Abstract
Background
PTPRU is an important signaling molecule that regulates a variety of cellular processes; however, the role of PTPRU in cancer development has remained elusive. Here, we report that PTPRU serves as a tumor suppressor that inhibits cancer stemness by attenuating Hippo/YAP signaling pathway.
Methods
Primary cancer cells and cell line cells were used in the study. The gene expression data were downloaded from R2 analysis and visualization platform and Kaplan–Meier analysis was performed to study the relationship between survival and PTPRU expression. qRT-PCR and Western blot were employed to study the expression of target genes in tissues and cells. Sphere and colony formation, proliferation, migration activities and the expression of stem cell and EMT markers were employed for characterizing the stemness. Gene manipulation was achieved by lentivirus-mediated gene delivery system. Luciferase reporter gene assay was used to study the transcriptional activity of the promoter, and ChIP-qPCR was employed to study the target binding sequence of the protein. Spearman correlation analysis was performed to study the correlation between two genes. Student’s t-test was used for determination of the significance between two experimental groups.
Results
PTPRU is downregulated in colorectal and gastric cancer tissues and cancer stem cells. High expression of PTPRU predicts poor prognosis. Overexpression of PTPRU attenuates the stemness of gastric cancer stem cells and knockdown of PTRPU improves the maintenance of the stemness of cancer stem cells. Mechanistic analysis showed that PTPRU inhibits Hippo/YAP signaling by suppressing the expression of YAP in a transcriptional level. Overexpression of YAP restored PTPRU-induced inhibited stemness of gastric cancer stem cells.
Conclusion
PTPRU serves as a tumor suppressor that inhibits the stemness of cancer stem cell by inhibiting Hippo/YAP signaling pathway.
Keywords: PTPRU, cancer stem cells, Hippo/YAP signaling, gastric cancer, colorectal cancer
Introduction
Cancer is a major cause of death worldwide. Although several therapeutic strategies have been developed, the incidence of solid tumor and leukemia is still increasing.1 Cancer stem cell is a key feature for cancer progression and is responsible for its survival.2–4 Stem cells are capable to self-renew and to differentiate to diverse cell types, and such capacities are comprised in the term stemness.5–7 Cancer stem cells are a small population of cells within tumor tissues maintaining stemness that sustain cancer progression.8–10 Thus, understanding the mechanisms underlying the maintenance of the stemness of cancer stem cells may provide more opportunities for effective therapies.
Protein tyrosine phosphatase receptor U (PTPRU, also known as PCP-2 or PTPλ), belongs to the protein tyrosine phosphatase (PTP) family.11–13 PTPs regulate a variety of important cellular processes including cell growth, differentiation and mitotic cycle.14,15 PTPRU represents a receptor-type PTP consisting of an extracellular region, a single transmembrane region and two tandem intracellular catalytic tyrosine phosphatase domains.16 PTPRU is known to be involved in cancer development17; however, the reports of the role of PTPRU expression in tumor progression are contradictory. For instance, PTPRU has been reported to inhibit cell proliferation and invasion by decreasing β-catenin tyrosine phosphorylation.18 In contrast, another study showed that endogenous PTPRU is required for glioma growth and motility, likely by activating β-catenin signaling and maintaining FA protein stability.19 It is also been reported that knockdown of PTPRU inhibits cell growth and motility of gastric cancer cells.20 These findings suggest that the role of PTPRU during cancer development depends on the cellular signaling context and cell type.
The objective of this study is to investigate whether PTPRU plays a role in cancer stem cells. In this report, we show that PTPRU serves as a tumor suppressor that inhibits cancer stemness by attenuating Hippo/YAP signaling pathway.
Methods
Cell Culture
SNU-16 cell was purchased from Chinese Academy of Sciences (Shanghai, China) and cultured in Roswell Park Memorial Institute (RPMI) 1640 media. Primary gastric and colorectal cancer cells were isolated from cancer tissues of patients with gastric and colorectal cancer and cultured in Dulbecco’s Modified Essential Media (DMEM) media. Cancer stem cells were isolated by suspension culture in ultra-low attachment multi-well plate (Corning Incorporated, NY, USA) with serum-free medium containing 100 µg/mL human bFGF, 20 µg/mL insulin and 10 ng/mL human EGF.
Human Samples
The gastric and colorectal cancer tissues were obtained from Renji Hospital Shanghai Jiao Tong University School of Medicine (Shanghai, China), which was approved by the Institutional Review Board. All patients provided written informed consent, and this was conducted in accordance with the Declaration of Helsinki.
