental evidence thus clearly points to a role of the NURF chromatin remodeling complex in regulating C. elegans sperm size.
The evolution of nurf-1 function in the LSJ lineage underlies laboratory adaptation to an axenic liquid medium (Large et al. 2016, 2017). Specifically, a key causal molecular variant underlying improved LSJ hermaphrodite reproduction in this environment (relative to N2) is the 60 bp nurf-1 deletion (Large et al. 2016), which we identified here as the (very likely) causal variant responsible for reduced male sperm size (Figure 6E). This deletion has been shown to have highly pleiotropic effects on hermaphrodite life history traits, including reproduction, growth rate, life span, and Dauer formation (Large et al. 2016). Given this demonstration that this nurf-1 variation specific to the LSJ lineage confers improved fitness of hermaphrodites, the evolution of reduced male sperm size in the LSJ lineage likely reflects a pleiotropic consequence stemming from selection on hermaphrodite function. Very possibly, the nurf-1 variation explaining reduced male sperm size is also responsible for the observed reduction of hermaphrodite sperm size in LSJ strains (Figure 5C); however, this remains to be experimentally confirmed. If true, evolution of reduced hermaphrodite sperm size in LSJ strains could thus be more directly linked to reported evolutionary changes of the LSJ lineage in both gametogenesis (Xu et al. 2019) and reproductive traits (Large et al. 2016). For example, production of smaller hermaphrodite sperm may go in hand with observed increases in sperm production (Figure 3C), which in turn, could explain observed increases in self-fertility (Large et al. 2016). More specifically, given that the different nurf-1 deletions occurring in LSJ strains and in the nurf-1(n4295) mutant differentially affect B and D isoforms (Xu et al. 2019), our findings that hermaphrodite sperm size is reduced in both n4295 mutant and LSJ strains (Figure 5C and Figure 6D) suggests that either NURF-D or both NURF-D and NURF-B affect sperm size determination. This could occur through a direct effect of NURF-1.D on genes regulating sperm size or as an indirect consequence of NURF-1.D acting on the timing of the sperm-oocyte decision. In the latter scenario, sperm size reduction may result as a by-product of increased sperm production as observed for LSJ hermaphrodites (Figure 3C).
C. elegans strains LSJ1 and LSJ2 were derived from the ancestral N2 strain by growing them in liquid culture for over 40 years (Large et al. 2016, 2017). Given that C. elegans males are generally presumed to be incapable of mating in liquid culture, laboratory evolution of the LSJ lineage likely occurred in the complete absence of outcrossing. Our finding of reduced male sperm size in the LSJ lineage thus illustrates how selection for improved C. elegans hermaphrodite function (Large et al. 2016) can affect a (likely unselected) male trait through a specific genetic variant. Consistent with the presumed absence of outcrossing over many hundreds of generations in liquid culture, LSJ1 showed significantly reduced male-male competitive ability relative to N2 (Figure 2, E–G). On the other hand, we found that LSJ1 male sperm function and mating ability with hermaphrodites remained largely preserved (Figure 2, A–D and Figure S1). These observations exemplify how essential male functions in C. elegans can be maintained despite absent or very rare outcrossing, i.e., strongly relaxed selection.
Acknowledgments
We thank Asher Cutter, Marie-Anne Félix, and Henrique Teotonio for discussion and comments on previous versions of the manuscript. We also thank three reviewers for their comments, which greatly helped to improve an earlier version of this article. Strains and materials were provided by the groups of Julie Ahringer, Cori Bargmann, Marie-Anne Félix, the C. elegans Natural Diversity Resource (Cook et al. 2017), and the Caenorhabditis Genetics Center, which is funded by National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440). C.B., C.G., and A.V. acknowledge financial support by the Centre National de la Recherche Scientifique. N.S.-S. was supported by an Erasmus mobility fellowship provided by the European Commission. S.Z. was supported by the National Institutes of Health Cell and Molecular Basis of Disease training grant (T32GM008061) and a Bernard and Martha Rappaport Fellowship. P.T.M. was supported by NIH grant R01GM114170. E.C.A. was supported by an American Cancer Society Research Scholar Grant (127313-RSG-15-135-01-DD).
