Abstract
A novel LIM-homeodomain gene, Lhx9, was isolated by degenerate RT-PCR followed by mouse embryonic library screening.Lhx9 cDNA encodes a protein that is most closely related to Drosophila apterous and rodent Lhx2 proteins. TheLhx9 spatiotemporal pattern of expression during embryogenesis was similar but distinct from Lhx2. Highest expression levels were found in the diencephalon, telencephalic vesicles, and dorsal mesencephalon. Domains of expression respected the proposed neuromeric boundaries (Puelles and Rubenstein, 1993).Lhx9 was also expressed in the spinal cord, forelimb and hindlimb mesenchyme, and urogenital system. AlthoughLhx9 expression was sustained in diencephalon and mesencephalon from embryonic day 10.5 (E10.5) to postnatal stages, it was transient in the future cerebral cortex, where it was turned off between E14.5 and E16.5. Lhx9 expression was highest if not exclusively located (depending on the region of interest) in the intermediate and mantle zones, as opposed to the mitotic ventricular zone. Lhx9 protein was tested for interaction with the recently discovered cofactors of LIM-homeodomain proteins and was found to interact strongly both with CLIM1 and CLIM2. The expression pattern and structural characteristics of Lhx9 suggest that it encodes a transcription factor that might be involved in the control of cell differentiation of several neural cell types. Furthermore, Lhx9 protein could act in a combinatorial manner with other LIM-homeodomain factors expressed in overlapping pattern.
Keywords: LIM-homeodomain, Lhx9, Lhx2, forebrain, development, neuromeres
A major issue in developmental neurobiology is the understanding of forebrain development and patterning. During embryogenesis, the anterior neural tube generates complex and highly organized structures, such as the cerebral cortex, the basal ganglia, and the thalamus. In adults, these structures are interconnected, almost always topographically.
Homeodomain genes play decisive roles in the establishment of cerebral structures and/or the generation of cell types. Among them, LIM-homeodomain (LIM-hd) genes encode developmentally expressed transcription factors that contain a homeodomain and two cysteine-rich LIM domains involved in protein–protein interactions (Sanchez-Garcia and Rabbitts, 1994; Dawid et al., 1995). In the current view, the LIM domains inhibit DNA binding by the homeodomain. After fixation of cofactors, transcription is activated, and synergism with other transcription factors is promoted. Indeed, cofactors for LIM-hd proteins have been isolated: NLI/Ldb1/CLIM2 and CLIM1 interact with LIM domains of LIM homeoproteins to potentiate transactivation of downstream genes (Agulnick et al., 1996; Jurata et al., 1996; Bach et al., 1997). These cofactors probably confer time and space specificity to the regulatory action of LIM-hd proteins.
LIM-hd genes were first implicated in cell fate determination in invertebrates; Caenorhabditis elegans mec-3 andlin-11 (Way and Chalfie, 1988; Freyd et al., 1990) andDrosophila apterous (Lundgren et al., 1995; Thor and Thomas, 1997) determine cell types and axonal pathfinding. In vertebrates, the spinal cord motor neurons give an example for similar roles of LIM-hd genes: motor neurons express a set of four LIM-hd genes, in a combinatorial manner that is correlated to the motor neuron position in the spinal cord and to the type of peripheral innervated target (Tsuchida et al., 1994). Thus, a LIM-hd combinatorial code might define pathfinding phenotypes. If similar combinations of LIM-hd genes were expressed in the forebrain, they could participate in the establishment of its highly organized circuits. Moreover,Drosophila islet governs aminergic phenotypes (Thor and Thomas, 1997). Forebrain LIM-hd genes could govern neurotransmitter expression in neurons as well.
Homeodomain genes expression patterns in the developing CNS have led to the theory of the prosomeric forebrain (Puelles and Rubenstein, 1993;Rubenstein et al., 1994; Puelles, 1995). In this model, regions and boundaries of expression of developmental factors are seen as landmarks for the determination of cerebral areas according to longitudinal and transverse subdivisions that form segmented structures. LIM-hd genes integrate this model well. Lhx1/lim-1 (Barnes et al., 1994),Lhx2/LH-2 (Xu et al., 1993), Lhx3/pLIM (Seidah et al., 1994; Bach et al., 1995), L3/Lhx8 (Matsumoto et al., 1996), Lhx4/Gsh4 (Li et al., 1994), Lhx5 (Sheng et al., 1997), Lhx6, and Lhx7 (Grigoriou et al., 1998) are all expressed in the developing rodent CNS, and their expression patterns respect neuromeric boundaries (present paper). The role of LIM-hd genes in cellular specification, combined with their apparent function in the coding of positional information, makes them important factors to be studied during brain regionalization and wiring.
In a search to isolate more LIM-hd involved in forebrain development, we hypothesized the existence of a subfamily of Lhx2-related LIM-hd genes. An RT-PCR cloning strategy allowed the isolation of mouseLhx9, which is similar to but distinct from Lhx2in sequence and expression pattern, the two genes thereby forming a new subfamily of forebrain LIM-hd genes.
MATERIALS AND METHODS
RT-PCR cloning. Total RNA from embryonic day 15 (E15) or E18 forebrain were reverse-transcribed to cDNA with avian myeloblastosis virus reverse transcriptase (Boehringer Mannheim, Mannheim, Germany) and used as templates for PCR reactions (Qiagen, Hilden, Germany) using the following degenerate oligonucleotide primers: 51, ATGCGIACITCITTYAARCAYCAYCARCT; 52, ATGAGRACITCITTYAARCAYCARCT; 31, MAYTTIGCIGCIGCRTTYTGRAACCA; and 32, MAYTTIGCYCTIGCRTTYTGRAACCA, where I is an inosine residue. 5′ (51 and 52) and 3′ (31 and 32) primers were designed to isolate genes of theLhx2 subfamily and were chosen to amplify a stretch of DNA in the homeodomain. Any combination of 3′ and 5′ primers led to the amplification of a single band of 160 bp as expected. After cloning of this 160 bp fragment into pGEM-T (Promega, Madison, WI), sequencing of 80 independent clones revealed the presence of three different fragments. One was mouse Lhx2, and the two others were unknown. We concentrated on one that we named Lhx9. The 160 bp DNA fragment was used to screen a λgt11 E14.5 mouse head cDNA library under high stringency. The Lhx9 cDNA was isolated using standard molecular biology techniques and sequenced on both strands with Amersham (Les Ulis, France) thermosequenase. The 1.016 kb PCR product of Lhx9 cDNA was subcloned into pGEM-T (Promega) for subsequent experiments.
Tissue preparation. Timed pregnant C57/Black6 mice were anesthetized with 0.1 ml/100 gm body weight pentobarbital. Embryos were harvested, fixed in 0.1 m 4-morpholinepropanesulfonic acid, pH 7.4, 2 mm EGTA, 1 mmMgSO4, and 3.7% formaldehyde at room temperature, cryoprotected in 30% sucrose, and frozen by immersion in isopentane on dry ice. Noon on the day after the night of mating was considered as E0.5. Postnatal day 1 (P1)–P7 brains were processed identically, except that sucrose was 20%. Whole embryos or dissected brains were cryostat-sectioned at 20 μm, and sections were stored at −80°C until use.
In situ hybridization and image processing. The pGEM-Lhx9 plasmid was linearized with NdEI orNcoI and used as template for RNA synthesis with T7 or SP6 polymerase in the presence of [35S]UTP (10 mCi/mmol; ICN Biochemicals, Costa Mesa, CA) for antisense and sense control probes, respectively. The probe thus included the 5′ noncoding region, the two LIM domains, the linker region, and the beginning of the homeodomain. The Bluescript-Lhx2 plasmid including the 1.1 kb Lhx2 insert was linearized with EcoRI, and T3 polymerase was used for antisense probe synthesis as described (Bach et al., 1997). The Bluescript-En2 3′ untranslated region plasmid was linearized with ClaI, and T7 was used to generate an 800 bp En-2 probe. Restriction enzymes were purchased from Appligene (Heidelberg, Germany) or Promega, and RNA polymerases were from Stratagene (La Jolla, CA). Briefly, slides were acetylated, dehydrated progressively, and hybridized 16 hr at 60°C in the presence of 5 × 105–106 cpm of probe/slide. They were then washed under high stringency, treated 1 hr with RNase A (20 μg/ml), and dehydrated. Sections were exposed 2–3 d to Eastman Kodak (Rochester, NY) Biomax MR film for autoradiography and then dipped in Kodak NTB-2 emulsion. Emulsions were developed after 10–12 d of exposure, and sections were counterstained with methylene blue. Photographs were taken on a Leica (Nussloch, Germany) DMBL microscope, scanned on a Canon (Tokyo, Japan) 2700 scanner, and mounted for figures with Adobe (Mountain View, CA) Photoshop. Images were corrected for color balance, contrast, brightness, or cropping, but no other corrections were made.