Plasmids
The coding sequence (CDs) regions of PTPRU and YAP were generated by reverse-transcription polymerase chain reaction (PCR). The shRNAs specifically against PPTRU were synthesized exogenously (Shenggong Biotech, Shanghai, China). The promoter region of YAP was also generated by reverse-transcription PCR. For overexpression, the CDs regions of PTPRU and YAP were cloned into pCDH lentivirus vectors. For knockdown assay, the shRNAs were cloned into pLKO.1 lentivirus vectors. For reporter gene assay, the promoter of YAP was cloned into PGL3 plasmid (Promega Corp., Madison, WI, USA). The sequences of PCR primer and shRNAs are listed in Supplementary table 1 and Supplementary table 2.
Antibodies And Reagents
The antibodies used in this study are listed in Supplementary table 3. All other reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Lentivirus Package, Transfection And Stable Cell Selection
Lentiviruses were generated by Lenti-X Packaging Single Shots (Clontech Laboratories Inc, Palo Alto, USA) according to the manufacturer’s manual. Media containing lentivirus particles were used to infect the cells. Stable cell line was selected by treatment with puromycin for 15–20 days. The efficiency was determined by Western blot assay.
Quantitative Real-Time Reverse-Transcription PCR
Total RNA was isolated by RNeasy Mini Kit (Qiagen, Hilden, Germany). cDNA was synthesized by SuperScript II reverse transcriptase (Thermo Fisher Scientific, Waltham, MA, USA). The qRT-PCR assay was then performed by TaqMan gene expression assay using specific primers (Supplementary Table 1). The results were expressed by comparative threshold cycle (Ct) method with GAPDH as a control.
Western Blot
The Western blot assay was performed as previously described.21–24 Briefly, samples were separated by electrophoresis using 4–12% NuPAGE Bis-Tris gels (Invitrogen, Carlsbad, CA). The samples were then transferred to nitrocellulose membranes. Membranes were blocked with 5% non-fat milk for 2 hrs at room temperature. Membranes were incubated with first antibodies (Supplementary Table 3) at 4ºC overnight. After washing in 0.05% Tween 20-TBS, membranes were incubated with the appropriate horseradish peroxidase-conjugated secondary antibodies for 1.5 hrs at room temperature. Immunoreactive bands were detected by chemiluminescence using SuperSignal chemiluminescent substrate (Pierce Biotechnology, Rockford, IL, USA).
Reporter Gene Assay
The cells were transfected with PGL3 vector containing the promoter of YAP and hRluc/TK vector. Luciferase activity was measured with Dual-Luciferases Reporter Assay kit (Promega Corp., Madison, WI, USA) according to the manufacturer’s manual.
Cell Proliferation
The cells were seeded into 96-well plates. After 2–3 days of incubation, the cells were washed and incubated with 10 μL of Cell Counting Kit-8 (CCK-8) solution (Dojindo Molecular Technologies Inc., Kumamoto, Japan). The absorbance at 450 nm was detected by a microplate reader (BioTek, Winooski, VT, USA).
Sphere Formation
The cells were cultured in ultra-low attachment multi-well plate (Corning Incorporated, NY, USA) with serum-free medium containing 100 µg/mL human bFGF, 20 µg/mL insulin and 10 ng/mL human EGF. After 10–15 days, the colonies were stained and counted.
Migration
The migration activities of the cells were measured by transwell assay (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s manual.
Statistics
Each experiment was performed at least three times. Data were represented as mean ± s.d. Student’s t-test was used for determination of the significance between two experimental groups.
Results
PTPRU Is Downregulated In Colorectal And Gastric Cancer Tissues And Stem Cells And Predicts Good Prognosis
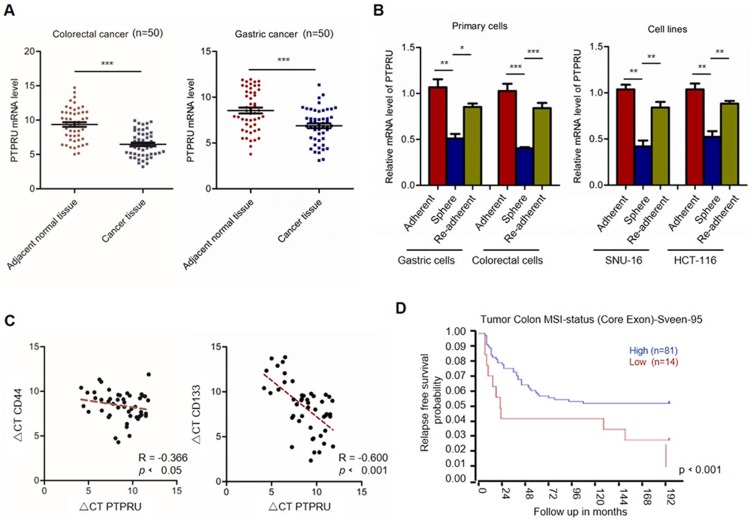
To investigate the effect of PTPRU in cancer development, we first examined the expression level of PTPRU in colorectal and gastric cancer tissues. As shown in Figure 1A, the results from qRT-PCR show that the mRNA level of PTPRU is downregulated in cancer tissues, which indicated that PTPRU may play a tumor suppressor role in cancer development. We then isolated the cancer stem cells from gastric and colorectal cancer tissues by suspension culture and tested the expression of PTPRU in adherent cells, suspension cells and re-adherent cells. As shown in Figure 1B (left), the mRNA level of PTPRU was downregulated in primary spheres and was recovered in re-adherent cells, which suggested that downregulation of PTPRU is associated with stemness maintenance of cancer stem cells. This result was subsequently confirmed in gastric and colorectal cancer cell lines (SNU-16 and HCT-116) (Figure 1B (right)).