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25386/genetics.9389255. One link to a second document is missing here, see comments for explanation.
Communicating editor: P. Wittkopp
Literature Cited
- Alcorn M. R., Callander D. C., Lopez-Santos A., Torres Cleuren Y. N., Birsoy B. et al. , 2016. Heterotaxy in Caenorhabditis: widespread natural variation in left-right arrangement of the major organs. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371: 20150404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhatib S. G., and Landry J. W., 2011. The nucleosome remodeling factor. FEBS Lett. 585: 3197–3207. 10.1016/j.febslet.2011.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen E. C., Lu X., and Horvitz H. R., 2006. C. elegans ISWI and NURF301 antagonize an Rb-like pathway in the determination of multiple cell fates. Development 133: 2695–2704. 10.1242/dev.02444 [DOI] [PubMed] [Google Scholar]
- Andersen E. C., Gerke J. P., Shapiro J. A., Crissman J. R., Ghosh R. et al. , 2012. Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat. Genet. 44: 285–290. 10.1038/ng.1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. L., Morran L. T., and Phillips P. C., 2010. Outcrossing and the maintenance of males within C. elegans populations. J. Hered. 101: S62–S74. 10.1093/jhered/esq003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A., Belicard T., Le Pen J., Sarkies P., Frezal L. et al. , 2013. A deletion polymorphism in the Caenorhabditis elegans RIG-I homolog disables viral RNA dicing and antiviral immunity. eLife 2: e00994 10.7554/eLife.00994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badenhorst P., Voas M., Rebay I., and Wu C., 2002. Biological functions of the ISWI chromatin remodeling complex NURF. Genes Dev. 16: 3186–3198. 10.1101/gad.1032202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi C., Viviano J., and Ellis R. E., 2011. A bias caused by ectopic development produces sexually dimorphic sperm in nematodes. Curr. Biol. 21: 1416–1420. 10.1016/j.cub.2011.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker D. M., 1994. Copulatory plugs and paternity assurance in the nematode Caenorhabditis elegans. Anim. Behav. 48: 147–156. 10.1006/anbe.1994.1221 [DOI] [Google Scholar]
- Barrière A., and Félix M.-A., 2005. High local genetic diversity and low outcrossing rate in Caenorhabditis elegans natural populations. Curr. Biol. 15: 1176–1184. 10.1016/j.cub.2005.06.022 [DOI] [PubMed] [Google Scholar]
- Barrière A., and Félix M.-A., 2007. Temporal dynamics and linkage disequilibrium in natural Caenorhabditis elegans populations. Genetics 176: 999–1011. 10.1534/genetics.106.067223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D., Mächler M., Bolker B., and Walker S., 2015. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67: 1–48. [Google Scholar]
- Birkhead T. R., and Moller A. P., 1998. Sperm Competition and Sexual Selection. Academic Press, New York. [Google Scholar]
- Birkhead T. R., Hosken D. J., and Pitnick S., 2009. Sperm Biology: An Evolutionary Perspective. Academic Press (Elsevier), Boston. [Google Scholar]
- Calhim S., Immler S., and Birkhead T. R., 2007. Postcopulatory sexual selection is associated with reduced variation in sperm morphology. PLoS One 2: e413 10.1371/journal.pone.0000413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasnov J. R., 2013. The evolutionary role of males in C. elegans. Worm 2: e21146 10.4161/Worm.21146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasnov J. R., and Chow K. L., 2002. Why are there males in the hermaphroditic species Caenorhabditis elegans? Genetics 160: 983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Shen Y., and Ellis R. E., 2014. Dependence of the sperm/oocyte decision on the nucleosome remodeling factor complex was acquired during recent Caenorhabditis briggsae evolution. Mol. Biol. Evol. 31: 2573–2585. 