In vitro protein–protein interaction assays. PCR products of full-length CLIM1a and CLIM2 were ligated in frame into theXhoI–XbaI sites of the bacterial expression vector pGEX-KG to yield a glutathione S-transferase (GST) fusion protein. The PCR product of the 1.016 kb Lhx9 cDNA was subcloned into the pGEM-T vector (Promega), and [35S]methionine-labeled Lhx9 protein was produced using the Promega in vitro transcription–translation kit, SP6 RNA polymerase, and [35S]methionine. Thein vitro protein–protein interaction assays were performed as described previously (Bach et al., 1995).
RESULTS
Cloning of Lhx9, a novel LIM homeodomain gene
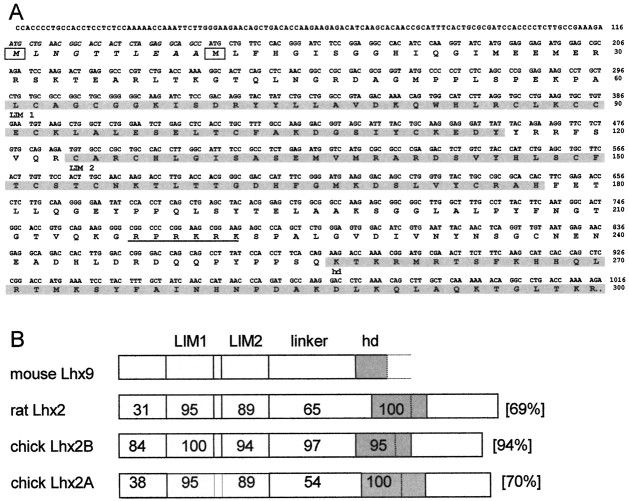
The RT-PCR strategy for cloning new LIM-hd genes expressed in the forebrain was based on the observation that the Lhx2 homeodomain was slightly different from other LIM-hd genes expressed in more posterior parts of the brain. An SFKHH amino acid sequence was found in the N terminus part of its homeodomain, instead of the often encountered TITAK motif. We hypothesized the existence of other genes of the same subfamily that could be specific for the forebrain. Degenerate PCR primers were thus designed to amplify a theoretical 160 bp DNA fragment spanning almost the entire homeodomain (with a 5′ primer at the level of the SFKHH motif and a 3′ primer at the level of the highly conserved VWFQN motif close to the C terminus of the homeodomain). Sequencing of the 160 bp RT-PCR fragment revealed a new sequence (19 nucleotides of 102 were different from Lhx2 but were conservative at the amino acid level). This PCR fragment was used to screen an E14.5 mouse head λgt11 cDNA library and allowed the isolation of Lhx9. The ∼1 kb cDNA contained 5′ noncoding sequences and, in the open reading frame, two LIM domains and a homeodomain (Fig.1A, shading). The C-terminal part and the stop codon were not included in this cDNA. Additional screens could not identify the 3′ end missing sequences. However, the 300 amino acids encoded by the cDNA are largely sufficient to allow for Lhx9 classification and riboprobe synthesis. There are two in-frame methionine residues (Fig. 1A, boxed) located 10 amino acids apart at the level of the translation initiation site. Based on similarity with Lhx2, we postulate that the second methionine will be used for translation start. Lhx9 shares 69% identity with rat Lhx2 at the amino acid level and 68% at the nucleotide level. The amino acid identity rises to 95% in the LIM domains (Fig. 1B) and to 100% in the homeodomain. The main differences between the two proteins are located between the second LIM domain and the homeodomain. The linker fragment between LIM2 and the homeobox is shorter in Lhx9 [79 amino acids (aa) in Lhx9 and 96 aa in Lhx2] and is more closely related in sequence and in length to chick Lhx2B [also 79 aa long (G. Tremml and T. M. Jessell, unpublished data; GenBank accession number L35566] (Nohno et al., 1997). A basic sequence (RPRKRK; Fig. 1A, underlined) that might serve as a nuclear translocation signal is conserved in the middle of the linker region. The sequence similarities between Lhx9 and Lhx2 strongly support the notion that they are two closely related family members (Fig.1B). Indeed, a comparison with other LIM-hd genes in GenBank showed that Lhx9 belongs to a growing subfamily including vertebrate Lhx2, Drosophila apterous, and C. elegans ttx-3 (Hobert et al., 1997). Moreover, the very high (overall 94%) identity between mouse Lhx9 and chick Lhx2 suggests that chick Lhx2 could rather be the avian homolog of rodent Lhx9 (Fig.1B).
Fig. 1.
Sequence of murine Lhx9.A, Nucleotide and deduced amino acid sequence ofLhx9. Amino acids are indicated by single-letter code. The two in-frame ATG start codons areboxed. The sequence between the two possible start codons is in italics solely to indicate that based on homology with Lhx2, the second one is more likely to be used. LIM1, LIM2, and the hd are indicated by shading. The putative nuclearization signal is underlined.B, Comparison of murine Lhx9 with rat and chick Lhx2. The percentage of identity in each domain between gene products is indicated. GenBank/EMBL accession numbers: L06804 (rat Lhx2), L35566(chick Lh2B), and AB005882 (chick Lh2A).
Expression of Lhx9 during embryogenesis of the nervous system
Whole-mount in situ hybridization showed that the onset of Lhx9 expression in the nervous system was at approximately E10.5 (data not shown). Thus, from E11.5 onward, the expression pattern was studied by in situ hybridization on sections (Figs. 2, 3; Table1). From E11.5 to E14.5, Lhx9expression was relatively widespread throughout the CNS alar neuroepithelium, with an increase between E11.5 and E12.5 (Fig. 2, compare A,B with C,D). Expression remained high in the following stages.
Fig. 2.
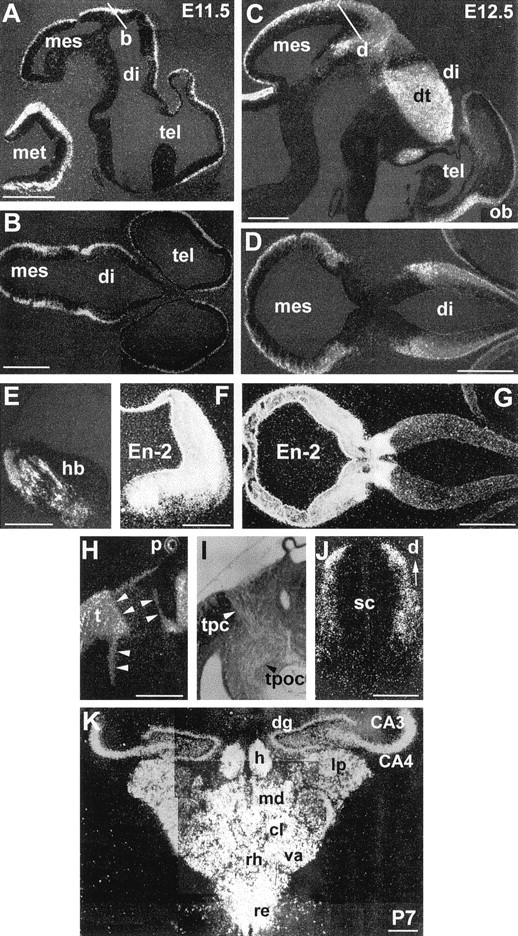
Lhx9 expression on brain sections from E11.5 (A, B), E12.5 (C–E, J), E13.5 (H, I), and P7 (K) embryos and comparison withEn-2 (F, G) on dark-field illumination photomicrographs. In this and subsequent figures, pictures are oriented so that anterior is right and dorsal is at thetop. A, C, Parasagittal sections through the entire brain at indicated stages. B, D, Horizontal sections (level and orientation indicated by white barsin A, C, respectively). G,En-2 expression on a section adjacent to that shown inD. E, F, Adjacent coronal hemisections showing the comparison of Lhx9 (E) and En-2 (F) expression in the anterior hindbrain. H, I, Bright-field (I) and dark-field (H) views of the same area showing the correlation between the position of the tpc and the tract of the postoptic commissure (tpoc) and Lhx9sharp boundaries of expression (arrowheads).J, Coronal section through the spinal cord.K, Coronal section at the level of the hippocampus and thalamus at P7, showing expression in CA3, CA4, and the dentate gyrus (dg) and the delineation of various thalamic nuclei by Lhx9 hybridization signal. cl, Central lateral; hr, habenula; lp, lateral posterior; md, mediodorsal; va, ventral anterior; re, reuniens; rh, rhomboid. See Table 1 for anatomical abbreviations. Scale bars:A–I, K, 100 μm; J, 50 μm.