Figure 1.
PTPRU is downregulated in cancer stem cells and predicts good prognosis. (A) The mRNA level of PTPRU in colorectal cancer and adjacent normal tissues. (B) The mRNA level of PTPRU in adherent, sphere and re-adherent primary gastric and colorectal cancer cells. (C) The correlation analysis of the expression of CD44 (left), CD133 (right) and PTPRU. (D) Kaplan–Meier analysis of the survival rate of colon cancer patients with high (blue) or low (red) PTPRU expression with gene expression dataset downloaded from R2 analysis and visualization platform (http://hgserver1.amc.nl/cgi-bin/r2/main.cgi). Data are represented as mean±SD; *P<0.05, **P<0.01, ***P<0.001; two-tailed Student’s t-test.
To confirm this hypothesis, we investigated the correlation between PTPRU and cancer stem cell markers (CD44 and CD133). The total mRNA was isolated from gastric cancer tissue and the expression of PTPRU, CD44 and CD133 was examined by qRT-PCR, and Spearman correlation analysis was performed. As shown in Figure 1C, the expression of PTPRU was negatively correlated with CD133 (R=−0.4074, P<0.001) and CD44 (R=−0.5706, P<0.001). Next, we downloaded the tumor gene expression and clinical data of colon cancer patients from R2 analysis and visualization platform (http://hgserver1.amc.nl/cgi-bin/r2/main.cgi) and performed Kaplan–Meier analysis. As shown in Figure 1D, high expression of PTPRU predicts good prognosis.
Taken together, these results suggested that downregulation of PTPRU may play an important role in the maintenance of the stemness of cancer stem cells.
Ectopic Expression Of PTPRU Attenuates The Stemness Of Cancer Stem Cells
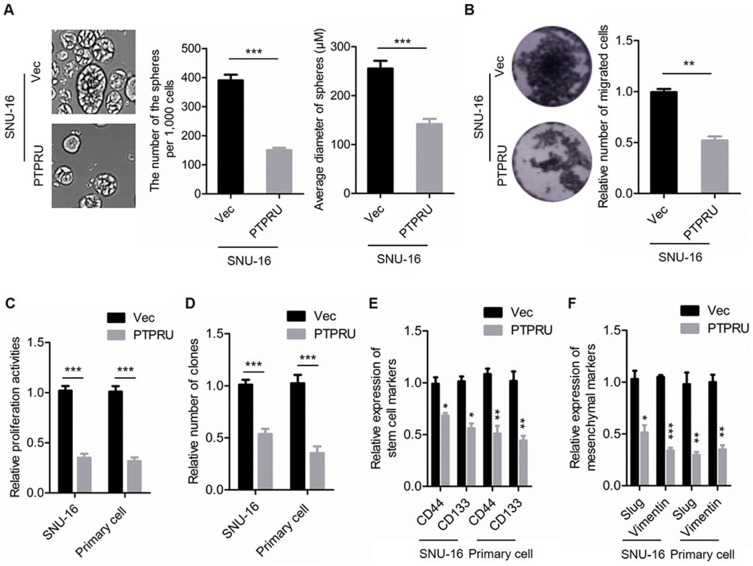
To confirm the role of PTPRU in cancer stem cells maintenance, we employed lentivirus-based gene delivery system to overexpress PTPRU in gastric cancer cell SNU-16 and primary gastric cancer cells (Supplementary Figure 1A). We first examined the activities of sphere formation, migration, proliferation and colony formation of the cells. As shown in Figure 2A (left), the number of the spheres is decreased by PTPRU overexpression. And the sphere size is also reduced by PTPRU overexpression (Figure 2A right). In addition, as shown in Figure 2B, the migration activities of the cells were attenuated by PTPRU overexpression. Moreover, ectopic expression of PTPRU decreased the number of the proliferated cells (Figure 2C). Furthermore, overexpression of PTPRU decreased the colony-formation activities of the cells (Figure 2D).
Figure 2.