10.1093/molbev/msu198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D. S., and Shakes D. C., 2013. Spermatogenesis. Adv. Exp. Med. Biol. 757: 171–203. 10.1007/978-1-4614-4015-4_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D. E., Zdraljevic S., Tanny R. E., Seo B., Riccardi D. D. et al. , 2016. The genetic basis of natural variation in Caenorhabditis elegans telomere length. Genetics 204: 371–383. 10.1534/genetics.116.191148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D. E., Zdraljevic S., Roberts J. P., and Andersen E. C., 2017. CeNDR, the Caenorhabditis elegans natural diversity resource. Nucleic Acids Res. 45: D650–D657. 10.1093/nar/gkw893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig H. L., Wirtz J., Bamps S., Dolphin C. T., and Hope I. A., 2013. The significance of alternative transcripts for Caenorhabditis elegans transcription factor genes, based on expression pattern analysis. BMC Genomics 14: 249 10.1186/1471-2164-14-249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter A. D., 2004. Sperm-limited fecundity in nematodes: how many sperm are enough? Evolution 58: 651–655 [PubMed] [Google Scholar]
- Demarco R. S., Eikenes A. H., Haglund K., and Jones D. L., 2014. Investigating spermatogenesis in Drosophila melanogaster. Methods 68: 218–227. 10.1016/j.ymeth.2014.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R. E., and Stanfield G. M., 2014. The regulation of spermatogenesis and sperm function in nematodes. Semin. Cell Dev. Biol. 29: 17–30. 10.1016/j.semcdb.2014.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endelman J. B., 2011. Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome 4: 250–255. 10.3835/plantgenome2011.08.0024 [DOI] [Google Scholar]
- Evans K. S., Zhao Y., Brady S. C., Long L., McGrath P. T. et al. , 2017. Correlations of genotype with climate parameters suggest Caenorhabditis elegans niche adaptations. G3 (Bethesda) 7: 289–298. 10.1534/g3.116.035162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix M.-A., and Braendle C., 2010. The natural history of Caenorhabditis elegans. Curr. Biol. 20: R965–R969. 10.1016/j.cub.2010.09.050 [DOI] [PubMed] [Google Scholar]
- Feng H., Craig H. L., and Hope I. A., 2012. Expression pattern analysis of regulatory transcription factors in Caenorhabditis elegans. Methods Mol. Biol. 786: 21–50. 10.1007/978-1-61779-292-2_2 [DOI] [PubMed] [Google Scholar]
- Fierst J. L., Willis J. H., Thomas C. G., Wang W., Reynolds R. M. et al. , 2015. Reproductive mode and the evolution of genome size and structure in Caenorhabditis nematodes. PLoS Genet. 11: e1005323 (erratum: PLoS Genet. 11: e1005497). 10.1371/journal.pgen.1005323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick J. L., and Baer B., 2011. Polyandry reduces sperm length variation in social insects. Evolution 65: 3006–3012. 10.1111/j.1558-5646.2011.01343.x [DOI] [PubMed] [Google Scholar]
- Geldziler B. D., Marcello M. R., Shakes D. C., and Singson A., 2011. The genetics and cell biology of fertilization. Methods Cell Biol. 106: 343–375. 10.1016/B978-0-12-544172-8.00013-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh R., Andersen E. C., Shapiro J. A., Gerke J. P., and Kruglyak L., 2012. Natural variation in a chloride channel subunit confers avermectin resistance in C. elegans. Science 335: 574–578. 10.1126/science.1214318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomendio M., Martin-Coello J., Crespo C., Magana C., and Roldan E. R. S., 2006. Sperm competition enhances functional capacity of mammalian spermatozoa. Proc. Natl. Acad. Sci. USA 103: 15113–15117. 10.1073/pnas.0605795103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J. M., Chavez D. R., and Stanfield G. M., 2015. COMP-1 promotes competitive advantage of nematode sperm. eLife 4: e05423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J., and Barnes T., 1991. More is not better: brood size and population growth in a self-fertilizing nematode. Proc. Biol. Sci. 246: 19–24. 10.1098/rspb.1991.0119 [DOI] [PubMed] [Google Scholar]