Fig. 3.
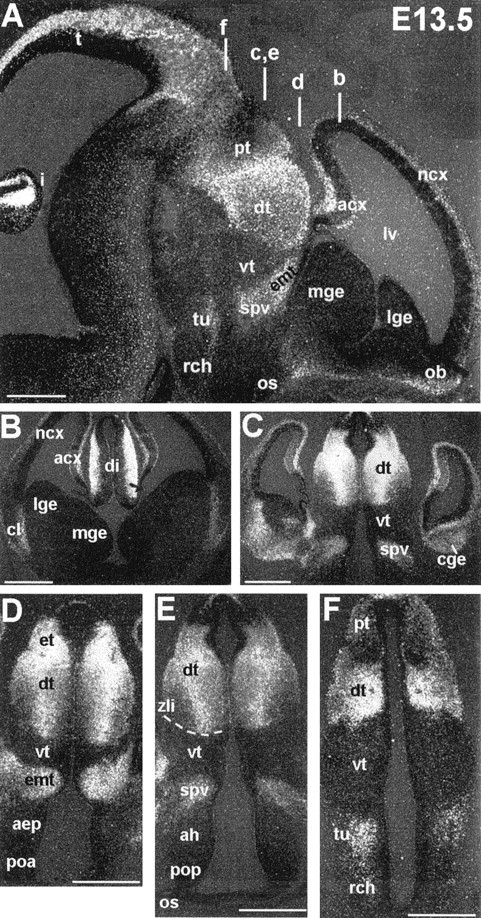
Lhx9 expression at E13.5.A, Sagittal section through the entire brain.B–F, coronal sections through the telencephalon and diencephalon (plan of section indicated by white bars inA) showing Lhx9 transverse boundaries in the rostrocaudal extension. The dotted line indicates the zli. Scale bars, 100 μm. See Table 1 for anatomical abbreviations.
Table 1.
List of anatomical abbreviations
| acx | Archicortex (hippocampus) |
| aep | Anterior entopeduncular area |
| ah | Anterior hypothalamus |
| cb | Cerebellum |
| cge | Caudal ganglionic eminence |
| ci, cs | Colliculus, inferior, superior |
| cl | Claustrum |
| cp | Choroid plexus |
| db | Diagonal band |
| di | Diencephalon |
| dt | Dorsal thalamus |
| e | Eye |
| emt | Eminentia thalami |
| et | Epithalamus |
| h | Habenula |
| hb | Hindbrain |
| hcc | Hypothalamic cell cord |
| is | Isthmus of the mesencephalon |
| lge | Lateral ganglionic eminence |
| lv | Lateral ventricle |
| ma | Mamillary area |
| mes | Mesencephalon |
| met | Metencephalon |
| mge | Medial ganglionic eminence |
| ncx | Neocortex |
| ob | Olfactory bulb |
| os | Optic stalk |
| p | Pineal gland |
| p1–p6 | Prosomeres 1–6 |
| pep | Posterior entopeduncular area |
| poa | Anterior preoptic area |
| pop | Posterior preoptic area |
| pt | Pretectum |
| rch | Retrochiasmatic area |
| rm | Retromammilary area |
| rp | Rathke’s pouch (pituitary) |
| sch | Suprachismatic area |
| se | Septum |
| spv | Supraoptic paraventricular area |
| t | Tectum |
| tel | Telencephalon |
| tpc | Tract of the posterior commissure |
| tu | Tuberal hypothalamus |
| vt | Ventral thalamus |
| zli | Zona limitans intrathalamica |
In the telencephalic vesicles Lhx9 mRNA was expressed in the future cerebral cortex, the hippocampus, the claustrum primordium (where it could correspond to a stream of migratory cells from the striatum to the cortex; see De Carlos et al., 1996), and the caudal ganglionic eminence (amygdaloid complex) (Figs.2A–C, 3A–C). It was also expressed in the olfactory bulb primordium (Figs. 2C, 3A). Interestingly, levels of expression in the telencephalic walls depended on the region examined: it was relatively weak in the parietal neocortex, high in the frontal neocortex (data not shown), and strongest in the archicortex (hippocampal field; Fig.3B,C).
The diencephalon showed the highest expression levels (Figs.2C,D, 3). Moreover, in this region, expression was observed in longitudinal and transverse bands, with sharp boundaries that respect the neuromeric regionalization proposed by Puelles and Rubenstein (1993) (Fig. 3). Indeed, from caudal to rostral (Fig.3A), Lhx9 was expressed in the pretectum (p1 prosomere) and in the dorsal thalamus (except for a thin ventral band close to the alar–basal boundary), the epithalamus, and the epiphysis in prosomere p2. Expression stopped abruptly at the zona limitans intrathalamica, showing no expression in the ventral thalamus (p3 prosomere; Fig. 3C–F). The supraoptic–paraventricular area and the eminentia thalami again formed a band of strong expression in the p4 prosomere that abuts on a negative region around the optic stalk (prosomeres p5 and p6; Fig.3A,D,E). A narrow band including the retrochiasmatic area and the tuberal hypothalamus expressed Lhx9 and represented the only domains of expression in the basal plate (Fig.3A,F; also see Fig. 8A). Interestingly, the boundaries of expression of Lhx9 at the diencephalic–mesencephalic junction delineated perfectly the borders of the trajectory used by pioneering tracts of the posterior and postoptic commissure (Fig. 2H,I).
Fig. 8.
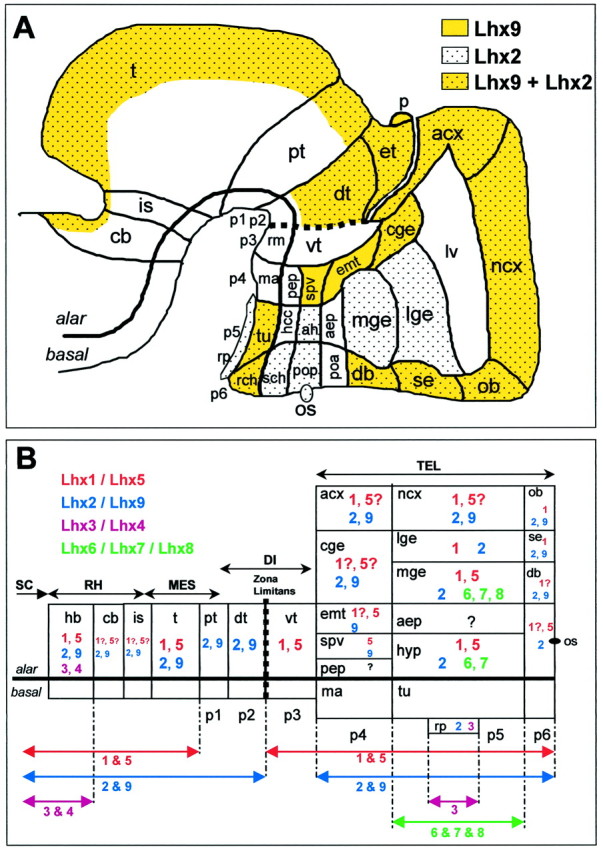
Summary of Lhx9 andLhx2 expression patterns in the context of the neuromeric model (A) and schematic representation of Lhx combinatorial expression in the developing brain (B). Adapted from Puelles and Rubenstein (1993).A, Schematic E13.5 brain shows similarities, differences, and boundaries of expression of Lhx9(yellow) and Lhx2(dotted). Expression in the ventral mesencephalon and metencephalon is not indicated because of its rather “nuclear-like” pattern. B, Compilation of expression data forLhx1/5 (red; Sheng et al., 1997), Lhx2/9 (blue; this paper),Lhx3/4 (pink; Li et al., 1994;Bach et al., 1995), and Lhx6/7/8(green; Matsumoto et al., 1996; Grigoriou et al., 1998) on a “flattened brain” adapted from Bulfone et al. (1993). Gene expression is indicated by color-coded numbers, and a possible Lhx combinatorial code clearly emerges. The anteroposterior extension of expression is recapitulated byarrows at the bottom. Basal plate expression is not indicated. The zli is indicated by a dotted line. p1–p6, Prosomeres 1–6. See Table 1 for anatomical abbreviations.