Ectopic expression of PTRPU attenuates the stemness of gastric cancer stem cells. (A) Sphere-formation activities of SNU-16 cells transfected with PTPRU and empty control vectors were detected by sphere-formation assay. (B) Migration activities of indicated cells were tested by transwell assay. (C) The proliferation activities of indicated cells were tested by CCK-8 assay. (D) The colony-formation activities of indicated cells were examined by colony-formation assay. (E, F) qRT-PCR analysis of the expression levels of stem cell markers (E) and mesenchymal markers (F) in indicated cells. Data are represented as mean±SD; *P<0.05, **P<0.01, ***P<0.001; two-tailed Student’s t-test.
To further investigate the relationship between PTPRU and cancer stem cells, we investigated the relationship between PTPRU and the regulation of the expression of cancer stem cell markers (CD44 and CD133). As shown in Figure 2E, the expression of stem cell markers was decreased in cells with PTPRU overexpression. In addition, the mRNA levels of epithelial–mesenchymal transition (EMT) markers (Slug and Vimentin) were also downregulated in PTPRU-overexpressing cells (Figure 2F).
Taken together, these results demonstrated that overexpression of PTPRU attenuates the stemness of cancer stem cells.
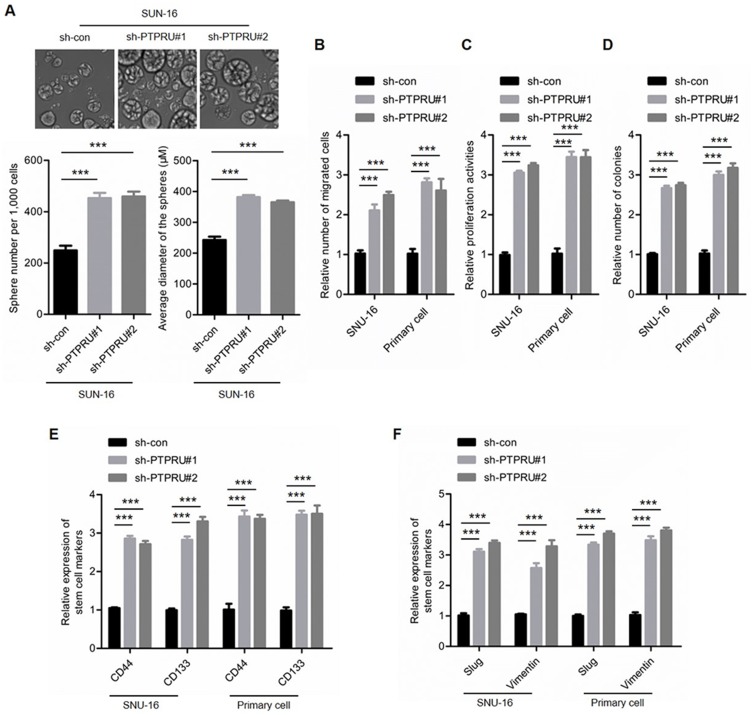
Knockdown Of PTPRU Improves The Maintenance Of Cancer Stem Cell Stemness
To confirm the suppressor role of PTPRU in the maintenance of cancer stem cell stemness, we next knocked down PTPRU in gastric cancer cells (SNU-16) and primary gastric cancer cells (Supplementary Figure 1B). As shown in Figure 3A, knockdown of PTPRU increased both the number and the size of spheres. In addition, the migration ability of the cells is also increased by PTPRU knockdown (Figure 3B). Moreover, the proliferation and the colony-formation activities were improved after PTPRU depletion (Figure 3C and D). Furthermore, the expression of stem cell markers (CD44 and CD133) as well as EMT markers (Slug and Vimentin) was elevated by knockdown of PTPRU (Figure 3E and F). Collectively, the above results demonstrated that PTPRU serves as a tumor suppressor that inhibits the maintenance of the stemness of cancer stem cells.
Figure 3.
Knockdown of PTRPU improves the stemness of gastric cancer stem cells. (A) Sphere formation activities of indicated cells were detected by sphere formation assay. (B) Migration activities of indicated cells were tested by transwell assay. (C) The proliferation activities of indicated cells were tested by CCK-8 assay. (D) The colony formation activities of indicated cells were examined by colony formation assay. (E,F) qRT-PCR analysis of the expression levels of stem cell markers (E) and mesenchymal markers (F) in indicated cells. Data are represented as mean±SD; ***P<0.001; two=tailed Student’s t-test.