- Hodgkin J., and Doniach T., 1997. Natural variation and copulatory plug formation in Caenorhabditis elegans. Genetics 146: 149–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immler S., Calhim S., and Birkhead T. R., 2008. Increased postcopulatory sexual selection reduces the intramale variation in sperm design. Evolution 62: 1538–1543. 10.1111/j.1558-5646.2008.00393.x [DOI] [PubMed] [Google Scholar]
- Joly D., Korol A., and Nevo E., 2004. Sperm size evolution in Drosophila: inter- and intraspecific analysis. Genetica 120: 233–244. 10.1023/B:GENE.0000017644.63389.57 [DOI] [PubMed] [Google Scholar]
- Jovelin R., Ajie B. C., and Phillips P. C., 2003. Molecular evolution and quantitative variation for chemosensory behaviour in the nematode genus Caenorhabditis. Mol. Ecol. 12: 1325–1337. 10.1046/j.1365-294X.2003.01805.x [DOI] [PubMed] [Google Scholar]
- Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R. et al. , 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. 10.1038/nature01278 [DOI] [PubMed] [Google Scholar]
- Kang H. M., Zaitlen N. A., Wade C. M., Kirby A., Heckerman D. et al. , 2008. Efficient control of population structure in model organism association mapping. Genetics 178: 1709–1723. 10.1534/genetics.107.080101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleven O., Laskemoen T., Fossøy F., Robertson R. J., and Lifjeld J. T., 2008. Intraspecific variation in sperm length is negatively related to sperm competition in passerine birds. Evolution 62: 494–499. 10.1111/j.1558-5646.2007.00287.x [DOI] [PubMed] [Google Scholar]
- Kwon S. Y., Xiao H., Wu C., and Badenhorst P., 2009. Alternative splicing of NURF301 generates distinct NURF chromatin remodeling complexes with altered modified histone binding specificities. PLoS Genet. 5: e1000574 10.1371/journal.pgen.1000574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMunyon C. W., and Ward S., 1995. Sperm precedence in a hermaphroditic nematode (Caenorhabditis elegans) is due to competitive superiority of male sperm. Experientia 51: 817–823. 10.1007/BF01922436 [DOI] [PubMed] [Google Scholar]
- LaMunyon C. W., and Ward S., 1998. Larger sperm outcompete smaller sperm in the nematode Caenorhabditis elegans. Proc. R. Soc. Lond. B Biol. Sci. 265: 1997–2002. 10.1098/rspb.1998.0531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMunyon C. W., and Ward S., 1999. Evolution of sperm size in nematodes: sperm competition favours larger sperm. Proc. R. Soc. Lond. B Biol. Sci. 266: 263–267. 10.1098/rspb.1999.0631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMunyon C. W., and Ward S., 2002. Evolution of larger sperm in response to experimentally increased sperm competition in Caenorhabditis elegans. Proc. R. Soc. Lond. B Biol. Sci. 269: 1125–1128. 10.1098/rspb.2002.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Large E. E., Xu W., Zhao Y., Brady S. C., Long L. et al. , 2016. Selection on a subunit of the NURF chromatin remodeler modifies life history traits in a domesticated strain of Caenorhabditis elegans. PLoS Genet. 12: e1006219 10.1371/journal.pgen.1006219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Large E. E., Padmanabhan R., Watkins K. L., Campbell R. F., Xu W. et al. , 2017. Modeling of a negative feedback mechanism explains antagonistic pleiotropy in reproduction in domesticated Caenorhabditis elegans strains. PLoS Genet. 13: e1006769 10.1371/journal.pgen.1006769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- L’Hernault, S.W., 2005 Spermatogenesis (February 20, 2006), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.85.1, http://www.wormbook.org.