Posteriorly, Lhx9 was expressed throughout the tectum in the mesencephalon (Figs. 2A–D, 3A), in the walls of the hindbrain (Fig. 2A,E), and in nuclei in the ventral midbrain and hindbrain (Fig.4I). The comparison with En-2, a major homeobox gene defining the mesencephalic–metencephalic region, showed that the expression of the two genes was quite different, neither totally overlapping nor strictly complementary (Fig. 2, compare D with G,E with F).
Fig. 4.
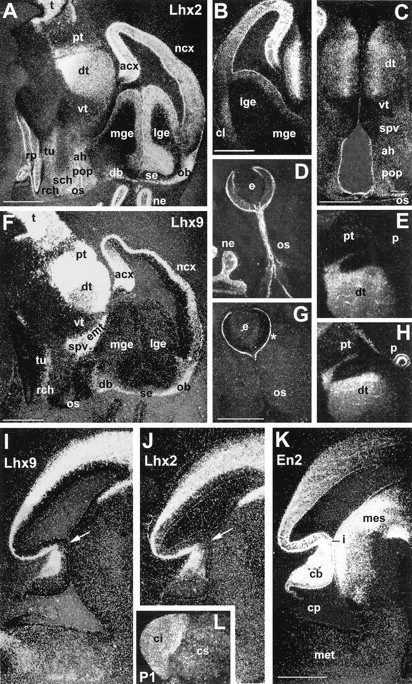
Comparison of Lhx2 (A–E, J),Lhx9 (F–I, L), and En-2(K) expression on sections. A, F, Parasagittal adjacent sections through the entire brain at E14.5, comparing expression of Lhx2 (A) and Lhx9 (F). B, C, Coronal sections at E13.5 showing Lhx2 expression in the telencephalon and diencephalon. Sections are adjacent to those shown in Figure 3, B and E, respectively, for comparison of transverse boundaries. D, G, Horizontal adjacent sections showing Lhx2 (D) and Lhx9 (G) expression in the eye and optic stalk at E14.5. The asterisk inG points to the nonspecific signal attributable to the presence of the eye pigmented epithelium. E, H, Adjacent parasagittal section showing Lhx2(E) and Lhx9(H) in the dorsal diencephalon and pretectum at E13.5. Note the absence of Lhx2 in the pineal gland.I–K, Adjacent parasagittal sections at E13.5 to compareLhx9 (I),Lhx2 (J), and En-2(K) expression through the mesencephalon and metencephalon. Arrows in I andJ show the thinning of expression at the isthmus.L, Sagittal section through the midbrain of a P1 animal to show the Lhx9 sharp boundary between the superior and the inferior colliculi. Compare with I (earlier stage). Scale bars, 100 μm. ne, Nasal epithelium. See Table 1for abbreviations.
In the spinal cord, Lhx9 was expressed as a gradient in the dorsal part of the neuroepithelium (Fig. 2J, corresponding to the region where commissural and association neurons differentiate) but not in motor neurons.
Between E14.5 and E16.5, a major change in Lhx9 expression occurred in the neocortical neuroepithelium, because it was turned off (Fig. 5; see below). However, expression persisted in the archicortex (in the dentate gyrus, CA3 and CA4) later on, as well as in the diencephalon, midbrain, and hindbrain. Indeed, at early postnatal stages, (P1–P7), expression was particularly abundant in the dorsal thalamus where it allowed the delineation of various nuclei (Fig. 2K). Lhx9 marked a sharp boundary between the very-high-expressing inferior colliculus and the high-expressing superior colliculus (Fig. 4L). A number of mesencephalic and pons nuclei, including deep cerebellar nuclei, still expressed Lhx9 at these postnatal stages (data not shown).
Fig. 5.
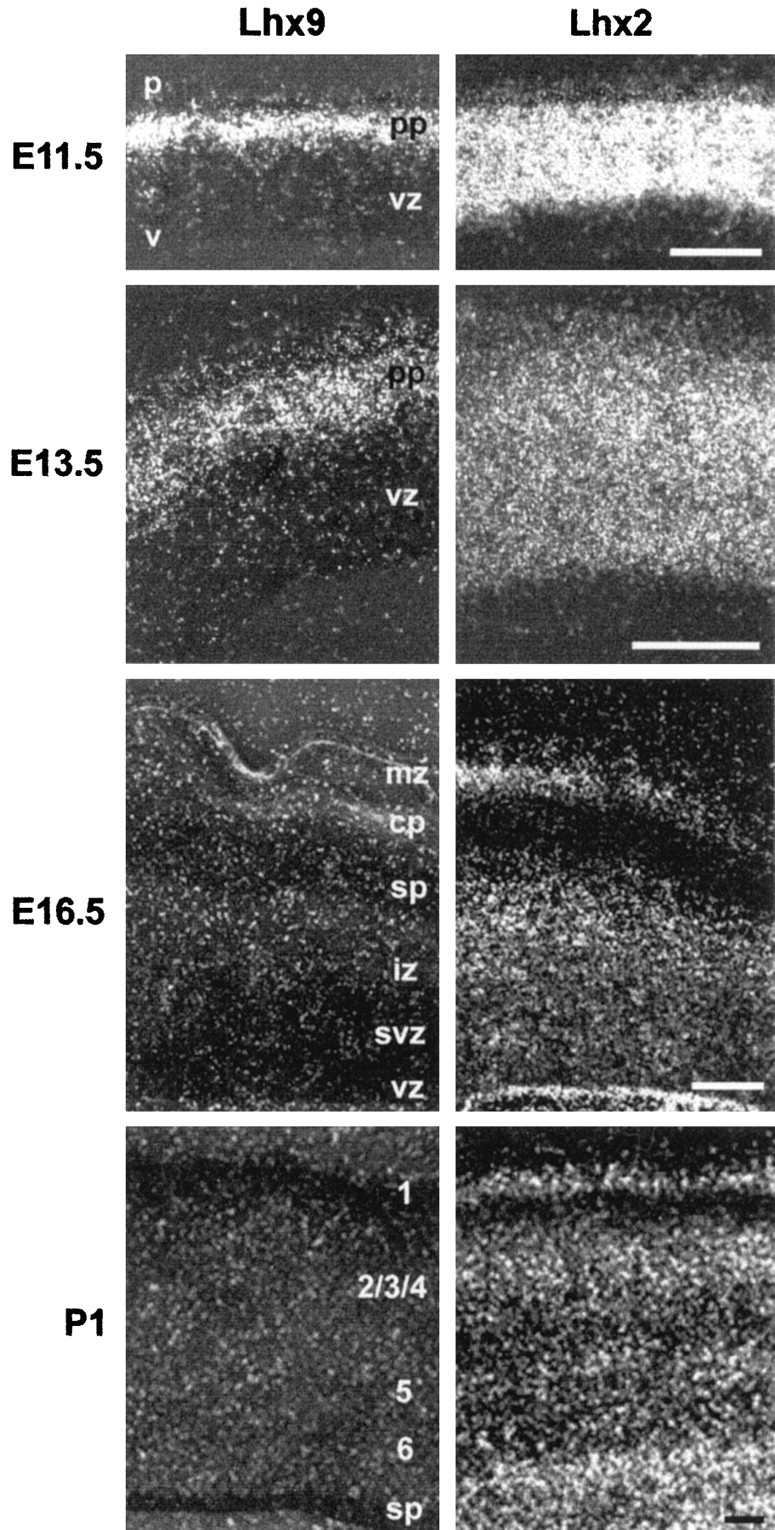
Comparison of Lhx9(left) and Lhx2 (right) expression in the neocortical epithelium at indicated stages. The pial surface (p) is at the top, and the ventricular surface (v) is at thebottom. Left and rightphotographs were taken on adjacent sections. cp, Cortical plate; iz, intermediate zone;mz, marginal zone; pp, preplate;sp, subplate; svz, subventricular zone;vz, ventricular zone. Scale bars, 100 μm.