PTPRU Inhibits Hippo/YAP Signaling Pathway
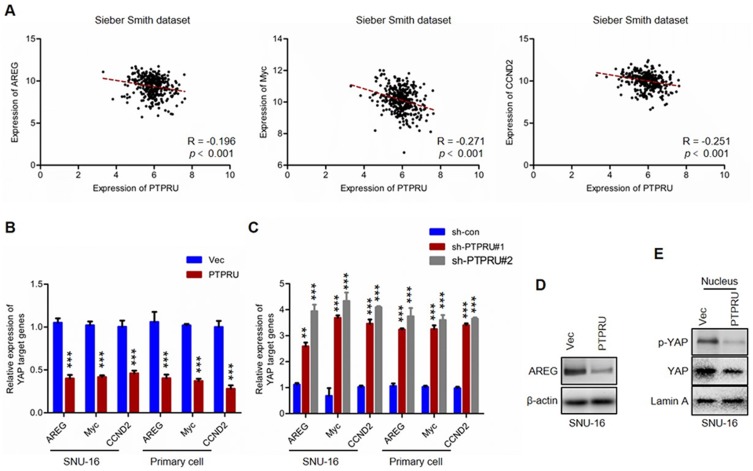
To investigate the underlying mechanisms for PTPRU inhibits the stemness of cancer stem cells, we performed bioinformatic analysis. The gene expression dataset was downloaded from R2 analysis and visualization platform (http://hgserver1.amc.nl/cgi-bin/r2/main.cgi). As shown in Figure 4A, results from Spearman correlation analysis showed that the expression of PTPRU was negatively correlated with YAP target genes (ARGE, Myc and CCND2). We subsequently performed qRT-PCR to confirm this phenomenon. As shown in Figure 4B and C, the results of qRT-PCR also show that the mRNA levels of YAP target genes were negatively regulated by PTPRU in gastric cancer cells (SNU-16). This result was subsequently confirmed by Western blot (Figure 4D). As YAP translocates into the nucleus to affect the transcription of its target genes, we then examined the abundance of nuclear YAP and phosphorylated YAP in the nucleus by Western blot. As shown in Figure 4E, the levels of both YAP and phosphorylated YAP in the nucleus were downregulated in PTPRU-overexpressing cells. These results thus demonstrated that PTPRU inhibits Hippo/YAP signaling pathway.
Figure 4.
PTPRU inhibits Hippo/YAP signaling in gastric cancer cells. (A) The correlation analysis of YAP target genes (AREG, Myc and CCND2) and PTPRU in colorectal cancer samples with gene expression dataset (Sieber Smith dataset) downloaded from R2 analysis and visualization platform (http://hgserver1.amc.nl/cgi-bin/r2/main.cgi). (B,C) qRT-PCR analysis of the expression of YAP target genes in indicated cells. (D) Western blot analysis of the expression of AREG in indicated cells. (E) Western blot analysis of the abundance of YAP and phosphorylated YAP in the nucleus of indicated cells. Data are represented as mean±SD; **P<0.01, ***P<0.001; two-tailed Student’s t-test.
Hippo/YAP Signaling Pathway Is Necessary For PTPRU Inhibiting The Stemness Of Cancer Cells
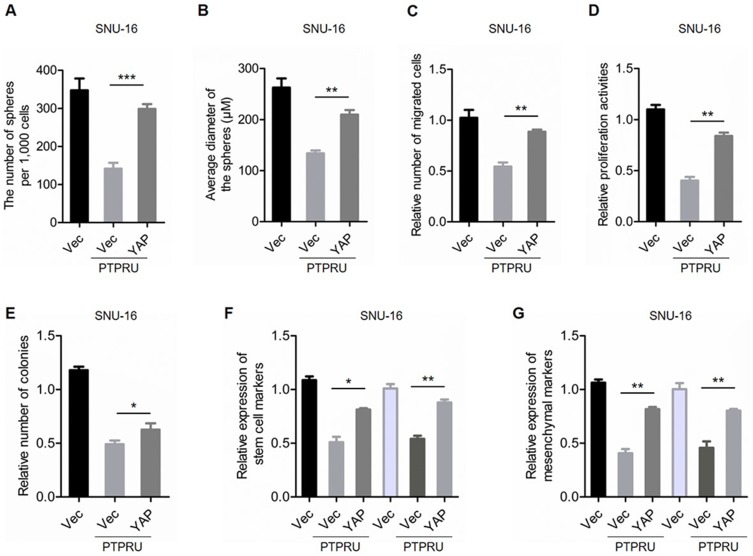
To confirm the role of Hippo/YAP signaling in PTPRU inhibiting the stemness of cancer cells, we overexpressed YAP in PTPRU-overexpressing cells (Supplementary Figure 1C). As shown in Figure 5A and B, overexpression of YAP restored the sphere-formation activity in both PTPRU-overexpressing SUN-16 and primary cells. In addition, the migration activity was also restored by YAP overexpression in PTPRU-overexpressing SUN-16 and primary cells (Figure 5C). Moreover, ectopic expression of YAP restored PTPRU-induced decreased proliferation and colony-formation activities in these cells (Figure 5D and E). Furthermore, the expression of cancer stem cell markers and EMT markers was also restored by YAP overexpression in PTPRU-overexpressing SNU-16 and primary gastric cancer cells (Figure 5F and G). These results thus demonstrated that Hippo/YAP signaling pathway is necessary for PTPRU inhibiting the stemness of cancer cells.