- Li H., 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. 10.1093/bioinformatics/btr509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long A., Mullaney S., Mackay T., and Langley C., 1996. Genetic interactions between naturally occurring alleles at quantitative trait loci and mutant alleles at candidate loci affecting bristle number in Drosophila melanogaster. Genetics 144: 1497–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath P. T., Rockman M. V., Zimmer M., Jang H., Macosko E. Z. et al. , 2009. Quantitative mapping of a digenic behavioral trait implicates globin variation in C. elegans sensory behaviors. Neuron 61: 692–699. 10.1016/j.neuron.2009.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath P. T., Xu Y., Ailion M., Garrison J. L., Butcher R. A. et al. , 2011. Parallel evolution of domesticated Caenorhabditis species targets pheromone receptor genes. Nature 477: 321–325. 10.1038/nature10378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G. T., Starmer W. T., and Pitnick S., 2003. Quantitative genetic analysis of among-population variation in sperm and female sperm-storage organ length in Drosophila mojavensis. Genet. Res. 81: 213–220. 10.1017/S0016672303006190 [DOI] [PubMed] [Google Scholar]
- Morran L. T., Cappy B. J., Anderson J. L., and Phillips P. C., 2009. Sexual partners for the stressed: facultative outcrossing in the self-fertilizing nematode Caenorhabditis elegans. Evolution 63: 1473–1482. 10.1111/j.1558-5646.2009.00652.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow E. H., and Gage M., 2001a Artificial selection and heritability of sperm length in Gryllus bimaculatus. Heredity 87: 356–362. 10.1046/j.1365-2540.2001.00921.x [DOI] [PubMed] [Google Scholar]
- Morrow E. H., and Gage M. J. G., 2001b Consistent significant variation between individual males in spermatozoal morphometry. J. Zool. (Lond.) 254: 147–153. 10.1017/S0952836901000656 [DOI] [Google Scholar]
- Murray R. L., Kozlowska J. L., and Cutter A. D., 2011. Heritable determinants of male fertilization success in the nematode Caenorhabditis elegans. BMC Evol. Biol. 11: 99 10.1186/1471-2148-11-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson G. A., and Ward S., 1980. Vesicle fusion, pseudopod extension and amoeboid motility are induced in nematode spermatids by the ionophore monensin. Cell 19: 457–464. 10.1016/0092-8674(80)90520-6 [DOI] [PubMed] [Google Scholar]
- Noble L. M., Chang A. S., McNelis D., Kramer M., Yen M. et al. , 2015. Natural variation in plep-1 causes male-male copulatory behavior in C. elegans. Curr. Biol. 25: 2730–2737. 10.1016/j.cub.2015.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palopoli M. F., Rockman M. V., Tinmaung A., Ramsay C., Curwen S. et al. , 2008. Molecular basis of the copulatory plug polymorphism in Caenorhabditis elegans. Nature 454: 1019–1022. 10.1038/nature07171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palopoli M. F., Peden C., Woo C., Akiha K., Ary M. et al. , 2015. Natural and experimental evolution of sexual conflict within Caenorhabditis nematodes. BMC Evol. Biol. 15: 93 10.1186/s12862-015-0377-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker G. A., and Begon M. E., 1993. Sperm competition games: sperm size and number under gametic control. Proc. Biol. Sci. 253: 255–262. 10.1098/rspb.1993.0111 [DOI] [PubMed] [Google Scholar]
- Pitnick S., Hosken D. J., and Birkhead T. R., 2009. Sperm morphological diversity, pp. 69–149 in Sperm Biology: An Evolutionary Perspective, edited by Birkhead T. R., Hosken D. J. and Pitnick S.. Academic Press (Elsevier), Boston. [Google Scholar]