In summary, Lhx9 expression was relatively widespread, mostly in the alar region of the neuroepithelium. In this respect, it is noteworthy that expression was absent from the large region of the ganglionic eminences in the ventral telencephalon (corresponding to the striatal–pallidal primordium).
Lhx9 and Lhx2 are expressed in overlapping but distinct patterns
Because Lhx9 and Lhx2 are closely related family members, we next compared directly the expression of the two genes.
Concerning regional expression, the two genes were expressed mainly in overlapping patterns, with few noticeable exceptions: Lhx2was expressed in the ganglionic eminences, the eyes, the optic stalk, and the neuromeres surrounding the optic stalk, whereas Lhx9was not (compare Figs. 4A,F, 3B,4B). In the prosencephalic region, the two mRNAs were expressed in complementary patterns, showing bands of expression with sharp boundaries, separated by expression-negative zones. More precisely, Lhx2 was not expressed in the eminentia thalami–supraoptic paraventricular areas, whereas Lhx9 was, but the Lhx9-negative zone at the optic stalk, preoptic area, and anterior hypothalamus was Lhx2 positive (compare Figs. 4A,F, 4D–G, 3E,4C). Expression of the two genes was perfectly overlapping in the dorsal thalamus and was absent for both genes in the ventral thalamus (Fig. 4, compare A–F). Thus, the addition of the two expression domains covered almost the entire diencephalic area with the exception of the ventral thalamus (for summary, see Fig. 8A).
In the tectal epithelium, the expression patterns of the Lhxgenes were overlapping in the marginal layers (Fig.4I–J). In contrast to engrailed,Lhx9 and Lhx2 were not expressed as a gradient. Their expression domains spanned the mesencephalic–metencephalic junction, slightly thinning out at the isthmus (Fig.4I–J, arrows). Furthermore, when compared withEn-2, they were not as largely expressed in the ventral mesencephalon (Fig. 4, compare I–K).
Interesting differences between the two family members were observed at the cellular level. In the telencephalic vesicles, Lhx9 mRNA was exclusively located in the differentiating layers of the neuroepithelium, as opposed to the mitotic ventricular zone, whereasLhx2 was expressed throughout the depth of the neocortical epithelium, including the ventricular and the differentiating mantle zones (Fig. 5). Concerning the timing of Lhx9 andLhx2 expression, a major difference was again found in the neocortex. Whereas Lhx9 expression stopped between E14.5 and E16.5, Lhx2 was still expressed in the neocortex until P1 (Fig. 5) and later (data not shown). Moreover, although Lhx9stopped in hippocampal fields CA1 and CA2 around birth (Fig.3K), Lhx2 was still expressed in the whole hippocampus (data not shown).
Thus, Lhx9 and Lhx2 are expressed in patterns that are compatible both for redundant and complementary roles during brain regionalization and neurogenesis.
Lhx9 expression outside the CNS
Lhx9 was also expressed in a few developing regions outside the CNS. This included the developing forelimb and hindlimb buds, where a gradient of expression was observed at early stages in the distal mesenchyme (Fig. 6B,C). Later on, expression was progressively restricted to the interdigit spaces corresponding to the region where programmed cell death happens for finger formation and to the region surrounding the cartilages (Fig.6D,E). Moreover, Lhx9 mRNA was also highly expressed in the caelomic cavity at the level of the urogenital ridge, including the gonads and parts of the pancreas and liver epithelium (Fig. 6A). As opposed to Lhx2,Lhx9 was expressed neither in the nasal epithelium nor in the pituitary (Fig. 4A).
Fig. 6.
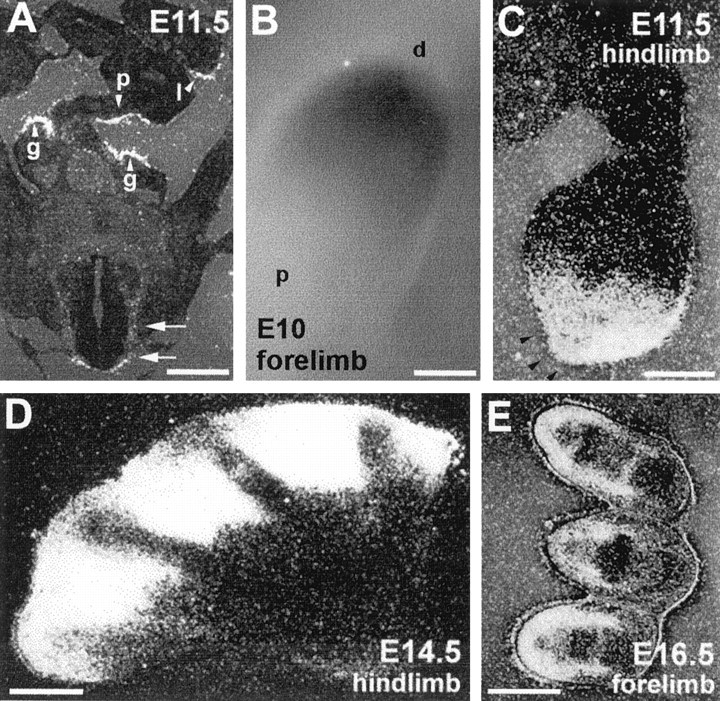
Lhx9 expression in limbs and urogenital ridge. A, Coronal section through the caelomic cavity and the spinal cord (double arrow) at E11.5. Arrowheads point to the developing gonads (g), pancreas (p), and liver (l). Because sex discrimination is not easy at this stage, it should be noted that the same expression pattern was found in all embryos examined. B, Expression in E10 forelimb after in toto in situ hybridization.p, Proximal; d, distal.C–E, Transverse sections through developing hindlimbs and forelimbs at indicated stages. Arrowheads inC indicate absence of expression in the apical ectodermal ridge. See Results for details. Scale bars: A, C–E, 50 μm; B, 200 μm.
Lhx9 protein interacts with CLIM1 and CLIM2 cofactors
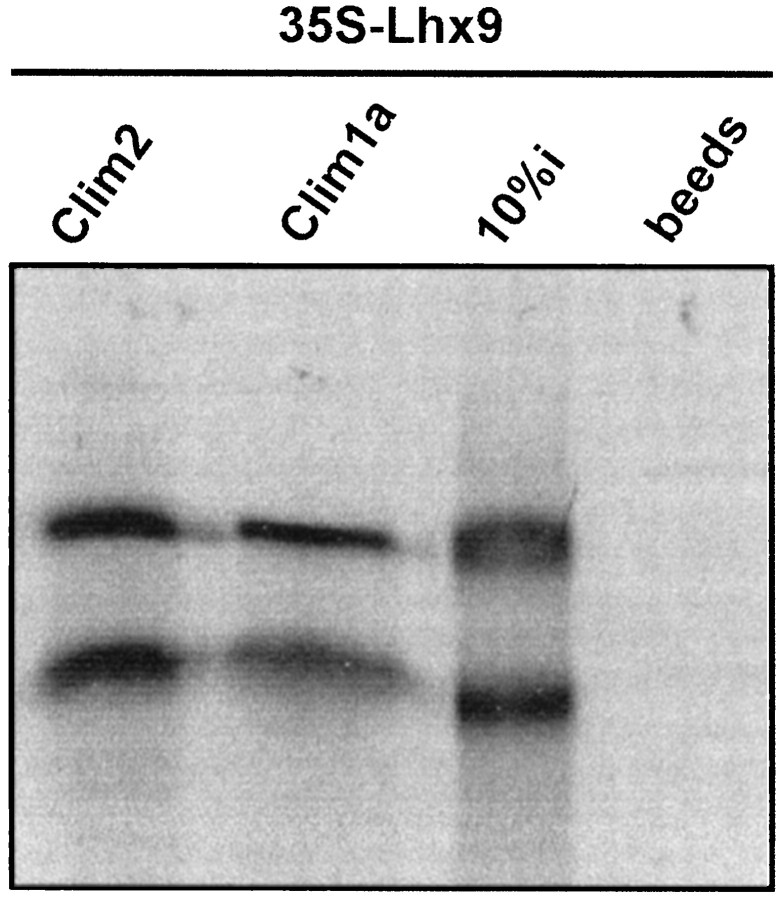
Because the Lhx9 expression pattern is partially overlapping with the expression of the recently isolated cofactors of LIM-hd proteins, we tested whether Lhx9 protein could physically interact with CLIM1 and CLIM2. GST pull-down assays were performed using [35S]Lhx9 and CLIM1 and CLIM2 GST fusion proteins. The results show that Lhx9 strongly interacts with both cofactors in vitro (Fig. 7), suggesting possible functional interactions in vivo.