Figure 5.
Hippo/YAP signaling is necessary for PTPRU inhibit the stemness of cancer cells. (A,B) The number (A) and size (B) of sphere generated from indicated cells were detected by sphere-formation assay. (C) Migration activities of indicated cells were tested by transwell assay. (D) The proliferation activities of indicated cells were tested by CCK-8 assay. (E) The colony-formation activities of indicated cells were examined by colony-formation assay. (F,G) qRT-PCR analysis of the expression levels of stem cell markers (F) and mesenchymal markers (G) in indicated cells. Data are represented as mean±SD; *P<0.05, **P<0.01, ***P<0.001; two-tailed Student’s t-test.
PTPRU Inhibits YAP Transcription By Directly Binding To YAP Promoter
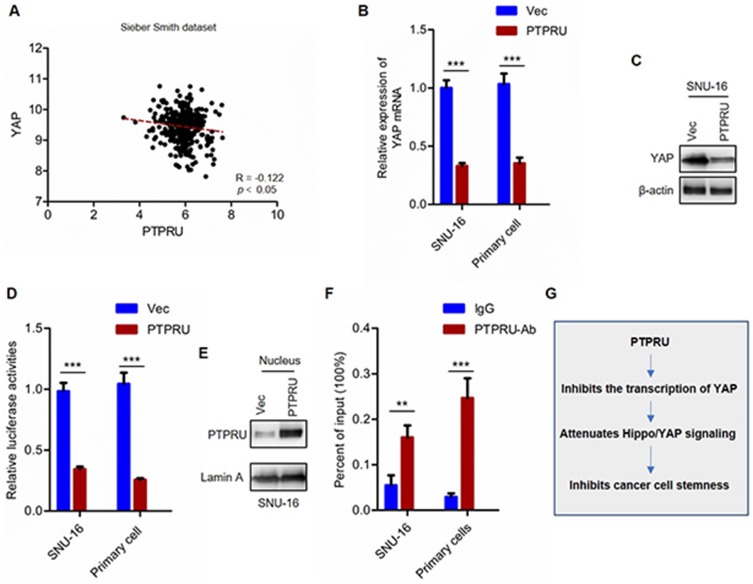
To investigate the mechanism underlying PTPRU inhibiting Hippo/YAP signaling pathway, we performed bioinformatic analysis to investigate whether there were key components in Hippo/YAP signaling pathway regulated by PTPRU. We found that the expression of YAP is negatively regulated by PTRPU (Figure 6A). This result was subsequently demonstrated by qRT-PCR and Western blot assay (Figure 6B and C). Next, to investigate whether PTPRU regulates the expression of YAP in a transcriptional level, we performed reporter gene assay. As shown in Figure 6D, the result from gene reporter assay shows that PTPRU inhibits the transcription activity of YAP promoter. These results suggested that PTPRU inhibits the expression of YAP at a transcriptional level.
Figure 6.
PTPRU inhibits YAP transcription by directly binding to YAP promoter. (A) The correlation analysis of YAP and PTPRU in colorectal cancer samples with gene expression dataset (Sieber Smith dataset) downloaded from R2 analysis and visualization platform. (B) qRT-PCR analysis of the expression of YAP in indicated cells. (C) Western blot analysis of the expression of YAP in indicated cells. (D) The transcriptional activity of YAP promoter was analyzed by reporter gene assay. (E) Western blot analysis of the abundance of nuclear YAP in indicated cells. (F) ChIP analysis the binding between PTPRU and YAP promoter. (G) The mechanism underlying PTPRU inhibits the stemness of cancer cells. Data are represented as mean±SD; **P<0.01, ***P<0.001; two-tailed Student’s t-test.
Next, we investigate whether PTPRU inhibits YAP transcription by directly binding to YAP promoter, and we performed Western blot assay and found that PTPRU does translocate into nucleus and its abundance in the nucleus was upregulated in PTPRU-overexpressing cells (Figure 6E). And the result from ChIP-qPCR showed that PTPRU directly bind to YAP promoter (Figure 6F). These results demonstrated that PTPRU inhibits YAP transcription by directly binding to YAP promoter.
In summary, our study demonstrated that PTPRU serves as a tumor suppressor that inhibits cancer stem cell stemness by inhibiting Hippo/YAP signaling pathway (Figure 6G).
Discussion
In this study, we revealed a novel mechanism underlying cancer stem cell stemness maintenance that PTPRU, as a tumor suppressor, inhibits the stemness of cancer stem cells.