- Poullet N., Vielle A., Gimond C., Ferrari C., and Braendle C., 2015. Evolutionarily divergent thermal sensitivity of germline development and fertility in hermaphroditic Caenorhabditis nematodes. Evol. Dev. 17: 380–397. 10.1111/ede.12170 [DOI] [PubMed] [Google Scholar]
- Poullet N., Vielle A., Gimond C., Carvalho S., Teotonio H. et al. , 2016. Complex heterochrony underlies the evolution of Caenorhabditis elegans hermaphrodite sex allocation. Evolution 70: 2357–2369. 10.1111/evo.13032 [DOI] [PubMed] [Google Scholar]
- Price A. L., Kryukov G. V., de Bakker P. I., Purcell S. M., Staples J. et al. , 2010. Pooled association tests for rare variants in exon-resequencing studies. Am. J. Hum. Genet. 86: 832–838. 10.1016/j.ajhg.2010.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramm S. A., Schärer L., Ehmcke J., and Wistuba J., 2014. Sperm competition and the evolution of spermatogenesis. Mol. Hum. Reprod. 20: 1169–1179. 10.1093/molehr/gau070 [DOI] [PubMed] [Google Scholar]
- Rasband, W.S., 1997–2014 ImageJ. National Institutes of Health, Bethesda, MD.
- Reece-Hoyes J. S., Shingles J., Dupuy D., Grove C. A., Walhout A. J. et al. , 2007. Insight into transcription factor gene duplication from Caenorhabditis elegans Promoterome-driven expression patterns. BMC Genomics 8: 27 10.1186/1471-2164-8-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg A. J., Li H., Milne T. A., Dewell S., McGinty R. K. et al. , 2011. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell 145: 692–706. 10.1016/j.cell.2011.03.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedl T., and Kimble J., 1988. fog-2, a germ-line-specific sex determination gene required for hermaphrodite spermatogenesis in Caenorhabditis elegans. Genetics 119: 43–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakes D. C., Wu J.-C., Sadler P. L., Laprade K., Moore L. L. et al. , 2009. Spermatogenesis-specific features of the meiotic program in Caenorhabditis elegans. PLoS Genet. 5: e1000611 10.1371/journal.pgen.1000611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons L. W., and Moore A. J., 2009. Evolutionary quantitative genetics of sperm, pp. 69–149 in Sperm Biology: An Evolutionary Perspective, edited by Birkhead T. R., Hosken D. J. and Pitnick S.. Academic Press (Elsevier), Boston. [Google Scholar]
- Sivasundar A., and Hey J., 2005. Sampling from natural populations with RNAI reveals high outcrossing and population structure in Caenorhabditis elegans. Curr. Biol. 15: 1598–1602. 10.1016/j.cub.2005.08.034 [DOI] [PubMed] [Google Scholar]
- Smith R. L., 1984. Sperm Competition and the Evolution of Animal Mating Systems. Academic Press, Orlando, FL. [Google Scholar]
- Snook R. R., 2005. Sperm in competition: not playing by the numbers. Trends Ecol. Evol. 20: 46–53. 10.1016/j.tree.2004.10.011 [DOI] [PubMed] [Google Scholar]
- Sterken M. G., Snoek L. B., Kammenga J. E., and Andersen E. C., 2015. The laboratory domestication of Caenorhabditis elegans. Trends Genet. 31: 224–231. 10.1016/j.tig.2015.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T., 2006. Maintenance of C. elegans (February 11, 2006), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.101.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teotónio H., Manoel D., and Phillips P. C., 2006. Genetic variation for outcrossing among Caenorhabditis elegans isolates. Evolution 60: 1300–1305. 10.1111/j.0014-3820.2006.tb01207.x [DOI] [PubMed] [Google Scholar]