Fig. 7.
Lhx9 interacts with CLIM1 and CLIM2. Autoradiogram of a representative GST pull-down assay in which35S-labeled, in vitro-translated Lhx9 was tested for its ability to bind bacterially expressed GST fusions of CLIM1 and CLIM2. Ten percent (10%i) of the total35S-labeled protein input and binding to beads alone are shown.
DISCUSSION
Lhx9 and Lhx2 form a subfamily of LIM-hd genes
We have isolated a new LIM-hd gene named Lhx9, thereby increasing the number of LIM-hd genes expressed in the developing CNS.Lhx9 is most closely related to rodent Lhx2 (Xu et al., 1993) and chick Lh2B-Lh2A (Nohno et al., 1997),Drosophila apterous (Lundgren et al., 1995), and C. elegans ttx-3 (Hobert et al., 1997). In particular, these proteins share a short but notable difference in their homeodomain when compared with other LIM-hd factors. Surprisingly, although the homology between mouse Lhx9 and chick Lh2B is very high (94%), the Lhx9expression pattern is more related to chick Lh2A, at least in limbs, in which chick Lh2A expression was thoroughly described (Nohno et al., 1997). The high degree of conservation betweenLhx9 and Lhx2 suggests that they belong to the same subfamily. Interestingly, in vertebrates, LIM-hd genes are often encountered in pairs of closely related homologs, such asLhx1/Lhx5, Lhx3/Lhx4, Lhx6/Lhx7/Lhx8, and now Lhx2/Lhx9. Generally, the two genes of the pair show similar expression patterns (Fig.8B). This redundancy among the gene family suggests critical and complementary (or synergistic) roles played by these transcription factors during brain formation.
In this respect, it is interesting to rediscuss the phenotype ofLhx2−/− embryos with the knowledge of the existence of Lhx9. Lhx2−/− embryos are anophtalmic, with an aplasic archicortex and hypoplasic neocortex and basal ganglia, but their diencephalon and mesencephalon look normal (Porter et al., 1997). Porter et al. (1997) hypothesized that, depending on the region examined, the more or less defective phenotype could result from functional compensation by an unknown relative gene.Lhx9 is not expressed in eyes and basal ganglia but is present in the neocortex, archicortex, diencephalon, and mesencephalon. Thus, functional compensation by Lhx9 might actually happen for the neocortex, diencephalon, and mesencephalon cases. It should be noted that the expression patterns of Lhx1 andLhx5 are also compatible for a partial functional redundancy at these levels, including basal ganglia (Sheng et al., 1997). In contrast, the eyeless phenotype ofLhx2−/− embryos suggests thatLhx2 is a major factor of this family in the eye. Finally, in the case of the hippocampus, it appears that althoughLhx9 is relatively strongly expressed in this area, it is not able to compensate for the lack of Lhx2 expression. This could mean that Lhx9 functions in the archicortex and neocortex are different. However, more detailed data on Lhx1and Lhx5 expression would help in understanding the possibleLhx combinations for regional specification. Overall, the very high expression levels of Lhx9 in the diencephalon and mesencephalon, combined with the lack of phenotype in these areas inLhx2−/− embryos, suggests thatLhx9 plays an important role in the specification of the thalamus and tectum.
Lhx9 respects neuromeric boundaries
Transverse and longitudinal subdivisions of the developing brain, corresponding to domains and boundaries of expression of a number of developmental factors, and coincident with morphological structures such as ventricular ridges and external furrows, have allowed the proposal of a neuromeric organization of the forebrain, as described in the model of Puelles and Rubenstein (e.g., Bulfone et al., 1993, 1995;Porteus et al., 1994). The detailed study of Lhx9 andLhx2 expression patterns shows that they respect the proposed neuromeric boundaries. This is summarized in Figure8A, and we would like to discuss the following points. (1) The expression patterns clearly corroborate the proposed demarcations for p4 prosomere and distinguish nicely both p3–p4 and p4–p5 boundaries, which is important because most gene markers do not distinguish the p3–p4 limit well. (2) In a more recent variation of their early model, Bulfone et al. (1995) suggest that the entire cerebral cortex (ncx and acx) could be either in p5 (implying that the lge–cortex boundary is longitudinal, and that the cge is the telencephalic portion of p4) or in p4, implying that the lge–cortex boundary is transverse (i.e., neuromeric). The expression patterns observed here are consistent with both views. From the Lhx9pattern, the cge clearly belongs to p4. In the telencephalon, however, the Lhx9 spatiotemporal pattern respects the putative transverse boundary between the archicortex (with strong and persistent expression) and the neocortex (with lower and transient expression), a pattern that is compatible with the early model with acx in p4 and ncx in p5. On the other end, as in many other genes, Lhx9 also distinguishes the cortex–lge boundary, a feature that is more in favor of the recent model in which this boundary is viewed as neuromeric. (3) In the diencephalon, Lhx2 and Lhx9 also give a sharp delineation of the ventral thalamus (in p3) that is free of signal between the adjacent prosomeres p4 and p2 (including the dorsal thalamus with massive expression and the epithalamus), thereby respecting the zona limitans intrathalamica. The other diencephalic subdivisions are well respected, with longitudinal and transverse divisions around the optic stalk region. At this level it is interesting to note that the Lhx9-negative optic stalk zone is complementary with Lhx2-positive areas. Also,Lhx9 and Lhx2 patterns respect a narrow negative band in the ventral part of the dorsal thalamus, close to the alar–basal boundary, that is similar to the Gbx-2expression pattern at this level (Bulfone et al., 1993). Interestingly, in the tectum, a sharp boundary appeared between the inferior and superior colliculi around birth and could be relevant to the establishment of optic versus auditory systems. Taken together, these results show that the Lhx9 pattern integrates well in the neuromeric model and could participate in the establishment of the identity of neuromeres.
The observation that Lhx9 (and Lhx2) expression borders the pathway of pioneering tracts at the dimesencephalic junction is also interesting. Indeed, it has been hypothesized that domains and boundaries of expression of a position-encoding homeodomain gene give landmarks for the establishment of the main axonal tracts in the brain (Figdor and Stern, 1993; Wilson et al., 1993;Macdonald et al., 1994; Rétaux et al., 1996). The two LIM-hd genes studied here might participate as local cues for the guidance of commissural axons at the dimesencephalic junction.
Lhx9 is transiently expressed in the marginal layers of the neocortical epithelium
Before E11, the cerebral walls are constituted of a pseudostratified ventricular epithelium in which progenitor proliferation occurs (for review, see Caviness et al., 1995). The onset of mouse cortical neurogenesis has been reported between E10 and E11 (Angevine and Sidman, 1961; Caviness, 1982), a time that corresponds almost exactly to the onset of Lhx9 expression. Moreover, as opposed to Lhx2 and Lhx1 (Sheng et al., 1997), whose expression spans the depth of the cortical neuroepithelium,Lhx9 expression is restricted to the outer layers. From its expression pattern in the cortical preplate where the first postmitotic cells are located, and from its absence in the mitotic ventricular zone, Lhx9 is a good candidate as a factor implicated in differentiation and phenotype acquisition. Moreover, neocorticalLhx9 expression is switched off between E14.5 and E16.5. The exact time of Lhx9 decrease in expression will have to be determined precisely, and the correlation with events related to neurogenesis will have to be studied. However, from birth-dating studies, this stage in mice (∼E16.5) corresponds approximately to the completion of generation of layer VI–V neurons, to the beginning of layer IV generation, and to the arrival of thalamic afferents (Caviness, 1982; Polleux et al., 1997). Thus, Lhx9 is expressed during the time deep layer (VI–V) cortical neurons are generated.