Cancer is still a major cause of death worldwide. Although therapeutic strategies for cancer management have been developed, the incidence of solid tumor and leukemia is still increasing.1 Identification of cancer stem cells (CSCs) might lead to a new thinking on cancer treatments as cancer stem cells are responsible for tumor initiation, growth, metastasis as well as therapeutic resistance.2–4 Similar to stem cells, cancer stem cells display high resistance to genotoxic agents.5–7 Conventional chemotherapy may shrink the tumor volume but cannot eliminate cancer.8–10 Therefore, eradication of cancer stem cells would be a novel therapeutic strategy. Thus, understanding the mechanisms underlying the maintenance of the stemness of cancer stem cells and identification of novel cancer stem cell drug targets is important for the development of novel cancer therapeutic strategies.
PTPRU belongs to the protein tyrosine phosphatase (PTP) family11–13 that regulates a variety of important cellular processes.14,15 The role of PTPRU in tumor progression is contradictory.17 PTPRU has been reported to inhibit cell proliferation by decreasing β-catenin tyrosine phosphorylation.18 In contrast, another study showed that endogenous PTPRU is required for glioma growth.19 It is also reported that knockdown of PTPRU inhibits cell growth and motility of gastric cancer cells.20 These findings suggested that the role of PTPRU during cancer development depends on the cellular signaling context and cell type. However, the role of PTPRU in cancer stemness has remained elusive.
In our study, we showed that PTPRU inhibits the stemness of cancer stem cells. The expression of PTPRU is downregulated in gastric and colorectal cancer stem cells (Figure 1B) and is negatively correlated with cancer stem cell markers (Figure 1C). In addition, overexpression of PTPRU inhibited the stemness of gastric cancer stem cells (Figure 2). Moreover, knockdown of PTPRU improved the stemness of gastric cancer stem cells (Figure 3). These results demonstrated that PTPRU inhibits the stemness of cancer stem cells.
Both Hippo/YAP and Wnt signaling pathways are important for regulation of the stemness of cancer stem cells, and they crosstalk in the cells.25,26 By bioinformatic analysis, we found that the expression of PTPRU is negatively correlated with YAP target genes (Figure 4A). The qRT-PCR and Western blot results confirmed this phenomenon (Figure 4B–D). To confirm that PTPRU inhibits Hippo/YAP signaling pathway, we found that the level of nuclear YAP is negatively correlated with PTPRU (Figure 4E), which demonstrated that PTPRU inhibits Hippo/YAP signaling pathway. The subsequent rescue experiment and ChIP study demonstrated the necessary role Hippo/YAP signaling and the mechanism that PTPRU inhibits YAP transcription by directly binding to YAP promoter (Figures 5 and 6). Our work thus revealed a novel mechanism of the regulation of the stemness of cancer stem cells.
Conclusion
In summary, this study revealed a novel mechanism underlying cancer stem cell stemness maintenance that PTPRU, as a tumor suppressor, inhibits the stemness of cancer stem cells.
Acknowledgment
This work was supported by Science and Technology Commission of Shanghai Municipality Fund (16ZR1420500).
Author Contributions
JQ, ZZ and TL designed the experiments and wrote the paper. JQ supervised the project. JQ, ZZ, XM, LY, JX, CT and KH collected clinical samples and performed most of the experiments with clinical samples. JG performed most of the experiments with cell line cells. TL performed bioinformatics analysis. All authors participated in conception and design, data acquisition, data analysis and interpretation, drafted the article, approved the final version to be published and agreed to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 2.Papaccio F, Paino F, Regad T, et al. Concise review: cancer cells, cancer stem cells, and mesenchymal stem cells: influence in cancer development. Stem Cells Transl Med. 2017;6(12):2115–2125. doi: 10.1002/sctm.17-0138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nassar D, Blanpain C. Cancer stem cells: basic concepts and therapeutic implications. Annu Rev Pathol. 2016;11:47–76. doi: 10.1146/annurev-pathol-012615-044438 [DOI] [PubMed] [Google Scholar]
- 4.Almendro V, Marusyk A, Polyak K. Cellular heterogeneity and molecular evolution in cancer. Annu Rev Pathol. 2013;8:277–302. doi: 10.1146/annurev-pathol-020712-163923 [DOI] [PubMed] [Google Scholar]
- 5.Mittal V. Epithelial mesenchymal transition in tumor metastasis. Annu Rev Pathol. 2018;13:395–412. doi: 10.1146/annurev-pathol-020117-043854 [DOI] [PubMed] [Google Scholar]
- 6.He S, Nakada D, Morrison SJ. Mechanisms of stem cell self-renewal. Annu Rev Cell Dev Biol. 2009;25:377–406. doi: 10.1146/annurev.cellbio.042308.113248 [DOI] [PubMed] [Google Scholar]