- Thomas C. G., Woodruff G. C., and Haag E. S., 2012. Causes and consequences of the evolution of reproductive mode in Caenorhabditis nematodes. Trends Genet. 28: 213–220. 10.1016/j.tig.2012.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., and Fire A., 1998. Specific interference by ingested dsRNA. Nature 395: 854 10.1038/27579 [DOI] [PubMed] [Google Scholar]
- Ting J. J., Woodruff G. C., Leung G., Shin N.-R., Cutter A. D. et al. , 2014. Intense sperm-mediated sexual conflict promotes gametic isolation in Caenorhabditis nematodes. PLoS Biol. 12: e1001915 10.1371/journal.pbio.1001915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting J. J., Tsai C. N., Schalkowski R., and Cutter A. D., 2018. Genetic contributions to ectopic sperm cell migration in Caenorhabditis nematodes. G3 (Bethesda) 8: 3891–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T., Daniel C., Tamkun J., and Wu C., 1995. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell 83: 1021–1026. 10.1016/0092-8674(95)90217-1 [DOI] [PubMed] [Google Scholar]
- Vielle A., Callemeyn-Torre N., Gimond C., Poullet N., Gray J. C. et al. , 2016. Convergent evolution of sperm gigantism and the developmental origins of sperm size variability in Caenorhabditis nematodes. Evolution 70: 2485–2503. 10.1111/evo.13043 [DOI] [PubMed] [Google Scholar]
- Ward P. I., 1998. Intraspecific variation in sperm size characters. Heredity 80: 655–659. 10.1046/j.1365-2540.1998.00401.x [DOI] [PubMed] [Google Scholar]
- Ward S., and Carrel J. S., 1979. Fertilization and sperm competition in the nematode Caenorhabditis elegans. Dev. Biol. 73: 304–321. 10.1016/0012-1606(79)90069-1 [DOI] [PubMed] [Google Scholar]
- Ward S., Argon Y., and Nelson G. A., 1981. Sperm morphogenesis in wild-type and fertilization-defective mutants of Caenorhabditis elegans. J. Cell Biol. 91: 26–44. 10.1083/jcb.91.1.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegewitz V., Schulenburg H., and Streit A., 2008. Experimental insight into the proximate causes of male persistence variation among two strains of the androdioecious Caenorhabditis elegans (Nematoda). BMC Ecol. 8: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J., Swigut T., Xiao H., Milne T. A., Kwon S. Y. et al. , 2006. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442: 86–90. 10.1038/nature04815 [DOI] [PubMed] [Google Scholar]
- Xu, W., L. Long, Y. Zhao, L. Stevens, R. E. Ellis et al., 2019 Evolution of Yin and Yang isoforms of a chromatin remodeling subunit results in the creation of two genes. bioRxiv doi:10.1101/616995 (Preprint posted April 24, 2019). [DOI] [PMC free article] [PubMed]
- Yin D., and Haag E. S., 2019. Evolution of sex ratio through gene loss. Proc. Natl. Acad. Sci. USA 116: 12919–12924. 10.1073/pnas.1903925116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin D., Schwarz E. M., Thomas C. G., Felde R. L., Korf I. F. et al. , 2018. Rapid genome shrinkage in a self-fertile nematode reveals sperm competition proteins. Science 359: 55–61. 10.1126/science.aao0827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdraljevic S., Strand C., Seidel H. S., Cook D. E., Doench J. G. et al. , 2017. Natural variation in a single amino acid substitution underlies physiological responses to topoisomerase II poisons. PLoS Genet. 13: e1006891 10.1371/journal.pgen.1006891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X., Hu Y., Li B., Abecasis G. R., and Liu D. J., 2016. RVTESTS: an efficient and comprehensive tool for rare variant association analysis using sequence data. Bioinformatics 32: 1423–1426. 10.1093/bioinformatics/btw079 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All raw data are provided in Additional File 1. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.9389255.