Although Lhx2 was first hypothesized to play a role in cortical neuron differentiation (Xu et al., 1993), it is probably also involved in precursor proliferation (Porter et al., 1997). As discussed above, Lhx9 is rather involved in differentiation. Other homeobox genes from the Emx and Otx families are expressed in interesting patterns in the cortical neuroepithelium:Emx2 is located exclusively in proliferating cells, whereasEmx1 is found in both proliferating and differentiated neurons (Gulisano et al., 1996). Otx1 is specifically expressed in dividing and differentiated layer V–VI neurons (Frantz et al., 1994). Genes from other families, such as T-Brain1, are expressed exclusively in postmitotic cortical cells (Bulfone et al., 1995). Thus, it seems possible that combinations of all these transcription factors interact to regulate cortical neurogenesis and layer formation (also see next section).
Lhx9 protein interacts with CLIM cofactors
In the pituitary, interaction between CLIM cofactors and LIM domains of the LIM-hd protein P-lim promotes synergistic interactions between P-lim and the homeobox-only protein P-otx on the glycoprotein hormone α subunit promoter (Bach et al., 1997). The CLIMs could physically and functionally interact as well with a number of LIM-hd proteins to confer time and space specificity to the transcriptional control they exert. CLIM1 and CLIM2 are widely expressed in the developing brain (Bach et al., 1997), including regions whereLhx9 is expressed. Moreover, we show that Lhx9 strongly interacts with both of them. In the cortical neuroepithelium, for example, CLIM1 is selectively expressed in the cortical plate, as well as Lhx9 until approximately E16.5.Otx1 is expressed in dividing and differentiating layer V–VI neurons (see above; Frantz et al., 1994). Making a parallel with the interactions described in the pituitary between P-lim, P-otx, and CLIMs, it is interesting to hypothesize that a trio of the same protein families, i.e., Lhx9, Otx1, and CLIM1, are candidates for functional interaction involved in the generation of deep layer cortical neurons.
LIM-hd genes acting in a combinatorial manner?
In the spinal cord, the combinatorial expression of the four LIM-hd genes, Islet-1, Islet-2, Lim-1, and Lim-3, distinguishes subclasses of motor neurons that select distinct axonal pathways and that occupy different columns in the spinal cord (Tsuchida et al., 1994). Although the complexity of the formation of structures and connections is probably degrees higher in the brain than in the spinal cord, the growing list of LIM-hd genes expressed in the forebrain suggests a possible similar role in the forebrain. In Figure 8B we present a tentative summary of the expression domains of Lhx1–9. A combinatorial pattern clearly emerges in the developing brain, with pairs of closely related homologs sharing comparable domains of expression and taking part in the determination of distinct parts of the brain (always respecting the prosomeric limits and particularly the zona limitans). For example, the richest combination is found in the mge, with a unique expression of Lhx6-7-8 that is restricted to p5. Of interest, Lhx1, Lhx2, Lhx5, and Lhx9 are all expressed in the diencephalon and in the cortex (Sheng et al., 1997; present work). If they act in a combinatorial mode, they could participate in the establishment of the highly topographic thalamocortical and/or corticothalamic projections (for review, see Molnar and Blakemore, 1995). Lhx9, highly expressed in median line and associative thalamic nuclei, could participate in the connections with the Lhx9-expressing frontal cortex. Noteworthy is the expression of Lhx9 in a number of limbic-related structures such as the archicortex, the olfactory bulb, the amygdala, the diagonal band, and the septum, pointing out a possible implication of Lhx9 in the development of the limbic system.
In summary, in the family of LIM-hd genes that are often expressed in pairs in the developing CNS, Lhx9 is one of the missing partners for Lhx2. Its expression pattern strongly suggests a role in forebrain development, both in the specification of brain subdivisions and in cellular determination.
Footnotes
This work was supported by the Association Franco-Israelienne pour la Recherche Scientifique et Technique, Fondation pour la Recherche Médicale, and Centre National de la Recherche Scientifique. We are grateful to Dr. A. Joyner for the En-2 in situ probe and J. L. Duband and Y. Bassaglia for discussions aboutLhx9 in the caelomic cavity.
Correspondence should be addressed to Dr. Sylvie Rétaux, Laboratoire de Neurochimie-Anatomie, Institut des Neurosciences, 9 quai St. Bernard, 75005 Paris, France.
REFERENCES
- 1.Agulnick AD, Taira M, Breen JJ, Tanaka T, Dawid IB, Westphal H. Interactions of the LIM-domain-binding factor Ldb1 with LIM homeodomain proteins. Nature. 1996;384:270–272. doi: 10.1038/384270a0. [DOI] [PubMed] [Google Scholar]
- 2.Angevine JB, Sidman RL. Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature. 1961;192:766–768. doi: 10.1038/192766b0. [DOI] [PubMed] [Google Scholar]
- 3.Bach I, Rhodes SJ, PearseII RV, Heinzel T, Gloss B, Scully KM, Sawchenko PE, Rosenfeld MG. P-Lim, a LIM homeodomain factor, is expressed during pituitary organ and cell commitment and synergizes with Pit-1. Proc Natl Acad Sci USA. 1995;92:2720–2724. doi: 10.1073/pnas.92.7.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bach I, Carriere C, Ostendorff HP, Andersen B, Rosenfeld MG. A family of LIM domain-associated cofactors confer transcriptional synergism between LIM and Otx homeodomain proteins. Genes Dev. 1997;11:1370–1380. doi: 10.1101/gad.11.11.1370. [DOI] [PubMed] [Google Scholar]
- 5.Barnes JD, Crosby JL, Jones MC, Wright CVE, Hogan BL. Embryonic expression of Lim-1, the mouse homolog of Xenopus XLIM-1, suggests a role in lateral mesoderm differentiation and neurogenesis. Dev Biol. 1994;161:168–178. doi: 10.1006/dbio.1994.1018. [DOI] [PubMed] [Google Scholar]
- 6.Bulfone A, Puelles L, Porteus MH, Frohman MA, Martin GR, Rubenstein JLR. Spatially restricted expression of Dlx-1, Dlx-2, Gbx-2, and Wnt-3 in the embryonic day 12.5 mouse forebrain defines potential transverse and longitudinal segmental boundaries. J Neurosci. 1993;13:3155–3172. doi: 10.1523/JNEUROSCI.13-07-03155.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bulfone A, Smiga SM, Shimamura K, Peterson A, Puelles L, Rubenstein JLR. T-brain-1:a homolog of Brachyury whose expression defines molecularly distinct domains within the cerebral cortex. Neuron. 1995;15:63–78. doi: 10.1016/0896-6273(95)90065-9. [DOI] [PubMed] [Google Scholar]
- 8.Caviness VS. Neocortical histogenesis in the normal and reeler mice: a developmental study based upon 3H-thymidine autoradiography. Dev Brain Res. 1982;4:293–302. doi: 10.1016/0165-3806(82)90141-9. [DOI] [PubMed] [Google Scholar]
- 9.Caviness VS, Takahashi T, Jr, Nowakowski RS. Numbers, time and neocortical neurogenesis: a general developmental and evolutionary model. Trends Neurosci. 1995;18:379–383. doi: 10.1016/0166-2236(95)93933-o. [DOI] [PubMed] [Google Scholar]
- 10.Dawid IB, Toyama R, Taira M. LIM domain proteins. C R Acad Sci III. 1995;318:295–306. [PubMed] [Google Scholar]