- 7.Dodge ME, Lum L. Drugging the cancer stem cell compartment: lessons learned from the hedgehog and Wnt signal transduction pathways. Annu Rev Pharmacol Toxicol. 2011;51:289–310. doi: 10.1146/annurev-pharmtox-010510-100558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lytle NK, Barber AG, Reya T. Stem cell fate in cancer growth, progression and therapy resistance. Nat Rev Cancer. 2018;18(11):669–680. doi: 10.1038/s41568-018-0056-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ratajczak MZ, Bujko K, Mack A, et al. Cancer from the perspective of stem cells and misappropriated tissue regeneration mechanisms. Leukemia. 2018;32(12):2519–2526. doi: 10.1038/s41375-018-0294-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Francesco EM, Sotgia F, Lisanti MP. Cancer stem cells (CSCs): metabolic strategies for their identification and eradication. Biochem J. 2018;475(9):1611–1634. doi: 10.1042/BCJ20170164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan HX, He YQ, Dong H, et al. Physical and functional interaction between receptor-like protein tyrosine phosphatase PCP-2 and beta-catenin. Biochemistry. 2002;41(52):15854–15860. doi: 10.1021/bi026095u [DOI] [PubMed] [Google Scholar]
- 12.Taniguchi Y, London R, Schinkmann K, et al. The receptor protein tyrosine phosphatase, PTP-RO, is upregulated during megakaryocyte differentiation and Is associated with the c-Kit receptor. Blood. 1999;94(2):539–549. [PubMed] [Google Scholar]
- 13.Cheng J, Wu K, Armanini M, et al. A novel protein-tyrosine phosphatase related to the homotypically adhering kappa and mu receptors. J Biol Chem. 1997;272(11):7264–7277. doi: 10.1074/jbc.272.11.7264 [DOI] [PubMed] [Google Scholar]
- 14.Tonks NK. Protein tyrosine phosphatases: from housekeeping enzymes to master-regulators of signal transduction. Febs J. 2013;280(2):346–378. doi: 10.1111/febs.12077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao S, Sedwick D, Wang Z. Genetic alterations of protein tyrosine phosphatases in human cancers. Oncogene. 2015;34(30):3885–3894. doi: 10.1038/onc.2014.462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar P, Munnangi P, Rammohan Chowdary KVS, et al. A human tyrosine phosphatase interactome mapped by proteomic profiling. J Proteome Res. 2017;16(8):2789–2801. doi: 10.1021/acs.jproteome.7b00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobs FM, van der Linden AJ, Wang Y, et al. Identification of Dlk1, Ptpru and Klhl1 as novel Nurr1 target genes in meso-diencephalic dopamine neurons. Development. 2009;136(14):2363–2373. doi: 10.1242/dev.037556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou R, Zhou X, Yin Z, et al. MicroRNA-574-5p promotes metastasis of non-small cell lung cancer by targeting PTPRU. Sci Rep. 2016;6:35714. doi: 10.1038/srep35714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu Z, Liu Y, Li K, et al. Protein tyrosine phosphatase receptor U (PTPRU) is required for glioma growth and motility. Carcinogenesis. 2014;35(8):1901–1910. doi: 10.1093/carcin/bgu123 [DOI] [PubMed] [Google Scholar]
- 20.Liu Y, Zhu Z, Xiong Z, et al. Knockdown of protein tyrosine phosphatase receptor U inhibits growth and motility of gastric cancer cells. Int J Clin Exp Pathol. 2014;7(9):5750–5761. eCollection 2014. [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Lang T, Jin B, et al. Luteolin inhibits colorectal cancer cell epithelial-to-mesenchymal transition by suppressing CREB1 expression revealed by comparative proteomics study. J Proteomics. 2017;161:1–10. doi: 10.1016/j.jprot.2017.04.005 [DOI] [PubMed] [Google Scholar]
- 22.Han K, Lang T, Zhang Z, et al. Luteolin attenuates Wnt signaling via upregulation of FZD6 to suppress prostate cancer stemness revealed by comparative proteomics. Sci Rep. 2018;8(1):8537. doi: 10.1038/s41598-018-26761-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lang T, Ding X, Kong L, et al. NFATC2 is a novel therapeutic target for colorectal cancer stem cells. Onco Targets Ther. 2018;11:6911–6924. eCollection 2018. doi: 10.2147/OTT.S169129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Z, Zhou T, Han X, et al. Mathematical models of amino acid panel for assisting diagnosis of children acute leukemia. J Transl Med. 2019;17(1):38. doi: 10.1186/s12967-019-1973-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kriz V, Korinek V. Wnt, RSPO and Hippo signalling in the intestine and intestinal stem cells. Genes (Basel). 2018;9(1):20. doi: 10.3390/genes9010020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papaspyropoulos A, Bradley L, Thapa A, et al. RASSF1A uncouples Wnt from Hippo signalling and promotes YAP mediated differentiation via p73. Nat Commun. 2018;9(1):424. doi: 10.1038/s41467-017-02786-5 [DOI] [PMC free article] [PubMed] [Google Scholar]