- 11.De Carlos JA, Lopez-Mascaraque L, Valverde F. Dynamics of cell migration from the lateral ganglionic eminence in the rat. J Neurosci. 1996;16:6146–6156. doi: 10.1523/JNEUROSCI.16-19-06146.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Figdor MC, Stern CD. Segmental organization of embryonic diencephalon. Nature. 1993;363:630–634. doi: 10.1038/363630a0. [DOI] [PubMed] [Google Scholar]
- 13.Frantz GD, Weimann JM, Levin ME, McConnell SK. Otx1 and Otx2 define layers and regions in developing cerebral cortex and cerebellum. J Neurosci. 1994;14:5725–5740. doi: 10.1523/JNEUROSCI.14-10-05725.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freyd G, Kim K, Horvitz HR. Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene. Nature. 1990;344:876–879. doi: 10.1038/344876a0. [DOI] [PubMed] [Google Scholar]
- 15.Grigoriou M, Tucker AS, Sharpe PT, Pachnis V. Expression and regulation of Lhx6 and Lhx7, a novel subfamily of LIM homeodomain encoding genes, suggests a role in mammalian head development. Development. 1998;125:2063–2074. doi: 10.1242/dev.125.11.2063. [DOI] [PubMed] [Google Scholar]
- 16.Gulisano M, Broccoli V, Pardini C, Boncinelli E. Emx1 and Emx2 show different patterns of expression during proliferation and differentiation of the developing cerebral cortex in the mouse. Eur J Neurosci. 1996;8:1037–1050. doi: 10.1111/j.1460-9568.1996.tb01590.x. [DOI] [PubMed] [Google Scholar]
- 17.Hobert O, Mori I, Yamashita Y, Honda H, Ohshima Y, Liu Y, Ruvkun G. Regulation of interneuron function in the C. elegans thermoregulatory pathway by the ttx-3 LIM homeobox gene. Neuron. 1997;19:345–357. doi: 10.1016/s0896-6273(00)80944-7. [DOI] [PubMed] [Google Scholar]
- 18.Jurata LW, Kenny DA, Gill GN. Nuclear LIM interactor, a rhombotin and LIM homeodomain interacting protein, is expressed early in neuronal development. Proc Natl Acad Sci USA. 1996;93:11693–11698. doi: 10.1073/pnas.93.21.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Witte DP, Branford WW, Aronow JB, Weinstein M, Kaur S, Wert S, Singh G, Schreiner CM, Whitsett JA, Scott WJ, Jr, Potter SS. Gsh-4 encodes a LIM-type homeodomain, is expressed in the developing central nervous system, and is required for early postnatal survival. EMBO J. 1994;12:2876–2895. doi: 10.1002/j.1460-2075.1994.tb06582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lundgren SE, Callahan CA, Thor S, Thomas JB. Control of neuronal pathway selection by the Drosophila LIM homeodomain gene apterous. Development. 1995;121:1769–1773. doi: 10.1242/dev.121.6.1769. [DOI] [PubMed] [Google Scholar]
- 21.Macdonald R, Xu Q, Barth A, Mikkola I, Holder N, Fjose A, Krauss S, Wilson SW. Regulatory gene expression boundaries demarcate sites of neuronal differentiation in the embryonic zebrafish forebrain. Neuron. 1994;13:1039–1053. doi: 10.1016/0896-6273(94)90044-2. [DOI] [PubMed] [Google Scholar]
- 22.Matsumoto K, Tanaka T, Furuyama T, Kashihara Y, Ishii N, Tohyama M, Kitanaka J, Takemura M, Mori T, Wanaka A. Differential expression of LIM-homeodomain genes in the embryonic murine brain. Neurosci Lett. 1996;211:147–150. doi: 10.1016/0304-3940(96)12749-x. [DOI] [PubMed] [Google Scholar]
- 23.Molnar Z, Blakemore C. How do thalamic axons find their way to the cortex? Trends Neurosci. 1995;18:389–397. doi: 10.1016/0166-2236(95)93935-q. [DOI] [PubMed] [Google Scholar]
- 24.Nohno T, Kawakami Y, Wada N, Ishikawa T, Ohuchi H, Sumihare N. Differential expression of the two closely related LIM-class homeobox genes LH-2A and LH-2B during limb development. Biochem Biophys Res Commun. 1997;238:506–511. doi: 10.1006/bbrc.1997.7320. [DOI] [PubMed] [Google Scholar]
- 25.Polleux F, Dehay C, Kennedy H. The timetable of laminar neurogenesis contributes to the specification of cortical areas in mouse isocortex. J Comp Neurol. 1997;385:96–116. [PubMed] [Google Scholar]
- 26.Porter FD, Drago J, Xu Y, Cheema SS, Wassif C, Huang SP, Lee E, Grinberg A, Massalas JS, Bodine D, Alt F, Westphal H. Lhx-2, a LIM homeobox gene, is required for eye, forebrain, and definitive erythrocyte development. Development. 1997;124:2935–2944. doi: 10.1242/dev.124.15.2935. [DOI] [PubMed] [Google Scholar]
- 27.Porteus MH, Bulfone A, Liu J-K, Puelles L, Lo L-C, John LR. DLX-2, MASH-1, and MAP-2 expression and bromodeoxyuridine incorporation define molecularly distinct cell populations in the embryonic mouse forebrain. J Neurosci. 1994;14:6370–6383. doi: 10.1523/JNEUROSCI.14-11-06370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puelles L. A segmental morphological paradigm for understanding vertebrate forebrain. Brain Behav Evol. 1995;46:319–337. doi: 10.1159/000113282. [DOI] [PubMed] [Google Scholar]
- 29.Puelles L, Rubenstein JLR. Expression patterns of homeobox and other putative regulatory genes in the embryonic forebrain suggest a neuromeric organization. Trends Neurosci. 1993;11:472–479. doi: 10.1016/0166-2236(93)90080-6. [DOI] [PubMed] [Google Scholar]
- 30.Rétaux S, McNeill L, Harris WA. Engrailed, retinotectal targeting, and axonal patterning in the midbrain during Xenopus development: an antisense study. Neuron. 1996;16:63–75. doi: 10.1016/s0896-6273(00)80024-0. [DOI] [PubMed] [Google Scholar]
- 31.Rubenstein JLR, Martinez S, Shimamura K, Puelles L. The embryonic vertebrate forebrain: the prosomeric model. Science. 1994;266:578–580. doi: 10.1126/science.7939711. [DOI] [PubMed] [Google Scholar]
- 32.Sanchez-Garcia I, Rabbitts TH. The LIM domain: a new structural motif found in zinc-finger-like proteins. Trends Genet. 1994;9:315–320. doi: 10.1016/0168-9525(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 33.Seidah NG, Barale JC, Marcinkiewicz M, Mattei MG, Day R, Chretien M. The mouse homeoprotein mLIM-3 is expressed early in cells derived from the neuroepithelium and persists in adult pituitary. DNA Cell Biol. 1994;13:1163–1180. doi: 10.1089/dna.1994.13.1163. [DOI] [PubMed] [Google Scholar]
- 34.Sheng HZ, Bertuzzi S, Chiang C, Shawlot W, Taira M, Dawid IB, Westphal H. Expression of murine Lhx-5 suggest a role in specifying the forebrain. Dev Dyn. 1997;208:266–277. doi: 10.1002/(SICI)1097-0177(199702)208:2<266::AID-AJA13>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 35.Thor S, Thomas JB. The Drosophila islet gene governs axon pathfinding and neurotransmitter identity. Neuron. 1997;18:397–409. doi: 10.1016/s0896-6273(00)81241-6. [DOI] [PubMed] [Google Scholar]
- 36.Tsuchida T, Ensini M, Morton SB, Baldassare M, Edlund T, Jessell TM, Pfaff SL. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79:957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- 37.Way JC, Chalfie M. mec-3, a homeobox-containing gene that specifies differentiation of the touch receptor neurons in C. elegans. Cell. 1988;54:5–16. doi: 10.1016/0092-8674(88)90174-2. [DOI] [PubMed] [Google Scholar]
- 38.Wilson SW, Placzek M, Furley AJ. Border disputes: do boundaries play a role in growth cone guidance? Trends Neurosci. 1993;16:316–323. doi: 10.1016/0166-2236(93)90108-x. [DOI] [PubMed] [Google Scholar]
- 39.Xu Y, Baldassare M, Fisher P, Rathbun G, Oltz EM, Yancopoulos GD, Jessell TM, Alt FW. LH-2: a LIM/homeodomain gene expressed in developing lymphocytes and neural cells. Proc Natl Acad Sci USA. 1993;90:227–231. doi: 10.1073/pnas.90.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]